Abstract
We performed two sets of in vitro selections to dissect the role of the −10 base sequence in determining the rate and efficiency with which Escherichia coli RNA polymerase-ς70 forms stable complexes with a promoter. We identified sequences that (i) rapidly form heparin-resistant complexes with RNA polymerase or (ii) form heparin-resistant complexes at very low RNA polymerase concentrations. The sequences selected under the two conditions differ from each other and from the consensus −10 sequence. The selected promoters have the expected enhanced binding and kinetic properties and are functionally better than the consensus promoter sequence in directing RNA synthesis in vitro. Detailed analysis of the selected promoter functions shows that each step in this multistep pathway may have different sequence requirements, meaning that the sequence of a strong promoter does not contain the optimal sequence for each step but instead is a compromise sequence that allows all steps to proceed with minimal constraint.
Gene expression is often regulated at transcription initiation. In the absence of gene regulatory proteins, the rate of initiation at a particular promoter depends on the concentration of RNA polymerase and the sequence of the promoter element. In both prokaryotes and eukaryotes, transcription initiation is a multistep process that begins with sequence-specific recognition and proceeds through formation of two or more intermediate RNA polymerase-promoter complexes prior to synthesis of the first phosphodiester bond of the product RNA (31). Subsequently, the initiated complex isomerizes into one that generates full-length RNA products (for exceptions, see references 18 and 34). The fractional occupancy of a promoter by RNA polymerase and the rate at which RNA polymerase proceeds through the initiation pathway determine, in part, the amount of RNA produced from a gene.
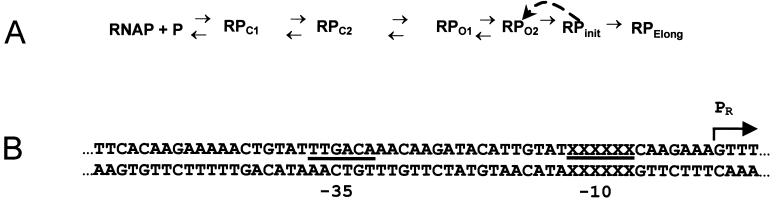
The pathway for transcription initiation is best understood in prokaryotes (Fig. 1A). In these organisms, RNA polymerase binds to promoter DNA and initially forms an unstable “closed” complex that dissociates with the addition of polyanionic competitors such as single-stranded DNA or heparin (31). The closed complex isomerizes through a number of intermediate complexes (21, 33) in which the DNA at the transcription start site remains base paired (Fig. 1A). Subsequently, these intermediates isomerize to form one or more competitor-resistant “open” complexes, in which the DNA strands surrounding the transcription start site display increasing separation (4, 36). RNA polymerase initiation complexes form after the DNA in the region between −11 and ca. +5 is fully denatured and the synthesis of small (8- to 12-base) “abortive” oligonucleotide products takes place and/or the polymerase escapes from the promoter, forming an elongation complex competent to synthesize full-length RNA transcripts.
FIG. 1.
Transcription pathway and sequence of the starting promoter. (A) Proposed transcription initiation pathway for E. coli RNA polymerase, based on evidence described in the text and reference 31. (B) Sequence of the modified bacteriophage 434 PR promoter that is the starting material for the selection experiments. The −35 and −10 regions in this promoter are underlined (2, 37).
In prokaryotes, both the affinity of RNA polymerase for DNA and the rate of isomerization of the RNA polymerase-DNA complexes to form the various intermediate species depend on the particular sequence of the promoter (12, 14). In addition, the rate of escape of RNA polymerase from the promoter to form full-length RNA transcripts may also be determined by promoter sequence (3, 15). Hence, the overall RNA polymerase occupancy of a promoter and the rate of RNA synthesis from that promoter depends on its sequence. It is not known, however, how promoter sequence determines the efficiency of the individual steps along the initiation pathway. Insight into this question is critical to understanding the interrelationship between promoter strength and the mechanisms by which gene regulatory proteins modulate transcription (19).
The consensus promoter utilized by Escherichia coli RNA polymerase-ς70 is comprised of two conserved, hexameric DNA sequence elements separated by ∼17 bp and located ∼35 and ∼10 bp upstream of the first transcribed nucleotide (11, 13, 23). The consensus sequences of these so-called −35 and −10 elements are TTGACA and TATAAT, respectively. In the RNA polymerase holoenzyme the ς70 subunit makes direct protein-DNA contacts with double-stranded DNA in the −35 region (7, 9, 35). In open complexes in which the DNA strands near the start point of transcription are separated, another portion of the ς70 subunit interacts with the bases on the nontemplate strand in the −10 region (25). Recent work (26) has examined the nature of the protein-DNA contacts made in the open promoter complex. Although they most certainly occur (1, 21, 33), the type of RNA polymerase-promoter DNA contacts made in the −10 region in the closed and the transitional intermediate promoter complexes has not yet been established.
The consensus sequence of the E. coli RNA polymerase-ς70 promoter has been deduced from sequence compilations (11, 13, 23), and the importance of these conserved promoter elements has been demonstrated by random (28) and site-directed (9, 17, 27) mutagenesis studies. Many of these studies focused on the effect of promoter sequence changes on transcriptional efficiency. While they provide useful information how sequence contributes to overall promoter efficiency, these methods provide only a composite measure of sequence effects on promoter function. More-recent studies examined the sequence dependence of the binding of single-stranded DNA (ssDNA) corresponding to the nontemplate strand of the −10 region to RNA polymerase (8, 25, 30). However, this type of experiment does not provide information about the relationship between promoter sequences and the efficiency of steps leading up to open-complex formation. Thus, these two types of studies are unable to examine the effects of promoter sequences on individual steps in the transcription initiation pathway.
As a first step to overcoming this limitation, we used E. coli RNA polymerase-ς70 holoenzyme to select promoter sequences from a pool of DNAs randomized at the −10 hexamer. Two types of selections were performed. We demanded that the promoter sequences either support rapid formation of heparin-resistant RNA polymerase-promoter complexes or support the formation of such complexes under conditions of very low RNA polymerase concentration. The first selection strategy should identify −10 sequences that allow RNA polymerase to advance rapidly through the isomerization steps that precede heparin-resistant complex formation. The second selection regimen should reveal sequences that allow RNA polymerase to productively bind promoter DNA with high affinity. Hence these selection protocols allow us to dissect the role of the −10 base sequence in determining, respectively, the rate and efficiency with which RNA polymerase forms stable complexes with a promoter. The starting material for these selections is derived from the PR promoter from the lambdoid bacteriophage 434 (Fig. 1B). This promoter bears a consensus −35 element sequence and is separated from the randomized −10 region by a 17-bp spacer.
MATERIALS AND METHODS
Construction of promoter DNA.
Bacteriophage 434 PR promoters bearing random sequences at the −10 region were constructed in two steps. We obtained (Integrated DNA Technologies) two pairs of oligonucleotides complementary to the upper and lower strands of 434 PR; in each pair one oligonucleotide contained six random bases at the position of the −10 region and the other was complementary to DNA 225 bp upstream or downstream of the 434 OR region in pJX (37). Two separate amplification reactions, one with each pair of oligonucleotides, resulted in DNA molecules that encoded the upstream and downstream halves, respectively, of the 434 OR region with a random sequence at the −10 region of PR. Subsequently, these two DNA molecules were annealed and the intact 434 OR region bearing a random −10 region was obtained by amplifying the annealed product with the upstream and downstream primers. DNA purified from this amplification reaction was labeled at the 5′ ends using T4 polynucleotide kinase and [γ-32P]ATP and was used directly in the selections.
In vitro selection.
For sequence selections based on affinity, initially 20 nM RNA polymerase was mixed with equimolar DNA in transcription buffer containing 100 mM KCl, 40 mM Tris (pH 7.9), 10 mM MgCl2, and 10 mM dithiothreitol (DTT) and incubated at 37°C for 15 min. This period is at least three times longer than the measured association and dissociation half-times of the DNA pool, ensuring that the mixture had reached equilibrium. Subsequently, 0.1 mg of heparin/ml was added and the mixture was incubated an additional 5 min at 37°C before the reaction products were fractionated on a 5% polyacrylamide gel at room temperature using TBE (89 mM Tris [pH 8.9], 89 mM boric acid, 1 mM EDTA) as the electrophoresis buffer. The RNA polymerase concentration was lowered to 5 nM at round 5 and to 1 nM at round 15, holding the DNA concentration constant. To select −10 sequences that bind rapidly to RNA polymerase, 20 nM RNA polymerase was mixed with equimolar DNA in transcription buffer and incubated for 15 s. Subsequently, 0.1 mg of heparin/ml was added and the mixture was incubated an additional 5 min at 37°C before the reaction products were fractionated on a 5% polyacrylamide gel.
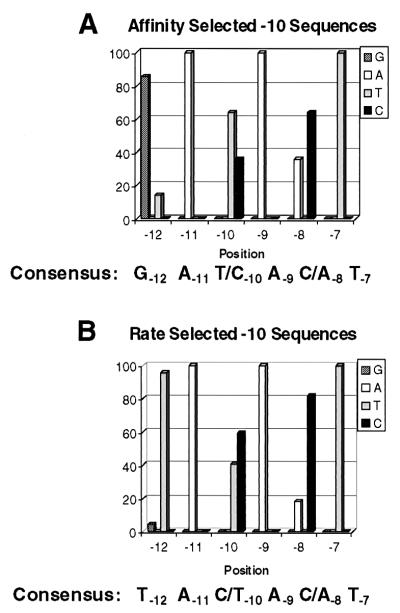
For both selection regimens, the gels were visualized on a PhosphorImager and the DNA in complex with RNA polymerase was recovered from the gel by crushing the isolated gel slice containing the complex and soaking the DNA out of the gel. The isolated DNA was amplified by PCR using the upstream and downstream primers. A portion of the amplified DNA was sequenced, and the remainder was used in subsequent selection rounds. Between selection rounds 17 and 22, the sequence of the selected DNAs stabilized, and the selected promoter DNAs were cloned into pEMBL8+ (5) and their sequences were determined. Multiple isolates of several different promoter sequences were obtained from both selection regimens, indicating that the population of selected promoter sequences was adequately sampled. Figure 2 depicts the frequency of occurrence of each base at the selected position as determined from the unique sequences from each selection regimen; 14 clones were analyzed in the case of the affinity-selected promoters and 22 clones were analyzed in the case of the rate-selected promoters.
FIG. 2.
The −10 sequences preferred by ς70 RNA polymerase for formation of heparin-resistant complexes. Sequences were selected and determined as described in Materials and Methods. Shown is the frequency distribution of the occurrence of a selected base sequence at each position in the selected promoter, as selected on the basis of high-affinity RNA polymerase binding (A) or rapid open-complex formation (B). The percent occurrence of a base pair at each position was derived from analysis of 14 and 22 cloned sequences for panels A and B, respectively.
Measurements of the rate and affinity of RNA polymerase-promoter binding.
The 450-bp DNA fragments containing selected promoters were separately isolated from the pEMBL8+ derivatives by cleavage with PvuII and HindIII and were labeled at their 3′ ends by incubation with [α-32P]dATP and the Klenow fragment of DNA polymerase I. Binding affinities were determined by mixing increasing concentrations of RNA polymerase with 0.1 pM promoter-containing DNA fragment and incubating at 37°C for 15 min. Subsequently, 0.1 mg of heparin/ml was added and the mixture was incubated an additional 5 min at 37°C before the reaction products were fractionated on a 5% polyacrylamide gel at room temperature. The incubation times were long enough to ensure that the reaction was at equilibrium (see above). Heparin was added to these mixtures to facilitate quantitation by removing RNA polymerase that was bound to DNA in non-sequence-specific complexes. Control experiments indicated that the amount of promoter-specific complex was unaffected by the concentration of heparin added or the length of the incubation time (data not shown). The gels were visualized on a PhosphorImager, and the amounts of complex and free DNA was determined. The apparent Kd was determined by fitting the bound counts/total counts fraction versus RNA polymerase concentration to a hyperbolic binding expression. For the rate measurements, 0.1 nM labeled DNA was separately mixed with 1, 2, 5, and 10 nM RNA polymerase. After incubation at 37°C for varying lengths of time, the reactions were quenched by adding 0.1 mg of heparin/ml before the reaction products were fractionated, visualized, and quantified as described above. The apparent association rate constant (ka) was determined from the slope of the plots of the pseudo-first-order rate constants determined from the progress curve at each RNA polymerase concentration versus RNA polymerase concentration. The apparent ka values reported are averages of three determinations.
In vitro transcription.
DNA fragments containing the selected promoters were isolated from the pEMBL8+ derivatives by cleavage with PvuII. Approximately 1 nM each fragment was separately incubated with increasing RNA polymerase concentrations for 15 min (for concentration dependence measurements, see Figures 3A and B) or with 10 nM RNA polymerase at increasing times at 37°C (for time dependence measurements, see Fig. 3C and D) in transcription buffer, prior to initiation of runoff transcription reactions by addition of ribonucleotide triphosphates together with 0.1 mg of heparin/ml. Heparin was added to these mixtures to ensure that only a single round of transcription occurred. Control experiments established that heparin addition did not affect the stability of open complexes formed by RNA polymerase on any of the promoters we used under these conditions. Hence, the time and RNA polymerase concentration dependence of transcript formation reveal the differential effects of promoter sequence, not variation in heparin competition. For the concentration dependence measurements, the RNA polymerase concentration was increased in twofold steps starting at 0.6 nM. All transcription reactions were terminated after 10 min by addition of a formamide-containing dye mixture and heating to 90°C for 5 min. Template DNA concentrations were determined by digitally comparing the fluorescent intensities of bands on ethidium bromide-stained gels of DNA samples of known and unknown concentrations. For these measurements, a standard curve was constructed from a DNA sample of known concentration that was identical in length and sequence composition to the unknown input template DNA. DNA concentrations determined in this fashion are reproducible with an error of ±5%.
FIG. 3.
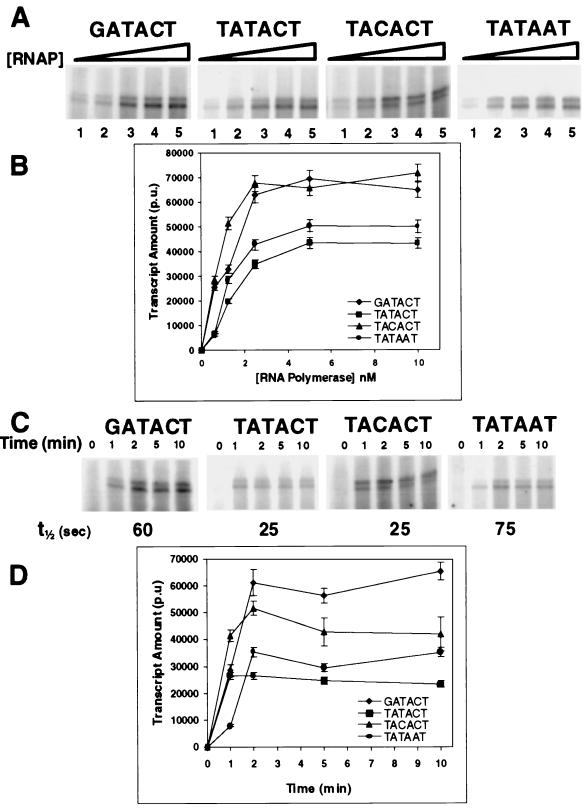
Runoff transcription directed by the rate- and affinity-selected promoters. Concentration-dependent (A and B) and time-dependent (C and D) runoff transcription directed by the rate- and affinity-selected promoters was determined in single-round runoff transcription assays, performed as described in Materials and Methods. The products of a typical transcription reaction are shown (A and C) as visualized by a PhosphorImager. Data from three experiments were quantified (B and D). Error bars, standard deviations. The amount of transcript is expressed in PhosphorImager units per microgram of input DNA (p.u.). Symbols in panels B and D represent promoters containing the −10 sequences.
KMnO4 footprinting.
DNA fragments containing the selected promoters were separately isolated from the pEMBL8+ derivatives by cleavage with PvuII and HindIII and were labeled on the template strand. These DNAs were incubated with 20 nM RNA polymerase for 15 min at 37°C, followed by addition of 15 mM KMnO4. After 2 min of further incubation at 37°C, the reaction was quenched by ethanol precipitation. The DNA was cleaved by incubation with piperidine at 90°C and was subsequently processed for electrophoresis and electrophoresed on an 8% polyacrylamide gel containing 7 M urea. The gels were visualized on a PhosphorImager, and the relative amount of KMnO4 reactivity was determined from the intensity of the bands corresponding to the entire −10 region, relative to the total intensity of the individual lane. These experiments were repeated three to five times, and the values obtained are reproducible within the errors of the measurement.
RESULTS AND DISCUSSION
Figure 2 shows that the preferred sequences obtained from the selections for a high rate of open-complex formation or a high-affinity RNA polymerase binding contain at least three (−7, −9, and −11) positions that match the consensus sequence. At these positions, RNA polymerase selects only the consensus base pair (Fig. 2). Work of others (30) showed that of positions −7, −9, and −11, only substitutions at the −11 position appear to substantially affect the affinity of ssDNA oligonucleotides encoding the nontemplate strand of the −10 region for RNA polymerase. Similarly, the identities of the bases at −7 and −11 have been shown to be critical for the binding of a “forked junction” template to RNA polymerase (26), a complex that has been proposed to mimic the open promoter complex (10, 32). Hence our findings extend these observations and suggest that in addition to −7 and −11, the identity of the base pair at position −9 is also critical to facilitating binding of the nontemplate DNA strand to RNA polymerase.
The consensus sequences of the selected −10 sequence promoters differ from the consensus −10 sequence (TATAAT) at as many as three (−8, −10, and −12) positions (Fig. 2). At these positions, RNA polymerase chooses between only two of the possible four base pairs. This finding suggests that the two base sequences not selected at each of these positions are incompatible with efficient open-complex formation. We find that the degree of preference for a particular base at a given position depends on the selection regimen (Fig. 2). Thus, the particular base sequence at positions −12, −10, and −8 may uniquely affect a particular step of the pathway towards open-complex formation.
To test this idea, we chose to analyze a subset of the promoters isolated during our selections. The promoter sequences were chosen to allow us to best examine the effect of base changes at the polymorphic positions −12, −10, and −8 on promoter function, both within identical sequence contexts and in the background of multiple sequence differences. However, since we obtained only a limited set of sequences in our selections, base changes at a particular site cannot be examined in the context of all base sequences at other positions. Nonetheless, among the sequences we chose is a promoter bearing the TATAAT sequence at its −10 region, facilitating comparisons with this naturally selected consensus sequence.
Position −8.
Although the preference of RNA polymerase for a C · G base pair at position −8 is stronger for the rate-based selection than for the affinity-based selection (80% versus 65%), the strong bias of RNA polymerase for this base pair over the consensus A · T, independent of the selection regimen (Fig. 2), is particularly striking. Compilations of E. coli RNA polymerase ς70 promoter sequences show that the consensus A · T pair is found in 55 to 60% (depending on spacer length) of known promoters (23). However, only 18 to 20% of known promoters contain C · G base pairs at this position, a frequency that is similar to that of the nonconsensus T · A base pairs at this position. Nonetheless, the preference of RNA polymerase for C · G base pairs at position −8 of the test promoter mirrors the positive effect that the presence of this base pair has on the function of several promoters (16, 27). Similarly, 50% of promoters that have been selected for high transcription levels have a C · G base pair at position −8 (28), and the consensus A · T base pair is not selected. Thus, a C · G base pair at position −8 contributes favorably to transcriptional efficiency of strong promoters. In their in vitro selection experiments, Gourse and colleagues find that the highest-affinity promoters for the E. coli RNA polymerase-ςs holoenzyme have either a C · G or an A · T base pair at this position (R. Gourse, personal communication). Thus, C · G pairs are well tolerated at position −8 in the high-affinity promoters of at least the related ς70 and ςs-RNA polymerase holoenzymes.
When viewed either in isolation or in the context of all selected promoter sequences, the selection results indicate that a C · G pair at position −8 may support open-complex formation better than do other base pairs at this position. This suggestion is confirmed by our finding that RNA polymerase forms open complexes on promoters containing a C · G base pair at position −8 12- to 50-fold faster than on promoters containing A · T base pairs at this position (Table 1) (note that the sequence of the promoter containing an A · T pair at position −8 is the consensus promoter sequence). Similarly, RNA polymerase binds to promoters containing a C · G pair at position −8 with up to 8-fold-higher affinity than it does to the promoter containing an A · T pair at this position (Table 1). However, consistent with the idea that the base at position −8 does not affect closed-complex formation (6), the specific effect of an isolated A · T→C · G change at this position on RNA polymerase affinity for the test promoter is less than fourfold. The observation that the C · G base pair has a greater effect on the rate of open-complex formation than it does on binding affinity is consistent with the selection results.
TABLE 1.
Characterization of rate- and affinity-selected −10 sequences
| Sequencea | Kd (nM)b | ka (M−1 s−1)b |
|---|---|---|
| GATACT | 1.58 ± 0.11 | (5.1 ± 0.54) × 108 |
| TATACT | 3.34 ± 0.2 | (2.0 ± 1.1) × 109 |
| TACACT | 6.7 ± 0.32 | (8.0 ± 0.65) × 108 |
| TATAAT | 12.8 ± 0.9 | (4.0 ± 2.1) × 107 |
From position −12 (leftmost) to position −7 (rightmost).
Determined as described in Materials and Methods. Values are means ± standard deviations from three independent determinations.
Position −10.
When promoters are selected solely on the basis of rapid open-complex formation, RNA polymerase prefers a C · G base pair at position −10, whereas if the selection is performed at a low enzyme concentration, RNA polymerase prefers T · A base pairs at this position (Fig. 2). The effect of selection regimen on position −10 base preference is not as large as that at position −8. The consensus T · A base pair is found at position −10 in ∼55% of known promoters. By contrast, a C · G base pair is found at position −10 in only 10 to 15% of naturally occurring promoters, a frequency at which the other nonconsensus base pairs are found at this position.
Consistent with the selection results, RNA polymerase binds to promoters bearing the TATACT sequence with a twofold-higher affinity than it does to promoters bearing the TACACT sequence (Table 1). However, despite its slightly higher preference for C · G pairs at position −10 in promoters selected for rapid open-complex formation, RNA polymerase forms open complexes with promoters bearing the TACACT sequence ∼3-fold more slowly than it does with promoters containing the TATACT sequence (Table 1). We do not understand why the effects of position −10 sequence on RNA polymerase binding affinity and the rate of open-complex formation by RNA polymerase do not accurately reflect the preferences uncovered by our selection regimen. It is possible that promoter sequence context alters the influence of position −10 substitution. However, because of the limited set of sequences obtained in our selections, we are unable to resolve the role of the base sequence context at positions −8 and −12 in the effect that substitution at position −10 has on RNA polymerase binding or the rate of open-complex formation by RNA polymerase. Alternatively, the rather small differences in binding and kinetic properties between promoters bearing a C · G or T · A pair at position −10 may not be resolvable by our selection regimens.
Despite the apparent inability of our selection regimens to distinguish between the similar properties of promoters bearing C · G or T · A pairs at position −10, it is clear from the selection results that only promoters bearing these sequences are capable of forming stable open complexes. Why, then, are these bases sequences preferred over G · C or A · T pairs? Studies performed in vitro under conditions similar to those of our selection experiments show that base substitutions at position −10 do not significantly affect either the binding of nontemplate ssDNA (8) or the affinity of binding of “fork junction” open-complex mimics to RNA polymerase (26). Hence, the identity of the base at position −10 may not be important in stabilizing the open complex. Nonetheless, in vivo studies show that T · A-to-C · G base substitutions decrease the relative activity of a promoter twofold (27). Hence, by deduction it appears that the base at position −10 affects predominantly the binding affinity of RNA polymerase. In view of our results, we suggest that the base at this position could affect the affinity of RNA polymerase for closed-complex formation, as well as the rate of isomerization steps that precede open-complex formation (see below).
Position −12.
Under conditions where RNA polymerase must rapidly form open complexes, the enzyme exhibits a strong preference for the consensus T · A base at position −12. If the selection is performed at a low enzyme concentration, RNA polymerase prefers the nonconsensus G · C base pair at this position. Consistent with the results of the selection, RNA polymerase binds to promoters bearing G · C base pairs at position −12 with twofold-higher affinity than it does to promoters bearing T · A base pairs at this position. Also consistent with the selection regimen, the apparent ka of RNA polymerase with T · A-containing promoters is ∼4-fold higher than that with G · C-containing promoters.
The modest magnitude of the effect of changing the −12 base pair on binding affinity and rate of association is similar to findings obtained by others (8, 16, 26, 30). However, these findings do not account for the importance of this position in determining promoter function. Although a G · C pair is found at the −12 positions of only 4 to 10% of naturally occurring promoters, many strong promoters, including λPR and T7A1 among others, contain this base pair at this position. Despite this observation, T · A base pairs, but not G · C base pairs, occur with high frequency at position −12 in the −10 regions of promoters selected for strong transcription (28). These observations suggest that the −12 position preferences may depend on the sequences of other bases in the promoter. Consistent with this suggestion, we find that the 75% of promoters containing C · G base pairs at −10 bear a G · C base pair at position −12 (data not shown). Although we have not examined the sequence dependence of −12 base identity on promoter function, others have shown that the presence of T · A and G · C base pairs differentially affects the relative strengths of two different sequence promoters (16).
Effect of −10 sequence on promoter function.
Previous studies have shown that increasing the correspondence of a promoter's sequence with the consensus does not always increase the rate or amount of mRNA product synthesized from the promoter (16). Since RNA transcript formation is the product of a sequential multistep reaction, the amount of active RNA polymerase-promoter complexes and the rate of RNA synthesis from these complexes can be limited by the affinity of RNA polymerase for the promoter as well as by the rates and efficiencies of steps that occur subsequent to binary complex formation. In an effort to determine whether our selected promoters efficiently mediate transcription initiation and whether the various promoter sequences differentially affect transcript formation, we measured amounts of runoff transcripts formed from promoters bearing changes at individual positions in time- and RNA polymerase concentration-dependent single-round runoff transcription assays. We also measured the extent of open-complex formation using KMnO4.
Promoters bearing C · G base pairs at position −8 form open complexes more rapidly than those containing an A · T base pair (Table 1). Consistent with this observation, at saturating RNA polymerase concentrations, the half-time for transcript formation directed by the promoter bearing the sequence TATACT is three times shorter than that for the promoter bearing the TATAAT sequence (Fig. 3C and D). Thus, a C · G base pair at position −8 favors transcript formation by enhancing the rate of open-complex formation. However, the rate enhancement induced by a C · G base pair at position −8 can be affected by the sequence at position −12 (see below).
Despite its effect on the rate of transcript formation, when only the position −8 sequence is considered, the identity of the base at this position has little effect on the amount of RNA synthesized from the promoter at high RNA polymerase concentrations. Nonetheless, under these conditions, the KMnO4 reactivity of the TATACT promoter is 1.8-fold greater than that of the TATAAT promoter (Fig. 4; compare lanes 3 and 4). These observations indicate that the presence of a C · G base pair at position −8 inhibits transcript formation by lowering the rate or efficiency of the steps subsequent to the formation of open complex. Hence, the presence of a C · G base pair at position −8 enhances the rate of steps prior to open-complex formation, but inhibits the rate of steps that follow open-complex formation. The opposite effects of C · G base substitution at position −8 on the various steps in the transcriptional initiation pathway are consistent with findings showing that introducing an abasic lesion or gap in the DNA strand at this position both enhances open-complex formation (22) and inhibits formation of the elongation complex (20).
FIG. 4.
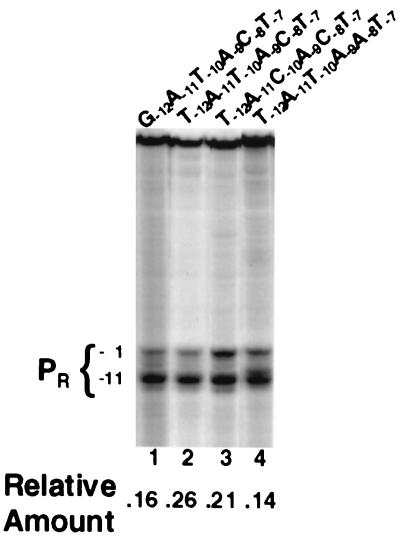
KMnO4 reactivity of selected promoters in the presence of saturating RNA polymerase concentrations. Labeled DNA fragments were incubated with RNA polymerase containing the indicated promoters and treated as described in Materials and Methods. The positions of the reactive thymines present on the template strand in the −10 region in all the selected promoters are indicated by the bracketed numbers. The relative KMnO4 reactivity of the −10 region is indicated below the gel and was determined as described in Materials and Methods.
A simple view of the transcription initiation pathway predicts that each RNA polymerase-open promoter complex should yield a full-length RNA transcript. Under the conditions of the transcription experiments for which results are shown in Fig. 3A, all the templates in the reaction mixture are completely occupied by RNA polymerase in a stable complex. In addition, KMnO4 probe experiments indicate that the extent of open-complex formation is essentially unaffected by the identity of the position −10 base pair (Fig. 4, lanes 2 and 3). Nonetheless, the TATACT promoter, bearing a T · A base pair at position −10, directs the synthesis of nearly twofold less runoff transcript than the promoter bearing a C · G base pair change (TACACT) at this position (Fig. 3A and B). This finding indicates that a smaller number of RNA polymerase-open promoter complexes formed on templates containing a T · A base at position −10 are capable of productive RNA synthesis. Thus, the identity of the position −10 base can regulate the activity of a promoter by fixing the RNA polymerase-promoter complex in an inactive state (18, 34) or inhibiting the rate of the transition of the RNA polymerase-open promoter complex to an elongation complex. Consistent with this idea, we find that the TACACT and TATACT promoters form transcripts at identical rates (Fig. 3C and D). This finding can be explained if the rate-limiting step for transcript formation by these promoters occurs after formation of an open complex. Since promoters bearing a T · A base pair at position −10 bind ∼2-fold better than those bearing C · G, we suggest that stronger protein-DNA contacts to bases at this position may inhibit the progression of RNA polymerase through the transcription cycle, an observation consistent with the findings of others (20, 22).
Considering only promoters whose base sequence differs at position −12, RNA synthesis initiates more rapidly on the promoter bearing a T · A base pair (TATACT) than it does on promoters bearing a G · C base pair at this position (GATACT) (Fig. 3C and D). The faster initiation of transcription by RNA polymerase on the TATACT promoter is qualitatively consistent with its ability to form heparin-resistant complexes more rapidly (Table 1). It should be noted that not all promoters containing a T · A base pair at position −12 form transcript rapidly. The half-time for transcript formation from the TATAAT-containing promoter is significantly longer than that for either of the other two promoters that contain a T · A base pair at −12 (Fig. 3C and D), an observation consistent with the low rate at which the TATAAT-containing promoter forms heparin-resistant complexes (Table 1). Since the other promoters containing T · A base pairs at −12 also bear base substitutions at positions −10 and −8, the effect of the position −12 base sequence is modulated by the identities of bases at other positions in the −10 region of the promoter.
Although transcripts initiate more rapidly from the TATACT promoter than from the GATACT promoter, at saturating RNA polymerase concentrations, the promoter bearing a G · C base pair at position −12 directs the synthesis of twofold more RNA than the promoter bearing a T · A base pair at this position (Fig. 3A and B). The increased amount of transcript is formed from a smaller number of open complexes, since the KMnO4 reactivity of complexes formed on the promoter bearing a G · C base pair at position −12 is 40% less than that of complexes formed on the promoter bearing a T · A base pair at this position (Fig. 4, lanes 1 and 2). Thus, a T · A base at position −12 either inhibits the transition from open complex to the initiation complex or prevents escape of the initiated RNA polymerase from abortive cycling. These findings suggest that the identity of the base at position −12 differentially affects multiple steps in the transcription initiation pathway. For example, the presence of a G · C base pair enhances the ability of RNA polymerase to form heparin-resistant complexes (Table 1). However, promoters bearing G · C base pairs at position −12 are inhibited in the rate at which these closed complexes isomerize to open complexes compared to the rate for the promoter bearing a T · A base pair at this position. Nonetheless, open complexes formed on G · C-containing promoters are much more efficient at proceeding through the steps in the initiation cycle that lead to RNA elongation than are those containing T · A base pairs.
Taken together, our results show that any individual position in the −10 region can affect the efficiency of one or more steps in the multistep pathway that precedes formation of an elongating RNA polymerase-DNA complex. More importantly, our findings demonstrate that the identical sequence at a particular position in the −10 region can have opposite effects on any given step in the transcriptional initiation pathway. Thus, the sequence of an individual promoter is unlikely to contain a sequence that is optimal for all steps in the transcription initiation pathway. Therefore, the activity of a promoter is a compromise between the rates of all the kinetic steps in the promoter, and the rate of each of these steps is determined by promoter sequence. This conclusion implies that the kinetic characteristics of naturally occurring promoters are precisely tailored by the individual sequence. Thus, in a manner similar to our selection regimens, the sequence of a particular naturally occurring promoter may have evolved to allow it to have the appropriate activity, for example, to strongly bind RNA polymerase such that it can effectively compete for scarce enzyme, or rapidly form open complexes to counterbalance the effects of repressors.
The failure to select particular base pairs at individual positions implies that the nonselected sequences are incompatible with formation of a heparin-resistant RNA polymerase-promoter complex. Hence the selection results suggest that the limited set of −10 sequences shown in Fig. 2 are those that permit efficient heparin-resistant complex formation. However, our failure to select a given base at any particular −10 position does not mean that the presence of that base is incompatible with the genesis of an active promoter. The −10 sequence selections were performed under equilibrium conditions, not transcription conditions. During transcription, the concentrations of individual RNA polymerase species in the transcriptional initiation pathway are at steady state and are determined by the rate of flux through the pathway. Although a given base may decrease the likelihood of heparin-resistant complex formation, it may also facilitate the isomerization of these complexes to a complex that is capable of RNA synthesis. Hence, under transcription conditions, a particular base sequence may drive the formation of such a complex. This realization may explain why bases that appear to be deleterious for heparin-resistant complex formation are found within functional promoters, and it suggests that −10 sequences selected for rapid formation of a transcription complex will differ from the sequences selected in this study.
An overarching conclusion of our study is that the base sequence at each position in the −10 region of the promoter can have differential effects on the individual steps that lead to formation of a stable RNA polymerase-promoter complex. Since both the selection regimens require that the promoter sequences support formation of a stable heparin-resistant RNA polymerase-promoter complex, we must be certain that the effects we observe are not due to sequence-dependent differences in the sensitivities of the selected RNA polymerase-promoter complexes to heparin addition (29). Several lines of evidence indicate that the sequence-dependent differences we observe are due only to sequence effects on promoter function. First, under the conditions of our rate and affinity measurements, heparin did not significantly alter the intensity of the band corresponding to the RNA polymerase-open promoter complexes, regardless of promoter sequence. Second, independent of promoter sequence, the addition of heparin and extended incubation of the complex with heparin did not affect the off-rate of the RNA polymerase-promoter complex. Third, heparin addition did not affect the amount of open complex detected by KMnO4 on any of our selected promoters.
Protein contacts to the nontemplate strand of the −10 region in the open complex have been proposed (24), and in one case, it has been demonstrated that Q437 of ς70 contacts the base at position −12 in the open complex (25). Our observation that −10 sequences containing a G · C pair at position −12 bind RNA polymerase with a higher affinity than do those bearing a T · A base pair at this position is consistent with this finding and suggests that protein-DNA contact in the open complex drives RNA polymerase −12 position preferences. Similarly, binding studies utilizing partial heteroduplex DNA fragments meant to mimic RNA polymerase–open-promoter complexes also show that promoters bearing a G · C pair at position −12 bind RNA polymerase with an affinity similar to that of those bearing a T · A base pair at this position (6). These earlier findings are not consistent with the more-recent experiments showing that the affinity of RNA polymerase for promoter DNAs containing mutations in the nontemplate strand of the −10 region is virtually unaffected by substitutions at position −12 (8, 26, 30). We also find inconsistencies between the recent binding results and the results of our selection experiments. Since the binding of the nontemplate strand of the −10 region to RNA polymerase is thought to be driven by the same protein-DNA contacts that occur in the open complex, these inconsistencies suggest that our selections are measuring effects of DNA sequence on steps that occur prior to open-complex formation. Thus, the selection methodology provides a method by which we can probe the role of sequence on steps prior to open-complex formation, a portion of the transcription initiation pathway that has heretofore been experimentally inaccessible.
ACKNOWLEDGMENTS
We thank P. Gollnick, V. J. Hernandez, and R. Gourse for critical reading of the manuscript.
This work was supported by PHS grant GM42138 from the National Institutes of Health.
REFERENCES
- 1.Buc H, McClure W R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 2.Bushman F D. The bacteriophage 434 right operator. Roles of OR1, OR2 and OR3. J Mol Biol. 1993;230:28–40. doi: 10.1006/jmbi.1993.1123. [DOI] [PubMed] [Google Scholar]
- 3.Choy H E, Adhya S. RNA polymerase idling and clearance in gal promoters: use of supercoiled minicircle DNA template made in vivo. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig M L, Suh W C, Record M T., Jr HO⋅ and DNase I probing of E sigma 70 RNA polymerase-lambda PR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 5.Dente L, Cesareni G, Cortese R. pEMBL: a new family of single-stranded plasmids. Nucleic Acids Res. 1983;11:1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dombroski A J. Recognition of the −10 promoter sequence by a partial polypeptide of ς70 in vitro. J Biol Chem. 1997;272:3487–3494. [PubMed] [Google Scholar]
- 7.Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription initiation factor ς70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 8.Fedoriw A M, Liu H, Anderson V E, DeHaseth P L. Equilibrium and kinetic parameters of the sequence-specific interaction of Escherichia coli RNA polymerase with nontemplate strand oligodeoxyribonucleotides. Biochemistry. 1998;37:11971–11979. doi: 10.1021/bi980980o. [DOI] [PubMed] [Google Scholar]
- 9.Gardella T, Moyle H, Susskind M M. A mutant Escherichia coli ς70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Gralla J D. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawley D K, McClure W R. Mechanism of activation of transcription initiation from the lambda PRM promoter. J Mol Biol. 1982;157:493–525. doi: 10.1016/0022-2836(82)90473-9. [DOI] [PubMed] [Google Scholar]
- 13.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley D K, McClure W R. The effect of a lambda repressor mutation on the activation of transcription initiation from the lambda PRM promoter. Cell. 1983;32:327–333. doi: 10.1016/0092-8674(83)90452-x. [DOI] [PubMed] [Google Scholar]
- 15.Hsu L M. Quantitative parameters for promoter clearance. Methods Enzymol. 1996;273:59–71. doi: 10.1016/s0076-6879(96)73006-9. [DOI] [PubMed] [Google Scholar]
- 16.Knaus R, Bujard H. PL of coliphage lambda: an alternative solution for an efficient promoter. EMBO J. 1988;7:2919–2923. doi: 10.1002/j.1460-2075.1988.tb03150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi M, Nagata K, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: effect of base substitutions in the promoter −35 region on promoter strength. Nucleic Acids Res. 1990;18:7367–7372. doi: 10.1093/nar/18.24.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T, Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- 19.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin J R, Blake J J, Ganunis R A, Tullius T D. The roles of specific template nucleosides in the formation of stable transcription complexes by Escherichia coli RNA polymerase. J Biol Chem. 2000;275:6885–6893. doi: 10.1074/jbc.275.10.6885. [DOI] [PubMed] [Google Scholar]
- 21.Li X Y, McClure W R. Characterization of the closed complex intermediate formed during transcription initiation by Escherichia coli RNA polymerase. J Biol Chem. 1998;273:23549–23557. doi: 10.1074/jbc.273.36.23549. [DOI] [PubMed] [Google Scholar]
- 22.Li X Y, McClure W R. Stimulation of open complex formation by nicks and apurinic sites suggests a role for nucleation of DNA melting in Escherichia coli promoter function. J Biol Chem. 1998;273:23558–23566. doi: 10.1074/jbc.273.36.23558. [DOI] [PubMed] [Google Scholar]
- 23.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malhotra A, Severinova E, Darst S A. Crystal structure of a ς70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 25.Marr M T, Roberts J W. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 26.Matlock D L, Heyduk T. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by E. coli RNA polymerase. Biochemistry. 2000;39:12274–12283. doi: 10.1021/bi001433h. [DOI] [PubMed] [Google Scholar]
- 27.Moyle H, Waldburger C, Susskind M M. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliphant A R, Struhl K. Defining the consensus sequences of E. coli promoter elements by random selection. Nucleic Acids Res. 1988;16:7673–7683. doi: 10.1093/nar/16.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeffer S R, Stahl S J, Chamberlin M J. Binding of Escherichia coli RNA polymerase to T7 DNA. Displacement of holoenzyme from promoter complexes by heparin. J Biol Chem. 1977;252:5403–5407. [PubMed] [Google Scholar]
- 30.Qiu J, Helmann J D. Adenines at −11, −9 and −8 play a key role in the binding of Bacillus subtilis E sigma(A) RNA polymerase to −10 region single-stranded DNA. Nucleic Acids Res. 1999;27:4541–4546. doi: 10.1093/nar/27.23.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. Escherichia coli RNA polymerase E (ς70) promoters and kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 32.Roberts C W, Roberts J W. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 33.Roe J H, Burgess R R, Record M T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 34.Sen R, Nagai H, Shimamoto N. Polymerase arrest at the lambda P(R) promoter during transcription initiation. J Biol Chem. 2000;275:10899–10904. doi: 10.1074/jbc.275.15.10899. [DOI] [PubMed] [Google Scholar]
- 35.Siegele D A, Hu J C, Walter W A, Gross C A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 36.Suh W C, Ross W, Record M T., Jr Two open complexes and a requirement for Mg2+ to open the lambda PR transcription start site. Science. 1993;259:358–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Koudelka G B. DNA-based positive control mutants in the binding site sequence of 434 repressor. J Biol Chem. 1998;273:24165–24172. doi: 10.1074/jbc.273.37.24165. [DOI] [PubMed] [Google Scholar]