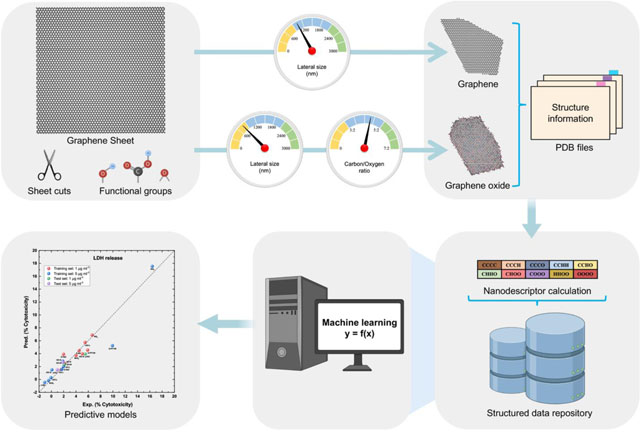
Abstract
Modern nanotechnology provides efficient and cost-effective nanomaterials (NMs). The increasing usage of NMs arises great concerns regarding nanotoxicity in humans. Traditional animal testing of nanotoxicity is expensive and time-consuming. Modeling studies using machine learning (ML) approaches are promising alternatives to direct evaluation of nanotoxicity based on nanostructure features. However, NMs, including two-dimensional nanomaterials (2DNMs) such as graphenes, have complex structures making them difficult to annotate and quantify the nanostructures for modeling purposes. To address this issue, we constructed a virtual graphenes library using nanostructure annotation techniques. The irregular graphene structures were generated by modifying virtual nanosheets. The nanostructures were digitalized from the annotated graphenes. Based on the annotated nanostructures, geometrical nanodescriptors were computed using Delaunay tessellation approach for ML modeling. The partial least square regression (PLSR) models for the graphenes were built and validated using a leave-one-out cross-validation (LOOCV) procedure. The resulted models showed good predictivity in four toxicity-related endpoints with the coefficient of determination (R2) ranging from 0.558 to 0.822. This study provides a novel nanostructure annotation strategy that can be applied to generate high-quality nanodescriptors for ML model developments, which can be widely applied to nanoinformatics studies of graphenes and other NMs.
Keywords: Nanotoxicity, Graphenes, Nanostructure annotation, Nanodescriptors, Machine learning
Graphical Abstract
1. Introduction
Modern nanotechnology is a critical technology for sustainable nanomaterial (NM) developments in both basic research and commercial applications [1–3]. The global nanotechnology market is expected to exceed $125 billion by 2024 [4]. Due to their diversity in structures, properties, and bioactivities, NMs have gained prominence in fields such as food security and safety processing [5–8], precision agriculture [9–11], clean energy [12], and clinical medicine [13, 14]. However, the increasing use of NM is coupled with concerns for nanotoxicity [15–21]. There is an urgent need for comprehensive risk assessments of both emerging and existing NMs. Traditional experimental methods to evaluate NM toxicity, which often use large numbers of animals, are expensive and time-consuming. With the development of Machine Learning (ML) approaches, computational modeling is emerging as an alternative for predicting the behavior of NMs in biological environments [22] and evaluating their nanotoxicity [23, 24]. Quantitative Nanostructure Activity Relationship (QNAR) modeling using ML approaches reveals the relationships between NMs’ structural features and biological activities such as toxicity in a quantitative manner [25]. In the Organization for Economic Co-operation and Development (OECD) 2022 report on risk assessment for NMs, ML-based modeling study was highlighted as a promising strategy for rapid toxicity screening of NMs [26].
ML has been successfully utilized in modeling studies of physicochemical properties and bioactivity for small molecules [27–31]. However, ML applications in computational nanotoxicology are limited because of lacking nanotoxicity data and the difficulties of nanostructure annotations [32]. Based on the EU-US Nanoinformatics Roadmap 2030 guidance for ML model development, the good performance of predictive nanotoxicity models relies heavily on the well-defined descriptors that tie nanostructures and physicochemical properties to the bioactivities of NMs [33]. Nanodescriptors represent NMs’ chemical and physical identities, intrinsic properties and extrinsic properties, which can be classified as experimental, empirical, and geometrical [34]. Experimental results of NMs’ morphological properties and physicochemical properties such as size [35], magnetic properties [36], and zeta potentials [37], can be used as descriptors for modeling purposes. However, these measured nanodescriptors vary significantly with different experimental conditions and may not be reliable without prior references. Empirical descriptors have been developed using molecular simulations and quantum chemistry [38–40], however, these descriptors need expertise for selecting appropriate force fields and calculation methods to generate descriptors. Geometrical descriptors provide more detailed information on nanostructures by annotating the important structural features such as molecular structures, mechanical properties, and electrical properties [23]. In our previous studies, novel geometrical nanodescriptors were developed by employing Delaunay tessellation and atomic properties. The nanodescriptors quantified nanostructures, by simulating the surface chemistry, to develop ML models for NMs such as metallic nanoparticles [41, 42]. However, for more complex NMs, such as two-dimensional nanomaterials (2DNMs) like graphenes, nanostructure annotations and simulations were not previously successful due to complex nanostructures.
As 2DNMs, graphenes are carbon-based NMs consisting of single- or few-layer atoms arranged in a planar honeycomb structure and are being widely applied in biomedicines, biosensors, and solar cells [43, 44]. Graphenes usually present planar structures with irregular edges, which is a key factor influencing their effects on cellular uptake and cytotoxicity [45–47]. Lateral size has long been shown to modulate the pathogenicity of graphenes [48, 49], but subtle surface modifications can also influence their bioactivity [46, 47, 50]. For example, primary endothelial cells develop more cytoplasmic protrusions and are more prone to losing their barrier function when exposed to increasingly oxidized graphene sheets [51]. The type of surfactant necessary to disperse the more hydrophobic graphenes and its concentration may also affect their cytotoxicity [52]. As graphene derivatives, graphene oxides have carbon frames oxidized with oxygen-containing functional groups on their edges and basal plane. The diverse graphene structures, especially the irregular edges, are difficult to annotate precisely through simulating experimental conditions, which prohibits the use of geometrical nanodescriptors in ML modeling.
In this study, we developed a novel structure annotation strategy by (1) simulating the nanostructures of synthesized graphenes; (2) developing geometrical nanodescriptors to characterize the structure features of graphenes; and (3) using the calculated nanodescriptors to develop ML models for various toxicity endpoints of graphenes. In a previous study, we synthesized and assessed the toxicities of graphenes and graphene-like inorganic 2DNMs [53], which generated a high-quality dataset for graphene toxicity modeling. The irregular structures of the graphenes and inorganic 2DNMs were constructed by modifying the number of vertices and edges on virtual nanosheets. For graphene oxides, functional groups including hydroxyl, epoxy, and carboxyl groups were added on the graphene surface to reach the carbon/oxygen (C:O) ratio experimentally measured. The annotated nanostructures were saved as Protein Data Bank (PDB) files. Novel geometrical nanodescriptors were computed from the annotated nanostructures using the Pauling electronegativity and the Delaunay tessellation approach. Partial least square regression (PLSR) models were developed for various nanotoxicity endpoints including LDH release, cell viability, oxidative stress, and apoptosis. This structure annotation strategy shows great potential in developing ML models for nanotoxicity predictions of NMs with complex nanostructures.
2. Material and methods
2.1. 2DNMs dataset
The 11 2DNMs used for ML modeling were synthesized and characterized in our previous study [53]. Graphene oxides were synthesized according to a modified Hummer’s method [54]. Reduced and partially reduced graphene oxides were synthesized by controlled reduction using L-ascorbic acid and size-sorted graphene oxide as the starting material. Graphene and inorganic 2DNMs were synthesized by liquid-phase exfoliation in the presence of sodium cholate hydrate (NaC) or Pluronic® F-108 (PF108). The toxicities of all the 2DNMs were investigated in a triculture model of small intestinal epithelium. LDH release, cell viability, and oxidative stress were tested after treating with 1 μg ml−1 and 5 μg ml−1 2DNMs and apoptosis was tested after treating with 5 μg ml−1 2DNMs. LDH release (plasma membrane damage) was calculated as % of LDH in lysed control. Cell viability (mitochondrial enzymatic activity) was expressed as % of activity (fluorescence) measured in cells treated with control fasting food model (FFM) digesta. Caspase 3/7 activity (apoptosis) was expressed as fold changes relative to that in cells exposed to FFM digesta. Oxidative stress (ROS generation) was expressed as fold changes relative to that in cells exposed to FFM digesta. The Pearson correlation analysis was implemented using the Python package SciPy 1.6.2 to test for results associations between two doses of 2DNMs in LDH release, cell viability, and oxidative stress.
2.2. Virtual 2DNMs library construction
The construction of virtual 2DNMs (v2DNMs) was performed by the new graphenes generation toolkit of VINAS, which took the target lateral size and C:O ratio as the input parameters. Briefly, a 2DNM sheet was first created with a lateral size larger than the target lateral size. Then, the irregular 2DNM was generated by scaling down the initial lateral size to the target and modifying the number of vertices and edges on the sheet. For GOs, rGOs, and prGO, the functional groups such as hydroxyl, epoxy, and carboxyl groups were randomly placed on the irregular graphene frame until the C:O ratio reached the target value. The structure information of annotated 2DNMs was then saved as PDB files. To avoid potential inconsistency of calculated nanodescriptor results due to different irregular shapes and randomly distributed functional groups, total 30 v2DNMs were generated for each 2DNMs in the dataset.
2.3. Nanodescriptor generation
Based on the generated v2DNMs, nanodescriptors were calculated using the new descriptor calculation toolkit of VINAS coded in Java 1.8.0_301. There are eight types of atoms in these v2DNMs: C (carbon), O (oxygen), H (hydrogen), N (nitrogen), S (sulfur), B (boron), Mo (molybdenum) and W (tungsten). Every four nearest atoms (e.g., CCCC, CCHO, etc.) that can form a trigonal planar, quadrangulation, or tetrahedron were identified from the v2DNM structures using the Delaunay tessellation approach. In 3D space, atoms within a distance cutoff of 2.0 Å and 2.5 Å played a key role in the physicochemical properties of GRMs and graphene-like inorganic 2DNMs, respectively [55–58]. Accordingly, the distance between any two atoms in a formed 3D tetrahedron fragment was set to within 2.0 Å in GRMs and within 2.5 Å in inorganic 2DNMs. In 2D planar, atoms within a distance cutoff of 2.5 Å were important for the 2D features of 2DNMs, the distance between any two atoms in a formed 2D trigonal planar or quadrangulation fragment was set to within 2.5 Å in all 2DNMs [59, 60]. The geometrical nanodescriptors were calculated without considering the atom order within a quadrangulation or tetrahedron (e.g., CCCO was the same as COCC). As described in our previous study [41], the procedure of geometrical nanodescriptors calculation can be summarized as follows: (1) the value of each quadrangulation or tetrahedron was the sum of the electronegativity of four atoms in this quadrangulation or tetrahedron. (2) The descriptor value in each v2DNM was computed as the value of the relevant quadrangulation or tetrahedron multiplied by its occurrences. (3) The final descriptor values of a 2DNM were averaged from results obtained from 30 v2DNMs. In this study, additional surfactant descriptors to describe the surfactants for experimental testing were also used.
2.4. Machine learning modeling
ML models were developed using the PLSR algorithm. PLSR is a method that combines principal component analysis and multiple regression [61]. PLSR performs a descriptor dimension reduction procedure and constructs a set of components that accounts for as much as possible of the total descriptors variance in the dataset, which can avoid multicollinearity and model overfitting [62, 63]. It is suitable for the modeling of small training sets using large sets of descriptors [64–66]. In this study, the generated geometrical nanodescriptors and surfactant descriptors were used to develop PLSR models for 2DNMs’ toxicities. The PLSR algorithm was implemented using scikit-learn 0.24.1 [67]. The original dataset was split into training and test sets with the ratio of 9:2 for cell viability, LDH release, and oxidative stress and 8:1 for apoptosis. The training sets were used to build models and relevant test sets were used for the prediction purpose. The leave-one-out cross-validation (LOOCV) procedure was performed to find the optimal number of components for modeling using the training set. Briefly, a single 2DNM was excluded from the training set and the remaining 2DNMs were used for model development. Then the developed models were used to predict the excluded 2DNM. This procedure was repeated until every 2DNM in the training set was used for prediction purpose one time. In the LOOCV procedure, the root mean square error (RMSE) and coefficient of determination (R2) were used as the parameters to identify the optimal number of components and the best PLSR model for each toxicity endpoint as follows:
| (1) |
| (2) |
where YEXP was the experimental results, YPRED was the predicted results, YMEAN was the mean value of the experimental results, n was the number of samples. Then, the external test set was used to evaluate the performance of selected PLSR models.
3. Results and Discussion
3.1. 2DNMs dataset overview
The information of the original 2DNMs dataset is summarized in Table 1. The dataset consisted of two types of 2DNMs: (1) eight graphene-related materials (GRMs) including two graphenes, three graphene oxides (GO), two reduced graphene oxides (rGO), and one partially reduced graphene oxide (prGO); (2) three graphene-like inorganic 2DNMs including one hexagonal boron nitride (h-BN), one molybdenum disulfide (MoS2), and one tungsten disulfide (WS2). Amphiphilic GOs were dispersed in water and the remaining 2DNMs, which are hydrophobic NMs, were dispersed in NaC or PF108 solutions for experimental testing. The toxicity data of the 2DNMs were summarized in Table S1. After treatment with 1 μg ml−1 2DNMs, the values for LDH release, cell viability, and oxidative stress range from 1.61% to 6.63%, 95.95% to 113.88%, and 0.81 to 2.23 (fold change) respectively. After treatment with 5 μg ml−1 2DNMs, the toxicity results of LDH release, cell viability, oxidative stress, and apoptosis range from −1.08% to 16.40%, 76.03% to 119.96%, 0.80 to 2.15 (fold change), and −2.50 to 0.55 (fold change) respectively. Pearson correlation analysis between the experimental results of 1 μg ml−1 and 5 μg ml−1 2DNMs were shown in Fig. S1. The resulted Pearson correlation coefficient r and two-tailed p-value showed that two doses 2DNMs have good correlation in oxidative results while they were not correlated in LDH release and cell viability results.
Table 1.
Key physicochemical properties of the 2DNMs used in this study.
| 2DNM | Lateral size (nm ± SD) | C:O ratio | Surfactant | |
|---|---|---|---|---|
| Graphene-related | Small-sized graphene in NaC (G-NaC) | 184 ± 23 | N/A | Na-Cholate |
| Small-sized graphene in PF108 (G-PF108) | 206 ± 51 | N/A | PF108 | |
| Small-sized graphene oxide (GO-S) | 271 ± 34 | 64:35 | Water | |
| Medium-sized graphene oxide (GO-M) | 462 ± 114 | 61:39 | Water | |
| Large-sized graphene oxide (GO-L) | 1560 ± 750 | 61:38 | Water | |
| Small-sized reduced graphene oxide (rGO-S) | 411 ± 79 | 78:22 | Na-Cholate | |
| Large-sized reduced graphene oxide (rGO-L) | 2015 ± 674 | 78:22 | Na-Cholate | |
| Partially reduced graphene oxide (prGO) | 357 ± 42 | 72:28 | Na-Cholate | |
| Inorganic | Hexagonal boron nitride (h-BN) | 149 ± 12 | N/A | Na-Cholate |
| Molybdenum disulphide (MoS2) | 428 ± 103 | N/A | Na-Cholate | |
| Tungsten disulphide (WS2) | 323 ± 28 | N/A | PF108 |
3.2. Annotating 2DNMs by constructing virtual 2DNMs
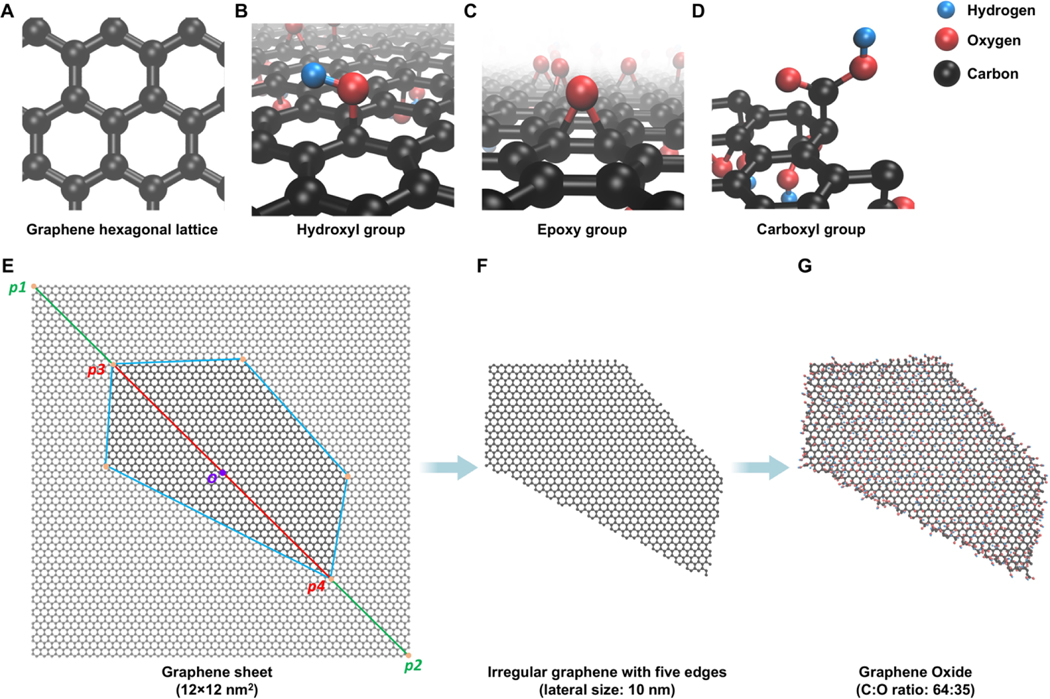
The in-house Virtual Nanostructure Simulations (VINAS) toolbox coded in Python 3.8 was used to construct all v2DNMs in this study with specified lateral size and C:O ratio as the input parameters. The key structure features of GRMs were shown in Fig. 1A-D. Pristine graphene represented a carbon frame arranged in a hexagonal lattice (Fig. 1A). Based on the Lerf-Klinowski-type structural models [68], hydroxyl groups were added randomly on the carbon frame (Fig. 1B); epoxy groups were placed on two adjacent connected carbon atoms (Fig. 1C); and carboxyl groups were (Fig. 1D) to form GO, rGO and prGO respectively. Each carbon atom on the graphene frame can be modified by adding one functional group and the functional groups can be added either above or below the carbon frame layer. The vGRMs generation procedure has three steps. First, a graphene sheet was created. The actual sheet’s lateral size, which was based on the experimental results, was marked as p3p4 on the diagonal line p1p2 (Fig. 1E). The virtual graphene sheet was generated by randomly forming other vertices with their distances to the sheet center o no larger than half of the target lateral size. Then these vertices were connected to form a polygon with an irregular shape, and the remaining sheet outside the polygon was deleted (Fig. 1F). To generate GO, rGO, and prGO, the functional groups were randomly added on the surface of the graphene to reach the target C:O ratio. For three inorganic 2DNMs in the dataset, the constructions of corresponding v2DNMs followed the same procedure except using different atoms and associated residues to generate the initial sheet. For example, h-BN had a lattice with B and N atoms in a hexagonal formation (Fig. 2). MoS2 and WS2 showed similar atom arrangements in which S and Mo or WWWW atoms were connected by covalent bonds as S-Mo-S and S-W-S respectively (Fig. 2). In the end, the constructed v2DNMs were saved as individual PDB files. All 11 2DNMs’ PDB files and bioactivity data are downloadable from in-house nanoinformatics portal (http://vinas-toolbox.com/explore_group/2DNMs).
Fig. 1.
Schematic workflow of v2DNMs development. The three-dimensional (3D) structures of graphene hexagonal lattice (A), hydroxyl group (B), epoxy group (C), and carboxyl group (D) are visualized by CPK drawing method in VMD. The v2DNMs generated in the workflow (E-G) are also shown by CPK drawing method in VMD. The construction of irregular graphene with 10 nm lateral size and five edges (F) is based on a 12×12 nm2 graphene sheet (E). Then, a graphene oxide with C:O ratio of 64:35 (G) is constructed based on the generated irregular graphene. The C, O, and H atoms are represented in black, red, and blue.
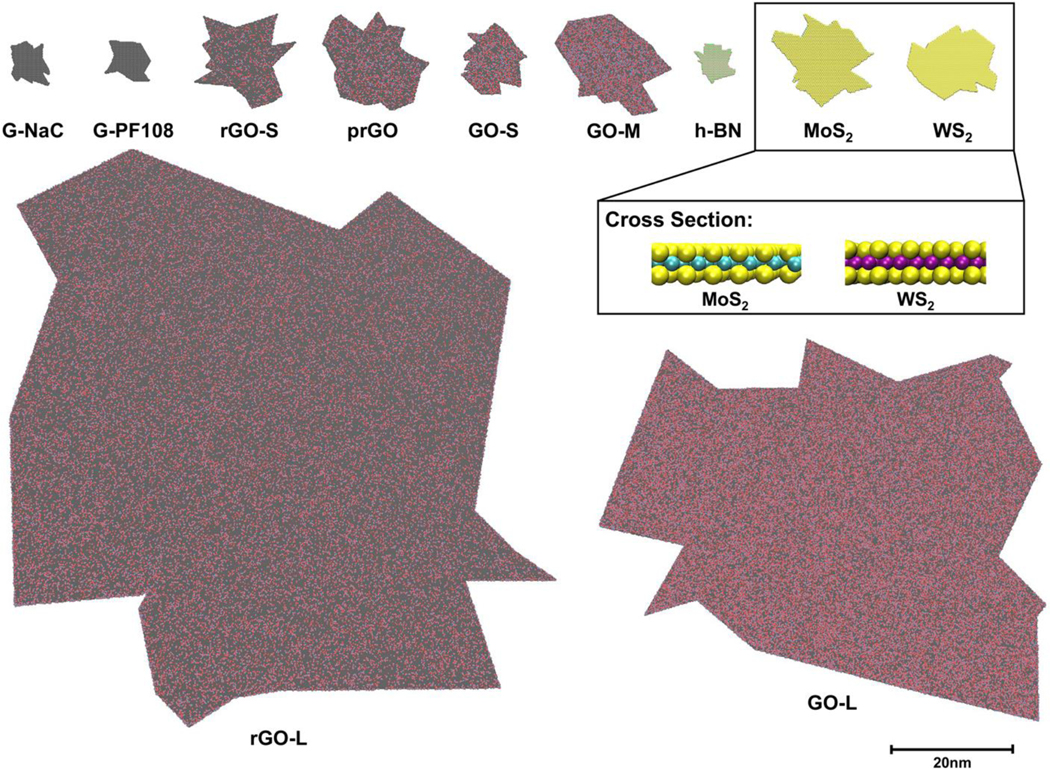
Fig. 2.
Visualization of the v2DNMs library. The 2DNMs of the dataset vary in shapes, lateral sizes, atom types, and surface groups. The 3D structures of the v2DNMs are rendered by VDW drawing method in VMD. The C, O, H, N, B, S, Mo, and W atoms are represented in black, red, blue, green, pink, yellow, cyan, and purple.
3.3. Nanostructure visualizations
Using the PDB files consisting of annotated 2DNMs, the nanostructures in the dataset can be rendered by visual molecular dynamics (VMD) [69] using van der Waals (VDW) method. All the GRMs and inorganic 2DNMs have irregular polygon structures that varied in sizes, edge numbers, atom types, and surface groups (Fig. 2). Specifically, G-NaC and G-PF108 are graphenes with different lateral sizes, which were constructed only with carbon atoms. GOs, rGOs, and prGO are GRMs oxidized with hydroxyl, epoxy, and carboxyl groups with C:O ratios ranging from 61:39 to 78:22. h-BN has a structure similar to graphene, where B and N atoms alternately constructed hexagonal lattices instead of carbons. MoS2 and WS2 showed a sandwich-like structure with two hexagonal planes of S atoms and a hexagonal plane of Mo or W atoms in the middle. All the annotated 2DNM structures were used to calculate nanodescriptors for modeling purposes.
3.4. Nanodescriptors generations
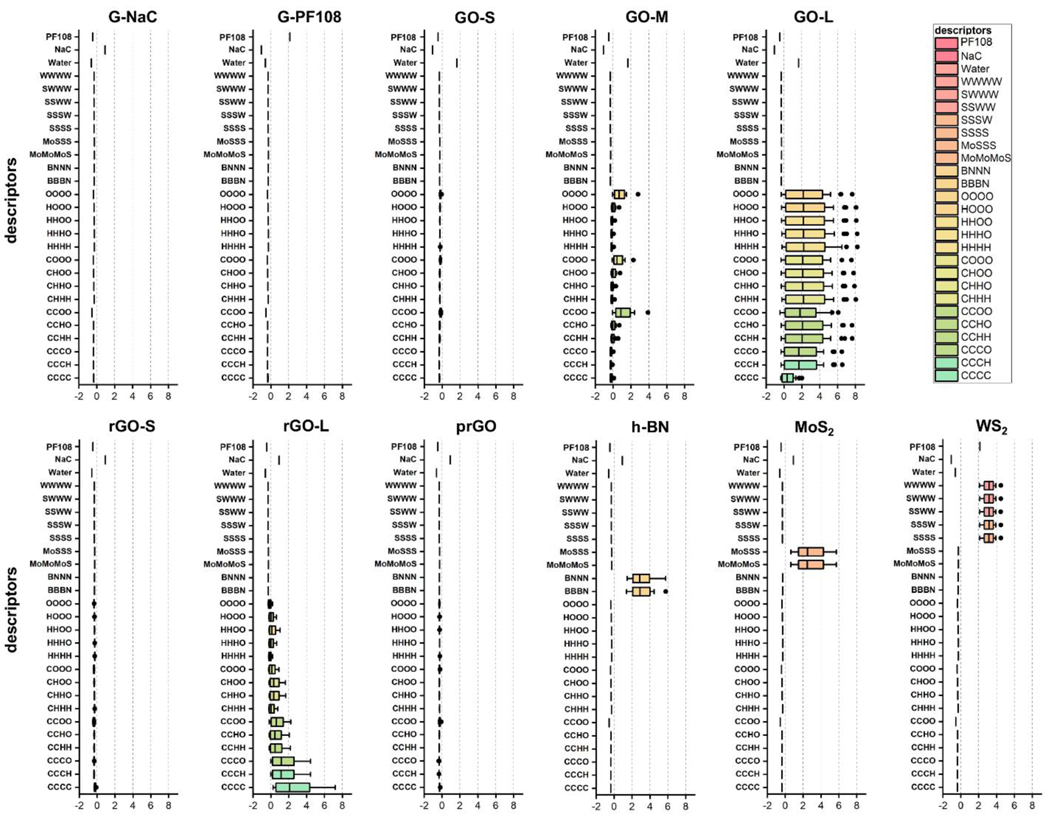
To account for the uncertainty of experial synthesis of 2DNMs, 30 v2DNMs were constructed for every 2DNM to mimic the diverse irregular structures of corresponding 2DNM [53]. The average lateral sizes of generated v2DNMs for each 2DNM are consistent to experimental conditions and were shown in Table 1. The designed nanodescriptors should be able to describe the diverse 2DNM and but not be sensitive to changes of irregular shapes of 2DNM due to the synthesis uncertainty. Based on the Delaunay tessellation approach, quadrangulations or tetrahedrons were generated for each four nearest neighboring atoms on v2DNM structure as nanodescriptors. The ientified quadrangulations or tetrahedrons can describe v2DNM structure features on the surface, which account for their properties, activities and toxicities. The value for each identified quadrangulation or tetrahedron was the sum of electronegativity values of the four atoms within this trigonal planar, quadrangulation or tetrahedron. For example, the value of CCCC was 10, obtained from the sum of four carbon electronegativity values (2.5 × 4). The atomic electronegativity values of all atoms were summarized in Table 2. Furthermore, a nanodescriptor value for a v2DNM was calculated as the value of each trigonal planar, quadrangulation, or tetrahedron electronegativity multiplied by its occurrences in this v2DNM. As mentioned above, 30 v2DNMs were constructed for every 2DNM. Thus, the nanodescriptors for a 2DNM were calculated by averaging descriptor values of 30 v2DNMs. Since surfactants can influence 2DNMs’ stability and surface chemistry [70–72], additional categorical descriptors were introduced to account for the effects of different surfactants on 2DNMs’ properties. Based on the surfactants used in the experimental testing (Table 1), the surfactant descriptors of the corresponding 2DNM were binary descriptors with “0” representing the absence and “1” representing the presence of the associated surfactant. Binary descriptor is widely used in ML modeling for drug discovery, such as MACCS keys fingerprints and PubChem fingerprints [73–76]. The results of calculated nanodescriptors for all 2DNMs were provided in Supporting Information Excel file E1. The standardized nanodescriptor values in each 2DNM were shown in Fig. 3. Some geometrical nanodescriptors have relatively high values for specific 2DNMs. Since the structures of G-NaC and G-PF108 were constructed with carbon atoms, the values of CCCC descriptor were higher than the other 2DNMs. In graphene lattice, six carbons located at the hexagon apexes with two sublattices, A and B. The carbon atom of one with three nearest neighbors of the other sublattice, which is one of the important features influencing graphene’s activity [59, 60]. As shown in Fig. S2, CCCC descriptors in graphene were developed using the trigonal planar geometry based on the relationship of sublattice A and B, which can reflect the structure feature of armchair edges (a and b)/zigzag (c), dangling atoms (d and e), and general atoms (f and g) in the plane. It can also extract the features of convex corner. For future explorations, descriptors reflecting 2DNMs’ frame structures can be divided into different subtypes. For example, CCCC descriptors reflecting the edge effect features can be separated from the general CCCC, and different weights can be given to these subtype descriptors for modeling purposes. Besides CCCC descriptor, surfactant descriptors NaC and PF108 showed high values in G-NaC and G-PF108, respectively (Fig. 3). These two surfactant descriptors were also used to further differentiate G-NaC and G-PF108. For GOs, rGOs and prGO, hydroxyl, epoxy and carboxyl groups were added on the graphene frame, so the values of nanodescriptors consisting of C, H, and O atoms were larger than the other nanodescriptors. h-BN, MoS2, and WS2 structures were constructed with inorganic atoms, so the nanodescriptors containing B, N, Mo, W, and S atoms showed higher values than others.
Table 2.
Atomic electronegativity values used in the calculation of nanodescriptors.
| Atoms | C | O | H | N | S | B | Mo | W |
|---|---|---|---|---|---|---|---|---|
| Electronegativity value | 2.5 | 3.5 | 2.2 | 3.1 | 2.4 | 2.0 | 2.2 | 2.4 |
Fig. 3.
Standardized nanodescriptors for 11 in-house 2DNMs in the dataset.
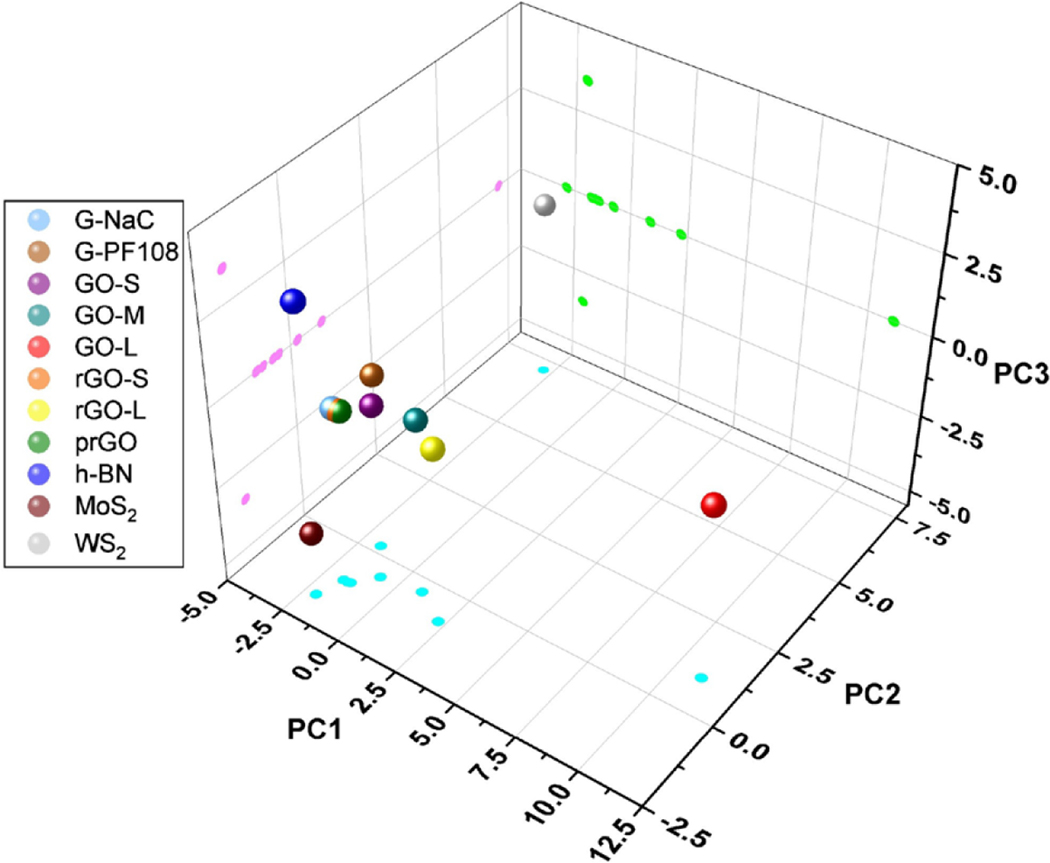
The calculated nanodescriptors can be used to show the chemical space of 2DNMs through principal component analysis (PCA). Both the top two and top three principal components were used to represent the distribution of all 2DNMs, which accounted for 73% and 81% of the total descriptor variance, respectively. As shown in Fig. 4 and Fig. S3, the 2DNMs in the dataset were structurally diverse due to various shapes, lateral sizes, atom types, surface chemistry, and surfactant types. The three GRMs, prGO, rGO-S, and G-NaC, are close to each other in both 2D and 3D chemical space, which mainly due to their similar lateral size and same surfactant. Although h-BN and, they locate differently in 3D chemical space. 3D chemical space retains more information of the original chemical space, which can better differentiate h-BN and MoS2. Compared to other GRMs, GO-L is a structure outlier mainly due to the large number of hydroxyl groups on its surface, which significantly increases the value of descriptor HHHH. To better analyze this type of GRMs, more graphene oxides with different sizes should be included in future studies.
Fig. 4.
Principal component analysis (PCA) of v2DNMs based on the nanodescriptors. Every 2DNMs is visualized in 3D chemical space. The projections of all 2DNMs on three planes (cyan, green, and pink) are also shown.
3.5. Computational modeling
Four nanotoxicity models were developed using the calculated nanodescriptors and PLSR approach for the 2DNMs in the dataset. When data in two doses (1 μg ml−1 and 5 μg ml−1) were available for LDH release and cell viability, both dose results were used in the model development (Table S1), so this effort resulted in models for low/high doses of these toxicity endpoints. Two medium-sized graphenes in the dataset, prGO and GO-M, were selected as test set, and the remaining nine 2DNMs were used as the training set for model development. For apoptosis dataset, which consists of nine 2DNMs total (Table S1), one medium-sized graphene (prGO) was selected for test purpose, and the remaining eight 2DNMs were used as training set for model development. The LOOCV procedure was used to evaluate the performance of developed nanotoxicity models within training sets.
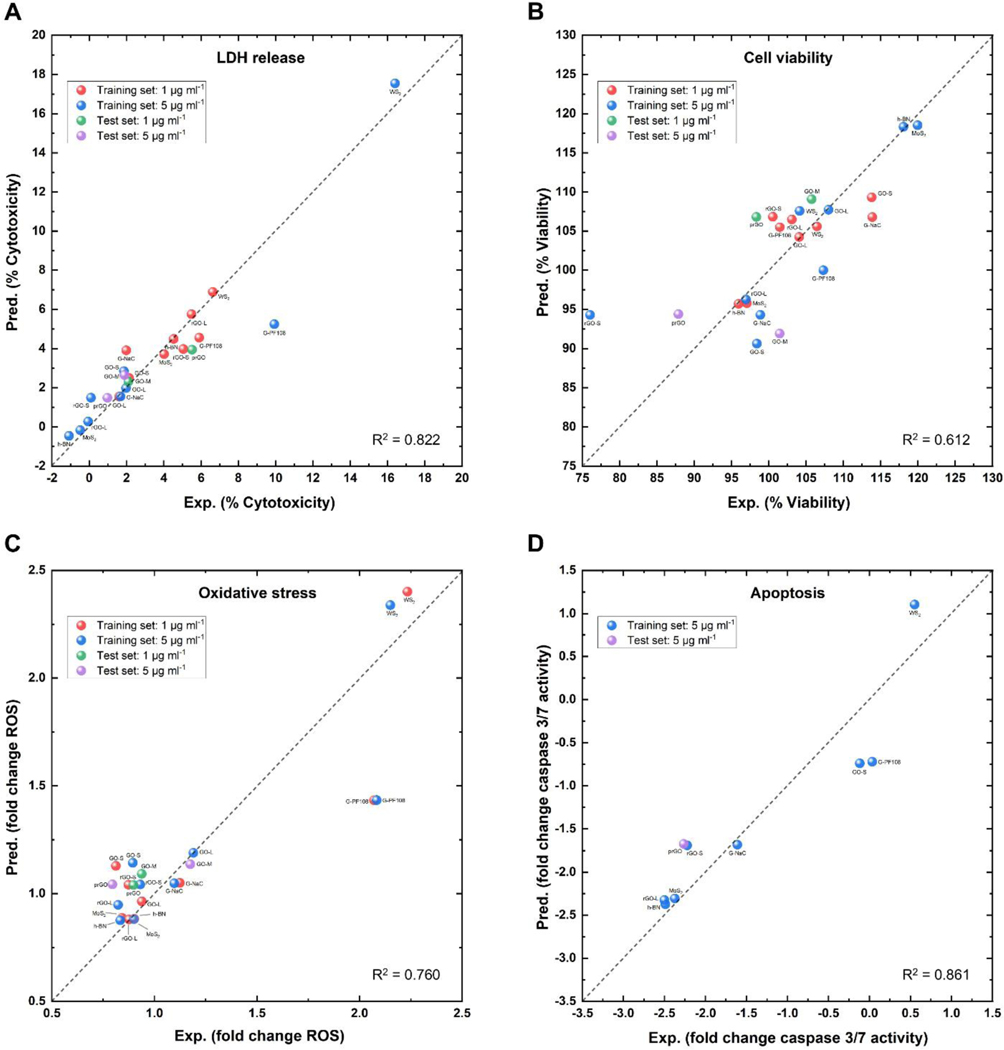
The correlations between experimental values and predicted values of the resulted four nanotoxicity models were shown in Fig. 5. The optimal number of components for developing the best PLSR model in the training set was obtained from the LOOCV procedure. The training set R2 values using the obtained optimal number of components ranged from 0.558 to 0.822 in corresponding PLSR models (Table 3). The RMSE values of training (test) set were 1.371 (0.916), 5.882 (7.361), 0.250 (0.164), and 0.447 (0.591) for the models of LDH release (%), cell viability (%), oxidative stress (fold change), and apoptosis respectively (fold change) (Table 3). The RMSE values between the training and test set of all four models are similar, indicating that the resulted nanotoxicity models are reliable for prediction purposes. Although most 2DNMs were correctly predicted, prediction errors still exist. For example, G-PF108 has relatively large prediction errors in the models of LDH release, oxidative stress, and apoptosis (Fig. 5 A, C and D). This issue was mainly due to the lack of other GRMs tested by using PF108 as the surfactant in the dataset. Compared to the other GRMs dispersed in NaC, G-PF108 was the only GRM dispersed in PF108. PF108 is a non-ionic surfactant while NaC is an ionic surfactant, which can influence the toxicity of 2DNMs [77, 78]. For the model of cell viability, 5 μg ml−1 rGO-S has a larger prediction error than others (Fig. 5 B and D). For the model of cell viability and apoptosis, 5 μg ml−1 rGO-S has a larger prediction error than others (Fig. 5 B and 5D). The reason for prediction errors of rGO-S was similar to G-PF108. rGO-S has its structural nearest neighbor as prGO. However, 5 μg ml−1 rGO-S and prGO have cell viability testing results as 76.03% and 87.88% respectively, and apoptosis testing results as −2.22 and −2.26 respectively. The issues are similar to the “activity cliff” of QSAR modeling studies [79]. There are only two types of rGO in the dataset, and the lateral size of rGO-L is at least 5 times larger than rGO-S. The training data used in this study are not sufficient to cover the structure diversity of rGO, so the model performance can be improved when new rGO with different lateral sizes are added into the training sets.
Fig. 5.
Correlations between experimental (Exp.) and predicted (Pred.) values for developed PLSR models in (A) LDH release, (B) cell viability, (C) oxidative stress, and (D) apoptosis. Red, blue, green, and purple dots are 1 μg ml−1 2DNMs in the training set, 5 μg ml−1 2DNMs in the training set, 1 μg ml−1 2DNMs in the test set and 5 μg ml−1 2DNMs in the test set, respectively. The coefficient of determination (R2) from the modeling results is also shown.
Table 3.
The optimal number of components, the coefficient of determination (R2) and root mean square error (RMSE) of PLSR models for four toxicity endpoints.
| Toxicity endpoint | The optimal number of components | R2 | RMSE for the training set | RMSE for the test set |
|---|---|---|---|---|
| LDH release (%) | 3 | 0.822 | 1.371 | 0.916 |
| Cell viability (%) | 3 | 0.612 | 5.882 | 7.361 |
| Oxidative stress (fold change) | 2 | 0.760 | 0.250 | 0.164 |
| Apoptosis (fold change) | 2 | 0.861 | 0.447 | 0.591 |
3.6. Mechanism analysis of 2DNMs-induced toxicities
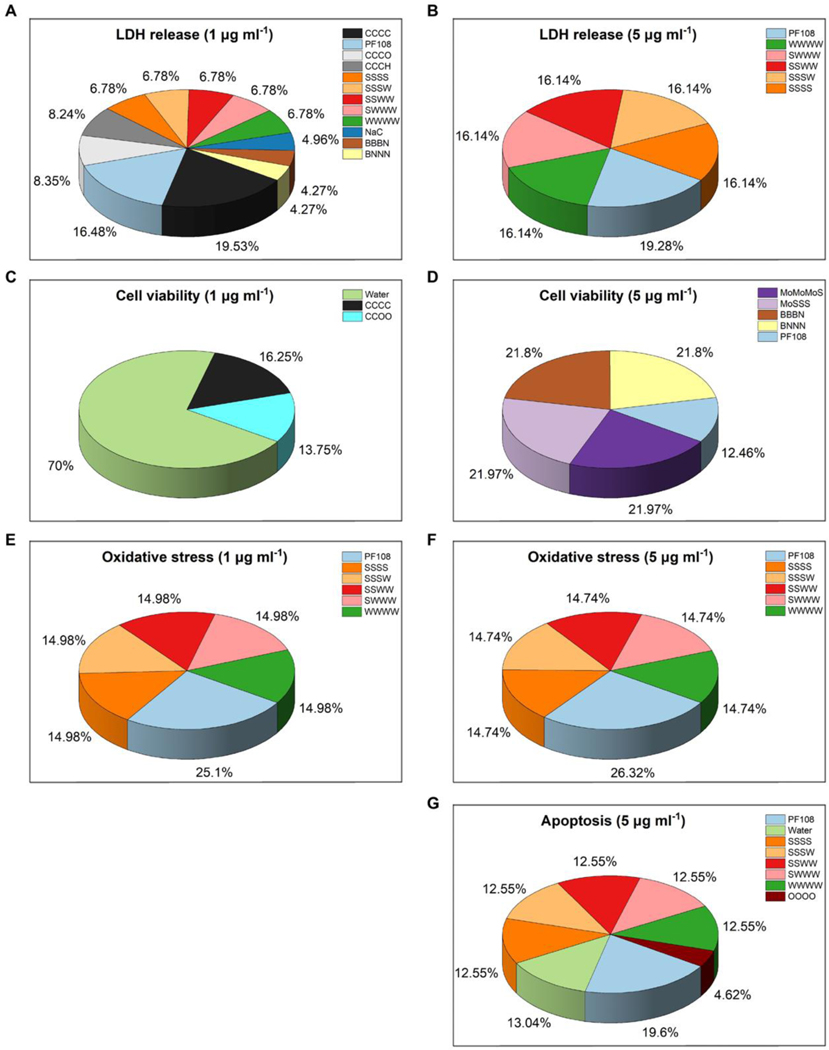
Analysis of the developed ML models allowed to identify nanostructure features responsible for nanotoxicity, which can be used to illustrate potential mechanisms of 2DNMs-induced toxicity and guide future 2DNMs design. The top ranked nanodescriptors were obtained from accepted PLSR models (Fig. 6). The high coefficient value of a descriptor indicates its critical contribution to the final models. The ranking was calculated based on the descriptors’ contributions to the models of 2DNMs. Descriptors with contributions greater than 4% of the total descriptor contributions were shown in Fig. 6. In a resulted ML model, the high frequency of a nanodescriptor utilization indicates its critical contribution to the associated nanotoxicity [41, 80].
Fig. 6.
Contributions of the top-𝑘 nanodescriptors from the PLSR modeling results in (A and B) LDH release, (C and D) cell viability, (E and F) oxidative stress, and (G) apoptosis. The descriptors contribution for both 1 μg ml−1 and 5 μg ml−1 2DNMs are shown from the model results. The percentage contributions of descriptors in all four models are harmonized.
To explore the mechanism of 2DNMs-induced toxicity, several descriptors were found to be critical for various toxicities of 2DNMs. Specifically, the descriptor CCCC was important for LDH release obtained by low concentration 2DNMs (1 μg ml−1) (Fig. 6 A), which mainly reflects the geometries of GRMs (Fig. S4 A). This result implies that geometries influence GRMs’ toxicities, which had been reported in previous studies [45, 81, 82]. Descriptors containing functional groups were also found to be important for 2DNMs’ toxicity in developed models (Fig. 6 A and G). For example, CCCO descriptor mainly reflects the neighbor relationship between a carboxyl group and a hydroxyl group (Fig. S4 B). This substructure can make GRMs interact with biomolecules by hydrogen bonding thus influencing GRMs’ toxicity [83, 84]. OOOO descriptor represents four oxygen atoms from neighbor hydroxyl groups (Fig. S4 C), which describes an oxidized region of GRMs. Intramolecular or Intermolecular hydrogen bonds can be formed with this substructure, which were important for GRMs’ bioactivity and toxicity [85, 86]. Descriptors, such as SSSS, SSSW, and WWWW, were found to be important for WS2-mediated toxicity in developed models (Fig. 6 A, B, E, F, and G). SSSS/WWWW descriptors capture structure features on sulfur/tungsten layers, and SSSW reflected the sublattice of WS2 (Fig. S4 D-F). Although the toxicity of WS2 is still under investigation, tungstate ion has been identified as a potentially toxicant against guppies and shows tumorigenicity and genotoxicity in vitro [87–89], which partially validate the importance of this descriptor. MoS2-related and h-BN-related descriptors, such as MoMoMoS, MoSSS, BBBN, and BNNN, showed positive contribution in cell viability models, which may reflect the low toxicity of MoS2 and h-BN (Fig. 6 D). Surfactant descriptors (i.e. PF108, Water and NaC) were also found to be important to nanotoxicity models, which increased the 2DNMs’ dispersibility and stabilized their dispersion under different mechanisms [77, 78].
3.7. Pitfalls and perspectives
In this study, nanodescriptors calculated from annotated 2DNMs structures afford the predictive modeling of 2DNM toxicity. Some pitfalls still exist due to limited data. For example, G-PF108 is a structural outlier and more similar 2DNMs needs to be tested to cover the relevant structure diversity. Furthermore, besides lateral size and surface chemistry, which are critical features influencing 2DNMs’ toxicity, thickness should also be considered as a potential feature in the modeling process, as investigated in previous studies [90]. Although the above issues exist, the current results showed the feasibility of using the current nanostructure annotation and modeling strategy to predict nanotoxicity in the future study.
4. Conclusions
In this study, novel structure annotation strategy and machine learning approach were integrated for computational modeling of nanotoxicities of 2DNMs with irregular shapes, which characterized the complex nanostructure features and enable the toxicity prediction of 2DNMs. A new computational approach was designed to construct virtual 2DNMs, which simulated 2DNMs’ irregular geometries and diverse functional modifications. The annotated 2DNMs structures were saved as PDB files and were further used for geometrical nanodescriptors calculations. Additional surfactant descriptors were also added. To prove the applicability of the nanostructure annotation and nanodescriptors calculation, a dataset containing diverse 2DNMs with different atom types, lateral sizes and surface chemistry was used to develop various nanotoxicity models. Good predictivities were shown in the resulted models for all available endpoints including LDH release, cell viability, oxidative stress, and apoptosis. This novel structure annotation strategy shows great potential to generate high-quality nanodescriptors for ML modeling purposes. Integration of structure annotation and machine learning approaches paves a road for the future development of NMs with complex structures.
Supplementary Material
Acknowledgments
This work was partially supported by the National Institute of Environmental Health Sciences (Grant R01ES031080) and National Science Foundation (Grant 2211489).
Footnotes
Declaration of Competing Interest
The authors declare no competing financial interest.
Tong Wang: Formal analysis, Investigation, Validation, Writing - Original Draft Daniel P. Russo: Methodology, Software Dimitrios Bitounis: Data Curation Philip Demokritou: Resources, Writing - Review & Editing Xuelian Jia: Visualization Heng Huang: Resources Hao Zhu: Conceptualization, Supervision, Funding acquisition, Writing - Review & Editing
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pokrajac L, Abbas A, Chrzanowski W, Dias GM, Eggleton BJ, Maguire S, Maine E, Malloy T, Nathwani J, Nazar L, Sips A, Sone J, van den Berg A, Weiss PS, Mitra S, Nanotechnology for a Sustainable Future: Addressing Global Challenges with the International Network4Sustainable Nanotechnology, Acs Nano 15(12) (2021) 18608–18623. [DOI] [PubMed] [Google Scholar]
- [2].Gottardo S, Mech A, Drbohlavova J, Malyska A, Bowadt S, Riego Sintes J, Rauscher H, Towards safe and sustainable innovation in nanotechnology: State-of-play for smart nanomaterials, NanoImpact 21 (2021) 100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nations United, Transforming our world: the 2030 agenda for sustainable development, Transforming our world: the 2030 agenda for sustainable development, United Nations, 2015. [Google Scholar]
- [4].Gharailou D, A review of market studies in different fields of nanotechnology, StatNano (2018) 0–15. [Google Scholar]
- [5].Eleftheriadou M, Pyrgiotakis G, Demokritou P, Nanotechnology to the rescue: using nano-enabled approaches in microbiological food safety and quality, Current opinion in biotechnology 44 (2017) 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aytac Z, Xu J, Raman Pillai SK, Eitzer BD, Xu T, Vaze N, Ng KW, White JC, Chan-Park MB, Luo Y, Enzyme-and Relative Humidity-Responsive Antimicrobial Fibers for Active Food Packaging, ACS Applied Materials & Interfaces 13(42) (2021) 50298–50308. [DOI] [PubMed] [Google Scholar]
- [7].Vaze N, Pyrgiotakis G, Mena L, Baumann R, Demokritou A, Ericsson M, Zhang Y, Bello D, Eleftheriadou M, Demokritou P, A nano-carrier platform for the targeted delivery of nature-inspired antimicrobials using Engineered Water Nanostructures for food safety applications, Food control 96 (2019) 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang R, Vaze N, Soorneedi A, Moore MD, Luo Y, Poverenov E, Rodov V, Demokritou P, A novel antimicrobial technology to enhance food safety and quality of leafy vegetables using engineered water nanostructures, Environmental Science: Nano 8(2) (2021) 514–526. [Google Scholar]
- [9].Zhao L, Lu L, Wang A, Zhang H, Huang M, Wu H, Xing B, Wang Z, Ji R, Nano-biotechnology in agriculture: use of nanomaterials to promote plant growth and stress tolerance, Journal of agricultural and food chemistry 68(7) (2020) 1935–1947. [DOI] [PubMed] [Google Scholar]
- [10].Xu T, Wang Y, Aytac Z, Zuverza-Mena N, Zhao Z, Hu X, g KWN, White JC, Demokritou P, Enhancing Agrichemical Delivery and Plant Development with Biopolymer-Based Stimuli Responsive Core–Shell Nanostructures, Acs Nano 16(4) (2022) 6034–6048. [DOI] [PubMed] [Google Scholar]
- [11].Zhao Z, Xu T, Pan X, Susanti JC White X. Hu Y. Miao P. Demokritou K.W. Ng, Sustainable Nutrient Substrates for Enhanced Seedling Development in Hydroponics, ACS Sustainable Chemistry & Engineering 10(26) (2022) 8506–8516. [Google Scholar]
- [12].Chen X, Li C, Gratzel M, Kostecki R, Mao SS, Nanomaterials for renewable energy production and storage, Chem Soc Rev 41(23) (2012) 7909–37. [DOI] [PubMed] [Google Scholar]
- [13].Bagheri AR, Aramesh N, Bilal M, Xiao J, Kim H-W, Yan B, Carbon nanomaterials as emerging nanotherapeutic platforms to tackle the rising tide of cancer–A review, Bioorganic & medicinal chemistry 51 (2021) 116493. [DOI] [PubMed] [Google Scholar]
- [14].Liu Y, Zhang B, Yan B, Enabling anticancer therapeutics by nanoparticle carriers: the delivery of Paclitaxel, Int J Mol Sci 12(7) (2011) 4395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Singh AV, Laux P, Luch A, Sudrik C, Wiehr S, Wild AM, Santomauro G, Bill J, Sitti M, Review of emerging concepts in nanotoxicology: opportunities and challenges for safer nanomaterial design, Toxicol Mech Methods 29(5) (2019) 378–387. [DOI] [PubMed] [Google Scholar]
- [16].Li Y, Zhang Y, Yan B, Nanotoxicity overview: nano-threat to susceptible populations, Int J Mol Sci 15(3) (2014) 3671–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang W, Wang L, Mettenbrink EM, DeAngelis PL, Wilhelm S, Nanoparticle Toxicology, Annu Rev Pharmacol Toxicol 61 (2021) 269–289. [DOI] [PubMed] [Google Scholar]
- [18].Sohal IS, O’Fallon KS, Gaines P, Demokritou P, Bello D, Ingested engineered nanomaterials: state of science in nanotoxicity testing and future research needs, Particle and fibre toxicology 15(1) (2018) 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pirela SV, Martin J, Bello D, Demokritou P, Nanoparticle exposures from nano-enabled toner-based printing equipment and human health: state of science and future research needs, Critical reviews in toxicology 47(8) (2017) 683–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cao X, Zhang T, DeLoid GM, Gaffrey MJ, Weitz KK, Thrall BD, Qian W-J, Demokritou P, Evaluation of the cytotoxic and cellular proteome impacts of food-grade TiO2 (E171) using simulated gastrointestinal digestions and a tri-culture small intestinal epithelial model, NanoImpact 17 (2020) 100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Parviz D, Bitounis D, Demokritou P, Strano M, Engineering two-dimensional nanomaterials to enable structure-activity relationship studies in nanosafety research, NanoImpact 18 (2020) 100226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Duan Y, Coreas R, Liu Y, Bitounis D, Zhang Z, Parviz D, Strano M, Demokritou P, Zhong W, Prediction of protein corona on nanomaterials by machine learning using novel descriptors, NanoImpact 17 (2020) 100207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jia Y, Hou X, Wang Z, Hu X, Machine Learning Boosts the Design and Discovery of Nanomaterials, ACS Sustainable Chemistry & Engineering 9(18) (2021) 6130–6147. [Google Scholar]
- [24].Brown KA, Brittman S, Maccaferri N, Jariwala D, Celano U, Machine learning in nanoscience: big data at small scales, Nano Letters 20(1) (2019) 2–10. [DOI] [PubMed] [Google Scholar]
- [25].Wang W, Yan X, Zhao L, Russo DP, Wang S, Liu Y, Sedykh A, Zhao X, Yan B, Zhu H, Universal nanohydrophobicity predictions using virtual nanoparticle library, Journal of cheminformatics 11(1) (2019) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].OECD, Important Issues on Risk Assessment of Manufactured Nanomaterials, Important Issues on Risk Assessment of Manufactured Nanomaterials, OECD, 2022. [Google Scholar]
- [27].Ciallella HL, Zhu H, Advancing Computational Toxicology in the Big Data Era by Artificial Intelligence: Data-Driven and Mechanism-Driven Modeling for Chemical Toxicity, Chem Res Toxicol 32(4) (2019) 536–-54.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao L, Ciallella HL, Aleksunes LM, Zhu H, Advancing computer-aided drug discovery (CADD) by big data and data-driven machine learning modeling, Drug Discov Today 25(9) (2020) 1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhu H, Big Data and Artificial Intelligence Modeling for Drug Discovery, Annu Rev Pharmacol Toxicol 60 (2020) 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jia X, Wen X, Russo DP, Aleksunes LM, Zhu H, Mechanism-driven Modeling of Chemical Hepatotoxicity Using Structural Alerts and an In Vitro Screening Assay, Journal of Hazardous Materials (2022) 129193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jia X, Ciallella HL, Russo DP, Zhao L, James MH, Zhu H, Construction of a virtual opioid bioprofile: a data-driven QSAR modeling study to identify new analgesic opioids, ACS sustainable chemistry & engineering 9(10) (2021) 3909–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Winkler DA, Role of Artificial Intelligence and Machine Learning in Nanosafety, Small 16(36) (2020) e2001883. [DOI] [PubMed] [Google Scholar]
- [33].Haase A, Klaessig F, EU US roadmap nanoinformatics 2030, (2018). [Google Scholar]
- [34].Yan X, Yue T, Zhu H, Yan B, Bridging the Gap Between Nanotoxicological Data and the Critical Structure–Activity Relationships, Advances in Toxicology and Risk Assessment of Nanomaterials and Emerging Contaminants, Springer 2022, pp. 161–183. [Google Scholar]
- [35].Liu R, Rallo R, George S, Ji Z, Nair S, Nel AE, Cohen Y, Classification NanoSAR development for cytotoxicity of metal oxide nanoparticles, Small 7(8) (2011) 1118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fourches D, Pu D, Tassa C, Weissleder R, Shaw SY, Mumper RJ, Tropsha A, Quantitative nanostructure-activity relationship modeling, Acs Nano 4(10) (2010) 5703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cho W-S, Duffin R, Thielbeer F, Bradley M, Megson IL, MacNee W, Poland CA, Tran CL, Donaldson K, Zeta potential and solubility to toxic ions as mechanisms of lung inflammation caused by metal/metal oxide nanoparticles, Toxicological Sciences 126(2) (2012) 469–477. [DOI] [PubMed] [Google Scholar]
- [38].Puzyn T, Rasulev B, Gajewicz A, Hu X, Dasari TP, Michalkova A, Hwang H-M, Toropov A, Leszczynska D, Leszczynski J, Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles, Nature nanotechnology 6(3) (2011) 175–178. [DOI] [PubMed] [Google Scholar]
- [39].Ahmed L, Rasulev B, Kar S, Krupa P, Mozolewska MA, Leszczynski J, Inhibitors or toxins? Large library target-specific screening of fullerene-based nanoparticles for drug design purpose, Nanoscale 9(29) (2017) 10263–10276. [DOI] [PubMed] [Google Scholar]
- [40].Wang W, Sedykh A, Sun H, Zhao L, Russo DP, Zhou H, Yan B, Zhu H, Predicting nano–bio interactions by integrating nanoparticle libraries and quantitative nanostructure activity relationship modeling, Acs Nano 11(12) (2017) 12641–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yan XL, Sedykh A, Wang WY, Zhao XL, Yan B, Zhu H, In silico profiling nanoparticles: predictive nanomodeling using universal nanodescriptors and various machine learning approaches, Nanoscale 11(17) (2019) 8352–8362. [DOI] [PubMed] [Google Scholar]
- [42].Yan XL, Sedykh A, Wang WY, Yan B, Zhu H, Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations, Nature Communications 11(1) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Garg M, Vishwakarma N, Sharma AL, Singh S, Different Types and Intense Classification of 2D Materials, Advanced Applications of 2D Nanostructures, Springer 2021, pp. 11–28. [Google Scholar]
- [44].Xu MS, Liang T, Shi MM, Chen HZ, Graphene-Like Two-Dimensional Materials, Chem Rev 113(5) (2013) 3766–3798. [DOI] [PubMed] [Google Scholar]
- [45].Li JL, Wang X, Mei KC, Chang CH, Jiang JH, Liu XS, Liu Q, Guiney LM, Hersam MC, Liao YP, Meng H, Xia T, Lateral size of graphene oxide determines differential cellular uptake and cell death pathways in Kupffer cells, LSECs, and hepatocytes, Nano Today 37 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ma Y, Wang J, Wu J, Tong C, Zhang T, Meta-analysis of cellular toxicity for graphene via data-mining the literature and machine learning, Sci Total Environ 793 (2021) 148532. [DOI] [PubMed] [Google Scholar]
- [47].Hu X, Zhou Q, Health and ecosystem risks of graphene, Chem Rev 113(5) (2013) 3815–3835. [DOI] [PubMed] [Google Scholar]
- [48].Bitounis D, Parviz D, Cao X, Amadei CA, Vecitis CD, Sunderland EM, Thrall BD, Fang M, Strano MS, Demokritou P, Synthesis and Physicochemical Transformations of Size‐Sorted Graphene Oxide during Simulated Digestion and Its Toxicological Assessment against an In Vitro Model of the Human Intestinal Epithelium, Small 16(21) (2020) 1907640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].de Luna LAV, Loret T, Fordham A, Arshad A, Drummond M, Dodd A, Lozano N, Kostarelos K, Bussy C, Lung recovery from DNA damage induced by graphene oxide is dependent on size, dose and inflammation profile, Particle and Fibre Toxicology 19(1) (2022) 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Parviz D, Bitounis D, Demokritou P, Strano M, Engineering Two-dimensional Nanomaterials to Enable Structure-Activity Relationship Studies in Nanosafety Research, NanoImpact 18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ardoña HAM, Zimmerman JF, Shani K, Kim S-H, Eweje F, Bitounis D, Parviz D, Casalino E, Strano M, Demokritou P, Differential modulation of endothelial cytoplasmic protrusions after exposure to graphene-family nanomaterials, NanoImpact 26 (2022) 100401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Coreas R, Castillo C, Li Z, Yan D, Gao Z, Chen J, Bitounis D, Parviz D, Strano MS, Demokritou P, Biological Impacts of Reduced Graphene Oxide Affected by Protein Corona Formation, Chemical Research in Toxicology 35(7) (2022) 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bazina L, Bitounis D, Cao XQ, DeLoid GM, Parviz D, Strano MS, Lin HYG, Bell DC, Thrall BD, Demokritou P, Biotransformations and cytotoxicity of graphene and inorganic two-dimensional nanomaterials using simulated digestions coupled with a triculture in vitro model of the human gastrointestinal epithelium, Environ Sci-Nano 8(11) (2021) 3233–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Parviz D, Strano M, Endotoxin‐Free Preparation of Graphene Oxide and Graphene‐Based Materials for Biological Applications, Current protocols in chemical biology 10(4) (2018) e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang H, Wu J, He J, Zhang Z, Effect of hole size on the fracture of graphene nanomesh, Proceedings of the 19th European Conference on Fracture, Kazan, Russia, 2012, pp. 26–31. [Google Scholar]
- [56].Chen L, Elibol K, Cai H, Jiang C, Shi W, Chen C, Wang HS, Wang X, Mu X, Li C, Direct observation of layer-stacking and oriented wrinkles in multilayer hexagonal boron nitride, 2D Materials 8(2) (2021) 024001. [Google Scholar]
- [57].Garcés E, Salas O, Magaña L, Optical absorption and reflectivity of four 2D materials: MoS2, MoP2, NbS2, and NbP2, Front. Mater. 8: 720768. doi: 10.3389/fmats (2021). [DOI] [Google Scholar]
- [58].Yang F, Wang K, Hu P, Wang X, Chang T, Chen Z, Deng J, Feng P, Volinsky AA, Detonation exfoliated mechanism of graphene-like MoS2 prepared by the intercalation-detonation method and promising exfoliation for 2D materials, Applied Surface Science 525 (2020) 145867. [Google Scholar]
- [59].Biro LP, Nemes-Incze P, Lambin P, Graphene: nanoscale processing and recent applications, Nanoscale 4(6) (2012) 1824–39. [DOI] [PubMed] [Google Scholar]
- [60].Mondal S, Basu S, Evolution of the Berry phase and topological properties of a band deformed Chern insulator, The European Physical Journal B 94(10) (2021) 1–8. [Google Scholar]
- [61].Wold S, Sjöström M, Eriksson L, PLS-regression: a basic tool of chemometrics, Chemometrics and intelligent laboratory systems 58(2) (2001) 109–130. [Google Scholar]
- [62].Abdi H, Partial least square regression (PLS regression), Encyclopedia for research methods for the social sciences 6(4) (2003) 792–795. [Google Scholar]
- [63].Tran L, Bañares MA, Rallo R, Modelling the toxicity of nanoparticles, Springer; 2017. [Google Scholar]
- [64].Qi R, Pan Y, Cao J, Yuan B, Wang Y, Jiang J, Toward comprehension of the cytotoxicity of heterogeneous TiO 2-based engineered nanoparticles: a nano-QSAR approach, Environmental Science: Nano 8(4) (2021) 927–936. [Google Scholar]
- [65].Buglak AA, Zherdev AV, Dzantiev BB, Nano-(Q) SAR for cytotoxicity prediction of engineered nanomaterials, Molecules 24(24) (2019) 4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Geladi P, Kowalski BR, Partial least-squares regression: a tutorial, Analytica chimica acta 185 (1986) 1–17. [Google Scholar]
- [67].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Scikit-learn: Machine learning in Python, the Journal of machine Learning research 12 (2011) 2825–2830. [Google Scholar]
- [68].Lerf A, He H, Forster M, Klinowski J, Structure of graphite oxide revisited, The Journal of Physical Chemistry B 102(23) (1998) 4477–4482. [Google Scholar]
- [69].Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics, Journal of molecular graphics 14(1) (1996) 33–38. [DOI] [PubMed] [Google Scholar]
- [70].Wajid AS, ram S Das.Irin F, Ahmed HT, Shelburne JL, Parviz D, Fullerton RJ, Jankowski AF, Hedden RC, Green MJ, Polymer-stabilized graphewastene dispersions at high concentrations in organic solvents for composite production, Carbon 50(2) (2012) 526–534. [Google Scholar]
- [71].Bari R, Parviz D, Khabaz F, Klaassen CD, Metzler SD, Hansen MJ, Khare R, Green MJ, Liquid phase exfoliation and crumpling of inorganic nanosheets, Physical Chemistry Chemical Physics 17(14) (2015) 9383–9393. [DOI] [PubMed] [Google Scholar]
- [72].Parviz D, Irin F, Shah SA, Das S, Sweeney CB, Green MJ, Challenges in liquid‐phase exfoliation, processing, and assembly of pristine graphene, Advanced Materials 28(40) (2016) 8796–8818. [DOI] [PubMed] [Google Scholar]
- [73].Durant JL, Leland BA, Henry DR, Nourse JG, Reoptimization of MDL keys for use in drug discovery, Journal of chemical information and computer sciences 42(6) (2002) 1273–1280. [DOI] [PubMed] [Google Scholar]
- [74].Kim S, Exploring chemical information in PubChem, Current Protocols 1(8) (2021) e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xu Y, Verma D, Sheridan RP, Liaw A, Ma J, Marshall NM, McIntosh J, Sherer EC, Svetnik V, Johnston JM, Deep dive into machine learning models for protein engineering, Journal of chemical information and modeling 60(6) (2020) 2773–2790. [DOI] [PubMed] [Google Scholar]
- [76].Saini V, Machine learning prediction of empirical polarity using SMILES encoding of organic solvents, Molecular Diversity (2022) 1–13. [DOI] [PubMed] [Google Scholar]
- [77].Ramalingam P, Pusuluri ST, Periasamy S, Veerabahu R, Kulandaivel J, Role of deoxy group on the high concentration of graphene in surfactant/water media, RSC advances 3(7) (2013) 2369–2378. [Google Scholar]
- [78].Chong JY, Mulet X, Waddington LJ, Boyd BJ, Drummond CJ, Steric stabilisation of self-assembled cubic lyotropic liquid crystalline nanoparticles: high throughput evaluation of triblock polyethylene oxide-polypropylene oxide-polyethylene oxide copolymers, Soft Matter 7(10) (2011) 4768–4777. [Google Scholar]
- [79].Maggiora GM, On outliers and activity cliffs why QSAR often disappoints, ACS Publications, 2006, pp. 1535–1535. [DOI] [PubMed] [Google Scholar]
- [80].Liu G, Yan X, Sedykh A, Pan X, Zhao X, Yan B, Zhu H, Analysis of model PM2. 5-induced inflammation and cytotoxicity by the combination of a virtual carbon nanoparticle library and computational modeling, Ecotoxicology and environmental safety 191 (2020) 110216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Akhavan O, Ghaderi E, Akhavan A, Size-dependent genotoxicity of graphene nanoplatelets in human stem cells, Biomaterials 33(32) (2012) 8017–8025. [DOI] [PubMed] [Google Scholar]
- [82].Gies V, Zou S, Systematic toxicity investigation of graphene oxide: evaluation of assay selection, cell type, exposure period and flake size, Toxicology research 7(1) (2018) 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Chen D, Feng H, Li J, Graphene oxide: preparation, functionalization, and electrochemical applications, Chem Rev 112(11) (2012) 6027–6053. [DOI] [PubMed] [Google Scholar]
- [84].He H, Klinowski J, Forster M, Lerf A, A new structural model for graphite oxide, Chemical physics letters 287(1–2) (1998) 53–56. [Google Scholar]
- [85].Sinclair RC, Coveney PV, Modeling nanostructure in graphene oxide: inhomogeneity and the percolation threshold, Journal of chemical information and modeling 59(6) (2019) 2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mittal S, Kumar V, Dhiman N, Chauhan LKS, Pasricha R, Pandey AK, Physico-chemical properties based differential toxicity of graphene oxide/reduced graphene oxide in human lung cells mediated through oxidative stress, Scientific reports 6(1) (2016) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Strigul N, Koutsospyros A, Christodoulatos C, Tungsten speciation and toxicity: acute toxicity of mono-and poly-tungstates to fish, Ecotoxicology and environmental safety 73(2) (2010) 164–171. [DOI] [PubMed] [Google Scholar]
- [88].Wasel O, Freeman JL, Comparative assessment of tungsten toxicity in the absence or presence of other metals, Toxics 6(4) (2018) 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Khan K, Tareen AK, Aslam M, Wang R, Zhang Y, Mahmood A, Ouyang Z, Zhang H, Guo Z, Recent developments in emerging two-dimensional materials and their applications, J Mater Chem C 8(2) (2020) 387–440. [Google Scholar]
- [90].Achawi S, Pourchez J, Feneon B, Forest V, Graphene-based materials in vitro toxicity and their structure–activity relationships: A systematic literature review, Chemical Research in Toxicology 34(9) (2021) 2003–2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.