Summary
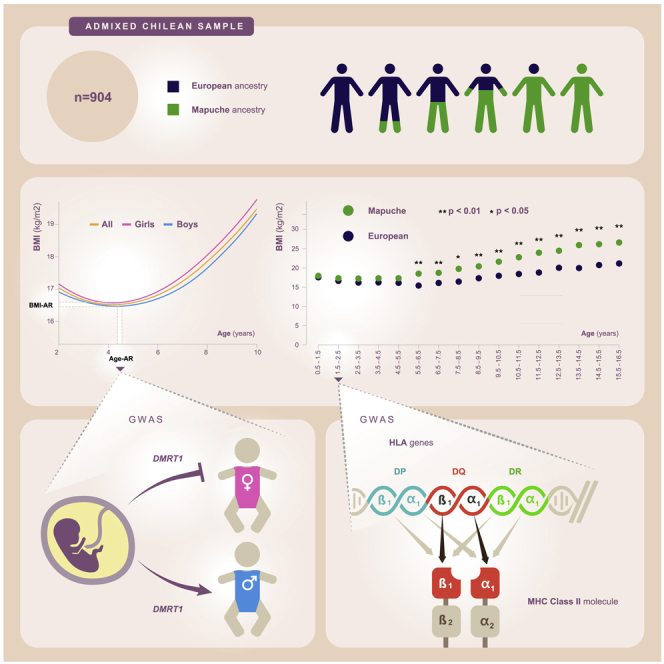
Body-mass index (BMI) is a hallmark of adiposity. In contrast with adulthood, the genetic architecture of BMI during childhood is poorly understood. The few genome-wide association studies (GWAS) on children have been performed almost exclusively in Europeans and at single ages. We performed cross-sectional and longitudinal GWAS for BMI-related traits on 904 admixed children with mostly Mapuche Native American and European ancestries. We found regulatory variants of the immune gene HLA-DQB3 strongly associated with BMI at years old. A variant in the sex-determining gene DMRT1 was associated with the age at adiposity rebound (Age-AR) in girls (P). BMI was significantly higher in Mapuche than in Europeans between 5.5 and 16.5 years old. Finally, Age-AR was significantly lower (P) by 1.94 years and BMI at AR was significantly higher (P) by 1.2 kg/, in Mapuche children compared with Europeans.
Subject areas: Health sciences, Human genetics, Genomics
Graphical abstract
Highlights
-
•
The genetics of childhood BMI and how ancestry affects it is poorly understood
-
•
BMI is higher but age at adiposity rebounds lower in Mapuche than in European children
-
•
GWAS captured variants of the immune gene HLA-DQB3 associated with BMI in toddlers
-
•
GWAS on age at adiposity rebound captured variant in sex-determining gene DMRT1
Health sciences; Human genetics; Genomics
Introduction
Childhood obesity is a major public health problem across the world. One well-studied indicator of obesity is the body mass index (BMI), whose alterations in children have been associated with risk of adult obesity, as well as related diseases, including type 2 diabetes1 and cardiometabolic diseases.2 Even though several environmental causes explain an important part of childhood obesity,3,4 there is still a lack of knowledge on the genetic factors underlying susceptibility to this disease.
BMI follows a nonlinear trajectory over time during childhood. In European children, BMI trajectory is characterized by three main phases: (1) a rapid increase with an adiposity peak (AP) at month of age; (2) a decline reaching its lowest value at years, also called the adiposity rebound (AR); and (3) a subsequent peak at early adulthood.5
The heritability of adult obesity is .6 Approximately of the adult BMI heritability is explained by 941 SNPs, according to a genome-wide association study (GWAS) meta-analysis performed in individuals of European ancestry.7 A few BMI GWAS performed on non-European populations such as 173,430 Japanese8 and a trans-ethnic population panel9 identified additional SNPs associated with BMI. Such studies have been mostly performed in adults, but for most of these loci it is unknown whether or not and to which extent they affect BMI in children. Some cross-sectional GWA studies on European populations have shown that several loci influence BMI in adulthood and childhood, whereas other loci seem to act only during adulthood or childhood. For instance, a GWAS found variants in the FTO and MC4R genes significantly associated with BMI in children ( 6 years old) as well as in adults, but a locus in MAF only associated in adults.10 A GWAS meta-analysis found significant associations with FTO, MC4R, TMEM18, SDCCAG8 and TNKS/MSRA loci, but associations for the later gene were limited to children and adolescents.11 Another GWAS meta-analysis identified 15 loci significantly associated with BMI (they considered a single BMI value at the oldest age between 2 and 10 years). Among them, 12 were previously associated with adult BMI or childhood obesity, suggesting that the 3 remaining loci, located near ELP3, RAB27B and ADAM23 genes, act only during childhood.12 A further GWAS meta-analysis identified 18 SNPs significantly associated with BMI in pediatric cohorts from diverse ancestries pooled together, although most of these loci only reached the genome-wide significance threshold in Europeans when same-ancestry cohorts were analyzed separately. Most of these loci had been previously associated with adult BMI, childhood BMI or childhood obesity.13
A few GWAS on European children have identified loci affecting BMI in distinct phases of child growth. A recent GWAS meta-analysis identified 4 loci significantly associated with childhood BMI-related traits: one LEPR/LEPROT gene variant associated with BMI at AP (BMI-AP); one FTO variant and one TFAP2B variant associated with Age at AR (Age-AR); and a GNPDA2 variant associated with BMI at AR (BMI-AR). Among them, the TFAP2B, GNPDA2 and FTO variants were also associated with adult BMI, whereas the LEPR/LEPROT variant was only associated with a BMI-related trait in the infant phase. The authors suggested that adult BMI-related variants start influencing BMI by the time of AR.5 A candidate gene study on a European multi-cohort found that a FTO locus is positively associated with BMI from 5.5 years onward, but is inversely associated below age 2.5 years old. They also confirmed this finding using a longitudinal linear mixed model.14 Warrington et al., 201515 performed a longitudinal GWAS to identify associations between genotypes and BMI trajectories across childhood. They found 4 loci previously associated with BMI in adults or children as well as a novel locus in theFAM120AOSgene significantly associated with BMI at 8 years of age. To our knowledge, Warrington et al., 2015 is the only longitudinal GWAS on childhood BMI.
As mentioned before, the vast majority of these GWA studies have been performed in Europeans, but we do not know to what extent these loci affect BMI in populations with other continental ancestries, in particular those with Native American ancestry. This is relevant, as obesity and obesity-related disorders are markedly affected by genetic ancestry.6 For instance, Latinos, which usually have mixed European, Native American, and African ancestries, are more susceptible to lipid-related disorders than any other US group, in part because of their Native-American genetic heritage.16
The aims of this study are two-fold: (1) To estimate how much Native American ancestry affects childhood BMI trajectory, Age-AR as well as BMI-AR; and (2) to identify genetic variants associated with these traits.
Results
Estimation of Age-AR and BMI-AR
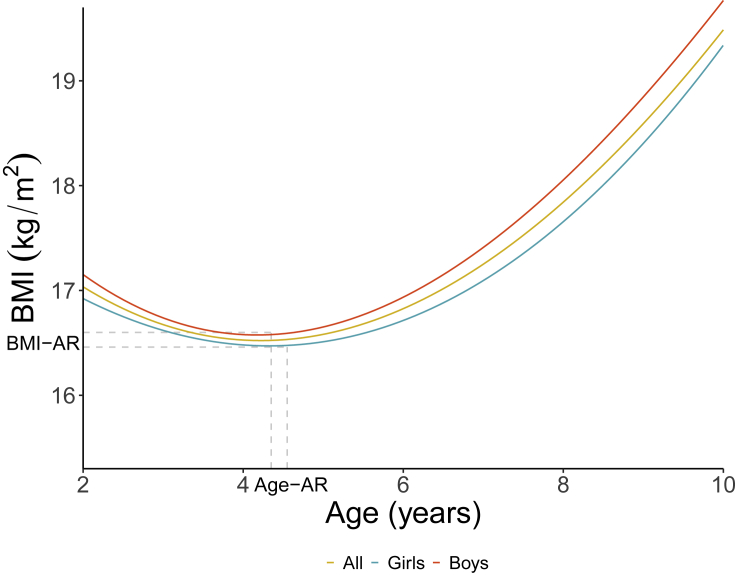
We first characterized BMI trajectories in 904 admixed Chilean children from the “Growth and Obesity Chilean Cohort Study” (GOCS),17 from age 2 years to age 10 years. We implemented longitudinal mixed models (see details in STAR Methods) in boys and girls pooled together as well as separately. Figure 1 shows the expected BMI trajectories for the three analyses. We observed a strong fit between observed and expected curves, as revealed by Pearson’s correlation coefficient of . Using the same longitudinal model, we estimated BMI-AR and Age-AR (see details in STAR Methods). The mean Age-AR was 4.5 (SD = 1.5) years in pooled children, 4.35 (SD = 1.35) years in boys and 4.55 (SD = 1.71) years in girls. The mean BMI-AR was 16.3 (SD = 1.4) kg/ in pooled children, 16.4 (SD = 1.29) kg/ in boys and 16.2 (SD = 1.42) kg/ in girls. Figure S1 shows examples of individual estimations of Age-AR and BMI-AR.
Figure 1.
Estimation of Age-AR and BMI-AR
Fitted BMI (kg/) curves over time (years) in the whole cohort as well as in boys and girls considered separately. Vertical and horizontal dotted lines pinpoint the observed mean Age-AR and mean BMI-AR in boys and girls.
Effect of global ancestry on BMI, Age-AR and BMI-AR
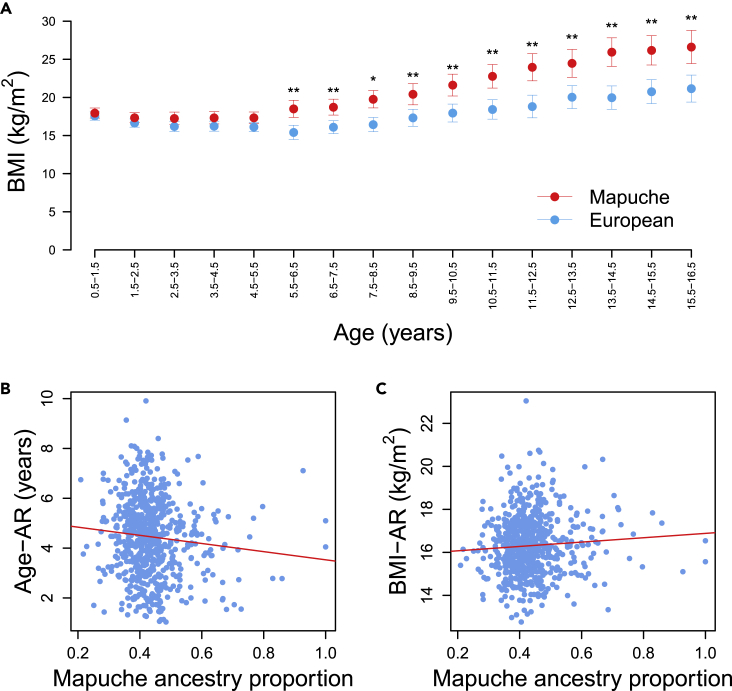
The children of this cohort have on average 52.1% European, 43.8% Mapuche Native American, 2.6% Aymara Native American, and 1.5% African global ancestry proportions18 (Figure S2). To quantify how much Mapuche and European ancestries affect BMI at each age stratum, we applied linear regressions with BMI as the dependent variable and global Mapuche ancestry as the independent variable. We estimated BMI values for hypothetical individuals with Mapuche ancestry and with European ancestry, hereafter referred to as “Mapuche” and “Europeans”. We adjusted for the maternal education level (MEL), as higher Mapuche ancestry proportions have been associated with a lower educational level and a lower socioeconomic status among Chileans.19Table S1 shows the individuals’ counts in each education category. When compared with Europeans, Mapuche individuals have significantly higher BMI at all age strata between 5.5 and 16.5 years old (P at all these strata except at 7.5–8.5 years old, where P; Wald test), but not before (Figure 2A and Data S1). Figure S3 shows that this effect is consistent for individuals grouped by different MEL categories (see details in STAR Methods). Furthermore, BMI differences between Mapuche and European individuals tend to increase with age (Figures 2A and S3). Similarly, global ancestry had a mild but significant effect in the whole longitudinal BMI trajectory (p = 0.045; effect size = 0.094; Wald test), as revealed by the mixed-effects model (see details in STAR Methods).
Figure 2.
Effect of global Mapuche and European ancestry on BMI trajectory, Age-AR and BMI-AR
(A) BMI (kg/) estimated at each age stratum for hypothetical individuals with 100% Mapuche (red dots) and 100% European global ancestry (blue dots). SE bars are included. ∗P; ∗∗P.
(B) Linear model of Age-AR with Mapuche ancestry and maternal educational level (MEL) as covariates. MEL Group 1 was chosen arbitrarily to illustrate this effect because all MEL categories behave similarly (Figure S3).
(C) Linear model of BMI-AR with Mapuche ancestry and MEL as covariates.
We explored whether individual Mapuche and European global ancestries have an effect on Age-AR and BMI-AR. When adjusting for MEL, our model predicted that Age-AR is significantly lower by 1.94 years in a Mapuche child (mean = 3.4 years) than in a European child (mean = 5.3 years; p = 0.004; Wald test; Figure 2B). In contrast, BMI-AR was significantly higher by 1.2 kg/ in a Mapuche child (mean = 17.0 kg/) than in a European child (mean = 15.8 kg/; p = 0.04; Wald test; Figure 2C). MEL did not have a significant effect on BMI at different age strata, Age-AR or BMI-AR.
Identification of variants associated with BMI
We performed GWAS on BMI at each age stratum by performing linear regression models (see details in STAR Methods; S2 Table shows the number of individuals in each stratum). Besides gender and age, we adjusted for the global ancestry proportion of each individual to account for population substructure as well as for local ancestry to account for the ancestry of each haplotype (i.e., SNP). Figures S4–S18 show the corresponding Manhattan plots. Figures S19 and S20 show the corresponding QQ-plots.
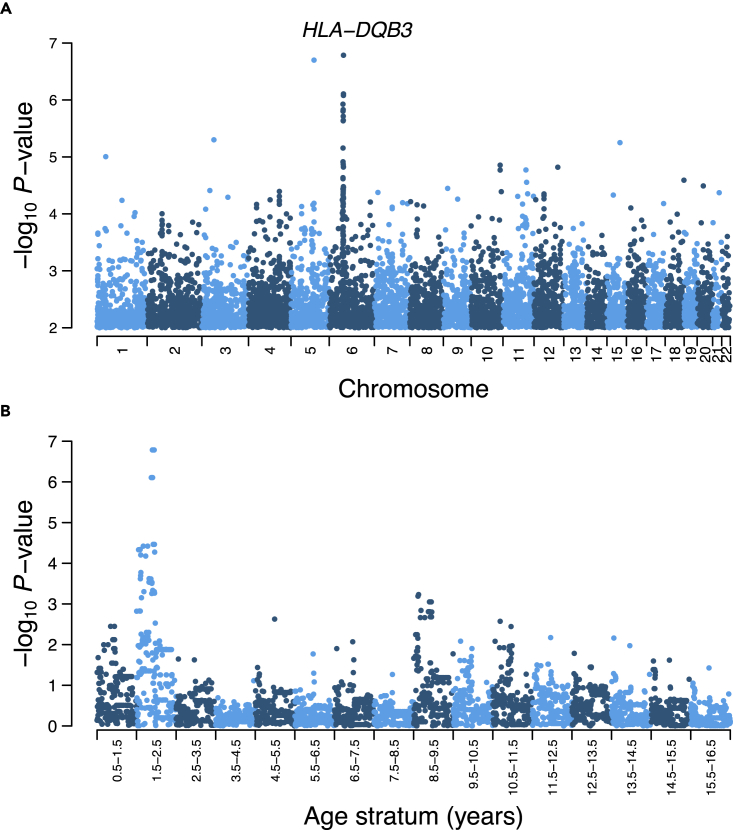
The strongest association was achieved by the intergenic variant rs269511 at the 1.5–2.5-year-old stratum (P; Table 1). The second strongest association (P) was achieved by a genetic signal at chromosome 6 harboring 3 variants in high linkage disequilibrium, as revealed by a clear peak in the Manhattan plot of Figure 3A. This genetic signal was apparent in the yearsold stratum, but not in other strata (Figure 3B). The strongest variants of this peak, namely, rs9275582, rs9275593 and rs9275595, map a promoter flanking region of the HLA-DQB3 pseudogene, which is part of the human leukocyte antigen (HLA) region (see discussion). We identified additional association peaks at chromosomes 4 and 20 (Figure S21). The strongest peak variant at chromosome 4, rs12501266, maps the gene SORCS2, whereas the strongest peak variant at chromosome 20, rs474169, maps the gene SNAP25-AS1. Of note, rs474169 is also associated with BMI at age stratum (Table 1). In contrast to the HLA peak, peak variants at chromosomes 4 and 20 show associations across several strata during childhood and/or adolescence (Figure S21). Table 1 shows the top 10 associated SNPs.
Table 1.
Top cross-sectional associations for BMI.
| SNP ID-Allele | Location | Consequence | Gene | -GT | P-GT | Age stratum | Frequency |
|---|---|---|---|---|---|---|---|
| rs269511-G | 5:114,688,671 | intergenic | – | −0.27 | 2.2E-7 | 1.5–2.5 | 0.4646 |
| rs9275582-T | 6:32,770,853 | promoter flanking region | HLA-DQB3 | 0.30 | 2.5E-7 | 1.5–2.5 | 0.2865 |
| rs9275593-A | 6:32,771,630 | promoter flanking region | HLA-DQB3 | 0.30 | 2.5E-7 | 1.5–2.5 | 0.2865 |
| rs9275595-C | 6:32,772,142 | promoter flanking region | HLA-DQB3 | 0.30 | 2.5E-7 | 1.5–2.5 | 0.2865 |
| rs7134291-A | 12:14,128,322 | intron | GRIN2B | −0.33 | 3.0E-7 | 8.5–9.5 | 0.1726 |
| rs7896870-C | 10:127,827,684 | intron | ADAM12 | 0.25 | 4.0E-7 | 0.5–1.5 | 0.4906 |
| rs474169-T | 20:10,081,800 | intron/NCT | SNAP25-AS1 | −0.29 | 4.8E-7 | 10.5–11.5 | 0.3031 |
| rs1495271-T | 15:101,962,723 | intron | PCSK6 | 0.26 | 6.0E-7 | 4.5–5.5 | 0.4060 |
| rs11244839-A | 10:127,834,046 | intron | ADAM12 | 0.25 | 6.7E-7 | 0.5–1.5 | 0.4751 |
| rs13257360-A | 8:6,117,252 | upstream | RP11-124B13.1 | 0.28 | 7.2E-7 | 14.5–15.5 | 0.3335 |
Shown are the top 10 associated variants. SNP rs ID with the associated allele, physical location, sequence ontology (SO) consequence type, gene name, effect size of the genotype , association p-value of the genotype (PGT), age stratum and frequency of the associated allele. NCT: noncoding transcript variant.
Figure 3.
Cross-sectional genome-wide associations for BMI
(A) GWAS at age stratum . Manhattan plot showing genome-wide per-SNP association p-values represented as-log10 p-value, along the 22 autosomes. Variants with-log10(p-value) between 0 and 2 are not shown.
(B) Association peak of chromosome 6 across age strata. The region shown has physical coordinates and was centered at the variant with the strongest association.
To assess the robustness of the rs269511 and HLA peak associations, we evaluated whether or not the following variables could affect the results: (1) MEL; (2) Using the first 10 genetic principal components (PCs) of each individual instead of global ancestry proportions, which is a more standard approach to account for population structure20; (3) The possibility that increasing the number of covariates could impact statistical power negatively. The four models are described in detail in (Equation 7), (Equation 8), (Equation 9), (Equation 10).
We observed that the rs269511 association strength essentially did not change in these 4 control regressions (P in analysis 1, P in analysis 2, P in analysis 3 and P in analysis 4). In the case of the 3 HLA peak variants, their association p-values were P in analysis 1, P in analysis 2, P in analysis 3 and P in analysis 4. Local ancestry inference in the HLA region can be challenging because of its high levels of polymorphisms.21 However, we observed that the strength of the associations is maintained when the local ancestry covariate is removed from the regression models (8) and (10). Moreover, we observed that the maternal education level did not have a significant effect in any of the genotype-phenotype associations (Data S1).
To further test the robustness of our associations, we simulated GWAS for different ranges of small p-values. We fed the simulation algorithm with the same number of SNPs, sample size, and parameter values of allele frequency distribution as the real data (see details in STAR Methods). The exponential distribution fit well the observed distribution, except for a slight shift toward frequencies (common variants) in the real data (Figure S22). This suggests that the simulations are well-powered to estimate random associations under the null hypothesis. In the case of the range between P and P, which harbors the strongest associations (Table 1), the simulations yielded 7, 95% CI [5, 8] random associations, whereas our GWAS on real data captured 11 associations (Table S3). This observation supports our hypothesis of non-random GWAS associations. Also, the associations of the top 10 SNPs from Table 1 have a statistical power of 0.73 on average for detecting p-values within the range between P and P (Table S4) (see discussion).
We assessed whether our associated variants have been associated with BMI in adults because there is no publicly available GWAS BMI data from pediatric cohorts. Thus, we mined GWAS data from the 2018 meta-analysis from the GIANT Consortium and the UK Biobank (GIANT-UKBB), which has BMI association results from 694,649 European adults.7 We found that 17 variants with association p-values of P in the GOCS cohort were significantly associated in GIANT-UKBB males and females considered together (Data S2), including the rs9275582 and rs9275595 variants from Table 1. Also, 5 variants were significantly associated in GIANT-UKBB females considered separately (Data S2).
To identify variants associated with the whole BMI trajectory during body growth, we performed a longitudinal GWAS on BMI (see details in STAR Methods). The strongest association was achieved by rs4655426 (P), which is an intron variant of the USH2A gene. Table S5 shows the top 10 associated variants and Data S3 shows the summary statistics of all associations. In addition, we found that 1 variant with an association P in the GOCS cohort was significantly associated in GIANT-UKBB males and females considered together as well as in females considered separately (Data S2).
Identification of variants associated with Age-AR and BMI-AR
To identify variants associated with Age-AR, we implemented a regression model described in the STAR Methods section. We detected one variant significantly associated between Age-AR and the interaction between genotype and female sex (P; Table 2 and Figure S23). This variant, namely rs445398, maps an intron of the DMRT1 gene. DMRT1 is a key gene involved in sex differentiation (see discussion). In addition, we identified 2 variants achieving nominal significance (P; Table 2). Similar to before, we tested the robustness of the associations between Age-AR and the interaction between rs445398 genotypes and female sex, by performing the 4 control regressions described in (Equation 18), (Equation 19), (Equation 20), (Equation 21). We found that the association remained genome-wide significant under these 4 scenarios (P in analysis 1, P in analysis 2, P in analysis 3 and P in analysis 4). Also, MEL did not have a significant effect on any of the genotype-phenotype associations (data not shown). Figure S25 (top panel) shows the QQ-plot of the GWAS on Age-AR.
Table 2.
Strongest associations for Age-AR.
| SNP ID-Allele | Location | Consequence | Gene | -GTxSex | P-GTxSex | Frequency |
|---|---|---|---|---|---|---|
| rs445398-T | 9:954,336 | intron | DMRT1 | 5.81 | 9.8E-9 | 0.165 |
| rs969092-A | 12:42,234,778 | intron/NCT | RP11-630C16.2 | 4.96 | 8.9E-7 | 0.238 |
| rs11066997-T | 12:114,661,524 | intergenic | – | 4.94 | 9.9E-7 | 0.358 |
| rs11070771-G | 15:50,669,890 | intergenic | – | −4.86 | 1.4E-6 | 0.327 |
| rs4775878-T | 15:50,672,420 | intergenic | – | −4.86 | 1.4E-6 | 0.326 |
| rs12542317-T | 8:70,715,243 | intron | SLCO5A1 | −4.81 | 1.9E-6 | 0.326 |
| rs3734266-C | 6:34,823,187 | intron | UHRF1BP1 | 4.66 | 3.9E-6 | 0.106 |
| rs13130318-G | 4:155,538,470 | upstream | FGG | 4.64 | 4.2E-6 | 0.133 |
| rs1397623-T | 4:13,887,783 | intron/NCT | RP11-341G5.1 | −4.60 | 5.2E-6 | 0.458 |
| rs7743724-A | 6:34,725,478 | intron | SNRPC | 4.55 | 6.4E-6 | 0.101 |
Shown are the top 10 associated variants. SNP rs ID with the associated allele, physical position in the chromosome, SO consequence type, gene name, effect size of the interaction between genotype and sex (GTxSex) with the corresponding association p-value (PGTxSex), and the frequency of the associated allele. NCT: noncoding transcript variant.
We found that 5 variants with association P in the GOCS cohort were significantly associated with Age-AR in GIANT-UKBB females alone as well as in males and females considered together (Data S2).
We also performed a GWAS for BMI-AR (see details in STAR Methods). The strongest association was achieved by the interaction between the rs2183606 variant and female sex (P). This variant is located within an intron of the GPC5 gene. Table S6 shows the top 10 associated variants. Figure S24 shows the Manhattan plot. Figure S25 (bottom panel) shows the corresponding QQ-plot. We did not find variants with association p-values of P in the GOCS cohort significantly associated in GIANT-UKBB males and females considered together or in females considered separately.
Discussion
In this study, we investigated the genetic architecture of BMI-related traits on admixed children with mainly Mapuche Native American and European ancestry. There are a few studies that have analyzed the genetic architecture of longitudinal growth traits during childhood and adolescence,10,11,12,13,15 and most of them have been performed in European populations. Previous studies have addressed the effect of Native American ancestry on traits such as adult height.22 However, to our knowledge, Vicuña et al., 202118 is the only study that has quantified the relationship between Native American genetic ancestry and growth traits in children and adolescents. The authors showed that the pubertal age where maximum height growth occurs (i.e., peak height velocity), is significantly older by 0.73 years in Europeans than in Mapuche adolescents on average. We are the first to quantify how Native American ancestry affects childhood growth traits. One of our most important findings is the observation that, when compared with European ancestry, increases in Mapuche ancestry are associated with significant increases in BMI between 5.5 and 16.5-year-old as well as with BMI trajectory between years old. Moreover, the difference in BMI between Mapuche and Europeans increases steadily after 5.5 years old. This makes sense because it is known that the contribution of heritable genetic variation to BMI increases from childhood (4 years) to young adulthood (19 years).23 We did not find significant differences at age stratum years old, possibly because of a lack of statistical power derived from the sample size of our cohort. Another important finding was the significantly lower Age-AR in the Mapuche compared with Europeans on average. Higher adiposity in the Mapuche during childhood could explain in part why this population is particularly susceptible to developing metabolic disorders such as insulin resistance, obesity, cholesterol gallstones, and metabolic syndrome during adulthood because all of these disorders are strongly associated with elevated lipid levels.24 Moreover, Mapuche ancestry among Chileans is distinctly associated with heart diseases, hypertension, and diabetes mellitus.19 Furthermore, the prevalence of type II diabetes and obesity among the Mapuche increased significantly following the change from a rural to an urban lifestyle,25 suggesting that genotype-environment interactions may lead to a higher genetic susceptibility to developing cardiometabolic diseases.
It is unknown whether the effect of Mapuche genetic ancestry on BMI-related traits was originated by random genetic drift in ancient Native American or European populations, by adaptation to selective pressures22,26 or by a combination of adaptation, drift and/or other evolutionary forces. It is also possible that environmental factors could partially contribute to such differences. One of such factors is the maternal education level (MEL). In this same cohort, mothers of boys with high Mapuche ancestry ( Mapuche last names: 4.8% of total boys) have a lower educational level than mothers of boys with low Mapuche ancestry (0 Mapuche last names: of boys),27 which in theory could have an effect on their son’s BMI. However, we did not observe a significant effect of MEL on BMI. Neither do we expect high variation in environmental factors such as nutrition, exposure to pollutants, or incidence of pathogenic diseases among these children because they belong to the same urban district in Santiago and to the same middle-lower socioeconomic group.18
The strongest association detected by our cross-sectional GWAS on BMI was for the intergenic variant allele rs269511-G at age stratum years. The second stronger association was for the rs9275582, rs9275593, and rs9275595 variants, which map a promoter flanking region of the HLA-DQB3 unprocessed pseudogene. Noteworthy, rs9275595 has been previously associated with BMI in adults (P; 0.016 kg/ increase).28 These three variants constitute the top of an association peak observed at age stratum years but not at other age strata. The HLA-DQB3 gene belongs to the human leukocyte antigen (HLA) gene cluster, which harbors hundreds of genes that are fundamental for immune function. HLA genes encode proteins of the major histocompatibility complex, which present antigenic peptides to immune cells, to distinguish between “self” and “non-self” agents.29 It is unknown why these HLA-DQB3 variants have such a distinct effect on BMI at years old, but the observation that they map a promoter flanking region suggests that they affect early BMI by regulating the expression of HLA-DQB3 or additional HLA genes in high linkage disequilibrium with HLA-DQB3. Future studies will be needed to validate this finding. It is worth noting that the HLA region shows high variability among ethnic groups30 and that the HLA-DR/DQ region is the major determinant of susceptibility to childhood type 1 diabetes.31 The strongest association of the longitudinal GWAS on BMI was for rs4655426, an intron variant of the USH2A gene. USH2A variants have been GWAS-associated with diverse phenotypes.32
We identified a variant of the DMRT1 gene significantly associated with Age-AR and the interaction between genotype and female sex. DMRT1 is a hallmark gene involved in sex differentiation by maintaining the fates of testes or ovaries in adult mammals.33 Indeed, deletion or inactivation of DMRT1 in humans causes XY male-to-female sex reversal.34 Thus, it is possible that genetic variation in this gene could affect endocrine mechanisms of infant growth.
The cross-sectional GWAS was well-powered to detect associations with p-values of P, as suggested by two lines of evidence. First, the power calculated for the variants from Table 1 was 0.73 on average (Table S4; see STAR Methods). However, this is probably an overestimate because the effect sizes used were estimated from within the sample, which is subject to winner’s curse. Second, the GWAS simulations indicate that the number of small p-values observed is larger than expected or it is within the expected number at several ranges of p-values (Table S3), including the range of P and P, which harbors the strongest associations (Tables 1 and S3).
Limitations of the study
We could not replicate our findings in an independent cohort because of the lack of longitudinal pediatric growth cohorts with enough longitudinal BMI measurements and relatively high Native American ancestry. To our knowledge, the Salvador-SCAALA cohort from Brazil35 and the COIPIS cohort from Mexico36 are the only pediatric cohorts with genome-wide genotype information and Native American admixture. However, these cohorts present severe limitations for replication. In the Salvador-SCAALA cohort, the mean Native American global ancestry is too low (6%), in contrast to their high mean African and European ancestries (51% and 46%, respectively).35 Also, in this cohort, only a single BMI measurement per individual was taken from a broad age window ( years old). Regarding the COIPIS cohort, although the level of Native American ancestry is relatively high (36%),36 it has a similar constraint, namely, that only a single BMI measurement per individual was taken in children aged years old.
Finally, the cross-sectional GWAS on BMI were affected by multiple testing of the 16 age strata. As always, associations should be considered cautiously, but nevertheless, we are reporting our top findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| GWAS Summary statistics | This study | NHGRI-EBI GWAS Catalog: GCST90132242 |
| Original code | This study | Mendeley Data: https://doi.org/10.17632/66rnn39z43.1 |
| Software and algorithms | ||
| Plink 1.9 | Purcell et al.37 | https://www.coggenomics.org/plink/ |
| RFmix 2.0 | Maples et al.38 | https://github.com/slowkoni/rfmix/blob/master/MANUAL.md |
| Shapeit2 | Delaneau et al.39 | https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html |
| Admixture 1.3.0 | Alexander et al.40 | https://dalexander.github.io/admixture/ |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Susana Eyheramendy (susana.eyheramendy@gmail.com)
Materials availability
This study did not generate new unique reagents.
Method details
Sample collection and genotyping
We analyzed individuals of the “Growth and Obesity Chilean Cohort Study” (GOCS),17 who were genotyped using the Infinium ® Multhi-Ethnic Global BeadChip (Illumina).18 Using Plink 1.9,37 we excluded the samples with call rate 0.98 (18 samples), gender mismatch (10 samples), and 1 sample from each pair of highly related individuals (IBS 0.2). We excluded variants with minor allele frequency (MAF) 0.01, as well as variants that fulfilled at least one of the following conditions: have heterozygous genotypes on the X chromosome in males, call genotypes on the Y chromosome in females, show high heterozigosity ( 3 SD from the mean), have 5% missing genotype data, have duplicated physical positions (one variant was kept from each duplicate pair) and show significant deviations from Hardy–Weinberg equilibrium (P). A-T and C-G transversions were removed to avoid inconsistencies with the reference human genome. Finally, we excluded 25 boys whose last BMI measurement was taken before they were 12 years old. After all these quality controls were applied, we obtained a filtered dataset of 904 individuals and 521,788 autosomal SNPs.
Weight and height measurements
Weight and height measurements were taken since 2006 at Instituto deNutrición y Tecnología de los Alimentos (INTA), Santiago, Chile, on children/adolescents in barefoot and light clothes by a trained dietitian following standardized protocols; ICC for all measurements was .41 Weight and height were measured twice at each individual visit, and the average was considered as the final value. Before and after puberty, weight and height were measured once per year; during puberty they were measured every 6 months. Weight was measured with a portable electronic scale (Seca 770), the precision of 0.1 kg, and height was measured with a portable stadiometer (Harpenden 603) to the nearest 0.1 cm. BMI (kg/) and Z-scores (for years old) were calculated based on the World Health Organization 2007 growth references. Weight and height dataprior to 2006 were retrieved from medical records.
Maternal education
Maternal education level was classified as: incomplete middle school, complete middle school, incomplete high school, complete high school, incomplete technical education, complete technical education, incomplete university education, complete university education, graduate studies, special education, does not know or does not reply, no studies, no classification and other. The count of individuals per category is shown in Table S1. Because at least half of these categories have individuals, all individuals were re-classified into 5 groups. Group 1 (n = 508) reflects basic levels of education. It includes no studies, incomplete middle school and complete middle school. Group 2 (n = 137) lies between middle and high school. It includes incomplete high school. Group 3 (n = 149) includes complete high school, incomplete technical education and complete technical education. Group 4 (n = 73) includes all kinds of university studies, namely, incomplete university education, complete university education and graduate studies. Group 5 (n = 17) includes categories without meaningful information, namely, special education, does not know or does not reply, no studies, no classification and other.
Principal component analysis
PCA was carried out with Plink 1.9,37 only including SNPs with minor allele frequency 0.01 and SNP calling rate 99%. We pruned SNPs using an independent pairwise approach with a window size of 50 kb, a step size of five SNPs, and an r2 cutoff threshold of 0.15. We used the first genetic principal components as covariates for the GWAS.
Local ancestry estimation
We used RFmix.v.2.038 to infer the local ancestry of each SNP allele from our Chilean sample. We used reference populations from the 1000 Genomes Project,42 namely, Yoruba (YRI, n = 108) for the African ancestry, Iberian Populations in Spain (IBS, n = 107) for the European ancestry, Peruvian in Lima Peru (PEL) individuals with Native American ancestry (n = 29). We excluded individuals with SNP missing rate. We inferred the gametic phase of individuals with Shapeit2,39 using the HapMap37 human genome build 37 recombination map. To obtain the local ancestry, we used the –forward-backward parameter, as recommended in the manual. Local ancestry estimation identified three continental ancestries: European, African, and Native American.
Global ancestry estimation
Global ancestry proportions of Chilean children were estimated with Admixture 1.3.040 in unsupervised mode. As reference populations, we used YRI (n = 108) for the African component and IBS (n = 107) for the European component. As reference populations for the Native American component, we used 11 Mapuche43 as well as 73 Aymara.44,45Global ancestry estimation identified four ancestry proportions corresponding to European, African, Aymara and Mapuche. We used the Mapuche proportion ancestry in the GWAS to identify its effect on the phenotypes. We used K = 4 ancestral populations, as this K value better distinguished the two main Native American subcomponents of Chileans, namely, Mapuche and Aymara (Figure S2). The cross-validation errors (CVs) of K = estimated with ADMIXTURE40 are shown in Figure S25. In order to distinguish Peruvian (PEL) individuals with Native American ancestry, we used K = 3 ancestral populations and the reference populations mentioned above.
Variant and gene annotations
Variant annotations [GRCh37 (hg19) assembly] were retrieved with the web tool Variant Effect Predictor (VEP) from Ensembl.46 Upstream and downstream variants were defined as those located 10 Kb upstream or downstream of the gene, respectively. Intergenic variants were defined as those located Kb upstream or downstream of the closest gene. Reported GWAS associations were retrieved from the NHGRI GWAS Catalog.32 When more than one variant in a gene has been associated with the same phenotype, we reported the strongest association.
Quantification and statistical analysis
Derivation of Age-AR and BMI-AR
Before modeling BMI trajectory over time, we applied some filters on the individuals. We excluded 51 individuals who had no sufficient measurements between 2 and 10 years old, as well as 156 individuals with all their measurements taken after 4.5 years old. We obtained a final dataset of 696 individuals with measurements between 2 and 10 years old. To estimate Age-AR, we implemented a longitudinal statistical model of BMI (in logarithm scale) on this filtered data. The model equation is:
| (Equation 1) |
where corresponds to the number of Age/BMI measurements for individual , and represents the gender of the individual (0 female, 1 male). The parameters for are fixed effects whereas the parameters for are random effects for each individual (i.e. ). The errors are assumed independent across the different individuals but dependent between observations for the same individual (index ) and are normally distributed with variance .
The predicted trajectory for individual can then be written as
| (Equation 2) |
where (hat) represents the estimators of the parameters. From this trajectory, the Age of Adiposity rebound (Age-AR) and the BMI at Age-AR are obtained by finding the Age at which the minimum BMI is found.
A local minimum of BMI is found by solving the following equation:
| (Equation 3) |
which has as its solution:
| (Equation 4) |
where the minimum obtained above needs to satisfy:
| (Equation 5) |
To identify BMI-AR, we replace the Age term from Equation 1 by the Age-AR value obtained from Equation 3.
Cross-sectional GWAS on BMI
For the cross-sectional GWAS, we analyzed individuals between 1.5 and 16.5 years old, across 16 strata of 1 year interval each (Table S2). BMI was expressed as kg/ and age in years. Individuals with no weight or height measurements in a particular stratum were removed. When measurement was taken in the same individual and time interval, we took the mean of the BMI measurements. To get a normal distribution of BMI values, we applied Box-Cox transformations ( values for each stratum are shown in Table S2). These transformed values were standardized to get mean = 0 and standard deviation = 1, obtaining zBMI scores.14,15 These zBMI scores were used to test for genome-wide associations. We applied the following linear regression model:
| (Equation 6) |
where represents the effect of the additive genotype (GT) of SNP ; represents the effect of gender (S); represents the effect of the global Mapuche Native American ancestry proportion (GA); and represents the effect of local Native American ancestry, where takes the values 0, 1 or 2 depending on the number of Native American alleles. Of note, while our global ancestry analysis is able to separately estimate the Mapuche and Aymara ancestral Native American subcomponents of Chileans, the local ancestry analysis only considers a general Native American population.
We also ran 4 new regressions between rs269511 genotypes and BMI: 1. With local ancestry, without global ancestry, and without the first 10 genetic PCs (see Equation 7); 2. Without local ancestry, with global ancestry, and without the first 10 genetic PCs (see Equation 8); 3. With local ancestry, without global ancestry, and with the first 10 genetic PCs (see Equation 9); 4. Without local ancestry, without global ancestry, and with the first 10 genetic PCs (see Equation 10). In all of these 4 new analyses, we included MEL as a covariate.
| (Equation 7) |
| (Equation 8) |
| (Equation 9) |
| (Equation 10) |
Power calculation for cross-sectional GWAS on BMI
We estimated the statistical power of the top associations of the cross-sectional GWAS on BMI (Table 1) following a reported method,47 which is based on the effect size of a particular SNP, the sample size N, a number M of SNPs and a significance level = 0.05/M. For example, let us calculate the statistical power of rs269511 at age stratum years old. Using Equation 6, we obtained an empirical effect size of = . We have N = 684 individuals, M = 521,788 SNPs and a significance level = 0.05/M. Using the R script Power-calculation.R available in https://github.com/lucas-vicuna/GWAS-BMI-2022, we obtain a statistical power = 0.655 (Table S4).
GWAS on longitudinal BMI data
To estimate longitudinal genotype-phenotype associations, we implemented a linear mixed model for BMI. We adjusted for gender, age, global ancestry, and local ancestry. We assumed an additive inheritance model. The model equation is as follows:
| (Equation 11) |
where corresponds to the number of Age/BMI measurements for individual , represents the gender of the individual (0 female, 1 male). Hereby, , and represent the random effects, while the remaining terms are fixed effects.
Because mixed models are computationally very expensive, we ran the model from Equation 11 in a subset of the SNPs. These SNPs were selected in the following way. We first ran the model from Equation 11 in a partition of five randomly chosen SNPs from the same chromosome; each subset of SNPs of a chromosome belonging to exactly one of the elements of the partition. In this way, we decreased to 20% the number of mixed models that we needed to estimate. Afterward, we selected only the 1065 SNPs with association p-values smaller than 0.001.
Similar to the cross-sectional GWAS, we evaluated whether the inclusion of MEL, the first 10 genetic principal components (PCs), and the number of covariates had a significant effect on the longitudinal GWAS results. Thus, we ran 4 new mixed models. 1. With local ancestry, without global ancestry, and without the first 10 genetic PCs (see Equation 12); 2. Without local ancestry, with global ancestry, and without the first 10 genetic PCs (see Equation 13); 3. With local ancestry, without global ancestry, and with the first 10 genetic PCs (see Equation 14); 4. Without local ancestry, without global ancestry, and with the first 10 genetic PCs (see Equation 15).
| (Equation 12) |
| (Equation 13) |
| (Equation 14) |
| (Equation 15) |
GWAS on Age-AR and BMI-AR
Similarly, as with BMI, we obtained standardized Age-AR and BMI-AR values by applying Box-Cox transformations followed by standardization. We obtained estimates of = 0.841 for Age-AR and = for BMI-AR. We performed genome-wide associations for standardized Age-AR, using the following linear regression model:
| (Equation 16) |
where represents the interaction effect between gender and genotype; = interaction effect between sex and a third degree polynomial of zBMI-AR. For the GWAS on BMI-AR, we used a similar model:
| (Equation 17) |
Similar to the previous GWAS, we evaluated whether the inclusion of MEL, the first 10 genetic principal components (PCs), and the number of covariates have a significant effect on the Age-AR GWAS results. Thus, we ran 4 new mixed models. 1. With local ancestry, without global ancestry, and without the first 10 genetic PCs (see Equation 18); 2. Without local ancestry, with global ancestry, and without the first 10 genetic PCs (see Equation 19); 3. With local ancestry, without global ancestry, and with the first 10 genetic PCs (see Equation 20); 4. Without local ancestry, without global ancestry, and with the first 10 genetic PCs (see Equation 21).
| (Equation 18) |
| (Equation 19) |
| (Equation 20) |
| (Equation 21) |
GWAS simulations
We simulated the GWAS for the 16 age strata on 521,788 SNPs, mimicking the real data. We estimated the distribution of the observed SNP’s allele frequencies through an exponential distribution with parameter = 3.041907 (Figure S22). Given that we filtered out frequencies , that the exponential distribution takes values between 0 and infinity, and that frequencies run between 0 and 1, in our simulations we discarded all values or . Due to the additive representation of the SNPs, we generated each SNP using a binomial distribution of size 2, with a probability obtained randomly from the exponential fit. For each SNP we ran 16 regressions, with the generated SNP as the independent variable and the observed zBMI as the dependent variable. Each regression yielded an association p-value. The whole process was performed 10 times to create confidence intervals of the mean.
Acknowledgments
This work was supported by the Fondo Nacional de Desarrollo Cientifico y Tecnologico (FONDECYT) [1200146 to S.E., L.V. and E.B.; 1190346 to V.M.; 1150416 and 1150486 to JL.S., 1190801 to C.M. ]. S.E., T.N. and L.V. were additionally supported by the Instituto Milenio de Investigación Sobre los Fundamentos de los Datos (IMFD). D.A. was supported by the UKRIMedical Research Council, grant number MC _UU_ 00002/5. Informed consent from participants was obtained from parents or guardians. Children agreed to participate when they turned 7 years old. This study was approved by the Scientific Ethics Committees of Instituto deNutrición y Tecnología en Alimentos (INTA) and Pontificia Universidad Católica deChile.
Author contributions
S.E. conceived the project. S.E. and L.V. designed experiments. E.B., L.V., T.N., C.M., D.A., and V.L. analyzed the data. A.P. and V.M. collected phenotype data. S.E., JL.S., and JC.G. raised funds for the genotyping data. L.V., S.E., and E.B. wrote the manuscript. All authors critically reviewed and accepted the final version.
Declaration of interests
The authors declare that there is no conflict of interest. The current affiliation of L.V. is the Department of Medicine, Genetics Section, University of Chicago, Chicago, USA.
Published: January 31, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106091.
Supplemental information
Data and code availability
-
•
The GWAS summary statistics have been deposited at the NHGRI-EBI GWAS Catalog server32 and are publicly available as of the date of publication. The accession number is listed in the key resources table. The genetic data used in this study are from adolescents, many of which are less than 18 years old. Thus, we are not allowed to publish or share their raw data.
-
•
All original code has been deposited at Mendeley Data and is publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Can B., Can B. Change in overweight from childhood to early adulthood and risk of type 2 diabetes. N. Engl. J. Med. 2018;378:2537–2538. doi: 10.1056/NEJMc1805984. [DOI] [PubMed] [Google Scholar]
- 2.Sovio U., Kaakinen M., Tzoulaki I., Das S., Ruokonen A., Pouta A., Hartikainen A.L., Molitor J., Järvelin M.R. How do changes in body mass index in infancy and childhood associate with cardiometabolic profile in adulthood? Findings from the Northern Finland Birth Cohort 1966 Study. Int. J. Obes. 2014;38:53–59. doi: 10.1038/ijo.2013.165. [DOI] [PubMed] [Google Scholar]
- 3.Sahoo K., Sahoo B., Choudhury A.K., Sofi N.Y., Kumar R., Bhadoria A.S. Childhood obesity: causes and consequences. J. Fam. Med. Prim. Care. 2015;4:187–192. doi: 10.4103/2249-4863.154628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malacarne D., Handakas E., Robinson O., Pineda E., Saez M., Chatzi L., Fecht D. The built environment as determinant of childhood obesity: a systematic literature review. Obes. Rev. 2022;23:133855–e13411. doi: 10.1111/obr.13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couto Alves A., De Silva N.M.G., Karhunen V., Sovio U., Das S., Taal H.R., Warrington N.M., Lewin A.M., Kaakinen M., Cousminer D.L., et al. GWAS on longitudinal growth traits reveals different genetic factors influencing infant, child, and adult BMI. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw3095. eaaw3095–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stryjecki C., Alyass A., Meyre D. Ethnic and population differences in the genetic predisposition to human obesity. Obes. Rev. 2018;19:62–80. doi: 10.1111/obr.12604. [DOI] [PubMed] [Google Scholar]
- 7.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M., GIANT Consortium Meta-analysis of genome-wide association studies for height and body mass index in 700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akiyama M., Okada Y., Kanai M., Takahashi A., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K., et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017;49:1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Rhodes L., Gong J., Haessler J., Franceschini N., Graff M., Nishimura K.K., Wang Y., Highland H.M., Yoneyama S., Bush W.S., et al. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture using Genomics and Epidemiology (PAGE) Study reveals evidence for multiple signals at established loci. Hum. Genet. 2017;136:771–800. doi: 10.1007/s00439-017-1787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyre D., Delplanque J., Chèvre J.C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proença C., et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 11.Scherag A., Dina C., Hinney A., Vatin V., Scherag S., Vogel C.I.G., Müller T.D., Grallert H., Wichmann H.E., Balkau B., et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felix J.F., Bradfield J.P., Monnereau C., van der Valk R.J.P., Stergiakouli E., Chesi A., Gaillard R., Feenstra B., Thiering E., Kreiner-Møller E., et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum. Mol. Genet. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradfield J.P., Vogelezang S., Felix J.F., Chesi A., Helgeland Ø., Horikoshi M., Karhunen V., Lowry E., Cousminer D.L., Ahluwalia T.S., et al. A trans-ancestral meta-analysis of genome-wide association studies reveals loci associated with childhood obesity. Hum. Mol. Genet. 2019;28:3327–3338. doi: 10.1093/hmg/ddz161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sovio U., Mook-Kanamori D.O., Warrington N.M., Lawrence R., Briollais L., Palmer C.N.A., Cecil J., Sandling J.K., Syvänen A.C., Kaakinen M., et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood:Tthe complex nature of genetic association through growth and development. PLoS Genet. 2011;7:e1001307–e1001313. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warrington N.M., Howe L.D., Paternoster L., Kaakinen M., Herrala S., Huikari V., Wu Y.Y., Kemp J.P., Timpson N.J., St Pourcain B., et al. A genome-wide association study of body mass index across early life and childhood. Int. J. Epidemiol. 2015;44:700–712. doi: 10.1093/ije/dyv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko A., Cantor R.M., Weissglas-Volkov D., Nikkola E., Reddy P.M.V.L., Sinsheimer J.S., Pasaniuc B., Brown R., Alvarez M., Rodriguez A., et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat. Commun. 2014;5:3983–4012. doi: 10.1038/ncomms4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira A., Garmendia M.L., González D., Kain J., Mericq V., Uauy R., Corvalán C. Breast bud detection: a validation study in the Chilean growth obesity cohort study. BMC Wom. Health. 2014;14:96–97. doi: 10.1186/1472-6874-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicuña L., Norambuena T., Miranda J.P., Pereira A., Mericq V., Ongaro L., Montinaro F., Santos J.L., Eyheramendy S. Novel loci and Mapuche genetic ancestry are associated with pubertal growth traits in Chilean boys. Hum. Genet. 2021;140:1651–1661. doi: 10.1007/s00439-021-02290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenzo Bermejo J., Boekstegers F., González Silos R., Marcelain K., Baez Benavides P., Barahona Ponce C., Müller B., Ferreccio C., Koshiol J., Fischer C., et al. Subtypes of Native American ancestry and leading causes of death: Mapuche ancestry-specific associations with gallbladder cancer risk in Chile. PLoS Genet. 2017;13:e1006756. doi: 10.1371/journal.pgen.1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uffelmann E., Posthuma D. Emerging methods and resources for biological interrogation of neuropsychiatric polygenic signal. Biol. Psychiatr. 2021;89:41–53. doi: 10.1016/j.biopsych.2020.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Meyer D., C Aguiar V.R., Bitarello B.D., C Brandt D.Y., Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. doi: 10.1007/s00251-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asgari S., Luo Y., Akbari A., Belbin G.M., Li X., Harris D.N., Selig M., Bartell E., Calderon R., Slowikowski K., et al. A positively selected FBN1 missense variant reduces height in Peruvian individuals. Nature. 2020;582:234–239. doi: 10.1038/s41586-020-2302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois L., Ohm Kyvik K., Girard M., Tatone-Tokuda F., Pérusse D., Hjelmborg J., Skytthe A., Rasmussen F., Wright M.J., Lichtenstein P., Martin N.G. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PLoS One. 2012;7:e30153. doi: 10.1371/journal.pone.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francisco J., Poblete M. Salud y enfermedad en nuestras poblaciones originarias: ¿Qué los hace ser más susceptibles o resistentes a ciertas enfermedades prevalentes? Rev. Chil. Cardiol. 2012;31:129–133. doi: 10.4067/S0718-85602012000200007. [DOI] [Google Scholar]
- 25.Pérez-Bravo F., Carrasco E., Santos J.L., Calvillán M., Larenas G., Albala C. Prevalence of type 2 diabetes and obesity in rural Mapuche population from Chile. Nutrition. 2001;17:236–238. doi: 10.1016/s0899-9007(00)00550-5. [DOI] [PubMed] [Google Scholar]
- 26.Neel J. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”. Am. J. Hum. Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández M., Pereira A., Corvalán C., Mericq V. Precocious pubertal events in Chilean children: ethnic disparities. J. Endocrinol. Invest. 2019;42:385–395. doi: 10.1007/s40618-018-0927-8. [DOI] [PubMed] [Google Scholar]
- 28.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lokki M.L., Paakkanen R. The complexity and diversity of major histocompatibility complex challenge disease association studies. HLA. 2019;93:3–15. doi: 10.1111/tan.13429. [DOI] [PubMed] [Google Scholar]
- 30.Degenhardt F., Wendorff M., Wittig M., Ellinghaus E., Datta L.W., Schembri J., Ng S.C., Rosati E., Hübenthal M., Ellinghaus D., et al. Construction and benchmarking of a multi-ethnic reference panel for the imputation of HLA class I and II alleles. Hum. Mol. Genet. 2019;28:2078–2092. doi: 10.1093/hmg/ddy443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilonen J., Kiviniemi M., Lempainen J., Simell O., Toppari J., Veijola R., Knip M., Finnish Pediatric Diabetes Register Genetic susceptibility to type 1 diabetes in childhood-estimation of HLA class II associated disease risk and class II effect in various phases of islet autoimmunity. Pediatr. Diabetes. 2016;17:8–16. doi: 10.1111/pedi.12327. [DOI] [PubMed] [Google Scholar]
- 32.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S., Ye L., Chen H. Sex determination and maintenance: the role of DMRT1 and FOXL2. Asian J. Androl. 2017;19:619–624. doi: 10.4103/1008-682x.194420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raymond C.S., Parker E.D., Kettlewell J.R., Brown L.G., Page D.C., Kusz K., Jaruzelska J., Reinberg Y., Flejter W.L., Bardwell V.J., et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 1999;8:989–996. doi: 10.1093/hmg/8.6.989. [DOI] [PubMed] [Google Scholar]
- 35.Scliar M.O., Sant'Anna H.P., Santolalla M.L., Leal T.P., Araújo N.M., Alvim I., Borda V., Magalhães W.C.S., Gouveia M.H., Lyra R., et al. Admixture/fine-mapping in Brazilians reveals a West African associated potential regulatory variant (rs114066381) with a strong female-specific effect on body mass and fat mass indexes. Int. J. Obes. 2021;45:1017–1029. doi: 10.1038/s41366-021-00761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costa-Urrutia P., Colistro V., Jiménez-Osorio A.S., Cárdenas-Hernández H., Solares-Tlapechco J., Ramirez-Alcántara M., Granados J., Ascencio-Montiel I.d.J., Rodríguez-Arellano M.E. Genome-wide association study of body mass index and body fat in Mexican-mestizo children. Genes. 2019;10:945. doi: 10.3390/genes10110945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 40.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kain J., Galván M., Taibo M., Corvalán C., Lera L., Uauy R. Evolution of the nutritional status of Chilean children from preschool to school age: anthropometric results according to the source of the data. Arch. Latinoam. Nutr. 2010;60:155–159. [PubMed] [Google Scholar]
- 42.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidal E.A., Moyano T.C., Bustos B.I., Pérez-Palma E., Moraga C., Riveras E., Montecinos A., Azócar L., Soto D.C., Vidal M., et al. Whole genome sequence, variant discovery and annotation in Mapuche-Huilliche Native South Americans. Sci. Rep. 2019;9:2132. doi: 10.1038/s41598-019-39391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindo J., Haas R., Hofman C., Apata M., Moraga M., Verdugo R.A., Watson J.T., Viviano Llave C., Witonsky D., Beall C., et al. The genetic prehistory of the Andean highlands 7000 years BP though European contact. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aau4921. eaau4921–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crawford J.E., Amaru R., Song J., Julian C.G., Racimo F., Cheng J.Y., Guo X., Yao J., Ambale-Venkatesh B., Lima J.A., et al. Natural selection on genes related to cardiovascular health in high-altitude adapted Andeans. Am. J. Hum. Genet. 2017;101:752–767. doi: 10.1016/j.ajhg.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abecasis G. 2010. Center for Statistical Genetics.https://genome.sph.umich.edu/wiki/Power_Calculations:_Quantitative_Traits [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The GWAS summary statistics have been deposited at the NHGRI-EBI GWAS Catalog server32 and are publicly available as of the date of publication. The accession number is listed in the key resources table. The genetic data used in this study are from adolescents, many of which are less than 18 years old. Thus, we are not allowed to publish or share their raw data.
-
•
All original code has been deposited at Mendeley Data and is publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.