Abstract
The numerous functions of the liver are controlled primarily at the transcriptional level by the concerted actions of a limited number of hepatocyte-enriched transcription factors (hepatocyte nuclear factor 1α [HNF1α], -1β, -3α, -3β, -3γ, -4α, and -6 and members of the c/ebp family). Of these, only HNF4α (nuclear receptor 2A1) and HNF1α appear to be correlated with the differentiated phenotype of cultured hepatoma cells. HNF1α-null mice are viable, indicating that this factor is not an absolute requirement for the formation of an active hepatic parenchyma. In contrast, HNF4α-null mice die during embryogenesis. Moreover, recent in vitro experiments using tetraploid aggregation suggest that HNF4α is indispensable for hepatocyte differentiation. However, the function of HNF4α in the maintenance of hepatocyte differentiation and function is less well understood. To address the function of HNF4α in the mature hepatocyte, a conditional gene knockout was produced using the Cre-loxP system. Mice lacking hepatic HNF4α expression accumulated lipid in the liver and exhibited greatly reduced serum cholesterol and triglyceride levels and increased serum bile acid concentrations. The observed phenotypes may be explained by (i) a selective disruption of very-low-density lipoprotein secretion due to decreased expression of genes encoding apolipoprotein B and microsomal triglyceride transfer protein, (ii) an increase in hepatic cholesterol uptake due to increased expression of the major high-density lipoprotein receptor, scavenger receptor BI, and (iii) a decrease in bile acid uptake to the liver due to down-regulation of the major basolateral bile acid transporters sodium taurocholate cotransporter protein and organic anion transporter protein 1. These data indicate that HNF4α is central to the maintenance of hepatocyte differentiation and is a major in vivo regulator of genes involved in the control of lipid homeostasis.
The adult liver executes numerous functions that are essential for metabolic homeostasis including plasma protein synthesis; carbohydrate, lipid, and amino acid metabolism; and xenobiotic metabolism. The majority of these functions are performed by hepatocytes. Hepatocyte-enriched transcription factors control transcription of genes that are preferentially expressed in liver (5). Our understanding of the role of liver-enriched transcription factors in gene expression has largely been developed using transfection assays in cultured cells. Cis-acting elements for transcription factor binding have been characterized using this approach in conjunction with in vitro DNA binding assays. However, many genes have regulatory elements for several transcription factors, and it becomes difficult to assess which factor predominates in vivo. Moreover, it is becoming increasingly clear that the in vitro regulation of a gene is not always reflected by the in vivo situation. For example, numerous hepatic genes are transactivated by hepatocyte nuclear factor 3β (HNF3β) in vitro but deletion of HNF3β in the mature hepatocyte using conditional gene targeting in mice has minimal consequences (54).
HNF4α is a highly conserved member of the nuclear receptor superfamily (NR2A1). It is expressed at the highest levels in the liver, kidney, intestine, and pancreas in mammals and in the homologous structures in invertebrates (39, 47, 69). This conserved expression pattern suggests that HNF4α fulfills an essential role in development, organogenesis, and maintenance of organ function. HNF4α can activate gene transcription in the absence of exogenous ligands (25, 48, 49). However, HNF4α binding activity may be modulated by fatty acyl-coenzyme A (CoA) thioesters, which may act as agonistic or antagonistic ligands depending on chain length and degree of saturation (17), and also by protein kinase A-mediated phosphorylation (57). This suggests that HNF4α may be responsive to and, by implication, important in the control of metabolic status. To add further weight to the proposal that HNF4α is involved in metabolic homeostasis, mutations in the HNF4α gene cause the disorder maturity onset diabetes of the young (MODY1) (67). Finally, a large number of putative HNF4α target genes have been identified, including those encoding several apolipoproteins, blood coagulation factors, and enzymes involved in lipid, amino acid, and glucose metabolism (47).
To determine the role of HNF4α in gene expression in an intact animal model and to circumvent the embryonic lethality of a standard gene knockout, conditional liver-specific disruption of the HNF4α gene was carried out using the Cre-loxP method with an albumin-Cre transgene. Here, data that strongly suggest a central role for hepatic HNF4α in the maintenance of lipid homeostasis are presented.
MATERIALS AND METHODS
Targeting of the HNF4α gene.
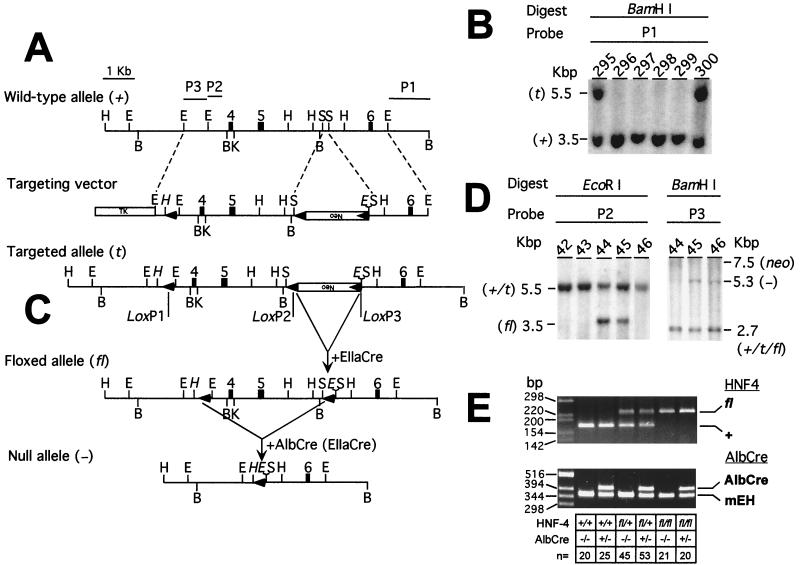
A mouse ES-129/SvJ genomic DNA bacterial artificial chromosome library (Genome Systems, St. Louis, Mo.) was screened with the rat HNF4α cDNA (a generous gift from Francis Sladek). A 5.5-kb EcoRI fragment containing exons 4, 5, and 6 of the mouse HNF4α gene was subcloned into a modified pGEM-3Z vector (Promega, Madison, Wis.) in which the polylinker from SacI to XbaI had been destroyed. A double-stranded oligonucleotide, 5′-TCGAATATAACTTCGTATAATGTATGCTATACGAAGTTATTAAGCTTCCCGGGG-3′ and 5′-TCGACCCCGGGAAGCTTAATAACTTCGTATAGCATACATTATACGAAGTTATAT-3′ (TCGA-loxP-HindIII-SmaI-SalI), was cloned into the SalI site of the polylinker (5′ of exon 4). Insertion and orientation of the linker were confirmed by sequencing. A 2-kb SacI fragment containing a neomycin resistance cassette flanked by two loxP motifs was cloned between the two SacI sites in intron 5 (deleting 178 bp of the intron and introducing an additional EcoRI site). This fragment was cloned such that all three loxP sites were in the same orientation. A blunt-ended, 0.75-kb EcoRI fragment from intron 3 (derived from a different subclone) was cloned into the SmaI site introduced with the first loxP oligonucleotide in order to ensure the inclusion of this loxP site during homologous recombination. Finally, a thymidine kinase gene cassette was cloned into the SalI site of the polylinker. Embryonic stem (ES) cells (RW4; Genome Systems) were electroporated with the linearized targeting construct, and 23 (out of 320 ES cells screened) homologous recombinants were identified using a flanking probe (probe 1; Fig 1). Of these, eight contained all three loxP sites. Clone 300 was injected into C57BL/6 blastocysts. The blastocysts were then inserted into foster NIH Swiss mice. Chimeric male mice were crossed with C57BL/6 females to generate heterozygous targeted/wild-type (t/+) mice. Deletion of the neomycin cassette in vivo was achieved by crossing the mice with an EIIaCre transgenic line (27) as described in Results.
FIG. 1.
Gene targeting and conditional deletion of exons 4 and 5 of the HNF4α gene. (A) Restriction maps of the wild-type allele, targeting vector, and targeted allele. P1, P2, and P3, probes used to assess recombination events; open boxes, neomycin (Neo) and thymidine kinase (TK) positive- and negative-selection cassettes, respectively; shaded boxes, exons, numbered as described by Taraviras et al. (55); arrowheads, position and orientation of loxP sites. The upside-down Neo shows that the Neo cassette is transcribed in an orientation opposite to that of the HNF4α gene. Restriction sites: E, EcoRI; H, HindIII; B, BamHI; K, KpnI; S, SacI. Sites introduced in the targeting vector are italicized. (B) Southern blot analysis of homologous recombination in ES cells electroporated with the targeting vector. Hybridizing fragments of wild-type (±) and targeted (t) alleles and their respective sizes are indicated. (C) Crosses between mice heterozygous for the targeted allele failed to yield targeted homozygotes (see text). In order to generate a viable, conditional knockout mouse line, heterozygous (t/+) males were crossed with EIIaCre females in order to generate both floxed (fl) and null (−) alleles. Shown is recombination between loxP sites 2 and 3 (although all recombination events, i.e., recombination between loxP sites 1 and 2, 2 and 3, and 1 and 3, were observed). (D) Southern blot analysis of tail DNA from pups derived from the cross described for panel C. Hybridizing fragments of wild-type (±), targeted (t), floxed (fl), null (-), and neomycin (neo) alleles and their respective sizes are indicated. The floxed, null, and neomycin alleles are the result of recombination between loxP sites 2 and 3, 1 and 3, and 1 and 2, respectively. (E) PCR genotyping of mice. (Top) HNF4α genotyping. Tail DNA was amplified using primers flanking loxP site 1 (A) in the floxed allele, yielding products of 241 and 180 bp for the floxed and wild-type alleles, respectively. (Bottom) Cre genotyping. mEH primers served as a positive control for amplification, yielding a fragment of 341 bp in all samples. The presence of the Cre transgene was indicated by the amplification of an additional band of 411 bp with Cre-specific primers.
Breeding scheme to produce animals with a liver-specific deletion of HNF4α.
Heterozygous animals carrying one floxed (fl; flanked by loxP) and one wild-type allele were crossed with animals hemizygous for the previously described albumin-Cre transgene (AlbCre) (66), kindly provided by Derek LeRoith. Heterozygous (fl/+) animals carrying one copy of the AlbCre transgene were then interbred with fl/+ littermates lacking Cre to generate HNF4α liver knockout mice (H4LivKO) and littermate control mice (H4Flox, AlbCre, and wild type, as described below in Results).
PCR genotyping of mice for HNF4α and AlbCre.
Genomic DNA was isolated from mouse tails as described previously (26). For HNF4α gene diagnostic PCR, approximately 50 ng of tail DNA was amplified in a 50-μl final reaction mixture containing 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 2.5 U of AmpliTaq, and 0.3 μM (each) HNF4α gene-specific primers prH4GT-LP1-F1 (5′-AGAATGACCCTGAAGCACCAGG-3′) and prH4GT-LP1-R1 (5′-GCCAGAGGTCTGTGAAACAAGG-3′). Cycling conditions were 94°C for 4 min and then 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a 10-min extension at 72°C. These primers amplify the region surrounding loxP site 1 (Fig. 1) in the flox allele, yielding products of 241 and 180 bp for the floxed and wild-type alleles, respectively.
For Cre/microsomal epoxide hydrolase (mEH) gene diagnostic PCR, approximately 50 ng of tail DNA was amplified in a 50-μl final reaction mixture containing 2.0 mM MgCl2, 2.5 U of AmpliTaq, 0.2 mM dNTPs, and 0.3 μM concentrations of the following primers: prCre-F (5′-AGGTGTAGAGAAGGCACTCAGC-3′), prCre-R (5′-CTAATCGCCATCTTCCAGCAGG-3′), prMEH-F (5′-AAGTGAGTTTGCATGGCGCAGC-3′), and prMEH-R (5′-CCCTTTAGCCCCTTCCCTCTG-3′). Cycling conditions were 94°C for 3 min and then 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by a 10-min extension at 72°C. mEH primers served as a positive control for amplification, yielding a fragment of 341 bp in all samples (40). An additional band of 411 bp was diagnostic for the presence of the Cre transgene.
Serum and organ collection.
Mice were anesthetized with 2.5% Avertin and decapitated, and trunk blood was collected in serum tubes. Serum was separated by centrifugation at 7,000 × g for 5 min and stored at −20°C prior to analysis. Serum was assayed for total cholesterol, high-density lipoprotein (HDL) cholesterol, triglycerides, and alanine aminotransferase using standard methodology (Anilytics Inc., Gaithersburg, Md.). Selected tissues were collected, weighed, and either fixed immediately for histological analysis or snap-frozen in liquid nitrogen for preparation of RNA or nuclei.
FPLC analysis of plasma lipids.
Total cholesterol and triglyceride (Sigma, St. Louis, Mo.), as well as free cholesterol and phospholipid (Wako, Osaka, Japan), concentrations were measured in 12-μl aliquots of plasma using commercial kits and the Hitachi 911 automated chemistry analyzer (Boehringer Mannheim, Indianapolis, Ind.). Plasma lipoproteins were analyzed by gel filtration on two Superose 6 columns in series (fast-protein liquid chromatography [FPLC]; Pharmacia, Piscataway, N.J.) at 0.3 ml/min in phosphate-buffered saline containing 0.1 mM EDTA and 0.02% sodium azide (28). Mouse apolipoproteins A-I, A-II, Cs, E, and B were identified by Western blotting in plasma and FPLC fractions using a mixture of polyclonal rabbit anti-mouse immunoglobulin G antisera raised against purified apolipoproteins (Biodesign, Saco, Maine).
Pathology.
Livers from 45-day-old representative H4LivKO and H4Flox control mice were fixed in 10% neutral buffered formalin and embedded in paraffin, and sections cut at a thickness of 4 to 6 μm were stained with hematoxylin and eosin (H & E). Selected livers were fixed in alcoholic formalin or frozen in Tissue-Tek O. C. T. compound (Miles, Inc., Elkhart, Ind.) for histochemistry of glycogen and fat by periodic acid-Schiff (PAS) and oil red O staining, respectively. Pieces of liver were fixed in 2.5% glutaraldehyde and postfixed in osmium tetroxide, and thin sections were stained with uranyl nitrate and lead citrate for ultrastructural studies.
Northern blot analysis.
Tissues were crushed on dry ice in a mortar and pestle, and total RNA was extracted with Ultraspec reagent (Biotecx, Houston, Tex.). RNA was separated on 1% agarose–0.22 M formaldehyde gels and transferred to GeneScreen Plus membranes (Dupont, Wilmington, Del.) by capillary blotting in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight. Blots were hybridized at 42°C in UltraHyb (Ambion Inc., Austin, Tex.) with random-primer-labeled cDNA probes. Probes for rat cDNAs were generously provided by Kiyoto Motojima (ApoA-I, -A-IV, -C-II, and -C-III) (41), Johann Auwerx (ApoB) (52), and Daniel Kelly (midchain acyl-CoA dehydrogenase [MCAD]) (11). All other probes were amplified from a mouse liver cDNA library using gene-specific primers and cloned into pCR TOPO II (Stratagene, La Jolla, Calif.). The probes were quality controlled by sequencing.
Western blot analysis.
Nuclei were prepared according to the method of Wu (65). Protein concentration was measured using the BCA kit (Pierce Chemical Co., Rockford, Ill.). Total nuclear protein was separated on a sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gel, transferred to nitrocellulose, and probed with an antibody raised to a C-terminal peptide of HNF4α (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). Equal loading of nuclear protein was demonstrated by reprobing the membrane with an antibody to histone H1 (a generous gift from Michael Bustin) (2). Bound antibody was detected with a horseradish peroxidase-conjugated secondary antibody (KPL, Gaithersburg, Md.), and immune complexes were developed with the ECL reagent (Pharmacia).
RESULTS
Generation of a conditional null allele of the HNF4α gene.
A triple-lox targeting vector was constructed; in this vector a phosphoribosyltransferase (neo) selection cassette flanked by loxP sites was introduced into intron 5 of the mouse HNF4α gene along with a third loxP site in intron 3 (Fig. 1A). Following standard electroporation and culture of ES cells, homologous recombinants were identified by Southern blotting of genomic DNA. All three loxP sites were incorporated into the HNF4α locus in 8 of 320 ES cells screened (Fig. 1B). A single clone (clone 300) was injected into C57BL/6 blastocysts to generate chimeric mice. The triple-lox, targeted allele (t) was transmitted through the germ line. However, extensive intercrosses between heterozygous (t/+) mice failed to yield homozygous targeted mice. Thus, the targeted allele was nonfunctional, most likely due to interference with HNF4α gene expression by the neo cassette.
In order to delete the neo cassette and thus rescue the mouse line, the targeted allele was crossed into a transgenic line expressing Cre under the control of the adenovirus EIIa promoter (EIIaCre) (27). EIIaCre-mediated recombination occurs early in development (two to eight cells) and at low levels, resulting in chimerism. Thus, it was hoped to selectively delete the neo cassette and thereby generate a heritable, active conditional null allele (Fig. 1C). Recombination events were analyzed by Southern blotting of tail DNA (isolated from pups from the cross HNF4αt/+ × EIIaCre+/−). This indicated the presence of all possible deletion patterns (Fig. 1D). Chimeric mice were selected according to the observed pattern of deletion and crossed to wild-type mice in order to generate lines carrying floxed (male 44) and null (male 45) alleles of the HNF4α gene. As predicted, the floxed allele, containing just small loxP sites flanking exons 4 and 5 of the HNF4α gene (Fig. 1C), was active and viable at homozygosity.
Deletion of exons 4 and 5 is predicted to result in the splicing of exon 3 to exon 6. This results in a frameshift, leading to a premature stop codon. Translation of the truncated mRNA results in an N-terminal peptide of just 128 amino acid residues (118 residues of HNF4α followed by 10 missense residues). While this protein retains both zinc finger motifs, it lacks both the A and T boxes necessary for high-affinity binding to DNA (14, 18), along with the ligand-binding and activation function 2 domains. Significantly, a mutant HNF4α protein containing 125 N-terminal residues fails to bind DNA at room temperature, although some low-affinity binding is observed at 4°C (18). The putative truncated protein also lacks sequences required for the recruitment of coactivator proteins CREB binding protein (CBP), SRC-1, and GRIP1 (10, 61, 68). Given that CBP-mediated acetylation of HNF4α is required for its retention in the nucleus (50), it is likely that any truncated protein produced would be quickly exported to the cytoplasm and therefore unable to interfere with transcription. While we cannot rule out subtle dominant-negative effects due to retention of some DNA binding, heterozygous animals were normal for all parameters measured (not shown), suggesting that this is not the case. Moreover, consistent with the embryonic lethality of germ line inactivation of HNF4α, crosses between the heterozygous null mice produced here (HNF4α+/−) failed to yield homozygous knockouts. Thus, the available evidence strongly suggests that deletion of exons 4 and 5 results in a null allele.
High-efficiency, liver-specific disruption of HNF4α.
To examine the role of HNF4α in the maintenance of hepatic gene expression and liver function, the active floxed allele was crossed into mice hemizygous for an albumin-Cre transgene (AlbCre). that express Cre exclusively in the postpartum liver (66). The (HNF4αfl/+; AlbCre+/−)F1 mice so generated were interbred with HNF4αfl/+ littermates lacking the AlbCre transgene. This breeding scheme yielded (HNF4αfl/fl; AlbCre+/−)F2 mice (designated H4LivKO) and three groups of littermate control mice, HNF4αfl/fl; AlbCre−/− (H4Flox), HNF4α+/+; AlbCre+/− (AlbCre), and HNF4α+/+; AlbCre−/− (wild type). Genotypes of all mice were assessed by PCR analysis of tail DNA (Fig. 1E). All alleles were inherited in a Mendelian fashion (Fig. 1E), suggesting that neither the floxed allele, the AlbCre transgene, nor the combination thereof resulted in any prenatal lethality.
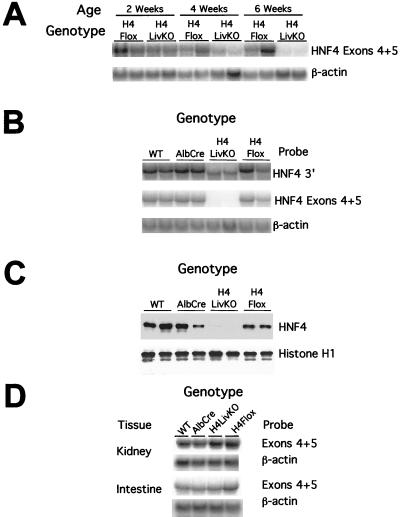
To assess the time course, extent, and specificity of Cre-mediated recombination at the HNF4α locus, Northern blots of total liver RNA from 2-, 4-, and 6-week-old H4LivKO and control animals were probed with a fragment of the HNF4α cDNA homologous to exons 4 and 5 (Fig. 2A). Deletion of the floxed exons of HNF4α was about 50% by 4 weeks of age, and, by 6 weeks of age, mRNA signal intensity in H4LivKO livers was below the limit of detection (Fig. 2A and B). Thus, intact HNF4α mRNA containing exons 4 and 5 is reduced to <10% that seen in control livers at 6 weeks of age. This time course is similar to that reported for a retinoid-X receptor-α (RXRα) floxed allele using the albumin promoter driving Cre expression (60). All further experiments were conducted using 45-day-old mice.
FIG. 2.
Exons 4 and 5 are deleted efficiently and specifically in the livers of H4LivKO mice. (A) Time course of Cre-mediated recombination at the HNF4α locus. Liver RNA from H4LivKO and control animals was subjected to Northern blotting and probed with a fragment of the HNF4α cDNA derived from the targeted exons (exons 4 and 5). (B to D) Tissues were isolated from 45-day-old wild-type (HNF4α +/+; AlbCre −/−), AlbCre (HNF4α +/+; AlbCre +/−), H4LivKO (HNF4α fl/fl; AlbCre +/−), and H4Flox (HNF4α fl/fl; AlbCre −/−) mice. (B) Northern blot analysis of 10 μg of total liver RNA. A truncated RNA species is evident in livers of H4LivKO mice; this species hybridizes to a 3′ fragment of the HNF4α cDNA but fails to hybridize to exons 4 and 5. (C) Western blot analysis of liver nuclei. Twenty micrograms of total nuclear protein was separated on a sodium dodecyl sulfate–10% polacrylamide gel electrophoresis gel and transferred to nitrocellulose. The membrane was probed with an antibody raised to a C-terminal peptide of HNF4α (Santa Cruz Biotechnology). Equal loading of nuclear protein was demonstrated by reprobing the membrane with an antibody directed against mouse histone H1. No full-length or truncated HNF4α protein was detected in livers of H4LivKO mice. (D) Northern blot analysis of total RNA isolated from control tissues of H4LivKO and control animals.
Northern blots of liver RNA probed with a 3′ fragment of the HNF4α cDNA revealed that H4LivKO mice expressed an mRNA that migrated with greater mobility than that from wild-type, AlbCre, or H4Flox mice (Fig. 2B). Reverse transcription-PCR of HNF4α cDNA from H4LivKO liver RNA failed to detect the full-length transcript. Sequencing the cloned, truncated cDNA revealed that the expected exon 3-to-exon 6 splice event had occurred (not shown). The steady-state level of the truncated HNF4α mRNA in H4LivKO hepatocytes is about 30% that seen for the undisrupted message in controls. This is consistent with the suggestion that HNF4α regulates its own expression (51) and/or that the abnormal transcript is less stable than the wild-type message.
Disruption of HNF4α RNA resulted in a corresponding loss of full-length protein as evidenced by Western blots of nuclear protein probed with an antibody raised to a C-terminal peptide of HNF4α (Fig. 2C). Equal loading of nuclear protein was demonstrated by reprobing the membrane with an antibody to histone H1. Thus, H4LivKO mouse liver nuclei contain less than 10% of the immunoreactive HNF4α protein present in controls.
To confirm that deletion of exons 4 and 5 was restricted to the liver, RNA from kidney and intestine, which also express HNF4α, was subjected to Northern blotting and probed with a probe consisting of exons 4 and 5. As expected, H4LivKO mice expressed full-length HNF4α mRNA in both tissues at levels similar to those found in wild-type, AlbCre, and H4Flox mice (Fig. 2D). Thus, HNF4α mRNA and protein were efficiently and specifically disrupted in the livers of adult H4LivKO mice by 45 days of age.
Hepatomegaly, hepatocyte hypertrophy, and abnormal glycogen and lipid deposition in livers of H4LivKO mice.
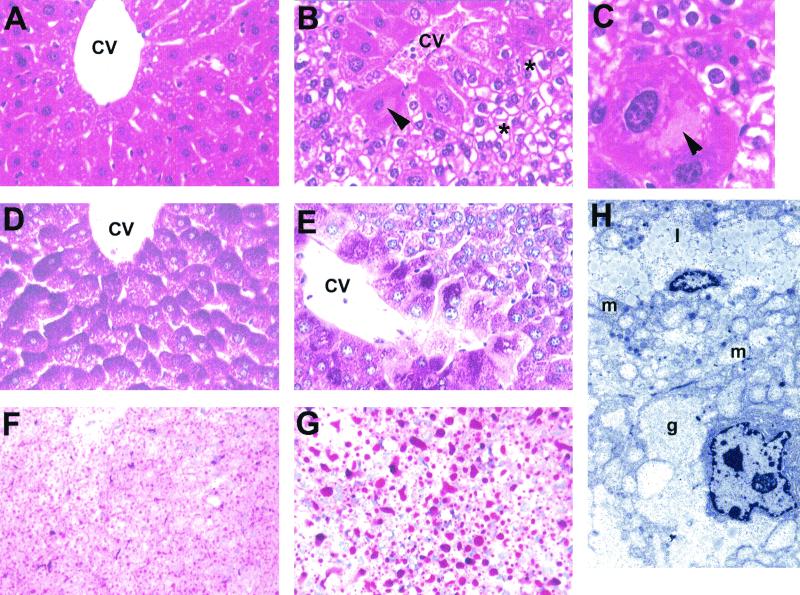
To begin to assess the influence of disruption of HNF4α on hepatic function, liver weights and pathology were examined. Livers from 45-day-old H4LivKO mice were significantly enlarged relative to those of controls (Table 1) and had a visibly gray, mottled appearance. Marked pathological lesions were evident in H4LivKO mouse livers but were absent in controls (Fig. 3). These lesions were more severe in male mice than in females. Centrilobular hepatocytes in H4LivKO mice were invariably hypertrophic, with pale eosinophilic intracytoplasmic inclusions (Fig. 3B and C). Hepatocytes throughout the liver lobule were markedly vacuolated (Fig. 3B). Although the material accumulating in the vacuoles had the typical morphology of glycogen, it did not stain PAS-positive in alcoholic formalin-fixed sections (Fig. 3D and E). Rather, the majority of this material stained positive for fat with oil red O stain (Fig. 3F and G).
TABLE 1.
Liver weights and serum chemistries of conditionally HNF4α-null mice and controlsa
| Parameter | Value for indicated mouse type
|
|||
|---|---|---|---|---|
| WT | AlbCre | H4LivKO | H4Flox | |
| Liver | ||||
| Wet wt (g) | 0.9 ± 0.1 | 0.9 ± 0.2 | 1.3 ± 0.3∗∗ | 0.8 ± 0.2 |
| % body wt | 4.0 ± 0.3 | 4.0 ± 0.2 | 7.3 ± 0.9∗∗ | 3.9 ± 0.1 |
| Serum chemistry | ||||
| Albumin (mg/dl) | 3.8 ± 0.4 | 3.8 ± 0.3 | 4.0 ± 0.4 | 4.0 ± 0.3 |
| ALT (U/liter) | 40 ± 20 | 40 ± 23 | 65 ± 7∗ | 40 ± 20 |
| Triglycerides (mg/dl) | 86 ± 23 | 76 ± 20 | 38 ± 7∗∗ | 80 ± 22 |
| Total cholesterol (mg/dl) | 119 ± 15 | 114 ± 14 | 48 ± 15∗∗ | 121 ± 28 |
| HDL cholesterol (mg/dl) | 93 ± 25 | 88 ± 22 | 37 ± 23∗∗ | 104 ± 24 |
| Free fatty acids (mg/dl) | 1.3 ± 0.4 | 1.3 ± 0.4 | 1.3 ± 0.2 | 1.8 ± 0.9 |
| Bile acids (μmol/liter) | 16 ± 7 | 17 ± 11 | 283 ± 61∗∗ | 10 ± 7 |
| Glucose (mg/dl) | 150 ± 32 | 145 ± 28 | 97 ± 39 | 127 ± 36 |
Mice (45 days old) were fasted for 4 h, anesthetized, and decapitated, and serum was prepared. All values are means ± standard deviations from at least four animals (not separated by sex). Statistics were performed using one-way analysis of variance followed by an unpaired Student t test if applicable. ∗, P < 0.05; ∗∗, P < 0.01. ALT, alanine aminotransferase; WT, wild type.
FIG. 3.
Histological analysis of the livers of H4LivKO mice. (A) Normal liver of a 45-day-old wild-type (+/+) mouse. Hepatocytes around the central vein have few vacuoles present. Shown is an H&E stain. Magnification, ×300. (B) Liver of H4LivKO mouse showing marked vacuolation of most hepatocytes (asterisks) except those around the central vein (CV). Those around the vein are eosinophilic, and one at the left side has a large intracytoplasmic pale eosinophilic inclusion (arrowhead; see panel C). Shown is an H&E stain. Magnification, ×300. (C) Centrilobular hepatocyte with large intracytoplasmic inclusion (arrowhead) in the liver of an H4LivKO mouse. Note the vacuolation of smaller hepatocytes. Shown is an H&E stain. Magnification, ×750. (D) PAS stain of normal wild-type mouse liver showing abundant PAS-positive glycogen (dark red color) in all hepatocytes around the central vein. Livers were fixed in alcoholic formalin. Magnification, ×300. (E) PAS stain of H4LivKO mouse liver showing decrease or loss of PAS-positive glycogen in most hepatocytes but abundant glycogen in centrilobular hepatocytes. Livers were fixed in alcoholic formalin. Magnification, ×300. (F) Frozen section of normal liver from a wild-type mouse, showing many small fat droplets in hepatocytes (red) throughout the liver. Shown is an oil red O stain. Magnification, ×150. (G) Frozen section of H4LivKO liver, showing abundant, large fat droplets in hepatocytes. Shown is an oil red O stain. Magnification, ×150. (H) Ultrastructure of the liver from a H4LivKO mouse showing abundant lipid (1) and mitochondria (m) in the hepatocyte at the top and areas of glycogen (g) in the hepatocyte at the bottom. Staining was with uranyl acetate and lead citrate. Magnification, ×6,000.
To further characterize these lesions, ultrastructure was analyzed. This revealed that most hepatocytes had abundant lipid droplets and large mitochondria (Fig. 3H, l and m, respectively), while others had both lipid droplets and abundant glycogen-like material (Fig. 3H, l and g, respectively). These abnormal cytoplasmic changes were not seen in control livers.
Liver-specific deletion of HNF4α alters serum lipid levels.
Consistent with the unusual fatty-liver phenotype observed in histological sections of H4LivKO livers, total cholesterol, HDL cholesterol, and triglyceride levels in sera from H4LivKO mice were dramatically reduced relative to those in sera from controls (Table 1). Conversely, serum bile acid concentrations were markedly elevated (Table 1). These alterations in serum lipid profiles could be due to either a generalized liver failure or to a more specific defect in lipid transport and metabolism. Mild liver dysfunction was indicated by a small elevation in the level of alanine aminotransferase in serum from H4LivKO mice (Table 1). However, the levels of albumin, nonesterified fatty acids, and glucose were indistinguishable from those in controls, suggesting the retention of many liver functions (Table 1). Thus, at this time point, disruption of HNF4α in the liver appears to induce a specific failure of normal hepatic lipid metabolism and/or transport rather than a generalized liver failure.
FPLC analysis of serum lipids.
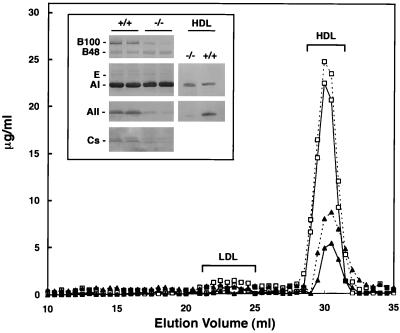
To further characterize the observed alterations in blood lipid composition, plasma from H4LivKO and H4Flox mice was subjected to FPLC analysis. Compared to controls, H4LivKO mice had significantly reduced plasma cholesterol (−61%) and phospholipids (−53%) (Fig. 4). Moreover, the elution profile of the lipids was altered (Fig. 4). Thus, both plasma low-density lipoprotein (LDL) and HDL cholesterol were dramatically decreased in H4LivKO mice compared to controls. Moreover, HDL cholesterol from H4LivKO plasma eluted later than that from controls and contained a significant amount of very-late-eluting, smaller HDL. This elution profile is indicative of production of unusually small, lipid-poor HDL particles. Western blot analysis of lipoprotein content in whole plasma from H4LivKO mice indicated that these mice exhibit reduced ApoB100, ApoA-II and ApoC content, while apolipoproteins A-I, E, and B48 are unaffected (Fig. 4, inset). Strikingly, HDL cholesterol from H4LivKO mice is virtually devoid of ApoA-II, with ApoA-I as the sole apolipoprotein component (Fig. 4, inset).
FIG. 4.
Serum lipoprotein profiles of control and H4LivKO mice. Lipoproteins were separated from 60 μl of pooled mouse plasma samples by FPLC (n = 4 for each group). Profiles from H4Flox (control; open squares) and H4LivKO (solid squares) mice are shown. y axis, concentrations of cholesterol (solid line) and phospholipids (dotted lines). (Inset) Immunoblot analysis of apolipoproteins B, E, A-I, A-II, and Cs in plasma- (left) and in HDL-eluted fractions (right) from control (+/+) and H4LivKO (−/−) mice.
HNF4α regulates hepatic genes involved in lipid metabolism and transport.
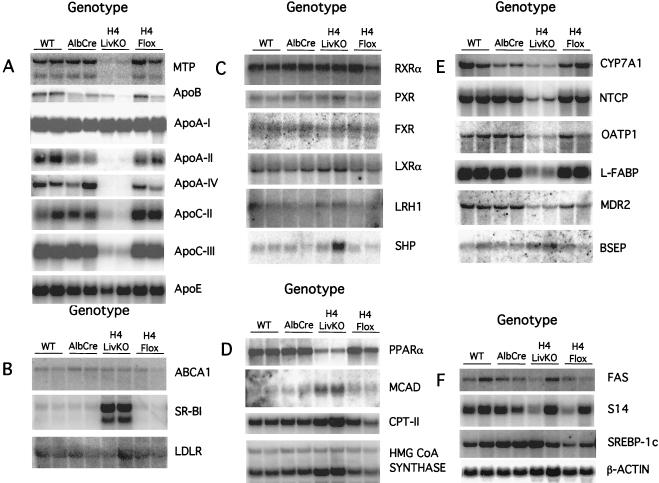
The HNF4α-mediated transactivation of numerous genes involved in lipid metabolism and transport has been demonstrated using cell culture systems. These include genes for ApoA-I (16, 34), ApoA-II (44), ApoA-IV (21), ApoB (37), ApoC-II (19), ApoC-III (38), and ApoE (9); MCAD (3, 4); microsomal triglyceride transfer protein (MTP) (15); and cholesterol 7α-hydroxylase (CYP7A) (8, 53). To examine whether the expression of these putative target genes was affected by disruption of the hepatic HNF4α gene, liver RNA from 45-day-old H4LivKO mice and controls was analyzed (Fig. 5). Strikingly, steady-state mRNA levels for apolipoproteins A-II, A-IV, C-II, and C-III and MTP (Fig. 5A) and CYP7A1 (Fig. 5E) were drastically reduced in H4LivKO livers compared to those in controls. In contrast, steady-state mRNA levels for ApoA-I and ApoE were relatively unaffected by disruption of the HNF4α gene (Fig. 5A), even though both of the genes are HNF4α responsive in transient transfection assays (9, 16, 42). Paradoxically, while MCAD is positively regulated by HNF4α in cultured cells (4), it is induced by disruption of the HNF4α gene (Fig. 5D). To further examine the influence of the deletion of hepatic HNF4α on lipid metabolism, the expression of numerous other key genes involved in lipid transport and metabolism was examined. Expression of the LDL receptor and ATP-binding cassette 1, both of which are essential lipid and cholesterol transporters, was unaffected, while expression of the major HDL receptor, SR-BI, was induced (Fig. 5B). The expression levels of several nuclear receptors implicated in the control of lipid homeostasis, RXRα, pregnane-X receptor, farsenoid-X receptor (FXR), oxysterol receptor-α, and liver receptor homologue 1, were unchanged by deletion of HNF4α (Fig. 5C). Expression of the nuclear receptor small heterodimer partner was variable (Fig. 5C). In contrast, expression of peroxisome proliferator-activated receptor α (PPARα) was lower in H4LivKO livers than in controls (Fig. 5D). Given this decrease in PPARα, it is interesting to note that expression of some PPARα target genes (carnitoyl-palmitoyl transferase-II, MCAD, and 3-hydroxy-3-methylglutaryl CoA synthase genes) is enhanced in H4LivKO livers relative to controls (Fig. 5D). The increased levels of serum bile acids suggested that these mice might be deficient in one or more pathways of bile acid trafficking. Consistent with this hypothesis, levels of sodium taurocholate cotransporter protein (Ntcp), organic anion transporter protein 1, liver fatty acid binding protein (L-FABP), and multidrug resistance protein 2 mRNA were markedly decreased in H4LivKO mice relative to controls (Fig. 5E). In contrast, the steady-state level of bile salt export pump (BSEP) mRNA was mildly elevated (Fig. 5E). Finally, expression of the key lipogenic genes encoding fatty acid synthase, sterol receptor element binding protein 1c, and spot-14 genes was unaffected in H4LivKO livers (Fig. 5F). Thus, consistent with the histological and serological alterations observed in these mice, disruption of hepatic HNF4α results in altered expression of genes involved in several pathways of lipid metabolism and transport. Conversely, the unchanged expression of other hepatic genes again argues that the observed dyslipidemia is due to disruption in a specific pathway(s) and not due to a generalized liver failure.
FIG. 5.
Northern blot analysis of liver RNA from H4LivKO and control animals. Total liver RNA (10 μg) was separated on 0.22 M formaldehyde–1% agarose gels and blotted to GeneScreen Plus membranes. Blots were hybridized in UltraHyb at 42°C for 16 h with the probes indicated. Blots were developed using a Storm 860 phosphorimager and quantified using ImageQuant software. Genes are grouped by function (see Discussion). (A) H4LivKO mice are deficient in expression of genes required for VLDL secretion and HDL synthesis. wt, wild type. (B) H4LivKO mice display increased expression of SR-BI, required for HDL uptake. ABCA1, ATP-binding cassette 1; LDLR, LDL receptor. (C) Expression levels of several nuclear receptors important for lipid homeostasis are unchanged by deletion of hepatic HNF4α. PXR, pregnane-X receptor; FXR, farsenoid-X receptor; LXRα, oxysterol receptor-α; LRH1, liver receptor homologue 1; SHP, small heterodimer partner. (D) Decreased expression of PPARα but increased expression of PPARα target genes in H4LivKO mice. CPT-II, carnitoyl-palmitoyl transferase-II; HMG, 3-hydroxy-3-methylglutaryl. (E) H4LivKO mice display gene expression patterns indicative of defective bile acid homeostasis. OATP1, organic anion transporter protein 1; MDR2, multidrug resistance protein 2. (F) Key genes in the fatty acid synthesis pathway are unaltered in H4LivKO mice. FAS, fatty acid synthase; S14, spot-14; SREBP-1c, sterol receptor element binding protein 1c.
Lethality associated with disruption of hepatic HNF4α.
To assess the influence of HNF4α deletion in liver on mouse development, mice were weighed at 10 days and 2 weeks of age and each week thereafter. The H4LivKO mice had no observable phenotype until they reached 5 weeks of age when they lost weight compared with the wild-type, AlbCre transgenic, and H4Flox mice (not shown). This weight loss coincided with the complete loss of HNF4α in the livers (between 4 and 6 weeks). To assess whether this weight loss was terminal, a cohort of mice was allowed to develop without further interference. In this group mortality reached >70% by 8 weeks of age. Thus, while we are as yet unable to explain the observed mortality at the functional level, this indicates that HNF4α activity is essential for the proper functioning of the liver.
DISCUSSION
The presence of functional HNF4α response elements in the promoters of numerous genes encoding metabolic enzymes suggests that HNF4α fulfills an essential role in cellular metabolism. Moreover, this nuclear receptor can respond to metabolic status through fatty acyl-CoA thioesters and cyclic AMP-dependent phosphorylation, suggesting that it may integrate signals from different pathways to fine tune the storage, import, and export of metabolic products (17, 57). Consistent with this hypothesis, the data presented here demonstrate that HNF4α is indispensable for the constitutive expression of several key hepatic genes involved in lipid transport and metabolism. This leads to profound decreases in cholesterol, HDL cholesterol, and triglycerides and a large increase in bile acids in the serum of mice lacking hepatic HNF4α. These data provide the first direct in vivo evidence for the role that HNF4α plays in controlling hepatic lipid metabolism and transport in the adult.
H4LivKO mice are deficient in expression of genes required for VLDL secretion.
Serum lipid levels are controlled in large part by the balance between hepatic cholesterol synthesis, degradation, export, and uptake. In this regard, it is salient to note that H4LivKO mice display decreased expression of two genes essential for very-low-density lipoprotein (VLDL) secretion (ApoB and MTP). While expression of ApoB in the H4LivKO mice is variable, expression of MTP is consistently and strongly decreased. Studies with hepatocyte-specific MTP knockout mice (6, 43) and MTP/ApoB double-heterozygous null animals (31) indicate that the concentration of MTP within the endoplasmic reticulum is the major determinant of hepatic VLDL secretion. Thus, it seems likely that the accumulation of lipid in hepatocytes of H4LivKO mice is due, at least partially, to a defect in VLDL secretion.
H4LivKO mice display increased expression of SR-BI, required for HDL uptake.
SR-BI appears to be the major mediator of selective cholesterol uptake from HDL by hepatocytes and, in combination with the LDL receptor, may also participate in cholesterol uptake from non-HDL cholesterol (20). SR-BI is subject to feedback regulation in response to changes in cellular cholesterol stores (20). However, most work on the regulation of SR-BI expression has been conducted with macrophages and steroidigenic tissues, and little is known about control of hepatic expression. Interestingly, expression of the macrophage homologue of SR-BI, CD36 (FAT/CLA-1), is induced by PPARα and -γ ligands (7). Given the increases in PPARα target gene expression observed here and the fact that HNF4α and PPARα compete for binding to a number of promoters (42, 45, 64), one may speculate that PPARα also regulates hepatic SR-BI expression. Experiments are under way to determine whether SR-BI is a direct target of HNF4α or whether the observed increase in expression is a secondary consequence of altered lipid homeostasis (e.g., due to changes in intracellular lipid levels). What is clear, however, is that expression of SR-BI is markedly increased in H4LivKO mice. Thus, it is likely that selective cholesterol uptake is increased in H4LivKO livers.
Given that disruption of HNF4α appears to have opposite effects on the expression of genes involved in VLDL secretion (down-regulation) and HDL cholesterol uptake (up-regulation), it is tempting to speculate that the overall effect of HNF4α may be to promote the net efflux of cholesterol from the liver. Direct measurements of VLDL secretion and HDL uptake in H4LivKO mice to definitively address these questions are under way.
H4LivKO mice display gene expression patterns indicative of defective bile acid homeostasis.
Expression of CYP7A1, which catalyzes the first, rate-limiting step in the neutral pathway of bile acid synthesis, is reduced in H4LivKO livers relative to controls. Thus, the in vivo data presented here are in close agreement with in vitro studies implicating HNF4α in the transcriptional regulation of CYP7A1 (8, 53). Similarly, in the absence of hepatic HNF4α, expression levels of both Ntcp and organic anion transporter protein 1 are markedly down-regulated. These proteins are the major mediators of hepatic basolateral bile acid uptake via sodium-dependent and -independent pathways, respectively (35). Consistent with the increased serum bile acid levels observed, it is likely that H4LivKO mice are unable to efficiently take up bile acids from the blood. Conversely, BSEP, the major canilicular bile acid export protein (12), is expressed at moderately elevated levels in H4LivKO mice. This suggests that clearance of bile acids from the liver into the gall bladder may be increased in H4LivKO mice. BSEP expression is induced by bile acids via the nuclear receptor FXR, while Ntcp expression is repressed by the same pathway (46). It is possible, therefore, that the altered expression of BSEP and Ntcp seen here may be due to elevated liver bile acid concentrations. However, several lines of evidence suggest that this is not the case. Thus, expression of oxysterol receptor-α, which is also induced by increased intracellular bile acid concentrations via FXR (46), is unchanged in H4LivKO livers relative to that in controls. Moreover, H4LivKO mice show none of the severe hepatotoxicity characteristic of elevated bile acid concentrations. Taken together, these observations suggest that the alterations in expression of bile acid transporters cannot be fully explained by indirect mechanisms and suggest that some of these genes may be novel targets for HNF4α.
Decreased expression of PPARα but increased expression of PPARα target genes in H4LivKO mice.
PPARα is consistently down-regulated in H4LivKO mouse liver. Although no measurements of glucocorticoids were performed, PPARα is known to be positively regulated by corticosterone (29, 30). Given that cholesterol is an essential precursor for the formation of glucocorticoids, it seems likely that the observed down-regulation is due to alterations in circulating hormone levels. Despite the decreased steady-state levels of PPARα mRNA, many prototypical PPARα target genes are induced in the H4LivKO mouse. Therefore, PPARα may not be limiting for target gene induction, and increases in the intracellular concentration of a PPARα agonist(s) such as free fatty acids may account for this effect.
H4LivKO mice show an atheroprotective serum lipid profile.
It is interesting to note that the overall effect of disruption of hepatic HNF4α activity is to reduce circulating lipid levels. This is consistent with the proposal that polyunsaturated fatty acids and saturated (C18:O) fatty acids act as antagonists of HNF4α, as diets rich in these fats also decrease blood lipid levels (13, 17). Similarly, dietary fats that increase lipid levels in blood were shown to act, via their acyl-CoA thiosters, as agonists for HNF4α (17). Thus, the data presented here support the proposal that the influence of dietary composition on circulating lipid levels may be mediated, at least in part, by HNF4α. By inference, this suggests that HNF4α may be important in the etiology of dyslipidemias.
The plasma lipid profile of H4LivKO mice is characterized by decreased plasma lipoproteins of all classes (ApoB containing as well as HDL). Overall, this profile may be regarded as atheroprotective, since (i) the atherogenic ApoB and LDL levels are reduced and (ii) the antiatherogenic ApoA-I-only HDL (LpA-I) but not the ApoA-I-plus-ApoA-II HDL (LpA-I/A-II) is found in HNF4α-null mice. The widely prescribed hypolipidemic fibrates actually decrease the ratio of LpA-I to LpA-I/A-II (36, 59). Thus, HNF4α antagonists may offer significant advantages over fibrates in the treatment of hyperlipidemias. Moreover, the observed decreases in ApoA-II, ApoA-IV, ApoC-III, and MTP found in the H4LivKO mice are consistent with the lower lipid levels in serum from the Apo-null (33, 62, 63) and MTP liver conditional-null (43) mice, all of which all display, to different degrees, decreased serum cholesterol levels.
HNF4α is essential for constitutive expression of genes with complex promoters.
The data presented here demonstrate that HNF4α is indispensable for the expression of Apo-AII, Apo-CII, Apo-CIII, L-FABP, and MTP. Interestingly, transcription factors in addition to HNF4α have been shown by transient transfection studies to play a role in regulating the corresponding promoters (19, 21, 24, 25, 38, 42, 44, 56, 58). For example, the ApoC-II promoter is activated by RXRα/T3Rβ and this increase is accentuated by the synergistic action of HNF4α and ARP-1 (19). Thus, while it remains possible that other transcription factors may contribute to the regulation of the genes under normal physiological conditions, HNF4α is absolutely required for their constitutive expression in the fully differentiated mouse liver. The dominant influence of HNF4α on the expression of some genes with complex promoters is consistent with the proposal that HNF4α-recruited chromatin-remodeling activity may be a prerequisite for the binding of other transcription factors (32). Alternatively, the dominant effect of HNF4α may be explained by the presence of a transcription factor cascade, whereby HNF4α controls the expression of other factors required for expression of a given gene. Indeed, HNF1α, a direct transcriptional target of HNF4α (23), has recently been shown to control the expression of L-FABP (1).
Dichotomy of HNF4α function in adult versus fetal hepatocytes?
The gene expression patterns reported here are broadly similar to those described recently for HNF4α-null livers produced by tetraploid rescue (32). However, significant differences are apparent. Disruption of HNF4α in the adult hepatocyte virtually abolishes ApoA-IV expression but does not significantly influence ApoA-I, while this pattern is reversed in the developing hepatocyte. Similarly, expression of pregnane-X receptor, which is abolished in fetal HNF4α-null livers (32), is unaffected by disruption of HNF4α in the fully differentiated liver. Thus, HNF4α appears to function differently in the maintenance of hepatic differentiation than in the establishment of the differentiated state. A similar disparity in the function of HNF3β in the developing liver and the fully differentiated hepatocyte has recently been published (54). It seems likely that more examples of this functional dichotomy may become apparent as conditional gene targeting is applied to examine the role of transcription factors in adult tissues.
Decreased hepatic HNF4α activity in MODY1 patients may explain decreases in serum triglyceride levels.
While the conditional HNF4α-null mouse, not surprisingly, does not resemble MODY1 subjects in terms of hyperglycemia, the evidence presented here suggests that the altered triglyceride levels observed in MODY1 subjects are a direct consequence of impaired hepatic HNF4α function. The demonstration that hepatic HNF4α deficiency in the mouse results in lower triglyceride levels in the serum suggests the possibility that triglyceride levels may be an early diagnostic test for the onset of diabetes in humans. It is still not clear whether diabetes associated with HNF4α haploinsufficiency in humans is due to decreased HNF4α activity in the β cell itself, particularly as this factor is expressed at very low levels in adult pancreas (39). Alternatively, perturbation of HNF4α activity in another organ such as the liver may lead to the exposure of the pancreas to a metabolite that gradually disrupts the function of the β cell. The phenotype of the conditional hepatic HNF4α-null mouse provides evidence that, at least in mice, lack of HNF4α activity in the liver does not produce the characteristic hyperglycemia of diabetes mellitus. To directly test the function of HNF4α in pancreatic β cells, experiments using the H4Flox line crossed with a β-cell Cre-expressing transgenic line are under way (22).
In conclusion, a mouse line carrying a conditionally null allele of the HNF4α gene was developed. Disruption of the HNF4α gene specifically in hepatocytes indicates that HNF4α is central to the maintenance of hepatic function and is a major in vivo regulator of genes involved in the control of lipid homeostasis. Further analysis of this model should aid in the elucidation of the role that HNF4α plays in the transcriptional control of liver function. The conditionally null allele described here should be useful in dissecting the role that HNF4α plays in gene regulation in other tissues and in the etiology of MODY1.
ACKNOWLEDGMENTS
We thank Roberta Smith, Keith Rogers, and Kunio Nagashina for assistance with the liver pathology and Derek LeRoith for providing the AlbCre mice. We also thank Johan Auwerx and Shioko Kimura for review of the manuscript and Chris Sinal and Taro Akiyama for useful discussions.
REFERENCES
- 1.Akiyama T E, Ward J M, Gonzalez F J. Regulation of the liver fatty acid-binding protein gene by hepatocyte nuclear factor 1alpha (HNF1alpha). Alterations in fatty acid homeostasis in HNF1alpha-deficient mice. J Biol Chem. 2000;275:27117–27122. doi: 10.1074/jbc.M004388200. [DOI] [PubMed] [Google Scholar]
- 2.Bustin M. Preparation and application of immunological probes for nucleosomes. Methods Enzymol. 1989;170:214–251. doi: 10.1016/0076-6879(89)70049-5. [DOI] [PubMed] [Google Scholar]
- 3.Carter M E, Gulick T, Moore D D, Kelly D P. A pleiotropic element in the medium-chain acyl coenzyme A dehydrogenase gene promoter mediates transcriptional regulation by multiple nuclear receptor transcription factors and defines novel receptor-DNA binding motifs. Mol Cell Biol. 1994;14:4360–4372. doi: 10.1128/mcb.14.7.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter M E, Gulick T, Raisher B D, Caira T, Ladias J A, Moore D D, Kelly D P. Hepatocyte nuclear factor-4 activates medium chain acyl-CoA dehydrogenase gene transcription by interacting with a complex regulatory element. J Biol Chem. 1993;268:13805–13810. [PubMed] [Google Scholar]
- 5.Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- 6.Chang B H, Liao W, Li L, Nakamuta M, Mack D, Chan L. Liver-specific inactivation of the abetalipoproteinemia gene completely abrogates very low density lipoprotein/low density lipoprotein production in a viable conditional knockout mouse. J Biol Chem. 1999;274:6051–6055. doi: 10.1074/jbc.274.10.6051. [DOI] [PubMed] [Google Scholar]
- 7.Chinetti G, Gbaguidi F G, Griglio S, Mallat Z, Antonucci M, Poulain P, Chapman J, Fruchart J C, Tedgui A, Najib-Fruchart J, Staels B. CLA-1/SR-BI is expressed in atherosclerotic lesion macrophages and regulated by activators of peroxisome proliferator-activated receptors. Circulation. 2000;101:2411–2417. doi: 10.1161/01.cir.101.20.2411. [DOI] [PubMed] [Google Scholar]
- 8.Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang J Y. Transcriptional activation of the cholesterol 7alpha-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res. 1998;39:2192–2200. [PubMed] [Google Scholar]
- 9.Dang Q, Walker D, Taylor S, Allan C, Chin P, Fan J, Taylor J. Structure of the hepatic control region of the human apolipoprotein E/C- I gene locus. J Biol Chem. 1995;270:22577–22585. doi: 10.1074/jbc.270.38.22577. [DOI] [PubMed] [Google Scholar]
- 10.Dell H, Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- 11.Disch D L, Rader T A, Cresci S, Leone T C, Barger P M, Vega R, Wood P A, Kelly D P. Transcriptional control of a nuclear gene encoding a mitochondrial fatty acid oxidation enzyme in transgenic mice: role for nuclear receptors in cardiac and brown adipose expression. Mol Cell Biol. 1996;16:4043–4051. doi: 10.1128/mcb.16.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann A F, Meier P J. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- 13.Grundy S M, Denke M A. Dietary influences on serum lipids and lipoproteins. J Lipid Res. 1990;31:1149–1172. [PubMed] [Google Scholar]
- 14.Hadzopoulou-Cladaras M, Kistanova E, Evagelopoulou C, Zeng S, Cladaras C, Ladias J A. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J Biol Chem. 1997;272:539–550. doi: 10.1074/jbc.272.1.539. [DOI] [PubMed] [Google Scholar]
- 15.Hagan D L, Kienzle B, Jamil H, Hariharan N. Transcriptional regulation of human and hamster microsomal triglyceride transfer protein genes. Cell type-specific expression and response to metabolic regulators. J Biol Chem. 1994;269:28737–28744. [PubMed] [Google Scholar]
- 16.Harnish D C, Malik S, Kilbourne E, Costa R, Karathanasis S K. Control of apolipoprotein AI gene expression through synergistic interactions between hepatocyte nuclear factors 3 and 4. J Biol Chem. 1996;271:13621–13628. doi: 10.1074/jbc.271.23.13621. [DOI] [PubMed] [Google Scholar]
- 17.Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 18.Jiang G, Sladek F M. The DNA binding domain of hepatocyte nuclear factor 4 mediates cooperative, specific binding to DNA and heterodimerization with the retinoid X receptor alpha. J Biol Chem. 1997;272:1218–1225. doi: 10.1074/jbc.272.2.1218. [DOI] [PubMed] [Google Scholar]
- 19.Kardassis D, Sacharidou E, Zannis V I. Transactivation of the human apolipoprotein CII promoter by orphan and ligand-dependent nuclear receptors. The regulatory element CIIC is a thyroid hormone response element. J Biol Chem. 1998;273:17810–17816. doi: 10.1074/jbc.273.28.17810. [DOI] [PubMed] [Google Scholar]
- 20.Krieger M. Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu Rev Biochem. 1999;68:523–558. doi: 10.1146/annurev.biochem.68.1.523. [DOI] [PubMed] [Google Scholar]
- 21.Ktistaki E, Lacorte J M, Katrakili N, Zannis V I, Talianidis I. Transcriptional regulation of the apolipoprotein A-IV gene involves synergism between a proximal orphan receptor response element and a distant enhancer located in the upstream promoter region of the apolipoprotein C-III gene. Nucleic Acids Res. 1994;22:4689–4696. doi: 10.1093/nar/22.22.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkarni R N, Bruning J C, Winnay J N, Postic C, Magnuson M A, Kahn C R. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 23.Kuo C J, Conley P B, Chen L, Sladek F M, Darnell J E, Jr, Crabtree G R. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 24.Lacorte J M, Ktistaki E, Beigneux A, Zannis V I, Chambaz J, Talianidis I. Activation of CAAT enhancer-binding protein delta (C/EBPdelta) by interleukin-1 negatively influences apolipoprotein C-III expression. J Biol Chem. 1997;272:23578–23584. doi: 10.1074/jbc.272.38.23578. [DOI] [PubMed] [Google Scholar]
- 25.Ladias J A, Hadzopoulou-Cladaras M, Kardassis D, Cardot P, Cheng J, Zannis V, Cladaras C. Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII, and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2, and EAR-3. J Biol Chem. 1992;267:15849–15860. [PubMed] [Google Scholar]
- 26.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert G, Amar M J, Martin P, Fruchart-Najib J, Foger B, Shamburek R D, Brewer H B, Jr, Santamarina-Fojo S. Hepatic lipase deficiency decreases the selective uptake of HDL- cholesteryl esters in vivo. J Lipid Res. 2000;41:667–672. [PubMed] [Google Scholar]
- 29.Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- 30.Lemberger T, Staels B, Saladin R, Desvergne B, Auwerx J, Wahli W. Regulation of the peroxisome proliferator-activated receptor alpha gene by glucocorticoids. J Biol Chem. 1994;269:24527–24530. [PubMed] [Google Scholar]
- 31.Leung G K, Veniant M M, Kim S K, Zlot C H, Raabe M, Bjorkegren J, Neese R A, Hellerstein M K, Young S G. A deficiency of microsomal triglyceride transfer protein reduces apolipoprotein B secretion. J Biol Chem. 2000;275:7515–7520. doi: 10.1074/jbc.275.11.7515. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Ning G, Duncan S A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda N, Li H, Lee D, Oliver P, Quarfordt S H, Osada J. Targeted disruption of the apolipoprotein C-III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J Biol Chem. 1994;269:23610–23616. [PubMed] [Google Scholar]
- 34.Malik S, Karathanasis S K. TFIIB-directed transcriptional activation by the orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol. 1996;16:1824–1831. doi: 10.1128/mcb.16.4.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier P J, Eckhardt U, Schroeder A, Hagenbuch B, Stieger B. Substrate specificity of sinusoidal bile acid and organic anion uptake systems in rat and human liver. Hepatology. 1997;26:1667–1677. doi: 10.1002/hep.510260641. [DOI] [PubMed] [Google Scholar]
- 36.Mellies M J, Stein E A, Khoury P, Lamkin G, Glueck C J. Effects of fenofibrate on lipids, lipoproteins, and apolipoproteins in 33 subjects with primary hypercholesterolemia. Atherosclerosis. 1987;63:57–64. doi: 10.1016/0021-9150(87)90082-7. [DOI] [PubMed] [Google Scholar]
- 37.Metzger S, Halaas J L, Breslow J L, Sladek F M. Orphan receptor HNF-4 and bZip protein C/EBP alpha bind to overlapping regions of the apolipoprotein B gene promoter and synergistically activate transcription. J Biol Chem. 1993;268:16831–16838. [PubMed] [Google Scholar]
- 38.Mietus-Snyder M, Sladek F M, Ginsburg G S, Kuo C F, Ladias J A, Darnell J E, Jr, Karathanasis S K. Antagonism between apolipoprotein AI regulatory protein 1, Ear3/COUP-TF, and hepatocyte nuclear factor 4 modulates apolipoprotein CIII gene expression in liver and intestinal cells. Mol Cell Biol. 1992;12:1708–1718. doi: 10.1128/mcb.12.4.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miquerol L, Lopez S, Cartier N, Tulliez M, Raymondjean M, Kahn A. Expression of the L-type pyruvate kinase gene and the hepatocyte nuclear factor 4 transcription factor in exocrine and endocrine pancreas. J Biol Chem. 1994;269:8944–8951. [PubMed] [Google Scholar]
- 40.Miyata M, Kudo G, Lee Y H, Yang T J, Gelboin H V, Fernandez-Salguero P, Kimura S, Gonzalez F J. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7, 12-dimethylbenz[a]anthracene. J Biol Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- 41.Motojima K, Passilly P, Peters J M, Gonzalez F J, Latruffe N. Expression of putative fatty acid transporter genes are (sic) regulated by peroxisome proliferator-activated receptor alpha and gamma activators in a tissue- and inducer-specific manner. J Biol Chem. 1998;273:16710–16714. doi: 10.1074/jbc.273.27.16710. [DOI] [PubMed] [Google Scholar]
- 42.Nishiyama C, Hi R, Osada S, Osumi T. Functional interactions between nuclear receptors recognizing a common sequence element, the direct repeat motif spaced by one nucleotide (DR- 1) J Biochem (Tokyo) 1998;123:1174–1179. doi: 10.1093/oxfordjournals.jbchem.a022058. [DOI] [PubMed] [Google Scholar]
- 43.Raabe M, Veniant M M, Sullivan M A, Zlot C H, Bjorkegren J, Nielsen L B, Wong J S, Hamilton R L, Young S G. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J Clin Investig. 1999;103:1287–1298. doi: 10.1172/JCI6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribeiro A, Pastier D, Kardassis D, Chambaz J, Cardot P. Cooperative binding of upstream stimulatory factor and hepatic nuclear factor 4 drives the transcription of the human apolipoprotein A-II gene. J Biol Chem. 1999;274:1216–1225. doi: 10.1074/jbc.274.3.1216. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez J C, Ortiz J A, Hegardt F G, Haro D. The hepatocyte nuclear factor 4 (HNF-4) represses the mitochondrial HMG- CoA synthase gene. Biochem Biophys Res Commun. 1998;242:692–696. doi: 10.1006/bbrc.1997.8032. [DOI] [PubMed] [Google Scholar]
- 46.Sinal C J, Tohkin M, Miyata M, Ward J M, Lambert G, Gonzalez F J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 47.Sladek F M. Hepatocyte nuclear factor 4. In: Tronche F, Yaniv M, editors. Liver gene expression. R. G. Austin, Tex: Landes Company; 1994. pp. 207–230. [Google Scholar]
- 48.Sladek F M, Ruse M D, Jr, Nepomuceno L, Huang S M, Stallcup M R. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4α1. Mol Cell Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sladek F M, Zhong W M, Lai E, Darnell J E., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 50.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 51.Spath G F, Weiss M C. Hepatocyte nuclear factor 4 expression overcomes repression of the hepatic phenotype in dedifferentiated hepatoma cells. Mol Cell Biol. 1997;17:1913–1922. doi: 10.1128/mcb.17.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staels B, van Tol A, Chan L, Will H, Verhoeven G, Auwerx J. Alterations in thyroid status modulate apolipoprotein, hepatic triglyceride lipase, and low density lipoprotein receptor in rats. Endocrinology. 1990;127:1144–1152. doi: 10.1210/endo-127-3-1144. [DOI] [PubMed] [Google Scholar]
- 53.Stroup D, Chiang J Y. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7alpha-hydroxylase gene (CYP7A1) J Lipid Res. 2000;41:1–11. [PubMed] [Google Scholar]
- 54.Sund N J, Ang S-L, Sackett S D, Shen W, Daigle N, Magnuson M A, Kaestner K H. Hepatocyte nuclear factor 3β (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol Cell Biol. 2000;20:5175–5183. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taraviras S, Monaghan A P, Schutz G, Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 56.Vergnes L, Taniguchi T, Omori K, Zakin M M, Ochoa A. The apolipoprotein A-I/C-III/A-IV gene cluster: ApoC-III and ApoA-IV expression is regulated by two common enhancers. Biochim Biophys Acta. 1997;1348:299–310. doi: 10.1016/s0005-2760(97)00071-4. [DOI] [PubMed] [Google Scholar]
- 57.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vorgia P, Zannis V I, Kardassis D. A short proximal promoter and the distal hepatic control region-1 (HCR-1) contribute to the liver specificity of the human apolipoprotein C-II gene. Hepatic enhancement by HCR-1 requires two proximal hormone response elements which have different binding specificities for orphan receptors HNF-4, ARP-1, and EAR-2. J Biol Chem. 1998;273:4188–4196. doi: 10.1074/jbc.273.7.4188. [DOI] [PubMed] [Google Scholar]
- 59.Vu-Dac N, Schoonjans K, Kosykh V, Dallongeville J, Fruchart J C, Staels B, Auwerx J. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. J Clin Investig. 1995;96:741–750. doi: 10.1172/JCI118118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan Y-J Y, An D, Cai Y, Repa J J, Chen T H-P, Flores M, Postic C, Magnuson M A, Chen J, Chien K R, French S, Mangelsdorf D J, Sucov H M. Hepatocyte-specific mutation establishes retinoid X receptor α as a heterodimeric integrator of multiple physiological processes in the liver. Mol Cell Biol. 2000;20:4436–4444. doi: 10.1128/mcb.20.12.4436-4444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J C, Stafford J M, Granner D K. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinstock P H, Bisgaier C L, Hayek T, Aalto-Setala K, Sehayek E, Wu L, Sheiffele P, Merkel M, Essenburg A D, Breslow J L. Decreased HDL cholesterol levels but normal lipid absorption, growth, and feeding behavior in apolipoprotein A-IV knockout mice. J Lipid Res. 1997;38:1782–1794. [PubMed] [Google Scholar]
- 63.Weng W, Breslow J L. Dramatically decreased high density lipoprotein cholesterol, increased remnant clearance, and insulin hypersensitivity in apolipoprotein A-II knockout mice suggest a complex role for apolipoprotein A-II in atherosclerosis susceptibility. Proc Natl Acad Sci USA. 1996;93:14788–14794. doi: 10.1073/pnas.93.25.14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winrow C J, Marcus S L, Miyata K S, Zhang B, Capone J P, Rachubinski R A. Transactivation of the peroxisome proliferator-activated receptor is differentially modulated by hepatocyte nuclear factor-4. Gene Expr. 1994;4:53–62. [PMC free article] [PubMed] [Google Scholar]
- 65.Wu C. Analysis of hypersensitive sites in chromatin. Methods Enzymol. 1989;170:269–289. doi: 10.1016/0076-6879(89)70052-5. [DOI] [PubMed] [Google Scholar]
- 66.Yakar S, Liu J L, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamagata K, Furuta H, Oda N, Kaisaki P J, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida E, Aratani S, Itou H, Miyagishi M, Takiguchi M, Osumu T, Murakami K, Fukamizu A. Functional association between CBP and HNF4 in trans-activation. Biochem Biophys Res Commun. 1997;241:664–669. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]
- 69.Zhong W, Sladek F M, Darnell J E., Jr The expression pattern of a Drosophila homolog to the mouse transcription factor HNF-4 suggests a determinative role in gut formation. EMBO J. 1993;12:537–544. doi: 10.1002/j.1460-2075.1993.tb05685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]