Abstract
It was previously demonstrated that the intestinal pathogen Vibrio cholerae could undergo an adaptive stress response known as the acid tolerance response (ATR). The ATR is subdivided into two branches, inorganic ATR and organic ATR. The transcriptional regulator ToxR, while not involved in inorganic ATR, is required for organic ATR in a ToxT-independent manner. Herein, we investigate the effect of organic acid stress on global protein synthesis in V. cholerae and show by two-dimensional gel electrophoresis that the stress response alters the expression of more than 100 polypeptide species. The expression of more than 20 polypeptide species is altered in a toxR strain compared to the wild type. Despite this, ectopic expression of the porin OmpU from an inducible promoter is shown to be sufficient to bypass the toxR organic ATR defect. Characterization of the effect of organic acid stress on ompU and ompT transcription reveals that while ompU transcription remains virtually unaffected, ompT transcription is repressed in a ToxR-independent manner. These transcript levels are similarly reflected in the extent of accumulation of OmpU and OmpT. Possible roles for OmpU in organic acid resistance are discussed.
Vibrio cholerae is the causative agent of the epidemic diarrheal disease cholera. After ingestion by a human host, passage through the gastric acid barrier, and colonization of the small intestine, this gram-negative bacterium produces cholera toxin and a subsequent profuse secretory diarrhea that is the hallmark of cholera (13). It was recently shown that V. cholerae is able to mount an adaptive stress response known as the acid tolerance response (ATR) (17). In addition, the acid-adapted V. cholerae was shown to be more virulent in a murine model of cholera than V. cholerae grown at neutral pH. These results have interesting implications for the V. cholerae ATR in the fitness of this pathogen in an individual host as well as in rapid epidemic spread.
The V. cholerae ATR consists of two branches: inorganic (low pH only) and organic (low pH plus organic acids). While some bacterial factors are required for both inorganic and organic ATR (17–19), some proteins are unique to the separate branches (17). One such protein is ToxR, which is necessary solely for the organic ATR, a division which suggests that different sets of regulatory factors mediate the ATR in response to inorganic versus organic acids. Notably, the toxR defect in organic ATR is toxT independent (17), implicating that additional ToxR-regulated factors are necessary for V. cholerae to mount a productive organic ATR.
ToxR is an inner membrane protein containing a cytoplasmic DNA binding domain that shows extensive similarity to those of the OmpR family of proteins (20). Working in conjunction with another inner membrane protein, ToxS, ToxR is responsible for sensing signals such as pH, temperature, osmolarity, and amino acid concentration in an as-yet undefined manner and then directly and indirectly regulating transcription of at least 17 different genes on the V. cholerae chromosome (reviewed in reference 28). This ToxR regulon has been further subdivided into two separate branches: toxT dependent and toxT independent (5). Within the toxT-dependent branch, ToxR and ToxS act synergistically with a homologous inner membrane signaling complex, TcpP and TcpH, to activate transcription of toxT. toxT encodes a transcriptional activator of the AraC family and is part of the V. cholerae pathogenicity island (8, 12, 14). Once produced, ToxT autoregulates its own expression as well as cholera toxin, the toxin coregulated pilus, and other factors that have been shown to be essential for full virulence of V. cholerae (reviewed in reference 28).
The toxT-independent branch of the toxR regulon includes two outer membrane porins (OMPs), OmpU and OmpT. These two porins are differentially regulated by ToxR in that ompU transcription is induced while ompT transcription is repressed (6, 15). This differential regulation leads to virtually exclusive expression of OmpU in wild-type V. cholerae and virtually exclusive expression of OmpT in a toxR strain when grown under standard laboratory conditions. Previous characterization of these OMPs has shown that OmpU composes 30 to 60% of the total outer membrane protein of V. cholerae, depending on the osmolarity of the growth medium. By analogy to homologous porins in Escherichia coli, both OmpU and OmpT are thought to function as trimers that are held together by hydrophobic interactions (4). The transport specificities of these porins are unknown, but the pore size of the OmpU channel has been shown to be on the order of 1.6 nm and that of OmpT to be smaller (4). In addition, OmpU has been hypothesized to function as a potential adhesion factor for V. cholerae (7, 27, 31, 32), though this has been disputed by other groups (21, 24). Most recently, it has been demonstrated that OmpU is important for survival of V. cholerae upon exposure to bile (24, 25); however, the role of OmpT in the V. cholerae life cycle, whether in the aquatic environment or within the host intestine, has yet to be elucidated.
Herein we investigate the nature of the organic ATR defect exhibited by a toxR strain and show by two-dimensional (2D) gel electrophoresis that while a large number of factors are differentially regulated upon exposure to low pH plus organic acids, only a subset of these factors are toxR regulated. Despite the fact that multiple factors are regulated by toxR upon exposure to low pH plus organic acids, ectopic expression of OmpU in a toxR strain was able to bypass the toxR defect in organic ATR. This suggests that the presence of OmpU in a toxR strain is sufficient to complement the loss of ToxR-regulated functions that are required for organic ATR. In addition, we characterize the expression of ompU and ompT in response to exposure to low pH and organic acids and show that while ompU is virtually unaffected by these changes, ompT expression is further repressed in a toxR-independent manner. This finding suggests that a novel regulator of ompT transcription exists.
MATERIALS AND METHODS
Strain and plasmid construction and growth conditions.
All strains, plasmids, and primers used in this study are listed in Tables 1 and 2. A 3,159-bp fragment carrying ompU was PCR amplified using primers K499 and K500 and was cloned into pBAD30 to create pCAM9. All strains were maintained at −80°C in Luria-Bertani (LB) broth containing 30% glycerol. All strains were grown at 37°C in LB broth. The pH of the medium was adjusted with HCl. Ampicillin (Ap) and streptomycin were used at concentrations of 100 μg ml−1. Induction of pBAD promoters was accomplished using l-arabinose at a concentration of 0.2% and was maintained in all steps subsequent to dilution of overnight cultures. RNA was harvested from strains grown in the following manner: overnight cultures of each test strain were grown in LB broth containing Ap and then diluted 1:150 into 30 ml of fresh medium plus Ap. The diluted cultures were grown with aeration until they reached an optical density (at 600 nm) of 0.16 to 0.20. At this point, cells were pelleted at 5,000 × g for 5 min at room temperature (RT), and the supernatants were removed by aspiration. Cells were resuspended in 1 ml of LB broth, pH 7.0, and then 10 and 90% of the cells were placed into two microcentrifuge tubes. The cells were pelleted at 12,000 × g for 1 min at RT, and the supernatants were removed by aspiration. The 10% cell pellet was resuspended in 1 ml of LB broth, pH 7.0, and the 90% cell pellet in 1 ml of LB broth, pH 5.7, plus organic acids, and these were transferred to culture tubes and grown at 37°C with aeration for 1 h. After 1 h, all of the pH 7.0 cells and half of the pH 5.7 cells were pelleted and then flash frozen in a −80°C isopropanol bath. The remainder of each pH 5.7 culture was resuspended in LB broth, pH 4.5, plus organic acids and was incubated at 37°C for 15 min. These cells were then pelleted and flash frozen as described above. Strains which were exposed to organic acids with LB broth at pH 5.7 or 4.5 were supplemented with 0.075× and 0.1× organic acid cocktail, respectively (1× cocktail was 87 mM acetic acid, 25 mM butyric acid, and 37 mM propionic acid). Cell pellets were then used for collection of total RNA.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| V. cholerae | ||
| C6709-1 | El Tor biotype, SmR | 26 |
| DSM-V468 | C6709-1 ΔtoxR | 17 |
| DSM-V705 | C6709-1 ΔtoxR, pCAM9 | This work |
| Plasmids | ||
| pBAD30 | Expression vector with inducible pBAD promoter | 10 |
| pCAM9 | pBAD30::ompU | This work |
TABLE 2.
Primer DNA sequences
| Primer | Sequence (5′ to 3′) |
|---|---|
| OmpUF | CTCTGAAAGATGGTAAGG |
| OmpUR | TTTTTCTGCGGCTGTGTT |
| OmpTF | ATTGGTTCTGGTTCTTCG |
| OmpTR | TTTGCATTATCTTCTGGA |
| K499 | CGCGGTACCCATAGCAATAACATCCACCAAG |
| K500 | GGAATTCGCTTACGTCGCACAAAAATC |
RNase protection assays and organic ATR.
RNase protection assays (RPAs) were conducted on total RNA isolated from V. cholerae strains AC-V168, DSM-V468, and DSM-V705 as previously described (17). A 287-bp ompU riboprobe and a 344-bp ompT riboprobe template were generated by PCR using Taq polymerase and primers OmpUF and OmpUR and OmpTF and OmpTR, respectively. The amplification products were ligated to pGemT (Promega), proper orientation was confirmed, and riboprobes were synthesized using the Maxiscript kit (Ambion) and 50 μCi of [32P]UTP (NEN) as previously described (17). RPAs were done by using the RPAII kit with 1 μg of RNA as described by the manufacturer (Ambion). The products of RNase protection were separated on 5% denaturing polyacrylamide gels and exposed to phosphor-screens (Kodak). Quantification and peak analysis of bands was conducted using a PhosphorImager and the ImageQuant program (Molecular Dynamics). Acid tolerance assays were conducted as previously described (17).
Protein expression and detection.
V. cholerae total proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% acrylamide gels and stained with Coomassie brilliant blue. Western blotting was performed with rabbit polyclonal antiserum against V. cholerae OmpU by using the ECL detection system (Amersham Pharmacia).
Protein samples used for 2D gel electrophoresis were isolated from strains grown exactly as described above for RNA isolation. The cell pellets were resuspended in 200 μl of osmotic lysis buffer (10 mM Tris [pH 7.4] and 0.3% SDS) containing nuclease (10× stock: 50 mM MgCl2, 100 mM Tris [pH 7.0], 500 μg of RNase A per ml, and 1,000 μg of DNase per ml) and protease inhibitors (100× stock: 20 mM AEBSF, 1 mg of leupeptin per ml, 0.36 mg of E-64 per ml, EDTA, and 5.6 mg of benzamidine per ml). The samples were mixed and allowed to stand on ice for 10 min before adding 200 μl of SDS boiling buffer (5% SDS, 10% glycerol, and 60 mM Tris [pH 6.8]) minus β-mercaptoethanol. Protein concentrations were determined using the bicinchoninic acid assay (30). The entire volume of each sample was lyophilized and then dissolved to 2.0 mg/ml in SDS boiling buffer with β-mercaptoethanol (5%). Two-dimensional electrophoresis was performed according to the method of O'Farrell (22) by Kendrick Labs, Inc. (Madison, Wis.) as follows. Isoelectric focusing was carried out in glass tubes with an inner diameter of 2.0 mm by using 2.0% pH 4 to 8 ampholines (BDH; Hoefer Scientific Instruments, San Francisco, Calif.) for 9,600 V · h. Fifty nanograms of an isoelectric focusing internal standard, tropomyosin protein, with a molecular weight (MW) of 33,000 and pI 5.2 was added to the samples. After equilibration for 10 min in buffer (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, and 0.0625 M Tris [pH 6.8]), the tube gel was sealed to the top of a stacking gel on top of a 10% acrylamide slab gel (0.75 mm thick). SDS slab gel electrophoresis was then carried out for 4 h at 12.5 mA. The slab gels were fixed in a solution of 10% acetic acid–50% methanol overnight. The following proteins (Sigma Chemical Co., St. Louis, Mo.) were added as MW standards to the agarose that was used to seal the tube gel to the slab gel: myosin (MW, 220,000), phosphorylase A (MW, 94,000), catalase (MW, 60,000), actin (MW, 43,000), carbonic anhydrase (MW, 29,000), and lysozyme (MW, 14,000). These standards appear as horizontal lines on the silver-stained 10% acrylamide slab gel. The gel was dried onto filter paper with the acidic edge to the left. Analysis of 2D gels for differentially expressed polypeptides was conducted by eye.
RESULTS
Differential protein expression in response to organic acid challenge.
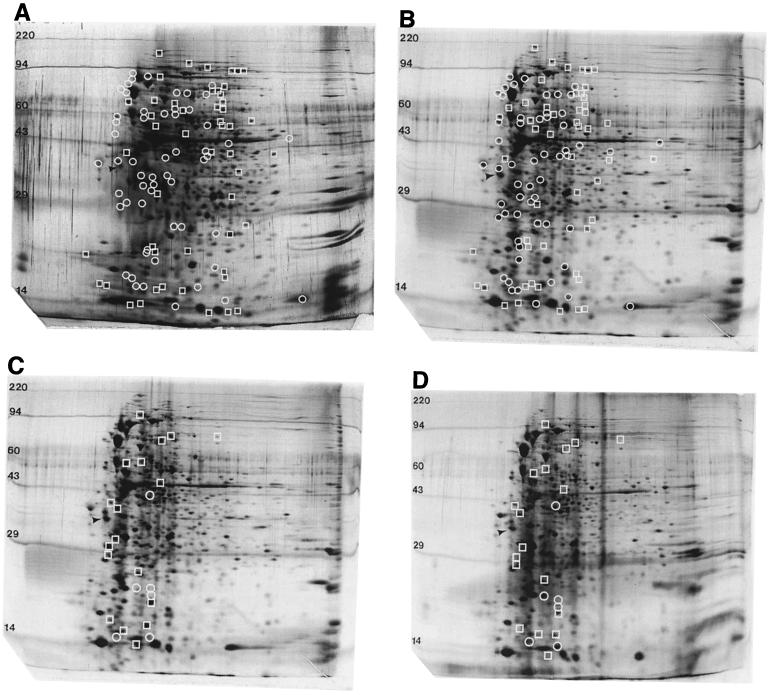
It has been previously shown that an important component of the ATR is the ability to induce the expression of a variety of different acid shock proteins (ASPs) (9). This ability has been studied fairly extensively in Salmonella enterica serovar Typhimurium, and it has been shown that approximately 50 different ASPs are induced upon exposure to acidic pH (9). It was previously demonstrated that the addition of protein synthesis inhibitors to V. cholerae cells blocked the ability of the bacterium to undergo a protective ATR (17), suggesting that, as with Salmonella, synthesis of ASPs is a key component of the V. cholerae ATR. In order to gain a better understanding of the global changes in protein expression that occur specifically when V. cholerae is exposed to organic acid challenge, 2D gel electrophoresis was conducted on total proteins collected from unadapted V. cholerae C6709-1 that had been exposed to pH 7.0 and proteins from adapted V. cholerae exposed to pH 5.7 plus a cocktail of organic acids common to the human intestinal tract (1). Comparison of the unadapted and adapted protein profiles revealed that the relative concentrations of a number of polypeptides is affected by exposure to organic acid stress. Approximately 60 different species were upregulated upon shift to adaptation conditions, showing that V. cholerae is able to induce the expression of a large number of organic ASPs. In addition, approximately 50 polypeptide species were downregulated (Fig. 1A and B).
FIG. 1.
Protein expression of V. cholerae C6709-1 during exposure to pH 7.0 and pH 5.7 plus organic acids. (A) Wild type at pH 7.0; (B and C) wild type at pH 5.7 plus organic acids; (D) ΔtoxR at pH 5.7 plus organic acids. Comparisons were made between panels A and B and panels C and D, respectively, to indicate differentially regulated polypeptide species. Circles indicate proteins whose expression is increased upon exposure to organic acid stress, while squares indicate proteins whose expression is decreased upon exposure to organic acid stress (panel B compared to panel A and panel C compared to panel D). The arrows indicate the internal isoelectric focusing standard, while numbers represent the relevant MWs (in kilodaltons) of polypeptide species.
We reasoned that we might identify the ToxR-regulated component responsible for the previously identified defect in organic ATR exhibited by a toxR strain by identification of differentially regulated polypeptides between the adapted wild type and mutant. Total protein was subsequently collected from the toxR strain DSM-V468 that had been adapted, and a comparison was made to wild-type V. cholerae exposed to identical conditions. To our surprise, 18 different polypeptide species were downregulated or lost in the toxR strain and 6 species were upregulated, indicating that a large number of factors are apparently regulated by ToxR under the conditions tested (Fig. 1C and D). Of note, the tentative identity of only two of these polypeptides, in addition to ToxR itself, could be assigned based on predicted pI and MW of known ToxR-regulated gene products. One of the downregulated species runs at the expected pI and MW of ToxS, which is transcribed within an operon with ToxR. This suggests that the constructed deletion of toxR results in a polar effect on downstream transcription of toxS. In addition, a tentative polypeptide assignment was made which corresponds to AldA, which is an aldehyde dehydrogenase (23). None of the other species correspond to the predicted pI and MW of any of the other known components of the ToxR regulon.
Expression of ompU is sufficient to bypass the defect in organic ATR of a toxR strain.
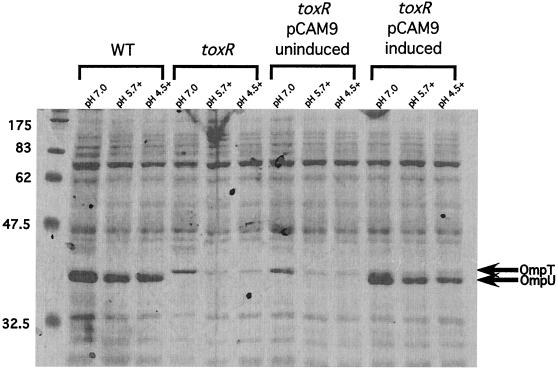
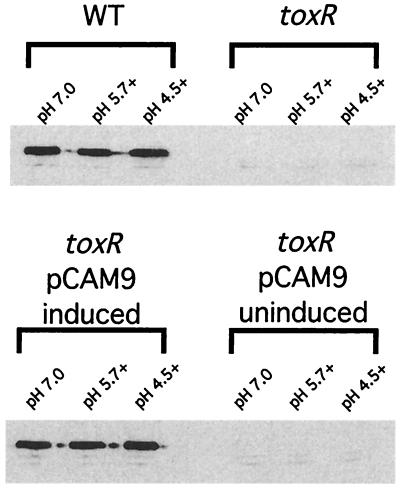
With so many polypeptides exhibiting aberrant regulation in the toxR strain upon exposure to organic acid challenge, we wished to understand which of these and perhaps other polypeptide species not resolved on the 2D gels were required for organic ATR. We first considered the known members of the toxT-independent branch of the ToxR regulon. ToxR positively and negatively regulates the expression of two OMPs, OmpU and OmpT, respectively, which are not resolved by the 2D gels shown in Fig. 1, as they lie outside of the effective pI range of the gels. In wild-type V. cholerae, almost exclusive expression of OmpU is seen, while the same is true of OmpT in a toxR strain (Fig. 2, WT and toxR strains at pH 7.0). Therefore, we reasoned that the previously observed defect in organic ATR might be due to the aberrant regulation of these two OMPs. In order to test this, we moved plasmid pCAM9, which contains the entire ompU coding sequence under the control of an l-arabinose inducible promoter, into the toxR strain DSM-V468, and this strain was designated DSM-V705. DSM-V705 was subsequently shown to express ompU upon induction with l-arabinose (Fig. 2, induced).
FIG. 2.
SDS-PAGE of total proteins from indicated V. cholerae strains. V. cholerae strains were grown at the indicated pHs as described in Materials and Methods. Equivalent optical density at 600 nm units of each strain were pelleted, and cell pellets were resuspended in 3× SDS buffer and boiled for 5 min. Cell debris was subsequently removed by centrifugation, and equivalent volumes of protein sample were separated on an SDS–12% PAGE gel and stained with Coomassie brilliant blue. WT, strain C6709-1; toxR, strain DSM-V468; and toxR pCAM9, strain DSM-V705 grown in the absence (uninduced) or presence (induced) of 0.2% l-arabinose. The relative positions of OmpT and OmpU are indicated by arrows. The presence of organic acids is indicated by a plus adjacent to the adaptation pH.
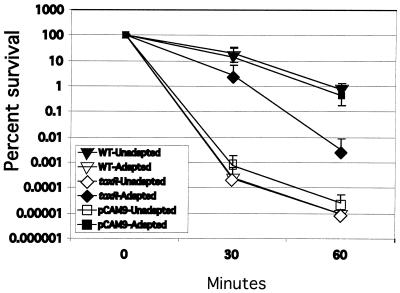
In order to determine the effect of ompU expression upon organic ATR in a toxR strain background, comparative organic ATR assays were conducted with wild-type V. cholerae, DSM-V468, and DSM-V705 expressing ompU. As expected, wild-type V. cholerae was able to mount a robust organic ATR (Fig. 3, compare organic acid-adapted wild type to organic acid-unadapted wild type). DSM-V468, while showing increased survival over organic acid-unadapted wild type, was attenuated in organic ATR, with approximately 100-fold-greater killing of the mutant at the 60-min time point than the wild type. Expression of ompU resulted in survival kinetics virtually identical to those of the wild type (Fig. 3). These results suggest that expression of ompU is sufficient to bypass the previously identified defect in organic ATR of the toxR strain. Of note, unadapted DSM-V705 was killed with similar kinetics to unadapted wild-type cells, suggesting that expression of ompU in the toxR background is not creating a V. cholerae strain that is inherently more resistant to organic acid stress. Instead, this result suggests that ectopically expressed OmpU is functioning within the organic ATR to promote survival.
FIG. 3.
Organic ATR assays of C6709-1 (WT), DSM-V468 (ΔtoxR), and DSM-V705 (ΔtoxR, pCAM9 plus l-arabinose). Strains were organic acid adapted or unadapted, and percent survival was calculated as a function of time after resuspension of bacteria in organic acid challenge medium as described in Materials and Methods. Data represent averages of three separate experiments. Standard deviations are represented by error bars.
ompU transcript and protein expressed ectopically in DSM-V705 resemble wild-type levels.
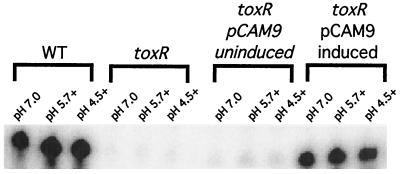
Since it is formally possible that overexpression of OmpU during acid stress conditions might result in a phenotype that mimics the wild-type organic ATR, we assessed the levels of transcription of ompU and OmpU protein accumulation in DSM-V705. RPAs were conducted using a riboprobe specific for ompU. As depicted in Fig. 4, wild-type V. cholerae shows a high steady-state level of ompU transcript, which is ToxR dependent (Fig. 4). Likewise, DSM-V705 grown in the absence of l-arabinose contains no detectable ompU transcript. However, upon induction with l-arabinose, ompU transcript accumulates to levels similar to, or slightly less than, that in the wild type under all conditions tested (Fig. 4).
FIG. 4.
RPA for ompU transcript in C6709-1 (WT), DSM-V468 (ΔtoxR), and DSM-V705 (ΔtoxR, pCAM9 plus [induced] and minus [uninduced] l-arabinose). Total RNA was prepared from bacteria grown at the indicated pH in the presence (indicated by a plus) or absence of organic acids as described in Materials and Methods.
To ensure that the accumulation of OmpU protein correlates with levels of ompU transcript seen using the RPA, SDS-PAGE and Western blot analysis were performed. As shown in Fig. 2, a band corresponding to the predicted size of OmpU appears in DSM-V705 lanes only after induction with l-arabinose. Western blot analysis using anti-OmpU antibody similarly revealed the presence of OmpU in DSM-V705 only after induction (Fig. 5). In both cases, levels of OmpU accumulation appear similar to that of the wild type. These results suggest that wild-type levels of OmpU protein are being produced in the induced DSM-V705 strain and support the idea that the presence of OmpU is sufficient to bypass the organic ATR defect exhibited by a toxR strain.
FIG. 5.
Western blot analysis of OmpU protein in C6709-1 (WT), DSM-V468 (ΔtoxR), and DSM-V705 (ΔtoxR, pCAM9 plus [induced] and minus [uninduced] l-arabinose). Total protein was collected as described in Materials and Methods. Samples were separated on SDS–12% PAGE gels, and Western blot analysis was performed with rabbit anti-OmpU polyclonal antibodies. The presence of organic acids is indicated by a plus adjacent to the adaptation pH.
Ectopic expression of ompU does not alter ompT expression.
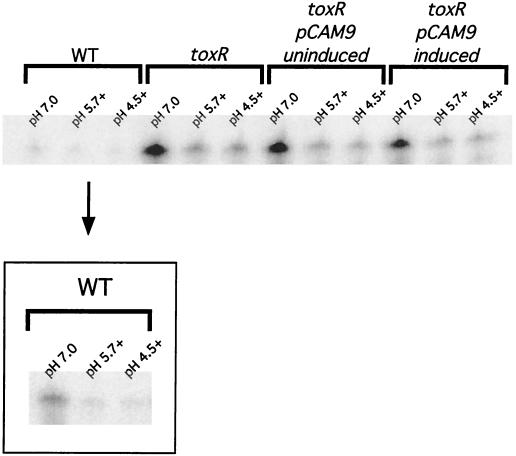
Since OmpU and OmpT are inversely regulated, these two proteins are not normally found to coexist at appreciable levels within the outer membrane. We considered the possibility that ectopic expression of ompU was able to complement the toxR organic ATR defect indirectly by altering the expression patterns of ompT. Specifically, the bypass effect of OmpU could be due to downregulation of ompT expression. This possibility was investigated by measuring the levels of ompT transcript produced in induced DSM-V705 compared to those of a toxR strain. As shown in Fig. 6, the steady-state levels of ompT transcript are similar between a toxR strain and strain DSM-V705 whether inducer has been added or not (compare pH 7.0 lanes). Likewise, similar levels of OmpT protein are produced (Fig. 2, compare lanes labeled toxR and toxR pCAM induced). Taken together, these results suggest that the complementation phenotype of DSM-V705 is due to the presence of OmpU and not to the loss of OmpT upon ectopic expression of the former.
FIG. 6.
RPA for ompT transcript in C6709-1 (WT), DSM-V468 (ΔtoxR), and DSM-V705 (ΔtoxR, pCAM9 plus [induced] and minus [uninduced] l-arabinose). Total RNA was prepared from bacteria grown at the indicated pH in the presence (indicated by a plus) or absence of organic acids as described in Materials and Methods. The three WT lanes offset by a box represent an additional ompT RPA conducted with a probe of approximately threefold-higher specific activity.
Organic acid stress represses ompT but not ompU transcription.
Since expression of ompU was sufficient to bypass the organic ATR defect exhibited by a toxR strain, and since 2D gel analysis revealed that multiple polypeptides were differentially regulated in response to organic acid stress, we wished to gain a better understanding of the effect of organic acid treatment on ompU and ompT transcript and protein levels. RPAs using a riboprobe specific for ompU show that the addition of organic acids results in very little, if any, change in the amount of ompU transcript (Fig. 4). This is similarly reflected in levels of OmpU protein which accumulate within organic acid-stressed cells (Fig. 5). These results demonstrate that OmpU is not an ASP per se but instead argue that the normal levels of OmpU are sufficient to mediate organic ATR in conjunction with other ToxR-independent ASPs.
In contrast to ompU, ompT transcription is affected by organic acid stress. In initial RPA experiments using ompT riboprobes with very high specific activity, we observed that the levels of ompT transcript were repressed upon exposure to organic acid stress (Fig. 6, WT box). This response was more readily seen in the toxR strain, where levels of ompT transcript are increased due to the loss of repression by ToxR. As shown in Fig. 6, ompT transcription is similarly repressed in a toxR strain upon exposure to organic acids. This demonstrates the existence of an organic acid-induced, toxR-independent repressor of ompT transcription. This repression of ompT transcription is similarly reflected in the reduced accumulation of OmpT protein in bacteria that are exposed to low pH (pH 5.7) plus organic acids (Fig. 2).
We attempted to determine the identity of the additional regulator of ompT by first investigating the only other known ompT regulator, cyclic AMP receptor protein (CRP). CRP was recently shown to act as a positive regulator of ompT (15) as well as a negative regulator of other V. cholerae virulence genes (29). Since CRP is able to function as both a repressor and an activator of different genes in E. coli (reviewed in reference 3), we considered the possibility that CRP might have the ability to serve as a dual regulator of ompT under different environmental conditions. RPAs of total RNA collected from a toxR crp double mutant revealed that the previously identified repression of ompT transcription upon exposure to organic acids was unaffected (data not shown). Therefore, CRP does not appear to be responsible for repression of ompT upon organic acid exposure.
We have additionally investigated the possibility that CadC, a recently identified member of the ToxR-like family of transcriptional regulators, might function as a repressor of ompT. CadC was shown to act as a positive transcriptional regulator of cadA and cadB, which code for a lysine decarboxylase and a lysine-cadaverine antiporter, respectively (17). CadC shows extensive homology to ToxR within its DNA binding domain and has been predicted to have a DNA binding site that exhibits a high degree of similarity to known ToxR binding sites. Indeed, it was previously noted that the predicted binding site of CadC within the cadB promoter region is strikingly similar to the consensus repeat sequence bound by ToxR within the ompT promoter (19). However, ompT expression levels in a cadC toxR strain were indistinguishable from those in a toxR strain (data not shown), indicating that CadC does not serve as the organic acid-induced repressor of ompT transcription. The identity of the factor involved in this phenomenon remains unknown.
DISCUSSION
The recent completion and annotation of the V. cholerae genome predicts that V. cholerae contains 3,885 open reading frames on its two circular chromosomes (11). Here, we have used 2D gel electrophoresis to show the altered expression of approximately 110 different polypeptide species in response to exposure to low pH plus organic acids. This represents approximately 3% of the predicted open reading frame products in the V. cholerae genome and, as many of the predicted polypeptides lie outside of the pI range of the 2D gels, is probably a conservative estimate. The altered expression of so many different polypeptides in response to organic acid stress is consistent with the fact that V. cholerae encounters such stress during the course of colonization of the small intestine and thus must be able to adapt in order to maximize its pathogenic potential.
Our comparative analysis of 2D gels of wild-type and toxR V. cholerae strains predicts that in addition to the 18 polypeptide species which are downregulated upon the loss of ToxR, 6 others are upregulated during organic ATR. ToxR is known to act as both an activator and a repressor of transcription of a number of different genes, collectively termed the ToxR regulon (reviewed in reference 28). To date, ompT is the only identified ToxR-repressed gene (TRG) (15), and to our knowledge, only one other TRG, which encodes a 58-kDa protein, has previously been shown to exist (33). The data presented here represent the first additional report suggesting the existence of additional TRGs. This, combined with the large number of polypeptides whose expression was downregulated with the loss of ToxR, implies that the ToxR regulon contains additional genes yet to be identified. Future elucidation of additional components of the ToxR regulon could provide valuable insight into the intricate nature of ToxR's ability to regulate not only ancestral genes (such as ompU and ompT) but also more recently acquired genes (such as those within the Vibrio pathogenicity island and CTXφ).
Ectopic expression of OmpU was able to bypass the organic ATR defect exhibited by a toxR V. cholerae strain. In addition, characterization of ompU and ompT transcript levels in response to organic acid stress conditions subsequently revealed that while the levels of ompU transcript remain unaffected, those of ompT are repressed in a ToxR-independent manner. That ompU transcription remains seemingly unaffected by organic acid stress is nevertheless somewhat surprising in light of recent data showing that treatment of V. cholerae with bile results in a ToxR-dependent increase in levels of ompU transcription (25). Provenzano and Klose further demonstrated that OmpU is involved in survival upon exposure to bile and other detergents that might be encountered by V. cholerae during intestinal colonization (24, 25). Perhaps, since V. cholerae is likely to be exposed to organic acid stress not only within the host small intestine but also during growth within the environment (as organic acids are produced as the bacteria undergo normal metabolic activities), levels of OmpU are consistently maintained in order to provide protection against general organic acid stress.
Additionally, the fact that ompU transcription levels are not altered upon exposure to organic acid suggests that the pathways and regulatory networks by which organic and bile stresses are received and then responded to are perhaps different. This is interesting when one considers that the only known regulator of ompU expression is ToxR, and it points to the fact that ToxR-mediated regulation in response to different environmental stimuli is a complex and multifactorial orchestration of signaling events. Gaining a better understanding of the mechanisms underlying the intricate complexities of ToxR regulation in V. cholerae should shed valuable insight into the mechanisms employed for gene regulation in a number of bacterial pathogens.
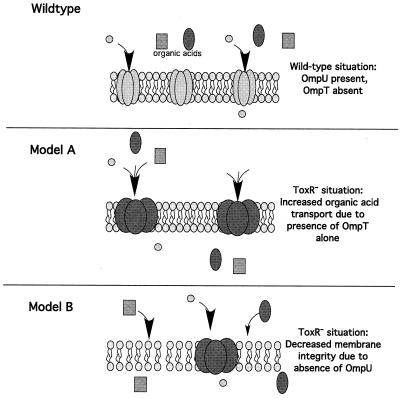
To our knowledge, OmpU represents the first porin identified as being involved in ATR. While OmpR, which regulates the OMPs OmpC and OmpF in S. enterica serovar Typhimurium, has been demonstrated to be crucial for regulation of stationary-phase ATR, this requirement is independent of OmpC and OmpF (2). What might be the role of OmpU in the protection of V. cholerae from organic acids? Data presented here support two of several possible models. In a toxR strain, OmpU, and thus OmpU porin activity, are lost from the V. cholerae outer membrane. At the same time, OmpT, and thus OmpT porin activity, are subsequently present. Model A predicts that there is increased transport of organic acids into the periplasm due to an alteration of porin activity (Fig. 7). Conceivably, the relevant alteration of porin activity could result from either loss of OmpU porin activity or gain of OmpT porin activity. The fact that coexpression of ompU and ompT results in regained resistance to organic acid stress suggests that it is OmpU porin activity which is the important factor. However, it is formally possible that coexpression of these two porins results in heterotrimers that disrupt OmpT porin activity, and thus OmpT porin activity may be the important factor after all. Our attempts to isolate such heterotrimeric species have, thus far, been unsuccessful. To investigate this possibility further, we overexpressed OmpT in a wild-type background and checked the organic ATR phenotype of these strains. For this strain background, we would hypothesize that increased levels of OmpT expression would drive the equilibrium towards increased numbers of homotrimeric OmpT and thus provide OmpT porin activity. If this OmpT porin activity is deleterious to the cell, we would expect that overexpression would result in an organic ATR defect that was similar to that of a toxR strain. This was not the case, as strains overexpressing OmpT behaved as wild type (data not shown), suggesting once again that it is the presence of OmpU which is the critical factor in ATR.
FIG. 7.
Two models for the effect of loss of OmpU on organic acid resistance. The outer membrane containing porin trimers is depicted. Arrows indicate the movement of organic acids or other unknown small molecules through either the porin (model A) or across the bacterial membrane in the absence of pore activity (model B). Model A suggests that the porin activity of OmpT and/or the loss of OmpU porin activity results in the transport of harmful organic acids across the bacterial membrane. Model B suggests that the loss of OmpU from the bacterial membrane results in an increase in membrane permeability and thus more movement of organic acids across the outer membrane.
Model B predicts that porin activity per se is not the important component for resistance to organic acid stress but that instead the loss of OmpU leads to an increase in outer membrane permeability to organic acids (Fig. 7). Since it has been shown that OmpU can compose 30 to 60% of the total outer membrane protein of V. cholerae, depending on the osmolarity of the growth medium, it would stand to reason that complete loss of OmpU could alter membrane permeability properties. This model does not go without precedent, as it has previously been shown that removal of a single protein from the outer membrane of Pseudomonas putida results in increased membrane permeability (16). In addition, when one considers the previous study by Chakrabarti et al. (4) that indicated that the actual pore size of OmpU was larger than that of OmpT, it seems inconsistent that the presence of a larger pore (OmpU) would result in the ability to survive exposure to organic acids. This having been said, it does remain formally possible that the pore sizes that were previously calculated using various carbohydrate solutes are not true indications of the relative permeability of the organic acids used in this study. Finally, overexpression of OmpT in the wild-type background resulted in wild-type ATR (data not shown). If coexpression of OmpU and OmpT results in the formation of heterotrimeric species that disrupt porin activity, one might hypothesize that overexpression of OmpT would not only result in the formation of OmpT homotrimers, as suggested above, but would also disrupt the number of OmpU homotrimers present, thus resulting in an ATR defect. This was not the case, supporting model B's prediction that it is the presence of OmpU and not porin activity per se which is the critical component of ATR. Both models taken into consideration, the exact nature of the requirement of OmpU for resistance to organic acid stress remains to be elucidated.
ACKNOWLEDGMENTS
This research was supported NIH grants AI 40262 and AI 45746 to A.C. and AI 19716 to J.B.K. and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928).
We thank C. C. Li, D. Provenzano, and K. Klose for helpful discussion. In addition, we thank E. Joyce for invaluable assistance with 2D gel electrophoresis, Michael Angelichio for statistical analysis, and David Hava for critical reading of the manuscript.
REFERENCES
- 1.Baik H S, Bearson S, Dunbar S, Foster J W. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology. 1996;142:3195–3200. doi: 10.1099/13500872-142-11-3195. [DOI] [PubMed] [Google Scholar]
- 2.Bang I S, Kim B H, Foster J W, Park Y K. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:2245–2252. doi: 10.1128/jb.182.8.2245-2252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botsford J L, Harman J G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti S R, Chaudhuri K, Sen K, Das J. Porins of Vibrio cholerae: purification and characterization of OmpU. J Bacteriol. 1996;178:524–530. doi: 10.1128/jb.178.2.524-530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 6.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 7.Das M, Chopra A K, Cantu J M, Peterson J W. Antisera to selected outer membrane proteins of Vibrio cholerae protect against challenge with homologous and heterologous strains of V. cholerae. FEMS Immunol Med Microbiol. 1998;22:303–308. doi: 10.1111/j.1574-695X.1998.tb01219.x. [DOI] [PubMed] [Google Scholar]
- 8.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster J W. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J Bacteriol. 1991;173:6896–6902. doi: 10.1128/jb.173.21.6896-6902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6874–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaper J B, Fasano A, Trucksis M. Toxins of Vibrio cholerae. In: Wachsmuth I K, Blake P A, Olsvik, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 145–176. [Google Scholar]
- 14.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C C, Crawford J A, Dirita V J, Kaper J B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 16.Llamas M A, Ramos J L, Rodriguez-Herva J J. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol. 2000;182:4764–4772. doi: 10.1128/jb.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrell D S, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 18.Merrell D S, Camilli A. Detection and analysis of gene expression during infection by in vivo expression technology. Philos Trans R Soc Lond B Biol Sci. 2000;355:587–599. doi: 10.1098/rstb.2000.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrell D S, Camilli A. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol. 2000;182:5342–5350. doi: 10.1128/jb.182.19.5342-5350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 21.Nakasone N, Iwanaga M. Characterization of outer membrane protein OmpU of Vibrio cholerae O1. Infect Immun. 1998;66:4726–4728. doi: 10.1128/iai.66.10.4726-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 23.Parsot C, Mekalanos J J. Expression of the Vibrio cholerae gene encoding aldehyde dehydrogenase is under control of ToxR, the cholera toxin transcriptional activator. J Bacteriol. 1991;173:2842–2851. doi: 10.1128/jb.173.9.2842-2851.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provenzano D, Klose K E. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci USA. 2000;97:10220–10224. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Provenzano D, Schuhmacher D A, Barker J L, Klose K E. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun. 2000;68:1491–1497. doi: 10.1128/iai.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts A, Pearson G D, Mekalanos J J. Proceedings of the 28th Joint Conference, U.S.-Japan Cooperative Medical Science Program on Cholera and Related Diarrheal Diseases. 1992. Cholera vaccine strains derived from a 1991 Peruvian isolate of Vibrio cholerae and other El Tor strains. [Google Scholar]
- 27.Sengupta D K, Sengupta T K, Ghose A C. Antibodies to outer membrane proteins of Vibrio cholerae induce protection by inhibition of intestinal colonization of vibrios. FEMS Microbiol Immunol. 1992;4:261–266. doi: 10.1111/j.1574-6968.1992.tb05004.x. [DOI] [PubMed] [Google Scholar]
- 28.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 29.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. . (Erratum, 163:279, 1987.) [DOI] [PubMed] [Google Scholar]
- 31.Sperandio V, Bailey C, Giron J A, DiRita V J, Silveira W D, Vettore A L, Kaper J B. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperandio V, Giron J A, Silveira W D, Kaper J B. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect Immun. 1995;63:4433–4438. doi: 10.1128/iai.63.11.4433-4438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]