Abstract
Association of motor proteins with organelles is required for the motors to mediate transport. Because axoplasmic organelles move on actin filaments, they must have associated actin-based motors, most likely members of the myosin superfamily. To gain a better understanding of the roles of myosins in the axon we used the giant axon of the squid, a powerful model for studies of axonal physiology. First, a ∼220 kDa protein was purified from squid optic lobe, using a biochemical protocol designed to isolate myosins. Peptide sequence analysis, followed by cloning and sequencing of the full-length cDNA, identified this ∼220 kDa protein as a nonmuscle myosin II. This myosin is also present in axoplasm, as determined by two independent criteria. First, RT-PCR using sequence-specific primers detected the transcript in the stellate ganglion, which contains the cell bodies that give rise to the giant axon. Second, Western blot analysis using nonmuscle myosin II isotype-specific antibodies detected a single ∼220 kDa band in axoplasm. Axoplasm was fractionated through a four-step sucrose gradient after 0.6 M KI treatment, which separates organelles from cytoskeletal components. Of the total nonmuscle myosin II in axoplasm, 43.2% copurified with organelles in the 15% sucrose fraction, while the remainder (56.8%) was soluble and found in the supernatant. This myosin decorates the cytoplasmic surface of 21% of the axoplasmic organelles, as demonstrated by immunogold electron-microscopy. Thus, nonmuscle myosin II is synthesized in the cell bodies of the giant axon, is present in the axon, and is associated with isolated axoplasmic organelles. Therefore, in addition to myosin V, this myosin is likely to be an axoplasmic organelle motor.
INTRODUCTION
Biochemical association between organelles and the microtubule motors, kinesin and cytoplasmic dynein, has provided some of the more convincing evidence that these motors play a role in intracellular transport (Schnapp and Reese, 1989; Schnapp et al., 1992; Yamazaki et al., 1995; Moreira et al., 1998). Strong evidence for a physiological association of kinesin with organelles was obtained by the retention of kinesin on organelles after extraction with potassium iodide (Schnapp et al., 1992). Immunogold labeling has been used to link specific motors with organelles (Yamazaki et al., 1995; Moreira et al., 1998).
The squid giant axon has provided a unique and powerful model system in which to study the physiology of axoplasmic transport. Indeed, the original observation of microtubule-based transport was made in squid axons (Brady et al., 1982), while biochemical identification of the first microtubule-based motor, kinesin, was achieved using squid optic lobe (Vale et al., 1985). Subsequently, squid axoplasmic organelles were shown to move on actin filaments, implicating myosins as additional transport motors (Kuznetsov et al., 1992; Bearer et al., 1993; Langford et al., 1994). Although the myosin superfamily is relatively well characterized, those myosins that associate with organelles have not yet been well defined.
In squid, only two myosins, myosin V and siphon muscle myosin II, have been previously identified by sequence analysis (Medeiros et al., 1998; Matulef et al., 1998; Molyneaux et al., 2000). One of these, myosin V, has been localized to the endoplasmic reticulum (ER), and ER movements are blocked by a peptide antibody raised against the tail domain of this myosin (Tabb et al., 1998; Molyneaux et al. 2000). Thus, myosin V appears to play a role in ER trafficking. In addition, there is evidence that at least one other myosin is involved with organelles. Our earlier studies demonstrated that an antimyosin antibody detected another protein, larger than myosin V, which copurified with organelles. (Bearer et al., 1993). This myosin antibody also labeled organelles by immunocytochemistry, suggesting that this second myosin, in addition to myosin V, is associated with axoplasmic organelles.
This second myosin has proven difficult to characterize for a number of reasons. First, the organelle fraction contains insufficient amounts of protein to obtain peptide sequences that could be useful in its identification. The relatively small amounts of protein in axons limit biochemical characterization. In fact, conventional squid kinesin, the founding member of this diverse family of motors, could not be obtained in quantity from axoplasm but had to be biochemically purified and characterized from optic lobe (Vale et al., 1985). Second, the antiscallop muscle myosin II antibody was not a reliable tool for the identification of this ∼220 kDa myosin. Although it recognized one prominent band of ∼220 kDa in a preparation of isolated organelles, in the axon it recognized at least five other bands by Western blot.
In this study, we applied a strategy similar to that used to isolate and characterize squid kinesin (Vale et al., 1985). First, we extracted myosins from optic lobe, which provides sufficient amounts of neural tissue; then we developed probes for these myosins that can be applied to the axon. A high molecular weight protein was obtained from optic lobe using a modification of an established myosin purification protocol. This protein was identified as a myosin through sequence analysis, and probes were generated to determine its presence in axoplasm and to study its distribution in the neuron.
MATERIALS AND METHODS
Materials
Squid (Loligo pealei) were obtained live from the Marine Resources Center, Marine Biological Laboratory, Woods Hole, MA. Oligonucleotides were synthesized by Integrated DNA Technologies, Coralville, IA. DNA sequencing of plasmid minipreps was performed by Davis Sequencing, Davis, CA. For all experiments, squid optic lobes and axons were dissected and stored in liquid nitrogen until use. Antineurofilament antibody was a generous gift from Philip Grant, National Institutes of Health, Bethesda, MD (Grant et al., 1995).
Purification of Squid Nonmuscle Myosin II
Squid nonmuscle myosin II was purified using a protocol modified from See and Metuzals (1976). Each step of the purification was carried out on ice or at 4°C. Squid optic lobes (100 g) were thawed in four volumes of ice-cold high-salt buffer (0.6 M KCl, 3 mM beta-mercaptoethanol, 5 mM MgCl2, 20 mM imidazole, pH 7.0) containing a protease inhibitor cocktail (10 mM benzamidine, 10 mM leupeptin, 10 mM pepstatin A, 10 mM aprotinin, and 10 mM phenanthroline), and the mixture was homogenized by hand using a glass dounce homogenizer. The homogenate was stirred for 30 min, followed by centrifugation at 30,000 × g for 15 min. The resulting supernatant was clarified by high-speed centrifugation at 100,000 × g for 90 min. The high-speed supernatant was diluted to a 0.1 M KCl final concentration by adding five volumes of ice-cold 2 mM MgCl2, and the pH of the solution was adjusted to 6.4 with 1 M potassium acetate buffer (pH 4.8). The sample was stirred for 15 min to precipitate actomyosin, and the precipitate was pelleted by centrifugation at 30,000 × g for 15 min. The pellet was resuspended in 5 ml S-500 buffer (25 mM HEPES, 600 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 2 mM DTT, pH 8.0) in the presence of 10 mM ATP and was dounced with a 10-ml homogenizer. The suspension was clarified at 100,000 × g for 1 h, and the resulting supernatant (S4) was placed over a 1.5 × 100 cm Sephacryl-500 gel filtration column (Amersham Pharmacia Biotech, Piscataway, NJ). Fractions (3 ml) were collected at a flow rate of 0.2 ml/min. Samples of each purification step were analyzed by Coomassie-stained SDS-PAGE. Peak fractions containing a ∼220 kDa myosin were pooled. Peptide sequences were obtained from the purified neural myosin by excising bands from SDS-PAGE gels, followed by limited proteolysis and Edman degradation as previously described (Medeiros et al., 1998).
Cloning and Sequencing of Squid Nonmuscle Myosin II
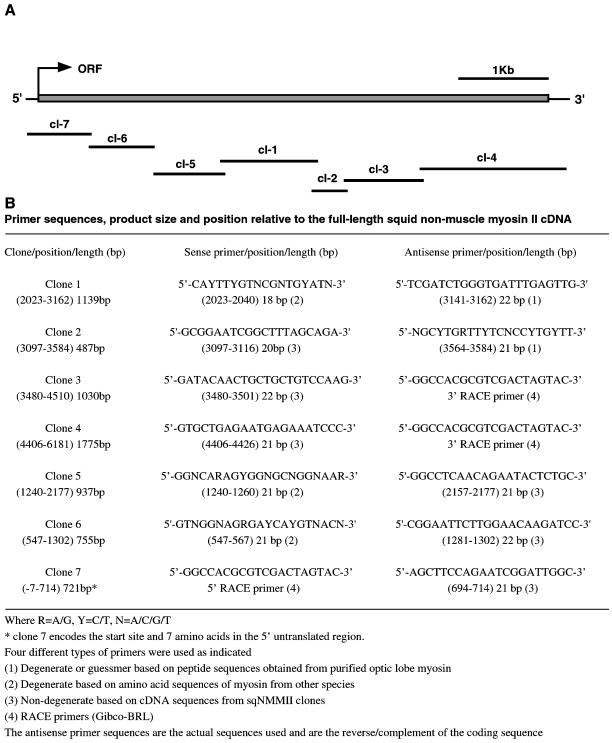
The full-length open reading frame of the squid nonmuscle myosin II cDNA was determined by seven overlapping clones obtained by PCR. PCR primers were designed to amplify squid nonmuscle myosin II transcripts. These primers were based on conserved sequences in the myosin ATP and actin-binding sites and on peptide sequences obtained from purified squid nonmuscle myosin II. Two primers 5′-CAYTTYGTNCGNTGYATN-3′ (sense) and 5′-TCGATCTGGGTGATTTGAGTTG-3′ (antisense) (Y = T/C, N = A/C/T/G) yielded a single band of ∼1.2 kb, as determined by ethidium bromide stained agarose gel electrophoresis using a 1-kb ladder as size standard (Life Technologies, Grand Island, NY). The PCR product was cloned using an Invitrogen TOPO-TA cloning kit (Invitrogen, Carlsbad, CA), and 10 of the resulting clones were screened for a 1.2-kb insert by Eco-RI digestion. Two positive clones were sequenced in both the forward and reverse directions using M13R and M13F primers that bind sequences internal to the vector and that flank the insertion site. Full-length sequencing of each of these inserts revealed two identical 1139 base-pair sequences. By fasta searches using gcg software, the insert sequences were determined to share sequence identity with other nonmuscle myosin IIs in the GenBank database. This initial cDNA was extended by a series of PCR reactions using a combination of gene specific primers, primers based on nonmuscle myosin II consensus sequences, and standard techniques in the rapid amplification of cDNA ends (RACE) (Life Technologies, Grand Island, NY). These primers are listed in Figure 2, and their position relative to the full-length squid nonmuscle myosin II cDNA sequence is shown in Figure 3.
Figure 2.
Cloning of squid nonmuscle myosin II. (A) Schematic representation of overlapping cDNA clones that encode the full-length squid nonmuscle myosin II cDNA nucleotide sequence (Accession No. 406790), and (B) table of sqNMMII PCR primers and their corresponding clones. Clones are numbered in the order that they were acquired (clones 1–7). The position along the full-length nonmuscle myosin II cDNA of each clone and primer is given, as well as their lengths in base pairs.
Figure 3.
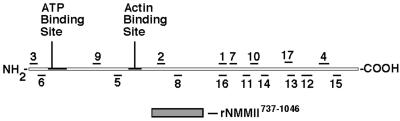
Position of sqNMMII peptide sequences and the αMII antibody domain along the full-length sqNMMII amino acid sequence. Peptide sequences are numbered (1–17) and refer to the p220 peptide sequences listed in Table 1. A polyclonal antibody was generated against a 309 amino acid sequence that maps to amino acids 737-1046 of squid nonmuscle myosin II (rNMMII737–1046).
Antibody Production
A 927 base-pair DNA fragment encoding a 309-aa sequence of the squid nonmuscle myosin II heavy chain (amino acids 737-1046) was ligated in frame into a Qiagen expression vector to create a His tag construct (Qiagen, Chatsworth, CA). The recombinant nonmuscle myosin II (rNMMII737–1046) was purified using a standard protocol (Qiagen). The rNMMII737–1046 was further purified by electrophoresis using a 12% SDS-PAGE curtain gel; the resulting protein band was excised from the gel and was used as the immunogen for antibody production in rabbits. Antisera were screened and affinity purified on rNMMII737–1046 affinity columns.
Antibodies to myosin V were also generated. A single peptide sequence, which was obtained from purified squid myosin V (DeGiorgis, Reese, and Bearer, unpublished results) that maps to the myosin V head domain (195KVLASNPIMESIGNAK211) was synthesized, coupled to ovalbumin, and used as an antigen to produce a polyclonal antibody in rabbit (Covance, Oakland, CA). Antisera were affinity purified against the peptide sequence by column chromatography. The affinity purified antimyosin V antibody (αMV) recognized 0.5 μg of purified myosin V at a 1:2000 dilution.
Preparation of Squid Optic Lobe Homogenates and Axoplasm for Western Blot Analysis
An optic lobe homogenate was prepared by homogenizing 5 g thawed squid optic lobes in six volumes ½× buffer (Schnapp et al., 1992) containing 10 mM each of the following protease inhibitors: benzamidine, leupeptin, pepstatin A, aprotinin, and phenanthroline. The homogenate was aliquoted into 1.5 ml Eppendorf tubes and centrifuged at 14,000 rpm for 10 min at 4°C. The resulting supernatant was drawn off and saved as the homogenate. Squid axoplasmic samples were prepared by extruding axoplasm from 10 thawed axons (∼ 50 μl) into five volumes of ½× buffer containing protease inhibitors. Gel sample buffer (6X) was added to each sample to a 1X final concentration (0.0625 M Tris (pH 6.8), 10% glycerol, 1% SDS, 1% beta-mercaptoethanol, 0.05% bromophenol blue), and the samples were boiled for 5 min before gel electrophoresis.
Coomassie Gels and Western Blot Analysis
For Western blots, proteins were transferred from 8.5% acrylamide gels onto nitrocellulose membranes. The membranes were blocked in 5% powdered milk in 1X Tris buffered saline (TBS) pH 7.4 for 1 h at room temperature. Primary antibodies were diluted to 1:500 in wash buffer (3% powdered milk, in TBS containing 0.2% Tween-20, pH 7.4), and the nitrocellulose blots were incubated for 90 min at room temperature in primary antibody solution. Blots were washed 3 × 10 min in wash buffer, then incubated in wash buffer containing 1:5000 alkaline phosphatase conjugated antirabbit IgG (Boehringer Mannheim, Indianapolis, IN). Blots were washed 3 × 10 min in TBS and developed with NBT/BCIP solution (Kirkgaard and Perry Laboratories, Gaithersburg, MD).
RT-PCR Assay for Squid Nonmuscle Myosin II Transcripts in Squid Neural Tissues
The expression of nonmuscle myosin II transcripts in squid neural tissues was assayed using tissue-specific cDNAs and gene specific primers. Total RNA was extracted from squid optic lobe and stellate ganglia using the Trizol Reagent method (Life Technologies, Grand Island, NY). RNA (2 μg) was treated with 2 U amplification grade DNAse I (15 min at 25°C) in 10 μl 1X reaction buffer (20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2) to remove potential genomic DNA contamination (Life Technologies, Grand Island, NY). The DNAse I was heat-inactivated at 65°C for 15 min in the presence of 2 mM EDTA. RNA (0.5 μg) was reverse transcribed with random hexamers and Superscript II reverse transcriptase at 42°C for 50 min, then treated with 1 μl RNAse H mix at 37°C for 30 min (Life Technologies, Grand Island, NY). The resultant cDNA was used as template for PCR reactions along with two squid, nonmuscle myosin II, gene specific primers. These primers correspond to a unique sequence in the squid nonmuscle myosin II tail domain (nt 4878–5239; 361 base-pair product) 5′-CTTGAACCAATTGTCTGAGCAACTG-3′ (sense) and 5′CCAACAGGTCTTCTAATTCGG-3′ (antisense). As a positive control, PCR reactions were carried out with gene specific primers for squid kinesin (nt 397–801; 404 base-pair product) 5′-ATATCGTCCTCAAACAACGCC-3′ (sense), 5′-CTCCA-AGTTTTCGTCCATTCC-3′ (antisense), and actin (608 base-pair product) 5′-GGAGAAGATCTGGCATCACACC-3′ (sense), 5′-GAAGTTCCTTCGAAACGAAAGG-3′ (antisense). Parallel PCR reactions were carried out for each primer set using cDNA in which reverse transcriptase was omitted from the first strand reaction. PCR reactions were performed in a Perkin Elmer-Cetus thermocycler at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min using 30 amplification cycles. PCR products (20 μl) were separated on 1% agarose gels containing ethidium bromide and photographed at f-22 for 1 s. Squid nonmuscle myosin II PCR products from both optic lobe and stellate ganglia were cloned using an Invitrogen TOPO TA cloning kit (Invitrogen, Carlsbad, CA), minipreped with Qiagen plasmid miniprep kit (Qiagen), and sequenced to verify their identity.
Axoplasmic Organelle Isolation
Squid axoplasm was extruded from 10 axons (∼50 μl) into 75 μl ½× buffer containing protease inhibitors (as above) and brought to 0.6 M KI with a 3 M KI stock. The solution was triturated 50 times with a wide bore pipette and placed on ice for 10 min to dissociate the cytoskeleton. The resulting homogenate was diluted 1:1 in ½× buffer, layered over a sucrose step gradient (100 μl 45% sucrose, 200 μl 15% sucrose, and 100 μl 12% sucrose in ½× buffer) and centrifuged at 35,000 rpm for 1.5 h at 4°C in a Beckman SW 55.1 rotor (Beckman, Fullerton, CA). The supernatant and each sucrose layer were removed with a syringe by puncturing the side of the centrifuge tube. Fractions were analyzed by SDS-PAGE and Western blots as well as by immunoelectron microscopy. Quantitative analysis of Western blots was carried out using a Bio-Rad Gel Doc and Quantity One software (Bio-Rad Laboratories, Hercules, CA). Protein concentration of axoplasmic sucrose fractions was determined using a standard Bradford protein assay.
Immunogold Labeling of Axoplasmic Organelles
Glow-discharged carbon-Formvar-coated grids were placed on droplets of KI-washed isolated axoplasmic organelles for 1 min to allow organelles to adhere to the coated surface. The grids were blocked in 10 mg/ml BSA (Sigma, St. Louis, MO) and 2% (vol/vol) fish gel (Tedd Pella, Redding, CA) in ½× buffer for 30 min, followed by incubation in 1:200 dilution of antisquid nonmuscle myosin II antibody (αMII) in blocking solution for 90 min. The grids were washed in blocking buffer (3 ×10 min) and were incubated in a 1:10 dilution of protein-A gold (Amersham Pharmacia Biotech, Piscataway, NJ) in blocking buffer for 1 h. Grids were washed 3 × 5 min in TBS and negative-stained in 1% uranyl acetate in water. Grids were processed in parallel, but in the absence of primary antibody, to determine nonspecific decoration by protein-A gold.
Six independent experiments were carried out using a variety of blocking agents. The combination of BSA and fish gel yielded the least amount of background labeling, and the resulting grids were used for statistical analysis. Areas of the grids were selected based on optimal negative stain and organelle density for visualization. Fields of organelles were photographed at 20,000 × in a JEOL CX 200, and gold particles were counted on randomly selected micrographs. Gold particles were considered to be associated with an organelle if the particle was on the surface of the organelle or was found within a 10-nm distance (diameter of one gold particle) of the organelle. The number of gold particles associated with each organelle and those in the background were counted, and the area occupied by organelles and background was determined.
RESULTS
Purification and Identification of Squid Nonmuscle Myosin II
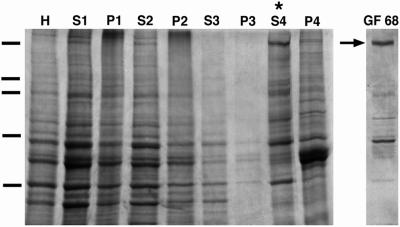
A protocol designed to purify myosin was used to extract a high molecular weight myosin from squid optic lobe (Figure 1) (See and Metuzals, 1976; Medeiros et al., 1998). By differential centrifugation a high-speed supernatant (Figure 1, S4) was obtained that was enriched for a protein (p220) migrating at ∼220 kDa, a molecular weight similar to that of other myosin heavy chains. This protein was further purified from the high-speed supernatant by gel filtration chromatography (Figure 1, GF68). From 100 g of squid optic lobes, ∼0.4 mg of p220 was obtained. The peak fraction contained p220 at a concentration of ∼100 μg/ml.
Figure 1.
Purification of a squid nonmuscle myosin II. Coomassie-stained 8.5% SDS-PAGE of optic lobe homogenate (H), supernatants (S) and corresponding pellets (P) of squid optic lobe samples obtained by a series of four centrifugation steps (S1-P4). The final high-speed supernatant, S4 fraction, (indicated by asterisks) is enriched in a high molecular weight protein (p220). The ∼220 kDa protein (arrow) was further separated from other proteins by gel filtration of the S4 fraction in the presence of ATP (GF68). Molecular weight markers indicated by dashes on left: 200, 116, 98, 68, 31 kDa.
Seventeen peptide sequences were obtained from purified p220 by Edman degradation sequencing (Table 1). These peptide sequences range in length from 7 to 25 amino acids and constitute a total of 210 residues. Eleven of the 17 peptide sequences match other myosin sequences in our myosin database by fasta search using gcg software (Medeiros et al., 1998). The other six peptides are not identifiable by BLAST of the NCBI databank or by fasta searches of our myosin directory.
Table 1.
Peptide sequences of purified p220
| Name | Amino acid sequence | Position | % Identity |
|---|---|---|---|
| Sequence #3 | KGDEVVVDVEDTGK | 59–72 | 100 |
| Sequence #6 | K*RTTFHRDDIQK | 72–83 | 100 |
| Sequence #9 | K*VGRDHVTK | 413–421 | 100 |
| Sequence #5 | K*LLGHHVNHPK | 558–568 | 90 |
| Sequence #2 | K*FEYEDLERK | 873–882 | 100 |
| Sequence #8 | KXAQVEEVQSQLARREEELQSALQK | 1075–1099 | 100 |
| Sequence #1 | KXELEVDNLK | 1172–1181 | 100 |
| Sequence #16 | LEVDNLK | 1175–1181 | 100 |
| Sequence #7 | K*QGENQALELRQK | 1189–1201 | 100 |
| Sequence #11 | K*MQTEYEQAV | 1285–1294 | 100 |
| Sequence #10 | K*VSNLQTDLA | 1310–1319 | 100 |
| Sequence #14 | K*NAETAESSRDNAEEELK | 1379–1396 | 100 |
| Sequence #17 | NELEGLVS | 1512–1519 | 100 |
| Sequence #13 | K*ELSGRDENADEIK | 1579–1592 | 100 |
| Sequence #12 | RAAQHDANTAF | 1666–1676 | 100 |
| Sequence #4 | LELRISELEDLLDEEQ | 1735–1750 | 100 |
| Sequence #15 | K*IASLEDQLDQD | 1823–1834 | 92 |
Where K* = assumed to be K based on the specificity of the enzyme endoproteinase Lys-C.
Where X = uninformative residues; S, H = mismatches.
Primers based on p220 peptide sequences and on conserved myosin consensus sequences were used to obtain a 1139 base-pair myosin fragment by RT-PCR of squid optic lobe total RNA (Figure 2, clone 1). This initial clone was extended in both the 5′ and 3′ directions by a series of PCR reactions using rapid amplification of cDNA ends techniques (RACE). Seven overlapping clones encode a 5892 base-pair open reading frame and 5′ and 3′ untranslated regions (Figure 2A). Comparison of the full-length deduced amino acid sequence to sequences in the NCBI databank identifies this protein as a squid nonmuscle myosin II heavy chain. This sequence most closely matches the sequences of the nonmuscle myosin II isoforms of Drosophila melanogaster (58.9%) (zipper gene, Accession No. A36014) and Caenorhabditis elegans (57.1%) (Accession No. T16416).
All 17 peptide sequences originally obtained from p220 were found to match squid nonmuscle myosin II (Figure 3). Four of the peptide sequences (No. 3, 5, 6, and 9) map to the myosin motor domain, two peptides (No. 2 and 8) map to the myosin neck domain, and the remaining 11 sequences map along the tail domain from amino acids 1175–1834. Those peptide sequences previously unidentifiable by BLAST and fasta searches each map to less conserved regions of the nonmuscle myosin II tail domain. These results verify that we have cloned the cDNA that encodes p220, and they identify p220 as a squid nonmuscle myosin II (sqNMMII).
Specificity of Nonmuscle Myosin II Antibody
A squid nonmuscle myosin II antibody (αMII) was generated against a unique recombinant 309 aa sequence (737–1046aa) of the sqNMMII heavy chain overexpressed in E. coli (rNMMII737–1046) (Figure 3). The rNMMII737–1046 includes part of the myosin head domain, the myosin neck domain which encompasses the IQ motifs, and part of the proximal tail in the region predicted to form a coiled coil. The rNMMII737–1046 does not contain the highly conserved ATP or actin-binding sites or other highly conserved regions of the myosin head or tail domains.
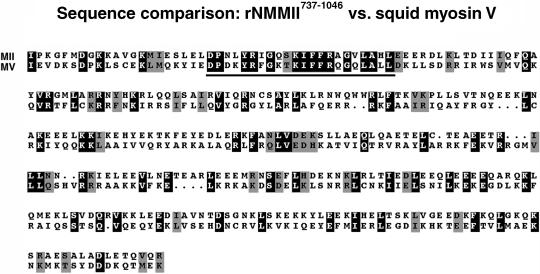
The region of sqNMMII we chose as an immunogen is significantly different from squid myosin V, the only other myosin known to be present in squid neural tissue (Figure 4; Table 2). Comparison of the rNMMII737–1046 sequence to squid myosin V (Accession No. AAF12809) reveals a sequence identity of only 23.3% (Table 2, upper panel). In contrast, a comparison to another nonmuscle myosin II from Drosophila reveals a 61.3% sequence identity (Table 2, upper panel). Sequences of rNMMII737–1046 and squid myosin V were compared to determine whether there are local regions of homology (Figure 4). Only a short sequence of 22 amino acids (758–779 aa) revealed any degree of similarity (63.3%) (Figure 4, underlined).
Figure 4.
Amino acid sequence comparison of the rNMMII737–1046 antigen sequence to the corresponding domain of the squid myosin V heavy chain sequence. Protein sequences were aligned with the gap command, using Wisconsin Sequence Analysis Package (Genetic Computer Group, Madison, WI). Identical residues are shaded in black and similar residues in gray (using BOXSHADE, available at: www.ch.embnet.org).
Table 2.
Distinguishing characteristics of nonmuscle myosin II compared with other myosins in the same species
| Name | Class | % Identity | % Similarity | Protein length (aa) | Protein AC# |
|---|---|---|---|---|---|
| rNMMII737–1046 | II | 1964 | AY040821 | ||
| Squid MM | II | 37.7 | 50.3 | 1935 | AAC24207 |
| Squid V | V | 23.3 | 33.0 | 1849 | AAF12809 |
| Dros. NM II (zip) | II | 61.3 | 71.0 | 1972 | A36014 |
| NM II (zip) | II | 1872 | A36014 | ||
| 1A | I | 21.5 | 32.1 | 1026 | S45573 |
| 1B | I | 23.0 | 33.7 | 1011 | S45574 |
| MHC | II | 34.8 | 51.6 | 1962 | PO5661 |
| Nina C | III | 20.5 | 35.6 | 1501 | P10676 |
| Myo V (Didum) | V | 23.0 | 33.0 | 1792 | T13939 |
| 95F (jargene) | VI | 25.3 | 40.4 | 1253 | Q01989 |
| ck (crinkled) | VII | 22.8 | 34.7 | 2167 | AAF44915 |
| VIIb 28b | VII | 26.3 | 38.5 | 2121 | AAF34810 |
As a further test that the rNMMII737–1046 encoded a domain unique to class II myosins, we also compared this domain to other myosin heavy chains. We could not do an extensive sequence comparison in squid as, in this species, only two myosin heavy chain genes have been previously sequenced. We therefore used the Drosophila database to compare the homologous rNMMII737–1046 domain of the Drosophila nonmuscle myosin II gene (zipper) to other myosins in the Drosophila genome. We reasoned that the degree of similarity between this domain of the zipper gene and other myosin sequences in the Drosophila genome would be similar to the degree of sequence homology between different myosin sequences within the genome of the squid. For instance, the sequence identity between rNMMII737–1046 and squid myosin V is 23.3% and to squid muscle myosin is 37.7%. This same domain in the Drosophila zipper gene is 23.0% identical to Drosophila myosin V and 34.8% identical to Drosophila muscle myosin II (MHCII) (Table 2). The Drosophila genome makes the sequence of the complete set of Drosophila myosins in the genome available (Yamashita et al., 2000). Comparison of the domain homologous to rNMMII737–1046 from Drosophila nonmuscle myosin II (zipper gene) with other myosins in the Drosophila genome shows that it has <27% sequence identity to any other unconventional myosin isoforms. Only the conventional myosins share any reasonable identity (34.8–37.7%) with the nonmuscle myosin II antibody domain (Table 2: lower panel). Thus, the rNMMII737–1046 domain provides a reasonably unique antigen for antibody production against nonmuscle myosin II isoforms.
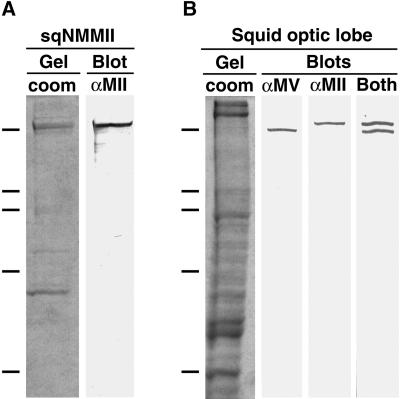
The resultant anti-nonmuscle myosin II antibody (αMII) recognized purified sqNMMII (Figure 5A) as well as a single band of the same molecular weight in optic lobe homogenates (Figure 5B). The fact that αMII recognized only a single band in optic lobe, the original source of purified sqNMMII, demonstrates that the antibody is specific for sqNMMII, since the optic lobe contains at least one other myosin isoform (myosin V; Tabb et al., 1998). The optic lobe would be expected to contain many, as yet unidentified, myosin isoforms, including myosins of glial cells as well as those of endothelial and of smooth muscle cells from blood vessels (Murakami and Elzinga, 1992).
Figure 5.
Antisquid nonmuscle myosin II antibody is specific for sqNMMII. Commassie-stained 8.5% SDS-PAGE gel of purified sqNMMII (A, sqNMMII/Gel, coom) and corresponding Western blot (A, sqNMMII/Blot, αMII) with αMII. Commassie stained 8.5% SDS-PAGE gel of optic lobe homogenate (B, squid optic lobe/Gel, coom) and corresponding Western blots (B, squid optic lobe/Blots) with αMV antibody (B, αMV), αMII antibody (B, αMII), and both antibodies (B, Both). Molecular weight markers indicated by dashes on left: 200, 116, 98, 68, 45 kDa.
The specificity of αMII was further tested by probing optic lobe homogenates in parallel with either αMII or αMV (antisquid myosin V) (Figure 5B). We also generated the αMV antibody against a unique peptide sequence that maps to the myosin V head domain (see MATERIALS AND METHODS). While αMII detects a band of ∼220 kDa in optic lobe homogenates, the αMV recognizes a lower band at ∼196 kDa. A lane probed with both antibodies shows two discrete bands (Figure 5B), demonstrating that these are different myosins and that each is recognized specifically by its respective antibody.
Expression of Nonmuscle Myosin II in the Stellate Ganglion
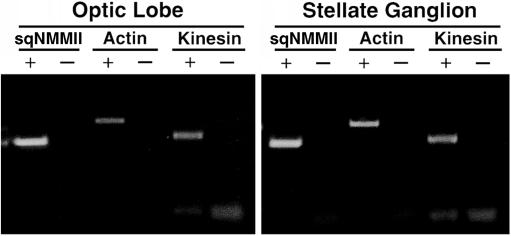
Proteins present in the giant axon are synthesized in the neuronal cell bodies of the stellate ganglion. PCR has been successfully applied to probe for myosin expression across tissue types in other organisms (Itoh and Adelstein, 1995). Thus, we used specific nonmuscle myosin II primers in RT-PCR reactions to assay for sqNMMII transcripts in the stellate ganglion. SqNMMII transcripts were detected in both the squid optic lobe and the stellate ganglion (Figure 6). Only samples prepared with reverse transcriptase produced products demonstrating that product was generated from message and not from genomic template. Each 361 base-pair product was cloned and sequenced to verify that it was sqNMMII. RT-PCR experiments using specific primers for kinesin and actin demonstrate that transcripts for each of these proteins are expressed in optic lobe and in the stellate ganglion.
Figure 6.
SqNMMII is expressed in the stellate ganglion. Ethidium bromide stained 1% agarose gel of PCR products using cDNA from optic lobe and stellate as template (as labeled). Primers are designed to amplify sqNMMII, actin and kinesin. PCR reactions were carried out using first strand cDNA generated in the presence (+) and absence (-) of reverse transcriptase.
Squid Nonmuscle Myosin II Copurifies with KI-stripped Axoplasmic Organelles
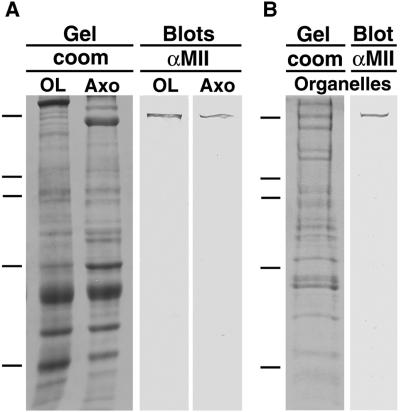
To determine whether sqNMMII is present in the axoplasm of the squid giant axon, Western blots of extruded axoplasm were probed with αMII (Figure 7). Squid optic lobe homogenate was electrophoresed and blotted in parallel with axoplasm. In both samples, a ∼220 kDa band was detected for sqNMMII (Figure 7A).
Figure 7.
SqNMMII is found in axoplasm and copurifies with axoplasmic organelles. Coomassie-stained 8.5% SDS-PAGE gel of optic lobe homogenate and axoplasm (A, Gel/coom OL, Axo) and corresponding Western blots with αMII antibody (A, Blots/αMII OL, Axo). Coomassie stained 8.5% SDS-PAGE gel of axoplasmic organelle fraction (B, Gel/coom, organelles) and corresponding Western blot with αMII antibody (A, Blots/αMII, organelles). Molecular weight markers indicated by dashes on left: 200, 116, 98, 68, 45 kDa.
To determine whether sqNMMII associates with axoplasmic organelles, we fractionated the axoplasm according to a protocol that involves incubation of axoplasm for 10 min in 0.6 M potassium iodide (KI) (Schroer et al., 1988). This KI treatment solubilizes the cytoskeleton, enabling the subsequent separation of organelles from other axoplasmic proteins by sucrose density gradient fractionation. These organelles have been shown to translocate toward the plus ends of microtubules (Schnapp et al., 1992), as well as along filamentous actin (Bearer et al., 1996a). The KI step is believed to strip cytoplasmic dynein and other loosely associated proteins from the organelles but does not remove kinesin (Schnapp et al., 1992). The αMII recognized a single band of ∼220 kDa in the sucrose fraction containing KI-stripped organelles (Figure 7B). This band is the same apparent molecular weight as the band recognized in both the axoplasmic and optic lobe samples. Thus, it appears that sqNMMII is present in all three samples.
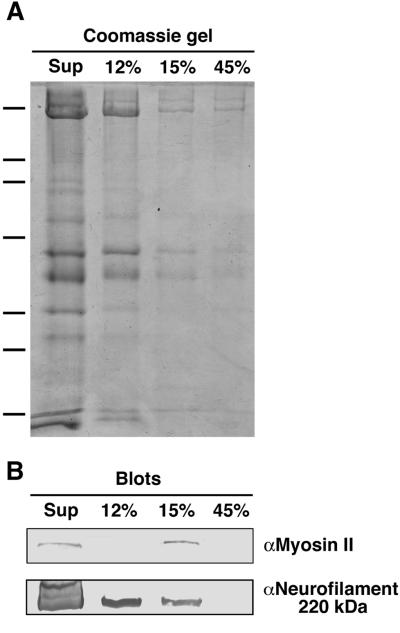
Quantitative analysis demonstrates that 43.2% of the total sqNMMII copurifies with organelles (Figure 8; Table 3). Equal volumes (10 μl) from each step in the gradient were separated by gel electrophoresis and were probed in parallel for either sqNMMII or neurofilament protein. The Western blot band intensities for each protein were measured by densitometry. The percentage of the total was calculated for each sucrose fraction, taking into account the fraction volume. SqNMMII was only detected in the supernatant and organelle fractions. The supernatant contains the remainder (56.8%) of the total sqNMMII. Protein concentration measurements show that the supernatant and the 12% fraction contain the majority of the total protein (92.8%) with only 1.6% in the organelle fraction. Thus, sqNMMII is enriched 15.5-fold in the organelle fraction compared with the supernatant fraction. In comparison, the intensity of the neurofilament band follows a pattern similar to that of the protein concentration, highest in the supernatant and decreasing in concentration down the gradient.
Figure 8.
SqNMMII preferentially fractionates with axoplasmic organelles. (A) Coomassie-stained 8.5% SDS-PAGE gel of axoplasmic sucrose density fractions and (B) corresponding Western blots with αMII antibody (top panel) and α-neurofilament antibody (bottom panel). Each lane is loaded with 10 μl each sucrose fraction, including supernatant (Sup), 12% sucrose fraction (12%), 15% sucrose fraction (15%), and 45% sucrose fraction (45%). The 15% fraction is highly enriched in organelles. Molecular weight markers indicated by dashes on left: 200, 116, 98, 68, 45, 31, 21 kDa.
Table 3.
Quantitative analysis of nonmuscle myosin II in axoplasmic fractions
| Sucrose fraction | Protein/ lane (μg)a | Proportion of total protein/lane (%)b | Proportion
of total antigen/fraction (%)cd
|
|
|---|---|---|---|---|
| sqNMMII | NF | |||
| Supernatant | 26.4 | 53.0 | 56.8 | 80.8 |
| 12% | 21.3 | 42.8 | 0.0 | 9.1 |
| 15% (organelles) | 1.3 | 2.6 | 43.2 | 10.1 |
| 45% | 0.8 | 1.6 | 0.0 | 0.0 |
| Total | 49.8 | 100 | 100 | 100 |
Total protein in each gel lane for Coomassie stain and Western blot analysis (10 μl each sucrose fraction).
Proportion of the total protein in each gel lane Coomassie stain and Western blot analysis.
Percent of nonmuscle myosin II (sqNMMII) and neurofilament (NF) in sucrose density fractions.
Percent of antigen/fraction has been adjusted to account for the total volume of each sucrose fraction.
Squid Nonmuscle Myosin II Associates with Isolated Axoplasmic Organelles
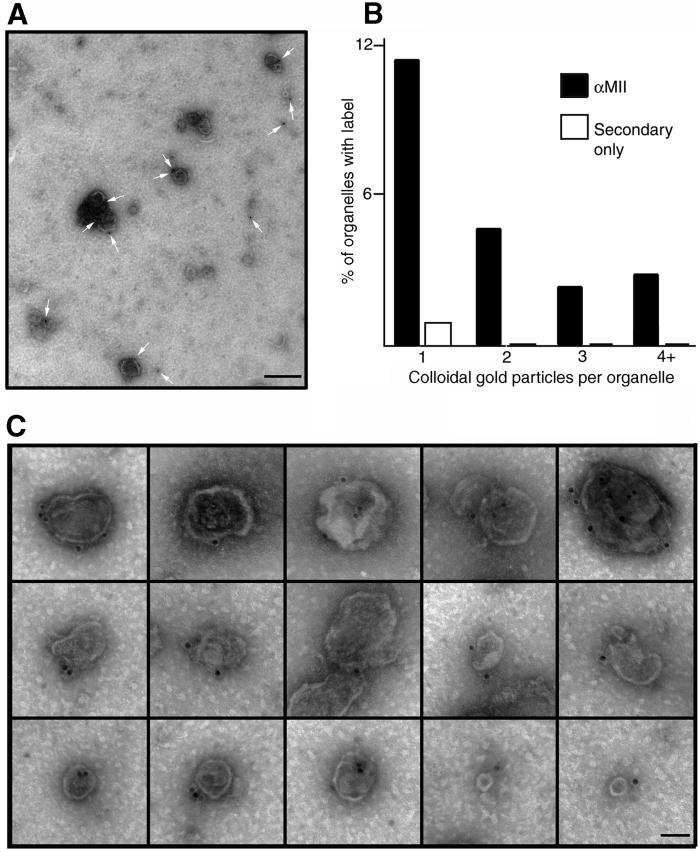
Cosedimentation with organelles suggests that this myosin is directly attached to organelles. To test whether sqNMMII was indeed associated with the axoplasmic organelles, organelles were stained with αMII/immunogold and examined by electron microscopy (Figure 9A, C). In ten fields, 11% of organelles were labeled with a single gold particle, while 10% were labeled with two or more particles (Figure 9B). Thus, 21% of all organelles were decorated by immunogold. Less than 1% of the organelles were labeled in grids stained with protein-A gold in the absence of primary antibody.
Figure 9.
SqNMMII associates with KI-washed axoplasmic organelles. (A) Low magnification field of KI-washed axoplasmic organelles labeled with αMII primary antibody and protein-A colloidal gold (scale bar = 200 nm), and (B) histogram of organelle labeling. Histogram shows the number of gold particles per organelle in the presence and absence of primary antibody (n = 581). Montage showing micrographs of representative organelles labeled with antisquid nonmuscle myosin II antibody and colloidal gold. (C) Organelles of various sizes are labeled with one or more gold particle (scale bar = 50 nm).
We quantified the number of particles in the area occupied by organelles compared with the number of particles occurring in the unoccupied (background) regions of the grids (Table 4). The frequency of particles associated with organelles was 6.01 particles/μm2. In contrast, the background area had only 0.24 particles/μm2. Thus, the area occupied by organelles had a 25-fold increase in gold particles compared with background. In the absence of primary antibody, no increase in association of gold particles with organelles was observed. With protein-A gold alone, the frequency of gold particle on organelles (0.32 particles/μm2) was similar to background levels (0.27 particles/μm2) (Table 4). Labeled organelles varied in size and shape, from the smallest vesicles in the preparation, measuring ∼50 nm, to the large mitochondria-like organelles, measuring ∼500 nm (Figure 9C). The surface contours of the labeled organelles also varied. Thus, sqNMMII appears to associate with a wide variety of organelles as determined by these morphological criteria.
Table 4.
Quantitative analysis of immunogold labeling of KI-washed axoplasmic organelles
| αMII & secondary
antibody
|
Secondary antibody alone
|
|||||
|---|---|---|---|---|---|---|
| Area (μm2)a | # Particlesb | Particles/μm2 | Area (μm2)a | Particlesb | Particles/μm2 | |
| Organelles | 19.3 | 116 | 6.01 | 22.1 | 7 | 0.32 |
| Background | 333.5 | 79 | 0.24 | 330.7 | 88 | 0.27 |
| Total | 352.8 | 195 | 352.8 | 95 | ||
Area on grid; see MATERIALS AND METHODS.
Gold particles counted.
DISCUSSION
The giant axon of the squid is a powerful model in which to study the physiology of axonal transport, but it poses significant difficulties for biochemical identification of the motor proteins involved. We initially reported that a myosin-like protein copurifies with motile organelles from the giant axon (Bearer et al., 1993), but until now, we have not been able to identify this protein definitively. Small amounts of this myosin could be obtained from axoplasm, but the protein is too large, and the yield is not sufficient for peptide sequencing (Bearer et al., 1996b). Identification by mass spectroscopy also was not feasible, as only two squid myosin sequences have been entered into the databank.
In this paper, we characterize this myosin and show that it is tightly associated with axoplasmic organelles. The present approach starts by obtaining proteins from the optic lobe, the largest structure of the squid CNS, with the expectation that proteins expressed in the optic lobe would include those also present in the giant axon. The optic lobe yielded a purified squid myosin (p220) that has provided peptide sequences (Table 1). Cloning and sequencing of the cDNA that encodes p220 allowed us to classify it as a nonmuscle myosin II (sqNMMII), according to the standard criteria for myosins, which is based on the amino acid sequence of the head domain (Goodson and Spudich, 1993; Cheney and Mooseker, 1993; Berg et al., 2001). The full-length sequence also allowed us to develop specific tools, including an antibody specific to nonmuscle myosin II and primers for PCR, to identify this myosin in other cells and tissues.
Is the Optic Lobe Myosin II Present in Axons?
Our data provide reasons to believe that this optic lobe nonmuscle myosin II is also present in the axon. First, RT-PCR demonstrates that the mRNA for sqNMMII is expressed in the cell bodies that give rise to the giant axon. This suggests but does not prove that the protein detected in the axon is sqNMMII. The proteins present in the axon are synthesized in the cell body, but some may not enter the axon proper. Some of the proteins expressed in neurons might remain solely in the cell body, or they may localize to other regions of the cell, such as dendritic processes. RT-PCR has been used in other species to detect tissue-specific expression of nonmuscle myosin II, and it has been found to be ubiquitously expressed (Itoh and Adelstein, 1995).
Second, by Western blot analysis, our antibody raised against a specific domain of sqNMMII (αMII) recognizes a single band in optic lobe homogenates as well as a band of the same molecular weight in squid axoplasm. This antibody was generated against a unique 309 amino acid domain that is highly specific for this myosin II, and the antibody does not cross-react with myosin V. Furthermore, this antibody is unlikely to recognize myosins of other classes, as these isoforms are equally or more divergent than myosin V from nonmuscle myosin IIs in the domain used for antibody production (aa 737-1046).
Even though the Drosophila genome, the closest to squid of the completed genomes, contains only a single nonmuscle myosin II gene, we cannot rule out the possibility that there are other nonmuscle myosin IIs in squid. There could also be minor molecular differences between the myosin II cloned from optic lobe and the protein present in axons. The axon could contain different splice forms, as myosin IIs are known to be alternatively spliced (Kelley and Adelstein, 1995), although whether splicing affects function is not clear. It has been difficult to differentiate splice forms by SDS-PAGE or Western blot even with peptide antibodies against spliced-in amino acid sequences. Other approaches will be necessary to determine whether small sequence variations exist in the axoplasmic nonmuscle myosin II isoform.
Guilt by Association?
Two lines of evidence provide strong support for a physiological association of sqNMMII with the cytoplasmic surface of axoplasmic organelles. First, 43.2% of the total sqNMMII remains associated with organelles even after stripping with 0.6 M KI. Similarly, the microtubule motor kinesin is not stripped with KI from organelles (Schnapp et al., 1992). Furthermore, by immunogold immunocytochemistry, sqNMMII is detected on the cytoplasmic surfaces of intact organelles, indicating that this myosin is exposed and thus available to serve as a motor and is not sequestered inside the organelle to be deployed later at some distant site. Such KI-stripped organelles are known to be motile on both actin filaments and microtubules (Schnapp et al., 1992; Bearer et al., 1996a).
This association between sqNMMII and axoplasmic organelles is strong evidence for a functional role in organelle transport. Association of motors with organelles, as demonstrated by various immunological and biochemical techniques, has proved to be a reliable predictor of function. Localization of kinesin after sequential extractions in cultured cells provided evidence of its role in vesicle transport (Morris and Hollenbeck, 1995). Immunofluorescence detection has served to identify the subcellular location of kinesin isoforms and thereby to differentiate their cellular function (Henson et al., 1992; Signor et al., 1999). Finally, immunogold labeling of axoplasmic and cellular organelles with antibodies specific for different kinesin isoforms has contributed to our understanding of which of this large superfamily are associated with organelles (Yamazaki et al., 1995; Moreira et al., 1998).
The strongest evidence of a role for sqNMMII in transport given the tools available is the biochemical association demonstrated here. Blocking antibodies and genetic knockout experiments can be used to determine whether a motor is involved in organelle transport (Yang et al., 2001; Sandberg et al., 2000; Doberstein et al., 1993). However, these approaches have significant drawbacks, especially in squid. For example, inhibitory antibodies are difficult to generate and antibody-blocking experiments can be hard to interpret. Although one myosin V antibody has been reported to block organelle movements in diluted axoplasm, another has no effect (Tabb et al., 1998). In the squid model system, genetic mutations are not yet an option. Now that myosin II has been found associated with organelles, studies in other organisms in which genetic approaches are possible can be initiated.
Could sqNMMII Be Membrane Associated?
That the myosin associated with organelles is a member of the myosin II subgroup comes as a surprise, as myosin IIs are generally thought to form multimeric antiparallel thick filaments that pull actin filaments against each other as in the classic Huxley model of contraction (Huxley and Simmons, 1971). The myosin II subgroup includes striated muscle, smooth muscle, and nonmuscle isoforms. The muscle myosins each have highly defined roles in muscle contraction, and it is well documented that nonmuscle myosin IIs are involved in a variety of contractile processes including cell division, cell motility, and chemotaxis (Straussman et al., 2001). However, a number of findings suggest that not all myosin IIs self-associate into thick filaments, and some may mediate functions other than contractility.
Evidence suggests that nonmuscle myosin II may take on a variety of structural conformations. The nonmuscle myosin II from Drosophila (zipper gene) has been shown to form short dumb-bells by rotary platinum shadow electron microscopy (Kiehart and Feghali, 1986). This myosin II is required for membrane movements during closure of the amnioserosa in Drosophila embryos. It could associate with membranes via the middle bar of the dumb-bell. In our early studies, electron microscopy of an axoplasmic myosin revealed “flowerettes,” or aggregates of myosins with all globular (head) domains at one end, held together by associations between the sinuous tails (Bearer et al., 1996a). This conformation of myosin multimers could easily support tail-mediated organelle association. Some of the sqNMMII described here could be associated with organelles via an interesting link with other myosin IIs in a thick filament conformation. However, evidence for such thick filaments was not found in our electron-microscopic examination of organelles by negative stain. Biochemical studies of nonmuscle myosin IIs have shown that thick filament formation is regulated by phosphorylation of the tail domain. For example, the phosphorylation of residues in the nonmuscle myosin IIB rod inhibits thick filament formation (Murakami et al., 1995). Phosphorylation mediates membrane association (Murakami et al., 1995). Thus, there may be different structural and functional forms of these myosin II motors, which could be generated by alternative splicing or posttranslational modifications.
Some Nonmuscle Myosin IIs are Membrane-associated Motors
Emerging evidence links nonmuscle myosin IIs with membrane dynamics in a wide range of species and tissue types. Myosin II knock-out mutations in Dictyostelium demonstrate that myosin II is required for agonist-induced rounding (Springer et al., 1994) and for surface membrane tension (Jay and Elson, 1992), as well as for cytokinesis in suspension (Springer et al., 1994). Actin-based vesicle movement in extracts of clam oocytes is blocked by myosin II-specific antibodies (Sandberg et al., 2000). Immunolocalization of myosin IIs in cultured neurons shows that myosin IIB is found at the leading edge of growth cones as well as in the organelle-rich central region at the end of extending axonal microtubule bundles (Cheng et al., 1992). In the present study, 43.2% of the nonmuscle myosin II in axoplasm is associated with KI-washed axoplasmic organelles. Thus, organelle-bound myosin represents a major component of the total nonmuscle myosin II in these cells.
Nonmuscle Myosin IIs May Play Other Roles in Neurons
In mature neurons, the role of nonmuscle myosin II is not well understood. Myosin II in the axon could be mediating vesicle transport on actin filaments. Alternatively, this organelle-associated myosin may be inactive while in transit to the synapse. Myosin II tails tagged by green fluorescent protein have been shown to localize to the cleavage furrow during cytokinesis. Thus, not all myosins reach their final intercellular destinations through their head domain motor activities (Zang and Spudich, 1998). Myosin IIs appear to perform necessary functions in the brains of vertebrates (Wylie and Chantler, 2001; Wylie et al., 1998; Chantler, 1997), where myosin IIB is required for normal brain development in mammals (Tullio et al., 2001). Myosin II could also be involved in maintaining axonal macrostructure through actomyosin contractions. Axons appear to be under tension, and it has been proposed that this tension is mediated by actomyosin interactions (Baas and Ahmad, 2001).
ACKNOWLEDGMENTS
We acknowledge Ben Greenfield for the original sqNMMII cDNA clone, Zhi Li for generating the αMII antibody, and Timna Onigman and Heather Davidson for characterizing the antibody on bacterially expressed protein. We thank Howard Jaffe for obtaining peptide sequences, and Jennifer Petersen and John Chludzinski for help in the laboratory. We are grateful for the work of Brown University undergraduates on this project, including Kendrick Jones, Eric Schneider, and Paul George. We thank Louie Kerr, at the Central Microscopy Facility, Marine Biological Laboratory, for advice and technical assistance and Jorge Moreira for advice on immunocytochemistry. We also thank Ed Enos and the staff of the Marine Resources Center, Marine Biological Laboratory, for the collection and maintenance of squid. Supported by National Institute of Neurological Disorders and Stroke (T.S.R.), the National Institutes of Health, GM-47368 (E.L.B.), and the National Science Foundation and GM-07601 (training grant to Graduate Program in Molecular Biology, Cell Biology, and Biochemistry).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–06–0315. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–06–0315.
REFERENCES
- Baas PW, Ahmad FJ. Force generation by cytoskeletal motor proteins as a regulator of axonal elongation and retraction. Trends Cell Biol. 2001;11:244–249. doi: 10.1016/s0962-8924(01)02005-0. [DOI] [PubMed] [Google Scholar]
- Bearer EL, DeGiorgis JA, Bodner RA, Kao AW, Reese TS. Evidence for myosin motors on organelles in squid axoplasm. Proc Natl Acad Sci USA. 1993;90:11252–11256. doi: 10.1073/pnas.90.23.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, DeGiorgis JA, Medeiros NA, Reese TS. Actin-based motility of axoplasmic organelles. Cell Motil Cytoskeleton. 1996a;30:50–66. doi: 10.1002/cm.970330202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, DeGiorgis JA, Jaffe H, Medeiros NA, Reese TS. An axoplasmic myosin with a calmodulin-like light chain. Proc Natl Acad Sci USA. 1996b;93:6064–6068. doi: 10.1073/pnas.93.12.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin consensus. Mol Biol Cell. 2001;12:795–808. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ST, Lasek RJ, Allen RD. Fast axonal transport in extruded axoplasm from squid giant axon. Science. 1982;218:1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- Chantler PD. A unique cellular myosin II exhibiting differential expression in the cerebral cortex. Biochem Biophys Res Commun. 1997;235:268. doi: 10.1006/bbrc.1997.6765. [DOI] [PubMed] [Google Scholar]
- Cheng TPO, Murakami N, Elzinga M. Localization of myosin IIB at the leading edge of growth cones from rat dorsal root ganglionic cells. FEBS letter. 1992;311:91–94. doi: 10.1016/0014-5793(92)81374-u. [DOI] [PubMed] [Google Scholar]
- Cheney RE, Mooseker MS. Phylogenetic analysis of the myosin superfamily. Cell Motil Cytoskeleton. 1993;24:215–223. doi: 10.1002/cm.970240402. [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Baines IC, Wiegand G, Korn ED, Pollard TD. Inhibition of contractile vacuole function in vivo by antibodies against myosin-I. Nature. 1993;365:841–843. doi: 10.1038/365841a0. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Spudich JA. Molecular evolution of the myosin family: relationships derived from comparisons of amino acid sequences. Proc Natl Acad Sci USA. 1993;90:659–663. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley AF, Simmons RM. Proposed mechanism of force generation of striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- Itoh K, Adelstein RS. Neuronal cell expression of inserted isoforms of vertebrate nonmuscle myosin heavy chain II-B. J Biol Chem. 1995;270:14533–14540. doi: 10.1074/jbc.270.24.14533. [DOI] [PubMed] [Google Scholar]
- Jay PY, Elson EL. Surface particle transport mechanism independent of myosin II in Dictyostelium. Nature. 1992;356:438–440. doi: 10.1038/356438a0. [DOI] [PubMed] [Google Scholar]
- Kelley CA, Adelstein RS. Characterization of myosin II isoforms containing insertions of amino acids in the flexible loop near the ATP-binding pocket. Biophys J. 1995;68:225S. [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophilamelanogaster. J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- Langford GM, Kuznetsov SA, Johnson D, Cohen DL, Weiss DG. Movement of axoplasmic organelles on actin filaments assembled on acrosomal processes: Evidence for a barbed-end-directed organelle motor. J Cell Sci. 1994;107:2291–2298. doi: 10.1242/jcs.107.8.2291. [DOI] [PubMed] [Google Scholar]
- Matulef K, Sirokman K, Perreault-Micale CL, Szent-Gyorgyi AG. Amino acid sequence of squid myosin heavy chain. J Muscle Res Cell Motil. 1998;19:705–712. doi: 10.1023/a:1005341416989. [DOI] [PubMed] [Google Scholar]
- Medeiros NA, Reese TS, Jaffee H, DeGiorgis JA, Bearer EL. Primary peptide sequences from squid muscle and optic lobe myosin IIs: a strategy to identify an organelle motor. Cell Biol Int. 1998;22:161–173. doi: 10.1006/cbir.1998.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Mulcahey MK, Stafford P, Langford GM. Sequence and phylogenetic analysis of squid myosin-V: a vesicle motor in nerve cells. Cell Motil Cytoskeleton. 2000;46:108–115. doi: 10.1002/1097-0169(200006)46:2<108::AID-CM3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Moreira JE, Dodane V, Reese TS. Immunoelectronmicroscopy of soluble and membrane proteins with a sensitive postembedding method. J Histochem Cytochem. 1998;46:847–854. doi: 10.1177/002215549804600708. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami N, Elzinga M. Studies on the distribution of cellular myosin with antibodies to isoform-specific synthetic peptides. Cell Motil Cytoskeleton. 1992;22:281–295. doi: 10.1002/cm.970220408. [DOI] [PubMed] [Google Scholar]
- Murakami N, Singh SS, Chauhan VPS, Elzinga M. Phospholipid Binging, phosphorylation by protein kinase C, and filament asembly of the COOH terminal heavy chain fragment of nonmuscle myosin II isoforms IIA and IIB. Biochemistry. 1995;34:11441–11451. doi: 10.1021/bi00049a019. [DOI] [PubMed] [Google Scholar]
- Sandberg L, Stafford P, Langford GM. Effects of myosin II antibody on actin-dependent vesicle transport in extracts of clam oocytes. Biol Bull. 2000;199:202–203. doi: 10.2307/1542898. [DOI] [PubMed] [Google Scholar]
- Schnapp BJ, Reese TS. Dynein is the motor for retrograde axonal transport of organelles. Proc Natl Acad Sci USA. 1989;86:1548–1552. doi: 10.1073/pnas.86.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp BJ, Reese TS, Bechtold R. Kinesin is bound with tight affinity to squid organelles that move to the plus-end of microtubules. J Cell Biol. 1992;119:389–399. doi: 10.1083/jcb.119.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA, Schnapp BJ, Reese TS, Sheetz MP. The role of kinesin and other soluble factors in organelle movement along microtubules. J Cell Biol. 1988;107:1785–1792. doi: 10.1083/jcb.107.5.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See YP, Metuzals J. Purification and characterization of squid brain myosin. J Biol Chem. 1976;251:7682–7689. [PubMed] [Google Scholar]
- Grant P, Tseng D, Gould RM, Gainer H, Pant HC. Expression of neurofilament proteins during development of the nervous system in the squid Loligo pealei. J Comp Neurol. 1995;356:311–326. doi: 10.1002/cne.903560212. [DOI] [PubMed] [Google Scholar]
- Springer ML, Patterson B, Spudich JA. Stage-specific requirement for myosin II during Dictyostelium development. Development. 1994;120:2651–2660. doi: 10.1242/dev.120.9.2651. [DOI] [PubMed] [Google Scholar]
- Straussman R, Enev L, Ravid S. Myosin II heavy chain isoforms are phosphorylated in an EGF-dependeny manner: involvement of protein kinase C. J Cell Sci. 2001;114:3047–3057. doi: 10.1242/jcs.114.16.3047. [DOI] [PubMed] [Google Scholar]
- Tabb JS, Molyneaux BJ, Cohen DL, Kuznetsov SA, Langford GM. Transport of ER vesicles on actin filaments in neurons by myosin V. J Cell Sci. 1998;111:3221–3234. doi: 10.1242/jcs.111.21.3221. [DOI] [PubMed] [Google Scholar]
- Tullio AN, Bridgman PC, Tresser NJ, Chan CC, Conti MA, Adelstein RS, Hara Y. Structural abnormalities develop in the brain after ablation of the gene encoding nonmuscle myosin II-B heavy chain. J Comp Neurol. 2001;433:62–74. doi: 10.1002/cne.1125. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SR, Chantler PD. Separate but linked functions of conventional myosins modulate adhesion and neurite outgrowth. Nat Cell Biol. 2001;3:88–92. doi: 10.1038/35050613. [DOI] [PubMed] [Google Scholar]
- Wylie SR, Wu PJ, Patel H, Chantler PD. A conventional myosin motor drives neurite outgrowth. Proc Natl Acad Sci USA. 1998;95:12967–12972. doi: 10.1073/pnas.95.22.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita RA, Sellers JR, Anderson JB. Identification and analysis of the myosin superfamily in Drosophila: a database approach. J Muscle Res Cell Motil. 2000;21:491–505. doi: 10.1023/a:1026589626422. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa NJ. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Xia C, Roberts EA, Bush K, Nigam SK, Goldstein LS. Molecular cloning and functional analysis of mouse C-terminal kinesin motor KifC3. Mol Cell Biol. 2001;21:765–770. doi: 10.1128/MCB.21.3.765-770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang JH, Spudich JA. Myosin II localization during cytokinesis occurs by a mechanism that does not require its motor domain. Proc Natl Acad Sci USA. 1998;23:13652–13657. doi: 10.1073/pnas.95.23.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]