Abstract
In this paper we describe the isolation of a second gene in the newly identified pyridoxine biosynthesis pathway of archaebacteria, some eubacteria, fungi, and plants. Although pyridoxine biosynthesis has been thoroughly examined in Escherichia coli, recent characterization of the Cercospora nicotianae biosynthesis gene PDX1 led to the discovery that most organisms contain a pyridoxine synthesis gene not found in E. coli. PDX2 was isolated by a degenerate primer strategy based on conserved sequences of a gene specific to PDX1-containing organisms. The role of PDX2 in pyridoxine biosynthesis was confirmed by complementation of two C. nicotianae pyridoxine auxotrophs not mutant in PDX1. Also, targeted gene replacement of PDX2 in C. nicotianae results in pyridoxine auxotrophy. Comparable to PDX1, PDX2 homologues are not found in any of the organisms with homologues to the E. coli pyridoxine genes, but are found in the same archaebacteria, eubacteria, fungi, and plants that contain PDX1 homologues. PDX2 proteins are less well conserved than their PDX1 counterparts but contain several protein motifs that are conserved throughout all PDX2 proteins.
Recent work in our laboratory with the filamentous, phytopathogenic fungus Cercospora nicotianae revealed that a highly conserved group of gene homologues found in eubacteria, archaebacteria, fungi, and plants play a role in a divergent pyridoxine (vitamin B6) biosynthesis pathway (8). PDX1 was originally identified as a gene required for resistance of this fungus to a singlet-oxygen-generating toxin, cercosporin, which it produces to parasitize plants (9, 10). During characterization of this gene, however, we discovered that it rescued both C. nicotianae and Aspergillus flavus pyridoxine auxotrophs to prototrophy (8). This observation was subsequently confirmed in Aspergillus nidulans, in which a PDX1 homologue, PYROA, also rescued pyridoxine auxotrophy (24). Interestingly, despite this direct evidence for the involvement of PDX1 homologues in pyridoxine synthesis, PDX1 shows no homology to any of the known Escherichia coli pyridoxine biosynthesis genes or to any gene in the completely sequenced E. coli genome. Database analysis determined that organisms with homologues to the E. coli genes (some eubacteria) lacked PDX1 homologues and that organisms with PDX1 homologues (other eubacteria, archaebacteria, fungi, and plants) lacked homologues to the E. coli genes. These data suggested that a divergence in the pyridoxine synthesis pathway occurred sometime during the evolution of the eubacteria (8).
The advent of genomic and other large-scale sequencing projects allows homology comparisons on an organismal level. Saccharomyces cerevisiae contains three unlinked PDX1 homologues, one of which (SNZ1, for snooze) was extensively studied because its expression increases dramatically during stationary phase (5). Analyses by Galperin and Koonin (12) uncovered that PDX1-containing organisms also contained a copy of a second homologous gene and that three of these organisms (S. cerevisiae and the eubacteria Bacillus subtilis and Haemophilus influenzae) all encode this second gene in close physical proximity to their PDX1 homologues. These researchers hypothesized that the protein encoded by this second open reading frame (ORF) (which they named SNZB) possesses glutamine amidotransferase activity. Clusters of orthologous groups of protein analysis performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/COG/) compares protein sequences encoded in 21 complete genomes from 17 major phylogenetic lineages. This analysis supports the supposition that PDX1 and SNZB are functionally linked because it indicates that there is only one other gene with the same organismal distribution pattern as PDX1, and that gene is SNZB (19, 29, 30). Padilla et al. (25) further characterized the yeast SNZB homologues, dubbing them SNO (Snz-proximal ORF), and showed that all three SNZ/SNO pairs were coordinately regulated during growth and nutrient limitation. They also expressed both proteins from one of the gene pairs using a yeast two-hybrid system and showed that the proteins interact.
Despite the above results, there is no direct evidence for a role of SNZB/SNO homologues in pyridoxine synthesis. We have five UV-generated mutant strains of C. nicotianae that are pyridoxine auxotrophs (8, 16, 18). Three of these strains can be restored at high frequency to prototrophy by transformation with PDX1, while the two remaining strains, CS2 (CS for cercosporin sensitive) and CS7, have wild-type PDX1 genes (9, 10), suggesting that they are mutant in a different gene in the biosynthesis pathway. In light of the above data, it seemed reasonable to isolate a C. nicotianae SNZB/SNO homologue and test its involvement in pyridoxine metabolism. Here we report the successful isolation of the C. nicotianae SNZB/SNO homologue and confirmation of its role in pyridoxine biosynthesis via both targeted gene disruption and the ability to rescue our C. nicotianae non-PDX1 pyridoxine mutants to prototrophy.
MATERIALS AND METHODS
Strains, cultural conditions, and fungal transformations.
The wild-type (ATCC 18366) and pyridoxine auxotrophic (CS2, CS7, CS8, and 9714-1) C. nicotianae strains used in this study were described previously (8, 16, 18). Briefly, CS2, CS7, and CS8 were generated by UV mutagenesis, while 9714-1 is a pdx1 null strain derived via targeted gene replacement. All strains were maintained on malt medium at 28°C. Experiments to determine pyridoxine auxotrophy or prototrophy used a minimal medium (17), bacteriological agar (Sigma), and supplementation, when necessary, with pyridoxine to a final concentration of 1 μg/ml. Cercosporin resistance was assayed by growth on CM medium (17) supplemented with 10 μM cercosporin. In both cases growth was assayed by transferring fungal mycelium as a toothpick point inoculation and measuring increase in colony diameter after 4 days of growth at 28°C. Cercosporin resistance assays were conducted in a lighted growth chamber (45 to 55 μEinsteins/m−2/s). C. nicotianae strains were transformed as previously described (9, 11).
Cloning of PDX2.
Degenerate primers for amplification of an internal portion of the C. nicotianae PDX2 gene were designed by the CODEHOP (consensus-degenerate hybrid oligonucleotide primers) program (http://blocks.fhcrc.org/blocks/codehop.html) (27), based on protein sequences from Neurospora crassa, Schizosaccharomyces pombe, and the three yeast homologues. The 5′ and 3′ primers were, respectively, GACCAACATAAACCTACTTGGGGTACNTGYGCNGG and GATCAATAATTTCTTCAATAACAGGAGCNCKDATRAA (see Fig. 4A for regions of the protein to which these correspond). Amplification was performed using Amplitaq Gold (PE Biosystems), an annealing temperature of 57°C, and 50 cycles of amplification. Because the amplification reaction also contained a high-molecular-weight fragment unlikely to correspond to the desired product, an approximately 200-bp band was gel purified and cloned in pGEM-T-Easy (Promega). Sequence analysis confirmed that the cloned fragment corresponded to the desired product.
FIG. 4.
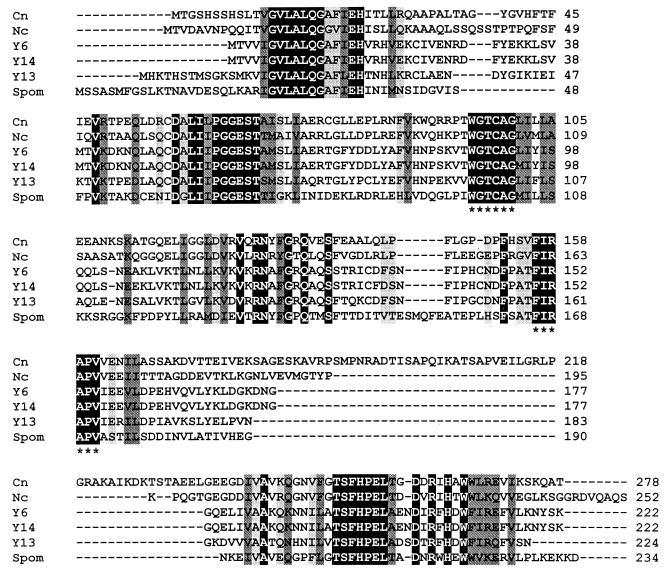
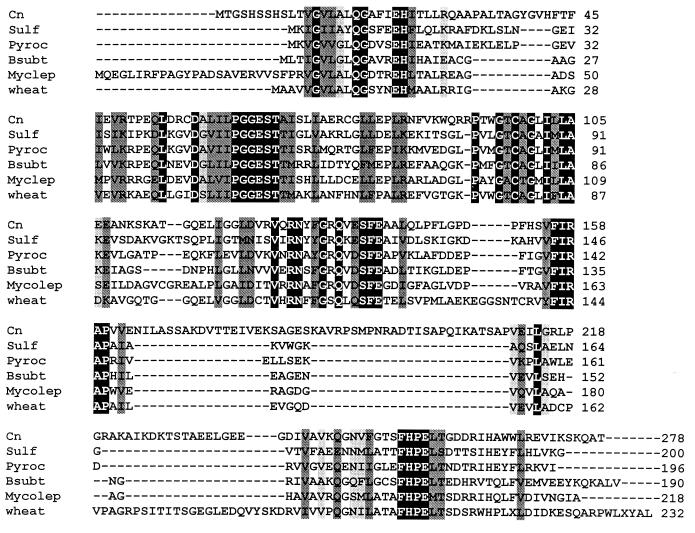
Comparison of the predicted amino acid sequences of PDX2 and homologues. (A) Comparison of all fungal PDX2 homologues. Species include C. nicotianae (Cn), Neurospora crassa (Nc), S. cerevisiae homologue from chromosome 6 (Y6), S. cerevisiae homologue from chromosome 14 (Y14), S. cerevisiae homologue from chromosome 13 (Y13), and S. pombe (Spom). The protein sequences from which the degenerate primers were derived are indicated by asterisks. (B) Comparison of the C. nicotianae (Cn) PDX2 protein sequence with representative sequences from other major taxa, including the archaebacteria Sulfolobus solfataricus (Sulf) and Pyrococcus horikoshii (Pyroc), the eubacteria Bacillus subtilis (Bsubt) and Mycobacterium leprae (Myclep), and the plant wheat (Triticum aestivum). The black-boxed residues with white letters indicate identical residues in all proteins, while the darker and lighter shading indicates, respectively, conserved substitutions and semiconserved substitutions.
The entire gene plus flanking regions was recovered using inverse PCR. Southern hybridization analysis was used to determine which restriction enzymes digested the C. nicotianae genome into fragments suitable for inverse PCR amplification. EcoRI-digested DNA was then self-ligated at dilute concentrations and used as a template for an inverse PCR amplification. When a fragment of the expected size was amplified, it was cloned into pGEM-T-Easy (Promega) for sequence analysis. After sufficient sequence was obtained to encompass the entire ORF plus upstream promoter and downstream sequences, primers were designed to amplify the intact gene from C. nicotianae genomic DNA using the high-fidelity DNA Pfu polymerase (Promega). After addition of an A residue to both ends, the amplification product was cloned into pGEM-T-Easy for sequence analysis. It was then recovered from pGEM-T-Easy for cloning into the fungal vector pBARKS1 (28) for transformation into C. nicotianae mutant strains.
Gene disruption.
To produce a gene disruption construct, the cloned inverse PCR product was recovered from pGEM-T-Easy and cloned into the fungal transformation vector pBARKS1. Because the PDX2 gene is located at either end of the recovered fragment, ligation into pBARKS1 produced a gene disruption construct with the entire vector interrupting the PDX2 ORF.
PCR amplification of PDX2 from mutant strains CS2 and CS7 and RT-PCR.
The PDX2 ORFs were amplified from CS2 and CS7 genomic DNA with the high-fidelity Pfu DNA polymerase using conditions described by the manufacturer (Promega). Reverse transcription (RT)-PCR was performed using the Access RT-PCR kit (Promega) according to the manufacturer's specifications.
Manipulations of nucleic acids.
Fungal genomic DNA was extracted as described (36), digested with restriction enzymes, and electrophoresed though 0.8% agarose prior to transfer to Magnagraph membrane (Osmonics). Probes were generated via PCR incorporation of digoxigenin-dUTP using either degenerate primers or, for the full-length PDX2 probe, the primers that span the start and stop codons, as shown in Fig. 1. Hybridizations were carried out in aqueous buffer at 65°C and washes at high stringency. Standard methods were used for the construction of plasmids and transformation into E. coli strain DH5α.
FIG. 1.
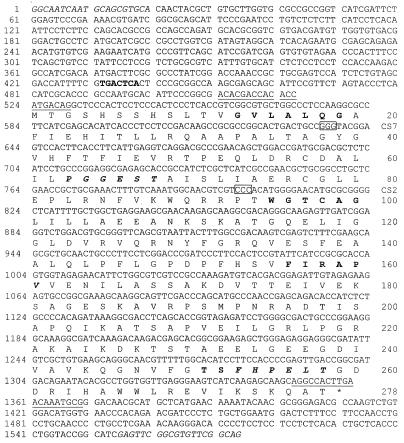
Nucleotide sequence of C. nicotianae PDX2 gene. The nucleotides shown are sufficient to complement the two non-pdx1 pyridoxine auxotrophs, CS2 and CS7. The nucleotides in italics at either end represent the primers used to amplify this sequence from C. nicotianae genomic DNA. The underlined nucleotides are the primers used to amplify the RT-PCR product and to label the coding sequence with digoxigenin-11-dUTP. The putative AP-1 recognition sequence is shown in bold. The boxed nucleotides represent the regions of the CS2 and CS7 pdx2 genes that are missing a single nucleotide. The bold italic amino acid residues are identical in all known PDX2 proteins, while the additional amino acid residues shown in bold type are completely conserved only among fungal PDX2 proteins.
DNA sequence analysis.
DNA sequence analysis was performed at the Molecular Genetics Facility at the University of Georgia, Athens. Homology searches were performed at the National Center for Biotechnology Information with the Blast network service (1). Both strands of the entire wild-type gene were sequenced, while one strand of RT-PCR products or PDX2 products from mutants was analyzed. Multiple alignments were performed using the European Bioinformatics Institute Clustal W Service (http://www2.ebi.ac.uk/clustalw) (33).
Nucleotide and protein sequence accession numbers.
The GenBank accession number for the C. nicotianae PDX2 gene is AF294268. The protein sequences shown in Fig. 4 include Neurospora crassa (GenBank AW713653), S. cerevisiae homologues (Swiss-Prot P53823, Swiss-Prot Q03144, and Swiss-Prot P43544), S. pombe (GenBank AL132798), Sulfolobus solfataricus (GenBank Y18930), Pyrococcus horikoshii (GenBank AP000006), Bacillus subtilis (Swiss-Prot P37527), Mycobacterium leprae (GenBank U00011), and wheat (Triticum aestivum) (GenBank BE217011).
RESULTS
Amplification of PDX2.
Using degenerate primers (see Materials and Methods and Fig. 4A), a fragment comparable in size to fragments amplified from both N. crassa and yeast SNO clones was amplified from C. nicotianae genomic DNA and cloned. After sequence analysis revealed it to encode a partial protein with strong homology to the SNO homologues of other fungi, the entire ORF and flanking regions were recovered using inverse PCR. Sufficient sequence was determined from the inverse PCR product for designing primers to amplify and clone a functional ca. 1,600-bp gene using the high-fidelity thermostable Pfu DNA polymerase (Fig. 1). Sequence analysis revealed that this fragment contains an uninterrupted ORF of 843 nucleotides encoding a putative polypeptide of 278 amino acid residues. This apparent lack of introns was confirmed by amplification, cloning, and sequence analysis of an RT-PCR product. No canonical promoter sequences are found in the region upstream of the PDX2 ORF. There is, however, an AP-1 response recognition site approximately 100 bp 5′ of the initiation codon (35).
Complementation of pyridoxine auxotrophs CS2 and CS7.
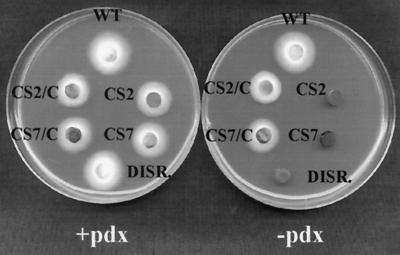
The entire PDX2 coding sequence plus flanking regions (Fig. 1) was cloned into the fungal transformation vector pBARKS1 (28) for transformation into the two C. nicotianae pyridoxine auxotrophic strains (CS2 and CS7) that are not pdx1 mutants. Ten and 25 bialaphos-resistant transformants of CS2 and CS7, respectively, were tested for ability to grow on minimal medium without pyridoxine (Fig. 2). Eighty percent of CS2 and 76% of CS7 transformants containing PDX2 grew to between 9 and 100% of wild-type growth on unsupplemented minimal medium. Neither of the parental strains nor any of the two CS2 or eight CS7 colonies transformed with the vector could grow on minimal medium. In previous work, we tested 40 CS2 and 46 CS7 strains transformed with the vector, and none of these vector-transformed strains exhibited even 1% of wild-type growth when tested for complementation (9).
FIG. 2.
Growth of wild-type (WT) C. nicotianae, pdx2 mutant strains CS2 and CS7, a pdx2 gene disruption mutant (DISR.), and pdx2 mutant strains complemented by transformation with the PDX2 gene (CS2/C and CS7/C) on minimal medium with (+pdx) and without (−pdx) pyridoxine. Fungal plugs (6 mm) were placed mycelium side down and incubated for 4 days at 28°C.
Targeted gene disruption of PDX2.
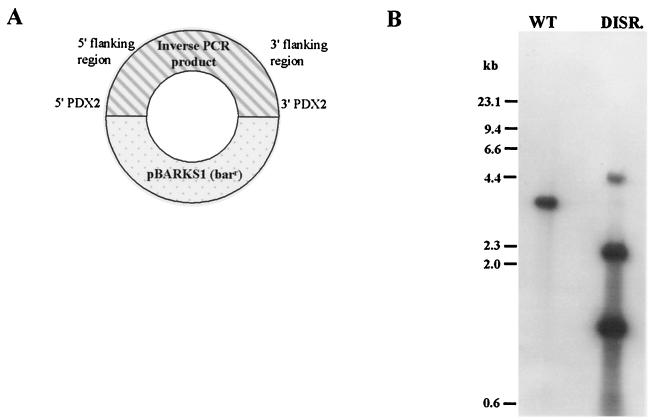
The inverse PCR product containing PDX2 was recovered from pGEM-T-Easy and cloned into pBARKS1. Standard gene disruption constructs that rely on double-crossover events for successful gene replacement generally contain an antibiotic resistance gene within the gene being disrupted. This plasmid (Fig. 3A) corresponds to a standard gene disruption construct except that the entire vector containing an antibiotic resistance marker interrupts PDX2 and its flanking regions. The disruption construct was transformed into the C. nicotianae wild-type strain, and bialaphos-resistant transformants were screened on minimal medium. One transformant of 758 tested was unable to grow on minimal medium, but could be rescued to wild-type growth by supplementation of the medium with pyridoxine (Fig. 2). Southern analysis (Fig. 3B) confirmed that this strain contained a disrupted pdx2 gene.
FIG. 3.
Disruption of PDX2. (A) Schematic describing the disruption plasmid. A clone was constructed in which the PDX2 inverse PCR product, which consists of the 5′ and 3′ ends of PDX2 separated by both 5′ and 3′ flanking sequences, was ligated to pBARKS1, a bialophos resistance-encoding vector. Standard gene disruption constructs that rely on double-crossover events for successful gene replacement generally contain an antibiotic resistance gene within the gene being disrupted. This plasmid corresponds to a standard gene disruption construct except that the entire vector containing the antibiotic resistance marker has been used to interrupt PDX2. (B) Southern hybridization analysis of a pdx2-disrupted strain. EcoRI-digested DNA from the wild-type (WT) and a pdx2-disrupted (DISR.) strains was probed with a PDX2-specific probe spanning the entire ORF. In the wild-type strain, the entire ORF is contained on a single EcoRI fragment, which is split into three parts in the gene disruption strain.
PDX2 protein sequence.
Figure 4 shows a comparison of the C. nicotianae PDX2 protein sequence and all available complete fungal homologues (Fig. 4A) and representative, homologues from other taxa (archaebacteria, eubacteria, and plants) (Fig. 4B). The C. nicotianae protein has a region towards the C terminus that is not found in any of the other proteins. Not surprisingly, the fungal proteins exhibit the strongest homology, while the other proteins are distinctly less well conserved. All regions conserved in the fungal proteins, however, are also found in the PDX2 protein sequences from more distantly related taxa, with the regions of homology tending towards conservative and semiconservative substitutions in the latter group. Two regions are highly conserved across all taxa, the PGGEST motif found at C. nicotianae residues 63 to 68 and the FHPE(LT) motif at C. nicotianae residues 253 to 258 (Fig. 1).
PDX2 sequences from mutants.
The gene ORFs from mutant strains CS2 and CS7 were amplified from each strain using the high-fidelity thermostable Pfu DNA polymerase and cloned for sequence analysis. Two independent amplification reactions were used to generate two independent clones from each strain. One strand of each clone was sequenced. The sequences from the two independent clones from each strain matched and showed that both the CS2 and CS7 pdx2 genes were missing a single nucleotide, leading to aberrant and truncated proteins of 13 and 12.4 kDa, respectively, in contrast to the wild-type protein of 30.1 kDa. The CS2 ORF is missing one of three cytosine residues at nucleotides 800 to 802, while the CS7 ORF is missing one of a guanosine residue triplet at nucleotides 635 to 637 (Fig. 1). The sequences of the resulting proteins diverge from the wild type at the altered codons (amino acid residues 38 and 93 for CS7 and CS2, respectively), converge with one another at amino acid residue 93, and terminate after 117 amino acid residues at the same stop codon. The CS7 protein contains only the first highly conserved domain (GVLALQGA), while the CS2 protein also contains the second (PGGEST) conserved domain.
Taxonomic distribution of PDX2 homologues.
Database searches were performed with the C. nicotianae PDX2 gene against GenBank and the finished and unfinished microbial databases available at the Institute for Genomic Research web site (http://www.tigr.org). As with PDX1, no organism containing homologues to the E. coli pyridoxine genes encodes a PDX2 homologue. A list of organisms encoding PDX2 homologues is shown in Table 1. PDX2 homologues are encoded by archaebacteria, fungi, plants, and eubacteria of the same groups that that encode PDX1 homologues. Nearly all of the organisms listed in Table 1 also have PDX1 homologues in the databases or are in the same genus as organisms with homologues. The only exceptions are a single plant (soybean, Glycine max) and a single eubacterium (Dehalococcoides ethenogenes), neither of which has been completely sequenced.
TABLE 1.
Occurrence of PDX2 homologues in organisms of diverse taxaa
| Plants |
| Arabidopsis thaliana |
| Glycine max |
| Hordeum vulgare |
| Lycopersicon esculentum |
| Medicago truncatula |
| Mesembryanthemum crystallinum |
| Oryza sativa |
| Triticum aestivum |
| Zea mays |
| Fungi |
| Aspergillus nidulans |
| Candida albicans |
| Neurospora crassa |
| Saccharomyces cerevisiae |
| Schizosaccharomyces pombe |
| Archaebacteria |
| Aeropyrum pernix |
| Archaeoglobus fulgidus |
| Methanobacterium thermoautotrophicum |
| Methanococcus jannaschii |
| Pyrococcus abyssi |
| Pyrococcus horikoshii |
| Sulfolobus solfataricus |
| Eubacteria |
| Bacillus anthracis |
| Bacillus stearothermophilus |
| Bacillus subtilis |
| Clostridium acetobutylicum |
| Corynebacterium diphtheriae |
| Dehalococcoides ethenogenes |
| Deinococcus radiodurans |
| Haemophilus ducreyi |
| Haemophilus influenzae |
| Mycobacterium avium |
| Mycobacterium bovis |
| Mycobacterium leprae |
| Mycobacterium tuberculosis |
| Staphylococcus aureus |
| Staphylococcus epidermidis |
| Streptococcus pneumoniae |
| Streptomyces coelicolor |
| Thermotoga maritima |
Organisms in boldface have been completely sequenced.
Transformation of pdx1 mutant strain CS8 and pdx1 null strain 9714-1 with PDX2.
The same construct used to transform and complement pdx2 mutant strains CS2 and CS7 was also used to transform two C. nicotianae pdx1 mutant strains. CS8 is the UV-derived strain (16, 18) whose complementation led to the identification of PDX1; strain 9714-1 was derived via targeted gene replacement and is null for PDX1 (10). In a previous study we showed that PDX1 can complement the two pdx2 mutant strains CS2 and CS7 at low frequency. Whereas pdx1 mutant strains were complemented at rates of 82 to 100%, CS2 and CS7 were complemented at rates of 35 and 11%, respectively (10). Because this study was conducted prior to our discovery that PDX1 was involved in pyridoxine synthesis, we used resistance to cercosporin as the measure of complementation. Pyridoxine is consumed during its quenching reaction with cercosporin-generated singlet oxygen, and therefore growth of a transformant in the presence of light and cercosporin indicates that the strain is capable of continued pyridoxine synthesis. In order to directly compare the effect of transformation of PDX2 into pdx1 mutant strains with the previously published converse experiment, CS8 and 9714-1 PDX2 transformants were tested for complementation on CM medium supplemented with 10 μM cercosporin. None of the 59 PDX2 transformants of the pdx1 null strain 9714-1 were able to grow in our assay. One of the 59 CS8 PDX2 transformants (1.7%) did grow in the presence of cercosporin, but only to 45% of the level of the wild-type strain.
DISCUSSION
In this work we provide evidence for a second gene involved in a recently described divergent pathway for pyridoxine biosynthesis. Using a PCR-based degenerate priming strategy, we isolated a C. nicotianae homologue of the yeast SNZB/SNO gene. Complementation of two C. nicotianae pyridoxine biosynthesis mutants with this gene demonstrated that it is required for pyridoxine biosynthesis. Sequencing of the pdx2 genes in these mutants revealed that both encode altered and truncated proteins, leading to their mutant phenotypes. Gene disruption experiments further corroborated the complementation studies and sequence analysis, and we have named this gene PDX2 in recognition of its newly defined role. Not surprisingly, database analysis revealed that PDX2 homologues exhibit the same distribution pattern as the PDX1 group of genes, the only other defined gene in this pathway. Additionally, while PDX2 proteins are not as well conserved as PDX1 proteins, several PDX2 domains are conserved across diverse phylogenetic groups.
In contrast to the situation in E. coli, very little is known about the PDX1/PDX2 biochemical pathway. 15N-labeling experiments suggested that the nitrogen of yeast pyridoxine originates with the amide moiety of glutamine (32), whereas in E. coli, glutamic acid provides the nitrogen (13, 14). Consistent with this, independent sequence motif analysis suggested that the PDX2 protein possesses glutamine amidotransferase activity (12). Data from Aspergillus nidulans, however, suggest that ammonium may be the source of the nitrogen atom (2, 3). Recent biochemical work with the eubacterium Rhizobium meliloti demonstrated that its pyridoxine biosynthesis pathway differs from that of E. coli (31). In E. coli, pyridoxine 5′-phosphate is formed via condensation of 4-phosphohydroxy-l-threonine and 1-deoxy-d-xylulose-5-phosphate (6, 7, 20), and 4-phosphohydroxy-l-threonine is formed by a four-step process from erythrose 4-phosphate. In R. meliloti, the latter compound is derived instead from condensation of glycine and glycoaldehyde. In E. coli, 4-hydroxy-l-threonine may be formed from glycine and glycoaldehyde, but only if glycoaldehyde is supplied. Unfortunately, no information is currently available on whether R. meliloti encodes the E. coli or PDX1-PDX2 homologues. We are currently engaged in experiments to determine if our C. nicotianae PDX1 and PDX2 genes and the E. coli pdxA and pdxJ genes (encoding the enzymes catalyzing the final condensation step between 1-deoxy-d-xylulose-5-phosphate and 4-phosphohydroxy-l-threonine) can functionally substitute for each other. In addition, we also intend to look at expression of pyridoxine genes in A. nidulans. Further data from these studies as well as from R. meliloti should provide valuable information on the actual roles of the PDX1 and PDX2 proteins and differentiate whether there are two completely unrelated pathways or the biochemical pathway is conserved and different proteins perform comparable enzymatic functions in different organisms.
It is currently unknown whether other genes are involved in C. nicotianae pyridoxine biosynthesis in addition to PDX1 and PDX2. Because genomic analyses strongly suggest that PDX1 and PDX2 are alone in their organismal distribution, it seems unlikely that there are other genes unique to this metabolic pathway. In B. subtilis and H. influenzae, the PDX1 and PDX2 homologues are in the same operon; however, the other flanking genes (e.g., seryl-tRNA synthetase and cytidylate kinase 2) have no obvious relationship to pyridoxine biosynthesis. It is possible that genes required for other metabolic pathways play a dual role in C. nicotianae pyridoxine synthesis, as is found in E. coli (21, 37). If so, a mutation in such a gene may be lethal. In a screen of over 11,000 UV-irradiated protoplasts regenerated on complete medium, only 5 pyridoxine auxotrophs were found (8, 16, 18), and all can be complemented to wild-type phenotype with either PDX1 or PDX2. Mutagenesis of any other gene in the pathway, even if also required for some other function, should nevertheless cause pyridoxine auxotrophy. All three yeast PDX2 homologues are found no further than 449 bp from their PDX1 cohorts, and the N. crassa PDX1 and PDX2 homologues are also closely linked (3a), and we initially attempted to isolate the C. nicotianae PDX2 by sequencing approximately 5 kb of DNA flanking either side of PDX1. Our sequencing efforts, however, have led us to conclude that the two known pyridoxine genes are unlinked in C. nicotianae, as they are in A. nidulans (http://www.gla.ac.uk/Acad/IBLS/molgen/aspergillus/). Gene clustering can be a useful tool for identifying pathway genes in fungi (15, 22, 26), but will not have utility in this case.
Previous complementation studies suggested that increasing PDX1 copy number can at least partially compensate for PDX2 mutation. The two pdx2 auxotrophs, CS2 and CS7, were transformed with complete and truncated versions of PDX1 and also with vector alone. Only intact copies of PDX1 could restore either CS2 or CS7 to prototrophy and only at a greatly reduced rate (9, 10). Altogether, 11 and 35% of CS7 and CS2 transformants, respectively, were complemented to pyridoxine prototrophy with PDX1, compared to an 82 to 100% complementation rate of three pdx1 mutant strains. This poorer rate of complementation and the lack of mutations in the CS2 and CS7 PDX1 genes led us to conclude these two strains were defective in a different part of the same pyridoxine pathway.
In this work we performed the corollary experiment, rescue of pdx1 mutant strains by transformation with PDX2, and showed that this reverse complementation does not work. Only a single PDX2 transformant of the mutant strain CS8 (1.7%) showed any rescue of the mutant phenotype, and that strain could only grow to 45% of wild-type levels. It is possible that increased amounts of PDX1 protein yield an enhanced level of substrate for partially functional PDX2 proteins. Alternatively, the two-hybrid analysis described by Padilla et al. (23) indicates that PDX1 and PDX2 proteins interact, suggesting that increased dosage of PDX1 stabilizes defective PDX2 proteins. The exact function of these proteins, however, has yet to be defined.
The sequence analysis of the pdx2 mutations in CS2 and CS7 revealed that both strains contain frameshift mutations leading to the translation of severely truncated proteins. While the wild-type protein contains 278 amino acid residues, both mutant strains are predicted to encode proteins of 117 amino acid residues. The CS2 protein diverges from the wild type at amino acid 93, while the CS7 protein diverges at residue 38. Nevertheless, the conserved domains that remain, one in the CS7 protein and two in the CS2 protein, must be sufficient for partial function. Otherwise, transformation of these strains with PDX1 would not result in rescue of their auxotrophy.
We have also found a response element for an AP-1-like transcription factor in both the C. nicotianae PDX1 and PDX2 genes. The yeast species S. cerevisiae, S. pombe, and Kluyveromyces lactis all encode AP-1-like transcription factors essential for their response to oxidative stress (4, 34, 35), and in S. pombe the PDX1 homologue is upregulated by overexpression of its AP-1-like transcription factor (M. W. Toone, personal communication). PDX1 and PDX2 were originally discovered during a search for genes involved in resistance to an active-oxygen-generating phototoxin, cercosporin. In organisms as diverse as rubbertree and S. cerevisiae, PDX1 homologue expression has also been linked to the presence of reactive oxygen species (5, 14, 21). The role of the AP-1 response pathway in cercosporin resistance is not yet clear. In yeast species, gene disruption and overexpression experiments have been valuable in delineating the biological role of their AP-1-like transcription factors. Comparable approaches should prove equally valuable in C. nicotianae to define the role of pyridoxine in oxidative stress response.
ACKNOWLEDGMENTS
We thank D. K. Wetzel for expert technical assistance and Margaret Werner-Washburne for providing clones for two of the yeast SNO genes and for the unpublished sequence of the N. crassa SNO gene.
This work was supported by grants from the National Science Foundation (MCB-9904746) and the USDA National Research Initiative Competitive Grants Program (96-35303-3204).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arst H N J. New evidence for old detective work. Microbiology. 1997;143:1481–1482. doi: 10.1099/00221287-143-5-1481. [DOI] [PubMed] [Google Scholar]
- 3.Arst H N J, Brownlee A G, Cousen S A. Nitrogen metabolite repression in Aspergillus nidulans: a farewell to tamA? Curr Genet. 1982;6:245–257. doi: 10.1007/BF00390345. [DOI] [PubMed] [Google Scholar]
- 3a.Bean L E, Dvorachek W H, Jr, Braun E L, Errett A, Saenz G S, Giles M D, Werner-Washburne M, Nelson M A, Natvig D O. Analysis of the pdx-1 (snz-1/sno-1) region of the Neurospora crassa genome. Correlation of pyridoxine-requiring phenotypes with mutations in two structural genes. Genetics. 2001;157:1067–1075. doi: 10.1093/genetics/157.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billard P, Dumond H, Bolotin-Fukuhara M. Characterization of an AP-1-like transcription factor that mediates an oxidative stress response in Kluyveromyces lactis. Mol Gen Genet. 1997;257:62–70. doi: 10.1007/s004380050624. [DOI] [PubMed] [Google Scholar]
- 5.Braun E L, Fuge E K, Padilla P A, Werner-Washburne M. A stationary-phase gene in Sacchromyces cerevisiae is a member of a novel, highly conserved gene family. J Bacteriol. 1996;178:6865–6872. doi: 10.1128/jb.178.23.6865-6872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cane D E, Du S, Robinson J K, Hsing Y, Spenser I D. Biosynthesis of vitamin B6: enzymatic conversion of 1-deoxy-d-xylulose-5-phosphate to pyridoxal phosphate. J Am Chem Soc. 1999;121:7722–7723. [Google Scholar]
- 7.Cane D E, Hsiung Y, Cormish J A, Robinson J K, Spenser I D. Biosynthesis of vitamin B6: the oxidation of 4-(phosphohydroxy)-l-threonine by PdxA. J Am Chem Soc. 1998;120:1936–1937. [Google Scholar]
- 8.Ehrenshaft M, Bilski P, Li M, Chignell C F, Daub M E. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA. 1999;96:9374–9378. doi: 10.1073/pnas.96.16.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrenshaft M, Chung K R, Jenns A E, Daub M E. Functional characterization of SOR1, a gene required for resistance to photosensitizing toxins in the fungus Cercospora nicotianae. Curr Genet. 1999;34:478–485. doi: 10.1007/s002940050423. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenshaft M, Jenns A E, Chung K R, Daub M E. SOR1, a gene required for photosensitizer and singlet oxygen resistance in Cercospora fungi is highly conserved in divergent organisms. Mol Cell. 1998;1:603–609. doi: 10.1016/s1097-2765(00)80060-x. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenshaft M, Jenns A E, Daub M E. Targeted gene disruption of carotenoid biosynthesis in Cercospora nicotianae reveals no role for cartenoids in photosensitizer resistance. Mol Plant-Microbe Interact. 1995;8:569–575. [Google Scholar]
- 12.Galperin M Y, Koonin E V. Sequence conservation of an exceptionally conserved operon suggests enzymes for a new link between histidine and purine biosynthesis. Mol Microbiol. 1997;24:443–445. doi: 10.1046/j.1365-2958.1997.3671706.x. [DOI] [PubMed] [Google Scholar]
- 13.Hill R E, Himmeldirks K, Kennedy I A, Pauloski R M, Sayer B G, Wolf E, Spenser I D. The biogenetic anatomy of vitamin B6: a 13C NMR investigation of the biosynthesis of pyridoxol in Escherichia coli. J Biol Chem. 1996;271:30426–30435. doi: 10.1074/jbc.271.48.30426. [DOI] [PubMed] [Google Scholar]
- 14.Hill R E, Spenser I D. Biosynthesis of vitamin B6. In: Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 695–703. [Google Scholar]
- 15.Hull E P, Green P M, Arst H N, Scazzocchio C. Cloning and physical characterization of the l-proline catabolism gene cluster of Aspergillus nidulans. Mol Microbiol. 1989;3:553–559. doi: 10.1111/j.1365-2958.1989.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 16.Jenns A E, Daub M E. Characterization of mutants of Cercospora nicotianae sensitive to the toxin cercosporin. Phytopathology. 1995;85:906–912. [Google Scholar]
- 17.Jenns A E, Daub M E, Upchurch R G. Regulation of cercosporin accumulation in culture by medium and temperature manipulation. Phytopathology. 1989;79:213–219. [Google Scholar]
- 18.Jenns A E, Scott D L, Bowden E F, Daub M E. Isolation of mutants of the fungus Cercospora nicotianae altered in their response to singlet-oxygen-generating photosensitizers. Photochem Photobiol. 1995;61:488–493. [Google Scholar]
- 19.Koonin E V, Tatusov R L, Galperin M Y. Beyond complete genomes: from sequence to structure and function. Curr Opin Struct Biol. 1998;8:355–363. doi: 10.1016/s0959-440x(98)80070-5. [DOI] [PubMed] [Google Scholar]
- 20.Laber B, Maurer W, Scharf S, Stepusin K, Schmidt F S. Vitamin B6 biosynthesis: formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-l-threonine and 1-deoxyxylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett. 1999;449:45–48. doi: 10.1016/s0014-5793(99)00393-2. [DOI] [PubMed] [Google Scholar]
- 21.Lam H M, Winkler M E. Metabolic relationships between pyridoxine (vitamin B6) and serine biosynthesis in Escherichia coli K-12. J Bacteriol. 1990;172:6518–6528. doi: 10.1128/jb.172.11.6518-6528.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamb H K, Hawkins A R, Smith M, Harvey I J, Turner G, Roberts C F. Spatial and biological characterization of the complete quinic acid utilization gene cluster in Aspergillus nidulans. Mol Gen Genet. 1990;223:17–23. doi: 10.1007/BF00315792. [DOI] [PubMed] [Google Scholar]
- 23.Mehdy M C, Sharma Y K, Sathasivan K, Bays N W. The role of activated oxygen species in plant disease resistance. Physiol Plant. 1996;98:365–374. [Google Scholar]
- 24.Osmani A, May G, Osmani S. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J Biol Chem. 1999;274:23565–23569. doi: 10.1074/jbc.274.33.23565. [DOI] [PubMed] [Google Scholar]
- 25.Padilla P A, Fuge E K, Crawford M E, Errett A, Werner-Washburne M. The highly conserved, coregulated SNO and SNZ gene families in Saccharomyces cerevisiae respond to nutrient limitation. J Bacteriol. 1998;180:5718–5726. doi: 10.1128/jb.180.21.5718-5726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne G A, Brown M P. Genetics and physiology of aflatoxin biosynthesis. Annu Rev Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- 27.Rose T M, Schultz E R, Henikoff J G, Pietrokovski S, McCallum C M, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly-related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straubinger B, Straubinger E, Wirsel S, Turgeon G, Yoder O C. Versatile fungal transformation vectors carrying the selectable bar gene of Streptomyces hygroscopicus. Fungal Genet Newsl. 1992;39:82–83. [Google Scholar]
- 29.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusov R L, Koonin E V, Lipman D J. A genomic perspective of protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 31.Tazoe M, Ichikawa K, Hoshino T. Biosynthesis of vitamin B6 in Rhizobium. J Biol Chem. 2000;275:11300–11305. doi: 10.1074/jbc.275.15.11300. [DOI] [PubMed] [Google Scholar]
- 32.Tazuya K, Adachi Y, Masuda K, Yamada K, Kumaoka H. Origin of the nitrogen atom of pyridoxine in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1244:113–116. doi: 10.1016/0304-4165(94)00205-c. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 35.Toone W M, Jones N. AP-1 transcription factors in yeast. Curr Opin Genet Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- 36.Woloshuk C P, Seip E R, Payne G A. Genetic transformation system for the aflatoxin-producing fungus Aspergillus flavus. Appl Environ Microbiol. 1989;55:86–90. doi: 10.1128/aem.55.1.86-90.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Zhao G, Man T K, Winkler M E. Involvement of the gapA-and epd (gapB)-encoded dehydrogenases in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. J Bacteriol. 1998;180:4294–4299. doi: 10.1128/jb.180.16.4294-4299.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]