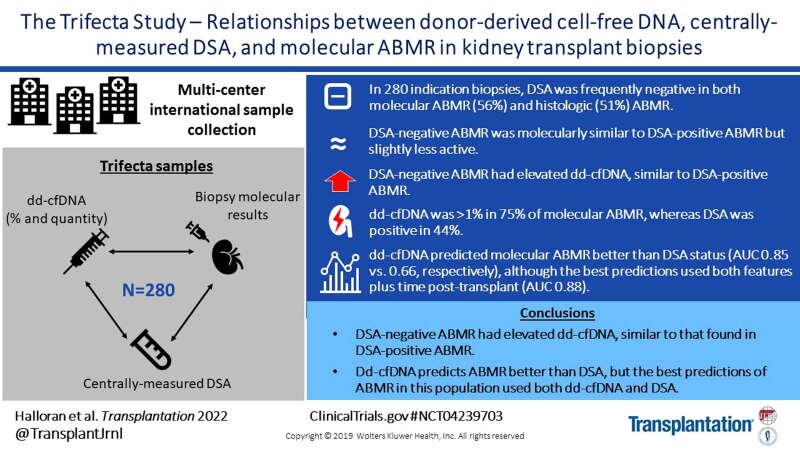
Background.
Trifecta (ClinicalTrials.gov #NCT04239703) is a prospective trial defining relationships between donor-derived cell-free DNA (dd-cfDNA), donor-specific antibody (DSA), and molecular findings in kidney transplant biopsies. Previous analyses of double results showed dd-cfDNA was strongly associated with rejection-associated molecules in the biopsy. The present study analyzed the triple results in 280 biopsies, focusing on the question of dd-cfDNA levels in DSA-negative antibody-mediated rejection (AMR).
Methods.
Molecular Microscope Diagnostic System biopsy testing was performed at Alberta Transplant Applied Genomics Centre, dd-cfDNA testing at Natera, Inc, and central HLA antibody testing at One Lambda Inc. Local DSA and histologic diagnoses were assigned per center standard-of-care.
Results.
DSA was frequently negative in both molecular (56%) and histologic (51%) AMR. DSA-negative AMR had slightly less molecular AMR activity and histologic peritubular capillaritis than DSA-positive AMR. However, all AMRs—DSA-positive or -negative—showed elevated %dd-cfDNA. There was no association between dd-cfDNA and DSA in biopsies without rejection. In AMR, %dd-cfDNA ≥1.0 was more frequent (75%) than DSA positivity (44%). In logistic regression, dd-cfDNA percent (area under the curve [AUC] 0.85) or quantity (AUC 0.86) predicted molecular AMR better than DSA (AUC 0.66). However, the best predictions incorporated both dd-cfDNA and DSA, plus time posttransplant (AUC 0.88).
Conclusions.
DSA-negative AMR has moderately decreased mean molecular and histologic AMR-associated features compared with DSA-positive AMR, though similarly elevated dd-cfDNA levels. In predicting AMR at the time of indication biopsies in this population, dd-cfDNA is superior to DSA, reflecting the prevalence of DSA-negative AMR, but the optimal predictions incorporated both dd-cfDNA and DSA.
INTRODUCTION
Plasma donor-derived cell-free DNA (dd-cfDNA) is elevated by the presence of rejection and injury in organ transplants,1-14 making it of interest as a noninvasive screening test for following transplant recipients. Dd-cfDNA is usually expressed as a fraction of the total cfDNA, but recent analyses indicate that using dd-cfDNA quantity also has value.15,16 Another test widely used to assess risk of rejection is the donor-specific HLA antibody (DSA) to predict antibody-mediated rejection (AMR), but this has been complicated by the recognition of DSA-negative AMR.17-20
We recently launched the Trifecta study to determine the relationships among 3 independent assessments done centrally at the time of the indication biopsy: dd-cfDNA, DSA, and the molecular phenotype of the biopsy as assessed by the Molecular Microscope Diagnostic System (MMDx).21 The initial Trifecta report analyzed the relationship between %dd-cfDNA and molecular biopsy results in the first 300 consecutive biopsies with both measurements.21 The case mix was similar to previous indication biopsy studies: 60% no rejection, 30% AMR, and 10% T cell–mediated rejection (TCMR) or mixed rejection. Dd-cfDNA was strongly related to active molecular rejection in the biopsies. The top genes in the biopsy correlating with %dd-cfDNA had all been previously correlated with AMR activity, including natural killer (NK) cell-expressed genes (eg, GNLY, CCL4, TRDC, and S1PR5) and IFNG-inducible genes (eg, PLA1A, IDO1, CXCL11, and WARS).22-25 The %dd-cfDNA was also high in active TCMR and in early biopsies with acute kidney injury. Multivariate random forests and logistic regression both showed that %dd-cfDNA was more strongly associated with molecular rejection than histologic rejection, confirming a previous study.5
The present study is the first analysis of the Trifecta triple relationships: dd-cfDNA versus DSA versus molecular biopsy findings. We studied 280 biopsies selected from the cohort of 300 previously reported,21 excluding only the 20 biopsies for which central HLA antibody testing could not be assessed. We evaluated the relative ability of DSA and dd-cfDNA blood tests at the time of indication biopsy for predicting molecular AMR in the biopsy. Given that AMR can be DSA-negative,19 we were particularly interested in the frequency of DSA-negative AMR and whether the dd-cfDNA levels were elevated in both DSA-negative and DSA-positive AMR.
Abbreviations and their definitions are shown in Table S1 (SDC, http://links.lww.com/TP/C545).
MATERIALS AND METHODS
Trifecta Study Population
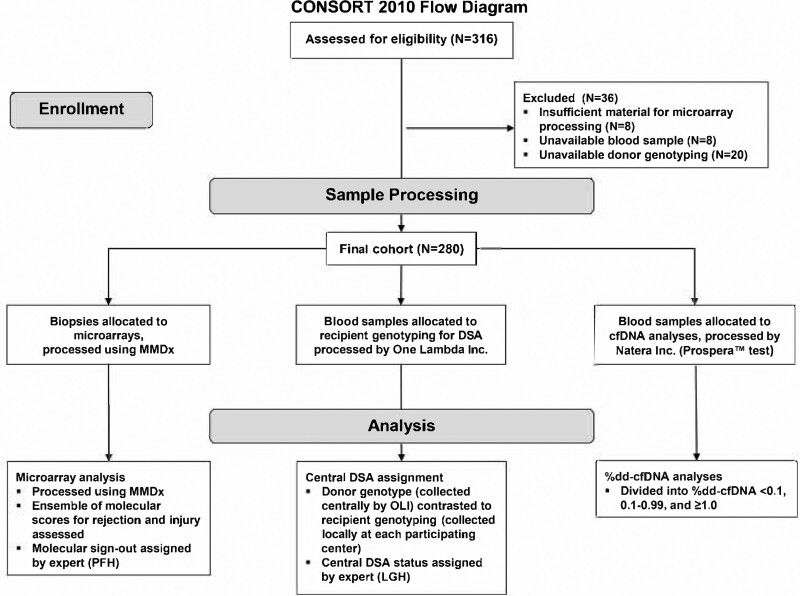
Trifecta (ClinicalTrials.gov #NCT04239703) is a prospective multicenter study of consenting patients involving 41 investigators from 18 transplant institutions (Table S2, SDC, http://links.lww.com/TP/C545) under local institutional review board–approved protocols as previously described.21 Of the 300 biopsies already reported,21 20 had to be excluded from the present report because DSA was not analyzable, leaving 280 with complete triple results. The study features are outlined in Figure 1. Central DSA/panel-reactive antibody (PRA) and dd-cfDNA (both quantity and percentage of total) were measured in blood drawn at time of biopsy per established protocols. Blood for dd-cfDNA was always taken before the biopsy to avoid detecting dd-cfDNA released by the procedure. MMDx results were immediately transmitted to the center. Central DSA and dd-cfDNA results were not made known to the center until many months postbiopsy to avoid influencing the decision to biopsy or patient management, although the centers did have access to their standard-of-care (SOC) DSA test results at the time of local diagnosis assignment.
FIGURE 1.
CONSORT flow diagram of the study design. CONSORT, Consolidated Standards Of Reporting Trials; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; OLI, One Lambda Inc.
Biopsy Sample, Data Collection, and Histologic Diagnoses
A portion of 1 core of each biopsy (mean length 3 mm)26 was immediately stabilized in RNAlater and shipped to the Alberta Transplant Applied Genomics Centre (http://atagc.med.ualberta.ca) at ambient temperature for RNA extraction and processing as previously described.26 Gene expression was measured using Affymetrix PrimeView microarrays (N = 49 495 probe sets). All molecular analyses and diagnoses were made without knowledge of the biopsy’s corresponding histology, clinical data, HLA antibody status, or dd-cfDNA results. MMDx reports were sent to the participating centers, usually within 2 working days of receiving the biopsy.
Of the 280 biopsies, 265 had available local histologic diagnoses assigned by the center (9 missing, 6 with inadequate material for assessment). Histologic and clinical data and DSA testing were collected at each center per SOC as approved by institutional review boards and submitted to the study as available. Central histology review was not permitted because it was not SOC. The histology findings were based on the local SOC opinion, following Banff 2019 guidelines. As detailed in the earlier report, histology diagnoses were entered into a database, interpreted as “no rejection,” “AMR,” “possible AMR,” “TCMR,” “possible TCMR,” and “mixed rejection” (with no knowledge of MMDx results) to compare the molecular and histology results using the same categories.
The Molecular Microscope Diagnostic System
The Molecular Microscope Diagnostic System (MMDx) is a central biopsy-based diagnostic system that measures genome-wide mRNA expression to assign molecular diagnoses. New samples are normalized against and compared with results in a large reference set (N = 1208). Automated output from machine-learning algorithms (classifiers) is used to make observer-independent archetype assignments.27-29 Archetypal analysis automatically assigns scores to each biopsy describing its relationship to each archetype group: no rejection, TCMR1 (active TCMR, often mixed), TCMR2 (less active TCMR), early stage AMR, fully developed AMR, and late-stage AMR, which is often relatively inactive.20,29 Archetypes recognize only the dominant phenotype and do not designate a separate mixed category.
In addition to automatic archetypes, the reports were given MMDx sign-outs by an expert reader based on the ensembles of molecular features without knowledge of DSA or dd-cfDNA: AMR, possible AMR, TCMR, possible TCMR, no rejection, and mixed rejection.
The %dd-cfDNA Assay
All blood samples for dd-cfDNA testing were collected immediately before the biopsy.30 Blood samples were drawn in two 10-mL quantities in DNA Streck tubes using 20- to 21-gauge needles and shipped immediately per established protocols to Natera (Natera Inc., Austin, TX) for analysis using the Prospera test. The Prospera test amplifies DNA by massively multiplexed-PCR targeting 13 926 single nucleotide polymorphisms designed to maximize the number of informative SNPs across ethnicities. This was followed by next-generation sequencing of the resultant amplicons on the Illumina NextSeq 500 on rapid run with an average of 8 million reads per sample.31 Samples were processed using standard operating procedures in the Clinical Laboratory Improvement Amendments–certified laboratory responsible for running the Prospera test.
For all samples, the dd-cfDNA fraction (analyzed as the percentage of total cfDNA, %dd-cfDNA) and quantity (genomic copies per milliliter; cp/mL) were measured. Most analyses in this study focused on the %dd-cfDNA, either as continuous numbers or using cutoffs (the historical diagnostic cutoff of ≥1%). However, because recent studies have suggested that actual dd-cfDNA quantity may also be diagnostically useful,15,32 in logistic regression, we assessed the predictive ability of the quantitative dd-cfDNA measurement (copies per mL), as well as %dd-cfDNA. All samples were analyzed, irrespective of time posttransplant and other potential confounders (eg, cancer), and thus the predictions are not intended to simulate the clinical results when a variety of exclusions are applied.
Central HLA Antibody Measurements and DSA Interpretation
Serum samples for HLA antibody testing by One Lambda Inc (OLI) were collected at the time of biopsy and shipped per standard protocols to OLI for centralized testing. HLA antibodies were tested using Luminex single antigen beads (OLI, Canoga Park, CA) according to the manufacturer’s recommendations. Donor and recipient HLA genotypes were provided by each center. HLA antibody test results were interpreted as DSA by a single expert (L.G.H.), blinded to other results.
HLA antibody specificities were interpreted as positive using a mean fluorescence intensity (MFI) threshold of 500. DSA was generally identified when the beads corresponding to the donor HLA type had ≥500 MFI. However, assignment of HLA antibody specificities did not depend solely on MFI cutoffs, rather they were assigned based on additional criteria, including patterns of epitope reactivity, avoidance of nonspecific bead reactivities, and assay background. Samples negative for all HLA antibodies were labeled PRA-negative (and by definition DSA-negative); samples positive for HLA antibodies but negative for DSA were labeled as DSA-negative-PRA-positive. Biopsies from PRA-positive patients with missing/unavailable donor phenotyping to assign DSA status were called PRA–high risk (PRAHR) and were analyzed here as DSA-positive.
As SOC, HLA antibody studies were also performed in each center and recorded. These results were available in the local centers at the time of the histology diagnoses.
Statistics
All analyses used version 4.1.0 of R.33 Logistic regression was used to predict AMR/mixed, with DSA (±), time of biopsy posttransplant, %dd-cfDNA, and donor cfDNA quantity as predictors. The latter 3 had skewed distributions and were therefore log-transformed for all analyses. Likelihood ratio tests were used to compare nested models. Akaike’s information criterion (AIC) is an estimate of model fit and is used to compare the relative quality of various predictive models. AICs were used to compare nonnested models (lower values are better, and differences of >2 are significant).
RESULTS
Demographics
Demographics of the 280 biopsies (Table S3, SDC, http://links.lww.com/TP/C545) were similar to those published for the published 300 biopsy cohort from which they were selected.21
Central DSA-negative Assessments Agree With Local DSA-negative Assessments
For 170 biopsies, DSA was tested locally as SOC. Agreement between central and local DSA-negativity calls was strong (Table 1): of 116 called DSA-negative locally, 113 of 116 (97%) were called DSA-negative centrally. Thus, the DSA-negative status was robust in 2 independent assessments.
TABLE 1.
Comparing local and central DSA and PRA in the 280-biopsy cohort
| Local DSA assessments | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Central DSA assessments | DSA | Class I | Class I/II | Class II | Positive/class not designated | Negative | Total recorded | Not done/missing | Total |
| Class I | 4 | 1 | 1 | 0 | 0 | 6 | 3 | 9 | |
| Class I/II | 0 | 0 | 5 | 0 | 0 | 5 | 1 | 6 | |
| Class II | 0 | 5 | 14 | 1 | 3 | 23 | 6 | 29 | |
| Negative | 3 | 2 | 13 | 2 | 108 | 128 | 92 | 220 | |
| PRAHRa | 0 | 1 | 2 | 0 | 5 | 8 | 8 | 16 | |
| Total | 7 | 9 | 35 | 3 | 116 | 170 | 110 | 280 | |
Shading indicates row and column totals.
Includes PRAHR. Biopsies from PRA-positive patients with missing/unavailable donor phenotyping to assign DSA status were called PRAHR in this study.
DSA, donor-specific antibody; PRA, panel-reactive antibody; PRAHR, panel-reactive antibody–high risk.
From this point on, “DSA” and “PRA” refer only to central test results.
Approximately Half of All AMR Is DSA-negative
There were 220 DSA-negative and 60 DSA-positive patients at the time of biopsy. Table 2 categorizes the biopsies by diagnosis. By MMDx sign-outs, automatically- assigned archetype groups, and histology diagnoses, DSA was increased in AMR compared with no rejection. However, 56% of MMDx sign-out AMRs, 63% of AMR-related archetypes, and 51% of histology AMRs were DSA-negative. Therefore, at least half of all AMRs in this prospective study were DSA-negative.
TABLE 2.
Diagnoses in 280 biopsies grouped by DSA status
| Biopsy groups | Number of biopsies | ||
|---|---|---|---|
| DSA-negative (N = 220) | DSA-positivea (N = 60) | Total | |
| MMDx diagnoses | |||
| No rejection | 147 | 17 | 164 |
| Possible TCMR | 6 | 0 | 6 |
| TCMR | 15 | 5 | 20 |
| Possible AMR | 7 | 3 | 10 |
| AMR | 36 | 25 | 61 |
| Mixed (AMR + TCMR) | 9 | 10 | 19 |
| All AMR (including mixed) (% of row total) | 45 | 35 | 80 |
| Automated rejection (K1208) archetype | |||
| No rejection | 150 | 19 | 169 |
| TCMR1 (many mixed) | 4 | 5 | 9 |
| TCMR2 | 15 | 6 | 21 |
| Early AMR | 21 | 8 | 29 |
| Full AMR | 16 | 20 | 36 |
| Late AMR | 14 | 2 | 16 |
| All AMR | 51 | 30 | 81 |
| Histology diagnoses | |||
| No rejection | 112 | 8 | 120 |
| Possible TCMR | 28 | 1 | 29 |
| TCMR | 22 | 7 | 29 |
| Possible AMR | 16 | 12 | 28 |
| AMR | 23 | 21 | 44 |
| Mixed (AMR + TCMR) | 7 | 8 | 15 |
| All AMR (including mixed) | 30 | 29 | 59 |
| Missing | 12 | 3 | 15 |
Bolding indicates rows with AMR, mixed. Bolding and italics indicate All AMR.
Includes PRAHR. Biopsies from PRA-positive patients with missing/unavailable donor phenotyping to assign DSA status were called PRAHR in this study and were analyzed as DSA-positive.
AMR, antibody-mediated rejection; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; PRA, panel-reactive antibody; PRAHR, panel-reactive antibody–high-risk biopsy, TCMR, T cell–mediated rejection.
DSA-negative AMR Has Lower AMR Activity Than DSA-positive
Table 3 shows the molecular scores for gene sets and classifiers in DSA-negative and DSA-positive AMR, as well as no rejection.
TABLE 3.
Molecular activity scores in 35 DSA-positive vs 45 DSA-negative MMDx AMR/mixed biopsies
| Transcript sets and classifiers | MMDx no rejection(N = 164) | MMDx mixed and AMR (N = 80) | |||
|---|---|---|---|---|---|
| DSA-negative(N = 45) | DSA-positivea(N = 35 ) | DSA-positive vs DSA-negative AMR/mixed(Wilcoxon test P) | |||
| AMR-related | AMR (AMRProb) classifier | 0.06 | 0.53 | 0.64 | 0.05 |
| Glomerular double contours (cg > 0Prob) classifier | 0.13 | 0.44 | 0.57 | 0.05 | |
| Glomerulitis (g > 0Prob) classifier | 0.14 | 0.58 | 0.66 | 0.06 | |
| Peritubular capillaritis (ptc > 0Prob) classifier | 0.13 | 0.66 | 0.78 | 0.004 | |
| DSA probability (DSAProb) classifier | 0.29 | 0.60 | 0.68 | 0.03 | |
| All rejection | Rejection (RejProb) classifier | 0.07 | 0.66 | 0.80 | 0.005 |
| TCMR-related | TCMR (TCMRProb) classifier | 0.02 | 0.09 | 0.15 | 0.37 |
| Interstitial infiltrate (i > 1Prob) classifier | 0.05 | 0.24 | 0.27 | 0.38 | |
| Tubulitis (t > 1Prob) classifier | 0.05 | 0.17 | 0.24 | 0.60 | |
| Macrophage-related | Constitutive macrophage-associated transcripts (QCMAT) | 0.32 | 0.74 | 0.76 | 0.67 |
| Alternative macrophage activation transcripts 1 (AMAT1) | 0.43 | 0.97 | 1.07 | 0.16 | |
| Recent injury | Injury/repair associated transcripts (human kidney) (IRRAT30) | 0.32 | 0.68 | 0.60 | 0.67 |
| Injury-repair induced transcripts, d 3 (IRITD3) | 0.04 | 0.16 | 0.13 | 0.95 | |
| Normal parenchymal | Kidney transcripts – set 1 (KT1) | –0.28 | –0.47 | –0.48 | 0.67 |
| Atrophy fibrosis | Interstitial fibrosis (ci > 1Prob) | 0.35 | 0.53 | 0.61 | 0.18 |
| Tubular atrophy (ct > 1Prob) | 0.29 | 0.45 | 0.52 | 0.31 | |
Bold indicates P < 0.05; bold and underline indicates P < 0.01.
Shading indicates rows with AMR-related variables.
Includes PRAHR biopsies. Biopsies from PRA-positive patients with missing/unavailable donor phenotyping to assign DSA status were called PRAHR in this study and were analyzed as DSA-positive.
AMR, antibody-mediated rejection; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; PRA, panel-reactive antibody; PRAHR, panel-reactive antibody–high risk, TCMR, T cell–mediated rejection.
All AMR-related gene sets and classifier scores were high in both DSA-positive and DSA-negative AMR compared with no rejection. However, despite being high in both, all AMR-related scores were slightly but significantly lower in DSA-negative AMR than in DSA-positive AMR. Scores related to TCMR, macrophages, recent injury, normal parenchyma, and atrophy-fibrosis were low in AMR and not different between DSA-negative and DSA-positive AMR. Additionally, DSA-negative AMR had lower histologic AMR activity, as represented by less peritubular capillaritis (lower ptc-score; Table S4, SDC, http://links.lww.com/TP/C545). These findings are similar to the findings in the INTERCOMEX study in a different population.20
DSA Positivity Is Associated With Higher %dd-cfDNA in the Whole Population
Positive DSA was associated with higher %dd-cfDNA levels across the whole 280-biopsy population: geometric mean of 0.45% in DSA-negative versus 1.28% in DSA-positive (P = 3.7 × 10–7).
dd-cfDNA Is High in Both DSA-negative AMR and DSA-positive AMR
Table 4 shows the %dd-cfDNA in DSA-negative versus DSA-positive biopsies, broken down by diagnosis (MMDx sign-outs, archetype group assignments, or histology diagnoses). Regardless of the diagnostic method, %dd-cfDNA was increased in both DSA-negative and DSA-positive AMR. The %dd-cfDNA was numerically higher in DSA-positive AMR than DSA-negative AMR (difference not significant).
TABLE 4.
Mean %dd-cfDNA in 280 biopsies grouped by DSA status
| Biopsy groups (AMR and mixed are bold) | Geometric mean %dd-cfDNA | P DSA-negative vs DSA-positiveb | ||
|---|---|---|---|---|
| DSA-negative (N = 220) | DSA-positivea(N = 60) | All (N = 280) | ||
| MMDx diagnoses | ||||
| No rejection | 0.31 | 0.40 | 0.32 | 0.40 |
| Possible TCMR | 0.20 | – | 0.20 | – |
| TCMR | 0.36 | 3.78 | 0.65 | 0.0004 (only 5 DSA-positive TCMR) |
| Possible AMR | 0.45 | 0.76 | 0.53 | 0.60 |
| AMR | 1.77 | 2.11 | 1.90 | 0.55 |
| Mixed (AMR + TCMR) | 1.77 | 1.88 | 1.83 | 0.90 |
| All AMR (including mixed) | 1.77 | 2.04 | 1.88 | 0.56 |
| Automated rejection (K1208) archetype | ||||
| No rejection | 0.31 | 0.49 | 0.33 | 0.18 |
| TCMR1 (many mixed) | 2.28 | 2.71 | 2.51 | 0.83 |
| TCMR2 | 0.39 | 2.46 | 0.66 | 0.005 |
| Early AMR | 1.57 | 1.84 | 1.64 | 0.74 |
| Full AMR | 2.29 | 2.08 | 2.17 | 0.79 |
| Late AMR | 0.34 | 0.52 | 0.36 | 0.33 |
| All AMR (including mixed) | 1.22 | 1.94 | 1.46 | 0.08 |
| Histology | ||||
| No rejection | 0.29 | 0.28 | 0.29 | 0.93 |
| Possible TCMR | 0.33 | 1.63 | 0.35 | Only 1 DSA-positive pTCMR |
| TCMR | 0.76 | 1.23 | 0.85 | 0.34 |
| Possible AMRc | 0.82 | 1.97 | 1.19 | 0.11 |
| AMR | 1.11 | 1.41 | 1.24 | 0.49 |
| Mixed (AMR + TCMR) | 1.82 | 3.00 | 2.38 | 0.41 |
| All AMR (including mixed) | 1.25 | 1.74 | 1.47 | 0.27 |
| Inadequate | 1.08 | – | 1.08 | – |
| Missing | 0.38 | 0.77 | 0.48 | 0.59 |
Bolding indicates rows with AMR and Mixed rejection, bolding and underlining indicate rows with all AMR.
Includes panel-reactive antibody–high risk (PRAHR) biopsies. Biopsies from PRA-positive patients with missing/unavailable donor phenotyping to assign DSA status were called PRAHR in this study and were analyzed as DSA-positive.
bTwo-tailed Welch t tests on logged %dd-cfDNA values.
Transplant glomerulopathy was included in possible AMR.
AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; PRA, panel-reactive antibody; TCMR, T cell–mediated rejection.
The %dd-cfDNA was increased in TCMR as previously reported.21 In biopsies with TCMR but no detectable AMR, DSA-positive TCMR did not have significantly higher %dd-cfDNA than DSA-negative TCMR, although there was a numerical trend. However, the relatively small number of TCMR biopsies limits the conclusions.
In biopsies with no rejection, DSA positivity was not associated with significantly increased mean %dd-cfDNA.
Details of Relationships Between DSA and dd-cfDNA
In Table 5, the rows stratify %dd-cfDNA at 1.0 and 0.1, and the columns show the DSA and PRA details. Dd-cfDNA percent ≥1.0 was more common in DSA-positive biopsies overall. Most instances of DSA positivity were anti-class II. High %dd-cfDNA (≥1.0%) was found in 29 of 44 DSA-positive and 8 of 16 DSA-negative PRAHR (total 37/60 or 62%) versus 67 of 220 (30%) DSA-negative biopsies. Among the 220 DSA-negative biopsies, the fraction of %dd-cfDNA positive was not significantly different in PRA-positive (45/127, 35%) versus PRA-negative (22/93, 24%). Very low %dd-cfDNA was uncommon in DSA-positive—2 of 60 (3%)—compared with DSA-negative—33 of 220 (16%).
TABLE 5.
%dd-cfDNA by DSA/PRA status in N = 280 biopsies
| Biopsies group by %dd-cfDNA | DSA-negative (N=220) | DSA-positive or PRAHRa (N = 60) | ||||||
|---|---|---|---|---|---|---|---|---|
| PRA-negative(N = 93) | PRA-positiveDSA-negative(N = 127) | All DSA-negative(N = 220) | I(N = 9) | II(N = 29) | I/II(N = 6) | PRAHR(N =16) | All DSA-positive or PRAHR(N = 60) | |
| %dd-cfDNA ≥1%(N = 104) | 22 | 45 | 67 | 5 | 21 | 3 | 8 | 37 |
| %dd-cfDNA 0.10–0.99(N = 138) | 57 | 60 | 117 | 3 | 8 | 3 | 7 | 21 |
| %dd-cfDNA <0.1%(N = 38) | 14 | 22 | 36 | 1 | 0 | 0 | 1 | 2 |
Bolding indicates all DSA-negative or all DSA-positive/PRAHR.
Biopsies from PRA-positive patients with missing/unavailable donor phenotyping to assign DSA status were called PRAHR in this study.
dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; PRA, panel-reactive antibody; PRAHR, panel-reactive antibody–high risk.
Visualizing Triple Relationships %dd-cfDNA in DSA-negative AMR
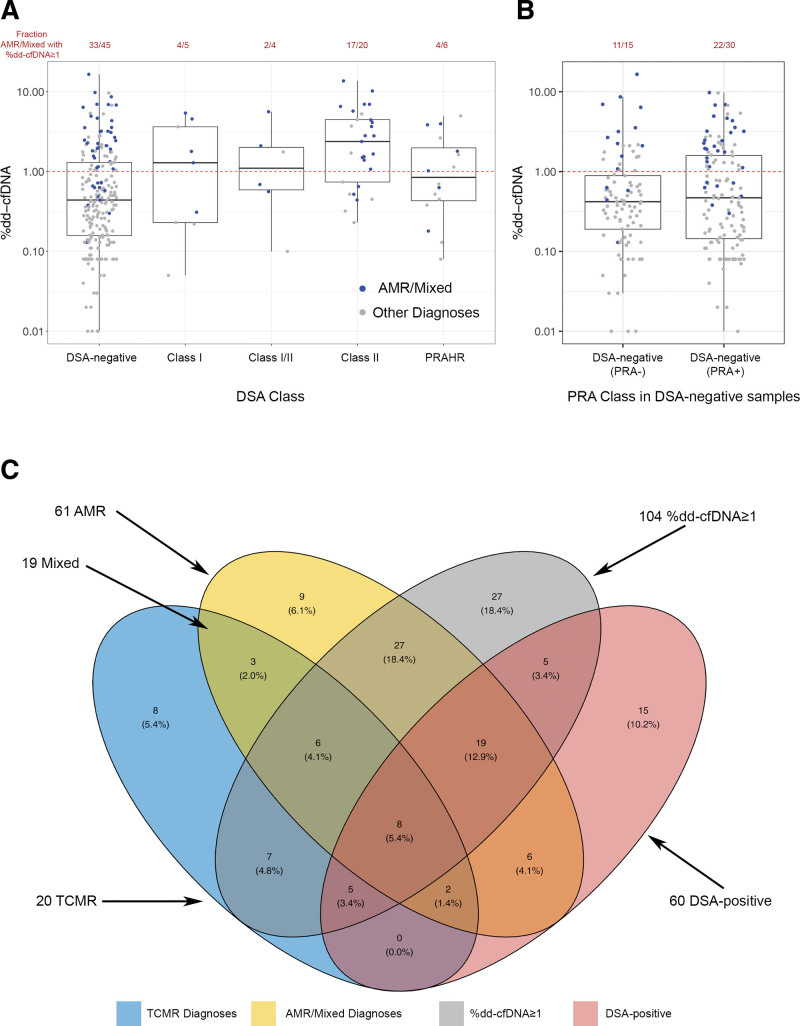
Biopsies are plotted in Figure 2A by their %dd-cfDNA (y-axis) and DSA class/status (x-axis). The %dd-cfDNA ≥1.0 cutoff is shown by the dashed red line, and biopsies called molecular AMR or mixed (by MMDx sign-outs) are represented as blue symbols.
FIGURE 2.
Relationships between MMDx rejection categories, DSA and %dd-cfDNA. A, Boxplots showing the distribution of %dd-cfDNA values across central DSA categories. Boxes represent the interquartile ranges and horizontal lines the medians. B, DSA-negative category split into PRA-positive and PRA-negative categories. C, Venn diagram showing overlap of samples with Molecular Microscope Diagnostic System TCMR, ABMR, %dd-cfDNA ≥1, and DSA positivity. Samples with mixed rejection are in the intersection between AMR and TCMR. One hundred thirty-three of the 280 samples have none of the 4 features and are therefore outside of the ellipses. AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; PRA, panel-reactive antibody; PRAHR, panel-reactive antibody–high risk; TCMR, T cell–mediated rejection.
Of 80 MMDx AMR/mixed biopsies, 60 (75%) had %dd-cfDNA ≥1.0, and 35 (44%) were DSA-positive (including PRAHR as DSA-positive).
Many DSA-negative and DSA-positive AMR/mixed biopsies had high dd-cfDNA. Of 45 DSA-negative AMR/mixed, 33 (73%) had %dd-cfDNA ≥1.0. Of 35 DSA-positive AMR, 27 (77%) had %dd-cfDNA ≥1.0.
DSA-negative AMR patients were usually sensitized: 30 of 45 (67%) were PRA-positive (Figure 2B). However, among DSA-negative AMRs, PRA positivity was not associated with higher dd-cfDNA: 11 of 15 (73%) PRA-negative versus 22 of 30 (73%) PRA-positive had %dd-cfDNA ≥1.0. Table 6 shows that, in DSA-negative AMR/mixed biopsies, the mean %dd-cfDNA level was similar in PRA-positive and PRA-negative biopsies.
TABLE 6.
Relationship of PRA status to %dd-cfDNA in AMR biopsies (including mixed)
| MMDx diagnoses | N | Arithmetic mean %dd-cfDNA | Geometric mean %dd-cfDNA |
|---|---|---|---|
| No rejection | 164 | 0.63 | 0.32 |
| All AMR (including mixed) | 80 | 3.07 | 1.88 |
| DSA-negative AMR (including mixed)a,b | 45 | 2.96 | 1.76 |
| PRA-negative DSA-negative AMR/mixed | 15 | 3.78 | 2.00 |
| PRA-positive DSA-negative AMR/mixed | 30 | 2.54 | 1.67 |
| DSA-positive AMR/mixed (including PRAHR) | 35 | 3.22 | 2.04 |
Shading indicates rows with PRA-negative DSA-negative AMR/mixed or PRA-positive DSA-negative AMR/mixed.
Bolding indicates rows with PRA-negative DSA-negative AMR/mixed or PRA-positive DSA negative AMR/mixed.
Within 45 biopsies called DSA-negative AMR (including mixed), %dd-cfDNA was not significantly different between PRA-positive and PRA-negative biopsies (P = 0.32).
%dd-cfDNA in DSA-negative (2.96) vs DSA-positive (3.22) in 80 MMDx AMR/mixed biopsies is not significantly different (P = 0.70).
AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; PRA, panel-reactive antibody; PRAHR, panel-reactive antibody–high risk.
Figure 2C shows details of the overlaps between the biopsies with MMDx AMR and TCMR, DSA positivity, and %dd-cfDNA ≥1.0.
AMR Was More Frequently dd-cfDNA-Positive Than DSA-positive
Table 7 compares the number (%) DSA-positive to the %dd-cfDNA-positive (≥1.0) biopsies in the 280-biopsy population. As stated above, within AMR/mixed, 44% were DSA-positive, and 75% were %dd-cfDNA-positive. There were more %dd-cfDNA-positive than DSA-positive biopsies in most categories, including in no rejection (18% versus 10%) and in TCMR (60% versus 25%), as expected because the 2 tests reflect different processes.
TABLE 7.
Number (% of 280) of DSA-positive and %dd-cfDNA positive biopsies by MMDx diagnoses
| MMDx diagnoses | Number | Number DSA-positive(% of row) | Number with %dd-cfDNA ≥1(% of row) |
|---|---|---|---|
| No rejection | 164 | 17 (10%) | 29 (18%) |
| Possible TCMR | 6 | 0 (0%) | 1 (17%) |
| TCMR | 20 | 5 (25%) | 12 (60%) |
| Possible AMR | 10 | 3 (30%) | 2 (20%) |
| Pure AMR | 61 | 25 (41%) | 46 (75%) |
| Mixed (AMR + TCMR) | 19 | 10 (53%) | 14 (74%) |
| All AMR including mixed | 80 | 35 (44%) | 60 (75%) |
| All AMR/mixed/TCMR | 100 | 40 (40%) | 72 (72%) |
Bolding indicates rows with pure AMR or Mixed rejection, bolding and italicization indicates rows with all AMR.
AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; TCMR, T cell–mediated rejection.
dd-cfDNA Is Better Than DSA for Predicting Molecular AMR
The DSA and %dd-cfDNA tests are used differently as predictors: DSA is used to assess the probability of AMR, and %dd-cfDNA is used to assess the probability of any active rejection, including TCMR (though it is also affected by AKI20). To compare dd-cfDNA and DSA directly, we used logistic regression to assess their relative ability to predict the same molecular process, namely, MMDx sign-outs of AMR including mixed rejection. (Histologic AMR that cannot be used for DSA is used as the target of the comparison part because the Banff guidelines for AMR use DSA as part of the diagnostic criteria for AMR.) We also included the dd-cfDNA quantity and time of biopsy posttransplant (TxBx) as predictor variables. No additional variables were used for this analysis because the goal was to compare the relative predictive ability of the 4 variables.
Logistic regression models are shown in Table 8. The areas under the curve for the 4 single variables in descending order were dd-cfDNA quantity (0.85), %dd-cfDNA (0.84), DSA status (0.66), and TxBx (0.61). Using AICs to compare models with single predictors, %dd-cfDNA predicted AMR/mixed better than DSA status (AIC 248.2 versus 308.4; smaller AICs are better, and differences >2 are usually considered to be significant). Dd-cfDNA quantity was slightly better than %dd-cfDNA but not significantly so (AICs 246.4 versus 248.2).
TABLE 8.
Predicting molecular antibody-mediated rejection/mixed considering only DSA, %dd-cfDNA, dd-cfDNA quantity, and TxBx
| Predictor variable(s) | P | AUC |
|---|---|---|
| TxBx | 4.4 × 10–3 | 0.61 |
| DSA | 3.2 × 10–8 | 0.66 |
| %dd-cfDNA | <1 × 10–16 | 0.84 |
| Quantity dd-cfDNA | <1 × 10–16 | 0.85 |
| %dd-cfDNA + DSA | <1 × 10–16 | 0.86 |
| %dd-cfDNA + DSA + quantity dd-cfDNA | <1 × 10–16 | 0.87 |
| %cfDNA + DSA + quantity dd-cfDNA + TxBx | <1 × 10–16 | 0.88 |
| Model comparison | P a | Interpretation |
| Quantity dd-cfDNA + %dd-cfDNA vs %dd-cfDNA | 0.007 | Quantity dd-cfDNA adds to %dd-cfDNA |
| %dd-cfDNA + quantity dd-cfDNA vs quantity dd-cfDNA | 0.01 | %dd-cfDNA adds to quantity dd-cfDNA |
| DSA + %dd-cfDNA vs %dd-cfDNA | 9.5 × 10–4 | DSA adds to %dd-cfDNA |
| DSA + %dd-cfDNA vs DSA | <1 × 10–16 | %dd-cfDNA adds to DSA |
| Quantity dd-cfDNA + %dd-cfDNA + DSA vs %dd-cfDNA + DSA | 0.0065 | Quantity dd-cfDNA adds slightly to %dd-cfDNA + DSA |
| TxBx + DSA + quantity dd-cfDNA + %dd-cfDNA vsDSA + quantity dd-cfDNA + %dd-cfDNA | 0.0018 | TxBx adds to DSA + quantity dd-cfDNA + %dd-cfDNA |
Likelihood ratio test.
AUC, area under the curve; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody;TxBx, time of biopsy posttransplant.
Considering models with 2 predictors, %dd-cfDNA added significantly to a model using DSA alone (P < 1 × 10–16). Adding dd-cfDNA quantity to %dd-cfDNA was better than either alone: quantity added more significantly to percent than vice versa (P = 0.007 versus P = 0.01, respectively). Quantity of dd-cfDNA or %dd-cfDNA each added greatly to DSA alone (P < 1 × 10–16 for each). DSA added slightly to %dd-cfDNA alone (P = 0.001). In each of these analyses, increased risk of AMR is associated with higher values of dd-cfDNA (quantity or %), longer time posttransplant, and DSA positivity.
Quantity dd-cfDNA adds slightly but significantly to %dd-cfDNA + DSA. Adding TxBx to quantity dd-cfDNA + %dd-cfDNA + DSA gave the best predictive model.
In summary, in predicting molecular AMR/mixed at the time of indication biopsy, dd-cfDNA (percent, quantity, or both) was superior to DSA alone. DSA added a small but significant predictive value to dd-cfDNA, and the best predictive model used dd-cfDNA quantity and percent, DSA, and time of the biopsy posttransplant.
DISCUSSION
Trifecta is a prospective multicenter study aimed at defining the relationship between blood tests performed at the time of indication biopsy and the findings in the biopsy, with all cases having the same independent, objective, and centrally performed tests. The present analysis addresses the triple relationship, DSA, dd-cfDNA, and molecular AMR in the biopsy, building on the double relationship between %dd-cfDNA and molecular AMR previously established.21 We were particularly interested in the frequency of DSA-negative AMR, its rejection activity relative to DSA-positive AMR, and its dd-cfDNA levels. DSA-negative status was robust in central and local assessment, with central testing confirming 97% of local SOC DSA-negative calls. In 280 biopsies with triple results from the published 300 biopsy cohort,21 about half of AMR was DSA-negative, whether molecular or histologic. AMR activity was slightly lower in DSA-negative than DSA-positive AMR, confirming previous results.20 DSA-negative AMR had high mean %dd-cfDNA, like DSA-positive AMR. The mean %dd-cfDNA was numerically slightly lower than in DSA-positive AMR (not significant), perhaps reflecting the lower AMR activity. In biopsies with AMR, %dd-cfDNA ≥1.0 was more frequently positive than DSA (75% versus 44%). In logistic regression, %dd-cfDNA or dd-cfDNA quantity both predicted AMR better than DSA, but DSA added modestly to predictions by dd-cfDNA alone. These findings are of particular interest given the rigorous trial design: a prospective, consented study with independent central assessments of HLA antibody, dd-cfDNA, and the molecular AMR diagnosis in all cases with no exclusions.
Across the population, dd-cfDNA was higher in DSA-positive cases, but this was because of the association of both with AMR. DSA positivity was not associated with higher dd-cfDNA in biopsies with no rejection.
The fact that elevated dd-cfDNA is a robust feature of DSA-negative, as well as DSA-positive, AMR adds to our understanding that DSA-negative AMR is essentially identical but slightly less active than DSA-positive AMR, as shown in previous studies.19,20,34 This impacts our discussion of the potential mechanisms operating in DSA-negative AMR. Possible explanations for DSA-negative AMR are usually considered to be anti-HLA DSA not detected by the current platforms, failed recognition of DSA in PRA-positive patients due to incomplete donor genotyping, DSA against non-HLA alloantigens, autoantibody, and NK recognition of missing self (discussed below). The explanations may differ in individual cases. Patients with DSA-negative AMR were usually sensitized (30/45 or 67% PRA-positive in the present study), but the PRA status did not affect the dd-cfDNA results, arguing against a major role for failed DSA assignments in PRA-positive patients as an explanation for DSA-negative AMR. Moreover, we offset this risk when genotyping was incomplete by analyzing DSA-negative PRA-positive high-risk biopsies as presumed DSA-positive. We believe that most DSA-negative AMR cases are truly negative for circulating HLA DSA as defined currently, not simply artifacts of incomplete genotyping.
The fact that most DSA-negative AMR patients were allo-sensitized (PRA-positive), and the high association of HLA antibody with risk of AMR in the transplant population, supports the argument that all AMRs—DSA-negative or -positive—reflect an adaptive alloimmune response increased by previous sensitization, that is, probably an alloantibody. The rarity of AMR in HLA identical transplants argues that the antibody is directed against HLA-related polymorphisms. Strong data supporting a role for recognition of “missing self” by NK cells in AMR have been presented,23,35,36 but the present analysis and other available data suggest that DSA-positive and DSA-negative AMRs are molecularly nearly identical.19,20 Missing self-recognition by NK cells probably plays a similar role in DSA-negative and DSA-positive AMR, not selective for DSA-negative AMR. As a working hypothesis to generate concepts for ongoing testing, we propose that most DSA-negative AMR is mediated by an HLA alloantibody not detected by the current platforms, augmented by missing self-recognition, and mechanistically identical in DSA-negative and DSA-positive AMR.
It follows that a key opportunity for improving the performance of DSA as a predictor of AMR is to find predictive antibodies in samples currently considered negative by existing cutoffs. DSA interpretation can differ between local laboratories and experts, with no central “gold standard,”37,38 but the agreement of central with local DSA-negative calls shows that interlaboratory variation cannot explain DSA-negative AMR. It is also possible that classifying the DSA-positive results in terms of de novo, complement binding, titer, immunoglobulin G subclass, and so on would enhance predictions, but Trifecta was not designed to assess these enhancements because there were no standardized criteria by which to judge the results.
The issue of non-HLA alloantibodies and autoantibodies is of great interest39-41 and is currently being addressed in the ongoing Sensitization in Transplantation: Assessment of Risk process.38 We are proceeding with central autoantibody measurements in the ongoing Trifecta study and should be able to clarify this issue in the future. Candidate antibodies to explain DSA-negative AMR must predict the presence of AMR, whether non-HLA alloantibodies or autoantibodies.
We are currently planning to reexamine the DSA-negative samples in the Trifecta study for HLA antibodies missed by the existing algorithms for interpreting Luminex results. In effect, we will see if the output of the HLA antibody assays can be recalibrated in a way that finds more HLA antibodies that predict AMR changes in patients currently called DSA-negative without an excessive number of false positives.
The predictive models at the time of an indication biopsy show that %dd-cfDNA or quantity of dd-cfDNA or both predict AMR activity much better than DSA status alone. This agrees with our calculations that the combination of percentage and quantity of dd-cfDNA offers slightly better predictions of AMR than either alone.16 For clinicians evaluating patients presenting with indications, considering quantity and percent dd-cfDNA and combining both with DSA (and possibly time posttransplant) using predictive models optimize the utility of these tests for the clinician.
Limitations of this study include the recruitment of indication biopsies, as this does not permit extrapolation of the dd-cfDNA or DSA results to patients without indications. Note also that there were relatively few early AMR cases before 3 months posttransplant in the population, suggesting that more of such cases need to be studied. Finally, the Trifecta biopsies are all relatively recent, limiting the availability of outcome data. Outcomes will be collected as the Trifecta study progresses. Validation of these results in a larger dataset is in progress.
It will be of interest to see how dd-cfDNA or DSA adds to clinical management as indices of response to treatment and perhaps even as guides for decisions regarding treatment. Should DSA-negative AMR be treated the same as DSA-positive AMR, and should the dd-cfDNA results impact therapy? The use of dd-cfDNA (quantity and %), as well as DSA, adds a new level of granularity that can potentially guide management of AMR and assess the effectiveness of treatment. For example, the prevalence of DSA-negative AMR that is nevertheless releasing dd-cfDNA suggests that a treatment that simply converts DSA-positive AMR to DSA-negative AMR may not necessarily be beneficial for long-term outcomes if dd-cfDNA levels remain high. On the other hand, rendering the dd-cfDNA negative could emerge as an indicator that AMR activity has been reduced if future outcome studies are supportive. Combining the dd-cfDNA and DSA tests, particularly with enhanced DSA testing, has the potential to advance patient management and inspire a new generation of treatment trials.
ACKNOWLEDGMENTS
The authors thank their valued clinicians in the Trifecta study group who partnered with us for this study by contributing biopsies and feedback. They also thank Dr Martina Mackova and Mrs Anna Hutton for biopsy processing for the microarray biopsy assessment component of this study and Natera, Inc, for blood sample processing and Prospera test results.
The Trifecta Investigators are Justyna Fryc, Beata Naumnik, Jonathan Bromberg, Matt Weir, Nadiesda Costa, Milagros Samaniego-Picota, Iman Francis, Anita Patel, Alicja Dębska-Ślizień, Joanna Konopa, Andrzej Chamienia, Andrzej Więcek, Grzegorz Piecha, Željka Veceric-Haler, Miha Arnol, Nika Kojc, Maciej Glyda, Katarzyna Smykal-Jankowiak, Ondrej Viklicky, Petra Hruba, Silvie Rajnochová Bloudíčkova, Janka Slatinská, Marius Miglinas, Marek Myślak, Joanna Mazurkiewicz, Marta Gryczman, Leszek Domański, Rajendra Balinga, Mahmoud Kamel, Agnieszka Perkowska-Ptasińska, Dominika Dęborska-Materkowska, Michał Ciszek, Magdalena Durlik, Leszek Pączek, Ryszard Grenda, Mirosław Banasik, Mladen Knotek, Ksenija Vucur, Zeljka Jurekovic, Thomas Müller, Thomas Schachtner, Andrew Malone, and Tarek Alhamad.
Supplementary Material
Footnotes
A full list of The Trifecta Investigators is included under ACKNOWLEDGMENTS.
The Trifecta study is an investigator-initiated study supported by a grant from Natera to Transcriptome Sciences Inc/Alberta Transplant Applied Genomics Centre. The Microarray biopsy assessment project is supported in part by a licensing agreement with One Lambda/Thermo Fisher Scientific.
CEL files are available on Gene Expression Omnibus (GSE192444).
P.F.H. reports having shares in Transcriptome Sciences Inc, a University of Alberta research company with an interest in molecular diagnostics, and is a consultant to Natera, Inc. K.S.M-T. is an employee of Transcriptome Sciences Inc. A.P., Z.D., P.G., and P.B. are all employees of Natera Inc., with stocks or options to buy stocks in the company. C.L. and D.L. are employees of One Lambda Inc. L.G.H. is a consultant for AdaptImmune Therapeutics. All remaining authors declare no conflicts of interest and have nothing to disclose.
Trifecta Study Clinical Trial Notation: ClinicalTrials.gov # NCT04239703.
P.F.H. is a principal investigator and contributed in article writing/reviewing, data interpretation, and study design. K.S.M-T. and J.R. contributed in article writing/reviewing, data analysis, and interpretation. P.G., P.B., Z.D., and A.P. contributed in article preparation, discussion of results, measurement of %dd-cfDNA, and interpretation. C.L. contributed in editing the article. D.L. is responsible for the HLA antibody measurements from OLI. L.G.H. interpreted the DSA status for central DSA assessment and edited the text. Trifecta Investigators contributed in biopsy collection and article reviewing.
Supplemental Visual Abstract; http://links.lww.com/TP/C598.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Huang E, Sethi S, Peng A, et al. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am J Transplant. 2019;19:1663–1670. [DOI] [PubMed] [Google Scholar]
- 2.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: results from a prospective observational study. Am J Transplant. 2019;19:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom RD, Bromberg JS, Poggio ED, et al. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigdel TK, Archila FA, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. 2018;8:E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. [DOI] [PubMed] [Google Scholar]
- 6.Agbor-Enoh S, Wang Y, Tunc I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. Ebiomedicine. 2019;40:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharon E, Shi H, Kharbanda S, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput Biol. 2017;13:e1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thongprayoon C, Vaitla P, Craici IM, et al. The use of donor-derived cell-free DNA for assessment of allograft rejection and injury status. J Clin Med. 2020;9:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataria A, Kumar D, Gupta G. Donor-derived cell-free DNA in solid-organ transplant diagnostics: indications, limitations, and future directions. Transplantation. 2021;105:1203–1211. [DOI] [PubMed] [Google Scholar]
- 10.Bloom RD, Augustine JJ. Beyond the biopsy: monitoring immune status in kidney recipients. Clin J Am Soc Nephrol. 2021;16:1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agbor-Enoh S, Shah P, Tunc I, et al. ; GRAfT Investigators. Cell-free DNA to detect heart allograft acute rejection. Circulation. 2021;143:1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinojosa RJ, Chaffin K, Gillespie M, et al. Donor-derived cell-free DNA May confirm real-time response to treatment of acute rejection in renal transplant recipients. Transplantation. 2019;103:e61. [DOI] [PubMed] [Google Scholar]
- 13.Filippone EJ, Farber JL. The monitoring of donor-derived cell-free DNA in kidney transplantation. Transplantation. 2021;105:509–516. [DOI] [PubMed] [Google Scholar]
- 14.Xiao H, Gao F, Pang Q, et al. Diagnostic accuracy of donor-derived cell-free DNA in renal-allograft rejection: a meta-analysis. Transplantation. 2021;105:1303–1310. [DOI] [PubMed] [Google Scholar]
- 15.Bunnapradist S, Homkrailas P, Ahmed E, et al. Using both the fraction and quantity of donor-derived cell-free DNA to detect kidney allograft rejection. J Am Soc Nephrol. 2021;32:2439–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. Combining donor-derived cell-free DNA fraction and quantity to detect kidney transplant rejection using molecular diagnoses and histology as confirmation. Transplantation. [Epub ahead of print. June 29, 2022]. doi:10.1097/TP.0000000000004212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senev A, Coemans M, Lerut E, et al. Histological picture of antibody-mediated rejection without donor-specific anti-HLA antibodies: clinical presentation and implications for outcome. Am J Transplant. 2019;19:763–780. [DOI] [PubMed] [Google Scholar]
- 18.Senev A, Callemeyn J, Lerut E, et al. Histological picture of ABMR without HLA-DSA: temporal dynamics of effector mechanisms are relevant in disease reclassification. Am J Transplant. 2019;19:954–955. [DOI] [PubMed] [Google Scholar]
- 19.Callemeyn J, Lerut E, de Loor H, et al. Transcriptional changes in kidney allografts with histology of antibody-mediated rejection without anti-HLA donor-specific antibodies. J Am Soc Nephrol. 2020;31:2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halloran PF, Madill-Thomsen KS, Pon S, et al. ; INTERCOMEX Investigators. Molecular diagnosis of AMR with or without donor-specific antibody in kidney transplant biopsies: differences in timing and intensity but similar mechanisms and outcomes. Am J Transplan. 2022;22:1976–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. The Trifecta study: comparing plasma levels of donor-derived cell-free DNA with the molecular phenotype of kidney transplant biopsies. J Am Soc Nephrol. 2022;33:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koenig A, Mezaache S, Callemeyn J, et al. Missing self-induced activation of NK cells combines with non-complement-fixing donor-specific antibodies to accelerate kidney transplant loss in chronic antibody-mediated rejection. J Am Soc Nephrol. 2021;32:479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koenig A, Chen CC, Marçais A, et al. Missing self triggers NK cell-mediated chronic vascular rejection of solid organ transplants. Nat Commun. 2019;10:5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venner JM, Hidalgo LG, Famulski KS, et al. The molecular landscape of antibody-mediated kidney transplant rejection: evidence for NK involvement through CD16a Fc receptors. Am J Transplant. 2015;15:1336–1348. [DOI] [PubMed] [Google Scholar]
- 25.Halloran PF, Venner JM, Madill-Thomsen KS, et al. Review: the transcripts associated with organ allograft rejection. Am J Transplant. 2018;18:785–795. [DOI] [PubMed] [Google Scholar]
- 26.Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: the INTERCOMEX study. Am J Transplant. 2017;17:2851–2862. [DOI] [PubMed] [Google Scholar]
- 27.Reeve J, Böhmig GA, Eskandary F, et al. ; INTERCOMEX MMDx-Kidney Study Group. Generating automated kidney transplant biopsy reports combining molecular measurements with ensembles of machine learning classifiers. Am J Transplant. 2019;19:2719–2731. [DOI] [PubMed] [Google Scholar]
- 28.Madill-Thomsen K, Perkowska-Ptasińska A, Böhmig GA, et al. ; MMDx-Kidney Study Group. Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant. 2020;20:1341–1350. [DOI] [PubMed] [Google Scholar]
- 29.Reeve J, Böhmig GA, Eskandary F, et al. ; MMDx-Kidney study group. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight. 2017;2:94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyeso Y, Bhalla A, Smith AP, et al. Donor-derived cell-free DNA kinetics post-kidney transplant biopsy. Transplant Direct. 2021;7:e703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altuğ Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation. 2019;103:2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osmanodja B, Akifova A, Budde K, et al. Absolute or relative quantification of donor-derived cell-free dna in kidney transplant recipients: case series. Transplant Direct. 2021;7:e778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The R Foundation. The R project for statistical computing. Available at http://www.r-project.org/. Accessed March 25, 2022.
- 34.Halloran PF, Famulski KS, Chang J. A Probabilistic approach to histologic diagnosis of antibody-mediated rejection in kidney transplant biopsies. Am J Transplant. 2017;17:129–139. [DOI] [PubMed] [Google Scholar]
- 35.Callemeyn J, Senev A, Coemans M, et al. Missing self-induced microvascular rejection of kidney allografts: a population-based study. J Am Soc Nephrol. 2021;32:2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callemeyn J, Lamarthée B, Koenig A, et al. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022;101:692–710. [DOI] [PubMed] [Google Scholar]
- 37.Reed EF, Rao P, Zhang Z, et al. Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA. Am J Transplant. 2013;13:1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tambur AR, Campbell P, Claas FH, et al. Sensitization in transplantation: assessment of risk (STAR) 2017 working group meeting report. Am J Transplant. 2018;18:1604–1614. [DOI] [PubMed] [Google Scholar]
- 39.Jackson AM, Wiebe C, Hickey MJ. The role of non-HLA antibodies in solid organ transplantation: a complex deliberation. Curr Opin Organ Transplant. 2020;25:536–542. [DOI] [PubMed] [Google Scholar]
- 40.Jackson AM, Delville M, Lamarthée B, et al. Sensitization to endothelial cell antigens: unraveling the cause or effect paradox. Hum Immunol. 2019;80:614–620. [DOI] [PubMed] [Google Scholar]
- 41.Jackson AM, Kuperman MB, Montgomery RA. Multiple hyperacute rejections in the absence of detectable complement activation in a patient with endothelial cell reactive antibody. Am J Transplant. 2012;12:1643–1649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.