Abstract
TFIID, a multiprotein complex comprising the TATA-binding protein (TBP) and TBP-associated factors (TAFs), associates specifically with core promoters and nucleates the assembly the RNA polymerase II transcription machinery. In yeast cells, TFIID is not generally required for transcription, although it plays an important role at many promoters. Understanding of the specific functions and physiological roles of individual TAFs within TFIID has been hampered by the fact that depletion or thermal inactivation of individual TAFs generally results in dissociation of the TFIID complex. We describe here C-terminally deleted derivatives of yeast TAF130 that assemble into normal TFIID complexes but are transcriptionally inactive in vivo. In vivo, these mutant TFIID complexes are dramatically reduced in their ability to associate with all promoters tested. In vitro, a TFIID complex containing a deleted form of TAF130 associates poorly with DNA, but it is unaffected for interacting with transcriptional activation domains. These results suggest that the C-terminal region of TAF130 is required for TFIID to associate with promoters.
TFIID is a multiprotein complex consisting of the TATA-binding protein (TBP) and at least 14 TBP-associated factors (TAFs) (5, 30, 42, 43, 47, 53). TBP specifically recognizes TATA elements (18, 19), which are present in most RNA polymerase II (Pol II) promoters, and it directly interacts with general transcription factors TFIIA (49) and TFIIB (34). As such, TBP is required to nucleate the assembly of the Pol II machinery on promoters. In the context of TFIID, certain TAFs contact the initiator and the downstream promoter elements and perhaps other sequences flanking the TATA element (4, 6, 37, 52, 54). In vitro, TAFs are important for transcription from promoters lacking TATA elements, but they are dispensable for basal TATA-dependent transcription. In addition, biochemical experiments suggest that TAFs play a role in the response to transcriptional activator proteins, perhaps by serving as direct targets of activation domains (53). However, transcriptional activation in vitro can occur in the absence of TAFs (20, 23, 38, 59).
In yeast cells, TBP is generally required for Pol II transcription (9), and the level of TBP occupancy of promoters is strongly correlated with transcriptional activity and with occupancy by TFIIA and TFIIB (25, 26, 27). In contrast, TAFs are significantly under-represented at many promoters, indicating that there are at least two forms of transcriptionally active TBP in vivo (25, 28). One form is presumably TFIID, whereas the other form lacks TAFs and corresponds either to TBP itself or to some other TBP complex. TFIID and the TAF-independent form of TBP have distinct promoter selectivities, which are in excellent accord with the TAF requirement for transcription in vivo. TFIID is the predominant (and perhaps exclusive) form of TBP at TAF-dependent promoters, whereas the TAF-independent form predominates at TAF-independent promoters (25, 28). However, TFIID is associated with most, and perhaps all, yeast promoters to some extent.
Because some TAFs are also present in the SAGA histone acetylase complex (12, 39, 47), elucidating TAF functions in the context of TFIID requires analysis of TFIID-specific TAFs. In yeast cells, depletion or thermal inactivation of TFIID-specific TAFs (TAFs 130, 67, 40 and 19) reduces transcription at only a subset of promoters, and it does not affect the response to many activator proteins (15, 32, 55). In addition, a TBP mutant defective for TFIID complex formation in vivo has selective effects on gene expression (40). Similarly, mutated derivatives of mammalian TAF250, the homolog of yeast TAF130, affect the transcription of only a subset of promoters in vivo (36, 48, 57). Individual depletion of yeast TFIID-specific TAFs results in a common effect on HIS3 TATA-element utilization and TRP3 transcription, indicating that these TAFs are required for the transcription from certain promoters lacking conventional TATA elements (32, 33). In addition, the TFIID-specific TAF130 is important for transcription of certain cell cycle and ribosomal protein genes in a manner that depends on the core promoter and not the enhancer (45, 50, 56).
Although the above genetic analyses demonstrate that TFIID plays an important role in core promoter function, they are not suitable for assigning specific functions to individual TAFs. First, depletion or thermal inactivation of an individual TAF often causes dissociation of the TFIID complex (33, 55), such that the state of TFIID varies in an unpredictable manner throughout the course of an experiment. In some cases, glutathione S-transferase (GST)-pulldown or coimmunoprecipitation experiments have shown that the mutated TAF derivatives can interact with TBP (10, 35, 50). However, since the N-terminal domains of TAF130 or TAF250 are sufficient for a stable interaction with TBP (22, 29), these assays are insufficient to demonstrate TFIID integrity. Second, some temperature-sensitive mutants of TAF130 may form normal TFIID complexes at the restrictive temperature (analysis was restricted to one additional TAF), but the limited molecular analysis of these mutants was insufficient to define specific functions of TAF130 (50). Third, although mutations that affect the acetylase or kinase activities of TAF250 have been described (10, 35), these and other biochemical assays have been performed on the isolated TAF derivatives and not in the context of TFIID. Fourth, although TFIID function in extracts prepared from temperature-sensitive TAF250 cell lines is reduced by heat treatment (48), TFIID integrity was not assessed. For these reasons, it is impossible to determine if the transcriptional phenotypes in vitro or in vivo are due to a specific function of the mutated TAF or to partial or complete disruption of the TFIID complex.
In order to identify specific and physiologically relevant functions of individual TAFs within the context of TFIID, it is essential to identify mutations of TFIID-specific TAFs that do not affect TFIID integrity. Furthermore, it is essential to analyze the corresponding mutant TFIID complexes (i.e., not isolated TAFs) for their transcriptional properties in vivo and biochemical functions in vitro. We describe here derivatives of TAF130 that assemble into a normal TFIID complex but are functionally defective. The mutant TFIID complexes interact poorly with promoters in vivo and in vitro but are unaffected for interacting with transcriptional activation domains. These results suggest that the C-terminal region of TAF130 is required for TFIID to interact with promoters.
MATERIALS AND METHODS
Isolation of TAF130 mutants.
To obtain dominant-negative mutants of TAF130, we started with plasmid pWC509, a derivative of URA3 centromeric plasmid pRS416 (46) that expresses hemagglutinin (HA)-tagged wild-type TAF130 from the GAL1 promoter (kindly provided by Michael Green). pWC509 was introduced into the Escherichia coli mutator strain XL1-Red, and pooled transformants were grown for 60 generations to obtain mutations at a level of approximately 1 per kb. The library of mutagenized plasmids was transformed into yeast strain FT4 (51), and transformants were grown for 2 days on medium containing 2% glucose and Casamino Acids lacking uracil. Colonies were then replica plated on comparable medium containing 2% galactose to identify cells that grew normally on glucose but poorly on galactose. Plasmids from the desired strains were isolated, and the TAF130-containing insert was recloned into the parental plasmid to confirm the phenotype.
Other TAF130 derivatives were obtained by standard procedures of DNA manipulation. TAF130-N787, -N690, and -N447 were made by digesting pWC509 with XbaI, EcoRI, and BsaBI, respectively, enzymes that cleave in the TAF130 coding region and in the terminator, followed by religation. TAF130-N569, -N333, -N202, and -N159 were made by PCR using appropriate oligonucleotides. TAF130-Δ16 and TAF130-Δ4 have been described previously (2) and were obtained from Tony Weil.
Immunoprecipitation of TFIID.
For analysis of the dominant-negative mutants, cells were grown in synthetic complete medium with 0.5% glucose and 1.5% galactose to allow expression of the TAF130 derivatives without impairing cell growth. For analysis of the other TAF130 derivatives, which were expressed from the native TAF130 promoter, cells were grown in comparable medium containing 2% glucose. In all cases, the strains contained untagged, wild-type TAF130 in the normal chromosomal location. HA or (HA)3 versions of wild-type and mutant TAF130 derivatives were immunoprecipitated from whole-cell extracts using the anti-HA monoclonal antibody 12CA5 as described previously (33). TFIID components were detected by Western blotting using antibodies against TBP and TAFs (kindly provided by Michael Green) and the Supersignal substrate (Pierce). Chromatin immunoprecipitation on strains containing (HA)3-tagged versions of wild-type or mutant TAF130 was performed with anti-HA F7 monoclonal antibody or anti-TBP polyclonal antibodies as described previously (25).
Transcriptional analysis.
ZMY117, the parental TAF130 depletion strain (32), was transformed with the vector (pRS313) or plasmids expressing (HA)3-tagged TAF130 or TAF130-Δ16 from the natural TAF130 promoter. Depletion of the untagged TAF130 was accomplished by adding 500 μM copper sulfate to the medium and then taking samples at 0, 2, and 4 h (32), and RNA was analyzed by S1 nuclease analysis using oligonucleotide probes (17). The oligonucleotide probes that have not been previously published are RPL9A (GGTGGAAAATTCGATA GTAACACCATCTCTAACTGGAACGTTTCTGATCTTCTTGTCACCAGGCGG) and RPL8A (GCCTAATTCGAACTTTCTCTTCTTTCTGAATTGAGCAC GCTTGGCACCGGAGAGAA).
Immobilized template assays.
Immobilized template assays were performed essentially as described previously (41), except that whole-cell extracts (58) were used instead of nuclear extracts. Whole-cell extracts were active as assayed by promoter-specific transcription in vitro. The HIS4 promoter, TATA-less HIS4 promoter derivative (mTATA), and promoterless DNAs were biotinylated by PCR and then incubated with streptavidin-linked magnetic beads (Dynabeads M280 Streptavidin; Dynal, Inc.) overnight at room temperature in buffer I (2 M NaCl, 10 mM Tris-HCl [pH 7.5], 0.01% NP-40). After incubation, the beads were washed three times with buffer I and three times with Tris-EDTA (TE) buffer and then stored in TE at 4°C (10-mg/ml final bead concentration). Before use, 1 mg of beads was incubated for 30 min with 1 ml of transcription buffer (25 mM HEPES-KOH [pH 7.5], 10 mM magnesium acetate, 5 mM EGTA, 125 mM potassium acetate, 10% glycerol, 2 mM dithiothreitol, 0.1% NP-40) containing 30 mg of bovine serum albumin and 5 mg of polyvinylpyrrolidone per ml. The immobilized template reactions consisted of 5 μl of DNA-bound beads, whole-cell extracts containing 80 μg of total protein, and 3 μg of the plasmid p(C2AT)19 (7) as competitor DNA in a total volume of 50 μl of transcription buffer. After the reactions were incubated at 4°C for 2 h in a rotator, the beads were washed four times in transcription buffer, and then 50 μl of sample buffer was added to the beads and the suspension was boiled for 5 min. Proteins were analyzed by Western blotting. Some reactions contained 200 ng of purified Gal4-VP16 activator protein (obtained from Steve Buratowski). Serial dilutions of the whole-cell extracts were used to compare the relative amounts of proteins detected after the immobilized template assay.
Activation domain interaction assay.
GST fusion proteins were expressed and bound to the glutathione beads as described previously (8), adjusted to a protein concentration of 1 mg/ml of bed, and stored at −80°C. The plasmid expressing GST fused to TADIV of Adr1 (24) was obtained from Clyde Denis. Beads were washed three times in transcription buffer prior to the assay. Binding assays were performed overnight at 4°C using 300 μg of whole-cell extract and 20 μl of beads in 200 μl of transcription buffer. Proteins bound to the beads were collected by centrifugation, washed four times with 0.5 ml of transcription buffer, and analyzed by Western blotting.
RESULTS
Isolation of dominant-negative TAF130 mutants that assemble into TFIID.
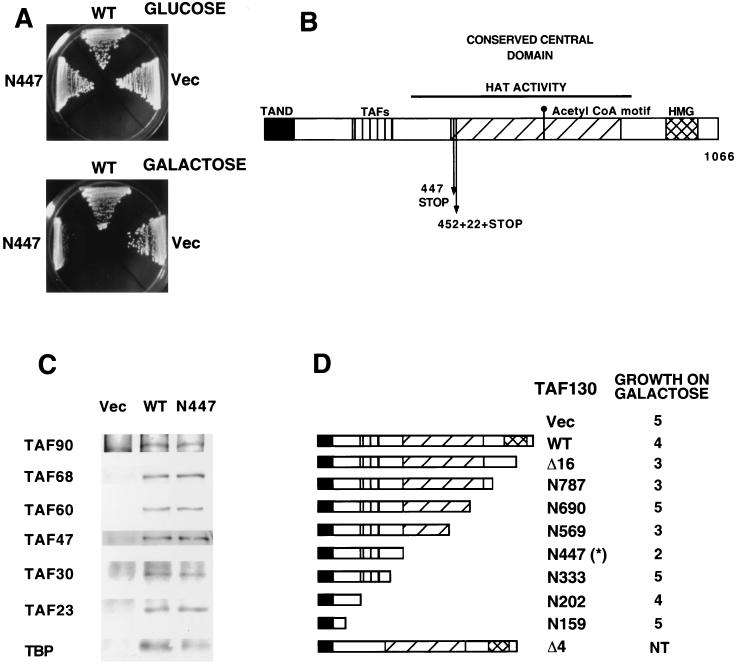
Functionally compromised TAF derivatives that assemble into TFIID complexes might behave in a dominant-negative fashion. Thus, we independently mutagenized HA-tagged versions of the TAF130, TAF17, TAF61, and TAF90 coding regions, expressed the libraries of mutant proteins from the GAL1 promoter, and searched for TAF mutants that specifically inhibit growth in galactose medium. Although this approach was unsuccessful for TAF17, TAF61, and TAF90, we obtained two TAF130 alleles that conferred a slow growth phenotype in galactose medium but not in glucose medium (Fig. 1A). These two alleles encode C-terminal deletions that lack most of the conserved central domain. One mutation introduces a termination codon at residue 448, whereas the other causes an insertion and reading frame shift at residue 453 (Fig. 1B). As expected from analysis of other TAF130 deletion mutants (2), these dominant-negative alleles are unable to support cell viability.
FIG. 1.
A dominant-negative TAF130 mutant that forms a normal TFIID complex. (A) Growth of yeast cells containing the plasmid vector or derivatives bearing wild-type TAF130 or the TAF130-N447 mutant expressed from the GAL1 promoter on plates containing glucose or galactose as the sole carbon source. (B) Location of dominant-negative mutations (vertical arrows) with respect to structural and functional features of TAF130. These features include a conserved central domain that has histone acetylase activity (31), a putative acetyl coenzyme A binding site (10), a putative HMG domain (44), an N-terminal inhibitory region (TAND) that strongly interacts with TBP (22, 29), an additional region interacting with TBP (2), and a region interacting with other TAFs (2). (C) TFIID complex formation. HA-tagged wild-type TAF130 or TAF130-N447 was immunoprecipitated from cell extracts with anti-HA antibodies, and components of the TFIID complex were detected by Western blotting with the indicated antibodies. (D) Dominant-negative effects due to overexpression of C-terminal deletions mutants of TAF130 (number indicates last residue of the protein). Growth phenotypes were determined on galactose medium and are defined in arbitrary units with a value of 5 indicating normal growth and a value of 0 indicating no growth. The structures of TAF130-Δ4 (lacks residues 208–303) and TAF130-Δ16 (lacks residues 913 to 1037) are also indicated. The asterisk denotes the N447 mutant isolated in the genetic screen.
We analyzed whether the TAF130-N447 derivative assembles into the TFIID complex by coimmunoprecipitation using antibodies against the HA tag (Fig. 1C). The TAF130-N447 derivative behaves indistinguishably from wild-type TAF130 with respect to its ability to coprecipitate all six TAFs tested. The mutant TAF130 efficiently coprecipitates TBP, although perhaps with a slightly reduced efficiency compared with wild-type TAF130. Although this analysis can not exclude the possibility that untested TAFs might be absent from the observed complex, the results strongly suggest that the TAF130-N447 derivative assembles into TFIID.
Since the dominant-negative effect of the mutant TAF is only evident upon overexpression, we considered the possibility that it might be caused by blocking of the TBP-DNA interaction by the N-terminal inhibitory domain on TAF130 (22, 29). However, analysis of a series of C-terminal deletion mutants (Fig. 1D) reveals that the dominant-negative phenotype is not observed in several derivatives that contain the N-terminal inhibitory region (e.g., the N159, N202, and N333 derivatives). All of these TAF derivatives were expressed, and the levels of the N202 and N447 derivatives were comparable (data not shown). In accord with our results, other laboratories have reported that overexpression of the N-terminal domain of TAF130 does not produce a dominant-negative phenotype (2).
TAF130 mutants do not associate with promoters in vivo.
Promoter association of the TAF130 derivatives in living yeast cells was analyzed by chromatin immunoprecipitation. Because the TAF130 mutants do not support viability, we used an approach previously described for inviable derivatives of TBP (11). (HA)3-tagged TAF130 mutants were expressed from the natural TAF130 promoter in a strain that contains an untagged wild-type copy of TAF130 at its normal chromosomal locus. In this way, we could directly assess the properties of the TAF130 derivative, and hence the mutant TFIID complex, in normally growing cells.
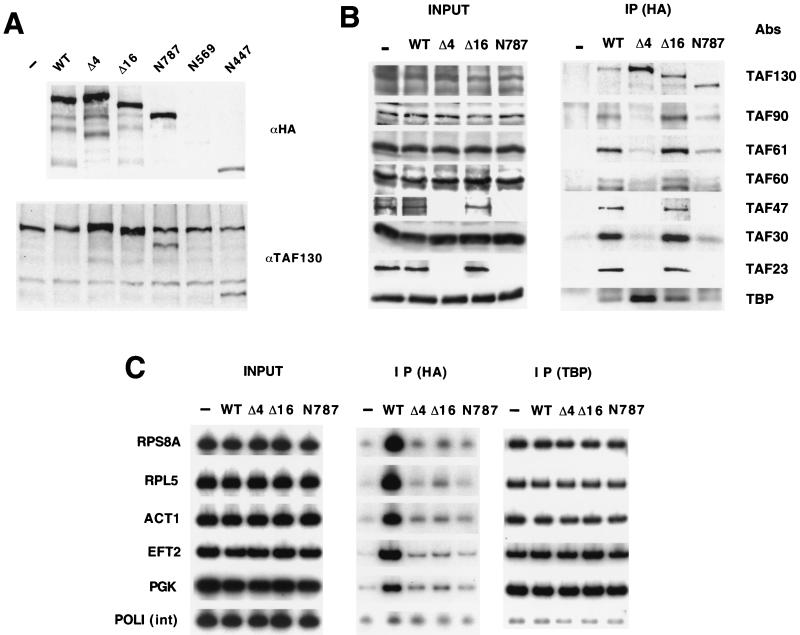
We examined several C-terminal deletions (Fig. 1D), as well as internal deletions (Δ4, which lacks residues 208 to 303, and Δ16, which lacks residues 913 to 1037) that are unable to support cell viability (2). The (HA)3-tagged Δ4, Δ16, and N787 derivatives are expressed at levels that are roughly comparable to the (HA)3-tagged wild-type protein but are ca. twofold lower than that of the genomic TAF130 (Fig. 2A). The N569 and N447 derivatives were expressed at significantly lower levels and thus were not analyzed further. Coimmunoprecipitation experiments (Fig. 2B) indicate that, as previously described (2), the Δ4 mutant interacts normally with TBP but fails to assemble with any of the TAFs tested. In contrast, the Δ16 derivative of TAF130 immunoprecipitates TBP and the five TAFs tested with an efficiency comparable to that of wild-type TAF130, strongly suggesting that it forms an otherwise normal TFIID complex. Consistent with its ability to form normal TFIID complexes, the Δ16 derivative of TAF130 confers a dominant-negative phenotype when overexpressed (Fig. 1D). The N787 derivative of TAF130 also assembles into a TFIID-like complex, although with a reduced efficiency. Importantly, wild-type TAF130 is not detected in complexes immunoprecipitated with the (HA)3-tagged Δ16 or N787 derivatives of TAF130 (Fig. 2B). This indicates that there is only one molecule of TAF130 in the TFIID complex and that the wild-type and mutant TFIID complexes coexist in physically distinct entities.
FIG. 2.
Promoter association of TAF130 derivatives in vivo. (A) Expression levels of the indicated (HA)3-tagged TAF130 derivatives expressed from the natural TAF130 promoter on centromeric plasmids as assayed by Western blotting using antibodies to the HA epitope or to TAF130. (B) TFIID complex formation by the indicated TAF130 derivatives. (HA)3-tagged TAF130 derivatives were immunoprecipitated, and components of the TFIID complex were detected with the indicated antibodies. (C) Cross-linked chromatin preparations from strains carrying the indicated (HA)3-tagged TAF130 derivatives or vector alone (the left panel represents the input sample) were immunoprecipitated with monoclonal antibodies against the HA tag (central panel) or polyclonal antibodies against TBP (right panel). PCR products corresponding to the indicated promoters or the POL1 structural gene are shown.
TAF occupancy at yeast promoters is not strictly correlated with transcriptional activity or with occupancy by TBP, TFIIA, or TFIIB, indicating that TFIID interacts preferentially with certain yeast promoters in vivo (25, 28). We therefore analyzed five promoters representing the full range of TFIID association: TFIID occupancy is high for ribosomal protein promoters RPS8A and RPL5, intermediate for the ACT1 and EFT2 promoters, and low for the PGK promoter (25). As expected, the levels of TBP occupancy for each promoter are indistinguishable in the five strains. However, at all five promoters, TAF130 occupancy is dramatically reduced for all the mutant derivatives tested (Fig. 2C). TAF130 association at these promoters is roughly comparable to that observed within the middle of the POL1 structural genes and hence is likely to be at or near the background level. Thus, all TAF130 derivatives, and therefore the TFIID complexes, tested are generally impaired in their association with yeast promoters in vivo. Of particular importance, the Δ16 mutant associates extremely poorly with promoters, even though it appears to form a normal TFIID complex, thereby highlighting the C-terminal region of TAF130 as playing an important functional role in the context of TFIID.
The Δ16 TAF130 mutant is transcriptionally nonfunctional in vivo.
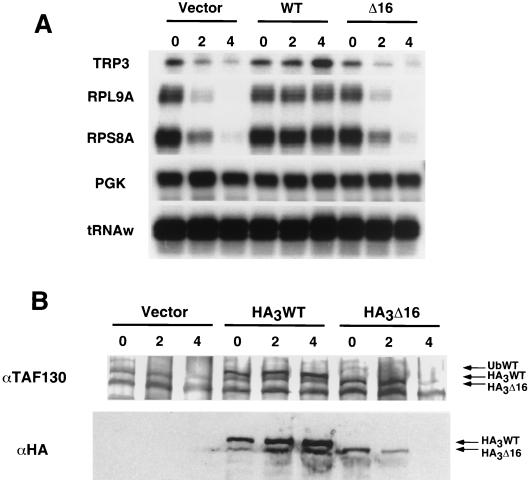
The transcriptional activity of the Δ16 derivative of TAF130 was analyzed in a strain where wild-type TAF130 was depleted by the copper-inducible, double-shutoff method (32). Upon copper induction, the parental strain and the strain bearing the Δ16 derivative of TAF130 had indistinguishable transcriptional patterns (Fig. 3A). Specifically, transcription of the three TAF130-dependent promoters (TRP3, RPS8A, and RPL9A) is significantly decreased after 2 h in copper is and virtually eliminated after 4 h, whereas the TAF-independent PGK and Pol III tRNAw promoters remain unaffected. Transcription of all genes tested was unaffected in a control strain containing a wild-type TAF130 allele.
FIG. 3.
The TAF130-Δ16 derivative is transcriptionally inactive. (A) RNA levels of the indicated genes in strains containing a copper-inducible TAF130-depletion allele and plasmids expressing (HA)3-tagged wild-type TAF130 or TAF130-Δ16 at 0, 2, and 4 h after the addition of copper. The TRP3, RPL9A, and RPS8A genes are dependent on TAF130 function, whereas the PGK and tRNAw genes are not (32, 45). (B) Levels of wild-type and mutant TAF130 proteins at the indicated times after addition of copper, as determined by Western blotting with antibodies to the HA epitope or to TAF130. The position of the ubiquitin-tagged wild-type TAF130 (UbWT), which is degraded upon copper addition is also indicated.
Unexpectedly, the level of the Δ16 mutant protein is modestly reduced 2 h after copper addition and drastically reduced after 4 h (Fig. 3B). This suggests that the mutant complex is undergoing degradation when a functional complex is eliminated and cells begin to die. The basis of this effect is unknown, but it suggests that the mutant TFIID complex is destabilized under conditions of severe cell stress and perhaps increased proteolytic activity. We doubt that transcriptional inactivity of the Δ16 derivative is simply due to protein degradation, because a considerable amount of the Δ16 protein remains at the 2-h time point, yet the expression of TAF130-dependent genes is indistinguishable from the control TAF130 depletion strain. Thus, in agreement with the chromatin immunoprecipitation experiments that are performed in wild-type cells, we conclude that the Δ16 derivative of TAF130 is transcriptionally nonfunctional in vivo.
The mutant TFIID complex containing TAF130-Δ16 is defective for promoter binding in vitro.
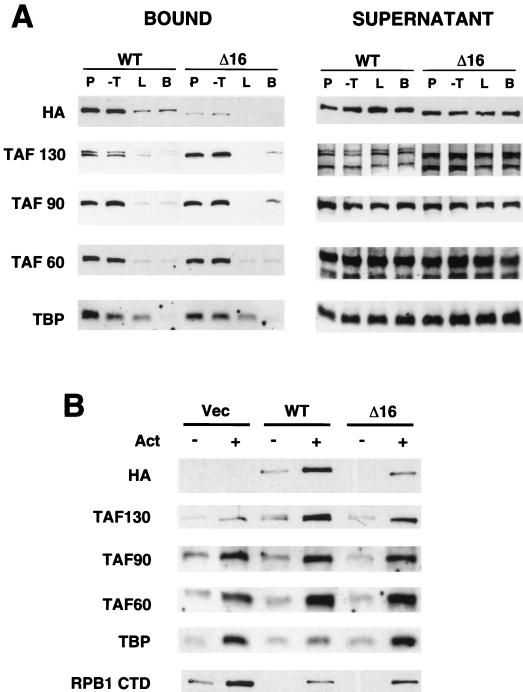
To compare biochemical properties of the mutant TFIID complex containing TAF130-Δ16 with those of wild-type TFIID, whole-cell extracts were incubated with immobilized DNA templates containing modified versions of the HIS4 promoter or a promoter-less fragment (41). As shown in Fig. 4A, wild-type TFIID binds to the DNAs in a promoter-dependent fashion, i.e., several TAFs and TBP efficiently associate with the HIS4 TATA-containing and TATA-lacking DNA fragments, whereas binding to the promoter-less DNA is reduced almost to the background level (beads alone). TAF association, and hence TFIID binding, is comparable on the TATA-containing and TATA-lacking fragments. A slight TATA dependence is observed for TBP, and this is likely to reflect the isolated subunit which exists in yeast extracts (14). The minimal TATA dependence in such immobilized template assays has been observed previously (41).
FIG. 4.
Promoter binding experiments. (A) Whole cell extracts containing (HA)3-tagged wild-type TAF130 or TAF130-Δ16 were incubated with paramagnetic bears containing a 500-bp fragment containing a derivative of the HIS4 promoter with one Gal4 binding site (P), the same HIS4 promoter fragment with several mutations at the TATA element (−T), a 280-bp promoterless fragment from the pBluescript vector present in the two previous constructs that contains the Gal4 binding site (L), and the beads without DNA (lane B). The left panel shows the indicated proteins bound to the DNA, whereas the right panel shows the analysis of 10% of the first supernatant after incubation. (B) Effect of the Gal4-VP16 activator on TFIID binding. The promoterless template containing the Gal4 binding site was incubated with extracts containing (HA)3-tagged wild-type TAF130 or TAF130-Δ16 in the absence (−) or presence (+) of Gal4-VP16.
In comparison to wild-type TFIID, the mutant complex containing TAF130-Δ16 binds poorly to the HIS4 promoter fragments. The weak binding of the mutant TFIID complex is significantly higher than that observed on the promoter-less fragment; hence, it is promoter dependent. The wild-type and mutant extracts are comparably active, as evidenced by equivalent levels of binding of nontagged TAF130, TAF90, TAF60, and TBP. In addition, the wild-type and mutant TFIID complexes are comparably stable during the procedure because equivalent amounts of intact TAFs remained in the supernatants after the binding. Taken together, these results indicate that the mutant TFIID complex is significantly impaired in DNA binding.
The Δ16 mutant complex is fully competent for interacting with activators.
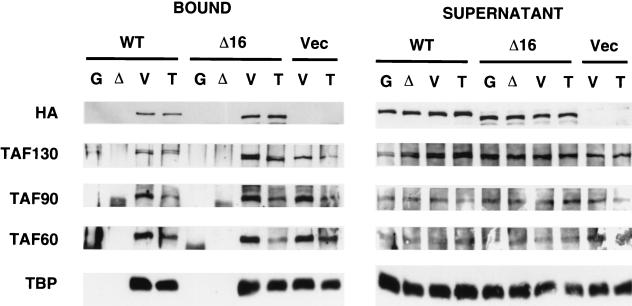
The immobilized template assay was also used to examine whether the Gal4-VP16 activator can recruit the mutant TFIID complex to DNA. This experiment employed the promoterless fragment containing one Gal4 binding site, because this fragment gave the highest activation ratio in previous studies (41). As shown in Fig. 4B, Gal4-VP16 conferred a sixfold stimulation in the association of TAF130-Δ16, and hence the mutant TFIID complex, to DNA. A similar six fold stimulation by Gal4-VP16 was observed for wild-type TFIID. The simplest interpretation of this result is that Gal4-VP16 interacts normally with the mutant TFIID complex to stimulate recruitment to DNA, but it cannot compensate for the initial defect in DNA binding. In accord with this interpretation, immobilized GST fusions to the VP16 or Adr1 (TAD4) activation domain (24) pulls down wild-type and mutant TAF130 derivatives, and hence TFIID complexes, to a comparable extent (Fig. 5). Both activators also pull down other TAFs and TBP, whereas no binding to TFIID components is observed for GST alone or a fusion to a transcriptionally inactive version of the VP16 activation domain. These experiments show that, in vitro, the Δ16 mutant TFIID complex is fully competent for interacting with activators, but it is specifically defective in a general DNA-binding function.
FIG. 5.
Interaction of TFIID with activation domains. Whole cell extracts containing (HA)3-tagged wild-type TAF130 or TAF130-Δ16 were incubated with glutathione-Sepharose beads bound to GST protein (G) or to GST fusions to a mutated form of the VP16 activation domain (Δ), the GST-VP16 activation domain (V), or the TADIV activation domain of Adr1 (T). The indicated TFIID components in the bound (left panel) or supernatant (right panel) fractions were detected by Western blotting with the indicated antibodies.
Evidence that the HMG domain within TAF130 has been mischaracterized.
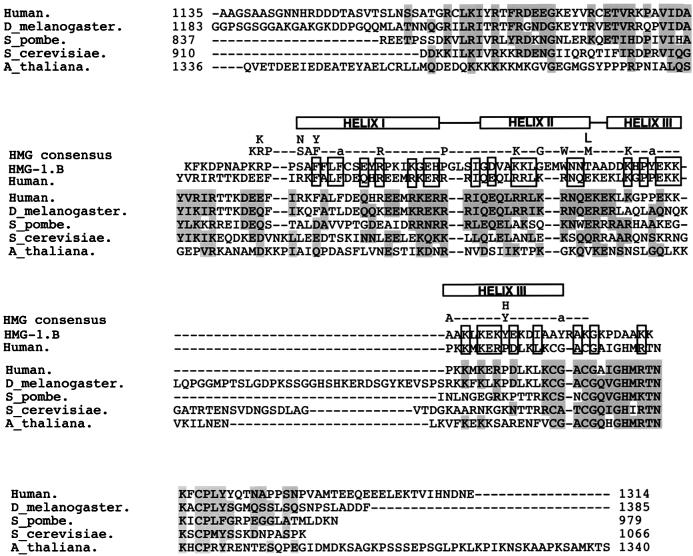
The region deleted in TAF130-Δ16 (residues 913 to 1037) corresponds to a region of human TAF250 that has been previously classified as being homologous to an HMG domain (44). This observation was intriguing because HMG domains are typically found in DNA-binding proteins, some of which bind without sequence specificity (13). However, an alignment between HMG proteins and TAF130 homologues (Fig. 6) strongly argues that the C-terminal region of TAF130 does not contain an HMG domain. There is very little correlation between the residues conserved in the TAFs and those similar between human TAF250 and the HMG-1.B. Only 2 of 18 residues in the HMG consensus are conserved across the known TAFs. Highly conserved aromatic residues in HMG proteins (Phe9, Phe12, Trp45, and Tyr56) that form a hydrophobic core that mediates the arrangement of helices II and III are not conserved among the TAF homologues, and only residues corresponding to Phe9 and Phe12 are present in the human TAF250. Furthermore, the TAF homologues contain a two-residue deletion of helix II and a large insertion in helix III with respect to HMG domains. These observations suggest that the C-terminal region of TAF130 (and presumably its homologues) interacts with DNA through a domain that is distinct from an HMG domain.
FIG. 6.
Analysis of the putative HMG domains of TAF130 homologues. Sequences corresponding to the HMG regions of several TAFs were aligned by using the program CLUSTAL W (EMBL Outstation) and compared to the original alignment between human TAF250 and the HMG-1.B (44) and to the HMG consensus (13). The three α-helices that form the HMG domain are depicted above the TAF sequences. Residues shaded in gray are conserved among the TAFs, and boxed residues are similar between human TAF250 and HMG-1.B.
DISCUSSION
A mutant TFIID complex that is defective for association with DNA.
Understanding the physiological roles of individual TAFs within TFIID requires the characterization of mutant TFIID complexes that have molecularly defined defects. Previous studies involving TAF depletion or thermal inactivation have been complicated by dissociation of the TFIID complex and/or by limited molecular analysis of the mutant TFIID complex (see the introduction). Here, we characterize TAF130 derivatives that assemble into TFIID complexes but are transcriptionally inactive in vivo. Specifically, TAF130-Δ16 and TAF130-N447 behave indistinguishably from wild-type TAF130 with respect to their abilities to coimmunoprecipitate TBP and all six TAFs tested. While we cannot exclude the possibility that some untested TAF is missing or substoichiometric, this observation strongly suggests that these TAF130 derivatives form TFIID complexes that are otherwise normal. As these mutant TFIID complexes were isolated from living cells and, since TFIID is unlikely to be assembled de novo in cell extracts, we believe that the mutant TFIID complexes exist in physiological conditions.
The N-terminal 447 residues of TAF130 (roughly 35% of the full-length protein) appear to define a minimal TFIID assembly domain. This TAF130 derivative includes determinants previously defined as being important for interactions with TBP and TAFs (2). Our results do not exclude other regions of TAF130 from contributing to TFIID stability and, in this regard, the TAF130-N787 mutant displays a minor defect in TFIID assembly. We do not fully understand why overexpression of TAF130-N447 and other TAF130 derivatives confers a dominant-negative phenotype. If the amounts of other TAFs are limiting in the cell, overexpression of such TAF130 derivatives might simply result in the preferential formation of mutant and transcriptionally incompetent TFIID complexes, thereby reducing the level of wild-type TFIID and causing a defect in cell growth. However, it is unclear why the genetically selected N447 and N452 derivatives confer a more severe dominant-negative phenotype than other TAF130 derivatives with less-extensive C-terminal deletions.
By epitope-tagging the TAF130 derivatives, we could directly assay the physiological and biochemical properties of the mutant TFIID complexes, even in the presence of wild-type TFIID. In vivo, the mutant TFIID complexes are severely defective for interacting with all promoters tested. Although many yeast promoters do not require TFIID-specific TAFs for transcription in vivo due to the existence of a TAF-independent form of transcriptionally active form of TBP, TFIID associates with TAF-independent promoters to a modest extent (25, 28). In accord with this general defect in promoter association, the mutant TFIID complexes appear to be transcriptionally inactive. However, there is a strong correlation between transcriptional activity and promoter association, because many components of the Pol II machinery are recruited together to promoters in vivo (26, 27). Thus, these in vivo experiments cannot determine whether the mutant TFIID complexes have a specific defect in promoter association or have some other functional defect (e.g., interaction with activators or a component of Pol II holoenzyme) that indirectly decreases promoter association in vivo.
Biochemical analysis of the mutant TFIID complex containing TAF130-Δ16 reveals a specific defect in promoter association. The observed defect is unlikely to be due to instability of the mutant TFIID complex in vitro because the conditions used for the immobilized template assays are similar to those used for the coimmunoprecipitation experiments that demonstrate TFIID integrity. Furthermore, the functional defect of the TAF130-Δ16 complex is not due to weakened interactions with TFIIB or Pol II holoenzyme, because TFIID binding to immobilized templates occurs independently of these components (41). Finally, the biochemical defect is specific in that the mutant TFID complex behaves indistinguishably from wild-type TFIID for its ability to interact with the VP16 and Adr1 (TAD4) activation domains under experimental conditions that are very similar to those used for promoter binding. Taken together, our results indicate that TAF130-Δ16 assembles into an otherwise normal TFIID complex that fails to associate with promoters in vitro and in vivo. To our knowledge, this represents the first physiological and biochemical analysis of a mutant TFIID complex with a defined biochemical defect.
The C-terminal region of TAF130 is required for general DNA binding by TFIID.
The biochemical and genetic properties of the mutant TFIID complex containing TAF130-Δ16 suggest that, in the context of TFIID, the C-terminal region of TAF130 is important for promoter association. The existence of this general DNA-binding function of TAF130 suggests that sequence-specific interactions between TBP and the TATA element (18, 19) and that certain TAFs with the initiator and downstream elements are not sufficient for efficient binding of TFIID to promoters (4, 6, 37, 52, 54). Thus, even though the isolated TBP subunit efficiently binds TATA elements, TBP in the context of TFIID requires the C-terminal region of TAF130 for efficient binding in vivo and in vitro. It seems likely that the contribution of the TAF130 C-terminal region toward TFIID binding is largely nonspecific for DNA sequence, although our results do not exclude some degree of sequence specificity.
Three molecular models could explain how the C-terminal region of TAF130 increases the association of TFIID with promoters. The simplest model is that this TAF130 region directly interacts with DNA. A direct interaction could increase the overall association of TFIID to promoter DNA, whether or not this interaction is sequence specific. In support of this idea, the C-terminal region of TAF130 is highly basic/ human TAF250 in the context of TFIID can be cross-linked to DNA in vitro, and a TAF250-TAF150 complex shows sequence-specific binding to initiator elements (6). However, the region of TAF250 that is cross-linked to DNA and the relative contributions of TAF250 and TAF150 to general and specific interactions to the initiator element are unknown. Other TAFs cross-link to promoter regions in vitro, suggesting that the promoter DNA wraps around TFIID (37), and a low resolution structure of TFIID reveals a horseshoe or clamp-shaped structure with considerable surface available for contacting promoter DNA (1, 3).
In a second model, the C-terminal region of TAF130 might not directly contact DNA but rather increase TFIID association by relieving inhibition of the TBP-TATA interaction by the N-terminal domain of TAF130. The N-terminal domain of TAF130 directly interacts with the DNA-binding surface of TBP and therefore acts as a significant inhibitor of TFIID binding to TATA elements (21, 22, 29). An intramolecular interaction between the N-terminal and C-terminal regions of TAF130 might relieve the inhibition of the DNA-binding surface of TBP, thereby increasing overall binding of TFIID to promoters. In a related model, the C-terminal region of TAF130 might alter the conformation of TFIID in a manner that displaces the N-terminal region of TAF130 from the DNA-binding surface of TBP.
In a third model, the C-terminal region of TAF130 might interact with TFIIA, thereby contributing to the formation or stability of the TFIID-TFIIA-DNA complex. In the immobilized template assay employed here, TFIIA stimulates TFIID association with promoter DNA (41). In vivo, the TFIIA-TBP occupancy ratio appears to be constant over many promoters, indicating that TBP and TFIIA co-occupy promoters in the context of physiological chromatin (25). Thus, by this model, loss of the TAF130 C-terminal region would weaken the TFIID-TFIIA-DNA complex and hence the observed interaction of the mutant TFIID complex with DNA. However, TAFs are not required for TFIIA to stimulate the TBP-TATA in reactions involving purified components (16) or crude extracts and immobilized templates (41), and the constant TFIIA-TBP occupancy ratio in vivo is independent of TAF occupancy (25). Models in which the TAF130 C-terminal region interacts with TFIIB or components of Pol II holoenzyme are unlikely because TFIID association with promoter DNA is unaffected by the presence or absence of these factors in the immobilized template assay (41).
These three models are not mutually exclusive, and indeed all may contribute to the association of TFIID with promoters in vivo and in vitro. Although the C-terminal region of TAF130 appears to be distinct from an HMG domain, this region is conserved among all known TAF130 homologues. It seems likely, therefore, that this region contributes to the association of TFIID complexes with promoters in other eukaryotic organisms.
ACKNOWLEDGMENTS
We thank Steve Buratowski, Eun-Jung Cho, and Oranart Matangkasombut for help with the immobilized template assays; Clyde Denis, Michael Green, and Tony Weil for DNAs; Michael Green for TAF antibodies; and Joseph Geisberg and Steve Hahn for useful comments on the manuscript.
This work was supported by a postdoctoral fellowship to M.M. from the Human Frontiers Science Program and by a research grant to K.S. from the National Institutes of Health (GM30186).
REFERENCES
- 1.Andel F, III, Ladurner A G, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIIA-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 2.Bai Y, Perez G M, Beechem J M, Weil P A. Structure-functional analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N-terminus of yeast TAFII130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286:2151–2153. doi: 10.1126/science.286.5447.2151. [DOI] [PubMed] [Google Scholar]
- 4.Burke T W, Kadonaga J T. The downstream core promoter element, DPE is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Chalkley G E, Verrijzer C P. DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho E J, Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J Biol Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- 8.Chou S, Struhl K. Transcriptional activation by TFIIB mutants that severely impair the interaction with promoter DNA and acidic activation domains. Mol Cell Biol. 1997;17:6794–6802. doi: 10.1128/mcb.17.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 10.Dunphy E L, Johnson T, Auerbach S S, Wang E H. Requirement for TAFII250 acetyltransferase activity in cell cycle progression. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisberg J V, Struhl K. TATA-binding protein mutants that increase transcription from enhancerless and repressed promoters in vivo. Mol Cell Biol. 2000;20:1478–1488. doi: 10.1128/mcb.20.5.1478-1488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. A subset of TBP-associated factors, TAFIIs are integral components of the SAGA complex that are required for nucleosomal acetylation and transcription stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 13.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S, Buratowski S, Sharp P A, Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: A gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 15.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 16.Imbalzano A N, Zaret K S, Kingston R E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 17.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP-TATA box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 21.Kokubo T, Gong D-W, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Drosophila 230-kd TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev. 1993;7:1033–1046. doi: 10.1101/gad.7.6.1033. [DOI] [PubMed] [Google Scholar]
- 22.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 24.Komarnitsky P B, Klebanow E R, Weil P A, Denis C L. ADR1-mediated transcriptional activation requires the presence of an intact TFIID complex. Mol Cell Biol. 1998;18:5861–5867. doi: 10.1128/mcb.18.10.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 26.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;389:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 27.Li X-L, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;389:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 28.Li X-Y, Bhaumik S R, Green M R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 29.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Solution structure of a TBP-TAFII230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 30.Matangkasombut O, Buratowski R M, Swilling N W, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 31.Mizzen C A, Yang X-Y, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 32.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;382:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 33.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 34.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien T, Tjian R. Functional analysis of the human TAFII250 N-terminal kinase domain. Mol Cell. 1998;1:905–911. doi: 10.1016/s1097-2765(00)80089-1. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien T, Tjian R. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc Natl Acad Sci USA. 2000;97:2456–2461. doi: 10.1073/pnas.97.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 38.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 39.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X-J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 40.Ranallo R T, Struhl K, Stargell L A. A TBP mutant defective in TFIID complex formation in vivo. Mol Cell Biol. 1999;19:3951–3957. doi: 10.1128/mcb.19.6.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranish J A, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reese J C, Zhang Z, Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem. 2000;275:17391–17398. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- 43.Sanders S L, Weil P A. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 44.Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol Cell Biol. 1991;11:3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen W C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 50.Tsukihashi Y, Miyake T, Kawaichi M, Kokubo T. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol Cell Biol. 2000;20:2385–2399. doi: 10.1128/mcb.20.7.2385-2399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 52.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 53.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 54.Verrijzer C P, Yokomori K, Chen J-L, Tjian R. Drosophila TAFII150: Similarly to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 55.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;382:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 56.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 57.Wang E H, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 58.Woontner M, Wade P A, Bonner J, Jaehning J A. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu S Y, Kershnar E, Chiang C M. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 1998;17:4478–4490. doi: 10.1093/emboj/17.15.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]