Abstract
On stably replicating episomes, transcriptional activation of the ɛ-globin promoter by the β-globin locus control region HS2 enhancer is correlated with an increase in nuclease sensitivity which is limited to the TATA-proximal nucleosome (N1). To elucidate what underlies this increase in nuclease sensitivity and the link between chromatin modification and gene expression, we examined the nucleoprotein composition and histone acetylation status of transcriptionally active and inactive promoters. Micrococcal nuclease digestion of active promoters in nuclei released few nucleosome-like nucleoprotein complexes containing N1 sequences in comparison to results with inactive promoters. We also observed that N1 DNA fragments from active promoters are of a subnucleosomal length. Nevertheless, chromatin immunoprecipitation experiments indicate that histones H3 and H4 are present on N1 sequences from active promoters, with H3 being dramatically hyperacetylated compared with that from inactive promoters and vector sequences. Strikingly, H3 in the adjacent upstream nucleosome (N2) does not appear to be differentially acetylated in active and inactive promoters, indicating that the nucleosome modification of the promoter that accompanies transactivation by HS2 is highly directed and specific. However, global acetylation of histones in vivo by trichostatin A did not activate transcription in the absence of HS2, suggesting that HS2 contributes additional activities necessary for transactivation. N1 sequences from active promoters also contain reduced levels of linker histone H1. The detection of a protected subnucleosomal sized N1 DNA fragment and the recovery of N1 DNA sequences in immunoprecipitations using anti-acetylated H3 and H4 antibodies argue that N1 is present, but in an altered conformation, in the active promoters.
Alterations of chromatin structure accompany the activation of gene transcription in vivo during development and in response to hormones and other stimuli. In the erythroid lineage, the five DNase I hypersensitive sites (HSs) (HS1 to -5), which comprise the locus control region (LCR) of the β-globin gene cluster, appear early in embryogenesis before transcription of the globin genes commences (19, 23, 41). The formation of these sites is temporally correlated with chromatin changes over the entire locus in erythroid cells characterized by an increase in general sensitivity to DNase I and a change in replication timing from early to late in S phase (17). Moreover, high-level expression of the individual members of the β-globin gene family during the process of development depends upon the LCR (3). During the developmental stages when the individual globin genes are being actively transcribed, a DNase I HS is detected in the promoter (21).
The appearance of DNase I HSs at the promoters of transcribed genes is indicative of altered chromatin structure associated with the binding of transcription factors to these regions (13). However, the alterations in chromatin structure at such sites may not be identical in every case. An early study of the chick β-globin gene showed that a short histone-free restriction fragment could be released from the promoter of the actively transcribing gene (30). In contrast, histones are present at the DNase I HSs of the induced mouse mammary tumor virus (MMTV) promoter (7, 40) and transcription factors and nucleosomes occupy the active albumin enhancer simultaneously (11). These observations raise the possibility that DNase I HSs are not necessarily devoid of nucleosomes (37).
An abundance of data implicates histone modification as a critical component in the control of gene expression through localized and higher-order alterations in chromatin structure. For example, there is a marked coincidence of hyperacetylation, as revealed by anti-acetyl-lysine antibodies, and nuclease sensitivity within the domain of the transcriptionally active chick β-globin locus (22). Other recent reports describe histone modification that is more localized. Hyperacetylated histones may be confined to the promoters of yeast genes activated by the acetyltransferase Gcn5, and increased H3 and H4 acetylation attributed to CREB-binding protein (CBP) (also called p300) was found in a region that could accommodate two or three nucleosomes surrounding the induced beta interferon promoter (26, 31). Conversely, deacetylation of H4 within a region potentially spanning two promoter nucleosomes accompanies repression of certain yeast genes (25, 27, 34). It is likely that modification of histones alters their interaction with each other and with DNA in vivo as it does in vitro, providing at least part of the basis for nuclease sensitivity (46).
Previous work in our laboratory using minichromosomes stably maintained in human cells indicated a series of positioned nucleosomes over the ɛ-globin gene. Transcription activation by the LCR HS2 enhancer alters the chromatin structure of the TATA-proximal N1 nucleosome in the promoter of the gene, creating a nuclease-sensitive site (20). However, the mechanism by which HS2 produces these effects has not been elucidated. To ask whether the promoter-proximal nucleosome is absent or altered under conditions of active transcription, we employed nucleoprotein electrophoresis and chromatin immunoprecipitation (ChIP) assays for linker histone H1 and for core histones H3 and H4 along with their acetylated variants. We also investigated the effect of trichostatin A (TSA)-mediated global histone acetylation on transcriptional activity of the ɛ-globin gene. These experiments indicate that N1 nucleoprotein complexes likely have altered properties, have reduced H1 levels compared to those of inactive promoters, and contain a subnucleosome-sized DNA component. Furthermore, a very specific and directed acetylation of H3 in the TATA-proximal nucleosome accompanies transcriptional activation. This modification requires a functional enhancer. Acetylation of H4 appears to be more widespread and is detected at two promoter nucleosomes spanning almost 400 bp of the 5′ ɛ-globin flanking sequence. Global histone acetylation in the absence of the HS2 enhancer is not sufficient for transcriptional activation, suggesting that enhancer function includes the recruitment of a specific acetylase activity responsible for high-level modification of H3 at the proximal promoter.
MATERIALS AND METHODS
Minichromosome construction, cell culture conditions, and transfection.
The construction of minichromosomes carrying the ɛ-globin gene with or without HS2 has been described elsewhere (20). The ɛ-globin gene was a 3.7-kb genomic EcoRI fragment (GenBank accession no. U01317, coordinates 17482 to 21233). LCR HS2 was a 374-bp HindIII-to-XbaI fragment (GenBank accession no. U01317, coordinates 8486 to 8860). Clustered point mutations were introduced into HS2 to eliminate binding of NF-E2. These mutations, the transfection conditions, the growth of K562 cells, and individual clones carrying minichromosomes have been described (20,29). A representative clone of each type was studied in this work. The minichromosome copy numbers were as follows: 20 for ɛ, 8 for ɛHS2, and 24 for ɛHS2(mut). ɛ-Globin RNA from the endogenous chromosomes was detected in all clones. Minichromosomal ɛ-globin RNA was detected only in the ɛHS2 clone.
Preparation of nuclei and nuclease digestion.
Nuclei of K562 cell clones (1 × 108 to 1.5 × 108 cells) were prepared as described elsewhere (20). The purified nuclei were suspended in 0.3 to 0.5 ml of wash buffer and digested with 0 or 200 U of micrococcal nuclease (MNase; Worthington Biochemical Corporation) per ml for 5 min at room temperature. EDTA was added to a final concentration of 10 mM, and the samples were centrifuged at 16,000 × g for 10 min at 4°C. Part of the supernatant (equivalent to 4 × 107 nuclei) was electrophoresed on a 1.5% agarose gel, and the nucleoproteins were transferred to Nytran by the Turboblot method (Schleicher & Schuell). A portion of the remaining supernatant was treated with proteinase K overnight at 37°C, and genomic DNA was purified using routine methods. The purified DNA was separated on an agarose gel and transferred to Nytran membranes. Southern blot hybridization was performed using Quickhyb solution (Stratagene). The probes were an ɛ-globin 197-bp BamHI-to-PvuII fragment (for N1) and a 263-bp XbaI-to-EcoRV fragment (for N2 and N3). Probes were labeled with [32P]dCTP by the random priming method. For restriction enzyme cleavage analyses, nuclei from 108 cells were incubated with restriction enzymes as indicated in the text and figures (0 to 800 U/ml of each enzyme) at 37°C for 30 min and the supernatant was collected as for the MNase digestions.
ChIP.
Immunoprecipitations were performed using modifications of published methods (22, 32). K562 cells (3 × 107) containing various minichromosomes were harvested and washed twice with phosphate buffered saline (PBS; pH 7.4), and the cell pellets were suspended in MN buffer {10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 0.4% NP-40, 10 mM butyrate, 0.1 mM benzamidine, 0.4 mM [4(2-aminoethyl)-benzene sulfonyl fluoride] hydrochloride [AEBSF]} for lysis. Following incubation on ice for 5 min, nuclei were digested with MNase (200 U/ml) at 37°C for 10 min. The digestion was stopped by adding 1/10 volume of stop solution (0.5 M NaCl, 50 mM EDTA), and the samples were placed on ice. The suspension was then centrifuged at 16,000 × g 30 s, and the supernatant (S1) was retained. The pellet was suspended in 0.5 times the original volume of MN buffer and incubated on ice for 5 min, followed by addition of a 1/20 volume of stop solution and centrifugation. The second supernatant (S2) was combined with S1. Aliquots of the combined supernatants were incubated at 4°C with buffer only or with 15 μg of specific antibodies, including anti-histone H4, anti-histone H3, anti-diacetylated histone H3, and anti-tetra-acetylated H4 antibodies (Upstate Biotechnology, Lake Placid, N.Y.). Each of the antibodies was raised against the N-terminal peptide of the corresponding histone. After 2 h of constant rocking at 4°C, 40 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) was added and the rocking continued overnight. The immunocomplexes were collected by centrifugation (700 × g for 2 min), and the pellets were washed five times with 1 ml of washing buffer (50 mM NaCl, 10 mM Tris-HCl [pH 7.4], 10 mM sodium butyrate, 5 mM EDTA). The bound material was released by resuspending the pellets in 150 μl of washing buffer containing 1.5% sodium dodecyl sulfate, followed by incubation at room temperature for 15 min and separation by centrifugation. The released chromatin was treated with RNase for 30 min at 37°C, followed by proteinase K digestion for 1 h at 37°C. DNA was further purified by standard extraction and precipitation procedures. When chromatin immunoprecipitation (ChIP) was performed using anti-histone H1 (Upstate Biotechnology), nuclei were prepared as described above, cross-linked with paraformaldehyde, digested with 100 u of MNase per ml for 5 min at room temperature, and fractionated on a CsCl gradient before immunoprecipitation was performed (12).
Analysis of coimmunoprecipitated DNA.
The concentration of the coimmunoprecipitated DNA was determined using a Fluorescent DNA Quantitation Kit (Bio-Rad). For PCR amplification, 2.5 to 5 ng of immunoprecipitated DNA was used as the template for a number of amplification cycles chosen to be in the linear response range. The PCR conditions were 94°C for 5 min (one cycle); 94°C for 30 s, 60°C for 30 s, and 72°C for 20 s (23 to 25 cycles); and 72°C for 7 min (one cycle). The primers for N1 were 5′-CACAG GTCAG CCTTG ACCAA TGACT-3′ (sense) and 5′-TTATT CTTTA CTGCC GAAGT TCTGG-3′ (antisense) and amplified a 75-bp region between the TATA box and the CACCC site. The primers for N2 (82-bp region) were 5′-TGAGA TTTGC TCCTT TATAT GAGGC-3′ (sense) and 5′-A CCCTC TTCAT CATCT TCCAA-3′ (antisense). The control primers amplified a 58-bp region in oriP of the minichromosome backbone. The sequences were 5′-GCACT CCCAA CTCTA CTACT GGGTA-3′ (sense) and 5′-TGCTA TCCTC ATGCA TATAC AGTC-3′ (antisense). The primers which amplified a 183-bp region of the ampicillin resistance gene were 5′-AGTGT TATCA CTCAT GGTTA TGGCA-3′ (sense) and 5′-AGTTC TGCTA TGTGG CGCGG TATTA-3′ (antisense). One primer of every primer pair was γ-32P labeled, and a trace amount of hot primer (0.1 μCi) was added to each PCR mixture. The amplification products were electrophoresed on 10% native polyacrylamide gels, and the signals of the amplified bands were quantitated with a PhosphorImager (Molecular Dynamics) using ImageQuant software.
TSA treatment, RNase protection assay, and histone analysis.
A K562 clone carrying ɛ minichromosomes was treated with TSA (150 ng/ml; Wako Chemicals), and RNA was prepared from 5 × 106 cells at various time points up to 48 h using PUREscript (Gentra). The episomal copy of the ɛ-globin gene is marked by a mutation in the 5′ untranslated region to distinguish its RNA transcripts from the endogenous ɛ-globin transcripts (20). RNase digestion and gel analyses were performed as suggested by the manufacturer of the reagents (Ambion). Histones were prepared at various time points during TSA treatment by modification of published methods (42). Cells (107) were harvested and washed twice with PBS, and the cells were lysed in PTB buffer (PBS containing 0.5 % Triton X-100, 10 mM sodium butyrate, and 0.4 mM AEBSF) on ice for 10 min. The lysate was centrifuged at 4°C at 16,000 × g for 10 min, and the pellet was resuspended in extraction buffer (0.4 N HCl, 10 mM dithiothreitol, 0.4 mM AEBSF, 10 mM sodium butyrate) and incubated overnight at 4°C. The acid extract was centrifuged at 16,000 × g at 4°C for 10 min, and the supernatant was retained. Histones were precipitated with 12 volumes of acetone. Following centrifugation at 1,200 × g and 4°C for 2 min, the pellets were washed twice with acetone–100 mM HCl (6:1) and three times with acetone. The histone preparation was vacuum dried and dissolved in water. Protein concentration was determined with Bio-Rad reagents by using commercial histones as the standard. Acid-extracted proteins were separated on sodium dodecyl sulfate–15% polyacrylamide gels and transferred to nitrocellulose membranes. Histones were detected with the same anti-histone antibodies used for ChIPs (Upstate Biotechnology) and enhanced chemiluminesence (Amersham).
RESULTS
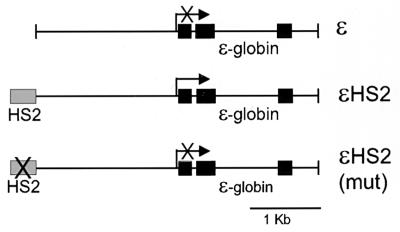
We previously reported the apparent loss or disruption of the TATA-proximal nucleosome (N1) in the human ɛ-globin gene when the gene was transcriptionally activated by the β-globin LCR HS2 on stably replicating minichromosomes in human erythroid K562 cells (20). To ascertain the fate of histones within promoter nucleosomes in vivo, we analyzed minichromosomes with and without functional HS2 linked to the ɛ-globin gene. Figure 1 illustrates the three minichromosomes used in these experiments, ɛ, ɛHS2, and ɛHS2(mut). ɛHS2 was constructed with the wild-type β-globin LCR HS2 fragment, while in ɛHS2(mut) the two NF-E2 binding sites in HS2 were destroyed by clustered point mutations. These NF-E2 sites are critical for the enhancer activity of HS2. Minichromosomes with the ɛ-globin gene alone, or with the NF-E2 sites of HS2 mutated, neither transcribe the gene nor remodel the N1 nucleosome (20). However, when HS2 is linked to the gene, the promoter-proximal nucleosome (N1) is altered such that it becomes sensitive to MNase, DNase I, and cleavage by NcoI and AvaII (20). The nucleosome positions in nontranscribed and actively transcribed minichromosomes, as determined by indirect end-labeling experiments, are shown in Fig. 2.
FIG. 1.
ɛ-Globin insertions into minichromosomes. Three constructs were inserted into the episomal vector p220.2 by blunt-end ligation into the unique SalI site (47). Each contained the 3.7-kb EcoRI human genomic ɛ-globin fragment. Coding regions of the gene are indicated by filled boxes. In addition, ɛHS2 contained HS2 of the β-globin LCR. In ɛHS2(mut) the tandem duplicated NF-E2 binding sites in HS2 have been mutated to destroy enhancer activity (×). The transcription start site and direction of transcription of the gene are indicated by a horizontal arrow. In the absence of a functional HS2, the ɛ-globin gene is not transcribed.
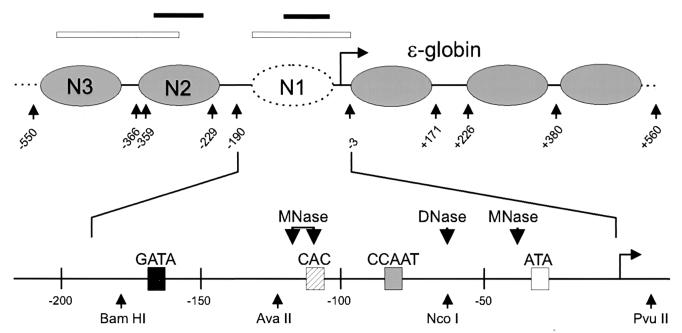
FIG. 2.
Structure of the ɛ-globin promoter and locations of positioned nucleosomes. Nucleosome positions in the region of the ɛ-globin promoter were previously determined by indirect end-labeling experiments (20). The dotted nucleosome is altered when the ɛ-globin gene is transcriptionally activated. An expanded view of the sequences underlying N1 illustrates regulatory sites within the promoter (shaded boxes), nuclease cleavage sites in actively transcribing promoters (large arrowheads), and restriction enzyme recognition sites (small arrowheads). Southern blot probes used in the experiments are indicated at the top by open bars over the corresponding sequences. The positions of amplified PCR products are indicated by filled bars.
Nucleoprotein complexes at N1.
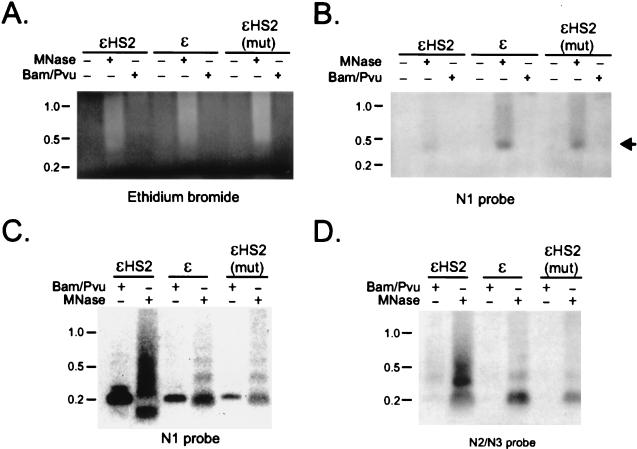
To investigate whether mononucleosomal nucleoprotein complexes could be isolated from particular nucleosomes in the proximal promoter region of the ɛ-globin promoter, nuclei from cell clones carrying different minichromosomes were digested with MNase or with BamHI plus PvuII, whose cutting sites are near the boundaries of N1 (197-bp expected fragment) (Fig. 2). Soluble chromatin obtained from 4 × 107 nuclei for each of the cell clones was electrophoresed on a 1% agarose gel, transferred to a nylon membrane, and hybridized with the N1 probe, indicated as an open bar over the nucleosome map in Fig. 2. Figure 3A shows the relevant portion of the ethidium bromide-stained gel, and Fig. 3B shows the hybridization pattern. There is a marked decrease in recovery of nucleoprotein complexes from the proximal promoter region (N1) of actively transcribing ɛ-globin genes linked to wild-type HS2, compared to that from inactive promoters either without HS2 or linked to a nonfunctional mutated HS2. However, naked DNA of mononucleosomal length is not visualized. Digestion of nuclei with the restriction enzymes BamHI plus NcoI (113-bp fragment) similarly failed to reveal nucleoprotein complexes or naked DNA of the predicted size from ɛHS2 (not shown).
FIG. 3.
Altered N1 nucleoprotein structure correlates with transcriptional activity. Nuclei from K562 clones containing the indicated minichromosomes were digested with 0 or 200 U of MNase per ml or with 800 U of BamHI and PvuII per ml. Soluble, recovered nucleoprotein was analyzed by agarose gel electrophoresis and Southern blotting. The probe used corresponded to sequences within N1 (Fig. 2). (A) Ethidium bromide-stained agarose gel. (B) Southern blot of the gel shown in panel A. (C) A part of each sample was deproteinized and analyzed by agarose gel electrophoresis and Southern blotting. The membrane was successively hybridized with the N1 (C) and N2-N3 (D) probes indicated in Fig. 2.
To ascertain the lengths of DNA fragments after MNase or restriction enzyme digestion and to confirm that enzymatic digestion had proceeded to completion, we treated the soluble chromatin fraction with proteinase K and purified the DNA. This DNA was electrophoresed on an agarose gel and Southern blotted to visualize DNA fragments from the proximal promoter region. Despite the apparent absence of MNase and restriction enzyme digestion products in nucleoprotein preparations of ɛHS2 nuclei (Fig. 3B), hybridization of extracted DNA with the N1 probe indicated cleavage of the expected 197-nucleotide BamHI-to-PvuII fragment in all three minichromosomes (Fig. 3C). Quantitatively more fragment was cleaved from the active promoter, commensurate with its greater accessibility to restriction enzymes (20). However, while MNase cleavage released a mononucleosome-sized DNA fragment from the minichromosomes when transcription was inactive (ɛ, ɛHS2mut), a subnucleosome-sized product was released from ɛHS2, which actively transcribes the ɛ-globin gene. This result is consistent with the cleavage sites for MNase mapped by indirect labeling (Fig. 2). The absence of full-length mononucleosomal fragments from N1 suggests that virtually all ɛHS2 episomal promoters undergo remodeling. Recovery of mononucleosome-sized fragments for all three minichromosome was observed when the blot was rehybridized with the N2-N3 probe, which does not detect the BamHI-PvuII product (Fig. 3D).
These results indicate that wild-type HS2 alters the chromatin structure of the N1 nucleosome of the ɛ-globin promoter such that internal cleavage by MNase occurs. There is also diminished recovery of N1 mononucleosomal nucleoprotein complex under the conditions of MNase digestion we employed. The 2.5- to 3-fold-lower copy number (see Materials and Methods) of ɛHS2 compared to those of ɛ and ɛHS2(mut) is insufficient to explain this result. It is unclear why we failed to detect either nucleoprotein complexes or naked DNA from the N1 region in crude preparations from ɛHS2 nuclei. Conceivably, the chromatin alterations at N1 resulted in complexes that were not sufficiently stable to survive electrophoresis (6). It is also possible that N1 of ɛHS2 was retained in large chromatin remodeling or transcription complexes and migrated poorly into the gel since we consistently observed some material hybridizing to the N1 probe in or near the wells of the gel.
Histones H3 and H4 at promoter-proximal nucleosomes.
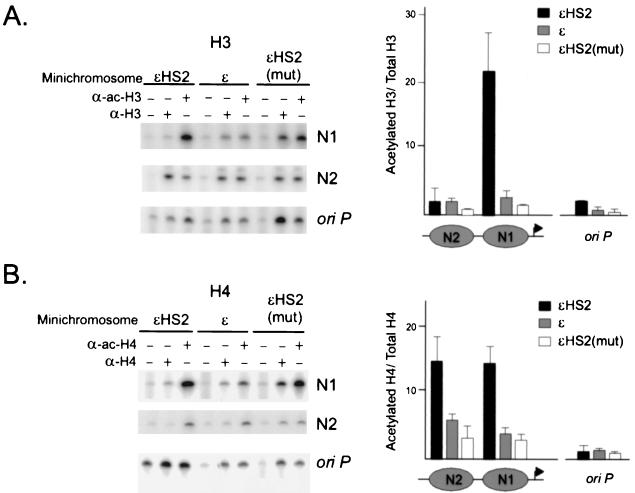
Although the MNase-resistant N1 DNA fragments released from ɛHS2 are not of canonical nucleosome length, much of the N1 sequence is clearly protected from MNase digestion. We hypothesized that such protection might occur if histones remained associated with the N1 DNA even after promoter remodeling and transcription activation. Therefore, we probed the nature of the N1 nucleoprotein complex, performing ChIPs with anti-histone H3 and H4 antibodies and antibodies to their acetylated forms. Soluble chromatin released by MNase digestion of nuclei from clonal cell lines carrying the different minichromosome constructs was precipitated with antibody and protein A/G-agarose beads. The resultant DNA was amplified by PCR with labeled primers and analyzed on acrylamide gels. Identical amounts of DNA were amplified for each of the primer pairs, and the number of PCR cycles was within the linear range of amplification. The positions of the amplified PCR fragments within particular nucleosomes are indicated above the nucleosome map in Fig. 2. A control primer pair amplified vector sequences within the Epstein-Barr virus origin of replication (oriP) in the minichromosome backbone, a region where the histone content and modification status is not expected to vary with the presence or absence of the HS2 enhancer.
Figure 4A and B depict representative examples of the results for H3 and H4, respectively. Similar exposures are presented, and the resultant amplified products were quantitated on a PhosphorImager. The ratios of amplified product obtained with anti-acetylated-H3 and anti-H3 antibodies (Fig. 4A) or with anti-acetylated-H4 and anti-H4 antibodies (Fig. 4B) were calculated for at least three independent experiments, and the results are presented in the accompanying bar graphs (± standard errors of the means [SEM]). As predicted, these ratios were very similar for the different minichromosomes when a primer pair amplifying oriP sequences was used. However, we observed that the ratio of acetylated H3 or acetylated H4 to total H3 or H4 was markedly elevated (H3, more than 20-fold; H4, 15-fold) in the TATA-proximal N1 for wild-type ɛHS2 minichromosomes in comparison with that in the same nucleosome in inactive promoters. Increased acetylation of H4 was seen in the adjacent upstream nucleosome (N2) of active promoters as well. However, N2 did not display differential levels of H3 acetylation in active and inactive promoters, indicating that the modification of H3 that accompanies transactivation by HS2 is highly directed and specific.
FIG. 4.
HS2 increases acetylation of H3 and H4 at N1. (A) Nuclei of K562 cells containing various minichromosomes were digested with MNase under conditions which yield primarily mononucleosomes (see Materials and Methods). The chromatin was immunoprecipitated with no added antibody (−lanes) or with antibodies to either H3 or acetylated H3 (α-H3 or α-ac-H3), and equal masses of the recovered, purified DNA was amplified by PCR for 23 cycles under identical conditions. The locations of sequences which are amplified by the N1 and N2 primer pairs are indicated by filled bars in Fig. 2. Representative PCR results are shown. The data from at least three independent experiments were quantitated on a PhosphorImager, and the ratios of signal obtained with anti-acetylated-H3 and anti-H3 antibodies were determined. The mean results of three or four experiments are plotted in the bar graph at the right of the panel. Error bars represent the SEM. (B) Nuclei of K562 cells were treated as described above except that immunoprecipitations were performed with anti-H4 or anti-acetylated-H4 antibody. PCR conditions and analysis were as described above.
Global histone acetylation does not activate an enhancerless ɛ-globin gene.
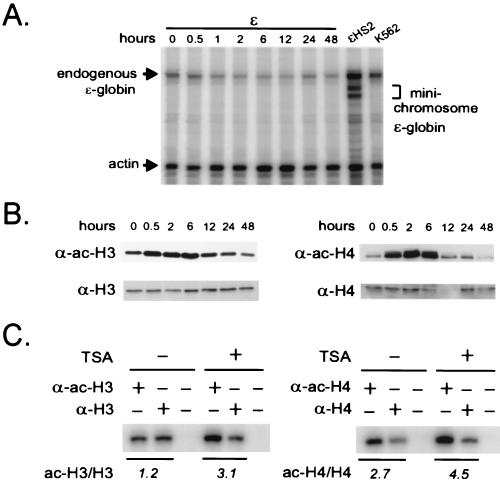
To ask whether histone acetylation is sufficient to transcriptionally activate the ɛ-globin promoter, we used the general deacetylase inhibitor TSA. K562 cell clones carrying the ɛ-globin minichromosome without HS2 are transcriptionally inactive (20). We therefore treated such a clone with 150 ng of TSA per ml over a time course of 48 h. RNA was prepared from equal numbers of cells for each time point and analyzed using a ribonuclease protection assay. ɛ-Globin transcripts from the episomal copy of the gene can be distinguished from endogenous transcripts because they are marked by a mutation in the 5′ untranslated region, resulting in a protected fragment(s) which is shorter than that from the endogenous transcripts (Fig. 5A, lane ɛHS2) (20).
FIG. 5.
Global histone acetylation in the absence of HS2 is insufficient to activate the ɛ-globin gene. (A) RNase protection. A K562 clone carrying ɛ minichromosomes was treated with TSA (150 ng/ml) for 48 h. RNA was prepared at various time points and analyzed by an RNase protection assay as described in Materials and Methods. The position of a band protected by endogenous ɛ-globin RNA transcripts in K562 cell RNA is indicated, as are the positions of bands protected by minichromosomal ɛ-globin RNA in ɛHS2 RNA. Actin RNA served as the load control. (B) Western analysis. Histones were acid extracted from the cells treated for 0 to 48 h with 150 ng of TSA per ml, and 2.5 μg of the acid-extracted proteins was electrophoresed on an acrylamide gel, transferred to a nitrocellulose membrane, and sequentially probed with either anti-H3 and anti-acetylated-H3 antibodies (α-H3 and α-ac-H3, respectively) or anti-H4 and anti-acetylated-H4 antibodies (α-H4 and α-ac-H4, respectively). (C) ChIP. After treatment of K562 cells containing ɛ minichromosomes with 150 ng of TSA per ml for 6 h, nuclei were prepared and digested with MNase and immunoprecipitation was carried out with anti-H3 and anti-acetylated-H3 antibodies or with anti-H4 and anti-acetylated-H4 antibodies as described in the legend to Fig. 4. Coprecipitated DNA was detected by PCR amplification using the N1 primers (Fig. 2).
The results in Fig. 5A show that TSA treatment does not activate transcription of the minichromosomal ɛ-globin gene in the absence of HS2. To ascertain that H3 and H4 became acetylated under the TSA conditions we used, bulk histones were isolated from cells at each time point and separated by electrophoresis and Western blots were prepared. The blots were probed sequentially with either anti-acetylated-H3 and anti-H3 or anti-acetylated-H4 and anti-H4 antibodies (Fig. 5B). Acetylation of bulk H3 and H4 occurred rapidly and reached a maximum of two to five times the untreated level at 6 h. By 12 h of incubation, acetylated H3 and H4 had returned to pre treatment levels, in agreement with the time course of histone acetylation in TSA-treated HeLa cells (8). Investigation of the acetylation status of H3 and H4 at N1 after TSA treatment for 6 h revealed a similar twofold increase in the ratio of amplified product from immunoprecipitation with anti-acetylated-H3 and anti-H3 antibodies or anti-acetylated-H4 and anti-H4 antibodies (Fig. 5C). These results suggest that HS2 may be required to recruit a specific acetylase activity to achieve high-level H3 and H4 acetylation at N1 and to result in remodeling and transcription activation of the ɛ-globin promoter. It also might be that specific acetylated lysines or combinations of acetylated lysines on the same or different histones are required (38).
Histone H1 on ɛ-globin sequences.
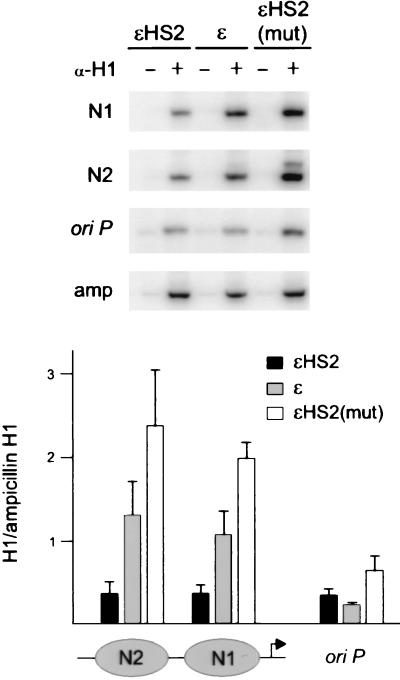
The detection of N1 DNA sequences in ChIPs using anti-acetylated-H3 and anti-acetylated-H4 antibodies argues that N1 is present but in an altered conformation at the active promoters. Since the linker histone H1 is thought to contribute to nucleosome stability and facilitate the formation of higher-order chromatin structures (46), removal of H1 may contribute to an altered and less stable N1 nucleosome. To examine whether H1 was associated with N1, K562 cells containing different minichromosomes were formaldehyde fixed and their nuclei were digested with MNase. Soluble chromatin was immunoprecipitated with an anti-H1 antibody, followed by PCR amplification of N1 and N2 sequences. As controls, oriP and ampicillin sequences in the vector backbone were amplified. We predicted that the ampicillin region, which is inactive in mammalian cells, would contain H1 but that the oriP region might be relatively depleted of H1. In these vector sequences the H1 status was not expected to vary among the different minichromosomes.
Figure 6 depicts representative examples of the results. Similar exposures are presented, and the amplified products were quantitated on a PhosphorImager. The results with the N1, N2, and oriP primers were normalized to the results with the ampicillin region primers and are shown in the bar graphs (± SEM). Actively transcribed ɛHS2 had H1 levels at both N1 and N2 which were comparable to those in oriP and which were reduced compared to those in the ampicillin region. In the minichromosomes where ɛ-globin was not transcribed H1 levels at N1 and N2 were two- to fourfold higher. The relative depletion of H1 at N1 for actively transcribing wild-type ɛHS2 was in agreement with previous observations demonstrating depletion of H1 on transcriptionally active MMTV promoters (7).
FIG. 6.
H1 is depleted at N1 and N2 of transcriptionally active ɛ-globin minichromosomes. Nuclei of K562 cells containing various minichromosomes were cross-linked with paraformaldehyde and digested with MNase (see Materials and Methods). The soluble chromatin was mock immunoprecipitated (−lanes) or was immunoprecipitated with antibodies to H1, and the recovered, purified DNA was amplified by PCR for 25 cycles with primer pairs which amplify N1, N2, or oriP sequences. Amplification of ampicillin sequences served as a control. Representative PCR amplifications are shown. The results were quantitated on a PhosphorImager, and the ratio of signal obtained with each of the test primer pairs to the ampicillin signal was determined. The mean results of three or four experiments are plotted in the bar graph on the right of the panel. Error bars are the SEM.
DISCUSSION
Transcriptional activation of the ɛ-globin gene by HS2 is accompanied by a chromatin structural change which is limited to the N1 nucleosome that overlies the TATA box. The altered structure is reflected in the increased sensitivity of N1 to nucleases and the generation of a DNase I HS, although the mechanism underlying this structural transition is unknown. We now find that at least some N1 sequences in promoters activated for transcription are still associated with histones H3 and H4 and are highly enriched for the acetylated forms of these histone proteins, particularly for H3. This specific, directed modification at the TATA-proximal nucleosome is consistent with a mechanism in which the acetylation of histones makes promoter sequences available to transcription factors that are necessary for high-level activity of the gene (27, 44). However, we find that global histone acetylation at the levels attainable in our experiments is insufficient to transcriptionally activate the ɛ-globin promoter in the absence of HS2. Transcription activation appears to require targeted modification of N1 mediated by the distant HS2 enhancer and perhaps other HS2-dependent activities as well.
Chromatin structure of a DNase I HS.
We previously reported that a functional HS2 enhancer was required for transcriptional activation of the ɛ-globin gene and the appearance of a DNase I HS at N1 (20). The presence of DNase HSs in genetic regulatory elements has often been assumed to indicate a region of histone-free DNA (45). An early study reported that a 114-bp restriction enzyme fragment of the chick β-globin promoter could be released from red blood cell nuclei as naked DNA (30). Using similar methodology, we were unable to detect naked DNA released from the N1 region of the actively transcribed human ɛ-globin promoter in K562 cell nuclei, even though N1 is more sensitive to nuclease digestion than in inactive promoters. However, when purified DNA was analyzed, MNase digestion revealed that a subnucleosome-sized N1 DNA fragment was released from actively transcribed promoters, while digestion of inactive promoters produced DNA fragments of nucleosomal length. Subnucleosomal fragments have been observed after hormone activation of the MMTV promoter in vivo (2). In our ChIP experiments, we readily detected N1 DNA from active promoters in association with H3 and H4, indicating that nuclease hypersensitivity can occur in the presence of histones. In contrast, we were unable to immunoprecipitate chromatin from HS2 using this methodology (data not shown). The differences among the ɛ-globin promoter, HS2, and the chick β-globin gene in nuclease digestion experiments support the notion that DNase I HSs may differ from one another in the extent of nucleosome disruption or the structure of the nucleoprotein complex.
Histone acetylation at a developmentally regulated promoter.
A number of activities have been demonstrated to be involved in nucleosome disruption and chromatin remodeling events, including the binding of transcription factors (15), histone acetylation or other posttranslational modifications, and ATP-dependent remodeling complexes such as SWI-SNF of Saccharomyces cerevisiae and NURF of Drosophila melanogaster (46). During development, transcriptional activation of globin genes is associated with the appearance of a DNase I HS at each promoter (21). Our data indicate that the increase in nuclease sensitivity of the transcriptionally activated ɛ-globin gene is associated with histone hyperacetylation, a situation which parallels that of the endogenous locus (see below). Others have also reported histone modification that is localized around promoters. Hyperacetylated histones are associated with the promoters of yeast genes activated by the acetyltransferase Gcn5, and increased H3 and H4 acetylation attributed to CBP was found in a region that could accommodate two or three nucleosomes surrounding the induced beta-interferon promoter (26, 31). We observe that hyperacetylation at the N1 nucleosome depends on linkage to a functional HS2 enhancer. The immunoglobulin H heavy-chain enhancer also mediates acetylation of histones in the promoter region of an episomal c-myc gene, although the effects are more widespread, extending over substantial distances up- and downstream from the promoter (28). Hence, enhancers may function at least in part by directing promoter histone acetylation.
Interestingly, the ratio of acetylated H3 to H3 at the active ɛ-globin promoter N1 nucleosome was much higher than the ratio of acetylated H4 to H4. In addition, we found increased H4 acetylation, but not H3 acetylation, in the adjacent upstream N2 nucleosome. Others have recently made complementary observations of the human globin locus. While general histone acetylation was observed across the open locus, hyperacetylation of H3 was found specifically at the LCR and at the β-globin gene. β-Globin promoter activity, which is LCR dependent, correlated with the localized H3 acetylation (35). In other studies, the transcriptionally active mb-1 promoter and the induced beta interferon promoter also showed a preferential acetylation of H3 relative to that of H4 (18, 31). Thus, an association exists between promoter chromatin remodeling, hyperacetylation of H3, and transcription activation. Our data along with that from the human globin locus and elsewhere (see above) indicate that H3 hyperacetylation may be targeted to highly restricted regions, in some cases as small as a single nucleosome.
Global histone acetylation and transcription activation.
TSA is a general inhibitor of histone deacetylases, and their inhibition results in elevated acetylation of histones in nuclear chromatin and altered expression of a variety of genes (43, 48). TSA treatment of cells carrying minichromosomes with the ɛ-globin gene alone resulted in a three- to fivefold increase in acetylated H3 and H4 as detected by Western blot analysis of bulk histones. ChIP assays revealed that acetylation of H3 and H4 at N1 increased by similar amounts. However, our studies indicate that treatment with TSA was insufficient to activate ɛ-globin transcription from minichromosomes in the absence of an enhancer. Interestingly, in the immunoglobulin H enhancer study cited above, global histone acetylation by TSA mimicked the effect of the enhancer on only one of the two c-myc promoters (28). H4 hyperacetylation via deacetylase inhibitors, while insufficient on its own, appeared to be a prerequisite for an additional signal necessary for activation of the chromosomal c-fos gene (1). These observations suggest that, while generalized histone acetylation may contribute to the regulation of transcription, additional regulatory events or targeted histone modifications are also likely to be necessary for the control of gene expression.
Accumulating evidence suggests that acetylation of the amino-terminal tails of H3 and H4 may be a principal regulator of transcription factor access to nucleosomal DNA (44). The N1 nucleosome spans recognition motifs for CACCC- and CCAAT-binding factors, the TATA box, and a GATA-1 site. Hence, acetylation of N1 H3 and H4 tails may play a key role in regulating the establishment of a transcription complex at the promoter. The role of the HS2 enhancer may be to recruit activities which are responsible for high-level, directed histone acetylation at N1. Such recruitment may occur via association with the transcriptional machinery, an erythroid-cell-specific transcriptional activator, or an activity independent of acetylation which is otherwise needed for transcriptional activation (33, 39). CBP is one candidate acetylase which plays important roles in hematopoietic cell differentiation (4). CBP has been shown to interact with the HS2-binding transcriptional activator NF-E2, with GATA-1, and with the human RNA polymerase II complex (5, 9, 10).
Depletion of histone H1 at remodeled N1.
Our experiments also reveal a relationship between a functional enhancer and promoter H1 content. Histone H1 binds to the linker region between nucleosome cores and is thought to limit nucleosome translational mobility and contribute to higher-order chromatin folding (16). However, little is known about how H1 affects transcription. Although H1 can be detected on an actively transcribing Balbiani ring gene, it also reduces the transcriptional efficiency of specific templates in vivo and transcription factor binding in vitro (14, 24, 36). Thus, depletion of H1 may facilitate both transcription initiation and elongation. We were able to detect H1 bound to active as well as inactive promoters. However, our ChIP experiments also indicated that H1 is depleted from N1 (and N2) sequences when transcription of the ɛ-globin gene is active. Furthermore, while the active promoter had H1 levels comparable to those of the replication origin oriP, the inactive promoters had H1 levels comparable to or slightly higher than those of the inactive ampicillin region. The depletion of H1 in combination with elevated H3 acetylation at N1 in active promoters might contribute to destabilization of the N1 nucleosome, perhaps as reflected in decreased detection of N1 nucleoprotein complexes (Fig. 2A). The data are consistent with a report that H1 is bound to the repressed MMTV promoter and depleted after hormone stimulation (7).
Protection of a subnucleosomal DNA fragment and detection of N1 DNA sequences in immunoprecipitations using anti-H3 and -H4 antibodies, combined with the diminished recovery of nucleoprotein complexes containing N1, argue that N1 is present but in an altered conformation in at least some of the active promoters. These results are consistent with a number of models: (i) the depletion of H1, H3 and H4 acetylation result in an N1 nucleosome that is packaged into chromatin but in such a way that it is no longer fully protected from nuclease attack, (ii) interaction of transcriptional activators and/or the transcriptional machinery with N1 DNA sequences result in loosening of histone-DNA contacts such that the resultant complex is not stable in electrophoresis, and (iii) histones at N1 are associated with other components of the transcription machinery and dynamically interact with N1 sequences. These possibilities are not mutually exclusive.
ACKNOWLEDGMENTS
We thank David Clark, Jurrien Dean, David Jackson, and Sharon Roth for discussions and critical review of the manuscript and Mark Groudine for communicating results before publication.
REFERENCES
- 1.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 2.Belikov S, Gelius B, Almouzni G, Wrange O. Hormone activation induces nucleosome positioning in vivo. EMBO J. 2000;19:1023–1033. doi: 10.1093/emboj/19.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender M A, Bulger M, Close J, Groudine M. β-Globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 4.Blobel G A. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 5.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyes J, Omichinski J, Clark D, Pikaart M, Felsenfeld G. Perturbation of nucleosome structure by the erythroid transcription factor GATA-1. J Mol Biol. 1998;279:529–544. doi: 10.1006/jmbi.1998.1783. [DOI] [PubMed] [Google Scholar]
- 7.Bresnick E H, Bustin M, Marsaud V, Richard-Foy H, Hager G L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W Y, Townes T M. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc Natl Acad Sci USA. 2000;97:377–382. doi: 10.1073/pnas.97.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng X, Reginato M J, Andrews N C, Lazar M A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirillo L A, Zaret K S. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 12.Cohen-Kaminsky S, Maouche-Chretien L, Vitelli L, Vinit M A, Blanchard I, Yamamoto M, Peschle C, Romeo P H. Chromatin immunoselection defines a TAL-1 target gene. EMBO J. 1998;17:5151–5160. doi: 10.1093/emboj/17.17.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elgin S C R. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1995;263:19259–19262. [PubMed] [Google Scholar]
- 14.Ericsson C, Grossbach U, Bjorkroth B, Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990;60:73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- 15.Fascher K D, Schmitz J, Horz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenfeld G, McGhee J D. Structure of the 30 nm chromatin fiber. Cell. 1986;44:375–377. doi: 10.1016/0092-8674(86)90456-3. [DOI] [PubMed] [Google Scholar]
- 17.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 18.Forrester W C, Fernandez L A, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–3014. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong Q H, McDowell J C, Dean A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the ɛ-globin gene in vivo by 5′ hypersensitive site 2 of the β-globin locus control region. Mol Cell Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groudine M, Kohwi-Shigematsu T, Gelinas R, Stamatoyannopoulos G, Papayannopoulou T. Human fetal to adult hemoglobin switching: changes in chromatin structure of the β-globin gene locus. Proc Natl Acad Sci USA. 1983;80:7551–7555. doi: 10.1073/pnas.80.24.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez G, Griffiths S D, Ford A M, Greaves M F, Enver T. Activation of the β-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci USA. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juan L J, Utley R T, Vignali M, Bohm L, Workman J L. H1-mediated repression of transcription factor binding to a stably positioned nucleosome. J Biol Chem. 1997;272:3635–3640. doi: 10.1074/jbc.272.6.3635. [DOI] [PubMed] [Google Scholar]
- 25.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 28.Madisen L, Krumm A, Hebbes T R, Groudine M. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell J C, Dean A. Structural and functional cross-talk between a distant enhancer and the epsilon-globin gene promoter shows interdependence of the two elements in chromatin. Mol Cell Biol. 1999;19:7600–7609. doi: 10.1128/mcb.19.11.7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGhee J D, Wood W I, Dolan M, Engel J D, Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- 31.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 32.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 34.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 35.Schubeler D, Francastel C, Cimbora D M, Reik A, Martin D I, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 36.Sera T, Wolffe A P. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Mol Cell Biol. 1998;18:3668–3680. doi: 10.1128/mcb.18.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steger D J, Workman J L. Remodeling chromatin structures for transcription: what happens to the histones? Bioessays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 38.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 39.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 40.Truss M, Bartsch J, Schelbert A, Hache R J, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuan D, Solomon W, Li Q, London I M. The “beta-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner B M, Fellows G. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur J Biochem. 1989;179:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- 43.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 44.Vitolo J M, Thiriet C, Hayes J J. The H3–H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Mol Cell Biol. 2000;20:2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallrath L L, Lu Q, Granok H, Elgin S C. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 46.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]