Abstract
Background
It seems that several members of intestinal gut microbiota like Streptococcus bovis, Bacteroides fragilis, Helicobacter pylori, Fusobacterium nucleatum, Enterococcus faecalis, Escherichia coli, Peptostreptococcus anaerobius may be considered as the causative agents of Colorectal Cancer (CRC). The present study used bioinformatics and immunoinformatics approaches to design a potential epitope-based multi-epitope vaccine to prevent CRC with optimal population coverage.
Methods
In this study, ten amino acid sequences of CRC-related pathogens were retrieved from the NCBI database. Three ABCpred, BCPREDS and LBtope online servers were considered for B cells prediction and the IEDB server for T cells (CD4+ and CD8+) prediction. Then, validation, allergenicity, toxicity and physicochemical analysis of all sequences were performed using web servers. A total of three linkers, AAY, GPGPG, and KK were used to bind CTL, HTL and BCL epitopes, respectively. In addition, the final construct was subjected to disulfide engineering, molecular docking, immune simulation and codon adaptation to design an effective vaccine production strategy.
Results
A total of 19 sequences of different lengths for linear B-cell epitopes, 19 and 18 sequences were considered as epitopes of CD4+ T and CD8+ cells, respectively. The predicted epitopes were joined by appropriate linkers because they play an important role in producing an extended conformation and protein folding. The final multi-epitope construct and Toll-like receptor 4 (TLR4) were evaluated by molecular docking, which revealed stable and strong binding interactions. Immunity simulation of the vaccine showed significantly high levels of immunoglobulins, helper T cells, cytotoxic T cells and INF-γ.
Conclusion
Finally, the results showed that the designed multi-epitope vaccine could serve as an excellent prophylactic candidate against CRC-associated pathogens, but in vitro and animal studies are needed to justify our findings for its use as a possible preventive measure.
Keywords: Colorectal cancer, Gut microbiota, Immunoinformatics, Vaccine design, In silico, Multi-epitope vaccine
Background
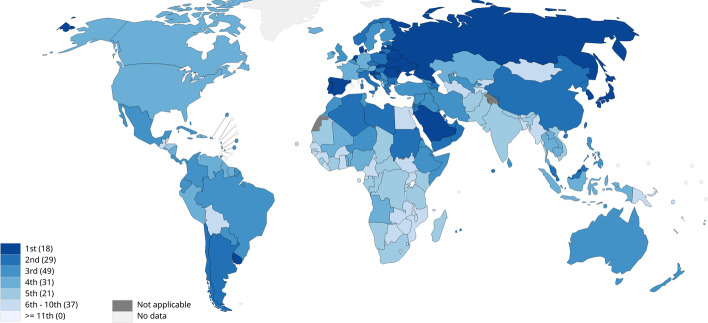
The human gut microbiome contains 1013–1014 bacterial cells which play important roles in health and disease prevention. These functions consist of providing an energy supply, balancing immune responses, preventing pathogens' colonization and maintenance of intestinal epithelium integrity [1]. Microbiome dysbiosis or any change in the composition of the human microbiome is the result of environmental factors [2] like diet, antibiotic treatment and recurrent infections which may lead to physiological and pathological alterations [3, 4]. Colorectal cancer (CRC) refers to a genetic disorder with uncontrolled proliferation of colorectal epithelial cells and may be triggered or exacerbated by microbiome dysbiosis. CRC has the first rank in terms of incidence and second in terms of mortality in both females and males among all cancers [5, 6]. Figure 1 shows the latest update (2020) of the CRC incidence rates according to the World Health Organization (WHO) report. CRC has a complicated etiology, while several cases of cancer have inherited and genetic backgrounds, most CRC cases arise due to predisposing environmental factors [1, 7]. According to the reports of the national cancer institute, other risk factors for CRC are personal history of colorectal adenomas, previous colorectal or ovarian cancer, familial adenomatous polyposis (FAP) and Lynch syndrome (hereditary nonpolyposis colorectal cancer [HNPCC]), personal history of long-term chronic ulcerative colitis or Crohn colitis, heavy alcohol consumption, smoking, special race/ethnicities and obesity [5, 8–13].
Fig. 1.
Incidence rank of CRC according to WHO reports (https://gco.iarc.fr/today)
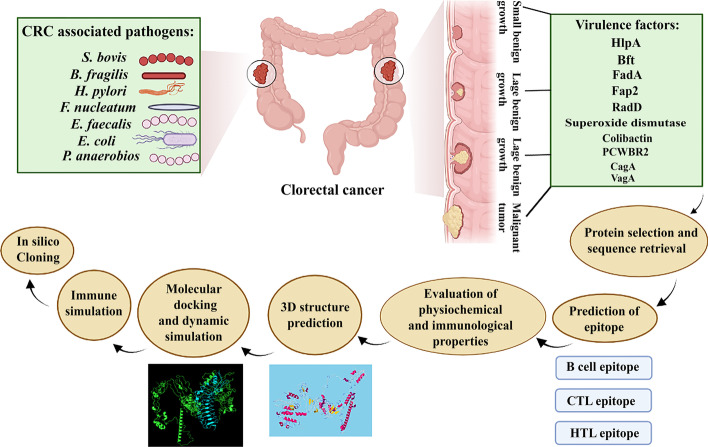
A remarkable result of intestinal dysbiosis is the replacement of commensal microorganisms with potential pathogens. Seven potential pathogens including Streptococcus bovis (S. bovis) [14, 15], Helicobacter pylori (H. pylori), Bacteroides Fragilis (B. Fragilis), Fusobacterium nucleatum (F. nucleatum) [16], Enterococcus faecalis (E. faecalis), Escherichia coli (E. coli) [17, 18], and Peptostreptococcus anaerobius (P. anaerobius) [19, 20] are eminent microorganisms involved in the occurrence of CRC [14].
The relationship between bacteria and malignancies is complex. In many cases, oral or intestine resident bacteria prevent the development of cancer by stimulating immune response and production of anti-inflammatory compounds [21] such as IL-10 and bacterial metabolites such as single chain fatty acids (SCFAs like butyrate and propionate) [22, 23], Lipopolysaccharide (LPS) in gram-negative bacteria [24] and ferrichrome in Lactobacillus casei [25]. On other hand, the importance of bacteria in inducing cancers such as H. pylori and gastric cancers is proven. H. pylori is carcinogenic by producing CagA and VacA toxins [26]. Inflammation processes triggered by the intestinal microorganisms can also cause cancer. These associations were observed in F. nucleatum and P. anaerobius, which colonize the oral cavity and can induce colorectal cancer by stimulating inflammation. Enterotoxigenic B. Fragilis (ETBF) stabilize intestinal colonization by biofilm formation and induce chronic inflammation and progression to cancers. E. faecalis and E. coli are transient members of the normal flora of the intestine, vagina and oral cavity. These species may cause CRC progression by producing virulence factors such as toxins and enzymes which cause chromosome instability in human chromosomes and cell cycle arresting in colon epithelial cells [27].
Histone-like protein A (HlpA) in S. gallolyticus, FadA, Fap2 and RadD in F. nucleatum [28], and PCWBR2 in P. anaerobius [1] are the main adhesins in CRC-related bacteria. Bacterial cell wall HlpA is the main surface immunogenic protein that enables S. gallolyticus to bind to Heparin sulfate proteoglycans (HSPGs) and stimulates a humoral immune response. Fap2 interacts and inactivates T lymphocytes in favor of tumor cell growth [29, 30]. RadD mediates biofilm formation and attachment of F. nucleatum cells to the host cells and the same as Fap2 supports the growth of tumor cells. It is also claimed that the interaction between the putative cell wall binding repeat 2 (PCWBR2) surface protein of P. anaerobius and α2/β1 integrin activates a signaling pathway associated with an increased risk of CRC [20]. Considering the potent immunogenic activity of the aforementioned proteins, they are potential vaccine candidates for the related bacteria causing CRCs.
Some bacterial protein toxins such as CagA and VacA produced by H. pylori [31–34], ETBF by B. fragilis and Cytolethal distending toxin (CDT) and Colibactin produced by E. coli are potent carcinogen promoters. They can elicit inflammatory reactions, interfere with signaling pathways and also may hamper cell cycles in favor of carcinogenesis. CagA and VacA toxins promote CRC during the reverse-feedback mechanism and hypergastrinemia and are known as the major factors of gastric cancer and possible inducers of CRC by affecting apoptosis and signal transduction systems of the cells, vacuolization and changing epithelial permeability, respectively [26, 35, 36]. ETBF is a zinc-dependent metalloprotease that cleaves E-cadherin molecules, and its interactions with intestinal epithelial cells lead to the activation of the STAT3 pathway. The toxin causes IL-2 reduction and IL-17 increase, which lead to the proliferation and survival of cancer cells [37, 38]. CDT, cytotoxic necrotizing factor (CNF), cycle inhibiting factor, Shiga toxin and subtilase toxin are important cyclomodulins which can alter the cell cycle in favor of bacterial invasion and colonization [39]. CDT and Colibactin arrest the cell cycle and damage the double-stranded structure of DNA by alkylating adenine bases [40]. Superoxide dismutase (SOD) as a virulence factor of E. faecalis induce damage in the DNA backbone and predisposes the colon epithelial cells to mutations and cancer [41–43]. To the best of our knowledge, no study is conducted on multi-epitope vaccines against different CRC-inducing pathogens, so the present study is the first report intending to design a multi-epitope vaccine based on in silico designing and immunoinformatics approach against the most important CRC-related bacterial pathogens.
Methods
In order to design multi-epitope vaccine against CRC-promoting bacteria, CD4+ and CD8+ T cell and B cell stimulating epitopes were selected. Then, validation, allergenicity, toxicity and physicochemical properties of all epitopes were performed using different web servers. Three linkers AAY, GPGPG and KK were used to connect cytotoxic T cell epitope (CTL), T-helper lymphocyte (HTL) and B-cell lymphocyte (BCL) epitopes, respectively. For assessment of the stability and binding affinity, the TLR4 receptor was docked by ligands using FireDock, PatchDock and ClusPro 2.0 servers. Finally, codon adaptation and in silico cloning studies were carried out. In addition, the C-ImmSim server was used to describe the humoral and cellular profile of the mammalian immune system against the designed vaccine. The workflow for this scientific study is shown in Fig. 2.
Fig. 2.
An overview of the steps of making a multi-epitope vaccine by in silico method in the present study
Retrieval of bacterial sequences associated with CRC
In this study, ten proteins including HlpA, BFT, (FadA, Fap2 and RadD), Superoxide, Colibactin, PCWBR2 and (CagA and VacA) were selected for design of multi-epitope vaccines against S. gallolyticus, ETBF, F. nucleatum, E. faecalis, E. coli, P. anaerobius and H. pylori, respectively. The related sequences were retrieved from the National Center for Biotechnology Information (NCBI) Protein Database (https://www.ncbi.nlm.nih.gov/protein/?term=) database (Table 1).
Table 1.
Details of sequences retrieved from CRC cancer related pathogens
| Organism | Protein | Accession numbers | Amino acids |
|---|---|---|---|
| Streptococcus gallolyticus | HlpA | KJF00052.1 | 91 aa |
| Bacteroides fragilis | Bft | AAB50410.2 | 389 aa |
| Fusobacterium nucleatum | FadA | AAY47043.1 | 129 aa |
| Fap2 | WP_059222898.1 | 3784 aa | |
| RadD | WP_238968484.1 | 3463 aa | |
| Enterococcus faecalis | Superoxide dismutase | EPI15045.1 | 202 aa |
| Escherichia coli | Colibactin | WP_193793145.1 | 3206 aa |
| Peptostreptococcus anaerobius | PCWBR2 | KXI10301.1 | 376 aa |
| Helicobacter pylori | CagA | P55980.1 | 1186 aa |
| VacA | AAU85846.1 | 746 |
Prediction of B cell epitopes
B cell epitopes play a pivotal role in the development of peptide vaccines, in the diagnosis of diseases as well as for allergy research [44]. Three servers, ABCpred (https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html), BCPREDS (http://ailab-projects1.ist.psu.edu:8080/bcpred/) and LBtope (https://webs.iiitd.edu.in/raghava/lbtope/protein.php), were used to predict B cell epitopes. We applied three different servers to obtain the best coverage of the predicted epitopes. ABCpred is developed based on a recursive neural network (machine-based technique) using a fixed length pattern and can predict epitopes with 65.93% accuracy using this network [44]. The BCPREDS server uses three developed methods, AAP, BCPred and FBCPred, to predict B cell epitopes [45, 46]. The server, on the other hand, uses a support vector machine (SVM) algorithm to predict B-cell (linear) epitopes. ABCpred and BCPREDS servers have variable epitope lengths (10–20) and (12–22) to predict B cell epitopes, respectively. The third server used to predict B cell was the LBtope server [47]. Due to the high accuracy of epitope prediction, we considered the cut-offs above 0.6 to predict B cell epitopes in this server.
MHC-I binding epitopes (CTL) prediction
Determination of peptide binding to major histocompatibility complex (MHC) class I is an important step in CTL detection methods for MHC class I peptide binding [48]. All 10 proteins in this study were screened for MHC-I (18 HLA-A, 32 HLA-B and 20 HLA-C) alleles based on the Immune Epitope Database server (IEDB; http://tools.iedb.org/mhci/) [49]. Length preferences can vary depending on the MHC allele but are generally limited to peptides of length 8–11 amino acids [50]. We considered 9-mer epitopes, NetMHCpan EL 4.1 method and a score above 0.5 to predict the desired epitopes.
MHC class II binding prediction
HLA class II molecules are expressed by human antigen-presenting cells (APCs) and are used directly to identify epitope candidates in infectious agents, allergens, cancer, and autoantigens [51]. The IEDB (http://tools.iedb.org/mhcii/) was used to predict HTL epitopes for 10 proteins. The IEDB parameters used for this study included a selection of peptide length 15 mer, IEDB recommended 2.22, and human Leukocyte Antigen (HLA) reference set (containing 27 alleles). The selection IEDB Recommended uses the Consensus approach, combining NN-align, SMM-align, CombLib and Sturniolo if any corresponding predictor is available for the molecule, otherwise, NetMHCIIpan is used [52]. Finally, to predict epitopes MHC class II with high binding power, we used adjusted rank < 2 to filter.
Vaccine construction
In the present study, CTL, HTL, and BCL were joined together with a suitable linker to make an effective multi-epitope vaccine. A total of three linkers, AAY, GPGPG, and KK were used to bind CTL, HTL and BCL epitopes, respectively. The reason for using these linkers is that they play a vital role in producing a wide conformation (flexibility), protein folding and separation of functional domains, and therefore they are able to make the protein structure more stable.
Prediction of various physicochemical properties
The study of physical and chemical properties reveals the functional and structural properties of a protein. ProtParam (http://web.expasy.org/protparam/) server was used to evaluate the physical and chemical properties of the final vaccine construct [53]. This server has various physical and chemical parameters of proteins such as amino acid composition, extinction coefficient, instability index, total hydropathic mean (GRAVY), aliphatic index, theoretical pI, atomic composition and molecular weight allowing us to understand the stability, activity and nature of proteins [54]. The instability index (II) of a protein indicates its stability of the protein. If the calculated protein instability index is less than 40, it was considered as a stable protein. GRAVY is used to indicate the hydrophobicity value of a peptide that calculates the sum of the hydropathic values of all amino acids divided by the length of the sequence [55]. In addition, the Aliphatic index (AI) is defined as the relative volume of protein occupied by its aliphatic side chains that are involved in the thermal stability of a protein.
Another important feature that should be considered in vaccine design is protein solubility, which is important in industrial and therapeutic applications. In this study, the Protein-Sol web server was used to predict protein solubility [56]. If our protein solubility score (scaled solubility value or QuerySol) was less than 0.45, it indicates that our protein is more soluble than the average soluble E. coli protein.
Identifying antigenicity, allergenicity and toxicity of protein sequences
The VaxiJen server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen_citation.html) is the first server for alignment-independent prediction of protective antigens [57]. This server examines bacterial, viral, and tumor protein datasets to predict protein antigenicity. In addition, it has shown a prediction accuracy of 70 to 89 percent [57]. In this study, our target organism was bacteria and other parameters were selected by default.
Allergy is a harmful consequence of a wrong immune response that has evolved to develop immunity to macroparasites [58]. The AllerTOPv.2 server (https://www.ddg-pharmfac.net/AllerTOP/) is used to predict allergenicity [59]. On the other hand, AllerTOP is known as the first suitable alignment-free server for in silico prediction of allergens based on the physicochemical properties of protein sequences. Version 2 of this server is a significant improvement over version 1 and has an accuracy of 88.7%. It is also highly sensitive (94%) compared to other allergenic prediction servers [59, 60]. ToxinPred (http://crdd.osdd.net/raghava/toxinpred/multi_submit.php) is a unique method in silicon that will be useful in predicting the toxicity of peptides/proteins, designing toxic peptides and detecting toxic regions in proteins [61].
Population coverage of epitopes
T cells detect a complex between a specific molecule of MHC and a specific pathogen-derived epitope [62]. Specific HLA alleles are expressed with very different frequencies in different ethnicities. Therefore, the IEDB population coverage server (http://tools.iedb.org/population/) was used in the design and development of T-cell epitope-based vaccines for population coverage analysis [62]. In this study, population coverage for a vaccine designed in both MHC Class I and MHC Class II types in different ethnicities was examined.
Secondary and tertiary structure prediction of the vaccine construct
One of the most important and challenging issues in the field of bioinformatics is the prediction of the secondary structure of the protein [63]. The secondary structure refers to the polypeptide backbone of local conformation proteins, which consists of three parts: regular secondary structure, α-helix and β-strand, and a type of irregular secondary structure, the coil region [63, 64]. In this study, PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) tool was used to predict the secondary structure. It is one of the most widely used servers that use two feed-forward neural networks to analyze the output obtained from PSI-BLAST [65].
The RaptorX server (http://raptorx.uchicago.edu/) was used to model the 3-dimensional (3D) structure. RaptorX offers high-quality structural models (5 models) for many purposes. It also takes the server about 35 min to complete the processing of a sequence of 200 amino acids. The server, on the other hand, is designed for protein secondary structure prediction, alignment quality assessment and sophisticated probabilistic alignment sampling [66].
Refinement, validation and quality assessment of the 3D structure
The importance of improving template-based model structures beyond the existing accuracy of template information in the structure prediction community is emphasized. For this reason, the GalaxyRefine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) was used to refine the structure of the protein model [67]. GalaxyRefine server has different parameters that include: global distance test-high accuracy (GDT-HA), root-mean-square deviation (RMSD), MolProbity, and Ramachandran favored score. MolProbity shows crystallographic resolution and typical scores for experimental structures range from 1 to 2. RMSD is the most commonly used quantitative measure of the similarity between two superimposed atomic coordinates [68]. A lower RMSD value indicates better stability, and an RMSD score between 0 and 1.2 Angstrom (Å) is usually acceptable.
UCLA-DOE LAB (https://saves.mbi.ucla.edu/) and ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) servers were used to evaluate the validity and quality of the selected 3D structure [69, 70]. The UCLA-DOE LAB server has various tools such as PROCHECK and ERRAT for 3D structure validation. Ramachandran diagram was analyzed using the PROCHECK section from the UCLA-DOE LAB server (http://molprobity.manchester.ac.uk/). The Ramachandran diagram shows the statistical distribution of the combination of the backbone dihedral angles φ and ψ, as well as the percentage and number of residues in the most favored, additional allowed, generously allowed, and disallowed region, which defines the quality of modeled structure [71].
ProSA-web server (https://prosa.services.came.sbg.ac.at/prosa.php) is a tool used to study 3D models of protein structures for possible errors [70]. One of the structural features derived from this server is the z-score. The z-score indicates overall model quality and measures the deviation of the total energy of the structure concerning an energy distribution derived from random conformations [72]. In addition, a plot of local quality scores points to problematic parts of the model which are also highlighted in a 3D molecule viewer to facilitate their detection [70].
Multi-epitope vaccine protein disulfide engineering
Disulfide bridges are formed between cysteine residues in peptides and proteins and are recognized as an essential element in the molecular architecture of proteins and peptides [73]. It is also believed that these bonds reduce conformational entropy and increase the free energy of the denatured state, thus increasing the stability of the protein structure [74]. In the present study, Disulfide by Design 2.0 (DbD2) (http://cptweb.cpt.wayne.edu/DbD2/) online server was used to detect disulfide bonds [75]. The server can provide refined 3D structures of the vaccine to identify residual pairs that can form disulfide bonds. When engineering the disulfide bonds, the intra-chain, inter-chain and Cβ for glycine residue, were selected and the v3 and Cα-Cβ-Sγ angles were kept at − 87° or + 97° ± 30 and 114.6° ± 10, respectively. Finally, an energy value of less than 1 kcal/mol was selected as the threshold for the remaining pair [75]. Because 90% of native disulfide bonds usually have an energy value of less than 2.2 kcal/mol [76].
Molecular docking of multi-epitope vaccine with TLR4
Docking is recognized as an important tool in computer-aided drug design. Protein–protein docking analysis was performed through the ClusPro 2.0 server [77]. This server requires two receptor and ligand files in the form of PDB. TLR-4 acts as a receptor for antigen recognition, which plays a role in immune activation and mediating cytokine induction [78]. The results obtained from this server include rigid body connection, clustering of the lowest energy structure, and structural refinement by minimizing energy. The vaccine-ligand complex was obtained based on the lowest energy and docking efficiency.
Docking analysis was again used by the PatchDock server (https://bioinfo3d.cs.tau.ac.il/PatchDock/) [79] to confirm the affinity of the vaccine structure designed with TLR4. The PatchDock server predicted potential complexity using three algorithm-molecular shape representations, surface patch matching, filtering, and scoring. Consequently, the top 10 results of the PatchDock server were evaluated using the FireDock (https://bioinfo3d.cs.tau.ac.il/FireDock/) server [80] to calculate the Global binding energy that consists of attractive and repulsive van der Waals (VdW) forces, atomic contact energy (ACE) and hydrogen bond. Before the docking process, the H2O molecules, ligands and polar hydrogens were removed while the Kollman charge was added. The structural coordinates of TLR4 were retrieved from the Protein Data Bank (PDB) (https://www.rcsb.org/) using the respective PDB ID: 2Z62. Finally, the visualization of complex vaccine-TLR interactions was performed by LigPlot + software.
Molecular dynamics simulation
The dynamic stability of the designed vaccine was investigated by performing a 100 ns molecular dynamics (MD) simulation. MD was performed using GROMACS package v2020, which provides a rich set of computational and analysis tools [81]. The parameters for MD simulation were derived from Amber sb99 force field and the system was the solvated by SPC/E water model. After electro-neutralization of the solvated simulation box the energy minimization was performed by the aim of steepest descend algorithm. Temperature and pressure were adjusted at 310 k and 1 bar respectively using a nose–hoover thermostat and a Parinello- Rahman barostat. All bonds were constrained by LINear Constraint Solver (LINCS) method. Both van der Waals and electrostatic non-bonded interactions were measured by the cutoff of 1 nm. In this regards the long range electrostatics were treated by PME method. Finally, a 100 ns MD simulation was carried out under the leap-frog algorithm.
Normal mode analysis (NMA)
The study of molecular dynamics (MD) is essential to evaluate the stability and physical motility of the vaccine-TLR4 docked complex in any in silico assay. Therefore, protein stability can be determined by comparing the dynamics of essential proteins with their normal modes [81, 82]. To perform the molecular dynamics simulation process, an iMODS server (http://imods.chaconlab.org/) based on a normal state analysis (NMA) conductor was used [82–84]. Then, the complex of vaccine construct-TLR was delivered to the iMODS server. This iMODS server evaluates the stability of a protein by calculating its NMA. The server also provides images of factor B-factor and deformability plots, covariance map, mode variance plot, eigenvalues and elastic networks.
Immune simulation
C-IMMSIM server (http://kraken.iac.rm.cnr.it/C-IMMSIM/index.php?page=1) is an agent-based simulator of the immune response that uses bioinformatics methods to predict T and B cell epitopes [85]. The C-ImmSim utilizes the Celada-Seiden model for describing both humoral and cellular profiles of a mammalian immune system against a designed vaccine. In summary, this server /C-IMMSIM displays images in which the major classes of cells of both the lymphoid [T helper lymphocytes (Th)], CTL, B lymphocytes, and antibody-producer plasma cells, PLB) and the myeloid lineage [macrophages (M) and dendritic cells] are represented [85]. The simulated parameters in this study included: (a) a vaccine without LPS, (b) considering three doses of vaccine (to create an efficient and long-lasting immune response) with time intervals of 1, 84 and 168 days, (c) the volume of the simulation and the simulation steps were adjusted to 10 and 1100, respectively. The other parameter "Random Seed" remains unchanged. It should be noted that one step of the simulation is equivalent to eight hours (8 h) of real-time, allowing immune response modeling for about 350 days [i.e. (1050 × 8 h)/(24 h)].
Codon-optimization and cloning for the design of multi-epitope vaccine
Today, we need a set of predictor servers to adapt the usage of the target gene codon for most sequenced prokaryotes and the eukaryotic gene expression hosts selected to improve heterologous protein production [86]. Java Codon Adaptation Tool (JCat) server (http://www.jcat.de/) was used to quantify the expression level of the multi-epitope vaccine in E. coli (strain K12). This server calculates two important outputs for the query sequence to ensure maximum expression. One of them is GC content and the other is Codon Adaptation Index (CAI) value [86]. CAI requires the definition of high-expression genes that allow a comparable value to be calculated for codon usage. Finally, the vaccine construct was cloned into plasmid pET-28a (+) using SnapGene software (version 5.2.3) (https://www.snapgene.com/).
Analysis of the vaccine MRNA
The Vienna RNA website is known as a comprehensive collection of tools for folding, designing and analyzing RNA sequences [87]. In this study, RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) web server was used to predict the secondary structure of MRNA. At this stage, after obtaining the optimized DNA sequence through the JCat server, for analysis of MRNA folding and vaccine secondary structure, first converted into a potential DNA sequence by DNA <—> RNA- > Protein at (http://biomodel.uah.es/en/lab/cybertory/analysis/trans.htm). Finally, the minimum free energy (MFE) score was important to us. MFE of ribonucleic acids (RNAs) increases at an apparent linear rate with sequence length. Simple indices, obtained by dividing the MFE by the number of nucleotides, have been used for a direct comparison of the folding stability of RNAs of various sizes [88].
Results
Retrieval of bacterial sequences associated with colorectal cancer
Ten protein sequences with different amino acid lengths from colorectal cancer-related pathogens were retrieved in the FASTA format.
Prediction of B cell epitopes
The reason for examining B-cell epitopes is their extraordinary ability to neutralize pathogenic molecules through the secretion of antibodies [89, 90]. ABCpred, BCPREDS and LBtope servers were used for B-cell prediction. Preliminary analysis showed that a total of 19 epitopes with criteria such as antigenic, non-allergenic and non-toxic were selected (Table 2). It should be noted that epitopes were considered for the final vaccine that overlapped at least two or three high-score servers.
Table 2.
Prediction of B cell epitopes based on ABCpred, LBtop and BCPREDS servers
| Protein | Length | Peptide | Start | ABCpred | LBtop | BCPREDS | Antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|---|---|
| HlpA | 12 | NFEVRERAARKG | 50 | 0.54 | – | 0.99 | 1.7361 | NON-ALLERGEN | Non-Toxin |
| BFT | 14 | SLKANPKAEGYDDQ | 278 | 0.74 | – | 0.905 | 1.2 | NON-ALLERGEN | Non-Toxin |
| 20 | TEYSCPSGNADEGLDGFTAS | 259 | 0.87 | 0.64 | – | 0.9369 | NON-ALLERGEN | Non-Toxin | |
| FadA | 12 | SQYQDLASKYED | 89 | 0.7 | 0.6 | – | 0.5765 | NON-ALLERGEN | Non-Toxin |
| 18 | LDAEYQNLANQEEARFNE | 32 | 0.78 | 0.61 | – | 0.7945 | NON-ALLERGEN | Non-Toxin | |
| Fap2 | 12 | DGASTNPDPNKL | 2518 | – | 0.77 | 0.999 | 1.033 | NON-ALLERGEN | Non-Toxin |
| 18 | EEVNLENSQVATREELKT | 42 | 0.87 | 0.66 | 0.928 | 1.1408 | NON-ALLERGEN | Non-Toxin | |
| RadD | 12 | EGTNNEVDHNTD | 1612 | 0.72 | 68.65 | 0.986 | 1.621 | NON-ALLERGEN | Non-Toxin |
| 14 | DLGTIDFNGDDGVG | 1222 | 0.76 | 70.12 | 0.974 | 1.3681 | NON-ALLERGEN | Non-Toxin | |
| Superoxide | 16 | HPELGEKSVEDLISDM | 46 | 0.81 | 0.62 | – | 0.5832 | NON-ALLERGEN | Non-Toxin |
| 20 | IPEDIRTAVRNNGGGHANHT | 64 | – | 0.61 | 0.882 | 1.0386 | NON-ALLERGEN | Non-Toxin | |
| Colibactin | 16 | LEAHQHEDDPSATGVR | 1503 | – | 0.64 | 1 | 1.3 | NON-ALLERGEN | Non-Toxin |
| 20 | QPPEGESNAPSPQPAVQTNT | 3163 | – | 0.78 | 1 | 1 | NON-ALLERGEN | Non-Toxin | |
| PCWBR2 | 12 | INKLNVSRISGK | 70 | 0.64 | 0.7 | – | 0.7269 | NON-ALLERGEN | Non-Toxin |
| 18 | RYETSVKVSDELQKMSSG | 83 | 0.78 | 0.65 | – | 0.9622 | NON-ALLERGEN | Non-Toxin | |
| CagA | 12 | NASKNPNKGVGA | 515 | – | 0.69 | 0.99 | 1.1717 | NON-ALLERGEN | Non-Toxin |
| 14 | ESSTKSFQKFGDQR | 108 | 0.74 | 0.65 | 0.89 | 0.6742 | NON-ALLERGEN | Non-Toxin | |
| VacA | 14 | VGGYKASLTTNAAH | 408 | – | 67.3 | 0.95 | 0.8495 | NON-ALLERGEN | Non-Toxin |
| 20 | NFEFKAGTDTKNGTATFNND | 472 | 0.87 | 0.67 | 0.95 | 1.5291 | NON-ALLERGEN | Non-Toxin |
MHC-I binding epitopes (CTL) prediction
The MHC-I binding epitopes (9 mer) predicted by the IEDB recommended method for 70 available alleles (including 18 HLA-A, 32 HLA-B, and 20 HLA-C) were performed by the IEDB server. Among a large number of MHC-I predicted epitopes, 18 epitopes were selected as vaccine candidates. The selection of epitopes based on characteristics such as high score (good binder), antigenic, non-allergenic and non-toxic is shown in Table 3.
Table 3.
Most probable predicted epitopes with MHC class I alleles from IEDB analysis tool
| Protein | Peptide sequence | Start | End | Allele | Score | Antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|---|
| HlpA | SAAAVDAVF | 22 | 30 | HLA-B*35:01 | 0.861527 | 0.5365 | NON-ALLERGEN | Non-Toxin |
| Bft | KANPKAEGY | 280 | 288 | HLA-A*30:02 | 0.830051 | 1.0961 | NON-ALLERGEN | Non-Toxin |
| HLA-B*15:01 | 0.665572 | |||||||
| HLA-B*57:01 | 0.615497 | |||||||
| HLA-B*58:01 | 0.596159 | |||||||
| YPGVMAHEL | 333 | 342 | HLA-B*35:01 | 0.819059 | 0.6723 | NON-ALLERGEN | Non-Toxin | |
| HLA-B*07:02 | 0.718513 | |||||||
| HLA-B*53:01 | 0.716264 | |||||||
| HLA-B*51:01 | 0.59173 | |||||||
| HLA-B*08:01 | 0.523581 | |||||||
| FadA | QVYNELSQR | 66 | 74 | HLA-A*68:01 | 0.957771 | 0.8312 | NON-ALLERGEN | Non-Toxin |
| HLA-A*31:01 | 0.795891 | |||||||
| HLA-A*11:01 | 0.657712 | |||||||
| HLA-A*33:01 | 0.58309 | |||||||
| HLA-A*03:01 | 0.508062 | |||||||
| Fap2 | KTISVTAEK | 1975 | 1983 | HLA-A*11:01 | 0.978654 | 0.7777 | NON-ALLERGEN | Non-Toxin |
| HLA-A*03:01 | 0.956606 | |||||||
| HLA-A*30:01 | 0.866036 | |||||||
| HLA-A*68:01 | 0.820562 | |||||||
| HLA-A*31:01 | 0.691021 | |||||||
| FADGLEQRY | 3426 | 3434 | HLA-A*01:01 | 0.972812 | 1.1206 | NON-ALLERGEN | Non-Toxin | |
| HLA-B*35:01 | 0.923357 | |||||||
| HLA-B*53:01 | 0.519186 | |||||||
| RadD | GANPSVEYW | 304 | 312 | HLA-B*58:01 | 0.991946 | 1.1284 | NON-ALLERGEN | Non-Toxin |
| HLA-B*57:01 | 0.987697 | |||||||
| HLA-B*53:01 | 0.73539 | |||||||
| KEQENISQM | 58 | 66 | HLA-B*40:01 | 0.954627 | 0.4345 | NON-ALLERGEN | Non-Toxin | |
| HLA-B*44:03 | 0.828703 | |||||||
| HLA-B*44:02 | 0.783902 | |||||||
| superoxide dismutase | YIDVETMHL | 18 | 26 | HLA-A*02:06 | 0.78953 | 1.3306 | NON-ALLERGEN | Non-Toxin |
| HLA-A*02:01 | 0.737224 | |||||||
| TPVLGLDVW | 158 | 166 | HLA-B*53:01 | 0.930286 | 1.9457 | NON-ALLERGEN | Non-Toxin | |
| HLA-B*35:01 | 0.50924 | |||||||
| Colibactin | YLDALAQQL | 2339 | 247 | HLA-A*02:01 | 0.981781 | 0.4388 | NON-ALLERGEN | Non-Toxin |
| HLA-A*02:06 | 0.926393 | |||||||
| HLA-A*02:03 | 0.777941 | |||||||
| KADLAQLRY | 970 | 978 | HLA-A*01:01 | 0.970875 | 0.9347 | NON-ALLERGEN | Non-Toxin | |
| HLA-A*30:02 | 0.650643 | |||||||
| HLA-B*58:01 | 0.611357 | |||||||
| PCWBR2 | YIIQNIYLV | 149 | 157 | HLA-A*02:06 | 0.914671 | 0.6827 | NON-ALLERGEN | Non-Toxin |
| HLA-A*02:01 | 0.884041 | |||||||
| HLA-A*02:03 | 0.70463 | |||||||
| HLA-A*68:02 | 0.587899 | |||||||
| RVDTAFAVY | 187 | 195 | HLA-A*01:01 | 0.860066 | 0.4398 | NON-ALLERGEN | Non-Toxin | |
| HLA-A*30:02 | 0.801369 | |||||||
| HLA-B*15:01 | 0.533769 | |||||||
| CagA | VPASLSAKL | 1050 | 1058 | HLA-B*07:02 | 0.925163 | 1.1446 | NON-ALLERGEN | Non-Toxin |
| HLA-B*35:01 | 0.668443 | |||||||
| HLA-B*53:01 | 0.618146 | |||||||
| HLA-B*51:01 | 0.60881 | |||||||
| GINPEWISK | 735 | 743 | HLA-A*11:01 | 0.904802 | 1.2858 | NON-ALLERGEN | Non-Toxin | |
| HLA-A*03:01 | 0.83302 | |||||||
| VacA | GEKLVIDEF | 600 | 608 | HLA-B*44:03 | 0.863097 | 0.4654 | NON-ALLERGEN | Non-Toxin |
| HLA-B*44:02 | 0.801589 | |||||||
| HLA-B*40:01 | 0.72177 | |||||||
| RVNNQVGGY | 456 | 464 | HLA-A*30:02 | 0.84623 | 1.5337 | NON-ALLERGEN | Non-Toxin | |
| HLA-B*15:01 | 0.597925 |
MHC class II binding prediction
MHC-II binding epitopes (15 mer) were examined for 27 alleles (including HLA-DR, HLA-DQ, and HLA-DP) using the IEDB-recommended method. From a large number of HTL epitopes, we selected 19 epitopes with a length of 15 amino acids, which are shown in Table 4. The criteria for selecting these epitopes were low adjusted rank (good binder), antigenic, non-allergenic and non-toxic properties.
Table 4.
Most probable predicted epitopes with MHC class II alleles from IEDB analysis tool
| Protein | Peptide sequence | Start | End | Allele | Score | Antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|---|
| HlpA | KKDSAAAVDAVFSAI | 19 | 33 | HLA-DQA1*04:01/DQB1*04:02 | 2.5 | 0.5391 | NON-ALLERGEN | Non-Toxin |
| Bft | SFILGDEFAVLRFYR | 94 | 108 | HLA-DPA1*01:03/DPB1*04:0 | 0.24 | 0.4059 | NON-ALLERGEN | Non-Toxin |
| HLA-DPA1*02:01/DPB1*01:01 | 0.29 | |||||||
| HLA-DPA1*01:03/DPB1*02:01 | 0.97 | |||||||
| HLA-DPA1*03:01/DPB1*04:02 | 1.3 | |||||||
| HGLKRFVNLHFVLYT | 244 | 258 | HLA-DRB1*15:01 | 0.33 | 0.4609 | NON-ALLERGEN | Non-Toxin | |
| HLA-DPA1*01:03/DPB1*04:01 | 1.4 | |||||||
| FadA | AVLAVSASAFAATDA | 8 | 22 | HLA-DQA1*03:01/DQB1*03:02 | 0.97 | 0.4315 | NON-ALLERGEN | Non-Toxin |
| HLA-DRB1*09:01 | 1.3 | |||||||
| HLA-DQA1*05:01/DQB1*03:01 | 1.5 | |||||||
| SLVGELQALDAEYQN | 24 | 38 | HLA-DQA1*05:01/DQB1*02:01 | 1.3 | 0.4966 | NON-ALLERGEN | Non-Toxin | |
| Fap2 | AVLVANNGANVEIAS | 1112 | 1126 | HLA-DRB1*13:02 | 0.01 | 0.7538 | NON-ALLERGEN | Non-Toxin |
| HLA-DRB3*02:02 | 0.15 | |||||||
| HLA-DQA1*01:02/DQB1*06:02 | 0.34 | |||||||
| EKIKNLRLELIQLME | 77 | 91 | HLA-DRB4*01:01 | 0.21 | 0.6094 | |||
| HLA-DPA1*03:01/DPB1*04:02 | 0.99 | |||||||
| HLA-DPA1*02:01/DPB1*01:01 | 1.5 | |||||||
| RadD | LVKFNINATKAIGIL | 599 | 613 | HLA-DRB3*02:02 | 0.02 | 0.4117 | NON-ALLERGEN | Non-Toxin |
| HLA-DRB1*07:01 | 0.21 | |||||||
| HLA-DRB1*13:02 | 0.89 | |||||||
| HLA-DRB1*09:01 | 1.4 | NON-ALLERGEN | Non-Toxin | |||||
| HLA-DPA1*02:01/DPB1*14:01 | 2 | |||||||
| AKLINNMNVTVGVDA | 2626 | 2640 | HLA-DRB1*13:02 | 0.15 | 0.806 | |||
| HLA-DRB3*02:02 | 0.3 | |||||||
| superoxide dismutase | ELPYAYDALEPYIDV | 7 | 21 | HLA-DRB3*01:0 | 0.93 | 0.4856 | NON-ALLERGEN | Non-Toxin |
| HLA-DQA1*05:01/DQB1*02:0 | 0.24 | |||||||
| HLA-DQA1*01:01/DQB1*05:01 | 1.7 | NON-ALLERGEN | Non-Toxin | |||||
| KAAFKTAATGRFGSG | 116 | 130 | HLA-DRB1*09:01 | 0.36 | 0.8373 | |||
| HLA-DPA1*02:01/DPB1*14:01 | 0.53 | |||||||
| Colibactin | QKGFRFSIAYALNYL | 425 | 438 | HLA-DPA1*02:01/DPB1*14:01 | 0.01 | 1 | NON-ALLERGEN | Non-Toxin |
| HLA-DRB1*07:0 | 0.14 | |||||||
| HLA-DRB3*02:02 | 0.33 | |||||||
| HLA-DRB5*01:01 | 0.37 | NON-ALLERGEN | Non-Toxin | |||||
| HLA-DPA1*01:03/DPB1*04:01 | 0.47 | |||||||
| HLA-DRB1*01:01 | 1.6 | |||||||
| HLA-DRB1*09:01 | 1.6 | |||||||
| ALPIAYLTAYYALVV | 2797 | 2811 | HLA-DRB1*01:01 | 0.2 | 0.6023 | |||
| HLA-DRB1*12:01 | 1.11 | |||||||
| HLA-DRB1*01:01 | 1.8 | |||||||
| HLA-DPA1*01:03/DPB1*04:01 | 0.56 | |||||||
| HLA-DPA1*01:03/DPB1*02:01 | 1.2 | |||||||
| PCWBR2 | DALTAGTMAAELEIP | 115 | 129 | HLA-DQA1*04:01/DQB1*04:02 | 0.61 | 0.8571 | NON-ALLERGEN | Non-Toxin |
| HLA-DQA1*01:02/DQB1*06:02 | 1.1 | |||||||
| HLA-DQA1*03:01/DQB1*03:02 | 1.6 | |||||||
| LEIPLLLTKSNKLPD | 126 | 140 | HLA-DRB1*15:01 | 0.84 | 0.4256 | |||
| CagA | FMEFLAQNNTKLDNL | 404 | 418 | HLA-DRB1*04:01 | 0.41 | 0.5405 | NON-ALLERGEN | Non-Toxin |
| HLA-DRB3*02:02 | 0.77 | |||||||
| HLA-DRB3*02:02 | 0.54 | 0.5348 | ||||||
| HLA-DRB1*13:02 | 0.7 | |||||||
| YKFNQLLIHNNALSS | 292 | 306 | HLA-DRB1*04:01 | 1.3 | NON-ALLERGEN | Non-Toxin | ||
| VacA | EYDLYKSLLSSKIDG | 97 | 111 | HLA-DRB1*01:01 | 0.19 | 0.6034 | NON-ALLERGEN | Non-Toxin |
| HLA-DRB1*07:01 | 0.69 | |||||||
| HLA-DRB1*04:05 | 0.72 | |||||||
| KLVIDEFYYSPWNYF | 602 | 616 | HLA-DRB1*04:01 | 0.94 | NON-ALLERGEN | Non-Toxin | ||
| HLA-DPA1*01:03/DPB1*04:01 | 0.61 | 0.6653 | ||||||
| HLA-DPA1*01:03/DPB1*02:01 | 2 |
Prediction of various physicochemical properties
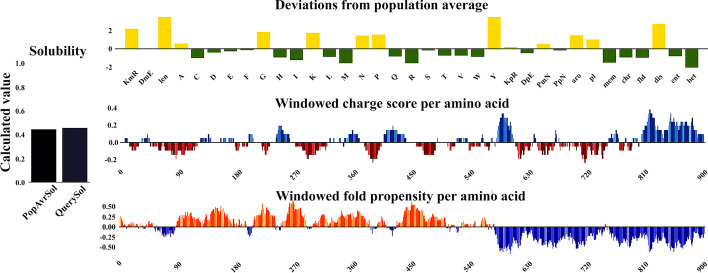
The final vaccine construct containing 924 amino acids and its molecular weight was determined based on ProtParam server 99 kDa. Since the final molecular weight of our final construct is less than 110 kDa, it can be considered a suitable vaccine [91]. The vaccine contained 105 (Arg + Lys) positively charged residues. The estimated half-life is 1.9 h (mammalian reticulocytes, in vitro), 20 h (yeast, in vivo), and more than 10 h (E. coli, in vivo). The vaccine construct was composed of 13,973 atoms, and its chemical formula was C4511H6895N1187O1370S10. The aliphatic index was 69.36 and the grand average hydropathicity index (GRAVY) was − 0.564, which reflects the vaccine’s polar nature and effective interaction with water, suggesting high solubility. The instability index was calculated at 23.00, which was < 40, classifying the vaccine as a stable protein (Table 5). The solubility of the vaccine construct was 0.460 according to QuerySol (Fig. 3).
Table 5.
Physicochemical properties of the final vaccine construct
| Characteristics | Assessment |
|---|---|
| Number of amino acids | 924 |
| Molecular weight | 99 KDa |
| Theoretical pI | 8.13 |
| Total number of positively charged residues (Arg + Lys) | 105 |
| Total number of atoms | 13,973 |
| Chemical formula | C4511H6895N1187O1370S10 |
| Estimated half-life (mammalian reticulocytes, in vitro), (yeast, in vivo), and (Escherichia coli, in vivo) | 1.9, 20 and 10 h |
| Aliphatic index | 69.36 |
| Instability index | 23.00 |
| Grand average of hydropathicity (GRAVY) | − 0.564 |
| Solubility | 0.460 |
| Antigenicity | 0.8952% |
| Allergenicity | Non-Allergen |
Fig. 3.
The solubility of the vaccine structure according to QuerySol was 0.460, which showed that it has good solubility
Evaluation of antigenicity, allergenicity and toxicity of protein sequences
The designed multi-epitope vaccine was evaluated for antigenic, non-allergenic and non-toxic properties. The antigenicity of the final vaccine construct was predicted at 0.8952% by VaxiJen at a 0.4% threshold for the bacterial model. Allergenicity and toxicity were evaluated to ensure that the candidate vaccine did not have any allergic reactions or toxic effects after entering the body. As predicted by AllerTOP 2.0 and ToxinPred web servers, the vaccine candidate was non-allergenic and non-toxic.
Multi-epitope vaccine construction
The multi-epitope vaccine construct was composed of a combination of 37 T cells (18 MHC-I and 19 MHC-II epitopes) and 19 linear B cell epitopes using AAY, GPGPG and KK linkers. All predicted epitopes were carefully selected and shown to be non-allergen, non-toxic, and highly antigenic.
Population coverage and conservancy of epitopes
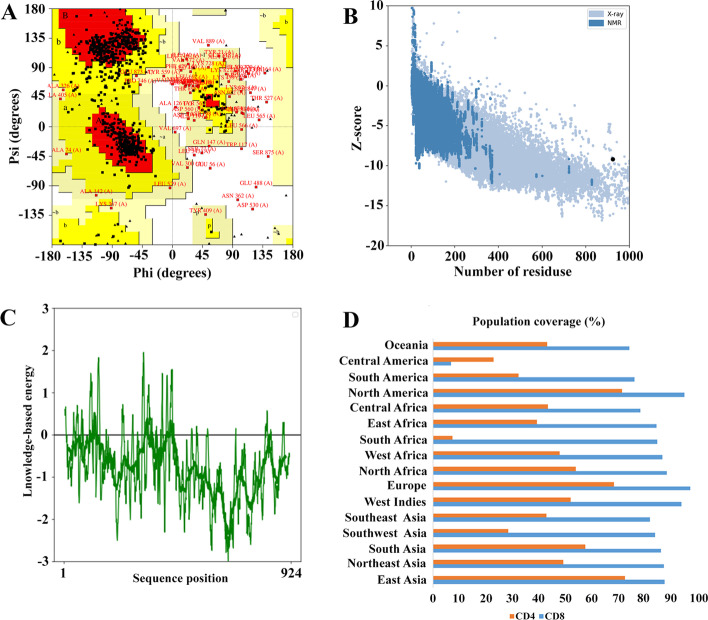
The potential efficacy of a potential vaccine can be determined by the frequency of distribution of HLA alleles in different ethnicities. In this study, population coverage in CD8+ and CD4+ T cells was investigated separately as well as their combined effect. The predicted T cell epitopes (CD8+ and CD4+) were exposed to population coverage in 16 different geographical regions of the world, as shown in Fig. 4D. Analyzes showed that among 18 CD8+ T cell epitopes, the highest coverage was in Europe (98.07%), North America (95.61%), and West India (94.69%). After that, North Africa (89.06%), East Asia (88.21%), Northeast Asia (88.03%), West Africa (87.42%), South Asia (86.82%), South Africa (85.42%), East Africa (85.26%), Southwest Asia (84.63%), Southeast Asia (82.77%), Central Africa (78.93%), Oceania (74.80%), South America (76.88%) provided other coverage. While the lowest coverage was related to the region Central America (7.01%).
Fig. 4.
Vaccine 3D Structure Validation by UCLA-DOE LAB and ProSA-web. A The statistics of the Ramachandran chart show the most favorable region, additionally allowed, generously and disallowed (outlier) area with 70.7%, 20.8%, 5.8%, and 2.7%, respectively. B Based on ProSA-web, the Z-score of the refined model is − 9.2. C The server also draws a plot to check the quality of the local model, which negative values indicating that there is no error in the structure of the model. D Worldwide population coverage rates based on CD8+ T cell epitopes and CD4+ T cell epitopes
On the other hand, the population coverage results for CD4+ T cell epitopes are shown in Fig. 4D. The highest coverage for CD4+ T-cell epitopes was found in East Asia (73.14%), North America (71.89%), and Europe (68.97%). Other results were reported in South Asia (58.10%), North Africa (54.47%), West Indies (52.58%), Northeast Asia (49.82%), West Africa (48.30%), Central Africa (43.89%), Oceania (43.57%), Southeast Asia (43.38%), East Africa (39.63%), and South America (32.67%). The lowest population coverage was for Southwest Asia (28.80%), Central America (23.09%) and South Africa (7.65%).
Secondary and 3D structure prediction of the vaccine construct
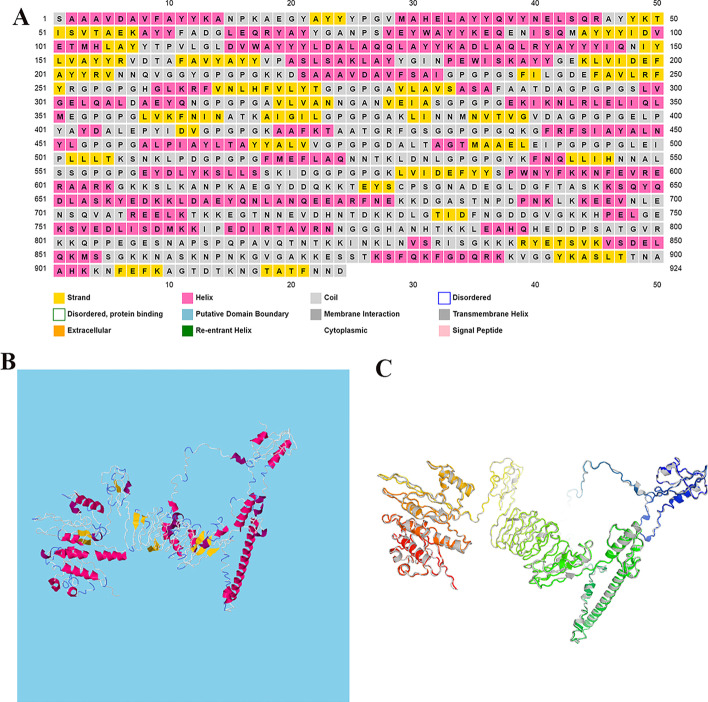
The secondary (i.e. α-helix, β-strand, and random coil) and 3D structure of the final vaccine construct were predicted by PSIPRED and RaptorX servers, respectively. According to the PSIPRED server, the final vaccine contained 34% of the amino acids in the α-helix structure and 16.66% and 49.34% of the amino acids in the β-strand and coil structures, respectively (Fig. 5A).
Fig. 5.
Displays the second and third structures of the final vaccine contracture. A In this Figure, the β-strands, the α-helix, and the random coils are shown in yellow, pink, and gray colors, respectively. B The 3D structure of a multi-epitope vaccine that was selected as the best model by the RaptorX server. β-strands, the α-helix, and the random coils are shown in yellow, red, and white-blue colors, respectively. C The 3D structure of multi-epitope vaccine after refinement
Five models were suggested by the RaptorX server for 3D structure in PDB format. Among the five models proposed by the RaptorX server, the structure of Model 3 (Fig. 5B) was selected.
Refinement, validation and quality assessment of the tertiary structure
The GalaxyRefine server was used to increase the overall and partial structural quality of the final vaccine construct. Among the 5 models proposed by this server, the best-refined model (5C) is shown in Fig. 5C with a GDT-HA score of 0.8888, an RMSD score of 0.571, a MolProbity score of 2.614, a Clash score of 32.2 and a Ramachandran score of 88.5 (Table 6). Therefore, it can be concluded that the quality of the refined model is high compared to the raw structure.
Table 6.
Quality scores of 5 models predicted by GalaxyRefine server
| Model | GDT-HA | RMSD | MolProbity | Clash score | Poor rotamers | Rama favored |
|---|---|---|---|---|---|---|
| Initial | 1.0000 | 0.000 | 5.202 | 318.7 | 89.4 | 77.7 |
| MODEL 1 | 0.8885 | 0.576 | 2.730 | 32.5 | 1.5 | 88.2 |
| MODEL 2 | 0.8861 | 0.587 | 2.586 | 32.8 | 1.0 | 88.5 |
| MODEL 3 | 0.8883 | 0.586 | 2.568 | 30.3 | 1.0 | 87.9 |
| MODEL 4 | 0.8899 | 0.588 | 2.581 | 31.1 | 1.0 | 87.7 |
| MODEL 5 | 0.8888 | 0.571 | 2.614 | 32.2 | 1.1 | 88.5 |
In validation, Ramachandran diagram analysis based on the PROCHECK server showed that 70.7%, 20.8%, 5.8% and 2.7% of protein residues were located in the most favored region, additional allowed, generously and disallowed (outlier) area of the final vaccine, respectively. (Fig. 4A). The quality and potential errors in the final vaccine 3D model were verified by ProSA-web. The Z-score, which indicates the overall quality of the model, was − 9.2 (Fig. 4B). However, a model with a lower Z-score is considered a higher-quality model. In addition, a plot was drawn to check the quality of the local model, where negative values indicate that there is no error in the model structure (Fig. 4C).
Protein disulfide bridging for vaccine stability
Disulfide engineering was applied to the multi-epitope vaccine construct refined model via DbD2. Four pairs of amino acids from the vaccine construct: 375GLY-377GLY, 642THR-645LYS, 668LEU-702SER, and 826ASN-844VAL, were selected for disulfide bond by the mutation because the bond energy had less than 1 kcal/mol. In addition, these mutations are shown in Fig. 6.
Fig. 6.
Disulfide engineering display in the final vaccine construct. The original form is shown on the left and the mutant form is on the right
Molecular docking of multi-epitope vaccine with TLR4 receptor
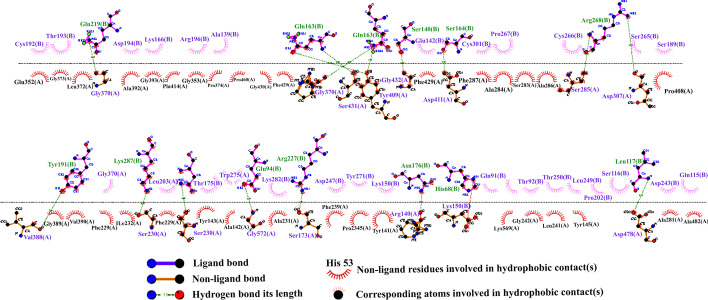
Molecular docking can evaluate the interactions between a ligand molecule and the receptor molecule to check the stability and binding affinity of their docked complex [84]. In this study, TLR4 was selected as the receptor for molecular binding. The energy scores obtained for best docking the vaccine-TLR4 complex from ClusPro v2.0 and PatchDock servers were − 1232.7 and − 32.40, respectively, which indicates a very good binding affinity. The score of the top models is shown in Table 7. These complexes were subjected to MD simulation to analyze their stability. In addition, a schematic diagram of the interaction between the vaccine structure and TLR4 was created by LigPlot + software (Fig. 7). Hydrogen bonds and salt bridge interactions were obtained by the DIMPLOT program. DIMPLOT was shown, Gly370, Arg428, Ser431, Tyr409, Gly432, Asp411, Ser285, Asp307, Val388, Gly233, Ser230, Gly572, Ser173, Arg140, Asp571, Asp478 residues from chain A of the vaccine were bound to Gln219, Gln188, Gln163, Ser140, Arg268, Tyr191, Lys278, Asn279, Glu94, Arg227, Asn176, His68, Leu117 residues from chain B by hydrogen bonds with bond lengths of 2.84 angstroms (Ǻ), 2.96 Ǻ, 3.26 Ǻ, 1.48 Ǻ, 2.94 Ǻ, 2.55 Ǻ, 2.54 Ǻ, 2.85 Ǻ, 3.21 Ǻ, 2.44 Ǻ, 2.66 Ǻ, 2.55 Ǻ, 1.5 Ǻ, 2.66 Ǻ, 3.33 Ǻ respectively. Also, the Asp571 residue from chain A of the vaccine binds to His68 residue from chain B of the TLR4 by salt bridge interaction.
Table 7.
Top models of docked complexes of designed vaccine with TLR4
| Cluster | Members | Representative | Weighted score |
|---|---|---|---|
| 0 | 147 | Center | − 843.1 |
| Lowest energy | − 1232.7 | ||
| 1 | 68 | Center | − 875.7 |
| Lowest energy | − 1098.3 | ||
| 2 | 65 | Center | − 950.8 |
| Lowest energy | − 1080.8 | ||
| 3 | 60 | Center | − 1241.8 |
| Lowest energy | − 1251.3 | ||
| 4 | 56 | Center | − 934.1 |
| Lowest energy | − 978.6 | ||
| 5 | 49 | Center | − 863.9 |
| Lowest energy | − 1174.1 | ||
| 6 | 26 | Center | − 856.9 |
| Lowest energy | − 926.3 | ||
| 7 | 22 | Center | − 844.5 |
| Lowest energy | − 949.0 | ||
| 8 | 18 | Center | − 993.8 |
| Lowest energy | − 993.8 | ||
| 9 | 17 | Center | − 935.4 |
| Lowest energy | − 935.4 | ||
| 10 | 16 | Center | − 856.4 |
| Lowest energy | − 946.8 |
Fig. 7.
Representation of interacting residues between vaccine docked with TLR4. The Gly370, Arg428, Ser431, Tyr409, Gly432, Asp411, Ser285, Asp307, Val388, Gly233, Ser230, Gly572, Ser173, Arg140, Asp571, Asp478 residues from chain A of the vaccine were bound to Gln219, Gln188, Gln163, Ser140, Arg268, Tyr191, Lys278, Asn279, Glu94, Arg227, Asn176, His68, Leu117 residues from chain B by hydrogen bonds
Molecular dynamic simulation
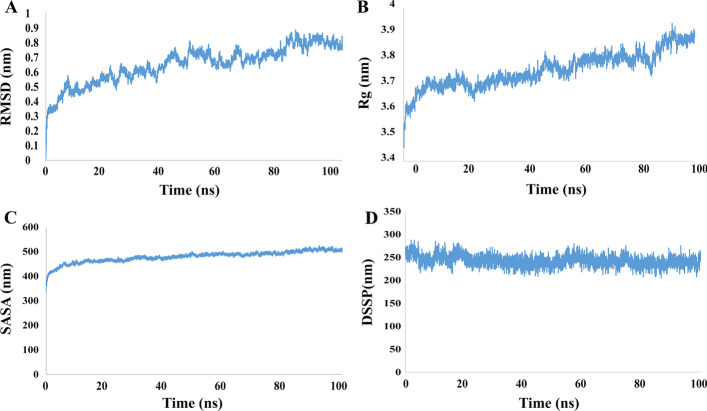
To evaluate the dynamic properties of the final vaccine, MD simulation was performed for 100 ns the results are represented in Fig. 8. At first, it is important to ensure that the simulation time is sufficient for a particular system. The best method for this is to measure the RMSD of the system during MD simulation. The results of this analysis is showed in Fig. 8A and as it is clear the protein reached its equilibrated state at the time of 50 ns was which followed by some fluctuations in diagram and this confirms that the simulation time is enough for this system. Also it can be seen that there is no sever fluctuation in RMSD diagram which is an index for structural stability of the as designed vaccine. Another issue that might be considered in order to vaccine retain its function is ensuring that the protein is not compressed and the epitopes are not inaccessible. Analyzing the radius of gyration (Rg) of protein is used in MD to evaluate time dependence changes in compactness of its structure. Figure 8b shows the changes in Rg for the designed vaccine during the simulation. As can be seen, the value of Rg is increased for the protein which indicated that its conformation is expanded after simulation. Another confirmation for this can be achieved by analyzing the value of solvent accessible surface area (SASA). The result of SASA analysis is reported in Fig. 8c and as it is clear in its diagram, the surface area is increased along the simulation time. Together with Rg analysis, these results suggest that the structure of protein did not undergo compactness and this prevent disabling the vaccine epitopes. Another important factor for vaccine Immuno-modulation is stability in its secondary structures which can be investigated by an analysis called DSSP. Figure 8d shows the changes in protein secondary structures during the simulation time. After 100 ns of MD simulation there is just less than 8 percent of protein residues which undergoes denaturation from their secondary structures. This predicts structural stability of the designed vaccine under the similar condition to which it may be assigned. In conclusion the results of MD simulations confirm that the as designed vaccine maintain its functional state in solution and can be tested for its Immuno-modulation ability in experimental.
Fig. 8.
The final construct of the molecular dynamics simulation vaccine with GROMACS software
NMA evaluation of the vaccine-receptor complex
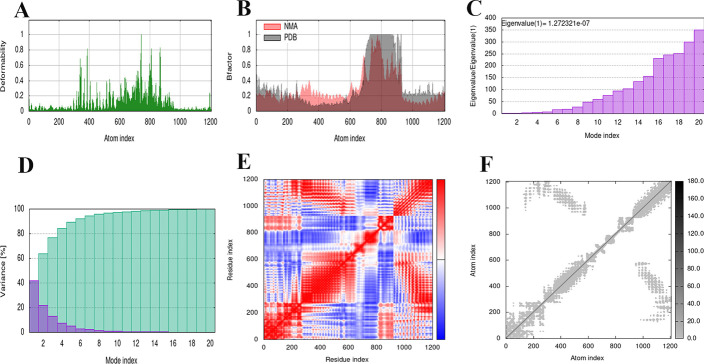
NMA was conducted to scrutinize protein stabilization and their large-scale mobility [92]. MD simulation of the vaccine candidate/TLR4 interactions is shown in Fig. 9. Figure 9A shows the deformation of the protein flexibility, which depends on the individual distortion of each residue depicted by the chain hinges. On the other hand, locations with hinges are areas with high deformability and illustrate a stable binding. The b-factor shows the relative amplitude of atomic displacement for the equilibrium position. According to Fig. 9B, few fluctuations of atomic displacement were observed for the TLR4-vaccine complex. Figure 9C showed the eigenvalue determined for the complex, which was 1.272 e-07. This Figure also showed that it has relatively least energy required to deform its structure based on the lowest eigenvalue. Figure 9D shows the variance plot of the complexes. In this diagram, the variance associated with the eigenvalue is inversely related to the individual variance shown by the blue-colored bands and the cumulative variance shown by the green bands (Fig. 9D). Figure 9E shows a covariance matrix map of the interaction between residue pairs of the proteins of a complex (red: correlated motion between a pair of residues, white: non-correlated motion, and blue: anti-correlated motion). Finally, the stiffness study of the protein complex was performed using elastic network analysis. As shown in Fig. 9F, the darker the gray dots, the greater the protein stiffness in certain sections.
Fig. 9.
The Molecular dynamics simulation of the vaccine–TLR4 complex. Six graphs including A Deformability index, B B-factor values calculated by normal mode analysis, C The eigenvalue of the docked complex, D The covariance matrix between pairs of residues, E The elastic network model are shown, F The darker the gray dots, the greater the protein stiffness in certain sections
Immune simulation
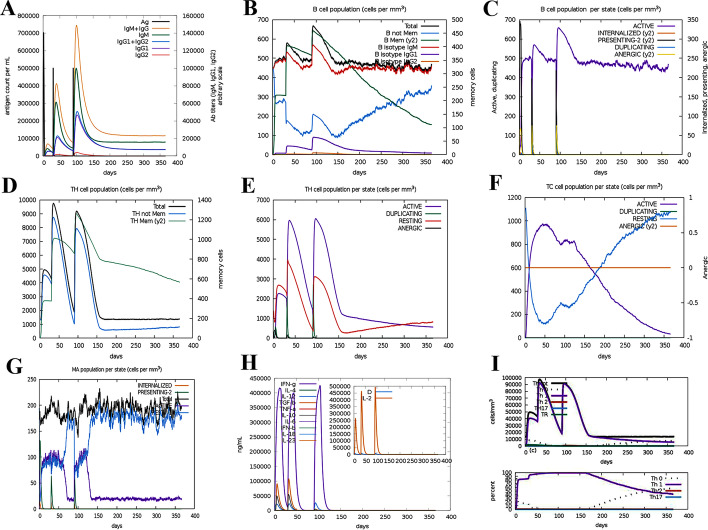
The multi-epitope vaccine designed to evaluate the specific immune response of the vaccine was submitted to the C-IMMSIM v10.1 server. Secondary and tertiary immune responses showed higher levels of antibodies (IgM + IgG, IgG1 + IgG2, IgM, IgG1) than the primary immune response, which coincided with a decrease in antigen levels (Fig. 10A). In addition, several long-lasting B cell isotypes were found, indicating possible isotype switching potentials and memory B cell formation (Fig. 10B). On the other hand, Fig. 10C shown the increase in cell proliferation in B cells as well as the presentation of antigens after vaccination. According to Fig. 10D–F, the levels of TH (helper) and TC (cytotoxic) cell populations also increased significantly in memory development. Increased macrophage activity and antigen presentation are shown in Fig. 10G. Figure 10H shows a significant increase in interferon-gamma titer as well as a moderate increase in interleukin 2 (IL-2) (Fig. 10H) after the third injection of the vaccine. Finally, we saw a significant increase in Th1 (Fig. 10I). All of these data suggest that our candidate multi-epitope vaccine can induce an effective immune response that can protect against pathogens.
Fig. 10.
Immunization simulation results by C-ImmSim of the construct of multi-epitope vaccine as an antigen. A Demonstration of immunoglobulin production in response to antigen injection after vaccine administration, shown as different color peaks. B B cell population after three vaccine injections, which indicates an increase in different types of B cells and their class-switching potential. C Displays the population results per state of B cell. D The evolution of T-helper cells. E Population per state of T-helper cell. F Production of cytotoxic-T cells after vaccine injection. G Macrophages population per state. H Induction of cytokines and interleukins (increased production of IFN-γ and IL-2) after vaccination. I Th1-mediated immune response
Codon adaptation and in silico cloning
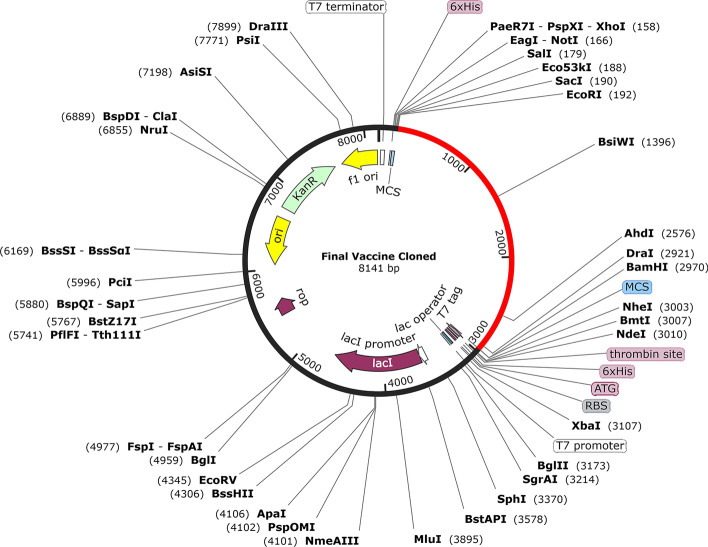
The CAI and GC content of the long nucleotide sequence of 2772 bp was evaluated to optimize the vaccine construct. Better expression (transcription and translation) in organisms requires a GC content between 30 and 70% to be optimal, while a CAI value should be higher than 0.8 to 1 [93, 94]. The GC content and CAI values of the optimized nucleotide sequence obtained from the Jcat server were 50.180% and 0.9913, respectively. EcoRI (GAATTC) and BamHI (GGATCC) restriction sites were added to the N and C terminals of the final vaccine codon sequence. Finally, SnapGene software was used to integrate the adapted DNA sequence to the pET-28a (+) vector, between the EcoRI and BamHI restriction sites (Fig. 11).
Fig. 11.
In silico cloning of the final vaccine construct into pET28a (+) expression vector. The vector was shown in black color, while the red color provided the gene coding for the vaccine to construct a protein. EcoRI and BamHI restriction enzyme sites have been proposed as cutting sites
MRNA prediction of the designed vaccine
The secondary structure of the vaccine MRNA sequence was predicted by the RNAfold server with a minimum free energy score of − 861.60 kcal/mol. A lower MFE indicates a higher thermodynamic stability of the MRNA secondary structure.
Discussion
CRC is known as the fourth cause of death among cancers and predisposing factors like different lifestyle, genetic and environmental risk reasons can promote the cancer [95]. CRC is induced by normal epithelium alteration into high-proliferative epithelial cells and results in the reorganization of intestinal epithelial cells and adenoma-carcinoma formation. This cancerous process metastasized to the colon and may progress to CRC [96]. There is a strong association between the presence of S. bovis, B. fragilis, H. pylori, F. nucleatum, E. faecalis, E. coli, and P. anaerobius and the incidence of CRC [1, 14, 95]. Even with antibiotic treatment, there is a high risk for the recurrence of disease and the emergence of antibiotic-resistant strains, so there is a need to develop novel methods like immunization via vaccines against pathogenic and toxigenic strains [97].
Peptide-based vaccines, especially those contain cocktail of several peptides, show a significant effect on the treatment outcome of patients with CRC [98]. These vaccines target either host proteins [99] or immunogen proteins of pathogens related to cancer [100].
A desirable multi-epitope vaccine should be consisting of peptides with capable of generating CTL, TH and B cells and triggering potential immune response against [101]. Today, the design of multi-epitope vaccines is recognized as an emerging area that is of considerable importance. However, vaccines designed with this approach have been shown in vivo efficacy with protective immunity, but have also entered phase I clinical trials [102–105].
This study aimed to design a multi-epitope and prophylactic vaccine against colorectal cancer-related pathogens based on the immunoinformatics approach. Considering the importance of the virulence factors, 10 proteins from different microorganisms were selected to predict effective epitopes.
A total of 56 epitopes (924 amino acids) including 19 epitopes for B cell, 19 epitopes for MHC I binding, and 18 epitopes for MHC II binding were considered. For the epitope to be effective and safe for the host, it must be antigenic, non-allergenic, non-toxic, and stable. All the selected epitopes were antigenic, non-allergenic and non-toxic.
The suitable molecular weight of the designed vaccine makes it easy for purification, so can be considered a suitable vaccine. Higher aliphatic index values indicate greater thermostability at several temperatures and negative GRAVY values indicate the hydrophilic nature of the candidate vaccine, so it can show strong interactions with water molecules. The vaccine instability index was calculated to be 23.00 and since it was less than 40.00, it was considered a stable protein. Also, the designed vaccine has good solubility.
PSIPRED and RaptorX web servers were used to evaluate the structure of the second and 3D candidate multi-epitope vaccines, respectively. Accordingly, the PSIPRED server predicted the α-helix, β-strand, and coil of the candidate vaccines to be 34%, 16.66%, and 49.34%, respectively.
In the final constriction refining, five proposed refined models were introduced and model 5 was selected as the best-refined model with GDT-HA 0.8722, an RMSD score of 0.586 a MolProbity score of 2.582, a Clash score of 32.1, and a Ramachandran score of 88.3. Studies have shown that RMSD < 2.0 Å corresponds to good docking solutions [106], and in the present study, our final RMSD construct was also in the best condition.
Because disulfide bonds play an important role in folding, stability, and protein function, if they are ignored, the stability of the target protein can be reduced [107]. For this reason, we saw 4 pairs of 375GLY-377GLY, 642THR-645LYS, 668LEU-702SER, and 826ASN-844VAL amino acids with less than one energy bond, which indicates more stability of the final construct.
Different frequencies of HLA type vary in different ethnicities around the world due to the high polymorphism of the MHC molecule. The selected alleles considered in this study proved to show sufficient population coverage a large scale (Fig. 4D). The highest population coverage in CD8+ T cells is in Europe (98.07%), North America (95.61%), and West India (94.69%). While the lowest CD8+ T cells population coverage is in Central America (7.01%). On the other hand, the largest population coverage in CD4+ T cells is in East Asia (73.14%), North America (71.89%), and Europe (68.97%). The lowest population coverage was for Southwest Asia (28.80%), Central America (23.09%) and South Africa (7.65%).
Two online servers, ClusPro 2.0 and PatchDock & FireDock were used for docking analysis to increase our forecast accuracy. These servers pointed to a strong interaction between the TLR4 and the designed vaccine. The energy scores obtained for binding the vaccine-TLR4 complex using these two servers indicated a very good binding affinity. In summary, the MD simulation findings obtained from the present study confirm that the designed vaccine molecule can interact optimally with the TLR4 protein. The C-IMMSIM server was then used to evaluate the ability of the candidate vaccine to initiate an immune response with an immune simulation. However, based on the results, enhancement of memory B cells and T cells was visible. Also, the secondary and tertiary immune responses showed higher levels of antibodies than the primary immune response. On the other hand, a significant increase in IFN-γ titer as well as a moderate increase in IL-2 was shown after the third injection of the vaccine. All these data suggest that our candidate multi-epitope vaccine can induce an effective immune response that can protect against pathogens.
Finally, to ensure the translation efficiency of the multi-epitope vaccine designed in a specific expression system, the vaccine MRNA was amplified using the JCAT. Adaptive DNA sequences between EcoRI (GAATTC) and BamHI (GGATCC) restriction enzyme cleavage sites were then added to N and C terminals, respectively, and subsequently cloned into pET28a (+), the expression vector. The codon adaptability index (0.98) and GC content (53.63%) were promising for the expression of high-level proteins in bacteria. On the other hand, during predicting the stability of the secondary structure of the vaccine mRNA, the RNAfold server produced less negative and less free energy, so it can be concluded that the predicted vaccine can be stable after in vivo transcription.
Conclusion
CRC is one of the most common cancers worldwide. Increasing evidence suggests that gut microbiota dysbiosis is closely related to CRC. Streptococcus bovis, Helicobacter pylori, Bacteroides Fragilis, Fusobacterium nucleatum, Enterococcus faecalis, Escherichia coli, and Peptostreptococcus anaerobius are the main microbial agents involved in CRC pathogenesis. Therefore, in the present study, an in silico vaccine was designed against their most important epitopes, then its effectiveness was evaluated through different immunoinformatics servers. The designed multi-epitope vaccine seems to act as an effective prophylactic candidate vaccine since the results showed an increase in antibodies, T lymphocytes, and its subtypes (such as helper T lymphocytes and cytotoxic T lymphocytes) as well as INF-γ levels. In general, the application of these results is pending validation in the wet lab experimental models.
Limitations
Each of these bioinformatics predictive servers has limitations that are not comparable to the experimental method. For example, the C-IMMSIM server simulator is limited because it does not have the disease layer and is unable to detect vaccine efficacy. On the other hand, NMA is probably the least computationally expensive method for studying the dynamics of macromolecules, but the MD method is more accurate than NMA.
A major limitation of this study is the lack of the experimental validation and evaluation of the safety and efficacy of the designed vaccine construct. However, major steps such as laboratory and animal studies are needed to justify our findings to determine safety, efficacy, and immunogenicity as a possible preventive measure. In general, the application of these results is pending validation in the wet lab experimental models.
Acknowledgements
None.
Author contributions
HM, MM, MS, SM, RA and AA: Conceptualizing, designing and study implementation, data collection and analysis, and writing the manuscript. HM, MM and PF: Cooperation in data collection, writing and editing of the manuscript. HM, RA and AA: Consulting in immunoinformatics. HM, RA and AA: Supervision and monitoring of the implementation of the study. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All the data supporting the findings are contained within the manuscript.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hamid Motamedi and Marzie Mahdizade Ari contributed equally to this work
Contributor Information
Amirhoushang Alvandi, Email: ah_alvandi@kums.ac.ir.
Ramin Abiri, Email: rabiri@kums.ac.ir.
References
- 1.Cheng Y, Ling Z, Li L. The intestinal microbiota and colorectal cancer. Front Immunol. 2020;11:615056. doi: 10.3389/fimmu.2020.615056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 3.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 4.Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74(16):2959–2977. doi: 10.1007/s00018-017-2509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang PS, Chen TY, Giovannucci E. Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis. Int J Cancer. 2009;124(10):2406–2415. doi: 10.1002/ijc.24191. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Liu J, Zheng X, Ren L, Yang Y, Li W, et al. Tumorigenic bacteria in colorectal cancer: mechanisms and treatments. Cancer Biol Med. 2021;19(2):147–162. doi: 10.20892/j.issn.2095-3941.2020.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale TF, Juluri R, Sherer EA, Glowinski EA, Johnson CS, Morelli MS. A risk index for advanced neoplasia on the second surveillance colonoscopy in patients with previous adenomatous polyps. Gastrointest Endosc. 2014;80(3):471–478. doi: 10.1016/j.gie.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 9.Mork ME, You YN, Ying J, Bannon SA, Lynch PM, Rodriguez-Bigas MA, et al. High prevalence of hereditary cancer syndromes in adolescents and young adults with colorectal cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2015;33(31):3544–3549. doi: 10.1200/jco.2015.61.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laukoetter MG, Mennigen R, Hannig CM, Osada N, Rijcken E, Vowinkel T, et al. Intestinal cancer risk in Crohn's disease: a meta-analysis. J Gastroint Surg Off J Soc Surg Aliment Tract. 2011;15(4):576–583. doi: 10.1007/s11605-010-1402-9. [DOI] [PubMed] [Google Scholar]
- 11.Fedirko V, Tramacere I, Bagnardi V, Rota M, Scotti L, Islami F, et al. Alcohol drinking and colorectal cancer risk: an overall and dose-response meta-analysis of published studies. Ann Oncol Off J Eur Soc Med Oncol. 2011;22(9):1958–1972. doi: 10.1093/annonc/mdq653. [DOI] [PubMed] [Google Scholar]
- 12.Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, van Ballegooijen M, Zauber AG, Jemal A. Contribution of screening and survival differences to racial disparities in colorectal cancer rates. Cancer Epidemiol Biomark Prevent. 2012;21(5):728–736. doi: 10.1158/1055-9965.epi-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koido S, Ohkusa T, Homma S, Namiki Y, Takakura K, Saito K, et al. Immunotherapy for colorectal cancer. World J Gastroenterol WJG. 2013;19(46):8531. doi: 10.3748/wjg.v19.i46.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seely KD, Morgan AD, Hagenstein LD, Florey GM, Small JM. Bacterial involvement in progression and metastasis of colorectal neoplasia. Cancers. 2022;14(4):1019. doi: 10.3390/cancers14041019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kambarev S, Caté C, Corvec S, Pecorari F. Draft genome sequence of erythromycin-resistant streptococcus gallolyticus subsp. gallolyticus NTS 31106099 Isolated from a patient with infective endocarditis and colorectal cancer. Genome Announc. 2015;3(2):e00370–e415. doi: 10.1128/genomeA.00370-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mainil J. Escherichia coli virulence factors. Vet Immunol Immunopathol. 2013;152(1-2):2–12. doi: 10.1016/j.vetimm.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Wassenaar TME. coli and colorectal cancer: a complex relationship that deserves a critical mindset. Crit Rev Microbiol. 2018;44(5):619–632. doi: 10.1080/1040841x.2018.1481013. [DOI] [PubMed] [Google Scholar]
- 19.Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus anaerobius Induces Intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152(6):1419–33.e5. doi: 10.1053/j.gastro.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Long X, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4(12):2319–2330. doi: 10.1038/s41564-019-0541-3. [DOI] [PubMed] [Google Scholar]
- 21.Akbar N, Khan N, Muhammad J, Siddiqui R. The role of gut microbiome in cancer genesis and cancer prevention. Health Sci Rev. 2022;2:100010. doi: 10.1016/j.hsr.2021.100010. [DOI] [Google Scholar]
- 22.Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, et al. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers. 2020 doi: 10.3390/cancers12061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei W, Sun W, Yu S, Yang Y, Ai L. Butyrate production from high-fiber diet protects against lymphoma tumor. Leuk Lymphoma. 2016;57:1–8. doi: 10.3109/10428194.2016.1144879. [DOI] [PubMed] [Google Scholar]
- 24.Hinrichs CS, Kaiser A, Paulos CM, Cassard L, Sanchez-Perez L, Heemskerk B, et al. Type 17 CD8+ T cells display enhanced antitumor immunity. Blood. 2009;114(3):596–599. doi: 10.1182/blood-2009-02-203935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J, et al. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat Comm. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strofilas A, Lagoudianakis EE, Seretis C, Pappas A, Koronakis N, Keramidaris D, et al. Association of helicobacter pylori infection and colon cancer. J Clin Med Res. 2012;4(3):172–176. doi: 10.4021/jocmr880w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seely KD, Morgan AD, Hagenstein LD, Florey GM, Small JM. Bacterial involvement in progression and metastasis of colorectal neoplasia. Cancers. 2022 doi: 10.3390/cancers14041019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46(4):1135–1140. doi: 10.1590/S1517-838246420140665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015;83(3):1104–1113. doi: 10.1128/iai.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butt J, Epplein M. Helicobacter pylori and colorectal cancer-A bacterium going abroad? PLoS Pathog. 2019;15(8):e1007861. doi: 10.1371/journal.ppat.1007861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapetanakis N, Kountouras J, Zavos C, Michael S, Tsarouchas G, Gavalas E, et al. Re: Helicobacter pylori infection and colorectal cancer risk: evidence from a large population-based case-control study in Germany. Am J Epidemiol. 2012;176(6):566–567. doi: 10.1093/aje/kws302. [DOI] [PubMed] [Google Scholar]
- 33.Navashenaq JG, Shabgah AG, Banach M, Jamialahmadi T, Penson PE, Johnston TP, et al. The interaction of Helicobacter pylori with cancer immunomodulatory stromal cells: new insight into gastric cancer pathogenesis. Semin Cancer Biol. 2021 doi: 10.1016/j.semcancer.2021.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Epplein M, Pawlita M, Michel A, Peek RM, Jr, Cai Q, Blot WJ. Helicobacter pylori protein-specific antibodies and risk of colorectal cancer. Am Soc Prevent Oncol. 2013;22(11):1964–1974. doi: 10.1158/1055-9965.EPI-13-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zumkeller N, Brenner H, Zwahlen M, Rothenbacher D. Helicobacter pylori infection and colorectal cancer risk: a meta-analysis. Helicobacter. 2006;11(2):75–80. doi: 10.1111/j.1523-5378.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- 36.Butt J, Varga MG, Blot WJ, Teras L, Visvanathan K, Le Marchand L, et al. Serologic response to helicobacter pylori proteins associated with risk of colorectal cancer among diverse populations in the United States. Gastroenterology. 2019;156(1):175–86.e2. doi: 10.1053/j.gastro.2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014;124(10):4166–4172. doi: 10.1172/jci72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8):782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 39.McCoy CS, Mannion AJ, Feng Y, Madden CM, Artim SC, Au GG, et al. Cytotoxic Escherichia coli strains encoding colibactin, cytotoxic necrotizing factor, and cytolethal distending toxin colonize laboratory common marmosets (Callithrix jacchus) Sci Rep. 2021;11(1):23093. doi: 10.1038/s41598-020-80000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019 doi: 10.1126/science.aar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132(2):551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 42.Alzahrani OM, Fayez M, Alswat AS, Alkafafy M, Mahmoud SF, Al-Marri T, et al. Antimicrobial resistance, biofilm formation, and virulence genes in enterococcus species from small backyard chicken flocks. Antibiotics. 2022;11(3):380. doi: 10.3390/antibiotics11030380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23(3):529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 44.Sudipto Saha GPS, Raghava, Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 45.EL-Manzalawy Y, Dobbs D, Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recogn Interdiscip J. 2008;21(4):243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Liu H, Yang J, Chou K-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33(3):423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 47.Singh H, Ansari HR, Raghava GP. Improved method for linear B-cell epitope prediction using antigen’s primary sequence. PLoS ONE. 2013;8(5):e62216. doi: 10.1371/journal.pone.0062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24(11):1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 49.Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:278. doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32(4):511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinf. 2010;11(1):1–12. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker JM. The proteomics protocols handbook. Springer; 2005. [Google Scholar]
- 54.Garg VK, Avashthi H, Tiwari A, Jain PA, Ramkete PW, Kayastha AM, et al. MFPPI–multi FASTA ProtParam interface. Bioinformation. 2016;12(2):74. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 56.Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J. Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8(1):1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palm NW, Rosenstein RK, Medzhitov R. Allergic host defences. Nature. 2012;484(7395):465–472. doi: 10.1038/nature11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2—a server for in silico prediction of allergens. J Mol Model. 2014 doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 60.Dimitrov I, Flower DR, Doytchinova I, editors. AllerTOP-a server for in silico prediction of allergens. BMC Bioinf; 2013. [DOI] [PMC free article] [PubMed]
- 61.Gupta S, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013;8(9):e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bui H-H, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7(1):1–5. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Y, Liu Y, Cheng J. Protein secondary structure prediction based on data partition and semi-random subspace method. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-28084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 65.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 66.Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7(8):1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heo L, Park H, Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41(W1):W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kufareva I, Abagyan R. Methods of protein structure comparison. Homology modeling. Springer; 2011. pp. 231–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucl Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laskowski R, MacArthur M, Moss D, Thornton J. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [PubMed] [Google Scholar]
- 72.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17(4):355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 73.Wiedemann C, Kumar A, Lang A, Ohlenschläger O. Cysteines and disulfide bonds as structure-forming units: insights from different domains of life and the potential for characterization by NMR. Front Chem. 2020;8:280. doi: 10.3389/fchem.2020.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dombkowski AA, Sultana KZ, Craig DB. Protein disulfide engineering. FEBS Lett. 2014;588(2):206–212. doi: 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 75.Craig DB, Dombkowski AA. Disulfide by design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinf. 2013;14(1):1–7. doi: 10.1186/1471-2105-14-S19-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prabhakar PK, Srivastava A, Rao KK, Balaji PV. Monomerization alters the dynamics of the lid region in Campylobacter jejuni CstII: an MD simulation study. J Biomol Struct Dyn. 2016;34(4):778–791. doi: 10.1080/07391102.2015.1054430. [DOI] [PubMed] [Google Scholar]
- 77.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Molteni M, Gemma S, Rossetti C. The role of toll-like receptor 4 in infectious and noninfectious inflammation. Med Inflam. 2016;2016. [DOI] [PMC free article] [PubMed]
- 79.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69(1):139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 81.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 82.López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Samad A, Ahammad F, Nain Z, Alam R, Imon RR, Hasan M, et al. Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach. J Biomol Struct Dyn. 2022;40(1):14–30. doi: 10.1080/07391102.2020.1792347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep. 2021;11(1):1–21. doi: 10.1038/s41598-021-81749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5(4):e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trotta E. On the normalization of the minimum free energy of RNAs by sequence length. PLoS ONE. 2014;9(11):e113380. doi: 10.1371/journal.pone.0113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jespersen MC, Mahajan S, Peters B, Nielsen M, Marcatili P. Antibody specific B-cell epitope predictions: leveraging information from antibody-antigen protein complexes. Front Immunol. 2019;10:298. doi: 10.3389/fimmu.2019.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fatoba AJ, Maharaj L, Adeleke VT, Okpeku M, Adeniyi AA, Adeleke MA. Immunoinformatics prediction of overlapping CD8+ T-cell, IFN-γ and IL-4 inducer CD4+ T-cell and linear B-cell epitopes based vaccines against COVID-19 (SARS-CoV-2) Vaccine. 2021;39(7):1111–1121. doi: 10.1016/j.vaccine.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baseer S, Ahmad S, Ranaghan KE, Azam SS. Towards a peptide-based vaccine against Shigella sonnei: a subtractive reverse vaccinology based approach. Biologicals. 2017;50:87–99. doi: 10.1016/j.biologicals.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Sayed SB, Nain Z, Khan M, Ahmed S, Abdulla F, Tasmin R, et al. Exploring lassa virus proteome to design a multi-epitope vaccine through immunoinformatics and immune simulation analyses. Int J Pept Res Ther. 2020;26(4):2089–2107. doi: 10.1007/s10989-019-10003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shey RA, Ghogomu SM, Esoh KK, Nebangwa ND, Shintouo CM, Nongley NF, et al. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019;9(1):1–18. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fathollahi M, Fathollahi A, Motamedi H, Moradi J, Alvandi A, Abiri R. In silico vaccine design and epitope mapping of New Delhi metallo-beta-lactamase (NDM): an immunoinformatics approach. BMC Bioinf. 2021;22(1):1–24. doi: 10.1186/s12859-021-04378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Int J Mol Sci. 2017 doi: 10.3390/ijms18061310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–2114. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 97.Pollard AJ, Bijker EM. A guide to vaccinology: from basic principles to new developments. Nat Rev Immunol. 2021;21(2):83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiao Y-F, Jie M-M, Li B-S, Hu C-J, Xie R, Tang B, et al. Peptide-based treatment: a promising cancer therapy. J Immunol Res. 2015;2015:1–13. doi: 10.1155/2015/761820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abdelmoneim AH, Mustafa MI, Abdelmageed MI, Murshed NS, Dawoud ED, Ahmed EM, et al. Immunoinformatics design of multiepitopes peptide-based universal cancer vaccine using matrix metalloproteinase-9 protein as a target. Immunol Med. 2021;44(1):35–52. doi: 10.1080/25785826.2020.1794165. [DOI] [PubMed] [Google Scholar]
- 100.Majid M, Andleeb S. Designing a multi-epitopic vaccine against the enterotoxigenic Bacteroides fragilis based on immunoinformatics approach. Sci Rep. 2019 doi: 10.1038/s41598-019-55613-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol. 2018;15(2):182–184. doi: 10.1038/cmi.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang P, Cai Y, Chen J, Ye X, Mao S, Zhu S, et al. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine. 2017;35(23):3096–3103. doi: 10.1016/j.vaccine.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 103.Lennerz V, Gross S, Gallerani E, Sessa C, Mach N, Boehm S, et al. Immunologic response to the survivin-derived multi-epitope vaccine EMD640744 in patients with advanced solid tumors. Cancer Immunol Immunother. 2014;63(4):381–394. doi: 10.1007/s00262-013-1516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Slingluff CL, Lee S, Zhao F, Chianese-Bullock KA, Olson WC, Butterfield LH, et al. A randomized phase II trial of multiepitope vaccination with melanoma peptides for cytotoxic T cells and helper T cells for patients with metastatic melanoma (E1602) multipeptide vaccine for advanced melanoma. Clin Cancer Res. 2013;19(15):4228–4238. doi: 10.1158/1078-0432.CCR-13-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toledo H, Baly A, Castro O, Resik S, Laferté J, Rolo F, et al. A phase I clinical trial of a multi-epitope polypeptide TAB9 combined with Montanide ISA 720 adjuvant in non-HIV-1 infected human volunteers. Vaccine. 2001;19(30):4328–4336. doi: 10.1016/S0264-410X(01)00111-6. [DOI] [PubMed] [Google Scholar]
- 106.Ramírez D, Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23(5):1038. doi: 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gao X, Dong X, Li X, Liu Z, Liu H. Prediction of disulfide bond engineering sites using a machine learning method. Sci Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-67230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data supporting the findings are contained within the manuscript.