Abstract
E2F is a family of transcription factors required for normal cell cycle control and for cell cycle arrest in G1. E2F4 is the most abundant E2F protein in many cell types. In quiescent cells, it is localized to the nucleus, where it is bound to the retinoblastoma-related protein p130. During entry into the cell cycle, the protein disappears from the nucleus and appears in the cytoplasm. The mechanism by which this change occurs has, in the past, been unclear. We have found that E2F4 is actively exported from the nucleus and that leptomycin B, a specific inhibitor of nuclear export, inhibits this process. E2F4 export is mediated by two hydrophobic export sequences, mutations in either of which result in export failure. Individual export mutants of E2F4, but not a mutant with inactivation of both export signals, can be efficiently excluded from the nucleus by forced coexpression of the nuclear export receptor CRM1. Similarly, CRM1 overexpression can prevent cell cycle arrest induced by the cyclin kinase inhibitor p16INK4a, an E2F4-dependent process. Taken together, these data suggest that nuclear export contributes to the regulation of E2F4 function, including its ability to regulate exit from G1 in association with a suitable pocket protein.
Members of the E2F family of transcription factors are important regulators of cellular proliferation (for reviews, see references 6 and 30). Binding sites for E2F proteins exist in the promoters of genes that are induced during the G1-to-S transition and are required for cell cycle progression and DNA synthesis (4). Deregulated expression of E2F can promote entry into the cell cycle, neoplastic transformation, and apoptosis (1). E2F activity is regulated by interactions with pRB, the product of the retinoblastoma susceptibility gene, and two related pocket proteins, p107 and p130. Complexes of pocket proteins and E2F act as transcriptional repressors with growth-suppressing activity (18). During the G1-to-S transition, the growth-inhibiting properties of pRB are inactivated by phosphorylation catalyzed by cyclin-dependent kinases (cdk's) (28). Cyclin-dependent kinase inhibitors, such as the tumor suppressor protein p16INK4a, inhibit cdk activity, thereby inducing cell cycle arrest in G1 (34). p16INK4a induces growth arrest only when cells synthesize functional pRB (13, 19, 25, 27) and at least one of the two pRB-related proteins p107 and p130 (3). Active E2F-dependent transcriptional repression is also required for p16INK4a-induced G1 arrest (40). Together, these data suggest that E2F is required not only for cell cycle progression but also for pocket protein-mediated growth inhibition.
E2F is a family of related proteins. High-affinity DNA binding requires their heterodimerization with a structurally related DP subunit. Six E2F proteins (E2F1 to -6) and two different DP subunits (DP1 and -2) have been identified (6). Conserved domains mediate DNA binding, heterodimerization, pocket protein binding, and transactivation. Based on homology and on certain functional characteristics, E2F proteins can be subgrouped into three distinct classes (6). The first group is composed of E2F1, -2, and -3. They bind exclusively to pRB and contain a cyclin A binding domain that mediates inhibition of DNA binding during entry into S phase. The second group is composed of E2F4 and -5. They associate preferentially with p107 and p130, although both can interact with pRB as well (29). Unlike E2F1, -2, and -3, E2F4 and -5 are synthesized constitutively. E2F4 accounts for the majority of the E2F proteins in many cell types. The last member of the E2F family, E2F6, is a transcriptional repressor that lacks pocket protein binding and transactivation domains (6).
Recent observations suggest that the cell cycle-promoting and -inhibiting activities of E2F are mediated by different family members. E2F1 and E2F3 are required for cellular proliferation (17, 22, 37). By contrast, in fibroblasts, E2F4 and -5 are dispensable for normal proliferation and for reentry into the cell cycle from G0 (11, 16, 23, 33). However, a common function of E2F4 and E2F5 is required for cell cycle arrest in G1 induced by the cyclin-dependent kinase inhibitor p16INK4a (11). Taken together, these data suggest a specific role for E2F4 and E2F5 in pocket protein-mediated G1 arrest.
While E2F1, -2, and -3 are constitutively located in the nucleus, a significant fraction of endogenous E2F4 is found in the cytoplasm, and ectopically overexpressed E2F4 is mainly cytoplasmic (24, 26). The protein can be translocated to the nucleus by forced coexpression of DP2 or pocket proteins, both nuclear localization sequence (NLS)-containing proteins. It was originally suggested that the cytoplasmic localization of E2F4 results from the lack of an NLS and that its subcellular localization is regulated mainly by its association with DP and pocket proteins.
During the cell cycle, the ratio of nuclear to cytoplasmic E2F4 changes. In quiescent cells, E2F4 is primarily nuclear, and it disappears from the nucleus during entry into S phase (24, 36). Nuclear E2F4 is mainly bound to p130 in G0 and to pRB and p107 during the G1-to-S transition. Very little free E2F4 is detected in the nucleus (36). The disappearance of E2F4 from the nucleus during cell cycle entry correlates well with the timing of pRB phosphorylation. It has been suggested that free, uncomplexed E2F4 is selectively lost from the nucleus after it is released from pRB (36). Taken together, these data suggest that loss of E2F4 from the nucleus contributes to the alleviation of transcriptional repression of E2F-dependent target genes during cell cycle entry.
It is unclear, however, whether regulated nuclear import, nuclear export, or nuclear degradation mediates disappearance of E2F4 from the nucleus. Here we report that E2F4 shuttles between the nucleus and the cytoplasm. Moreover, it is exported from the nucleus in a process that depends on the nuclear export receptor, CRM1. Additional evidence suggests that the G1 arrest activity of E2F4, a property of certain pocket protein-E2F4 complexes (11), is negatively regulated by its controlled nuclear export.
MATERIALS AND METHODS
Cell culture.
Cells were cultivated at 37°C in a 10% CO2-containing atmosphere. U2-OS cells and HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS; HyClone). NIH 3T3 cells were maintained in DMEM supplemented with 10% bovine calf serum (GIBCO).
Plasmids.
E2F4 deletion and point mutants (E2F4[1-265], E2F4[266-416], E2F4[68,70A], E2F4[Δ84-105], E2F4[Δ169-199], E2F4[Δ383-388] and E2F4[Δ404-416]) were generated by standard cloning techniques using pCDNA-HA-E2F4 as a template. Details of the construction of these mutants are available upon request. The identities of the mutants were confirmed by DNA sequencing. Other expression plasmids have been described elsewhere: pCDNA3-HA-E2F4 (24), pCMV-HA-E2F1 (20), pCDNA-HA-E2F6 (12), pCMV-HA-DP2 (39), pCDNA-mycNPc-NLS (32), and pCDNA-HA-crm1 (9).
Transfection of U2-OS cells, leptomycin B treatment, and immunostaining.
U2-OS cells were plated onto coverslips in 30-mm-diameter cell culture dishes and transfected with 1 μg of each expression plasmid and 3 μl of Fugene 6 (Roche). Twenty-four hours later, cells were fixed in 3% paraformaldehyde and 2% sucrose in phosphate-buffered saline (PBS) for 10 min at room temperature. Where indicated, cells were incubated with 10 ng of leptomycin B (kind gift of B. Wolff, Vienna, Austria) per ml for 3 h before fixation. Cells were then permeabilized with 0.2% Triton X-100 in PBS for 5 min and incubated with primary antibody at room temperature for 1 h. The following primary antibodies were used: anti-E2F4 C108 (Santa Cruz), antihemagglutinin (anti-HA), anti-CRM1 (kind gift of G. Grosveld, St. Jude, Memphis, Tenn.), and anti-c-myc (9E10). To detect endogenous E2F4 with monoclonal antibody LLF4, fixation and permeabilization were performed as described elsewhere (36). Secondary antibody conjugated to rhodamine or fluorescein isothiocyanate was used where indicated (Roche). Nuclei were counterstained with 1 μg of Hoechst 33258 (Sigma) per ml in PBS. Immunostaining was visualized using the 60× objective of a Microphot SA fluorescence microscope (Nikon).
Heterokaryon fusion assay.
To detect nucleocytoplasmic shuttling, a heterokaryon fusion assay was performed essentially as described previously (32). HeLa cells (2 × 105) were plated on glass coverslips in 3-cm-diameter dishes. Twenty-four hours later, expression plasmids were transfected with 5 μl of Lipofectamine (GIBCO) in 1 ml of serum-free DMEM. Five hours later, 1 ml of DMEM–20% FCS was added, and the cells were incubated at 37°C for ≈16 h and then washed with serum-free medium and refed with DMEM–10% FCS. One hour later, 106 NIH 3T3 cells were plated onto the HeLa cells. After 3 h, the cells were treated with 75 μg of cycloheximide (Sigma) per ml for 1 h. The cells on the coverslip were then overlaid with a solution of 50% polyethylene glycol 8000 (Sigma) in DMEM for 2 min at 37°C to induce cell fusion. The cells were then washed twice with PBS and transferred back to DMEM–10% FCS containing 75 μg of cycloheximide per ml. One hour later, cells were fixed and stained as described above.
Analysis of entry into S phase by BrdU incorporation.
Aliquots of 4 × 105 U2-OS cells were plated onto coverslips in 60-mm-diameter cell culture dishes. Sixteen to 20 h later, cells were transfected with 0.5 μg of pCDNA-p16, pCDNA-p21, and/or pCDNA-HA-crm1 and 2 μl of Fugene 6 (Roche). A green fluorescent protein expression vector (0.2 μg at 10 ng/ml) was cotransfected to allow identification of transfected cells. Twenty-four hours later, cells were pulse-labeled with 50 μM bromodeoxyuridine (BrdU) for 1 h. Cells were then stained with an anti-BrdU antibody (Becton Dickinson) as described elsewhere (12). Transfected cells were identified by their green fluorescence, and the number of BrdU-positive cells was determined.
RESULTS
E2F4 shuttles between the nucleus and the cytoplasm.
To determine whether E2F4 is exported from the nucleus, we performed a heterokaryon fusion assay (32). In this assay, the ability of a protein to shuttle from one nucleus to a foreign one is analyzed in a cycloheximide-treated heterokaryon, a cell containing two different nuclei. Apparent transport from one nucleus to another would depend upon export from the former.
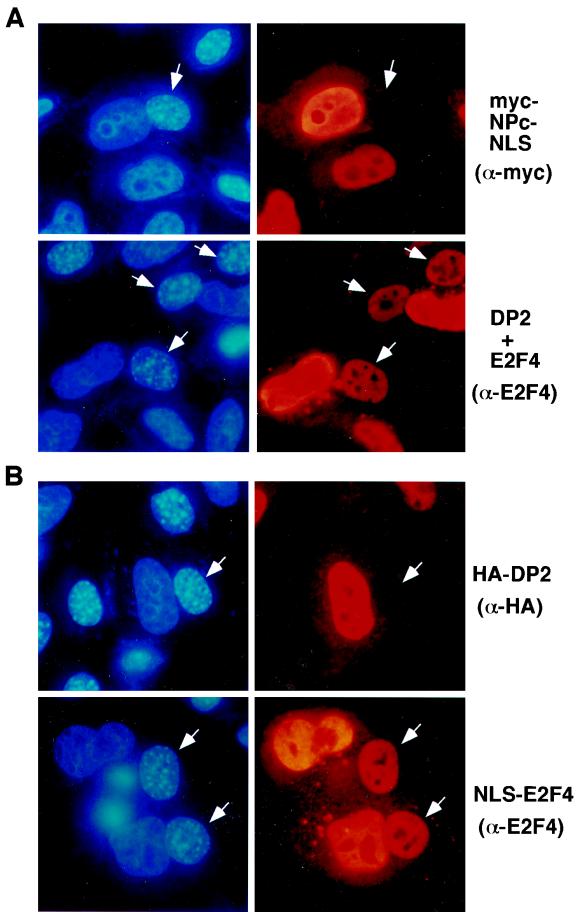
HeLa cells were transfected with an E2F4 expression vector and with DP2 to promote E2F4 import into the HeLa nucleus (24, 26). The transfected HeLa cells were fused to NIH 3T3 cells in the presence of cycloheximide to prevent further synthesis of E2F4, which would confuse the results. One hour after fusion, the culture was fixed, and the localization of the ectopically expressed protein was analyzed by immunostaining. E2F4 was repeatedly detected in NIH 3T3 nuclei of the resulting heterokaryons (Fig. 1A, bottom), indicating that it had been exported from a HeLa nucleus and then retransported into the NIH 3T3 nucleus. Thus, E2F4 can shuttle between the nucleus and the cytoplasm. A myc-tagged nucleoplasmin core fragment fused to an NLS (myc-NPc-NLS) was studied as a negative control. myc-NPc-NLS does not shuttle from the nucleus to the cytoplasm (32), and as expected, it was readily detected in HeLa but not NIH 3T3 nuclei after cell fusion (Fig. 1A, top).
FIG. 1.
(A) E2F4 shuttles between the nucleus and the cytoplasm. HeLa cells were transfected with expression plasmid for myc-NPc-NLS or E2F4 and DP2. After fusion with NIH 3T3 cells and incubation for 1 h, cells were fixed and stained using antibody 9E10 (α-myc) or C-108 (α-E2F4) and rhodamine-conjugated secondary antibodies (right panels). Nuclei were stained with Hoechst no. 33258 to identify murine nuclei in the fusion by their distinctive dot-like pattern (left panels). White arrows indicate NIH 3T3 nuclei in transfected heterokaryons. (B) E2F4 does not require DP2 to shuttle between the nucleus and the cytoplasm. HeLa cells were transfected with HA-tagged DP2 or NLS-E2F4, fused to NIH 3T3 cells as described for panel A, and stained with antibody 12CA5 (α-HA) or with C-108 (α-E2F4).
To find out whether E2F4 nuclear export is a specific property of HeLa cells, we fused E2F4-transfected monkey kidney (CV-1) cells to NIH 3T3 cells under the same conditions used in the HeLa-NIH 3T3 cell fusion experiments. E2F4 was again detected in the NIH 3T3 nuclei in these fused cells (data not shown). Therefore, nuclear export of E2F4 is not cell type specific and, given the human papillomavirus-infected property of HeLa cells, does not require human papillomavirus oncoproteins to support this process.
In these experiments, E2F4 was coexpressed with a heterodimeric binding partner, DP2, to concentrate it in the HeLa or CV-1 nucleus prior to the fusion (24, 26). Thus, nuclear export of E2F4 could, in principle, be mediated by DP2. However, when expressed alone, DP2 did not accumulate in NIH 3T3 nuclei in HeLa-NIH 3T3 heterokaryons. This result implies that DP2 was not exported from the nucleus and is not required for export of E2F4 per se (Fig. 1B, top). We also tested E2F4 fused to an NLS (NLS-E2F4) in these experiments. As shown previously, NLS-E2F4 localized in the nucleus of transfected HeLa cells without coexpression of DP2 (24). In HeLa-NIH 3T3 heterokaryons, NLS-tagged E2F4 was also detected in NIH 3T3 nuclei (Fig. 1B, bottom). These data reconfirm the view that E2F4 nuclear export is a DP2-independent process.
Leptomycin B inhibits nuclear export of E2F4.
In search of additional experimental evidence that E2F4 is exported from the nucleus, we utilized leptomycin B, a specific inhibitor of nuclear export. Exportin, or CRM1, has been identified as a receptor that is responsible for nuclear export of proteins that contain specific nuclear export sequences (NES) (8, 10, 31). Leptomycin B binds to CRM1 and inhibits its interaction with NES-containing proteins (21, 38).
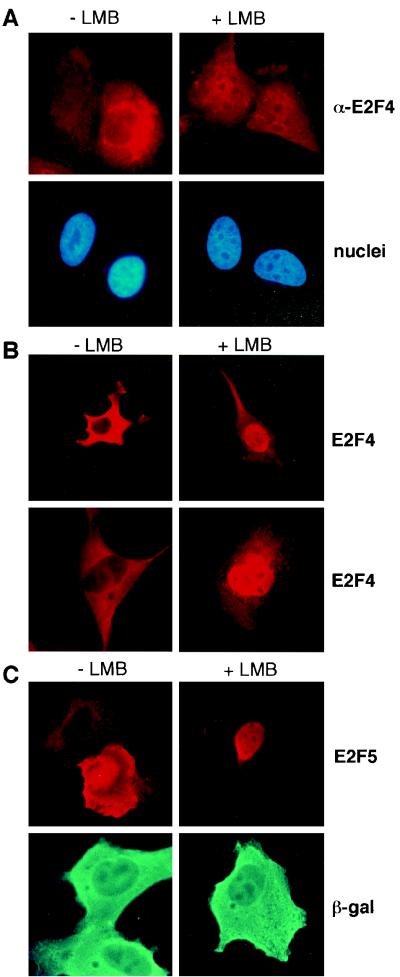
Consistent with previous studies, endogenous E2F4 was predominantly cytoplasmic in untreated U2-OS cells, as reflected by immunostaining with a monoclonal anti-E2F4 antibody (Fig. 2, −LMB). However, after incubation for 3 h with leptomycin B, E2F4 was detected in the nuclei of most cells in the culture (Fig. 2A, +LMB). These data further support the view that E2F4 is exported from the nucleus and suggest that this process is CRM1 dependent.
FIG. 2.
Nuclear export of E2F4 is inhibited by leptomycin B. (A) Asynchronously growing U2-OS cells were treated with carrier (−LMB) or 10 ng of leptomycin B per ml (+LMB) for 3 h and then stained with a monoclonal anti-E2F4 antibody (LLF4) (top). Nuclei were counterstained with Hoechst no. 33258. E2F4 was cytoplasmic in the majority of asynchronously growing U2-OS cells. In leptomycin B-treated cells E2F4 is no longer excluded from the nucleus. (B) U2-OS cells (top) or NIH 3T3 cells (bottom) were transfected with an expression plasmid for E2F4. Twenty-four hours after transfection, cells were mock treated (−LMB) or exposed to 10 ng of leptomycin B per ml (+LMB) for 3 h and then stained with anti-E2F4 (C-108) antibody. (C) U2-OS cells were transfected with expression plasmids for HA-E2F5 (top) or β-galactosidase (bottom). Cells were treated as described for panel B and stained with anti-HA (12CA5) or with anti-β-galactosidase antibodies.
To address the possibility that the cytoplasmic localization of ectopically overexpressed E2F4 is also a consequence of efficient nuclear export, we transiently expressed E2F4 and treated the cells with leptomycin B. The localization of E2F4 was then analyzed by immunofluorescence. As expected, ectopically overexpressed E2F4 was cytoplasmic in untreated NIH 3T3 and U2-OS cells (Fig. 2B, −LMB). However, after treatment with leptomycin B for 3 h, E2F4 efficiently accumulated in the nucleus (Fig. 2B, +LMB), suggesting that in the absence of drug, high levels of E2F4 accumulate in the cytoplasm as a result of efficient nuclear export.
To test whether the closely related protein E2F5 is also exported from the nucleus, we transiently transfected HA-tagged E2F5 into U2-OS cells, which were then exposed to leptomycin B. These cells were then immunostained with anti-HA antibody. Like E2F4, E2F5 was cytoplasmic in untreated cells but accumulated in the nucleus after leptomycin B treatment (Fig. 2C), indicating that E2F5 is also exported from the nucleus and that nuclear export of E2F5 is also mediated by CRM1. Importantly, leptomycin B had no effect on the cytoplasmic localization of transiently expressed β-galactosidase (Fig. 2C, bottom). Thus, the closely related proteins E2F4 and -5 are both targets of CRM1-mediated nuclear export.
E2F is an unstable protein, and its turnover is controlled by the ubiquitin-proteasome degradation system (14, 15). It has been suggested that regulated nuclear degradation contributes to the selective loss of nuclear E2F4 during entry into the cell cycle. By contrast, we observed that proteasome inhibitors had no effect on the cytoplasmic localization of E2F4, suggesting that proteasome-dependent nuclear degradation is not responsible for the cytoplasmic concentration of E2F4 (data not shown).
E2F4 contains two NES.
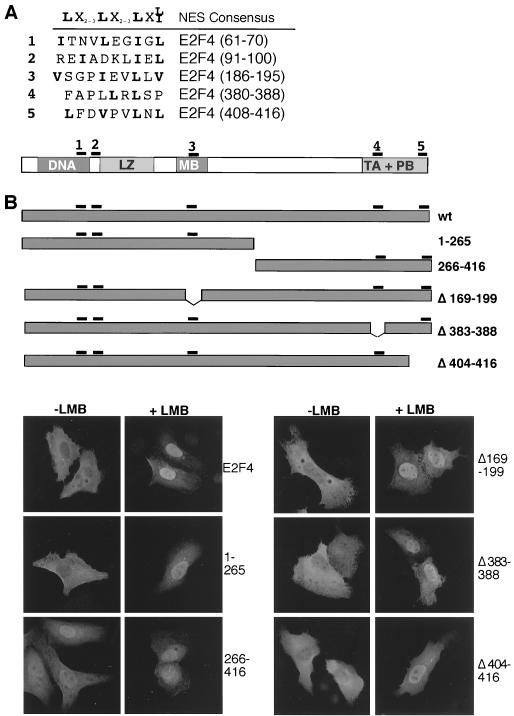
CRM1-dependent NES are characterized by short leucine- or isoleucine-rich hydrophobic regions (2). E2F4 contains five such sequences (Fig. 3A). To determine whether one or more of them are involved in its nuclear export, we generated a number of E2F4 deletion and missense mutants (Fig. 3B and C). Given that ectopically expressed E2F4 is exported from the nucleus (Fig. 2), we asked whether these mutants behaved similarly in transiently transfected cells. The data show that a fragment containing the amino-terminal 265 residues of E2F4 (E2F4[1-265]) was largely cytoplasmic in untreated cells but accumulated in the nucleus after leptomycin B treatment (Fig. 3B). In contrast, the carboxy-terminal portion of the protein, E2F4[266-416], was nuclear, even in the absence of leptomycin B. Thus, nuclear export appears to be a function of the amino-terminal region of E2F4. Consistent with these results, deletion of two potential export motifs in the carboxy terminus of otherwise intact E2F4 had no effect on its cytoplasmic localization (Fig. 3B, mutants E2F4[Δ383-388] and E2F4[Δ404-416]). Likewise, deletion of residues 169 to 199 did not affect the cytoplasmic localization of E2F4, suggesting that this region is also not involved in nuclear export (Fig. 3B, E2F4[Δ169-199]). Importantly, leptomycin B treatment resulted in nuclear accumulation of those mutants that, like wild-type protein, were cytoplasmic, indicating that the relevant mutation did not alter the ability of E2F4 to be imported into the nucleus (Fig. 3B, right panels).
FIG. 3.
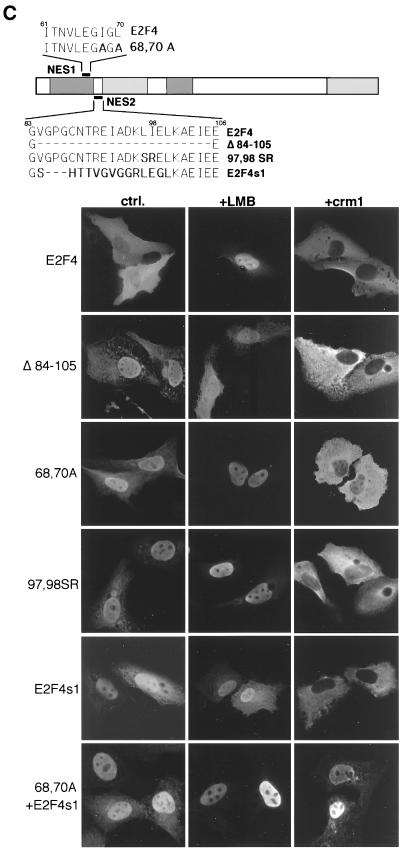
E2F4 contains two NES. (A) NES-like sequences in E2F4 were compared to a consensus NES (2). The location of the NES-like sequences in E2F4 is shown. (B) (Top) Schematic representation of E2F4 deletion mutants. (Bottom) E2F4 mutants were expressed in U2-OS cells. Their localization before and after treatment with 10 ng of leptomycin B per ml for 3 h was assessed by immunostaining with polyclonal anti-E2F4 antibody (C-108). (C) (Top) Schematic representation of NES1 and NES2 mutants. (Bottom) The E2F4 mutants shown were expressed in U2-OS cells, and their subcellular localization was evaluated by immunostaining with C-108. Where indicated, cells were treated with 10 ng of leptomycin B per ml for 3 h before fixation (+LMB). CRM1 was coexpressed where indicated (+crm1). The expression of CRM1 was confirmed by immunostaining with a polyclonal anti-CRM1 antibody (not shown). Nuclei were counterstained with Hoechst no. 33258 (not shown). ctrl., control.
Among the potential NES motifs in E2F4, the most amino terminal NES-like sequence (residues 61 to 70) most closely resembles a CRM1-dependent nuclear export motif (Fig. 3A). Indeed, conversion of two hydrophobic residues, isoleucine 68 and leucine 70, to alanine resulted in nuclear accumulation of E2F4 (Fig. 3C). Analogous changes in export motifs of other proteins also inhibited (CRM1-dependent) export (2). This result suggests that residues 61 to 70 function as a CRM1-dependent NES. Surprisingly, deletion of the second hydrophobic motif in the N-terminal region (residues 84 to 105) also resulted in nuclear accumulation of the mutant protein (Fig. 3C). These residues of E2F4 make up the short spacer region between the DNA binding and dimerization domains. When this segment (residues 84 to 99) was replaced by the corresponding sequence of E2F1 (residues 186 to 198), the resulting mutant, E2F4s1, was also nuclear in the absence of leptomycin B treatment (Fig. 3C). Thus, the E2F1 sequence did not function as a NES in this assay. Given the limited structural similarity of the second export motif to a consensus NES, we next asked whether hydrophobic amino acids in this region were needed for the cytoplasmic localization of E2F4. To address this possibility, we replaced the two hydrophobic residues, 97 (leucine) and 98 (isoleucine), with serine and arginine, respectively. The resulting mutant, E2F4[97,98SR], was nuclear in the absence of leptomycin B (Fig. 3C), indicating that these residues are likely required for the cytoplasmic localization of E2F4. These data suggest that this motif is also a hydrophobic NES. Consequently, we have referred to the two elements of the NES of E2F4 as NES1 (residues 61 to 70) and NES2 (residues 91 to 100) (Fig. 3C).
CRM1 overexpression results in exclusion of nuclear export mutants of E2F4 from the nucleus.
Because there are two, discrete NES in E2F4, we wondered whether single NES1 and/or NES2 mutants could still be exported from the nucleus by CRM1. It has been demonstrated that overproduction of CRM1 can result in exclusion of NES-containing proteins from the nucleus (41). As expected, overproduction of CRM1 did not change the cytoplasmic localization of wild-type E2F4 (Fig. 3C). However, it efficiently relocated the single NES1 and NES2 mutants E2F4[68,70A], E2F4[Δ84-105], and E2F4s1 from the nucleus to the cytoplasm (Fig. 3C), suggesting that there is a remaining CRM1-responsive NES in each of these mutant proteins. Consistent with this notion, the double mutant E2F4[68,70A+s1], with mutations in both hydrophobic regions, was nuclear in the majority of cells that overproduced CRM1 (Fig. 3C). Since neither NES1 nor NES2 is sufficient to direct nuclear export under normal conditions, they may represent the two parts of a bipartite export signal that are separated by a 20-amino-acid spacer.
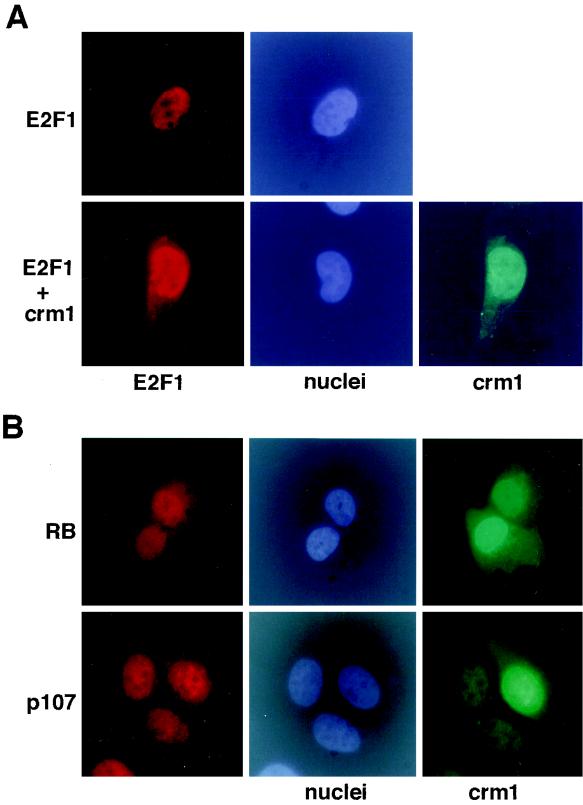
E2F1, -2, and -3 and pocket proteins are not excluded from the nucleus by CRM1.
Although the NES1 sequence is conserved among all E2F proteins, E2F1, -2, and -3 are constitutively localized to the nucleus (36). Given that ectopically expressed CRM1 resulted in exclusion of single NES mutants of E2F4 from the nucleus, we asked whether coexpression of CRM1 had a similar effect on the localization of E2F1. However, when E2F1 was expressed together with CRM1, E2F1 remained nuclear in the presence of CRM1 (Fig. 4A), suggesting that it is not normally exported from the nucleus. Nuclear localization of overexpressed E2F2, -3, and -6 was also unchanged by overexpression of CRM1 (data not shown). Taken together, these data imply that among the E2F proteins, nuclear export is a specific property of E2F4 and -5. On the other hand, one cannot rule out the formal possibility that E2F1, -2, and -3 do contain a functional NES but its experimental detection is masked by a powerful effect of the NLS present in these proteins.
FIG. 4.
Overexpression of CRM1 does not result in cytoplasmic localization of E2F1, pRB, and p107. (A) HA-E2F1 was expressed alone or coexpressed with CRM1 in U2-OS cells, as indicated. The subcellular localization of HA-E2F1 was determined by immunostaining with an anti-HA antibody. Expression of CRM1 was confirmed by immunostaining with a polyclonal anti-CRM1 antibody. (B) The subcellular localization of p107 and pRB in U2-OS cells expressing CRM1 was analyzed by immunostaining. For p107 a mixture of monoclonal antibodies, SD6 and SD9, was used. pRB was detected with a monoclonal antibody, 245. Expression of CRM1 was confirmed by immunostaining with a polyclonal anti-CRM1 antibody.
It has been suggested that association with pocket proteins plays a role in the regulation of the subcellular localization of E2F4 (24, 36). To address the question of whether pocket proteins are also exported from the nucleus, we overproduced CRM1 in U2-OS cells and then analyzed their subcellular localization by immunostaining. pRB and p107 were nuclear in the presence of ectopically overexpressed CRM1, implying that they are not exported from the nucleus by CRM1 (Fig. 4B). We were unable to detect p130 in asynchronously growing U2-OS cells by immunostaining. Together with the fact that E2F4 contains two NES, these data strongly suggest that nuclear export of E2F4 is not mediated by association with pocket proteins and is a function of E2F4 itself.
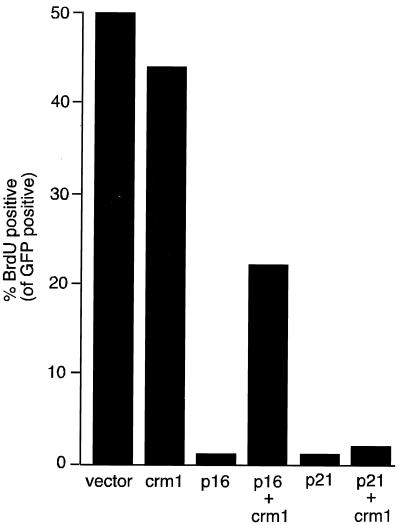
CRM1 overcomes p16INK4a-induced G1 arrest.
Cell cycle arrest induced by the cdk4 inhibitor p16INK4a requires either E2F4 or E2F5, consistent with a role for E2F4 and -5 in pocket protein-mediated cell cycle arrest in G1 (11). Because CRM1 can affect E2F4 intracellular localization (Fig. 3C), we wondered whether overexpression of CRM1 had any effect on G1 arrest induced by the cyclin kinase inhibitor p16INK4a. To address this possibility, we transiently expressed p16INK4a in U2-OS cells and analyzed entry into S phase by BrdU incorporation. As expected, p16INK4a reduced the number of cells in S phase by about 90% compared to the number of such cells transfected with a control plasmid (Fig. 5). Remarkably, the ability of p16INK4a to induce G1 arrest was inhibited when CRM1 was coexpressed. In contrast, when CRM1 was expressed alone, it had no significant effect on entry into S phase. Importantly, p16INK4a expression levels were unaffected by CRM1, and p16INK4a remained in the nucleus in cells expressing CRM1, as analyzed by immunostaining (not shown). Moreover, CRM1 had no effect on p21WAF1-induced growth arrest (Fig. 5), which is a pocket protein-independent process (5).
FIG. 5.
CRM1 overcomes p16INK4a-mediated cell cycle arrest. U2-OS cells were transfected with a control vector (pCDNA3) or with expression plasmids for CRM1, p16INK4a, or p21waf1 as indicated. A green fluorescent protein (GFP) expression plasmid was cotransfected to allow identification of transfected cells. Twenty-four hours after transfection, cells were labeled with BrdU for 1 h, fixed, and then stained with an anti-BrdU antibody. Transfected cells were identified by their green fluorescence, and the number of BrdU positive cells was determined. The experiment was repeated four times with similar results. Results of a typical experiment are shown.
Taken together, these data suggest that CRM1 inactivates one or more components of the pocket protein G1 arrest pathway. Since E2F4 and E2F5, and no other E2F family members or pocket proteins, are exported from the nucleus by CRM1, these data suggest that CRM1-dependent nuclear export regulates the G1 arrest function of E2F4 and E2F5. This model is consistent with our earlier finding that a shared function of either E2F4 or E2F5 is required for p16INK4a-mediated cell cycle arrest (11).
DISCUSSION
The activity of E2F family members is regulated by a variety of mechanisms. Given the results reported here, the list likely includes changes in subcellular localization.
It has been suggested that the predominant cytoplasmic localization of E2F4 results from the lack of an intrinsic NLS and that timely nuclear localization of the protein was achieved by association with pocket and/or DP proteins (24, 26). While it is possible that complex formation with these proteins contributes to E2F4 nuclear localization, data reported here show that E2F4 shuttles between the nucleus and the cytoplasm and that incubation with the export inhibitor leptomycin B results in its nuclear accumulation. Thus, the relatively low nuclear levels of ectopically overexpressed E2F4 and of the endogenous protein in asynchronous cultures are maintained by efficient, CRM1-dependent nuclear export.
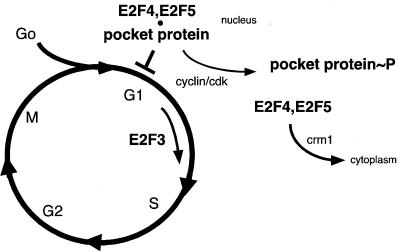
This mechanism could well explain the prior observation that free, uncomplexed E2F4 is selectively lost from the nucleus (36). E2F4 is largely nuclear in G0 cells and becomes progressively cytoplasmic as cells emerge into G1 and S (22, 33). The disappearance of E2F4 from the nucleus during entry into the cell cycle correlates well with the known timing of pocket protein phosphorylation, suggesting that E2F4 is exported from the nucleus once it is released from a phosphorylated pocket protein(s). Thus, our data suggest that the newly described E2F4 NES motifs contribute to the cell cycle-dependent changes in its subcellular localization. Moreover, given the fact that CRM1 overproduction overrode a p16INK4a-induced G1 block, which is an E2F4- and -5-dependent process (11), our results are consistent with the hypothesis that nuclear export of E2F4 contributes to cell cycle progression (Fig. 6).
FIG. 6.
Model for the role of CRM1-dependent nuclear export in G1 control (see text). E2F4 and E2F5 are required for pocket protein-mediated arrest in G1 (11). During entry into the cell cycle, pocket proteins are phosphorylated (∼P) by cdk and E2F is released. E2F4 (and probably E2F5) is exported from the nucleus by CRM1 after release from a relevant pocket protein. Other E2Fs, such as E2F3, are not exported, remain nuclear, and presumably contribute to the transcriptional activation of E2F-responsive genes (17, 22).
Several lines of evidence suggest that E2F4 and E2F5 are the components of the pocket protein pathway that are specifically inactivated by nuclear export. First, among the E2F proteins, nuclear export is a specific behavior of E2F4 and E2F5. Other E2F family members are constitutively nuclear and were not excluded from the nucleus by CRM1 overproduction (Fig. 4 and data not shown). The nuclear localization of p16INK4a and of pocket proteins was also not affected by CRM1 expression. Thus, of the different components tested, only E2F4 and E2F5 appear to be exported from the nucleus in a CRM1-dependent manner.
This model is consistent with recent observations that E2F4 and -5 serve primarily as negative regulators of cell cycle progression (11, 35). While they are not necessary for normal proliferation of embryonic fibroblasts, they exhibit a shared function that is required for cell cycle arrest induced by the cdk inhibitor p16INK4a. Since G1 arrest by pocket proteins depends upon their ability to form E2F-containing transcription-repressing complexes (40), these observations further suggest that E2F4 and E2F5 mediate pocket protein-dependent transcriptional repression in G1. We assume that E2F proteins that are likely not exported, such as E2F1 and E2F3, remain in the nucleus during cell cycle entry and contribute to the activation of E2F-responsive genes. Consistent with that notion, E2F3 is required for reentry into the cell cycle from G0 and plays a major role in promoting the G1-to-S transition of cycling cells (17, 22).
Intriguingly, the export function of certain transcription factor NES is regulated by the timely phosphorylation of specific neighboring residues (7). It is noteworthy that the state of phosphorylation of endogenous E2F4 was different depending upon whether it was nuclear (in G0/early G1) or not (in late G1 and S phase) (unpublished observations). Furthermore, the phosphorylation pattern of pRB-bound E2F4 differs from that of free E2F4 (11). One wonders whether specific phosphorylation of a serine or threonine contributes to regulation of its nuclear export.
ACKNOWLEDGMENTS
We thank Stefanie Hauser, Ulrike Kutay, Fabio Martelli, Pamala Silver, and our laboratory and divisional colleagues for many helpful conversations. We thank Sara Nakielny and G. Dreyfus for sending us the nucleoplasmin expression plasmid and for important advice on the use of the heterokaryon fusion assay. We also thank G. Grosveld (St. Jude Children's Hospital) for the polyclonal CRM1 antiserum and CRM1 expression plasmid and B. Wolff (Novartis, Vienna, Austria) for her gift of leptomycin B.
This work was supported by grants from the NIH-NCI to D.M.L., by fellowships from the European Molecular Biology Organization and the Leukemia and Lymphoma Society to S.G., and by a fellowship from the National Health and Medical Research Council of Australia to G.J.L.
REFERENCES
- 1.Adams P D, Kaelin W G., Jr The cellular effects of E2F overexpression. Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce J L, Hurford R K, Clason M, Koh J, Dyson N. Requirements for cell cycle arrest by p16INK4a. Mol Cell. 2000;6:737–742. doi: 10.1016/s1097-2765(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 4.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimri G P, Nakanishi M, Desprez P Y, Smith J R, Campisi J. Inhibition of E2F activity by the cyclin-dependent protein kinase inhibitor p21 in cells expressing or lacking a functional retinoblastoma protein. Mol Cell Biol. 1996;16:2987–2997. doi: 10.1128/mcb.16.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 7.Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 11.Gaubatz S, Lindeman G L, Ishida S, Jakoi L, Nevins J R, Livingston D M, Rempel R E. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 12.Gaubatz S, Wood J G, Livingston D M. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan K L, Jenkins C W, Li Y, Nichols M A, Wu X, O'Keefe C L, Matera A G, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 14.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway—regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann F, Martelli F, Livingston D M, Wang Z Y. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 16.Humbert P O, Rogers C G S, Landsberg R L T J M, Dandapani S, Brugnara C, Erdman S, Schrenzel M, Bronson R T, Lees J A. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol Cell. 2000;6:281–291. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 17.Humbert P O, Verona R, Trimarchi J M, Rogers C, Dandapani S, Lees J A. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 18.Kaelin W G., Jr Functions of the retinoblastoma protein. Bioessays. 1999;21:950–958. doi: 10.1002/(SICI)1521-1878(199911)21:11<950::AID-BIES7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 19.Koh J, Enders G H, Dynlacht B D, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 20.Krek W, Livingston D M, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 21.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 22.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindeman G J, Dagnino L, Gaubatz S, Xu Y, Bronson R T, Warren H B, Livingston D M. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 1998;12:1092–1098. doi: 10.1101/gad.12.8.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindeman J G, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 26.Magae J, Wu C L, Illenye S, Harlow E, Heintz N H. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- 27.Medema R H, Herrera R E, Lam F, Weinberg R A. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 29.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 31.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 32.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between the nucleus and the cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 33.Rempel R E, T. S-R M, Storms R, Morham S, Ishida S, Engel A J L, Melhem M F, Pipas J M, Smith C, Nevins J R. Loss of E2F4 activity leads to abnormal development of multiple lineages. Mol Cell. 2000;6:293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 34.Sherr C J, Roberts J M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi Y, Rayman J B, Dynlacht B D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 36.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z M, Yang H, Livingston D M. Endogenous E2F-1 promotes timely G0 exit of resting mouse embryo fibroblasts. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 39.Wu C-L, Zukerberg L R, Ngwu C, Harlow E, Lees J A. In vivo association of E2F and DP family proteins. Mol Cell Biol. 1995;15:2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H S, Postigo A A, Dean D C. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]