Abstract
Cyclins A and E and their partner cyclin-dependent kinases (Cdks) are key regulators of DNA synthesis and of mitosis. Immunofluorescence studies have shown that both cyclins are nuclear and that a proportion of cyclin A is localized to sites of DNA replication. However, recently, both cyclin A and cyclin E have been implicated as regulators of centrosome replication, and it is unclear when and where these cyclin-Cdks can interact with cytoplasmic substrates. We have used live cell imaging to study the behavior of cyclin/Cdk complexes. We found that cyclin A and cyclin E are able to regulate both nuclear and cytoplasmic events because they both shuttle between the nucleus and the cytoplasm. However, we found that there are marked differences in their shuttling behavior, which raises the possibility that cyclin/Cdk function could be regulated at the level of nuclear import and export. In the course of these experiments, we have also found that, contrary to published results, mutations in the hydrophobic patch of cyclin A do affect Cdk binding and nuclear import. This has implications for the role of the hydrophobic patch as a substrate selection motif.
INTRODUCTION
Successive waves of cyclin-dependent kinase (Cdk) activity control progress through the eukaryotic cell cycle. Cdks are activated by binding a member of the cyclin family and phosphorylation by Cdk-activating kinase. The cyclin-Cdks that have been most strongly implicated in controlling entry into, and progress through, DNA replication are cyclins A and E. Both cyclins bind to Cdk2 (Elledge and Spottswood, 1991; Tsai et al., 1991; Koff et al., 1992), and the levels of both cyclins are strictly regulated throughout the cell cycle, by transcriptional and proteolytic mechanisms. Cyclin E levels are primarily dictated by the rate of its transcription because it is an unstable protein that is rapidly degraded by two different pathways of ubiquitin-dependent proteolysis (Clurman et al., 1996; Won and Reed, 1996; Singer et al., 1999; Wang et al., 1999; Winston et al., 1999; Nakayama et al., 2000). In contrast, cyclin A is stable until cells enter mitosis. Cyclin A levels first start to increase at the beginning of S phase and continue to rise throughout S and G2 phases until prometaphase, when they are rapidly degraded by ubiquitin-dependent proteolysis (Pines and Hunter, 1990; Hunt et al., 1992).
There is extensive evidence to indicate that the activation of cyclin E/Cdk2 leads to the initiation of DNA replication. Cyclin E/Cdk2 activity is maximal at G1/S (Koff et al., 1992), replication of DNA in vitro is dependent on cyclin E/Cdk2 activity (Jackson et al., 1995; Strausfeld et al., 1996; Krude et al., 1997) and, in vivo, cyclin E is essential for DNA replication in Drosophila (Sauer et al., 1995). Cyclin A/Cdk2 has also been demonstrated to promote DNA replication (Girard et al., 1991; Pagano et al., 1992; Zindy et al., 1992; Strausfeld et al., 1996). However, cyclin A may be more significant in regulating progression through S phase (Connell-Crowley et al., 1998), perhaps through mediating the nuclear export and degradation of Cdc6 (Saha et al., 1998; Petersen et al., 1999). Furthermore, in Drosophila embryos at the cellular blastoderm stage, cyclin E can promote S phase without cyclin A (Knoblich et al., 1994), and cyclin A itself is located in the cytoplasm in S phase and only becomes nuclear during prophase (Lehner and O'Farrell, 1989).
There are also data indicating that at least cyclin A and perhaps cyclin E interact directly with the DNA replication machinery. Immunofluorescence studies have shown that both cyclin A and cyclin E are nuclear proteins (Girard et al., 1991; Pines and Hunter, 1991; Ohtsubo et al., 1995) and that cyclin A can be detected at sites of DNA replication in cells (Cardoso et al., 1993; Ohtsubo et al., 1995) and bound to the origin recognition complex in vitro (Romanowski et al., 2000). Therefore, proteins at DNA replication origins are probably physiological substrates of either cyclin A/Cdk2 or cyclin E/Cdk2 (Hua and Newport, 1998). Thus, nuclear localization is crucial to the function of both cyclin A/Cdk2 and cyclin E/Cdk2; indeed, cyclin E has a classical nuclear localization sequence (NLS) that targets it to the nucleus via the well characterized importin-α/importin-β nuclear import pathway (Moore et al., 1999).
More recently, cyclin A- and cyclin E-dependent kinases have been implicated in the control of centrosome replication. Mammalian somatic cell extracts require cyclin A-dependent kinase activity to duplicate centrosomes (Meraldi et al., 1999), whereas Xenopus oocytes and early embryos require cyclin E-associated kinase activity (Hinchcliffe et al., 1999; Lacey et al., 1999). Cyclin E-associated kinase also phosphorylates nucleophosmin (Okuda et al., 2000; Tokuyama et al., 2001), a component of the centrosome, causing it to dissociate from the centrosome, which may allow the centrosome to duplicate. Thus, both cyclin E and cyclin A may regulate cell cycle events in both the nucleus and the cytoplasm, but they appear to be exclusively nuclear by immunofluorescence. This raises the question of whether the cyclin-Cdks are more dynamic in living cells or whether their cytoplasmic substrates must first enter the nucleus to be recognized by the kinases.
The behavior of the major mitotic cyclin-Cdk, cyclin B1-Cdk1, sets a precedent for dynamic behavior of cell cycle regulators. Cyclin B1 appears to be exclusively cytoplasmic by immunofluorescence, but this is a reflection of its slow nuclear import being counteracted by more rapid nuclear export. The slow nuclear import of cyclin B1 appears to be mediated by a direct association between cyclin B1 and importin-β, one of the most abundant nuclear import carriers. Cyclin B1 is exported by exportin 1, the primary protein export receptor. It binds directly to exportin 1, and its export is blocked by treating cells with leptomycin B (LMB), which covalently inactivates exportin 1 (Kudo et al., 1999).
In contrast to cyclins E and B1, much less is known about how cyclin A is targeted to the nucleus. Neither cyclin A nor its partner kinase, Cdk2, has consensus NLSs. Using a series of deletion mutants, Nigg and colleagues demonstrated that the nuclear localization of cyclin A correlated with its ability to bind to a Cdk (Maridor et al., 1993). In turn, the cyclin A/Cdk complex is known to bind a variety of proteins with a recognizable NLS, such as p107, E2F1, and the p21 and p27 proteins of the Kip/Cip family. Thus, it was suggested that cyclin/Cdk complexes might “piggy-back” into the nucleus by binding NLS-containing proteins (for discussion, see Gallant et al., 1995). A possible precedent for this is the observation that overexpressing members of the Kip/Cip family increase the amount of cyclin D1 imported into nuclei (Diehl and Sherr, 1997; LaBaer et al., 1997). However, cyclin A is still correctly localized to the nucleus in cells lacking p21CIP1 (Deng et al., 1995).
In this study, we used live cell imaging to show that cyclin A/Cdk2 and cyclin E/Cdk2 continuously shuttle between the nucleus and the cytoplasm. However, in contrast to cyclin B1/Cdk1, cyclin E and cyclin A are exported more slowly than they are imported, and their export is not inhibited by LMB. Using an in vitro import assay, we show that cyclin A/Cdk2 import depends on binding to Cdk2 and that it does not piggy-back into the nucleus using NLS-containing proteins. Furthermore, the nuclear import of cyclins A, B1, and E are distinct, raising the possibility that each can be separately regulated at the level of nuclear transport.
MATERIALS AND METHODS
Construction of Chimeras
Chimaeras were created between a modified form of green fluorescent protein (GFP)-MmGFP (Zernicka-Goetz et al., 1996) and cyclin A, cyclin E, cyclin B1, and Cdk2. In each case, MmGFP was attached at the carboxyl terminus of the protein. For recombinant baculoviral constructs, the cyclin A and cyclin E fusion proteins were constructed by polymerase chain reaction. The stop codon was removed from the cyclin and replaced with an EcoRI site. This was ligated in frame to an EcoRI site 5′ to the first residue of MmGFP, creating a glutamic acid-phenylalanine linker. Similarly, a four-amino acid linker (CPEF) was created between CDK2 and MmGFP. Cyclin B1 5E-GFP cloned into the pGEX2T expression vector had been previously described (Hagting et al., 1998). Cyclin B1 with MKAIL mutation was fused at the C terminus via a AGAQF linker to the second amino acid of MmGFP. The cDNAs encoding the cyclin A and E fusion proteins were tagged at the 5′ with an XbaI site and at the 3′ with a BglII site and cloned into XbaI-BglII–digested pAcAB4 (Belyaev and Roy, 1993). The cDNA encoding cyclin B1-GFP was tagged with a HindIII site at the 5′ and XbaI site at the 3′ end and ligated into pCDNA3. The initiating methionine codon of CDK2-MmGFP cDNA was altered to a NcoI site (CCATGG) and the cDNA was cloned in frame with the (his)6 tag of pRSET B (Invitrogen, Breda, The Netherlands). To construct the cyclin ER130A and cyclin ER130K mutants, the cDNA for human cyclin ER130A (a kind gift of by Dr. B. Clurman, FHRC, Seattle, WA) or human cyclin ER130K (E. Sahai and J.P., unpublished results) was altered by polymerase chain reaction to include a 5′ HindIII site and 3′ EcoRI site as above and used to replace the wild-type sequence in a pCMX-cyclin E-MmGFP construct. All constructs were sequenced by an ABI automated sequencer (Department of Biochemistry, University of Cambridge, Cambridge, UK).
Protein Expression and Purification
Sf9 cells were infected and coinfected with recombinant baculovirus encoding human (his)6-cyclin A, (his)6-cyclin A-GFP, cyclin E-GFP, and human (his)6-Cdk2K33R, harvested 48 h postinfection, and lysed as described by Krude et al. (1997). Supernatants taken following centrifugation at 100,000 × g were partially purified via their (his)6- tag using nickel-nitrilotriacetic acid Superflow (Qiagen, Crawley UK). Proteins were then dialyzed against 25 mM HEPES/KOH, pH 7.1, 150 mM NaCl, 5 mM β-mercaptoethanol in a slide-a-lyzer 10-kDa dialysis cassette (Pierce Chemical, Rockford, IL) and then purified by fast-performance liquid chromatography using a Mono-Q ion exchange column (Amersham Pharmacia Biotech UK, Little Chalfont, Buckinghamshire, UK). Cyclin A-GFP and cyclin E-GFP, with or without coexpressed Cdk2, were eluted from the Mono-Q column with 200 mM NaCl. Proteins were then concentrated to a final concentration of ∼150 mM using Vivaspin concentrators, 50,000 molecular weight cut-off (Vivascience, Lincoln, UK) and stored in liquid N2. (his)6-Cdk2-GFP and (his)6-Cks1 was expressed in Escherichia coli BL21 DE3(pLysS) at 30°C and purified by Ni2+ affinity chromatography followed by gel filtration through a Superdex 200 column. Glutathione S-transferase-cyclin B1 5E-GFP was expressed and purified as described by Hagting et al. (1998). Rch1, nucleoplasmin, and nucleoplasmin core domain, all tagged at the C terminus with (his)6, were expressed and purified as described by Görlich et al. (1994). Human importin-β was expressed from a pQE60 vector (Qiagen) and purified from a bacterial lysate by anion exchange and affinity chromatography on an immobilized importin-β–binding domain of Xenopus importin-α. Ran/TC4 was expressed with an N-terminal (his)6-tag and purified as described by Görlich et al. (1994). Importin-binding domain (IBB) from Xenopus importin-α (gift of D. Görlich) was expressed and purified as described by Gorlich et al. (1996). Xenopus importin-β Δ N 44 (gift of D. Görlich) was expressed and purified as described by Kutay et al. (1997). Ran/TC4 Q69L (gift of D. Görlich) was expressed and purified as described by Görlich et al. (1997) but with an additional final gel filtration purification step using a Superdex 200 column (Pharmacia Amersham Biotech). Proteins were analyzed using 15% SDS-PAGE, and concentrations were determined using the Coomassie Plus Protein Assay Reagent (Pierce). Fluorescein isothiocyanate-conjugated (FITC)-labeled M9 domain and transportin were kindly supplied by D. Görlich. Xenopus extract was prepared as described by Kubota et al. (1995). HeLa cell cytosolic extract was prepared by swelling HeLa S3 cells in hypotonic buffer, 20 mM HEPES/KOH, pH 7.6, 20 mM NaCl, 1 mM EDTA, 5 mM β-mercaptoethanol, and protease inhibitor cocktail (Boehringer-Mannheim, Indianapolis, IN) and lysing by Dounce homogenization. After lysis, NaCl in the lysate was immediately adjusted to 150 mM before spinning at 15,000 × g for 15 min. The resulting supernatant was spun at 100,000 × g for 1 h to give a supernatant with a final protein concentration of 8.7 mg/ml, aliquots of which were frozen in liquid nitrogen.
Transfection
Cyclin A-GFP, cyclin E-GFP, and cyclin ER130A-GFP were expressed from the cytomegalovirus early promoter in the pCMX eukaryotic expression vector. Cyclin A and cyclin B1 constructs were expressed from the pCDNA3 expression vector. All transfections were carried out as described by Jackman et al. (1995). Subcellular localizations of transfected cyclins were visualized 24 h after transfection using confocal laser microscopy or by time-lapse fluorescence and differential interference contrast (DIC) microscopy. Subcellular localization of myc-cyclin A was carried out using methanol/acetone fixation as described by Pines and Hunter (1991) using the anti-myc 9310 mAb (1:200, 0.2 mg/ml, Santa Cruz Biotechnology, Santa Cruz, CA).
Nuclear Import Assays
Nuclear import assays were carried out as described by Görlich et al. (1994). Import reactions were started by adding digitonin-permeabilized HeLa cells to the following reaction mixture: 20 mM HEPES-KOH, pH 7.5, 80 mM potassium acetate, 3 mM magnesium acetate, 250 mM sucrose, an energy-regenerating mixture (1 mM ATP, 100 mM GTP, 10 mM creatine phosphate, 50 mg/ml creatine kinase), 1 mM importin-α(Rch1), 200 nM importin-β, 3 mM RanGDP, 5 mg/ml bovine serum albumin (BSA), and 2 mg/ml nucleoplasmin core (to out-compete nonspecific core binding) or Xenopus extract or HeLa cell extract supplemented with energy-regenerating mixture. Nuclear import substrates were used at the following concentrations: cyclin A-GFP/Cdk2K33R and cyclin E-GFP/Cdk2K33R, 5 mM; cyclin A-GFP, cyclin E-GFP and cyclin B15E-GFP, 4 mM; GFP and GFP-Cdk2K33R, 10 mM; tetramethylrhodamine B isothiocyanate (TRITC)-nucleoplasmin, 2 mM. Nuclei were incubated for 30 min at 22 or 37°C, fixed with 4% paraformaldehyde, and centrifuged through a 30% sucrose cushion onto polylysine-coated coverslips. In experiments using importin-β Δ N 44, wheat germ agglutinin, RanQ69L, or IBB, nuclei were preincubated with, or without, these inhibitors in whole-reaction mixtures on ice for 15 min before import substrates were added, and the nuclei warmed to room temperature. Nuclear import was assayed by laser scanning confocal microscopy using a 1024 confocal microscope (Bio-Rad, Hercules, CA), and the relative amounts of fluorescent substrate imported were measured using the Laser Sharp program (Bio-Rad). One hundred cells were counted per time point, and the mean of at least three independent experiments was calculated.
Nuclear Export Assays
To assay nuclear-cytoplasmic shuttling, HeLa cells were microinjected with purified proteins and incubated in DMEM supplemented with 10% calf serum at 37°C/10% CO2. Cells were fused for 5 min with 50% polyethylene glycol (PEG) 3350/DMEM prewarmed to 37°C, washed five times with phosphate-buffered saline, and analyzed by time-lapse DIC-fluorescence microscopy as described previously (Hagting et al., 1998). To study shuttling in the presence of LMB, cells were pretreated with 20 nM LMB for 1 h, fusions were carried out as above, and LMB was included in the postfusion incubation.
Absorption of Cyclin A/CDK2 onto Cks1
(His) 6-tagged Cks1 was purified by Ni2+-affinity chromatography and coupled onto Ultralink biosupport medium (Pierce). HeLa cells transfected with either pcDNA3-myc-cyclin A or with pcDNA3-myc-cyclin A M210A, L214A, W217A M210A were lysed in 0.1% Nonidet P40, 50 mM Tris pH 8.0, 150 mM NaCl, 0.1 mM EDTA and a protease inhibitor cocktail (Roche Diagnostics, Lewes, East Sussex, UK). Myc-cyclin A was absorbed from a 10,000 × g supernatant by adding 20 μl of Cks1-linked beads to a 1-ml lysate of 1.5 × 107 cells, incubated for 1 h at 4°C, and then washed four times with lysis buffer. Material absorbed onto beads was resolved by SDS-PAGE and myc-cyclin A associated with Cdk was detected by Western blotting with the anti-myc epitope antibody 9E10.
RESULTS
Both Cyclin A and Cyclin E Shuttle between the Nucleus and the Cytoplasm
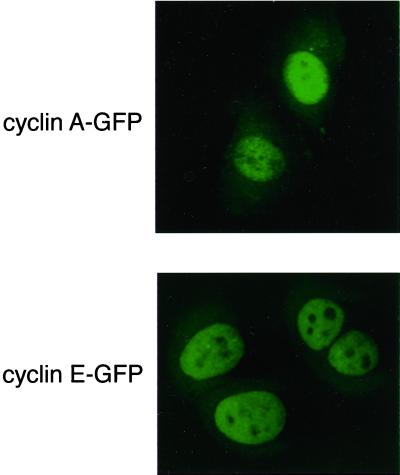
Although cyclins A and E appear to be nuclear by immunofluorescence, their localization might be dynamic when analyzed in real time. To study this, we generated fusion proteins between human cyclin A and E and the GFP of Aequorea victoria. We have previously described our cyclin A-GFP fusion protein (den Elzen and Pines, 2001); like cyclin A-GFP, the cyclin E fusion protein binds and activates its partner Cdk (Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results) and localizes to the nucleus when expressed in tissue culture cells (Figure 1). Therefore, the GFP-fusion proteins can be used as markers for the behavior of the wild-type cyclins.
Figure 1.
Cyclin A-GFP and cyclin E-GFP chimaeras are correctly localized to the nucleus of living cells. Shown are representative fluorescence confocal microscopy images of HeLa cells transfected with cyclin A-GFP (top) or cyclin E-GFP (bottom).
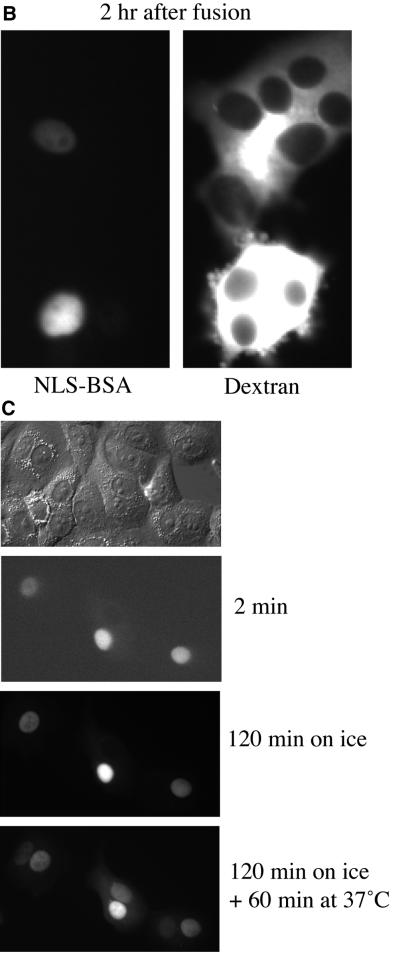
To analyze whether the nuclear localization of cyclin A- and cyclin E-CDK complexes revealed by immunofluorescence was accurate, or masked more dynamic behavior in living cells, we used a cell fusion assay (Borer et al., 1989) and time-lapse imaging. We injected cyclin A-GFP or cyclin E-GFP into the nucleus of a cell and then fused the cell to its neighbors using PEG (see MATERIALS AND METHODS). As a control we injected TRITC-labeled BSA tagged with an NLS that should not be exported from the nucleus. Any proteins that shuttled between the nucleus and the cytoplasm would enter an uninjected nucleus that shared the cytoplasm with a labeled nucleus. We found that both cyclin A-GFP and cyclin E-GFP visibly began to enter uninjected nuclei ∼20 min after cell fusion (Figure 2A), but NLS-BSA remained only in injected nuclei for at least 2 h following fusion (Figure 2B). Cyclin export was not by diffusion because it was blocked by incubating the cells on ice after fusion (Figure 2C). Thus, both cyclin A and E shuttled, and their apparent nuclear localization was a reflection of their nuclear import being more rapid than their export.
Figure 2.
Cyclin A and cyclin E actively shuttle between the nucleus and cytoplasm in a temperature-dependent manner. (A) Cyclins A and E shuttle between nuclei. HeLa cells were injected in the nucleus with 25 mg/ml cyclin A-GFP//CDK2K33R (top) or 35 mg/ml cyclin E-GFP/CDK2K33R (bottom). Cells were fused with PEG 1 h after microinjection. The GFP-chimaeras were detected by fluorescence imaging before and at the indicated time points after injection. (B) NLS-BSA does not shuttle. FITC-conjugated NLS-BSA was injected into HeLa cell nuclei 60 min after cell fusion. Texas Red-conjugated dextran (MW 70,000; 1 mg/ml) was injected at the end of the experiment to show that cells injected with FITC-conjugated NLS-BSA had fused with their neighbors. (C) Cyclin A does not shuttle at 4°C. HeLa cells were fused with PEG, injected in the nucleus with cyclin A-protein, and immediately placed on ice. After 2 h cells were warmed to 37°C. Cyclin A-GFP was detected by fluorescence imaging at the indicated time points after injection
Cyclin A and Cyclin E Export Is Insensitive to LMB
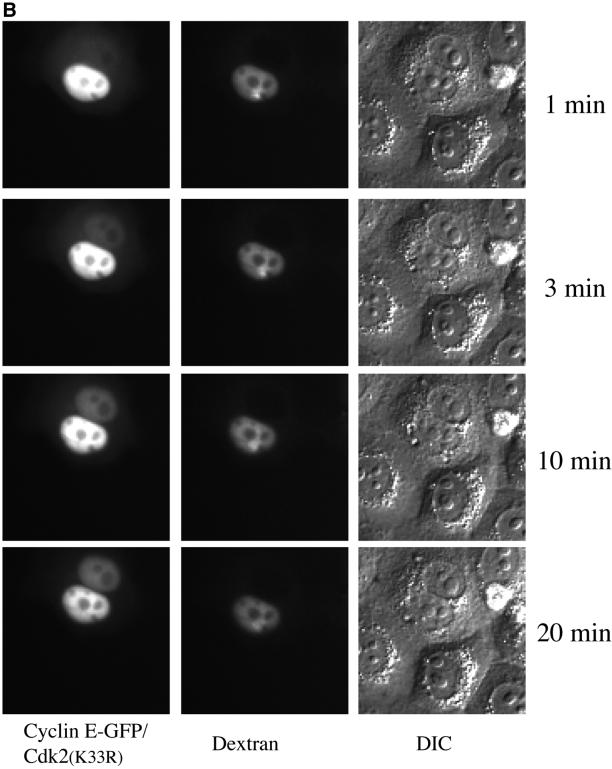
A large number of proteins exported from the nucleus, including cyclin B1, were shown to depend, directly or indirectly, on the nuclear export receptor, exportin 1 by inactivating exportin 1 with LMB (Kudo et al., 1999). Therefore, we analyzed cyclin A- and E-GFP nuclear export by the cell fusion assay in the presence of 20 nM LMB. This showed that both cyclin A and cyclin E were still exported to the surrounding nuclei (Figure 3 and supplementary Figure 9) and indicated that cyclins A and E, unlike cyclin B1 (Hagting et al., 1998; Toyoshima et al., 1998; Yang et al., 1998; Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results), do not depend on exportin 1 for their nuclear export. In parallel experiments the same concentration of LMB inhibited the nuclear export of cyclin B1 (supplementary Figure 8).
Figure 3.
Shuttling of cyclin A and cyclin E is insensitive to LMB. HeLa cells were fused and 20 nM LMB was added 40 min later. Nuclei were injected with 35 mg/ml cyclin A-GFP (A) or coinjected with 35 mg/ml cyclin E-GFP/CDK2K33R and 1 mg/ml Texas Red-conjugated dextran (MW 70,000) 20 min after addition of LMB (B). Fluorescence and DIC images were taken at the indicated time points after injection.
Cyclin A/Cdk2 Is Actively Imported into Nuclei without Proteins with a Recognizable NLS
The similarities in nuclear export of cyclins A and E raised the possibility that they might also be imported in a similar manner. Cyclin E had been shown to be transported into nuclei by the classical importin-α/β receptors (Moore et al., 1999), whereas cyclin B1 bound directly to importin-β and, unusually, was transported into nuclei independently of the small GTPase, Ran (Takizawa et al., 1999). The cyclin A sequence did not have a classical NLS, and its nuclear localization had been correlated with the ability to bind to its CDK. Thus, it was suggested that the cyclin A-CDK complex piggy-backed into the nucleus by binding to a protein with a NLS (Maridor et al., 1993). To test this hypothesis we analyzed cyclin A import in vitro. We expressed recombinant cyclin A-GFP with, and without, Cdk2, in baculovirus-infected Sf9 cells, and purified the proteins to homogeneity. We assayed the ability of the purified proteins to be imported into nuclei using the well-characterized in vitro nuclear transport system based on digitonin-permeabilized HeLa cells (Adam et al., 1992). Nuclei were incubated with an energy regenerating mix, plus Ran, NTF2, importin-β and the most abundant subtype of importin-α, Rch1. To eliminate any possible effects of Cdk2 kinase activity on nuclear membrane integrity, we routinely used a kinase deficient mutant of Cdk2, Cdk2K33R. We measured the amount of protein imported into nuclei by confocal fluorescence microscopy and immunoblotted nuclei that had imported the GFP-fusion proteins with an anti-GFP antibody to show that the nuclear fluorescence was from the full-length GFP-fusion proteins.
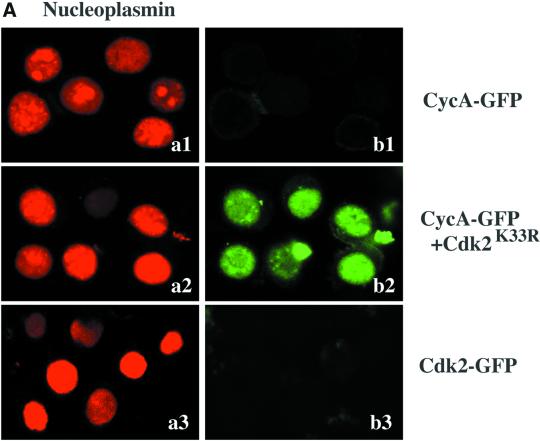
Using the in vitro import assay, we found that cyclin A-GFP/Cdk2K33R was imported into nuclei in a temperature-dependent and energy-dependent manner (Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results, but see Figure 5A), indicating that cyclin A-GFP/Cdk2K33R did not passively diffuse into nuclei. Most importantly, cyclin A/Cdk2 could be nuclearly imported without exogenous NLS-containing proteins, such as p107 or p21 (Figure 4A). To demonstrate that cyclin A-GFP/Cdk2 was imported via nuclear pore complexes, we treated nuclei with a dominant-negative mutant of importin-β (importin-β Δ N 44; Kutay et al., 1997). This mutant binds to the nuclear pore but cannot be released. It strongly blocks facilitated translocation of molecules through the NPC but only mildly inhibits passive diffusion (see Ribbeck and Gorlich, 2001). Importin-β (Δ N 44) blocked the nuclear import of cyclin A-GFP/Cdk2K33R (Figure 4A).
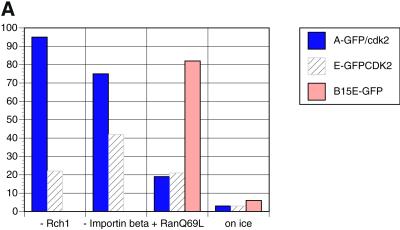
Figure 5.
Cyclin A-GFP/Cdk2(K33R) import is not dependent on Rch1/importin-β but is blocked by the IBB. (A) Permeabilized HeLa cells were incubated with cyclin A-GFP/Cdk2K33R, cyclin E-GFP/Cdk2K33R, or cyclin B15E-GFP. Control levels of nuclear import with exogenous Rch1, importin-β, and Ran GDP after 30 min of incubation at 22°C were set to 100%. Import was measured in parallel experiments, without exogenous Rch1, without importin-β, with 60 μM RanQ69L, or under standard control conditions but with incubation on ice instead of at 22°C. For each experimental condition, the average amount of each substrate imported into 100 nuclei was calculated and the mean of at least two independent experiments is shown. (B) Permeabilized HeLa cells were incubated in import buffer containing Rch1, importin-β, Ran GDP, and an energy-regenerating mixture, with TRITC-labeled nucleoplasmin (top), cyclin A-GFP/Cdk2K33R (middle), or Texas Red-labeled M9 domain (bottom) in the absence (left) or presence (right) of IBB (15 μM final concentration). In the experiments using TRITC-labeled M9 domain (a3 and b3), the protein was incubated under standard nuclear import assay conditions but with 300 nM transportin instead of Rch1 and importin-β in the reaction mixture. After 30 min of incubation at 22°C, nuclear import was visualized by confocal fluorescence microscopy. Transportin was unable to stimulate cyclin A-GFP/Cdk2K33R import (not shown).
Figure 4.
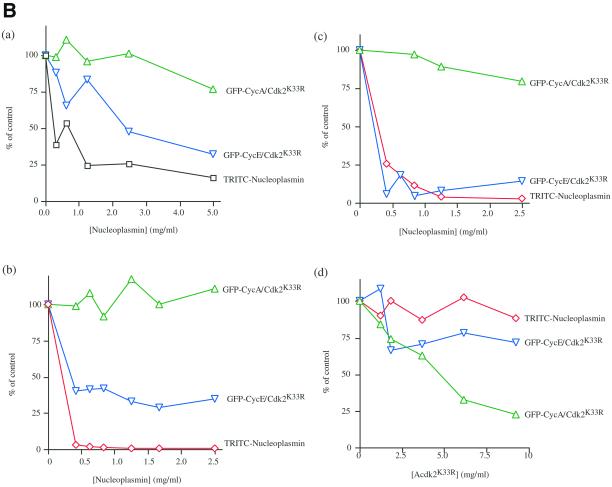
Nucleoplasmin and cyclin E-GFP/Cdk2K33R but not cyclin A-GFP/Cdk2K33R compete with one another for their nuclear import. Cyclin A/Cdk2K33R is imported into nuclei in vitro. Permeabilized HeLa cells were coincubated with Rch1, importin-β, Ran GDP, an energy-regenerating mixture, and TRITC-labeled nucleoplasmin (a1 and a2) and cyclin A GFP/Cdk2K33R (b1 and b2). Nuclear uptake in the absence (a1 and b1) or presence of 2.5 μM importin-β ΔN44 (a2 and b2) after 30 min of incubation at 22°C was visualized by confocal fluorescence microscopy. (B) Competition experiments among cyclin A, cyclin E, and nucleoplasmin. Permeabilized HeLa cells were coincubated in HeLa cell extract (4.4 mg/ml [a]) or Xenopus egg extract (3.3 mg/ml [b]), with an energy-regenerating mixture, TRITC-labeled nucleoplasmin, and cyclin A-GFP/Cdk2K33R or cyclin E-GFP/Cdk2K33R and increasing concentrations of unlabeled nucleoplasmin. After 30 min of incubation in HeLa extract at 37°C or Xenopus egg extract at 22°C, nuclear uptake of TRITC-labeled nucleoplasmin, cyclin A-GFP/Cdk2K33R, and cyclin E-GFP/Cdk2K33R were visualized by confocal fluorescence microscopy. c and d, permeabilized HeLa cells were incubated under standard conditions using purified proteins in place of cell extracts, with TRITC-labeled nucleoplasmin, cyclin A-GFP/Cdk2K33R, or cyclin E-GFP/Cdk2K33R in the presence of increasing amounts of unlabeled nucleoplasmin (c) or increasing amounts of unlabeled cyclin A/Cdk2(K33R) (d). The average amount of each substrate imported into 100 nuclei was calculated in at least two independent experiments and the mean of these values are shown.
Nuclear Import of Cyclin A Differs from That of Cyclins E and B1
Given that both cyclin A and cyclin E were predominantly nuclear, but both shuttled between the nucleus and the cytoplasm in the presence of LMB, we wished to determine whether they were imported into nuclei in a similar manner. The nuclear import of cyclin E/Cdk2 (and of cyclin E alone) had been shown to require importin-α/β (Moore et al., 1999); therefore, we compared its import in vitro with that of cyclin A. We used purified nuclei in the presence of an energy-regenerating mixture and Xenopus extract or HeLa cell extracts to provide all the components necessary for nuclear transport. We assayed the nuclear import of cyclin A-GFP/Cdk2K33R and cyclin E-GFP/Cdk2K33R in the presence of increasing amounts of unlabeled nucleoplasmin, a good importin-α/β cargo, to act as a competitor substrate. TRITC-labeled nucleoplasmin was coincubated with cyclin A-GFP/Cdk2K33R or cyclin E-GFP/Cdk2K33R in the assay to indicate when importin-α/β-dependent nuclear import had been saturated. Increasing the amount of nucleoplasmin in HeLa cell extract or Xenopus extract significantly reduced the nuclear import of cyclin E-GFP/Cdk2K33R but did not affect the import of cyclin A-GFP/Cdk2K33R (Figure 4B, a and b). These results were confirmed in competition experiments using purified import components in place of cell extracts; unlabeled nucleoplasmin competed with cyclin E-GFP/Cdk2K33R but did not significantly inhibit the nuclear import of cyclin A GFP/Cdk2K33R (Figure 4B). Moreover, unlabeled cyclin A/Cdk2K33R inhibited the import of cyclin A-GFP/Cdk2K33R but not that of nucleoplasmin or cyclin E-GFP/Cdk2K33R (Figure 4B, d).
These competition assays showed that cyclin A did not enter nuclei in the same manner as cyclin E. Therefore, we compared the nuclear import of cyclins A and E and tested whether it differed from that published for cyclin B1. Wild-type cyclin B1 had been shown to be predominantly cytoplasmic in interphase because it was rapidly exported from the nucleus. Thus, we assayed the import of a mutant form of cyclin B1–5E in which its nuclear export was compromised (Hagting et al., 1998; Takizawa et al., 1999). The nuclear import of cyclin B1–5E was unusual because it did not appear to require GTP hydrolysis by the small GTP-binding protein Ran (Takizawa et al., 1999), nor was it blocked by the Ran Q69L dominant-negative mutant of Ran (Takizawa et al., 1999; Figure 5A). Cyclin B1 import was stimulated by importin-β but insensitive to IBB, the amino-terminal domain of importin-α that binds importin-β and inhibits importin-α–mediated import α/β (Moore et al., 1999; Takizawa et al., 1999; Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results). In contrast, we found that cyclin A-GFP/Cdk2K33R import was blocked by RanQ69L and was not stimulated by exogenous importin-β (Figure 5A). Furthermore, IBB almost completely inhibited cyclin A/Cdk2K33R nuclear import in the presence of either purified nuclear import factors (Figure 5B) or cytosol (Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results). In these experiments, IBB acted as a specific inhibitor because it blocked the import of the importin-α/β substrate, nucleoplasmin, but not the transportin-dependent import of M9 (Figure 5B). Thus, cyclin A-CDK2 was imported in a manner distinct from either cyclin E or cyclin B1.
Cyclin A Needs to Bind to Cdk2 for Nuclear Import In Vitro
Previous studies using mutant forms of cyclin A transfected into cells showed a correlation between the ability of cyclin A to bind Cdk2 and its import into the nucleus (Maridor et al., 1993). However, this could either have reflected a requirement to bind Cdk2 or have been because the mutant cyclin A was also unable to be recognized by the nuclear import machinery. Therefore, we assayed the import of cyclin A-GFP in the absence of Cdk2K33R. We confirmed that cyclin A-GFP had not copurified with significant amounts of insect cell Cdk2 by immunoblotting the protein preparations with an anti-PSTAIRE antibody that recognized insect Cdk1 and Cdk2. We found that cyclin A-GFP could not be imported into nuclei in the absence of Cdk2 and that we could induce cyclin A-GFP import by adding purified Cdk2 (Figure 6A). A Cdk2K33R-GFP (Figure 6A) fusion protein nor GFP alone was not actively imported into nuclei, indicating that cyclin A-GFP/Cdk2K33R was not imported because of a NLS on Cdk2.
Figure 6.
Cyclin A needs to bind a Cdk for nuclear import in vitro and in vivo. (A) In vitro import. Permeabilized HeLa cells were coincubated with Rch1, importin-β, Ran GDP, an energy-regenerating mixture, and TRITC-labeled nucleoplasmin (a1–3), cyclin A-GFP (b1), or cyclin A-GFP that had been preincubated with purified Cdk2K33R for 60 min at 37°C (b2) or a Cdk2 K33R-GFP fusion protein (b3). After 30 min of incubation at 22°C, nuclear import was visualized by confocal fluorescence microscopy. (B) In vivo import. HeLa cells were transfected with non-Cdk–binding cyclin mutants; cyclin AR211K-GFP (top left), cyclin ER130A-GFP (top right), wild-type cyclin B1-GFP (bottom left), or cyclin B1R202K-GFP (bottom right) under the control of the cytomegalovirus promoter. Subcellular localizations of the cyclin-GFP chimaeras were visualized 24 h later in the living cells by fluorescence microscopy. Thirty minutes before observation, 20 nM LMB was added to HeLa cells transfected with cyclin B1-GFP constructs to allow the nuclear import of cyclin B1-GFP to be visualized.
Cyclins A and B1, but Not Cyclin E, Need to Bind to Cdk2 for Nuclear Import In Vivo
To confirm this result in vivo, we generated GFP-fusion proteins from mutants of cyclin A that cannot bind to a Cdk. The most well characterized of these have been point mutations of arginine 211 in the sequence MRAIL, part of helix 1 of the cyclin box, that has been shown to form a buried salt bridge with aspartic acid 240 to stabilize helices 1 and 2 (Jeffrey et al., 1995). Mutating R211 to K or A prevented cyclin A from binding Cdk2 in vitro (Kobayashi et al., 1992) and, in agreement with our in vitro results, this caused cyclin A to remain in the cytoplasm (Figure 6B). Unlike cyclin A-GFP, cyclin E-GFP did not need to bind its Cdk to become nuclear; cyclin E was imported into the nucleus in the absence of Cdk2 (Figure 6B), and a cyclin E mutant unable to bind Cdk2, cyclin ER130A (Clurman et al., 1996) was imported into nuclei in vivo (Figure 6B). Cyclin B1 resembled cyclin A in that it was not imported into nuclei in the absence of its Cdk in vivo; mutating R202 in the MRAIL motif to lysine or alanine markedly reduced its import into nuclei of interphase cells treated with LMB (Figure 6B).
Mutations in the Hydrophobic Patch Alter the Binding between Cyclin A and Cdk2
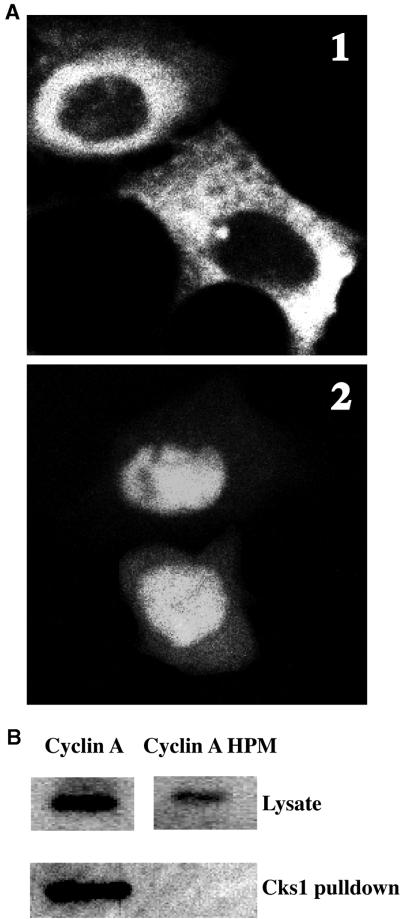
We were anxious to exclude the possibility that altering R211 might have had effects other than preventing Cdk binding. In particular, we thought it might alter the “hydrophobic patch” region on the other face of the MRAIL helix that was identified as a potential substrate interaction motif (Schulman et al., 1998; Cross et al., 1999) and bind to substrates with a “Cy” motif (consensus RxL). Therefore, we attempted to distinguish the effects of altering the hydrophobic patch from those due to preventing Cdk binding. We made three mutations in the hydrophobic patch, M210A, L214A, and W217A (Schulman et al., 1998), tagged the hydrophobic patch-mutated protein with the myc epitope, transfected it into tissue culture cells, and analyzed its localization by immunofluorescence. To our surprise, we found that the M/L/W→A hydrophobic patch mutant of cyclin A was cytoplasmic (Figure 7A); as was a GFP-tagged version of this mutant (Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results). The hydrophobic patch mutant of cyclin A was originally described as being nuclear (Schulman et al., 1998), but in that paper the authors had cotransfected the mutant with Cdk2. Therefore, we cotransfected Cdk2 with the hydrophobic patch mutant and found that there was a substantial increase in the amount of cyclin A in the nucleus (Figure 7A).
Figure 7.
Mutations in the hydrophobic patch of cyclin A alter both its binding to Cdk2 and its localization. (A) Localization. HeLa cells were transfected with myc epitope-tagged cyclin A (M210A, L214A, W217A) alone (top) or with CDK2 (bottom). Cells were fixed and processed for immunofluorescence using the anti-myc epitope antibody. (B) Cdk binding. Myc epitope-tagged cyclin A and cyclin A hydrophobic patch mutants were transfected into HeLa cells. Eighteen hours after transfection, cell lysates were incubated with p9Cks1-Sepharose beads. The amounts of myc-cyclin A and myc-cyclin A HPM bound to p9Cks1 was compared by immunoblotting using an anti-myc epitope antibody. Top, the amounts of myc-cyclin A and myc-cyclin A hydrophobic patch mutant expressed in each cell lysate.
These results indicated either that the hydrophobic patch mutations had blocked nuclear import or that they had interfered with cyclin A binding to its Cdk. Therefore, we tested the ability of the cyclin A hydrophobic patch mutant to bind to Cdk2. In agreement with its intracellular localization, we found that the M/L/W→A hydrophobic patch mutant had a markedly reduced affinity for Cdk2 when transfected into cells (as determined by assaying its ability to bind to CDKs using a p9Cks1 affinity column; Figure 7B).
DISCUSSION
Cyclin A-, B1-, and E-Cdk Complexes All Shuttle between the Nucleus and the Cytoplasm
In this paper, we have shown that all the cyclin/Cdk complexes that are essential for S phase and for mitosis are highly dynamic in living cells. The S phase cyclin/Cdks, cyclin E/Cdk2 and cyclin A/Cdk2, are constantly shuttling between the nucleus and the cytoplasm, although they appear to be nuclear by immunofluorescence. The major mitotic cyclin/Cdk complex, cyclin B1/Cdk1, also shuttles but is predominantly cytoplasmic (Hagting et al., 1998; Toyoshima et al., 1998; Yang et al., 1998). Remarkably, the localizations of the different cyclins appear to be mediated by distinct pathways. The nuclear export of cyclin B1 is more rapid than its import, whereas the converse is true for cyclin A/Cdk2 and cyclin E/Cdk2. In this respect, it is probably significant that the export of cyclins A and E is not sensitive to LMB, in contrast to that of cyclin B1 (Hagting et al., 1998; Toyoshima et al., 1998; Yang et al., 1998). Cyclin A- and cyclin E-Cdk complexes are not the only proteins whose export is not blocked by LMB. Proteins involved in RNA processing and transport, such as hnRNPs with an M9 motif that bind to transportin (Nakielny and Dreyfuss, 1999), the hnRNP K protein (Michael et al., 1997), and RNA helicase A (Tang et al., 1999), are also insensitive to LMB. We have not yet identified the region(s) of cyclins A and E that are necessary for their export. However, cyclin A, cyclin E, and CDK2 have no M9 motif or a “K nuclear shuttling domain,” and neither cyclin A nor CDK2 have been found associated with RNA, although cyclin E does bind components of the pre-mRNA–splicing machinery (Seghezzi et al., 1998). Recently, β-catenin has been demonstrated to be exported via two distinct pathways, one of which is insensitive to LMB (Eleftheriou et al., 2001; Wiechens and Fagotto, 2001)
Our observation that both cyclin A- and cyclin E-Cdk complexes are able to shuttle between the nucleus and the cytoplasm means that they could directly phosphorylate both nuclear and cytoplasmic substrates in their roles as regulators of both DNA and centrosome duplication (Hinchcliffe et al., 1999; Lacey et al., 1999; Meraldi et al., 1999). Indeed, cyclin A has been detected on centrosomes in prophase cells by immunofluorescence (den Elzen and Pines, 2001; Pagano et al., 1992).
Cyclins A, B1, and E All Have Different Nuclear Import Characteristics
Despite the similarities in their nuclear export, cyclin A/Cdk2 and cyclin E/Cdk2 are imported by different pathways, and both of these are distinct from the pathway that imports cyclin B1. Cyclin E binds to importin-α/β, and cyclin B can bind directly to importin-β (Moore et al. 1999), whereas cyclin A/Cdk2 is transported by a pathway that does not require exogenous importin-α or -β but is energy and temperature dependent and inhibited by both importin-β (Δ 78 44) and wheat germ agglutinin. Moreover, cyclin A/Cdk2K33R does not require additional NLS-containing carrier proteins to piggy-back into the nucleus via the importin-α/β pathway.
In the course of these studies, we also found that mutations in the hydrophobic patch of cyclin A have profound effects on Cdk binding and on the subcellular localization of the cyclin. These mutations have been thought to be specific for a substrate selection region on the cyclin (Schulman et al., 1998; Cross et al., 1999). Thus, it will be important to control for effects on Cdk binding and localization in future studies that use hydrophobic patch mutants to investigate substrate selection.
In vivo, mutants of both cyclin A and mammalian cyclin B1 that cannot bind their Cdk cannot be imported into nuclei. Non-Cdk–binding mutants of chicken cyclin B3 also remain in the cytoplasm (Gallant and Nigg, 1994). However, our results with cyclin B1 appear to be at variance with studies by Moore et al. (1999), who found that Xenopus cyclin B1 does not need Cdk1 for nuclear import. Moore et al. (1999) used cyclin B1 lacking the first 127 amino acids, whereas we used full-length cyclin B1. Thus, it is possible that the amino terminus of cyclin B1 obscures the region to which importin-β binds until the cyclin binds to its Cdk.
The nuclear import of cyclin A-GFP/Cdk2 is not stimulated by additional importin-α, importin-β, or transportin (Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results). Thus, cyclin A-GFP/Cdk2K33R is transported via receptors that remain associated with preparations of HeLa cell nuclei. Although this could be importin-β itself, our data indicate that it is likely to be another member of the importin-β receptor family (Görlich et al., 1997), because cyclin A-GFP/Cdk2K33R does not compete for import with either nucleoplasmin or NLS-BSA under conditions in which importin-β is limiting. If cyclin A/CDK complexes do use importin-β, then the pathway is markedly different from that used to import cyclin B1, because we found that cyclin A/Cdk2 import is blocked by Ran Q69L and IBB, both of which fail to inhibit cyclin B1 transport (Takizawa et al., 1999). We do not think that the receptor is transportin, because cyclin A/Cdk2 import is not stimulated by exogenous transportin, and it is inhibited by levels of IBB at which M9 substrates are still imported.
Alternatively, importin-β might still be the receptor for cyclin A/Cdk2 import, but an adaptor protein, distinct from Rch1 may be required. Such an adaptor would have to remain associated with our preparations of nuclei. We have excluded the importin-α subtype NPI-1, which can import classical NLS-substrates and Stat1 via separate domains (Sekimoto et al., 1997), because a dominant-negative mutant of NPI-1 (NPI-1 78–538, kind gift of T. Sekimoto) does not block cyclin A-GFP/Cdk2 import (Jackman, Kubota, Den Elzen, Hagting, and Pines, unpublished results). A third importin-α subfamily member (importin-α3/Qip1/hSRPα; Kohler et al., 1997; Miyamoto et al., 1997; Nachury et al., 1998) is unlikely to be the import receptor for cyclin A/Cdk2 because it has a highly tissue-specific distribution (Nachury et al., 1998).
Cyclin/Cdk Regulation by Nuclear Import and Export
Our results raise the intriguing possibility that different cyclin/Cdks could be regulated by modulating their import and export from the nucleus. In addition to controlling their localization, this might also affect their abundance. For example, cyclin D1 is imported into the nucleus in G1 phase, possibly by piggy-backing on the p21Cip1/p27Kip1 Cdk-inhibitors (Diehl and Sherr, 1997; LaBaer et al., 1997), and is then exported in S phase concomitant with phosphorylation by GSK3 and subsequent proteolysis (Diehl et al., 1998). We have not yet determined whether the shuttling of short-lived proteins such as cyclin E and cyclin D1 or of cyclins A and B1 that are unstable in G1 phase cells is also relevant to their proteolysis.
In the early Drosophila embryo, cyclin A remains in the cytoplasm throughout S phase and only translocates into the nucleus at prophase (Lehner and O'Farrell, 1989). It will be fascinating to discover whether this behavior reflects changes in the recognition of cyclin A by the nuclear import machinery or a change in the cyclin A/Cdk import machinery itself. Studies of other proteins involved in the G1/S transition, such as the E2F family members (Muller et al., 1997; Verona et al., 1997), as well as components of the DNA replication machinery such as Cdc6 (Saha et al., 1998; Petersen et al., 1999) and the MCM proteins (Chen et al., 1992; Yan et al., 1993), have also revealed strong regulation at the level of their nuclear localization. The identification of the nuclear import mechanisms that are used by specific cyclins, and the elucidation of whether and how these pathways are altered in different phases of the cell cycle, may illuminate a novel aspect of cell cycle control.
Figure.
Supplemental Figure 8. LMB treatment of fused cells inhibits cyclin B1 export but not cyclin A shuttling. (A) HeLa cells were injected with an expression construct for cyclin B1-YFP and fused as described in MATERIALS AND METHODS 3 h later. Twenty minutes after fusion, 20 nM LMB was added to the cells. Fluorescence pictures were taken at different time points after LMB addition (top). (B) HeLa cells were fused, and after 20 min, 20 nM LMB was added. Cyclin A-GFP protein was injected into the nucleus 30 min after the addition of LMB, and fluorescence pictures were taken at different time points after injection.
Figure.
Supplemental Figure 9. Graphs show two separate experiments quantitating loss and appearance of cyclin A-GFP (fluorescence, arbitrary units, and corrected for background) in nuclei of binucleate fused cells. Black lines with squares show loss of cyclin A-GFP from nuclei into which it was injected. Lines with filled circles show increase in cyclin A-GFP in nuclei not injected with cyclin A-GFP. Movement of cyclin A-GFP between nuclei in two fused cells without LMB treatment (A) and with LMB treatment (B).
ACKNOWLEDGMENTS
We thank Dirk Görlich for his generosity in supplying reagents and for helpful suggestions. We thank Helena Pinwica-Worms for supplying recombinant baculovirus encoding human Cdk2K33R, Bruce Clurman for the cyclin ER130A mutant, and Brenda Schulmann for cDNA encoding the cyclin A hydrophobic patch mutant. We are also grateful to Lucy Perkins for maintaining insect cell cultures and to Christina Karlsson for constructing the cyclin E-GFP fusion protein. Y.K. was supported by a Research Fellowship and by a Cancer Research Campaign grant to Ron Laskey, SP1961/O502, N.d.E. was a Commonwealth Universities scholar, and A.H. was supported by an EU Training and Mobility Research network grant. M.J. and J.P. were supported by Cancer Research Campaign grant SP2143/0103 to J.P.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc. 01–07–0361. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–07–0361.
REFERENCES
- Adam SA, Sterne-Marr R, Gerace L. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 1992;219:97–110. doi: 10.1016/0076-6879(92)19013-v. [DOI] [PubMed] [Google Scholar]
- Belyaev AS, Roy P. Development of baculovirus triple and quadruple expression vectors: co-expression of three or four bluetongue virus proteins and the synthesis of bluetongue virus-like particles in insect cells. Nucleic Acid Res. 1993;21:1219–1223. doi: 10.1093/nar/21.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–90. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hennessy KM, Botstein D, Tye BK. CDC46/MCM5, a yeast protein whose subcellular localization is cell cycle-regulated, is involved in DNA replication at autonomously replicating sequences. Proc Natl Acad Sci USA. 1992;89:10459–10463. doi: 10.1073/pnas.89.21.10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by Cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1790. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L, Elledge SJ, Harper JW. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr Biol. 1998;8:65–68. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- Cross FR, Yuste-Rojas M, Gray S, Jacobson MD. Specialization and targeting of B-type cyclins. Mol Cell. 1999;4:11–39. doi: 10.1016/s1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–36. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Sherr CJ. A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftheriou A, Yoshida M, Henderson BR. Nuclear export of human beta-catenin can occur independent of CRM1 and the adenomatous polyposis coli tumor suppressor. J Biol Chem. 2001;276:25883–25888. doi: 10.1074/jbc.M102656200. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Spottswood MR. A new human p34 protein kinase, CDK2, identified by complementation of a cdc28 mutation in Saccharomyces cerevisiae, is a homolog of Xenopus Eg1. EMBO J. 1991;10:2653–2659. doi: 10.1002/j.1460-2075.1991.tb07808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P, Fry AM, Nigg EA. Protein kinases in the control of mitosis: focus on nucleocytoplasmic trafficking. J Cell Sci Suppl. 1995;19:21–28. doi: 10.1242/jcs.1995.supplement_19.3. [DOI] [PubMed] [Google Scholar]
- Gallant P, Nigg EA. Identification of a novel vertebrate cyclin: cyclin B3 shares properties with both A- and B-type cyclins. EMBO J. 1994;13:595–605. doi: 10.1002/j.1460-2075.1994.tb06297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard F, Strausfeld U, Fernandez A, Lamb NJC. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on Cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Luca FC, Ruderman JV. The requirements for protein synthesis and degradation, and the control of destruction of cyclins A and B in the meiotic and mitotic cell cycles of the clam embryo. J Cell Biol. 1992;116:707–724. doi: 10.1083/jcb.116.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Stewart E, Poon R, Adamczewski JP, Gannon J, Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32Cdk2 protein kinase subunits. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-Cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Krude T, Jackman MR, Pines JN, Laskey RA. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;87:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S-i, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. Expression and function of Drosophila cyclin A during embryonic cell cycle progression. Cell. 1989;56:957–968. doi: 10.1016/0092-8674(89)90629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maridor G, Gallant P, Golsteyn R, Nigg E. Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with CDK catalytic subunits. J Cell Sci. 1993;106:535–544. doi: 10.1242/jcs.106.2.535. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- Moore JD, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Ryder UW, Lamond AI, Weis K. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, et al. Targeted disruption of skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclins A and B are differentially located in the cell and undergo cell cycle dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–3130. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Marr J, Madine MA, Rowles A, Blow JJ, Gautier J, Laskey RA. Interaction of Xenopus Cdc2-cyclin A1 with the origin recognition complex. J Biol Chem. 2000;275:4239–4243. doi: 10.1074/jbc.275.6.4239. [DOI] [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, Lawlis SJ, Hou ZH, Hendricks M, Parvin JD, Dutta A. Human CDC6/Cdc18 associates with Orc1 and cyclin-Cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Knoblich JA, Richardson H, Lehner CF. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 1995;9:1327–1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Descombes P, Chevalier S, Rempel RE, Adamczewski J, Maller JL, Hunt T, Blow JJ. Both cyclin A and cyclin E have S phase promoting (Spf) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- Takizawa CG, Weis K, Morgan DO. Ran-independent nuclear import of cyclin B1-Cdc2 by importin beta. Proc Natl Acad Sci USA. 1999;96:7938–7943. doi: 10.1073/pnas.96.14.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, McDonald D, Middlesworth T, Hope TJ, Wong-Staal F. The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain. Mol Cell Biol. 1999;19:3540–3550. doi: 10.1128/mcb.19.5.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr199 by cyclin-dependent kinase 2-cyclin E, and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai L-H, Harlow E, Meyerson M. Isolation of the human Cdk2 gene that encodes the cyclin A and adenovirus E1A-associated p33 kinase. Nature. 1991;353:174–177. doi: 10.1038/353174a0. [DOI] [PubMed] [Google Scholar]
- Verona R, Moberg K, Estes S, Starz M, Vernon JP, Lees JA. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Penfold S, Tang X, Hattori N, Riley P, Harper JW, Cross JC, Tyers M. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr Biol. 1999;9:1191–1194. doi: 10.1016/S0960-9822(00)80024-X. [DOI] [PubMed] [Google Scholar]
- Wiechens N, Fagotto F. CRM1- and Ran-independent nuclear export of beta-catenin. Curr Biol. 2001;11:18–27. doi: 10.1016/s0960-9822(00)00045-2. [DOI] [PubMed] [Google Scholar]
- Winston JT, Chu C, Harper JW. Culprits in the degradation of cyclin E apprehended. Genes Dev. 1999;13:2751–2757. doi: 10.1101/gad.13.21.2751. [DOI] [PubMed] [Google Scholar]
- Won KA, Reed SI. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- Yan H, Merchant AM, Tye BK. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- Yang J, Bardes ES, Moore JD, Brennan J, Powers MA, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Pines J, Ryan K, Siemering KR, Haseloff J, Gurdon JB. An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development. 1996;122:3719–3724. doi: 10.1242/dev.122.12.3719. [DOI] [PubMed] [Google Scholar]
- Zindy F, Lamas E, Chenivesse X, Sobczak J, Wang J, Fesquet D, Henglein B, Brechot C. Cyclin A is required in S phase in normal epithelial cells. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]