Significance
V-ATPases are powerful proton pumps required for acidification of numerous intracellular compartments. The enzyme comprises a catalytic V1 region that hydrolyzes ATP and a membrane-embedded VO region that translocates protons. The VO complex is assembled in the endoplasmic reticulum (ER) before it is transported to the Golgi where V1 binds. Formation of intact V-ATPase activates proton pumping through a rotary catalytic mechanism. We used high-resolution cryoEM to show how the proteins Vma12p, Vma21p, and Vma22p work together for both V-ATPase assembly and quality control. The resulting structures show how a sequence of coordinated interactions and conformational changes ensures that only properly assembled VO leaves the ER and proton pumping into the neutral ER is avoided.
Keywords: V-ATPase; assembly; structure; Vma12p, Vma22p, Vma21p; cryoEM
Abstract
Vacuolar-type adenosine triphosphatases (V-ATPases) are rotary proton pumps that acidify specific intracellular compartments in almost all eukaryotic cells. These multi-subunit enzymes consist of a soluble catalytic V1 region and a membrane-embedded proton-translocating VO region. VO is assembled in the endoplasmic reticulum (ER) membrane, and V1 is assembled in the cytosol. However, V1 binds VO only after VO is transported to the Golgi membrane, thereby preventing acidification of the ER. We isolated VO complexes and subcomplexes from Saccharomyces cerevisiae bound to V-ATPase assembly factors Vma12p, Vma21p, and Vma22p. Electron cryomicroscopy shows how the Vma12-22p complex recruits subunits a, e, and f to the rotor ring of VO while blocking premature binding of V1. Vma21p, which contains an ER-retrieval motif, binds the VO:Vma12-22p complex, “mature” VO, and a complex that appears to contain a ring of loosely packed rotor subunits and the proteins YAR027W and YAR028W. The structures suggest that Vma21p binds assembly intermediates that contain a rotor ring and that activation of proton pumping following assembly of V1 with VO removes Vma21p, allowing V-ATPase to remain in the Golgi. Together, these structures show how Vma12-22p and Vma21p function in V-ATPase assembly and quality control, ensuring the enzyme acidifies only its intended cellular targets.
Vacuolar-type adenosine triphosphatases (V-ATPases) are membrane-embedded enzyme complexes that couple ATP hydrolysis to proton translocation across a membrane (1). V-ATPases are ubiquitous in eukaryotic cells where they acidify intracellular compartments such as vacuoles, lysosomes, endosomes, and the trans-Golgi network (2, 3). V-ATPases in the plasma membrane of specialized cells, including osteoclasts (4), kidney intercalated cells (5), and some cancer cells (6), acidify the extracellular environment. Due to their numerous roles, V-ATPase malfunction can lead to various diseases such as osteopetrosis (7, 8), distal renal tubular acidosis (9), and neurodegenerative disorders (10, 11).
V-ATPases consist of a soluble V1 region that hydrolyzes ATP and a membrane-embedded VO region that translocates protons (1, 3). Within V1, a hexamer of subunits A and B catalyzes ATP hydrolysis, causing conformational changes that drive rotation of a central rotor (subunits D, F, and d) that is attached to a membrane-embedded c ring (subunits c8, c′, c″, and Voa1p) within the VO region (12–14). Rotation of the c ring against the membrane-embedded C-terminal domain of subunit a drives proton translocation. The membrane-embedded subunits e and f, which have unknown functions, bind the C-terminal domain of subunit a. During rotation of the c ring, protons enter a cytosolic half channel between subunit a and the c ring, protonating conserved glutamate residues on each of the c, c′, and c″ subunits. Continued rotation of the ring carries protons through the lipid bilayer to a luminal half channel in subunit a where they exit the complex on the other side of the membrane (12). Three pairs of E and G subunits form peripheral stalk structures that interact with the soluble subunits C and H, as well as the cytosolic N-terminal domain of subunit a, holding subunit a stationary relative to the rotating c ring. As a result of its rotary catalytic mechanism, electron cryomicroscopy (cryoEM) of intact V-ATPase shows the enzyme in three main conformations, known as rotational “State 1,” “State 2,” and “State 3” (14, 15).
The V1 and VO regions of V-ATPase can undergo reversible dissociation and reassembly to regulate enzyme activity in response to changes in cellular conditions (16, 17). Glucose depletion in the growth medium of the yeast Saccharomyces cerevisiae causes V1 and VO to separate, with the V1 and VO complexes adopting inhibited conformations and subsequent dissociation of subunit C from the V1 complex (18, 19). On restoration of glucose to the growth medium, the enzyme complex known as RAVE (regulator of the ATPase of vacuoles and endosomes) mediates V-ATPase reassembly, likely by bringing V1 and VO into close proximity and inducing conformational changes needed for their interaction (20–22). When separated from the V1 region, the VO complex adopts rotational State 3 (12), with the isolated intact V1 complex adopting rotational State 2 but sampling all three rotational states after separation of subunit C (19).
The initial assembly of the VO complex in the endoplasmic reticulum (ER) membrane of yeast requires the assembly factors Vma12p, Vma21p, and Vma22p (23–25). These proteins, which are homologous with mammalian TMEM199, VMA21, and CCDC115, respectively, are necessary for V-ATPase activity in cells but are not found in the mature enzyme. The mammalian proteins are required for functional lysosomes (26, 27), with their deficiency associated with X-linked myopathy with excessive autophagy (27), follicular lymphoma (28), and congenital disorders of glycosylation (29–32). Vma21p is an integral membrane protein that participates in VO assembly but also accompanies fully assembled VO out of the ER (24, 33). Following assembly of V1 with VO, a C-terminal ER-retrieval motif on Vma21p allows it to be transported back to the ER to participate in additional rounds of VO assembly (24, 33). Vma12p, which contains a transmembrane region, forms a complex with the soluble protein Vma22p (the Vma12-22p complex) that stabilizes subunit a and mediates its assembly with the c ring (23, 25, 34).
To understand the structural basis by which Vma21p and the Vma12-22p complex facilitate VO assembly, we used cryoEM to determine structures of yeast VO assembly intermediates bound to both species. The structures show that the Vma12-22p complex binds both a partially assembled VO complex that lacks subunits a, e, and f, as well as fully assembled VO with its c ring in a different rotational state than found in mature VO. In contrast, Vma21p binds complexes that include mature VO, the assembly intermediate that contains Vma12-22p, and a small population of particles that appear to consist of loosely packed c rings bound to the uncharacterized proteins YAR027W and YAR028W. Together, these structures suggest how Vma12-22p and Vma21p carry out their functions and the sequence of events in VO assembly.
Results
Vma12-22p Forms Complexes with VO and VO Lacking Subunits a, e, and f.
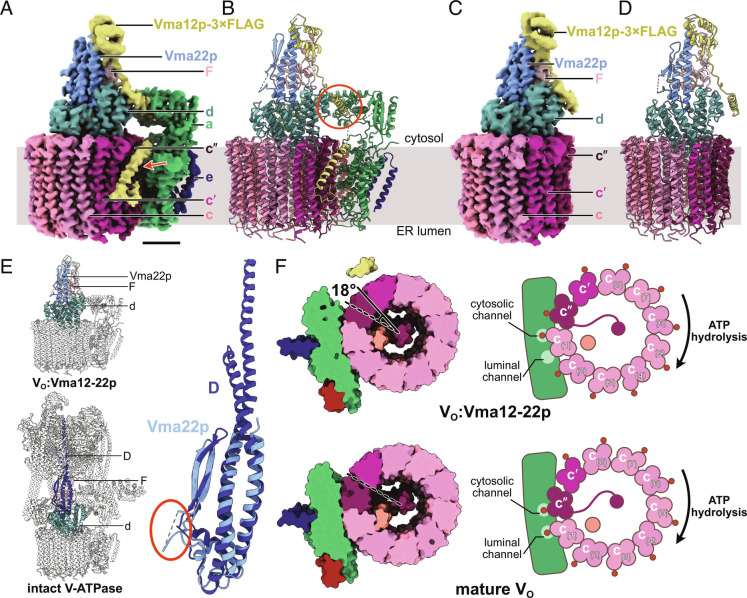
To understand how the Vma12-22p complex facilitates VO assembly, we integrated DNA encoding a 3×FLAG tag into the yeast chromosome 3′ of the gene for Vma12p. Following yeast growth, cell membranes were isolated, solubilized with detergent, and protein was purified by anti-FLAG affinity chromatography. The resulting protein preparation was subjected to structural analysis by cryoEM (SI Appendix, Fig. S1A). This preparation gave rise to two distinct 3D structures. The first structure resembles the VO complex but with additional proteins bound and has a nominal overall resolution of 2.6 Å, with local resolutions ranging from 2.3 to 4.8 Å EMD-27984 (Fig. 1A and SI Appendix, Fig. S1B blue curve, SI Appendix, Fig. S1 C and D). The map enabled construction of an atomic model PDB 8EAS (Fig. 1B and SI Appendix, Table S1 and Fig. S2A), which included all the known yeast VO subunits and subunit F from the V1 region. The map also allowed fitting and refinement of models of Vma12p and Vma22p from AlphaFold (35) (Fig. 1 A and B, yellow and blue densities and models). Consequently, this complex is designated the VO:Vma12-22p complex. In this structure, the folds predicted for domains in Vma12p and Vma22p fit the map with high fidelity, while two extended sequences within Vma12p were predicted poorly. Side chain orientations for both proteins also required substantial refinement. The second structure, which was obtained from ~27% of the particle images, appears as a partial VO complex lacking subunits a, e, and f but with Vma12-22p bound EMD-27985 (Fig. 1C). This map was refined to 3.1 Å resolution, with local resolutions ranging from 2.6 to 6.1 Å (SI Appendix, Fig. S1B, green curve, SI Appendix, Fig. S1 E and F). The map enabled construction of an atomic model consisting of the c ring (subunits c8, c′, c″, and Voa1p), subunit d, the Vma12-22p complex, and subunit F PDB 8EAT (Fig. 1D and SI Appendix, Table S1 and Fig. S2B). Based on its subunit composition, we designate this assembly the VO∆aef:Vma12-22p complex.
Fig. 1.
Structure of the VO:Vma12-22p complex. (A) CryoEM map of the VO:Vma12-22p complex. (Scale bar, 25 Å.) (B) Atomic model for the VO:Vma12-22p complex. (C) CryoEM map of the VO∆aef:Vma12-22p complex. (D) Atomic model for the VO∆aef:Vma12-22p complex. (E) Structural comparison of Vma22p and subunit D from the V1 region of intact V-ATPase. (F) Comparison of cross-sections through the VO:Vma12-22p complex and the mature VO complex (PDB: 6O7T) shows a difference in rotation of the c ring.
Interaction between Vma12-22p complex and subunit a is known to be transient (34). However, we were still concerned that the presence of VO∆aef:Vma12-22p in the dataset could be due to dissociation of subunits a, e, and f from VO:Vma12-22p during cryoEM specimen preparation, rather than representing a species that exists in the ER membrane. A structure of VO∆aef has not been described in previous cryoEM analysis of VO complexes (12, 13, 36, 37), but it was not clear whether these broken complexes were removed during image analysis in those studies. Therefore, we reprocessed a published dataset of cryoEM images of VO complex (36) that had been purified and frozen in conditions equivalent to those used for the Vma12p-3×FLAG preparation. The results showed only intact VO (SI Appendix, Fig. S3A), indicating that subunits a, e, and f are unlikely to separate from VO during grid freezing, and suggesting that the VO∆aef:Vma12-22p complex is present in the cell and is not an artifact of cryoEM specimen preparation. However, we cannot exclude the possibility that the interaction between subunit a and the c ring in VO:Vma12-22p is less stable than it is the mature VO complex, possibly owing to Vma12-22p destabilizing the complex and causing subunits a, e, and f to dissociate at the air-water interface during grid freezing. While this interpretation is possible, previous work has suggested that Vma12p and Vma22p stabilize the interaction of subunit a with the c ring rather than destabilize it (34).
Vma22p Mimics Subunit D from V1 and blocks V1 Binding.
In both the VO:Vma12-22p and VO∆aef:Vma12-22p complexes, Vma22p sits on top of subunit d in a site that is occupied by subunit D from the V1 region in intact V-ATPase (14, 38) (Fig. 1E, Left). Vma22p consists primarily of a twisted pair of α helices abutted by two antiparallel β strands (Fig. 1E, Right, light blue). This fold resembles the fold of subunit D (Fig. 1E, Right, dark blue), which shares 18% amino acid sequence similarity and 10% amino acid sequence identity with Vma22p (SI Appendix, Fig. S3B). The α helical structure in Vma22p is less than half the length of the corresponding α helical region in subunit D, making Vma22p too short to serve the same role as subunit D in the V1 complex. Thus, the structure of Vma22p suggests that it would block V1 from binding subunit d, which would prevent premature attachment of V1 to partially assembled VO complexes in the ER membrane. Blocking V1 from binding VO would prevent proton pumping into the ER lumen, which is maintained at near neutral pH (39, 40). Compared to subunit D, Vma22p possesses an additional disordered loop of unknown function comprising 35 residues C-terminal to its two β strands (Fig. 1E, Right, circled in red).
Surprisingly, V-ATPase subunit F is found interacting with Vma12-22p (Fig. 1 A–D, light pink). Subunit F is a small protein that normally binds subunit D in both intact V-ATPase and the dissociated V1 complex, but is not part of the dissociated VO complex. The role of subunit F when bound to subunit D is not known. However, both the VO:Vma12-22p and VO∆aef:Vma12-22p structures show the N-terminal domain of subunit F in contact with Vma22p, while its C-terminal α helix interacts with both Vma12p and Vma22p (SI Appendix, Fig. S3C). This interaction with both Vma12p and Vma22p suggests that subunit F may help mediate association of the two proteins. Deletion of the gene for subunit F leads to reduced levels of VO subunits a and c in yeast, with normal levels of subunit d but reduced association of subunit d with the vacuolar membrane (41). This phenotype is consistent with the association between subunit F and Vma12-22p being necessary for function of the assembly factors.
Vma12p Helps Attach Subunit a to the c Ring.
Vma12p possesses a compact folded N-terminal domain comprising three α helices and two β strands (Fig. 1 A–D, yellow). In both the VO:Vma12-22p complex and the VO∆aef:Vma12-22p complex, this N-terminal domain interacts with subunit F and Vma22p. Following the N-terminal domain of Vma12p in the VO:Vma12-22p structure, there is an extended linker and an α helix that contacts both a loop (residues 48 to 53) from subunit a and two short α helices (residues 27 to 35 and 39 to 48) from subunit d (Fig. 1B, red circle, SI Appendix, Fig. S3D). This α helix from Vma12p is followed by a second extended linker and a membrane-embedded α helix that is tilted relative to the plane of the lipid bilayer (Fig. 1A, red arrow). Vma12p is predicted to have two C-terminal transmembrane α helices, with its C terminus on the cytosolic side of the membrane (25, 42). However, apparent flexibility in this region of the protein prevents the second transmembrane α helix from being resolved in the cryoEM map. Neither of the two transmembrane α helices from Vma12p could be resolved in the VO∆aef:Vma12-22p map (Fig. 1C). To ensure that flexibility in the C-terminal transmembrane α helices from Vma12p is not due to the C-terminal 3×FLAG tag introduced in the protein, we constructed a second yeast strain with the affinity tag at the C terminus of Vma22p instead. CryoEM of protein purified from this second strain resulted in maps of VO:Vma12-22p and VO∆aef:Vma12-22p that are indistinguishable from the maps obtained from the Vma12p-3×FLAG preparation (SI Appendix, Fig. S3 E and F). This finding indicates that flexibility in the C-terminal transmembrane α helices of Vma12p is not due to the incorporation of the affinity tag. The equivalence of the two structures obtained from the Vma12p-3×FLAG and Vma22p-3×FLAG preparations also shows that Vma12p and Vma22p form an obligate heterodimer. It appears that interaction between Vma12p and the N-terminal domain of subunit a reduces mobility in one of the transmembrane α helices from Vma12p, allowing it to be resolved in the cryoEM map. This transmembrane α helix from Vma12p does not interact directly with the transmembrane C-terminal domain of subunit a or the c ring. Therefore, other than anchoring Vma12-22p to the membrane, the role of the tilted transmembrane domain in Vma12p is unclear. In contrast, the interaction between subunit a, subunit d, and the short α helix N-terminal of the transmembrane domain in Vma12p is consistent with Vma12-22p’s proposed role of stabilizing subunit a and recruiting it to VO (34).
The c Ring Rotation Differs between VO:Vma12-22p and the Mature VO Complex.
The c ring has an asymmetric distribution of proton-carrying Glu residues, with the protonatable Glu residue of subunit c″ found on the second ring-forming α helix, while the protonatable Glu residues of the eight c subunits and one c′ subunit are located on their fourth ring-forming α helices (12) (Fig. 1F, Right). Previous structures of yeast VO found the complex in rotational State 3 with Glu108 from subunit c″ aligned with the cytosolic half channel (12, 13, 36, 37), while Glu137 from subunit c(1) is aligned with the luminal half channel (Fig. 1F, Lower). Compared to these structures of mature VO, the VO:Vma12-22p complex shows an ~18° rotation of the c ring in the ATP-hydrolysis direction (Fig. 1F, Upper). As a result, in the VO:Vma12-22p complex, Glu137 from subunit c(1) is aligned with the cytosolic half channel and Glu137 from subunit c(2) is far from the luminal half channel (Fig. 1F, Upper Right). A previous study of yeast VO complexes by cryoEM detected a similar ~14° rotation of the ring in a 3D class from ~5% of particle images (43). Both the small population of particle images in the class and molecular dynamic simulations indicated that this conformation is less energetically favorable than VO in rotational State 3 (43). Comparison of the VO:Vma12-22p and mature VO structures shows that even with the ~18° rotation of the c ring relative to the C-terminal domain of subunit a, the interface between subunit d and N-terminal domain of subunit a remains unchanged (SI Appendix, Fig. S3 G, i, as shown previously (43). Further, rotation of the c ring from its VO:Vma12-22p conformation to the mature VO conformation would cause a clash between Vma12p and subunit a that would prevent Vma12-22p from binding to VO (SI Appendix, Fig. S3 G, ii, orange circle). Therefore, the structure suggests that after Vma12p recruits subunits a, e, and f to the c ring with subunit d, relaxation of the c ring into its energetically favored conformation disrupts the interaction between Vma12-22p and VO, releasing Vma12-22p and resulting in the mature VO structure. However, this release of Vma12-22p from VO must occur either slowly or with other proteins involved, or else it would not be possible to isolate the VO:Vma12-22p complex biochemically. The rate of Vma12-22p release may also be slower in detergent micelles compared to the lipid bilayer of the ER membrane.
Vma21p Binds the VO Complex in Multiple Positions around the c Ring.
To understand how the activity of Vma21p relates to the VO assembly process, we next investigated VO complexes that contain Vma21p. As with Vma12-22p, we constructed a yeast strain with DNA for a 3×FLAG tag integrated in the chromosome 3′ of the VMA21 gene. When grown on medium containing zinc, the Vma21p-3×FLAG strain exhibited growth comparable to wild-type yeast, while a ∆Vma21p strain showed the expected VMA− V-ATPase deficiency phenotype (SI Appendix, Fig. S4), indicating that despite Vma21p’s C-terminal Lys-Lys-Glu-Asp ER-retrieval motif (24) the C-terminal 3×FLAG tag does not prevent assembly of functional V-ATPase complexes.
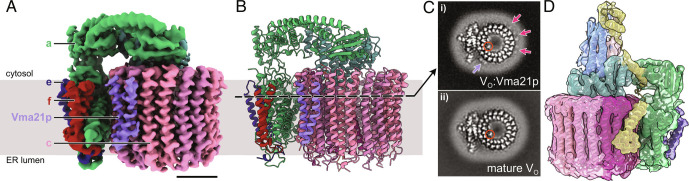
CryoEM of protein purified from the Vma21p-3×FLAG strain led to a 3D map resembling the mature VO complex in rotational State 3 that we designate the VO:Vma21p complex EMD-27986 (Fig. 2A and SI Appendix, Fig. S5). The nominal overall resolution of the map is 3.1 Å with local resolutions ranging from 2.2 to 7.5 Å (SI Appendix, Fig. S5B, orange curve, SI Appendix, Fig. S5C). The map enabled construction of an atomic model through a combination of rigid body fitting and refinement of a published VO model (36) and a model for Vma21p predicted by AlphaFold (35) PDB 8EAU (Fig. 2B and SI Appendix, Table S1 and Fig. S6). Again, AlphaFold predicted the overall fold of Vma21p accurately, but the orientation of the protein’s α helices and side chains required refinement. Clearly defined density accommodated a single copy of Vma21p (Fig. 2 A and B, purple). No conformational differences could be detected between the VO part of VO:Vma21p and a mature VO structure (SI Appendix, Fig. S7A). In the structure, Vma21p forms a transmembrane α helical hairpin wedged between two adjacent c subunits near the luminal half channel of subunit a. The N-terminal α helix from Vma21p interacts with the second α helix of subunit c(2) and the fourth α helix of subunit c(3), while the C-terminal α helix from Vma21p contacts the second and fourth α helices of subunit c(2) (SI Appendix, Fig. S7B).
Fig. 2.
Structure of the VO:Vma21p complex. (A) CryoEM map of the VO:Vma21p complex. (Scale bar, 25 Å.) (B) Atomic model for the VO:Vma21p complex. (C) A slice through the VO:Vma21p map (i) showing additional protein densities surrounding the c ring compared to the mature VO complex (accession code EMD-0644) (ii). Purple arrow, strong density for Vma21p. Magenta arrows, weaker density for Vma21p. Orange circle, density for Voa1p. (D) A subset of the particle images from the Vma21p-3×FLAG dataset gave a map of VO:Vma12-22p complex, shown here with the atomic model of VO:Vma12-22p from the Vma12p-3×FLAG dataset fitted.
The well-defined single Vma21p binding site in VO:Vma21p involves only two of the eight c subunits and does not appear to be unique within the c ring. To understand Vma21p’s apparent preference for a single binding site, cross-sections through the VO:Vma21p map were inspected and compared to cross-sections through a map of mature VO complex (Fig. 2C). The two α helices from the well-resolved Vma21p are clear in the cross-section (Fig. 2 C, i, purple arrow). However, features corresponding to additional similar but lower density α helices can be seen surrounding the c ring (Fig. 2 C, i, magenta arrows). These additional densities, which are not seen in a map of the mature VO complex determined by equivalent methods (36) (Fig. 2 C, ii) suggest that multiple copies of Vma21p bind around the c ring but with a low binding occupancy in all but one site. In its preferred site, Vma21p does not interact with the nearby subunit a, and it is not clear why this site has a high occupancy while the others sites have a low occupancy. It is worth noting that in the VO:Vma12-22p complex this Vma21p would be in contact with subunit a. However, the VO:Vma12-22p map does not show density for Vma21p at this position. Cross-sections through the map also show density for the C-terminal transmembrane α helix from Voa1p (Fig. 2 C, i, orange circle), which co-precipitates with Vma21p (44) and can be seen in maps of VO∆aef:Vma12-22p and VO:Vma12-22p (Fig. 1F, orange), as well as mature VO complex (Fig. 2 C, ii, orange circle) (12, 13, 36).
The VO:Vma21p structure appears to show the assembled mature VO complex that is escorted from the ER by Vma21p. A Vma21p mutant with a defective ER-retrieval motif was previously found to interact with intact V-ATPase (33), and V1 subunits were detected in mass spectrometry of the Vma21p-3×FLAG protein preparation (Dataset S1). However, we did not identify particle images from the Vma21p-3×FLAG dataset that show intact V-ATPase, suggesting any interaction between Vma21p and intact V-ATPase is unstable or transient.
Vma21p Binds a VO:Vma12-22p Complex and a Complex Containing YAR027W and YAR028W.
3D classification of the cryoEM dataset from Vma21p-3×FLAG tagged protein identified two additional populations of particle images. The first allowed calculation of a 3D map at 4.4 Å resolution that was indistinguishable from the VO:Vma12-22p complex EMD-27988 (Fig. 2D and SI Appendix, Fig. S5B, green curve, SI Appendix, Fig. S5 E and F). Density for Vma21p could not be detected in this map, suggesting that again Vma21p interacts with the c ring, but with variable positions that leads to incoherent averaging in 3D reconstruction. The VO:Vma12-22p map from the Vma21p-3×FLAG sample was calculated from 17,884 particle images, compared to 127,922 particle images for the VO:Vma21p map from the same sample. Therefore, Vma21p appears to bind a small population of VO complex before release of Vma12-22p, while most of the VO complexes it binds have already released Vma12-22p.
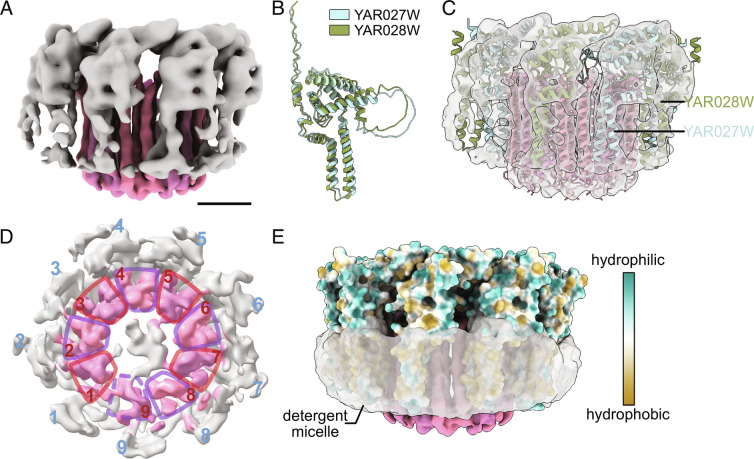
The second unique class of particle images isolated with 3×FLAG tagged Vma21p allowed calculation of a 3D map that resembles a c ring with additional protein densities attached EMD-27987 (Fig. 3A). Only 19,849 out of 607,746 particle images from the dataset contribute to this structure, leading to a poor overall resolution of 5.7 Å, with local resolutions between 4.6 and 9.2 Å (SI Appendix, Fig. S5B, pink curve, SI Appendix, Fig. S5 G and H). To identify the additional protein density seen in this map, we inspected the list of proteins identified in the preparation by mass spectrometry of tryptic fragments (Dataset S1) and attempted to fit models of candidate proteins predicted by AlphaFold into the map. This process identified two candidate proteins, YAR027W and YAR028W. The amino acid sequences for these two putative integral membrane proteins are 58% identical and 73% similar (SI Appendix, Fig. S8A), with AlphaFold predicting nearly indistinguishable folds for the two proteins (Fig. 3B). Due to the low resolution of the cryoEM map and the similarity in the predicted folds of YAR027W and YAR028W, the two proteins could not be distinguished in the map (Fig. 3C) PDB 8EAV. A view of the map perpendicular to the plane of the detergent micelle shows nine copies of YAR027W or YAR028W surrounding a ring that appears to consist of nine loosely packed c subunits (Fig. 3D, dashed circles). Therefore, we designated this assembly the YAR027W/028W:c9 complex. Due to the limited resolution of the map, the c, c′, and c″ subunits could not be distinguished in the ring. Examination of the fold and surface properties of YAR027W and YAR028W suggests that they each form two N-terminal transmembrane α helices that are buried in the detergent micelle, with a C-terminal soluble domain likely on the cytosolic side of the ring (Fig. 3E, blue and yellow surfaces).
Fig. 3.
Structure of YAR027W and YAR028W binding to a loosely packed c ring. (A) A subset of the particle images from the Vma21p-3×FLAG dataset produced a map that resembles a c ring with additional protein densities attached. (Scale bar, 25 Å.) (B) Comparison of the AlphaFold predicted structures of YAR027W and YAR028W. (C) Fitting of the c subunit structure and AlphaFold predicted structures of YAR027W and pYAR028W into the map PDB 8EAV. (D) A view of the map perpendicular to the plane of the detergent micelle shows nine copies of YAR027W or YAR028W surrounding a structure that resembles nine loosely packed c ring protomers. (E) Surface hydrophobicity of YAR027W and YAR028W compared with the position of the detergent micelle in the map.
The YAR027W and YAR028W genes are members of the S. cerevisiae DUP240 multigene family, which contains non-essential membrane-associated proteins (45, 46). The two proteins have been shown in high-throughput studies to interact with each other, as well as multiple other proteins, and YAR028W has been shown to interact with V-ATPase subunit c″ (47, 48). YAR027W, also known as UIP3, was reported to localize to the nuclear envelope (46). A recent study showed that yeast cells that were resistant to killer toxin K28 became sensitive to the toxin when the gene YAR028W was deleted (49). While it is tempting to speculate that YAR027W and YAR028W are somehow involved in c ring assembly, the resolution of the map precludes determining the precise locations of YAR027W, YAR028W, c, c′, or c″ in the complex. To examine whether the two proteins are essential for V-ATPase function, we generated a haploid yeast strain with both the YAR027W and YAR028W genes deleted. When grown on medium containing zinc, the strain did not exhibit the VMA− V-ATPase deficiency phenotype, indicating that YAR027W and YAR028W are not essential for V-ATPase activity (SI Appendix, Fig. S8B). However, numerous questions related to YAR027W and YAR028W still require investigation. For example, it remains unknown if most of the YAR027W and YAR028W in the cell are bound to V-ATPase c rings with Vma21p, or if this population is a minor portion of the YAR027W and YAR028W. It is also unclear if YAR027W and YAR028W interact with other proteins besides the components of the c ring and Vma21p and if these interactions are somehow related to their interaction with V-ATPase. A high-resolution structure of YAR027W, YAR028W, the c ring, and Vma21p may help answer some of these questions. While extensive further study is clearly required, the results presented here indicate that Vma21p interacts with a complex that involves c subunits, YAR027W, and YAR028W.
The C-terminal Lys-Lys-Glu-Asp ER-retrieval motif of Vma21p allows the protein to travel back to the ER membrane after escorting the fully assembled VO complex out of the ER. Mutating the motif to Gln-Gln-Glu-Arg results in Vma21p’s mis-localization to the vacuolar membrane (24). A recent study showed that removal of this retrieval motif on human VMA21 results in the protein becoming enriched in lysosomes, eventually leading to activation of autophagy (28). The structures obtained from the Vma21p-3×FLAG sample suggest that Vma21p binds all VO subcomplexes that contain c rings. By binding to these complexes, the ER-retrieval motif on Vma21p would return any partially assembled VO complexes that are transported to the Golgi back to the ER membrane. Therefore, removal of Vma21p is required for the mature VO complex to remain in the Golgi or travel further to its designated cellular location.
Discussion
Unsurprisingly, owing to their importance for VO assembly, mutations in the human homologues of Vma12p, Vma22p, and Vma21p have been linked to disease. The results presented here provide a structural context for understanding these mutations. Mutations of the human homologue of Vma22p, CCDC115, are found in patients affected by congenital disorders of glycosylation (30, 31). The human mutation D11Y maps onto a region at the N terminus of Vma22p that is not resolved in the structure. However, L31S in CCDC115 maps onto Leu34 in the yeast Vma22p structure, where mutation could affect the binding of subunit F (SI Appendix, Fig. S9A, blue circle with cyan fill). Mutations in TMEM199, the human homologue of Vma12p, are also found in the same disease (32) and map to the N-terminal domain of Vma12p (32) (SI Appendix, Fig. S9A, yellow circles with cyan fill). These residues do not appear to be involved in protein–protein interactions, but their mutation may disrupt the fold of Vma12p. Mutations of the human gene VMA21 have been reported in X-linked myopathy with excessive autophagy (27) and follicular lymphoma (28). Three of the reported mutations (equivalent to Thr35, Pro36, and Thr38 in the yeast Vma21p) are found in the luminal loop of the protein (SI Appendix, Fig. S9B). The remaining reported mutations map to the transmembrane region of yeast Vma21p where it interacts with c subunits and could partially disrupt this interaction (28, 29) (SI Appendix, Fig. S9B). In yeast, loss of function of Vma12p, Vma21p, or Vma22p leads to the conditionally lethal VMA− phenotype, suggesting that loss of function of CCDC115, VMA21, or TMEM199 would be embryonic lethal in humans. Therefore, disease-linked mutations in the assembly factors likely only weaken interaction with VO subunits or reduce the stability of the assembly factors. Mutations that are far from where the assembly factors interact with VO subunits may affect interaction with other, currently unidentified, binding proteins.
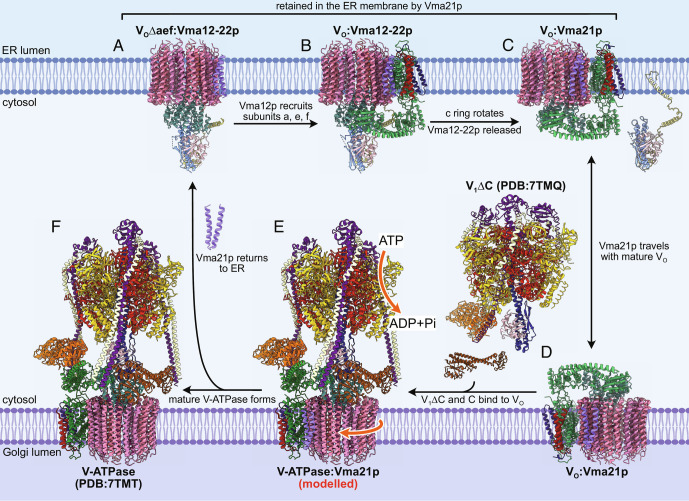
The structures described above suggest the sequence of events that occur during VO assembly in the ER membrane and subsequent binding of V1 in the Golgi (Movie S1 and Fig. 4). The structure consisting of a partial c ring with YAR027W and YAR028W purified via Vma21p-3×FLAG may or may not be on the assembly pathway and requires further investigation. The c ring assembles around Voa1p (13, 36), with binding of subunit d to the c8c′c″Voa1p ring (33) masking the ER-retrieval motif of Voa1p (13). The VO∆aef:Vma12-22p structure indicates that the Vma12-22p complex binds this fully assembled ring prior to interaction with subunits a, e, and f (Fig. 4A). Vma12-22p helps recruit and secure the interaction of subunit a with the c ring (34) through interaction of Vma12p with subunits a and d (Fig. 4B). During recruitment of subunits a, e, and f, Vma22p mediates the interaction between Vma12p and subunit d and prevents premature attachment of the V1 complex to the partially assembled VO complex. Subunit F does not directly participate in the assembly process but mediates the association of Vma12p and Vma22p. The fully assembled VO complex with Vma12-22p can then transition to the energetically favored conformation of the c ring, disrupting the Vma12-22p binding site on VO and removing Vma12-22p from the complex (Fig. 4C). This release of Vma12-22p is required for the subsequent assembly of V1 with VO. The structures presented here suggest that Vma21p binds all assembly intermediates that contain a c ring, with the ER-retrieval motif from Vma21p ensuring that partially assembled complexes are returned to the ER membrane (Fig. 4 A–C).
Fig. 4.
Sequence of events in VO assembly. (A) Vma12-22p binds a complex containing the c ring, subunit d, and Vma21p. (B) Vma12p recruits subunits a, e, and f to the rest of VO. (C) The c ring in VO relaxes to its preferred orientation, releasing Vma12-22p. Vma21p ensures VO assembly intermediates are retrieved to the ER membrane. (D) Vma21p accompanies the fully assembled VO to the Golgi membrane. (E) V1∆C and subunit C bind VO and initiate rotary catalysis. (F) Rotation of the c ring releases Vma21p, which travels back to the ER membrane, leaving functional V-ATPase in the Golgi membrane (Movie S1).
Despite the ER-retrieval motif of Vma21p, the assembly factor is found in a subpopulation of VO complexes on coat protein complex II-coated transport vesicles (33), which facilitate VO transport from the ER membrane to the Golgi membrane (Fig. 4D). In the absence of Vma12-22p, this VO:Vma21p complex is capable of interacting with V1, presumably with the RAVE complex mediating binding of VO, V1∆C, and subunit C (19, 22) (Fig. 4E). The attachment of V1 would initiate rotary catalysis, so that rotation of the c ring drives dissociation of Vma21p (Fig. 4F). The removal of Vma21p allows intact V-ATPase to remain in the Golgi membrane or travel further to the vacuole, while free Vma21p returns to the ER membrane owing to its ER-retrieval motif. This substitution of Vma21p with V1 is consistent with the simultaneous loss of Vma21p and binding of V1 to VO observed previously (33).
Overall, the data presented here provide a structural basis for understanding disease-causing mutations in CCDC115, TMEM199, and VMA21. Furthermore, the structures suggest how Vma12-22p and Vma21p coordinate their known roles in the assembly of the VO complex and formation of an active intact V-ATPase after departure of VO from the ER membrane. They also reveal an unexpected protein complex involving the V-ATPase c ring, Vma21p, and the uncharacterized proteins YAR027W and YAR028W. Importantly, the structures illustrate how Vma21p and Vma12-22p play central roles in both V-ATPase assembly and quality control.
Methods
Yeast Strains.
S. cerevisiae strains SABY125, SABY129, and SABY130 were prepared by homologous recombination to integrate DNA sequence for a 3×FLAG tag downstream of the VMA12, VMA21, and VMA22 genes, respectively, in the background strain BJ2168 (MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 gal2), respectively. Plasmid pJT1 (38) was used to amplify DNA sequence encoding the 3×FLAG tag followed by a URA3 marker. The PCR products were then used to transform BJ2168 by standard lithium acetate transformation. Transformed cells were selected by growth on minimal media lacking uracil, and successful transformants were confirmed by PCR. The ∆Vma21p yeast strain YSC6273-201936318 (vma21::KanMX MATa his3Δ1 leu2Δ0 met15Δ0) was purchased from Horizon Discovery. The S. cerevisiae strain SABY149 was prepared from strain W303-1A by simultaneous deletion of the genes YAR027W and YAR028W. These genes are adjacent to each other on chromosome I and were deleted by homologous recombination with a linear DNA fragment encoding the URA3 marker. Primer sequences used for yeast strain construction and confirmation are listed in SI Appendix, Table S2.
Protein Purification.
Yeast strains were grown in 11 L yeast extract peptone dextrose media (20 g/L peptone, 20 g/L glucose, 10 g/L yeast extract) supplemented with 100 μg/mL ampicillin and 0.02% antifoam in a Microferm fermenter (New Brunswick Scientific) at 30 °C for 1 d (>22 h), with aeration of 34 cubic feet per hour and stirring at 300 rpm. All subsequent steps were performed at 4 °C. Cells were harvested by centrifugation at 4,000 × g for 15 min and resuspended in 1 mL/g lysis buffer (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, 80 g/L sucrose, 20 g/L sorbitol, 20 g/L glucose, 5 mM 6-aminocaproic acid, 5 mM benzamidine hydrochloride, 5 mM ethylenediaminetetraacetic acid, 10 mg/L phenylmethylsulfonyl fluoride, pH 7.4). Cells were lysed with 0.5 mm glass beads in a bead beater (BioSpec), with six cycles of 1 min of bead beating and 1 min of cooling. Cell debris was removed by centrifugation at 4,000 × g for 15 min. Membranes were then collected by ultracentrifugation (Beckman L-90K, Ti70 rotor) at 145,000 × g for 40 min, resuspended in 0.5 mL/g lysis buffer using a Dounce homogenizer, and stored at −80 °C prior to protein purification.
Frozen membranes were thawed at room temperature, and all the purification steps were performed at 4 °C. Membranes were solubilized with 1% (w/v) n-dodecyl β-D-maltoside (DDM; Anatrace) and mixed for 30 min. Insoluble material was removed by ultracentrifugation at 158,000 × g for 1h (Beckman L-90K, Ti70 rotor), and membranes were filtered with a 0.45 μm syringe filter and applied to a 0.5 mL anti-FLAG M2 affinity gel in a column (Millipore Sigma) pre-equilibrated in DDM Tris-buffered saline (DTBS; 50 mM Tris-HCl, 150 mM NaCl, 0.02% [w/v] DDM, pH 7.4). The column was washed with 10 column volumes of DTBS, and protein was eluted with three column volumes of DTBS containing 150 μg/mL of 3×FLAG peptide and one column volume of DTBS without peptide. Protein was then concentrated to ~200 μL with a 100 kDa molecular weight cutoff (MWCO) Amicon Ultracentrifugal filter (Millipore Sigma) at 1,000 × g, diluted with 4 mL glyco-diosgenin Tris-buffered saline (50 mM Tris-HCl, 150 mM NaCl, 0.004% [w/v] glyco-diosgenin [Anatrace], pH 7.4) and concentrated to ~200 μL in the same concentrator. For Vma12p-3×FLAG and Vma22p-3×FLAG, samples were further concentrated to ~1 to 2 mg/mL with a 100 kDa MWCO Vivaspin 500 centrifugal concentrator (Sartorius) at 12,000 × g. For Vma21p-3×FLAG, the sample was concentrated at 1,000 × g. Protein concentration was determined by bicinchoninic acid assay (Pierce).
CryoEM Specimen Preparation and Data Collection.
Samples were applied to homemade nanofabricated holey gold grids with regular arrays of ~2 μm holes (50, 51). Grids were glow-discharged in air for 2 min, and 1.5 μL of sample was applied to the grids, blotted for 2 s at 4 °C and 80% relative humidity, and frozen rapidly in liquid ethane with a Leica EM GP2 freezing device. Sample screening was performed with a FEI Tecnai F20 electron microscope operating at 200 kV and equipped with a Gatan K2 Summit direct detector device camera. Images were collected as movies at a magnification of 25,000×, with 30 fractions at 5 e−/pixel/s and a calibrated pixel size of 1.45 Å. High-resolution data collection was performed with a Titan Krios G3i electron microscope (Thermo Fisher) operating at 300 kV and equipped with a Falcon 4 camera (Vma22p dataset) or a prototype Falcon 4i camera (Vma12p and Vma21p datasets). Automated data collection was performed with the EPU software package. Movies consisting of 30 exposure fractions were collected at a nominal magnification of 75,000×, corresponding to a calibrated pixel size of 1.03 Å.
Image Analysis.
Movie alignment with patch-based motion correction and estimation of contrast transfer function (CTF) parameters were performed with cryoSPARC Live. All other image analysis steps were performed with cryoSPARC v3 (52). After removing movies with undesirable CTF fit, ice thickness, or motion, 4,032, 4,417, and 4,179 movies from the Vma12p-×FLAG, Vma21p-×FLAG, and Vma22p-×FLAG datasets were selected for further processing, respectively. Templates for particle selection were generated from 2D classification of manually selected particle images. Individual particle motion correction was performed (53), and datasets were cleaned with multiple rounds of 2D classification and ab initio 3D classification. This process provided 692,432 particle images for the Vma12p-×FLAG dataset, 607,746 particle images for the Vma21p-×FLAG dataset, and 462,692 particle images for the Vma22p-×FLAG dataset.
Ab initio 3D classification and heterogeneous refinement were applied to all the datasets. For the Vma21p dataset, this procedure allowed identification of classes corresponding to VO:Vma21p, VO:Vma12-22p, and the YAR027W/028W:c9 ring structure. For both the Vma12p and Vma22p datasets, the process led to identification of VO:Vma12-22p and VO∆aef:Vma12-22p structures. The classes corresponding to VO:Vma21p from Vma21p dataset and VO:Vma12-22p and VO∆aef:Vma12-22p from Vma12p dataset were refined with nonuniform refinement (54), followed by two rounds of local and global CTF refinement, and nonuniform refinement.
Atomic Model Building.
The previously published VO model 6O7T (36), AlphaFold models of Vma12p, Vma21p, and Vma22p (35), and subunit F from previously published V-ATPase model 7TMQ (19) were used for rigid body fitting into the maps of VO:Vma12-22p, VO∆aef:Vma12-22p, and VO:Vma21p with University of California San Francisco (UCSF) Chimera (55). Atomic models were constructed by manual model building in Coot (56), followed by refinement with ISOLDE (57) and real space refinement with Phenix (58). Figures were rendered with UCSF Chimera (55) and UCSF ChimeraX (59).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Model for the sequence of events in VO assembly.
Acknowledgments
We thank Samir Benlekbir for assistance with cryoEM data collection, Spencer Freeman and Sergio Grinstein for discussions about ER-retrieval, and Patricia Kane for a critical reading of the manuscript. J.L.R. was supported by the Canada Research Chairs program. This research was supported by Canadian Institutes of Health Research grant PJT166152 (J.L.R.). CryoEM data were collected at the Toronto High-Resolution High-Throughput cryoEM facility, supported by the Canada Foundation for Innovation and Ontario Research Fund. Mass spectrometry data were collected at the Network Biology Collaborative Center at the Lunenfeld-Tanenbaum Research Institute.
Author contributions
H.W. purified the protein samples and performed cryoEM and associated analysis; S.A.B. prepared the Vma12p-, Vma21p-, and Vma22p-3×FLAG yeast strains, performed proof-of-principle protein purifications, and prepared the ∆YAR027W∆YAR028W yeast strain; J.L.R. conceived the study and supervised the research; and H.W. and J.L.R. wrote the manuscript and prepared figures.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. J.A.L. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
Electron microscopy maps are available from the electron microscopy databank with accessioncodes EMD-27984, EMD-27985, EMD-27986, EMD-27987, and EMD-27988 (60–64), and atomic models are available from the protein databank with accession codes 8EAS, 8EAT, 8EAU, and 8EAV (65–68).
Supporting Information
References
- 1.Forgac M., Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Hinton A., Bond S., Forgac M., V-ATPase functions in normal and disease processes. Pflüg. Arch. Eur. J. Physiol. 457, 589–598 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Vasanthakumar T., Rubinstein J. L., Structure and roles of V-type ATPases. Trends Biochem. Sci. 45, 295–307 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Qin A., et al. , V-ATPases in osteoclasts: Structure, function and potential inhibitors of bone resorption. Int. J. Biochem. Cell Biol. 44, 1422–1435 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Brown D., Paunescu T. G., Breton S., Marshansky V., Regulation of the V-ATPase in kidney epithelial cells: Dual role in acid–base homeostasis and vesicle trafficking. J. Exp. Biol. 212, 1762–1772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stransky L., Cotter K., Forgac M., The function of V-ATPases in cancer. Physiol. Rev. 96, 1071–1091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frattini A., et al. , Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat. Genet. 25, 343–346 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Kornak U., Mutations in the a3 subunit of the vacuolar H+-ATPase cause infantile malignant osteopetrosis. Hum. Mol. Genet. 9, 2059–2063 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Karet F. E., et al. , Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat. Genet. 21, 84–90 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Colacurcio D. J., Nixon R. A., Disorders of lysosomal acidification—The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korvatska O., et al. , Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS). Hum. Mol. Genet. 22, 3259–3268 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazhab-Jafari M. T., et al. , Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature 539, 118–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roh S.-H., et al. , The 3.5-Å cryoEM structure of nanodisc-reconstituted yeast vacuolar ATPase VO proton channel. Mol. Cell 69, 993–1004.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Benlekbir S., Rubinstein J. L., Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 521, 241–245 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Abbas Y. M., Wu D., Bueler S. A., Robinson C. V., Rubinstein J. L., Structure of V-ATPase from the mammalian brain. Science 367, 1240–1246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane P. M., Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem. 270, 17025–17032 (1995). [PubMed] [Google Scholar]
- 17.Sumner J.-P., et al. , Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270, 5649–5653 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Parra K. J., Keenan K. L., Kane P. M., The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275, 21761–21767 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Vasanthakumar T., Keon K. A., Bueler S. A., Jaskolka M. C., Rubinstein J. L., Coordinated conformational changes in the V1 complex during V-ATPase reversible dissociation. Nat. Struct. Mol. Biol. 29, 430–439 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Jaskolka M. C., Tarsio M., Smardon A. M., Khan M., Kane P. M., Defining steps in RAVE-catalyzed V-ATPase assembly using purified RAVE and V-ATPase subcomplexes. J. Biol. Chem. 296, 100703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seol J. H., Shevchenko A., Shevchenko A., Deshaies R. J., Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3, 384–391 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Smardon A. M., Tarsio M., Kane P. M., The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 277, 13831–13839 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Hill K. J., Stevens T. H., Vma22p is a novel endoplasmic reticulum-associated protein required for assembly of the yeast vacuolar H+-ATPase complex. J. Biol. Chem. 270, 22329–22336 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Hill K. J., Stevens T. H., Vma2lp is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H+-ATPase complex. Mol. Biol. Cell 5, 12 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson D. D., Stevens T. H., VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J. Biol. Chem. 272, 25928–25934 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Miles A. L., Burr S. P., Grice G. L., Nathan J. A., The vacuolar-ATPase complex and assembly factors, TMEM199 and CCDC115, control HIF1α prolyl hydroxylation by regulating cellular iron levels. eLife 6, e22693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramachandran N., et al. , VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. (Berl.) 125, 439–457 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Wang F., et al. , Follicular lymphoma-associated mutations in the V-ATPase chaperone VMA21 activate autophagy creating a targetable dependency. Autophagy 18, 1–19 (2022), 10.1080/15548627.2022.2050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cannata Serio M., et al. , Mutations in the V-ATPase assembly factor VMA21 cause a congenital disorder of glycosylation with autophagic liver disease. Hepatology 72, 1968–1986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girard M., et al. , CCDC115-CDG: A new rare and misleading inherited cause of liver disease. Mol. Genet. Metab. 124, 228–235 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Jansen J. C., et al. , CCDC115 deficiency causes a disorder of Golgi homeostasis with abnormal protein glycosylation. Am. J. Hum. Genet. 98, 310–321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen J. C., et al. , TMEM199 deficiency is a disorder of Golgi homeostasis characterized by elevated aminotransferases, alkaline phosphatase, and cholesterol and abnormal glycosylation. Am. J. Hum. Genet. 98, 322–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malkus P., Graham L. A., Stevens T. H., Schekman R., Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell 15, 5075–5091 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham L. A., Hill K. J., Stevens T. H., Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 142, 39–49 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasanthakumar T., et al. , Structural comparison of the vacuolar and Golgi V-ATPases from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 116, 7272–7277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keon K. A., Benlekbir S., Kirsch S. H., Müller R., Rubinstein J. L., Cryo-EM of the yeast VO complex reveals distinct binding sites for macrolide V-ATPase inhibitors. ACS Chem. Biol. 17, 619–628 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Benlekbir S., Bueler S. A., Rubinstein J. L., Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol. 19, 1356–1362 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Casey J. R., Grinstein S., Orlowski J., Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Kim J. H., et al. , Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl. Acad. Sci. U.S.A. 95, 2997–3002 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham L. A., Hill K. J., Stevens T. H., VMA7 encodes a novel 14-kDa subunit of the Saccharomyces cerevisiae vacuolar H(+)-ATPase complex. J. Biol. Chem. 269, 25974–25977 (1994). [PubMed] [Google Scholar]
- 42.Hirata R., et al. , VMA12 is essential for assembly of the vacuolar H(+)-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J. Biol. Chem. 268, 961–967 (1993). [PubMed] [Google Scholar]
- 43.Roh S.-H., et al. , Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of VO complex. Sci. Adv. 6, eabb9605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan M., Graham L. A., Stevens T. H., Voa1p Functions in V-ATPase Assembly in the Yeast Endoplasmic Reticulum. Mol. Biol. Cell 19, 5131–5142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poirey R., et al. , Functional analysis of the Saccharomyces cerevisiae DUP240 multigene family reveals membrane-associated proteins that are not essential for cell viability. Microbiology 148, 2111–2123 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Takahashi Y., Mizoi J., Toh-e A., Kikuchi Y., Yeast Ulpl, an Smt3-specific protease, associates with nucleoporins. J. Biochem. (Tokyo) 128, 723–725 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Miller J. P., et al. , Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. 102, 12123–12128 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu H., et al. , High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreev I., KTD1 is a yeast defense factor against K28 killer toxin. http://biorxiv.org/lookup/doi/10.1101/2021.10.25.465803 (2021) 10.1101/2021.10.25.465803 (Accessed 14 June 2022). [DOI]
- 50.Marr C. R., Benlekbir S., Rubinstein J. L., Fabrication of carbon films with ∼500nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Russo C. J., Passmore L. A., Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Rubinstein J. L., Brubaker M. A., Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Pettersen E. F., et al. , UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis: UCSF ChimeraX visualization system. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO in complex with Vma12-22p. EMBL's European Bioinformatics Institute. https://www.ebi.ac.uk/emdb/EMD-27984. Deposited 29 August 2022.
- 61.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO missing subunits a, e, and f in complex with Vma12-22p. EMBL's European Bioinformatics Institute. https://www.ebi.ac.uk/emdb/EMD-27985. Deposited 29 August 2022.
- 62.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO in complex with Vma21p. EMBL's European Bioinformatics Institute. https://www.ebi.ac.uk/emdb/EMD-27986. Deposited 29 August 2022.
- 63.Wang H., Bueler S. A., Rubinstein J. L., YAR027W and YAR028W in complex with c subunits from yeast VO complex. EMBL's European Bioinformatics Institute. https://www.ebi.ac.uk/emdb/EMD-27987. Deposited 29 August 2022.
- 64.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO in complex with Vma12-22p purified via Vma21p-3xFLAG. EMBL's European Bioinformatics Institute. https://www.ebi.ac.uk/emdb/EMD-27988. Deposited 29 August 2022.
- 65.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO in complex with Vma12-22p. Protein Data Bank. https://www.rcsb.org/structure/8EAS. Deposited 29 August 2022.
- 66.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO missing subunits a, e, and f in complex with Vma12-22p. Protein Data Bank. https://www.rcsb.org/structure/8EAT. Deposited 29 August 2022.
- 67.Wang H., Bueler S. A., Rubinstein J. L., Yeast VO in complex with Vma21p. Protein Data Bank. https://www.rcsb.org/structure/8EAU. Deposited 29 August 2022.
- 68.Wang H., Bueler S. A., Rubinstein J. L., YAR027W and YAR028W in complex with c subunits from yeast VO complex. Protein Data Bank. https://www.rcsb.org/structure/8EAV. Deposited 29 August 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Model for the sequence of events in VO assembly.
Data Availability Statement
Electron microscopy maps are available from the electron microscopy databank with accessioncodes EMD-27984, EMD-27985, EMD-27986, EMD-27987, and EMD-27988 (60–64), and atomic models are available from the protein databank with accession codes 8EAS, 8EAT, 8EAU, and 8EAV (65–68).