Abstract
The cAMP response element binding protein (CREB) is a bifunctional transcription activator, exerting its effects through a constitutive activation domain (CAD) and a distinct kinase inducible domain (KID), which requires phosphorylation of Ser-133 for activity. Both CAD and phospho-KID have been proposed to recruit polymerase complexes, but this has not been directly tested. Here, we show that the entire CREB activation domain or the CAD enhanced recruitment of a complex containing TFIID, TFIIB, and RNA polymerase II to a linked promoter. The nuclear extracts used mediated protein kinase A (PKA)-inducible transcription, but phosphorylation of CRG (both of the CREB activation domains fused to the Gal4 DNA binding domain) or KID-G4 did not mediate recruitment of a complex, and mutation of the PKA site in CRG abolished transcription induction by PKA but had no effect upon recruitment. The CREB-binding protein (CBP) was not detected in the recruited complex. Our results support a model for transcription activation in which the interaction between the CREB CAD and hTAFII130 of TFIID promotes the recruitment of a polymerase complex to the promoter.
The cAMP response element (CRE) mediates both constitutive and cAMP-induced transcription activation of many genes in a variety of cell types (8, 30, 40, 43, 50, 52). The CRE-binding protein, CREB, a member of the basic leucine zipper family of transcription factors, binds constitutively to the CRE in the promoter of the target gene (51) and can activate constitutive transcription in the absence of hormonal stimuli (3, 29, 50). Extracellular stimuli that activate protein kinases can lead to phosphorylation of CREB on Ser-133, e.g., by cAMP-activated protein kinase A (PKA), resulting in a further enhancement of transcriptional activation (3, 16, 50). Mutation of the Ser-133 PKA phosphorylation site in CREB to an alanine abolishes kinase-inducible activation (17, 50) but not constitutive activation (3, 29, 50). We and others have shown that these constitutive and kinase-inducible activities map to two separate and independently acting transcription activation domains: a constitutive activation domain (CAD) responsible for activating constitutive transcription and a kinase-inducible domain (KID) that mediates activation in response to cAMP-activated PKA (3, 29, 50) and several other kinases (11, 15, 26, 63, 68). However, the exact mechanism of action of these domains in stimulating constitutive and kinase-inducible transcription has not been defined.
Transcription of a class II gene by RNA polymerase II requires the assembly of general transcription factors and coactivators around the transcription start site in the gene's promoter (reviewed in references 22 and 44). The general transcription factors (TFIID, TFIIA, TFIIB, TFIIF-pol II, TFIIE, and TFIIH) were initially identified as the basic nuclear components required to reconstitute in vitro transcription by RNA polymerase II (9, 54–56, 66, 67). These general factors are required for accurate and optimal positioning of RNA polymerase II at the transcriptional start site, melting the template and facilitating promoter clearance to allow synthesis of an mRNA transcript (reviewed in references 21, 44 and 72). Much work has focused on the role of activators in mediating recruitment of these essential factors, which is a necessary first step in transcription initiation. In particular, many activators interact with TFIID (5, 12–14, 23, 28, 38, 47, 61, 62, 65) or with TFIIB (7, 25, 36, 57). Recent work has demonstrated that the general factors, RNA polymerase II, and coactivators often exist as macromolecular complexes in cells rather than as isolated factors (18, 22). Thus, transcription activators must recruit and modify the activity of complexes for promoter recognition (TFIID) (1, 48, 49) and mRNA synthesis (Pol II holoenzyme) (32, 33, 39), processes which are often facilitated by coactivators (2, 19, 27, 34, 59, 70, 71).
Although recruitment of a holoenzyme complex is essential to transcription in vivo, the polymerase still must be positioned properly at the start site in the promoter of the target gene to accurately initiate the synthesis of a transcript. Recruitment of the TFIID complex represents a crucial first target in assembly of a functional polymerase complex. Early studies of in vitro transcription showed that binding of TFIID to a promoter enhanced the association of other polymerase complex components, which did not readily exchange with other promoters in template challenge assays (66). This suggested a processive mechanism for assembly of a functional polymerase complex. Even though it is now recognized that general factors are found in complexes rather than as isolated factors, the same type of mechanism may operate in vivo, where TFIID and holoenzyme exist as distinct complexes in cells. Support for this idea comes from experiments in which VP16 mutants that cannot bind to TFIID are defective for holoenzyme recruitment (31). In addition, several lines of evidence have suggested that CREB plays a role in recruitment of TFIID and RNA polymerase II, as discussed below.
Early evidence for the recruitment activity of CREB came from the demonstration that inclusion of activating transcription factor (ATF)/CRE sites upstream of the adenovirus major late promoter resulted in extension of the footprint downstream of the TATA region to include the transcription initiation site (20). Although the ATF/CREB family member responsible for this activity was not determined in those studies, it was found that the complex responsible for the expanded footprint included RNA polymerase II and TFIIB (24). Those studies were done without phosphorylation of ATF/CREB proteins. Subsequent studies provided evidence that the CAD in CREB binds TFIID through its TATA binding protein-associated factors (TAFs) (69) and that this interaction occurs through hTAF130 or dTAF110 in TFIID (12, 13, 64) and is mediated by a conserved set of hydrophobic residues in the CREB CAD (12). Phosphorylation of the KID in CREB promotes binding of the CREB-binding protein (CBP) (34, 53), which has also been shown to bind TFIIB and RNA helicase A associated with holoenzyme (34, 41, 42). Thus, both of the CREB activation domains, the CAD and the KID, have been proposed to mediate recruitment of the polymerase complex (Fig. 1) (12, 34, 41).
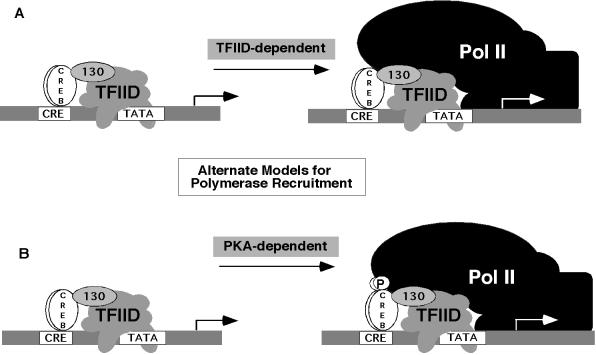
FIG. 1.
Models for transcription activation by the two activation domains of CREB. (A) A sequential model for CAD activation involves interaction between CAD and the hTAF130 subunit of TFIID to recruit TFIID to the promoter, thereby increasing the rate of formation of the polymerase complex and the overall rate of transcription (12, 13). (B) An alternative model involves bivalent recruitment of TFIID by CAD and holoenzyme by P-KID interaction with CBP and RNA helicase A (41, 42).
We have suggested that the mechanism of activation of the CREB CAD involves recruitment of TFIID through interaction with hTAFII130, followed by association of the polymerase complex through its interaction with promoter-bound TFIID (Fig. 1A; 12). Similar mechanisms have been proposed based on the interactions between other transcription activators and TAF components of TFIID (14, 37, 38, 61, 65). On the other hand, Nakajima and coworkers showed that phospho-CREB bound to purified holoenzyme preparations in vitro and that the CAD (Q2) domain bound to TFIID (42). Based on these results, a mechanism for CREB activation was proposed in which each domain, CAD and P-KID, recruited individual components of the transcription machinery, TFIID and holoenzyme, independently (Fig. 1B). However, the CAD can activate transcription on its own (3, 29, 50), which suggests that the CAD is able to reach the end point of polymerase recruitment through its recruitment of TFIID. Neither of these hypotheses has been directly tested by measuring recruitment of RNA polymerase II to a template.
To further address the question of polymerase complex recruitment by the CREB activation domains, we designed an assay to determine the contributions of the constitutive and kinase-inducible activation domains in CREB to recruitment of an RNA polymerase II-containing complex to promoter DNA. We adapted an agarose electrophoretic mobility shift assay (Ag-EMSA) (35), previously used to show recruitment of purified TATA-binding protein (TBP) by the factor Zta, to detect the formation of polymerase complexes on promoter DNA. A template containing five Gal4 sites upstream of a minimal promoter was incubated with purified proteins containing either or both of the CREB activation domains fused to the Gal4 DNA binding domain (CRGs) and rat liver nuclear extract (RLNE) to determine which CRGs could promote complex formation on the promoter. These binding reactions were done under conditions where CRG stimulated in vitro transcription, which was enhanced by phosphorylation of Ser- 133 in CREB with PKA. In addition, a sensitive procedure was developed to determine whether specific components, such as TBP or RNA polymerase II, are present in the recruited complex. We also tested whether phosphorylation of CRG contributes to this recruitment process, by testing a PKA phosphorylation site mutant of CRG and by phosphorylation of CRG or KID by preincubation with protein kinase A. Our results show that the CAD in CREB mediates recruitment of a complex containing TBP, RNA polymerase II, and TFIIB and that this process is unaffected by protein kinase A-mediated phosphorylation of the CREB activation domain.
MATERIALS AND METHODS
Expression and purification of CRG proteins.
Coding regions for the various CRG proteins (CRG, G4-DBD, CAD-G4, KID-G4, and CRG-S133A) were subcloned into the pBacPak baculovirus expression vector, and Sf9 insect cells in culture were infected using the BaculoGold baculovirus system (Pharmingen). The CRG proteins are identical to those described previously except that they contain amino acids (aa) 4 to 94 rather than aa 4 to 147 of the Gal4 DNA binding domain (50). All constructs include CREB aa 1 to 8 to provide appropriate translation initiation signals. In addition, the constructs include the following amino acids of the CREB protein: CRG, aa 1 to 276; CAD-G4, aa 165 to 252; and KID-G4, aa 98 to 142. CRG proteins were purified from Sf9 nuclear extracts using DNA affinity chromatography. Oligonucleotides containing five Gal4 binding sites and cohesive ends for HindIII were ligated for an average length of a 10-mer and then ligated to a biotinylated linker. The resulting biotinylated DNA template was bound to streptavidin-coated Dynabeads (Dynal) in Tris-EDTA (TE), pH 7.5, plus 1 M KCl at room temperature overnight. Beads were immobilized with a magnet and washed once with an equal volume of TE and 1 M KCl and then twice with DAP-A (20 mM HEPES, pH 7.9, 1 mM EDTA, 15% glycerol, 0.05% NP-40, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) plus 100 mM KCl. The affinity support was then incubated with the Sf9 nuclear extracts (0.5 mg of protein/1 pmol of bound DNA) in DAP-A plus 70 mM (NH4)2SO4 at room temperature for 15 min. Bound proteins were washed three times in an equal volume of DAP-A and 100 mM KCl and then eluted from the affinity support with 0.2 volumes of DAP-A plus 1 M KCl at room temperature for 15 min. Protein was tested for purity on a silver-stained gel and stored at −80°C.
Production of RLNE.
Livers from four 125- to 150-g freshly killed rats were homogenized in 80 ml of homogenization buffer (10 mM HEPES, pH 7.9, 25 mM KCl, 0.5 mM spermidine, 0.15 mM spermine, 1 mM EDTA, 2 M sucrose, 10% glycerol, 0.5 mg of leupeptin/ml, 1 mg of pepstatin/ml, 1 mM benzamidine, 0.5 mM PMSF) at 4°C, filtered, and centrifuged through a homogenization buffer gradient at 24,000 rpm in an SW27 rotor for 30 min at 4°C to pellet nuclei. Nuclei were lysed in a nuclear lysis buffer (10 mM HEPES, pH 7.9, 100 mM KCl, 3 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 0.5 mg of leupeptin/ml, 1 mg of pepstatin/ml, 1 mM benzamidine, 0.5 mM PMSF) with a Dounce homogenizer, and chromatin was precipitated from the lysate by the addition of a 4 M (NH4)2SO4 solution, pH 7.9, to a 400 mM final concentration. Precipitated chromatin was removed by centrifugation at 35,000 rpm in a Ti60 rotor at 4°C for 1 h. Nuclear proteins were precipitated with the addition of solid (NH4)2SO4 to 0.3 g/ml followed by centrifugation at 35,000 rpm in a Ti60 rotor for 25 min at 4°C. Pelleted proteins were resuspended in dialysis buffer (25 mM HEPES, pH 7.9, 40 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol) and were dialyzed against the same buffer four times for 30 min each time at 4°C. Protein extracts were concentrated by centrifugation through a Millipore concentrating filter in a tabletop centrifuge at 3,200 rpm for 5 to 6 h at 4°C. Nuclear extracts were stored in small aliquots at −80°C.
In vitro transcription assay.
In vitro transcription was performed as previously described (51), with the following modifications. In vitro reactions contained 50 μg of nuclear extract, 150 fmol of 5XGT-Luc template (750 fmol of Gal4 sites), and 750 fmol of CREB-Gal4 protein. The mRNA synthesized was quantitated by primer extension analysis. Where indicated, 0.5 U of PKA plus 0.5 uM okadaic acid were added to phosphorylate P-KID-G4 and P-CRG, after which 0.5 U of PKI was added to inhibit further PKA activity. Equivalent results were obtained for using two different preparations of purified proteins and nuclear extracts.
Ag-EMSA.
Probe DNA (150 bp) with five Gal4 binding sites (5×GT) and a minimal TATA-containing promoter (see Fig. 4A) was end labeled with [α-32P]dATP using Klenow. Five femtomoles of probe was incubated with 10 fmol of purified protein and 3 μg of RLNE (see above) in 1× Ag-EMSA binding buffer (12.5 mM HEPES, pH 7.9, 12.5% glycerol, 5 mM MgCl2, 70 mM KCl, 0.2 mM EDTA, 10 mM 2-mercaptoethanol, 0.5 mg of bovine serum albumin/ml, 40 μg of poly-dI/dC/ml) for 15 min on a nutator at 4°C. Reactions were run on a 1% Seakem agarose gel (FMC) in 1× buffer G (45 mM Tris, pH 8.3, 45 mM boric acid, 0.5 mM EDTA, 5 mM MgAc) at 100 V for 2 h. The gel was then placed on filter paper, dried overnight at room temperature under vacuum, and exposed to film to detect complexes. For antibody supershifting experiments, 1 μl of primary antibody was incubated with 3 μg of RLNE in 1× Ag-EMSA binding buffer for 1 h on a nutator at 4°C. Ten femtomoles of purified protein, 2 μl of biotinylated secondary antibody, and 3 μl of streptavidin-coated Dynabeads were added, and the samples were incubated for an additional 1 h on a nutator at 4°C. Five femtomoles of probe was then added, and the samples were incubated for an additional 15 min at 4°C. Reactions were run on a 1% Seakem agarose gel, and complexes were visualized as described above.
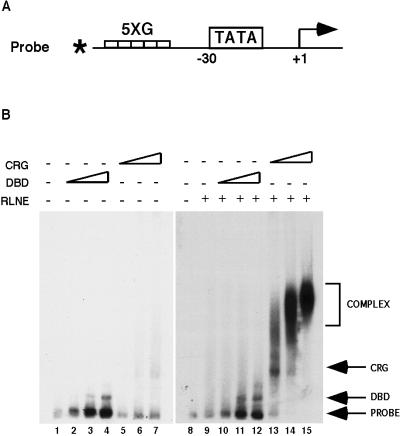
FIG. 4.
CRG stimulates the formation of a large complex at the promoter in an Ag-EMSA. (A) The Ag-EMSA method uses a 32P-end-labeled, 150-bp DNA probe that contains five Gal4 binding sites (5×G) and a TATA-containing minimal promoter. The probe is incubated with purified protein and RLNE under conditions that allow DNA binding (see Materials and Methods). (B) Ag-EMSA binding reactions included 5 fmol of probe incubated with 1, 5, or 10 fmol of purified G4-DBD or CRG protein and 3 μg of RLNE where indicated. Complexes were resolved and visualized as indicated in Materials and Methods.
RESULTS
CRG recruits a large complex to the promoter DNA in an Ag-EMSA.
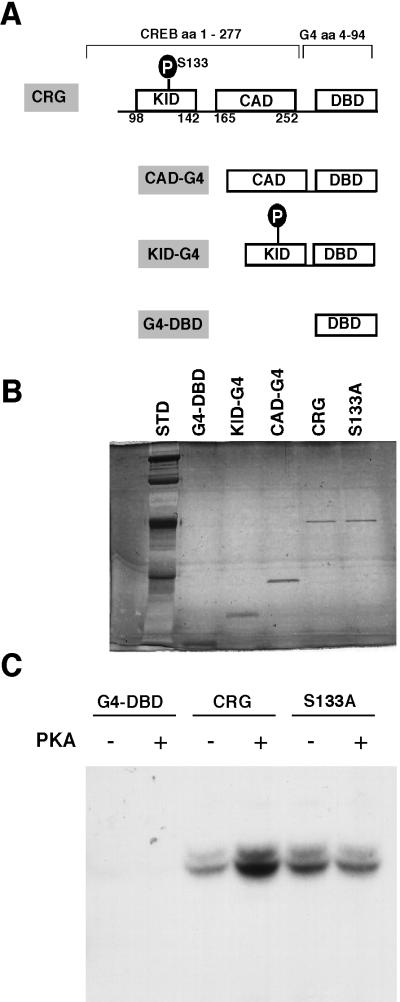
To obtain functional CRG proteins containing either or both of the functional CREB activation domains (Fig. 2A) for our assays, we expressed recombinant baculovirus in Sf9 insect cells and purified the expressed CRGs, using DNA affinity chromatography. The eluted CRG proteins were separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and silver stained (Fig. 2B), which shows that the CRG proteins were purified to near homogeneity. The recombinant CRG proteins and RLNEs used in our binding studies were also tested for their ability to support constitutive and PKA-induced transcription controlled by a minimal promoter with five Gal4 binding sites (5×GT), which was also used for the binding studies (see Fig. 4A). As illustrated in Fig. 2C, CRG increased transcription above the background level obtained with the Gal4 DNA binding domain (G4-DBD) alone, and this transcription was enhanced significantly by phosphorylation of CRG with PKA. In addition, the CRG-S133A PKA phosphorylation site mutant provided stimulation of constitutive but not PKA-inducible activity, indicating that the constitutive and PKA-inducible CRG activities previously described in transfected cells (50) are reproduced in the in vitro transcription system.
FIG. 2.
Purification of CRG proteins and their function in an in vitro transcription assay. (A) The schematic shows the domains of the CRG (CREB-Gal4) proteins used, including the amino acids of CREB included in each. (B) CRG proteins were expressed in Sf9 insect cells using a baculovirus system. Proteins were purified from nuclear extracts using DNA affinity chromatography. Purified protein was separated on an SDS–15% PAGE gel and was silver stained by standard methods to visualize the protein bands. (C) Purified G4-DBD and CRG proteins were tested for activation in an in vitro transcription system with and without PKA added to the reaction as indicated.
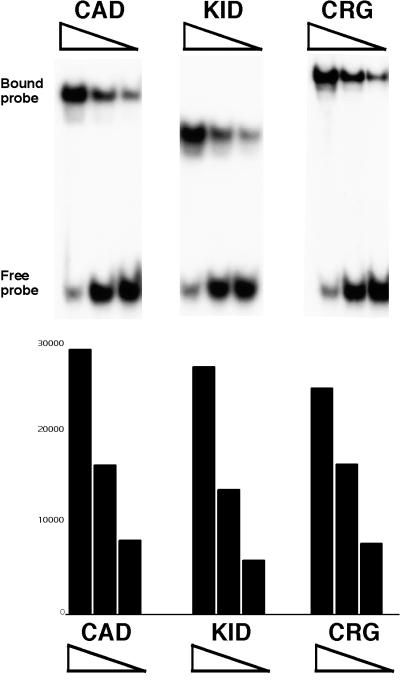
The concentrations of purified CRG proteins shown in Fig. 2B were determined empirically by testing their binding to a DNA probe encoding a Gal4 binding site in a classic EMSA. The results from these experiments (Fig. 3) showed that the purified proteins retain DNA binding activity, and the correlation between amount of protein added and amount of probe bound was used to estimate protein concentrations.
FIG. 3.
Mobility shift analysis of purified CRG proteins with a Gal4 probe. Affinity purified G4-CAD, G4-KID and CRG were diluted 1:20, 1:50, and 1:100 and bound to 10 fmol of 32P-labeled Gal4 probe in a standard EMSA. Bound and free probes were separated on a 5% polyacrylamide mobility shift gel. Densitometry was used to quantify the relative amounts of probe bound by the various protein samples. A representative experiment is shown.
To test for stimulation of complex formation on promoter DNA by CRG, we adapted an Ag-EMSA that had been previously used to separate free promoter from Zta/TBP/TFIIA-bound promoter (35). Agarose gels are used because their larger matrix makes it possible to resolve much larger complexes, such as those formed by RNA polymerase II (megadaltons), than those that can be resolved (kilodaltons) on the polyacrylamide gels normally used for gel shift assays. In our method (see Materials and Methods), a 32P-end-labeled DNA probe (Fig. 4A), which contains five Gal4 binding sites ligated to a TATA-containing minimal promoter (5×GT), was incubated with purified CRG proteins and RLNE to provide the general transcription factors under conditions identical to those used for in vitro transcription. Bound and free probes were separated on a 1% Seakem agarose gel (Fig. 4B). The binding of G4-DBD or CRG alone produced only a slight decrease in the mobility of the probe in these gels (Fig. 4B, lanes 2 to 4 and 5 to 7).
In the presence of RLNE, the addition of increasing amounts of CRG protein to the binding reactions produced an increase in the formation of a large, slowly migrating complex on the DNA probe (Fig. 4B, lanes 13 to 15). No such effect was observed with similar amounts of DBD protein (Fig. 4B, lanes 10 to 12), and complex formation was dependent on the presence of RLNE in the reaction (in Fig. 4B, compare lanes 5 to 7 with lanes 13 to 15). These results indicate that CRG bound at the promoter was specifically stimulating the recruitment of a large complex of proteins from the extract to the promoter DNA.
The complex recruited to the promoter by CRG contains TFIID, TFIIB, and RNA polymerase II.
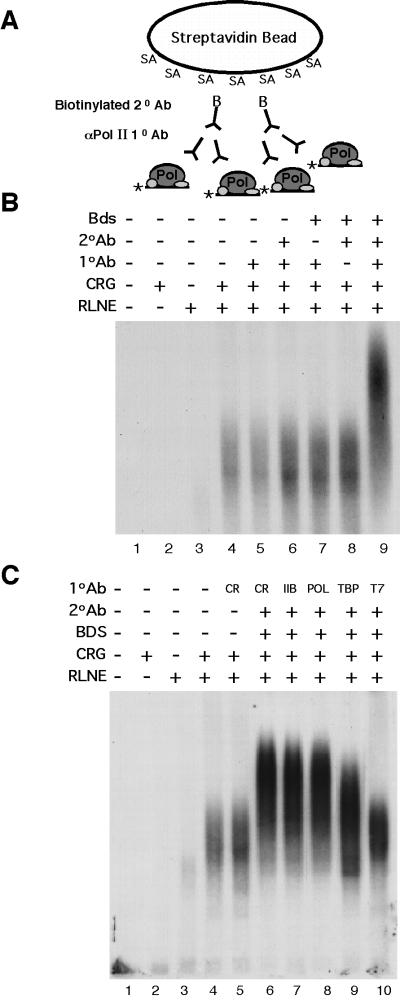
To determine whether the complex recruited by CRG has components expected to be found in an RNA polymerase II complex, we developed a supershift assay to test the complexes for the presence of specific components, such as RNA polymerase II. The method, illustrated in Fig. 5A, makes use of a multiorder binding array to cross-link the recruited complexes and derives component specificity from the use of primary antibodies specific for CREB or for proteins known to be required in the RNA polymerase II complex, including TFIIB, TBP of TFIID, and the large subunit of pol II. Because the complexes formed in this assay are extremely large (in the megadalton range), the addition of a specific primary antibody (kDa) alone is not sufficient to produce a detectable increase in the size of the complex (mDa) (Fig. 5B, lane 5), even in the presence of secondary antibody (anti-CREB; Fig. 5B, lane 6). It was necessary to include a biotinylated secondary antibody specific for the primary antibody along with streptavidin-coated Dynabeads (Fig. 5B, lane 9) to cross-link the complexes into larger “supercomplexes” that could then be detected through their shift to a higher molecular weight on the agarose gel. Supershifting was not observed when any of the components (primary antibody, secondary antibody, streptavidin beads) were omitted from the reaction (Fig. 5B, lanes 6 to 8). The addition of a primary antibody specific for CREB, together with the other supershifting reagents, resulted in a visible shift of the complex to a higher molecular weight on the gel (Fig. 5C, lane 4 versus lane 6). Likewise, antibodies specific for TFIIB or the large subunit of RNA polymerase II (Pol) produced a comparable shift in the molecular weight of the complex (Fig. 5C, lanes 7 and 8). Antibody against the TBP subunit of TFIID also retarded the complex, but to a somewhat lesser extent (Fig. 5C, lane 9), which may be attributable to reduced accessibility of TBP within the TFIID complex or interference of the antibody with further growth of the complex. Antibodies specific for an irrelevant T7 epitope did not supershift the complex (Fig. 5C, lane 10).
FIG. 5.
The complex recruited to the promoter by CRG contains components of an RNA polymerase II complex, as indicated by an antibody supershift assay. (A) The method uses rabbit primary antibodies specific for components of the polymerase complex, a biotinylated goat anti-rabbit secondary antibody, and streptavidin-conjugated Dynabeads to bind the complexes into larger “supercomplexes.” (B) Binding reactions contained primary antibody (1° Ab) directed against CREB, secondary antibody (2° Ab), and/or beads, as indicated, with 5 fmol of probe, 10 fmol of purified CRG, and 3 μg of RLNE. (C) Primary antibodies specific for CREB, TFIIB, the large subunit of RNA polymerase II, TBP, or an irrelevant T7 antibody were included with secondary antibody and streptavidin beads where indicated, as in panel B.
CAD, but not KID, is sufficient to recruit the complex to the promoter DNA.
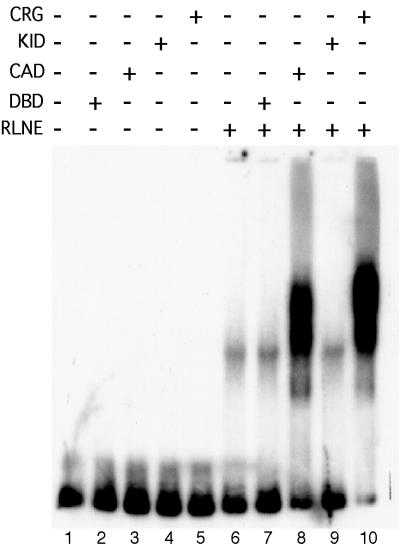
To test whether stimulation of complex formation was occurring specifically through one or both of the two activation domains of CREB, purified KID-G4 or CAD-G4 proteins (Fig. 2B) were included in Ag-EMSA binding reactions. The CAD was able to stimulate significant complex formation on the promoter DNA (Fig. 6, lane 8), while the level observed with KID-G4 was no different from that with G4-DBD alone (Fig. 6, lane 9 versus lane 7). This indicates that the CAD, but not the KID, of CREB is sufficient to recruit the complex to the promoter DNA. However, the KID would not be expected to mediate recruitment in the absence of phosphorylation by PKA, because CBP has been shown to bind specifically to the KID in CREB only when the KID is phosphorylated (6, 34, 53).
FIG. 6.
The CAD but not the KID of CREB is sufficient to recruit a complex to the promoter. Ag-EMSA binding reactions included 5 fmol of probe DNA, 10 fmol of purified G4-DBD, KID-G4, CAD-G4, or CRG, and 3 μg of RLNE where indicated.
Recruitment of the complex to the promoter by CRG is independent of CRG phosphorylation by PKA.
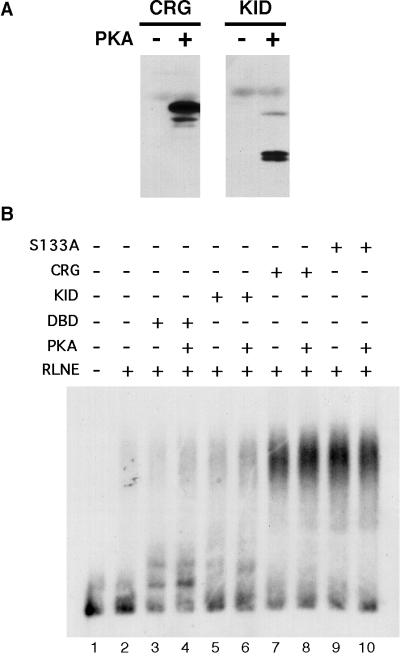
To determine whether phosphorylation of CRG by PKA had an effect on recruitment of a complex to the promoter DNA, we tested P-CRG and P-KID-G4. To avoid artifacts due to phosphorylation of nuclear extract proteins by PKA, we incubated CREB-Gal4 proteins with PKA and terminated the reaction with an inhibitor of the kinase before the addition of phosphorylated factors to the binding reaction. Phosphorylation of KID-G4 and CRG by PKA and the maintenance of the phosphorylation state under the conditions of the Ag-EMSA was confirmed by Western blot using an antibody specific for CREB that is phosphorylated on the Ser-133 PKA phosphorylation site (Fig. 7A). The extent of phosphorylation was assessed by including [γ-32P]ATP of known specific activity in an in vitro kinase assay. Under the conditions used in these experiments, CRG and KID-G4 were phosphorylated stoichiometrically by PKA. However, phosphorylation of CRG by PKA had no effect on the amount of complex formed on the promoter DNA (Fig. 7B, lane 7 versus lane 8). Likewise, phosphorylation of KID-G4 did not allow it to stimulate complex formation (Fig. 7B, lane 6). This is in contrast to the ability of PKA to stimulate CRG-mediated transcription with the same reagents in the in vitro transcription assay (Fig. 2C). To determine whether inadvertent phosphorylation of CRG by endogenous PKA in the extracts was having an unrecognized effect on complex recruitment by CRG, we tested CRG-S133A, a PKA phosphorylation site mutant, in the Ag-EMSA. Complex recruitment by CRG-S133A was indistinguishable from recruitment by wild-type CRG (Fig. 7B, lane 9 versus lane 7) and was also equivalent in the presence of PKA (Fig. 7B, lane 9 versus lane 10). The result with CRG S133A provides further evidence that PKA phosphorylation is not playing a role in recruitment of the complex to the promoter DNA by CRG. These results indicate that recruitment by CRG is independent of PKA phosphorylation and provide direct evidence that phosphorylation of the KID does not allow it to recruit the polymerase complex in this system, whether the KID acts alone or in concert with the CAD in CRG.
FIG. 7.
Recruitment of the complex by CRG is independent of CRG phosphorylation by PKA. (A) To test for phosphorylation of CRG and KID-G4 by PKA in the Ag-EMSA system, we incubated 100 fmol of each protein without and with 5 U of PKA under the same conditions used in the Ag-EMSA binding reactions. Samples were separated on an SDS–15% PAGE gel and transferred to a membrane, and a Western blot was performed with a primary antibody specific for Ser-133 phosphorylated CREB. (B) Ten femtomoles of each purified protein was included with 5 fmol of probe and 3 μg of RLNE in Ag-EMSA binding reactions as indicated. Five units of PKA were included where shown.
CBP antibody does not supershift the complex recruited by CRG.
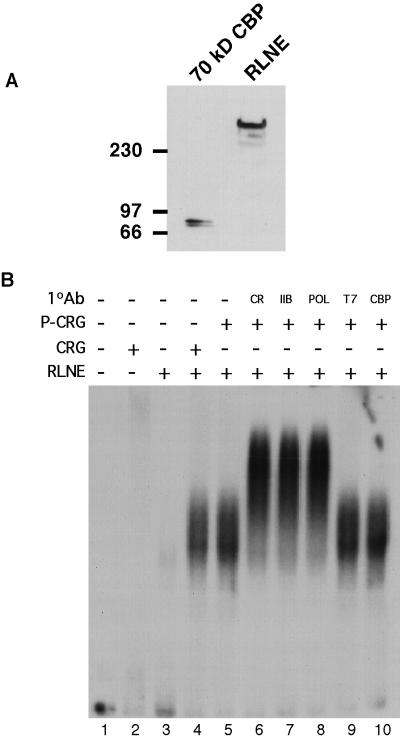
Because interaction with CBP has been proposed to mediate activation of transcription by phosphorylated CREB, we were interested to see if an antibody specific for CBP could supershift the complex recruited to the promoter by CRG. Endogenous CBP was detected with the CBP antibody in the RLNE (Fig. 8A), demonstrating that it is present in significant amounts in the recruitment assay. We are also able to immunoprecipitate CBP from nuclear extracts with this antibody, indicating that it recognizes native CBP (data not shown). Using this same CBP antibody, we tested PKA-phosphorylated CRG to see if inclusion of CBP in the recruited complex was dependent on phosphorylation of CRG. Phosphorylation of CRG still did not allow supershifting of the complex by the CBP primary antibody (Fig. 8B, lane 10), indicating either that CBP is not present in the complex recruited to the promoter by CRG, regardless of the CRG phosphorylation state, or that the amino terminal CBP epitope is masked in the complex. In addition, two other antibodies directed against amino and carboxy terminal epitopes in CBP were tested alone and together, but they also failed to produce a supershift (data not shown). The maintenance of phosphorylation of CRG under the conditions of the assay was again confirmed by Western blot, as described above (Fig. 7A).
FIG. 8.
Phosphorylated CRG does not recruit CBP to the complex. (A) To confirm the presence of CBP in the RLNE, a Western blot was performed using an antibody specific for CBP. RLNE (250 μg) was separated on an SDS–5% PAGE gel and transferred to a membrane, and a Western blot was performed. A 70-kDa N-terminal CBP fragment was synthesized in a coupled in vitro transcription-translation reaction and included on the gel as a positive control for the antibody. (B) A supershift assay was performed similar to that shown in Fig. 5C, except 5 U of PKA was included to phosphorylate the CRG protein. The indicated primary antibodies were included along with biotinylated secondary antibody and streptavidin beads. The CBP antibody was the same as that described in the Fig. 7A legend.
DISCUSSION
Based on previous results showing an interaction between the CREB CAD and the dTAF110 or hTAF130 subunit of TFIID (12, 13, 60, 69), we proposed a model for CAD activation through sequential recruitment of the TFIID and polymerase complexes. We developed the Ag-EMSA method reported here to test this hypothesis directly. The results of our Ag-EMSA experiments demonstrate the following: (i) CRG is able to recruit a polymerase complex to the promoter; (ii) this complex contains TFIID, TFIIB, and RNA polymerase II; (iii) the CAD, but not the KID, of CREB is sufficient to mediate recruitment; (iv) recruitment is independent of the phosphorylation state of CRG; and (v) CBP was not detected in the recruited complex, even when CRG was phosphorylated.
CRG stimulation of the formation of a polymerase complex required the CREB activation domain, as no increase in complex formation was detected with G4-DBD and RLNE. The incubation of G4-DBD or CRG with the probe DNA showed only a very minor shift in the mobility of the probe in the agarose gel (Fig. 4B, lanes 2 to 7), corresponding to the binding of the purified proteins to the Gal4 sites, as shown in Fig. 4. The addition of increasing amounts of CRG to RLNE produced a corresponding increase in the amount of complex with very slow mobility in the gel. This indicates that a factor binding to the DNA probe is specifically stimulated by the presence of CRG and is not simply a function of Gal4 protein binding to the probe, consistent with a role of the CREB activation domains in active recruitment of such a complex.
It is apparent in some experiments that incubation of RLNE with a promoter DNA probe in an Ag-EMSA binding reaction produced a modest level of complex formation on the promoter DNA in the absence of activator. This effect is particularly evident in Fig. 6 and 7. This minor complex formation likely represents the low level of polymerase complex formation and in vitro transcription that can occur in the absence of an activator (4). The formation of this complex was consistent and was unchanged by the addition of purified G4-DBD to the binding reaction (Fig. 6), suggesting that it did indeed reflect spontaneous, activator-independent binding.
That the complex recruited to the promoter by CRG contains factors expected to be found in an RNA polymerase II complex was demonstrated by our antibody supershifting experiments (Fig. 5). Through the use of a multiorder binding array, we were able to detect further shifts in the molecular weights of the large complexes formed on the DNA, which were specific for the CREB activation domain and for the general transcription factors known to be required in the polymerase complex. The multicomponent supershifting method utilized in these studies retains the specificity provided by the primary antibody, as in the single component supershifting used in a classic polyacrylamide gel shift assay. The method we report here is far more sensitive than previously described procedures utilizing Western blotting (36, 58). Based on our results with CRG, we can readily detect a supershift with ∼5% of the protein needed for detection with an avid antibody by Western blot. An advantage of using a combination of a selective Gal4-targeted activation domain together with heterogeneous nuclear extracts that support transcription activation is that it is possible to test for the inclusion of specific factors recruited to the complex. As expected, supershifting of the complex was observed with a CREB primary antibody, due to the presence of CRG bound to the probe. Comparable supershifting was also seen with primary antibodies specific for the general transcription factor TFIIB or the large subunit of RNA polymerase II. Antibody against the TBP subunit of TFIID produced a smaller supershift, which may indicate that the antibody produced a steric block to full complex formation or that access to TBP was limited. It has been shown that a complex containing TFIID, TFIIB, and RNA polymerase II-TFIIF is sufficient to transcribe a supercoiled template (46). Thus, the presence of TFIIB, RNA polymerase II, and TBP in the complex recruited to the promoter by CRG (Fig. 5C), together with the ability of CRG to stimulate transcription under identical conditions (Fig. 2C), suggests that the recruited complex is functional.
The recruitment activity of CRG is mediated through the CAD in CREB, as evidenced by the recruitment of the complex by purified CAD-G4 but not by KID-G4 (Fig. 6). Although the full-length activation domain of CRG was more effective than CAD-G4 in promoting recruitment, this is not due to phosphorylation of KID, because CRG-S133A is as effective as CRG in recruitment. This result is consistent with previous studies demonstrating that the CAD interacts with the general transcription machinery through the dTAF110 or hTAF130 subunit of TFIID (12, 13, 60, 69). Mammalian cells contain a homologue of dTAF110, hTAF130, in which the domains that interact with TAF250 in TFIID and with the CREB or Sp1 activation domains are highly conserved (60, 64). A yeast screen for TAF110 interaction-defective CAD mutants mapped the CREB amino acids required for interaction with TAF110 to a central hydrophobic cluster of amino acids in the CAD (12). As will be demonstrated elsewhere, the interaction between the CAD and TAF is necessary and sufficient for constitutive transcription (Felinski and Quinn, unpublished data). Mutations in these hydrophobic residues which abolished interaction in the yeast two-hybrid system also abolished the following: (i) direct interaction between the CAD and the TAF, (ii) recruitment of the RNA polymerase II complex in the assay described here, and (iii) transcription activation by these mutated CAD-G4 proteins in vivo. This establishes a genetic link between CAD-TAF interaction, recruitment, and transcription. Taken together, our results provide strong support for the idea that the mechanism by which the CAD activates transcription involves binding and recruitment of TFIID by the CREB CAD, which in turn facilitates recruitment of the polymerase complex (Fig. 1A).
To determine whether the KID had potential recruiting activity that was simply masked by the strong activity of the CAD, we tested the KID alone. Even when phosphorylated, KID-G4 had no effect on recruitment in this assay. In addition, recruitment of the polymerase complex by CRG was not affected by phosphorylation of CRG by PKA or by a mutation, CRG-S133A, in KID that prevents phosphorylation by PKA. Identical results were obtained when we employed a functional assay (single-round transcription) to assess recruitment; neither KID alone nor PKA phosphorylation of the entire CREB activation domain had any effect upon recruitment (29). Thus, results from two independent assays, which measured distinct end points, involving direct assessment of recruitment or the presence of a functional polymerase complex (29), indicate that KID alone is incapable of recruiting a polymerase complex, even when phosphorylated. An important difference between the experiments reported here and those of Nakajima et al. (41) showing that CBP mediated interaction between phosphorylated CREB and holoenzyme is that we included template in the recruitment reaction, whereas Nakajima et al. supplied template and other factors subsequent to the interaction assay. Thus, a strict interpretation of that result is that CBP can mediate interaction between CREB and holoenzyme. However, phosphorylated CREB would have to interact with holoenzyme in order to affect any step in transcription initiation, so the demonstration of such an interaction does not indicate that recruitment is the step affected. When recruitment was assayed independently here, we saw no effect of phosphorylation of CRG on this process.
The coactivator CBP has been demonstrated to interact with CREB when it is phosphorylated on Ser-133 (6, 34). Thus, the fact that we were unable to detect the presence of CBP in the complex, even when CRG was phosphorylated, was somewhat of a surprise. CBP has been suggested to mediate recruitment of the polymerase complex by simultaneously interacting with phosphorylated CREB and RNA helicase A of the holoenzyme complex to enhance activation by CREB (41, 42, 45). The observation that CBP was not detected in the complex, even in the presence of PKA (Fig. 8B), suggests either that phosphorylation of CRG is not sufficient to recruit CBP under these conditions, that CBP is present in the complex but that the CBP epitopes required for antibody recognition are masked by interaction with another factor, or that CBP association is not stable to electrophoresis. We did confirm by Western blot that CBP is present in significant quantity in our RLNE and thus is available in the Ag-EMSA binding reactions (Fig. 8A). Regardless of whether CBP is absent or masked, the observation that PKA does not influence recruitment by CRG under conditions where it stimulates transcription in a phosphorylation site-dependent manner indicates that recruitment of CBP or of the polymerase complex in general may not be required for phosphorylated CREB to augment induction of transcription. Immunodepletion of CBP from nuclear extracts had no effect upon induction by PKA (data not shown), but we were unable to deplete CBP by more than ∼75%, even when using large quantities of a combination of amino- and carboxy-directed antibodies. We cannot exclude the possibility that the residual CBP, which is inaccessible to antibodies recognizing amino- and carboxy-terminal epitopes, may be in a functional complex required for transcription activation by phosphorylated CREB.
In summary, the results presented here demonstrate that recruitment of a complex containing TBP, TFIIB, and polymerase II can be facilitated by the presence of CRG bound to an upstream enhancer. This recruitment activity is solely a function of the CAD in CREB and was unaffected by phosphorylation of KID-G4, whether acting as a distinct domain or in concert with the CAD in CRG. The data presented here show that CAD-G4 effectively recruits an RNA polymerase II complex to the promoter but do not support a role for P-CREB–CBP in recruitment of a polymerase complex. Work reported elsewhere showed that CAD-G4 but not KID-G4 mediated single-round transcription, a functional measure of recruitment, whereas phosphorylation of CRG stimulated later steps in the reaction: isomerization, as estimated by abortive initiation, and promoter clearance and/or reinitiation, as estimated by multiple-round transcription (29). Taken together, our results are consistent with the idea that the CREB CAD can interact with the TAF130 subunit of TFIID, leading to recruitment of a polymerase complex and the establishment of constitutive transcription (Fig. 1A). The experiments reported here are in excellent agreement with experiments recently reported in which artificial recruitment of TFIID to a promoter could establish transcription in mammalian cells, whereas recruitment of holoenzyme could not (10). Finally, the assay described here should be useful, at least in principle, in the identification of specific factors recruited to a variety of protein-nucleic acid complexes under different conditions.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health, R01DK43871.
The authors thank Dongying Cui for unpublished results with anti-CBP antibodies and David Spector and Anita Hopper for critical reading of the manuscript.
REFERENCES
- 1.Albright S R, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Brindle P, Linke S, Montminy M. Protein-kinase-A-dependent activator in transcription factor CREB reveals new role for CREM repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 4.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Chrivia J, Kwok R, Lamb N, Hagiwara M, Montminy M, Goodman R. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 7.Colgan J, Ashali H, Manley J L. A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol Cell Biol. 1995;15:2311–2320. doi: 10.1128/mcb.15.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delegeane A M, Ferland L H, Mellon P L. Tissue-specific enhancer of the human glycoprotein hormone α-subunit gene: dependence on cyclic AMP-inducible elements. Mol Cell Biol. 1987;7:3994–4002. doi: 10.1128/mcb.7.11.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;104:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 10.Dorris D R, Struhl K. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 12.Felinski E A, Quinn P G. The CREB constitutive activation domain interacts with TATA-binding protein-associated factor 110 (TAF110) through specific hydrophobic residues in one of the three subdomains required for both activation and TAF110 binding. J Biol Chem. 1999;274:11672–11678. doi: 10.1074/jbc.274.17.11672. [DOI] [PubMed] [Google Scholar]
- 13.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor SP1 contacts the dTAFII 110 component of the drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginty D D, Bonni A, Greenberg E. Nerve growth factor activates a ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez G A, Yamamoto K K, Fischer W H, Karr D, Menzel P, Biggs W I, Vale W W, Montminy M R. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 18.Greenblatt J. RNA polymerase II holoenzyme and transcriptional regulation. Curr Opin Cell Biol. 1997;9:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 19.Guermah M, Malik S, Roeder R G. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-kappaB and Spl. Mol Cell Biol. 1998;18:3234–3244. doi: 10.1128/mcb.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hai T W, Horikoshi M, Roeder R G, Green M R. Analysis of the role of the transcription factor ATF in the assembly of a functional preinitiation complex. Cell. 1988;54:1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- 21.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 23.Hoey T, Weinzierl R O J, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 24.Horikoshi M, Hai T, Lin Y S, Green M R, Roeder R G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 25.Ing N H, Beekman S Y, Tsai S Y, Tsai M J, O'Malley B W. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 26.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H J, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 28.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Lu J-F, Quinn P. Distinct cAMP response element binding protein (CREB) domains stimulate different steps in a concerted mechanism of transcription activation. Proc Natl Acad Sci USA. 2000;97:11292–11296. doi: 10.1073/pnas.97.21.11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim K S, Lee M K, Carroll J, Joh T H. Both the basal and inducible transcription of the tyrosine hydroxylase gene are dependent upon a cAMP response element. J Biol Chem. 1993;268:15689–15695. [PubMed] [Google Scholar]
- 31.Kobayashi N, Horn P J, Sullivan S M, Triezenberg S J, Boyer T G, Berk A J. DA-complex assembly activity required for VP16C transcription activity. Mol Cell Biol. 1998;18:4023–4031. doi: 10.1128/mcb.18.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 33.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 34.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 35.Lieberman P, Berk A. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991;5:2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 37.May M, Mengus G, Carre L, Chambon P, Davidson I. Human TAFII28 promotes transcriptional stimulation by the activation function 2 of the retinoid X receptors. EMBO J. 1997;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- 38.Mengus G, May M, Carre L, Chambon P, Davidson I. Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 39.Meyer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 40.Montminy M R, Bilezikjian L M. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 43.Nilson J H, Bokar J A, Andersen B, Bohinski R, Kennedy G, Keri R A, Farmerie T A, Fenstermaker R A. CRE-binding proteins interact cooperatively to enhance placental-specific expression of the glycoprotein hormone alpha-subunit gene. Ann N Y Acad Sci. 1989;564:77–85. doi: 10.1111/j.1749-6632.1989.tb25889.x. [DOI] [PubMed] [Google Scholar]
- 44.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 45.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parvin J, Sharp P. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 47.Petty K J, Krimkevich Y I, Thomas D. A TATA binding protein-associated factor functions as a coactivator for thyroid hormone receptors. Mol Endocrinol. 1996;10:1632–1645. doi: 10.1210/mend.10.12.8961272. [DOI] [PubMed] [Google Scholar]
- 48.Pugh B F, Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992;267:679–682. [PubMed] [Google Scholar]
- 49.Pugh B F, Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 50.Quinn P G. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 51.Quinn P G, Granner D K. Cyclic AMP-dependent protein kinase regulates transcription of the phosphoenolpyruvate carboxykinase gene but not binding of nuclear factors to the cyclic AMP regulatory element. Mol Cell Biol. 1990;10:3357–3364. doi: 10.1128/mcb.10.7.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn P G, Wong T W, Magnuson M A, Shabb J B, Granner D K. Identification of basal and cyclic AMP regulatory elements in the promoter of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1988;8:3467–3475. doi: 10.1128/mcb.8.8.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 54.Reinberg D, Horikoshi M, Roeder R G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987;262:3322–3330. [PubMed] [Google Scholar]
- 55.Reinberg D, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 56.Reinberg D, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J Biol Chem. 1987;262:3331–3337. [PubMed] [Google Scholar]
- 57.Roberts S G, Ha I, Maldonado E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 58.Roberts S G E, Choy B, Walker S S, Lin Y-S, Green M R. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- 59.Ryu S, Zhou S, Ladurner A G, Tjian R. The transcriptional cofactor CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 60.Saluja D, Vassallo M, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sauer F, Hansen S K, Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 62.Sauer F, Wassarman D, Rubin G, Tjian R. TAFII's mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 63.Sun P, Enslen H, Myung P S, Maurer R A. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 64.Tanese N, Saluja D, Vassallo M F, Chen J L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thut C J, Chen J, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–103. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 66.Van Dyke M W, Sawadogo M, Roeder R G. Stability of transcription complexes on class II genes. Mol Cell Biol. 1989;9:342–344. doi: 10.1128/mcb.9.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weil P A, Luse D S, Segall J, Roeder R G. Selective and accurate transcription of the Ad2 major late promoter in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979;18:469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- 68.Xie H, Rothstein T L. Protein kinase C mediates activation of nuclear cAMP response element-binding protein (CREB) in B lymphocytes stimulated through surface Ig. J Immunol. 1995;154:1717–1723. [PubMed] [Google Scholar]
- 69.Xing L, Gopal V K, Quinn P G. cAMP response element-binding protein (CREB) interacts with transcription factors IIB and IID. J Biol Chem. 1995;270:17488–17493. doi: 10.1074/jbc.270.29.17488. [DOI] [PubMed] [Google Scholar]
- 70.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W, Freedman L P. 20-Epi analogues of 1,25-dihydroxyvitamin D3 are highly potent inducers of DRIP coactivator complex binding to the vitamin D3 receptor. J Biol Chem. 1999;274:16838–16845. doi: 10.1074/jbc.274.24.16838. [DOI] [PubMed] [Google Scholar]
- 72.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]