Summary
This study describes a novel, neutralizing monoclonal antibody (mAb), 11D7, discovered by mouse immunization and hybridoma generation, against the parental Wuhan‐Hu‐1 RBD of SARS‐CoV‐2. We further developed this mAb into a chimeric human IgG and recombinantly expressed it in plants to produce a mAb with human‐like, highly homogenous N‐linked glycans that has potential to impart greater potency and safety as a therapeutic. The epitope of 11D7 was mapped by competitive binding with well‐characterized mAbs, suggesting that it is a Class 4 RBD‐binding mAb that binds to the RBD outside the ACE2 binding site. Of note, 11D7 maintains recognition against the B.1.1.529 (Omicron) RBD, as well neutralizing activity. We also provide evidence that this novel mAb may be useful in providing additional synergy to established antibody cocktails, such as Evusheld™ containing the antibodies tixagevimab and cilgavimab, against the Omicron variant. Taken together, 11D7 is a unique mAb that neutralizes SARS‐CoV‐2 through a mechanism that is not typical among developed therapeutic mAbs and by being produced in ΔXFT Nicotiana benthamiana plants, highlights the potential of plants to be an economic and safety‐friendly alternative platform for generating mAbs to address the evolving SARS‐CoV‐2 crisis.
Keywords: COVID‐19, Monoclonal antibody (mAb), Variants of Concern, Plant‐made antibody, Neutralization synergy, Antibody cocktail
Introduction
Coronavirus disease 2019 (COVID‐19) is caused by infection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (Azkur et al., 2020; Chen et al., 2020; Chou et al., 2022; Cotugno et al., 2021). Vaccine development against SARS‐CoV‐2 proceeded at an unprecedented pace and has resulted in multiple effective products, even against emerging variants (Carreño et al., 2021; Garcia‐Beltran et al., 2022; Hager et al., 2022; Heath et al., 2021; El Sahly et al., 2021; Thomas et al., 2021). However, immunocompromised and elderly individuals may still benefit from the continued development of prophylactic and therapeutic monoclonal antibodies (mAbs), given that these subpopulations may not develop a robust and durable response from vaccination or prior infection (Brockman et al., 2022; Obeid et al., 2022; Ryan et al., 2022).
The cellular receptor mediating SARS‐CoV‐2 infection in humans is angiotensin‐converting enzyme 2 (ACE2) (Qiao et al., 2020; Schurink et al., 2022; Q. Wang et al., 2020). The spike (S) protein of the virus interacts with ACE2 and can be broadly broken down into two domains. S1 contains the N‐terminal domain (NTD) and the receptor‐binding domain (RBD), while S2 houses the fusion peptide, necessary for the structural rearrangements leading to membrane fusion between virus and host (Lan et al., 2020; Xu et al., 2021). The RBD is flexible, being found mostly in an ‘up and open’ or a ‘down and closed’ conformation, with a transitionary conformation between the two also being observed (Pramanick et al., 2021; Walls et al., 2020). It is upon the binding of ACE2 with one ‘up and open’ RBD, that structural changes in the S protein result in viral entry (Cai et al., 2020). The interaction between ACE2 and RBD makes the RBD a dominant target for antibodies with neutralizing capability and indeed, accounts for most of the neutralizing antibodies in a polyclonal response (Greaney et al., 2021; Piccoli et al., 2020; Schmidt et al., 2021).
Monoclonal antibodies are useful therapeutics for preventing viral disease by neutralizing viral particles, accomplishing the same results as the polyclonal response to immunization or natural infection (Both et al., 2013). Many groups have identified and developed potently neutralizing mAbs against SARS‐CoV‐2 in the last 2 years, with some being advanced to clinical use via Emergency Use Authorization (EUA) (Dong et al., 2021; Loo et al., 2022; Rogers et al., 2020; Shi et al., 2020; Wu et al., 2020; Zost et al., 2020). The mechanism by which many mAbs interfere with viral infection has been elucidated by structural studies and can be broadly categorized into four classes based on the mAb's epitope and which RBD conformation it recognizes (Barnes et al., 2020; Greaney et al., 2021; Gruell et al., 2022). Class 1 mAbs overlap with the ACE2 binding site of the RBD in the ‘up and open’ conformation, while Class 2 mAbs overlap with the ACE2 binding site in both ‘up and open’ and ‘down and closed’ conformations of the RBD. By directly interfering with viral binding to ACE2, these classes tend to be potent neutralizers. Class 3 mAbs also bind to both conformations of the RBD, with the epitopes laying outside of the ACE2 binding site, yet many mAbs of this class are also potent neutralizers. Class 4 mAbs bind further away from the ACE2 binding site and are generally less potent neutralizers (Huo et al., 2020). Characterizing mAbs using these classes helps inform which mAbs may be useful in mAb cocktails, where inclusion of multiple classes with non‐overlapping epitopes can reduce immune evasion (Ku et al., 2021). Indeed, the urgent need for continued development of mAb cocktails is driven by the emergence of variants of concern accumulating mutations in the RBD, causing changes in binding to ACE2 (Ozono et al., 2021; Wang et al., 2022). This will potentially reduce neutralizing potency of host antibody response against the new variants (Greaney et al., 2021; Hoffmann et al., 2021) and, in some cases, abolish the utility of previously potent therapeutic mAbs (Chen et al., 2021; Starr et al., 2021). Several mAbs that received EUA from the Food and Drug Administration (FDA) have been amended or revoked as they lose therapeutic activity against emerging variants, specifically the B.1.1.529 (Omicron) and other variants of this lineage (Focosi et al., 2022). This necessitates the development of mAbs that have potential synergy in cocktails to help reduce immune escape by SARS‐CoV‐2.
Discovery of new mAbs against antigens of interest can be accomplished in several ways. Single‐cell antigen‐specific sorting and sequencing (Gilchuk et al., 2020) and phage display techniques (Kumar et al., 2019) are among the leading technologies used for mAb generation and screening. The method of mouse immunization followed by hybridoma generation (Köhler and Milstein, 1975; Mitra and Tomar, 2021) is the classic method of obtaining full‐length, monoclonal IgGs. The hybridoma method has benefits over single‐cell and phage techniques, in that it does not require sophisticated instrumentation and preserves the process of in vivo affinity maturation, respectively. Moreover, variable region gene sequences of mAbs from lead hybridoma candidates can be rescued and humanized for recombinant production.
Plants are a versatile system for recombinant protein production and have made substantial contributions in developing countermeasures against the current COVID‐19 pandemic. For example, a plant‐based vaccine based on virus‐like particles (VLP) displaying a modified S protein of SARS‐CoV‐2 was developed. Results from clinical trials demonstrated that this vaccine was safe and had 78.8% efficacy against moderate‐to‐severe disease caused by the homologous strain and 69.5% overall efficacy against five circulating variants of concern (VOCs) (Hager et al., 2022). This plant‐made vaccine is currently licensed for human use by Canada Health (Health Canada, 2022). Several other plant‐made COVID‐19 vaccine candidates are currently being evaluated in human clinical trials (Stander et al., 2022). Plant‐made ACE2 has also been explored as a prophylactic/therapeutic to prevent or treat SARS‐CoV‐2 infection (Siriwattananon et al., 2021). An innovative approach of using ACE2 is the use of an ACE2‐formulated chewing gum to trap SARS‐CoV‐2 in the saliva, blocking transmission of the virus among individuals in the population (Daniell, 2022). Studies have shown that chewing gums containing plant‐made cholera‐toxin B (CTB)‐ACE2 fusion protein significantly reduced salivary SARS‐CoV‐2 load in vitro (Daniell, 2022). This approach is being evaluated in a Phase I/II clinical trial and will have a broad impact on the prevention of other respiratory viral diseases, as well as on the development of orally deliverable drug platforms that do not require cold chain.
Plants have also been demonstrated as a favourable system for mAb development and production. Traditional mammalian expression platforms for mAb production require sterile facilities and can be cost‐intensive. However, producing mAbs in plants has potential to significantly reduce overall costs, increase safety by reducing the possibility of introducing human pathogens during production and increase efficacy by allowing for facile and homogeneous manipulation of N‐linked glycans (Chen, 2016, 2022; Loos and Steinkellner, 2014; Nandi et al., 2016). In addition, transient production of mAbs in plants is accomplished through simple cloning of mAb genes and agroinfiltration, resulting in rapid, high‐level accumulation of recombinant mAbs within a short period of time (Chen et al., 2013; Leuzinger et al., 2013). Many examples of mAbs produced in plants also show that plant‐made pharmaceuticals have potential to be viable alternatives to mammalian‐made counterparts (Jugler et al., 2021; Kallolimath et al., 2021; Lai et al., 2010; Rattanapisit et al., 2020; Shanmugaraj et al., 2020; Sun et al., 2021, 2023).
In the current study, we describe the development of an original mAb against the RBD of SARS‐CoV‐2 by hybridoma generation, followed by chimerization and expression in plants. We also provide evidence that this mAb, namely 11D7, neutralizes variants of concern, including B.1.617.2 (Delta) and B.1.1.529 (Omicron). 11D7 has a unique RBD‐binding site that is not overlapping with those of several other EUA‐authorized, SARS‐CoV‐2‐neutralizing mAbs in Classes 1, 2 and 3 and displays neutralizing synergy with these classes of therapeutic mAbs in neutralizing the Omicron and other variants. This study highlights the utility of non‐ACE2‐competing mAbs in forming effective cocktails and of plant‐based expression systems in developing novel mAbs against the ever‐evolving SARS‐CoV‐2 pandemic.
Results
Expression of a novel Anti‐SARS‐CoV‐2 RBD monoclonal antibody in Nicotiana benthamiana
BALB/c mice were immunized with recombinant Wuhan‐Hu‐1 RBD (BEI Resources, NR‐52309) on a prime and boost regimen followed by splenocyte isolation and hybridoma generation. Hybridomas were screened for reactivity to the immunogen (Wuhan‐Hu‐1 RBD). Several strongly reactive antibodies were identified, and one, 11D7, was chosen for variable region gene rescue. The murine variable regions of the light and heavy chains were grafted onto human kappa and human gamma constant regions, respectively. The resulting chimeric mAb was codon‐adapted for plant‐based expression and transiently expressed in ΔXFT N. benthamiana mutant, carrying N‐glycans lacking plant‐specific xylose and fucose residues (Strasser et al., 2008). Plant‐made 11D7 (p11D7) reached peak expression of 131 μg of mAb per gram of fresh leaf weight (FLW) 5 days after transgene delivery (Figure 1). When total plant protein extract containing the p11D7 mAb was subjected to Protein A affinity chromatography, the recombinant mAb was homogenously purified in a manner similar to a mammalian‐made mAb purified by the same method (Figure 2a). Furthermore, the bands observed from SDS‐PAGE analysis were confirmed to be the expected heterotetrameric IgG (Figure 2b) and its constitutive light chain (Figure 2c) and heavy chain (Figure 2d). We next investigated the glycosylation status of p11D7 and 11D7 produced in mouse hybridoma (m11D7) by mass spectrometry analysis. Our p11D7 was found to carry 91.1% bi‐ or mono‐antennary N‐acetylglucosamine (GnGn, MGn), mammalian‐like glycans typical for mAbs produced in ΔXFT plants, compared with the more heterogeneous, mouse hybridoma‐produced m11D7 (Table 1).
Figure 1.
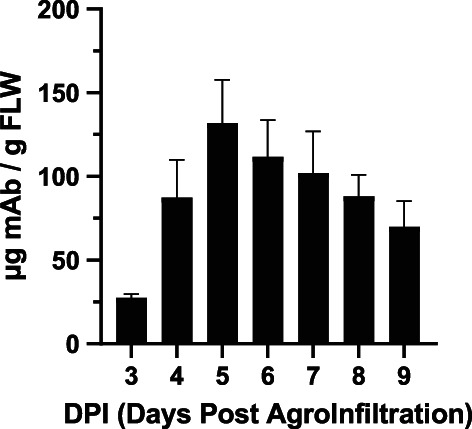
Recombinant expression of 11D7 in ΔXFT Nicotiana benthamiana. ΔXTF N. benthamiana leaves were infiltrated with 11D7 gene constructs and total soluble leaf proteins were extracted 3–9 days post agroinfiltration (DPI). The expression levels of p11D7 were analysed by a sandwich ELISA that detects only fully assembled IgG. Mean ± SEM are shown from two independent experiments performed in technical duplicates.
Figure 2.
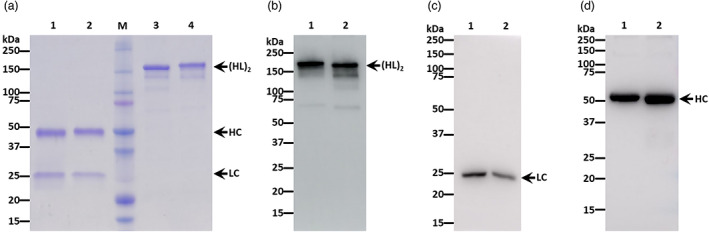
Biochemical characterization of p11D7. Protein A‐purified, p11D7 and an IgG isotype control were subjected to SDS‐PAGE under reducing (a, Lanes 1 and 2) or non‐reducing conditions (a, Lanes 3 and 4) and total protein content was stained with Coomassie blue. In parallel, SDS‐PAGE‐separated proteins under non‐reducing (b) or reducing conditions (c and d) were transferred to PVDF and probed for human kappa light (b and c) or for human gamma chain (d). Lanes 1 and 3: IgG isotype control. Lanes 2 and 4: p11D7. HC: heavy chain. LC: light chain. (HL)2: assembled heterotetrameric form of IgG. One representative result of multiple experiments is shown.
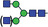
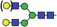
Table 1.
Glycan analysis of m11D7 and p11D7
| Major N‐glycan species | Schematic presentation | m11D7 (%) | p11D7 (%) |
|---|---|---|---|
| GnGn/MGn |
|
5.7 | 91.1 |
| AGn/AA |
|
12.3 | |
| GnGnF6/MGnF6 |
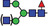
|
25.7 | |
| AMF6 |
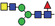
|
3.3 | |
| AGnF6 |
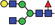
|
33.4 | |
| AAF6 |
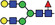
|
19.5 | |
| Man7‐9 |
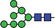
|
0.1 | 8.9 |
Heavy chains of either hybridoma‐made 11D7 (m11D7) or 11D7 produced in ΔXFT Nicotiana benthamiana plants (p11D7) were extracted after SDS‐PAGE, trypsin digested and analysed by LC‐ESI‐MS. Glycopeptide peaks were identified using FreeStyle 1.8 and assigned percentages based on approximate molar ratios from the peak heights. Consortium for Functional Glycomics nomenclature was used.
 Mannose;
Mannose;  N‐acetylglucosamine;
N‐acetylglucosamine;  Fucose;
Fucose;  Galactose.
Galactose.
p11D7 binds to the RBD of the B.1.1.529 (Omicron) variant
We next assessed the functionality of the mAb by testing the binding to the antigen of interest, the SARS‐CoV‐2 RBD. ELISA analysis with the original WA1/2020 strain RBD showed specific binding of p11D7 in a dose‐dependent manner, as expected (Figure 3a). Furthermore, a dissociation constant (KD) for p11D7 of 0.15 nM was determined from the ELISA curve, suggesting that p11D7 has a relatively high affinity for the WA1/2020 RBD. However, given the continuing emergence of variants of SARS‐CoV‐2 with novel mutations in the RBD that may impact mAb binding, we also tested the binding of p11D7 to the RBD of the B.1.1.529 (Omicron) variant of concern to assess the practical utility of this mAb against emerging variants. We observed specific binding of p11D7 to the Omicron RBD at higher concentrations than that of the WA1/2020, indicating a reduced affinity (Figure 3b). By contrast, a Class 1 (CB6) and a Class 4 (CR3022) mAb against the RBD of the WA1/2020 variant showed no binding activity to the Omicron RBD (Figure 3b). As expected, the IgG isotype negative control did not show specific binding to the RBD (Figure 3a). In spite of the reduced affinity, our novel p11D7 mAb still retained recognition of the Omicron RBD, suggesting it may be able to neutralize emerging variants of concern.
Figure 3.
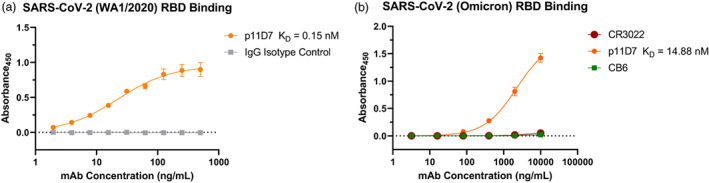
p11D7 recognition of RBD from various SARS‐CoV‐2 variants. Various dilutions of p11D7 and reference mAbs were incubated with the WA1/2020 RBD (a) or the B.1.1.529 (Omicron) RBD (b) that was immobilized on ELISA plates. Specific binding of the RBD‐mAb complex was detected with a horseradish peroxidase (HRP)—conjugated secondary antibody. The absorbance at 450 nm is plotted and is representative of at least two independent experiments performed with technical duplicates. Error bars represent SEM.
p11D7 neutralizes multiple variants of SARS‐CoV‐2
After validating the binding of the chimeric p11D7 to its target antigen, the neutralizing potency of the mAb was assessed. Utilizing authentic SARS‐CoV‐2 in a foci‐forming assay (FFA), a neutralization curve generated against the ancestral WA1/2020 strain resulted in a half‐maximal inhibitory concentration (IC50) of 25.37 μg/mL (Figure 4a). The ongoing emergence of new variants of concern also urged us to test p11D7 against the B.1.617.2 (Delta) variant, where an IC50 of 59.52 μg/mL was observed (Figure 4b), and the B.1.1.529 (Omicron) variant, giving an IC50 of 948.7 μg/mL (Figure 4c). The overall neutralization data showed that the neutralizing ability of p11D7 decreased with the accumulation of mutations in the variants derived from the ancestral strain, but nevertheless still neutralizes variants of concern of SARS‐CoV‐2.
Figure 4.
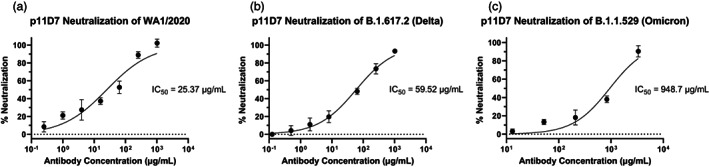
Neutralization of SARS‐CoV‐2 variants by p11D7. Serial dilutions of p11D7 were mixed with SARS‐CoV‐2 WA1/2020 (a), Delta (b), or Omicron (c) before adding to Vero E6 (a and b) or Vero‐hACE2‐TMPRSS2 (c) cells. Cells were then fixed, permeabilized and stained for SARS‐CoV‐2 spike protein (a and b) or nucleocapsid protein (c). Foci were counted, per cent neutralization determined, and IC50 calculated using GraphPad Prism 9. Error bars are SD and curves represent at least two independent experiments performed in technical triplicates.
Synergy of therapeutic mAbs with p11D7 in neutralizing omicron
Next, we investigated whether our novel p11D7 mAb could be categorized into a mAb class against the RBD of SARS‐CoV‐2 as described previously (Barnes et al., 2020; Greaney et al., 2021) by performing a competitive binding assay with well‐characterized mAbs of all RBD‐binding classes. In an ELISA assay using the WA1/2020 RBD and p11D7 conjugated to horseradish peroxidase (HRP), we observed that CR3022 binding to the RBD abolished the ability of p11D7 to bind to the RBD, while several other EUA therapeutic mAbs (CB6, tixagevimab and cilgavimab) did not inhibit the binding of p11D7 to the RBD (Figure 5). This was the expected outcome for CR3022 and CB6, given that we have previously provided evidence of this (Jugler et al., 2022), validating that p11D7 has at least a partially overlapping epitope on the RBD with CR3022 (a Class 4 mAb) but not with CB6 (also known as etesevimab, a Class 1 mAb). Similarly, the two mAbs, tixagevimab (also known as COV2‐2196, a Class 1/2 mAb) and cilgavimab (COV2‐2130, a Class 3 mAb) did not compete with the binding of p11D7 to the RBD. The unique binding site of p11D7 encouraged us to evaluate potential synergy of p11D7 with anti‐RBD mAbs in other classes as a dual and triple mAb cocktails. After empirically determining the IC50 and IC25 for p11D7, tixagevimab and cilgavimab in the FFA, we combined these mAbs at the IC50 and IC25 concentration and measured per cent neutralization of the mAb cocktail against the B.1.1.529 (Omicron) variant. We also calculated the predicted per cent neutralization for the cocktail by importing the individually measured per cent neutralizations of each mAb into SynergyFinder and analysed them with the highest single agent (HSA) and the Loewe models. The calculated values from the models indicate the per cent neutralization where there is no synergistic interaction between mAbs in the cocktail. In all the combinations, the cocktails' observed per cent neutralization is consistently higher than those calculated by prediction models. Specifically, we observed strong neutralization synergy for the p11D7 + tixagevimab combination and for the tixagevimab + cilgavimab combination at their respective IC50's (Table 2), and with the highest per cent neutralization for the triple mAb cocktail at both IC50's and IC25's (Table 2 and Table 3). This indicates that there is neutralizing synergy between p11D7 and tixagevimab and cilgavimab, both as dual combinations, as well as a triple mAb cocktail.
Figure 5.
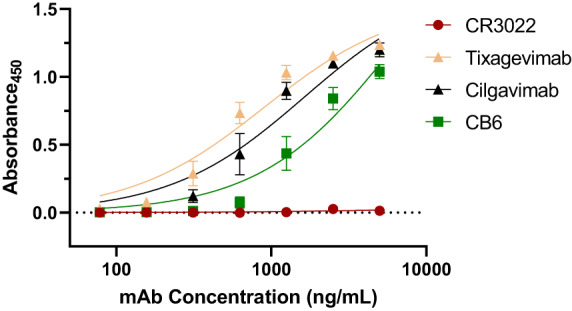
Competitive binding between p11D7 and other anti‐RBD mAbs to the WA1/2020 RBD. Dilutions of each reference mAb were coated on 96‐well plates followed by addition of 2 μg/mL of the WA1/2020 RBD. After washing, the plates were incubated with HRP‐conjugated p11D7 to detect either non‐competitive binding (detection of a signal) or overlapping binding (absence of a signal) of p11D7 with each reference mAb to RBD. Error bars represent SD and at least two independent experiments with technical triplicates were performed.
Table 2.
Neutralizing synergy of p11D7 with plant‐made tixagevimab and plant‐made cilgavimab against the B.1.1.529 variant
| mAb Combination | Observed Neutralization (%) | HSA Predicted Neutralization (%) | Loewe Predicted Neutralization (%) |
|---|---|---|---|
| p11D7 + tixagevimab | 91.97 ± 2.60 | 35.58 ± 7.15 | 40.89 ± 11.29 |
| p11D7 + cilgavimab | 68.21 ± 13.60 | 38.31 ± 12.57 | 48.98 ± 1.90 |
| tixagevimab + cilgavimab | 89.63 ± 2.26 | 47.50 ± 3.38 | 50.34 ± 0.55 |
| p11D7 + tixagevimab + cilgavimab | 96.59 ± 0.64 | 47.85 ± 3.04 | 66.22 ± 5.40 |
Per cent neutralization was obtained via FFA for each individual and combination of mAbs with each mAb at the concentration that corresponds to the IC50. The per cent neutralization data from each individual mAb was used to calculate the predicted per cent neutralization of the cocktail with the highest single agent (HSA) and Loewe models in SynergyFinder.org, assuming there is no neutralization synergy between mAbs in the cocktail. If the empirically determined observed per cent neutralization value of the cocktail is higher than that predicted by the models, it indicates synergistic interaction of the mAbs in the cocktail. At least two independent experiments with technical triplets were performed for each mAb and mAb combination with SD presented.
Table 3.
Neutralizing synergy of p11D7 with plant‐made tixagevimab and cilgavimab at IC25 concentrations against the B.1.1.529 variant
| mAb Combination | Observed Neutralization (%) | HSA Predicted Neutralization (%) | Loewe Predicted Neutralization (%) |
|---|---|---|---|
| p11D7 + tixagevimab | 43.96 ± 9.59 | 18.84 ± 3.96 | 26.08 ± 4.84 |
| p11D7 + cilgavimab | 34.76 ± 27.73 | 29.18 ± 14.99 | 48.90 ± 1.98 |
| tixagevimab + cilgavimab | 65.03 ± 2.62 | 35.14 ± 9.02 | 38.33 ± 7.02 |
| p11D7 + tixagevimab + cilgavimab | 93.92 ± 1.88 | 35.14 ± 9.02 | 50.45 ± 7.46 |
FFA data for each individual and combination of mAbs were performed at the IC25 and per cent neutralization was analysed with the highest single agent (HSA) and Loewe models in SynergyFinder.org. Predicted values represent the expected per cent neutralization if no synergy between mAbs was occurring, while the observed per cent neutralization was the empirically determined value of the combination at the IC25. An observed value higher than a predicted value is inferred to have synergetic interaction. At least two independent experiments with technical triplets were performed with SD values presented.
Discussion
The continuing pandemic caused by SARS‐CoV‐2 urges the ongoing development of novel mAb therapeutics using platforms that have potential to optimize utility and safety. This study describes a novel, SARS‐CoV‐2‐neutralizing mAb discovered in a traditional hybridoma system and further developed in a plant‐based expression system. Initial screening of mAbs secreted by hybridomas generated from RBD‐immunized mice was performed by ELISA with both biotinylated and non‐biotinylated RBD. This ensured that RBD‐binding mAbs were not overlooked whose epitope may have been excluded by direct adhering of antigen to the plate. Of several positive hits from the screening process, one mAb, 11D7, was chosen for further development and recombinant expression in plants due to its ability to efficiently neutralize the WA1/2020 strain in the FFA. The chimeric 11D7 transiently expressed in ΔXFT N. benthamiana plants peaked within a week after agroinfiltration to approximately 131 μg of p11D7 per gram of FLW. This level of expression is similar to other mAbs recently produced by our group using the same expression vector based on the geminiviral bean yellow dwarf virus (Jugler et al., 2022) and allowed accumulation of milligram levels necessary for in vitro and future in vivo studies. However, this level of expression is lower than those of mAbs we produced using more optimized geminiviral vectors (Diamos et al., 2020) or other plant‐made proteins such as griffithsin (GRFT) (Fuqua et al., 2015) that have shown potent anti‐viral activity against SARS‐CoV‐2 (Alsaidi et al., 2021; Palmer, 2022). The yield of p11D7 can be increased by using optimized versions of expression vectors (Diamos et al., 2020) and/or via co‐expression with chaperons (Margolin et al., 2020) to make it cost‐effective for commercial production. Our result also showed that recombinant p11D7 assembled correctly into an IgG, as determined by the Western blot analysis, without any observable degradation or truncation of the heterotetrametric protein. The correct assembly was further validated by observing specific binding of the mAb to its target antigen, the WA1/2020 RBD. The dissociation constant determined from the ELISA was 0.15 nM, suggesting that our novel mAb has a relatively high affinity for the SARS‐CoV‐2 RBD as this is similar to the dissociation constant for other neutralizing mAbs against SARS‐CoV‐2, CA1 and CB6, produced in plants (Jugler et al., 2022). Interestingly, although the affinity is close to that of these neutralizing mAbs, the IC50 for p11D7 in neutralizing the WA1/2020 strain, is much higher at 25.37 μg/mL, indicating that p11D7 is not a potently neutralizing mAb by itself. However, taken together with its unique binding site on the RBD, we speculate that p11D7 may neutralize viral particles by a mechanism other than interfering with ACE2 binding. For example, its binding may change the overall conformation of the S protein and hinder the post‐binding steps required for viral entry.
The continuous emergence of viral variants with accumulating mutations in the spike protein must be considered when developing therapeutic mAbs. The S protein that is responsible for host cell entry is under high selective pressure due to its wide use as a vaccine and therapeutic target. The variant of concern that emerged at the end of 2021, B.1.1.529, colloquially termed Omicron, has overtaken all previous variants, with the parental Omicron variant having accumulated 15 mutations in the RBD region, a prominent neutralizing target (Hu et al., 2022; Kuchipudi et al., 2022). As a result, it is important that novel mAbs being developed for therapeutic purposes retain recognition to circulating variants that have outcompeted prior variants. We assessed the capability of p11D7 to bind to the B.1.1.529 RBD and observed specific binding of p11D7, with an approximately 100‐fold reduction in affinity compared with the WA1/2020 RBD. Of note, CR3022, a Class 4 non‐neutralizing mAb against SARS‐CoV‐2 (Barnes et al., 2020), derived from the original SARS‐CoV outbreak (ter Meulen et al., 2006) did not recognize the Omicron RBD. This is of particular interest because we validated in this study that p11D7 has an overlapping epitope with CR3022 (Jugler et al., 2022), strongly suggesting that p11D7 may itself be a Class 4 RBD‐binding mAb. However, even with an overlapping epitope, the undetermined epitope of p11D7 appears to be more conserved on the RBD than that of CR3022, due to retained binding with accumulating RBD mutations. Furthermore, we show that p11D7 retained neutralizing capacity against the B.1.617.2 (Delta) variant, albeit with an approximately 2.3‐fold reduction in the IC50, as well as the Omicron variant with a further approximately 15.9‐fold reduction in IC50 when compared to the Delta variant. Ultimately, this is a 37.4‐fold reduction in neutralizing ability from the WA1/2020 strain to the Omicron variant that emerged nearly 2 years later. The evidence that p11D7 retains recognition of the Omicron RBD at all is significant, given that many other more potent neutralizing mAbs that were developed and used clinically now do not bind or neutralize what is now the dominant variant of SARS‐CoV‐2 (Planas et al., 2021; VanBlargan et al., 2022). From a structural and mechanistic perspective, this is not surprising, given that many of the anti‐RBD mAbs receiving an EUA early belong to Class 1 or 2, with epitopes on the RBD that overlap with the ACE2 binding site, making them particularly sensitive to mutations that impact ACE2 binding (Deshpande et al., 2021). However, Class 4 mAbs bind to epitopes on the RBD outside and distant from the ACE2 binding site (Deshpande et al., 2021), suggesting they have a neutralization mechanism that is different from ACE2‐competition like Class 1 and 2 mAbs. In addition, the Class 4 mAbs tend to have more conserved epitopes on the RBD, making them more resistant to mutations that occur more often within or near the ACE2 binding site (Gruell et al., 2022a). This suggests that p11D7, which overlaps with a Class 4 mAb, is a good candidate to complement other classes of mAbs in a cocktail to provide synergistic potency against multiple variants. Hence, we sought to use it as a cocktail member with other, more potent mAbs that are still active against the Omicron variant. Our plant‐made mAbs derived from the parental mAbs COV2‐2130 (commercialized into cligavimab or AZD1061) and COV2‐2196 (commercialized into tixagevimab or AZD8895) (Dong et al., 2021; Zost et al., 2020), which are well‐characterized mAbs designated to be Class 3 and Class 1/2, respectively (Barnes et al., 2020; Greaney et al., 2021), do not interfere with p11D7 binding to the WA1/2020 RBD, as determined by a competitive ELISA. We also performed the competition assay with the Class 1 CB6, knowing that it should not interfere with p11D7 binding to the RBD (Jugler et al., 2022). These observations further support the hypothesis that p11D7 is likely a Class 4 mAb against the RBD and that p11D7, not having overlapping epitopes with cilgavimab and tixagevimab, would be a complementary cocktail partner for these two mAbs that are still functionally active against Omicron variants (Planas et al., 2021; VanBlargan et al., 2022).
The synergy analysis performed with dual and triple combinations of p11D7 with plant‐made cilgavimab and tixagevimab at IC50 concentration, do indeed suggest that there is neutralizing synergy of both these mAbs with p11D7 against the Omicron variant. Synergy was also observed at the lower IC25 concentration as dual combinations of p11D7 with tixagevimab and tixagevimab with cilgavimab, and as an triple combination. Notably, the triple mAb combination showed a greater increase in neutralizing activity than all of the dual combinations, especially at low mAb concentrations. Interestingly, the combination of cilgavimab and tixagevimab, used clinically as Evusheld™, still retains neutralizing potency against highly divergent SARS‐CoV‐2 variants, such as BA.4/5, highlighting the continuing clinical utility of this mAb cocktail (Wang et al., 2022; Yamasoba et al., 2022). Previously, we have demonstrated that m11D7 has synergy with the Class 1 EUA mAb, etesevimab (CB6) in neutralizing the WA1/2020 strain (Jugler et al., 2022). Combined with the results reported in this study, the cumulative observations suggest that p11D7 has potential to enhance the neutralizing potency of partner mAbs against multiple variants of concern in a cocktail setting. Therefore, efforts to continue to evaluate p11D7 against emerging variants will inform whether this unique mAb may have potential to increase the neutralizing synergy of an already effective mAb cocktail by a different mechanism to reduce the occurrence of viral immune escape.
We further developed this mAb by generating it as a murine/human chimeric IgG in plants to take advantage of the benefits mAb production in plants offers. Specifically, plant‐based production utilizes inexpensive facilities and growth media for easily scalable mAb generation (Chen and Davis, 2016), as well as potential economic advantages in upstream production (Buyel et al., 2017; Nandi et al., 2016). This, in combination with homogenous N‐linked glycosylation achieved with ΔXFT N. benthamiana plants, allows for development of mAbs with potentially greater safety and efficacy at lower costs than mAbs produced in typical mammalian systems (Dent et al., 2016; Kallolimath et al., 2021; Lai et al., 2010; Strasser et al., 2008; Sun et al., 2021). Indeed, the glycan analysis performed on p11D7 confirmed that 91.1% of the recombinant mAb carried the mammalian‐like glycoform of GnGn or a partially processed MGn, the human‐like intermediate for more complex N‐linked glycans (Chen, 2016). This is regarded as necessary by regulatory agencies for potential human therapeutics made in plants to eliminate immunogenicity of plant‐specific glycans (Schähs et al., 2007). In addition, the N‐linked glycan pattern of IgGs located in the constant heavy 2 (CH2) region at asparagine 297 (N297) is necessary for Fc effector functions mediated through Fc gamma receptors (FcγRs) (Jefferis and Lund, 2002). Fc effector functions are responsible for pathogen clearance through cell‐mediated mechanisms, as well as enhancing antigen presentation (Lu et al., 2017). It has also been demonstrated that Fc function is necessary for more favourable therapeutic protection against SARS‐CoV‐2 (Winkler et al., 2021). The lack of core‐fucose residues is of interest here, in that fucosylated IgGs weaken antibody‐dependent cellular cytotoxicity (ADCC) activity by decreasing affinity to the FcγRIIIa (Wang et al., 2018). Furthermore, ADCC activity of IgGs against SARS‐CoV‐2 has been observed in plasma containing both neutralizing and non‐neutralizing IgGs (Tso et al., 2021), indicating that there may be a protective role for weakly neutralizing or non‐neutralizing mAbs that still bind to SARS‐CoV‐2 through an Fc‐mediated mechanism. Although p11D7 is not as potent a neutralizer compared with other therapeutically developed mAbs for SARS‐CoV‐2 (Gruell et al., 2022b; Jugler et al., 2022; Shi et al., 2020; VanBlargan et al., 2022; Wu et al., 2020; Zost et al., 2020), the plant‐made mAb developed in this study, has a highly homogenous (91.1%) glycan population without detectable core fucose, compared with the 81.9% of the hybridoma‐made mAb that contains core fucose. This suggests that p11D7 may have increased in vivo efficacy by means other than neutralizing potency, namely through Fc‐mediated mechanisms such as ADCC, and will be the focus of future studies.
Conclusion
To our knowledge, this is the most comprehensive study describing a novel mAb produced in plants that binds to a unique epitope on the RBD, exhibiting neutralizing potency against multiple variants of concern by itself, and shows synergy in neutralizing the Omicron variant of SARS‐CoV‐2. As variants continue to emerge, there is a need to proceed to develop therapeutic mAbs that maintain antigen recognition, as well as continue to neutralize the virus, which can be most efficiently accomplished with synergizing antibody cocktails. Our study specifically highlights the utility of mAbs that do not directly hinder ACE2 binding to the RBD in forming effective cocktail components, as well as plants as a mAb expression platform and the potential of this platform to contribute to therapeutic development against the ever‐evolving SARS‐CoV‐2 pandemic.
Methods
Generation of anti‐RBD monoclonal antibodies and variable gene rescue
Generation of the hybridoma as the source of the mAb 11D7 was described previously (Jugler et al., 2022). Briefly, BALB/c mice were immunized subcutaneously with RBD protein (NR‐52309 obtained from BEI) under protocol #22‐1881 T approved by the Institutional Animal Care and Use Committee (IACUC) of Arizona State University. Splenocytes were isolated to establish hybridomas using a standard method (Köhler and Milstein, 1975). Hybridomas were screened for reactivity to both biotinylated and non‐biotinylated RBD (Wuhan‐Hu‐1 RBD) to ensure the recovery of all RBD‐binding mAbs including those with epitopes that may be excluded by direct adhering of antigen to the ELISA plate (Jugler et al., 2022). cDNA was generated from cells of the 11D7 hybridoma using a RNeasy kit for total RNA isolation (Qiagen), followed by first‐strand generation by the Reverse Transcription System (Promega). The coding sequences of mAb variable regions of were generated by PCR using degenerate primers (Wang et al., 2000), cloned into the CloneJET PCR Cloning Kit (Thermo Scientific), and sequenced by Sanger sequencing as described previously (Jugler et al., 2022).
Variable gene plant codon adaptation and plant expression
Variable region sequences rescued and determined from the 11D7 hybridoma and the variable regions for two mAbs, COV2‐2130 (cilgavimab) and COV2‐2196 (tixagevimab) from the literature (Dong et al., 2021; Loo et al., 2022; Zost et al., 2020) were codon‐adapted for plant‐based expression using GeneDesigner2.0. and synthesized by Integrated DNA Technologies (IDT). Synthesized fragments were further grafted onto a human kappa constant sequence that was codon‐adapted for plant expression of the light chain and a human gamma constant sequence codon‐adapted for plant expression of the heavy chain. Subsequent light and heavy chain genes were inserted into bean yellow dwarf virus geminiviral vector and transiently expressed in N. benthamiana plants by agroinfiltration (Chen et al., 2013; Jugler et al., 2021, 2022; Leuzinger et al., 2013).
Antibody extraction and purification
The extraction and purification of mAbs from plants followed our published protocols (Jugler et al., 2020; Lai et al., 2010). Briefly, N. benthamiana leaves expressing mAbs were harvested 5 days after infiltration and homogenized in extraction buffer (1X PBS pH 5.2 with 10 mg/mL Na‐L‐ascorbate, 2 mM PMSF and 1 mM EDTA) and filtered through cheesecloth. Total plant protein extract was then centrifuged several times and incubated at least 4 h at 4 °C at pH 5.2 to precipitate host proteins. Further centrifugation was performed, and the clarified protein extract was filtered through a 0.2‐micron vacuum filter. Protein extract containing recombinant mAbs was then subjected to gravity flow Protein A purification prior to further analysis.
SDS‐PAGE and western blot analysis
SDS‐PAGE and Western blot analysis were performed as described previously (Hurtado et al., 2020). Briefly, purified mAbs were subjected to SDS‐PAGE analysis under reducing and non‐reducing conditions on 4%–20% acrylamide gels and total protein content was stained with Coomassie Blue R‐250. For Western blots, proteins were separated on 12% acrylamide gels under reducing conditions or on 4%–20% acrylamide gels under non‐reducing conditions. After separation, proteins were transferred to PVDF membranes, blocked with 5% milk in 1X PBST and probed with goat anti‐human kappa chain conjugated to horseradish peroxidase (HRP) (Southern Biotech) or goat anti‐human IgG‐HRP (Southern Biotech). Pierce West Pico Western blotting substrate (Thermo Scientific) was added to the membranes and images were taken with an Amersham ImageQuant instrument.
p11D7 temporal expression in plants
Expression of 11D7 in plants was performed utilizing an ELISA assay as described previously (Dent et al., 2016). Briefly, 96‐well plates were coated with goat anti‐human IgG (Southern Biotech) at 2 μg/mL overnight at 4 °C and blocked with 5% milk in 1X PBST. All wash steps between incubations were performed three times with 1X PBST. N. benthamiana leaves expressing recombinant p11D7 were harvested at 24‐h intervals ranging from 3 to 9 days post agroinfiltration. Leaves were homogenized as described above and the extract clarified by centrifugation. After blocking, various dilutions of each time point was added to the plate for 1 h at 37 °C, alongside a control IgG of known concentration. Recombinant, p11D7 was then detected by adding goat anti‐human kappa chain‐HRP diluted 1:4000 in 5% milk in 1X PBST. TMB substrate (SeraCare Life Sciences Inc.) was then added and developed for 5 min before using a 1 M H2SO4 stop solution. Absorbance was read at 450 nm and GraphPad Prism was used to calculate microgram of recombinant 11D7 per gram of fresh leaf weight.
Glycan analysis
The N‐glycosylation profiles of purified plant‐made 11D7 and hybridoma‐made 11D7 were determined by mass spectrometry (MS) as described previously (Esqueda and Chen, 2023; Kallolimath et al., 2021; Sun et al., 2021). In brief, heavy chains were extracted from an SDS‐PAGE, trypsin digested and analysed with an LC‐ESI‐MS system (Orbitrap Exploris 480, Thermo Scientific). Glyco‐peptides were identified as sets of peaks consisting of the peptide and the attached N‐glycan moiety, varying in the number of N‐acetylhexosamine (HexNAc) units, hexose (mannose, galactose, glucose, etc.), deoxyhexose (fucose) and pentose (xylose) residues. Using FreeStyle 1.8 (Thermo Scientific), manual glycopeptide searches were performed, and deconvolution was done using the Extract function. Heights of peaks roughly reflect the molar ratios of the glycoforms. Glycan nomenclature according to Consortium for Functional Glycomics (http://www.functionalglycomics.org) was used.
RBD‐specific binding of mAbs
Indirect ELISAs were used to show specific binding to the WA1/2020 or the B.1.1.529 RBD using a previously described method (He et al., 2021). Briefly, 2 μg/mL (WA1/2020) or 5 μg/mL (B.1.1.529) of RBD was coated on 96‐well plates overnight at 4 °C and blocked with 5% milk in 1X PBST. All wash steps between incubations were performed three times with 1X PBST. Dilutions of mAbs were made in 5% milk in 1X PBST and incubated 1 h at 37 °C, followed by detection with goat anti‐human IgG‐HRP. TMB substrate was then added, and the plate developed for 5 min before using a 1 M H2SO4 stop solution. Absorbance was read at 450 nm and GraphPad Prism was used to generate graphs and calculate approximate dissociation constants (KD) of p11D7 with one‐site specific‐binding model.
Competitive ELISA
An ELISA was used to assess RBD‐binding competition between 11D7 and other RBD‐binding mAbs as previously described (Jugler et al., 2022). Dilutions of each mAb (all produced in plants) were coated on 96‐well plates overnight at 4 °C and blocked with 5% milk in 1X PBST. All wash steps between incubations were performed three times with 1X PBST. The WA1/2020 RBD was incubated at 2 μg/mL for 1 h at 37 °C. Plant‐made 11D7 was conjugated to HRP using EZ Link Plus Activated Peroxidase Kit (Thermo Scientific) and added to the plate at a 1:1000 dilution. TMB substrate was then added, and the plate developed for 5 min before using a 1 M H2SO4 stop solution. Absorbance was read at 450 nm and GraphPad Prism was used to generate binding curves.
Focus‐forming assay
The focus‐forming assay was used to assess neutralization of mAbs as described previously (Case et al., 2020; Jugler et al., 2022), with some modifications. Briefly, 25 000 Vero E6 cells or Vero‐hACE2‐TMPRSS2 cells (for infection with Omicron) were plated in 100 μL of DMEM +10% FBS 1 day prior to the assay in 96‐well clear, flat bottom plates. On the day of the assay, mAbs were diluted to various concentrations and SARS‐CoV‐2 stocks were diluted to 2000 plaque forming units (PFU) and mixed with the mAb dilutions for 1 h at 37 °C prior to adding to the plated cells for an additional hour at 37 °C. Dilutions were performed in DMEM +2% FBS. A 100 μL per well MEM:methylcellulose overlay was then added and infected cells were incubated for 24 h (WA1/2020 and Omicron) or 40 h (Delta). The overlay was then removed, and cells were fixed with 4% paraformaldehyde. Cells were further washed six times with 0.1% saponin and 0.1% BSA in 1X PBS. Cells infected with WA1/2020 or Delta strains were stained with plant‐made CR3022 at 2 μg/mL followed by detection with goat anti‐human IgG‐HRP (Sigma). Omicron‐infected cells were stained with a SARS‐CoV‐2 nucleocapsid antibody (GeneTex) and goat anti‐rabbit IgG‐HRP (Sigma). KPL TrueBlue substrate (SeraCare Life Sciences Inc.) was added, and foci were quantified with an AID Spot Reader. Data were analysed with GraphPad Prism 9 and per cent neutralization was calculated as (average number of foci in virus‐only wells—number of foci in antibody‐treated well)/average number of foci in virus‐only wells. Each antibody was tested in triplicate and at least two independent experiments were performed.
Neutralization synergy
Neutralizing synergy of mAb combinations was performed as previously described (Jugler et al., 2022). In brief, mAbs were diluted to their corresponding IC25 and IC50, followed by FFA experiments with mAbs alone, as dual combinations, or as a triple combination of three mAbs. Per cent neutralization was then calculated as described for the FFA, and data were imported into SynergyFinder and analysed with the highest single agent (HSA) or Loewe models to calculate predicted per cent neutralization at each concentration tested (Berenbaum, 1989; Ianevski et al., 2020; Loewe, 1953). Predicted neutralization was then directly compared with empirically determined per cent neutralization of each combination and concentration. Synergy was inferred when the observed per cent neutralization was higher than that predicted by both models.
Conflict of interest
M.F. is an employee of Halberd Corporation, a company that may have interest in 11D7 for possible commercial development.
Author Contributions
Q.C. conceptualized research. HY.S., C.J. and H.S. designed experiments. HY.S., K.N., C.J. and R.P. performed experiments and analysed data. M.F. provided funding resources. C.J. drafted the manuscript and Q.C. revised the paper with helpful suggestions by H.S., HY.S, R.P. and M.F. All authors have reviewed and agreed to the submitted version of the manuscript.
Supporting information
Table S1 Neutralization potency (IC90) of p11D7 against various SARS‐CoV‐2 variants. Serial dilutions of p11D7 were mixed with the indicated SARS‐CoV‐2 variant before adding to Vero E6 (WA1/2020 and Delta) or Vero‐hACE2‐TMPRSS2 (Omicron) cells. Cells were then fixed, permeabilized and stained for SARS‐CoV‐2 protein. Foci were counted, per cent neutralization determined and IC90 calculated using GraphPad Prism 9. At least two independent experiments were performed with technical triplicates.
Acknowledgements
The authors thank Dr. K. Kibler for technical assistance, Dr. Y. Li for sharing SARS‐CoV‐2 B.1.1.529 stock and H. Lai for sharing the recombinant RBD proteins of various SARS‐CoV‐2 variants. We also thank Dr. D. Lake and F. Grill for their advice in gene rescue and contributions described in Jugler et al., 2022, and Clemens Gruenwald‐Gruber (Core Facility Mass Spectrometry, BOKU VIENNA) for MS analyses. This research was supported by a sponsored research agreement (FP00026198) funding to Q. Chen Lab via Arizona State University by the Halberd Corporation. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS‐Related Coronavirus 2, Isolate USA‐WA1/2020, NR‐52281. The following reagent was obtained through BEI Resources, NIAID, NIH: SARS‐Related Coronavirus 2, Isolate hCoV‐19/USA/PHC658/2021 (Lineage B.1.617.2; Delta Variant), NR‐55611, contributed by Dr. Richard Webby and Dr. Anami Patel. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS‐Related Coronavirus 2, Isolate hCoV‐19/USA/HI‐CDC4359259‐001/2021 (Lineage B.1.1.529; Omicron Variant), NR‐56475, contributed by Centers for Disease Control.
References
- Alsaidi, S. , Cornejal, N. , Mahoney, O. , Melo, C. , Verma, N. , Bonnaire, T. , Chang, T. et al. (2021) Griffithsin and carrageenan combination results in antiviral synergy against SARS‐CoV‐1 and 2 in a pseudoviral model. Mar. Drugs 19, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azkur, A.K. , Akdis, M. , Azkur, D. , Sokolowska, M. , Veen, W. , Brüggen, M. , O'Mahony, L. et al. (2020) Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy 75, 1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, C.O. , Jette, C.A. , Abernathy, M.E. , Dam, K.‐M.A. , Esswein, S.R. , Gristick, H.B. , Malyutin, A.G. et al. (2020) SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum, M.C. (1989) What is synergy? Pharmacol. Rev. 41, 93–141. [PubMed] [Google Scholar]
- Both, L. , Banyard, A.C. , van Dolleweerd, C. , Wright, E. , Ma, J.K.C. and Fooks, A.R. (2013) Monoclonal antibodies for prophylactic and therapeutic use against viral infections. Vaccine 31, 1553–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman, M.A. , Mwimanzi, F. , Lapointe, H.R. , Sang, Y. , Agafitei, O. , Cheung, P.K. , Ennis, S. et al. (2022) Reduced magnitude and durability of humoral immune responses to COVID‐19 mRNA vaccines among older adults. J Infect Dis 225, 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyel, J.F. , Twyman, R.M. and Fischer, R. (2017) Very‐large‐scale production of antibodies in plants: The biologization of manufacturing. Biotechnol. Adv. 35, 458–465. [DOI] [PubMed] [Google Scholar]
- Cai, Y. , Zhang, J. , Xiao, T. , Peng, H. , Sterling, S.M. , Walsh, R.M. , Rawson, S. et al. (2020) Distinct conformational states of SARS‐CoV‐2 spike protein. Science 369, 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño, J.M. , Alshammary, H. , Tcheou, J. , Singh, G. , Raskin, A. , Kawabata, H. , Sominsky, L. et al. (2021) Activity of convalescent and vaccine serum against SARS‐CoV‐2 Omicron. Nature 602, 682–688. [DOI] [PubMed] [Google Scholar]
- Case, J.B. , Bailey, A.L. , Kim, A.S. , Chen, R.E. and Diamond, M.S. (2020) Growth, detection, quantification, and inactivation of SARS‐CoV‐2. Virology 548, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Wu, D. , Guo, W. , Cao, Y. , Huang, D. , Wang, H. , Wang, T. et al. (2020) Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 130, 2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. (2016) Glycoengineering of plants yields glycoproteins with polysialylation and other defined N‐glycoforms. Proc. Natl. Acad. Sci. U. S. A. 113, 9404–9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. (2022) Development of plant‐made monoclonal antibodies against viral infections. Curr. Opin. Virol. 52, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. and Davis, K.R. (2016) The potential of plants as a system for the development and production of human biologics [version 1; referees: 3 approved]. F1000Research 5(F1000 Faculty Rev), 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Lai, H. , Stahnke, J. , Hurtado, J. , Leuzinger, K. and Dent, M. (2013) Agroinfiltration as an effective and scalable strategy of gene delivery for production of pharmaceutical proteins. Advanced Techniques in Biology & Medicine 1, 1–9. 10.4172/atbm.1000103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.E. , Zhang, X. , Case, J.B. , Winkler, E.S. , Liu, Y. , VanBlargan, L.A. , Liu, J. et al. (2021) Resistance of SARS‐CoV‐2 variants to neutralization by monoclonal and serum‐derived polyclonal antibodies. Nat. Med. 27, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, J. , Thomas, P.G. and Randolph, A.G. (2022) Immunology of SARS‐CoV‐2 infection in children. Nat. Immunol. 23, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotugno, N. , Ruggiero, A. , Bonfante, F. , Petrara, M.R. , Zicari, S. , Pascucci, G.R. , Zangari, P. et al. (2021) Virological and immunological features of SARS‐CoV‐2‐infected children who develop neutralizing antibodies. Cell Rep. 34, 108852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. (2022) A phase 1/2, randomized double‐blind placebo‐controlled trial to test the safety and antiviral activity of ACE2 chewing gum on SARS‐CoV‐2 viral load (COVID 19). https://clinicaltrials.gov/ct2/show/NCT05433181. (Accessed Nov 28, 2022).
- Dent, M. , Hurtado, J. , Paul, A.M. , Sun, H. , Lai, H. , Yang, M. , Esqueda, A. et al. (2016) Plant‐produced anti‐dengue virus monoclonal antibodies exhibit reduced antibody‐dependent enhancement of infection activity. J. Gen. Virol. 97, 3280–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, A. , Harris, B.D. , Martinez‐Sobrido, L. , Kobie, J.J. and Walter, M.R. (2021) Epitope classification and RBD binding properties of neutralizing antibodies against SARS‐CoV‐2 variants of concern. Front. Immunol. 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamos, A.G. , Hunter, J.G.L. , Pardhe, M.D. , Rosenthal, S.H. , Sun, H. , Foster, B.C. , DiPalma, M.P. et al. (2020) High level production of monoclonal antibodies using an optimized plant expression system. Front. Bioeng. Biotechnol. 7, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, J. , Zost, S.J. , Greaney, A.J. , Starr, T.N. , Dingens, A.S. , Chen, E.C. , Chen, R.E. et al. (2021) Genetic and structural basis for SARS‐CoV‐2 variant neutralization by a two‐antibody cocktail. Nat. Microbiol. 6, 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esqueda, A. and Chen, Q. (2023) Producing Biologics with Defined N‐Glycosylation in Plants. In Chemokine‐Glycosaminoglycan Interactions: Methods and Protocols( Lucas, A.R. , ed), pp. 235–250. New York, NY: Springer US. 10.1007/978-1-0716-2835-5_17 [DOI] [PubMed] [Google Scholar]
- Focosi, D. , McConnell, S. , Casadevall, A. , Cappello, E. , Valdiserra, G. and Tuccori, M. (2022) Monoclonal antibody therapies against SARS‐CoV‐2. Lancet Infect. Dis. 22, e311–e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, J.L. , Hamorsky, K. , Khalsa, G. , Matoba, N. and Palmer, K.E. (2015) Bulk production of the antiviral lectin griffithsin. Plant Biotechnology 13, 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Beltran, W.F. , Denis, K.J.S. , Hoelzemer, A. , Lam, E.C. , Nitido, A.D. , Sheehan, M.L. , Berrios, C. et al. (2022) mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell 185, 457–466 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk, P. , Bombardi, R.G. , Erasmus, J.H. , Tan, Q. , Nargi, R. , Soto, C. , Abbink, P. et al. (2020) Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nature Biomedical Engineering 4, 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney, A.J. , Loes, A.N. , Crawford, K.H.D. , Starr, T.N. , Malone, K.D. , Chu, H.Y. and Bloom, J.D. (2021) Comprehensive mapping of mutations in the SARS‐CoV‐2 receptor‐binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney, A.J. , Starr, T.N. , Barnes, C.O. , Weisblum, Y. , Schmidt, F. , Caskey, M. , Gaebler, C. et al. (2021) Mapping mutations to the SARS‐CoV‐2 RBD that escape binding by different classes of antibodies. Nat. Commun. 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell, H. , Vanshylla, K. , Weber, T. , Barnes, C.O. , Kreer, C. and Klein, F. (2022a) Antibody‐mediated neutralization of SARS‐CoV‐2. Immunity 55, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager, K.J. , Marc, G.P. , Gobeil, P. , Diaz, R.S. , Heizer, G. , Llapur, C. , Makarkov, A.I. et al. (2022) Efficacy and safety of a recombinant plant‐based adjuvanted Covid‐19 vaccine. New England Journal of Medicine 386, 2084–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J. , Lai, H. , Esqueda, A. and Chen, Q. (2021) Plant‐produced antigen displaying virus‐like particles evokes potent antibody responses against west nile virus in mice. Vaccine 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Canada . (2022). Health Canada authorizes Medicago COVID‐19 vaccine for adults 18 to 64 years of age. https://www.canada.ca/en/health‐canada/news/2022/02/health‐canada‐authorizes‐medicago‐covid‐19‐vaccine‐for‐adults‐18‐to‐64‐years‐of‐age.html. (Accessed October 25, 2022).
- Heath, P.T. , Galiza, E.P. , Baxter, D.N. , Boffito, M. , Browne, D. , Burns, F. , Chadwick, D.R. et al. (2021) Safety and efficacy of NVX‐CoV2373 Covid‐19 vaccine. New England Journal of Medicine 385, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Arora, P. , Groß, R. , Seidel, A. , Hörnich, B.F. , Hahn, A.S. , Krüger, N. et al. (2021) SARS‐CoV‐2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184, 2384–2393.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Peng, P. , Cao, X. , Wu, K. , Chen, J. , Wang, K. , Tang, N. et al. (2022) Increased immune escape of the new SARS‐CoV‐2 variant of concern Omicron. Cell. Mol. Immunol. 19, 293–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, J. , Zhao, Y. , Ren, J. , Zhou, D. , Duyvesteyn, H.M.E. , Ginn, H.M. , Carrique, L. et al. (2020) Neutralization of SARS‐CoV‐2 by destruction of the prefusion spike. Cell Host Microbe 28, 445–454.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado, J. , Acharya, D. , Lai, H. , Sun, H. , Kallolimath, S. , Steinkellner, H. , Bai, F. et al. (2020) In vitro and in vivo efficacy of anti‐chikungunya virus monoclonal antibodies produced in wild‐type and glycoengineered Nicotiana benthamiana plants. Plant Biotechnol. J. 18, 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski, A. , Giri, A.K. and Aittokallio, T. (2020) SynergyFinder 2.0: visual analytics of multi‐drug combination synergies. Nucleic Acids Res. 48(W1), W488–W493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis, R. and Lund, J. (2002) Interaction sites on human IgG‐Fc for FcγR: current models. Immunol. Lett. 82, 57–65. [DOI] [PubMed] [Google Scholar]
- Jugler, C. , Grill, F.J. , Eidenberger, L. , Karr, T.L. , Grys, T.E. , Steinkellner, H. , Lake, D.F. et al. (2022) Humanization and expression of igg and igm antibodies in plants as potential diagnostic reagents for valley fever. Front. Plant Sci. 13, 925008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugler, C. , Joensuu, J. and Chen, Q. (2020) Hydrophobin‐protein A fusion protein produced in plants efficiently purified an anti‐west nile virus monoclonal antibody from plant extracts via aqueous two‐phase separation. Int. J. Mol. Sci. 21, 2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugler, C. , Sun, H. and Chen, Q. (2021) SARS‐CoV‐2 spike protein‐induced interleukin 6 signaling is blocked by a plant‐produced anti‐interleukin 6 receptor monoclonal antibody. Vaccine 9, 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugler, C. , Sun, H. , Grill, F. , Kibler, K. , Esqueda, A. , Lai, H. , Li, Y. et al. (2022) Potential for a plant‐made SARS‐CoV‐2 neutralizing monoclonal antibody as a synergetic cocktail component. Vaccine 10, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallolimath, S. , Sun, L. , Palt, R. , Stiasny, K. , Mayrhofer, P. , Gruber, C. , Kogelmann, B. et al. (2021) Highly active engineered IgG3 antibodies against SARS‐CoV‐2. Proc. Natl. Acad. Sci. 118, e2107249118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, G. and Milstein, C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497. [DOI] [PubMed] [Google Scholar]
- Ku, Z. , Xie, X. , Davidson, E. , Ye, X. , Su, H. , Menachery, V.D. , Li, Y. et al. (2021) Molecular determinants and mechanism for antibody cocktail preventing SARS‐CoV‐2 escape. Nat. Commun. 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchipudi, V. , Kapur, V. , Jacob, J. , Ghimire, D. , Han, Y. and Lu, M. (2022) Structural plasticity and immune evasion of SARS‐CoV‐2 spike variants. Viruses 14, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R. , Parray, H.A. , Shrivastava, T. , Sinha, S. and Luthra, K. (2019) Phage display antibody libraries: A robust approach for generation of recombinant human monoclonal antibodies. Int. J. Biol. Macromol. 135, 907–918. [DOI] [PubMed] [Google Scholar]
- Lai, H. , Engle, M. , Fuchs, A. , Keller, T. , Johnson, S. , Gorlatov, S. , Diamond, M.S. et al. (2010) Monoclonal antibody produced in plants efficiently treats West Nile virus infection in mice. Proc. Natl. Acad. Sci. 107, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, J. , Ge, J. , Yu, J. , Shan, S. , Zhou, H. , Fan, S. , Zhang, Q. et al. (2020) Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature 581, 215–220. [DOI] [PubMed] [Google Scholar]
- Leuzinger, K. , Dent, M. , Hurtado, J. , Stahnke, J. , Lai, H. , Zhou, X. and Chen, Q. (2013) Efficient agroinfiltration of plants for high‐level transient expression of recombinant proteins. J. Vis. Exp. 77, e50521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, S. (1953) The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3, 285–290. [PubMed] [Google Scholar]
- Loo, Y.‐M. , McTamney, P.M. , Arends, R.H. , Abram, M.E. , Aksyuk, A.A. , Diallo, S. , Flores, D.J. et al. (2022) The SARS‐CoV‐2 monoclonal antibody combination, AZD7442, is protective in non‐human primates and has an extended half‐life in humans. Sci. Transl. Med. 14, eabl8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos, A. and Steinkellner, H. (2014) Plant glyco‐biotechnology on the way to synthetic biology. In Frontiers in Plant Science 5, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L.L. , Suscovich, T.J. , Fortune, S.M. and Alter, G. (2017) Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, E. , Oh, Y.J. , Verbeek, M. , Naude, J. , Ponndorf, D. , Meshcheriakova, Y.A. , Peyret, H. et al. (2020) Co‐expression of human calreticulin significantly improves the production of HIV gp140 and other viral glycoproteins in plants. Plant Biotechnol. J. 18, 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, S. and Tomar, P.C. (2021) Hybridoma technology; advancements, clinical significance, and future aspects. Journal of Genetic Engineering & Biotechnology 19, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, S. , Kwong, A.T. , Holtz, B.R. , Erwin, R.L. , Marcel, S. and McDonald, K.A. (2016) Techno‐economic analysis of a transient plant‐based platform for monoclonal antibody production. MAbs 8, 1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid, M. , Suffiotti, M. , Pellaton, C. , Bouchaab, H. , Cairoli, A. , Salvadé, V. , Stevenel, C. et al. (2022) Humoral Responses Against Variants of Concern by COVID‐19 mRNA Vaccines in Immunocompromised Patients. JAMA Oncol. 8, e220446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono, S. , Zhang, Y. , Ode, H. , Sano, K. , Tan, T.S. , Imai, K. , Miyoshi, K. et al. (2021) SARS‐CoV‐2 D614G spike mutation increases entry efficiency with enhanced ACE2‐binding affinity. Nat. Commun. 12, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, K. E. (2022) PREVENT‐COVID‐19: A Q‐Griffithsin Intranasal Spray, https://clinicaltrials.gov/ct2/show/NCT05122260 (Accessed Oct 25, 2022).
- Piccoli, L. , Park, Y.J. , Tortorici, M.A. , Czudnochowski, N. , Walls, A.C. , Beltramello, M. , Silacci‐Fregni, C. et al. (2020) Mapping neutralizing and immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell 183, 1024–1042.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas, D. , Saunders, N. , Maes, P. , Guivel‐Benhassine, F. , Planchais, C. , Buchrieser, J. , Bolland, W.H. et al. (2021) Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature 602, 671–675. [DOI] [PubMed] [Google Scholar]
- Pramanick, I. , Sengupta, N. , Mishra, S. , Pandey, S. , Girish, N. , Das, A. and Dutta, S. (2021) Conformational flexibility and structural variability of SARS‐CoV2 S protein. Structure 29, 834–845 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y. , Wang, X.M. , Mannan, R. , Pitchiaya, S. , Zhang, Y. , Wotring, J.W. , Xiao, L. et al. (2020) Targeting transcriptional regulation of SARS‐CoV‐2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. U. S. A. 118, e2021450118. 10.1073/PNAS.2021450118/SUPPL_FILE/PNAS.2021450118.SAPP.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanapisit, K. , Shanmugaraj, B. , Manopwisedjaroen, S. , Purwono, P.B. , Siriwattananon, K. , Khorattanakulchai, N. , Hanittinan, O. et al. (2020) Rapid production of SARS‐CoV‐2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci. Rep. 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, T.F. , Zhao, F. , Huang, D. , Beutler, N. , Burns, A. , He, W.T. , Limbo, O. et al. (2020) Isolation of potent SARS‐CoV‐2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, A. , Lee, Y.B. , Wong, S.Y. , Yi, L. , Chai, A. , Lee, S.C. , Lee, M.X. et al. (2022) Efficacy of covid‐19 vaccines in immunocompromised patients: systematic review and meta‐analysis. BMJ 376, e068632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sahly, H.M. , Baden, L.R. , Essink, B. , Doblecki‐Lewis, S. , Martin, J.M. , Anderson, E.J. , Campbell, T.B. et al. (2021) Efficacy of the mRNA‐1273 SARS‐CoV‐2 vaccine at completion of blinded phase. New England Journal of Medicine 385, 1774–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schähs, M. , Strasser, R. , Stadlmann, J. , Kunert, R. , Rademacher, T. and Steinkellner, H. (2007) Production of a monoclonal antibody in plants with a humanized N‐glycosylation pattern. Plant Biotechnol. J. 5, 657–663. [DOI] [PubMed] [Google Scholar]
- Schmidt, F. , Weisblum, Y. , Rutkowska, M. , Poston, D. , da Silva, J. , Zhang, F. , Bednarski, E. et al. (2021) High genetic barrier to SARS‐CoV‐2 polyclonal neutralizing antibody escape. Nature 600, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurink, B. , Roos, E. , Vos, W. , Breur, M. , van der Valk, P. and Bugiani, M. (2022) ACE2 protein expression during childhood, adolescence, and early adulthood. Pediatr. Dev. Pathol. 25, 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugaraj, B. , Rattanapisit, K. , Manopwisedjaroen, S. , Thitithanyanont, A. and Phoolcharoen, W. (2020) Monoclonal antibodies B38 and H4 produced in nicotiana benthamiana neutralize SARS‐CoV‐2 in vitro. Front. Plant Sci. 11, 589995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, R. , Shan, C. , Duan, X. , Chen, Z. , Liu, P. , Song, J. , Song, T. et al. (2020) A human neutralizing antibody targets the receptor‐binding site of SARS‐CoV‐2. Nature 584, 120–124. [DOI] [PubMed] [Google Scholar]
- Siriwattananon, K. , Manopwisedjaroen, S. , Kanjanasirirat, P. , Purwono, P.B. , Rattanapisit, K. , Shanmugaraj, B. , Smith, D.R. et al. (2021) Development of plant‐produced recombinant ACE2‐Fc fusion protein as a potential therapeutic agent against SARS‐CoV‐2. Front. Plant Sci. 11, 604663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stander, J. , Mbewana, S. and Meyers, A.E. (2022) Plant‐derived human vaccines: Recent developments. BioDrugs 36, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, T.N. , Greaney, A.J. , Addetia, A. , Hannon, W.W. , Choudhary, M.C. , Dingens, A.S. , Li, J.Z. et al. (2021) Prospective mapping of viral mutations that escape antibodies used to treat COVID‐19. Science 371, 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, R. , Stadlmann, J. , Schähs, M. , Stiegler, G. , Quendler, H. , Mach, L. , Glössl, J. et al. (2008) Generation of glyco‐engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human‐like N‐glycan structure. Plant Biotechnol. J. 6, 392–402. [DOI] [PubMed] [Google Scholar]
- Sun, H. , Lecio, J. , & Chen, Q. (2023). Development of antibody‐based therapeutics against West Nile virus in plants. In: Nile West Virus: Methods and Protocols ( Bai, F. ed) pp. 211‐225. New York, NY: Springer US. 10.1007/978-1-0716-2760-0_19 [DOI] [PubMed] [Google Scholar]
- Sun, L. , Kallolimath, S. , Palt, R. , Stiasny, K. , Mayrhofer, P. , Maresch, D. , Eidenberger, L. et al. (2021) Increased in vitro neutralizing activity of SARS‐CoV‐2 IgA1 dimers compared to monomers and IgG. Proc. Natl. Acad. Sci. 118, e2107148118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen, J. , van den Brink, E.N. , Poon, L.L.M. , Marissen, W.E. , Leung, C.S.W. , Cox, F. , Cheung, C.Y. et al. (2006) Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 3, e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S. J. , Moreira Edson D., Jr., Kitchin, N., Absalon, J. , Gurtman, A. , Lockhart, S. , Perez, J. L. , Marc, G. P ., Polack, F. P. , Zerbini, C. , Bailey, R , Swanson, K. A. , Xu, X. , Roychoudhury, S. , Koury, K. , Bouguermouh, S. , Kalina, W. V. , Cooper, D. , Robert W. Frenck, Jr , Jansen, K. U. (2021). Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine through 6 months. New England Journal of Medicine, 385, 1761‐1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso, F.Y. , Lidenge, S.J. , Poppe, L.K. , Peña, P.B. , Privatt, S.R. , Bennett, S.J. , Ngowi, J.R. et al. (2021) Presence of antibody‐dependent cellular cytotoxicity (ADCC) against SARS‐CoV‐2 in COVID‐19 plasma. PLOS ONE 16, e0247640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan, L.A. , Errico, J.M. , Halfmann, P.J. , Zost, S.J. , Crowe, J.E. , Purcell, L.A. , Kawaoka, Y. et al. (2022) An infectious SARS‐CoV‐2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 28, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A.C. , Park, Y.J. , Tortorici, M.A. , Wall, A. , McGuire, A.T. and Veesler, D. (2020) Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 181, 281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Chung, C.Y. , Chough, S. and Betenbaugh, M.J. (2018) Antibody glycoengineering strategies in mammalian cells. Biotechnol. Bioeng. 115, 1378–1393. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Guo, Y. , Iketani, S. , Nair, M.S. , Li, Z. , Mohri, H. , Wang, M. et al. (2022) Antibody evasion by SARS‐CoV‐2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Raifu, M. , Howard, M. , Smith, L. , Hansen, D. , Goldsby, R. and Ratner, D. (2000) Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3’ to 5’ exonuclease activity. J. Immunol. Methods 233, 167–177. 10.1016/S0022-1759(99)00184-2 [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Zhang, Y. , Wu, L. , Niu, S. , Song, C. , Zhang, Z. , Lu, G. et al. (2020) Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell 181, 894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Liu, C. , Zhang, C. , Wang, Y. , Hong, Q. , Xu, S. , Li, Z. et al. (2022) Structural basis for SARS‐CoV‐2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat. Commun. 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, E.S. , Gilchuk, P. , Yu, J. , Bailey, A.L. , Chen, R.E. , Chong, Z. , Zost, S.J. et al. (2021) Human neutralizing antibodies against SARS‐CoV‐2 require intact Fc effector functions for optimal therapeutic protection. Cell 184, 1804–1820.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Wang, F. , Shen, C. , Peng, W. , Li, D. , Zhao, C. , Li, Z. et al. (2020) A noncompeting pair of human neutralizing antibodies block COVID‐19 virus binding to its receptor ACE2. Science 368, 1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Wang, Y. , Liu, C. , Zhang, C. , Han, W. , Hong, X. , Wang, Y. et al. (2021) Conformational dynamics of SARS‐CoV‐2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo‐EM. Science Advances 7, eabe5575. 10.1126/SCIADV.ABE5575/SUPPL_FILE/ABE5575_SM.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba, D. , Kosugi, Y. , Kimura, I. , Fujita, S. , Uriu, K. , Ito, J. and Sato, K. (2022) Neutralisation sensitivity of SARS‐CoV‐2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect. Dis. 22, 942–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost, S.J. , Gilchuk, P. , Case, J.B. , Binshtein, E. , Chen, R.E. , Nkolola, J.P. , Schäfer, A. et al. (2020) Potently neutralizing and protective human antibodies against SARS‐CoV‐2. Nature 584, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Neutralization potency (IC90) of p11D7 against various SARS‐CoV‐2 variants. Serial dilutions of p11D7 were mixed with the indicated SARS‐CoV‐2 variant before adding to Vero E6 (WA1/2020 and Delta) or Vero‐hACE2‐TMPRSS2 (Omicron) cells. Cells were then fixed, permeabilized and stained for SARS‐CoV‐2 protein. Foci were counted, per cent neutralization determined and IC90 calculated using GraphPad Prism 9. At least two independent experiments were performed with technical triplicates.


