Significance
The remarkably heterogenous nature of microglia has impeded efforts to understand their contribution to Alzheimer’s disease (AD) pathology. We show that OPN production divides CD11c+ microglia into pathogenic (CD11c+OPN+) and protective (CD11c+OPN−) subsets. OPN production promotes a microglial proinflammatory phenotype and may inhibit Aβ uptake and consolidation through the TREM2/TAM-lysosomal phagocytic pathway. Genetic ablation of OPN or administration of anti-OPN mAb reduces proinflammatory microglia and Aβ plaque pathology, and improves cognition. Analysis of brain tissue of AD patients reveals a strong correlation between OPN+CD11c+ microglial levels with dementia severity and neuropathology. These findings suggest that OPN represents a promising and tractable target for the development of potentially effective immunotherapy to halt or reverse AD progression.
Keywords: Osteopontin, integrins, microglia, neuroinflammation, Alzheimer’s disease
Abstract
Alzheimer’s disease (AD) is the most common form of incurable dementia and represents a critical public health issue as the world’s population ages. Although microglial dysregulation is a cardinal feature of AD, the extensive heterogeneity of these immunological cells in the brain has impeded our understanding of their contribution to this disease. Here, we identify a pathogenic microglial subset which expresses the CD11c surface marker as the sole producer of Osteopontin (OPN) in the 5XFAD mouse model of AD. OPN production divides Disease-Associated Microglia (DAM) into two functionally distinct subsets, i.e., a protective CD11c+OPN− subset that robustly ingests amyloid β (Aβ) in a noninflammatory fashion and a pathogenic CD11c+OPN+ subset that produces proinflammatory cytokines and fails to ingest significant amounts of Aβ. Genetic ablation of OPN or administration of monoclonal anti-OPN antibody to 5XFAD mice reduces proinflammatory microglia, plaque formation, and numbers of dystrophic neurites and results in improved cognitive function. Analysis of brain tissue from AD patients indicates that levels of OPN-producing CD11c+ microglia correlate strongly with the degree of cognitive deficit and AD neuropathology. These findings define an OPN-dependent pathway to disease driven by a distinct microglial subset, and identify OPN as a novel therapeutic target for potentially effective immunotherapy to treat AD.
Although microglial dysregulation is a cardinal feature of Alzheimer’s disease (AD) (1), the heterogeneity of this immune cell type has impeded efforts to understand its contribution to disease development and pathology. Transcriptomic studies of microglia in mouse models and AD patients (2–5) have indicated that the Spp1 gene, which encodes Osteopontin (OPN), is strongly up-regulated by microglia in both mouse models of AD and the human disease (2, 3, 6–8).
OPN, a cytokine expressed by a fraction of activated peripheral dendritic cells and macrophages, regulates innate and adaptive immune responses through interactions with its integrin receptors (9–13). Although OPN has been implicated in neuroinflammatory disorders (14, 15), the potential contribution of OPN-producing microglia to AD is unknown.
Here, we describe a microglial subset that promotes disease development in the 5XFAD mouse model. This subset accounts for <10% of total microglia, expresses the CD11c receptor and is the sole producer of OPN. Genetic deletion of OPN in 5XFAD mice results in a sharp reduction in proinflammatory microglia, amyloid β (Aβ) plaques and dystrophic neurites, and a marked improvement in cognitive function. In situ analysis of periplaque areas reveals that OPN production differentiates the disease-associated microglia (DAM) into two distinct subpopulations, i.e., a protective CD11c+OPN− (single positive, SP) subset that robustly takes up Aβ in a noninflammatory manner and a pathogenic CD11c+OPN+ (double positive, DP) subset that produces proinflammatory cytokines and failed to ingest Aβ fibrils.
Analysis of the underlying molecular mechanism suggests that two OPN-dependent pathways drive disease development in 5XFAD mice. An interaction between OPN and its αVβ3 integrin receptor that induces TNF-α production and inflammasome activation may convert homeostatic microglia into proinflammatory microglia that do not efficiently ingest Aβ. OPN-dependent inhibition of the TREM2/TAM phagocytic pathway may also inhibit lysosomal activation and destruction of Aβ fibrils. Proof-of-concept studies indicate that administration of monoclonal anti-OPN Ab to 5XFAD mice at an advanced stage of disease (6 mo) efficiently inhibits disease development, as judged by reductions in proinflammatory microglia and Aβ plaque pathology.
Analysis of human brain tissue confirmed that levels of the CD11c+OPN+ microglial subset correlate strongly with both dementia severity and AD neuropathology (neuritic plaques and neurofibrillary tau tangles) in clinically and neuropathologically characterized AD patients and healthy controls. Collectively, these studies define a pathogenic microglial subset marked by CD11c and OPN production that drives AD pathology by promotion of neuroinflammation and inhibition of Aβ plaque condensation. These findings refine current understanding of DAM and suggest a tractable target for immunotherapy of AD.
Results
OPN Contributes to AD Pathology and Cognitive Impairment in 5XFAD Mice.
To identify the cellular source of OPN during disease progression in brains of 5XFAD mice, we measured OPN expression by astrocytes, neurons, and microglia at 3-, 6-, and 9 mo of age. Astrocytes and neurons did not produce detectable amounts of OPN, while microglial production of OPN was robust and increased with disease progression (SI Appendix, Fig. S1A). There was a 10- to 20-fold increase in OPN mRNA and a twofold to threefold increase in OPN protein by 9-mo-old 5XFAD microglia compared with age-matched B6-WT controls (SI Appendix, Fig. S1 B and C), suggesting microglial OPN production closely parallels disease development.
Microglia that coexpress CD11c represent the sole producer of OPN in brains of healthy mice (16). We observed that microglial OPN production in 5XFAD mice is also confined to this CD11c+ microglial subset, which increased dramatically during the initial 6 mo of disease progression compared with relatively low levels in age-matched healthy (B6-WT) controls (Fig. 1 A and B).
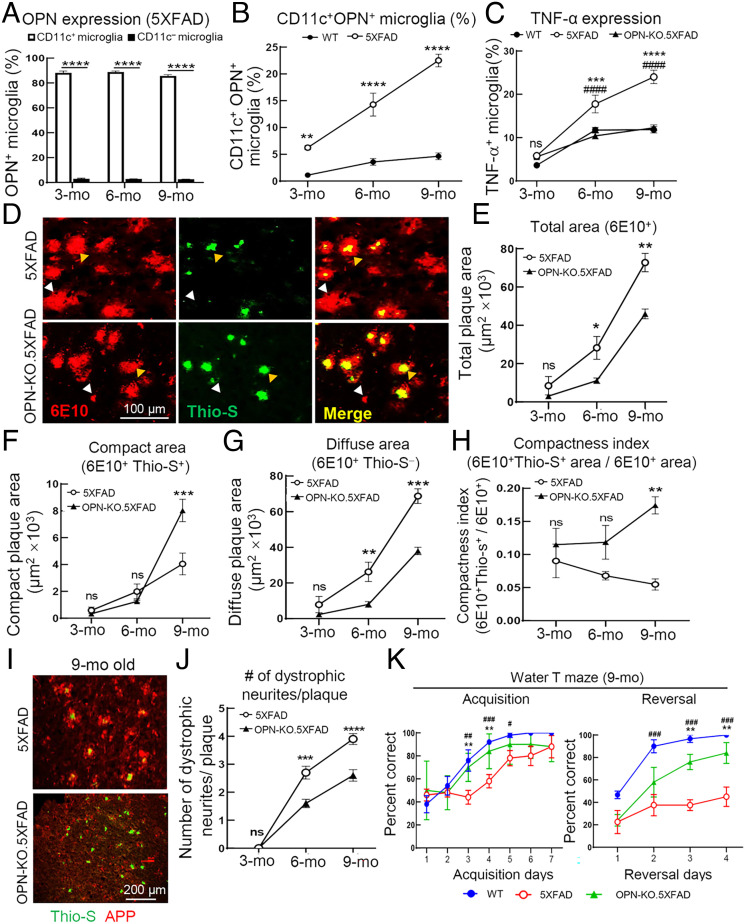
Fig. 1.
OPN deletion diminishes AD pathology and rescues cognitive deficits in 5XFAD mice. (A) Flow cytometric analysis of OPN expression in CD11c+ and CD11c− microglia from 5XFAD mice (n=6) at different disease stages. ****P < 0.0001 by two-way ANOVA with Bonferroni’s multiple comparisons test. (B) Flow cytometric analysis showing the increased percentage in CD11c+ OPN+ microglia from 5XFAD mice compared with age-matched WT mice during disease progression (n=5 to 6). ****P < 0.0001, **P < 0.01 by two-way ANOVA with Bonferroni’s multiple comparisons test. (C) Ex vivo analysis of microglial tumor necrosis factor-alpha (TNF-α) expression in WT, 5XFAD, and OPN-KO.5XFAD mice (n=6) during disease progression according to flow cytometry. *: WT vs. 5XFAD, ***P < 0.001, ****P < 0.0001; #: 5XFAD vs. OPN-KO.5XFAD, ####P < 0.0001, ns: not significant by two-way ANOVA with Bonferroni’s multiple comparisons test. (D) Representative immunofluorescent images of brain sections from 9-mo-old 5XFAD mice and OPN-KO.5XFAD mice stained with 6E10 (red) and Thioflavin-S (green) displaying different forms of Aβ plaques. Total plaques were defined as 6E10+. White arrows indicate diffuse plaques (6E10+ Thio-S−), while compact plaques (6E10+ Thio-S+) are indicated by yellow arrows. (Scale bar, 100 μm.) (E–G) Quantification of total area (6E10+), compact area (6E10+Thio-S+), and diffuse area (6E10+Thio-S−) of Aβ plaques in 3-, 6-, and 9-mo-old 5XFAD mice and OPN-KO.5XFAD mice (n= 54 fields from 3 mice/group). ***P < 0.001, **P < 0.01, *P < 0.05, ns: not significant by two-way ANOVA with Bonferroni’s multiple comparisons test. (H) The compactness index of Aβ plaques in 3-, 6-, and 9-mo-old 5XFAD mice and OPN-KO.5XFAD mice. The compactness index was calculated as 6E10+Thio-S+ area/ 6E10+ area (n =54 fields from three mice/group). **P < 0.01, ns: not significant by two-way ANOVA with Bonferroni’s multiple comparisons test. (I and J) Representative immunofluorescent images and quantification of dystrophic neurites (labeled with N terminus APP) per plaque (labeled with Thioflavin-S) in brain sections of 5XFAD and OPN-KO.5XFAD mice. The number of APP+ dystrophic neurites was quantified within 25 µm area of each plaque (n=30 to 40 plaques from three mice per group). ****P < 0.0001, ***P < 0.001, ns: not significant by two-way ANOVA with Bonferroni’s multiple comparisons test. (Scale bar, 200 μm.) (K) Spatial learning and memory (acquisition trials) and cognitive flexibility (reversal trials) were assessed by water T maze in 9-mo-old 5XFAD and OPN-KO.5XFAD mice. Age-matched WT mice were included as controls (n=3 to 5, #: WT vs. 5XFAD, #P < 0.05, ##P < 0.01, ###P < 0.001; *: 5XFAD vs. OPN-KO.5XFAD, **P < 0.01). Statistical analysis was performed by two-way ANOVA with Bonferroni’s multiple comparisons test. All data are presented as mean ± SEM.
We analyzed the impact of OPN on AD pathology using OPN-KO.5XFAD mice after crossing 5XFAD transgenic mice with Spp1flstop (OPN-KO) mice (17) and confirmed OPN deletion at both the genetic and protein levels (SI Appendix, Fig. S2). Genetic deletion of OPN in 5XFAD mice resulted in reduction in microglial production of TNF-α to levels similar to age-matched healthy (B6-WT) controls (Fig. 1C).
To directly test the potential contribution of OPN to microglial processing of Aβ plaques, we costained mouse brain cryosections with anti-Aβ mAb 6E10 (to identify both neurotoxic diffuse form and nontoxic condensed forms of Aβ plaques) and Thioflavin-S (which identifies only β-sheet+ Aβ condensed plaques) (18). We found that the 40 to 60% decrease in total plaque area in OPN-KO.5XFAD mice noted in Fig. 1 D and E was accompanied by an increase in compact plaque (6E10+Thio-S+) area and a decrease in diffuse plaque (6E10+Thio-S−) area (Fig. 1 F and G), as judged from a striking increase in plaque compactness index (6E10+Thio-S+ area/6E10+ area) (Fig. 1H).
Neuritic dystrophy, a central element of 5XFAD disease pathology consisting of swollen, bulbous-shaped neurites comprised of dysfunctional axons and terminals expressing the amyloid precursor protein (APP) (19), correlates with the clinical severity of dementia (20, 21). Since neurotoxic diffuse plaques positively correlate with dystrophic neurites (18), we analyzed the impact of OPN on neuritic dystrophy. We noted that the numbers of dystrophic neurites per plaque were reduced by approximately 50% in OPN-KO.5XFAD mice (Fig. 1 I and J). Given that increased numbers of dystrophic neurites correlate closely with cognitive decline (22, 23), we measured the impact of OPN expression on cognitive function of 9-mo-old 5XFAD mice using the water T maze to assess spatial learning and memory (acquisition trials) and cognitive flexibility (reversal trials). OPN deletion markedly improved these cognitive functions, as indicated by a 40 to 50% increase in correct choices made by OPN-KO.5XFAD mice in both the acquisition and reversal phases of these trials (Fig. 1K). Collectively, these data suggest that OPN deficiency substantially decreases microglial proinflammatory responses, total and diffuse Aβ plaques, dystrophic neurites, and improves cognitive function in 5XFAD mice.
OPN Promotes a CD11c + Microglial Proinflammatory Response via an Interaction with the αVβ3 Integrin Receptor and May Inhibit Aβ Plaque Compaction through Suppression of the TREM2/TAM−Lysosomal Phagocytic Pathway.
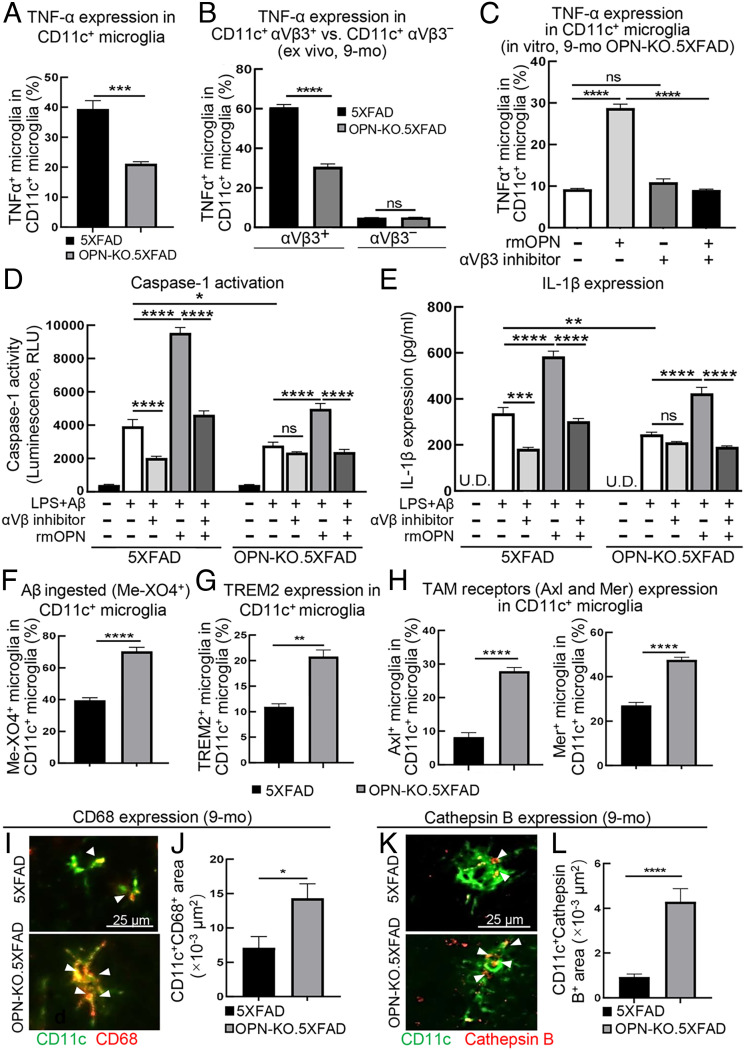
Since genetic deletion of OPN reduces production of TNF-α by total microglia to levels found in nondiseased mice (Fig. 1C), we examined the potential mechanistic relationship between OPN and the production of TNF-α by microglia. Microglia that expressed CD11c displayed a 50% reduction in TNF-α production compared with OPN-WT (5XFAD) mice (Fig. 2A). Expression of the canonical αVβ3 integrin receptor for OPN (16, 24) increased from 5 to 20% of microglia, while other OPN receptors, CD44 and αVβ5, were not detectable (SI Appendix, Fig. S3). We noted that a) CD11c+ microglia lacking the OPN receptor (αVβ3− CD11c+ microglia) did not produce detectable TNF-α and b) OPN-deficiency led to reduced proportion of TNF-α+ cells in αVβ3+ CD11c+ microglia (Fig. 2B). These findings suggest that OPN might promote TNF-α production via engagement of its αVβ3 receptor on CD11c+ microglia. This notion was supported by the observation that induction of microglial TNF-α expression by recombinant mouse OPN (rmOPN) was abolished by treatment with a cyclic RGD compound (αVβ3 inhibitor) that blocks the interaction between OPN and the αVβ3 integrin (Fig. 2C).
Fig. 2.
OPN promotes CD11c+ microglial proinflammatory responses via an interaction with the αVβ3 integrin receptor and impairs Aβ plaque compaction via suppression of the TREM2/TAM-lysosomal phagocytic pathway. (A) Flow cytometric analysis of CD11c+ microglial TNF-α production in 9-mo-old 5XFAD and OPN-KO.5XFAD mice (n=6). ***P < 0.001 by two-tailed Student’s t test. (B) Expression of TNF-α in CD11c+ αVβ3+ vs. CD11c+ αVβ3− microglia in 9-mo-old 5XFAD and OPN-KO.5XFAD mice (n=3). ****P < 0.0001, ns: not significant by two-tailed Student’s t test. (C) The impact of OPN−αVβ3 interaction on CD11c+ microglial production of TNF-α. Up-regulated TNF-α production induced by rmOPN was fully abrogated by αVβ3 inhibitor in CD11c+ microglia of 9-mo-old OPN-KO.5XFAD mice (n=3). ****P < 0.0001, ns: not significant by one-way ANOVA with Bonferroni’s multiple comparisons test. (D and E) The impact of OPN−αVβ3 interaction on microglial caspase-1 activation and IL-1β production. Activation of caspase-1 and IL-1β by provision of rmOPN was blocked following provision of αVβ3 inhibitor to microglia from 9-mo-old 5XFAD and OPN-KO.5XFAD mice (n=3). ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns: not significant by one-way ANOVA with Bonferroni’s multiple comparisons test. U.D. undetectable. (F) Aβ engulfment (Me-XO4+) by CD11c+ microglia in 9-mo-old 5XFAD and OPN-KO.5XFAD mice was analyzed by flow cytometry. Increased Me-XO4+ microglia were observed in OPN-deficient 5XFAD mice compared with 5XFAD mice (n=6). ****P < 0.0001 by two-tailed Student’s t test. (G) Flow cytometric analysis of TREM2 expression by CD11c+ microglia from 9-mo-old 5XFAD mice and OPN-KO.5XFAD mice (n=3). **P < 0.01 by two-tailed Student’s t test. (H) Flow cytometric analysis of Axl and Mer expression by CD11c+ microglia from 9-mo-old 5XFAD mice and OPN-KO.5XFAD mice (n=6). ****P < 0.0001 by two-tailed Student’s t test. (I and J) Representative immunofluorescent images and quantification of CD68+ area in CD11c+ microglia located within 25 μm of Aβ plaques (defined as plaque-associated CD11c+ microglia) in 9-mo-old 5XFAD and OPN-KO.5XFAD mice. (n=20 ROIs from 3 mice/group). *P < 0.05 by two-tailed Student’s t test. (Scale bar, 25 μm.) (K and L) Representative immunofluorescent images and quantification of cathepsin B+ area in plaque-associated CD11c+ microglia of 9-mo-old 5XFAD and OPN-KO.5XFAD mice (n=22 ROIs from 3 mice/group). ****P < 0.0001 by two-tailed Student’s t test. (Scale bar, 25 μm.) All data are presented as mean ± SEM.
Activation of the NLRP3 (NLR family pyrin domain containing 3) inflammasome represents a major neuroinflammatory component of AD pathology in both human and mouse models of AD (25). We analyzed the impact of OPN on microglial inflammasome activation in vitro according to caspase-1 activity and IL-1β production (25). Microglial intracellular caspase-1 activity was determined using a bioluminescent assay, which specifically detects caspase-1 activation, confirmed by a selective caspase-1 inhibitor (SI Appendix, Fig. S4). We observed that rmOPN substantially increased caspase-1 activity and IL-1β production in LPS+Aβ-stimulated microglia from both 9-mo 5XFAD and OPN-KO.5XFAD mice. This OPN-dependent promotion of inflammasome activation was fully blocked by the αVβ3 inhibitor (Fig. 2 D and E), suggesting that OPN-dependent promotion of microglial inflammasome activation reflects its interaction with αVβ3 integrin.
We then examined Aβ uptake by CD11c+ microglial subset using methoxy-X04 (Me-XO4) [which labels fibrillar Aβ (25, 26)] in 5XFAD and OPN-KO.5XFAD mice followed by flow cytometric analysis of Me-XO4+ microglia. OPN-deficiency was associated with a twofold increase in Me-XO4+ CD11c+ microglia (Fig. 2F). TREM2 facilitates microglial uptake of Aβ (27, 28) as a receptor for Aβ and contributes to microglial condensation of diffuse plaques (29, 30). More recently, TAM receptors (including Axl and Mer), which may work downstream of TREM2, may also regulate microglial engulfment of Aβ plaques and promote lysosomal degradation/compaction of ingested Aβ material, leading to microglial extrusion of inert condensed Aβ at the expense of diffuse neurotoxic forms of Aβ plaque in APP/PS1 mice (18).
We therefore asked whether the decrease in diffuse plaques noted in OPN-deficient 5XFAD brains (Fig. 1G) might reflect increased activation of this TREM2/TAM−lysosomal phagocytic pathway. We found that CD11c+ microglia from OPN-KO.5XFAD expressed increased TREM2, Axl and Mer, compared with control 5XFAD mice (Fig. 2 G and H). Analysis of CD11c+ microglia from OPN KO.5XFAD mice revealed enhanced lysosomal activation compared with age-matched control 5XFAD mice, as judged by increased expression of the CD68 lysosomal activation glycoprotein (Fig. 2 I and J) and the cathepsin B cysteine protease, which may cleave and degrade Aβ (31, 32) (Fig. 2 K and L).
Taken together, these data suggest that the reduced proportion of diffuse plaques (6E10+Thio-S−) within total plaques (6E10+) in OPN-deficient 5XFAD mice (Fig. 1G) may reflect de-repression of this TREM2/TAM−lysosomal Aβ compaction pathway. We further tested the contribution of OPN to lysosomal Aβ degradation in vitro. We found that rmOPN inhibited lysosomal activation, as judged by reduced CD68 expression, and this reduction could be reversed by addition of anti-OPN Ab (SI Appendix, Fig. S5 A and B). Suppression of lysosomal activation was accompanied by a substantial reduction in Aβ degradation [(Aβ MFI1h – Aβ MFI24h)/Aβ MFI1h] in the lysosomes of CD11c+ microglia that was fully reversed by addition of anti-OPN Ab (SI Appendix, Fig. S5C). Collectively, these data suggest that OPN-dependent inhibition of the TREM2/TAM−lysosomal phagocytic pathway, and associated Aβ plaque compaction may underpin a portion of OPN-dependent promotion of 5XFAD disease.
OPN Production Divides CD11c+ Microglia into a Pathogenic and Protective Subset.
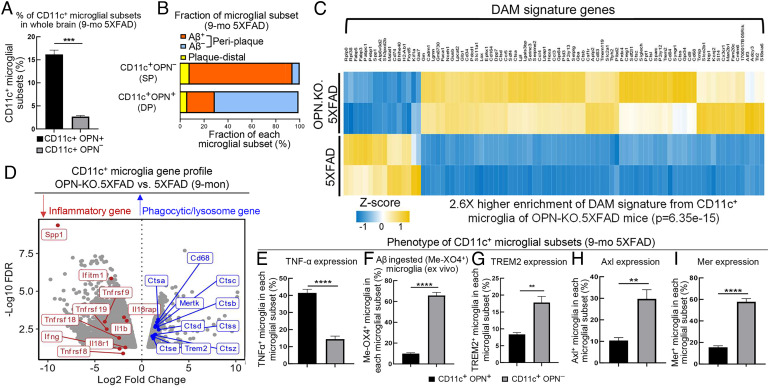
Both Spp1 (encoding OPN) and Itgax (encoding CD11c) are genetic hallmarks of the protective DAM phenotype (2). However, our findings suggest that OPN production inhibits Aβ uptake and promotes an inflammatory microglial phenotype. Flow cytometric analysis of microglia that coexpress OPN and CD11c indicates that about 15 to 20% of microglia express both CD11c and OPN (DP), and 3 to 5% express the CD11c+OPN− (SP) in brain tissue from 9-mo-old 5XFAD mice (Fig. 3A).
Fig. 3.
OPN production divides CD11c+ microglia into a pathogenic and protective subset. (A) The percentage of CD11c+ OPN+ and CD11c+ OPN− microglial subsets in whole brain of 9-mo-old 5XFAD mice was determined by flow cytometry and calculated as the number of each subset within the number of total microglia of the brain (n=3). ***P < 0.001 by two-tailed Student's t test. (B) Bar plot showing the fraction of CD11c+ OPN+ and CD11c+ OPN− microglial subsets in periplaque area (within 25 μm of Aβ plaque core) and the plaque-distal area (>25 μm of Aβ plaque core) in the brain of 9-mo-old 5XFAD mice. Aβ ingested or noningested cells of each microglial subset was calculated as the number of Aβ engulfed (Aβ+) or Aβ nonengulfed (Aβ−) microglia within the total number of each microglial subset resided in the periplaque area (33 ROIs from three mice were analyzed). The bar plot was a representative result of three independent experiments. (C) Enrichment of DAM gene signature was performed by mapping previously reported DAM signature genes with our CD11c+ microglial differentially expressed genes (DEGs) from OPN-KO.5XFAD compared with 5XFAD mice. Heatmap displaying OPN-deficient CD11c+ microglia highly resembled DAM-like gene signature with a 2.6-fold higher enrichment score (P=6.35e-15) compared with CD11c+ microglia from 5XFAD mice. Gene expression was considered significant if log2FC > 1 or log2FC < −1 with a FDR-adjusted P value < 0.05. (D) Volcano plot displaying CD11c+ microglial DEGs from 9-mo-old OPN-KO.5XFAD mice compared with 5XFAD mice. OPN deletion in CD11c+ microglia showed downregulation of a series of proinflammatory response-related genes (highlighted in red, i.e., Tnfrsf9, Il1b, Ifng) and upregulation of phagocytic/lysosomal activation-related genes essential for Aβ uptake and lysosomal degradation (highlighted in blue, i.e., Mertk, Trem2, Cd68, Ctsb). Gene expression was considered significant if log2FC > 1 or log2FC < −1 with a FDR-adjusted P value < 0.05. (E) Flow cytometric analysis of TNF-α expression in CD11c+ OPN+ and CD11c+ OPN− microglia from 9-mo-old 5XFAD mice (n=6). ****P < 0.0001 by two-tailed Student’s t test. (F) Flow cytometric analysis of Aβ engulfment (Me-XO4+) by CD11c+ OPN+ and CD11c+ OPN− microglial cell subsets from 9-mo-old 5XFAD mice (n=6). ****P < 0.0001 by two-tailed Student’s t test. (G) Flow cytometric analysis of TREM2 expression in CD11c+ OPN+ and CD11c+ OPN− microglia from 9-mo-old 5XFAD mice (n=3). **P < 0.01 by two-tailed Student’s t test. (H and I) Flow cytometric analysis of Axl and Mer expression by CD11c+ OPN+ and CD11c+ OPN− microglial subsets from 9-mo-old 5XFAD mice (n=6). ****P < 0.0001, **P < 0.01 by two-tailed Student’s t test. All data are presented as mean ± SEM.
Since DAM cells almost locate exclusively in plaque areas, we determined the plaque-associated fraction for each microglial subset by in situ analysis of brain cryosections from 9-mo-old 5XFAD mice. The specificity of CD11c-specific immunofluorescent signal was validated by several controls to confirm the validity of CD11c+OPN+ and CD11c+OPN− subset division (SI Appendix, Fig. S6 A and B). Analysis of microglia that resided within periplaque areas (within 25 μm of the Aβ plaque core) (33) and plaque-distal areas indicated that virtually all CD11c+ (both DP and SP) microglia were located within periplaque areas (Fig. 3B). Analysis of Aβ ingestion by periplaque DP (OPN+CD11c+) microglia revealed that less than 20% contained ingested Aβ. In contrast, over 90% periplaque SP (CD11c+ OPN−) microglia contained ingested Aβ material (Fig. 3B), indicating the phagocytic and potentially protective DAM phenotype may primarily reflect SP (CD11c+ OPN−) microglia.
This conclusion was supported by RNA-seq analysis. Since FACS-sorting of CD11c+OPN+ vs. CD11c+OPN− microglia depends on intracellular staining of OPN, we sorted CD11c+ microglia from 9-mo-old 5XFAD mice and OPN-KO.5XFAD mice according to the CD11c surface marker. We validated the FACS gating strategy of CD11c+ microglia in 5XFAD mice by flow cytometric analysis, as previously described (16) (SI Appendix, Fig. S7). We utilized DAM signature genes (2) as a reference gene set to analyze enrichment of a DAM-like signature by CD11c+ microglia from 5XFAD and OPN-deficient (OPN-KO).5XFAD mice. We noted that CD11c+ microglia from OPN-KO.5XFAD mice strongly resembled the DAM-like signature, as judged by a 2.6-fold enrichment (P=6.35e-15) of DAM signature genes compared with 5XFAD mice (Fig. 3C). Unbiased analysis of differentially expressed genes (DEGs) also revealed that OPN-deficient CD11c+ microglia expressed a reduced proinflammatory phenotype along with an enhanced phagocytic/lysosomal activation phenotype, as indicated by down-regulation of a series of proinflammatory genes (highlighted in red) and up-regulation of genes that contribute to Aβ uptake and lysosomal compaction/degradation (highlighted in blue, e.g., Mertk, Trem2, Cd68, Ctsb) (Fig. 3D and Dataset S1). These findings support the view that the OPN-deficient CD11c+ microglial subset may represent the protective DAM-like microglial subset.
We then characterized the phenotypes of CD11c+OPN+ and CD11c+OPN− microglial subsets by flow cytometry. The CD11c+OPN− (SP) microglia expressed low levels of TNF-α (Fig. 3E) and a sevenfold increase in Aβ ingestion (Me-XO4+ microglia) compared with the CD11c+OPN+ (DP) subset (Fig. 3F). Analysis of TREM2 and TAM expression revealed that CD11c+OPN−microglia displayed increased expression of TREM2 (twofold) and Axl and Mer (threefold) compared with CD11c+OPN+ microglia (Fig. 3 G–I). These data lend further support to the view that OPN production differentiates between the pathogenic and protective phenotype of CD11c+ microglia.
Collectively, these findings define the distinct contributions of two microglial subsets to 5XFAD disease. Although virtually all CD11c+OPN+ and CD11c+OPN− microglia locate within plaque areas, OPN production by these two CD11c+ microglial subsets accounts for their distinct functional periplaque phenotypes. The CD11c+OPN+ microglial subset produces high levels of TNF-α and less than 15% contain ingested Aβ, and reduced periplaque levels of TREM2/Axl/Mer, consistent with their pathogenic contribution to disease. In contrast, CD11c+OPN− microglia do not produce significant levels of TNF-α, engulf Aβ, and express higher levels of the TREM2/TAM proteins, consistent with increased lysosomal activation.
Increased OPN-Producing CD11c+ Microglia Correlate with Severity of Cognitive Impairment and Neuropathology in AD Patients.
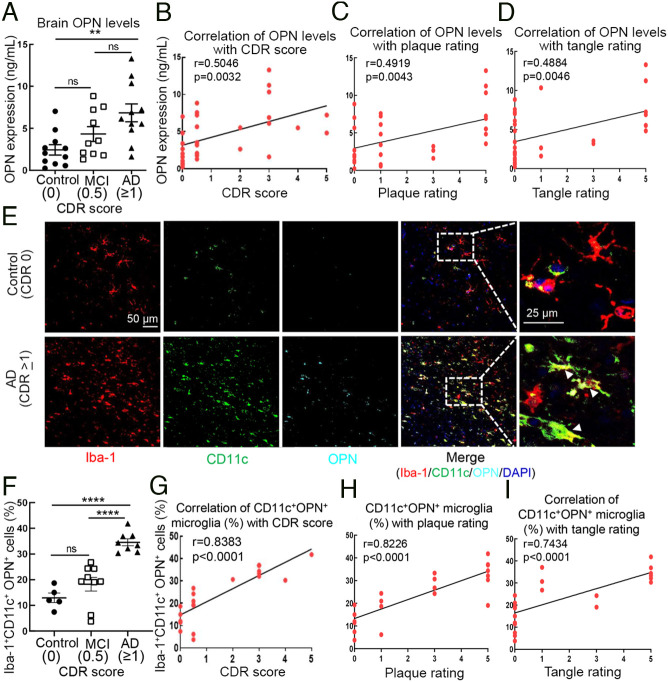
We extended these findings to an analysis of human AD using brain tissues from clinically and neuropathologically confirmed AD patients (cohort characterization in SI Appendix, Tables S1 and S2) whose clinical dementia rating (CDR) was ≥1 at death, as well as patients with mild cognitive impairment (MCI, CDR = 0.5) and cognitively normal controls (CDR = 0). Measurements of OPN protein level in middle frontal gyrus homogenates revealed a threefold increase in OPN expression in AD patient brains compared with that of cognitively normal control subjects. Increased OPN expression in AD patient brains compared with brains of patients with MCI was also noted but did not reach statistical significance (Fig. 4A). A correlative analysis of OPN expression in brain tissues and CDR score indicated that increased OPN expression correlated positively with dementia severity (r=0.5046, P=0.0032) (Fig. 4B). We also determined the potential correlation between OPN levels with neuritic plaques and neurofibrillary tangle density ratings that were determined postmortem by a neuropathologist before sample storage in the Mount Sinai brain repository as described (34). We found that higher brain OPN levels correlated with higher neuritic plaque levels (r=0.4919, P=0.0043) and neurofibrillary tangle ratings (r=0.4884, P=0.0046) (Fig. 4 C and D).
Fig. 4.
Increased microglial OPN production correlates with AD severity in human AD brains. (A) Analysis of OPN protein expression in brain (middle frontal gyrus) homogenates from AD patients (CDR ≥ 1, n=11) with mild cognitive impairment (MCI, CDR 0.5, n=10) and cognitively normal controls (CDR 0, n=11). **P < 0.01, ns: not significant by one-way ANOVA with Bonferroni’s multiple comparisons test. (B) Pearson’s correlation analysis of OPN expression levels in brain tissues from AD patients with CDR scores (CDR ≥ 1, n=11), MCI patients (CDR 0.5, n=10), and controls (CDR 0, n=11). r=0.5046, P=0.0032. (C and D) Pearson’s correlation analysis of brain OPN concentrations correlated with neuritic plaque rating (r=0.4919, P=0.0043) and tangle rating (r=0.4884, P=0.0046). (E and F) Representative immunofluorescent images from middle frontal gyrus of AD patients and normal controls stained for Iba-1 (microglia, red), CD11c (green) and OPN (cyan) are indicated by white arrows. Percent of CD11c+OPN+ microglia in brains of AD patients (CDR ≥1, n=8) compared with MCI (CDR 0.5, n=9) and control subjects (CDR 0, n=5). Each dot represents the average percentage of CD11c+OPN+ microglia over total Iba-1+ cells in 10 to 12 fields of each brain sample. (Scale bar, 25 μm.) ****P < 0.0001, ns: not significant by one-way ANOVA with Bonferroni’s multiple comparisons test. (G) Pearson’s correlation analysis of the percentage of CD11c+OPN+ microglia in human brain tissues correlated with CDR scores in AD patients (CDR ≥1, n=8), MCI patients (CDR 0.5, n=9) and controls (CDR 0, n=5). r=0.8383, P < 0.0001. (H and I) Pearson’s correlation analysis of the percentage of CD11c+OPN+ microglia in human brain tissues correlated with neurite plaque rating (r=0.8226, P < 0.0001) and tangle rating (r=0.7434, P < 0.0001). All data are presented as mean ± SEM.
To better resolve the potential correlation between OPN and AD pathology, we then determined the percentage of CD11c+ OPN+ microglia by immunofluorescent analysis of human brain sections from the middle frontal gyrus (35). Normal controls and MCI subjects displayed similar proportions of CD11c+ OPN+ microglia (CD11c+OPN+Iba-1+). We observed a threefold increase in CD11c+ OPN+ microglia in brain sections from AD patients compared with sections from normal controls (Fig. 4 E and F and Movies S1 and S2). Moreover, the percentage of CD11c+ OPN+ microglia from AD patients was significantly higher than that from MCI patients (Fig. 4F), suggesting that the percentage of CD11c+ OPN+ microglia represents a more sensitive parameter for differentiation of AD patients from patients who suffer from MCI than levels of OPN alone. This conclusion was further supported by the very strong correlation of the percentage of CD11c+ OPN+ microglia with CDR scores (r=0.8383, P < 0.0001) (Fig. 4G). Similar to the associations found with dementia severity, the percentage of CD11c+OPN+ (DP) microglia strongly correlated with the density of neuritic plaques (r=0.8226, P < 0.0001) and neurofibrillary tangles (r=0.7434, P < 0.0001) (Fig. 4 H and I). These findings highlight the strong correlation of this pathogenic microglial subset with disease severity in AD patients.
Administration of Anti-OPN mAb Inhibits Microglial Proinflammatory Response and Ameliorates Aβ Plaque Pathology in 5XFAD Mice.
Our analyses of OPN-KO.5XFAD mice suggest that OPN may contribute to AD pathology; therefore, we asked whether Ab-mediated blockade of OPN might alter progression of 5XFAD disease. The monoclonal anti-OPN antibody (clone: MPIIIB10, mouse IgG1) binding specificity was validated in a competition ELISA assay: Preincubation of plate-bound rmOPN with graded concentrations of AF808 anti-OPN Ab (goat IgG) resulted in a dose-dependent reduction in MPIIIB10 anti-OPN mAb binding (SI Appendix, Fig. S8A). Anti-OPN mAb (MPIIIB10) also displayed a dose-dependent reduction in in vitro TNF-α production by CD11c+ microglia from 9-mo-old 5XFAD mice (SI Appendix, Fig. S8B).
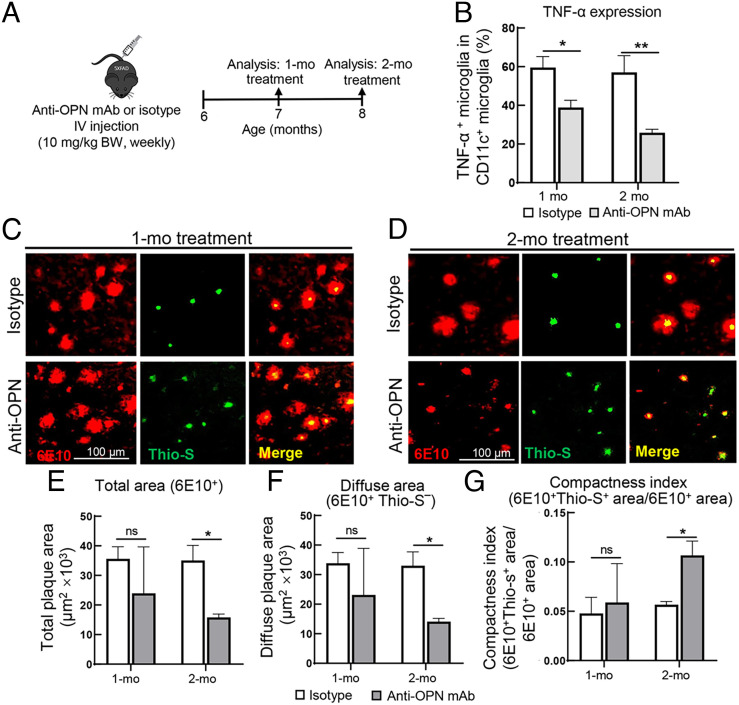
To test the impact of anti-OPN mAb (MPIIIB10) on the progression of 5XFAD disease, we performed weekly intravenous (I.V.) injections of anti-OPN mAb (10 mg/kg) into 6-mo-old 5XFAD mice for 1 or 2 mo followed by analysis of microglial proinflammatory responses and Aβ plaque pathology (Fig. 5A). Analysis of the impact of anti-OPN mAb on microglial proinflammatory responses revealed a 35% reduction in TNF-α+ CD11c+ microglia after 1 mo and 55% by 2 mo (Fig. 5B). Although anti-OPN mAb did not alleviate Aβ plaque pathology after 1-mo treatment (Fig. 5 C, E–G), administration for 2 mo resulted in a 55 to 60% decrease in total plaque (6E10+Thio-S+) and diffuse plaque (6E10+Thio-S−) areas, and a substantial increase in the plaque compactness index (6E10+Thio-S+ area/6E10+ area) (Fig. 5 D–G). These data support the hypothesis that OPN targeting by monoclonal antibody represents a potentially novel and effective mechanism-based approach to treat AD.
Fig. 5.
Administration of anti-OPN mAb inhibits microglial proinflammatory responses and ameliorates Aβ plaque pathology. (A) Schematic outline of anti-OPN mAb administration. Weekly intravenous injections (10 mg/kg) of anti-OPN mAb or isotype control (mouse IgG1) into 5XFAD mice starting at 6 mo of age for 1 or 2 mo, as described in Materials and Methods. Microglial proinflammatory responses and Aβ plaque pathology were analyzed after 1-mo and 2-mo treatment. (B) The percentage of TNF-α+CD11c+ microglia was determined by flow cytometric analysis following treatment of age-matched 5XFAD mice for 1or 2 mo with anti-OPN mAb or isotype-matched IgG1 is shown (n=3 to 4/group). **P < 0.01, *P < 0.05 by two-tailed Student’s t test. (C and D) Representative immunofluorescent images of condensed and diffuse forms of Aβ plaque distinguished by anti-Aβ 6E10 (red) and Thioflavin-S (green) staining in brain sections from 5XFAD mice after 1-mo or 2-mo treatment with anti-OPN mAb or isotype control. (Scale bar, 100 μm.) (E–G) Quantitation of total plaque area (6E10+), diffuse plaque area (6E10+Thio-S–) and compactness index of plaques in 5XFAD mice after 1-mo or 2-mo treatment with anti-OPN mAb compared with isotype treated mice (n=3). The compactness index is calculated as 6E10+Thio-S+ area/ 6E10+ area (n= 54 fields from 3 mice/group). *P < 0.05, ns: not significant by two-tailed Student’s t test. All data are presented as mean ± SEM.
Discussion
Microglial dysregulation characterized by a sustained inflammatory response and impaired Aβ plaque processing may contribute to neuritic dystrophy and cognitive decline in the setting of AD (1, 36). We show that OPN production by CD11c+ microglia promotes a proinflammatory microglial phenotype and impairs TREM2/TAM−dependent Aβ plaque consolidation in 5XFAD mice. Analysis of human brain tissue revealed that increased levels of the CD11c+ microglial subset may represent a sensitive and robust indication of AD-associated dementia and neuropathology. Proof-of-concept studies indicate that administration of anti-OPN mAb reduces microglial proinflammatory responses and ameliorates Aβ plaque pathology. These findings suggest that targeting the microglial OPN response may represent an effective therapeutic strategy for this disease.
Microglial heterogeneity has impeded a clear definition of its contribution to AD (2, 18). The view that DAM represent a protective microglial subset comes from observations that DAM locate exclusively within plaque areas and contain ingested Aβ material (2). Both Spp1 and Itgax are part of the genetic signature of the DAM subset (2). Our analysis of CD11c and OPN expression by microglia that surround Aβ plaques indicates a significant functional heterogeneity within the DAM description. Although virtually all CD11c+ microglia (both CD11c+OPN+ and CD11c+OPN−) reside in the periplaque area, OPN production distinguishes pathogenic from protective CD11c+ microglia. The CD11c+OPN− microglial subpopulation robustly ingests Aβ without concomitant TNF-α production, while CD11c+OPN+ microglia produce substantial amounts of TNF-α but do not efficiently ingest Aβ and express relatively low levels of TREM2 and TAM receptors. RNA-seq analysis of CD11c+ microglia sorted from 5XFAD and OPN-KO.5XFAD mice confirms that the OPN-deficient fraction of CD11c+ microglia expressed a DAM-like gene signature. These findings refine our understanding of the DAM phenotype and suggest that the protective vs. pathogenic contribution of DAM (2) may reflect the production of OPN by the latter microglial subpopulation.
The TREM2 receptor may allow microglia to surround and alter Aβ plaque structure into more compact forms, perhaps reflecting a TREM2-dependent phagocytic pathway of Aβ internalization and digestion by activated lysosomes (26, 37, 38). As a consequence, TREM2 deficiency in both mouse models and AD patients may increase Aβ burden, plaque diffusion, dystrophic neurites, and cognitive impairment (20, 26, 30, 39). Lemke and colleagues find that TAM receptors, which may act downstream of the TREM2 pathway, also represent critical molecular components of this microglial confinement mechanism (18). Up-regulation of TAM by plaque-associated microglia facilitates robust engulfment, degradation, and compaction of Aβ plaques by activated microglial lysosomes (18). Interestingly, TAM receptors also play a central role in noninflammatory phagocytosis of apoptotic cells (ACs), termed efferocytosis, by macrophages and microglia (40, 41). This confinement mechanism may be analogous to macrophage-dependent formation of granulomas of tuberculosis (42), that limits the dissemination of toxic Aβ oligomers throughout the brain. We find that OPN inhibits expression of the key molecular components of the TREM2/TAM pathway by CD11c+ microglia and that genetic ablation of OPN increases microglial lysosomal activation and plaque condensation, perhaps reflecting inhibition of the TREM2/TAM pathway. Transcriptomic analysis of OPN-deficient microglia also indicates that OPN may inhibit the TREM2/TAM–lysosomal pathway (Mertk, Trem2, Cd68, and Ctsb).
Until recently, clinical trials of AD therapeutics aimed at reducing Aβ plaque load have largely failed to improve cognition, possibly reflecting the indiscriminate targeting of both forms of Aβ plaques (43). Since microglial condensation of Aβ fibrils into dense plaques may represent a neuroprotective mechanism (18, 44), antibodies that target Aβ may disaggregate condensed nontoxic Aβ fibrils into oligomers that increase neurotoxicity may not be effective in rescuing cognitive impairment (43, 45). Our mechanistic analysis suggests that depletion of OPN may favorably alter the ratio of proinflammatory to phagocytic microglia leading to increased TAM-associated noninflammatory Aβ uptake and conversion of neurotoxic Aβ material into nontoxic dense-core plaques, that reduce neurite destruction and associated cognitive impairment.
A proof-of-concept analysis of the effects of administration of anti-OPN mAb for 2 mo indicated substantial reduction in both CD11c+ microglial proinflammatory responses and plaque pathology in 5XFAD mice at a relatively advanced stage of disease (6 mo). This therapeutic approach may convert the proinflammatory microglia to noninflammatory microglia equipped with enhanced ability to uptake and destroy Aβ fibrils. Optimization of this antibody-based regimen and modification of anti-OPN antibody to enhance brain penetration and reduce Fc–FcγR interactions may enhance the therapeutic effects of mAb-based OPN targeting.
Not all findings from AD mouse models are applicable to human AD. However, the findings reported here appear to apply to the human disease. Although elevated OPN levels in CSF and plasma samples have been reported in AD patients (46), the relationship between brain OPN levels and disease severity of AD patients has not been clear. Consistent with our findings in 5XFAD mice, brain OPN levels and especially the proportion of CD11c+ OPN+ microglia correlate strongly with plaque pathology according to neuritic plaque ratings of brain samples and correlate directly with dementia severity according to CDR scores of AD patients and controls. Neurofibrillary tangles, consisting mainly of aggregated hyperphosphorylated tau protein, represent an additional pathological feature of AD. We noted that brain OPN levels and the percent of CD11c+OPN+ microglia also correlate closely with neurofibrillary tangle ratings, suggesting that OPN may also contribute to tauopathy.
NLRP3 inflammasome activation contributes to the pathogenesis of tauopathy in an AD mouse model (47). Our transcriptomic analysis revealed a down-regulation of Il1b gene (encoding IL-1β, a key member of NLRP3 inflammasome pathway) in OPN-deficient CD11c+ microglia, and a marked increase in caspapse-1 activation and IL-1β production induced by rmOPN further supports the potential contribution of OPN to tauopathy. These findings suggest that OPN may regulate multiple disease-escalating factors that contribute to AD pathology. Although generalized increase in microglial proliferation has been described in the brains of AD patients (48), the majority of plaque-associated microglia do not proliferate (49), suggesting that the CD11c+ OPN+ microglial subset we define here may be distinct from the microgliosis response. CD11c+OPN+ microglia reside predominantly in periplaque areas, supporting the view that increased CD11c+OPN+ microglia in human brain tissue may represent a sensitive and reliable indicator of premortem cognitive impairment and severity of AD neuropathology.
In sum, these studies define a pathogenic CD11c+ OPN+ microglial subset that drives AD pathology via OPN-dependent promotion of proinflammatory responses and inhibition of a protective TREM2/TAM−lysosomal Aβ phagocytosis/compaction pathway. This two-pronged OPN-dependent mechanism of action, the strong correlation between levels of this OPN-producing CD11c+ microglial subset and human disease severity, and the disease-ameliorating effects of mAb-mediated OPN blockade suggest OPN as a promising therapeutic target for the treatment of AD.
Materials and Methods
Mice.
C57BL/6 (B6) and B6. Cg-Tg (APPSwFlLon, PSEN1*M146L*L286V)6799Vas/Mmjax (5XFAD) mice were obtained from the Jackson Laboratory (MMRRC). B6. Spp1flstop (OPN-KO) mice were previously generated by our lab (17). OPN-KO.5XFAD mice were generated by crossing B6. Spp1flstop mice with 5XFAD mice as a previously described strategy for breeding genetic-modified AD mouse models featuring amyloid pathology (5, 19, 39, 50). Spp1 gene mutation and 5XFAD transgene were validated by PCR genotyping. All mice were on a C57BL/6 background. Age-matched and sex-balanced mice were used for all experiments. Experimental cohorts of mice were cohoused from birth to control for microbiota (51, 52). All mice were housed in pathogen-free conditions with a 12-12-h light–dark cycle with ad libitum access to food and water. All experiments were performed in compliance with federal laws and institutional guidelines as approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute (DFCI).
Microglia Isolation.
Single-cell suspensions were prepared using a cold isolation protocol to avoid ex vivo artifacts as previously described (16, 53). Briefly, mice were anesthetized with isoflurane and transcardially perfused using ice-cold Phosphate Buffered Saline (PBS). Brains were quickly dissected and minced on ice using a scalpel followed by Dounce homogenization in ice-cold Hank’s balanced salt solution (HBSS). Cell suspensions were passed through a 70-μm cell strainer followed by spinning at 500 g for 5 min at 4°. Myelin and debris were removed using 10 mL ice-cold 40% Percoll (Sigma), spun for 30 min at 500 g before washing with 10 mL ice-cold HBSS, and spun again for 5 min at 500 g at 4°. All samples were then resuspended in ice-cold FACS buffer (0.5% BSA, 1mM EDTA, in 1× PBS) for staining. This isolation protocol has been used for all experiments unless stated otherwise.
Flow Cytometry.
The Ghost dye Violet 510 (1:1,000, Tonbo Biosciences) and anti-CD16/CD32 antibody (2.4G2, 1:100, BD Biosciences) were used to exclude dead cells and to avoid nonspecific staining, respectively. Microglia were stained with appropriate surface markers, including anti-CD11b (M1/70, 1:100 Biolegend), anti-CD45 (30-F11, 1:100 Biolegend), anti-CD11c (N418, 1:50 Biolegend), anti-αV integrin (RMV-7, 1:50, Biolegend), anti-β3 integrin (2C9.G2, 1:50, Biolegend), anti-β5 integrin (KN52, 1:50, eBioscience), anti-CD44 integrin (IM7, 1:100, BD Bioscience), anti-TREM2 (237920, 1:20, Novus Biologicals), anti-Axl (MAXL8DS, 1:50, eBioscience), anti-Mer (DS5MMER, 1:100, eBioscience) followed by fixation and permeabilization with an Intracellular Fixation & Permeabilization buffer kit (eBioscience) for the subsequent intracellular staining with anti-OPN (IC808, 1:10, R&D Systems), anti-TNF-α (MP6-XT22, 1:50, Biolegend), and anti-CD68 (FA-11, 1:100 Biolegend). Data were acquired on CytoFLEX LX (Beckman Coulter) and analyzed with FlowJo (Tree Star).
For detection of OPN expression in major brain cell types from 5XFAD mice at different stages of disease development, magnetic-activated cell sorting (MACS) was performed before flow cytometric analysis. Briefly, after generation of brain single-cell suspensions, MACS buffer (PBS pH 7.2, 2 mM EDTA and 0.5% BSA) was used to resuspend pelleted cells. Anti-mouse CD11b microbeads (Miltenyi) were used for magnetic isolation of total microglial populations, before anti-mouse ACSA-2 microbeads (Miltenyi) were used for subsequent isolation of astrocytes. Neurons were negatively enriched using anti-CD11b, anti-ACSA-2, and anti-O4 microbeads (Miltenyi) to deplete microglia, astrocytes, or oligodendrocytes following the manufacturer’s instructions. Cells were then incubated with Ghost dye Violet 510 (1:1,000, Tonbo Biosciences) and anti-CD16/CD32 antibody (2.4G2, 1:100, BD Biosciences) before staining with appropriate markers for further validation of cell types after magnetic isolation: cell surface staining of anti-CD11b (M1/70, 1:100, Biolegend) for microglia and intracellular staining of anti-GFAP (2E1.E9, 1:100, Biolegend) for astrocytes and anti-MAP2 (SMI52, 1:50, Biolegend) for neurons followed by intracellular staining with anti-OPN (IC808, 1:10, R&D Systems).
Validation of microglial CD11c expression was performed as previously described (16). Briefly, brain single-cell suspensions from 9-mo-old 5XFAD mice were stained with Ghost dye Violet 510 (1:1,000, Tonbo Biosciences) and anti-CD16/CD32 antibody (2.4G2, 1:100, BD Biosciences) followed by incubation with anti-CD11b (M1/70, 1:100, Biolegend), anti-CD45 (30-F11, 1:100, Biolegend), anti-CD11c (N418, 1:50, Biolegend), anti-TMEM119 (106-6, 1:200, ABCAM), anti-CCR2 (SA203G11, 1:100, Biolegend). CD11b+ cells were gated from single/live cells followed by subsequent gating of CD11b+ CD45low as microglia and CD11b+CD45high as macrophage. The microglial-specific marker Tmem119 (54) and CCR2 that expressed by blood-derived macrophage, but not expressed by microglia were included to distinguish microglia and macrophage (55, 56). CD11b+CD45high cells that expressed CCR2 but not Tmem119 were confirmed as macrophages, while CD11b+ CD45low cells that express Tmem119, but do not express CCR2 were confirmed as microglia. Fluorescence minus one negative controls were included to confirm the specificity of CD11c staining in CD11b+CD45low microglial populations. Brain CD45− cells that mainly contain nonimmune cells that do not express CD11c were also included as negative controls to further validate staining specificity.
RNA-Seq Analysis of CD11c Microglia.
Fluorescence-activated cell sorting for RNA-seq analysis.
Ghost dye Violet 510 (1:1,000, Tonbo Biosciences) and anti-CD16/CD32 antibody (2.4G2, 1:100, BD Biosciences) were used to exclude dead cells and to block Fc receptors, respectively. Single-cell suspensions were then stained with anti-CD11b (M1/70, 1:100, Biolegend), anti-CD45 (30-F11, 1:100, Biolegend), and anti-CD11c (N418, 1:50, Biolegend) antibodies for 20 min on ice. Samples were washed with ice-cold FACS buffer, spun down for 5 min at 500 g, and cell pellets were resuspended in 5 mL FACS buffer before sorting on a BD FACS Aria II using the 70-μm nozzle with purity mode and a sorting speed of ~10,000 events per second. After sorting, each sample was spun down, and cell pellets were immediately stored at −80° until further processing.
Due to the small population size of CD11c+ microglia and low yield of microglia by cold isolation in general, two biological samples were prepared for each group. For 9-mo-old 5XFAD mice, each CD11c+ microglial sample was pooled from 22 mice (11 male + 11 female) for a total of 44 mice (22 male + 22 female). For 9-mo-old OPN-KO.5XFAD mice, one CD11c+ microglia sample was pooled from 13 mice (6 male + 7 female) and the other sample was pooled from 14 mice (7 male + 7 female) for a total of 27 mice (13 male +14 female).
RNA extraction, library preparation, and HiSeq sequencing.
Total RNA was extracted from FACS-sorted cell pellets using the RNeasy Plus Universal Mini Kit following the manufacturer’s instructions (QIAGEN).
Library preparations for sequencing reactions were conducted at GENEWIZ, LLC. as follows: Concentration of extracted RNA was quantified using Qubit 2.0 Fluorometer (Life Technologies) and RNA integrity was determined using Agilent TapeStation 4200 (Agilent Technologies). Due to very limited cell numbers of CD11c+ microglia that can be obtained from adult mice, microglial samples of 9-mo-old 5XFAD and OPN-KO.5XFAD mice were processed with the SMART-Seq v4 Ultra Low Input Kit for sequencing for full-length complementary DNA (cDNA) synthesis and amplification (Clontech), and Illumina Nextera XT library for sequencing library preparation. Briefly, cDNA was fragmented and adaptor was added using transposase, followed by limited-cycle PCR to enrich and add index to the cDNA fragments. The final library was assessed with Agilent TapeStation.
Sequencing libraries clustered on flowcell lanes were loaded on the Illumina HiSeq 4000 according to the manufacturer’s instructions. Samples were sequenced using a 2×150-bp paired end (PE) configuration. Image analysis and base calling were conducted by HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and demultiplexed using Illumina's bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
RNA-Seq data analysis.
Mapping and gene counting were performed by GENEWIZ (South Plainfield, NJ, USA). After quality check of raw data, sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Mus musculus reference genome available on ENSEMBL using the STAR aligner v.2.5.2b, a splice aligner that detects splice junctions and incorporates them to help align entire read sequences, resulting in generation of BAM files. Unique gene hit counts were calculated using feature counts from the Subread package v.1.5.2. Only unique reads that fell within exon regions were counted.
Analysis of differentially expressed genes (DEGs) was performed by a negative binomial model implemented in the R package edgeR (Bioconductor) (57, 58), comparing OPN-KO.5XFAD to 5XFAD mice using a one-factor design and default parameters. The false discovery rate (FDR)–adjusted P value was estimated using the Benjamini–Hochberg correction. Gene expression was considered significantly up-regulated if log2FC > 1 or down-regulated if log2FC < −1 with FDR-adjusted P value < 0.05. Microglia similarity to the protective DAM phenotype as previously identified (2) was performed using a custom gene-set made from all DAM signature genes (2) input into the pathfindR R library (Bioconductor) using otherwise default settings. Significant DEGs from this gene-set were shown as a z-score-adjusted heatmap, with the overall enrichment score shown generated from the entire gene-set. Volcano plots were generated from the edgeR DEG output filtered on an FDR-adjusted P-value < 0.05.
In Vivo Aβ Phagocytosis Assay.
In vivo Aβ phagocytosis assay was performed as previously described (25, 26). Briefly, 5XFAD and OPN-KO.5XFAD mice were injected intraperitoneally with methoxy-X04 (Tocris Bioscience) at 10 mg/kg in 50% DMSO/50% NaCl (0.9%), pH 12. Brains were isolated 3 h after injection and made into single-cell suspensions followed by analysis of frequencies of methoxy-X04+ microglia by flow cytometry using a CytoFLEX LX (Beckman Coulter) followed by analysis using FlowJo (Tree Star).
Quantitative Real-Time PCR (qRT-PCR).
Microglial RNA was extracted using the RNeasy Plus Universal Mini Kit per the manufacturer’s instructions (QIAGEN). cDNA was then reverse transcribed using 100 ng RNA with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. Real-time quantitative PCR for Spp1 was performed with the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems) using 5 μL cDNA, 4.92 μL PowerUp™ SYBR™ Green Master Mix (Applied Biosystems), and 0.08 μL primer (Integrated DNA Technologies (IDT), working concentration: 200 nM) per reaction. Gene expression levels of Spp1 were compared using the ΔΔCt method normalized to β-actin.
Water T-Maze.
Animal cognitive testing was conducted in the NeuroBehavior Laboratory, Harvard Institute of Medicine (Boston, MA). The water T-maze (WTM) behavioral paradigm assesses spatial learning and memory by training mice to use spatial cues in a room to navigate to a hidden platform to escape from the water. This test also measures cognitive flexibility through a reversal learning procedure in which mice must learn a new location of the hidden platform. These tests were performed as previously described (59). Briefly, testing apparatus, consisting of a (14 cm × 4.6 cm) clear plexiglass maze with arms designated as north (N), south (S), east (E), or west (W), is divided to block off an arm to train the mouse to choose only E or W to escape. Mice are then placed in the N or S arms, in a semirandom order, at the start of each trial. The maze is then filled with (25 to 26 °C) water, and the escape platform is placed on the E side of the maze submerged 1 cm below the surface of the water. To ensure that mice cannot see the submerged platform, nontoxic white paint is added to water to increase opacity. At the start of the trial, the divider is placed to block off the appropriate arm, and mice are carried to the relevant start point. The experimenter scores correct and incorrect responses for each trial, and mice are allowed to stay on the platform for 10 s before being removed. Mice were given 10 trials per day, and the percent of correct responses was calculated by averaging correct responses across the 10 trials for each day. The platform is then moved to the opposite side, and the procedure is repeated for the reversal trial until mice learn the new position of the platform and scored as above.
Immunofluorescence.
Following transcardial perfusion, mouse brains were removed and fixed in 4% paraformaldehyde solution (paraformaldehyde (PFA), Electron Microscopy Sciences) at 4 °C overnight. After rinsing with PBS, fixed brains were dehydrated in 30% sucrose at 4 °C overnight before brains were embedded in optimal cutting temperature (OCT) compound (Sakura Finetek) and serial sagittal frozen sections (10 μm) were cut using a Cryostat (CM3050S, Leica). Brain cryosections were permeabilized with PBS containing 0.1% Triton X100 (PBS-T) for 1 h. After incubation for 1 h in a blocking solution containing 5% normal donkey serum (Jackson ImmunoResearch Lab) in PBS-T to prevent nonspecific binding, sections were incubated for 24 h with the appropriate primary antibodies: rabbit anti-Iba-1 (1:1,000, WAKO), anti-Amyloid β (6E10, 1:1,000, Biolegend), Biotin anti-mouse CD11c (N418, 1:50, Biolegend), goat anti-OPN (AF808, 1:100, R&D), anti-APP (A4, 1:1,000, Millipore), rat anti-CD68 (FA-11, 1:100, Bio-Rad), goat anti-Cathepsin B (AF965, 1:100, R&D). To amplify CD11c signal, the sections were washed with PBS and incubated for 5 min with the Alexa Fluor 488 streptavidin Tyramide SuperBoost Kit (Invitrogen), according to the manufacturer’s instructions. Sections were then incubated for 1 h with appropriate secondary antibodies: Alexa-594-conjugated donkey anti-rabbit IgG or Alexa-488-conjugated donkey anti-mouse IgG (1:500; Invitrogen). Brain cryosections incubated without anti-CD11c primary Ab or tyramide signal amplification (TSA) were included as negative controls to exclude nonspecific immunofluorescent signals. DAPI (Invitrogen) was used as a nuclear counterstain (10-min incubation). For Thioflavin-S staining, brain cryosections were incubated in filtered 1% aqueous Thioflavin-S for 10 min at room temperature and washed 2× for 3 min in 80% ethanol and 3 min in 95% ethanol, prior to adding blocking solution. Samples were analyzed under Olympus fluorescence microscope. Quantification of fluorescent microscopy images was performed using ImageJ (NIH). Detailed analysis methods of immunofluorescent staining data for each experiment are stated in the corresponding figure legend. Preparation of mouse brain section samples and image acquisition/quantification procedures were performed by independent investigators in a fully blinded manner.
In Vitro Primary Microglia Culture Assays.
Microglia isolation by MACS.
Adult mice were anesthetized with isoflurane followed by transcardial perfusion using ice-cold PBS. Since isolation of microglia from adult mice using a cold isolation protocol may lead to a low cell yield, which is not optimal for in vitro cell culture assays, we used a well-established enzymatic microglial isolation approach to increase cell yield along with subsequent MACS-based cell isolation (51, 60) for in vitro assays with some modifications (e.g., minimized enzyme dissociation time). Briefly, brains were minced and subjected to enzymatic dissociation using Collagenase type IV (300 U/mL, Worthington) or Papain (20 U/mL, Worthington) and DNase I (60 U/mL, Worthington) at 37° for 15 min, before myelin was removed by 30% Percoll gradient. MACS buffer (PBS pH 7.2, 2 mM EDTA and 0.5% BSA) was used to resuspend pelleted cells, and anti-mouse CD11b microbeads (Miltenyi) were used for magnetic isolation of total microglial populations (purity >95%), according to the instructions, for subsequent culture assays.
The impact of OPN−αVβ3 interaction on CD11c+ microglial production of TNF-α.
Microglia isolated from 9-mo-old OPN-KO.5XFAD mice were seeded into 12-well plates at 3×105 cells/well in a conventional microglia culture medium (DMEM-F12 with 10% fetal bovine serum + 1% penicillin/ streptomycin + 10 ng/mL recombinant mouse M-CSF). Microglia were preincubated with 10 μM of a selective αVβ3 inhibitor (Cilengitide, Selleck, Cat. S6387) for 1 h followed by the addition of 12.5 μg/mL rmOPN and cultured for 24 h. BD GolgiPlug with brefeldin A (1 μL/mL, BD Biosciences) was added into culture for the last 4 h before analysis of CD11c+ microglial expression of TNF-α by flow cytometry (CytoFLEX LX, Beckman Coulter).
The impact of OPN−αVβ3 interaction on microglial inflammasome activation.
Microglia isolated from 9-mo-old 5XFAD and OPN-KO.5XFAD mice were seeded into 96-well plates at 6×104 cells/well in a conventional microglia culture medium (DMEM-F12 with 10% fetal bovine serum + 1% penicillin/ streptomycin + 10 ng/mL recombinant mouse M-CSF). To induce inflammasome activation, microglia were primed with 100 ng/mL LPS for 3 h followed by stimulation with 10 μM Aβ1-42 fibrils overnight in the presence or absence of 12.5 μg/mL rmOPN. The αVβ3 inhibitor (Cilengitide, Selleck, 10 μM) was added into culture 1 h before the addition of rmOPN. Microglial intracellular caspase-1 activity was analyzed by the bioluminescent Caspase-Glo® 1 Inflammasome Assay Kit (Promega) per the manufacturer’s instruction. The detection specificity of caspase-1 activity was validated using a selective caspase-1 inhibitor (Ac-YVAD-CHO, 1 μM) included in the kit. Culture medium was collected for quantitative determination of microglial IL-1β production by the mouse IL-1β DuoSet ELISA kit (R&D Systems) according to the manufacturer’s instructions.
The impact of OPN on lysosome activation and Aβ degradation.
Microglia enriched from 9-mo-old 5XFAD and OPN-KO.5XFAD mice were seeded into 12-well plates at 3×105 cells/well in a conventional microglia culture medium (DMEM-F12 with 10% fetal bovine serum + 1% penicillin/ streptomycin + 10 ng/mL recombinant mouse M-CSF). Two batches of microglial cultures were set up and preincubated with anti-OPN Ab (10 μg/mL) for 1 h followed by incubation with rmOPN (12.5 μg/mL) overnight. Microglia were then incubated with FAM-Aβ1–42 peptide (1 μM) for 1 h followed by washing with cold PBS. Surface bound FAM-Aβ1–42 was quenched by incubation with 0.2% trypan blue in PBS (pH 4.4) for 1 min. Then one batch of microglia were harvested, and the mean fluorescence intensity (MFI) of FAM-Aβ1–42 in lysosomes of CD11c+ microglia (CD11c+CD68+) was determined by flow cytometry and defined as Aβ MFI1h. The other batch of microglia remained in culture for 24 h in the presence or absence of anti-OPN Ab (10 μg/mL) and/or rmOPN (12.5 μg/mL) before analysis of CD11c+ microglial expression of CD68 (MFI) and MFI of retained FAM-Aβ1–42 in CD11c+CD68+ microglia (defined as Aβ MFI24h) by flow cytometry (CytoFLEX LX, Beckman Coulter). Aβ degradation rate was calculated as (Aβ MFI1h - Aβ MFI24h) / Aβ MFI1h.
Anti-OPN mAb Binding Specificity assay.
The anti-OPN mAb (clone: MPIIIB10, isotype: mouse IgG1) was produced using hybridoma cells [Cat. MPIIIB10 (1)] from the Developmental Studies Hybridoma Bank (DSHB). Antibody scale-up and purification were performed by Bio-X-Cell. The binding specificity of anti-OPN mAb to rmOPN was determined by competitive ELISA immunoassay. Plates coated with 2 μg/mL rmOPN (R&D System) were preincubated with a well-defined polyclonal anti-OPN Ab (clone: AF808, goat IgG, R&D System) or its isotype goat IgG (R&D System) at graded concentrations (0, 0.625, 1.25, 2.5, 5, 10, or 20 nM) at room temperature (RT) for 2 h followed by incubation with 200 nM anti-OPN mAb (clone: MPIIIB10, DSHB). Binding of anti-OPN mAb (clone: MPIIIB10, DSHB) to rmOPN was detected using HRP-anti-mouse IgG (1:250, Invitrogen) and developed in TMB solution. The binding level was evaluated as the absorbance at 450 nm measured by a microplate reader (BioTek).
The impact of anti-OPN mAb on microglial proinflammatory responses.
Microglia isolated from 9-mo-old 5XFAD mice were incubated with anti-OPN mAb (clone: MPIIIB10, DSHB) at increasing concentrations (5, 10, 20 μg/mL) for 24 h. BD GolgiPlug with brefeldin A (1 μL/mL, BD Biosciences) was added to cultures for the last 4 h before flow cytometric analysis of TNF-α production by CD11c+ microglia. Incubation of microglia with isotype control (mouse IgG1) was included as a negative control.
In Vivo Administration of anti-OPN mAb.
To test the potential therapeutic effect of anti-OPN mAb (clone: MPIIIB10, DSHB), 6-mo-old 5XFAD mice received weekly intravenous injections (IV; 10 mg/kg body weight) of anti-OPN mAb or isotype control (mouse IgG1) were performed for 1 or 2 mo. Microglial proinflammatory responses and Aβ plaque pathology were analyzed at each end point. Microglial proinflammatory responses were evaluated according to the percentage of TNF-α+CD11c+ microglia by flow cytometry (CytoFLEX LX, Beckman Coulter). Aβ plaque pathology was assessed by immunofluorescent costaining of anti-Amyloid β (6E10, 1:1,000, Biolegend) and Thioflavin-S in mouse brain cryosections. Quantification of total plaque area (6E10+), diffuse area (6E10+Thio-S−), and compactness index (calculated as 6E10+Thio-S+ area/ 6E10+ area) were performed by ImageJ (NIH).
Human Brain Sample Processing and Assessment.
Brain sample processing.
Clinical and postmortem diagnoses, brain harvesting procedures, tissue processing, and assessment of AD pathology were performed and described in detail elsewhere (35). Deidentified brain tissues of the middle frontal gyrus (a region affected early by AD) (35) of clinically and neuropathologically characterized AD patients and controls were obtained from the Mount Sinai Brain Bank through the Neurobiobank (https://neurobiobank.nih.gov/). Three groups of decedents were included in this study: cognitively normal (n=11), mild cognitive impairment (MCI, n=10), and AD patients (n=11). SI Appendix, Tables S1 and S2 describe this cohort in detail. Cognitive status at death was ascertained using the clinical dementia rating (CDR) scale (61), which assesses cognitive and functional impairments associated with dementia and provides specific severity criteria for classifying subjects as nondemented (CDR = 0), questionably demented (CDR = 0.5), or with increasing levels of severity of dementia from CDR ≥ 1. Participants with an acute neurological condition such as stroke or traumatic brain injury were excluded.
Plaque and tangle assessment in human brain tissue.
Neuritic plaque and neurofibrillary tangle assessments were conducted at the Mount Sinai Brain Bank, according to the Consortium to Establish a Registry for Alzheimer's Disease protocol (62). Sections from paraffin-embedded blocks were variably stained with hematoxylin and eosin, modified Bielschowski, modified thioflavin S, anti-β amyloid (4G8), and anti-tau (AD2). All neuropathology data regarding the extent and distribution of neuropathologic lesions were collected in a blinded fashion relative to the subject's dementia status. Each case was assigned a Braak AD-staging score for progression of neurofibrillary neuropathology (63, 64). In addition, quantitative data regarding the density of neuritic plaques were collected as described (34).
OPN ELISA of human tissues.
For quantification of brain OPN concentrations in human brain samples, 10 mg frozen tissue was homogenized in 0.3 mL lysis buffer [20 mM Tris-Hcl pH8, 130 mM NaCl, 1% triton X100 and protease inhibitor cocktail (Roche)], kept on ice for 45 min and centrifuged at 13,000 g at 4 °C for 20 min. Protein concentrations of brain lysate were measured using a BCA kit (ThermoFisher Scientific), and 10 µg protein was loaded into each well of a human OPN Quantikine ELISA plate (R&D Systems), according to the manufacturer's instructions.
Human brain staining for CD11c+OPN+ microglia.
Paraffin-embedded brain tissue sections, from the middle frontal gyrus, were deparaffinized in xylene, rehydrated in graded ethanol, and washed with PBS containing 0.1% Triton X100 (PBS-T). Brain sections were boiled in 10 mM citric acid buffer (pH 6) for antigen retrieval and washed with PBS-T. The endogenous peroxidase activity of samples was quenched using 3% hydrogen peroxide solution for 60 min, followed by washing and incubation with Zyblack (Zytovision, BS-0002-8) for 30 min to reduce auto fluorescence. Endogenous biotin was blocked with the Biotin-Blocking Kit (Invitrogen, E21390). Staining was performed by multiplexing three Tyramide SuperBoost kits (Invitrogen, B40936, B40912, B40923). First, sections were blocked using blocking buffer for 60 min, then incubated with primary antibodies [biotinylated anti-OPN 1:50 (R&D systems, BAF1433), anti CD11c 1:150 (Novus, NBP2-44598) and anti Iba-1 1:500 (Wako, 019-19741)] at 4 °C overnight. After washing, sections were incubated with HRP-conjugated streptavidin for 60 min, washed and incubated with Alexa Fluor™ 647 Tyramide streptavidin reagent for 10 min, followed by reaction stop solution for 5 min. This was followed by superboost kits Alexa Fluor™ 488 tyramid anti-mouse and Alexa Fluor™ 555 tyramid anti-rabbit. Cell nuclei were marked with Hoechst, and slides were mounted with the Immu-Mount medium (Thermofisher Scientific).
Brain slices of control (n=5), MCI (n=9) and AD (n=8) were coded and 10 to 12 images were captured under a confocal microscope (Leica DMi8), under the same conditions of laser intensities, at three separate sequences; laser 408 nm at 5%, lasers 488 nm at 0.1%, laser 638 nm at 6.5%, and laser 552 nm at 3% exposures using magnification of X400. Triple-positive cells expressing Iba-1, CD11c, and OPN were counted, as well as total Iba-1-positive cells from each slice using ImageJ software (NIH). The percent of CD11c+OPN+ microglia (CD11c+OPN+Iba-1+) out of total Iba-1-positive cells was calculated for each slice, and an average was calculated for each brain sample. Blinded analysis was performed by an independent investigator.
Quantification and Statistical Analysis.
Data were presented as mean ± SEM. Statistical analysis was performed using GraphPad Prism software version 9.0. Analysis for comparing multiple groups was performed by one-way or two-way ANOVA with Bonferroni’s test. Comparisons between two groups were analyzed with two-tailed Student’s t test. The correlation analysis was performed using Pearson correlation. P < 0.05 was considered to denote significance and expressed as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The statistical details, including the specific statistical tests used in different analyses, as well as the respective sample sizes, were given in the respective figure legends.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Confocal 3D images of human brain section stained for CD11c+OPN+Iba-1+ microglia from a normal control. Microglia shown in the images were stained for Iba1 (red), CD11c (green) and OPN (cyan). 3D images consist of 34 Z stacks, each slice 0.29 μm thick accumulating to a total of 9.85 μm in depth for the entire image. The image was acquired at a magnification of ×63 with a zoom in effect.
Confocal 3D images of human brain section stained for CD11c+OPN+Iba-1+ microglia from an AD patient. Microglia represented in the images were stained for Iba1 (red), CD11c (green) and OPN (cyan). The 3D image consists of 29 Z stacks, each slice 0.29 μm thick accumulating to a total of 8.36 μm in depth for the entire image. The image was acquired at a magnification of ×63 with a zoom in effect.
Acknowledgments
We thank S. Carrillo and O. Oseghali for mouse genotyping and A. Angel for manuscript and figure preparation. The hybridoma MPIIIB10 (1) was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health (NIH) and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported in part by research grants from the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH under award numbers R01AI037562 and R01AI048125 (H.C.), the Edward N. & Della L. Thome Memorial Foundation Awards Program in Alzheimer’s Disease Drug Discovery Program and a gift from the LeRoy Schecter Research Foundation (H.C.).
Author contributions
Y.Q., X.S., S.L.-Z., I.C., M.S.B., and H.C. designed research; Y.Q., X.S., O.R., D.A., D.R., and S.L.-Z. performed research; Y.Q., X.S., A.E.W., H.-J.K., S.L.-Z., I.C., M.S.B., and H.C. analyzed data; and Y.Q., X.S., and H.C. wrote the paper.
Competing interests
The authors have patent filings to disclose. Provisional US patent applications relating to targeting OPN for immunotherapy of AD were filed on 5/31/2022 (63/347,113) and 1/18/2023 (63/480,398).
Footnotes
Reviewers: M.L.S., Duke University School of Medicine; and L.S., Stanford University.
Data, Materials, and Software Availability
RNA-seq data for CD11c+ microglia from 9-mo-old 5XFAD and 9-mo-old OPN-KO.5XFAD mice have been deposited to NCBI-Gene Expression Omnibus (GEO) under accession number GSE191118 (65). Data are available upon reasonable request from the authors.
Supporting Information
References
- 1.Heneka M. T., et al. , Neuroinflammation in Alzheimer’s disease. Lancet. Neurol. 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keren-Shaul H., et al. , A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Mathys H., et al. , Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grubman A., et al. , A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y., et al. , Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q., et al. , Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safaiyan S., et al. , White matter aging drives microglial diversity. Neuron 109, 1100–1117.e10 (2021), 10.1016/j.neuron.2021.01.027. [DOI] [PubMed] [Google Scholar]
- 8.Sala Frigerio C., et al. , The major risk factors for Alzheimer’s disease: Age, sex, and genes modulate the microglia response to Abeta Plaques. Cell Rep. 27, 1293–1306.e1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patarca R., et al. , Structural and functional studies of the early T lymphocyte activation 1 (Eta-1) gene. Definition of a novel T cell-dependent response associated with genetic resistance to bacterial infection. J. Exp. Med. 170, 145–161 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashkar S., et al. , Eta-1 (osteopontin): An early component of Type 1(cell-mediated) immunity. Science 287, 860–864 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Inoue M., Shinohara M. L., Cutting edge: Role of osteopontin and integrin alphav in T cell-mediated anti-inflammatory responses in endotoxemia. J. Immunol. 194, 5595–5598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danzaki K., Kanayama M., Alcazar O., Shinohara M. L., Osteopontin has a protective role in prostate tumor development in mice. Eur. J. Immunol. 46, 2669–2678 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal N., et al. , Secreted osteopontin from CD4(+) T cells limits acute graft-versus-host disease. Cell Rep. 37, 110170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hur E. M., et al. , Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat. Immunol. 8, 74–83 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Steinman L., New targets for treatment of multiple sclerosis. J. Neurol. Sci. 274, 1–4 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Shen X., Qiu Y., Wight A. E., Kim H. J., Cantor H., Definition of a mouse microglial subset that regulates neuronal development and proinflammatory responses in the brain. Proc. Natl. Acad. Sci. U.S.A. 119, e2116241119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavenworth J. W., Verbinnen B., Yin J., Huang H., Cantor H., A p85alpha-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nat. Immunol. 16, 96–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y., et al. , Microglia use TAM receptors to detect and engulf amyloid beta plaques. Nat. Immunol. 22, 586–594 (2021), 10.1038/s41590-021-00913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulrich J. D., et al. , ApoE facilitates the microglial response to amyloid plaque pathology. J. Exp. Med. 215, 1047–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKee A. C., Kosik K. S., Kowall N. W., Neuritic pathology and dementia in Alzheimer’s disease. Ann. Neurol. 30, 156–165 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Nelson P. T., et al. , Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. 71, 362–381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terry R. D., et al. , Physical basis of cognitive alterations in Alzheimer’s disease: Synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 30, 572–580 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Koffie R. M., Hyman B. T., Spires-Jones T. L., Alzheimer’s disease: Synapses gone cold. Mol. Neurodegener. 6, 63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber G. F., et al. , Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J. Leukoc. Biol. 72, 752–761 (2002). [PubMed] [Google Scholar]
- 25.Heneka M. T., et al. , NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., et al. , TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y., et al. , TREM2 is a receptor for beta-amyloid that mediates microglial function. Neuron 97, 1023–1031.e1027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinberger G., et al. , TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6, 243ra286 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Wang S., et al. , Anti-human TREM2 induces microglia proliferation and reduces pathology in an Alzheimer’s disease model. J. Exp. Med. 217, e20200785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan P., et al. , TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe Axonal dystrophy. Neuron 92, 252–264 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Chapman H. A., Riese R. J., Shi G. P., Emerging roles for cysteine proteases in human biology. Annu Rev. Physiol. 59, 63–88 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Mueller-Steiner S., et al. , Antiamyloidogenic and neuroprotective functions of cathepsin B: Implications for Alzheimer’s disease. Neuron 51, 703–714 (2006). [DOI] [PubMed] [Google Scholar]
- 33.McAlpine C. S., et al. , Astrocytic interleukin-3 programs microglia and limits Alzheimer’s disease. Nature 595, 701–706 (2021), 10.1038/s41586-021-03734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haroutunian V., et al. , Regional distribution of neuritic plaques in the nondemented elderly and subjects with very mild Alzheimer disease. Arch. Neurol. 55, 1185–1191 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Haroutunian V., Katsel P., Schmeidler J., Transcriptional vulnerability of brain regions in Alzheimer’s disease and dementia. Neurobiol. Aging 30, 561–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarlus H., Heneka M. T., Microglia in Alzheimer’s disease. J. Clin. Invest. 127, 3240–3249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi P., et al. , TREM2 modulates differential deposition of modified and non-modified Abeta species in extracellular plaques and intraneuronal deposits. Acta Neuropathol. Commun. 9, 168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parhizkar S., et al. , Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nat. Neurosci. 22, 191–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., et al. , TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fourgeaud L., et al. , TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zagorska A., Traves P. G., Lew E. D., Dransfield I., Lemke G., Diversification of TAM receptor tyrosine kinase function. Nat. Immunol. 15, 920–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemke G., Huang Y., The dense-core plaques of Alzheimer’s disease are granulomas. J. Exp. Med. 219, e20212477 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panza F., Lozupone M., Logroscino G., Imbimbo B. P., A critical appraisal of amyloid-beta-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 15, 73–88 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Baik S. H., Kang S., Son S. M., Mook-Jung I., Microglia contributes to plaque growth by cell death due to uptake of amyloid beta in the brain of Alzheimer’s disease mouse model. Glia 64, 2274–2290 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Long J. M., Holtzman D. M., Alzheimer disease: An update on pathobiology and treatment strategies. Cell 179, 312–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y., et al. , Elevated osteopontin levels in mild cognitive impairment and Alzheimer’s disease. Mediators Inflammation 2013, 615745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ising C., et al. , NLRP3 inflammasome activation drives tau pathology. Nature 575, 669–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez-Nicola D., Fransen N. L., Suzzi S., Perry V. H., Regulation of microglial proliferation during chronic neurodegeneration. J. Neurosci. 33, 2481–2493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boekhoorn K., Joels M., Lucassen P. J., Increased proliferation reflects glial and vascular-associated changes, but not neurogenesis in the presenile Alzheimer hippocampus. Neurobiol. Dis. 24, 1–14 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Hong S., et al. , Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulland T. K., et al. , TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 170, 649–663.e613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stappenbeck T. S., Virgin H. W., Accounting for reciprocal host-microbiome interactions in experimental science. Nature 534, 191–199 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Hammond T. R., et al. , Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett M. L., et al. , New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. U.S.A. 113, E1738–E1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizutani M., et al. , The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J. Immunol. 188, 29–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wlodarczyk A., et al. , A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36, 3292–3308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy D. J., Chen Y., Smyth G. K., Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic. Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ofengeim D., et al. , RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, E8788–E8797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Y., et al. , Overexpressing low-density lipoprotein receptor reduces tau-associated neurodegeneration in relation to apoE-linked mechanisms. Neuron 109, 2413–2426.e2417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marin D. B., et al. , Reliability and validity of a chronic care facility adaptation of the Clinical Dementia Rating scale. Int. J. Geriatr. Psychiatry 16, 745–750 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Mirra S. S., et al. , The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–486 (1991). [DOI] [PubMed] [Google Scholar]
- 63.Braak H., Braak E., Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 64.Braak H., Alafuzoff I., Arzberger T., Kretzschmar H., Del Tredici K., Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112, 389–404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu Y., Shen X., Kim H.-J., Cantor H., Osteopontin-producing microglia contribute to Alzheimer's Disease(AD) pathology and severity. NCBI GEO: Expression profiling by high throughput sequencing. Accession No. GSE191118. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE191118.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Confocal 3D images of human brain section stained for CD11c+OPN+Iba-1+ microglia from a normal control. Microglia shown in the images were stained for Iba1 (red), CD11c (green) and OPN (cyan). 3D images consist of 34 Z stacks, each slice 0.29 μm thick accumulating to a total of 9.85 μm in depth for the entire image. The image was acquired at a magnification of ×63 with a zoom in effect.
Confocal 3D images of human brain section stained for CD11c+OPN+Iba-1+ microglia from an AD patient. Microglia represented in the images were stained for Iba1 (red), CD11c (green) and OPN (cyan). The 3D image consists of 29 Z stacks, each slice 0.29 μm thick accumulating to a total of 8.36 μm in depth for the entire image. The image was acquired at a magnification of ×63 with a zoom in effect.
Data Availability Statement
RNA-seq data for CD11c+ microglia from 9-mo-old 5XFAD and 9-mo-old OPN-KO.5XFAD mice have been deposited to NCBI-Gene Expression Omnibus (GEO) under accession number GSE191118 (65). Data are available upon reasonable request from the authors.