Abstract
The levels of molecular chaperones and folding enzymes in the endoplasmic reticulum (ER) are controlled by a transcriptional induction process termed the unfolded protein response (UPR). The mammalian UPR is mediated by the cis-acting ER stress response element (ERSE), the consensus sequence of which is CCAAT-N9-CCACG. We recently proposed that ER stress response factor (ERSF) binding to ERSE is a heterologous protein complex consisting of the constitutive component NF-Y (CBF) binding to CCAAT and an inducible component binding to CCACG and identified the basic leucine zipper-type transcription factors ATF6α and ATF6β as inducible components of ERSF. ATF6α and ATF6β produced by ER stress-induced proteolysis bind to CCACG only when CCAAT is bound to NF-Y, a heterotrimer consisting of NF-YA, NF-YB, and NF-YC. Interestingly, the NF-Y and ATF6 binding sites must be separated by a spacer of 9 bp. We describe here the basis for this strict requirement by demonstrating that both ATF6α and ATF6β physically interact with NF-Y trimer via direct binding to the NF-YC subunit. ATF6α and ATF6β bind to the ERSE as a homo- or heterodimer. Furthermore, we showed that ERSF including NF-Y and ATF6α and/or β and capable of binding to ERSE is indeed formed when the cellular UPR is activated. We concluded that ATF6 homo- or heterodimers recognize and bind directly to both the DNA and adjacent protein NF-Y and that this complex formation process is essential for transcriptional induction of ER chaperones.
Two mammalian proteins with molecular masses of 78 and 94 kDa identified in the mid-1970s were named glucose-regulated proteins (GRP78 and GRP94, respectively) due to the marked increases in their levels on depletion of glucose from media for cell culture (24). Subsequent studies indicated that this phenomenon is part of the cellular response to the accumulation of unfolded proteins in the endoplasmic reticulum (ER) (8, 18). Not only glucose deprivation but also various physiological and environmental stress conditions (ER stress) cause unfolding or misfolding of proteins in the ER, where newly synthesized secretory and transmembrane proteins fold and assemble. As only correctly folded molecules are allowed to move along the secretory pathway, eukaryotic cells must deal with protein misfolding in the ER promptly and appropriately to avoid malfunction and/or mislocalization of proteins that are synthesized on membrane-bound ribosomes and translocated into the ER. A typical cellular strategy to cope with unfolded proteins in the ER is induction of molecular chaperones and folding enzymes localized in the lumen of the ER, such as BiP/GRP78 and GRP94, resulting in augmentation of the folding capacity of the ER. This homeostatic response is achieved by a transcriptional induction process coupled with signaling from the ER to the nucleus, now known as the unfolded protein response (UPR).
The transcriptional apparatus responsible for the mammalian UPR was poorly understood until the cis-acting ER stress response element (ERSE), necessary and sufficient for the induction, was identified as a sequence of 19 nucleotides, the consensus of which is CCAAT-N9-CCACG (20, 26). Based on our recent results (5, 27) and the previous finding reported by Li et al. (13) that the CCAAT part of the ERSE is constitutively occupied at least in the GRP78 promoter, we proposed that the ER stress response factor (ERSF), a transcription factor responsible for the mammalian UPR, is a heterologous protein complex composed of a constitutive component that binds to CCAAT and an inducible component that binds to CCACG. The constitutive component of ERSF is almost unambiguously NF-Y (CBF), a general transcription factor involved in transcription of numerous genes (16), as essentially all cellular binding activity to the CCAAT sequence in ERSE present in nuclear extracts was supershifted by anti-NF-Y antiserum on electrophoretic mobility shift assays (EMSAs) (20, 27). We recently identified the basic leucine zipper (bZIP)-type transcription factor ATF6 as an inducible component of ERSF based on its DNA-binding specificity and tightly regulated mechanism of activation (5, 27).
It was previously reported (28) that mammalian cells express another bZIP protein closely related to ATF6, which is encoded by the cyclic AMP response element binding protein-related protein (CREB-RP) gene (17) (also called the G13 gene [9]). Our recent study revealed that this gene product also functions as an inducible component of ERSF, and we thus proposed calling the ATF6 gene product ATF6α and the CREB-RP (G13) gene product ATF6β (K. Haze, T. Okada, H. Yoshida, H. Yanagi, T. Yura, M. Negishi, and K. Mori, submitted for publication). Both ATF6α and ATF6β are constitutively synthesized as type II transmembrane glycoproteins embedded in the ER and subjected to proteolytic processing in response to ER stress, allowing translocation into the nucleus of N-terminal fragments liberated from the ER membrane. The N-terminal regions of ATF6α and ATF6β contain all the hallmarks required for an active transcription factor, i.e., DNA binding, dimerization, and activation domains, and bind to the CCACG part of ERSE when the CCAAT part is bound to NF-Y. Thus, the soluble forms of ATF6α and ATF6β produced in ER-stressed cells (designated p50ATF6α and p60ATF6β, respectively) activate transcription of mammalian UPR target genes in collaboration with NF-Y.
The CCACG part of ERSE appears to be a half-site of an E box (CANNTG)-like palindromic sequence, to which transcription factors containing a basic region as a DNA-binding domain are known to bind (6, 15).
We therefore examined in this study whether ATF6α and ATF6β bind to ERSE as monomers or as dimers. Interestingly, CCAAT and CCACG parts are separated by a spacer of 9 bp in all ERSE-like sequences found in the promoter regions of various ER stress-inducible genes (26), and we showed that this spacing is critical for both ATF6-binding and transcription-inducing activities of ERSE (27). These results prompted us to examine whether ATF6α and ATF6β physically interact with NF-Y. We further investigated whether ER stress indeed induces formation of ERSF that contains both NF-Y and endogenous p50ATF6α or p60ATF6β.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics (100 U of penicillin and 100 μg of streptomycin per ml). Cells were maintained at 37°C in a humidified 5% CO2–95% air atmosphere. Transfection was carried out by the standard calcium phosphate method (21) as described in our previous report (26).
Construction of plasmids.
Recombinant DNA techniques were performed according to standard procedures (21). Expression plasmids were constructed on the basis of mammalian expression vectors pcDNA3.1(+) (Invitrogen, Carlsbad, Calif.), pBIND (Promega, Madison, Wis.), and pACT (Promega). Various subregions of ATF6α and ATF6β were amplified by PCR and inserted into appropriate restriction enzyme sites of appropriate vectors after their sequences had been confirmed. In the case of N-terminal deletion mutants, the consensus sequence for efficient initiation of translation (10) was introduced at the N terminus together with an initiation codon.
Two-hybrid assays in mammalian cells.
pBIND-based bait plasmid and pACT-based prey plasmid were transfected into HeLa cells cultured in 96-well plates together with reporter plasmid pG5luc containing five Gal4p binding sites in a minimal promoter upstream of the firefly luciferase gene (Promega). pBIND carried the Renilla luciferase gene to normalize transfection efficiency. Luciferase activities were determined as described (27), and relative luciferase activity was defined as the ratio of firefly luciferase activity to Renilla luciferase activity.
Pull-down assays.
Recombinant NF-Y trimer composed of T7-tagged NF-YA, T7-tagged NF-YB, and histidine-tagged NF-YC was prepared as described previously (1, 27). Various subregions of ATF6α and ATF6β cloned in pcDNA3.1(+) were translated in vitro using the TNT T7 quick coupled transcription-translation system (Promega) in the presence or absence of [35S]methionine using EXPRE35S35S protein labeling mix (DuPont, Wilmington, Del.) according to the manufacturer's instructions.
Recombinant NF-Y trimer (10 pmol) was rotated with 10 μl of Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen, Munich, Germany) in binding buffer I (20 mM HEPES [pH 7.9], 100 mM KCl, 10% glycerol, 1 mM MgCl2, 1 mM 2-mercaptoethanol, 0.1% Tween 20, and 20 mM imidazole) at 4°C for 1 h to immobilize NF-Y trimer through binding of the histidine-tagged NF-YC subunit to nickel resin. After washing with binding buffer I six times, various subregions of ATF6α or ATF6β labeled with [35S]methionine during in vitro translation were rotated with NF-Y-immobilized resin in binding buffer I at 4°C for 1 h. After washing with binding buffer I six times, resin was suspended in sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 10% glycerol) followed by boiling for 5 min. After brief centrifugation, supernatants were subjected to SDS–12% polyacrylamide gel electrophoresis (PAGE). Bound materials were visualized by exposure to X-ray film.
Histidine-tagged NF-YA, histidine-tagged NF-YB, and histidine-tagged NF-YC were expressed in Escherichia coli cells and purified separately as described previously (1). Each purified protein migrated as a single band on SDS-PAGE, and their protein concentrations were determined using the Bio-Rad protein assay kit. Each of the NF-Y subunits (10 pmol) was also immobilized on 10 μl of Ni-NTA-agarose and used for pull-down assays as described for NF-Y trimer.
EMSAs.
Synthetic double-stranded oligonucleotides containing GRP78-ERSE1 and its flanking nucleotides (AGGGCCTTCACCAATCGGCGGCCTCCACGACGGGGCT; nucleotides matching the ERSE consensus are underlined) were radiolabeled using the Klenow fragment of DNA polymerase I and [α-32P]dCTP (222 TBq/mmol; DuPont) and purified by centrifugation through ProbeQuant G50 microcolumns (Amersham Pharmacia Biotech). 32P-labeled oligonucleotide probes (0.1 pmol, ∼9,000 cpm) were incubated with various mutant forms of ATF6α(∼800 fmol) or ATF6β (∼400 fmol) translated in vitro in the presence of recombinant NF-Y trimer (10 fmol) in binding buffer E (20 mM HEPES [pH 7.9], 100 mM KCl, 10% glycerol, 1 mM MgCl2, 1 mM 2-mercaptoethanol, 0.1% Tween 20, 20 ng of poly[dI-dC]:poly[dI-dC] per μl) at 4°C for 1 h. Electrophoresis was carried out essentially as described previously (27).
Immunoblotting analysis.
Immunoblotting was carried out according to the standard procedure (21) as described previously (5) using an enhanced chemiluminescence Western blotting detection system kit (Amersham Pharmacia Biotech). Rabbit anti-NF-YA antibody and mouse anti-KDEL monoclonal antibody (clone 10C3) were obtained from Rockland (Gilbertsville, Pa.) and StressGen Biotechnologies (Victoria, British Columbia, Canada), respectively. Anti-ATF6α and anti-ATF6β antibodies were prepared as described previously (5, 27).
Pull-down assays for ERSF.
Nuclear extracts were prepared as described previously (2) from HeLa cells untreated or treated with 300 nM thapsigargin for 4 h. Synthetic double-stranded oligonucleotides containing wild-type or mutant GRP78-ERSE1 (see Fig. 8A for their sequences) were biotinylated and immobilized on UltraLink resin (Pierce, Rockford, Ill.) by streptavidin-biotin interaction. ERSE-coupled resin was rotated with nuclear extracts in binding buffer B (20 mM HEPES [pH 7.9], 100 mM KCl, 10% glycerol, 1 mM MgCl2, 1 mM 2-mercaptoethanol, 0.1% Tween 20, and 0.1% bovine serum albumin) at 4°C for 1 h, washed with binding buffer B six times, and then suspended in SDS sample buffer, followed by boiling for 5 min. After brief centrifugation, supernatants were subjected to SDS–12% PAGE and analyzed by immunoblotting using anti-ATF6α or anti-ATF6β antibodies.
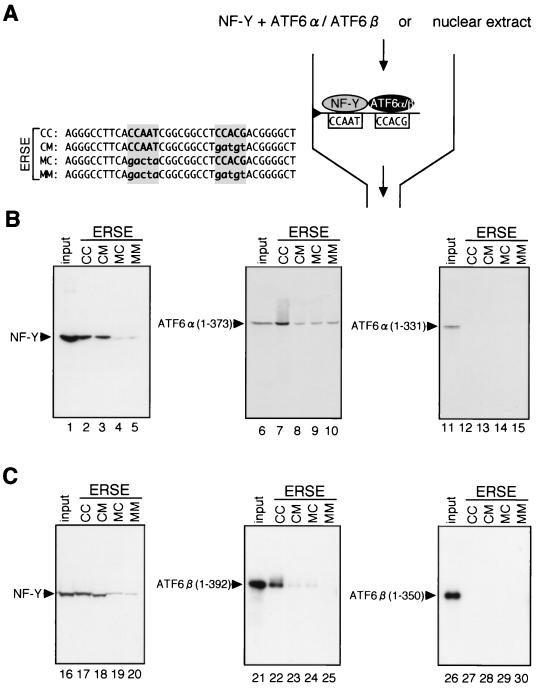
FIG. 8.
In vitro reconstitution of ERSF consisting of NF-Y and ATF6α or ATF6β. (A) Nucleotide sequences of oligonucleotides immobilized on resin. A 37-bp sequence containing GRP78-ERSE1 and its flanking nucleotides is referred to as ERSE-CC. Two critical regions of ERSE (shaded) were mutated separately or simultaneously (indicated by lowercase letters) to generate ERSE-CM, ERSE-MC, and ERSE-MM. Each of the synthetic double-stranded oligonucleotides was immobilized on resin. Principles of assays to analyze formation of ERSF are shown schematically on the right. (B and C) Binding of ATF6α(B) and ATF6β (C) in the presence of NF-Y to resin carrying ERSE-CC. Various subregions of ATF6α and ATF6β labeled with [35S]methionine during in vitro translation were mixed with recombinant NF-Y trimer and applied to resin to which one of the four oligonucleotides (ERSE-CC,-CM,-MC, or -MM) had been immobilized. After washing, bound materials were eluted and subjected to SDS–12% PAGE together with 10% aliquots of input material. Eluted NF-Y was detected by immunoblotting using anti-NF-YA antibody, whereas eluted ATF6α and ATF6β were visualized by exposure to X-ray film.
RESULTS
ATF6α and ATF6β bind to ERSE as a homo- or heterodimer.
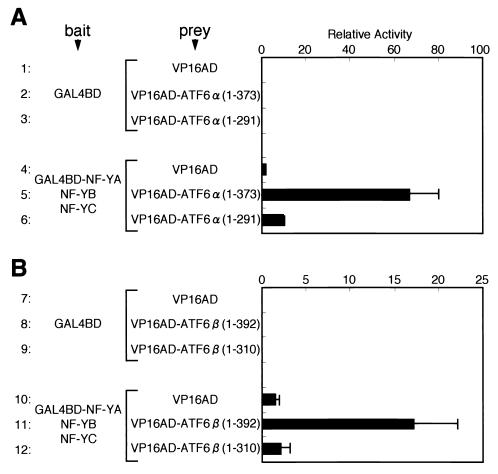
As both p50ATF6α and p60ATF6β, soluble and active forms of ATF6α and ATF6β, respectively, are bZIP-type transcription factors localized in the nucleus, we performed two-hybrid assays to determine whether ATF6α and ATF6β can dimerize through their leucine zippers. HeLa cells were transfected with pBIND-based bait plasmid and pACT-based prey plasmid together with reporter plasmid pG5luc, and then reporter luciferase activities constitutively expressed in transfected cells were determined. pBIND carried the DNA-binding domain of yeast transcriptional activator Gal4p (GAL4BD), whereas pACT carried the activation domain of VP16 (VP16AD). ATF6α(1-373) and ATF6β(1-392), representing p50ATF6α and p60ATF6β, respectively, were fused to VP16AD to generate VP16AD-ATF6α(1-373) and VP16AD-ATF6β(1-392), respectively, whereas GAL4BD was fused to ATF6α(171-373), which lacked the activation domain present in the N-terminal region (see schematic structures depicted in Fig. 1).
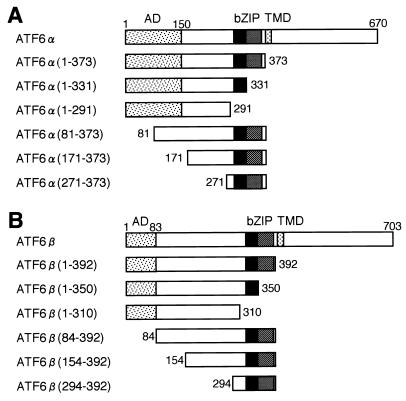
FIG. 1.
Schematic structures of full-length and mutant forms of ATF6α(A) and ATF6β (B). Numbers indicate amino acid positions from the N terminus. The locations of the activation domain (AD), basic leucine zipper region (bZIP), and transmembrane domain (TMD) are marked.
Reporter expression in cells expressing GAL4BD-ATF6α(171-373) and VP16 alone (Fig. 2A, line 4) was as low as that in cells expressing GAL4BD alone and VP16AD alone (line 1), indicating no interaction between ATF6α(171-373) and VP16AD. Interaction between GAL4BD and ATF6α(1-373) was also marginal (compare line 2 with line 1). In contrast, coexpression of GAL4BD-ATF6α(171-373) with VP16AD-ATF6α(1-373) markedly enhanced luciferase expression (line 5). Importantly, GAL4BD-ATF6α(171-373) did not interact with VP16AD-ATF6α(1-291), lacking the leucine zipper (line 6). Similarly, GAL4BD-ATF6α(171-373) interacted with VP16AD-ATF6β(1-392), and this interaction was dependent on the leucine zipper region of ATF6β (Fig. 2B). These results suggested that ATF6α and ATF6β can homo- and heterodimerize via their leucine zipper regions.
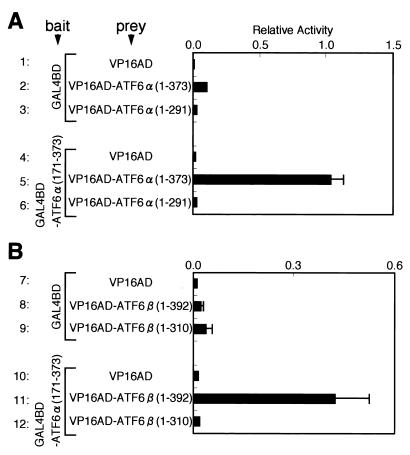
FIG. 2.
Formation of homodimers of ATF6α(A) or heterodimers between ATF6α and ATF6β (B) detected by two-hybrid assays. Mammalian expression plasmids pBIND and pACT carried the DNA-binding domain of yeast transcriptional activator Gal4p (GAL4BD) and the activation domain of VP16 (VP16AD), respectively. ATF6α(171-373) was fused with GAL4BD in pBIND to generate GAL4BD-ATF6α(171-373), which was used as bait, whereas ATF6α(1-373), ATF6α(1-291), ATF6β(1-392), and ATF6β(1-310) were fused with VP16AD in pACT and used as prey (see schematic structures depicted in Fig. 1). HeLa cells were transiently transfected with a combination of bait plasmid (100 ng) and prey plasmid (100 ng), as indicated, together with the reporter plasmid pG5luc (100 ng), containing five GAL4BD binding sites upstream of the firefly luciferase gene. Twenty-four hours after transfection, relative luciferase activity constitutively expressed in transfected cells was determined, and averages from four independent experiments are presented with standard deviations (bars).
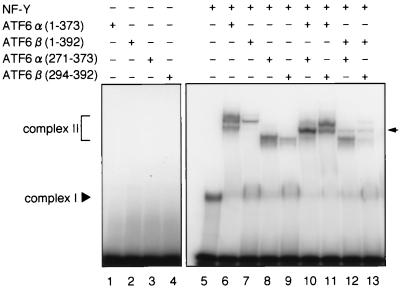
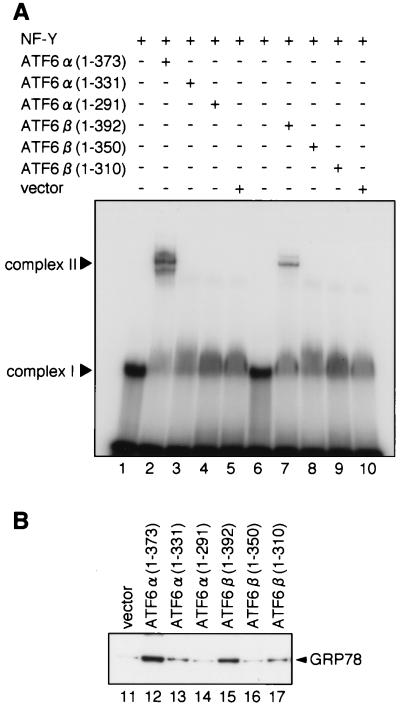
To visualize dimer formation using EMSAs, we analyzed truncated forms of ATF6α and ATF6β, ATF6α(271-373) and ATF6β(294-392), respectively (Fig. 1), in addition to p50ATF6α-like mutant ATF6α(1-373) and p60ATF6β-like mutant ATF6β(1-392). As reported previously (27; Haze et al., submitted) and reproduced in Fig. 3, neither in vitro-translated ATF6α(1-373) alone (lane 1) nor ATF6β(1-392) alone (lane 2) could bind to 32P-labeled ERSE, whereas recombinant NF-Y trimer alone bound to the probe, resulting in formation of a DNA-protein complex designated complex I (lane 5). However, in vitro-translated ATF6α(1-373) and ATF6β(1-392) bound to ERSE in the presence of NF-Y trimer, resulting in formation of a DNA-protein complex designated complex II (lanes 6 and 7, respectively). Similarly, both ATF6α(271-373) and ATF6β(294-392) translated in vitro formed complex II only in the presence of NF-Y trimer (compare lanes 8 and 9 with lanes 3 and 4), which migrated faster than complex II composed of NF-Y trimer and ATF6α(1-373) or ATF6β(1-392) bound to 32P-labeled ERSE (lane 6 or 7). Under these conditions, simultaneous translation in vitro of ATF6α(1-373) and ATF6α(271-373) and subsequent incubation with 32P-labeled ERSE in the presence of NF-Y trimer resulted in formation of complex II (lane 10) that migrated at a position intermediate between those of ATF6α(1-373)-containing complex II (lane 6) and ATF6α(271-373)-containing complex II (lane 8). Similarly, heterodimer formation was observed between ATF6α(1-373) and ATF6β(294-392) (lane 11), ATF6β(1-392) and ATF6α(271-373) (lane 12), and ATF6β(1-392) and ATF6β(294-392) (lane 13). These results clearly demonstrated that ATF6α and ATF6β bind to ERSE as dimers.
FIG. 3.
Formation of homo- and heterodimers between ATF6α and ATF6β detected by EMSA. Various subregions of ATF6α and ATF6β were translated in vitro individually or together with other subregions, as indicated, and then incubated with 32P-labeled oligonucleotide probe GRP78-ERSE1 in the presence or absence of NF-Y trimer. DNA-protein complexes formed were analyzed by EMSA. The positions of complexes I and II are indicated. The arrow marks the positions of complex II composed of NF-Y and heterodimer of ATF6α and/or ATF6β bound to 32P-labeled GRP78-ERSE1.
ATF6α and ATF6β directly interact with NF-Y trimer through binding to NF-YC subunit.
To gain insight into possible interactions of ATF6α and ATF6β with NF-Y, we performed two-hybrid assays, as shown in Fig. 4. As NF-Y is a heterotrimer composed of NF-YA, NF-YB, and NF-YC, each subunit was fused to GAL4BD to generate GAL4BD-NF-YA, GAL4BD-NF-YB, and GAL4BD-NF-YC, respectively. In addition, GAL4BD-NF-YA was expressed together with full-length NF-YB and NF-YC to allow heterotrimer formation in transfected cells. Subregions of ATF6α and ATF6β fused with VP16AD were used as prey.
FIG. 4.
Interaction between NF-Y and ATF6α(A) or ATF6β (B) detected by two-hybrid assays. Full-length NF-YA was fused with GAL4BD in pBIND to express GAL4BD-NF-YA fusion protein, whereas unfused full-length NF-YB or NF-YC was expressed from a pcDNA3.1(+)-based plasmid. Various subregions of ATF6α and ATF6β were fused with VP16AD in pACT to express various VP16AD fusion proteins, as indicated. HeLa cells were transiently transfected with a combination of bait plasmid (total, 100 ng) and prey plasmid (100 ng) together with reporter plasmid pG5luc (100 ng). Relative luciferase activity was determined and is presented as in Fig. 2.
Coexpression of VP16AD-ATF6α(1-373) with GAL4BD-NF-YA, GAL4BD-NF-YB, or GAL4BD-NF-YC did not significantly affect luciferase expression (data not shown). Thus, no interaction could be detected between ATF6α and any of the three subunits of NF-Y by two-hybrid assays. Importantly, however, coexpression of VP16AD-ATF6α(1-373) with GAL4BD-NF-YA in the presence of full-length NF-YB and NF-YC resulted in marked enhancement of luciferase expression (Fig. 4, line 5) compared to the control (line 4). Essentially identical results were obtained with ATF6β (lines 10 and 11 and data not shown). These results suggested that p50ATF6α and p60ATF6β interact with NF-Y trimer.
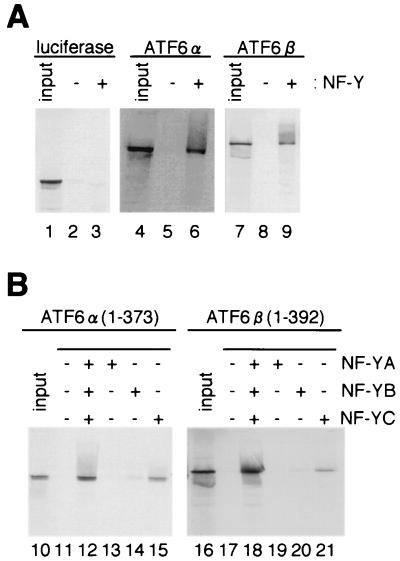
We next performed pull-down assays. In vitro-translated and [35S]methionine-labeled ATF6α(1-373) or ATF6β(1-392) was mixed with resin to which recombinant NF-Y trimer had been immobilized, and then bound materials were eluted and subjected to SDS-PAGE. As shown in Fig. 5A, approximately 10% of ATF6α (lanes 4 to 6) and ATF6β (lanes 7 to 9) applied to resin bound to NF-Y trimer, whereas binding of luciferase to NF-Y trimer was negligible (lanes 1 to 3). These results further confirmed the interaction of ATF6α and ATF6β with NF-Y. We then examined whether direct binding of ATF6α and ATF6β with any of the three subunits of NF-Y could be detected by pull-down assays. As shown in Fig. 5B, in vitro-translated and [35S]methionine-labeled ATF6α(1-373) and ATF6β(1-392) bound to resin to which the NF-YC subunit (lanes 15 and 21) but not the NF-YA (lanes 13 and 19) or NF-YB (lanes 14 and 20) subunit had been immobilized, albeit much less efficiently than to NF-Y trimer (compare with lanes 12 and 18). These results were inconsistent with those of the two-hybrid assays described above, in which we failed to detect interaction of ATF6α or ATF6β with any of the three NF-Y subunits. However, we found in two-hybrid assays that reporter luciferase expression was not enhanced when VP16AD-ATF6α(1-373) or VP16AD-ATF6β(1-392) was coexpressed with GAL4BD-NF-YC in the presence of full-length NF-YA and NF-YB (data not shown), in marked contrast to the results shown in Fig. 4, where VP16AD-ATF6α(1-373) or VP16AD-ATF6β(1-392) was coexpressed with GAL4BD-NF-YA in the presence of full-length NF-YB and NF-YC. Thus, the discrepancy between the two-hybrid and pull-down assays could be explained by the inability of the GAL4BD-NF-YC fusion protein to bind to ATF6α or ATF6β despite the ability of unfused NF-YC to bind to ATF6α or ATF6β both in vivo and in vitro. From these results, we concluded that ATF6α and ATF6β interact directly with NF-Y trimer through binding to the NF-YC subunit.
FIG. 5.
Interaction between NF-Y and ATF6α or ATF6β detected by pull-down assays. (A) Pull-down assays using immobilized NF-Y trimer. In vitro-translated and [35S]methionine-labeled luciferase, ATF6α(1-373), or ATF6β(1-392) was mixed with resin to which recombinant NF-Y trimer had been immobilized (lanes +) or control resin (lanes −). After washing, bound materials were eluted, subjected to SDS-12% PAGE, and visualized by exposure to X-ray film. Aliquots (10%) of input materials were run on the same gel for comparison (lanes input). (B) Pull-down assays using immobilized NF-Y subunit. In vitro-translated and [35S]methionine-labeled ATF6α(1-373) or ATF6β(1-392) was mixed with resin to which NF-Y trimer (lanes 12 and 18) or one of the three subunits of NF-Y (NF-YA, NF-YB, and NF-YC, lanes 13 to 15 and lanes 19 to 21) had been immobilized, or control resin (lanes 11 and 17). Bound materials and 10% aliquots of input materials were analyzed as in panel A.
bZIP regions of ATF6α and ATF6β are sufficient for interaction with NF-Y.
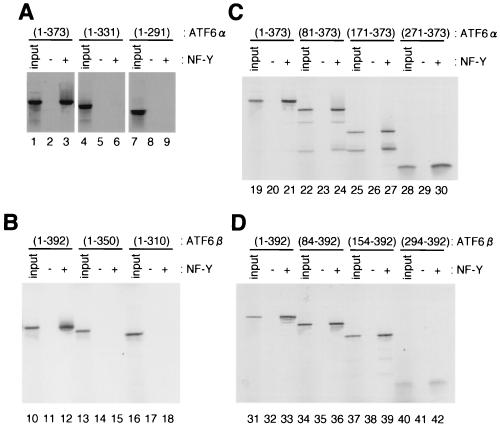
As ATF6α and ATF6β bind to ERSE as dimers, we determined whether dimerization is required for interaction with NF-Y trimer. As shown in Fig. 6A and B, we found that deletion of the leucine zipper made ATF6α and ATF6β unable to interact with NF-Y trimer; neither ATF6α(1-331) (lanes 4 to 6) nor ATF6β(1-350) (lanes 13 to 15) could be retained in NF-Y trimer-immobilized resin. Consistent with these results, deletion of the leucine zipper from ATF6α and ATF6β abolished their association with NF-Y trimer as detected by two-hybrid assays (Fig. 4, lines 6 and 12, respectively). We then examined the effects of deleting the leucine zipper regions on the ERSE-binding activities of ATF6α and ATF6β using EMSA. As shown in Fig. 3, in vitro-translated ATF6α(1-373) and ATF6β(1-392) bound to ERSE only in the presence of NF-Y trimer, resulting in formation of complex II (Fig. 7A, lanes 2 and 7, respectively). In marked contrast, ATF6α(1-331) and ATF6β(1-350) containing the DNA-binding basic region but lacking the leucine zipper failed to form complex II even in the presence of NF-Y trimer (lanes 3 and 8, respectively). ATF6α and ATF6β lacking the leucine zipper were inactive in activating transcription of UPR target genes (Fig. 7B); overexpression of ATF6α(1-373) and ATF6β(1-392) led to a constitutively enhanced level of GRP78 protein (lanes 12 and 15, respectively), as reported previously (5, 27; Haze et al., submitted), but overexpression of ATF6α(1-331) and ATF6β(1-350) failed to do so (lanes 13 and 16, respectively). It should be noted that not only ATF6α(1-373) and ATF6β(1-392) but also ATF6α(1-331) and ATF6β(1-350) accumulated in the nuclei of transfected cells (Haze et al., submitted). Thus, leucine zipper region-mediated dimerization of ATF6α and ATF6β appeared not to be critical for nuclear localization but to be indispensable for their interaction with NF-Y, binding to ERSE, and transcriptional activator activity.
FIG. 6.
Identification of the regions in ATF6α(A and C) and ATF6β (B and D) important for interaction with NF-Y. Interactions of NF-Y trimer with various subregions of ATF6α and ATF6β translated in vitro and labeled with [35S]methionine were determined by pull-down assays as described for Fig. 5A.
FIG. 7.
Effects of deleting leucine zipper regions on the activities of ATF6α and ATF6β. (A) Effects on ERSE-binding activity. Various C-terminal deletion mutants of ATF6α and ATF6β translated in vitro were incubated with 32P-labeled oligonucleotide probe GRP78-ERSE1 in the presence of NF-Y trimer. Rabbit reticulocyte lysate that had been incubated with vector alone was used as a control. DNA-protein complexes formed were analyzed by EMSA. The positions of complexes I and II are indicated. (B) Effects on transcriptional activity. HeLa cells were transiently transfected with 10 μg of pcDNA3.1(+) alone (vector) or one of the pcDNA3.1(+)-based plasmids to express various C-terminal deletion mutants of ATF6α or ATF6β, as indicated. Forty-eight hours after transfection, cells were lysed with phosphate-buffered saline containing 1% SDS and boiled for 5 min. Samples (6 μg of protein) were subjected to SDS–10% PAGE and analyzed by immunoblotting with anti-KDEL antibody, which recognizes GRP78.
To identify the region(s) important for the interaction with NF-Y trimer, we constructed N-terminal deletion mutants of ATF6α(1-373) and ATF6β(1-392) (see Fig. 1 for their schematic structures). Binding of various subregions of ATF6α(1-373) and ATF6β(1-392) to NF-Y trimer was analyzed by pull-down assays. As shown in Fig. 6C and D, deletion of N-terminal regions from ATF6α(1-373) or ATF6β(1-392) did not significantly affect the binding activities; both ATF6α(271-373) (lanes 28 to 30) and ATF6β(294-392) (lanes 40 to 42) containing almost only the bZIP region bound to NF-Y trimer as efficiently as ATF6α(1-373) (lanes 19 to 21) and ATF6β(1-392) (lanes 31 to 33). These results provided the basis for our finding that both ATF6α(271-373) and ATF6β(294-392) were able to bind to 32P-labeled ERSE in the presence of NF-Y (see Fig. 3). Thus, the bZIP regions of ATF6α and ATF6β were sufficient for their interaction with NF-Y trimer.
There are two explanations for our observations. NF-Y may directly interact with ATF6α and ATF6β via the leucine zipper region, although a computer-based search revealed no leucine zipper-like motifs in the amino acid sequences of NF-YA, NF-YB, or NF-YC. Alternatively, NF-Y may directly bind to the basic regions of ATF6α and ATF6β only when they are homo- or heterodimerized. Currently, we are unable to distinguish between these two possibilities because the leucine zipper regions of ATF6α and ATF6β alone are too small to be analyzed by two-hybrid or pull-down assays. In any event, our results indicated that direct interaction between NF-Y and ATF6α or ATF6β is mediated through binding of the NF-YC subunit to the bZIP region of ATF6α or ATF6β.
ERSF including NF-Y trimer and ATF6α and/or ATF6β is formed in response to ER stress.
Previously, we failed to demonstrate ER stress-induced formation of complex II (ERSF composed of NF-Y trimer and p50ATF6α or p60ATF6β bound to ERSE) by means of EMSA because of the low levels of p50ATF6α and p60ATF6β produced in ER-stressed cells (27). We therefore examined whether ERSF formation could be detected by pull-down assays. To ensure that the system used was reliable, we attempted to reconstitute complex II in vitro as shown in Fig. 8. Double-stranded oligonucleotides encoding GRP78-ERSE1 (referred to here as ERSE-CC) were immobilized on resin using biotin-avidin interaction. To confirm binding specificity, two critical regions of ERSE were mutated separately or simultaneously to generate ERSE-CM, ERSE-MC, and ERSE-MM (see Fig. 8A for their sequences), and mutant ERSEs were similarly immobilized on resin.
In vitro-translated and [35S]methionine-labeled ATF6α(1-373) (Fig. 8B) or ATF6β(1-392) (Fig. 8C) was mixed with NF-Y trimer and applied to resin to which one of the four ERSEs had been immobilized. Bound materials were eluted and analyzed by immunoblotting for NF-Y or autoradiography for ATF6α and ATF6β. As NF-Y recognizes the CCAAT part of ERSE, NF-Y bound to and eluted from resin carrying ERSE-CC or ERSE-CM but not ERSE-MC or ERSE-MM (Fig. 8B, lanes 1 to 5, and Fig. 8C, lanes 16 to 20), as expected. In contrast, ATF6α(1-373) and ATF6β(1-392) were enriched only in the eluate from resin carrying ERSE-CC (Fig. 8B, lanes 6 to 10, and Fig. 8C, lanes 21 to 25, respectively). Importantly, leucine zipper-mediated dimerization was required for this enrichment, as expected from the above results; neither ATF6α(1-331) nor ATF6β(1-350) was retained by any of the resins under the same conditions (Fig. 8B, lanes 11 to 15, and Fig. 8C, lanes 26 to 30). These results strongly indicated that binding of ATF6α and ATF6β to ERSE requires both interaction with NF-Y trimer and intactness of the CCACG part of ERSE.
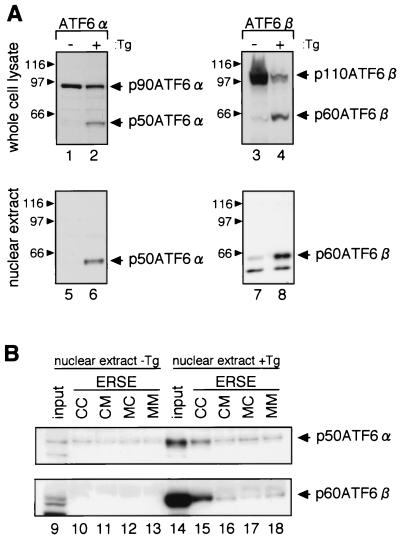
As the ERSE-CC-immobilized resin but not other resins efficiently trapped ERSF in our pull-down system, we then analyzed nuclear extracts prepared from HeLa cells that had been untreated or treated for 4 h with thapsigargin, a potent inducer of ER stress (12). As expected from previous reports (5; Haze et al., submitted), thapsigargin treatment induced proteolytic processing of both ATF6α and ATF6β (Fig. 9A, lanes 1 to 4). Importantly, p50ATF6α and p60ATF6β but not p90ATF6α or p110ATF6β were recovered in nuclear extracts (lanes 5 to 8), consistent with their subcellular localization. When nuclear extract of thapsigargin-treated cells was subjected to the pull-down assays described above, both p50ATF6α and p60ATF6β were enriched in the eluate from resin carrying ERSE-CC (Fig. 9B, lane 15), whereas no such enrichment was observed when nuclear extract of untreated cells was applied (lane 10). From these results, we concluded that p50ATF6α and p60ATF6β produced by ER stress-induced proteolysis indeed form a complex with NF-Y on the ERSE in the nucleus and that the resulting ERSF activates transcription of UPR target genes.
FIG. 9.
Formation of ERSF in nuclear extract of ER-stressed cells. (A) Effects of thapsigargin treatment on the processing of ATF6α and ATF6β. HeLa cells were untreated (−Tg) or treated with 300 nM thapsigargin for 4 h (+Tg). The cells were lysed directly in 50 μl of 1× Laemmli's SDS sample buffer and boiled for 5 min. Aliquots (5 μl) of the samples were subjected to SDS-PAGE and analyzed by immunoblotting with anti-ATF6α or anti-ATF6β antibody. Nuclear extracts were also prepared as described in Materials and Methods and analyzed similarly by immunoblotting. The positions of p90ATF6α, p50ATF6α, p110ATF6β, and p60ATF6β are indicated by arrowheads. The positions of prestained SDS-PAGE molecular size standards (Bio-Rad, Hercules, Calif.) are also shown (in kilodaltons). (B) ER stress-induced formation of ERSF. Nuclear extract of HeLa cells prepared as in panel A was mixed with resin to which one of the four oligonucleotides (ERSE-CC,-CM,-MC, or -MM) had been immobilized, as indicated. After washing, bound materials were eluted and subjected to SDS–12% PAGE together with 10% aliquots of input materials. Eluted ATF6α and ATF6β were detected by immunoblotting using anti-ATF6α and anti-ATF6β antibodies, respectively.
DISCUSSION
Identification of the cis-acting ERSE responsible for the mammalian UPR as 19 nucleotides of CCAAT-N9-CCACG not only provided the opportunity to search for transcription factors specific to the mammalian UPR but also delineated the nature of the expected binding protein(s) (20, 26). The expected proteins should bind to the CCACG part of ERSE, because the general transcription factor NF-Y binds to the CCAAT part. The characteristics of the expected proteins should explain why the binding site is always located 9 bp downstream of the NF-Y-binding site; altering the spacing from 9 bp to 8 or 10 bp inactivated ERSE regardless of whether the ERSE was present in the natural XBP-1 promoter (27) or transplanted upstream of the heterologous simian virus 40 promoter (26). Furthermore, the transcriptional activity of the expected proteins should be specifically regulated, because the UPR is activated only in response to the accumulation of unfolded proteins in the ER. The bZIP proteins ATF6α and ATF6β were promising candidate transcription factors because they bind to CCACG in ERSE in an NF-Y-dependent manner and their transcriptional activities are tightly regulated by ER stress-induced proteolysis (5, 27; Haze et al., submitted). In this study, we determined why the CCAAT and CCACG parts of ERSE must be separated by a spacer of 9 bp. Homo- or heterodimers of ATF6α and ATF6β physically interact with NF-Y trimer through direct association with the NF-YC-subunit, as depicted in Fig. 10. Thus, ATF6α and ATF6β recognize and bind to both NF-Y and ERSE simultaneously to activate transcription, and this bipartite interaction requires optimal spacing. Interestingly, the bZIP regions of ATF6α and ATF6β alone were sufficient for association with the NF-Y trimer (Fig. 6) and binding to ERSE (Fig. 3), consistent with our unpublished observation that ATF6α(271-373) and ATF6β (294-392), both lacking the activation domain, exhibited strong dominant negative effects on the UPR.
FIG. 10.
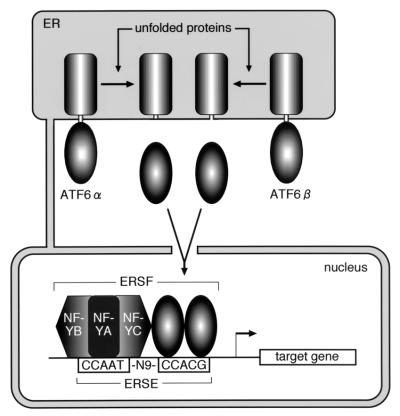
Model for mammalian UPR. Under normal growth conditions, the general transcription factor NF-Y constitutively occupies the CCAAT part of ERSE. On the other hand, both ATF6α and ATF6β are sequestered from the CCACG part of ERSE due to their anchoring in the ER membrane. Upon accumulation of unfolded proteins in the ER, both ATF6α and ATF6β are cleaved, allowing entrance of the resulting N-terminal fragments into the nucleus, where homo- and/or heterodimers of ATF6α and ATF6β bind to both NF-YC and CCACG. ER stress-induced formation of the transcription factor complex ERSF composed of NF-Y and ATF6α/β culminates in induced transcription of UPR target genes.
NF-Y is a trimer composed of three subunits, NF-YA, NF-YB, and NF-YC (16). Among these, NF-YB and NF-YC contain regions homologous to each other and similar to the histone fold motif. NF-YB and NF-YC form a stable heterodimer, and this heterodimerization allows association of the third subunit, NF-YA, resulting in the formation of a heterotrimer capable of binding to the CCAAT motif (16). NF-Y not only controls basal transcription of many genes by itself but also participates in inducible transcription by interacting with other specific transcription factors. One such example is interaction of NF-Y with regulatory factor X (RFX), involved in gamma interferon-induced transcription of the major histocompatibility complex (MHC) class II genes (14, 19). The promoters of the MHC class II genes contain the RFX binding site (X box) approximately 20 bp upstream of the NF-Y binding site (Y box), and both RFX and NF-Y can bind independently to their respective target sequences. Interestingly, the binding of RFX to the X box was enhanced severalfold when the Y box was bound to NF-Y, and the amounts of RFX–NF-Y–DNA complex, rather than those of RFX-DNA or NF-Y–DNA complex, correlated well with the promoter activity, suggesting that the RFX–NF-Y interaction is critical for transcriptional activation of the MHC class II genes. This cooperative binding and subsequent activation of the promoter required correct alignment between RFX and NF-Y because formation of the RFX–NF-Y–DNA complex as well as promoter activation were not affected by altering the spacing by one helical turn (insertion or deletion of 10 bp) but were abolished by changing the spacing by a half turn (insertion or deletion of 5 bp).
NF-Y is also involved in sterol-regulated transcription of several genes, such as those encoding farnesyl diphosphate synthase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, and squalene synthase, by interacting with sterol regulatory element-binding protein (SREBP) (3, 4, 7, 22). In the promoter of the farnesyl diphosphate synthase gene, the SREBP binding site is located 20 bp downstream of the NF-Y binding site, and weak binding of SREBP to its target sequence was enhanced over 20-fold by simultaneous incubation with NF-Y (4). Insertion of 4 bp between the SREBP and NF-Y binding sites abolished the cooperative binding of SREBP and NF-Y as well as the activation of the promoter for the farnesyl diphosphate synthase gene in response to depletion of sterol (4). In the promoter of the 3-hydroxy-3-methylglutaryl coenzyme A synthase gene, the SREBP binding site is located 17 bp downstream of the NF-Y binding site, and insertion of 5 or 10 bp between the SREBP and NF-Y binding sites made the promoter insensitive to sterol depletion (3). Consistent with these observations, it was shown that spacing of 16 to 20 bp between the SREBP and NF-Y binding sites is optimal for sterol-regulated transcription (7). Thus, to our knowledge, nothing is as strict as the interaction between NF-Y and ATF6α or ATF6β; insertion or deletion of 1 bp is sufficient to inactivate transcriptional activity of ERSE by abolishing binding of ATF6α or ATF6β to ERSE. These results indicated that the transcriptional activation by ATF6α or ATF6β definitely requires the presence of both a precise DNA sequence and NF-Y trimer at an optimal distance. Interestingly, SREBP was found to interact directly with NF-Y only when NF-Y is trimerized (3), whereas we showed that ATF6α and ATF6β associate with NF-Y trimer through direct binding to the NF-YC subunit. Because binding of ATF6α and ATF6β to the NF-YC subunit is much less efficient than that to the NF-Y trimer (Fig. 5), ATF6α and ATF6β translocated into the nucleus would form a functional complex with NF-Y trimer on the ERSE (see below) without being trapped by free NF-YC subunit nonproductively, as proposed for the case of SREBP–NF-Y interaction (3).
Wang et al. conducted binding-site selection experiments using the bZIP domain of ATF6α tagged with a polyhistidine that had been expressed and purified from bacterial cells (25). As a result, the consensus binding sequence, designated the ATF6 site, was deduced to be TGACGTGG/A, which contains a partially palindromic sequence (GACGTG), suggesting that ATF6α binds to the ATF6 site as a dimer. The ATF6 site conferred strong ER stress inducibility on the c-fos minimal promoter. A point mutation in the 3′ half-site from G to T (TGACGTTG; the mutated nucleotide is underlined) blocked not only its binding activity to ATF6α but also its transcriptional response to ER stress. It is therefore possible that ATF6α activates transcription of its target genes via direct binding to the ATF6 site even in the absence of the NF-Y binding site nearby, as the authors proposed, although the ATF6 site has not yet been identified in natural promoters of ER stress-inducible genes. Interestingly, the CCACG part of ERSE is entirely complementary to the 3′ half-site of the ATF6 site (TGACGTGG). We showed in this study that ATF6α and ATF6β bind to ERSE as dimers in the presence of NF-Y. Thus, the half-site of the palindromic sequence appears to be sufficient for recognition and binding when the ATF6 dimer is physically associated with NF-Y.
Li et al. recently analyzed the mechanism of activation of ATF6α by ER stress (11) using anti-ATF6α antibody raised by Zhu et al. (28). They found that the level of p90ATF6α decreased 2 h after thapsigargin treatment, consistent with our previous results (5), but they failed to detect p50ATF6α, contrary to our results. This discrepancy could be due, at least in part, to the region of ATF6α used to immunize rabbits; the anti-ATF6α antibody produced by Zhu et al. was raised against ATF6α(155-345), whereas our anti-ATF6α antibody was raised against ATF6α(6-307). We also had difficulty in detecting p50ATF6α using their anti-ATF6α antibody (data not shown). Four hours after thapsigargin treatment, they observed the recovery of p90ATF6α together with the appearance of a new band migrating slightly faster than p90ATF6α. Although the amounts of these doublet protein bands increased over time from 4 to 12 h after thapsigargin treatment, it is not yet clear whether this increase is required for the induction of GRP78 mRNA, a target of the mammalian UPR. Perhaps the most critical difference between their results and our results was that they recovered both p90ATF6α and the band migrating slightly faster than p90ATF6α in nuclear extracts of HeLa cells (11), whereas we recovered only the cleaved product, p50ATF6α, in HeLa nuclear extracts, as shown in Fig. 9A. We have no explanation for this discrepancy because we could not find a difference significant enough between their methods (23) and our methods (2) for preparing nuclear extracts from HeLa cells to account for the discrepancy. Nevertheless, it is very difficult to see how a protein containing a transmembrane domain can be extracted as a soluble nuclear protein as they reported. Although they speculate that alternative splicing might be able to produce a soluble form of ATF6α without proteolysis by removing the short hydrophobic stretch acting as a transmembrane domain from p90ATF6α, such specific changes at the mRNA level have not been observed in ER-stressed HeLa cells (26).
Li et al. also reported that ATF6α was coimmunoprecipitated with YY1, a ubiquitous transcription factor, but not with NF-Y from ATF6α-overproducing COS cells (11). However, the physiological significance of their findings remains unclear, as they overexpressed full-length ATF6α containing the transmembrane domain that anchors ATF6α in the ER membrane, and they performed immunoprecipitation after solubilization of cell extracts with detergent (0.5% NP-40); ATF6α may have been dissociated from NF-Y under these conditions. Thus, the most important finding of this study was the ER stress-induced formation of a transcription factor complex ERSF in the nucleus including NF-Y and p50ATF6α and/or p60ATF6β that binds to ERSE and activates transcription of ER chaperone genes. Although the amounts of this complex were too low to be detected by EMSA, as we reported previously (27), the complex was detected in the nuclear extract of ER-stressed cells by pull-down assays in the present study (Fig. 9). These results, together with our previous observations (5, 27; Haze et al., submitted), unambiguously established that ER stress-induced activation of ATF6α and ATF6β through proteolytic processing plays a major role in the mammalian UPR. The next important issue to be resolved is the mechanism of regulated proteolysis that activates ATF6α and ATF6β.
ACKNOWLEDGMENTS
We are grateful to Ron Prywes (Columbia University) for providing us with anti-ATF6α antibody. We thank Masako Nakayama, Seiji Takahara, and Tomoko Yoshifusa for technical assistance.
This work was supported in part by Research for the Future Program of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Bellorini M, Zemzoumi K, Farina A, Berthelsen J, Piaggio G, Mantovani R. Cloning and expression of human NF-YC. Gene. 1997;193:119–125. doi: 10.1016/s0378-1119(97)00109-1. [DOI] [PubMed] [Google Scholar]
- 2.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooley K A, Millinder S, Osborne T F. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 4.Ericsson J, Jackson S M, Edwards P A. Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J Biol Chem. 1996;271:24359–24364. doi: 10.1074/jbc.271.40.24359. [DOI] [PubMed] [Google Scholar]
- 5.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst H. Protein profile-transcription factors 1: bZIP proteins. Vol. 2. London, U.K: Academic Press; 1995. [PubMed] [Google Scholar]
- 7.Inoue J, Sato R, Maeda M. Multiple DNA elements for sterol regulatory element-binding protein and NF-Y are responsible for sterol-regulated transcription of the genes for human 3-hydroxy-3-methylglutaryl coenzyme A synthase and squalene synthase. J Biochem. 1998;123:1191–1198. doi: 10.1093/oxfordjournals.jbchem.a022060. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman R J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 9.Khanna A, Campbell R D. The gene G13 in the class III region of the human MHC encodes a potential DNA-binding protein. Biochem J. 1996;319:81–89. doi: 10.1042/bj3190081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, Lee A S. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W W, Alexandre S, Cao X, Lee A S. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- 13.Li W W, Sistonen L, Morimoto R I, Lee A S. Stress induction of the mammalian GRP78/BiP protein gene: in vivo genomic footprinting and identification of p70CORE from human nuclear extract as a DNA-binding component specific to the stress regulatory element. Mol Cell Biol. 1994;14:5533–5546. doi: 10.1128/mcb.14.8.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linhoff M W, Wright K L, Ting J P. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: the invariant chain promoter as a model. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Littlewood T, Evan G. Protein profile-transcription factors 2: helix-loop-helix. Vol. 2. London, U.K: Academic Press; 1995. [PubMed] [Google Scholar]
- 16.Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 17.Min J, Shukla H, Kozono H, Bronson S K, Weissman S M, Chaplin D D. A novel Creb family gene telomeric of HLA-DRA in the HLA complex. Genomics. 1995;30:149–156. doi: 10.1006/geno.1995.9891. [DOI] [PubMed] [Google Scholar]
- 18.Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 19.Reith W, Siegrist C A, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci USA. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy B, Lee A S. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 1999;27:1437–1443. doi: 10.1093/nar/27.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 24.Shiu R P, Pouyssegur J, Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteins in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci USA. 1977;74:3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman R J, Prywes R. Activation of ATF6 and an ATF6 DNA Binding Site by the Endoplasmic Reticulum Stress Response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins; involvement of basic-leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Mori K. ATF6 activated by proteolysis directly binds in the presence of NF-Y (CBF) to the rcis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]