Abstract
Two efficient feather-degrading bacteria were isolated from honeybee samples and identified as Bacillus sonorensis and Bacillus licheniformis based on 16S rRNA and genome sequencing. The strains were able to grow on chicken feathers as the sole carbon and nitrogen sources and degraded the feathers in a few days. The highest keratinase activity was detected by the B. licheniformis CG1 strain (3800 U × mL−1), followed by B. sonorensis AB7 (1450 U × mL−1). Keratinase from B. licheniformis CG1 was shown to be active across a wide range of pH, potentially making this strain advantageous for further industrial applications. All isolates displayed antimicrobial activity against Micrococcus luteus; however, only B. licheniformis CG1 was able to inhibit the growth of Mycobacterium smegmatis. In silico analysis using BAGEL and antiSMASH identified gene clusters associated with the synthesis of non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKSs) and/or ribosomally synthesized and post-translationally modified peptides (RiPPs) in most of the Bacillus isolates. B. licheniformis CG1, the only strain that inhibited the growth of the mycobacterial strain, contained sequences with 100% similarity to lichenysin (also present in the other isolates) and lichenicidin (only present in the CG1 strain). Both compounds have been described to display antimicrobial activity against distinct bacteria. In summary, in this work, we have isolated a strain (B. licheniformis CG1) with promising potential for use in different industrial applications, including animal nutrition, leather processing, detergent formulation and feather degradation.
Keywords: Bacillus licheniformis, honey, bee, keratin, antibiotic, lichenicidin, feed, biodegradation
1. Introduction
Around 1 million tons of chicken feathers are produced as waste products annually by the poultry industry around the world [1]. In general, all types of feathers have a protein content of around 90%, which indicates that this waste could have great potential as a source of protein and amino acids for animal feed or many other applications [2]. However, the most abundant protein in feathers is keratin, an insoluble protein that is difficult to degrade due to the presence of hydrogen bonds, disulphide bonds and hydrophobic interactions [3]. Commonly, the discarded feathers are either dumped in landfills or incinerated. Sometimes, feathers are converted to feather meal by steam pressure and/or chemical treatment and utilized on a limited basis as a dietary protein supplement for animal feedstuffs [4]. However, this approach is unusual due to the poor digestibility of keratin by animals and the high cost of the production process. In fact, in its native state, keratin is not degradable by common proteolytic enzymes such as trypsin, pepsin or papain. Therefore, prior to usage, the feathers are steam pressure cooked or chemically treated to make them more digestible [5]. An alternative to these mechanical and chemical approaches is proteolysis of keratin by enzymatic and/or microbiological methods, thereby improving the nutritional value of feather waste by avoiding the destruction of certain amino acids, such as methionine, tryptophan and lysine [6].
Keratin degradation requires a specific class of proteases known as keratinase [4]. Although several microorganisms, including fungi and bacteria, are known to degrade keratin, Bacillus species are considered as the most effective keratin degraders [7]. Lin et al. [8] were the first to successfully isolate and characterize a keratinase enzyme from a keratin-degrading bacterium, Bacillus licheniformis PWD-1, that they previously isolated. Recently, we have also characterized a keratin-degrading strain that, in addition to feathers, can degrade other type of wastes, such as bioplastics [9]. The strain, initially referred to as Bacillus pumilus B12 [10], was reclassified as Bacillus altitudinis B12 after genomic analyses [9]. Other strains with keratinolytic activity have also been reported and, in some cases, highlighted as promising candidates for the development of cost-effective and eco-friendly platforms for the conversion of feathers into value-added products, especially in the form of livestock feed [2,5,6,7]. To develop a biotechnological application for that purpose, it would be of great interest to characterize a strain that, in addition to its keratinolytic activity, would exhibit an important antimicrobial potential. This feature would inhibit certain pathogenic microorganisms present in the feathers, thereby decreasing the risk of causing diseases (such as gastroenteritis or tuberculosis) in animals fed with these wastes [11,12,13]. There is currently a global antibiotic resistance crisis. New antibiotics are rarely discovered, and the number of resistant microbes is increasing worldwide.
At present, scientists are searching for new antimicrobial weapons and therapies against microbial infections in natural products and unexplored ecological niches. For decades, soil has been used as the main source for the identification of novel antibiotics. However, there is an urgent need to explore other ecological niches as the screening of soil samples has become overexploited. Interestingly, ecological niches in which symbionts work together to defend themselves by producing bioactive compounds are relatively underexploited. This is the case in beehives. Some authors have found antimicrobial metabolites in honeybee products, such as pollen and honey, where a significant variety of different bacteria derive from plants and bees [14,15]. Honeybees manufacture honey, bee bread (pollen) and royal jelly using secretions from their stomach, salivary glands and hypopharynx glands, and these natural products are continuously exposed to the environment [15]. Honeybees transfer microorganisms vertically (mother to daughter) or horizontally, via trophallaxis (mouth to mouth feeding) and coprophagy (anus to mouth). These microorganisms have a very important function in the digestion of plant polymers and in the preservation of honey and bee pollen by controlling the growth of pathogens [14,15]. Therefore, honeybee products are promising sources for isolating novel antibiotic-producing microorganisms. In previous work, we have shown that raw honeybee products provide antibiotic-producing isolates (Streptomyces) and lactic acid bacteria (LAB) with high antimicrobial potential and probiotic features, respectively [14,15]. Among the antibiotic producers isolated, Streptomyces species stand out as most interesting, as they are responsible for production of over half of the antibiotics discovered so far [16,17,18]. On the other hand, the isolated LAB (Lactobacillus and Pediococcus) can potentially exhibit good probiotic properties in animals and/or humans as they are able, among other capabilities, to activate type-I interferon production [15]. In another study, both probiotic and antimicrobial properties have been described for some Bacillus strains isolated from stingless bee honey collected across Malaysia [19]. Therefore, the products that honeybees manufacture are a very valuable source for the isolation of antibiotic-producing bacteria. Whether these bacteria derive as symbionts of honeybees or environmental contaminants is a very important question that remains to be elucidated.
The aim of this study was to isolate novel Bacillus strains with both high keratinolytic activity to degrade feather wastes in an efficient manner and antimicrobial potential to inhibit major pathogens, such as Mycobacterium. To this end, studies were carried out on antibiotic-producing and probiotic strains previously isolated from natural honeybee products collected from 15 apiaries across Southeast England [20]. Genome sequencing analysis of the isolates resulted in the identification of novel strains. Furthermore, phenotypic characterization was performed in a systematic manner to select for the most promising candidate with potential in industrial applications.
2. Materials and Methods
2.1. Microbial Growth Conditions and Growth Determination
Bacillus cells were grown in a mineral salt medium with feathers as the sole carbon and nitrogen sources, with the following composition (g/L): KH2PO4 (0.7), K2HPO4 (1.4), MgSO4 (0.1), NaCl (0.5), ZnSO4·7H20 (0.05), FeSO4·7H20 (0.015) and feathers (10). pH of the medium was maintained at 7, nevertheless, growth of cells was also determined across a range of pH (6–9). Chicken feathers provided by a poultry farm in Valladolid, Spain, were first washed with tap water and Triton-X to get rid of any debris, followed by a final cleaning step with distilled water. Then, clean feathers were dried at 60 °C overnight and manually cut into small pieces. Cultures were performed in either 120 mL or 2.2 L bottles, depending on the experiment. In all experiments, cultures of the respective strain grown until the mid-exponential phase (i.e., OD600~1.5) were inoculated into the defined media at 2% (v/v). The bottles containing the inoculated cultures were hermetically closed with an isoprene rubber and an aluminum crimp seal and incubated at 37 °C with shaking (150 rpm) for several days. Determination of bacterial cell growth was performed by measuring O2 consumption and CO2 production in the glass bottles using a Gas Chromatograph (Varian Bruker 430-GC, Middelburg, The Netherlands) equipped with a thermal conductivity detector (GC-TCD), as previously described [9]. Abiotic controls (without bacterial inoculum) were also maintained under the same conditions to monitor O2 and CO2 concentrations in the bottles, which should remain invariable.
2.2. Experimental Design, Sampling, DNA Extraction and 16S rRNA Sequencing
Honeybee samples were collected from 15 apiaries across Southeast England. Sample collection took place between mid-June and mid-August and targeted several habitats and soils in 4 different counties, as well as different beehives, some of which were located within the same apiary [20]. Samples were collected directly from honeycomb frames. Ten grams each of bee product sample were collected in sterile swab tubes and containers respectively, which were then immersed in liquid nitrogen and stored at −80 °C. To ensure full representation of the whole beehive, samples were collected from different parts of the honeycomb.
To enrich the presence of Bacillus strains in the honeybee products, the samples were incubated at 80 °C for 10 min to select for endospore-forming bacteria. The samples were serially diluted and seeded onto Tryptone Soya Agar (TSA) medium to favor Bacillus growth. After incubation, isolates with stronger antimicrobial properties were selected and further analyzed using 16S rRNA sequencing to select strains belonging to the Bacillus genus.
DNA extraction was carried out from cultures in Tryptone Soya Broth (TSB) using the EZNA bacterial DNA kit according to the manufacturer’s instructions (Omega Bio-Tek, Doraville, CA, USA) and as previously reported [18]. 16S rRNA gene amplicon sequencing was performed on an Illumina MiSeq sequencer using universal 16S rRNA bacterial primers for V3–V4 regions. Among the isolates, three were found to be Bacillus sonorensis, one was B. licheniformis, one was B. subtilis and one was an unclassified Bacillus species that had 100% sequence similarity to Bacillus sp. BAB-4886.
2.3. Genome Sequencing, Assembly and Annotation
DNA sequencing was performed by MicrobesNG (University of Birmingham, United Kingdom) using the Illumina MiSeq platform as previously described [21]. Briefly, the library was prepared with the 250 NexteraTM XT Library Prep Kit Genome sequencing and the quality of the generated reads was trimmed with Trimmomatic [22]. The generated contigs were assembled from the paired-end reads using Shovill v.1.0.41 with SPAdes 3.13.0 [23] and the resulting genome assemblies were verified by N50 and L50 using Quast v.4.5 [24] and annotated with Prokka v.1.13 [25]. The secretome for each of the strains was inferred by predicting the presence of signal peptides using SignalP-5.0 [26]. Sequencing reads, genome assemblies and metadata have been uploaded onto Genbank in BioProject PRJNA907715, with the next genome accession numbers: JAPQWB000000000 (Z1), JAPQWC000000000 (CG1), JAPQWD000000000 (X1), JAPQWE000000000 (X2), JAPQWF000000000 (AB6) and JAPQWG000000000 (AB7).
2.4. Taxonomic Analysis
For the taxonomy analysis, the web-based tool JSpeciesWS [27] was employed (https://jspecies.ribohost.com/; accessed on 3 November 2022). An initial search for the closest relative of each of the isolates was carried out using Tetra Correlation Search (TCS) [28], followed by a more systematic comparison with B. subtilis, B. sonorensis and B. licheniformis species using pairwise Average Nucleotide Identity with BLAST (ANIb) calculations [29]. The phylogenetic analysis was performed using the dendrogram function in SciPy. A distance between species was defined as 100 minus the obtained ANI values and the resulting matrix of distances was used to construct the dendrogram.
2.5. Keratinase Activity Detection Assay
Keratinase activity was determined following previous existing methods [8,30]. Liquid samples of 0.7 mL were daily extracted of each culture and centrifuged 20 min at 13,000 rpm to obtain a final 0.5 mL aliquot of supernatant. With the centrifugation step, cells and feather debris are removed. The 0.5 mL aliquot was mixed with 0.5 mL of 100 mM glycine-NaOH (pH 10) containing 1% casein and incubated at 37 °C for 20 min. In this reaction, tyrosine is released after the breaking of casein by the keratinases, among other enzymes. The reaction was stopped by addition of 0.5 mL of trichloroacetic acid (20% w/v) and incubation for 15 min at room temperature. Following the inactivation of the enzymes, the samples were centrifuged during 15 min at 13,000 rpm and the supernatant collected and measured with a spectrophotometer at 280 nm. A standard curve was performed using solutions of 0–700 mg L−1 of tyrosine. One keratinase unit (U) was defined as the amount of enzyme required to increase the absorbance by 0.01 (OD280nm) in one minute under the assay conditions employed.
2.6. Growth Inhibition Bioassay
To study the growth inhibition of Micrococcus luteus and Mycobacterium smegmatis, aliquots of 5 µL of each of the Bacillus isolates were added onto the surface of TSA plates containing 108 CFU × mL−1 of the respective indicator strain. Plates were incubated at 30 °C until visible halos of inhibition appeared.
2.7. Analysis of Gene Clusters with Antimicrobial Potential
The genomes of Bacillus isolates were analyzed to identify gene clusters associated with the biosynthesis of antimicrobial compounds. The secondary metabolites clusters were identified using antiSMASH v.6.0 [31] and BAGEL v.3 [32].
3. Results
3.1. Isolation and Selection of Bacillus Strains from Raw Honeybee Products
Natural Bacillus strains with antimicrobial potential were isolated using a screening approach from samples of raw honey, bee bread (referred as to pollen henceforth) and royal jelly collected from 15 apiaries across Southeast England [20]. Colonies with stronger antimicrobial properties against Micrococcus luteus ATCC4698 were selected and further analyzed using 16S rRNA sequencing to select for Bacillus species. We isolated six different Bacillus strains (Z1, CG1, AB6, AB7, X1 and X2) from either raw honey or royal jelly samples (see Table 1).
Table 1.
Strains isolated from honeybee samples in South England and characterized in this and previous studies.
| Strain | Organism | Source | Location | BioSample | References |
|---|---|---|---|---|---|
| O29 | Lactobacillus kunkeei | Bee pollen | Verney Junction, BUCKINGHAMSHIRE | SAMN11831833 | [15] |
| E1 | Pediococcus acidilactici | Raw honey | Saffron Walden, ESSEX | SAMN11831832 | [15] |
| AD1 | Streptomyces drozdowiczii | Raw honey | Woking, SURREY | SAMN20207146 | [14] |
| AD2 | Streptomyces griseoaurantiacus | Raw honey | Woking, SURREY | SAMN20207147 | [14] |
| AN1 |
Streptomyces
albus |
Bee pollen | West Byfleet, SURREY | SAMN20207148 | [14] |
| Z1 |
Bacillus
subtilis |
Raw honey | Woking, SURREY | SAMN31988539 | This study |
| CG1 | Bacillus licheniformis | Raw honey | Shere, SURREY | SAMN31988540 | This study |
| AB6 | Bacillus sonorensis | Raw honey | West Byfleet, SURREY | SAMN31988543 | This study |
| AB7 | Bacillus sonorensis | Raw honey | West Byfleet, SURREY | SAMN31988544 | This study |
| X1 | Bacillus sonorensis | Royal jelly | Woking, SURREY | SAMN31988541 | This study |
| X2 | Bacillus sonorensis | Royal jelly | Woking, SURREY | SAMN31988542 | This study |
3.2. Taxonomy of the Bacillus Isolates and General Features of Their Genomes
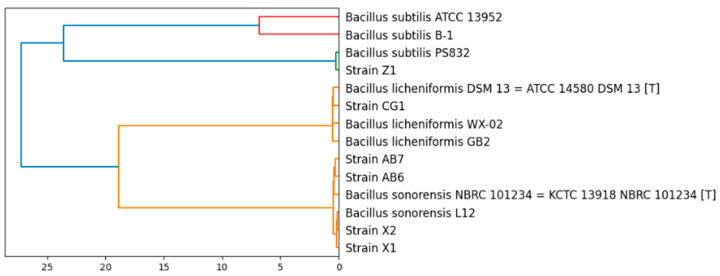
The 16S rRNA sequencing analysis informed that the six isolates were Bacillus, and more specifically B. subtilis, B. licheniformis and B. sonorensis, depending on the strain (see Table 1). To have a better insight about these strains, their genomes were sequenced. The sequenced genomes were used to classify the isolated strains based on whole genome similarities (to confirm or correct the taxonomy obtained with 16S rRNA sequencing). A pairwise ANIb calculation was carried out between all the possible combinations of the isolated strains and several strains of B. subtilis, B. sonorensis and B. licheniformis contained in the JSpeciesWS database [27]. The constructed dendrogram (Figure 1) clearly shows that the newly sequenced strains belong to the species B. subtilis (Z1), B. licheniformis (CG1) and B. sonorensis (AB6, AB7, X1 and X2).
Figure 1.
Phylogenetic relations of the isolated strains with strains of the species B. subtilis, B. sonorensis and B. licheniformis.
Table 2 summarizes the predicted general features of each of the Bacillus genomes as well as the proteins that each of the strains might secrete according to RAST and SignalP-5.0 analyses.
Table 2.
Data obtained from the genomes of the six Bacillus strains isolated in this study.
| Strain | Genome Accession | Bases | Contigs | Encoded Proteins | Secreted Proteins |
|---|---|---|---|---|---|
|
Bacillus subtilis Z1 |
JAPQWB000000000 | 4316935 | 305 | 4280 | 399 |
| Bacillus licheniformis CG1 | JAPQWC000000000 | 4228198 | 208 | 4252 | 407 |
| Bacillus sonorensis AB6 | JAPQWF000000000 | 4767461 | 595 | 4532 | 441 |
| Bacillus sonorensis AB7 | JAPQWG000000000 | 4580150 | 233 | 4440 | 437 |
| Bacillus sonorensis X1 | JAPQWD000000000 | 4903862 | 502 | 4770 | 447 |
| Bacillus sonorensis X2 | JAPQWE000000000 | 4809962 | 323 | 4807 | 445 |
3.3. Antimicrobial Assays
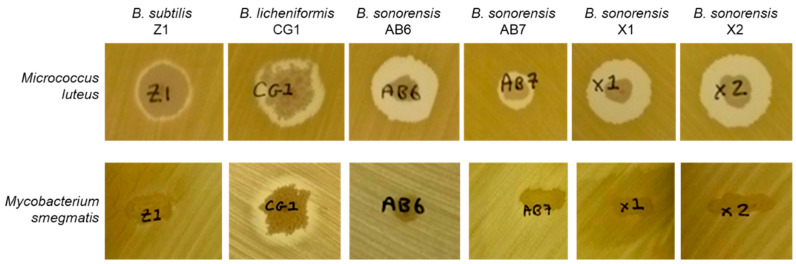
After the initial antimicrobial screening against M. luteus (see Figure 2), the six Bacillus isolates were also tested against other pathogenic bacteria, such as Paenibacillus alvei DSM29 (B), Staphylococcus aureus NCTC8325 (C) and Mycobacterium smegmatis MC2155. Antagonistic activity was only seen by B. licheniformis CG1 against M. smegmatis (Figure 2). Therefore, while M. luteus was susceptible to the six strains, M. smegmatis was only susceptible to the CG1 strain. As Mycobacterium species are quite resistant to antibiotic treatments, this result highlights the B. licheniformis CG1 isolate as an interesting antibiotic-producing strain.
Figure 2.
Antimicrobial assays of Bacillus isolates against Micrococcus luteus ATCC4698 and Mycobacterium smegmatis MC2155.
3.4. Characterization of the Keratin-Degrading Activity of the Isolated Bacillus Strains
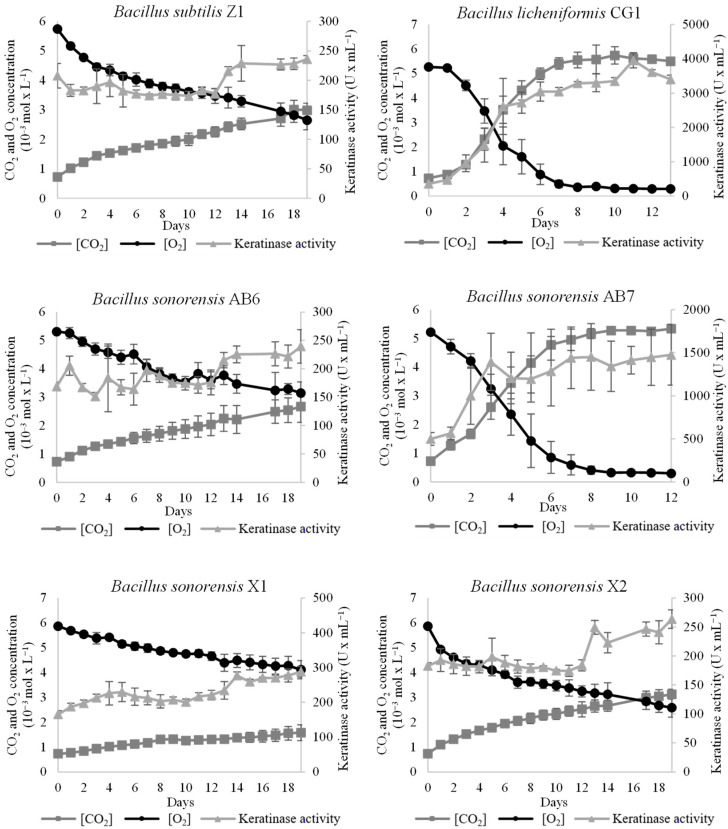
To study the keratin-degrading activity of the Bacillus isolates over keratin-rich wastes, the strains were assessed on their ability to grow on a medium with chicken feathers as the sole carbon and nitrogen source, as well as on their production of keratinase. The results showed in Figure 3 indicated that the strains with better degrading capabilities were B. licheniformis CG1 and B. sonorensis AB7. Both strains were able to grow and produce keratinases until the oxygen (O2) was depleted from the sealed bottes. Keratinase activity was more than 2.6 times higher in B. licheniformis CG1 in comparison to B. sonorensis AB7, and almost 16 times higher than the rest of isolates (see Table 3), which makes this strain a promising keratinase producer under minimal nutritional requirements.
Figure 3.
Growth and keratinase activity of the Bacillus isolates on chicken feathers as sole carbon and nitrogen source. Samples for growth (CO2 production and O2 consumption) and keratinase activity were collected every 24 h until O2 was depleted from the bottles or until when 20 days were reached. Vertical error bars correspond to the standard error of the mean of three replicated experiments.
Table 3.
Summary of the antimicrobial and keratinolytic activities obtained for each of the strains.
| Strain | Anti- Micrococcus Activity | Anti- Mycobacterium Activity | Max. Keratinase Activity (U/mL) |
|---|---|---|---|
| Bacillus subtilis Z1 | Positive | Negative | 240 |
| Bacillus licheniformis CG1 | Positive | Positive | 3800 |
| Bacillus sonorensis AB6 | Positive | Negative | 240 |
| Bacillus sonorensis AB7 | Positive | Negative | 1450 |
| Bacillus sonorensis X1 | Positive | Negative | 290 |
| Bacillus sonorensis X2 | Positive | Negative | 260 |
3.5. Gene Cluster Associated with the Biosynthesis of Putative Antimicrobial Metabolites
Gene clusters associated with the synthesis of non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKSs) and/or ribosomally synthesized and post-translationally modified peptides (RiPPs) were identified in most of the Bacillus isolates using both BAGEL3 and antiSMASH_v.6.0 software (see Supplementary Table S1). BAGEL3 was unable to identify NRPS, PKSs or RiPPs in AB7 and X2 strains. Nevertheless, antiSMASH_v.6.0 analysis predicted gene clusters for the production of lichenysin in all strains, with the exception of B. subtilis Z1. This metabolite has been shown to display antimicrobial activity against Gram-positive bacteria [33] and it could account for the inhibition of M. luteus growth by all Bacillus isolates tested. Surfactin [34], another predicted compound, would account for the antimicrobial activity displayed by B. subtilis Z1 (see Supplementary Table S1).
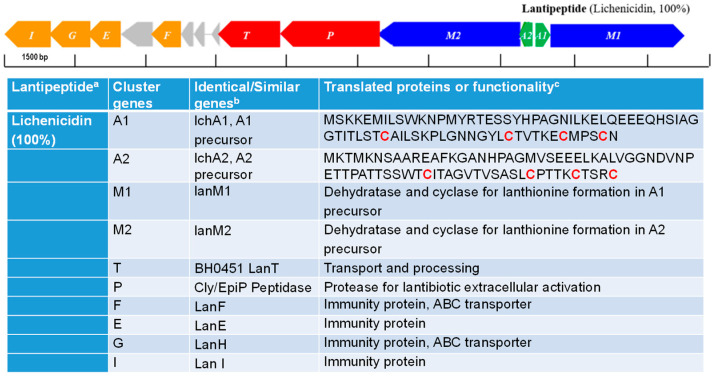
Interesting, B. licheniformis CG1, the only strain that inhibited the growth of the mycobacterial strain (see Figure 2), contained (in addition to the sequences with 100% similarity to lichenysin) another sequence with 100 % similarity to lichenicidin (see Figure 4). Lichenicidin is a lantibiotic compound with antimicrobial activity against a wide range of Gram-positive bacteria [35], which has been previously shown to be produced by another B. licheniformis strain, i.e., DSM13 [36]. This antibiotic might account for the antimicrobial activity against M. smegmatis observed by the CG1 strain; however, this hypothesis would require further testing.
Figure 4.
Gene cluster associated with the synthesis of Lichenicidin identified in the genome of B. licheniformis CG1. The color of the arrows indicate the nomenclature for genes encoding for: hypothetical precursor peptides (green arrows), posttranslational modification enzymes (blue arrows), transport/processing proteins (red arrows), immunity proteins (orange arrows) and hypothetical proteins (grey arrows). The hypothetical RiPPs are indicated next to each cluster, on the right-hand side, along with their corresponding identity alignment scores when compared to similar RiPPs (in brackets). In the Table: a percentage of similarity (indicated in brackets) with described lichenicidin; b Similarity with proteins reported previously; c cysteines involved in the lanthionine biosynthesis (indicated in red).
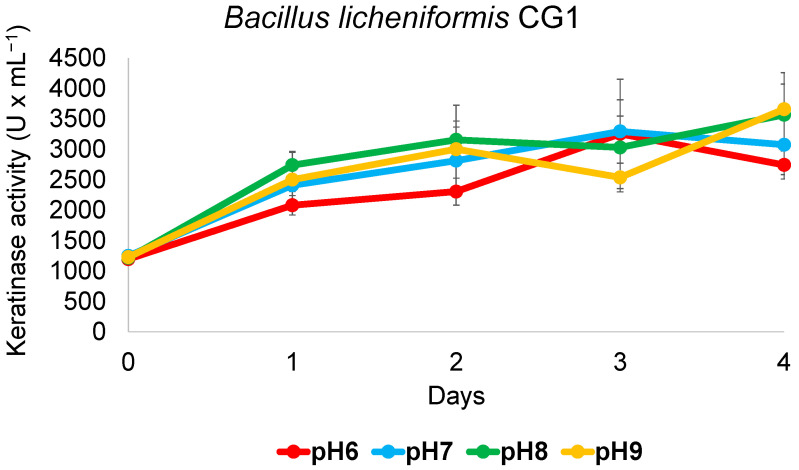
3.6. B. licheniformis CG1 Keratinase Activity Detected across a Wide Range of pH
Factors such as microbial strain, medium composition, pH and temperature play major roles in the production of highly active keratinase enzyme. As we have shown, B. licheniformis CG1 displays the most interesting antimicrobial potential and the highest keratinase activity among the Bacillus isolates. Therefore, this strain would be a good candidate for biotechnological applications in feather valorization. For industrial approaches, it is important that the employed strain remains active across a range of pH, among other parameters. To study this, keratin degradation using smaller bottles and less volume of medium (20 mL instead of 200 mL) at pH values from 6 to 9 was performed. As is shown in Figure 5, the strain showed a similar keratinase activity with all pH values tested, reaching around 3000 units per mL in just 2–4 days, which might provide an advantage for further industrial applications.
Figure 5.
Keratinase activity of B. licheniformis CG1 across a range of pH. Cultures grown at 37 °C and 150 rpm for 4 days in 120 mL bottles hermetically closed showed similar keratinase activity in a medium adjusted to varying pH values and where chicken feathers were the sole carbon and nitrogen source. Vertical error bars correspond to the standard error of the mean of three replicated experiments.
4. Discussion
Famine and lack of resources are a problem in certain parts of the world, whereas waste management is a problem in developed countries, which results in environmental problems and diseases. In this sense, a more sustainable use of natural resources is of vital importance. This especially applies to those wastes that can serve as food, either for humans or for animals. A clear example is the feather residues generated by the poultry industry, which are usually discarded into the environment, generating ecological problems [37]. Biodegradation is a process carried out by organisms that transform waste (and other compounds) into novel products that can be of interest for the industry. Therefore, this natural process could play a key role in the creation of viable end-products using wastes of a different nature in a circular and environmentally friendly approach.
Feathers serve as an example of cheap waste with potential nutritional value [38]. Poultry farming produces thousands of tons of chicken feathers in every year [4]. This waste product cannot be digested by most animals; therefore, it has to be degraded in some form to be usable. Several microorganisms with keratinolytic activity have been reported in the last few decades [7]. In this study, we have isolated and characterized two Bacillus strains with significant keratinolytic activity. The highest keratinolytic activity was achieved by B. licheniformis CG1 (3800 U × mL−1). In previous work, Bacillus altitudinis B12 displayed maximal keratinolytic activity of 1500 U × mL−1 [9]. In this study, B. sonorensis AB7 exhibited a comparatively similar magnitude of keratinolytic activity (1450 U × mL−1). This means that B. licheniformis CG1 shows a keratinolytic activity more than 2.6 times higher in comparison to the other two strains. Our results correlate well with previous studies. For example, in a study with B. licheniformis strain PWD-1, a similar magnitude of keratinolytic activity was reported (i.e., 3750 U × mL−1) [8]. Similarly, Hmidet et al. [39], after an in-depth optimization of the testing conditions, reported B. licheniformis strain NH1 with a maximal keratinase activity of 3960 U × mL−1. On the other hand, many studies have shown that the Bacillus strains analyzed (or other genera of bacteria) reached a maximal keratinolytic activity of 1000 U × mL−1 [40]. For example, Bacillus pumilus AR57 was found to be the most potent keratinase producer out of 39 isolates. However, the maximum keratinase activity reached by this strain was around 700 U × mL−1 [41], more than five times lower than B. licheniformis CG1, PWD-1 or NH1. In summary, our work corroborates previous studies that report that B. licheniformis species constitute one of the most important and effective keratin-degrading microorganisms discovered to date [8,39]. These observations highlight B. licheniformis species as promising candidates for feather degradation and keratinase production approaches. Keratinases are highly employed in different industrial applications, including animal nutrition, leather processing, detergent formulation and biofertilizer development [42]. In addition, as the bacterium grows on feathers as the sole carbon, nitrogen, sulfur and energy source, the utilization of feathers by these strains represents a method for replacing the commercial and more expensive substrates normally employed, therefore cheapening the cost of the culturing conditions. Thus, the utilization of feathers as substrate could result in a cost-effective process suitable for large-scale production of commercial enzymes produced by this species, such as those used in laundry detergent formulations [42]. The production of any other valuable metabolite that the bacterium could produce (either intrinsically or heterologously) would also be cheapened. Moreover, the absence of rapidly metabolizable carbohydrates in chicken feathers could also be beneficial for avoiding carbon catabolite repression [43], which is often observed in the production of extracellular proteases, including the well-known alkaline proteases produced by this bacterium [44].
This study has found that honeybee products, including honey and royal jelly, can contain Bacillus species. For centuries, it has been known that honey is an effective wound-healing agent and can even heal wounds that are unable to be healed by conventional treatments [45,46]. Studies into the antimicrobial activity of honey believed its effects were due to high osmolality of honey, and the presence of hydrogen peroxide [47]. However, more recent studies have shown that bacterial isolates from honey are able to exhibit antimicrobial activity against several foodborne pathogens, which suggests that the bacteria within honey contribute to its antimicrobial effects [48]. For example, it was shown that some Bacillus species, present in the gut of honeybees, were able to kill the pathogenic Paenibacillus larvae, which is the causative agent of foulbrood disease in bees, providing further indications that the bacteria within the honeybee microbiome may exhibit antimicrobial properties [49]. Actually, Bacillus species are an important source of antibiotics [50]. Some of these antimicrobial compounds are lipopeptides, which are produced by NRPS encoded in clusters on the bacterial genome [50]. Lipopeptides, that act through the disruption of membranes, are a particularly important source of antimicrobials, as it is less likely that resistance will develop towards them compared to other antibiotics. However, because of their toxicity, their use as therapeutics has been limited to mainly topical agents [51]. Well-known lipopeptides produced from Bacillus species include surfactin, fengysin and lichenysin, which have powerful antibacterial and antiviral properties [52]. Additionally, secondary metabolites of Bacillus can also be produced by PKS, also encoded within gene clusters on the bacterial genome. There are three categories of PKS: type I, type II and type III [53]. PKS are categorized in terms of structure; where the catalytic domains of type I PKS enzymes are within a single protein, the catalytic domains of type II and type III PKS enzymes are on separate proteins [54]. The possession of PKS genes is very common in Bacillus species, including in B. subtilis and B. licheniformis. In this study, and others, it has been shown that B. licheniformis is able to synthesize different antimicrobial compounds that can inhibit the growth of specific bacteria, which could be useful to tackle certain diseases associated with animals and plants in downstream applications with the strain [55]. The B. licheniformis CG1 isolate was predicted to have secondary metabolite clusters associated with the synthesis of lichenysin and lichenicidin, which is consistent with the literature [56,57]. Clusters for lichenysin were also present in the B. sonorensis isolates from this study. Of the six isolates found in the honeybee samples, four were identified as B. sonorensis. To our knowledge, this is the first report that describes the presence of B. sonorensis in honeybee products. Two B. sonorensis strains (X1 and X2) were isolated from royal jelly samples, which has never been reported before. The high nutritional value of royal jelly could make this substance as a good growth medium for these bacteria [58]. Alternatively, the presence of Bacillus strains in the royal jelly samples could be the result of a contamination, since bees mix royal jelly with pollen and nectar to produce worker jelly [59].
In summary, B. licheniformis has been defined, not only as one of the most important bacterial organisms employed in industry for enzyme production, but also as a promising probiotic organism for animal feed and as an efficient biofertilizer to prevent diseases and promote growth in plants [60]. Therefore, the B. licheniformis CG1 strain characterized in this study constitutes a promising candidate for use in different industrial applications based on keratin wastes utilization.
Acknowledgments
We thank Charlotte Doidge for useful discussion on the analysis of the secondary metabolite cluster predictions and other results.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020456/s1, Table S1: Antimicrobial Potential. References [57,61,62,63,64,65,66,67,68,69,70,71,72,73,74] are cited in the supplementary materials.
Author Contributions
F.S.-B. conceptualized the study, analyzed and interpreted the results. J.G.-M. and S.S. isolated the Bacillus strains, sequenced the strains and performed the antimicrobial assays. D.M.-G. performed the keratinolytic experiments. S.B. analyzed the genome sequencing data and deposited the genomes in GenBank. D.M.-G. wrote an initial draft of the manuscript. F.S.-B. wrote and edited the final manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequenced genomes have been deposited at GenBank under the accession numbers: JAPQWB000000000 (Z1), JAPQWC000000000 (CG1), JAPQWD000000000 (X1), JAPQWE000000000 (X2), JAPQWF000000000 (AB6) and JAPQWG000000000 (AB7); BioProject: PRJNA907715.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
We thank the FEDER program (TCUE 2021–2023) for allowing Fernando Santos-Beneit to obtain the project with reference number: 067/229111.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Converting Poultry Feathers Discarded Waste into Valuable Raw Materials. [(accessed on 9 October 2022)]. Available online: https://cordis.europa.eu/article/id/418077-converting-poultry-feathers-from-discarded-waste-into-valuable-raw-materials.
- 2.Lyndall M.P., Kurtböke D.I. Development of an Environmentally Friendly Biofertilizer with Keratin Degrading and Antiobiotic Producing Actinomycetes. Actinomycetologica. 2004;18:34–42. [Google Scholar]
- 3.Zouari-Fakhfakh N., Haddar A., Hmidet N., Frikha F., Nasri M. Application of statistical experimental design for optimization of keratinases production by Bacillus pumilus A1 grown on chicken feather and some biochemical properties. Process Biochem. 2010;45:617–626. doi: 10.1016/j.procbio.2009.12.007. [DOI] [Google Scholar]
- 4.Verma A., Singh H., Anwar S., Chattopadhay A., Tiwari K.K., Kaur S., Dhilo G.S. Microbial keratinases: Industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 2017;37:476–491. doi: 10.1080/07388551.2016.1185388. [DOI] [PubMed] [Google Scholar]
- 5.Reddy M.R., Reddy K.S., Chouhan R., Bee H., Reddy G. Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour. Tehcnol. 2017;243:254–263. doi: 10.1016/j.biortech.2017.06.067. [DOI] [PubMed] [Google Scholar]
- 6.Tamreihao K., Mukherjee S., Khunjamayum R., Devi L.J., Asem R.S., Ningthoujam D.S. Feather degradation by keratinolytic bacteria and biofertilizing potential for sustainable agricultural production. J. Basic Microbiol. 2019;59:4–13. doi: 10.1002/jobm.201800434. [DOI] [PubMed] [Google Scholar]
- 7.Dariot D.J., Brandelli A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2013;34:372–384. doi: 10.3109/07388551.2013.794768. [DOI] [PubMed] [Google Scholar]
- 8.Lin X., Lee C.G., Casale E.S., Shih J.C. Purification and Characterization of a Keratinase from a Feather-Degrading Bacillus licheniformis Strain. Appl. Environ. Microbiol. 1992;58:3271–3275. doi: 10.1128/aem.58.10.3271-3275.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordel S., Martín-González D., Muñoz R., Santos-Beneit F. Genome sequence analysis and characterization of Bacillus altitudinis B12, a polylactic acid- and keratin-degrading bacterium. Mol. Genet. Genom. 2022 doi: 10.1007/s00438-022-01989-w. [DOI] [PubMed] [Google Scholar]
- 10.Bonifer K.S., Wen X., Hasim S., Phillips E.K., Dunlap R.N., Gann E.R., DeBruyn J.M., Reynolds T.B. Bacillus pumilus B12 degrades polylactic acid and degradation is affected changing nutrient conditions. Front. Microbiol. 2019;10:2548. doi: 10.3389/fmicb.2019.02548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golden C.E., Rothrock M.K., Mishra A. Comparison Between Random Forest and Gradient Boosting Machine Methods for Predicting Listeria spp. Prevalence in the Environment of Pastured Poultry Farms. Int. Food Res. J. 2019;122:47–55. doi: 10.1016/j.foodres.2019.03.062. [DOI] [PubMed] [Google Scholar]
- 12.Velasquez C.G., Macklin K.S., Kumar S., Bailey M., Ebner P.E., Oliver H.F., Martin-Gonzalez F.S., Singh M. Prevalence and antimicrobial resistance patterns of Salmonella isolated from poultry farms in southeastern United States. Poult. Sci. 2018;97:2144–2152. doi: 10.3382/ps/pex449. [DOI] [PubMed] [Google Scholar]
- 13.Neoigi S.B., Islam M.M., Islam S.K.S., Akhter A.H.M.T., Sikder M.M.H., Yamasaki S., Kabir S.M.L. Risk of multi-drug resistant Campylobacter spp. and residual antimicrobials at poultry farms and live bird markets in Bangladesh. BMC Infect. Dis. 2020;20:278. doi: 10.1186/s12879-020-05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos-Beneit F., Ceniceros A., Nikolaou A., Salas J.A., Gutierrez-Merino J. Identification of Antimicrobial Compounds in Two Streptomyces sp. Strains Isolated From Beehives. Front. Microbiol. 2022;13:742168. doi: 10.3389/fmicb.2022.742168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez-Merino J., Isla B., Combes T., Martinez-Estrada F., Maluquer De Motes C. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microbes. 2020;11:771–788. doi: 10.1080/19490976.2019.1707015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos-Beneit F., Roberts D.M., Cantlay S., McCormick J.R., Errington J. A mechanism for FtsZ-independent proliferation in Streptomyces. Nat. Commun. 2017;8:1378. doi: 10.1038/s41467-017-01596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández-Martínez L.T., Santos-Beneit F., Martín J.F. Is PhoR-PhoP partner fidelity strict? PhoR is required for the activation of the pho regulon in Streptomyces coelicolor. Mol. Genet. Genom. 2012;287:565–573. doi: 10.1007/s00438-012-0698-4. [DOI] [PubMed] [Google Scholar]
- 18.Santos-Beneit F. Genome sequencing analysis of Streptomyces coelicolor mutants that overcome the phosphate-depending vancomycin lethal effect. BMC Genom. 2018;19:457. doi: 10.1186/s12864-018-4838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amin F.A.Z., Sabri S., Ismail M., Chan K.W., Isamail N., Esa N.M., Lila M.A.M., Zawawi N. Probiotic Properties of Bacillus Strains Isolated from Stingless Bee (Heterotrigona itama) Honey Collected across Malaysia. Int. J. Environ. Res. Public Health. 2020;17:278. doi: 10.3390/ijerph17010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santorelli L.A., Wilkinson T., Abdulmalik R., Rai Y., Creevey C.J., Huws S., Gutierrez-Merino J. Beehives possess their own distinct microbiomes. Environ. Microbiome. 2023;18:1. doi: 10.1186/s40793-023-00460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stedman A., Vilet A.H.M., Chambers M.A., Gutierrez-Merino J. Gut commensal bacteria show beneficial properties as wildlife probiotics. Ann. N. Y. Acad. Sci. 2020;1467:112–132. doi: 10.1111/nyas.14302. [DOI] [PubMed] [Google Scholar]
- 22.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 27.Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teeling H., Meyerdierks A., Bauer M., Amann R., Glöckner F.O. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 2004;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- 29.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kembhavi A.A., Kulkarni A., Pant A. Salt-tolerant and thermostable alkaline protease from Bacillus subtilis NCIM no. 64. Appl. Biochem. Biotechnol. 1993;38:83–92. doi: 10.1007/BF02916414. [DOI] [PubMed] [Google Scholar]
- 31.Blin K., Shaw S., Kloosterman A.M., Charlop-Powers Z., van Wezel G.P., Medema M.H., Weber T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Heel A.J., de Jong A., Montalbán-López M., Kok J., Kuipers O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41:W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudiña E.J., Teixeira J.A. Bacillus licheniformis: The unexplored alternative for the anaerobic production of lipopeptide biosurfactants? Biotechnol. Adv. 2022;60:108013. doi: 10.1016/j.biotechadv.2022.108013. [DOI] [PubMed] [Google Scholar]
- 34.Chen X., Lu Y., Shan M., Zhao H., Lu Z., Lu Y. A mini-review: Mechanism of antimicrobial action and application of surfactin. World J. Microbiol. Biotechnol. 2022;38:143. doi: 10.1007/s11274-022-03323-3. [DOI] [PubMed] [Google Scholar]
- 35.Barbosa J.C., Silva Í.C., Caetano T., Mösker E., Seidel M., Lourenço J., Süssmuth R.D., Santos N.C., Gonçalves S., Mendo S. Assessing the potential of the two-peptide lantibiotic lichenicidin as a new generation antimicrobial. World J. Microbiol. Biotechnol. 2022;38:18. doi: 10.1007/s11274-021-03196-y. [DOI] [PubMed] [Google Scholar]
- 36.Dischinger J., Josten M., Szekat C., Sahl H.G., Bierbaum G. Production of the Novel Two-Peptide Lantibiotic Lichenicidin by Bacillus licheniformis DSM 13. PLoS ONE. 2009;4:e6788. doi: 10.1371/journal.pone.0006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta R., Ramnani P. Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechnol. 2006;70:21–33. doi: 10.1007/s00253-005-0239-8. [DOI] [PubMed] [Google Scholar]
- 38.Chatuvedi V., Agrawal K., Verma P. Chicken feathers: A treasure cove of useful metabolites and value-added products. J. Sustain. 2021;4:231–243. [Google Scholar]
- 39.Hmidet N., Ali Nel H., Zouari-Fakhfakh N., Haddar A., Nasri M., Sellemi-Kamoun A. Chicken feathers: A complex substrate for the co-production of alpha-amylase and proteases by B. licheniformis NH1. J. Ind. Microbiol. Biotechnol. 2010;37:983–990. doi: 10.1007/s10295-010-0792-8. [DOI] [PubMed] [Google Scholar]
- 40.Yahaya R.S.R., Normi Y.M., Phang L.Y., Ahmad S.A., Abdullah J.O., Sabri S. Molecular strategies to increase keratinase production in heterologous expression systems for industrial applications. Appl. Microbiol. Biotechnol. 2021;105:3955–3969. doi: 10.1007/s00253-021-11321-y. [DOI] [PubMed] [Google Scholar]
- 41.Jagadeesan Y., Meenakshisundaram S., Saravanan V., Balaiah A. Sustainable production, biochemical and molecular characterization of thermo-and-solvent stable alkaline serine keratinase from novel Bacillus pumilus AR57 for promising poultry solid waste management. Int. J. Biol. Macromol. 2020;163:135–146. doi: 10.1016/j.ijbiomac.2020.06.219. [DOI] [PubMed] [Google Scholar]
- 42.Anna S., Veronika L., Svetlana T., Alexander O. Patented Keratinolytic Enzymes for Industrial Application: An Overview. Recent Pat. Biotechnol. 2022 doi: 10.2174/1872208317666221212122656. [DOI] [PubMed] [Google Scholar]
- 43.Nair A., Sarma S.J. The impact of carbon and nitrogen catabolite repression in microorganisms. Microbiol. Res. 2021;251:126831. doi: 10.1016/j.micres.2021.126831. [DOI] [PubMed] [Google Scholar]
- 44.Gupta R., Beg Q.K., Lorenz P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- 45.Efem S.E. Clinical observations on the wound healing properties of honey. Br. J. Surg. 1988;75:679–681. doi: 10.1002/bjs.1800750718. [DOI] [PubMed] [Google Scholar]
- 46.Subrahmanyam M. Topical application of honey in treatment of burns. Br. J. Surg. 1991;78:497–498. doi: 10.1002/bjs.1800780435. [DOI] [PubMed] [Google Scholar]
- 47.Wahdan H.A. Causes of the antimicrobial activity of honey. Infection. 1998;26:26–31. doi: 10.1007/BF02768748. [DOI] [PubMed] [Google Scholar]
- 48.Lashani E., Davoodabadi A., Soltan Dallal M.M. Some probiotic properties of Lactobacillus species isolated from honey and their antimicrobial activity against foodborne pathogens. Vet. Res. Forum. 2020;11:121–126. doi: 10.30466/vrf.2018.90418.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabaté D.C., Carrillo L., Audisio M.C. Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res. Microbiol. 2009;160:193–199. doi: 10.1016/j.resmic.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Clements-Decker T., Kode M., Khan S., Khan W. Underexplored bacteria as reservoirs of novel antimicrobial lipopeptides. Front. Chem. 2022;10:1025979. doi: 10.3389/fchem.2022.1025979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antonioli Júnior R., Poloni J.F., Pinto É.S.M., Dorn M. Interdisciplinary Overview of Lipopeptide and Protein-Containing Biosurfactants. Genes. 2022;14:76. doi: 10.3390/genes14010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Díaz P.R., Torres M., Petroselli G., Erra-Balsells R., Audisio M.C. Antibacterial activity of Bacillus licheniformis B6 against viability and biofilm formation of foodborne pathogens of health importance. World J. Microbiol. Biotechnol. 2022;38:181. doi: 10.1007/s11274-022-03377-3. [DOI] [PubMed] [Google Scholar]
- 53.Wang H., Fewer D.P., Holm L., Rouhiainen L., Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA. 2014;111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aleti G., Sessitsch A., Brader G. Genome mining: Prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput. Struct. Biotechnol. J. 2015;13:192–203. doi: 10.1016/j.csbj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramirez-Olea H., Reyes-Ballesteros B., Chavez-Santoscoy R.A. Potential application of the probiotic Bacillus licheniformis as an adjuvant in the treatment of diseases in humans and animals: A systematic review. Front. Microbiol. 2022;13:993451. doi: 10.3389/fmicb.2022.993451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madslien E.H., Rønning H.T., Lindbäck T., Hassel B., Andersson M.A., Granum P.E. Lichenysin is produced by most Bacillus licheniformis strains. J. Appl. Microbiol. 2013;115:1068–1080. doi: 10.1111/jam.12299. [DOI] [PubMed] [Google Scholar]
- 57.Begley M., Cotter P.D., Hill C., Ross R.P. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 2009;75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagameri L., Baci G.M., Dezmirean D.S. Royal Jelly as a Nutraceutical Natural Product with a Focus on Its Antibacterial Activity. Pharmaceutics. 2022;14:1142. doi: 10.3390/pharmaceutics14061142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson K.E., Sheehan T.H., Mott B.M., Maes P., Snyder L., Schwan M.R., Walton A., Jones B.M., Corby-Harris V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera) PLoS ONE. 2013;8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.James N., Umesh M., Sarojini S., Shanmugam S., Nasif O., Alharbi S.A., Lan Chi N.T., Brindhadevi K. Unravelling the potential plant growth activity of halotolerant Bacillus licheniformis NJ04 isolated from soil and its possible use as a green bioinoculant on Solanum lycopersicum L. Environ. Res. 2023;216:114620. doi: 10.1016/j.envres.2022.114620. [DOI] [PubMed] [Google Scholar]
- 61.Yakimov M.M., Timmis K.N., Wray V., Fredrickson H.L. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol. 1995;61:1706–1713. doi: 10.1128/aem.61.5.1706-1713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eppelmann K., Doekel S., Marahiel M.A. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 2001;276:34824–34831. doi: 10.1074/jbc.M104456200. [DOI] [PubMed] [Google Scholar]
- 63.May J.J., Wendrich T.M., Marahiel M.A. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- 64.Coburn P.S., Gilmore M.S. The Enterococcus faecalis cytolysin: A novel toxin active against eukaryotic and prokaryotic cells. Cell Microbiol. 2003;5:661–669. doi: 10.1046/j.1462-5822.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 65.Koumoutsi A., Chen X.H., Henne A., Liesegang H., Hitzeroth G., Franke P., Vater J., Borriss R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 2004;186:1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wescombe P.A., Tagg J.R. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 2003;69:2737–2747. doi: 10.1128/AEM.69.5.2737-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umezawa H., Aoyagi T., Nishikiori T., Okuyama A., Yamagishi Y., Hamada M., Takeuchi T. Plipastatins: New inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. I. Taxonomy, production, isolation and preliminary characterization. J. Antibiot. 1986;39:737–744. doi: 10.7164/antibiotics.39.737. [DOI] [PubMed] [Google Scholar]
- 68.Hegemann J.D., Zimmermann M., Xie X., Marahiel M.A. Lasso peptides: An intriguing class of bacterial natural products. Acc. Chem. Res. 2015;48:1909–1919. doi: 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- 69.Allenby N.E., Watts C.A., Homuth G., Pragai Z., Wipat A., Ward A.C., Harwood C.R. Phosphate starvation induces the sporulation killing factor of Bacillus subtilis. J. Bacteriol. 2006;188:5299–5303. doi: 10.1128/JB.00084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babasaki K., Takao T., Shimonishi Y., Kurahashi K. 1985. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: Isolation, structural analysis, and biogenesis. J. Biochem. 1985;98:585–603. doi: 10.1093/oxfordjournals.jbchem.a135315. [DOI] [PubMed] [Google Scholar]
- 71.Patel P.S., Huang S., Fisher S., Pirnik D., Aklonis C., Dean L., Meyers E., Fernandes P., Mayerl F. Bacillaene, a novel inhibitor of procaryotic protein synthesis produced by Bacillus subtilis: Production, taxonomy, isolation, physico-chemical characterization and biological activity. J. Antibiot. 1995;48:997–1003. doi: 10.7164/antibiotics.48.997. [DOI] [PubMed] [Google Scholar]
- 72.Rogers H.J., Newton G.G., Abraham E.P. Production and purification of bacilysin. Biochem. J. 1965;97:573–578. doi: 10.1042/bj0970573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arima K., Kakinuma A., Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968;31:488–494. doi: 10.1016/0006-291X(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 74.Paik S.H., Chakicherla A., Hansen J.N. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 1998;273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequenced genomes have been deposited at GenBank under the accession numbers: JAPQWB000000000 (Z1), JAPQWC000000000 (CG1), JAPQWD000000000 (X1), JAPQWE000000000 (X2), JAPQWF000000000 (AB6) and JAPQWG000000000 (AB7); BioProject: PRJNA907715.