Significance
Previous studies, mostly in animal models, suggest that microRNAs (miRNAs) play an important role in type 2 diabetes (T2D). Yet, we have limited knowledge of miRNA expression in human pancreatic islets (HPIs), a T2D-relevant tissue. Here, we present the largest sequencing-based analysis of miRNA expression in HPIs to date and evaluate islet miRNA expression in the context of T2D. We identify genetic effects that drive miRNA expression and show that highly heritable islet miRNAs are largely regulated by trans-effects. We also identify 14 miRNAs associated with T2D and 4 with a polygenic score for glycated hemoglobin. This study provides key insights into HPI miRNA biology and future pathways for investigating T2D pathophysiology.
Keywords: pancreatic islets, diabetes, microRNA, eQTL
Abstract
Genetic studies have identified ≥240 loci associated with the risk of type 2 diabetes (T2D), yet most of these loci lie in non-coding regions, masking the underlying molecular mechanisms. Recent studies investigating mRNA expression in human pancreatic islets have yielded important insights into the molecular drivers of normal islet function and T2D pathophysiology. However, similar studies investigating microRNA (miRNA) expression remain limited. Here, we present data from 63 individuals, the largest sequencing-based analysis of miRNA expression in human islets to date. We characterized the genetic regulation of miRNA expression by decomposing the expression of highly heritable miRNAs into cis- and trans-acting genetic components and mapping cis-acting loci associated with miRNA expression [miRNA-expression quantitative trait loci (eQTLs)]. We found i) 84 heritable miRNAs, primarily regulated by trans-acting genetic effects, and ii) 5 miRNA-eQTLs. We also used several different strategies to identify T2D-associated miRNAs. First, we colocalized miRNA-eQTLs with genetic loci associated with T2D and multiple glycemic traits, identifying one miRNA, miR-1908, that shares genetic signals for blood glucose and glycated hemoglobin (HbA1c). Next, we intersected miRNA seed regions and predicted target sites with credible set SNPs associated with T2D and glycemic traits and found 32 miRNAs that may have altered binding and function due to disrupted seed regions. Finally, we performed differential expression analysis and identified 14 miRNAs associated with T2D status—including miR-187-3p, miR-21-5p, miR-668, and miR-199b-5p—and 4 miRNAs associated with a polygenic score for HbA1c levels—miR-216a, miR-25, miR-30a-3p, and miR-30a-5p.
Type 2 diabetes (T2D) is a leading contributor to global morbidity and mortality (1) and is characterized by reduced insulin response in insulin-sensitive tissues and pancreatic islet beta cell dysfunction (2, 3). However, despite our knowledge of the role of insulin in the disease, the underlying molecular and cellular mechanisms driving T2D are not wholly understood. Recent studies investigating circulating biomarkers of T2D identified several microRNAs (miRNAs) with altered levels in diabetic patients (4–9), raising the possibility that miRNAs play an important role in human T2D pathophysiology.
Mature miRNAs are small (~22 nucleotides long), non-coding RNA molecules that typically function as post-transcriptional gene regulators by tethering the miRNA-induced silencing complex to target messenger RNAs (mRNAs), leading to mRNA silencing and degradation (10–12). Several seminal studies have implicated miRNAs in pancreatic islet development and function (13–24). However, the bulk of these studies are in model systems (e.g., animal models, cell lines), so our knowledge of miRNAs in human pancreatic islets (HPIs) remains limited. To date, most studies examining miRNA expression in HPIs have tested only a small number of miRNAs (25–28). Of the two more comprehensive sequencing-based studies of HPI miRNAs (29, 30), both have been limited in sample size (n ≤ 7).
In addition, incorporating genetic information in the analysis of miRNA expression in HPIs may help identify miRNAs with a causal role in T2D pathophysiology. To date, ≥240 loci have been reported to be associated with T2D risk, yet the majority of these signals lie in non-coding regions of the genome, masking the underlying molecular mechanisms (31, 32). Previous studies have successfully integrated genetic and RNA-seq data in HPIs to characterize the genetic drivers of mRNA expression and nominate candidate causal genes for T2D by identifying shared genetic signals between loci associated with islet gene expression quantitative trait loci (eQTLs) and T2D risk (33–37). However, such integrative analyses have yet to be performed for miRNAs in HPIs.
Here, we present data from 63 individuals, the largest sequencing-based analysis of miRNA expression in HPIs to date (Fig. 1 and SI Appendix, Table S1; 57 with genotypes). Using genotype information (n = 57), we estimate the SNP-based heritability for each miRNA (i.e., the proportion of variation in expression that can be explained by additive effects of common genetic variants) and find 84 highly heritable miRNAs. To perform a comparative analysis of heritability between miRNAs and mRNAs, we also estimate the SNP-based heritability of mRNAs using a subset of islets with genotype and mRNA expression data from this study (n = 39) and a previous study (n = 189) (33). We show that regulation of heritable miRNAs is primarily driven by trans-acting genetic effects, whereas heritable mRNAs are regulated by a combination of cis-acting and trans-acting genetic effects. Given the importance of islet dysfunction in T2D pathophysiology, we apply several different strategies to assess the role of miRNA expression in diabetes. First, we map miRNA-eQTLs and colocalize the five identified miRNA-eQTLs with genetic signals from association studies of T2D and glycemic traits. Next, we identify single nucleotide polymorphisms (SNPs) from these genetic studies that are in the 99% credible sets (i.e., variants that are most likely to be the causal variant at each independent genetic association signal) and lie in miRNA seed regions or predicted target sites. Finally, using all miRNA expression data (n = 63), we identify miRNAs associated with T2D status and polygenic scores (PGSs) of T2D-related traits, as well as sex, age, and body mass index (BMI).
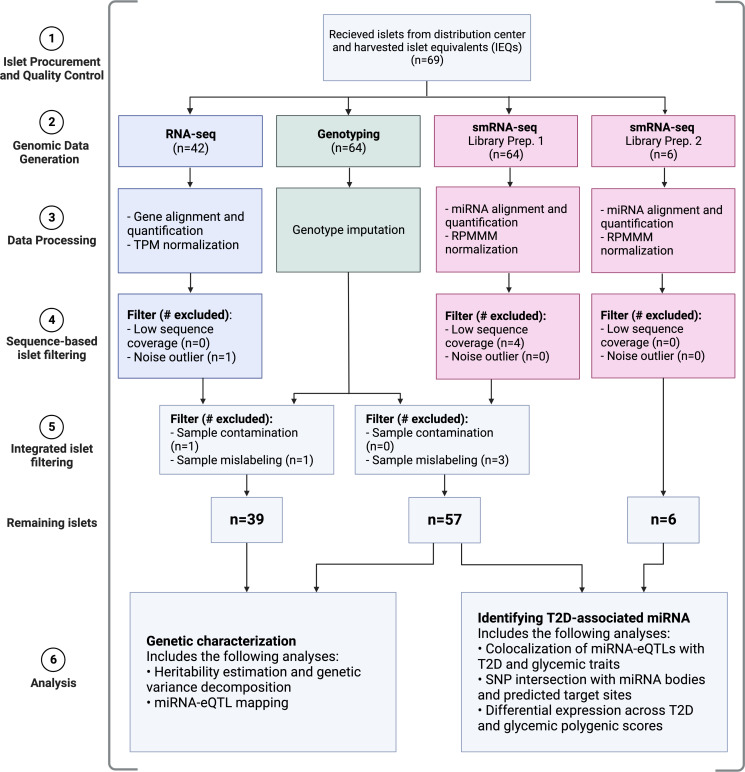
Fig. 1.
Study overview. Colors correspond to data modalities. TPM: transcripts per million; RPMMM: reads per million mapped to miRNAs. Created with BioRender.com.
Results
Data Overview.
We procured 69 HPI samples and performed small RNA-sequencing (smRNA-seq), RNA-sequencing (RNA-seq), and genotyping, retaining 63 samples with smRNA-seq, 39 samples with RNA-seq, and 57 samples with genotypes after quality control steps (Materials and Methods; Fig. 1 and SI Appendix, Fig. S1 and Table S1). We note that not all samples were subject to each assay (Fig. 1). For the smRNA-seq, we generated data through two experiments: library preparation 1 (LP1; 57 samples) and library preparation 2 (LP2; 6 samples). In LP1, we generated an average of 38.87 million read pairs per sample (±14.45 SD; minimum read count ≥19.98 million), with a mean read length of 23.24 nucleotides (SI Appendix, Fig. S2A). In LP2, we generated an average of 64.36 million read pairs per sample (±4.18 SD; minimum read count ≥58.61 million), with a mean read length of 22.62 nucleotides (SI Appendix, Fig. S2B). Across both LPs, we found that the miRNAs identified by previous mouse and human studies (17, 29, 38) to be most abundant in islets (e.g., miR-375) were also the most abundant miRNAs in the data generated in this study (SI Appendix, Fig. S4). In total, we identified 2,959 unique miRNAs, including 1,989 miRNA isoforms (isomiRs). Of these, 2,279 are either canonical (reference) miRNAs or isomiRs with nucleotide shifts ≤2 in either direction, which we referred to as “high confidence.” For the RNA-seq, we generated an average of 57.51 million read pairs per sample (±16.48 SD; minimum read count ≥21.49 million). Similar to smRNA-seq, we found that highly abundant mRNAs (e.g., PRSS1, REG1A) from previous HPI studies (35, 37) were also highly abundant in the islet mRNA data generated in this study (SI Appendix, Fig. S5), underscoring the data quality.
Heritability and Genetic Architecture of miRNA Expression.
Studies in mice hepatic and lung tissues (39, 40) suggest that the genetic architecture of miRNA expression is primarily driven by trans-acting factors, much more than mRNA which has a substantial cis-acting genetic component; however, to date, a comparison of miRNA and mRNA regulatory trends has yet to be explored in human islets. To compare the trends in genetic regulation of miRNA and mRNA species, we estimated the SNP-based heritability () for miRNA and mRNA transcripts using imputed genotypes of common SNPs (Materials and Methods). We found that the fraction of heritable miRNA transcripts ( ≥ 0.9) was substantially smaller than the fraction of heritable mRNA transcripts for both all miRNA transcripts (P = 8.70 × 10−4, Chi-squared test; Fig. 2A and SI Appendix, Fig. S6A) and high-confidence miRNA transcripts (P = 1.56 × 10−3, Chi-squared test; Fig. 2A and SI Appendix, Fig. S6A), suggesting that miRNAs are under stronger selection pressure than mRNAs (SI Appendix, Note S1).
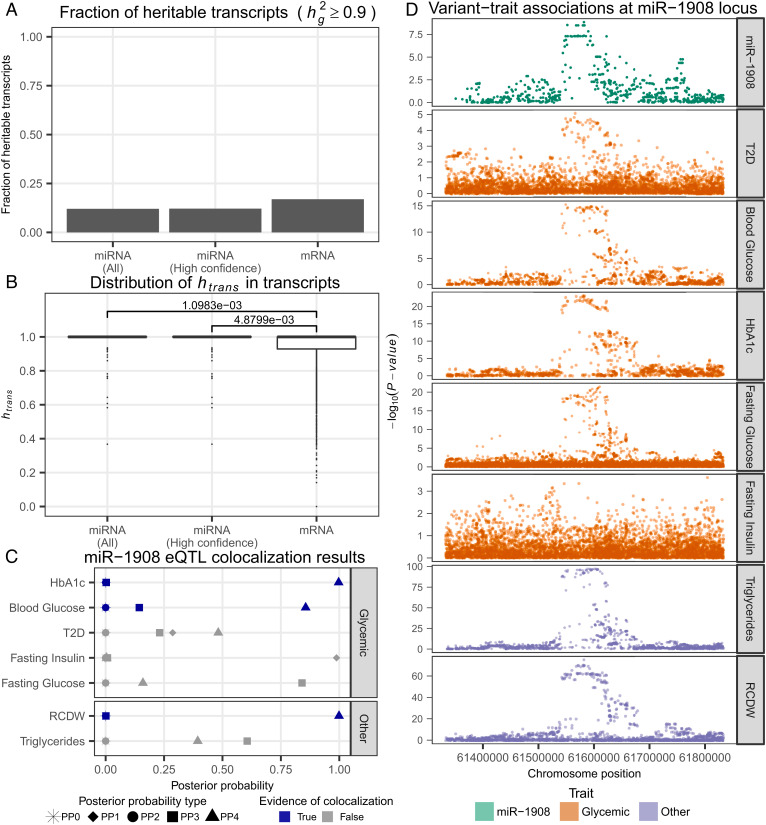
Fig. 2.
Genetic analysis of miRNA expression in HPIs. (A) Fraction of heritable transcripts ( ≥ 0.9) across miRNAs and mRNAs. (B) Distribution of variance explained by trans-acting genetic effects in the expression of heritable transcripts. P-value calculated using Mann–Whitney U test. (C) Evidence of genetic colocalization between miR-1908 eQTL and disease/trait genetic association signals (Materials and Methods). Posterior probability definitions: PP0 – neither trait is associated, PP1 – miRNA-eQTL is associated, PP2 – Genome-wide association study (GWAS) is associated, PP3 – both traits are associated with different causal variants, PP4 – both traits are associated and share a single causal variant. (D) For each disease/trait used in the colocalization analysis, Manhattan plots for all variants within a 250 kb window on either side of the mature miR-1908 transcript.
We sought to understand the genetic architecture of heritable transcripts and decomposed the variance in transcript expression into cis- or trans-acting genetic effects, assuming that proximal SNPs [i.e., SNPs found within 250 kb of the mature miRNA transcript or mRNA transcription start site (TSS)] function primarily via cis-acting mechanisms and distal SNPs (i.e., outside of a 250 kb window) function primarily via trans-acting mechanisms, as has been shown previously (41). We found that the variance explained by trans-effects (htrans) was greater in heritable miRNA transcripts than in heritable mRNA transcripts (P = 1.10 × 10−3, Mann–Whitney U test; Fig. 2B and SI Appendix, Fig. S6B). These trends held true when just considering high-confidence miRNA transcripts (P = 4.88 × 10−3, Mann–Whitney U test; Fig. 2B and SI Appendix, Fig. S6B) as well as a larger 20 Mb cis window size, to account for the fact that some miRNA TSSs may be quite far away from the miRNA body (P = 1.00 × 10−2, Mann–Whitney U test; SI Appendix, Fig. S8) (42). To ensure these results were not driven by the limited sample size of the mRNA dataset (n = 39), we repeated the analysis using an independent dataset of 189 HPI samples with mRNA expression and genetic data from a previous study (33) and replicated these findings (SI Appendix, Fig. S9 A and B; P = 2.75 × 10−14, Chi-squared test for comparative heritability analysis; P = 3.75 × 10−2, Mann–Whitney U test for trans/cis variance decomposition). Combined, these results suggest the genetic regulatory architecture of miRNAs differs from mRNAs as miRNA genetic regulation is primarily driven by trans-effects while mRNA is driven by a combination of trans- and cis-effects. Such a model is consistent with previous studies in rodents (39, 40).
Identification of Genetic Associations with miRNA Expression.
We tested for genetic associations with miRNA expression in order to identify miRNA-eQTLs (Materials and Methods). At a false discovery rate (FDR) of ≤5%, we found five miRNA-eQTLs (SI Appendix, Table S3). We performed colocalization analysis between these miRNA-eQTLs and genetic loci associated with T2D and glycemic traits, including fasting blood glucose levels, blood glucose levels, fasting insulin levels, and glycated hemoglobin (HbA1c; Materials and Methods). We found no colocalizations with T2D. However, for one miRNA-eQTL, an eQTL for miR-1908 tagged by rs174559, we found evidence of colocalization with HbA1c and blood glucose levels (Fig. 2 C and D). Since rs174559 is also strongly associated with triglycerides (TG) (43) and red blood cell distribution width (RCDW, a metric representing the heterogeneity of red blood cell volume) (44), we also performed colocalization with these traits. We found no evidence of colocalization with TG, but evidence of colocalization with RCDW (Fig. 2C), possibly suggesting pleiotropic effects of this locus across tissues.
Focusing on the islet-relevant colocalization with glycemic traits, we looked for i) cis-effects of rs174559 on the transcription of nearby protein coding genes in HPIs and ii) trans-effects of rs174559 on protein coding genes throughout the genome to identify candidate target transcripts of miR-1908. We performed colocalization analysis between the miRNA-eQTL signal for miR-1908 and genetic associations for gene and exon expression identified in an islet eQTL study spanning 420 individuals (33). We found evidence of colocalization with several FADS1 exons and with gene level FADS1 expression (SI Appendix, Fig. S11A). Although miR-1908 is located within a FADS1 exon (there are several FADS1 isoforms), we found that the strongest colocalization signal was for variants associated with an exon ~4.3 kb away from miR-1908. We performed Mendelian randomization and meditation analyses to test for a potential causal relationship between miR-1908 and FADS1 (or vice versa; Materials and Methods). We found no evidence for a causal relationship between miR-1908 and FADS1 expression (SI Appendix, Fig. S11B). Next, we looked for trans-associations between rs174559 and protein coding genes in 228 samples [39 samples from this study and 189 samples from an independent dataset (33)] as trans-eQTL summary statistics from previous islet eQTL studies were not available (Materials and Methods). We found no associations, which may be driven by no signal or the lack of power to detect trans-eQTL effects at this sample size.
Previous studies have mapped miRNA-eQTLs in blood (45, 46). To understand the tissue specificity of miRNA genetic effects, we compared the effect sizes of SNP-miRNA pairs from HPI miRNA-eQTLs to blood miRNA-eQTLs (Materials and Methods). We found that although some effects are replicated across datasets [e.g., the HPI rs174559 miR-1908 eQTL is also found in the study by Sonehara et al. (46)], the HPI effects were generally not correlated with the genetic effects observed in blood (SI Appendix, Fig. S12; maximum Spearman’s rho = 0.07), suggesting that genetic effects on miRNAs may to some extent be tissue/cell type specific.
Identification of T2D-Related Genetic Effects that May Perturb miRNA Function.
To identify genetic variants associated with T2D and T2D-related phenotypes that may affect islet miRNA function (as opposed to overall expression levels), we overlapped 99% credible set SNPs from genetic studies for T2D and other glycemic traits with i) mature miRNA genomic coordinates (which include areas outside of the seed region and may affect the ability of the miRNA to bind to targets) and ii) genomic coordinates for predicted miRNA target regions (i.e., the miRNA binding site on the mRNA transcript). To limit the analysis to islet-relevant results, we retained SNPs where the miRNA and target mRNA were highly expressed in HPI data presented in this study (Materials and Methods; SI Appendix, Table S4). Consistent with previous work (47), we did not identify any SNPs within mature miRNA coordinates. However, we did identify 16 T2D, 8 HbA1c, and 1 blood glucose credible set SNPs within predicted miRNA target regions. Because SNPs within miRNA target regions would exert their effects on the target transcript via cis-acting mechanisms, we further explored the effects of these SNPs using an islet cis-eQTL study spanning 420 individuals (33). We found one SNP, rs1464569—in the 99% credible set for an HbA1c signal—that lies within NICN1 at a miR-532-3p target site and is in high linkage disequilibrium (1000GENOMES phase_3 FIN = 1.00, CEU = 1.00, GBR = 1.00) with the tag SNP, rs4955440, for the gene-level NICN1 eQTL. To explore whether rs1464569 functions as an eQTL for NICN1 by perturbing miRNA binding, we tested for an interaction effect by modeling NICN1 expression with rs1464569 and miR-532-3p expression using 33 samples with mRNA expression, miRNA expression, and genetic data (Materials and Methods). We found no evidence of an interaction effect (P > 0.05), leaving the mechanism driving the genetic association at this locus unresolved.
Differentially Expressed miRNAs.
Next, we sought to identify miRNAs associated with sex, age, BMI, T2D disease status, and PGSs for T2D and glycemic traits (Materials and Methods).
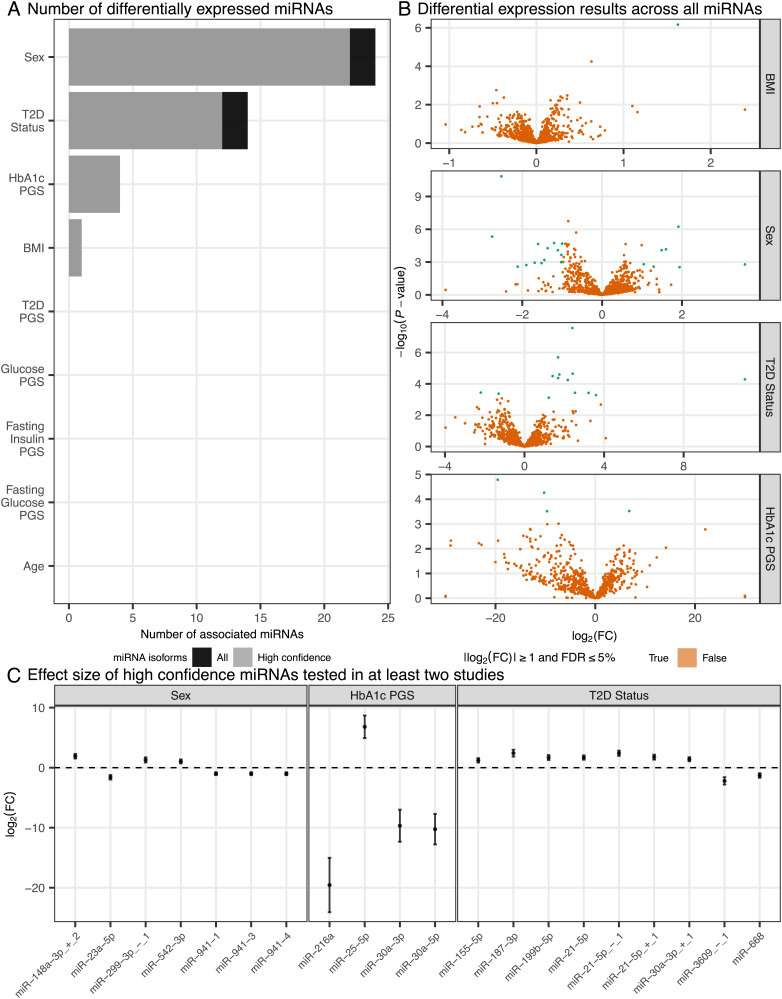
For sex, age, and BMI, we performed a meta-analysis of differential expression results from this study (n = 63 total from two LPs) and a previous smRNA-seq study of HPIs (n = 7) (29). We identified 1 BMI-associated and 24 sex-associated miRNAs [FDR ≤ 5% and |log2(fold change (FC)]| ≥ 1; Fig. 3 and SI Appendix, Figs. S13 and S14). The large number of sex-associated miRNAs is consistent with a previous study describing substantial genomic differences between male and female HPIs (28). Notably, all but two (miR-542-3p and miR-767-5p) of the sex-associated miRNAs occurred on autosomes.
Fig. 3.
Differentially expressed miRNAs across diseases/traits. (A) Number of differentially expressed miRNAs (FDR ≤ 5% and |log2(FC)| ≥ 1) across each disease/trait. (B) Volcano plots describing the log2(FC) (x axis) and -log10(P-value) (y axis) of miRNAs across BMI, sex (male vs. female), T2D status, and HbA1c PGS. (C) Forest plot describing the effect of high-confidence differentially expressed miRNAs (FDR ≤ 5% and |log2(FC)| ≥ 1). Effect sizes are from meta-analysis.
Similarly, for T2D status, we compared T2D (n = 4) to normal glucose-tolerant (NGT, n = 59) donors and meta-analyzed the differential expression results from this study with a previous study (29) consisting of 7 islet samples (n = 4 T2D, n = 3 NGT). We identified 14 miRNAs with altered expression in T2D (FDR ≤ 5% and |log2(FC)| ≥ 1), 13 of which were not identified previously in the study by Kameswaran et al. (Fig. 3 and SI Appendix, Fig. S15), including 12 high-confidence miRNAs such as miR-21-5p, miR-187-3p, miR-199b-5p, miR-668, and miR-4497.
For the PGSs, we used all miRNA samples with genotypes (n = 57) and meta-analyzed differential expression results from individuals of European (n = 42), African (n = 7), and Hispanic/Latino (n = 6) ancestries. We identified four miRNAs associated with the HbA1c PGS (FDR ≤ 5% and |log2(FC)| ≥ 1; Fig. 3 and SI Appendix, Fig. S16)—miR-216a, miR-25-5p, miR-30a-5p, and miR-30a-3p.
Across all of the phenotypes considered, we compared the overlap of associated miRNAs (SI Appendix, Fig. S17). We found that one miRNA, miR-122-5p, was associated with both T2D and BMI.
To establish mRNAs that may be regulated by the miRNAs identified by our differential expression and genetic analyses, we tested for associations between miRNA expression and the mRNA expression of i) all genes and ii) only the predicted target genes (from TargetScan) using 33 samples with mRNA and miRNA expression data (Materials and Methods). At an FDR ≤ 5%, we identified no associations, likely driven by limited power with only 33 samples.
Finally, previous studies describe cell type heterogeneity as a possible confounder for bulk tissue gene expression studies (48, 49). To assess the contribution of cell type heterogeneity to the results described in this study, we estimated the fraction of alpha and beta cells, the primary cell types in processed HPIs (50–52), using miRNA profiles of sorted alpha and beta cells (Materials and Methods; SI Appendix, Fig. S20) (29). For both the miRNA-eQTL (SI Appendix, Fig. S21A) and differential expression analyses (SI Appendix, Fig. S21 B–E), we observed little change in the results of the models, suggesting cell type heterogeneity is not a primary driver of the results reported in this study.
Discussion
MiRNAs are small, non-coding RNA molecules that serve as important post-transcriptional gene regulators in human health and disease [reviewed in the study by Singh and Storey (53)]. The critical contribution of miRNAs to proper pancreatic islet development and function as well as the dysregulation of miRNAs in the context of T2D is well documented by previous studies using rodent models and cell-based systems (13–24). Despite these advances, our knowledge of miRNA expression and activity in human islets remains limited.
In this study, we present the largest sequencing-based analysis of miRNA expression in HPIs to date (n = 63). We describe the genetic regulation of HPI miRNA expression and provide insight into the role of miRNAs in T2D pathophysiology.
In our genetic analysis of HPIs, we show that the expression of highly heritable miRNAs is driven primarily by trans-acting genetic effects, whereas the expression of highly heritable mRNAs is driven by a combination of cis- and trans-acting genetic effects. This model is supported by previous studies in rodents that compare the heritability of miRNAs and mRNAs (39, 40) and suggests that miRNAs may be under greater selective pressure than mRNAs, as has been postulated by previous studies in humans (47, 54). Such selection pressures imply that to map miRNA-eQTLs thoroughly, large sample sizes will be required. Although this study replicates a previous finding, it is important to note that the heritability estimates of miRNAs and mRNAs will be affected by differences in signal-to-noise dynamics, making a direct comparison between the two features challenging (55) (SI Appendix, Note S1). We attempt to minimize the effects of noise by using noisyR (56) to remove low-abundance, noisy transcripts (Materials and Methods). However, in order to confidently compare the heritability between two biological phenomena measured by two different assays, future studies using repeat sampling of replicates will be required (55, 57).
Among the five identified miRNA-eQTLs, we find one miRNA-eQTL (miR-1908 tagged by rs174559) that colocalizes with genetic signals for multiple physiological traits—blood glucose, HbA1c, and RCDW. This signal also colocalizes with a genetic association for gene- and exon-level expression of FADS1, an essential enzyme for long-chain polyunsaturated fatty acid synthesis that has been implicated in a range of diseases, including diabetes and related glycemic traits (58–62). However, we highlight three limitations. First, given the sample size of this study and the number of expected recombination events, our power to detect colocalization events is limited, so the lack of colocalizations for five miRNA-eQTLs does not rule out possible colocalizations at larger sample sizes (63). Second, it is unclear whether the identified miR-1908 effects are mediated by miR-1908, FADS1, both, or neither. We perform an analysis to disentangle directionality between the two transcripts, but we find no evidence of a causal relationship between miR-1908 and FADS1, suggesting that the genetic effect may be independent. Further experiments to understand the molecular effects at this locus will be required. Finally, it is unclear whether the HbA1c and blood glucose effect is mediated by islets, blood, or some other tissue type. On the one hand, given i) the rs174559 miR-1908 eQTL also occurs in blood (46), ii) the RCDW effect at this locus, and iii) extensive studies that document how erythrocytic (i.e., red blood cell related) genetic effects can mediate HbA1c levels (64–68), it is plausible that observed effect on HbA1c is mediated by blood. On the other hand, an erythrocytic mediated effect would not explain the observed blood glucose genetic effect. In addition, at this locus, a study partitioned HbA1c genetic associations into erythrocytic and glycemic effects (69). Using a SNP in high linkage disequilibrium with rs174559 (1000GENOMES phase_3 FIN = 0.67, CEU = 0.70, GBR = 0.71) (70, 71), this study determined that the effect at rs174559 on HbA1c was driven by glycemic effects rather than erythrocytic effects which would suggest that the observed effects in islets may indeed be relevant.
As part of our genetic analysis, we also intersect 99% credible set SNPs with the genomic coordinates of predicted miRNA target sites. We find one SNP, rs1464569, in the 99% credible set for an HbA1c signal that lies in the NICN1 target site for miR-532-3p and is in high linkage disequilibrium with the tag SNP for the islet NICN1 eQTL. This finding may indicate that the eQTL functions by perturbing miR-532-3p regulation of NICN1. However, when we tested for an interaction effect in NICN1 expression, we did not find evidence of an interaction effect as would be expected if the NICN1 eQTL functioned by perturbing miR-532-3p binding. NICN1 is a tumor suppressor gene that has been shown to promote cell differentiation in a cell line derived from nasopharyngeal carcinomas (72); to date, little is known about the function of this gene in HPIs—a topic for future studies.
In addition, we identify several HPI miRNAs differentially expressed across BMI, sex, T2D status, and HbA1c genetic scores. Of the miRNAs identified, 26 (61.9%) have been previously implicated at some level as being associated with a related phenotype (38, 73–89). Focusing on the miRNAs associated with T2D status and HbA1c genetic scores, these findings provide critical human islet-based confirmation of previous studies in rodents and cell lines. For example, we find HPI levels of miR-216a and miR-25 to be negatively and positively correlated with HbA1c genetic scores, respectively. In previous work, the miR-216a knockout mouse was reported to have defects in insulin secretion in the context of chronic high-fat diet (90), and miR-25 has been described as a negative regulator of insulin synthesis in INS-1 cells, a rat insulinoma cell line (75). As another example, we show that miR-199b-5p is associated with T2D status, consistent with a previous study in murine beta cells where miR-199b-5p was shown to be highly responsive to glucose stimulation (91). We also find that miR-21-5p is strongly associated with T2D status. This observation comports with a recent study in which overexpression of miR-21-5p led to reduced glucose-stimulated insulin secretion in mice and human beta cells (92). We report that increased HPI expression of one miRNA, miR-122-5p, is associated with both increased BMI and T2D status. These findings are consistent with a previous study that reports that increased expression of miR-122-5p in blood is associated with T2D status and increased circulating insulin levels (81). As a final example, we identify the expression of miR-668, previously reported to be associated with T2D in human skeletal muscle (87), as a newly identified T2D association in HPIs.
Across all of the miRNAs identified by our genetic analysis and differential expression analysis, we tested for associations with potential target mRNAs, but found no associations (FDR ≤ 5%), likely driven by the small sample size of samples with paired miRNA and mRNA data (n = 33). Future studies with multiomic readouts or targeted miRNA perturbations paired with mRNA sequencing will be required to understand the target genes of the identified miRNAs.
Unraveling the genetic factors that contribute to pancreatic islet function and T2D is a high priority for the diabetes research community. However, much of the focus has been on mRNA and chromatin. Here, we add data on another important source of molecular regulation. While this is the largest sequencing-based analysis of HPI miRNA expression to date, larger studies will be required to probe the genetics of HPI miRNAs more comprehensively and to uncover more subtle associations between miRNA expression and phenotypes of interest. Of particular importance is increasing the sample size of islets from T2D patients, as the current sample size of T2D patients analyzed (n = 8) is still small. Nonetheless, the findings and associations identified in this study are a step toward understanding HPIs in the context of diabetes and will help guide the prioritization of miRNAs for future mechanistic studies.
Materials and Methods
A detailed description of computational and experimental analyses is provided in SI Appendix, Materials and Methods. Briefly, we performed smRNA-seq on 63 HPI samples and RNA-seq on 39 HPIs—quantifying miRNA and mRNA expression. Using 63 HPIs with genotypes, we estimated the SNP-based heritability for miRNA (n = 57) and mRNA (n = 39) transcripts and decomposed the variance in the expression of heritable transcripts into cis- and trans-acting genetic components using LIMIX v3.0.4. We tested for genetic variants associated with miRNA expression (miRNA-eQTLs) using LIMIX v3.0.4. To identify T2D-relevant miRNAs, we performed a colocalization analysis with miRNA-eQTLs and genetic loci associated with T2D and glycemic traits using coloc v3.1. To identify potential gene targets of miRNAs that colocalized with T2D or a glycemic trait, we also performed a colocalization analysis with miRNA-eQTLs and genetic loci associated with exon and gene expression in HPIs. We overlapped 99% credible set SNPs from genetic studies for T2D and glycemic traits with miRNA mature transcripts and target sites to identify variants that may alter miRNA function. Finally, we used DESeq2 v1.32.0 to identify miRNAs differentially expressed across T2D status, PGSs of T2D and glycemic traits, and other common phenotypes (i.e., sex, age, and BMI).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This research was supported in part by United States’ NIH grants ZIA-HG000024 (to F.S.C.), Gates Cambridge Trust (to H.J.T.), the NIH Oxford-Cambridge scholars program (to H.J.T.), and American Diabetes Association Pathway Award 1-16-ACE-47 (to P.S.). We thank Peter S. Chines and Chad Krilow for supporting the work presented in this study. We also thank the investigators of the Integrated Network for Systematic analysis of Pancreatic Islet RNA Expression consortium for their support in sharing data, with special thanks to Dr. Anna Gloyn and Dr. Mark McCarthy.
Author contributions
H.J.T., Y.-H.H., P.S., F.S.C., and D.L.T. designed research; H.J.T., Y.-H.H., M.R.E., A.J.S., L.L.B., P.S., F.S.C., and D.L.T. performed research; H.J.T., Y.-H.H., N.N., M.K., T.Y., and C.M.G. analyzed data; P.S., F.S.C., and D.L.T. supervised the study; and H.J.T., Y.-H.H., N.N., P.S., F.S.C., and D.L.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: H.L., The University of Texas MD Anderson Cancer Center; E.K.S., Indiana University School of Medicine; and O.S., EMBL-European Bioinformatics Institute.
Contributor Information
Praveen Sethupathy, Email: pr46@cornell.edu.
Francis S. Collins, Email: francis.collins@nih.gov.
Data, Materials, and Software Availability
Anonymized data deposits: the individual-level genotype, RNA-seq, and LP1 smRNA-seq data generated in this study are available in the database of Genotypes and Phenotypes (dbGap) with accession no. phs001188.v2.p1 (93); data are accessible through dbGaP’s standard data access request procedures. The individual-level LP2 smRNA-seq data from this study are available in the Gene Expression Omnibus data repository with accession no. GSE196797 (94). Summary statistics for the differential expression and miRNA-eQTL analyses are available in the Zenodo data repository with accession no. 7516377 (95).
Supporting Information
References
- 1.Khan M. A. B., et al. , Epidemiology of type 2 diabetes - global burden of disease and forecasted trends. J. Epidemiol. Glob. Health 10, 107–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galicia-Garcia U., et al. , Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 21, 6275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn S. E., Cooper M. E., Del Prato S., Pathophysiology and treatment of type 2 diabetes: Perspectives on the past, present, and future. Lancet 383, 1068–1083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C., et al. , Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci. Rep. 6, 20032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rong Y., et al. , Increased microRNA-146a levels in plasma of patients with newly diagnosed type 2 diabetes mellitus. PLoS One 8, e73272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belongie K. J., et al. , Identification of novel biomarkers to monitor β-cell function and enable early detection of type 2 diabetes risk. PLoS One 12, e0182932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiménez-Lucena R., et al. , A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: From the CORDIOPREV study. Exp. Mol. Med. 50, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghai V., et al. , Circulating RNAs as predictive markers for the progression of type 2 diabetes. J. Cell. Mol. Med. 23, 2753–2768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farr R. J., Joglekar M. V., Taylor C. J., Hardikar A. A., Circulating non-coding RNAs as biomarkers of beta cell death in diabetes. Pediatr. Endocrinol. Rev. 11, 14–20 (2013). [PubMed] [Google Scholar]
- 10.O’Brien J., Hayder H., Zayed Y., Peng C., Overview of microrna biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 9, 402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dexheimer P. J., Cochella L., Micrornas: From mechanism to organism. Front. Cell Dev. Biol. 8, 409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel D. P., Metazoan microRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poy M. N., et al. , miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U.S.A. 106, 5813–5818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latreille M., et al. , MicroRNA-7a regulates pancreatic β cell function. J. Clin. Invest. 124, 2722–2735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalis M., et al. , Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One 6, e29166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melkman-Zehavi T., et al. , miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J. 30, 835–845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poy M. N., et al. , A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226–230 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Xu G., Chen J., Jing G., Shalev A., Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat. Med. 19, 1141–1146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belgardt B.-F., et al. , The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 21, 619–627 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Roggli E., et al. , Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes 61, 1742–1751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Q., et al. , Obesity-induced miR-455 upregulation promotes adaptive pancreatic β-cell proliferation through the CPEB1/CDKN1B pathway. Diabetes 71, 394–411 (2022), 10.2337/db21-0134. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F., et al. , Obesity-induced overexpression of miR-802 impairs insulin transcription and secretion. Nat. Commun. 11, 1822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joglekar M. V., Joglekar V. M., Hardikar A. A., Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns 9, 109–113 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Joglekar M. V., Parekh V. S., Mehta S., Bhonde R. R., Hardikar A. A., MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev. Biol. 311, 603–612 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Klein D., et al. , MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS One 8, e55064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolmeson C., et al. , Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochem. Biophys. Res. Commun. 404, 16–22 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Ofori J. K., et al. , Human islet microRNA-200c is elevated in type 2 diabetes and targets the transcription factor ETV5 to reduce insulin secretion. Diabetes 71, 275–284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall E., et al. , Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 15, 522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kameswaran V., et al. , Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 19, 135–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Bunt M., et al. , The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS One 8, e55272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan A., et al. , Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vujkovic M., et al. , Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 52, 680–691 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viñuela A., et al. , Genetic variant effects on gene expression in human pancreatic islets and their implications for T2D. Nat. Commun. 11, 4912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonso L., et al. , TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 37, 109807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fadista J., et al. , Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. U.S.A. 111, 13924–13929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Bunt M., et al. , Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 11, e1005694 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varshney A., et al. , Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc. Natl. Acad. Sci. U.S.A. 114, 2301–2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karagiannopoulos A., et al. , Human pancreatic islet miRNA-mRNA networks of altered miRNAs due to glycemic status. iScience 25, 103995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Que E., et al. , Genetic architecture modulates diet-induced hepatic mRNA and miRNA expression profiles in Diversity Outbred mice. Genetics 218, 241–259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Rutledge H., et al. , Identification of microRNAs associated with allergic airway disease using a genetically diverse mouse population. BMC Genomics 16, 633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.GTEx Consortium et al. , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethupathy P., Illuminating microRNA transcription from the epigenome. Curr. Genomics 14, 68–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinnott-Armstrong N., et al. , Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 53, 185–194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan-UKB team, Pan-UK Biobank. https://pan.ukbb.broadinstitute.org (2020) (March 18, 2021).
- 45.Huan T., et al. , Genome-wide identification of microRNA expression quantitative trait loci. Nat. Commun. 6, 6601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonehara K., et al. , Genetic architecture of microRNA expression and its link to complex diseases in the Japanese population. Hum. Mol. Genet. 31, 1806–1820 (2021), 10.1093/hmg/ddab361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saunders M. A., Liang H., Li W.-H., Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. U.S.A. 104, 3300–3305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor D. L., et al. , Integrative analysis of gene expression, DNA methylation, physiological traits, and genetic variation in human skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 116, 10883–10888 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruning O., et al. , Confounding factors in the transcriptome analysis of an in-vivo exposure experiment. PLoS One 11, e0145252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabrera O., et al. , The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl. Acad. Sci. U.S.A. 103, 2334–2339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brissova M., et al. , Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 53, 1087–1097 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Ionescu-Tirgoviste C., et al. , A 3D map of the islet routes throughout the healthy human pancreas. Sci. Rep. 5, 14634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh G., Storey K. B., MicroRNA cues from nature: A roadmap to decipher and combat challenges in human health and disease? Cells 10, 3374 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rotival M., et al. , Population variation in miRNAs and isomiRs and their impact on human immunity to infection. Genome Biol. 21, 187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge T., Holmes A. J., Buckner R. L., Smoller J. W., Sabuncu M. R., Heritability analysis with repeat measurements and its application to resting-state functional connectivity. Proc. Natl. Acad. Sci. U.S.A. 114, 5521–5526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moutsopoulos I., et al. , noisyR: Enhancing biological signal in sequencing datasets by characterizing random technical noise. Nucleic Acids Res. 49, e83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ponzi E., Keller L. F., Bonnet T., Muff S., Heritability, selection, and the response to selection in the presence of phenotypic measurement error: Effects, cures, and the role of repeated measurements. Evolution 72, 1992–2004 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Reynolds L. M., et al. , FADS genetic and metabolomic analyses identify the ∆ desaturase (FADS1) step as a critical control point in the formation of biologically important lipids. Sci. Rep. 10, 15873 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao M., et al. , Polymorphisms of rs174616 in the FADS1-FADS2 gene cluster is associated with a reduced risk of type 2 diabetes mellitus in northern Han Chinese people. Diabetes Res. Clin. Pract. 109, 206–212 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Yuan S., Larsson S. C., Association of genetic variants related to plasma fatty acids with type 2 diabetes mellitus and glycaemic traits: A Mendelian randomisation study. Diabetologia 63, 116–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S.-W., et al. , Polymorphisms in FADS1 and FADS2 alter plasma fatty acids and desaturase levels in type 2 diabetic patients with coronary artery disease. J. Transl. Med. 14, 79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bazinet R. P., Layé S., Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 15, 771–785 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Giambartolomei C., et al. , Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lacy M. E., et al. , Association of sickle cell trait with hemoglobin a1c in african americans. JAMA 317, 507–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paré G., et al. , Novel association of HK1 with glycated hemoglobin in a non-diabetic population: A genome-wide evaluation of 14,618 participants in the Women’s Genome Health Study. PLoS Genet. 4, e1000312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soranzo N., et al. , Common variants at 10 genomic loci influence hemoglobin A1 (C) levels via glycemic and nonglycemic pathways. Diabetes 59, 3229–3239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen P., et al. , Multiple nonglycemic genomic loci are newly associated with blood level of glycated hemoglobin in East Asians. Diabetes 63, 2551–2562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen P., et al. , A study assessing the association of glycated hemoglobin A1C (HbA1C) associated variants with HbA1C, chronic kidney disease and diabetic retinopathy in populations of Asian ancestry. PLoS One 8, e79767 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wheeler E., et al. , Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 14, e1002383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howe K. L., et al. , Ensembl 2021. Nucleic Acids Res. 49, D884–D891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fairley S., Lowy-Gallego E., Perry E., Flicek P., The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 48, D941–D947 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hou D., et al. , Cloning and characterization of the NPCEDRG gene promoter. Mol. Cell. Biochem. 346, 1–10 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Jones A., et al. , miRNA signatures of insulin resistance in obesity. Obesity (Silver Spring) 25, 1734–1744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang P., et al. , miR-216a-targeting theranostic nanoparticles promote proliferation of insulin-secreting cells in type 1 diabetes animal model. Sci. Rep. 10, 5302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Setyowati Karolina D., Sepramaniam S., Tan H. Z., Armugam A., K., Jeyaseelan, miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 10, 1365–1378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim J. W., et al. , miRNA-30a-5p-mediated silencing of Beta2/NeuroD expression is an important initial event of glucotoxicity-induced beta cell dysfunction in rodent models. Diabetologia 56, 847–855 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Kwekel J. C., et al. , Sex and age differences in the expression of liver microRNAs during the life span of F344 rats. Biol. Sex Differ. 8, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ziats M. N., Rennert O. M., Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 19, 848–852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ameling S., et al. , Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med. Genomics 8, 61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lizarraga D., et al. , miRNAs differentially expressed by next-generation sequencing in cord blood buffy coat samples of boys and girls. Epigenomics 8, 1619–1635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mononen N., et al. , Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci. Rep. 9, 8887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.López-Beas J., et al. , miR-7 modulates hESC differentiation into insulin-producing beta-like cells and contributes to cell maturation. Mol. Ther. Nucleic Acids 12, 463–477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Locke J. M., da Silva Xavier G., Dawe H. R., Rutter G. A., Harries L. W., Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia 57, 122–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sato-Kunisada R., Yoshida N., Nakamura S., Uchiyama H., Matsumoto H., Enhanced expression of miR-199b-5p promotes proliferation of pancreatic β-Cells by down-regulation of MLK3. Microrna 5, 57–65 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Lakhter A. J., et al. , Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 61, 1124–1134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ding L., et al. , Identification of the differential expression of serum microRNA in type 2 diabetes. Biosci. Biotechnol. Biochem. 80, 461–465 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Gallagher I. J., et al. , Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jankauskas S. S., Gambardella J., Sardu C., Lombardi A., Santulli G., Functional role of miR-155 in the pathogenesis of diabetes mellitus and its complications. Noncoding RNA 7, 39 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cui C., et al. , Identification and analysis of human sex-biased microRNAs. Genomics Proteomics Bioinf. 16, 200–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Erener S., et al. , Deletion of pancreas-specific miR-216a reduces beta-cell mass and inhibits pancreatic cancer progression in mice. Cell Rep. Med. 2, 100434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Werneck-de-Castro J. P., Blandino-Rosano M., Hilfiker-Kleiner D., Bernal-Mizrachi E., Glucose stimulates microRNA-199 expression in murine pancreatic β-cells. J. Biol. Chem. 295, 1261–1270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ibrahim S., et al. , β-Cell pre-mir-21 induces dysfunction and loss of cellular identity by targeting transforming growth factor beta 2 (Tgfb2) and Smad family member 2 (Smad2) mRNAs. Mol. Metab. 53, 101289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ibrahim M. R., et al. , Data from “The Finland-United States Investigation of NIDDM Genetics (FUSION) Study - Islet Expression and Regulation by RNAseq and ATACseq. database of Genotypes and Phenotypes.” https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001188.v2.p1. Accessed 21 August 2020.
- 94.Hung Y.-H., Kanke M., from Data “microRNA Expression in Normal and Diabetic Pancreatic Islets.” Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196797. Accessed 21 August 2020.
- 95.Taylor H. J., et al. , Human pancreatic islet microRNAs implicated in diabetes and related traits by large-scale genetic analysis. Zenodo. https://zenodo.org/record/7516377#.Y9IZOslBy5c. Deposited 9 January 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Anonymized data deposits: the individual-level genotype, RNA-seq, and LP1 smRNA-seq data generated in this study are available in the database of Genotypes and Phenotypes (dbGap) with accession no. phs001188.v2.p1 (93); data are accessible through dbGaP’s standard data access request procedures. The individual-level LP2 smRNA-seq data from this study are available in the Gene Expression Omnibus data repository with accession no. GSE196797 (94). Summary statistics for the differential expression and miRNA-eQTL analyses are available in the Zenodo data repository with accession no. 7516377 (95).