Abstract
The yeast cadmium factor (Ycf1p) is a vacuolar ATP binding cassette (ABC) transporter required for heavy metal and drug detoxification. Cluster analysis shows that Ycf1p is strongly related to the human multidrug-associated protein (MRP1) and cystic fibrosis transmembrane conductance regulator and therefore may serve as an excellent model for the study of eukaryotic ABC transporter structure and function. Identifying intramolecular interactions in these transporters may help to elucidate energy transfer mechanisms during transport. To identify regions in Ycf1p that may interact to couple ATPase activity to substrate binding and/or movement across the membrane, we sought intragenic suppressors of ycf1 mutations that affect highly conserved residues presumably involved in ATP binding and/or hydrolysis. Thirteen intragenic second-site suppressors were identified for the D777N mutation which affects the invariant Asp residue in the Walker B motif of the first nucleotide binding domain (NBD1). Two of the suppressor mutations (V543I and F565L) are located in the first transmembrane domain (TMD1), nine (A1003V, A1021T, A1021V, N1027D, Q1107R, G1207D, G1207S, S1212L, and W1225C) are found within TMD2, one (S674L) is in NBD1, and another one (R1415G) is in NBD2, indicating either physical proximity or functional interactions between NBD1 and the other three domains. The original D777N mutant protein exhibits a strong defect in the apparent affinity for ATP and Vmax of transport. The phenotypic characterization of the suppressor mutants shows that suppression does not result from restoring these alterations but rather from a change in substrate specificity. We discuss the possible involvement of Asp777 in coupling ATPase activity to substrate binding and/or transport across the membrane.
The yeast cadmium factor protein (Ycf1p) is a vacuolar membrane protein involved in heavy metal and drug detoxification in Saccharomyces cerevisiae. It is an ATP-dependent pump able to transport organic glutathione S (GS) conjugates (32), GS-metal complexes (18, 31), glutathione (41, 42), and other compounds, like unconjugated bilirubin (39). Ycf1p belongs to the ATP binding cassette (ABC) superfamily of transporters that includes the yeast a-factor transporter Ste6p (28), the Pfmdr-1 of Plasmodium falciparum which is associated with antimalarial drug resistance (17), and the human proteins P glycoprotein (16) and multidrug-associated protein (MRP1) (10) involved in multidrug resistance or the cystic fibrosis transmembrane conductance regulator, in which mutations cause cystic fibrosis (44). There are many sequence and mechanistic similarities between ABC transporters (21, 22), and they have a common evolutionary origin (11). Structural homology among ABC transporters reflects functional similarity in some cases, since MRP1 is able to suppress the Cd2+ hypersensitivity of a yeast Δycf1 mutant (57) and Ycf1p can transport the physiological substrate of MRP1, the leukotriene LTC4 (15, 43). Ycf1p may thus be an excellent model for examining structure-function issues relating to human MRP1 and eukaryotic ABC transporters in general. Secondary-structure predictions suggest that Ycf1p is formed by two transmembrane domains (TMDs) and two nucleotide binding domains (NBDs) (55), as are nearly all members of the ABC superfamily. In addition, it possesses two subfamily-specific domains: a putative regulatory domain common to the MRP and cystic fibrosis transmembrane conductance regulator subfamilies and a third N-terminal TMD present only in the MRP subfamily (58). The most characteristic feature of ABC transporters is the NBDs that contain the highly conserved Walker A (GXXGXGKS/T [X, any amino acid]), Walker B (Rx6-8 hyd4D) (59), and ABC signature (LSXGXK/R) (25) motifs. Walker A and Walker B are common to a wide variety of nucleotide binding proteins, whereas the ABC signature sequence, just upstream of the Walker B motif, is distinctive to the ABC family. ATP binding and hydrolysis at these domains are essential for subsequent substrate transport, and ATPase activity stimulation by substrate binding in several systems has been demonstrated (12, 34, 50, 54). Coupling of ATPase activity to substrate binding and transport involves interactions between the distinct domains of the protein, but the exact nature of the intramolecular interactions that underlie these effects is not known. One approach to detecting structural and functional interactions is to screen for second-site mutations that compensate for a primary defect in a gene. In this study, we performed an intragenic suppression analysis of five ycf1 mutations located in highly conserved motifs of the NBDs, all of them involved in binding and/or hydrolysis of ATP (7, 37, 47) and characterized in a previous report (15). We successfully isolated intragenic suppressors for one of the five mutations, D777N, in the Walker B motif of NBD1. The positions of the suppressors indicate that NBD1 functionally interacts with NBD2 and both TMDs. We discuss the possibility of a direct involvement of Walker B region in coupling ATPase activity and substrate binding and/or transport.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
A Δycf1 derivative of S. cerevisiae strain W303-1A (MATa ycf1Δ::URA3 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1) (15) was used. Escherichia coli strains XL1-Blue and XL1-Red (mutD5 mutS mutT) (Stratagene) were used for plasmid amplification and mutagenesis, respectively. The centromeric plasmid pRS315 (53) and the episomal plasmid pRS425 (9) were used for expressing wild-type and mutant YCF1 alleles. In all cases, YCF1 possessed the nine-amino-acid 12CA5 epitope sequence from human influenza hemagglutinin (HA) protein immediately before the termination codon (15). In all experiments, growth of yeast cells was at 30°C in SD medium (0.7% yeast nitrogen base without amino acids [US Biological], 2% glucose, pH 5.5) supplemented with the appropriate auxotrophic requirements (100 μg/ml). SD medium for resistance assays was supplemented with drop-out mix (BIO 101).
Mutagenesis and selection of revertants
Chemical mutagenesis and propagation of the cloned gene into a mutator E. coli strain were used to introduce random mutations in the different mutant ycf1 alleles. For chemical mutagenesis, the plasmid containing the ycf1 allele with the primary mutation was treated with 0.5 M hydroxylamine and 1 mM EDTA, pH 6, for 4 h at 70°C. The hydroxylamine was removed by ethanol precipitation. For in vivo mutagenesis, the plasmid was transformed into Epicurian E. coli XL1-Red competent cells by following the manufacturer's instructions (Stratagene). After growth of transformants for 24 h at 37°C, 2 × 103 to 3 × 103 colonies were picked from the transformation plates and pooled. Mutated plasmid DNA was isolated from the pooled transformants and transformed into XL1-Blue strain cells for DNA amplification. Randomly mutated plasmid DNA obtained by any of the two procedures was used to transform the yeast Δycf1 strain by using the lithium acetate procedure (26). Revertants were selected by replica plating onto 50 μM CdCl2 plates for transformants carrying centromeric plasmids or 150 μM CdCl2 plates for those carrying episomal plasmids. Plasmid DNA was rescued from revertants and recovered in E. coli cells (46). Yeast transformation and selection of revertants were repeated with the recovered plasmid as described above.
Mapping and sequencing of revertant mutants.
To determine whether the reversion occurred at the site of the original mutation, the DNA regions that include the primary mutation were sequenced in the plasmids rescued from revertants of G663V, G756D, and G1306E mutations. The D777N mutation introduces an MseI restriction endonuclease site that is absent in the wild-type allele. In this case, a 0.6-kb DNA fragment was amplified by PCR using plasmid DNA rescued from revertants as template and subjected to restriction analysis with MseI. The presence of the primary mutation in the rescued plasmids would indicate the existence of a second mutation in YCF1 able to suppress the inactive primary mutation. The second-site mutations were located within specific YCF1 fragments by single-strand conformation polymorphism analysis of PCR-amplified fragments (40). Fifteen 0.3- to 0.44-kb overlapping PCR fragments were generated from each ycf1 suppressor allele so that, when put together, they included DNA encoding the entire Ycf1p. The PCR mixture contained 5 ng of plasmid DNA, 10 pmol of each primer, 5 nmol of each of the four deoxynucleotides, 2.5 mM MgCl2, 1 μCi of [α-33P]dATP (2,000 Ci/mmol) (10 mCi/ml) (Amersham), and 2.5 U of Ampli Taq DNA polymerase (Perkin-Elmer) in 25 μl of the buffer supplied by the manufacturer. One reaction cycle was performed at 95°C for 3 min, 30 cycles were performed at at 95, 58, and 72°C for 0.5, 0.5, and 1 min, respectively, and one cycle was performed at 72°C for 7 min, using a GeneAmp PCR System 2400 (Perkin-Elmer). Electrophoresis of the PCR products on nondenaturing 0.35× mutation detection enhancement (MDE) (FMC BioProducts) gels was performed as previously described (40), at 6 W in a 4°C cold room for 16 h. The revertant ycf1 regions corresponding to PCR fragments with altered mobility were sequenced using fluorescence-labeled dideoxynucleotides in an ABI Prism 377 DNA Sequencer. Once the second-site mutation was identified, a 1.3-kb BsmI-StuI fragment for V543I, F565L, and S674L suppressors or a 1.8-kb NdeI-SalI fragment for the remainder of the suppressors was excised from the revertant and exchanged with the corresponding fragment in pRS425-ycf1D777N-HA. The entire restriction fragment exchanged was sequenced to exclude the possibility of other mutations.
Construction of mutant ycf1 alleles containing only second-site mutations.
The plasmids containing both the original D777N and the second-site mutations were digested with BsmI and NcoI for V543I, F565L, and S674L suppressors or with NdeI and SalI for the remaining suppressors. The excised fragments, 1.2 and 1.8 kb, respectively, were exchanged with the corresponding fragment in pRS315-YCF1-HA. The resultant plasmids, containing the ycf1 variants with the isolated suppressor mutations, were sequenced to verify the presence of the second-site mutation and the absence of the D777N change.
Cadmium and diamide resistance assays.
Qualitative and quantitative determinations were performed. Cells were cultured for 2 days on SD plates and suspended in water to an optical density at 660 nm (OD660) of 0.4 (2.4 × 107 cells/ml) to be used as inoculum. For qualitative assays, 5-μl samples were dropped on plates with CdCl2 or diamide at the indicated concentrations. Growth was scored after 2 to 3 days of incubation. For quantitative determination of the MIC, flat-bottom 96-well microtiter plates containing medium with concentrations ranging from 0 to 1 mM CdCl2 or 0 to 3 mM diamide were inoculated to a final cell density of 6 × 105 cells/ml. Inoculum-free wells were also included. The OD595 of each well was determined after a 24-h incubation in the case of diamide or a 2-day incubation in the case of CdCl2. Data were fitted to a sigmoidal dose-response equation by using Prism 2.0 GraphPad Software. The MIC is defined as the lowest concentration at which prominent inhibition of cell growth (90 to 95%) is observed.
Isolation of vacuolar membrane vesicles.
Intact vacuoles were isolated by flotation centrifugation of spheroplast lysates on Ficoll 400 step gradients, as previously described (15). The resulting vacuole fraction was vesiculated in 5 mM MgCl2–25 mM KCl–10 mM Tris-MES (morpholineethanesulfonic acid) (pH 6.9), pelleted by centrifugation (37,000 × g, 25 min), and resuspended in buffer (1.1 M glycerol, 2 mM dithiothreitol, 1 mM EGTA, 5 mM Tris-MES, pH 7.6). All buffers used contained a protease inhibitor mixture (1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 1 mM phenylmethylsulfonyl fluoride).
Measurement of [3H]LTC4 uptake.
Standard uptake experiments were performed at 30°C in TS buffer (250 mM sucrose, 25 mM Tris-MES, pH 8.0) containing 10 mM ATP, 10 mM MgCl2, 10 mM creatine phosphate, 20 U of creatine kinase/ml, and 50 nM [3H]LTC4 (13 nCi/pmol) in a final volume of 55 μl. Uptake was initiated by addition of vesicles (1 to 5 μg of protein). LTC4 uptake into vacuolar vesicles increased linearly with the amount of vacuolar membrane protein, at least to 10 μg. Five aliquots (10 μl) were removed at times between 0 and 1.5 min, diluted in 1 ml of ice-cold TS buffer, immediately filtered through nitrocellulose filters (pore size, 0.45 μm; Millipore) presoaked in TS buffer, and washed twice with 5 ml of ice-cold TS buffer. The retained radioactivity was counted using liquid scintillation fluid. Initial rates were calculated from the first 1 min of uptake.
Protein analysis.
Protein concentration was measured by the Bradford method (8), using the Bio-Rad protein assay reagent and bovine immunoglobulin G as standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 7% gels was performed as previously described (30). Sample solubilization and Western blot analysis were performed as previously described (45, 51). Reversible protein staining with Ponceau S (1) and immunodetection of Ycf1p-HA using mouse anti-HA monoclonal antibody and a second antibody coupled to alkaline phosphatase (Bio-Rad) was as previously described (5).
Chemicals.
[3H]LTC4 (165 Ci/mmol) was obtained from DuPont NEN. Unlabeled LTC4 was from Sigma. All other reagents were analytical grade and purchased from Sigma, Roche, Pharmacia, or US Biological.
RESULTS
Isolation of intragenic suppressors of mutations in NBDs.
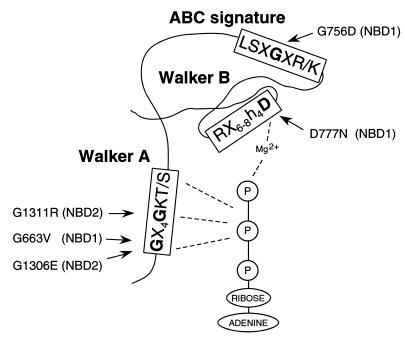
For the intragenic suppressor analysis of mutations localized in the NBDs, we selected G663V, G756D, D777N, G1306E, and G1311R changes (Fig. 1), all of which affect residues in highly conserved motifs. Residues Gly663 in NBD1 or Gly1306 and Gly1311 in NBD2 are located in the Walker A motif, which is postulated to form a flexible loop that interacts with bound nucleotide phosphate groups in the catalytic site of nucleotide-binding proteins (7). Asp777 in the Walker B motif of NBD1 is presumed to play an essential role in Mg2+ binding during ATP hydrolysis (47). Gly756 is located in the ABC signature motif of NBD1, which is thought to participate in nucleotide binding and hydrolysis (37). We previously reported on the effect of these amino acid replacements on yeast Cd2+ tolerance, Ycf1p biogenesis, and transport activity, showing that all result in severely impaired Ycf1p-dependent Cd2+ tolerance and transport function without altering the amount of protein in the vacuolar membrane (15).
FIG. 1.
Ycf1p mutations chosen for the intragenic suppression analysis. The amino acid substitutions and their position in the conserved motifs of Ycf1p NBDs, NBD1 and NBD2, are indicated in a schematic representation of the predicted topology of an NBD (3, 13, 23, 24, 61). The consensus sequences for Walker A, Walker B, and ABC signature motifs are indicated. Interactions of bound ATP with Walker A and Walker B regions are represented by dashed lines.
Centromeric or episomal plasmids carrying each of the five ycf1 mutant alleles selected for suppression analysis were mutagenized in vitro with hydroxylamine or in vivo by propagating the plasmids into a mutator E. coli strain as described in Materials and Methods (summarized in Table 1). The mutagenized plasmids were introduced into the Δycf1 strain, and transformants were screened for growth on Cd2+ plates. Mutagenesis of plasmids containing the G663V, G756D, D777N, and G1306E mutations resulted in transformants that grew in the presence of Cd2+, whereas mutagenesis of a plasmid containing the G1311R allele did not (Table 1). To disprove that mutations in the yeast genome could contribute to the Cd2+-tolerant phenotype, plasmids of revertants were rescued and reintroduced into Δycf1 cells to ascertain their activity. The isolated plasmids were then tested for retention of the original substitutions as described in Materials and Methods. All revertants of G663V, G756D, and G1306E mutants were full revertants of the initial mutation, and 30 out of 83 revertants isolated for the D777N mutant were due to a second-site suppressor mutation.
TABLE 1.
Summary of suppressor isolation
| Mutagenized ycf1 allele | Plasmid | Mutagenesis | No. of transformants screened | No. of revertants
|
|
|---|---|---|---|---|---|
| Full revertants | Second-site suppressor mutants | ||||
| G663V | Episomal | Mutator strain | 16 × 104 | 18 | 0 |
| G756D | Centromeric | Chemical | 5 × 104 | 0 | 0 |
| Episomal | Mutator strain | 5 × 104 | 2 | 0 | |
| Chemical | 3 × 104 | 15 | 0 | ||
| D777N | Episomal | Mutator strain | 16 × 104 | 53 | 30 |
| G1306E | Centromeric | Chemical | 3 × 104 | 1 | 0 |
| Mutator strain | 5 × 104 | 4 | 0 | ||
| G1311R | Centromeric | Chemical | 3 × 104 | 0 | 0 |
| Episomal | Chemical | 3 × 104 | 0 | 0 | |
Mapping and sequencing of suppressors of D777N mutant.
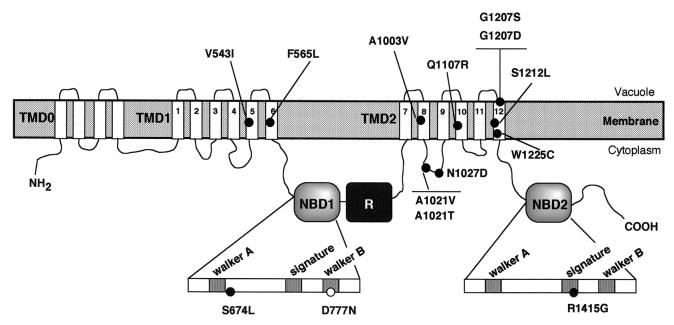
The suppressing mutations were mapped by single-strand conformation polymorphism (see Materials and Methods). Once the region bearing the intragenic suppressor mutation had been narrowed down to 300 to 450 bp, it was sequenced. The DNA sequence changes and predicted amino acid alterations are shown in Table 2. To test whether or not the mutations identified were sufficient to confer Cd2+ tolerance, all changes were reconstructed into the original ycf1D777N by exchange of different restriction fragments (see Materials and Methods). In those cases in which two amino acid changes were found, the mutation responsible for the restoration of Cd2+ tolerance was identified by subcloning the mutations separately. Identification of the mutational changes in the suppressing alleles revealed 13 different amino acid changes that suppress D777N mutation. In each case, the change identified was sufficient to confer suppression. Certain mutations were detected more than once; these included A1021V (four times), A1021T (three), G1207D (two), G1207S (two), and W1225C (seven). In these cases, identification of the same nucleotide substitution in independent clones suggests that these mutations emerged from amplification of a single mutagenic event during growth of the mutator strain. Nevertheless, the fact that A1021 and G1207 residues were targeted more than once but with different amino acids each time and that two distinct codons were found for the W1225C substitution indicates that these mutations arose from independent mutagenic events and suggests that these residues may play an important role in suppressing the defect of the D777N mutant. The second-site mutations mapped to four different domains of Ycf1p (Fig. 2). Two mutations (V543I and F565L) were localized in TMD1, nine substitutions (A1003V, A1021T, A1021V, N1027D, Q1107R, G1207D, G1207S, S1212L, and W1225C) were found within TMD2, one mutation (S674L) was localized in NBD1, and another (R1415G) was in NBD2.
TABLE 2.
Suppressors of D777N ycf1 mutant
| Suppressor no. | Codon change(s)a | Amino acid change(s)b |
|---|---|---|
| 1 | TTC→CTC | F565L |
| 2 | —c | — |
| 3 | GCC→GTC | A1021V |
| 4 | GTT→ATT | V5431 |
| 5 | GGT→GAT | G1207D |
| 6 | GCC→ACC | A1021T |
| 7 | GCC→ACC | A1021T |
| 8 | TTA→CTA, TGG→TGC | L677L, W1225C |
| 9 | — | — |
| 10 | TGG→TGC | W1225C |
| 11 | GCC→GTC | A1021V |
| 12 | TGG→TGC | W1225C |
| 13 | GCC→GTC | A1021V |
| 14 | CAG→CGG, TTA→TTG | Q1107R, L1418L |
| 15 | TCA→TTA | S674L |
| 16 | TGG→TGT, ACT→GCT | W1225C, T1454A |
| 17 | TGG→TGT | W1225C |
| 18 | GGT→GAT | G1207D |
| 19 | — | — |
| 20 | TGG→TGC | W1225C |
| 21 | GCC→ACC | A1021T |
| 22 | — | — |
| 23 | GCA→GTA | A1003V |
| 24 | GGT→AGT | G1207S |
| 25 | AGA→GGA | R1415G |
| 26 | GGT→AGT | G1207S |
| 27 | TCA→TTA | S1212L |
| 28 | GCC→GTC | A1021V |
| 29 | AAC→GAC | N1027D |
| 30 | TGG→TGC | W1225C |
The nucleotide changes are underlined.
The mutation responsible for the restoration of Cd2+ tolerance is underlined when there are two changes.
—, No mobility shift was detected in any of the PCR-amplified fragments analyzed. These suppressors were not studied further.
FIG. 2.
Location of the suppressor mutations found in revertants of the D777N mutant. The positions of the amino acid substitutions are included in the predicted model for the domain structure of Ycf1p based on the structural model that was previously proposed (58). In the additional TMD0 domain, four TM segments have been represented, but it is predicted to contain four to six segments. The highly conserved regions of the NBDs, Walker A, ABC signature, and Walker B, are indicated, as well as the position of the original D777N mutation.
Next, we characterized several aspects of Ycf1p function in the revertants from the D777N mutant. This characterization included the capacity to confer resistance to different toxic substrates in vivo, the protein expression level, and the transport capacity in vacuolar membrane vesicles.
Phenotypic characterization of revertants from D777N mutant.
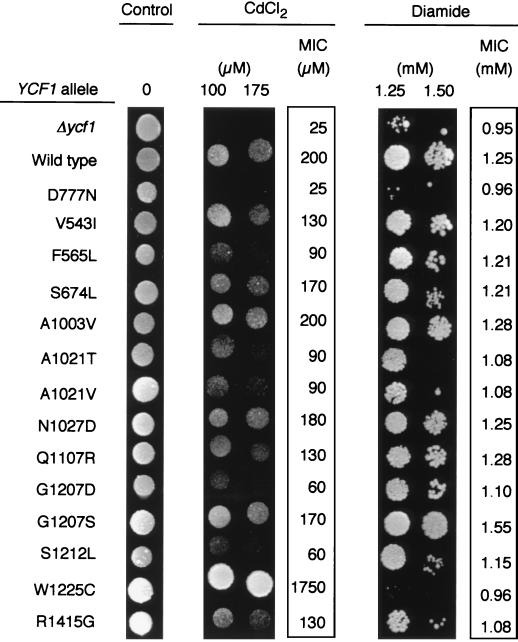
The ability of the second-site mutations to suppress the Cd2+ sensitivity of the D777N mutant was tested on plates containing 100 or 300 μM CdCl2 and further quantified by determining the MICs of the compound for these mutants (Fig. 3). Only one of these mutants, bearing a Trp1225-to-Cys change as well as the original change, was considerably more active than the wild type. The remaining mutants showed partial restoration of Cd2+ tolerance to various degrees. Ycf1p is also involved in tolerance to diamide, an oxidative stress agent (60). To determine whether the suppressor mutations affected Ycf1p-dependent resistance to other toxic substrates, we examined growth on diamide of the suppressed mutants in comparison with the wild type and D777N mutant. Δycf1 and D777N mutant strains were hypersensitive to diamide relative to the resistance shown by the wild-type strain (Fig. 3). The majority of the suppressor mutations improved to different extents the growth on diamide of the D777N mutant. Although the original mutant failed to grow on 1.5 mM diamide, 11 suppressed mutants grew at this concentration, of which 5 grew at an even higher concentration (V543I, F565L, Q1107R, G1207D, G1207S). On the contrary, mutations W1225C and R1415G did not restore the growth defect of the D777N mutant on diamide. In a detailed comparison of the ability to grow on Cd2+ and diamide for each of the suppressors, a lack of correlation was apparent between the increase in Cd2+ tolerance and the correction of the defect on diamide. Suppressor W1225C not only restored the growth defect of the D777N mutant but, as mentioned above, produced a gain-of-function phenotype for Cd2+ resistance, whereas it was completely unable to grow on 1.5 mM diamide. The mutant R1415G did not grow at all on diamide but showed a nearly twofold increase in its ability to grow on Cd2+ compared to the mutant D777N. On the other hand, among the group of mutants that corrected the Cd2+ defect to a similar extent (V543I, F565L, A1003V, A1021T, A1021V, Q1107R, G1207D, G1207S, and S1212L), only five (V543I, F565L, Q1107R, G1207D, and G1207S) were able to grow on 2 mM diamide (Fig. 3). These data indicate a change in the substrate specificity of some of the suppressors.
FIG. 3.
Resistance profile of the revertants of the D777N mutant. Cells of yeast strain Δycf1 were transformed with the episomal plasmid pRS425 (Δycf1), pRS425-YCF1-HA (wild type), or pRS425-ycf1-HA (revertant listed) and grown on SD plates. Drops of each diluted strain (see Materials and Methods) were placed onto SD drop-out plates containing the indicated CdCl2 or diamide concentrations, grown for 48 h (diamide) or 72 h (CdCl2) at 30°C, and photographed. For quantitative determination of CdCl2 and diamide tolerance, MIC measurement was performed as described (see Materials and Methods) after growth of each strain at 30°C on microtiter plates containing medium with different concentrations of the compounds. Values are the means of independent duplicate experiments.
To determine whether the increased growth capacity on Cd2+ or diamide of the suppressors was due to an increment in the amount of the mutant protein, the relative amount of Ycf1p in the vacuolar membrane of the suppressor strains was estimated by immunoassay. The mutant proteins were expressed at similar levels, or at least not higher, when compared with the expression of the wild-type and D777N controls (Fig. 4A). As expected, no Ycf1p was detectable in the Δycf1 strain transformed with the empty vector. Similar results were obtained in total membranes (data not shown). These results indicate that the greater resistance of the suppressors is not due to an increase of mutant Ycf1p in the vacuolar membrane.
FIG. 4.
Expression levels of wild-type and mutant Ycf1p in yeast vacuolar membranes. (A) Vacuolar membrane vesicles of the Δycf1 strain transformed with pRS425 (Δycf1), pRS425-YCF1-HA (wild type), or related plasmids encoding each of the revertant mutant enzymes were isolated as described (see Materials and Methods), subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (2.5 μg of protein/lane), and immunodetected with an anti-HA monoclonal antibody and a second antibody coupled to alkaline phosphatase. (B) Vacuolar membrane vesicles of the Δycf1 strain transformed with pRS315 (Δycf1), pRS315-YCF1-HA (wild type), or related plasmids encoding each of the isolated suppressor mutant enzymes were prepared, electrophoresed (4 μg of protein/lane), and immunodetected as described for panel A.
To shed some light on the suppression mechanism of the mutants, the kinetic parameters of LTC4 transport were determined in vacuolar membrane vesicles from each revertant. As mentioned above, Asp777 is proposed to be the residue that interacts with magnesium ion during ATP hydrolysis (24, 47). Accordingly, D777N mutant protein exhibits a strong defect in the apparent affinity for ATP and maximal activity of LTC4 transport (15). These defects in the kinetic parameters of the transporter could be the basis for its low tolerance to different Ycf1p transport substrates in vivo. The suppressor alleles provide a tool to test this proposal. In the case of the W1225C suppressor mutation, these kinetic parameters could not be determined since this mutant exhibited no detectable transport activity for LTC4. Table 3 shows that, contrary to what might be expected, none of the mutants showed significant changes in the apparent affinity for ATP, and minor increases in the Vmax compared with that of D777N were detected in only three cases. Two of the suppressors even showed a decrease in the maximal activity. The majority of the mutants had 68 to 120% of the maximal activity of the D777N mutant enzyme. The highest activity was observed in mutants N1027D (147%) and G1207S (138%), whereas mutants F565L (33%) and G1207D (47%) had the lowest. These data suggest that neither the low affinity for ATP nor the Vmax defect, as measured with LTC4, of the D777N mutant is the primary defect responsible for the inability of this mutant protein to support cell growth in the presence of metal ions or other toxins.
TABLE 3.
Effect of the D777N suppressor mutations on the kinetic parameters of LTC4 uptake in vacuolar membrane vesicles
| Mutant | Km (ATP)a | Vmaxa |
|---|---|---|
| Wild type | 0.05 ± 0.01 | 4.40 ± 0.31 |
| D777N | 1.42 ± 0.18 | 1.69 ± 0.24 |
| D777N/V543I | 1.09 ± 0.28 | 1.15 ± 0.04 |
| D777N/F565L | 1.44 ± 0.30 | 0.58 ± 0.04 |
| D777N/S674L | 1.37 ± 0.26 | 1.21 ± 0.05 |
| D777N/A1003V | 1.55 ± 0.46 | 1.64 ± 0.13 |
| D777N/A1021T | 1.14 ± 0.20 | 1.28 ± 0.12 |
| D777N/A1021V | 1.71 ± 0.39 | 1.47 ± 0.15 |
| D777N/N1027D | 1.16 ± 0.18 | 2.49 ± 0.34 |
| D777N/Q1107R | 1.07 ± 0.09 | 1.48 ± 0.13 |
| D777N/G1207D | 1.06 ± 0.36 | 0.8 ± 0.06 |
| D777N/G1207S | 1.12 ± 0.27 | 2.33 ± 0.14 |
| D777N/S1212L | 1.39 ± 0.12 | 1.36 ± 0.13 |
| D777N/W1225C | —b | — |
| D777N/R1415G | 1.65 ± 0.44 | 2.02 ± 0.27 |
To determine the apparent Km for ATP (mM), the initial rate of LTC4 uptake in vacuolar membrane vesicles was assayed with 50 nM LTC4 and ATP concentrations ranging from 0.035 to 6 mM (see Materials and Methods). The Vmax (nmol · min−1 · mg−1) was determined with 10 mM ATP and LTC4 concentrations ranging from 0.1 to 3 μM. Data were fitted to the Michaelis-Menten equation using Prism 2.0 GraphPad Software. Values are the average ± standard deviation of duplicate determinations in two independently isolated vacuolar membrane preparations.
No detectable transport activity for LTC4.
Generation and phenotypic characterization of mutants containing only secondary mutations.
One way to get further information is the study of the mutants carrying ycf1 alleles with the second-site mutation alone. If the substituted amino acid forms a critical interaction with another residue, it might be expected that mutagenesis of this residue in an otherwise wild-type background would yield a defective phenotype. For this reason, we separated the suppressors from the original D777N mutation and tested their properties by analyzing their expression level and their ability to grow on plates containing CdCl2 or diamide and determining the MICs of these substrates. The mutants expressed Ycf1p at wild-type levels (Fig. 4B), but only three of them, S674L, A1003V, and N1027D, showed a wild-type phenotype when their sensitivity to Cd2+ and diamide was tested (Fig. 5). The resistance phenotypes of cells expressing the remaining 10 suppressor mutations could be grouped according to their different phenotypes. First, mutants A1021T, A1021V, G1207D, S1212L, and R1415G were more sensitive than the wild-type strain to inhibition by both Cd2+ and diamide, with 35 to 80% decreases in the MICs of these substrates when compared to those for the wild type. Second, mutants V543I, F565L, and Q1107R grew on diamide medium as much as the wild-type control, whereas growth on Cd2+ was clearly reduced (MICs ranging from 37 to 60% of that for the control). Third, mutant G1207S displayed a Cd2+ resistance similar to that of the wild-type cells but an enhanced tolerance to diamide. Finally, cells with mutant W1225C Ycf1p tolerated a Cd2+ concentration ninefold higher than the wild-type cells did, but their growth on diamide-containing medium was indistinguishable from that of the Δycf1 strain. Thus, the resistance phenotype analysis of the second-site mutants showed a group of nonfunctional mutants for both substrates, A1021T, A1021V, G1207D, S1212L, and R1415G, indicating that Ala1021, Glu1207, Ser1212, and Arg1415 are functionally important residues in Ycf1p. In addition, it revealed a group of suppressor mutants, V543I, F565L, Q1107R, G1207S, and W1225C, with a switch in their resistance profile indicating that Val543, Phe565, Gln1107, Gly1207, and Trp1225 are involved in determination of substrate specificity.
FIG. 5.
Resistance profile of the second-site mutants. Cells of the Δycf1 yeast strain were transformed with the centromeric plasmid pRS315 (Δycf1), pRS315-YCF1-HA (wild type), or pRS315-ycf1-HA (revertant listed) and grown on SD plates. Drops of each diluted strain (see Materials and Methods) were placed onto SD drop-out plates containing the indicated CdCl2 or diamide concentrations, grown for 48 h (diamide) or 72 h (CdCl2) at 30°C, and photographed. For quantitative determination of CdCl2 and diamide tolerance, MIC measurement was performed as described (see Materials and Methods) after growth of each strain at 30°C on microtiter plates containing medium with different concentrations of the compounds. Values are the mean of independent duplicate experiments.
DISCUSSION
Existing models for the transport cycle of ABC transporters suggest a close interaction between the NBDs and the TMDs. Nevertheless, the precise mechanism of signaling after substrate binding from the TMDs to the NBDs for stimulation of ATP hydrolysis remains poorly understood. In the same way, the mechanism by which ATP hydrolysis at the NBDs causes a structural change in the protein, presumably in the TMDs, to originate substrate transport across the membrane is unknown. This study was designed to gain insight into these problems and to identify interacting regions of the protein that could potentially be involved in the conformational changes produced in the catalytic sites during the transport cycle.
We performed a revertant analysis of five mutations located in the NBDs of Ycf1p, namely G663V, G756D, D777N, G1306E, and G1311R. The altered residues are completely conserved among ABC transporters and apparently involved in ATP binding and/or hydrolysis, as they are located in the Walker A, Walker B, and ABC signature motifs (48). Using this genetic approach, we isolated 13 different second-site mutations that suppress, to various degrees, the high sensitivity of the D777N mutant to Cd2+ and diamide.
We were unable to identify any intragenic suppressors of the other four alleles, G663V, G756D, G1306E, and G1311R. There are a number of reasons that suppressor mutations may have gone undetected, including a limited number of transformants screened, or mutagenesis that was not entirely random due to mutational hot spots and specificity of the mutagenic agent used. We used a combination of mutagenesis procedures and tested a large number of transformants. One of the mutagenic protocols, the use of a mutator strain of E. coli, is described as largely unbiased (19). On the basis of these observations and of the isolation, for the D777N mutant, of independent suppressor mutations in 13 residues, we believe that there is a severe limitation on the number of single-amino-acid alterations that will suppress these mutations. Moreover, the NBD1 mutations G663V and G756D were not suppressed when combined with some of the suppressors isolated for D777N (see below). Intragenic suppressors of four mutations located in the Walker A motif in the β-subunit of the yeast mitochondrial ATPase have been sought (52). The lack of suppressors for one mutation and the identification of only one suppressor for each of the other three in that study also argues in favor of our interpretation.
The suppressors isolated for the D777N mutant are located in four domains: TMD1, TMD2, NBD1, and NBD2. The location of second-site revertants within TMD1, TMD2, and NBD2 supports the structural and/or functional interaction between these domains and NBD1.
Eleven of the 13 suppressors isolated are located in the TMDs, not only in the predicted intracytoplasmic loops (A1021T, A1021V, and N1027D) but included in the membrane (V543I, F565L, A1003V, Q1107R, S1212L, and W1225C) or even facing the vacuolar lumen (G1207D and G1207S). This localization suggests intimate interaction between NBD1 and both TMDs. This is in agreement with present structural models that are based on studies on several ABC transporters in which NBD accessibility to proteases and biotinylated reagents from both sides of the membrane was investigated (4, 6, 20, 49). These models propose that part of the NBDs may span the lipid bilayer and be exposed to the noncytoplasmic surface through the pore formed by the TMDs.
Currently, based on the crystal structure of the ATPase subunits of several ABC transporters, two conflicting models for the dimeric arrangement of the NBDs are emerging. One of these models proposes that the two nucleotide binding sites are facing away from each other (24). In the second, the ABC signature motif of one NBD completes the ATP binding site (Walker A and B motifs) of the other (3, 23, 27, 61). In the case of Ycf1p, it is noteworthy that one of the second-site D777N suppressor mutations, R1415G, affects a highly conserved Arg residue in the ABC signature motif of NBD2, suggesting a model in which interaction of the two NBDs occurs.
To unravel the mechanism by which the suppressor mutations might act, they were studied in isolation. The results show that a significant fraction of the suppressors,V543I, F565L, A1021T, A1021V, Q1107R, G1207D, S1212L, and R1415G, are deficient for Ycf1p function in cadmium detoxification, pointing to a specific suppression mechanism. The specificity of the suppression is further supported by the fact that neither the W1225C nor R1415G mutations were able to suppress the defective growth or Ycf1p transport function of the other NBD1 primary mutations, namely G663V and G756D (data not shown). These results suggest that the suppressor mutations do not suppress by bypassing the function of Asp777 in Ycf1p and rather indicate that the Val543, Phe565, Ala1021, Gln1107, Gly1207, Ser1212, and Arg1415 residues are involved in intramolecular interactions that are relevant for the connection with the Walker B region of the NBD. In contrast, mutations G1207S and W1225C produce an enhanced Ycf1p function even in the absence of D777N mutation, indicating a different mechanism of suppression that involves a change in Ycf1p substrate specificity. Finally, the suppressor effect of S674L, A1003V, and N1027D mutations is probably due to nonspecific compensating structural alterations, since these mutations exhibited wild-type behavior in the absence of the primary mutation.
Phenotypic characterization of the suppressor mutants separated from the primary mutation showed that five of them, namely V543I, F565L, Q1107R, G1207S, and W1225C, exhibit individual different responses to the substrates tested (Fig. 5). This behavior concurs with their localization in the TMDs. Previous mutational analysis of several ABC transporters showed that changes introduced into the TMDs (2, 14, 29, 33, 56) can affect substrate specificity. In fact, all of them map in TM segments for which contribution to specific binding sites has been documented, namely TM5, TM6, TM10, and TM12 (35, 36, 38). W1225C in TM12 showed a drastic specificity shift. This mutant appeared to completely disrupt Ycf1p-substrate interactions except for Cd2+ since no resistance to diamide or LTC4 transport could be detected. The W1225C mutant deserves further investigation, since it may provide useful insights into Ycf1p transport substrate specificity.
The use of suppression genetics to enlighten structure and function studies is based on the premise that an existing altered function allele can be restored to wild-type function by a second mutational change. Thus, by definition, the suppressor mutations reverse the critical defect of the starting mutant. In addition, the suppressors can confirm or deny that a property observed in vitro is the critical one in vivo. The D777N mutant shows in vivo a greatly reduced resistance to Cd2+, and in vitro kinetic analysis of the mutant protein revealed that it has an apparently wild-type Km for LTC4 but lowered ATP affinity and Vmax for LTC4 transport (15). The results presented here show that the suppressor mutants recovered the ability to grow on Cd2+ whereas none of them restored the Vmax for LTC4 or improved the affinity for ATP. On the contrary, some of them share an alteration in substrate specificity, suggesting that this feature underlies the suppression mechanism. These findings indicate that the growth defect of the D777N mutant does not derive from the kinetic defects detected in vitro. The crucial functional defect may rather be due to another essential function of the Asp777 residue in addition to ATP binding and/or hydrolysis, such as coupling these processes to substrate binding and/or transport. The identified second-site mutations could have restored the interactions involving the substrate and ATP binding sites, which are required for transport to occur and presumably disrupted in the D777N mutant protein. Consistent with this proposed dual function for the NBD1 invariant Asp residue in the Walker B region are the results for some of the known NBD structures. In ArsA, the ATPase subunit of the ArsAB pump of E. coli, two aspartic residues involved in coordination of Mg2+ are in close proximity to the allosteric metal binding site (61) and are proposed to participate in signal transmission between metal and nucleotide binding sites. In addition, the high-resolution crystal structure of HisP, the ATPase subunit of the histidine permease of Salmonella enterica serovar Typhimurium, places the homologous residue to Asp777 in the particularly strategic position connecting most residues that contact ATP with those that may interact with the TM subunits (24). The location and phenotype of the suppressor mutations may thus be interpreted if substrate union to its binding site is directly connected with the NBD1 Walker B region to stimulate ATPase activity. Further characterization of the suppressor mutants will allow a deeper understanding of the intramolecular interactions that are important for Ycf1p transport activity and may be shared by other ABC transporters.
ACKNOWLEDGMENTS
This study was supported by Fondo de Investigaciones Sanitarias Grant 98/1279 and by the Fundación Sira Carrasco para Ayuda a la Fibrosis Quística.
We thank J. Martín for the anti-HA antibody, F. Portillo for critical reading of the manuscript and helpful discussions and suggestions, and Eulalia Morgado for technical assistance in some experiments.
REFERENCES
- 1.Aebersold R H, Leavitt J, Saavedra R A, Hood L E, Kent S B H. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M P, Gregory R J, Thompson S, Souza D W, Paul S, Mulligan R C, Smith A E, Welsh M J. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong S, Tabernero L, Zhang H, Hermodson M, Stauffacher C. Powering the ABC transporter: the 2.5 Å crystallographic structure of the ABC domain of RBSA. Pediatr Pulmonol. 1998;26:91–92. [Google Scholar]
- 4.Baichwal V, Liu D, Ames F L. The ATP-binding component of a prokaryotic traffic ATPase is exposed to the periplasmic (external) surface. Proc Natl Acad Sci USA. 1993;90:620–624. doi: 10.1073/pnas.90.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake M S, Johnston K H, Russell-Jones G H, Gotschlich E C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 6.Bliss J M, Silver P S. Evidence that KpsT, the ATP-binding component of an ATP-binding cassette transporter, is exposed to the periplasm and associates with polymer during translocation of the polysialic acid capsule of Escherichia coli K1. J Bacteriol. 1997;179:1400–1403. doi: 10.1128/jb.179.4.1400-1403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, Deeley R G. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 11.Croop J M. Evolutionary relationships among ABC transporters. Methods Enzymol. 1998;292:101–116. doi: 10.1016/s0076-6879(98)92010-9. [DOI] [PubMed] [Google Scholar]
- 12.Davidson A L, Shuman H A, Nikaido H. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci USA. 1992;89:2360–2364. doi: 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egner R, Rosenthal F E, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falcón-Pérez J M, Mazón M J, Molano J, Eraso P. Functional domain analysis of the yeast ABC transporter Ycf1p by site-directed mutagenesis. J Biol Chem. 1999;274:23584–23590. doi: 10.1074/jbc.274.33.23584. [DOI] [PubMed] [Google Scholar]
- 16.Fojo A T, Whang-Peng J, Gottesman M M, Pastan I. Amplification of DNA sequences in human multidrug-resistant KB carcinoma cells. Proc Natl Acad Sci USA. 1985;82:7661–7665. doi: 10.1073/pnas.82.22.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foote S J, Thompson J K, Cowman A F, Kemp D J. Amplification of the multidrug resistance gene in some chloroquine-resistant isolates of P. falciparum. Cell. 1989;57:921–930. doi: 10.1016/0092-8674(89)90330-9. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh M, Shen J, Rosen B P. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greener A, Callahan M. XL1-Red: a highly efficient random mutagenesis strain. Strategies. 1994;7:32–34. [Google Scholar]
- 20.Gruis D B, Price E M. The nucleotide binding folds of the cystic fibrosis transmembrane conductance regulator are extracellularly accessible. Biochemistry. 1997;36:7739–7745. doi: 10.1021/bi9701585. [DOI] [PubMed] [Google Scholar]
- 21.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 22.Holland I B, Blight M A. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner K-P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 24.Hung L-W, Wang I X, Nikaido K, Liu P-Q, Ames G F-L, Kim S-H. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 25.Hyde S C, Emsley P, Hartshorn M J, Mimmack M M, Gileadi U, Pearce S R, Gallagher M P, Gill D R, Hubbard R E, Higgins C F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 26.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P M, George A M. Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol Lett. 1999;179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuchler K, Sterne R E, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwan T, Gros P. Mutational analysis of the P-glycoprotein first intracellular loop and flanking transmembrane domains. Biochemistry. 1998;37:3337–3350. doi: 10.1021/bi972680x. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Li Z-S, Lu Y-P, Zhen R-G, Szczypka M, Thiele D J, Rea P A. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z-S, Szczypka M, Lu Y-P, Thiele D J, Rea P A. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 33.Linsdell P, Evagelidis A, Hanrahan J W. Molecular determinants of anion selectivity in the cystic fibrosis transmembrane conductance regulator chloride channel pore. Biophys J. 2000;78:2973–2982. doi: 10.1016/S0006-3495(00)76836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C E, Liu P-Q, Ames G F-L. Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter) J Biol Chem. 1997;272:21883–21891. doi: 10.1074/jbc.272.35.21883. [DOI] [PubMed] [Google Scholar]
- 35.Loo T W, Clarke D M. Identification of residues in the drug-binding site of human P-glycoprotein using a thiol-reactive substrate. J Biol Chem. 1997;272:31945–31948. doi: 10.1074/jbc.272.51.31945. [DOI] [PubMed] [Google Scholar]
- 36.Loo T W, Clarke D M. Defining the drug-binding site in the human multidrug resistance P-glycoprotein using methanethiosulfonate analog of verapamil, MTS-verapamil. J Biol Chem. 2001;276:14972–14979. doi: 10.1074/jbc.M100407200. [DOI] [PubMed] [Google Scholar]
- 37.Manavalan P, Dearborn D G, McPherson J M, Smith A E. Sequence homologies between nucleotide binding regions of CFTR and G-proteins suggest structural and functional similarities. FEBS Lett. 1995;366:87–91. doi: 10.1016/0014-5793(95)00463-j. [DOI] [PubMed] [Google Scholar]
- 38.Morris D I, Greenberger L M, Bruggemann E P, Cardarelli C, Gottesman M M, Pastan I, Seamon K B. Localization of the forskolin labeling sites to both halves of P-glycoprotein: similarity of the sites labeled by forskolin and prazosin. Mol Pharmacol. 1994;46:329–337. [PubMed] [Google Scholar]
- 39.Petrovic S, Pascolo L, Gallo R, Cupelli F, Ostrow J D, Goffeau A, Tiribelli C, Bruschi C V. The products of YCF1 and YLL015w (BPT1) cooperate for the ATP-dependent vacuolar transport of unconjugated bilirubin in Saccharomyces cerevisiae. Yeast. 2000;16:561–571. doi: 10.1002/(SICI)1097-0061(200004)16:6<561::AID-YEA551>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 40.Ravnik-Glavac M, Glavac D, Dean M. Sensitivity of single-strand conformation polymorphism and heteroduplex method for mutation detection in the cystic fibrosis gene. Hum Mol Genet. 1994;3:801–807. doi: 10.1093/hmg/3.5.801. [DOI] [PubMed] [Google Scholar]
- 41.Rebbeor J F, Connolly G C, Dumont M E, Ballatori N. ATP-dependent transport of reduced glutathione in yeast secretory vesicles. Biochem J. 1998;334:723–729. doi: 10.1042/bj3340723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebbeor J F, Connolly G C, Dumont M E, Ballatori N. ATP-dependent transport of reduced glutathione on YCF1, the yeast orthologue of mammalian multidrug resistance associated proteins. J Biol Chem. 1998;273:33449–33454. doi: 10.1074/jbc.273.50.33449. [DOI] [PubMed] [Google Scholar]
- 43.Ren X-Q, Furukawa T, Chen Z-S, Okumura H, Aoki S, Sumizawa T, Tani A, Komatsu M, Mei X-D, Akiyama S. Functional comparison between YCF1 and MRP1 expressed in Sf21 insect cells. Biochem Biophys Res Commun. 2000;270:608–615. doi: 10.1006/bbrc.2000.2483. [DOI] [PubMed] [Google Scholar]
- 44.Riordan J R, Rommens J M, Kerem B-S, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, Drumm M L, Iannuzzi M C, Collins F S, Tsui L-C. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 45.Romero I, Maldonado A M, Eraso P. Glucose-independent inhibition of yeast plasma-membrane H+-ATPase by calmodulin antagonists. Biochem J. 1997;322:823–828. doi: 10.1042/bj3220823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose M D, Winston F, Hieter P, editors. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 128–129. [Google Scholar]
- 47.Schlichting I, Almo S C, Rapp G, Wilson K, Petratos K, Lentfer A, Wittinghofer A, Kabsch W, Pai E F, Petsko G A, Goody R S. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature. 1990;345:309–315. doi: 10.1038/345309a0. [DOI] [PubMed] [Google Scholar]
- 48.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 49.Schneider E, Hunke S, Tebbe S. The MalK protein of the ATP-binding cassette transporter for maltose of Escherichia coli is accessible to protease digestion from the periplasmic side of the membrane. J Bacteriol. 1995;177:5364–5367. doi: 10.1128/jb.177.18.5364-5367.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senior A E, Al-Shawi M K, Urbatsch I L. ATP hydrolysis by multidrug-resistance protein from Chinese hamster ovary cells. J Bioenerg Biomembr. 1995;27:31–36. doi: 10.1007/BF02110328. [DOI] [PubMed] [Google Scholar]
- 51.Serrano R, Montesinos C, Roldán M, Garrido G, Ferguson C, Leonard K, Monk B C, Perlin D S, Weiler E W. Domains of yeast plasma membrane and ATPase-associated glycoprotein. Biochim Biophys Acta. 1991;1062:157–164. doi: 10.1016/0005-2736(91)90387-n. [DOI] [PubMed] [Google Scholar]
- 52.Shen H, Sosa-Peinado A, Mueller D M. Intragenic suppressors of P-loop mutations in the β-subunit of the mitochondrial ATPase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271:11844–11851. doi: 10.1074/jbc.271.20.11844. [DOI] [PubMed] [Google Scholar]
- 53.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun H, Molday R S, Nathans J. Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J Biol Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 55.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 56.Taguchi Y, Kino K, Morishima M, Komano T, Kane S E, Ueda K. Alteration of substrate specificity by mutations at the His61 position in predicted transmembrane domain 1 of human MDR1/P-glycoprotein. Biochemistry. 1997;36:8883–8889. doi: 10.1021/bi970553v. [DOI] [PubMed] [Google Scholar]
- 57.Tommasini R, Evers R, Vogt E, Mornet C, Zaman G J, Schinkel A H, Borst P, Martinoia E. The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor 1. Proc Natl Acad Sci USA. 1996;93:6743–6748. doi: 10.1073/pnas.93.13.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tusnády G E, Bakos E, Váradi A, Sarkadi B. Membrane topology distinguishes a subfamily of the ATP-binding cassette (ABC) transporters. FEBS Lett. 1997;402:1–3. doi: 10.1016/s0014-5793(96)01478-0. [DOI] [PubMed] [Google Scholar]
- 59.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wemmie J A, Moye-Rowley W S. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol Microbiol. 1997;25:683–694. doi: 10.1046/j.1365-2958.1997.5061868.x. [DOI] [PubMed] [Google Scholar]
- 61.Zhou T, Radaev S, Rosen B P, Gatti D L. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]