Abstract
The most widely used genome editing toolkit is CRISPR (clustered regularly interspaced short palindromic repeats). It provides the possibility of replacing and modifying DNA and RNA nucleotides. Furthermore, with advancements in biological technology, inhibition and activation of the transcription of specific gene(s) has become possible. Bioinformatics tools that target the evolution of CRISPR-associated protein 9 (Cas9) turn this protein into a vehicle that is specific for a DNA or RNA region with single guide RNA (sgRNA). This toolkit could be used by researchers to investigate the function of stem cell gene(s). Here, in this review article, we cover recent developments and applications of this technique in stem cells for research and clinical purposes and discuss different CRISPR/Cas technologies for knock-out, knock-in, activation, or inhibition of gene expression. Additionally, a comparison of several deliveries and off-target detecting strategies is discussed.
Keywords: CRISPR, genome editing, stem cells, cell therapy, bioinformatics
1. Introduction
Stem cells are heterogeneous and unspecialized cells that are the foundation of every organ and cell in our body [1]. Depending on their origin, stem cells are categorized into embryonic stem cells (ESCs), which exist in the inner cell mass (ICM) at an early stage of development; adult stem cells (ASC), which are found in specific tissues and act as a source to repair the damage of their specific tissue; and induced pluripotent stem cells (iPSC), which are adult stem cells that are reprogrammed into another type of adult stem cells and one of the most important cells that can be used for medical purposes. Additionally, there are prenatal stem cells which come from the fetal membrane, umbilical cord, and amniotic fluid extra-embryonic cells, and mesenchymal stem cells (MSC) which are adult stem cells originating from bone marrow, liver, and muscles [2,3]. For a cell to be considered a stem cell, two criteria must be met. First, stem cells must possess an unlimited capacity for self-renewal in order to produce descendants that are identical to the original cell. Second, they must possess the ability to differentiate into other healthy specialized cells of the body [4]; this specialization can occur depending on the physiological needs of tissue and organs at different times [5].
These properties of stem cells make them highly valuable, especially for medical purposes. Much research on these cells has shown the potential usage of stem cells in treating many diseases, such as cancer [6,7,8]. Regenerative medicine is one of the therapeutic applications of stem cells which can help to restore damaged organs or tissues in patients suffering from chronic diseases or injuries [9]. Another application of stem cells is their use in testing new drugs prior to animal and human trials [10]. Today, with the advancement of technology, various tools have been provided to researchers, among which we can mention genome editing techniques, which have given us the ability to change and manipulate the genome sequence at the desired point. By using this technology in stem cells, an important tool has been provided for researchers in the production of drugs and advanced treatments. In the following section, we introduce CRISPR, which is one of the most important tools in genome editing.
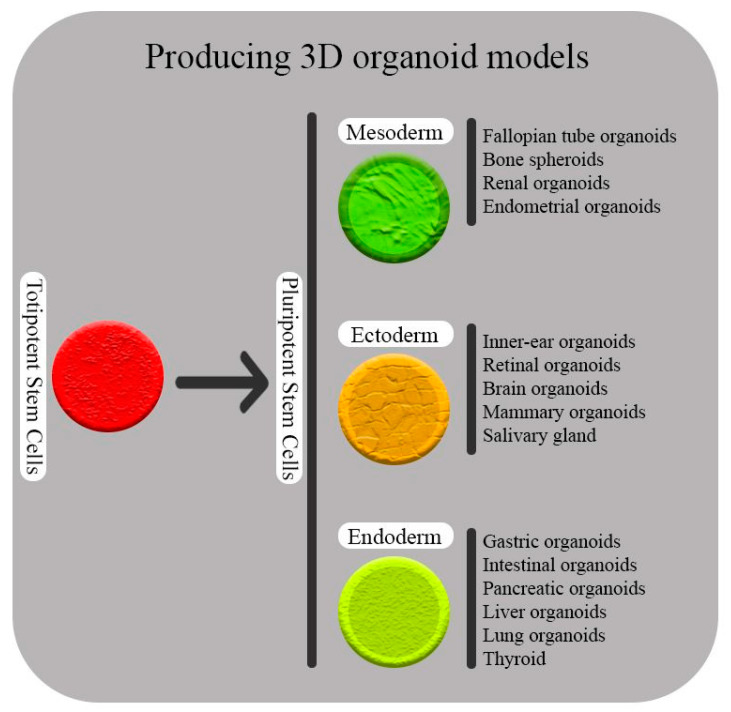
The cluster regulatory interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR-Cas9) system consists of a short and repetitive nucleotide that was first discovered in the genomes of bacteria and archaea that act as adoptive immune systems. It works by removing exogenous genetic elements that assemble with Cas proteins [11]. The deactivation of the endogenous genetic elements consists of three steps. First, a few endogenous short nucleotides might integrate with the host’s CRISPR loci as new spacers. Then, a crRNA/Cas complex is created by the transcription of CRISPR RNAs (crRNAs). Finally, under the base complementation pairing rule, the complexes will inactivate the exogenous element [12]. Most of the RNase and DNase activities of Cas proteins are predicted with bioinformatics tools. New-generation sequencing has led to the discovery of a large number of Cas proteins; thus, new classification based on sequence information is necessary. The new classification is various and has evolved fast. The new classification has three categories: type 1, type 2 (Cas9 is included in this type and is based on the presence of the HNH domain), and type 3; each type contains a large number of Cas proteins. Despite this classification, some subtypes of Cas proteins are still unclassified. This issue would be improved by further studies in the topic [13,14]. The type II CRISPR-Cas9 immune system stands out among them because it uses RNase III for cleaving the transcript into mature crRNAs and only needs one Cas9 protein to form a crRNA/Cas9 complex. This technology is simple, fast, cheap, and applicable. These characteristics make it a strong candidate for the development of a completely new genome-editing tool for biological and medical research (explained in Section 2) [15]. Gene editing using CRISPR/Cas9 technology has been widely implemented in biological and biomedical research, and stem cell-mediated cell treatment and gene therapy are recognized as essential elements in human medicine because of their capacity for tissue repair and regeneration [16]. Some different stem cell types have so far been successfully used in clinical studies and have received scientific approval. Besides viral genome modification, more and more publications are confirming that CRISPR/Cas9 genome editing is a potent technique that can greatly advance biomedicine, such as in virus genome editing and in stem cell research [11,17,18,19]. Organoids are interesting new systems that help us improve our knowledge of diseases’ mechanisms, development, evolution, homeostasis, and therapy. They can derive from ESs, ASCs, and PSCs (or even differentiate cells) (Figure 1). Organoids form three-dimensional (3D) cell culture models (in vivo-like morphogenesis), and our understanding of signaling pathways (cascades) that result in this formation is important. Indeed, utilizing the CRISPR/Cas9 system for editing in organoids increases our understanding of different disorders (e.g., human digestive diseases). This is mostly through removal of genes of interest and investigating their role in such conditions. Genome-wide CRISPR screens are another applicable tool that help researchers to find mutations and genes involved in organ regeneration, tumorigenesis, metastasis mechanisms and off-target analyses (this topic is discussed in Section 2.2) [20,21,22].
Figure 1.
The zygote cell and its initial divisions (embryonic cells), which are considered totipotent stem cells, have the capacity to give rise to fully developed living organisms (body and placenta). Pluripotent stem cells, which can generate all cells that make up a live organism’s body following totipotent stem cells, are the next stage (mesoderm, endoderm, and ectoderm). Organoids are the new era for disease modeling, homeostasis, and development studies. Organoids derived from mesoderm, endoderm, and ectoderm are considered a new field of interest in biomedical research.
Stem Cell Application in Medicine
Stem cell therapy has the potential to cure a wide range of aggressive and deadly human diseases. Physiological, morphological, and developmental subjects, in addition to formation, regeneration, and repair of tissues have all benefited from stem cell research during the last decade [23,24,25]. Recent advancements in stem cell technology have allowed scientists to use ex vivo and in vivo stimulation to differentiate stem cells into functional offspring for therapeutic purposes [26,27]. Many studies have shown that particular stem cells can be used to treat a variety of human pathological conditions [28,29,30]. Additionally, engineered stem cells could be employed to cure and reverse inherited genetic abnormalities [31]. Stem cells have been found to be a promising therapeutic tool for treating pathological conditions like cancer by transferring altered genes to the injured organ/tissue [32,33]. NSCs, for instance, have been demonstrated to migrate via the central nervous system and reach the extra cranial neoplastic location [32,34,35]. Despite all the advantages of stem cells, there are several disadvantages that have limited their routine clinical use until now. Some of these limitations include their heterogenicity and instability (stem cell malignant transformation), long term immune rejection, and limited access to ASCs; these are some of the reasons which have resulted in few confirmations for their clinical application [36,37,38,39]. Different immunological behaviors were observed from stem cells derived from different tissues. For example, hiPSC-derived retinal pigment epithelial (RPE) cells were tolerated in different locations, but autologous integration-free hiPSCs have shown immune rejection [40]. In another example, researchers noticed that fetal liver-derived MSCs are more immunosuppressive and immunoregulatory than bone marrow-derived MSCs [41]. Researchers have been attempting to circumvent these limitations by inventing and discovering new technologies such as genome editing tools like CRISPR-Cas9 and CRISPR/Cas12. By using these technologies, we can delete gene(s); insert or delete nucleotide(s), genes, or gene clusters; and inhibit or activate gene expression. Also we can manipulate RNAs with the CRISPR/Cas13 method [42]. This system has been used to evaluate the function of genes in stem cells and to optimize their function. In the following sections, these functions will be discussed in detail.
2. CRISPR/Cas
2.1. CRISPR/Cas9 and CRISPR/Cas12
CRISPR is a technique that is designed for targeted modification of specific DNA or RNA sequences; nevertheless, it sometimes produces unwanted or unexpected changes in DNA [43]. In bacteria and archaebacteria, this mechanism works as an adaptive defense against invading nucleic acids (phages) [44]. CRISPR/streptococcus pyogenes CRISPR associated protein 9 (spCas9), the most well-known CRISPR system, is derived from the Streptococcus pyogenes bacterium [45,46,47]. In Streptococcus pyogenes, this system is made up of two primary components. An RNA (containing two distinct RNAs named CRISPR RNA (crRNA) and trans-activator RNA (tracrRNA)) and the Cas9 endonuclease protein, which targets crRNA and tracrRNA [48].
Bioinformatics methods were used to create the functional system-related RNA of this bacterium, and instead of two independent components (crRNA and tracrRNA), it has become a single guide RNA (sgRNA). The 20 nucleotides in terminal 5’ of the sgRNA are designed to complement the DNA target location. The presence of a sequence called protospacer adjacent motif (PAM), consisting of 5′-NGG-3′, on the target sites at the end of three of these 20 nucleotides is required for system function [49,50]. In terms of length and nucleotides, this sequence differs amongst bacteria [51,52,53].
The Cas9 protein identifies this region after the 5’ sgRNA ends of 20 nucleotides bind to the target site. The Cas9 protein then makes a blunt end double-strand break in DNA [51,52,53]. It can take one of two paths after breaking the DNA code—the cell either commits suicide or fixes the damage (Figure 2) [54]. NHEJ (non-homologous end joining) and HDR (homology directed repair) are the two main processes for repairing double-stranded breaks in DNA [55]. In mammals, the NHEJ mechanism is the most common mode of repair, and most cells use it to mend double-stranded breaks. A few nucleotides are inserted/removed, or maybe larger deletions occur quite often. Moreover, in certain reports, the loss of chromosome arms or deletions of thousands of kb have been reported in the cut region during NHEJ repair (INDEL mutation) [56,57]. A frameshift mutation occurs when the insertion and deletion of nucleotides do not multiply by three, resulting in loss of gene function (knock-out) [58]. The capacity to target several DNA loci concurrently (multiplexing) is one of the system’s features [59]. The knock-out method has been widely utilized to assess the function of genes involved in stem cell stemness and differentiation. Using this technology, the histopathological role of mutations and diseases mechanisms were assessed with the help of stem cells. For example, paired-like homeodomain 2 (PITX2) (a transcription factor involved in atrial fibrillation (AF) disease), beta-2-microglobulin (B2M) (a serum protein that is associated with human leukocyte antigen (HLA) class I), and galactosidase alpha (GLA) (to produce a cell line model for Fabry disease (an X-linked inherited disease)) which is related to polysaccharides, glycoproteins, and glycolipids cleavage, were targeted to find and model the roles of those genes in development of disease mechanisms within the stem cells, which in turn provided new insights for designing new therapeutic approaches [60,61,62,63]. Some of the other studies are addressed in Table 1. CRISPR/Cas12 is another form of this system. It was first discovered in Acidaminococcus and Lachnospiraceae bacteria, and afterwards in additional bacteria in various forms. Double-strand sticky end breaks in DNA, DNA cut distance from PAM, longer binding-site-to-target region, shorter sgRNA length, smaller Cas protein size than CRISPR/Cas9, and AT-rich PAM (unlike CRISPR/Cas9) are all features of this system [49,64,65]. A large number of Cas12 (Cas12a to k) have been identified so far [66], but the most studied and practical system for stem cells is still Cas12a. Base editing (BE) and prime editing (PE) represent the next generation of CRISPR technology. Using BE, we can change single nucleotides (in RNAs (temporary) or DNAs (permanent)). If BE efficiency, precision, and specificity are improved, it has high potential for being used in personalized medicine [67]. PE is one of the latest technologies derived from Cas9. Modifications have been applied to this technology to create changes in DNA that have the highest efficiency and are the least off-target. Several generations of PEs are being developed. The latest generations, PE4 and PE5, have two-fold increased efficiency compared to the previous one (PE3). In these generations (PE4 and PE5), an inhibitory mismatch repair (MMR) protein has been added to the Cas9 nickase protein and it has proper editing efficiency in-vitro [68]. Furthermore, many changes are being made to increase the efficiency of this technology. For example, by adding different domains to the Cas9 nickase protein and making changes in peg RNA, the efficiency of this technology has been greatly improved [69,70,71]. Spinal muscular atrophy (SMA) is an autosomal recessive disease that is caused by mutations in SMN-2 and SMN-1 genes and causes problems in motor neurons. Mutations in the SMN-1 gene are fatal to fetuses, but adults who have mutations in the SMN-1 gene have the disease. Researchers removed intronic splicing silencer-N1 (ISS-N1) by PE technology in SMA-patient specific iPSCs (SMA-iPSCs) (24/7 efficiency) [72]. In another study, editing hiPSC-derived cardiomyocytes of Moloney murine leukemia virus with split and non-split reverse transcriptase (RT) deleted the RNase H domain; MMLV-RTΔRH showed 1.4% to 16.7% editing efficiency in all four locations [73].
Table 1.
Application of CRISPR knock-out system in studying the function of stem cell genes (all of the knock-outs done by CRISPR-Cas9 system).
| Stem Cell | Gene(s) Knock-Out | Gene Function | Results |
|---|---|---|---|
| Pancreatic cancer stem cell | PolymeraseII-associated factor 1 (PAF1) | Regulates stem cell features (i.e., decreased the orthotopic pancreatic tumors capacity to expand and evolution in mice and cancer stem cells) | The PAF1 interaction with PHF5A (PHD Finger Protein 5A), DDX3 (DEAD-box RNA helicase 3) genes to regulate stemness with NANOG expression [74]. |
| Mouse Embryonic stem cell | Recombination activating 1 gene (RAG-1) | Associated in immunoglobulin V-D-J recombination activation | Efficiently create RAG1 biallelic homozygous and compound heterozygous indel mutations (92%) [75]. |
| hESCs | Fos Proto-Oncogene, AP-1 Transcription Factor Subunit (FOS) | Involved in cell differentiation, proliferation and transformation regulator | HESCs line production for hematopoietic differentiation assay [76]. |
| Pluripotent stem cell | WW Domain Containing Transcription Regulator 1 (WWTR1) | Associated with signal transduction, differentiation, and hippo signaling regulation | WWTR1 knock-out does not have any effect on karyotype, phenotype, and differentiation [77]. |
| hESCs | RAP1 (Ras-proximate-1) | A small cytosolic GTPase that is vital for signal transduction | RAP1 deficiency enhance self-renewal and delay cellular senescence. Therefore, It has a role in hESCs homeostasis (telomeric and non-telomeric role) [78]. |
| hESCs | Histone deacetylase 6 (HDAC6) | A transcription repressor | HDAC6 homozygote knockout in hESCs does not have any effect on karyotype, differentiation, and pluripotency [79]. |
| Spermatogonia stem cell | EPH receptor B2 (Ephb2) | Tyrosine kinase receptor that has role in differentiation, division and motility | The Ephb2 knockout cells showed less colonies compared to the wild type cells, which demonstrated the role of this gene in pluripotency [80]. |
| hESCs | Poly (ADP-Ribose) Polymerase 1 (PARP1) | A chromatin-associated enzyme that has associated with tumor transformation, proliferation, differentiation and cell damage recovery | PARP1 knock-out cell lines showed normal differentiation ability, karyotype and stem cell markers expression [81]. |
| mESCs | The Methyltransferase-like 3 and 14 (METTL3, METTL14) | Complex of methyltransferase that are sequence-specific DNA adenine methyltransferase (in unpaired and single-strand DNA) | m6 A RNA methylation as a way to restrict ERVs [82]. |
| hMSCs | NAD-dependent deacetylase sirtuin-3 (SIRT3) | Is a histone deacetylases that has widespread effects in nuclear gene expression control | SIRT3 knock-out resulted in the detachment of genomic lamina-associated domains (LADs) from the nuclear lamina, chromatin accessibility increases and enhance cell senescence [83]. |
| hMSCs | Receptor activator of nuclear factor kappa-Β ligand (RANKL) | An apoptosis regulator that is involved in immune system, bone remodeling, regeneration and controls cell proliferation | The mesenchymal stem cells showing capacity of bone formation is immortalized [84]. |
| mESCs | Telomeric repeat binding factor 2 (TRF2) | A key gene for telomeres protectection | TRF2 knock out showed that it is dispensable for the proliferation and survival of mouse embryonic stem cells [85]. |
| Myeloma cells | V-Set Pre-B Cell Surrogate Light Chain 1 (VPREB1) | Involved in early stages of B cell development | Knock out of VPREB1 effective in inhibition of primary myeloma grows [86]. |
| hESCs | Acidic nuclear phosphoprotein 32 family member A (ANP32A) |
A RNA binding protein that is associated with nucleocytoplasmic transport | The knock out cells shows the normal karyotype and typical stem cell morphology, in accordance with high expression of pluripotent genes and the differentiation potential in-vitro [87]. |
| Adult epithelial stem cells | Interferon Regulatory Factor 2 (IRF2) | An interferon regulatory factor | IRF2 is an antagonist of stemness. With the knock-down of this gene in Keratinocytes, migration, self-renewal and epidermis formation increases [88]. |
HDR repair is used much less in human cells. DNA is employed as a template in this sort of repair, and the repair is precise. As a result, the repaired DNA will be identical to the original DNA. With this repair we can change nucleotide(s) in the target DNA, or nucleotides/gene fragments can insert into this region utilizing this method of repair (knock-in) [89,90].
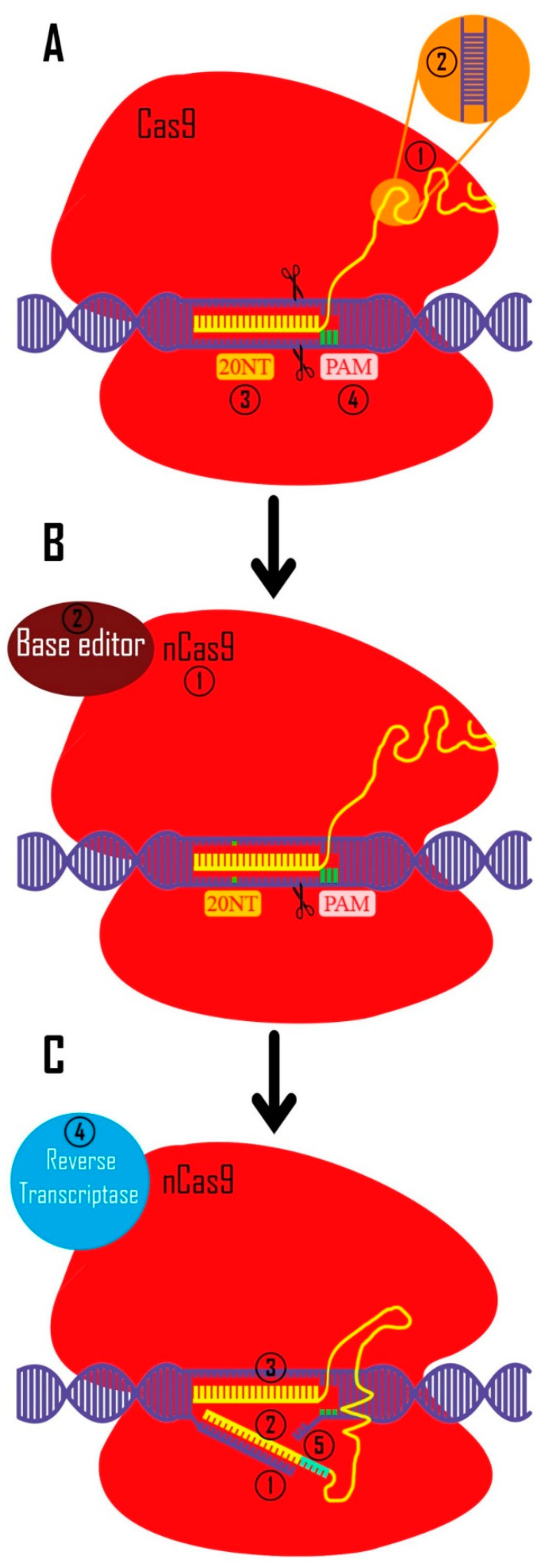
Figure 2.
CRISPR toolkit. (A). CRISPR technology was originally used to create double-strand breaks in eukaryotic DNA (with a bacterial origin (Streptococcus pyogenes)). 1. In bacteria, crRNA and tracrRNA guide Cas9 to target the intended region. These RNAs are artificially synthetized as a unique sgRNA to be more applicable in other creatures (yellow) 2. crRNA and tracrRNA are widely used in multiple experimental systems (e.g., mouse embryo microinjections, RNP electroporation into mammalian cell lines, etc.) [91,92] 3. Twenty nucleotides complementary to the target site are used to identify the target area (these nucleotides are designed in a targeted manner). 4. Before these 20 nucleotides, there are three PAM nucleotides (5′-NGG-3′ in Streptococcus pyogenes Cas9 system) which are necessary for CRISPR/Cas9 function. (B). 1. In order to modify the bases in a targeted way, the Cas9 protein was altered to cut only one strand of DNA by changing one amino acid in Cas9 protein (nickase Cas9 [nCas9]). 2. Additionally, they coupled the different base editor domains to the Cas9 protein. (C). 1. Prime editing, the subsequent iteration of this technique, cuts a DNA strand by creating a cut at the intended location. 2 and 3. The sgRNA is made in such a way that its 3′-end complements the two sides of cut site, and its 5′-end can recognize the target site. 4. The reverse transcriptase enzyme turns 3′ sgRNA into cDNA using this 3′ end as a primer. 5. In the cut region, bases are designed for knock-in to produce highly accurate results.
2.2. Genome-Wide CRISPR/Cas Knock-Out
The genome-scale CRISPR knock-out (GeCKO) method was first developed to examine genes in the human genome [93]. With the addition of non-coding areas as new targets for the CRISPR library system, the technology was improved to GeCKO V2 (GeCKO Version 2) [94]. This method (genome-wide screening) has been frequently utilized to identify key and fundamental genes involved in diseases [95,96]. This technique was employed in a study assessing loss of function in iPSC-derived macrophages. The function of macrophages in chronic inflammation, neurological disorders, cancer progression, and immune response was revealed for the first time in this study [97]. Another study on ESCs looked at metabolic signals for transitions in embryonic cell fate and discovered that inhibiting the Tfe3 gene causes ESC differentiation [98]. This method was also utilized to explore the medication resistance of pancreatic cancer stem cells, which led to the recommendation of autoimmune medicines as a novel treatment option for pancreatic cancer patients [99].
2.3. Dead/Deactivated Cas9 (CRISPR i or a (Inhibition/Activation))
Researchers discovered that D10 (aspartic acid 10) in the RuvC domain and H840 (histidine 840) in the HNH domain are the two amino acids responsible for Cas9 protein double strand DNA cleavage [49]. The Cas9 protein loses its capacity to create cuts in the target region when certain amino acids (D10A (aspartic acid 10 alanine) and H840A (histidine 840 alanine)) are changed. Dead/deactivated Cas9 (dCas9) is a modified protein that can only connect to the target site (directed by sgRNA) [100]. This protein guides functional domains to the target region chosen by sgRNA by transposing protein functional domains into dCas9. For example, to increase and reduce mRNA expression, dCas9-binding transcription-activator and inhibitor domains are utilized (CRISPR a and I (CRISPR activator and inhibitor)). The sgRNA was created with transcriptional regulatory areas in mind. The functional domain of raising or reducing mRNA transcription is transferred to this area by dCas9. In fact, dCas9 is a carrier of specific functional domains [101,102,103] (e.g., epigenetic regulatory protein domains (acetylase, methylase, deacetylase, demethylase) [104], green fluorescence protein (GFP) to identify the DNA locus in the cell [105], and base editors to modify the nucleotide without cutting in DNA (Figure 2) [106]) and has proved to be a powerful tool of this technology.
This technology has been widely utilized to investigate the impact of changing gene expression levels on stem cell activity. In mouse ESCs, dCas9-Kruppel associated box (KRAB) inducible with doxycycline knock-in was used to study gene expression and repression, cell differentiation, or reprogramming [107]. In another investigation, in MSCs, CRISPR/dCas9-KRAB (CRISPRi) and CRISPR/dCas9- herpes simplex virus-based transcriptional activator VP64 domain (VP64) (CRISPRa) inducible with doxycycline (it has an inducible promoter that becomes activated with doxycycline) were used to control the expression of alkaline phosphatase (ALP) gene. In this setting, the osteoblast differentiation capacity of MSCs was enhanced or inhibited in vitro [108]. Multiple activation with CRISPRa in adipose stem cells leads to the expression and production of nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and glial cell line-derived neurotrophic factor (GDNF). Designed sheets with NGF, BDNF and GDNF neurotrophic factors related to gene activation in the rats induced Schwann cells in vitro to improve migration, proliferation, and neurite extension and promoted nerve reinnervation, regeneration, and efficient in vivo recovery [109]. Table 2 has more examples of this platform application.
Table 2.
The effect of CRISPR a/i on the gene expression regulation in the stem cell assay.
| Stem Cell | CRISPR System | Result |
|---|---|---|
| Adult epithelial stem cells | Interferon Regulatory Factor 2 (IRF2) | |
| Rat bone marrow-derived mesenchymal stem cell (rBMSC) | Endogenous SOX9 activation/peroxisome proliferator-activated receptor gamma (PPAR-γ) repression (dCas9 by modules with MS2 coat protein [MCP]-p65- heat shock factor 1 [HSF1] (MPH) as activation complex and Com-(Krüppel-associated box) KRAB (CK) as repression complex used fot this study) | This system in 2D culture arouses chondrogenesis and suppressed adipogenesis. However, the formation of manipulated cartilage and recovery of calvarial bone healing are enhanced in 3D culture system [110]. |
| hiPSCs derived from Parkinson disease | CRISPR/dCas9-DNA-methylation (DNA methyltransferase 3A [DNMT3A]) | CRISPR/dCas9-DNA-methylation (DNMT3A) designed for alpha-synuclein gene (SNCA) intron 1. As a result, 30% decrease in SNCA mRNA and protein expression were observed [111]. |
| iPSCs | dCas9- KRAB repressor | Identify genes that differentiate iPSCs into Cardiomyocyte [112]. |
2.4. RNA Editing
Another kind of this system is CRISPR/Cas13. The only difference is that it breaks RNA rather than DNA [113]. Cas13a, b, c, and d are four different types of this system, each with its own set of characteristics [113,114,115]. In CRISPR/Cas13, the area known as PAM in CRISPR/Cas9 and CRISPR/Cas12 is termed as PFS (protospacer flanking site) [115]. However, other Cas13 varieties (such as Cas13d) do not require PFS [116]. In RNA, this mechanism causes a break [117]. Changes to the amino acids in the Cas13 protein have been made, similarly to the CRISPR/Cas9 and CRISPR/Cas12 systems, so that the Cas13 protein binds to the target site only with crRNA guidance. dCas13 gains the ability to effect modifications at the RNA level by binding particular functional domains [118,119,120]. In one study, scientists created an exon-specific isoform expression reporter system (EXSISERS) to determine the type of isoform expressed in cells. With this method, a link between protein and polypeptide is established by intein (a protein that mediates the split-Cas9 system) without affecting the protein, and it is efficient for detection of RNA isoforms. This method was used to evaluate exon 10 of tau protein in iPSCs derived from a patient and the effect of targeting effectors for the specificity of isoforms by use of Cas13. This showed that the method was sensitive and applicable for this purpose [121].
2.5. Off-Target
Dedicated sgRNA design is normally carried out using web-based applications [122,123,124]. The most crucial component of a basic design is selecting the optimal sgRNA, created by the software, that has the maximum performance and specificity. Following design, sgRNAs are examined for practical performance in in vitro and in vivo settings, as well as the ability to make cuts (INDEL mutations) using methods such as next-generation sequencing, T7E1, or SURVEYOR kits [125,126]. Once the best sgRNA has been determined, it will be used for further research. The risk of sgRNA binding to places other than the target site and producing cuts in those regions (off-target) is one of the issues with the CRISPR system [127]. Various approaches are employed to prevent these off-target cuts. Double nickase is one of these techniques. The method employs Cas9 systems that have been engineered to cleave a strand of DNA. One dCas9 cuts the sense strand, and the second dCas9 cuts the antisense strand at the same time, resulting in a sticky end double-strand break [128]. The off-target cuts are reduced by 50 to 1500 times using this strategy [50]. This technique has been utilized to limit the generation of off-target stem cells. R201H has been reported to increase intracellular cGMP production in hPSCs cells utilizing this technology in the guanine nucleotide-binding protein alpha gene stimulating activity polypeptide 1 (GNAS) gene. These cells can be utilized to figure out how the GNAS gene works [129]. To test the function of the genes RB1 (retinoblastoma 1) and immune-reactive antigen domain containing 1 (OCIAD1) in iPSCs, this method was employed to generate heterozygous knock-out iPSCs [130,131].
Other approaches include shortening the sgRNA target region to 17 nucleotides to lower the chance of mismatches, sgRNA and Cas9 protein engineering, and the use of particular medicines [132,133,134]. In human stem cells however, off-target mutations are rare [135].
2.6. Knock-In
The knock-in method can be accomplished by a number of ways. All of these techniques are based on using donor DNA to make nucleotide modifications or add a gene or gene cluster to the target site [136]. A DNA fragment with a length of 100 to 200 bp can be used to make modifications in several bases. The changed nucleotides are placed in the ssODN middle where the DNA is cut, and the 3′ and 5′ sides of these single stranded oligodeoxynucleotides (ssODNs) are complementary to the two sides of the CRISPR system’s target site. HDR repair is accomplished after DNA cleavage, and the desired nucleotides are altered [137,138]. Another option is to use prime editing to make these changes. The reverse transcriptase enzyme is coupled to the nickase Cas (nCas) protein in this approach. The sgRNA is likewise built so that its 5′ end is complemented by two sides of the cut area, with the nucleotides we want to modify placed in the middle (at the cut site).
A 5′-strand sgRNA complement is inserted into the target site once the sgRNA is bound to the target area (the DNA acts as a primer, and the reverse transcriptase enzyme synthesizes DNA from the RNA). This DNA serves as a donor template for the cell. The HDR repair system repairs the target spot, and KI occurs with great accuracy (Figure 2) [139].
We can use longer homology arms on either side of the insert site to insert chunks longer than the donor DNA [140,141]. Drugs with different methods (e.g., Nocodazole (G2 and M phase cell cycle arrest), RS1 (binding homologous recombination-binding protein (RAD51) to the cut DNA area facilitators), and nu7441 molecules (NHEJ inhibitor)) considerably boost HDR repair and hence KI efficiency [142]. They monitor long-term expression changes by attaching reporter genes (KI) to the c-terminus of stemness-related genes [143,144,145,146]. In Wilson’s disease hiPSCs, the mutation in R778L (arginine 778 lysine) in ATPase copper transporting beta (ATP7B) was induced; this has application in drug screening and finding disease model mechanisms for Wilson disease [147]. In iPSCs derived from a patient with global development delay, the c.1730T>A mutation was induced in the mental retardation autosomal dominant 7 (MRD7) gene [148]. In iPSCs-derived motor neurons amyotrophic lateral sclerosis, the G4C2 hexanucleotide repeat expansion (HRE) was replaced with a normal region (HRE complete correction) by CRISPR-Cas9 and homology-directed repair [149]. Urinary-induced iPSC-derived monomeric cardiomyocytes red fluorescence protein, firefly luciferase (Fluc) for bioluminescence and herpes simplex virus, thymidine kinase for positron emission tomography (PET) imaging Cells imaging, approved these cells’ enhanced cardiac function in infarcted heart [150]. Table 3 shows examples of knock-in and its uses in stem cells.
Table 3.
Knock-in and application of this technology in stem cells (all the knock-ins done by using CRISPR/Cas9 technology).
| Stem Cell | Gene | Result |
|---|---|---|
| hiPSCs | Pancreatic And Duodenal Homeobox 1 (PDX1) [a transcription activator for several genes] | EGFP receptor introduced in PDX1 c-terminal gene by CRISPR/Cas9 knock-in in KSCBi005-A-3 which is used to monitor PDX1 expression during B-cell differentiation in live cells [151]. |
| hiPSCs | Activity-regulated cytoskeleton-associated light inducible (Arc light) [a kind of genetically-encoded voltage indicators] |
Arc light stable expression from Adeno-Associated Virus Integration Site 1 (AAVS1) locus. This hiPSCs is useful for cardiac development studies [145]. |
| Ovarian cancer stem cells | Green fluorescence protein (GFP) [a fluorescent protein] | GFP knock-in in NANOG gene is performed to study NANOG and androgen receptor (AR) expression and co-localization [152]. |
| Recessive dystrophic epidermolysis bullosa-specific-iPSCs | Drug preserve selection cassette (i.e., a drug-resistance gene, for positive cells selection) | Properly corrected a pathogenic mutation in Autosomal recessive dystrophic EB (RDEB)-specific iPSCs [153]. |
| iPSCs | Tetratricopeptide repeat domain 3 (TTC3) [ubiquitin-dependent protein catabolic mechanism and associated in protein K48-linked ubiquitination] | A cell line (p.S1038C) is used for the risk of late onset Alzheimer’s assessment [154]. |
| hESCs | Akaluc (a sensitive luciferase reporter) | Insert Akaluc into the AAVS1 locus to generate human embryonic stem cell lines capable of being traceable with near-infrared emission light [155]. |
| ESCs | GFI1-tag | Generating cells with GFI1-tag that can be identified via western blot and immunohistochemistry [156]. |
| hESCs | RYBP (RING1 And YY1 Binding Protein) gene with Flag-HA | Normal morphology and karyotype, while it maintains pluripotency to differentiate into three germ layers [157]. |
3. CRISPR Delivery Methods
Delivery methods are classified into two categories: viral vectors and non-viral vectors [158]. Safety, low immunogenicity, specific function, minimal toxicity, and high efficiency are all critical properties of a good vector [159,160]. Adenoviruses, adeno-associated viruses (AAVs), and lentiviruses are the most common viral vectors. These viruses are being used in a number of clinical investigations [159]. Lentiviral vectors can transport larger amounts of DNA, and the third generation of these viruses is undergoing clinical trials [161,162]. Additionally, these viruses have the ability to penetrate cells efficiently, produce a large number of in vitro viruses, and they have a high cell transduction rate [163,164,165]. However, one of the most essential characteristics of viruses is their capacity to integrate into host DNA. AAVs were found, in a 10-year long-term study, to have the ability to integrate into DNA and induce cancer in dogs with hemophilia who have been treated with the virus [166]. The virus can enter the genome through the CRISPR system’s cut area and remain there for a long time [167,168]. To overcome this problem, virus-like particles that do not have the ability to integrate their genome into the host’s genome are used. This delivery method is used for both in vitro and in vivo editing. These viruses are non-proliferative and have good delivery efficiency [169]. The PiggyBac transposon system is the other technology for gene(s) or cluster delivery. By this technology, dCas9-VPR and dCas9-KRAB transgenes and sgRNAs are delivered to hPSCs to change TCF4 gene expression (a transcription factor for NSCs regulation). Data has shown that this method is capable of stable delivery [170].
Immunogenicity is another issue with these viruses. The immune system responds to these viruses in the early stage of their admission into the body, and the body develops immunity to their re-entry [171].
Mechanical procedures such as electroporation and microinjection are examples of non-viral carriers [172,173]. Liposomes and nanoparticles are the two primary kinds of chemical techniques (metal nanoparticles and lipid nanoparticles). These approaches are generally safe, have a low immunogenicity, and are primarily employed in ex vivo studies [174,175,176].
4. Conclusions
CRISPR-based stem cell genome editing is a novel field in regenerative medicine. This technology has the ability to make precise changes in DNA and RNA that could ameliorate stem cell therapy. Knocking-out, knocking-in, base editing, and changing RNA expression are all some of the possibilities with this technology. Researchers employed this technique to examine and improve stem cell function by better understanding the involvement of important genes and biological cascades. This technology is expected to dramatically expand our understanding of stem cells and facilitate the use of these cells in therapy.
Author Contributions
All the authors participated in the article preparation, wrote the draft, and helped revise the draft. S.J. collected the documentation materials. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
Dr. Sina Naserian is the CEO of CellMedEx company. The rest of the authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding Statement
The research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zakrzewski W., Dobrzynski M., Szymonowicz M., Rybak Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019;10:68. doi: 10.1186/s13287-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mummery C., van de Stolpe A., Roelen B.A.J., Clevers H., editors. Stem Cells. 2nd ed. Academic Press; Boston, MA, USA: 2014. Chapter 5—Origins and Types of Stem Cells: What’s in a Name? pp. 101–129. [Google Scholar]
- 3.Razazian M., Khosravi M., Bahiraii S., Uzan G., Shamdani S., Naserian S. Differences and similarities between mesenchymal stem cell and endothelial progenitor cell immunoregulatory properties against T cells. World J. Stem Cells. 2021;13:971–984. doi: 10.4252/wjsc.v13.i8.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biehl J.K., Russell B. Introduction to Stem Cell Therapy. J. Cardiovasc. Nurs. 2009;24:98–103. doi: 10.1097/JCN.0b013e318197a6a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh V.K., Saini A., Kalsan M., Kumar N., Chandra R. Describing the Stem Cell Potency: The Various Methods of Functional Assessment and In silico Diagnostics. Front. Cell Dev. Biol. 2016;4:134. doi: 10.3389/fcell.2016.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown C., McKee C., Bakshi S., Walker K., Hakman E., Halassy S., Svinarich D., Dodds R., Govind C.K., Chaudhry G.R. Mesenchymal stem cells: Cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 2019;13:1738–1755. doi: 10.1002/term.2914. [DOI] [PubMed] [Google Scholar]
- 7.Kobolak J., Dinnyes A., Memic A., Khademhosseini A., Mobasheri A. Mesenchymal stem cells: Identification, phenotypic characterization, biological properties and potential for regenerative medicine through biomaterial micro-engineering of their niche. Methods. 2016;99:62–68. doi: 10.1016/j.ymeth.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida G.J., Saya H. Therapeutic strategies targeting cancer stem cells. Cancer Sci. 2015;107:5–11. doi: 10.1111/cas.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahla R.S. Stem Cells Applications in Regenerative Medicine and Disease Therapeutics. Int. J. Cell Biol. 2016;2016:6940283. doi: 10.1155/2016/6940283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saleem U., Van Meer B.J., Katili P.A., Yusof N.A.N.M., Mannhardt I., Garcia A.K., Tertoolen L., De Korte T., Vlaming M.L.H., McGlynn K., et al. Blinded, Multicenter Evaluation of Drug-induced Changes in Contractility Using Human-induced Pluripotent Stem Cell-derived Cardiomyocytes. Toxicol. Sci. 2020;176:103–123. doi: 10.1093/toxsci/kfaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenti M.T., Serena M., Carbonare L.D., Zipeto D. CRISPR/Cas system: An emerging technology in stem cell research. World J. Stem Cells. 2019;11:937–956. doi: 10.4252/wjsc.v11.i11.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunin V., Sorek R., Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova K., Wolf Y., Koonin E. Classification and nomenclature of CRISPR-cas systems: Where from here? CRISPR J. 2018;1:325–336. doi: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J.J., Charpentier E., Horvath P., Moineau S., Mojica F.J.M., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Genet. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Zhang Y., Gao F., Han S., Cheah K.S., Tse H.-F., Lian Q. CRISPR/Cas9 Genome-Editing System in Human Stem Cells: Current Status and Future Prospects. Mol. Ther.-Nucleic Acids. 2017;9:230–241. doi: 10.1016/j.omtn.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 17.Jamehdor S., Pajouhanfar S., Saba S., Uzan G., Teimoori A., Naserian S. Principles and Applications of CRISPR Toolkit in Virus Manipulation, Diagnosis, and Virus-Host Interactions. Cells. 2022;11:999. doi: 10.3390/cells11060999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamehdor S., Zaboli K.A., Naserian S., Thekkiniath J., Omidy H.A., Teimoori A., Johari B., Taromchi A.H., Sasano Y., Kaboli S. An overview of applications of CRISPR-Cas technologies in biomedical engineering. Folia Histochem. Cytobiol. 2020;58:163–173. doi: 10.5603/FHC.a2020.0023. [DOI] [PubMed] [Google Scholar]
- 19.Jamehdor S., Naserian S., Teimoori A. Enhanced High Mutation Rate and Natural Selection to Produce Attenuated Viral Vaccine with CRISPR Toolkit in RNA Viruses especially SARS-CoV-2. Infect Genet Evol. 2022;97:105188. doi: 10.1016/j.meegid.2021.105188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lancaster M.A., Huch M. Disease modelling in human organoids. Dis. Model. Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahu S., Sharan S.K. Translating Embryogenesis to Generate Organoids: Novel Approaches to Personalized Medicine. Iscience. 2020;23:101485. doi: 10.1016/j.isci.2020.101485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii M., Clevers H., Sato T. Modeling Human Digestive Diseases With CRISPR-Cas9–Modified Organoids. Gastroenterology. 2019;156:562–576. doi: 10.1053/j.gastro.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 23.Lindvall O., Kokaia Z., Martinez-Serrano A. Stem cell therapy for human neurodegenerative disorders–how to make it work. Nat. Med. 2004;10:S42–S50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 24.Bryder D., Rossi D.J., Weissman I.L. Hematopoietic Stem Cells: The Paradigmatic Tissue-Specific Stem Cell. Am. J. Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balaji S., King A., Crombleholme T.M., Keswani S.G. The Role of Endothelial Progenitor Cells in Postnatal Vasculogenesis: Implications for Therapeutic Neovascularization and Wound Healing. Adv. Wound Care. 2013;2:283–295. doi: 10.1089/wound.2012.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schäffler A., Büchler C. Concise Review: Adipose Tissue-Derived Stromal Cells—Basic and Clinical Implications for Novel Cell-Based Therapies. STEM CELLS. 2007;25:818–827. doi: 10.1634/stemcells.2006-0589. [DOI] [PubMed] [Google Scholar]
- 27.Oh S., Witek R.P., Bae S., Zheng D., Jung Y., Piscaglia A.C., Petersen B.E. Bone Marrow–Derived Hepatic Oval Cells Differentiate Into Hepatocytes in 2-Acetylaminofluorene/Partial Hepatectomy–Induced Liver Regeneration. Gastroenterology. 2007;132:1077–1087. doi: 10.1053/j.gastro.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Mimeault M., Hauke R., Batra S.K. Stem Cells: A Revolution in Therapeutics—Recent Advances in Stem Cell Biology and Their Therapeutic Applications in Regenerative Medicine and Cancer Therapies. Clin. Pharmacol. Ther. 2007;82:252–264. doi: 10.1038/sj.clpt.6100301. [DOI] [PubMed] [Google Scholar]
- 29.Weiss M.L., Troyer D.L. Stem cells in the umbilical cord. Stem Cell Rev. 2006;2:155–162. doi: 10.1007/s12015-006-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohban R., Pieber T.R. Mesenchymal Stem and Progenitor Cells in Regeneration: Tissue Specificity and Regenerative Potential. Stem Cells Int. 2017;2017:5173732. doi: 10.1155/2017/5173732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Zhang R., Qiao H., Zhang H., Wang Y., Yuan H., Liu Q., Liu D., Chen L., Pei X. Generation of insulin-producing cells from PDX-1 gene-modified human mesenchymal stem cells. J. Cell. Physiol. 2007;211:36–44. doi: 10.1002/jcp.20897. [DOI] [PubMed] [Google Scholar]
- 32.Aboody K.S., Bush R.A., Garcia E., Metz M.Z., Najbauer J., Justus K.A., Phelps D.A., Remack J.S., Yoon K.J., Gillespie S., et al. Development of a Tumor-Selective Approach to Treat Metastatic Cancer. PLoS ONE. 2006;1:e23. doi: 10.1371/journal.pone.0000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J.J., Sun X., Yuan X., Lee J.-W., Snyder E.Y., Yu J.S. Immunomodulatory neural stem cells for brain tumour therapy. Expert Opin. Biol. Ther. 2006;6:1255–1262. doi: 10.1517/14712598.6.12.1255. [DOI] [PubMed] [Google Scholar]
- 34.Müller F.J., Snyder E.Y., Loring J.F. Gene therapy: Can neural stem cells deliver? Nat. Rev. Neurosci. 2006;7:75–84. doi: 10.1038/nrn1829. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt N.O., Przylecki W., Yang W., Ziu M., Teng Y., Kim S.U., Black P.M., Aboody K.S., Carroll R.S. Brain Tumor Tropism of Transplanted Human Neural Stem Cells Is Induced by Vascular Endothelial Growth Factor. Neoplasia. 2005;7:623–630. doi: 10.1593/neo.04781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anasetti C., Logan B.R., Lee S.J., Waller E.K., Weisdorf D.J., Wingard J.R., Cutler C.S., Westervelt P., Woolfrey A., Couban S., et al. Peripheral-Blood Stem Cells versus Bone Marrow from Unrelated Donors. New Engl. J. Med. 2012;367:1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.J., Park J.-S. Usage of Human Mesenchymal Stem Cells in Cell-based Therapy: Advantages and Disadvantages. Dev. Reprod. 2017;21:1–10. doi: 10.12717/DR.2017.21.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mertens J., Reid D., Lau S., Kim Y., Gage F.H. Aging in a Dish: iPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu. Rev. Genet. 2018;52:271–293. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naserian S., Leclerc M., Shamdani S., Uzan G. Current Preventions and Treatments of aGVHD: From Pharmacological Prophylaxis to Innovative Therapies. Front. Immunol. 2020;11:607030. doi: 10.3389/fimmu.2020.607030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao T., Zhang Z.-N., Westenskow P., Todorova D., Hu Z., Lin T., Rong Z., Kim J., He J., Wang M., et al. Humanized Mice Reveal Differential Immunogenicity of Cells Derived from Autologous Induced Pluripotent Stem Cells. Cell Stem Cell. 2015;17:353–359. doi: 10.1016/j.stem.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Y., Valderrama A.V., Han Z., Uzan G., Naserian S., Oberlin E. Human fetal liver MSCs are more effective than adult bone marrow MSCs for their immunosuppressive, immunomodulatory, and Foxp3+ T reg induction capacity. Stem Cell Res. Ther. 2021;12:138. doi: 10.1186/s13287-021-02176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh V., Dhar P.K. Genome Engineering Via CRISPR-Cas9 System. Academic Press; Cambridge, MA, USA: 2020. [Google Scholar]
- 43.Yan F., Wang W., Zhang J. CRISPR-Cas12 and Cas13: The lesser known siblings of CRISPR-Cas9. Cell Biol. Toxicol. 2019;35:489–492. doi: 10.1007/s10565-019-09489-1. [DOI] [PubMed] [Google Scholar]
- 44.Marraffini L.A., Sontheimer E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Rhun A., Escalera-Maurer A., Bratovič M., Charpentier E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019;16:380–389. doi: 10.1080/15476286.2019.1582974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou Z., Zhang Y., Propson N.E., Howden S.E., Chu L.-F., Sontheimer E.J., Thomson J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tóth E., Weinhardt N., Bencsura P., Huszár K., Kulcsár P.I., Tálas A., Fodor E., Welker E. Cpf1 nucleases demonstrate robust activity to induce DNA modification by exploiting homology directed repair pathways in mammalian cells. Biol. Direct. 2016;11:46. doi: 10.1186/s13062-016-0147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Featherstone C., Jackson S.P. DNA double-strand break repair. Curr. Biol. 1999;9:R759–R761. doi: 10.1016/S0960-9822(00)80005-6. [DOI] [PubMed] [Google Scholar]
- 55.Scully R., Panday A., Elango R., Willis N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutin J., Cappellen D., Rosier J., Amintas S., Dabernat S., Bedel A., Moreau-Gaudry F. ON-Target Adverse Events of CRISPR-Cas9 Nuclease: More Chaotic than Expected. CRISPR J. 2022;5:19–30. doi: 10.1089/crispr.2021.0120. [DOI] [PubMed] [Google Scholar]
- 58.You Y., Ramachandra S.G., Jin T. A CRISPR-based method for testing the essentiality of a gene. Sci. Rep. 2020;10:14779. doi: 10.1038/s41598-020-71690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim Y.-K., Yu J.H., Min S.-H., Park S.-W. Generation of a GLA knock-out human-induced pluripotent stem cell line, KSBCi002-A-1, using CRISPR/Cas9. Stem Cell Res. 2019;42:101676. doi: 10.1016/j.scr.2019.101676. [DOI] [PubMed] [Google Scholar]
- 61.Mun D., Kang J.-Y., Chun Y., Park D.-S., Kim H., Yun N., Lee S.-H., Joung B. Generation of two PITX2 knock-out human induced pluripotent stem cell lines using CRISPR/Cas9 system. Stem Cell Res. 2022;65:102940. doi: 10.1016/j.scr.2022.102940. [DOI] [PubMed] [Google Scholar]
- 62.Rudraraju R., Gartner M.J., Neil J.A., Stout E.S., Chen J., Needham E.J., See M., Mackenzie-Kludas C., Lee L.Y.Y., Wang M., et al. Parallel use of pluripotent human stem cell lung and heart models provide new insights for treatment of SARS-CoV-2. bioRxiv. 2022. preprint . [DOI] [PMC free article] [PubMed]
- 63.Thongsin N., Wattanapanitch M. CRISPR/Cas9 Ribonucleoprotein Complex-Mediated Efficient B2M Knockout in Human Induced Pluripotent Stem Cells (iPSCs) Methods Mol. Biol. 2021;2454:607–624. doi: 10.1007/7651_2021_352. [DOI] [PubMed] [Google Scholar]
- 64.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I., et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Genet. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Porto E.M., Komor A.C., Slaymaker I.M., Yeo G.W. Base editing: Advances and therapeutic opportunities. Nat. Rev. Drug Discov. 2020;19:839–859. doi: 10.1038/s41573-020-0084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen P.J., Hussmann J.A., Yan J., Knipping F., Ravisankar P., Chen P.-F., Chen C., Nelson J.W., Newby G.A., Sahin M., et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–5652.e29. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson J.W., Randolph P.B., Shen S.P., Everette K.A., Chen P.J., Anzalone A.V., An M., Newby G.A., Chen J.C., Hsu A., et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022;40:402–410. doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen P.J., Liu D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2022. Online ahead of print . [DOI] [PMC free article] [PubMed]
- 71.Song M., Lim J.M., Min S., Oh J.-S., Kim D.Y., Woo J.-S., Nishimasu H., Cho S.-R., Yoon S., Kim H.H. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat. Commun. 2021;12:5617. doi: 10.1038/s41467-021-25928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou M., Tang S., Duan N., Xie M., Li Z., Feng M., Wu L., Hu Z., Liang D. Targeted-Deletion of a Tiny Sequence via Prime Editing to Restore SMN Expression. Int. J. Mol. Sci. 2022;23:7941. doi: 10.3390/ijms23147941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grünewald J., Miller B.R., Szalay R.N., Cabeceiras P.K., Woodilla C.J., Holtz E.J.B., Petri K., Joung J.K. Engineered CRISPR prime editors with compact, untethered reverse transcriptases. Nat. Biotechnol. 2022. Online ahead of print . [DOI] [PMC free article] [PubMed]
- 74.Karmakar S., Rauth S., Nallasamy P., Perumal N., Nimmakayala R.K., Leon F., Gupta R., Barkeer S., Venkata R.C., Raman V., et al. RNA Polymerase II-Associated Factor 1 Regulates Stem Cell Features of Pancreatic Cancer Cells, Independently of the PAF1 Complex, via Interactions With PHF5A and DDX3. Gastroenterology. 2020;159:1898–1915.e6. doi: 10.1053/j.gastro.2020.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehravar M., Shirazi A., Mehrazar M.M., Nazari M., Banan M., Salimi M. Efficient Production of Biallelic RAG1 Knockout Mouse Embryonic Stem Cell Using CRISPR/Cas9. Iran. J. Biotechnol. 2019;17:45–53. doi: 10.21859/ijb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li C., Wang Q., Peng Z., Lin Y., Liu H., Yang X., Li S., Liu X., Chen J. Generation of FOS gene knockout lines from a human embryonic stem cell line using CRISPR/Cas9. Stem Cell Res. 2019;39:101479. doi: 10.1016/j.scr.2019.101479. [DOI] [PubMed] [Google Scholar]
- 77.Lorthongpanich C., Jiamvoraphong N., Supakun P., Damkham N., Terbto P., Waeteekul S., U-Pratya Y., Laowtammathron C., Issaragrisil S. Generation of a WWTR1 mutation induced pluripotent stem cell line, MUSIi012-A-1, using CRISPR/Cas9. Stem Cell Res. 2019;41:101634. doi: 10.1016/j.scr.2019.101634. [DOI] [PubMed] [Google Scholar]
- 78.Zhang X., Liu Z., Liu X., Wang S., Zhang Y., He X., Sun S., Ma S., Shyh-Chang N., Liu F., et al. Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell. 2019;10:649–667. doi: 10.1007/s13238-019-0610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie L., Huang J., Li X., Dai L., Lin X., Zhang J., Luo J., Zhang W. Generation of a homozygous HDAC6 knockout human embryonic stem cell line by CRISPR/Cas9 editing. Stem Cell Res. 2019;41:101610. doi: 10.1016/j.scr.2019.101610. [DOI] [PubMed] [Google Scholar]
- 80.N’Tumba-Byn T., Yamada M., Seandel M. Loss of tyrosine kinase receptor Ephb2 impairs proliferation and stem cell activity of spermatogonia in culturedagger. Biol. Reprod. 2020;102:950–962. doi: 10.1093/biolre/ioz222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Y., Li J., Zhou T., Pan G., Huang K. Generation of PARP1 gene knockout human embryonic stem cell line using CRISPR/Cas9. Stem Cell Res. 2021;53:102288. doi: 10.1016/j.scr.2021.102288. [DOI] [PubMed] [Google Scholar]
- 82.Chelmicki T., Roger E., Teissandier A., Dura M., Bonneville L., Rucli S., Dossin F., Fouassier C., Lameiras S., Bourc’His D. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature. 2021;591:312–316. doi: 10.1038/s41586-020-03135-1. [DOI] [PubMed] [Google Scholar]
- 83.Diao Z., Ji Q., Wu Z., Zhang W., Cai Y., Wang Z., Hu J., Liu Z., Wang Q., Bi S., et al. SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 2021;49:4203–4219. doi: 10.1093/nar/gkab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gross T., Jeney C., Halm D., Finkenzeller G., Stark G.B., Zengerle R., Koltay P., Zimmermann S. Characterization of CRISPR/Cas9 RANKL knockout mesenchymal stem cell clones based on single-cell printing technology and Emulsion Coupling assay as a low-cellularity workflow for single-cell cloning. PLoS ONE. 2021;16:e0238330. doi: 10.1371/journal.pone.0238330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Markiewicz-Potoczny M., Lobanova A., Loeb A.M., Kirak O., Olbrich T., Ruiz S., Denchi E.L. TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature. 2020;589:110–115. doi: 10.1038/s41586-020-2959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khaled M., Moustafa A.S., El-Khazragy N., Ahmed M.I., Elkhalek M.A.A., El Salahy E.M. CRISPR/Cas9 mediated knock-out of VPREB1 gene induces a cytotoxic effect in myeloma cells. PLoS ONE. 2021;16:e0245349. doi: 10.1371/journal.pone.0245349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang C., Wei Y., Zhang Y., Zhang J., Xing Q., Zhou M., Shan Y. Establishment of a CRISPR/Cas9-mediated ANP32A homozygous knockout human embryonic stem cell line. Stem Cell Res. 2021;52:102234. doi: 10.1016/j.scr.2021.102234. [DOI] [PubMed] [Google Scholar]
- 88.Mercado N., Schutzius G., Kolter C., Estoppey D., Bergling S., Roma G., Keller C.G., Nigsch F., Salathe A., Terranova R., et al. IRF2 is a master regulator of human keratinocyte stem cell fate. Nat. Commun. 2019;10:4676. doi: 10.1038/s41467-019-12559-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He X., Li Y.-X., Feng B. New Turns for High Efficiency Knock-In of Large DNA in Human Pluripotent Stem Cells. Stem Cells Int. 2018;2018:9465028. doi: 10.1155/2018/9465028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumita W., Sato K., Suzuki Y., Kurotaki Y., Harada T., Zhou Y., Kishi N., Sato K., Aiba A., Sakakibara Y., et al. Efficient generation of Knock-in/Knock-out marmoset embryo via CRISPR/Cas9 gene editing. Sci. Rep. 2019;9:12719. doi: 10.1038/s41598-019-49110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S., et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 92.Ma X., Chen C., Veevers J., Zhou X., Ross R.S., Feng W., Chen J. CRISPR/Cas9-mediated gene manipulation to create single-amino-acid-substituted and floxed mice with a cloning-free method. Sci. Rep. 2017;7:42244. doi: 10.1038/srep42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 96.Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Navarro-Guerrero E., Tay C., Whalley J.P., Cowley S.A., Davies B., Knight J.C., Ebner D. Genome-wide CRISPR/Cas9-knockout in human induced Pluripotent Stem Cell (iPSC)-derived macrophages. Sci. Rep. 2021;11:4245. doi: 10.1038/s41598-021-82137-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Villegas F., Lehalle D., Mayer D., Rittirsch M., Stadler M.B., Zinner M., Olivieri D., Vabres P., Duplomb-Jego L., De Bont E.S., et al. Lysosomal Signaling Licenses Embryonic Stem Cell Differentiation via Inactivation of Tfe3. Cell Stem Cell. 2018;24:257–270.e8. doi: 10.1016/j.stem.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 99.Lytle N.K., Ferguson L.P., Rajbhandari N., Gilroy K., Fox R.G., Deshpande A., Schürch C.M., Hamilton M., Robertson N., Lin W., et al. A Multiscale Map of the Stem Cell State in Pancreatic Adenocarcinoma. Cell. 2019;177:572–586.e22. doi: 10.1016/j.cell.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brezgin S., Kostyusheva A., Kostyushev D., Chulanov V. Dead Cas Systems: Types, Principles, and Applications. Int. J. Mol. Sci. 2019;20:6041. doi: 10.3390/ijms20236041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.MacLeod R.S., Cawley K.M., Gubrij I., Nookaew I., Onal M., O’Brien C.A. Effective CRISPR interference of an endogenous gene via a single transgene in mice. Sci. Rep. 2019;9:17312. doi: 10.1038/s41598-019-53611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schoger E., Argyriou L., Zimmermann W.-H., Cyganek L., Zelarayán L.C. Generation of homozygous CRISPRa human induced pluripotent stem cell (hiPSC) lines for sustained endogenous gene activation. Stem Cell Res. 2020;48:101944. doi: 10.1016/j.scr.2020.101944. [DOI] [PubMed] [Google Scholar]
- 104.Pulecio J., Verma N., Mejía-Ramírez E., Huangfu D., Raya A. CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell. 2017;21:431–447. doi: 10.1016/j.stem.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen B., Huang B. Imaging Genomic Elements in Living Cells Using CRISPR/Cas9. Methods Enzymol. 2014;546:337–354. doi: 10.1016/b978-0-12-801185-0.00016-7. [DOI] [PubMed] [Google Scholar]
- 106.Eid A., AlShareef S., Mahfouz M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li R., Xia X., Wang X., Sun X., Dai Z., Huo D., Zheng H., Xiong H., He A., Wu X. Generation and validation of versatile inducible CRISPRi embryonic stem cell and mouse model. PLOS Biol. 2020;18:e3000749. doi: 10.1371/journal.pbio.3000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen L., Shi K., Qiu W., Aagaard L., Kassem M. Generation of Inducible CRISPRi and CRISPRa Human Stromal/Stem Cell Lines for Controlled Target Gene Transcription during Lineage Differentiation. Stem Cells Int. 2020;2020:8857344. doi: 10.1155/2020/8857344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hsu M.-N., Liao H.-T., Truong V.A., Huang K.-L., Yu F.-J., Chen H.-H., Nguyen T.K.N., Makarevich P., Parfyonova Y., Hu Y.-C. CRISPR-based Activation of Endogenous Neurotrophic Genes in Adipose Stem Cell Sheets to Stimulate Peripheral Nerve Regeneration. Theranostics. 2019;9:6099–6111. doi: 10.7150/thno.36790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Truong V.A., Hsu M.-N., Nguyen N.T.K., Lin M.-W., Shen C.-C., Lin C.-Y., Hu Y.-C. CRISPRai for simultaneous gene activation and inhibition to promote stem cell chondrogenesis and calvarial bone regeneration. Nucleic Acids Res. 2019;47:e74. doi: 10.1093/nar/gkz267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tagliafierro L., Ilich E., Moncalvo M., Gu J., Sriskanda A., Grenier C., Murphy S.K., Chiba-Falek O., Kantor B. Lentiviral Vector Platform for the Efficient Delivery of Epigenome-editing Tools into Human Induced Pluripotent Stem Cell-derived Disease Models. J. Vis. Exp. 2019 doi: 10.3791/59241-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nishiga M., Qi L.S., Wu J.C. CRISPRi/a Screening with Human iPSCs. Methods Mol. Biol. 2021;2320:261–281. doi: 10.1007/978-1-0716-1484-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Molecular cell. 2018;70:327–339.e325. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Granados-Riveron J.T., Aquino-Jarquin G. CRISPR–Cas13 Precision Transcriptome Engineering in Cancer. Cancer Res. 2018;78:4107–4113. doi: 10.1158/0008-5472.CAN-18-0785. [DOI] [PubMed] [Google Scholar]
- 118.Li J., Chen Z., Chen F., Xie G., Ling Y., Peng Y., Lin Y., Luo N., Chiang C.-M., Wang H. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020;48:5684–5694. doi: 10.1093/nar/gkaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shimizu Y., Bandaru S., Hara M., Young S., Sano T., Usami K., Kurano Y., Lee S., Kumagai-Takei N., Sano S., et al. An RNA-immunoprecipitation via CRISPR/dCas13 reveals an interaction between the SARS-CoV-2 5’UTR RNA and the process of human lipid metabolism. Res. Sq. 2021. Preprint . [DOI] [PMC free article] [PubMed]
- 120.Crunkhorn S. Expanding the RNA-editing toolbox. Nat. Rev. Drug Discov. 2019;18:667. doi: 10.1038/d41573-019-00128-2. [DOI] [PubMed] [Google Scholar]
- 121.Truong D.-J.J., Phlairaharn T., Eßwein B., Gruber C., Tümen D., Baligács E., Armbrust N., Vaccaro F.L., Lederer E.-M., Beck E.M., et al. Non-invasive and high-throughput interrogation of exon-specific isoform expression. Nature. 2021;23:652–663. doi: 10.1038/s41556-021-00678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D., Zhou C., Zhu C., Chen K., Duan B., et al. DeepCRISPR: Optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Keough K.C., Lyalina S., Olvera M.P., Whalen S., Conklin B.R., Pollard K.S. AlleleAnalyzer: A tool for personalized and allele-specific sgRNA design. Genome Biol. 2019;20:167. doi: 10.1186/s13059-019-1783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu H., Wei Z., Dominguez A., Li Y., Wang X., Qi L.S. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation: Figure 1. Bioinformatics. 2015;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dong Y., Li H., Zhao L., Koopman P., Zhang F., Huang J.X. Genome-Wide Off-Target Analysis in CRISPR-Cas9 Modified Mice and Their Offspring. G3 Genes Genomes Genet. 2019;9:3645–3651. doi: 10.1534/g3.119.400503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zischewski J., Fischer R., Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017;35:95–104. doi: 10.1016/j.biotechadv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 127.Zhang X.-H., Tee L.Y., Wang X.-G., Huang Q.-S., Yang S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Watanabe K., Nakamura T., Onodera S., Saito A., Shibahara T., Azuma T. A novel GNAS-mutated human induced pluripotent stem cell model for understanding GNAS-mutated tumors. Tumor Biol. 2020;42:1010428320962588. doi: 10.1177/1010428320962588. [DOI] [PubMed] [Google Scholar]
- 130.Shetty D.K., Kalamkar K.P., Inamdar M.S. OCIAD1 controls electron transport chain complex I activity to regulate energy metabolism in human pluripotent stem cells. Stem Cell Rep. 2018;11:128–141. doi: 10.1016/j.stemcr.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tu J., Huo Z., Liu M., Wang D., Xu A., Zhou R., Zhu D., Gingold J., Shen J., Zhao R., et al. Generation of human embryonic stem cell line with heterozygous RB1 deletion by CRIPSR/Cas9 nickase. Stem Cell Res. 2018;28:29–32. doi: 10.1016/j.scr.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jain S., Xun G., Abesteh S., Ho S., Lingamaneni M., Martin T.A., Tasan I., Yang C., Zhao H. Precise Regulation of Cas9-Mediated Genome Engineering by Anti-CRISPR-Based Inducible CRISPR Controllers. ACS Synth. Biol. 2021;10:1320–1327. doi: 10.1021/acssynbio.0c00548. [DOI] [PubMed] [Google Scholar]
- 133.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang J.-P., Li X.-L., Neises A., Chen W., Hu L.-P., Ji G.-Z., Yu J.-Y., Xu J., Yuan W.-P., Cheng T., et al. Different Effects of sgRNA Length on CRISPR-mediated Gene Knockout Efficiency. Sci. Rep. 2016;6:28566. doi: 10.1038/srep28566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low Incidence of Off-Target Mutations in Individual CRISPR-Cas9 and TALEN Targeted Human Stem Cell Clones Detected by Whole-Genome Sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Banan M. Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. J. Biotechnol. 2019;308:1–9. doi: 10.1016/j.jbiotec.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 137.Bialk P., Rivera-Torres N., Strouse B., Kmiec E.B. Regulation of Gene Editing Activity Directed by Single-Stranded Oligonucleotides and CRISPR/Cas9 Systems. PLoS ONE. 2015;10:e0129308. doi: 10.1371/journal.pone.0129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Okamoto S., Amaishi Y., Maki I., Enoki T., Mineno J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 2019;9:4811. doi: 10.1038/s41598-019-41121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang B., Li K., Wang A., Reiser M., Saunders T., Lockey R.F., Wang J.-W. Highly efficient CRISPR/HDR-mediated knock-in for mouse embryonic stem cells and zygotes. Biotechniques. 2015;59:201–208. doi: 10.2144/000114339. [DOI] [PubMed] [Google Scholar]
- 142.Smirnikhina S.A., Anuchina A.A., Lavrov A.V. Ways of improving precise knock-in by genome-editing technologies. Hum. Genet. 2018;138:1–19. doi: 10.1007/s00439-018-1953-5. [DOI] [PubMed] [Google Scholar]
- 143.Lee Y., Choi H.Y., Kwon A., Park H., Park M., Kim Y.-O., Kwak S., Koo S.K. Generation of a NESTIN-EGFP reporter human induced pluripotent stem cell line, KSCBi005-A-1, using CRISPR/Cas9 nuclease. Stem Cell Res. 2019;40:101554. doi: 10.1016/j.scr.2019.101554. [DOI] [PubMed] [Google Scholar]
- 144.Li S., Xue H., Wu J., Rao M.S., Kim D.H., Deng W., Liu Y. Human Induced Pluripotent Stem CellNEUROG2Dual Knockin Reporter Lines Generated by the CRISPR/Cas9 System. Stem Cells Dev. 2015;24:2925–2942. doi: 10.1089/scd.2015.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun Y.-H., Kao H.K., Chang C.-W., Merleev A., Overton J.L., Pretto D., Yechikov S., Maverakis E., Chiamvimonvat N., Chan J.W., et al. Human induced pluripotent stem cell line with genetically encoded fluorescent voltage indicator generated via CRISPR for action potential assessment post-cardiogenesis. STEM CELLS. 2019;38:90–101. doi: 10.1002/stem.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yoshimatsu S., Sato T., Yamamoto M., Sasaki E., Nakajima M., Nakamura M., Shiozawa S., Noce T., Okano H. Generation of a male common marmoset embryonic stem cell line DSY127-BV8VT1 carrying double reporters specific for the germ cell linage using the CRISPR-Cas9 and PiggyBac transposase systems. Stem Cell Res. 2020;44:101740. doi: 10.1016/j.scr.2020.101740. [DOI] [PubMed] [Google Scholar]
- 147.Kim D., Kim S.-B., Ryu J.L., Hong H., Chang J.-H., Yoo T.-J., Jin X., Park H.-J., Han C., Lee B.H., et al. Human Embryonic Stem Cell-Derived Wilson’s Disease Model for Screening Drug Efficacy. Cells. 2020;9:872. doi: 10.3390/cells9040872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ma L., Wu Z., Tang Q., Ji X., Mei Y., Peng T., Xu Q., Zhou W., Xiong M. Generation of an induced pluripotent stem cell line from a patient with global development delay carrying DYRK1A mutation (c.1730T > A) and a gene correction isogenic iPSC line. Stem Cell Res. 2021;53:102305. doi: 10.1016/j.scr.2021.102305. [DOI] [PubMed] [Google Scholar]
- 149.Ababneh N.A., Scaber J., Flynn R., Douglas A., Barbagallo P., Candalija A., Turner M.R., Sims D., Dafinca R., Cowley S.A., et al. Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair. Hum. Mol. Genet. 2020;29:2200–2217. doi: 10.1093/hmg/ddaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao Y., Wu S., Pan J., Zhang K., Li X., Xu Y., Jin C., He X., Shi J., Ma L., et al. CRISPR/Cas9-edited triple-fusion reporter gene imaging of dynamics and function of transplanted human urinary-induced pluripotent stem cell-derived cardiomyocytes. Eur. J. Nucl. Med. 2020;48:708–720. doi: 10.1007/s00259-020-05087-0. [DOI] [PubMed] [Google Scholar]
- 151.Lee Y., Choi H.Y., Kwon A., Park H., Park M.H., Kim J.W., Kim M.J., Kim Y.-O., Kwak S., Koo S.K. Generation of a PDX1-EGFP reporter human induced pluripotent stem cell line, KSCBi005-A-3, using the CRISPR/Cas9 system. Stem Cell Res. 2019;41:101632. doi: 10.1016/j.scr.2019.101632. [DOI] [PubMed] [Google Scholar]
- 152.Ling K., Jiang L., Liang S., Kwong J., Yang L., Li Y., Yin P., Deng Q., Liang Z. Nanog interaction with the androgen receptor signaling axis induce ovarian cancer stem cell regulation: Studies based on the CRISPR/Cas9 system. J. Ovarian Res. 2018;11:36. doi: 10.1186/s13048-018-0403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Itoh M., Kawagoe S., Tamai K., Nakagawa H., Asahina A., Okano H.J. Footprint-free gene mutation correction in induced pluripotent stem cell (iPSC) derived from recessive dystrophic epidermolysis bullosa (RDEB) using the CRISPR/Cas9 and piggyBac transposon system. J. Dermatol. Sci. 2020;98:163–172. doi: 10.1016/j.jdermsci.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 154.Laverde-Paz M.J., Nuytemans K., Wang L., Vance J.M., Pericak-Vance M.A., Dykxhoorn D.M., Cukier H.N. Derivation of stem cell line UMi028-A-2 containing a CRISPR/Cas9 induced Alzheimer’s disease risk variant p. S1038C in the TTC3 gene. Stem Cell Res. 2021;52:102258. doi: 10.1016/j.scr.2021.102258. [DOI] [PubMed] [Google Scholar]
- 155.Zhou M., Xing Q., Zhang D., Zhang C., Zhang Y., Zhang J., Shan Y. Generation of an Akaluc knock-in human embryonic stem cell reporter line using CRISPR-Cas9 technology. Stem Cell Res. 2021;56:102532. doi: 10.1016/j.scr.2021.102532. [DOI] [PubMed] [Google Scholar]
- 156.Kang B., Cong Z., Duan J., Liu K., Wang Y. Generation of a GFI1-flag knock-in human embryonic stem cell line using CRISPR-Cas9 technology. Stem Cell Res. 2022;60:102724. doi: 10.1016/j.scr.2022.102724. [DOI] [PubMed] [Google Scholar]
- 157.Zhang J., Zhou T., Shan Y., Pan G. Generation of RYBP FLAG-HA knock-in human embryonic stem cell line through CRISPR/Cas9-mediated homologous recombination. Stem Cell Res. 2022;62:102803. doi: 10.1016/j.scr.2022.102803. [DOI] [PubMed] [Google Scholar]
- 158.Sung Y.K., Kim S.W. Recent advances in the development of gene delivery systems. Biomater. Res. 2019;23:8. doi: 10.1186/s40824-019-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021;6:53. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ramamoorth M., Narvekar A. Non viral vectors in gene therapy- an overview. J. Clin. Diagn Res. 2015;9:GE01-6. doi: 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Loza L.I.M., Yuen E.C., McCray J.P.B. Lentiviral Vectors for the Treatment and Prevention of Cystic Fibrosis Lung Disease. Genes. 2019;10:218. doi: 10.3390/genes10030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Negre O., Eggimann A.V., Beuzard Y., Ribeil J.A., Bourget P., Borwornpinyo S., Hongeng S., Hacein-Bey S., Cavazzana M., Leboulch P., et al. Gene Therapy of the beta-Hemoglobinopathies by Lentiviral Transfer of the beta(A(T87Q))-Globin Gene. Hum. Gene Ther. 2016;27:148–165. doi: 10.1089/hum.2016.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ghaleh H.E.G., Bolandian M., Dorostkar R., Jafari A., Pour M.F. Concise review on optimized methods in production and transduction of lentiviral vectors in order to facilitate immunotherapy and gene therapy. Biomed. Pharmacother. 2020;128:110276. doi: 10.1016/j.biopha.2020.110276. [DOI] [PubMed] [Google Scholar]
- 164.Ricobaraza A., Gonzalez-Aparicio M., Mora-Jimenez L., Lumbreras S., Hernandez-Alcoceba R. High-Capacity Adenoviral Vectors: Expanding the Scope of Gene Therapy. Int. J. Mol. Sci. 2020;21:3643. doi: 10.3390/ijms21103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Westhaus A., Cabanes-Creus M., Rybicki A., Baltazar G., Navarro R.G., Zhu E., Drouyer M., Knight M., Albu R.F., Ng B.H., et al. High-Throughput In Vitro, Ex Vivo, and In Vivo Screen of Adeno-Associated Virus Vectors Based on Physical and Functional Transduction. Hum. Gene Ther. 2020;31:575–589. doi: 10.1089/hum.2019.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kaiser J. Virus used in gene therapies may pose cancer risk, dog study hints. Science. 2020;10 doi: 10.1126/science.aba7696. [DOI] [Google Scholar]
- 167.Hanlon K.S., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Volak A., Spirig S.E., Muller A., Sousa A.A., Tsai S.Q., Bengtsson N.E., et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019;10:4439. doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nelson C., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D., Robinson-Hamm J.N., Bulaklak K., Rivera R.M.C., Collier J.H., et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Banskota S., Raguram A., Suh S., Du S.W., Davis J.R., Choi E.H., Wang X., Nielsen S.C., Newby G.A., Randolph P.B., et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell. 2022;185:250–265.e16. doi: 10.1016/j.cell.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hazelbaker D.Z., Beccard A., Angelini G., Mazzucato P., Messana A., Lam D., Eggan K., Barrett L.E. A multiplexed gRNA piggyBac transposon system facilitates efficient induction of CRISPRi and CRISPRa in human pluripotent stem cells. Sci. Rep. 2020;10:635. doi: 10.1038/s41598-020-57500-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shirley J.L., de Jong Y.P., Terhorst C., Herzog R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020;28:709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Abe T., Inoue K.-I., Furuta Y., Kiyonari H. Pronuclear Microinjection during S-Phase Increases the Efficiency of CRISPR-Cas9-Assisted Knockin of Large DNA Donors in Mouse Zygotes. Cell Rep. 2020;31:107653. doi: 10.1016/j.celrep.2020.107653. [DOI] [PubMed] [Google Scholar]
- 173.Alghadban S., Bouchareb A., Hinch R., Hernandez-Pliego P., Biggs D., Preece C., Davies B. Electroporation and genetic supply of Cas9 increase the generation efficiency of CRISPR/Cas9 knock-in alleles in C57BL/6J mouse zygotes. Sci. Rep. 2020;10:17912. doi: 10.1038/s41598-020-74960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Inglut C.T., Sorrin A.J., Kuruppu T., Vig S., Cicalo J., Ahmad H., Huang H.-C. Immunological and Toxicological Considerations for the Design of Liposomes. Nanomaterials. 2020;10:190. doi: 10.3390/nano10020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mout R., Ray M., Tonga G.Y., Lee Y.-W., Tay T., Sasaki K., Rotello V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Thi T., Suys E., Lee J., Nguyen D., Park K., Truong N. Lipid-Based Nanoparticles in the Clinic and Clinical Trials: From Cancer Nanomedicine to COVID-19 Vaccines. Vaccines. 2021;9:359. doi: 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]