Significance
Using DNA dendrons as a platform for vaccine design, we investigate the structure-function relationships that dictate molecular vaccine efficacy. By exploring how adjuvant and antigen placement and attachment chemistry affect biological properties, we uncovered key design parameters that impact cellular uptake, immune activation, and the ability to raise a specific immune response against a cancer target, in vitro and in vivo. Indeed, DNA dendrons successfully treated a mouse cervical cancer model where vaccine structure defined its overall efficacy, with one of the architectures dramatically reducing tumor burden and maintaining 100% animal survival throughout the duration of the study. Taken together, this work establishes the DNA dendron as a potentially generalizable approach to probe concepts in rational therapeutic design with molecular precision.
Keywords: DNA dendrons, vaccines, DNA therapeutics
Abstract
A foundational principle of rational vaccinology is that vaccine structure plays a critical role in determining therapeutic efficacy, but in order to establish fundamental, effective, and translatable vaccine design parameters, a highly modular and well-defined platform is required. Herein, we report a DNA dendron vaccine, a molecular nanostructure that consists of an adjuvant DNA strand that splits into multiple DNA branches with a varied number of conjugated peptide antigens that is capable of dendritic cell uptake, immune activation, and potent cancer killing. We leveraged the well-defined architecture and chemical modularity of the DNA dendron to study structure-function relationships that dictate molecular vaccine efficacy, particularly regarding the delivery of immune-activating DNA sequences and antigenic peptides on a single chemical construct. We investigated how adjuvant and antigen placement and number impact dendron cellular uptake and immune activation, in vitro. These parameters also played a significant role in raising a potent and specific immune response against target cancer cells. By gaining this structural understanding of molecular vaccines, DNA dendrons successfully treated a mouse cervical human papillomavirus TC-1 cancer model, in vivo, where the vaccine structure defined its efficacy; the top-performing design effectively reduced tumor burden (<150 mm3 through day 30) and maintained 100% survival through 44 d after tumor inoculation.
Immunotherapy has emerged as a powerful approach to treat cancer due to its ability to train the immune system to attack specific cancer cells and reduce off target effects (1–5). Cancer vaccines function via the delivery of two components: an adjuvant (an immune system activator) and an antigen (an immune system target) (6, 7). Nanomaterials are particularly attractive agents for the delivery of adjuvant and antigen components because they provide unprecedented control over vaccine chemistry and structure and hence their properties and functions (3, 6–13). Indeed, the rational design of nanoscale therapeutics allows one to tune the biodistribution, codelivery, kinetics of component processing, and temporal degradation of a vaccine, all of which are critical factors that determine its efficacy (14–19). These findings are the foundation for rational vaccinology (20) and underscore the importance of vaccine structure and architecture in dictating vaccine function.
The spherical nucleic acid (SNA), a nanoparticle core functionalized with a dense shell of radially oriented DNA, has emerged as a powerful platform to study concepts in rational vaccinology because it enables control over vaccine structure (20–25). Due to the chemical modularity of the SNA, key parameters such as adjuvant and antigen ratios, (25) antigen attachment chemistry, (20, 22), and SNA stability (23) can be modified independently, enabling the exploration of the impact of structure on function. As with many nanoparticle-based systems however, achieving molecularly defined structural control over the SNA is a challenge due to the inherent chemical heterogeneity of many nanoparticle cores and the inability to control oligonucleotide placement on the surface of the particle with atom-scale precision. As a result, SNAs afford only moderate structural control, specifically with regard to DNA surface coverage, antigen conjugation, and nanoparticle size (26).
DNA dendrons are molecularly well-defined and can be used to access SNA-like properties as a result of the highly oriented, dense packing of DNA on the dendron branches (27, 28). These dendritic architectures typically consist of a DNA stem that splits into multiple DNA branches. Similar to SNAs, these multivalent DNA constructs undergo rapid cellular uptake, are resistant to degradation, and can elicit an enhanced therapeutic effect of conjugated cargo (27). Herein, we hypothesize that by utilizing the DNA dendron as the basis for a cancer vaccine, structure-function relationships could be probed in a molecularly defined manner, providing novel insights into vaccine design and function.
Accordingly, we report the design, synthesis, and evaluation of DNA dendron-based molecular vaccines as cancer immunotherapies. Specifically, we investigated how adjuvant placement and attachment chemistry impact dendron uptake and adjuvant potency. By conjugating an antigenic peptide, derived from cervical cancer due to human papillomavirus (HPV), to different positions on the dendron architecture, we used this system to investigate how antigen placement impacts vaccine cellular uptake and immune activation. Finally, we utilized these structures to train immune cells to target and kill cancer cells, both in vitro and in vivo, demonstrating that vaccine efficacy is highly structure-dependent. Taken together, the data establish the DNA dendron as a powerful tool to study the fundamental structure-function relationships that govern cancer vaccine efficacy. Furthermore, the observation that DNA dendrons themselves can behave as potent vaccines has important implications in the development of a new class of cancer immunotherapeutics.
Results
Design, Uptake, and Efficacy of Immune-Stimulating DNA Dendrons.
The adjuvant is a key component of a vaccine because it leads to immune activation and cellular processing of the antigen target (29). Previous work has shown that increasing the valency of DNA dendrons (and thereby the number of adjuvant sequences) expectedly leads to increased immune activation; (30) however, important structural questions, such as the effects of steric hindrance and DNA hybridization are yet to be explored and can have a significant impact on adjuvant function (vide infra). When considering multivalent adjuvant delivery and efficacy, steric hindrance can play a crucial role in determining adjuvant potency.
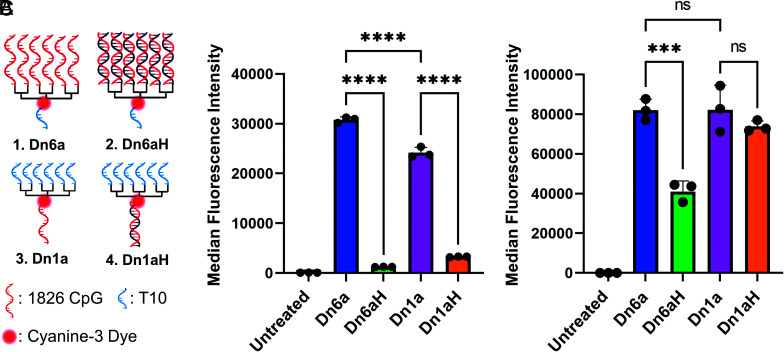
To that end, we designed four different six-branched DNA dendrons that contain adjuvant CpG (cytosine-phosphate-guanine) DNA sequences that behave as potent toll-like receptor 9 (TLR-9) agonists: 1) a T10 sequence stem that branches into six CpG sequences (termed Dn6a, for dendron with six adjuvant strands); 2) a T10 sequence stem that branches into six CpG complementary sequences that hybridize the CpG sequences (termed Dn6aH, for dendron with six adjuvant strands hybridized); 3) a CpG sequence stem that branches into six T10 sequences (termed Dn1a, for dendron with one adjuvant strand); and 4) a CpG complement strand that can hybridize the CpG strand and branches into six T10 sequences (termed Dn1aH, for dendron with one adjuvant strand hybridized) (Fig. 1A). T10 sequences were chosen due to synthetic ease and lack of secondary structure formation (31–33). These designs enabled us to probe steric effects in detail. By comparing data from Dn6a and Dn1a, we can deduce whether multivalency improves or impairs adjuvant function as a result of steric hindrance from dense DNA packing on the branches. Comparing hybridized (Dn6aH and Dn1aH) and unhybridized structures (Dn6a and Dn1a) reveals whether the dendron molecule itself sterically inhibits proper TLR-9 binding and processing, and if a supramolecular dendron design is therefore necessary for potent activation.
Fig. 1.
Cellular uptake of immune-stimulating DNA dendrons. (A) Four different DNA dendron designs were investigated: 1) six CpG sequences as the branches and a T10 stem (Dn6a); 2) six CpG sequences hybridized to the branches and a T10 stem (Dn6aH); 3) T10 branches and a CpG stem (Dn1a); 4) T10 branches and a CpG sequence hybridized to the stem (Dn1aH). All DNA dendrons have a cyanine 3 fluorescent dye in the middle of the molecule. T10 sequences were chosen due to synthetic ease and lack of secondary structure formation. (B) Cellular uptake after 1 h incubation at 250 nM shows the unhybridized structures have preferential access to cells. (C) Cellular uptake after 15 h incubation at 1 μM shows this trend holds, with hybridized branches having lower cellular uptake. The MFI ± SD is shown for n = 3. ns = not significant; ***P < 0.001; ****P < 0.0001. Not all significances are shown for clarity.
For in vitro measurements via flow cytometry, all DNA dendrons were synthesized with a cyanine 3 (Cy3) fluorescent dye in the middle of the structure, which not only allowed for a quantitative assessment of dendron uptake, but also allowed for the facile tracking of the dendron throughout its synthesis and purification. The DNA dendrons were synthesized through solid-phase automated synthesis, purified by polyacrylamide gel electrophoresis (PAGE), and characterized by PAGE and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) (SI Appendix, Table S1). To form the hybridized structures, CpG strands were mixed with Dn6aH and Dn1aH at stoichiometric amounts, annealed from 90 to 20 °C over 1 h, and characterized by native PAGE (SI Appendix, Fig. S1).
First, we determined how the structure of each dendron impacted the cellular uptake efficiency. To test this, murine bone marrow-derived dendritic cells (BMDCs) were treated with 250 nM fluorophore-labeled DNA dendrons (1:1 ratio of dendron molecule to fluorescent tag) for 1 h in serum-containing media. Cellular uptake was assessed by measuring the median fluorescence intensity (MFI) of the Cy3 fluorophore in treated cells, via flow cytometry. After 1 h, we observed that Dn6aH and Dn1aH had significantly lower cellular uptake efficiencies compared to Dn6a and Dn1a (Fig. 1B), presumably due to a decrease in DNA-scavenger receptor A recognition. The non-hybridized structures had MFIs over two orders of magnitude greater than those of the hybridized structures, indicating that dendron uptake is maximized when the branches and stems are single-stranded DNA. The Dn6a-treated cells also had 1.2-fold greater MFI than those treated with the Dn1a. This observation can be attributed to the fact that longer DNA sequences can lead to increased uptake efficiencies (SI Appendix, Fig. S2) (27). We repeated this experiment with 1 μM of fluorescently labeled DNA dendron and an incubation time of 15 h. Even at this longer timepoint, we observe that the non-hybridized structures enter cells in greater quantity than the hybridized forms. Specifically, the Dn6aH structure suffers from low uptake indicating that the double-stranded dendron branches significantly inhibit cellular uptake (Fig. 1C).
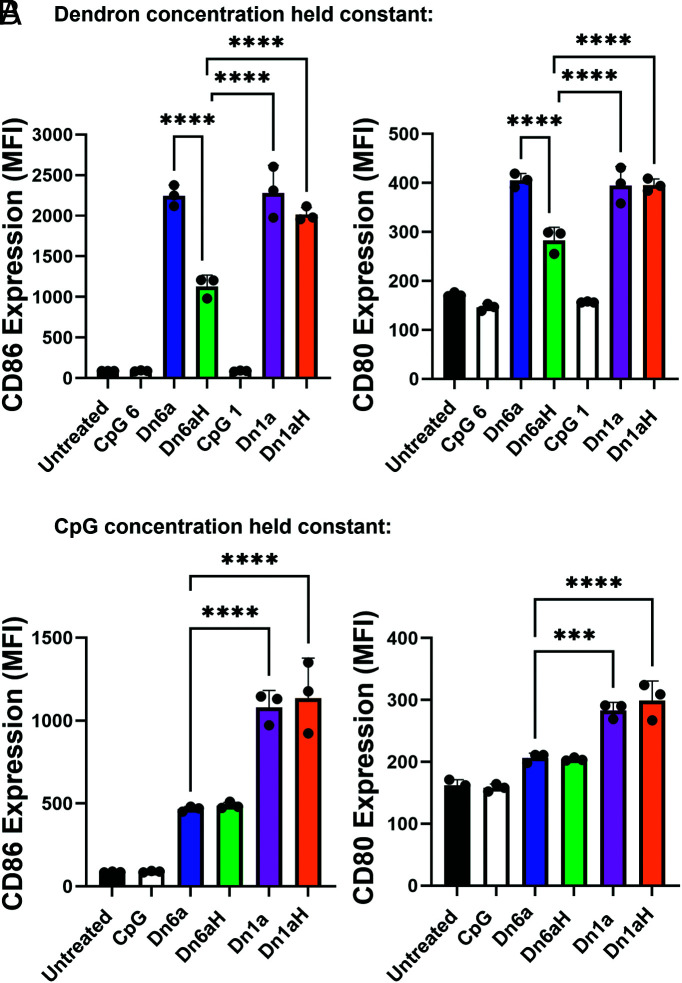
Next, BMDCs were treated with either linear CpG controls or one of the four dendron designs and immune activation was measured after 15 h incubation. Upon TLR-9 activation by CpG adjuvant DNA, immune costimulatory markers, such as cluster of differentiation (CD) 86 and CD80, become up-regulated and presented on the dendritic cell surface (34). We can assess which dendrons elicit the greatest immune response by measuring the amount of CD80 and CD86 expressed. To address the fact that the Dn6a and Dn6aH structures deliver sixfold more CpG per dendron than the Dn1a and Dn1aH structures, this experiment was conducted in two different ways. First, cells were treated such that the total dendron concentration was held constant, meaning that the Dn6a- and Dn6aH-treated cells received sixfold more CpG than the Dn1a- and Dn1aH-treated cells but that the amount of dendron delivered was the same for each group (Fig. 2A). Due to the two different CpG concentrations between the Dn1a and Dn6a structures, two linear CpG controls were used to match those concentrations (CpG 1, to match Dn1a and Dn1aH, and CpG 6, to match Dn6a and Dn6aH). Expectedly, the linear CpG controls at either concentration had negligible cellular uptake and immune activation compared to the dendron treatment groups due to the rapid degradation of linear DNA sequences. For cells treated with DNA dendrons, we observed trends in CD86 and CD80 expression that match the uptake profile of the different groups—the Dn6a, Dn1a, and Dn1aH had no significant differences in CD86 and CD80 expression, while the Dn6aH structure produced significantly less CD86 and CD80 expression among murine CD11c+ dendritic cells. This trend held when the percent of cells positive for CD86 and CD80 expression was measured. Specifically, Dn6a, Dn1a, and Dn1aH induced CD86 expression in nearly 100% of cells, while the Dn6aH structure induced expression in only 80% of cells (SI Appendix, Fig. S3). Expression of CD80 followed a similar trend. These observations can be attributed to the lower cellular uptake of the Dn6aH, as discussed above. Furthermore, these results indicated that although the Dn6a dendron delivered sixfold more CpG than the Dn1a and Dn1aH dendrons, the total immune activation was nearly identical.
Fig. 2.
Immune activation after treatment of murine bone marrow-derived dendritic cells with immune-stimulating DNA dendrons. (A) MFI of CD86 (Left) and CD80 (Right) expression when the total dendron concentration was held constant. A similar trend was observed as that in the uptake experiment, whereby the unhybridized structures perform better than the hybridized constructs with no significant differences between Dn6a, Dn1a, and Dn1aH despite the sixfold increase in CpG delivered by the Dn6a dendron. CpG 1 and CpG 6 represent CpG concentrations that correspond to Dn1a and Dn6a, respectively. (B) MFI of CD86 (Left) and CD80 (Right) expression when the total CpG concentration was held constant. Results indicate that the Dn1a dendron was the most efficient design to deliver functional DNA adjuvant. The MFI ± SD is shown for n = 3. ***P < 0.001; ****P < 0.0001. Not all significances are shown for clarity.
To further investigate this observation, cells were treated such that the CpG concentration was held constant across treatment groups. Therefore, cells treated with Dn6a and Dn6aH received 1/6th the amount of dendron molecule as those treated with Dn1a and Dn1aH (Fig. 2B and SI Appendix, Fig. S3B). Interestingly, when the CpG concentration was held constant, a different trend in immune activation emerged. The Dn6a and Dn6aH structures resulted in significantly lower immune activation, whereby CD86 and CD80 levels were reduced by approximately 50% and 33% compared to the Dn1a and Dna1H responses, respectively (Fig. 2B). These data indicate that although the Dn6a dendron presented CpG multivalently, this multivalency did not lead to increased CpG processing and cell activation. This suggests that dendron valency does not impart the proper spatial requirements necessary for multivalent interactions with TLR-9 (35). Taken together, these data reveal that the Dn1a and Dn1aH designs are the most effective for adjuvant delivery. Due to synthetic ease and molecular precision, the non-hybridized Dn1a structure was used in subsequent experiments. Moreover, since the branches of the DNA dendron are no longer used as the CpG sequence, they can be leveraged as orthogonal handles to improve cellular uptake and immune activation (i.e., G-rich sequences) (SI Appendix, Fig. S4) or as secondary functional sequences (e.g., alternative TLR agonist sequences).
Antigen Conjugation and Its Effects on Immune Activation.
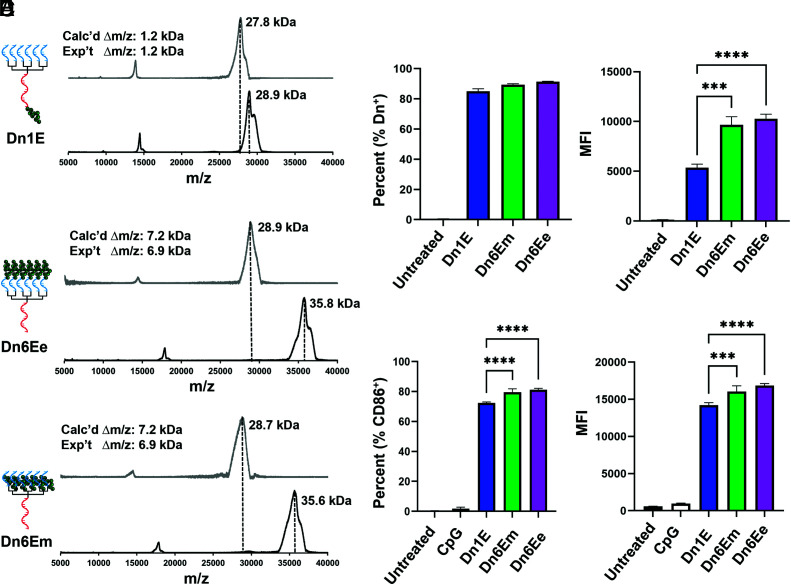
Having established design rules for dendron-mediated adjuvant delivery, we next sought to understand how the placement of an antigenic peptide affects dendron vaccine efficacy. We hypothesized that antigen placement on the dendron would significantly impact dendron uptake and overall vaccine efficacy due to the hydrophobic nature of many peptide antigens (36). We further hypothesized that the antigen:adjuvant ratio would significantly affect vaccine efficacy (25). To begin exploring the impact of antigen placement, three dendrons were designed based on the Dn1a architecture. The first contained a primary amine on the 3′ end of the dendron stem such that a single antigen could be added to each structure (Dn1E, dendron with 1 epitope, Fig. 3A). The second design included primary amines on the 5′ end of the dendron, thereby producing dendrons with six peptides on the ends of the branches (one peptide/branch) (Dn6Ee, dendron with six epitopes on the end, Fig. 3B). To test whether peptide conjugation to the 5′ end impacted dendron uptake (due to a potential loss of scavenger receptor A binding interactions), the third design had primary amines located in the middle of each branch, enabling the conjugation of six antigen molecules without blocking the multivalent DNA branches (Dn6Em, dendron with six epitopes in the middle, Fig. 3C). Interestingly, these dendrons on their own, without conjugated antigens, had distinct uptake properties. Specifically, the dendrons with more primary amine functional groups achieved nearly twofold increases in cellular uptake (SI Appendix, Fig. S5). This result is likely due to the positive charge of the primary amine under biological conditions, which can facilitate cellular uptake (37). Nevertheless, upon peptide conjugation, the amine is converted to an amide and the positive charge is negated.
Fig. 3.
Antigenic peptide conjugation and its impact on cellular uptake and immune activation. (A–C) MALDI-TOF MS of dendron-peptide conjugates, post purification, reveal expected mass shifts for each of the synthesized conjugates: Dn1E, Dn6Ee, and Dn6Em. (D) Cellular uptake of the dendron-peptide conjugates after 1 h at 250 nM. An increase in cellular uptake for the conjugates that contain a greater number of hydrophobic antigenic peptides is observed. (E) Immune activation of dendritic cells after 15 h treatment with dendron-peptide conjugates at 1 μM. The trends observed are similar to that seen in the cellular uptake experiments; the Dn6E structures produced a stronger immune response than did the Dn1E structure. The MFI ± SD is shown for n = 3. ***P < 0.001; ****P < 0.0001. Not all significances are shown for clarity.
We utilized the peptide antigen E649–58 (V10C, sequence: VYDFAFRDLC, mw = 1.25 kDa, SI Appendix, Table S2) derived from the E6 protein, which is found in cervical cancer cells caused by HPV. The peptide contains a single terminal cysteine amino acid which is conjugated to the dendron through a reducible disulfide bond. Specifically, the heterobifunctional crosslinker, succinimidyl 3-(2-pyridyldithio)propionate (SPDP), was used, which contains an activated ester on one end, employed for primary amine functionalization, and a pyridyldithiol group on the other end, used for sulfhydryl functionalization. First, the activated ester was reacted with the primary amines on the DNA dendrons, producing pyridyldithiol functionalized dendrons. Second, the E6 peptide was reacted with the pyridyldithiol to form peptide conjugates. All conjugates were purified by PAGE. MALDI-TOF MS results indicate that for each of the dendron designs, the expected product was synthesized and purified successfully because the expected mass shifts between unconjugated and conjugated DNA dendrons were observed (Fig. 3 A–C, Top and Bottom, respectively). Specifically, the Dn1E structure shows a mass shift of 1.2 kDa, while the Dn6E structures show a mass shift of 6.9 kDa, closely matching the expected changes in mass for dendrons conjugated with one or six peptides, respectively.
After synthesizing the dendron-peptide conjugates, we investigated how peptide placement affects cellular uptake. We hypothesized that the increased hydrophobicity of the conjugated peptides would facilitate uptake and lead to increased uptake frequency and fluorescence intensity. Using flow cytometry, we found that a similar percentage of cells were positive for each of the structures (% Dn+) (Fig. 3 D, Left); however, cells treated with dendrons with a greater number of conjugated peptides (Dn6Em and Dn6Ee) exhibited a nearly twofold higher MFI compared to those treated with Dn1E (Fig. 3 D, Right), demonstrating that more Dn6Em and Dn6Ee entered each cell when compared to the mono-conjugated Dn1E dendron.
A similar trend was observed when measuring immune activation via CD86 expression (Fig. 3E). We observed a statistically significant increase in the frequency of cells expressing CD86 (% CD86+) when Dn6Em or Dn6Ee was used when compared to Dn1E (75 to 80%). When measuring the amount of CD86 expressed (MFI), we observed a similar trend as seen in the uptake experiments, wherein the Dn6E structures led to the expression of more CD86 than the Dn1E dendron (Fig. 3 E, Right). A similar trend was also observed when measuring the CD80 costimulatory marker (SI Appendix, Fig. S6). These results confirm our hypothesis that increasing the number of peptides attached to the dendron leads to increased cellular uptake and immune activation. Due to the similarity in uptake and activity for the Dn6Em and Dn6Ee constructs, we chose to continue experimenting with the Dn6Em structure for synthetic reasons, finding that Dn6Em peptide conjugation consistently had a 30% yield while the Dn6Ee structure had a 15% yield.
We further explored the uptake mechanism of these materials by selectively inhibiting cellular uptake pathways (SI Appendix, Fig. S7). Specifically, cells were pretreated with either fucoidan (a scavenger receptor A inhibitor), methyl-β-cyclodextrin (cholesterol/lipid raft depletion), or incubated at 4 °C (which inhibits all active transport mechanisms). We observed that inhibiting active transport through incubation at 4 °C resulted in the greatest decrease in uptake for each of the peptide conjugates, indicating that these peptides were not facilitating significant passive transport across the cell membranes. After treatment with methyl-β-cyclodextrin, which abstracts hydrophobic cholesterol and lipid rafts from the cell surface, we observed a decrease in cellular uptake, which is likely due to decreased interactions between hydrophobic peptides and hydrophobic components of the cell membrane. Finally, we observed that by inhibiting scavenger receptor A mediated endocytosis using fucoidan, the most significant drop (at least 50% in each treatment group) in uptake was observed, indicating that the primary pathway of dendron uptake was through scavenger receptor A, regardless of where the antigenic peptide was located on the dendron. We further investigated if peptide conjugation impacts the complexation of serum proteins to the dendron, which may explain the observed differences in cellular uptake efficiencies. To test this assertion, Dn1a, Dn1E, and Dn6Em were incubated in fetal bovine serum and subsequently analyzed by gel electrophoresis. Based on shifts in electrophoretic mobility, the results suggest that there are mild differences in serum protein complexation to the dendrons depending on the amount of conjugated peptide (SI Appendix, Fig. S8). These differences in protein complex formation may contribute to the observed uptake properties.
Investigating Vaccine Efficacy In Vitro and In Vivo.
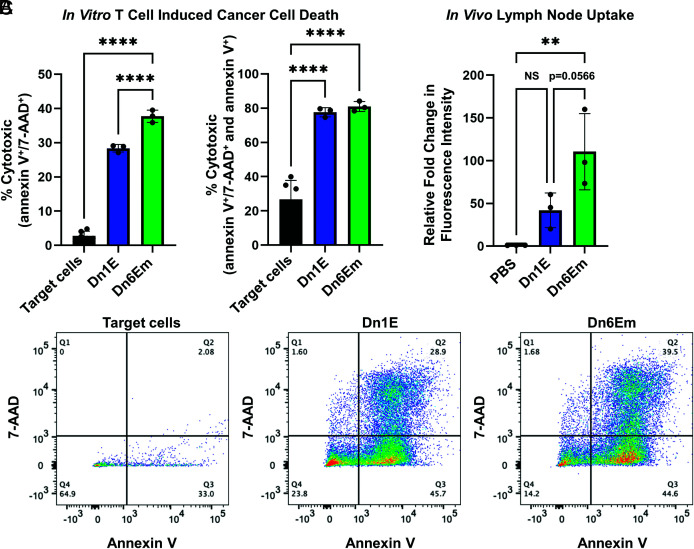
Before studying vaccine efficacy in vivo, we first confirmed that the DNA dendron vaccines elicited a potent and specific immune response against cervical cancer cells. Mouse peripheral blood mononuclear cells (PBMCs) were treated with the different vaccines. The resulting raised T cells were isolated from the PBMCs and co-cultured separately with TC-1 mouse cervical cancer cells at a 10 to 1 ratio of T cells to cancer cells. After incubation, cancer cell death was quantified by measuring 7-AAD (necrosis) and Annexin V (early apoptosis). We observed that all tested DNA dendrons produced a potent immune response against the target cancer cells. Specifically, ~30% of the cells treated with Dn1E and ~40% of the cells treated with Dn6Em induced TC-1 expression of markers for both apoptosis and necrosis (Fig. 4 A, Left). However, if the target cells that were expressing only apoptotic markers were included (indicating that they are undergoing early apoptosis but not yet full necrosis), both vaccines killed approximately 80% of the target cells (Fig. 4 A, Right). Since early apoptosis occurs prior to necrosis, these results suggest that the vaccines produce an immune response at different rates. Indeed, the Dn6Em vaccine seems to result in a potent immune response earlier than the Dn1E vaccine, leading to a greater number of necrotized target cells. This observation is likely due to the increase in antigen loading on the Dn6Em structure, as well as its increased cellular uptake. Representative scatter plots are provided in Fig. 4C.
Fig. 4.
Ex vivo vaccine efficacy and in vivo vaccine uptake. (A) Murine PBMCs were treated with the dendron vaccines, raised T cells were then isolated and incubated in the presence of target TC-1 cancer cells at a ratio of 5:1, and cell death was quantified in terms of expression of both necrosis and early apoptosis markers (double positive, Left) or early apoptosis markers (double and single positive, Right). (B) Dendron vaccines were injected subcutaneously into female C57BL/6 mice (n = 3), and fluorescence intensity of the lymph nodes was measured after 4 h, using IVIS. Results indicate that the Dn6Em structure was taken up significantly more than the Dn1E structure in vivo. (C) Representative scatter plots that correspond to the data reported in A. ns = not significant; **P < 0.01; ****P < 0.0001. Not all significances are shown for clarity.
To further assess the potential in vivo efficacy of the dendron vaccines, we studied how the dendron architecture facilitates effective uptake into the lymph nodes, the biological hub for immune cells responsible for producing immune responses (38). Female C57BL/6 mice (n = 3 per group) received either a saline control [phosphate-buffered saline (PBS)], Dn1E (6 nmol) or Dn6Em (6 nmol), subcutaneously. After 4 h, the mice were sacrificed, the lymph nodes were collected, and Cy3 fluorescence was quantified using an in vivo imaging system (IVIS). We observed that both vaccines localized to one or both of the proximal lymph nodes (SI Appendix, Fig. S9) and that the Dn6Em structure was taken up by the lymph nodes more than the Dn1E vaccine (P = 0.0566) (Fig. 4B). Negligible fluorescence was observed in the lymph nodes of the PBS treated mice, and importantly, there is no statistically significant difference between the fluorescence intensities of PBS and Dn1E treated mice. This indicates that peptide location and amount on the dendron structure can significantly impact vaccine accumulation at the desired immune centers when delivered in vivo. Previous research has shown that the hydrophobicity of the therapeutic can significantly enhance lymph node uptake (39–41). Since the Dn6Em structure has sixfold more peptides per dendron, those hydrophobic moieties likely play an important role in mediating vaccine trafficking and uptake.
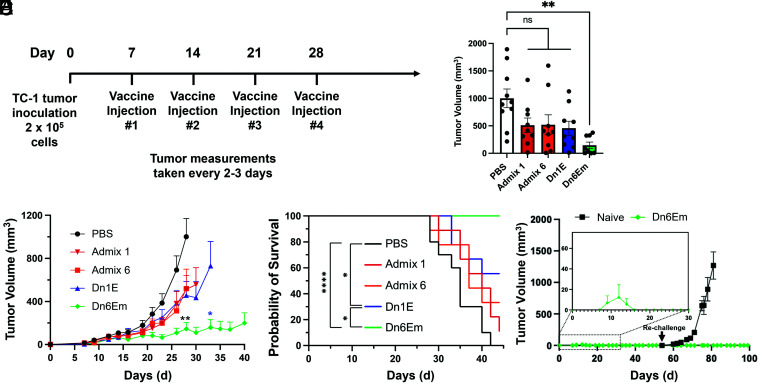
To evaluate the in vivo therapeutic efficacy, we treated TC-1 tumor-bearing female mice (n = 8 to 10) with the different dendron architectures and evaluated tumor growth and animal survival. TC-1 tumor cells (2 × 105) were inoculated subcutaneously into the right flank of C57BL/6 mice and allowed to grow to ∼50 mm3 before the first of four treatments (schedule provided in Fig. 5A). Specifically, animals were treated once per week with Dn1E, Admix 1 (simple mixture corresponding to Dn1E adjuvant/antigen ratio), Dn6Em, Admix 6 (simple mixture corresponding to Dn6Em adjuvant/antigen ratio), or PBS. Tumor growth was measured every 2 to 3 d, while survival was quantified; animals were sacrificed when tumor burden reached 1,500 mm3. Mice that were treated with Dn6Em exhibited potent suppression of tumor growth (Fig. 5 B and C and SI Appendix, Fig. S10). In fact, tumor growth inhibition was observed in 100% of animals treated with Dn6Em (on average <150 mm3 through day 30), and they were protected from death through the 44 d study as their tumors did not reach the 1,500 mm3 cutoff during this timeframe (Fig. 5D). In contrast, mice treated with the Dn1E dendron failed to exhibit a significant improvement in tumor burden, and only 56% survived to day 44. As expected, animals treated with saline all perished by the conclusion of the study (median survival—35 d), and animals treated with either of the admix controls were unable to mount a sufficient immune response to fight off tumor burden effectively. Spider plots for all groups are provided in SI Appendix, Fig. S10. This study illustrates the impact of precise structural changes to the dendron architecture on its ability to dramatically impact animal survival and tumor burden in vivo. At the conclusion of the study (after 44 d), mice with no measurable tumors were re-challenged with TC-1 tumor cells (Fig. 5E). Mouse health and tumor growth were monitored and measured for an additional 60 d. Naïve control mice experienced rapid increases in tumor size and required euthanasia by day 80 (36 d post challenge). Nevertheless, the Dn6Em re-challenged mice were protected from the cancer cells, showing no signs of tumor growth through the conclusion of the study (Day 100). These results demonstrate that the DNA dendron vaccines can impart an adaptive immune response capable of recognizing targets for several weeks post-immunization.
Fig. 5.
Treating tumor-bearing mice that were inoculated with cervical cancer cells. (A) Mice were treated with 2 × 105 cancer cells on day 0 and received treatment once/week for 4 wk, starting on day 7. Tumor measurements were taken every 2 to 3 d. (B) Tumor volumes at day 28, which demonstrate the significant tumor growth inhibition that results from Dn6Em treatment over the other groups. (C) Plots of the average tumor volumes show that the Dn6Em structure is the most effective vaccine for inhibiting tumor growth. (D) This inhibition in tumor growth led to a 100% survival rate for Dn6Em throughout the study (44 d). ns = not significant; *P < 0.05; **P < 0.01; ****P < 0.0001. Not all significances are shown for clarity. (E) Mice with no measurable tumors at the end of the study were re-challenged with TC-1 cancer cells. Mice that received the Dn6Em vaccine were protected against tumor growth.
Discussion
By leveraging the chemical addressability and structural modularity of the DNA dendron, this work explores concepts of rational vaccinology with molecular precision that underscore the importance of a well-defined vaccine architecture to elicit a potent immune response. Importantly, while these studies only explore a subset of all possible dendron valencies and adjuvant:antigen ratios, this investigation yielded several fundamental observations that should be broadly applicable to any DNA dendron-based vaccine. First, we learned that the placement and attachment chemistry of the adjuvant CpG sequence within the dendron was critical for eliciting efficient cellular uptake and a potent immune response. Specifically, the DNA dendron that contained a single CpG sequence as the stem proved to be the most effective design for the delivery of functional adjuvant to cells, while the small size of the DNA dendron prevented any benefit from adjuvant multivalency and adjuvant hybridization significantly inhibited cellular uptake. Next, we discovered that the number and placement of antigenic peptides along the dendron architecture affect dendron uptake, activation, and vaccine efficacy in vitro. Specifically, by increasing the amount of antigen conjugated to the dendron, cellular uptake and immune activation increased. It was also determined that the location of peptide placement was less important than the overall number in affecting vaccine properties. We observed that the number of peptide antigens conjugated to the dendron impacted the rate at which dendron vaccines prompted an immune response against TC-1 ovarian cancer cells. Indeed, Dn6Em resulted in an approximately 30% increase in the amount of tumor cells that were both apoptotic and necrotic, likely a result of the dendrons increased uptake, immune activation, and antigen presentation. Translation of these vaccine designs in vivo was supported by the observed trend that increased peptide conjugation results in Dn6Em preferentially draining to murine lymph nodes to a greater extent than the mono-functionalized Dn1E architecture. Finally, we observed that vaccine structure affects the overall immunogenicity of a cancer vaccine in vivo. Indeed, while highly immunogenic Dn6Em effectively reduced tumor burden (<150 mm3 through day 30), an additive mixture of its component adjuvant and antigen parts, as well as the Dn1E vaccine, failed to provide any significant benefit. While we expect the conclusions drawn from this work to be broadly applicable to DNA dendron-based molecular vaccines, extensive structure-function relationships will be required to adapt the DNA dendron for other therapeutic applications. Parameters such as receptor distribution in the cell and processing of therapeutic components must be considered. Nevertheless, by utilizing the DNA dendron as a platform to study such concepts, structure-function relationships can be determined with molecular precision. Furthermore, although dendritic DNA architectures have been employed in the past for the delivery of therapeutic agents, many of these approaches are heterogenous and lack structural modularity. This DNA dendron-based vaccine consists of a single molecule, made using common oligonucleotide synthesis methods, with an easily modifiable structure. Taken together, this work establishes the DNA dendron as a powerful tool for studying fundamental structure-function relationships that govern vaccine efficacy with molecular precision, underscoring the importance of rational vaccinology and fine-tuned therapeutic design in the development of next generation medicines. Due to the modularity of this molecular vaccine, these design rules can be applied to develop vaccines that treat a variety of cancers.
Materials and Methods
Materials.
TC-1 cells were kindly provided by Dr. Bin Zhang. All animals were used in accordance with approved protocols of the Institutional Animal Care and Use Committee of Northwestern University. Animals (female C57BL/6, 8 to 12 wk old) were obtained from Jackson Laboratories.
Synthesis of Oligonucleotides.
Oligonucleotide synthesis, purification, and characterization were conducted as reported previously. (27) Briefly, reagents and solid-phase supports were purchased from Glen Research. Linear oligonucleotides were synthesized using a MerMade 12 synthesizer (Bio Automation) on controlled pore glass (CPG) beads (Universal UnyLinker Support (1,000 Å)), using conditions recommended by the manufacturer. DNA dendrons were synthesized using a modified coupling protocol reported previously (27). Specifically, they were synthesized using an ABI synthesizer on a dT CPG (2,000 Å) with 2× phosphoramidite concentration for bases on the branches of the dendron. Linear oligonucleotides were purified using reverse-phase high-performance liquid chromatography (RP-HPLC; Agilent), while the DNA dendrons were purified using denaturing PAGE. The samples were characterized using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF; AutoFlex-III, Bruker) mass spectrometry (matrix: dihydroxyacetone phosphate). A complete list of synthesized oligonucleotides can be found in SI Appendix, Tables S1 and S2. The concentrations of DNA dendrons and DNA-containing templates were determined by measuring the solution absorbance at λ = 260 nm (Cary 5000 UV–vis spectrophotometer, Varian) and using the extinction coefficients calculated by the OligoAnalyzer tool (Integrated DNA Technologies).
Cellular Uptake and Immune Activation of DNA Dendrons and Conjugates.
BMDCs were harvested from C57BL/6 mice. Cells were flushed from inside the bone and then collected by centrifugation (1,200 rpm, 5 min). Cells were lysed using 2 mL of ACK lysing buffer for 4 min at room temperature to selectively lyse red blood cells. The remaining cells were washed once with PBS and cultured by incubating in dishes with Gibco Roswell Park Memorial Institute 1640 Medium (RPMI) containing 10% heat inactivated fetal bovine serum, 1% penicillin/streptomycin (P/S), and 40 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) for 5 d. Media was added after 3 d to maintain appropriate nutrients. Cells were collected from the plate and transferred to microtiter tubes prior to addition of treatment.
Cells described above were treated with DNA for 1 h at 50 nΜ or 250 nM, or for 15 h at 1,000 nM (by DNA) and stored in a 37 °C/5% CO2 incubator for the specified amount of time. At the completion of the timepoint, cells were washed with PBS and spun at 1,200 rpm for 5 min to remove supernatant and incubated with fluorophore-conjugated antibodies (Fixable Live/Dead—DAPI, Invitrogen #L34969; CD11c—Alexa Fluor 647, Biolegend #117312 and BV421, Biolegend #117330; CD80—FITC, Biolegend #104706; CD86—PerCP cy5.5, Biolegend #105028) for 15 min at 4 °C. Then, cells were washed with PBS, resuspended in fixation buffer (Biolegend #420801), and incubated at 4 °C for at least 15 min or until analyzed by flow cytometry. All experiments have been conducted in triplicate and with at least three experimental replicates.
DNA Dendron Uptake Mechanism Experiments.
Dendron uptake mechanisms were tested by repeating the previously described uptake experiment in dendritic cells with minor modifications. The cells were pretreated with 50 µg/mL of fucoidan from fucus vesiculous (Sigma F8190), methyl-beta-cyclodextran (12.5 mg/mL), or at 4 °C for 30 min prior to DNA treatment. DNA was added to the cells at concentrations of either 50 nM or 250 nM (by DNA) and incubated for 1 h and 6 h at either 37 °C/5% CO2 or 4 °C. At the completion of the timepoints, cells were washed with PBS and spun at 1,200 rpm for 5 min to remove the supernatant and incubated with fluorophore-conjugated antibodies (Fixable Live/Dead—Ultra-Violet, Invitrogen #L34961and CD11c—Alexa Fluor 647, Biolegend #117312) for 15 min at 4 °C. Then, cells were washed with PBS, resuspended in fixation buffer (Biolegend #420801), and incubated at 4 °C for at least 15 min or until analyzed by flow cytometry (up to 3 d after collection).
Lymph Node Uptake.
Female C57BL/6 mice (8 to 12 wk old) were administered a single subcutaneous injection into the abdomen. Treatment dose was maintained at 6 nmol. After 4 h, mice were euthanized and the skin containing the lymph nodes was resected. Fluorescence was assessed using an IVIS 200 Spectrum (PerkinElmer) IVIS with a narrow band excitation of 535 and emission of 580. Quantitative analysis was performed using Living Image software.
Ex Vivo T Cell Killing.
Murine PBMCs (BioIVT) were thawed from storage in liquid nitrogen, and 1 × 104 cells were added to a 96-well round bottom plate in 1000 μL volume. Treatment was added to each well in 450 μL volume, and cells were left in a 37 °C in a 5% CO2 incubator for 48 h. The following procedures were modified from a previously established protocol (24). Briefly, CD8+ T cells were magnetically isolated using a murine CD8a Positive Selection Kit II (StemCell Technologies). Concurrently, TC-1 cells were collected from a passage and were stained with efluor450 (eBioscience) following the manufacturer’s protocol. TC-1 cells were counted after staining and were plated in a new 96-well round bottom plate with 5,000 cells per well in a volume of 100 μL. Cells recovered while T cells were isolated and counted. T cells were added to be at a final ratio of 5:1 with TC-1 target cells in the 96-well plate. Cells were co-cultured together for ~24 h in a 37 °C/5% CO2 incubator. After the incubation time, all cells were collected into microtiter tubes using trypsin to detach adherent cells and media to neutralize the trypsin. Samples were washed once with PBS, centrifuged at 1,200 rpm for 5 min, aspirated, and resuspended in a solution of 100 μL Annexin V binding buffer (BioLegend) containing 0.5 μL each of 7-AAD (Fisher, 50169259) and Annexin V (BioLegend, 640906). This staining solution was left with cells for 15 min at room temperature in the dark prior to flow cytometry.
In Vivo Therapeutic Efficacy.
Female C57BL/6 mice aged 8 to 12 wk (Jackson Laboratory) were inoculated with 2 × 105 TC-1 tumor cells subcutaneously into the right flank and were allowed to grow to ∼50 mm3 (7 d) prior to treatment. Treatments were administered at a dose of 6 nmol (60 μM in 100 μL volume) by subcutaneous injection into the abdomen once per week, following the schedule provided. Tumor growth was measured every 2 to 3 d, and volume was calculated using the following equation: tumor volume = length × width2 × 0.5. Animals were euthanized when tumor volumes reached 1,500 mm3 or when animal health necessitated them to be sacrificed for humane reasons.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This material is based upon work supported by the Polsky Urologic Cancer Institute of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University at Northwestern Memorial Hospital, the Air Force Office of Scientific Research under award FA9550-17-1-0348, and the Lefkofsky Family Foundation. This research was also supported by the National Cancer Institute of the NIH under awards P50CA221747 and R01CA257926. M.H.T. acknowledges support from Northwestern University’s Cancer Nanotechnology Training Program supported by the National Cancer Institute of the NIH award T32CA186897 and support from Edward Bachrach. M.E. was partially supported by the Alexander S. Onassis Public Benefit Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Dr. Bin Zhang for supplying the TC-1 cervical cancer cells. This work made use of the IMSERC MS facility at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-2025633), the State of Illinois, and the International Institute for Nanotechnology (IIN). Imaging work was performed at the Northwestern University Center for Advanced Molecular Imaging generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Some figures were made using BioRender.
Author contributions
M.E.D., J.P.C., M.H.T., and C.A.M. designed research; M.E.D., J.P.C., M.H.T., and M.E. performed research; M.E.D., J.P.C., and C.A.M. contributed new reagents/analytic tools; M.E.D., J.P.C., M.H.T., M.E., and C.A.M. analyzed data; and M.E.D., J.P.C., M.H.T., and C.A.M. wrote the paper.
Competing interest
C.A.M. and M.H.T. have financial interests in SNAP Therapeutics Inc which could potentially benefit from the outcomes of this research.
Footnotes
Reviewers: D.J.M., Harvard University; and N.F.S., University of California, San Diego.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Ashley C. E., et al. , The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat. Materials 10, 389–397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coulie P. G., Van den Eynde B. J., Van Der Bruggen P., Boon T., Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Irvine D. J., Swartz M. A., Szeto G. L., Engineering synthetic vaccines using cues from natural immunity. Nat. Materials 12, 978–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil D. N., Smith E. L., Brentjens R. J., Wolchok J. D., The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 13, 273–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L., et al. , Nanoparticles in medicine: Therapeutic applications and developments. Clin. Pharmacol. Ther. 83, 761–769 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Lin S., Wang X.-Y., Zhu G., Nanovaccines for cancer immunotherapy. WIREs Nanomed. Nanobiotechnol. 11, e1559 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu G., Zhang F., Ni Q., Niu G., Chen X., Efficient nanovaccine delivery in cancer immunotherapy. ACS Nano 11, 2387–2392 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Couvreur P., Nanoparticles in drug delivery: Past, present and future. Adv. Drug Del. Rev. 65, 21–23 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kemp J. A., Shim M. S., Heo C. Y., Kwon Y. J., “Combo” nanomedicine: Co-delivery of multi-modal therapeutics for efficient, targeted, and safe cancer therapy. Adv. Drug Del. Rev. 98, 3–18 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Wong K. K., Li W. A., Mooney D. J., Dranoff G., Advances in therapeutic cancer vaccines. Adv. Immunol. 130, 191–249 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Anselmo A. C., Mitragotri S., Nanoparticles in the clinic: An update post COVID-19 vaccines. Bioeng. Transl. Med. 6, e10246 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sallam M. A., Prakash S., Kumbhojkar N., Shields C. W. IV, Mitragotri S., Formulation-based approaches for dermal delivery of vaccines and therapeutic nucleic acids: Recent advances and future perspectives. Bioeng. Transl. Med. 6, e10215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahdev P., Ochyl L. J., Moon J. J., Biomaterials for nanoparticle vaccine delivery systems. Pharm. Res. 31, 2563–2582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu L., Mooney D. J., Biomaterials and emerging anticancer therapeutics: Engineering the microenvironment. Nat. Rev. Cancer 16, 56–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshy S. T., Mooney D. J., Biomaterials for enhancing anti-cancer immunity. Curr. Opin. Biotechnol. 40, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzeng A., et al. , Temporally programmed CD8α+ DC activation enhances combination cancer immunotherapy. Cell Rep. 17, 2503–2511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rincon-Restrepo M., et al. , Vaccine nanocarriers: Coupling intracellular pathways and cellular biodistribution to control CD4 vs CD8 T cell responses. Biomaterials 132, 48–58 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Irvine D. J., Aung A., Silva M., Controlling timing and location in vaccines. Adv. Drug Del. Rev. 158, 91–115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta N. K., et al. , Pharmacokinetic tuning of protein–antigen fusions enhances the immunogenicity of T-cell vaccines. Nat. Biomed. Eng. 4, 636–648 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S., et al. , Rational vaccinology with spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 116, 10473–10481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radovic-Moreno A. F., et al. , Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 112, 3892–3897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skakuj K., et al. , Conjugation chemistry-dependent T-cell activation with spherical nucleic acids. J. Am. Chem. Soc. 140, 1227–1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meckes B., Banga R. J., Nguyen S. T., Mirkin C. A., Enhancing the stability and immunomodulatory activity of liposomal spherical nucleic acids through lipid-tail DNA modifications. Small 14, 1702909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teplensky M. H., et al. , Spherical nucleic acid vaccine structure markedly influences adaptive immune responses of clinically utilized prostate cancer targets. Adv. Healthcare Mater. 10, 2101262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teplensky M. H., et al. , Spherical nucleic acids as an infectious disease vaccine platform. Proc. Natl. Acad. Sci. U.S.A. 119, e2119093119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Irvine D. J., Guiding principles in the design of molecular bioconjugates for vaccine applications. Bioconjugate Chem. 26, 791–801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Distler M. E., et al. , DNA dendrons as agents for intracellular delivery. J. Am. Chem. Soc. 143, 13513–13518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng H. F., et al. , Nanoparticle superlattices through template-encoded DNA dendrimers. J. Am. Chem. Soc. 143, 17170–17179 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Reed S. G., Bertholet S., Coler R. N., Friede M., New horizons in adjuvants for vaccine development. Trends Immunol. 30, 23–32 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Kawamoto Y., et al. , Enhanced immunostimulatory activity of covalent DNA dendrons. ChemBioChem 23, e202100583 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Kim S. H., Lee T.-H., Conformational dynamics of poly (T) single-stranded DNA at the single-molecule level. J. Phys. Chem. Lett. 12, 4576–4584 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang X., Kuhn H., Frank-Kamenetskii M. D., Monitoring single-stranded DNA secondary structure formation by determining the topological state of DNA catenanes. Biophys. J. 90, 2877–2889 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Ziv R., Libchaber A., Effects of DNA sequence and structure on binding of RecA to single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 9068–9073 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanier L. L., et al. , CD80 (B7) and CD86 (B70) provide similar costimulatory signals for T cell proliferation, cytokine production, and generation of CTL. J. Immunol. 154, 97–105 (1995). [PubMed] [Google Scholar]
- 35.Comberlato A., Koga M. M., Nüssing S., Parish I. A., Bastings M. M. C., Spatially controlled activation of toll-like receptor 9 with DNA-based nanomaterials. Nano Lett. 22, 2506–2513 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowell D., et al. , TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc. Natl. Acad. Sci. U.S.A. 112, E1754–E1762 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fröhlich E., The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 7, 5577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sainte-Marie G., The lymph node revisited: Development, morphology, functioning, and role in triggering primary immune responses. The Anat. Rec. 293, 320–337 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Ali Khan A., Mudassir J., Mohtar N., Darwis Y., Advanced drug delivery to the lymphatic system: Lipid-based nanoformulations. Int. J. Nanomed. 8, 2733–2744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawley A. E., Davis S. S., Illum L., Targeting of colloids to lymph nodes: Influence of lymphatic physiology and colloidal characteristics. Adv. Drug Del. Rev. 17, 129–148 (1995). [Google Scholar]
- 41.Liu J., et al. , Targeting colloidal particulates to thoracic lymph nodes. Lung Cancer 51, 377–386 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.