Abstract
Phosphatidylinositol transfer proteins (PITPs) regulate the interface between signal transduction, membrane-trafficking, and lipid metabolic pathways in eukaryotic cells. The best characterized mammalian PITPs are PITPα and PITPβ, two highly homologous proteins that are encoded by distinct genes. Insights into PITPα and PITPβ function in mammalian systems have been gleaned exclusively from cell-free or permeabilized cell reconstitution and resolution studies. Herein, we report for the first time the use of genetic approaches to directly address the physiological functions of PITPα and PITPβ in murine cells. Contrary to expectations, we find that ablation of PITPα function in murine cells fails to compromise growth and has no significant consequence for bulk phospholipid metabolism. Moreover, the data show that PITPα does not play an obvious role in any of the cellular activities where it has been reconstituted as an essential stimulatory factor. These activities include protein trafficking through the constitutive secretory pathway, endocytic pathway function, biogenesis of mast cell dense core secretory granules, and the agonist-induced fusion of dense core secretory granules to the mast cell plasma membrane. Finally, the data demonstrate that PITPα-deficient cells not only retain their responsiveness to bulk growth factor stimulation but also retain their pluripotency. In contrast, we were unable to evict both PITPβ alleles from murine cells and show that PITPβ deficiency results in catastrophic failure early in murine embryonic development. We suggest that PITPβ is an essential housekeeping PITP in murine cells, whereas PITPα plays a far more specialized function in mammals than that indicated by in vitro systems that show PITP dependence.
INTRODUCTION
Phosphatidylinositol transfer proteins (PITPs) are operationally defined by their ability to catalyze the transfer phosphatidylinositol (PtdIns) or phosphatidylcholine (PtdCho) monomers between membrane bilayers in vitro (Cleves et al., 1991a; Wirtz, 1991). Both yeast and metazoan PITPs contain one phospholipid-binding site that exhibits a dual headgroup specificity (Sha et al., 1998; Phillips et al., 1999; Yoder et al., 2001). Thus, PITPs accomodate two dissimilar phospholipid substrates in a mutually exclusive binding reaction.
From a biological perspective, little is known about the genuine physiological functions of PITPs. The best-studied case is in yeast where PITP function has been subjected to genetic analysis. Sec14p, the major yeast PITP, is essential for protein transport from the Golgi complex (Bankaitis et al., 1989, 1990). A dissection of how Sec14p translates PtdIns/PtdCho transfer activity into biological function is driven by analyses of mutations that relieve cells of the essential Sec14p requirement for Golgi function and cell viability (Cleves et al., 1989, 1991b; Fang et al., 1996; Kearns et al., 1997; Rivas et al., 1999; Li et al., 2000; Xie et al., 2001). Such “bypass Sec14p” mutants identify novel interfaces between Golgi function, lipid metabolism, and the actions of novel proteins whose functions are only now beginning to be understood. Precise execution points for individual PITPs in mammalian cells remain unknown, although reduced PITP function leads to specific neurodegenerative defects in flies and in the vibrator mouse (Hamilton et al., 1997; Milligan et al., 1997).
There are at least three soluble mammalian PITPs and these are designated PITPα, PITPβ, and rdgBβ (Dickeson et al., 1989; Tanaka and Hosaka, 1994; Fullwood et al., 1999). The best characterized of these are PITPα and PITPβ. These two isoforms share 77% sequence identity and are encoded by distinct genes. PITPβ is distinguished from PITPα in that it catalyzes the in vitro transfer of PtdIns, PtdCho, and sphingomyelin (SM), whereas PITPα is unable to catalyze SM transfer (van Tiel et al., 2000). PITPα reconstitutes as a cytosolic factor required for 1) Ca2+-activated discharge of secretory granule cargo in permeabilized neuroendocrine cells (Hay and Martin, 1993), 2) budding of both constitutive secretory vesicles and immature secretory granules from the trans-Golgi network of neuroendocrine cells (Ohashi et al., 1995), 3) budding of constitutive secretory vesicles from hepatocyte trans-Golgi network (Jones et al., 1998), and 4) hydrolysis of phosphatidylinositol bisphosphate mediated by various G protein-coupled phospholipase C isoforms (Cunningham et al., 1996). The former two reconstitution systems implicate a role for mammalian PITP in regulation of vesicle trafficking, an activity analogous to Sec14p function in yeast.
In all of these in vitro systems, PITPs stimulate the synthesis of various phosphoinositides that play critical roles in facilitating the reactions that are being reconstituted. Moreover, PITPβ and yeast Sec14p can fully replace PITPα in each of these reconstituted reactions. This is a rather remarkable result given that yeast Sec14p and mammalian PITPs share no primary sequence homology nor structural similarity at all (Skinner et al., 1993; Sha et al., 1998; Yoder et al., 2001). Yet, the interchangeability of PITPs in the reconstituted in vitro systems is significantly at odds with the results of biological experiments. Saccharomyces cerevisiae expresses five distinct Sec14p-like PITPs, but none of these PITPs shares perfect physiological redundancy with the others, and each regulates a distinct step in phospholipid metabolism (Li et al., 2000; Wu et al., 2000). Moreover, mammalian PITPα exerts potent dominant-negative effects when expressed in lieu of a homologous Drosophila PITP that harbors the same biochemical properties as does PITPα in vitro (Milligan et al., 1997). These data strongly suggest that individual mammalian PITPs also execute specific in vivo functions. Furthermore, these data indicate that the physiological specificity of mammalian PITPs is unavailable for experimental scrutiny in present in vitro systems.
In this report, we describe our use of genetic approaches to gain insight into the physiological functions of PITPα and PITPβ in murine cells. Our data clearly indicate that disruption of both copies of the PITPα structural gene in murine cells fails to compromise their growth or vigor, and that ablation of PITPα function has no significant effect on bulk phosphoinositide or phospholipid metabolism. Moreover, PITPα −/− cells are competent for protein trafficking through the constitutive secretory pathway, are normal with regard to endocytic pathway function, are fully competent for both biogenesis and agonist-induced exocytosis of dense core secretory granules (DSGs), are responsive to bulk growth factor stimulation, and retain their pluripotency. Thus, PITPα does not uniquely execute an essential housekeeping function in murine embryonic stem (ES) cells. In contrast, we were unable to evict both PITPβ alleles from murine ES cells, and we demonstrate that PITPβ deficiency leads to early failure in murine embryonic development. We suggest that PITPβ is an essential housekeeping PITP in murine cells, whereas PITPα plays a far more specialized function in mammals than what is indicated by in vitro systems that demonstrate PITP dependence.
MATERIALS AND METHODS
Cell Culture and Antibodies
R1 and AB1 ES cell lines have been described (McMahon and Bradley, 1990; Nagy et al., 1993) and were maintained in standard media consisting of DMEM (4.5 g/l glucose), 1 mM sodium pyruvate, 1× nonessential amino acids, 2 mM l-glutamine, a 50-μg/ml penicillin/streptomycin cocktail, 15% fetal bovine serum (Invitrogen, Carlsbad, CA), 2-mercaptoethanol, and leukemia inhibitory factor (LIF). Media used for differentiating ES cells to embryoid bodies were identical except LIF and 2-mercaptoethanol were omitted. ES cells were plated on feeder layers consisting of either mitomycin C-treated murine embryonic fibroblast feeder cells or gelatin-coated tissue culture dishes. When necessary, cells were dispersed with a 0.25% trypsin/0.1% EDTA solution.
Isolation of Genomic PITPα and PITPβ Clones
A mouse 129SVJ genomic λ-phage library was screened by standard filter-lift plaque hybridization methods by using random primed 32P-radiolabeled rat PITPα or PITPβ cDNAs as probes. Hybridized filters were washed in 2× SSPE, 0.5% SDS at room temperature followed by two washes in 0.2× SSPE, 0.5% SDS at 65°C. From such screens, λ-phage clones containing 12- and 15-kb genomic insert fragments of PITPα and PITPβ were recovered. Iterative cycles of physical mapping and nucleotide sequence analysis confirmed the identities of the genomic fragments, and identified the precise nature and physical positions of the exons contained in each genomic fragment.
Transfection and Selection of ES Cell Recombinants
All experiments were carried out with ES cells grown on embryonic murine fibroblast feeder cells. For electroporation, ES cells (80% confluence) were harvested by trypsinization, centrifuged through culture medium, and resuspended in fresh culture media. A 20-μg load of linearized targeting vector was electroporated into 2 × 107 cells via a single discharge (300 V, 250 μF) delivered by a single cell electroporator (Bio-Rad, Hercules, CA). Immediately after electroporation the cells were distributed onto 100-mm embryonic mouse fibroblast feeder plates and allowed to recover for 24 h before challenge with 0.5 mg/ml G418 and 2 mM gangcyclovir. Selection media were exchanged every 3–4 d, for a total of 12 d, after which individual colonies were picked and expanded.
DNA Hybridization
DNA from each clonal ES cell line was prepared using the Wizard genomic DNA purification kit marketed by Promega (Madison, WI). Purified DNA (10 μg) was subsequently digested with either EcoRI or XbaI, electrophoresed through a 1% agarose gel, transferred to Hybond N nylon membrane (Amersham Biosciences, Piscataway, NJ), and hybridized with probe 1 (Figure 1A) under stringent nonaqueous conditions (Southern, 1975). Hybridized products were washed at 68°C in 1× SSC/0.1% SDS and 0.2× SSC/0.1% SDS.
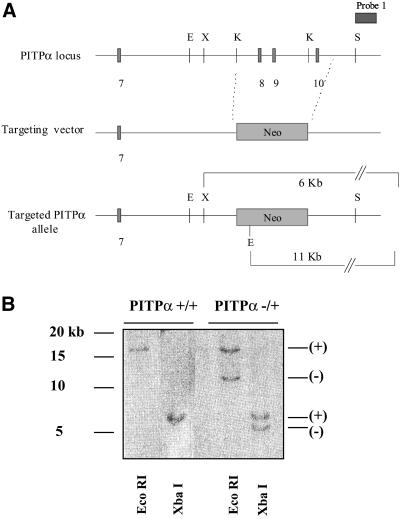
Figure 1.
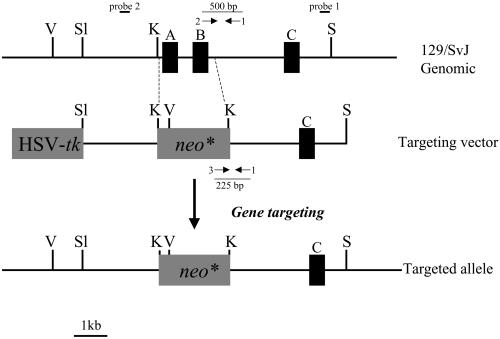
(A) Strategy for targeting of the PITPα locus. An HSV-tk expression cassette from pPNT (Tybulewicz et al., 1991) was engineered so that it flanks a 12-kb mouse genomic PITPα clone bracketed by SstI restriction sites. A 0.4-kb DNA fragment was released from the extreme 3′ end of the genomic insert so that it is not represented in the targeting vector. This fragment was subsequently used as a diagnostic probe, designated probe 1, in Southern hybridization analyses. PITPα exons 8–10 were exchanged for a selectable neo expression cassette derived by polymerase chain reaction from pMC1NEO (Stratagene, La Jolla, CA). As described in the text, this amplified cassette contains an uncharacterized mutation that attenuates, but does not abolish, activity of the gene product. (B) Identification of targeted ES cell clones by Southern hybridization. The PITPα-targeting vector was linearized, introduced into ES cells by electroporation, and recombinants were selected on the basis of acquired resistance to G418 and gangcyclovir (see MATERIALS AND METHODS). Genomic DNA was prepared from +/+ controls and candidate targeted (−/+) ES cells, digested to completion with either restriction endonuclease EcoRI or XbaI (as indicated at bottom), transferred to nitrocellulose, and hybridized with radiolabeled probe generated by random priming of probe 1 DNA. Diagnostic signatures of the wild-type (+) and targeted (−) PITPα alleles are indicated at right. Approximately 5% of total R1- and AB1-derived recombinants exhibited physical signatures consistent with one wild-type and one targeted PITPα allele as shown.
Quantitative Enzyme-linked Immunosorbent Assay (ELISA)
Total ES cell protein (20 μg) or 50 and 100 ng of PITPβ C-terminal peptide was affixed onto coated ELISA plates (Falcon Plastics, Oxnard, CA) by baking overnight at 60°C. Wells were incubated with 0.5% Tween 20 in phosphate-buffered saline (PBS) (30 min at 37°C), flushed three times with PBS, and blocked with 2% bovine serum albumin (BSA) for 90 min at 37°C. PITPβ-specific rabbit polyclonal antibody was added to wells and incubated at 37°C for 3 h. After extensive washing with PBS, secondary goat anti-chicken antibody was added to wells, incubated at 37°C for 3 h, and again washed with PBS. ELISA signal was developed with the OPD reagent system marketed by Sigma (St. Louis, MO). Signal was quantified as absorbance at 405 nm by using a Biospec Products plate reader.
Lipid Chromatography
ES cells were plated onto 100-mm tissue culture dishes at a density of 1 × 106 cells/plate and labeled in standard media containing 10 μCi/ml of [3H]inositol and supplemented with serum (final concentration of 15%) dialyzed to remove all serum components with molecular mass of <1000 Da. After 72 h, cells were harvested by washing in cold PBS followed by scraping into glass tubes. Samples were then extracted with 1.88 ml of CHCl3/methanol/HCl and incubated for 10 min on ice. Subsequently, 0.625 ml of CHCl3 and 0.625 ml of 0.1 M HCl were sequentially added, and the phases separated by centrifugation for 10 min at 160 × g. The organic phase was collected and dried under a stream of nitrogen gas. Samples were resuspended in a small volume of CHCl3 and loaded onto a dried silica gel 60 thin layer chromatography (TLC) plate and resolved in a CHCl3/methanol/H2O/concentrated ammonia (48:40:7:5) solvent system. The TLC plates were exposed to film, inositol lipid species were identified, and individual species were quantified by scraping from the TLC plate and scintillation counting.
SM, PtdCho, and water-soluble choline metabolite analyses were performed by plating and ES cells as described above with the exception that cells were radiolabeled with 1.5 μCi/ml of [14C]choline chloride. Cells were subsequently washed in ice-cold PBS and scraped off the plates into 1 ml of 5% trichloroacetic acid. Cells were pelleted, washed in PBS, resuspended in 1 ml of polar extraction solvent (15 ml of H2O/15 ml of 95% ethanol/5 ml of diethylether/1 ml of pyridine/36 μl of NH4OH/75 μl of 5% butylated hydroxytoluene in CHCl3/MeOH [2:1]), and incubated at 60°C for 20 min. Subsequently, 0.5 ml of H2O and 5 ml of CHCl3/methanol/butylated hydroxytoluene (2:1:0.005) were added to the samples and the mixtures were incubated at 4°C for 60 min. Samples were centrifuged for 5 min at 160 × g to separate the aqueous (choline, phosphorylcholine, and cytidine-diphosphocholine-choline [CDP]–containing) and organic (PtdCho- and SM-containing) phases. These phases were individually collected and evaporated to dryness under nitrogen gas.
SM and PtdCho were further fractionated by deacylation of PtdCho upon addition of 0.1 N KOH to the lipid film, and incubation at 37°C for 1 h. After addition of CHCl3/balanced salt solution/EDTA, the organic (SM-containing) and aqueous (PtdCho-derived, glycerophosphocholine-containing) phases were collected and dried. SM was resolved on silica gel TLC plates with a CHCl3/methanol (1:1) solvent system. Water-soluble choline metabolites were separated on silica gel TLC plates by using a methanol/aqueous 0.5% NaCl/NH4OH (100:100:4) solvent system. Individual choline-containing species were detected by autoradiography and quantified by scraping and scintillation counting.
Ratiometric Calcium Measurements
ES cells were grown on feeder layers in 100-mm dishes to 80% confluence in complete media, seeded onto gelatinized coverslips at a very low cell density, and allowed to grow for 14 h. Cells were incubated in serum-free media for 2 h before loading in saline solution with fura 2-acetoxymethylester (Teflabs, Austin, TX) for 40 min at a final concentration of 5 μM fura (Manning and Sontheimer, 1997). Cells were transferred to a Series 20 Microperfusion chamber on the stage of a Nikon Diaphot 200 inverted epifluorescence microscope and kept under constant perfusion with HEPES buffer supplemented with 2 mM Ca2+. Immediately before stimulation the chamber was flushed with Ca2+-free HEPES buffer and cells were stimulated with serum (3 or 10%) or LPA. Fura was alternately excited at 340 and 380 nm with a single-wavelength monochromator and fluorescence ratio obtained every 6 s. Emitted fluorescence >520 nm was captured with an intensified charge-coupled device camera, digitized, and analyzed using ImageMASTER software. The ratio of the two images (340/380 nm) was calculated and converted to absolute calcium concentrations (Grynkiewicz et al., 1985).
Transferrin Receptor (TfR) Recycling
Steady-state distribution of TfR at 37°C was performed by the method of Odorizzi et al. (1994). Cells were plated in triplicate wells (24-well plate) and 24 h later incubated first in serum-free media for 1 h then with 4-μg/ml 125I-transferrin (Tf) in 0.1% BSA in PBS for 1 h at 37°C. The labeling media were removed, the cells rinsed three times in 0.1% BSA in PBS, and cells were then washed twice for 3 min with 0.5 ml of 0.2 M acetic acid, 0.5 M NaCl, pH 2.4, to remove surface-bound 125I-Tf. Cells were lysed with 0.1 M NaOH to monitor intracellular 125I-Tf. Radioactivity in the acid washes and the cell lysates was quantified and a ratio obtained.
Internalization assays used the IN/SUR method (Wiley and Cunningham, 1982; Kang et al. 1998). Externalization assays have been described (Jing et al., 1990).
Generation of Mast Cells
ES cells were cultured (10% CO2 atmosphere) on gelatin-coated culture dishes for 48 h in Iscove's modified Dulbecco's medium (IMDM) with 15% fetal bovine serum (Invitrogen), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 2 mM l-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, 100 μM monothioglycerol, and 10 ng/ml LIF. Cells were harvested using trypsin-EDTA and resuspended as single cells in IMDM.
Cells were seeded in methylcellulose-based medium (Stem Cell Technologies (Vancouver, British Columbia, Canada), supplemented as described above but without LIF and containing 40 ng/ml murine stem cell factor, at a density of 500 cells/ml (phase 1 medium) and cultured for up to 21 d. At various times after day 5, the embryoid bodies were disrupted using 0.5% collagenase (Sigma Chemical) and the cell suspension was passed through a 21-gauge needle three times. The hematopoietic progenitors were reseeded in methylcellulose medium supplemented as before but also including 1% BSA, 10 μg/ml insulin, 200 μg/ml transferrin, 30 ng/ml murine interleukin-3, and 30 ng/ml human interleukin-6 (phase 2 medium). Cultures were maintained for up to 40 d and were fed weekly with the same medium. At various times, cultures were scored for surface FcεRI by using fluorescein isothiocyanate (FITC)-labeled mouse IgE and for morphology by electron microscopy.
IgE Labeling and Fluorescence-activated Cell Sorting (FACS) Analysis
Cells were harvested from methylcellulose cultures and treated with collagenase as described above to obtain a single cell suspension. Cells were “rested” for 12 h in IMDM supplemented with 5% fetal bovine serum, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 2 mM l-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 100 μM monothioglycerol. FITC-labeled IgE (gift of B. Wilson, University of New Mexico, Albuquerque, NM) was applied to cells (final concentration 5 nM). After 4 h, cells were collected and fixed. FACS analysis was performed on BD FACSCalibur and Coulter Epics Elite instruments.
Regulated Exocytosis Measurements
Permeabilized Cells.
Calcium/EGTA buffers were prepared by mixing solutions, made up at identical concentrations adjusted to pH 6.8, of EGTA and endpoint-titrated Ca-EGTA (Gomperts and Tatham, 1993). Cells, suspended in an iso-osmotic salts buffer solution [137 mM NaCl, 2.47 mM KCl, 1 mM MgCl2, 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8] supplemented with 1 mg/ml BSA, were then incubated with metabolic inhibitors (0.6 mM 2-deoxyglucose and 10 μM antimycin A) for 5 min (37°C) and cooled on ice. Streptolysin O (SLO, final concentration 1.6 IU/ml in the same buffer) was then added. After 5 min, unbound SLO was washed away by dilution and centrifugation. Rundown was initiated by transferring cells to prewarmed (37°C) iso-osmotic buffer supplemented with 0.3 mM Ca-EGTA (to regulate pCa8) and 1 mM MgATP in wells of 96-microtiter plates. Cell load was ∼10,000/well. After allowing predetermined times for rundown, the cells were stimulated to secrete by adding prewarmed solutions containing Ca-EGTA buffer (3 mM final) formulated to regulate pCa5 (or pCa7 as control) and guanosine-5′-O-(3-thio)triphosphate to a final concentration of 100 μM. After 10 min, the incubations were quenched by addition of an equal volume of ice-cold iso-osmotic buffer supplemented with 5 mM EGTA. The cells were sedimented by centrifugation and the supernatants sampled for secreted hexosaminidase as described (Gomperts and Tatham, 1993; Pinxteren et al., 1998). Secretion is expressed as percentage of total hexosaminidase released relative to total content release evoked by cell permeabilization with 0.2% Triton X-100.
Intact Cells.
Cells were harvested from methylcellulose cultures, treated with collagenase, rested as described above, transferred to fresh IMDM (supplemented as described above) containing 1 or 10 μg/ml antidinitrophenol (DNP) IgE (Sigma Chemical), and incubated for 4 h under standard conditions (Buckley and Coleman, 1992). After three washes in PBS (with Ca2+ and Mg2+) 1% BSA, cells were transferred to 96-well microtiter plates. Human serum albumin conjugated to dinitrophenol (Sigma Chemical) was applied at 0, 1, 10, or 100 μg/ml. Cells were incubated (30 min, 37°C), pelleted, and sampled as described above.
Preparation of Mast Cells for Electron Microscopy
Mast cells were washed in PBS and placed in primary fixative consisting of 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 2 h at 4°C. Cells were rinsed three times in 0.1 M cacodylate buffer for 5 min and incubated in 1% osmium tetroxide for 1 h. After three rinses with 0.1 M cacodylate cells were stained in 0.5% tannic acid for 10 min.
Before embedding, cells were rinsed three times with 0.1 M cacodylate buffer, two times with water, and dehydrated by serial 5-min incubations in 50, 75, and 95% ethanol followed by three 5-min incubations in 100% ethanol. Samples were infiltrated overnight in a 1:1 mixture of ethanol/Spurr's resin followed by incubation in 100% Spurr's resin for 2 h. Samples were finally polymerized in fresh resin by overnight incubation at 60°C. Thin sections were prepared and stained with alcoholic uranyl acetate and Reynold's lead citrate before imaging by electron microscopy.
RESULTS
Targeting of PITPα Mutation into Murine Embryonic Stem Cell Genome
Data from permeabilized cell systems suggest that PITPα plays critical roles in a number of constitutive and regulated membrane trafficking events, and in the optimal activation of signaling pathways that involve G protein-coupled receptors (Cunningham et al., 1996). We therefore expected that PITPα would be essential for the viability of eukaryotic cells. To test this prediction, we used homologous gene targeting methods to assess the cellular consequences of loss of PITPα function. To this end, we recovered a 12-kb fragment of genomic DNA derived from a 129SVJ mouse strain (see MATERIALS AND METHODS). As diagrammed in Figure 1A, this genomic fragment harbors exons 7–10 of the PITPα gene. These exons collectively encode amino acids 126–257 of the 278-residue PITPα polypeptide. From this genomic DNA, we constructed a homologous recombination vector carrying a mutant PITPα allele where exons 8–10 are replaced with a neo∗ cassette (Figure 1A). This mutation (PITPαΔ::neo∗) deletes amino acid residues 162–257 from PITPα, and we previously established that this region of the protein is critical for PITPα activity (Alb et al., 1995). In the targeting vector, a total of 9 kb of PITPα genomic flanking sequences is available for directing replacement of a wild-type PITPα allele by PITPαΔ::neo∗ via homologous recombination.
The targeting vector was linearized by restriction in the vector backbone and introduced by electroporation into ES cell lines derived from R1 and AB1 mouse strains (see MATERIALS AND METHODS). Recombinants were selected using a positive-negative selection strategy of dual resistance to neomycin and gangcyclovir (Mansour et al., 1988). In these experiments, 120 and 76 clones derived from R1 and AB1 ES cells, respectively, survived the double selection and were screened for legitimate recombination events. The screening procedure used Southern hybridization to monitor the genomic arrangement of sequences flanking both ends of the integrated PITPαΔ::neo∗ allele. These analyses demonstrated that six and three clones derived from parental R1 and AB1 ES cell lines, respectively, exhibited the signatures of PITPαΔ::neo∗/+ heterozygotes (Figure 1B). These data indicate a targeting efficiency at the PITPα locus of ∼5%.
PITPα Is Nonessential for Viability of Murine Embryonic Stem Cells
The neo∗ cassette used to generate the PITPαΔ::neo∗ allele contains a missense mutation that reduces the activity of the neomycin acetyltransferase gene product such that a single genomic copy of the neo∗ cassette supports only low-level resistance to 400 μg/ml G418. Two or more genomic copies of the neo∗ cassette, however, endow cells with resistance to high concentrations of G418 (2 mg/ml). This property of neo∗ offers a facile method for generating PITPα −/− ES cell lines provided cells remain viable when challenged with PITPα deficiency.
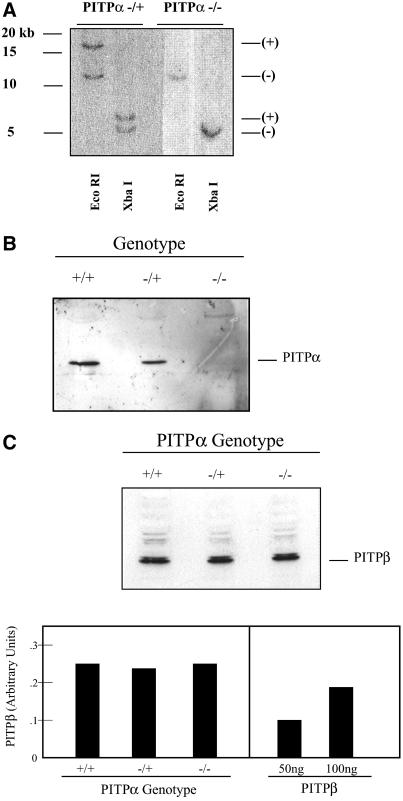
To determine whether ES cells tolerate loss of PITPα, three of the −/+ R1 ES cell lines and two of the −/+ AB1 ES cell lines were challenged with a high concentration of G418 (2 mg/ml) in attempts to convert PITPαΔ::neo∗ to a homozygous state. Surprisingly, we recovered survivors from the high G418 selection that bore the genetic signatures of −/− ES cells. Of the 98 and 72 −/+ R1 and AB1 ES cell-derived cell lines that passed the 2 mg/ml G418 selection, 6 and 3 clones exhibit Southern hybridization profiles diagnostic of a homozygous PITPαΔ::neo∗ genotype (Figure 2A). The authenticity of these −/− clones was confirmed by immunoblotting by using an isoform-specific PITPα antibody. No PITPα antigen is detected in the candidate −/− ES cell lines (Figure 2B).
Figure 2.
(A) Genetic diagnosis of PITPα −/− clones. R1- and AB-1–derived PITPα −/+ ES cells were challenged with 2 mg/ml G418 and surviving clones were isolated. Genomic DNA was prepared from −/+ ES cell controls and high G418-resistant ES cell clones and analyzed as detailed in Figure 1B. Diagnostic signatures of the wild-type (+) and targeted (−) PITPα alleles are indicated at right. Approximately 5% of total R1- and AB1-derived −/+ ES cells that passed the high G418 selection exhibited physical signatures consistent with two targeted PITPα alleles as shown. (B) Immunoblot analysis of candidate PITPα −/− clones. ES cell strains were grown to confluence on gelatin-coated tissue culture dishes, harvested, and lysed by repeated trituration through a tuberculin syringe (Pierce Chemical). A sample load of 150 μg of total cell lysate was resolved by SDS-PAGE, and displayed proteins were transferred to nitrocellulose membranes. Membranes were decorated with PITPα-specific antibodies and developed with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence detection (Amersham Biosciences). PITPα genotypes are indicated at top. Chicken antibodies specific for PITPα were raised against a peptide corresponding to the C-terminal 15 PITPα amino acids. (C) Measurements of PITPβ levels. Top, an immunoblot experiment performed as described in A with 150 μg of total cell lysate per lane and PITPβ-specific antibodies as probe. The PITPβ-specific antibodies were rabbit polyclonal antibodies directed against the C-terminal 18 residues of this protein and were generously provided by Bruce Hamilton (University of California, San Diego, CA). PITPα genotypes are indicated at top. Bottom, quantitative ELISA assay (see MATERIALS AND METHODS) was used to measure levels of PITPβ in isogenic PITPα +/+, −/+, and −/− ES cell lines (left, PITPα genotype indicated at bottom). For comparison, the signals recorded for the indicated amounts of PITPβ C-terminal peptide against which the PITPβ-specific antibodies were raised is given (right).
Quantitative ELISA experiments indicated that PITPα and PITPβ constitute 0.065 and 0.060% of total ES cell protein, respectively (our unpublished data). We therefore considered the possibility that expression of the closely related PITPβ may increase in PITPα −/− ES cells as a compensatory mechanism for loss of PITPα function. Immunoblotting experiments indicate that PITPβ expression is not increased in PITPα −/− ES cells (Figure 2C), and quantitative ELISA experiments independently confirm that PITPβ levels are comparable in PITPα +/+, −/+, and −/− ES cells (Figure 2C). Thus, PITPβ expression is not increased in PITPα −/− cells as part of some adaptive response to PITPα deficiency. Finally, analysis of chromosome spreads demonstrated that the both the R1- and AB1-derived PITPα −/− ES cell lines harbor the normal murine diploid count of 40 chromosomes (our unpublished data), indicating that aneuploidy is not associated with survival of PITPα −/− ES cells. We conclude that PITPα is nonessential for ES cell viability.
Choline Phospholipid Metabolism in PITPα-deficient Cells
Functional analyses of yeast Sec14p reveal a role for this protein in regulating the interface between PtdCho metabolism and Golgi function (Cleves et al., 1991b; Skinner et al., 1995; Xie et al., 1998, 2001). A role for PITPα in regulating metabolism of choline phospholipids in mammalian cells has also been proposed. Antisense approaches suggest that modest reductions in PITPα levels result in significant alterations in PtdCho and SM metabolism in one breast cancer cell line (Monaco et al., 1998).
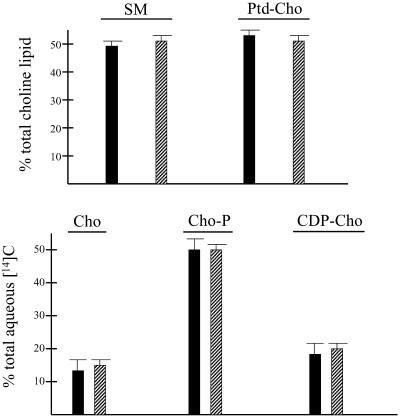
To test whether PITPα regulates choline phospholipid metabolism, +/+ and −/− ES cells were radiolabeled to steady state with [14C]choline, lipids were extracted, and radiolabeled choline phospholipids were resolved by TLC. As shown in Figure 3A, there were no significant differences in steady-state levels of bulk SM or PtdCho between +/+ and −/− ES cells. Moreover, intracellular levels of soluble choline-containing compounds that serve as precursors of PtdCho synthesized via the CDP-choline pathway were unaffected by PITPα deficiency (Figure 3B).
Figure 3.
(A) Bulk PtdCho and SM profiles of PITPα +/+ and −/− ES cell lines. ES cells were labeled to steady state with [14C]choline, and choline-containing phospholipids (SM and PtdCho) were resolved from water-soluble choline-containing precursors of PtdCho in a two-phase organic/aqueous solvent system. (A) Choline-containing lipids SM and PtdCho were distinguished by deacylation of the lipid fraction and separation of the reaction products by organic/aqueous partitioning in a two-phase system. SM and deacylated PtdCho (glycerophosphocholine) were resolved from the organic and aqueous fractions by TLC, respectively (see MATERIALS AND METHODS). Data are from three independent experiments, and are presented as percentage of total choline lipid recovered as SM and PtdCho, respectively, in isogenic PITPα +/+ (black bars) and PITPα −/− ES cell lines (hatched bars). All ES cell lines exhibited similar incorporation of [14C]choline into phospholipid. (B) Choline-containing biosynthetic precursors of PtdCho in PITPα +/+ and −/− ES cell lines were resolved by TLC. The data are given as the contribution of each indicated choline compound to total water-soluble radioactivity. Cho, choline; Cho-P, phosphorylcholine; CDP-Cho, cytidine-diphosphocholine. Values represent averages of three independent experiments, and the solid and hatched bars represent data from PITPα +/+ and −/− ES cell lines, respectively.
Bulk Phosphoinositides in PITPα-deficient Cells
Speculations regarding the physiological function of PITPs in mammalian cells have focused on the role of these proteins in regulating phosphoinositide synthesis. To determine the consequences of PITPα deficiency on phosphoinositide levels in ES cells, isogenic PITPα +/+ and −/− ES cells were radiolabeled to steady-state with [3H]inositol. The inositol lipids were then extracted and resolved by TLC.
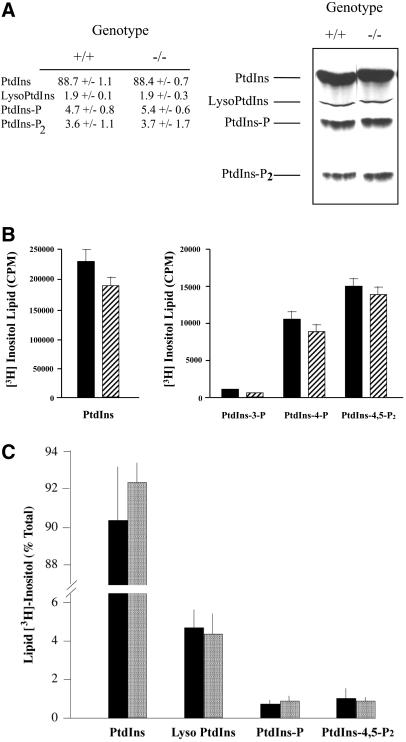
The steady-state inositol lipid profiles of +/+ and −/− ES cells are essentially identical (Figure 4A). As expected, PtdIns was the major labeled lipid recovered from both +/+ and −/− ES cells (88.7 ± 1.1 and 88.4 ± 0.7% of inositol lipid, respectively). No differences were detected in the steady-state levels of monophosphorylated PtdIns species (i.e., combined levels of PtdIns-3-phosphate and PtdIns-4-phosphate with the latter representing by far the predominant species) in +/+ versus −/− ES cells (4.7 ± 0.8 and 5.4 ± 0.6% of inositol lipid, respectively). Similarly, bis-phosphorylated PtdIns derivatives (i.e., combined levels of PtdIns-4,5-P2, PtdIns-3,4-P2, and PtdIns-3,5-P2 with PtdIns-4,5-P2 representing by far the predominant species) were also indistinguishable in +/+ versus −/− cells (3.6 ± 1.1 and 3.7 ± 1.7% of inositol lipid, respectively). These results were verified by deacylating the inositol lipid fraction and resolving the glycerophosphoinositide derivatives by anion-exchange chromatography. No differences in bulk steady-state levels of PtdIns, PtdIns-3-phosphate, PtdIns-4-phosphate, or PtdIns-4,5-P2 were observed (Figure 4B). Moreover, the deacylation data were in quantitative agreement with the TLC data.
Figure 4.
Steady-state levels of inositol phospholipids. (A) ES cells were labeled to steady state with [3H]inositol, and phospholipids were extracted and resolved by TLC (see MATERIALS AND METHODS). A representative chromatogram is given (right) with identification of lipid species (left) and PITPα genotype (top). At left are given the quantitative data obtained from the average of three independent experiments. (B) ES cells were labeled to steady state with [3H]inositol, phosphoinositides were extracted, deacylated in the presence of methylamine, and the corresponding glycerophospho-derivatives were resolved by anion exchange chromatography (Stolz et al., 1998). The left panel presents the deacylated PtdIns data for PITPα +/+ (solid bars) and −/− cells (hatched bars), whereas the right panel shows data for deacylated PtdIns-3-P, PtdIns-4-P, and PtdIns-4,5-P2. All values represent averages of at least three independent experiments. (C) ES cells were pulse radiolabeled for 1 h with 100 μCi/ml of [3H]inositol. Lipids were extracted and resolved by TLC, identified by autoradiography, scraped, and quantified by scintillation counting. Data are expressed as percentage of total lipid [3H]inositol incorporated into each lipid species identified at bottom. Values from PITPα +/+ (solid bars) and −/− cells (stippled bars) are given. PtdIns-P represents the sum of PtdIns-4-P and PtdIns-3-P values. Data represent the averages of three independent experiments.
To assess rates of phosphoinositide synthesis in PITPα +/+ and −/− ES cells, cells were pulse-radiolabeled with [3H]inositol and radiolabeled inositol lipids were then extracted, resolved, and quantified. As shown in Figure 4C, the relative rates of PtdIns, PtdIns-phosphate (primarily PtdIns-4-phosphate), and PtdIns-4,5-P2 were comparable in these isogenic ES cell lines. To compare efficiencies of phosphoinositide resynthesis in PITPα +/+ and −/− ES cells after stimulation, cells were radiolabeled with [3H]inositol to steady state. Radiolabeled cells were then serum starved, stimulated with serum for various times, and labeled inositol phospholipids were extracted and quantified. The inositol phospholipid profiles of isogenic PITPα +/+ and −/− cell lines as a function of time were again indistinguishable (our unpublished data). We conclude that there are no major reductions in rates of phosphoinositide resynthesis in PITPα-deficient ES cells.
Because PITPα is concentrated in the nucleus, PITPα −/− cells might exhibit alterations in nuclear phosphoinositide levels. To address this issue, a detergent method was used to purify nuclei from isogenic +/+ and −/− ES cells labeled to steady state with [3H]inositol. Nuclear phosphoinositide levels were then analyzed. Again, we were unable to discern any significant differences in the identities of nuclear phosphoinositide species or in their steady-state levels between +/+ and −/− ES cells (our unpublished data). These data indicate that PITPα deficiency does not manifest itself in discernible changes in steady-state levels of either bulk membrane or nuclear phosphoinositide pools.
Bulk Signaling in PITPα-deficient Cells
Although the steady-state data suggest that PITPα does not play a quantitatively obvious role in regulating bulk or nuclear phosphoinositide pools, these measurements are potentially insensitive to regulated fluctuations in specific pools. In that regard, PITPα is proposed to be a critical regulator of the metabolism of PIP pools that sustain signaling via receptors localized at the plasma membrane (Kauffmann-Zeh et al., 1995). Thus, we used several independent assays to measure responsiveness of +/+ and −/− ES cells to serum stimulation. First, isogenic ES cells were labeled to steady state with [3H]inositol, cells were serum starved, and serum-deprived cells were stimulated with either undialyzed serum or lysophosphatidic acid. Inositol-1,4,5-trisphosphate was resolved and quantified from samples collected at various times poststimulation. These time course experiments failed to reveal obvious differences in bulk inositol-1,4,5-trisphosphate metabolism in −/− ES cells relative to isogenic +/+ cells (our unpublished data).
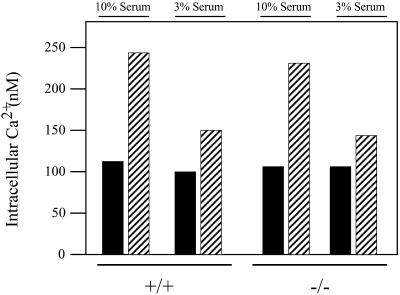
A second independent approach involved monitoring evoked Ca2+ mobilization in cells. We measured no significant difference between +/+ and −/− ES cells in the overall increase in cytosolic Ca2+ when serum-deprived cells were stimulated with 10% serum. Under these stimulation conditions, virtually all of the cells imaged responded to serum by rapidly increasing cytosolic Ca2+ to levels ∼2.5-fold greater than those recorded for resting cells (Figure 5). Because stimulation with high serum might saturate cell surface receptors, and thereby obscure more subtle signaling defects in −/− cells, we repeated the experiments using a 3% serum stimulation. As seen in Figure 5, this low-serum condition represents a suboptimal stimulation as the amplitude of the response was reduced relative to that scored for the 10% serum condition. Moreover, only ∼30% of the wild-type cells responded to the low-serum stimulus. Yet, even under these conditions, the latency of the Ca2+ response, the number of cells responding to the serum stimulus, and the response amplitude were all comparable for +/+ and −/− ES cells. Similar results were obtained when serum-deprived cells were stimulated with LPA alone (our unpublished data). The data indicate that PITPα is dispensable for bulk cellular responses to serum stimulation, and that plasma membrane signaling through at least the LPA receptor is unaffected by PITPα deficiency.
Figure 5.
Bulk signaling properties of PITPα +/+ and −/− cells. ES cells were serum starved for 2 h, loaded with fura 2, and stimulated with serum (3 or 10%). Ratios of fura 2 excitation at 340 and 380 nm were determined and converted to absolute calcium concentrations. These concentrations are presented as a function of serum concentration used to stimulate cells (top) and PITPα genotype (bottom). Values represent the averages and SDs of cytosolic Ca2+ concentrations measured for 69 PITPα +/+ and 67 PITPα −/− cells in the 10% serum stimulation condition, and 48 PITPα +/+ and 45 PITPα −/− cells in the 3% serum stimulation condition, respectively.
Constitutive Secretory Pathway Function in PITPα-deficient Cells
The availability of isogenic +/+ and −/− cell lines allowed a survey of the involvement of PITPα in defined cellular functions. Several lines of evidence obtained from biochemical reconstitution systems suggest that PITPα regulates constitutive pathways for membrane trafficking through the Golgi complex (Paul et al., 1998), and from the trans-Golgi network to the cell surface (Jones et al., 1998). To assess constitutive secretory pathway function in PITPα-deficient cells, we monitored the synchronized transport of the mutant vesicular stomatitis virus glycoprotein (VSV-G) tsO45 protein from the endoplasmic reticulum (ER) to the Golgi complex and the cell surface. Incubation of both +/+ and −/− cells at 39.5°C results in accumulation of core glycosylated VSV-G tsO45 in the ER (Aridor and Balch, 1996). Chase of VSV-G tsO45 from the ER was initiated by addition of cycloheximide to cells and shifting the infected cells to 32°C to permit proper folding of the mutant protein.
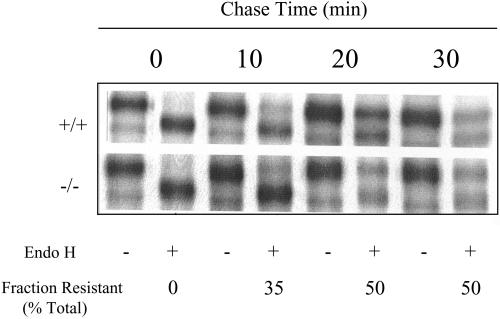
In both +/+ and −/− cells, ∼35% of the VSV-G tsO45 was exported from the ER to the Golgi complex by 15 min of chase at 32°C as scored by the remodeling of VSV-G glycosyl chains to a form where these are resistant to endoglycosidase H (EndoH) treatment (Figure 6). By 45 and 60 min of chase, 50% of the VSV-G tsO45 had acquired EndoH resistance in the +/+ and the −/− cells (Figure 6). Longer chase times did not result in additional export of VSV-G tsO45 from the ER (our unpublished data). The 50% efficiency of VSV-G tsO45 export from the ER is in agreement with values reported by others (Bi et al., 1997). These data indicate that VSV-G tsO45 transport from the ER to the medial-Golgi (i.e., where EndoH resistance is acquired) occurs with similar kinetics and efficiencies in +/+ and −/− cells. Surface labeling methods with VSV-G antibodies revealed robust plasma membrane labeling in intact +/+ and −/− cells within 30 min of release of the 39.5°C block (our unpublished data). The surface-labeling data, although qualitative, suggest that the rates of VSV-G tsO45 transit from the ER to the cell surface are similar in PITPα +/+ and −/− cells.
Figure 6.
Trafficking of VSV-G from the ER to medial-Golgi in PITPα +/+ and −/− cells. The mutant VSV-G tsO45 protein was accumulated in the ER of infected cells (genotypes at left) at 39.5°C and chase was initiated by adding cycloheximide and reducing culture temperature to 32°C (t = 0 min). VSV-G tsO45 was chased from the ER for various times (top). Samples were collected and either challenged with endoglycosidase H (+) or not (−) as indicated at bottom. EndoH resistance signifies arrival in medial-Golgi, and the fraction of EndoH-resistant VSV-G tsO45 increases with chase. EndoH sensitivity is scored by increased migration of VSV-G tsO45 after EndoH challenge. Quantification of the fraction of VSV-G tsO45 that has acquired EndoH resistance at each time point is given at bottom. One set of quantifications is given because values were essentially identical for +/+ and −/− cells.
Constitutive Pathways for Endocytosis Operate Normally in PITPα −/− ES Cells
Operation of the constitutive endocytic cycle is strongly perturbed by agents such as wortmannin that inhibit pathways for synthesis of 3-phosphorylated inositol lipids and inositides (Corvera et al., 1999). We therefore tested whether PITPα deficiency compromises endocytic pathway activity. In these experiments, we used the constitutively recycling TfR as a sensitive reporter for function of the endocytic pathway in +/+ and −/− ES cells. Because TfR is a long-lived protein that recycles rapidly through the endosomal pathway, even subtle defects in TfR internalization from the cell surface, or in TfR recycling from endosomes back to the cell surface, exert large effects on steady-state TfR distribution. Using an 125I-transferrin binding assay to register surface-exposed TfR, we found that the steady-state distributions of TfR are similar in +/+ and −/− cells irrespective of whether cells are incubated in 10 or 3% serum. In all cases, 60% of the TfR is present in intracellular compartments, whereas 40% of the TfR resides at the cell surface (Figure 7A). These steady-state distributions of TfR are in close agreement with those recorded for other cell types (Jing et al., 1990; Cotlin et al., 1999).
Figure 7.
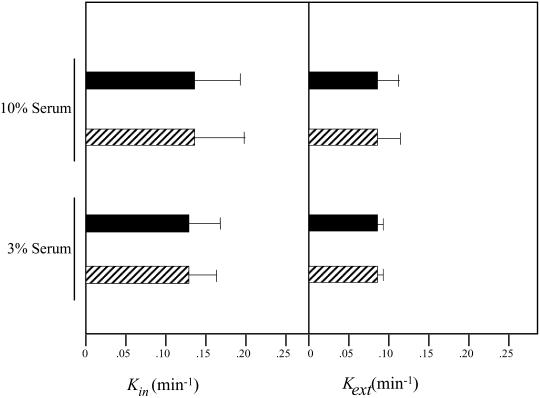
(A) Steady-state distribution of TfR on the cell surface of PITPα +/+ and −/− cells. Steady-state distribution of TfR was performed as described previously at 37°C (Odorizzi et al., 1994; see MATERIALS AND METHODS) with the modification that PITPα +/+ and −/− ES cells were plated in triplicate in standard media containing either 10 or 3% serum. The percentage of total 125I-Tf (and therefore TfR) located at the cell surface (solid bars) and within the cell (hatched bars) is given for each ES cell PITPα genotype (+/+ or −/−; bottom) and for each serum concentration tested (3 and 10%; top). (B) Kinetics of TfR internalization and recycling in PITPα +/+ and −/− cells. For internalization assays, PITPα +/+ (solid bars) and −/− cells (hatched bars) were preincubated in serum-free media at 37°C and prewarmed 125I-Tf was added for various times up to 10 min. At each time point, the ratio of internalized and surface 125I-Tf was distinguished by an acid strip of the cell surface, and Tf(internal)/Tf(surface) ratio was plotted as a function of time. The slope of each curve yields the TfR internalization constant kin, and the data represent the mean and SDs derived from at least three independent determinations. To measure the recycling constant kext, cells were incubated with 125I-Tf for 1 h at 37°C and chilled. Surface-bound 125I-labeled Tf was removed by chelation with deferosamine at 4°C, cells were warmed to 37°C to initiate chase of 125I-Tf from endocytic compartments to the cell surface, and measurements of cell-associated and -released radioactivity were taken at various times of chase. The Tf(secreted)/Tf(internal) ratio was plotted against time, and the slope of each curve yields kext. The data represent the mean and SE derived from at least three independent determinations.
The normal TfR distribution between the plasma membrane and intracellular pools in −/− ES cells suggests either that rates of TfR internalization and recycling are unadulterated in PITPα-deficient cells, or that any alterations in internalization or recycling rates are compensated by alterations in the opposing arm of the TfR trafficking pathway. To measure TfR internalization rates (expressed as kin), we used the IN/SUR method to monitor the rate at which surface 125I-transferrin–TfR complexes acquired resistance to surface stripping with a low-pH wash. As shown in Figure 7B, +/+ cells exhibit a kin = 0.123 ± 0.075 when incubated in 10% serum and 0.125 ± 0.064 when the assay is performed in cells cultured in 3% serum. Similarly, TfR kin values are 0.129 ± 0.078 and 0.133 ± 0.042 in −/− cells incubated in 10 and 3% serum, respectively. Thus, PITPα deficiency does not impose a defect on rate of TfR internalization into cells.
Finally, the data show that the rate of TfR recycling from intracellular compartments to the cell surface is also refractory to PITPα dysfunction. By monitoring the rate at which internalized 125I-transferrin–TfR complexes reach the cell exterior, rates of TfR recycling (kext) in +/+ and −/− cells were calculated. The recycling rates are essentially the same in +/+ and −/− cell lines irrespective of whether cells are incubated at 3% serum (kext = 0.085 ± 0.007 vs. 0.085 ± 0.007 min−1) or 10% serum (kext = 0.075 ± 0.021 vs. 0.080 ± 0.028 min−1; Figure 7B). These collective data indicate that the constitutive endocytic cycle operates normally in PITPα-deficient cells.
Regulated Exocytosis in PITPα-deficient Mast Cells
Because PITPα has been implicated in catalyzing formation and priming of secretory granules for agonist-induced exocytosis (Hay and Martin, 1993; Ohashi et al., 1995; Fensome et al., 1996; Pinxteren et al., 1998, 2001), we investigated the consequence of PITPα deficiency to the activity of the regulated exocytotic pathway in ES cell-derived mast cells. Initially, we tested whether −/− ES cells could be induced to become functional mast cells. We subjected isogenic +/+ and −/− ES cell lines to conditions that induce ES cells to differentiate into the mast cell lineage (see MATERIALS AND METHODS). At various times during a 40-d period of maturation in phase 2 medium, the differentiating cell cultures were harvested and subjected to two tests for acquisition of mast cell properties. First, FACS was used monitor expression of FcεRI, a mast cell marker, on the cell surface. Second, we used to electron microscopy to visualize formation of mast cell-like DSGs in the differentiating cell population. Mature cells (>30 d in phase 2) were then used in established assays that measure secretory capacity directly.
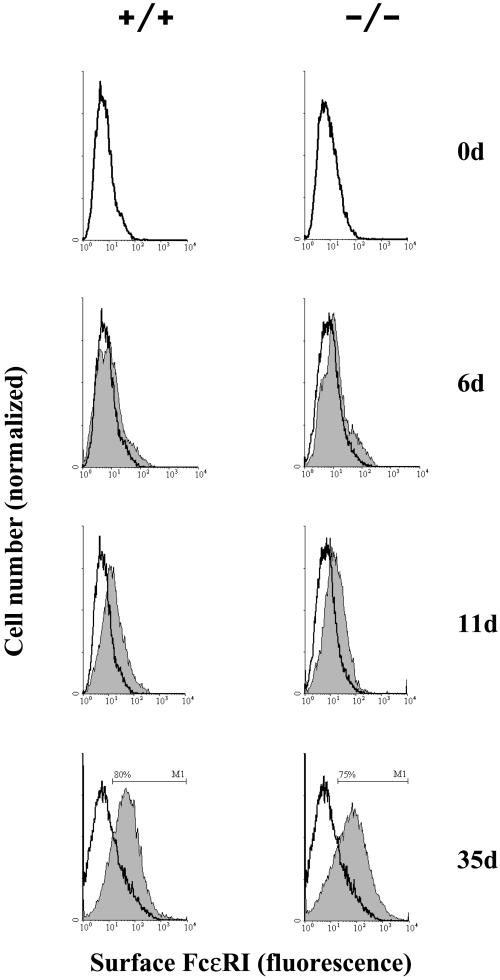
As shown in Figure 8A, both +/+ and −/− ES cells express the high-affinity receptor for IgE (FcεRI) equally well and the efficiency of differentiation to mast cells is dependent on the length of culture incubation in phase 1 medium. The optimum incubation was 9 d, with 75–80% of the cells expressing FcεRI after 35 d in phase 2 (Figure 8A, bottom). Shorter or longer periods in phase 1 medium gave a lower yield of mast cells, e.g., 50% (5 d) and 58% (21 d). No further increase in the expression of FcεRI was observed after 35 d in phase 2 (our unpublished data).
Figure 8.
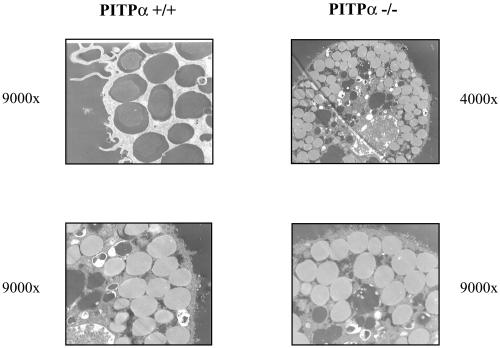
(A) Expression of IgE binding sites on the surface of +/+ and isogenic PITPα −/− ES cells during phase 2 culture. Cells were treated with FITC-labeled IgE (5 nM) and surface IgE-FITC was quantified by flow cytometry. Shaded histograms represent populations from phase 2 cultures with ages in days indicated at right. Results from −/− cells are in the right-hand panels. Control cells (open histograms) represent passage-matched cells maintained for the same period exclusively in phase 1 medium. Results are representative of three independent experiments. (B) Mast cells from isogenic PITPα +/+ and −/− ES cells (genotype at top) were visualized by thin-section electron microscopy. Magnifications for each panel are given. PITPα +/+ and −/− ES cells yielded mast cells packed with morphologically correct dense core secretory granules.
Visualization of differentiated cell populations by electron microscopy confirmed that both +/+ and −/− ES cells efficiently manufacture DSGs, and that these granules are morphologically indistinguishable from those present in authentic mast cells. In cells derived from both +/+ and −/− lineages, the mast-like cells were packed with DSGs (Figure 8B). Classification of these granules as DSGs is further supported by the demonstration that the granule-bearing cells express both the mast cell-specific marker tryptase and histamine. By these criteria, ∼80% of the +/+ and −/− ES cells adopted the morphological characteristics of mast cells. We conclude that both DSG biogenesis and mast cell differentiation proceed efficiently in the face of PITPα deficiency.
To characterize the secretory competence of these differentiated cells, we stimulated intact cells to exocytose. Both +/+ and −/− ES cell-derived mast cells are perfectly able to secrete granule contents after loading of the cell surface with anti-DNP IgE and stimulating the loaded cells with a DNP-conjugated human serum albumin agonist (Table 1). These data demonstrate that the physiologically relevant regulated exocytotic machinery is in place in PITPα-deficient cells.
Table 1.
IgE-dependent secretion from peritoneal mouse mast cells and ES cell-derived mast cells The values (%) express the secretion (± SEM, n = 4 separate determinations) elicited by applying human serum albumin-DNP to anti-DNP IgE-loaded cells at the indicated concentrations and under the conditions as described in MATERIALS AND METHODS. Genotypes of the cells are indicated at left. Authentic peritoneal mast cells (PMCs) were us as positive controls and were isolated by peritoneal lavage of 2–10-wk-old male C57 Bl/6J mice. Mast cells were purified (>95%) by centrifugation through a Percoll cushion, essentially as previously described (Pinxteren et al., 1998).
| IgE (μg/ml) HSA-DNP (μg/ml) | 1
|
10
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 10 | 100 | 0 | 1 | 10 | 100 | |
| PMCs | 1.1 ± 0.3 | 19.6 ± 1.8 | 21.1 ± 1.8 | 20.5 ± 1.4 | 2.2 ± 0.7 | 11.2 ± 2.7 | 11.4 ± 1.5 | 12.9 ± 2.4 |
| +/+ | 1.5 ± 0.2 | 19.6 ± 2.0 | 18.8 ± 0.7 | 19.5 ± 2.8 | 0.7 ± 0.2 | 11.2 ± 1.3 | 9.0 ± 2.1 | 8.7 ± 0.7 |
| −/− | 1.2 ± 0.4 | 18.6 ± 1.7 | 20.0 ± 1.4 | 20.2 ± 2.3 | 0.9 ± 0.5 | 10.5 ± 0.9 | 10.2 ± 0.9 | 10.5 ± 0.4 |
Rundown in Permeabilized PITPα-deficient Mast Cells
Agonist-induced exocytotic events have been reconstituted in permeabilized mast cells, and this reaction exhibits “rundown.” Rundown is the progressive decrease in exocytotic efficiency that is recorded as the time interval between cell permeabilization and addition of secretory stimulus increases, and reflects the progressive destruction of phosphoinositides in the permeabilized cell ghosts (Pinxteren et al., 1998, 2001). Addition of either cytosol or purified PITPα to the permeabilized cells strongly diminishes rundown rate, suggesting that PITPα may represent the key cytosolic factor that stimulates the reconstituted exocytotic event (Pinxteren et al., 1998, 2001).
If PITPα is indeed a major factor in synthesis of phosphoinositide pools required for agonist-induced exocytosis, significant differences in the rates of rundown in permeabilized PITPα +/+ versus PITPα −/− mast cells should be apparent. As shown in Table 2, application of 100 μM guanosine-5′-O-(3-thio)triphosphate at pCa5 to SLO-permeabilized peritoneal mast cells or to +/+ and −/− ES cell-derived mast cells elicited the massive degranulation normally recorded upon such treatment, and the corresponding values recorded for each cell type were set at 100%. As reported previously (Pinxteren et al., 1998, 2001), the exocytotic efficiency recorded for peritoneal mast cells was inversely proportional to time of preincubation of permeabilized cells before stimulation. This rate of peritoneal mast cell rundown closely resembles that measured for permeabilized PITPα +/+ ES cell-derived mast cells. These data further demonstrate that ES cell-derived mast cells exhibit the properties of genuine mast cells. Rundown in the PITPα-deficient system was not accelerated relative to the peritoneal mast cell and PITPα +/+ ES cell-derived mast cell controls (Table 2). These data indicate that PITPα is not a major factor in the resynthesis of phosphoinositide pools whose integrity is scored in the reconstituted reaction. It remains possible that PITPβ is the physiological PITP isoform that is dedicated to this reaction. Quantitative ELISA experiments indicated that PITPβ is more abundant in mast cells than is PITPα (our unpublished data).
Table 2.
Secretion from permeabilized peritoneal- and ES cell-derived mast cells Peritoneal mast cells (PMCs; isolated as described in the legend to Table 1) and isogenic +/+ and −/− ES cell-derived mast cells, pretreated with SLO at 0°C were permeabilized by transfer to a buffer (pH 6.8) containing 1 mM MgATP and 0.1 mM EGTA to reduce free [Ca2+] to below 10−8M. This treatment induces a time-dependent rundown of secretion efficiency that is a function of progressive loss of a phosphoinositide pool required for the exocytotic event (Pinxteren et al., 1998, 2001). At the indicated rundown times, cells were stimulated by a secretagogue cocktail containing 100 μM GTPγS and 3 mM Ca-EGTA formulated to regulate pCa5. After a 10-min incubation, the systems were quenched with an ice-cold salts buffer containing 5 mM EGTA. The cells were sedimented and the supernatants were sampled and assayed for secreted hexosaminidase. Data points (%) represent mean values ± SEM (n = 4) normalized to maximum secretion (100%) determined in absence of rundown (time 0). PMCs serve as positive control.
| Time of rundown | 0 min | 2 min | 10 min | 30 min |
|---|---|---|---|---|
| PMCs | 100 | 57.7 ± 2.8 | 25.3 ± 2.0 | 2.6 ± 0.3 |
| +/+ | 100 | 55.7 ± 12.1 | 23.7 ± 3.0 | 4.0 ± 1.5 |
| −/− | 100 | 51.2 ± 3.6 | 22.6 ± 3.1 | 2.6 ± 1.1 |
Growth and Pluripotency of PITPα-deficient ES Cells in Nude Mice
PITPα-deficient ES cells are robust by the definition that such cells exhibit wild-type growth rates in culture when presented with a range of fetal calf serum concentrations (15, 10, and 3%; our unpublished data). We considered the possibility that −/− ES cell lines might exhibit growth defects when placed into an environment where these cells must compete vigorously with wild-type cells for growth factors. To this end, isogenic R1-derived +/+ and −/− ES cell lines and isogenic AB1-derived +/+ and −/− ES cell lines were introduced into adult athymic nude mice via a subcutaneous injection route. The abilities of these ES cell lines to form tumors in nude mice were then quantified.
In all cases, palpable tumors were produced within 3 wk of injection, and no significant differences were recorded between +/+ and −/− ES cell lines with regard to average size of tumors (Table 3). In the R1 ES cell genetic background, all seven mice injected with +/+ ES cells elaborated tumors with an average tumor mass of 0.88 ± 0.56 g. Similarly, all 10 mice injected with −/− cells elaborated tumors, and these tumors exhibited a mass of 0.86 ± 0.43 g. Consistent results were obtained when +/+ and −/− ES cells of the AB1 genetic background were analyzed. The average tumor mass measured from four independent +/+ AB1-derived ES cell experiments was 1.27 ± 0.13 g. Two separate experiments, each involving the injection of five mice with isogenic AB1-derived PITPα −/− ES cells, yielded average tumor masses of 1.66 ± 0.36 and 1.28 ± 0.55g, respectively (Table 3). Thus, PITPα deficiency does not compromise either R1 or AB1 ES cell vigor when this property is assayed in a physiological context.
Table 3.
Tumorigenic potential of PITPα +/+ and −/− ES cells Athymic nude mice were injected subcutaneously in the hind flank with 107 ES cells suspended in 0.2 ml of PBS/glucose. The tumors that formed were surgically harvested after 3 wk and weighed. Values represent the mean and standard error derived from analyses of all tumors in a given set.
| Injected cells | Mice with tumors/ mice injected | Tumor mass (g) |
|---|---|---|
| R1 PITPα +/+ | 7 /7 | 0.88 ± 0.56 |
| R1 PITPα −/− | 10 /10 | 0.86 ± 0.43 |
| AB1 PITPα +/+ | 4 /4 | 1.27 ± 0.13 |
| AB1 PITPα −/− | 10 /10 | 1.48 ± 0.45 |
Finally, no obvious distinctions were apparent between the histology of tumors derived from +/+ and −/− ES cell lines. Tumors from both genetic backgrounds represented complex teratomas consisting of neural, respiratory, muscle, gastrointestinal, developing spinal chord, and collagenous tissues. Moreover, inner masses of −/− ES cell-derived tumors often exhibit complex structures, e.g., glandular structures lined with ciliated epithelia (our unpublished data). Thus, PITPα deficiency fails to compromise ES cell vigor and is of no obvious consequence for ES cell differentiation into multiple lineages and into intricate multicellular structures. The fundamental dispensability of PITPα for murine development is confirmed by our finding that PITPα −/− mice are born alive at standard Mendelian frequencies (Alb and Bankaitis, unpublished data).
Targeting of PITPβ Mutation into Murine Embryonic Stem Cell Genome
The dispensability of PITPα for housekeeping functions in murine cells is consistent with the possibility that PITPβ and PITPα are largely redundant and contribute rather equally to essential housekeeping functions. Alternatively, PITPβ might represent the essential mammalian housekeeping PITP, whereas PITPα serves a very specific and as-yet-unidentified function. We therefore used gene-targeting methods to assess the cellular consequences of PITPβ dysfunction. We isolated a 5-kb fragment of 129SVJ mouse genomic DNA that harbors three exons of the PITPβ gene. These exons encode amino acids 66–121 of the 278 residue PITPβ polypeptide, and the homologous recombination vector derived from this fragment carries a mutant PITPβ allele where exons A and B are replaced with a neo∗ cassette (Figure 9A). This mutation (PITPβΔ::neo∗) deletes residues 66–99 from PITPβ. Based on our functional analyses of PITPα (Alb et al., 1995), PITPβΔ::neo∗ almost certainly represents a null mutation.
Figure 9.
(A) Targeting the PITPβ locus. A 5-kb SacI-SacI genomic fragment was excised from a cloned 15-kb genomic DNA fragment and determined to carry three PITPβ exons (designated A, B, and C) that encode residues 66–152 of the mouse PITPβ. Southern analyses of the adjacent 10-kb genomic fragment did not reveal any exons. From this 10-kb DNA, a 4-kb SacI/EcoRV fragment that lies adjacent to the 5-kb SacI-SacI genomic DNA was cloned to generate a cassette containing 9 kb of total homology to the PITPβ. Exons A and B were replaced with a neo∗ cassette described in Figure 1A, and HSV-tk was introduced at one end of the cloned PITPβ DNA. The targeting vector was linearized, introduced into ES cells by electroporation, and recombinants that acquire resistance to both G418 and gangcyclovir were selected. Positions of probes 1 and 2 for diagnosis of wild-type and targeted PITPβ alleles by Southern hybridization are shown. In hybridization experiments, EcoRV digestion was routinely used as a diagnostic. The wild-type PITPβ locus generates a 10.5-kb EcoRV restriction fragment, and the ends of this fragment both lie outside the targeting region. NEO∗ introduces a new EcoRV site into the PITPβ locus so that the targeted allele yields an 8.0- and a 2.7-kb fragment revealed with probes 1 and 2, respectively. (B) Distribution of PITPβ genotypes in the F1. The 78 live-born F1 progeny analyzed from intercross of PITPβ −/+ mice were scored by PCR. The positions of the three primers for PCR diagnosis of wild-type and targeted PITPβ alleles are shown. The wild-type allele was diagnosed using primer 1 (PITPβ-1045-R; 5′-GCGAACAACCTGTGAGCC CAG-3′) and primer 2 (PITPβ-545-F; 5′-GTGAACAAGTACCAGCAGC-GC-3′). The PITPβ null allele was diagnosed using primer 1 (PITPβ-1045-R; 5′-GCGAACAACCTGTG A-GCCCAG-3′) and primer 3 (3-NEO1; 5′-GCCAGA-GGCCACTTGTGTAGCGCC-3′). Polymerase chain reaction products derived from the wild-type PITPβ and targeted PITPβ alleles are 500 and 225 base pairs, respectively. The number of live births obtained for each PITPβ genotype (bottom) is indicated. (C) PITPβ genotypes in F1 embryos derived from intercross of PITPβ −/+ mice. Sixty-six F1 embryos derived from intercross of PITPβ −/+ mice were harvested in an E7.5-E11.5 window. The number of embryos obtained for each PITPβ genotypic class (bottom) is given. Embryos were genotyped by polymerase chain reaction as outlined in B.
The targeting vector was linearized and electroporated into murine ES cell lines derived from the AB1 mouse strain. Recombinants were again selected using a selection for recombinants exhibiting dual resistance to neomycin and gangcyclovir. Of clones that survived the double selection, 50 were screened for legitimate recombination events. One ES cell line exhibited the genetic signatures of a PITPβΔ::neo∗/+ heterozygote (Figure 9B). We challenged this cell line with high G418 in an attempt to generate PITPβ −/− ES cells and recovered 150 clones resistant to high concentrations of G418. In contrast to the results obtained with PITPα −/+ ES cells, genotypic analysis of these G418-resistant ES cell clones failed to identify PITPβ −/− ES cell clones (our unpublished data).
Consequences of PITPβ Deficiency in Mice
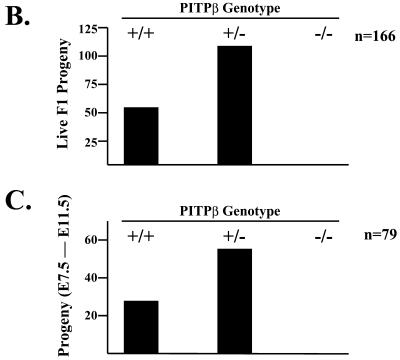
Our failure to recover PITPβ −/− ES cells via the high G418 resistance selection may be the direct consequence of ablating an essential activity of PITPβ at the cellular level, or may simply reflect some trivial difficulty in obtaining such clones that is unrelated to loss of PITPβ function. If PITPβ is the sole, or even the major, executor of an essential cellular function, PITPβ deficiency should manifest itself in an early embryonic lethal phenotype. If PITPβ is not an essential cellular housekeeping PITP then PITPβ −/− mice should exhibit significant developmental progress before failure is realized. To distinguish these possibilities, the PITPβΔ::neo∗ allele was passed through the murine germline and a colony of PITPβ −/+ mice was generated (see MATERIALS AND METHODS). We then analyzed the F1 progeny generated by intercross of PITPβ −/+ heterozygotes for −/− mice or embryos. From a set of 166 live births we recovered a ratio of 58 +/+:108 −/+ F1 progeny that closely conformed to the 1:2 ratio predicted by Mendel's rules (Figure 9C). Yet, we failed to recover any −/− progeny in the F1 at all. Equal numbers of −/− and +/+ F1 progeny are expected if PITPβ deficiency is not associated with a cell- or embryonic-lethal phenotype. Thus, PITPβ −/− mice do not survive to term.
To further assess the consequences of PITPβ deficiency for murine development, we performed a series of timed mating experiments and surveyed a total of 79 F1 embryos harvested in an E7.5-E11.5 developmental window. Again, a Mendelian distribution of 24 +/+:58 −/+ F1 embryos was recorded, but no −/− embryos were recovered in the F1 progeny (Figure 9D). In these experiments, we failed to detect any unusual signs of embryo resorption. These data indicate that PITPβ deficiency minimally results in early embryonic lethality of the developing mouse.
DISCUSSION
Genetic experiments in yeast have demonstrated that specific PITPs, such as the budding and fission yeast Sec14ps, execute essential cellular housekeeping functions (Bankaitis et al., 1989, 1990; Nakase et al., 2001). Other PITP isoforms, such as the nonclassical SFH PITPs of S. cerevisiae and the one known Sec14p of the dimorphic yeast Yarrowia lipolytica, perform specific nonessential functions (Lopez et al., 1994; Li et al., 2000; Wu et al., 2000). The genetic approaches used in studies of yeast PITP function, and also in the function of a nonessential Drosophila PITP (Milligan et al., 1997), have revealed striking specificities of physiological function. These specificities, at least to this point, have been invisible in the in vitro systems used to study mammalian PITP function (reviewed in Kearns et al., 1998).
Herein, we report the outcome of genetic experiments that for the first time characterize the physiological consequences associated with functional ablation of PITPα and PITPβ isoforms in murine ES cells. Our results clearly demonstrate that, in two distinct ES cell lines, specific genetic ablation of PITPα function does not compromise ES cell viability. Detailed analyses of inositol- and choline-phospholipid metabolism failed to reveal any significant aberrations in PITPα-deficient cells. In this regard, our data do not support a previous report that PITPα is required for normal metabolism of PtdCho and SM in living cells (Monaco et al., 1998). The data also fail to support proposals, based solely on overexpression data, that PITPα plays a major physiological role in stimulating deacylation of PtdIns to lyso-PtdIns in vivo (Snoek et al., 1999).
Of special note was the surprising indifference of bulk phosphoinositide metabolism to loss of PITPα function. Although interpretations of bulk lipid analyses are subject to the caveat that PITPα may play a significant role in synthesis/maintenance of a phosphoinositide pool(s) that makes only a quantitatively minor contribution to the bulk membrane phosphoinositide content, our present in vivo data fail to reveal the existence of such a PITPα-dependent pool. For example, we have not detected enhanced sensitivity of PITPα −/− cells to inhibitors of phosphoinositide synthesis such as wortmannin (our unpublished data). Moreover, PITPα deficiency has no significant effect on the activity of cellular functions that have documented phosphoinositide requirements. These include 1) constitutive protein trafficking, 2) internalization of TfR from the plasma membrane and its recycling from endocytic compartments back to the cell surface, and 3) the biogenesis and agonist-induced fusion of DSGs with the mast cell plasma membrane. In addition, no deficiencies in growth factor signaling were evident either in PITPα −/− cells stimulated in culture, or in efficiency of tumor formation in vivo. Thus, PITPα-deficient ES cells compete robustly for growth factors in situ, these cells respond normally to growth factors in the competitive context of the whole animal, and the pluripotency of ES cells is not compromised by loss of PITPα function. Finally, the dispensability of PITPα for cell viability and fetal murine development is amply demonstrated by our finding that PITPα −/− mice develop to term (Alb and Bankaitis, unpublished data). These mice fail to thrive, however. Detailed analysis of PITPα −/− mice will be presented elsewhere.
Although the data demonstrate PITPα does not significantly contribute to essential housekeeping functions in murine ES cells, our results suggest that PITPβ does. Starting with a PITPβ −/+ cell line, we failed to recover PITPβ −/− ES cells even though we used a positive selection for such mutants. Although the failure to isolate PITPβ −/− ES cells is by itself a negative result, our demonstrations that PITPβ −/− mice do not survive to term, and that PITPβ deficiency results in early embryonic lethality, are consistent with incompatibility of PITPβ dysfunction with cell viability. The timing and mechanism of PITPβ-less lethality in the mouse remain to be determined.
In closing, the collective data allow us to discard the possibility that PITPα and PITPβ are perfectly redundant PITPs that share equal responsibilities in executing essential cellular housekeeping functions. We are presently left with two interpretations of the data: 1) PITPβ is a major PITP isoform, whereas PITPα is a functionally redundant minor isoform; or 2) PITPα and PITPβ are not functionally redundant PITPs and PITPα is a highly specialized isoform dedicated to functions that apply only to very specific cell types. Demonstrations that PITPα and PITPβ defects evoke strikingly different phenotypic consequences, when coupled with PITPα and PITPβ being biochemically distinct proteins that localize to different subcellular compartments (de Vries et al., 1996; our unpublished data), lead us to favor the interpretation that PITPα and PITPβ are not functionally redundant PITPs. We suggest that the PITPα is a highly specialized isoform dedicated to functions that apply only to very specific cell types, whereas PITPβ executes essential housekeeping functions in many cell types. Extension of the genetic approaches described herein will permit direct tests of these possibilities, and will contribute to our understanding of PITPα and PITPβ function in mammalian cells.
ACKNOWLEDGMENTS
We thank Allan Bradley and Tim Townes for providing the AB1 and R1 ES cell lines, respectively. The technical assistance of A. Pizzey (University College London) and L. Millican (University of Alabama at Birmingham) is also acknowledged. This work was supported by National Institutes of Health grant NS37723 (to V.A.B.) J.P. was sponsored by a Bogue Fellowship from the University College London.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09–0457. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09–0457.
REFERENCES
- Alb JG, Jr, Gedvilaite A, Cartee RT, Skinner HB, Bankaitis VA. Mutant rat phosphatidylinositol/phosphatidylcholine transfer proteins specifically defective in phosphatidylinositol transfer: implications for the regulation of phospholipid transfer activity. Proc Natl Acad Sci USA. 1995;92:8826–8830. doi: 10.1073/pnas.92.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Balch WE. Principles of selective transport: coat complexes hold the key. Trends Cell Biol. 1996;6:315–320. doi: 10.1016/0962-8924(96)10027-1. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K, Roth MG, Ktistakis NT. Phosphatidic acid formation by phospholipase D is required for protein transport from the endoplasmic reticulum to the Golgi complex. Curr Biol. 1997;7:301–307. doi: 10.1016/s0960-9822(06)00153-9. [DOI] [PubMed] [Google Scholar]
- Buckley MG, Coleman JW. Cycloheximide treatment of mouse mast cells inhibits serotonin release. Evidence of a requirement for newly synthesized protein in the exocytotic response. Biochem Pharmacol. 1992;44:659–664. doi: 10.1016/0006-2952(92)90400-d. [DOI] [PubMed] [Google Scholar]
- Cleves AE, McGee TP, Bankaitis VA. Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1991a;1:30–34. doi: 10.1016/0962-8924(91)90067-j. [DOI] [PubMed] [Google Scholar]
- Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991b;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvera S, D'Arrigo A, Stenmark H. Phosphoinositides in membrane traffic. Curr Opin Cell Biol. 1999;11:460–465. doi: 10.1016/S0955-0674(99)80066-0. [DOI] [PubMed] [Google Scholar]
- Cotlin LF, Siddiqui MA, Simpson F, Collawn JF. Casein kinase II activity is required for transferrin receptor endocytosis. J Biol Chem. 1999;274:30550–30556. doi: 10.1074/jbc.274.43.30550. [DOI] [PubMed] [Google Scholar]
- Cunningham E, Tan SK, Swigart P, Hsuan J, Bankaitis VA, Cockcroft S. The yeast and mammalian isoforms of phosphatidylinositol transfer protein can all restore phospholipase C-mediated inositol lipid signaling in cytosol-depleted RBL-2H3 and HL-60 cells. Proc Natl Acad Sci USA. 1996;93:6589–6593. doi: 10.1073/pnas.93.13.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries KJ, Westerman J, Bastiaens PIH, Jovin TM, Wirtz KWA, Snoek GT. Fluorescently labeled phosphatidylinositol transfer protein isoforms (α and β), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp Cell Res. 1996;227:33–39. doi: 10.1006/excr.1996.0246. [DOI] [PubMed] [Google Scholar]
- Dickeson SK, Lim CN, Schuyler GT, Dalton TP, Helmkamp GM, Jr, Yarbrough LR. Isolation and sequence of cDNA clones encoding rat phosphatidylinositol transfer protein. J Biol Chem. 1989;264:16557–16564. [PubMed] [Google Scholar]
- Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MKY, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- Fensome A, Cunningham E, Prosser S, Tan SK, Swigart P, Thomas G, Hsuan J, Cockcroft S. ARF and PITP restore GTP gamma S-stimulated protein secretion from cytosol-depleted HL60 cells by promoting PIP2 synthesis. Curr Biol. 1996;6:730–738. doi: 10.1016/s0960-9822(09)00454-0. [DOI] [PubMed] [Google Scholar]
- Fullwood Y, dos Santos M, Hsuan JJ. Cloning and characterization of a novel human phosphatidylinositol transfer protein, rdgBβ. J Biol Chem. 1999;274:31553–31558. doi: 10.1074/jbc.274.44.31553. [DOI] [PubMed] [Google Scholar]
- Gomperts BD, Tatham PER. Regulated exocytotic secretion from permeabilized cells. In: Rothman JE, editor. Reconstitution of Intracellular Transport. New York: Academic Press; 1993. [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Hamilton BA, et al. The vibrator mutation causes neurodegeneration via reduced expression of PITPα: positional complementation cloning and extragenic suppression. Neuron. 1997;18:711–722. doi: 10.1016/s0896-6273(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Hay JC, Martin TFJ. Phosphatidylinositol transfer protein is required for ATP-dependent priming of Ca2+-activated secretion. Nature. 1993;366:572–575. doi: 10.1038/366572a0. [DOI] [PubMed] [Google Scholar]
- Jing S, Spencer T, Miller K, Hopkins C, Trowbridge IS. Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J Cell Biol. 1990;110:283–294. doi: 10.1083/jcb.110.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Alb JG, Jr, Phillips SE, Bankaitis VA, Howell KE. A phosphatidylinositol 3-kinase and phosphatidylinositol transfer protein act synergistically in formation of constitutive transport vesicles from the trans-Golgi network. J Biol Chem. 1998;273:10349–10354. doi: 10.1074/jbc.273.17.10349. [DOI] [PubMed] [Google Scholar]
- Kang S, Liang L, Parker CD, Collawn JF. Structural requirements for major histocompatibility complex class II invariant chain endocytosis and lysosomal targeting. J Biol Chem. 1998;273:20644–20652. doi: 10.1074/jbc.273.32.20644. [DOI] [PubMed] [Google Scholar]
- Kauffmann-Zeh A, Thomas GM, Ball A, Prosser S, Cunningham E, Cockcroft S, Hsuan JJ. Requirement for phosphatidylinositol transfer protein in epidermal growth factor signaling. Science. 1995;268:1188–90. doi: 10.1126/science.7761838. [DOI] [PubMed] [Google Scholar]
- Kearns BG, Alb JG, Jr, Bankaitis VA. Phosphatidylinositol transfer proteins: the long and winding road to physiological function. Trends Cell Biol. 1998;8:276–282. doi: 10.1016/s0962-8924(98)01281-1. [DOI] [PubMed] [Google Scholar]
- Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. An essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Routt S, Xie Z, Cui X, Fang M, Kearns MA, Bard M, Kirsch D, Bankaitis VA. Identification of a novel family of nonclassical yeast PITPs whose function modulates activation of phospholipase D, and Sec14p-independent cell growth. Mol Biol Cell. 2000;11:1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MC, Nicaud J-M, Skinner HB, Vergnolle C, Kader JC, Bankaitis VA, Gaillardin C. A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J Cell Biol. 1994;125:113–127. doi: 10.1083/jcb.125.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning TJ, Jr, Sontheimer H. Bovine serum albumin and lysophosphatidic acid stimulate calcium mobilization and reversal of cAMP-induced stellation in rat spinal cord astrocytes. Glia. 1997;20:163–172. doi: 10.1002/(sici)1098-1136(199706)20:2<163::aid-glia8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Milligan SC, Alb JG, Jr, Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco ME, Alexander RJ, Snoek GT, Moldover NH, Wirtz KW, Walden PD. Evidence that mammalian phosphatidylinositol transfer protein regulates phosphatidylcholine metabolism. Biochem J. 1998;335:175–179. doi: 10.1042/bj3350175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y, Nakamura T, Hirata A, Routt SM, Skinner HB, Bankaitis VA, Shimoda C. The Schizosaccharomyces pombe spo20+ gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol Biol Cell. 2001;12:901–917. doi: 10.1091/mbc.12.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi CG, Trowbridge IS, Xue L, Hopkins CR, Davis CD, Collawn JF. Sorting signals in the MHC class II invariant chain cytoplasmic tail and transmembrane region determine trafficking to an endocytic processing compartment. J Cell Biol. 1994;126:317–330. doi: 10.1083/jcb.126.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M, de Vries KJ, Frank R, Snoek G, Bankaitis V, Wirtz K, Huttner WB. A role for phosphatidylinositol transfer protein in secretory vesicle formation. Nature. 1995;377:544–547. doi: 10.1038/377544a0. [DOI] [PubMed] [Google Scholar]
- Paul KS, Bogan AA, Waters MG. Phosphatidylinositol transfer protein (PITPα) stimulates in vitro intra-Golgi transport. FEBS Lett. 1998;431:91–96. doi: 10.1016/s0014-5793(98)00722-4. [DOI] [PubMed] [Google Scholar]
- Phillips SE, et al. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol Cell. 1999;4:187–197. doi: 10.1016/s1097-2765(00)80366-4. [DOI] [PubMed] [Google Scholar]
- Pinxteren JA, Gomperts BD, Phillips SE, Rogers D, Tatham PER, Thomas GMH. Biochem J. 2001;356:287–296. doi: 10.1042/0264-6021:3560287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinxteren JA, O'Sullivan AJ, Tatham PER, Gomperts BD. Regulation of exocytosis from rat peritoneal mast cells by G protein beta gamma-subunits. EMBO J. 1998;17:6210–6218. doi: 10.1093/emboj/17.21.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to ‘bypass Sec14p’ and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- Skinner HB, Alb JG, Jr, Whitters EA, Helmkamp GM, Jr, Bankaitis VA. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. 1993;12:4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner HB, McGee TP, McMaster C, Fry MR, Bell RM, Bankaitis VA. Phosphatidylinositol transfer protein stimulates yeast Golgi secretory function by inhibiting choline-phosphate cytidyltransferase activity. Proc Natl Acad Sci USA. 1995;92:112–116. doi: 10.1073/pnas.92.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek GT, Berrie CP, Geijtenbeek TB, van der Helm HA, Cadee JA, Iurisci C, Corda D, Wirtz KW. Overexpression of phosphatidylinositol transfer protein alpha in NIH3T3 cells activates a phospholipase A. J Biol Chem. 1999;274:35393–35399. doi: 10.1074/jbc.274.50.35393. [DOI] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. Inp51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-sensitive phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Hosaka K. Cloning of a cDNA encoding a second phosphatidylinositol transfer protein from rat brain by complementation of the yeast sec14 mutation. J Biochem. 1994;115:981–984. doi: 10.1093/oxfordjournals.jbchem.a124448. [DOI] [PubMed] [Google Scholar]
- van Tiel CM, Luberto C, Snoek GT, Hannun YA, Wirtz KWA. Rapid replenishment of sphingomyelin in the plasma membrane upon degradation by sphingomyelinase in NIH3T3 cells overexpressing the phosphatidylinositol transfer protein beta. Biochem J. 2000;346:537–543. doi: 10.1042/0264-6021:3460537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley HS, Cunningham DD. The endocytotic rate constant. a cellular parameter for quantitating receptor-mediated endocytosis. J Biol Chem. 1982;257:4222–4229. [PubMed] [Google Scholar]
- Wirtz KWA. Phospholipid transfer proteins. Annu Rev Biochem. 1991;60:73–99. doi: 10.1146/annurev.bi.60.070191.000445. [DOI] [PubMed] [Google Scholar]
- Wu W-I, Routt S, Bankaitis V, Voelker DR. A new gene involved in the transport dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the phosphatidylinositol/phosphatidylcholine transfer protein, S.E.C14. J Biol Chem. 2000;275:14446–14456. doi: 10.1074/jbc.275.19.14446. [DOI] [PubMed] [Google Scholar]
- Xie Z, Fang M, Rivas MP, Faulkner A, Sternweis PC, Engebrecht J, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci USA. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Fang M, Topalof L, Bankaitis VA. Evidence for an intrinsic toxicity of phosphatidylcholine to Sec14p-dependent protein transport from the yeast Golgi complex. Mol Biol Cell. 2001;12:1117–1129. doi: 10.1091/mbc.12.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MD, Thomas LM, Tremblay JM, Oliver RL, Yarbrough LR, Helmkamp GM., Jr Structure of a multifunctional protein: mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J Biol Chem. 2001;276:9246–9252. doi: 10.1074/jbc.M010131200. [DOI] [PubMed] [Google Scholar]