Abstract
In mammalian cells reiterated binding sites for Sp1 and two overlapping and inverted E2F sites at the transcription start site regulate the dhfr promoter during the cell growth cycle. Here we have examined the contributions of the dhfr Sp1 and E2F sites in the repression of dhfr gene expression. In serum-starved cells or during serum stimulation, the Chinese hamster dhfr gene was not derepressed by trichostatin A (TSA), an inhibitor of histone deacetylases (HDAC). Immunoprecipitation experiments showed that HDAC1 and hypophosphorylated retinoblastoma protein (pRb) are associated with Sp1 in serum-starved CHOC400 cells. In transfection experiments, reporter plasmids containing the reiterated dhfr Sp1 sites were stimulated 10-fold by TSA, while a promoter containing four dhfr E2F sites and a TATA box was responsive to E2F but was completely unaffected by TSA. HDAC1 did not coprecipitate with p130-E2F DNA binding complexes, the predominant E2F binding activity in cell extracts after serum starvation, suggesting that p130 imposes a TSA-insensitive state on the dhfr promoter. In support of this notion, recruitment of GAL4-p130 to a dihydrofolate reductase-GAL4 reporter rendered the promoter insensitive to TSA, while repression by GAL4-pRb was sensitive to TSA. Upon phosphorylation of pRb and p130 after serum stimulation, the Sp1-pRb and p130-E2F interactions were lost while the Sp1-HDAC1 interaction persisted into S phase. Together these studies suggest a dynamic model for the cooperation of pRb and p130 in repression of dhfr gene expression during withdrawal from the cell cycle. We propose that, during initial phases of cell cycle withdrawal, the binding of dephosphorylated pRb to Sp1-HDAC1 complexes and complexes of E2F-1 -to -3 with DP results in transient, HDAC-dependent suppression of dhfr transcription. Upon withdrawal of cells into G0, recruitment of p130 to E2F-4–DP-1 complexes at the transcription start site results in a TSA-insensitive complex that cooperates with Sp1-HDAC-pRb complexes to stably repress dhfr promoter activity in quiescent cells.
E2F plays an important role in the regulation of genes required for cell cycle progression and entry into the S phase (reviewed in references 2, 12, and 19). The mammalian E2F protein family contains at least six members (E2F-1 to -6) that can associate with the two members of the DP protein family (DP-1 and DP-2), resulting in multiple heterodimers capable of binding E2F sequences. Control of E2F activity during the cell cycle is complex: expression of E2F-1 to -3 is low in quiescent cells and increases during growth stimulation, while E2F-4 and E2F-5 accumulate in quiescent cells or during differentiation (2, 12, 19). Various E2F-DP heterodimers also preferentially bind different members of the pRb tumor suppressor protein family: E2F-1 to -3 tend to associate with pRB, while E2F-4 and -5 are most often found in complexes with p107 and p130 (12). E2F-pRb family member complexes are dynamic during the cell cycle, with association and dissociation regulated by cyclin-dependent and other protein kinases. Moreover, members of the E2F and pRb families have been reported to be controlled by nuclear localization, phosphorylation, protein stability, and transcriptional regulation by E2F itself (reviewed in references 2, 12, and 19).
Of interest is the functional relationship between specific E2F-DP-pRb family member complexes and the regulation of specific E2F-dependent genes. In some instances, E2F appears more important for repression of gene expression during early G1 than for activation after the G1 restriction point (reviewed in references 2, 12, and 19). In contrast, E2F appears to play a role in both repression and activation of the major dhfr gene promoter (14). The dhfr major promoter is highly conserved in humans, mice, and hamsters; the core promoters lack obvious TATA and CAAT sequences but contain an 18-bp conserved sequence at the transcription start site that includes two overlapping and inverted E2F sites (reviewed in references 40 and 41). Upstream of the overlapping E2F sites are multiple binding sites for Sp1, a ubiquitous transcription factor found in association with E2F sites in several promoters that are regulated in late G1 (reviewed in reference 2). Transcription of dhfr is low in quiescent cells and is activated after pRb protein family phosphorylation during growth stimulation (4, 15, 23, 42, 51, 52). In some settings the overlapping dhfr E2F sites alone are sufficient for induction of transcription after the G1 restriction point (42). Other studies suggest that the reiterated Sp1 sites, which are required for selection of transcriptional start sites and which regulate basal levels of expression (reviewed in references 41 and 42), contribute to activation of dhfr during growth stimulation (3, 23, 36). Mutational analysis of the E2F-1 activation domain indicates that E2F-dependent activation of dhfr transcription is correlated with the binding of CREB binding protein (CBP), a transcriptional coactivator with histone acetylase activity (14). Proper control of dhfr requires the E2F sites to be precisely located relative to the Sp1 sites (15), suggesting that stereospecific interactions between proteins tethered to DNA through the Sp1 and E2F sites regulate dhfr transcription.
Cooperation between Sp1 and E2F in regulation of dhfr gene expression also is supported by observations that members of these two protein families interact with one another. For example, Sp1 has been reported to interact directly with p107 (9), pRb (35), and E2F-1 to -3 (24, 26, 38), as well as pRb binding protein histone deacetylase 1 (HDAC1) (11). The role of pRb in the expression of dhfr, however, has proven difficult to assess. Cotransfection of pRb expression plasmids with dhfr reporter genes stimulated expression in some studies (25, 47, 48), perhaps by titration of a negative repressor (8), while in others pRb repressed dhfr promoter activity in a manner dependent on the overlapping E2F sites (10). In serum-stimulated mouse embryo fibroblasts lacking various combinations of pRb family members, dhfr was regulated normally in cells lacking pRb and was up-regulated slightly earlier in G1 in cells lacking p130 and p107 (21), suggesting that p130 and p107 function in repression of dhfr in a manner that is not entirely redundant with that in which pRb functions.
Here we have examined the effects of trichostatin A (TSA), a nonreversible inhibitor of HDAC activity (46), on activation and repression of the dhfr major promoter under various growth conditions. Our results support a dynamic model in which Sp1-HDAC1-pRb and E2F-4–DP-1–p130 complexes cooperate to establish stable repression of dhfr gene expression in quiescent cells.
MATERIALS AND METHODS
Cell culture and cell synchrony.
CHOC 400 and U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). For arrest in G0/G1 by serum deprivation, 3 × 105 cells were plated in 60-mm-diameter culture dishes. After 24 h cells were washed and incubated in DMEM with 0.2% FBS for 72 h. Cells were stimulated to reenter the cell cycle with DMEM containing 10% FBS. As indicated in the Figure legends, 100nM TSA or 400 μM mimosine [β-N(3-hydroxy-4-pyridone)-α-aminoproprionic acid] or both were added in culture medium. Cell cycle progression was monitored by flow cytometry as described previously (52). Cell culture supplies were from Life Technologies; mimosine and TSA were purchased from Sigma.
Plasmids.
The [E2F]×4 luciferase reporter, which contains four E2F sites with the sequence TTTCGCGC and the TATA box from the adenovirus E1b gene, was generated by subcloning the promoter from [E2F]×4 chloramphenical acetyltransferase (20) into reporter plasmid pGL3-Basic (Promega). [E2F]×4 luciferase was a gift from F. Dick and N. Dyson (MGH Cancer Center). The dihydrofolate reductase (DHFR)-GAL4 reporter (14) and the GAL4-CBP constructs were a gift gift from C. Fry and P. Farnham. GAL4-pRb and GAL4-p130 plasmids (32) were a gift from J. Nevins.
For the wild-type (WT) dhfr reporter construct, a fragment encompassing nucleotides −230 to −16 (relative to the translational start site) was generated by PCR with oligonucleotide primers containing terminal 5′ SacI and 3′ BglII restriction enzyme sites. The fragment was gel purified, digested with SacI and BglII, and cloned into the SacI and BglII sites of pGL3-Basic. To generate plasmids with altered or missing E2F sites, a fragment encompassing nucleotides −230 to −85 (which contains GC boxes I to IV) was generated by PCR with oligonucleotides containing terminal 5′ SacI and 3′ NheI restriction sites. The fragment was gel purified, digested with SacI and NheI, and cloned into the SacI and NheI sites of pGL3-Basic to generate a reporter lacking E2F sites (pGL3-DHFR-SP1). To introduce various E2F sites, double-stranded oligonucleotides with compatible overhangs were cloned into pGL3-DHFR-SP1 that had been digested with NheI and BglII and treated with calf intestinal phosphatase. In order to maintain the same distance between the E2F and Sp1 sites, the first T in the WT DHFR site, DHFR 2 site, or DHFR mutant 2 site test sequence was placed at the same position as the first T in the TTTCGCGC E2F CG site in the native dhfr promoter. The WT and reconstructed WT (2 site) plasmids were designed such that no sequence alterations associated with cloning occurred within the 30-bp genomic footprint that encompasses the highly conserved 18-bp overlapping E2F sites (51). For the CG and GG test plasmids, single E2F sites with the same E2F core sequences as the dhfr CG and GG sites but with different flanking sequences (51, 52) were used. The CG site, which is derived from the adenovirus EIIa enhancer, was placed on the top strand in the same position as the dhfr CG site. The GG site was oriented on the bottom strand in the same position as the dhfr GG site. As a control for the cloning strategy, the expression vector containing the WT promoter from −230 to −16 (pGL3-DHFR-WT) was compared to pGL3-DHFR-2 SITE, which contains the WT overlapping E2F sites cloned into pGL3-DHFR-SP1. No differences in activity between the two plasmids were observed. The oligonucleotides used to generate the indicated reporter plasmids in pGL3-DHFR-SP1 were as follows (E2F sequences are in boldface): pGL3-DHFR-2 SITE, 5′-CCGGGCGAATGCAATTTCGCGCCAAACTTGGGGGAAGC-3′; pGL3-DHFR MUTANT 2 SITE, 5′-CCGGGCGAATGCAATTTCTCGCCAAACTTGGGGGAAGC-3′; pGL3-DHFR CG SITE, 5′-CCGCCTCTAGAAGTTTTCGCGCTTAAATCTAGACCAGC-3′; pGL3-DHFR GG SITE, 5′-CCGGGTCTAGATTTAAGCGCCAAAACTTCTAGAGGGC-3′. The sequence of each reporter plasmid was confirmed by DNA sequencing. Plasmid DNA was prepared for transfection by banding in CsCl gradients as described previously (30).
Transfection and reporter gene assays.
CHOC 400 or U2OS cells were plated at 3 × 105 cells per 60 mm-diameter culture dish in DMEM with 10% FBS. After 16 h, cells were fed fresh medium and transfected with a total of 7.2 μg of DNA containing 2.4 μg of reporter plasmid and 4.8 μg of salmon sperm carrier DNA by calcium phosphate coprecipitation as described previously (29, 30). Cells were washed three times with phosphate-buffered saline 20 h after transfection to remove the DNA precipitates. Except for experiments with DHFR-GAL4, cotransfection with pCMV-GFP was used to evaluate transfection efficiency, which routinely exceeded 35% in both CHOC 400 and U2OS cells. For experiments with DHFR-GAL4, the cells were cotransfected with pCVM-β-gal as the control and β-galactosidase activity was measured as described previously (29). In synchronization experiments, transfected cells were washed after 20 h and arrested in early G1 by serum deprivation by incubation in DMEM with 0.2% FBS for 60 to 72 h. Serum-starved transfected cells were stimulated to reenter the cell cycle by incubation in DMEM with 10% FBS.
For luciferase assays, cells were washed twice with ice-cold phosphate-buffered saline, scraped into Eppendorf tubes, and pelleted by centrifugation in a microcentrifuge for 1 min. The cell pellet was resuspended in 200 μl of reporter lysis buffer (Promega) and incubated on ice for 10 min, and the sample was subjected to centrifugation at 13,000 × g for 1 min. Twenty microliters of supernatant was assayed for luciferase activity using the Promega luciferase assay system and a Berthold Lumat LB 9501 luminometer. Luciferase assays were performed with duplicate samples from at least two separate experiments; individual determinations from duplicate samples normally varied less than 3%. The statistical significance of differences between groups was assessed by analysis of variance.
Northern blot analysis.
Total RNA was isolated with Trizol reagent as described by the manufacturer (Life Technologies). Total RNA (10 μg per sample) was resolved by electrophoresis on 1% agarose gels containing formaldehyde, transferred to Hybond-N membranes (Amersham), and hybridized with 32P-labeled probes derived from DHFR and/or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNAs. After the washing, hybridization signals were first visualized by exposure to Kodak X-Omat film and then quantified with a Bio-Rad GS250 molecular imager.
Immunoblotting.
Cells were scraped from plates into Eppendorf tubes, lysed in 2 × sodium dodecyl sulfate (SDS) sample buffer (4% SDS, 20% glycerol, 200 mM dithiothreitol, 120 mM Tris-HCl [pH 6.8], and 0.002% bromphenol blue), and heated to 95°C for 10 min, and 10 to 20 μg of protein sample was resolved on 6, 8, or 12% denaturing polyacrylamide gels, depending on the size of the protein to be detected. Total protein was determined using Bio-Rad DC protein assays as described by the manufacturer. After transfer to Immobilon membranes (Millipore), immunoblots were incubated sequentially with primary antibodies and horseradish peroxidase-coupled secondary antibodies as described previously (30). Signals were generated by chemiluminescence with ECL substrate (Amersham) and visualized by exposure to Kodak X-Omat film.
Immunoprecipitations (IPs).
Nuclear extracts were prepared as described for electrophoretic gel mobility shift assays (44). Extract from approximately 3 × 106 cells (100 μl containing 200 μg of protein) was diluted to 1 ml in whole-cell extraction buffer (25 mM HEPES [pH 7.4), 1 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM NaF, 2 μg of phenylmethylsulfonyl fluoride per ml, 0.1 μg of aprotinin and leupeptin per ml, and 1 mM dithiothreitol and incubated with 3 μl of antibody for 2 h at 4°C with gentle shaking. After addition of 50 μl of protein A-agarose beads (Santa Cruz Biotechnology), the suspension was incubated another 2 h at 4°C. The beads were pelleted by centrifugation, washed three times with 1.5 ml of cold extraction buffer, and resuspended in 50 μl of 2 × SDS sample buffer. After the suspension was heated to 95°C for 10 min, 20-μl samples were resolved on denaturing SDS-polyacrylamide gels, transferred to membranes, and probed for the presence of specific proteins by immunoblotting as described above.
Antibodies.
Polyclonal rabbit antibodies to E2F-4 (C-108), pRb (C-15), p107 (C-18), and p130 (C-18) and p300 were purchased from Santa Cruz Biotechnology. Polyclonal rabbit antibodies to Sp1 were purchased from Geneka and Santa Cruz Biotechnology (pep2). Rabbit polyclonal antibodies to HDACI were purchased from Upstate Biotechnology and Santa Cruz Biotechnology (H-51).
Electrophoretic mobility shift assays.
Gel shift assays were performed with nuclear extracts prepared as described above and a 32P-labeled double-stranded DNA oligonucleotide probe representing the overlapping dhfr E2F sites as described previously (51, 52).
RESULTS
Effects of TSA on expression of the endogenous CHOC 400 dhfr genes.
The promoter for the cyclin-dependent kinase inhibitor p21, which contains five Sp1 binding sites upstream of a TATA box, is markedly induced by TSA and other inhibitors of HDACs (7, 34, 39, 43). The dhfr gene promoter is similar to that of p21 but contains overlapping and inverted E2F sites downstream of the Sp1 sites rather than a TATA box. In some studies, inhibitors of HDACs have been reported to induce expression of dhfr (14), while in others HDAC inhibitors had no effect (6, 39). These differences may be due to cell type, growth and treatment conditions, or indirect effects of TSA on other processes.
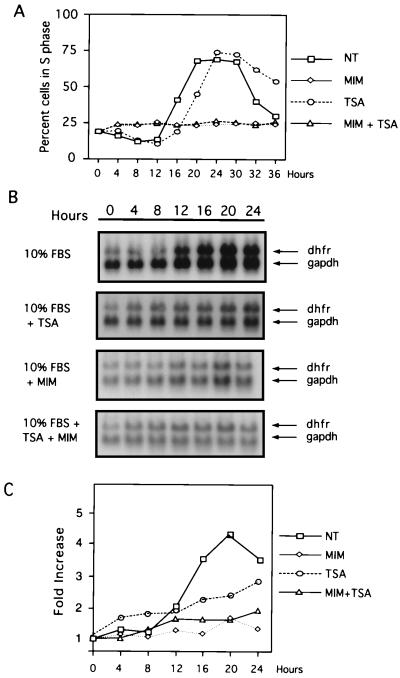
To test the effect of TSA on repression of the CHO dhfr gene, serum-starved CHOC 400 cells were incubated with medium containing 10% FBS with or without TSA (100 nM), cells were harvested at 4-h intervals, and dhfr mRNA levels were measured by Northern blotting (Fig. 1). Progression through G1 and entry into the S phase was assessed by flow cytometry. Under these synchronized growth conditions, addition of medium with 10% FBS caused entry into the S phase by 16 to 18 h (Fig. 1A) and 5- to 10-fold increases in dhfr mRNA levels at the G1/S boundary (Fig. 1B and C). In serum-stimulated cells treated with TSA, S-phase entry was delayed (Fig. 1A) and dhfr mRNA levels never reached the levels observed in control cultures (Fig. 1B and C). However, a reproducible 1.5- to 2-fold increase in dhfr mRNA was observed in three individual experiments 4 to 8 h after serum stimulation in the presence of 100 nM TSA (Fig. 1B and C; data not shown), suggesting that TSA can partially derepress the dhfr gene prior to phosphorylation of pRb, p130, and p107 at the G1 restriction point.
FIG. 1.
Cell cycle kinetics and dhfr gene expression in the presence of mimosine and TSA. CHOC 400 cells were arrested in G0/G1 by incubation in 0.2% FBS for 60 h (zero time). Cells were then induced to reenter the cell cycle by the addition of fresh medium with 10% FBS (NT), 10% FBS plus TSA (TSA), 10% FBS plus mimosine (MIM), or 10% FBS plus TSA and mimosine (MIM + TSA). (A) Cultures were harvested at 4-h intervals, and the percentages of cells in S phase were determined by flow cytometry. (B) Total RNA was prepared from replicate cultures and probed simultaneously for dhfr and gapdh mRNA by Northern blotting. (C) Northern blot hybridization signals from two replicate experiments were quantified with a phosphorimaging system and are expressed as average increases in expression relative to that at time zero.
Cells treated with mimosine, a plant amino acid that alters nucleotide pools (16), induces the expression of the cyclin-dependent kinase inhibitor p21 (1), and stalls cells prior to S phase (33), did not progress into S phase (Fig. 1A) or display increases in dhfr mRNA relative to GAPDH mRNA during serum stimulation (Fig. 1B and C). In addition, mimosine ablated cell cycle progression and the induction of dhfr mRNA in the presence of TSA (Fig. 1).
Phosphorylation of pRb family members is delayed by TSA.
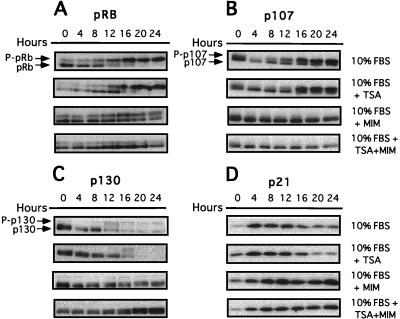
Because E2F is regulated by its association with pRb, p130, and p107, we examined the effects of TSA on the expression and phosphorylation of pRb family members and on E2F DNA binding activity in serum stimulated cells. In serum-starved CHOC 400 cells, phosphorylation of pRb family members occurred at 12 to 16 h after addition of medium with 10% FBS (Fig. 2A to C). After phosphorylation of pRb family members, cyclin-dependent kinase inhibitor p21 decreased in abundance (Fig. 2D). Mimosine blocked the phosphorylation of all three pRb family members completely, either in the presence or absence of TSA, likely by inducing expression of p21 (Fig. 2D). Thus, phosphorylation of pRb family members appears to be an obligate step for induction of dhfr in serum-stimulated cells.
FIG. 2.
Effect of TSA and mimosine on phosphorylation of pRb family members. CHOC 400 cells were arrested in G0/G1 by incubation in low serum for 60 h and then were stimulated to reenter the cell cycle in medium with 10% FBS, with or without TSA and/or mimosine (MIM) as for Fig. 1. Cultures were harvested at 4-h intervals, total cellular protein was resolved by electrophoresis under denaturing conditions, and the abundances and/or phosphorylation states of pRB (A), p107 (B), p130 (C), and p21 (D) were examined by immunoblotting. The phosphorylation patterns of pRb family members suggests that the G1 restriction point occurs 12 to 16 h after the addition of 10% FBS.
However, phosphorylation of pRb family members alone did not appear to be sufficient for full activation of dhfr. Although TSA delayed phosphorylation of pRb family members approximately 2 to 4 h compared to that in the untreated control cells (Fig. 2 A to C), an effect that was particularly evident for the phosphorylation and degradation of p130 (Fig. 2C), by 20 h after serum stimulation in TSA the phosphorylation status of pRb, p130, and p107 and levels of p21 were similar to those for control cells. Yet under these conditions at 20 h the levels of dhfr mRNA in TSA-treated cells were less than 50% of control levels (Fig. 1).
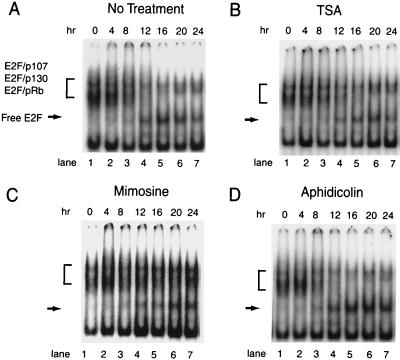
Gel mobility shift experiments indicated that TSA did not block the dissolution of E2F-pRb family member complexes (Fig. 3B), whereas inhibition of the phosphorylation of pRb, p130, and p107 by mimosine prevented the release of E2F DNA binding activity from pRb, p130, and p107 (Fig. 3C). This change in E2F DNA binding complexes was not a consequence of failure to enter the S phase, because aphidicolin blocked S-phase entry without blocking dissolution of E2F-pRb, E2F-p130, and E2F-p107 DNA binding complexes (Fig. 2D) or the accumulation of dhfr mRNA (data not shown).
FIG. 3.
TSA does not block the dissolution of E2F-pRb and E2F-p130 complexes after serum stimulation. CHOC 400 cells were arrested in G0/G1 and induced to reenter the cell cycle in the presence of 10% FBS (A), 10% FBS plus TSA (B), 10% FBS plus mimosine (C), or 10% FBS plus aphidicolin (D). Nuclear extracts were prepared at 4-h intervals and examined for E2F DNA binding activity by gel mobility shift assays using the overlapping E2F sites from the dhfr promoter as a probe. Brackets, E2F-pRb family member complexes; arrows, E2F not bound to pRb, p130, or p107 (free E2F).
Together these experiments support other studies that indicate that repression of dhfr is relatively insensitive to inhibitors of HDACs (6, 39) and indicate that both relief of repression and activation of the dhfr promoter are required for full induction in serum-stimulated cells. Preliminary experiments indicate that dhfr levels fail to reach control levels in serum-stimulated cells treated with TSA because accumulation of CBP and p300 coactivator complexes in the nucleus is impaired (see below).
Differential sensitivity of the dhfr Sp1 and E2F sites to TSA.
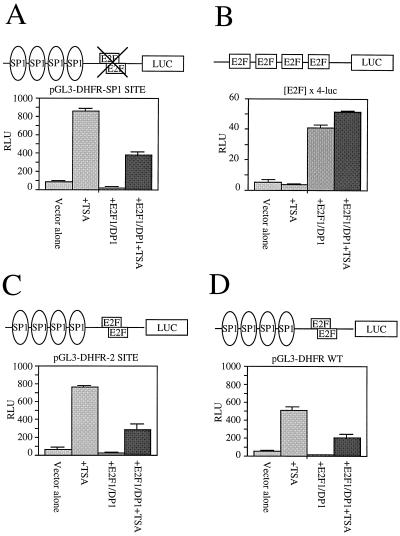
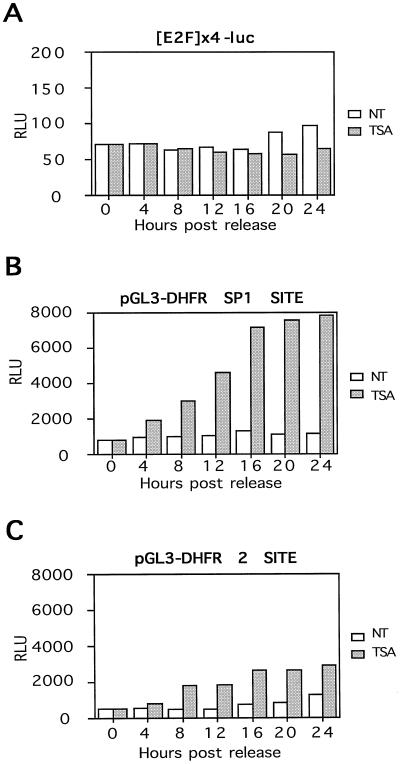
The insensitivity of the dhfr promoter to TSA suggested that the overlapping E2F sites may mask the sensitivity of the Sp1 sites to inhibitors of HDACs. To investigate the role of E2F and Sp1 sites in the sensitivity of the dhfr promoter to TSA, we used a series of model promoters linked to luciferase reporter genes. Plasmids containing the WT promoter (DHFR-WT), a reconstructed WT promoter (DHFR 2 Site), the four upstream Sp1 sites only (DHFR-SP1), or four TTTCGCGC E2F sites only ([E2F]×4) were transfected into CHOC 400 cells, and luciferase activity was measured after 20 h (Fig. 4). Under standard growth conditions constructs containing the reiterated dhfr Sp1 sites were about 10-fold more active for luciferase expression than the [E2F]×4 reporter (compare Fig. 4A, C, and D to B), supporting work that indicates that the Sp1 sites are critical for initiation of transcription of dhfr. Exposure of the transfected cells to 100 nM TSA for 20 h after the removal of DNA resulted in a 10-fold induction of luciferase activity from reporter constructs containing the DHFR Sp1 sites but had no effect on the [E2F]×4 construct (Fig. 4B).
FIG. 4.
Stimulation of dhfr promoter activity by TSA requires Sp1 binding sites. CHOC 400 cells were transfected with the indicated luciferase reporter constructs with or without cotransfection of pCMV-E2F-1 and pCMV-DP-1. Twelve hours after the addition of DNA, cultures were washed and incubated for 20 h in medium with 10% FBS with or without TSA as indicated. Luciferase activity was measured in cell extracts and is expressed in relative light units (RLU) (103) as an average of duplicate transfection samples. Error bars, standard errors of the means.
The failure of the [E2F]×4 reporter to respond to TSA was not due to an inability to recruit E2F to the plasmid construct. When the [E2F]×4 reporter was cotransfected with expression vectors for E2F-1 and DP-1, luciferase activity from it increased 10-fold, showing that the construct was responsive to E2F (Fig. 4B). TSA had no effect on the level of induction by E2F-1 and DP-1 (Fig. 4B). In contrast, cotransfection with 50 ng of pCMV expression vectors for E2F-1 and DP-1 suppressed the activity of all dhfr reporters that contained Sp1 sites, including the pGL3-DHFR-SP1 reporter, which lacks an E2F site (Fig. 4 and data not shown). TSA partially reversed the effects of E2F-1 and DP-1 on promoters containing reiterated Sp1 sites (Fig. 4).
We then transfected CHOC 400 cells with a panel of reporter plasmids, arrested the cells in G0/G1 by serum deprivation, and assayed luciferase expression after serum stimulation. In serum stimulation experiments, TSA did not enhance expression of luciferase from the [E2F]×4 reporter at any time (Fig. 5A). Over 24 h of serum stimulation, the reporter containing only the reiterated Sp1 sites again was stimulated 10-fold by TSA, with a 2-fold induction in expression evident as early as 4 h after the addition of FBS (Fig. 5B). Introduction of the WT dhfr E2F sites into the DHFR-Sp1 site reporter to generate the pGL3-DHFR-2 SITE plasmid resulted in a promoter that was much less sensitive to TSA in serum-stimulated cells at all time points after induction (compare Fig. 5B to C). With all reporters mimosine suppressed activation by TSA at least 50% (data not shown).
FIG. 5.
Induction of dhfr reporter genes by TSA during serum stimulation requires Sp1 sites. CHOC 400 cells were transfected with the indicated reporter constructs. After 20 h the cells were washed free of DNA and incubated in medium containing 0.2% FBS for 60 h. The cells then were treated with medium containing 10% FBS (NT) or 10% FBS plus TSA (TSA). At the indicated times duplicate plates were harvested and extracts were assayed for luciferase activity. Relative light units (RLU) (103) for the averages of duplicate samples are presented.
Interactions of Sp1 with HDAC1 and pRb during the cell cycle.
Here the isolated dhfr Sp1 sites were markedly responsive to TSA, yet the endogenous dhfr gene and the [E2F]×4 reporter were not, suggesting that E2F may mask the effect of TSA on gene regulation by Sp1. In addition, conditions that blocked or delayed phosphorylation of pRb family members impaired dhfr gene expression (Fig. 1 to 3), while in certain other studies cotransfection of expression vectors for pRb stimulated expression from dhfr reporter plasmids (8, 25, 35). In an attempt to resolve the role of pRb and other regulatory factors in dhfr gene expression, we examined the interactions between endogenous Sp1, E2F, pRb family members, and HDAC1 during the cell cycle by IP experiments.
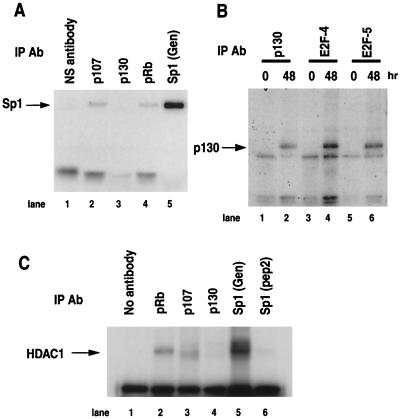
HDAC1 has been reported to interact with all three members of the pRb protein family (5, 13, 22, 27, 31), as well as Sp1 (11). Antibodies to p107 and pRb were able to coprecipitate Sp1 from nuclear extract, whereas antibodies to p130 did not (Fig. 6A). Approximately 8 to 10% of the total Sp1 in nuclear extract could be precipitated by a rabbit polyclonal antibody to full-length human Sp1 (Fig. 6A and data not shown). Although p130–E2F-4 and p130–E2F-5 complexes could be readily recovered from nuclear extract from serum-starved CHOC 400 cells with antibodies to p130, E2F-4, or E2F-5 (Fig. 6B), none of the p130 or p130-E2F complexes contained HDAC1 (Fig. 6C and data not shown). In contrast, IP of either pRb or Sp1 resulted in recovery of HDAC1 (Fig. 6C, lanes 2, 5, and 6). HDAC1 was more abundant in the Sp1 IPs than in the pRb IPs (Fig. 6C), perhaps reflecting the large difference in the abundances of the two proteins in Chinese hamster cells and the cell cycle-dependent interaction of HDAC1 with pRb (18, 49) (see below). Little HDAC1 was found in association with p107 (lane 3, Fig. 6C). HDAC1 was also recovered from Sp1 IPs performed with nuclear extracts from serum-starved U2OS cells (data not shown), indicating that the binding of HDAC1 to Sp1 is not unique to Chinese hamster cells.
FIG. 6.
HDAC1 binds Sp1 and pRb but not p130 in Chinese hamster cell extracts. IPs were performed with nuclear extracts prepared from log phase CHOC 400 cells (zero time) or CHOC 400 cells incubated in 0.2% FBS for 48 h. (A) Antibodies (Ab) to the indicated proteins were used in the primary IP reaction, and the immunoprecipitates were probed for Sp1. Nonspecific rabbit serum (NS) was used as the control. (B) Antibodies to p130, E2F-4, or E2F-5 were used for the primary IP, and the immunoprecipitates were then probed by immunoblotting for p130. (C) Nuclear extracts from cells incubated in 0.2% FBS for 48 h were subjected to IP with the indicated antibodies, and the immunoprecipitates were then probed for HDAC1.
Binding of pRb but not HDAC1 to Sp1 is regulated during the cell cycle.
The interactions of endogenous Sp1 with pRb proteins were examined during transition from G0/G1 to the S phase, when dhfr transcription is activated. Nuclear extracts were prepared from serum-starved cells or cells stimulated with serum for various periods of time, Sp1 was immunoprecipitated, and the proteins recovered in the Sp1 IPs were examined by immunoblotting. Immunoblotting of total-cell lysates and nuclear extract was used to assess the recovery, phosphorylation state, and cellular distribution of each protein during the IP experiments.
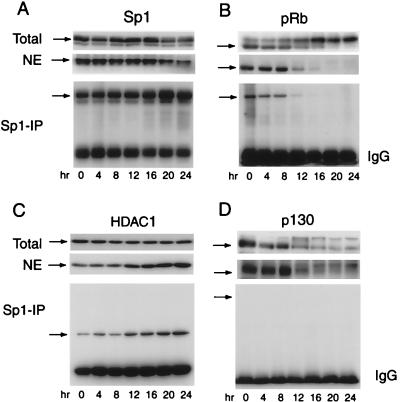
During serum starvation (data not shown) and stimulation there was little change in the total amount of cellular Sp1, the amount of Sp1 in nuclear extract, or the amount of Sp1 recovered in IP experiments (Fig. 7A). As shown above, immunoblotting of total cellular protein revealed the progressive phosphorylation of pRb in G1 after serum stimulation (Fig. 7B). Only hypophosphorylated pRb was detected in nuclear extract, and only hypophosphorylated pRb was recovered in Sp1 IPs (Fig. 7B). In contrast, both unphosphorylated and phosphorylated p130 was found in nuclear extract (Fig. 7D), and no form of p130 was found in association with Sp1. The amount of HDAC1 increased slightly in nuclear extract and in the Sp1 IP after serum stimulation (Fig. 7C). We were unable to detect E2F-1 in Sp1 IPs of nuclear extracts at any time after serum stimulation of CHOC 400 cells (data not shown).
FIG. 7.
A cell cycle-regulated interaction of pRb with Sp1. Total cell lysate (total) and nuclear extract (NE) from serum-starved (zero time) cells or cells stimulated with 10% FBS for 4 to 24 h were probed for the presence of Sp1 (A), pRb (B), HDAC1 (C), and p130 (D) by immunoblotting (top two sections). Sp1 was immunoprecipitated from nuclear extract, and the IP reaction mixtures were probed for Sp1, pRb, HDAC1, and p130 (bottom section) by immunoblotting. In panels B and D the arrows indicate the hypophosphorylated forms of pRb and p130, respectively. Note that only hypophosphorylated pRb is found in nuclear extract or in association with Sp1. IgG, immunoglobulin G.
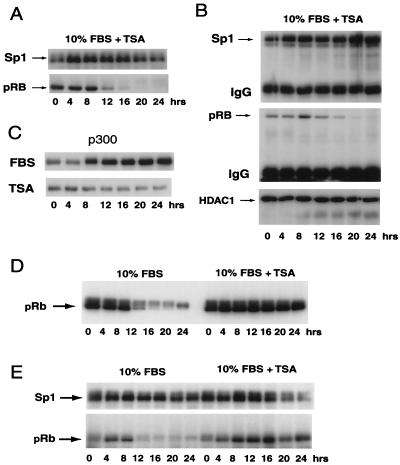
The recovery of hypophosphorylated pRb in Sp1 IPs in a cell cycle-dependent manner from CHOC 400 cells suggested that phosphorylation of pRb disrupts the Sp1-pRb interaction. We therefore examined the effect of TSA on the pRb-Sp1 interaction in IPs of nuclear extract from CHOC 400 and U2OS cells (Fig. 8). Immunoblotting CHOC 400 nuclear extract confirmed that TSA delays phosphorylation of pRb, leading to its retention in nuclear extract (Fig. 8A). In contrast, TSA had no effect on the amount of HDAC in nuclear extract (data not shown) or the association of HDAC1 with Sp1 (Fig. 8B). TSA markedly reduced the accumulation of p300 in nuclear extract (Fig. 8C), suggesting that the failure to fully activate the dhfr gene in serum-stimulated cells treated with TSA (Fig. 1) may result from defects in the assembly of coactivator complexes.
FIG. 8.
Effects of TSA on the interaction of pRb and HDAC1 with Sp1. (A) Nuclear extracts from serum-stimulated CHOC 400 cells treated with TSA were probed for Sp1 and pRb. (B) Sp1 was immunoprecipitated from nuclear extracts from serum-stimulated CHOC 400 cells treated with TSA, and the immunoprecipitates were probed for Sp1, pRb, and HDAC1. Note that TSA delays the loss of pRb from nuclear extract after serum stimulation (compare panel B to Fig. 6B). (C) Nuclear extracts from CHOC 400 cells stimulated with 10% FBS (FBS) or 10% FBS and TSA (TSA) were probed for p300 by immunoblotting. (D) TSA inhibits phosphorylation of pRb in serum-stimulated U2OS cells. Serum-starved U2OS cells were incubated with medium containing 10% FBS or medium with 10% FBS and TSA. Nuclear extracts were prepared and probed for pRb by immunoblotting. (E) Inhibition of pRb phosphorylation by TSA and delayed dissociation of pRb from Sp1 in U2OS cells. Sp1 was immunoprecipitated from nuclear extracts prepared from cells stimulated with serum or serum plus TSA as for panel D, and the immunoprecipitates were probed for Sp1 and pRb. In cells treated with TSA, hypophosphorylated pRb was recovered in association with Sp1 throughout the course of the experiment. IgG, immunoglobulin G.
Like several other human tissue culture cell lines (7, 34, 39), serum-stimulated U2OS cells treated with 100 nM TSA do not progress into S phase (data not shown) or phosphorylate pRb (Fig. 8D). As for CHOC 400 cells, the amounts of Sp1 in nuclear extract or recovered in Sp1 IPs from extracts of U2OS cells prepared at different times after serum stimulation were similar at all time points (Fig. 8E). In U2OS cells, only hypophosphorylated pRb was found associated with Sp1, and the Sp1-pRb interaction was lost by 12 h after serum stimulation, a time coinciding with pRb phosphorylation (Fig. 8E). In serum-stimulated U2OS cells treated with TSA, pRb was not phosphorylated and the Sp1-pRb interaction persisted for at least 24 h. Mimosine also blocked phosphorylation of pRb and its dissociation from Sp1 (data not shown), suggesting that inhibition of phosphorylation of pRb prevents the release of pRb from the Sp1 IP complex. In contrast, TSA had no effect on the interaction of HDAC1 with Sp1.
Establishment of repression is inhibited by TSA.
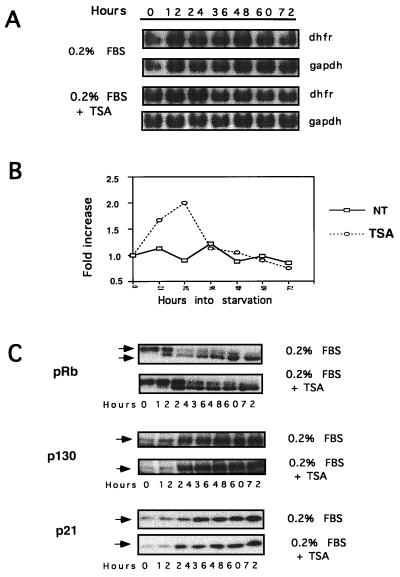
We then examined the effect of TSA on repression of dhfr gene expression during serum deprivation. CHOC 400 cells were plated in medium containing 0.2% FBS with or without TSA, RNA was isolated at 12-h intervals, and dhfr mRNA levels were measured by Northern blotting. GAPDH mRNA was used as a control. In 0.2% FBS dhfr levels dropped slowly over 48 to 72 h, whereas in medium with TSA dhfr levels rose about twofold during the first 24 h of serum deprivation and then dropped to control levels immediately thereafter (Fig. 9A and B). In low-serum conditions TSA transiently delayed the dephosphorylation of pRb but did not affect the accumulation of p130 or p21 by 24 h or thereafter (Fig. 9C).
FIG. 9.
TSA transiently stimulates endogenous dhfr promoter activity during serum withdrawal. Subconfluent CHOC 400 cells were incubated in fresh medium with 0.2% FBS (NT) or 0.2% FBS plus TSA (TSA) as indicated. At 12-h intervals cultures were harvested and total cellular RNA was examined for dhfr and gapdh mRNA levels by Northern blotting. (A) Autoradiographic signals of Northern blots for dhfr and gapdh mRNA as a function time in medium with 0.2% FBS with or without TSA. (B) The hybridization signals in panel A were quantified by phosphorimaging. dhfr mRNA levels first were normalized to gapdh mRNA levels and then plotted as a function of the percentage increase in signal relative to the levels of dhfr mRNA at zero time. (C) Total cell lysates from cells treated with or without TSA during serum deprivation were probed for pRb, p130, and p21 by immunoblotting.
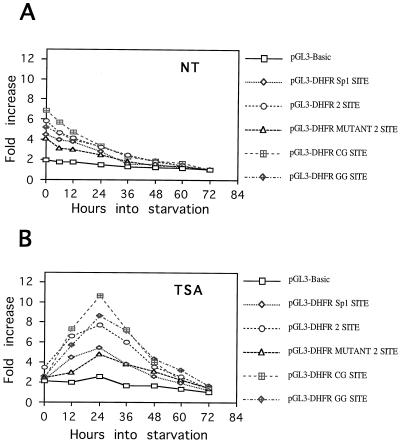
The effect of TSA on dhfr reporter gene expression was also examined during serum deprivation. CHOC 400 cells were transfected with the panel of dhfr reporter plasmids, the DNA was removed 20 h later, and the cells then were incubated in medium with 0.2% FBS with or without TSA. Except for the pGL3 vector control, each reporter lost activity as cells approached quiescence (Fig. 10A). In medium with 0.2% FBS and TSA, the reporter constructs containing WT, CG, or GG E2F sites were activated nearly 10-fold after 24 h, and then declined thereafter (Fig. 10B). The constructs containing the Sp1 sites only or the mutant version of the overlapping E2F sites showed less than a fivefold induction by TSA during serum withdrawal (Fig. 10B).
FIG. 10.
E2F sites may contribute to transient stimulation of dhfr promoter activity by TSA during serum withdrawal. The indicated luciferase reporter plasmids were transfected into CHOC 400 cells. After being washed, the cultures were incubated in fresh medium containing 0.2% FBS (A) or 0.2% FBS plus TSA (B). Cultures were harvested at 12-h intervals, and duplicate samples were assayed for luciferase activity. The level of luciferase activity is plotted as a function of time relative to the 72-h value.
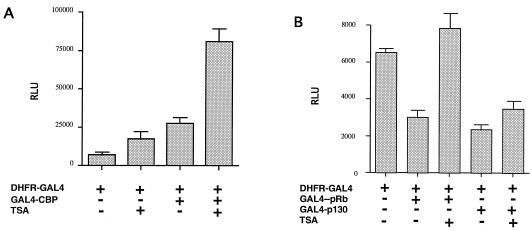
While these trends were maintained in three independent experiments, the differences between reporters with or without functional E2F sites were not statistically significant in all three experiments. The insensitivity of the endogenous dhfr gene and transfected dhfr reporter plasmids with functional E2F sites to TSA after 24 h, a time when p130 was expressed, suggested that p130 may render the dhfr gene insensitive to TSA. To test this possibility, we cotransfected DHFR-GAL4, a dhfr reporter plasmid in which a GAL4 DNA binding site was substituted for the E2F sites, with expression plasmids for GAL4-pRb and GAL-p130 fusion proteins. As a positive control, we also included GAL4-CBP in these experiments.
As reported elsewhere (14), expression of luciferase activity from DHFR-GAL4 was stimulated about fourfold by GAL4-CBP (Fig. 11A). Interestingly, expression was increased nearly 15-fold by GAL4-CBP and TSA (Fig. 11A). In contrast to expression of GAL4-CBP, expression of either GAL4-pRB or GAL4-p130 repressed luciferase expression from DHFR-GAL4 by greater than 50% after 24 h (Fig. 11B). Although both fusion proteins repressed expression to similar extents, TSA restored expression to control levels in the presence of GAL4-pRb but had no effect on repression by GAL4-p130 (Fig. 11B). In this instance, the difference between expression in the presence and absence of TSA for GAL4-pRb was statistically significant (P = 0.011), whereas the difference between TSA treatment and the control for GAL4-p130 was not (P = 0.414). Neither GAL4-pRb nor GAL4-p130 had any significant effect on expression of the pCVM-β-gal control plasmid (not shown).
FIG. 11.
p130 renders the dhfr promoter insensitive to TSA. CHOC 400 cells were transfected with 0.5 μg of DHFR-GAL4, a luciferase reporter plasmid in which the E2F sites were replaced with a GAL4 DNA binding site, per plate and pCMV-β-gal (1 μg) as the control, with or without expression plasmids for GAL4-CBP (20 μg) (A) or GAL4-pRB (2 μg) and GAL4-p130 (2 μg) (B). After removal of DNA, cells were incubated for 24 h in fresh medium containing 10% FBS or 10% FBS with 100 nM TSA as indicated. Luciferase activity was determined in cell extracts, was normalized to expression of β-galactosidase, and is expressed in relative light units (RLU) as averages of duplicate samples. Error bars, standard errors of the means. GAL4-CBP, GAL4-pRb, and GAL4-p130 influenced expression of pCMV-β-gal less than 5% under all conditions tested.
DISCUSSION
While dhfr is considered a typical E2F-dependent gene (i.e., it is repressed in quiescent cells and activated late in G1 after the phosphorylation of pRb family members), it is one of the few E2F-dependent genes in which E2F appears to play a prominent role in both repression and activation of transcription (14, 40, 41). For example, in serum starvation and stimulation experiments with mouse embryo fibroblasts lacking p130 and p107, the dhfr gene was not derepressed in G0-G1 as were cdc2, b-myb, and several others but rather was the only gene examined that showed premature induction after serum stimulation (21). The behavior of dhfr in these and other experiments likely results from the unique arrangement of Sp1 and E2F sites in the dhfr promoter. However, assigning specific roles for Sp1 and E2F in regulation of dhfr gene expression has generated considerable debate.
For instance, several studies have suggested different roles for Sp1 and E2F in activation of dhfr after growth stimulation. Experiments with the mouse promoter suggest that the reiterated Sp1 sites are not sufficient for activation after serum stimulation (42), while the experiments here support other studies that indicate that Sp1 sites alone respond to serum stimulation (23, 36). These and other experiments have inspired discussions about the relative contributions of E2F to repression and/or activation of the gene under various growth conditions. Here the DHFR-Sp1 reporter gene containing the four hamster dhfr Sp1 sites was 10-fold more active under standard growth conditions than the [E2F]×4 reporter, showed 2- to 4-fold increases in activity after serum stimulation, and displayed 10-fold increases after serum stimulation with TSA. In contrast, the [E2F]×4 reporter gene, although responsive to E2F, had weak basal activity, showed less than a twofold increase in activity after serum stimulation, and was completely unresponsive to TSA.
Importantly, no reporter construct employed here, including those with the WT promoter, precisely recapitulated the behavior of the endogenous dhfr gene. For example, elevated levels of E2F-1 and DP-1 expressed from tetracycline-responsive promoters in stable cell lines accentuate activation of dhfr gene expression, but only after the G1 restriction point (28) (data not shown). When cotransfected with various reporter plasmids bearing either viral or cellular promoters, including those used here, E2F-1 reduced expression in a dose-dependent manner by transcriptional squelching (29). While under the same conditions the [E2F]×4 plasmid responds to E2F, it contains four consensus E2F binding sites upstream of a TATA box, a situation not yet encountered in nature.
Despite these limitations, transient transfection assays indicate that TSA is able to stimulate gene expression through the reiterated dhfr Sp1 sites, as has been observed for the Sp1 sites associated with the p21/WAF1/Cip1 promoter (7, 34, 43). Others have shown that the effect of HDAC inhibitors on Sp1 does not involve changes in Sp1 DNA binding activity (34, 43). Direct interactions between specific domains of Sp1 and HDAC1 (11) and Sp1 and E2F-1 to -3 (24, 26, 38) have been detected in vitro and in transfected cells. The interactions of Sp1 with HDAC1 and E2F are mutually exclusive, suggesting that expression of E2F-1 in late G1 might displace HDAC1 from Sp1 and thereby relieve repression of gene expression (11).
Here HDAC1 was readily recovered in immunoprecipitates of Sp1, and this interaction persisted in serum-starved cells (Fig. 7), supporting the notion that Sp1 plays a role in repression of dhfr through HDAC1. However, our results suggest that activity of HDAC1 during the cell cycle may be regulated in a manner other than displacement from Sp1 by E2F-1. Our IP results indicate that HDAC1 is associated with Sp1 during all of G1 in CHO and U2OS cells and that the association of pRb with Sp1 is regulated by pRb phosphorylation. The interaction of hypophosphorylated pRb with Sp1 may be mediated by HDAC1 because Sp1 and pRb do not interact directly in vitro (27, 49), and phosphorylation of pRb by cyclin D-cdk4 has been shown to disrupt pRb-HDAC1 complexes (18). Sp1 is also phosphorylated in mid-G1 in C-terminal regions (3) that may be involved in binding HDAC1 (11). While we have not tested for Sp1 phosphorylation, no change in the binding of HDAC1 to Sp1 was observed in extracts from serum-stimulated cells. Thus it also appears that phosphorylation of Sp1 does not disrupt the Sp1-HDAC1 interaction during G1. While it remains to be determined how (or if) the binding of pRb to the Sp1-HDAC1 complex affects HDAC activity, it is clear that TSA does not disrupt Sp1-HDAC1 interactions. Thus, we suggest that phosphorylation of pRb during the transition through G1 releases pRb from the Sp1-HDAC1 complex and that this event contributes to relief of repression of the dhfr gene.
Our work also supports a role for E2F in activation of dhfr transcription. After phosphorylation of pRb and p130 at the G1 restriction point, the genomic footprint of the E2F sites becomes occupied (51) and binding of E2F-1 to -3 to the promoter is detected by chromatin IP (ChIP) assays (50). Activation by E2F may occur by recruitment of CBP or p300 to the E2F transactivation domain (14), leading to modification of chromatin in the vicinity of the promoter by the HDAC activity of CBP, p300, or other E2F coactivators. ChIP experiments show that E2F-dependent transcriptional activity for several E2F-dependent genes is accompanied by histone acetylation (45), supporting a role for chromatin modification in activation of dhfr. Here TSA inhibited the accumulation of p300 in nuclear extract (Fig. 8C), providing a possible explanation for failure to fully activate the dhfr gene in serum-stimulated cells treated with TSA (Fig. 1). If recruitment of CBP or p300 is compromised by TSA, the levels of expression of dhfr in TSA at the G1/S boundary (about 50% that in control cells) may represent that fraction of expression due solely to relief of repression.
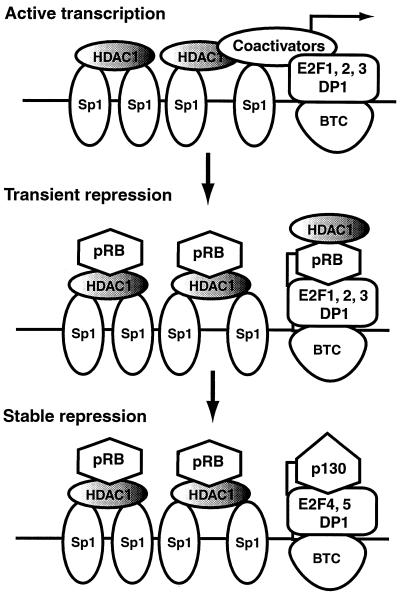
Our experiments suggest a dynamic role for the E2F sites in the establishment of stable repression of dhfr during withdrawal from the cell cycle (Fig. 12). During the first 24 h of serum deprivation, we propose that dephosphorylated pRb accumulates and binds both Sp1-HDAC1 complexes and complexes of E2F-1 to -3 with DP, thereby dampening the transactivation capacity of both types of complexes. Note that during the early phases of cell cycle withdrawal, prior to expression of p130, both endogenous dhfr genes and dhfr reporter constructs containing functional E2F sites were susceptible to transient activation by TSA (Fig. 10 and 11), indicating that HDAC activity is important for repression during this time. In support of this interpretation, others have shown that ectopic expression of pRb from a tetracycline-regulated promoter for 24 h down-regulates dhfr transcription in cycling U2OS cells and that this inhibition is relieved by TSA (27).
FIG. 12.
A dynamic model for cooperative repression of the dhfr promoter by Sp1 and E2F. In cycling cells, Sp1 complexes and complexes of and E2F-1 to -3 with DP drive transcription of the dhfr gene. During the first 24 h of serum withdrawal, dephosphorylation of pRb results in the binding of pRb to Sp1-HDAC1 complexes and complexes of E2F-1 to -3 with DP, thereby dampening transcription. During the initial phases of serum deprivation, the dhfr promoter is sensitive to TSA, suggesting that HDAC activity associated with pRb is involved in repression at this time. After 24 h a TSA-insensitive state that correlates with the expression of p130 and E2F-4 is achieved, suggesting that E2F-4–DP–p130 complexes cooperate with Sp1-HDAC1-pRb complexes to establish stable repression of the promoter. Upon growth stimulation, repression is relieved by phosphorylation of pRb and p130 at the G1 restriction point. Activation occurs thereafter by recruitment of complexes of E2F-1 to -3 with DP and associated coactivators to the E2F sites. HDAC1 activity may be regulated by association with pRb or other mechanisms. The model posits that Sp1 and E2F cooperate in a dynamic fashion during the establishment of repression, relief of repression during G1, and activation of transcription after the G1 restriction point. Based on genomic footprinting experiments, the model suggests that a basal transcription complex (BTC) is present at the E2F sites throughout the cell cycle. The components of the BTC are not known.
After 24 h, increases in expression of p130 and E2F-4 lead to occupation of the dhfr sites by an E2F-4–DP-1–p130 complex, an interaction that has been documented by both gel mobility shift and ChIP experiments (46, 50). Recruitment of E2F-4–p130 complexes at E2F sites without displacement of hypophosphorylated pRb from Sp1 may explain why no E2F-dependent promoters other than dhfr and TK were recovered by pRB ChIP assays in serum-starved cells even though pRb was bound to chromatin (45, 50). In contrast to recruitment of pRb, recruitment of p130 to the dhfr promoter results in TSA-insensitive repression, perhaps by the binding of the E1A C-terminal binding protein (CtBP) corepressor complex to p130 (32). Thus, rather than a model of repression that invokes either HDAC-dependent or HDAC-independent activities, we suggest that interactions of pRb with Sp1 and p130 with E2F upon withdrawal from the cell cycle lead to inactivation of both transcription factors by complementary mechanisms. Because these mechanisms of repression are not redundant, they ensure that the promoter is stably repressed over long periods of time in quiescent cells.
One major issue remains unresolved for regulation of dhfr. Genomic footprinting shows that there is a large protein complex spanning 30 bp that is bound to the overlapping E2F and transcription initiation start sites during the entire cell cycle (51). Although the constituents of the basal complex at the E2F sites are not known, ChIP experiments indicate that E2F-4 and E2F-5 are bound to the dhfr promoter during the entire cell cycle (50), presenting the interesting possibility that E2F-4 (or E2F-5) is a constituent of the basal transcription complex for the dhfr gene. The next important step in understanding the regulation of dhfr will be to identify the components of the basal complex and to determine if these components include E2F and how this complex differs from those at TATA boxes during the cell cycle.
We note that our IP results differ from those of others who have reported interactions between HDAC1 and p130. After treatment of HaCaT cells with transforming growth factor β (TGF-β), an interaction between endogenous HDAC1 and dephosphorylated p130 was observed (22), an interaction that we did not detect. Yet other studies have inferred an interaction between HDAC1 and p130 (13, 27, 53). Several factors may contribute to differences between our results and those from others. Use of different conditions for extract preparation or different antibodies in the IP experiments may account for some differences. However, we expect that the most important consideration in comparing results between systems is the likelihood that the interplay between regulatory proteins, which depends on numerous factors, including relative abundances of individual proteins, subcellular localization, and posttranslational modifications such as acetylation and phosphorylation, differs in various cell types and under different growth conditions. For example, treatment of cells with TGF-β may result in distinct changes in the phosphorylation state of p130 that result in recruitment of HDAC1.
Finally, in transient transfection experiments, expression of pRb has been shown to superactivate several genes, including c-fos, c-myc, and dhfr, through Sp1 binding sites (25, 35, 47). Paradoxically, superactivation of Sp1 transcriptional activity by pRb required regions of pRb involved in growth suppression (48). One mechanism for superactivation involves titration of a negative regulator of Sp1 DNA binding activity by pRb (8). In addition to the observations that pRb stimulates Sp1-mediated transcription in transfection experiments, it has been reported that pRb does not repress the transcriptional activity of Sp1 tethered to an artificial promoter through a GAL4 DNA binding domain (27, 49). Yet in other studies pRb repressed dhfr reporter gene activity in a manner dependent on E2F sites (10). Here we have documented a cell cycle-regulated interaction between pRb and Sp1. This complex was present in early G1 when dhfr transcription was low, and conditions that inhibited pRb phosphorylation also inhibited dhfr expression and dissolution of the Sp1-pRb complex. Together these data support a model for repression of dhfr that involves cooperation between pRb-HDAC1-Sp1 and p130–E2F-4–DP-1 complexes. We expect that the highly conserved architecture of the dhfr promoter reflects requirements for spacing, alignment, and orientation of proteins that are important for the assembly and maintenance of a stereospecific transcription complex. At other E2F-dependent promoters, different arrangements of regulatory elements may permit E2F and pRb family members to participate in other mechanisms of repression and activation.
ACKNOWLEDGMENTS
We thank N. Dyson, F. Dick, M. Classon, C. Fry, P. Farnham, and J. Nevins for plasmids and other reagents, J. Wells and P. Farnham for discussing unpublished results, and the Vermont Cancer Center DNA sequencing facility for DNA sequencing.
This work was supported by grant GM54726 from the NIH to N.H. Y.-C.C. was supported in part by a J. Walter Juckett Postdoctoral Fellowship from the Vermont Cancer Center.
REFERENCES
- 1.Alpan R S, Pardee A B. p21/WAF1/CIP1/SDI1 is elevated through a p53-independent pathway by mimosine. Cell Growth Differ. 1996;7:893–901. [PubMed] [Google Scholar]
- 2.Black A R, Azizkhan-Clifford J. Regulation of E2F: a family of transcription factors involved in proliferation control. Gene. 1999;237:281–302. doi: 10.1016/s0378-1119(99)00305-4. [DOI] [PubMed] [Google Scholar]
- 3.Black A R, Jensen D, Lin S-Y, Azizkhan J C. Growth/cell cycle regulation of Sp1 phosphorylation. J Biol Chem. 1999;274:1207–1215. doi: 10.1074/jbc.274.3.1207. [DOI] [PubMed] [Google Scholar]
- 4.Blake M C, Azizkhan J C. Transcription factor E2F is required for efficient expression of the dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Buchmann A M, Swaminathan S, Thimmapaya B. Regulation of cellular genes in a chromosomal context by the retinoblastoma tumor suppressor protein. Mol Cell Biol. 1998;18:4565–4576. doi: 10.1128/mcb.18.8.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buquet-Fagot C, Lallemand F, Charollais R-H, Mester J. Sodium butyrate inhibits the phosphorylation of the retinoblastoma gene product in mouse fibroblasts by a transcription-dependent mechanism. J Cell Physiol. 1996;166:631–636. doi: 10.1002/(SICI)1097-4652(199603)166:3<631::AID-JCP18>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y-H F, Grunwald S, Chui R. The reintoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta P K, Raychaudhuri P, Bagchi S. Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription. Mol Cell Biol. 1995;15:5444–5452. doi: 10.1128/mcb.15.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick F A, Sailhamer E, Dyson N J. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol Cell Biol. 2000;20:3715–3727. doi: 10.1128/mcb.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry C J, Pearson A, Malinowski E, Bartley S M, Greenblatt J, Farnham P J. Activation of the murine dihydrofolate reductase promoter by E2F1. A requirement for CBP recruitment. J Biol Chem. 1999;274:15883–15891. doi: 10.1074/jbc.274.22.15883. [DOI] [PubMed] [Google Scholar]
- 15.Fry C J, Slansky J, Farnham P J. Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol Cell Biol. 1997;17:1966–1976. doi: 10.1128/mcb.17.4.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert D M, Neilson A, Miyazawa H, DePamphilis M, Burhans W C. Mimosine arrests DNA synthesis at replication forks by inhibiting deoxyribonucleotide metabolism. J Biol Chem. 1995;270:9597–9606. doi: 10.1074/jbc.270.16.9597. [DOI] [PubMed] [Google Scholar]
- 17.Good L, Dimri G P, Campisi J, Chen K Y. Regulation of dihydrofolate reductase gene expression and E2F components in human diploid fibroblasts during growth and senescence. J Cell Physiol. 1996;168:580–588. doi: 10.1002/(SICI)1097-4652(199609)168:3<580::AID-JCP10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Harbour J W, Luo R X, Dei Santi A, Postigo A A, Dean D C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 19.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 20.Helin K, Wu C-L, Fattaey A, Lees J, Dynlacht B, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative transactivation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 21.Hurford R, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 22.Iavarone A, Massague J. E2F and histone deacetylase mediate transforming growth factor β repression of cdc25A during keratinocye cell cycle arrest. Mol Cell Biol. 1999;19:916–922. doi: 10.1128/mcb.19.1.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen D E, Black A R, Swick A G, Azizkhan J C. Distinct roles for Sp1 and E2F sites in the growth/cell cycle regulation of the DHFR promoter. J Cell Biochem. 1997;67:24–31. doi: 10.1002/(sici)1097-4644(19971001)67:1<24::aid-jcb3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 24.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S-J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S-Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 28.Magae J, Illenye S, Chang Y-C, Mitsui Y, Heintz N H. Association with E2F-1 governs intracellular trafficking and polyubiquitination of DP-1. Oncogene. 1999;18:593–605. doi: 10.1038/sj.onc.1202345. [DOI] [PubMed] [Google Scholar]
- 29.Magae J, Illenye S, Tejima T, Chang Y-C, Mitsui Y, Tanaka K, Omura S, Heintz N H. Transcriptional squelching by ectopic expression of E2F-1 and p53 is alleviated by proteosome inhibitors MG-132 and lactacystin. Oncogene. 1997;15:759–769. doi: 10.1038/sj.onc.1201251. [DOI] [PubMed] [Google Scholar]
- 30.Magae J, Wu C-L, Illenye S, Harlow E, Heintz N H. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- 31.Magnaghi J L, Groisman R, Naguibneva I, Robin P, Lorain S, Le V J, Troalen F, Trouche D, Harel B A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 32.Meloni A R, Smith E J, Nevins J R. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc Natl Acad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosca P J, Dijkwel P A, Hamlin J L. The plant amino acid mimosine may inhibit initiation at origins of replication in Chinese hamster cells. Mol Cell Biol. 1992;12:4375–4383. doi: 10.1128/mcb.12.10.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano K, Mizuno T, Sowa Y, Orita T, Yoshino T, Okuyama Y, Fujita T, Ohtani-Fujita N, Matsukawa Y, Tokino T, Yamagishi H, Oka T, Nomura H, Sakai T. Butyrate activates WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J Biol Chem. 1997;272:22199–22206. doi: 10.1074/jbc.272.35.22199. [DOI] [PubMed] [Google Scholar]
- 35.Noe V, Alemany C, Chasin L A, Ciudad C J. Retinoblastoma protein associates with SP1 and activates the hamster dihydrofolate reductase promoter. Oncogene. 1998;16:1931–1938. doi: 10.1038/sj.onc.1201718. [DOI] [PubMed] [Google Scholar]
- 36.Noe V, Chen C, Alemany C, Nicolas M, Caragol I, Chasin L A, Ciudad C J. Cell-growth regulation of the hamster dihydrofolate reductase gene promoter by transcription factor Sp1. Eur J Biochem. 1997;249:13–20. doi: 10.1111/j.1432-1033.1997.00013.x. [DOI] [PubMed] [Google Scholar]
- 37.Noe V, Ciudad C J, Chasin L A. Effect of polyadenylation and cell growth phase on dihydrofolate reductase mRNA stability. J Biol Chem. 1999;274:27807–27814. doi: 10.1074/jbc.274.39.27807. [DOI] [PubMed] [Google Scholar]
- 38.Rotheneder H, Geymayer S, Haidweger E. Transcription factors of the Sp1 family: interaction with E2F and regulation of the murine thymidine kinase promoter. J Mol Biol. 1999;293:1005–1015. doi: 10.1006/jmbi.1999.3213. [DOI] [PubMed] [Google Scholar]
- 39.Sambucetti L C, Fischer D D, Zabludoff S, Kwon P O, Chamberlin H, Trogani N, Xu H, Cohen D. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 40.Schilling L J, Farnham P J. Transcriptional regulation of the dihydrofolate reductase/rep-3 locus. Crit Rev Eukaryot Gene Expr. 1994;4:19–53. doi: 10.1615/critreveukargeneexpr.v4.i1.20. [DOI] [PubMed] [Google Scholar]
- 41.Slansky J E, Farnham P J. Transcriptional regulation of the dihydrofolate reductase gene. Bioessays. 1996;18:55–62. doi: 10.1002/bies.950180111. [DOI] [PubMed] [Google Scholar]
- 42.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sowa Y, Orita T, Hiranabe-Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through Sp1 sites. Ann N Y Acad Sci. 1999;886:195–199. doi: 10.1111/j.1749-6632.1999.tb09415.x. [DOI] [PubMed] [Google Scholar]
- 44.Staal F J, Roederer M, Herzenberg L A, Herzenberg L A. Intracellular thiols regulate activation of nuclear factor kappa B and transcription of human immunodeficiency virus. Proc Natl Acad Sci USA. 1990;87:9943–9947. doi: 10.1073/pnas.87.24.9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi Y, Rayman B, Dynlacht B D. Analysis of promoter binding by E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 46.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 47.Udvadia A J, Rogers K T, Higgins P D R, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udvadia A J, Templeton D J, Horowitz J M. Functional interactions between retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc Natl Acad Sci USA. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 50.Wells J, Boyd K E, Fry C J, Bartley S M, Farnham P J. Target gene specificity of E2F and pocket protein family members in living cells. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells J, Held P, Illenye S, Heintz N H. Protein-DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the Chinese hamster cell cycle. Mol Cell Biol. 1996;16:634–647. doi: 10.1128/mcb.16.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells J, Illenye S, Magae J, Wu C-L, Heintz N H. Accumulation of E2F-4.DP-1 DNA binding complexes correlates with induction of dhfr gene expression during the G1 to S phase transition. J Biol Chem. 1997;272:4483–4492. doi: 10.1074/jbc.272.7.4483. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H S, Gavin M, Dahiya A, Postigo A A, Ma D, Luo R X, Harbour J W, Dean D C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]