Abstract
The opportunistic human pathogenic fungus Penicillium marneffei is dimorphic and is thereby capable of growth either as filamentous multinucleate hyphae or as uninucleate yeast cells which divide by fission. The dimorphic switch is temperature dependent and requires regulated changes in morphology and cell shape. Cdc42p is a Rho family GTPase which in Saccharomyces cerevisiae is required for changes in polarized growth during mating and pseudohyphal development. Cdc42p homologs in higher organisms are also associated with changes in cell shape and polarity. We have cloned a highly conserved CDC42 homolog from P. marneffei named cflA. By the generation of dominant-negative and dominant-activated cflA transformants, we have shown that CflA initiates polarized growth and extension of the germ tube and subsequently maintains polarized growth in the vegetative mycelium. CflA is also required for polarization and determination of correct cell shape during yeast-like growth, and active CflA is required for the separation of yeast cells. However, correct cflA function is not required for dimorphic switching and does not appear to play a role during the generation of specialized structures during asexual development. In contrast, heterologous expression of cflA alleles in Aspergillus nidulans prevented conidiation.
In a multicellular organism, the regulation of polarized growth and division allows the development of morphologically varied cell types. Fungal growth and development are characterized by regulated changes in polarized growth. During germination cells undergo a change from isotropic growth to polarized growth (15). Polarized growth of a hypha, which extends and branches, is achieved by the continual transport of vesicles containing cell wall material along the cytoskeleton to the hyphal apex (17). At a later stage of growth, hyphal cells can differentiate to undergo asexual development. Specialized cell structures are generated by the sequential budding of apical cells, culminating in the production of conidiospores, a process which also requires changes in polarized growth (28). Although a number of studies of filamentous fungi have been conducted, the molecular mechanisms of how polarity is generated and maintained are not fully understood (22, 29).
Hyphae are partitioned at regular intervals after mitosis into multinucleate cellular compartments during septation, a process analogous to cytokinesis in other organisms. Septation is controlled by signals derived from the positioning of nuclei, mitosis, and actin localization (19, 39). Aspergillus nidulans temperature-sensitive mitotic and nuclear migration mutants fail to form septa and can display hyphal cells with various lengths (30, 39). Septation is inhibited when actin localization is abolished by the actin polymerization inhibitor cytochalasin A (19). In A. nidulans, temperature-sensitive screens have identified three classes of septation mutants which are affected during either early or late stages of septum formation (19). One of these mutants, sepA, is also associated with morphological defects. Deletion of sepA results in abnormally wide hyphae with excessive branching and abnormally branched conidiophore stalks with split vesicles. sepA encodes a member of the formin homology family, which comprises proteins thought to facilitate actin localization by serving as molecular scaffolds (21). Other sep genes, such as sepB, play a role in monitoring chromosomal DNA metabolism (20). In the analogous process in the fission yeast Schizosaccharomyces pombe, cytokinesis mutants (sid [septation initiation defective]) which form an actinomysin contractile ring at the presumptive site of septation but which fail to form a septum have been isolated. The cells continue to grow and undergo nuclear division, giving rise to elongated, multinucleate cells (9).
Saccharomyces cerevisiae cytokinesis mutants include several cell division cycle (CDC) genes, and these mutants exhibit unbudded and multinucleate phenotypes (1). This phenotype can also be caused by mutations in genes which encode proteins involved in organizing the actin cytoskeleton and linking cytoskeletal organization with the mitotic cycle (BNI4, CLA4, GIN4, and CDC42) (5, 16, 35). Cdc42p is a member of the Rho family of GTPases, which in S. cerevisiae coordinate changes in polarized growth and regulate cytokinesis, mitotic budding, pseudohyphal growth, and polarized growth toward pheromones. These GTPases cycle between an active GTP-bound state and an inactive GDP-bound state. The active state of the protein is regulated by guanine exchange factors (GEFs) and GTPase-activating proteins (reviewed in reference 10). Dominant-negative mutations in the GDP-to-GTP exchange domain leading to an inactive protein which is unable to exchange GDP for GTP but which can still interact with the GEF (CDC24p) have been characterized. Dominance arises from GEF sequestration by the mutant protein. A dominant-activated phenotype arises from a mutation in the GTP binding domain, which results in the production of a constitutively activated protein (14). In S. cerevisiae, Cdc42p is proposed to control polarized growth via the activation of mitogen-activated protein kinase (MAPK) cascades, in addition to localizing the MAPK components, actin, and actin-associated proteins to the site of polarized growth. Cdc42p also localizes proteins required for cytokinesis and links growth to the cell cycle by activating cyclin-dependent kinase Clb2-Cdc28p and localizing the Cla4p p21-activated kinase (reviewed in references 4 and 37). Similarly in S. pombe, CDC42 is required for polarized growth and cytokinesis (27). In other organisms, CDC42 mutations result in a loss of polarized growth and morphology (reviewed in references 18 and 25).
Fungal dimorphism provides an amenable model system for the study of regulated changes in morphology and polarized growth during development. Dimorphic fungi have the ability to grow either as filamentous multinucleate hyphae or as uninucleate yeast cells. The switch from one growth state to the other requires regulated changes in cellular polarization and morphology. Penicillium marneffei is a dimorphic ascomycete which, like many other dimorphic fungi, is also an opportunistic pathogen (12, 23). P. marneffei exhibits a temperature-dependent dimorphic switch. At 25°C, P. marneffei grows as a multinucleate, septate mycelium which produces asexual conidia. At 37°C, the hyphae lay down double septa, which consequently promote cell separation to produce single uninucleate yeast cells (arthroconidia) which divide by fission. It is this growth form which is pathogenic.
As Cdc42p is involved in cytokinesis and polarized growth, we have investigated the role of the homologous protein in P. marneffei growth and development. The cloning of the P. marneffei CDC42 homolog named cflA (cdc42-like gene A) has allowed the generation of dominant-activated and dominant-negative mutations equivalent to those characterized in S. cerevisiae. We show that correct P. marneffei CflA function is required for polarized growth of the vegetative mycelium at 25°C and for correct cellular morphology of yeast cells at 37°C. Interestingly, CflA does not appear to be involved in development or in dimorphic switching.
MATERIALS AND METHODS
Molecular techniques.
Plasmid DNA was isolated using the Roche high-purity plasmid kit. Genomic DNA from P. marneffei and A. nidulans was isolated as previously described (7). RNA was prepared using the FastRNA kit from Bio101 as previously described (6). Southern and Northern blotting was performed with Amersham Hybond N+ membranes according to the manufacturer's instructions. Filters were hybridized using [α-32P]dATP-labeled probes by standard methods (36).
A P. marneffei genomic library, constructed by ligating partially Sau3AI-digested P. marneffei genomic DNA into BamHI-digested λGEM-11, was screened at low stringency (50% formamide, 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]; 37°C) for CDC42 homologs using A. nidulans modA (CDC42 homolog), supplied by S. Harris (pML5). Three positive clones were isolated from the screen, and a 4.6-kb hybridizing genomic SacI fragment was cloned into pBluescript II SK+, generating pKB4288. Subclones of pKB4288 were generated for sequencing. Sequencing was performed by the Australian Genome Research Facility and analyzed using Sequencher 3.1.1 (Gene Codes Corporation). Database searches and a sequence comparison were performed using the Australian National Genomic Information Service. A cDNA library, constructed using the SMART cDNA library construction kit (Clontech Laboratories Inc.) and RNA isolated from vegetative mycelia grown at 25°C (6) were probed with a 1.0-kb HindIII fragment of cflA at high stringency (50% formamide, 0.1× SSC; 37°C) to isolate cDNAs. Reverse transcriptase PCR was performed using the Superscript II kit from Life Technologies, primers F66 (5′ CTG GTA CCG ATT CGA GAG AAA CGT TG 3′) and F67 (5′ TAG GAT CCT TAT AGA ACA ACG CAC 3′), and with mRNA isolated from a vegetative mycelium grown at 25°C using the Qiagen Oligtex kit. Mutagenesis was performed using the GeneEditor in vitro site-directed mutagenesis kit with mutagenic primers G14V (5′ CGA CCG CAA CGT CACA CAA C 3′) and D120A (5′ CTC GCA AAG CGG TCT GGG 3′) (base mismatches are underlined). This was performed on a cflA PstI/SacII pBluescript II SK+ subclone (pKB4469) to generate pKB4478 (cflAG14V) and pKB4479 (cflAD120A).
To make the cflA::gfp construct, PCR with the T7 universal primer and F33 (5′ CAC CAT GGA TAG AAC AAC GCA CTT G 3′) was used to amplify the coding region of cflA and 946 bp of the promoter. The F33 primer incorporated a unique NcoI site, removed the cflA stop codon, and added an additional base to make an in-frame fusion. The NcoI- and PstI-digested PCR product was ligated into the NcoI/PstI sites of gfp-containing vector pAA4110 (A. Andrianopoulos, unpublished data). The PstI/SacII cflA::gfp fragment was then inserted into pAA4707, which contains the A. nidulans pyrG gene in pBluescript II, to generate pKB4705.
The alcA(p)::cflA, alcA(p)::cflAG14V, and alcA(p)::cflAD120A constructs were made by ligating a KpnI/BamHI fragment from a PCR product using primers F66 (5′ CTG GTA CCG ATT CGA GAG AAA CGT TG 3′) and F67 (5′ TAG GAT CCT TAT AGA ACA ACG CAC 3′), spanning 29 bases before the cflA start codon to the stop codon, into KpnI- and BamHI-digested pAL3 [alcA(p)] (26) to generate pKB4698.
Fungal strains and media.
The P. marneffei type strain, FRR2161, was obtained from J. Pitt (CSIRO Food Industries, Sydney, Australia). Strain SPM3 was isolated as a chlorate-resistant sector of FRR2161 and cannot grow on nitrate as the sole nitrogen source (7). Strain SPM4 was isolated as an SPM3 sector resistant to 5-fluoroorotic acid and is unable to grow in the absence of uridine (5 mM) and uracil (5 mM) (7). Transformation was carried out using the previously described protoplast method (7). The cflAD120A and cflAG14V strains (KBS086-089 and KBS090-093) were generated by cotransformation of strain SPM4 with pAB4342 (pyrG in pBluescript II SK+) and pKB4478 (cflAG14V) or pKB4479 (cflAD120A). The alcA(p)::cflA strains (KBS094-097) were produced by transformation of SPM4 with pKB4698, selecting for pyrG+. The cflA::gfp strains were generated by transformation of strain SPM4 with pKB4705 (KBS098-102), selecting for pyrG+.
At 25°C strains were grown on yeast synthetic dextrose (SD) medium (2) or on Aspergillus nidulans medium (ANM) supplemented with 10 mM γ-amino butyric acid (GABA) as the sole nitrogen source (13). At 37°C strains were grown on brain heart infusion (BHI) medium or on yeast SD medium (2). Dimorphic-switching experiments were performed in liquid SD medium. Strains were grown in liquid medium for 2 days at 25°C and transferred to 37°C. Strains were also grown for 10 days at 37°C and transferred to 25°C. Samples were obtained from both cultures after 1, 2, 3, 5, and 7 days. alcA(p)::cflA, alcA(p)::cflAD120A, and alcA(p)::cflAG14V strains were grown on carbon-free ANM(13)–GABA–fructose (noninduced) or on carbon-free ANM–GABA–fructose–cyclopentanone (induced).
Microscopy.
P. marneffei strains were grown on slides covered with thin layers of solid medium resting in liquid medium (6). cflA::gfp strains were grown at 25°C on SD slides and grown in liquid SD at 37°C. alcA(p)::cflA overexpression strains were grown in liquid ANM with 0.1% glucose plus GABA for 2 days at 25°C and induced by replacement of the medium with cyclopentanone (0.1 M) and 0.1% fructose for 2 days. alcA(p)::cflA overexpression strains were grown on BHI slides in BHI liquid medium for 4 days at 37°C before replacement of the medium with cyclopentanone (0.1 M) and 0.1% fructose for 2 days. Slides were examined using differential interference contrast or epifluorescence optics for green fluorescent protein fluorescence and staining with fluorescent brightener 28 (Calcofluor) and 4′,6-diamidino-2-phenylindole (DAPI) and viewed on a Reichart Jung Polyvar II microscope. Images were captured using a SPOT charge-coupled device camera (Diagnostic Instruments Inc.) and processed in Adobe Photoshop, version 4.0.
Nucleotide sequence accession number.
The GenBank accession number of the P. marneffei cflA gene is AF330694.
RESULTS
cflA encodes a highly conserved P. marneffei CDC42 homolog.
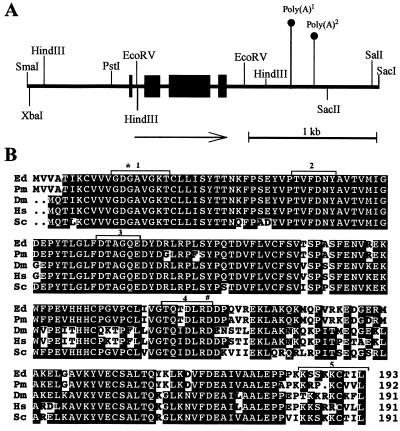
The P. marneffei CDC42 homolog was cloned by low-stringency hybridization using the CDC42 homolog from A. nidulans (modA) provided by S. Harris. The gene was named cflA and encodes a highly conserved protein homologous to the Rho family of small GTPases and specifically to the CDC42 class (Fig. 1). cflA is predicted to encode a 192-amino-acid protein containing the amino acid sequence TQXD at positions 114 to 117. This sequence is present in all Cdc42-like proteins and is NKXD in other related non-CDC42 GTPase proteins (11). This sequence is located in the domain required for association of the protein with effectors and is proposed to be required for interactor specificity (Fig. 1). The cflA gene spans an open reading frame of 819 bp and, based on the predicted amino acid sequence, consists of four exons and three introns. To confirm the positions of the predicted introns, reverse transcriptase PCR was performed on poly(A) mRNA produced from a vegetative mycelium grown at 25°C and the PCR product was sequenced. Database searches and sequence comparisons show that the cflA gene is most closely related to the homologous gene from Exophiala dermatitidis (AF162788.1 [40]) which is 83% identical at the DNA level and 96% identical in predicted protein sequence (Fig. 1). The predicted protein sequence contains the conserved domains previously shown to be necessary for Cdc42p and Rho1p GTPase function in S. cerevisiae. These include the GTP binding and hydrolysis domains and the domain required for GDP-to-GTP exchange and interaction with effector proteins (Fig. 1) (24). The predicted protein also contains a CAAX motif (A indicates an aliphatic amino acid) at the C terminus (positions 189 to 192), which in other related proteins allows prenylation and subsequent association of the protein with the plasma membrane (32).
FIG. 1.
(A) Gene structure of cflA. The gene is composed of four exons (filled boxes) and three introns. Arrow, direction of transcription. The two alternative polyadenylation sites and restriction enzyme recognition sites are also indicated. (B) Protein sequence alignment of the Cdc42p homologs from E. dermatitidis (Ed; AF162788.1), P. marneffei (Pm), D. melanogaster (Dm; P40793), Homo sapiens (Hs; P21181), and S. cerevisiae (Sc; Q01112). Identical amino acids are shaded black, similar amino acids are shaded grey, and nonconserved amino acids are unshaded. Amino acids mutated in these studies are indicated (asterisk, G14V; hash sign, D120A). The domains important for function are indicated as follows: GTP binding, 1; interaction with effectors, 2; GTP hydrolysis, 3; GDP-to-GTP exchange, 4; targeting to the plasma membrane, 5.
cflA produces two differentially expressed transcripts.
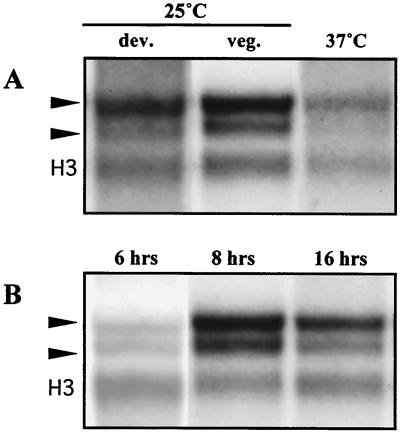
RNA was isolated from vegetative hyphal cells grown in liquid medium for 2 days at 25°C, from conidiating cultures grown on solid medium at 25°C for 4 days, and also from yeast cells grown at 37°C in liquid medium for 8 days. Northern blot analysis showed that cflA is expressed under all conditions as two differentially expressed transcripts, approximately 1.3 and 1.5 kb in size (Fig. 2A). Two cflA partial cDNA clones were isolated from a vegetative-hyphal-cell library. Sequence analysis revealed two alternative polyadenylation sites 200 bp apart in the 3′ untranslated region, accounting for the two cflA transcripts (Fig. 1A). Both cflA transcripts were expressed during vegetative growth and development at 25°C and during yeast-like growth at 37°C. However, the larger 1.5-kb transcript was expressed at a higher level than the smaller 1.3-kb transcript under all conditions tested (Fig. 2). The levels of cflA transcripts did not significantly differ between cultures grown at 25 or 37°C, compared to that of the histone H3 control. RNA was also isolated from P. marneffei during the yeast-to-hypha dimorphic switch. Yeast cells grown at 37°C for 8 days were transferred to 25°C in liquid culture and incubated for 6, 18, and 24 h in order to determine if cflA expression is regulated in response to the yeast-to-hyphal growth transition. cflA expression was upregulated during switching from yeast to hyphal growth, with the levels of both of the transcripts increasing upon transfer (Fig. 2B).
FIG. 2.
(A) Northern blot analysis of cflA expression. Total RNA was isolated from asexually developing cultures at 25°C (dev.), vegetatively growing mycelia at 25°C (veg.), and yeast cells (37°C). Northern blots were probed with cflA and a histone H3 probe (H3) as an RNA loading control. Arrowheads, the two cflA transcripts of 1.3 and 1.5 kb. (B) Northern blot analysis using total RNA isolated during the dimorphic switching from 37 to 25°C. Yeast cells were transferred from 37 to 25°C, and total RNA was isolated at 6, 18, and 24 h posttransfer. Northern blots were probed with cflA and a histone H3 probe as an RNA loading control. Arrowheads, the two cflA transcripts of 1.3 and 1.5 kb.
Overexpression of cflA does not affect morphogenesis.
Development depends on the accurate timing of polarized growth. To investigate whether increased levels of CflA are detrimental to P. marneffei development, cflA was overexpressed by placing it under the control of the alcohol-inducible A. nidulans alcA promoter (26). The alcA(p)::cflA overexpression construct (pKB4698) was transformed into strain SPM4, and transformants were isolated by direct selection for pyrG+. Transformants containing the pKB4698 construct were confirmed by Southern blot analysis of genomic DNA and probing with cflA (data not shown). alcA(p)::cflA strains (KBS094-097), which ranged in plasmid copy number from three to six copies, were grown on medium which induced expression of cflA and examined at the colonial level during vegetative growth at 25°C, during asexual development at 25°C, and during yeast growth at 37°C. The colonies of the alcA(p)::cflA strains displayed no change in phenotype on inducing medium at either 25 (Fig. 3B) or 37°C (data not shown). alcA(p)::cflA strains were also grown on slides for 2 days at 25°C or for 4 days at 37°C and then induced for an additional 2 days (see Materials and Methods). Microscopic examination of these cultures stained for nuclei (DAPI) or cell walls (Calcofluor) showed that the morphology, septation, and nuclear distribution of the alcA(p)::cflA strains were the same as those of the SPM4 control strain (data not shown).
FIG. 3.
Colonial morphology of the P. marneffei and A. nidulans dominant-activated cflAG14V and dominant-negative cflAD120A transformants. (A) Colonial morphology of the P. marneffei cflAG14V and cflAD120A transformants compared to that of the parental SPM4 control strain (cflA+). Strains were grown on ANM-GABA-uridine-uracil (5 mM) medium at 25°C for 8 (25°C vegetative) or 12 days (25°C developmental) or on SD-uridine-uracil (5 mM) medium for 6 days at 37°C (37°C vegetative). (B) Colonial morphology of the P. marneffei and A. nidulans alcA(p)::cflA, alcA(p)::cflAD120A, and alcA(p)::cflAG14V transformants. Strains were grown at 25°C for 12 days on noninducing medium (carbon-free medium [CF]-GABA-fructose medium [noninduced]) or on inducing medium (CF-GABA-fructose-cyclopentanone [induced]).
cflA mutant transformants display defects at the colonial level.
Dominant-negative cflAD120A and dominant-activated cflAG14V mutant alleles were generated to examine the role of CflA during growth and development. The cflAD120A mutation was located in the position equivalent to that of the CDC42D118A allele of S. cerevisiae, which leads to a temperature-sensitive dominant-negative phenotype. Likewise, the cflAG14V mutation was located in the position equivalent to that of the CDC42G12V allele of S. cerevisiae, which results in a dominant-activated phenotype (14). SPM4 was cotransformed with the mutated cflA constructs, cflAD120A and cflAG14V (pKB4479 and pKB4478), and the pAB4342 (pyrG) selectable plasmid. Cotransformants were isolated, and the presence of the nonselected plasmids was confirmed and the copy number was determined by Southern blot analysis of genomic DNA using a cflA probe (data not shown). Four representative cotransformants of each mutant cflA construct, ranging in copy number from four to eight, were analyzed further. Comparison of the growth rates of transformants with the determined copy numbers showed that high-copy-number transformants had a severely decreased growth rate. Colonial growth at 25°C for the dominant-negative cflAD120A and dominant-activated cflAG14V transformants was slower than that of the wild-type SPM4 control strain (Fig. 3A). cflAD120A transformants exhibited a more compact colony morphology than the wild type, and cflAG14V transformants displayed fewer aerial hyphae. Both kinds of transformants conidiated at the same stage as the control strain. At 37°C, both transformants formed yeast-like colonies which were identical to that of the wild type (Fig. 3A).
CflA affects the rate of conidial germination.
One hypothesis for the reduced growth rate observed in the cflA mutant transformants is a decrease in the rate of conidial germination. To investigate this further, the cflA mutant transformants were analyzed at different time points during germination for polarity and growth defects. Conidia were germinated on slides, stained for nuclei (DAPI) and cell walls (Calcofluor), and observed microscopically. The rates of germination were measured by counting the numbers of nonpolarized (spherical spores 10 ± 1 μm in diameter), polarized (ellipsoidal cells 10 ± 1 μm in diameter), and germinated conidia (polarized conidia with a visible germ tube) in a population of 100 at a number of different time points.
Wild-type P. marneffei asexual conidia were activated after 6 h of incubation at 25°C and began isotropic cell growth. After 8 h the conidia became polarized, and a germ tube emerged after approximately 12 h. The first septum was laid down after 15 h of growth, and the second germ tube emerged after 18 h of growth. After 12 h in the SPM4 control strain 14% of spores showed isotropic growth, 36% were polarized, and 50% had extended the primary germ tube. The cflAD120A transformants exhibited a severe decrease in germination rates, and the mutant conidia required a longer time to become polarized and consequently extend a germ tube. After 12 h, 52% of the conidia were still growing isotropically, only 28% had become polarized, and only 20% had extended a primary germ tube. Despite the germination delay, the spore volume failed to increase, suggesting that the delay in germination was caused by a growth rather than morphogenetic defect. In contrast, the cflAG14V transformants germinated at a faster rate than wild-type conidia. Only 6% of conidia showed isotropic growth, 24% were polarized, and 70% had extended a primary germ tube after 12 h. The morphology and nuclear distribution of both of the cflAD120A and cflAG14V transformant germlings were normal compared to those of the SPM4 control strain.
To investigate whether the cflA mutant transformants possessed polarity defects during germination, the polarity of germ tube emergence was measured. The angles of secondary germ tube emergence (relative to the primary germ tube) were measured by classifying whether the secondary germ tube had emerged proximal (≤90°) or distal (>90°) to the primary germ tube in a population of 100 germinating conidia grown on slides for 18 h. In the wild type, the second germ tubes exhibited random emergence, with germ tubes extended either proximally or distally at roughly equal proportions. The polarity of germ tube emergence in the cflAD120A and cflAG14V transformants did not significantly differ from that in the SPM4 control strain (data not shown).
cflA is required for correct cellular polarization during vegetative growth.
Early during postgermination stages (1 to 2 days), the SPM4 control strain grows as polarized hyphae which branch and are septate at regular intervals. Hyphal cells are approximately 40 ± 5 μm in length and contain a single nucleus. Tip cells contain multiple nuclei and can range from 50 to 100 μm in length. To further investigate the role CflA plays during vegetative growth, the cflA mutant transformants were grown on a solid surface at 25°C and stained for nuclei (DAPI) and cell walls (Calcofluor), viewed microscopically, and observed for nuclear distribution, growth, and septation defects.
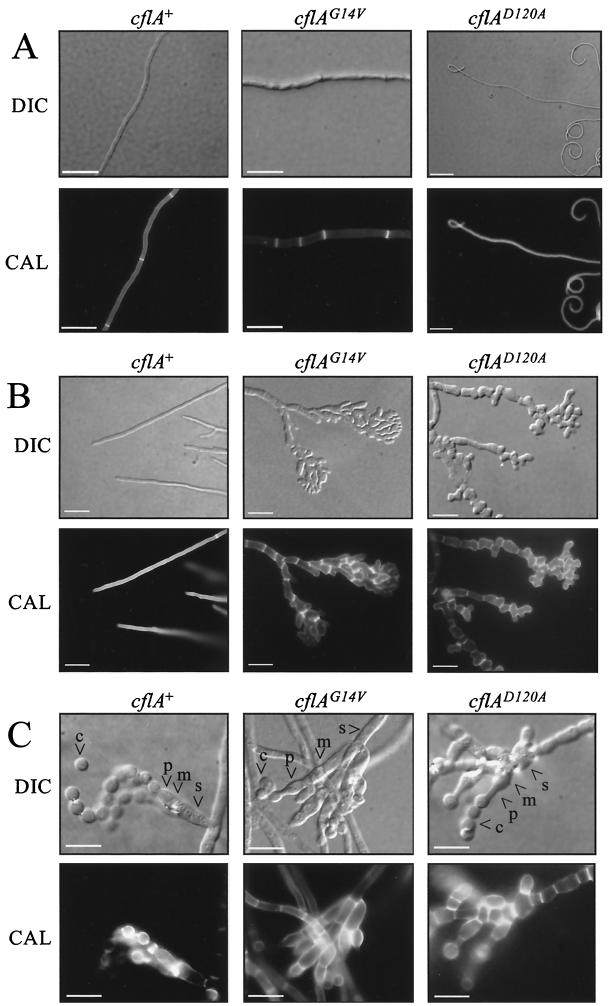
During early hyphal growth, the cflAD120A transformants possessed hyphae displaying misdirectional growth. The hyphae were curled in both anticlockwise and clockwise directions (Fig. 4A). The nuclear distribution, septation, and cell length did not differ from those for the SPM4 control strain. Unlike the cflAD120A transformants, the cflAG14V transformants did not show misdirectional growth. Although the nucleus-to-cell ratio did not differ from that for the SPM4 control strain (data not shown), the cell size was smaller, suggesting an increase in septation. The lengths of cells varied along a hypha, with some cells reduced in length to 20 μm (Fig. 4A). This is in contrast to the wild type, in which septa are regularly spaced along a hypha.
FIG. 4.
cflA mutant transformants at 25°C show numerous morphological defects. (A) Early vegetative hyphae. Strains were grown for 2 days at 25°C on SD solid medium. The cflAG14V transformants showed increased septation compared to the wild-type strain. The cflAD120A transformants possessed curled hyphal tip cells. Scale bars, 50 μm. (B) Vegetative hyphae at a later stage. Strains were grown for 5 days at 25°C on SD solid medium. The cflAG14V and cflAD120A transformants displayed a variety of morphological abnormalities including branched and swollen hyphae, which also exhibited increased septation. Apical cells show significant loss of polarization leading to fused, lysed, and aberrant cells. Scale bars, 50 μm. (C) Conidiation. Strains were grown at 25°C for 5 days on ANM-GABA solid medium. Conidiation proceeds normally in the cflA mutant transformants. Scale bars, 20 μm. Arrowheads, conidiophore structures (s, stalk; m, metulae; p, phialides; c, conidia). Images were captured using differential interference contrast (DIC) or under epifluorescence to observe Calcofluor-stained cell walls (CAL).
Later during hyphal growth (5 days) the basal mycelium in the wild type shows increased septation and hyphal cells are reduced in length to approximately 20 ± 2 μm. Both mutant transformants displayed aberrant hyphal morphology when analyzed at a later stage of growth at 25°C (Fig. 4B). The hyphal cells of the cflAD120A transformants became swollen and consequently appeared greatly reduced in length (10 ± 5 μm). Cell wall staining showed that septa appeared to be more numerous than in the wild type. Also observed were multiple subapical and apical hyphal cells, which were clumped together to produce a mass of deformed hyphae, which were occasionally fused. Random patches of depolarization were observed within cells along a hypha. Similar to what was found for the wild type, the older hyphal cells remained uninucleate and branched and newer cells were multinucleate (data not shown).
The cflAG14V transformants also had morphological defects after 5 days at 25°C (Fig. 4B). The hyphal cells were also swollen but to a greater degree than the cflAD120A transformants and appeared greatly reduced in length (7 ± 5 μm). In addition, unlike what was found for the cflAD120A transformants, uniform depolarization of entire cells was observed. The hyphal cells were also frequently lysed, releasing cellular contents, and consequently appeared anucleate. Multiple subapical and apical cells were clumped together and occasionally fused. The older hyphal cells remained uninucleate and branched, and newer cells were multinucleate (data not shown).
Asexual development in P. marneffei does not require correct cflA function.
The cflAD120A and cflAG14V transformants displayed wild-type conidiation patterns at the colonial level (Fig. 3A). In order to examine whether the conidiophores were also normal in morphology, the cflAD120A and cflAG14V transformants were grown on a solid surface for 5 days and the nuclei (DAPI) and cell walls (Calcofluor) were stained. When observed microscopically, all conidiophore structures, including the metulae, phialides, and conidia, were present in the mutant strains and appeared wild type in morphology and arrangement compared with those for the SPM4 control strain (Fig. 4C). Furthermore, the onset of conidiation was not affected by mutations in cflA.
To investigate whether the apparent lack of function of CflA during asexual development was conserved in other fungi, the mutant cflA alleles were placed behind the alcohol-inducible alcA promoter and transformed into both A. nidulans and P. marneffei and transformants were isolated by direct selection for pyrG+ (see Materials and Methods). At 25°C, the P. marneffei transformants containing the alcA(p)::cflAD120A and alcA(p)::cflAG14V alleles displayed no change in phenotype on both noninducing and inducing medium (Fig. 3B). Both the alcA(p)::cflAD120A and alcA(p)::cflAG14V P. marneffei transformants were capable of undergoing asexual conidiation. The A. nidulans transformants, containing alcA(p)::cflAD120A and alcA(p)::cflAG14V alleles, displayed no change in phenotype on noninducing medium. Interestingly, A. nidulans alcA(p)::cflA transformants exhibited a decrease in conidiation under inducing conditions and alcA(p)::cflAD120A and alcA(p)::cflAG14V transformants displayed a complete lack of conidiation under inducing conditions (Fig. 3B). To investigate this further, the alcA(p)::cflAD120A and alcA(p)::cflAG14V P. marneffei and A. nidulans strains were grown at 25°C on agar-coated slides for 2 days and then induced for an additional 2 days (see Materials and Methods). Microscopic examination of the P. marneffei strains showed that, upon induction, the vegetative morphological defects were the same as previously described for the cflAD120A and cflAD120A pyrG+ cotransformants. Conidiophore structures (metulae, phialides, and conidia) were observed in the P. marneffei transformants (data not shown). Microscopic examination of both the alcA(p)::cflAD120A and alcA(p)::cflAG14V A. nidulans transformants revealed hyphae with morphological abnormalities under inducing conditions, including uniformly depolarized hyphal cells. However, in contrast to those of the P. marneffei transformants, the tip cells of the A. nidulans transformants appeared normal in morphology and a complete lack of conidiation was observed (data not shown).
cflA mutants produce yeast cells with aberrant morphology.
To examine the role of CflA during yeast growth, the cflAD120A and cflAG14V transformants were grown on a solid surface for 4 days at 37°C, stained, and viewed microscopically (see Materials and Methods). In wild-type P. marneffei, yeast cells are produced at 37°C from an arthroconidiating mycelium after 4 days and are uninucleate, are approximately 20 ± 3 μm in length, and divide by fission. In a 37°C culture, two kinds of yeast cells were observed: short chains of cells (two to five cells) which break away from the arthroconidiating mycelium and true uninucleate yeast cells dividing by fission. After 6 days, true yeast cells are predominantly observed.
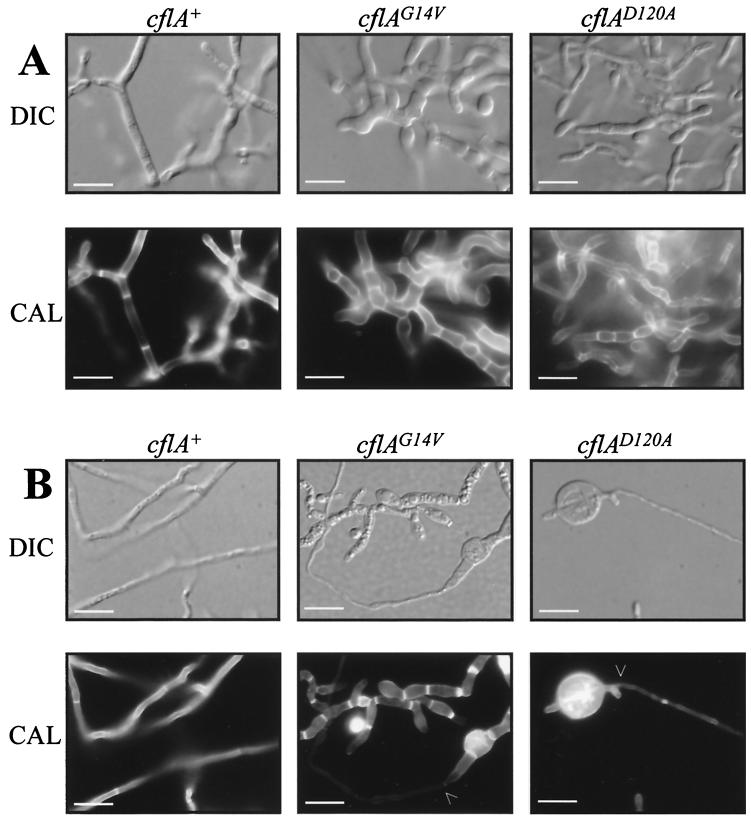
The cflAD120A transformants were capable of producing yeast cells at 37°C (Fig. 5). Yeast cells detached from the arthroconidiating mycelium but possessed a variety of morphological defects and failed to separate. True yeast cells produced by fission could not be identified. Yeast cells produced by the cflAD120A transformants had the overall appearance of yeast-like cells which had broken away from the arthroconidiating mycelium. The cells could be branched, unseparated, and irregularly shaped or swollen and ranged in size from 40 to 100 μm. The nucleus-to-septum ratio (one nucleus per cell) remained identical to that for vegetative mycelia (data not shown).
FIG. 5.
cflAG14V and cflAD120A transformants produce yeast cells with aberrant morphology. Cultures were incubated at 37°C for 4 days in BHI medium. The yeast cells produced by the cflAG14V transformants were swollen and septate. Some septa appeared thicker than those of dividing SPM4 control yeast cells (arrowhead), and some cells did not contain a nucleus. The cflAD120A transformants produced yeast cells which were branched, swollen, and septate. Scale bars, 20 μm. Images were captured using differential interference contrast (DIC) or under epifluorescence to observe Calcofluor cell wall stain (CAL).
Yeast cells produced by the cflAG14V transformants were also aberrant in morphology (Fig. 5). The cells detached from the arthroconidiating mycelium, but unlike the cflAD120A transformants true yeast cells were observed. The yeast cells were approximately 30 ± 10 μm in size. The yeast cells were swollen but to a greater extent than the cflAD120A transformants, and random patches of depolarization were observed within cells. The yeast cells appeared bumpy, misshapen, and swollen but were not branched like the cflAD120A transformants. The cflAG14V cells were also septate (unseparated), and some septa appeared thicker than those for the SPM4 control strain (Fig. 5). Anuclear cells were also observed; however, the majority (94%) of cells contained a single nucleus (data not shown).
Correct cflA function is not required for dimorphic switching.
The role of cflA during the dimorphic transition was investigated by examining the cflAD120A and cflAG14V transformants when switched from 37 to 25°C and vice versa.
In the wild-type SPM4 control, a liquid culture at 25°C consisted of septate hyphal filaments which were less branched than when grown on solid medium. Hyphal cells were approximately 40 ± 2 μm in length and uninucleate. After being transferred to 37°C for 1 day, the hyphae became highly septate and cells decreased to approximately 20 ± 5 μm in length (data not shown). After 3 days, chains of yeast cells broke away from the arthroconidiating mycelium. The culture produced consisted of cells which clump together and which were composed of chains of two or three uninucleate cells (10 ± 2 μm in length) (Fig. 6A).
FIG. 6.
cflAG14V and cflAD120A dominant-negative and dominant-activated transformants are able to undergo the dimorphic temperature-dependent switch, but resulting cell types display aberrant morphology. (A) cflA mutant transformants grown at 25°C for 2 days and transferred to 37°C are able to produce chains of yeast-like cells after 3 days; however, these cells are swollen, highly septate, and unseparated. Scale bars, 20 μm. (B) cflAG14V and cflAD120A transformants extend narrow hyphae from swollen yeast cells when grown at 37°C for 10 days and transferred to 25°C for 3 days (arrowheads). Scale bars, 20 μm. Images were captured using differential interference contrast (DIC) or under epifluorescence to observe Calcofluor cell wall stain (CAL).
When transferred from 25 to 37°C, liquid cultures of both cflAD120A and cflAG14V transformants showed dimorphic switching. Like those of the wild type, the hyphae became more septate within 1 day. After 3 days both cultures were composed of chains of uninucleate cells; however, these cells exhibited uniform depolarization and could be swollen, fat, or misshapen and appeared shorter in length (10 ± 5 μm) than those of the control strain (Fig. 6A). The cells did not separate to produce the two- or three-cell hyphal fragments observed in the wild type, even at a later time point.
In the reciprocal experiment, wild-type yeast cells transferred from 37 to 25°C for 1 day elongated and became multinucleate to produce hyphae. The new hyphae did not become septate at regular intervals until day 3, after which cells became uninucleate (40 ± 2 μm in length) and branching occurred (Fig. 6B).
After 1 day, the yeast cells of the cflAD120A and cflAG14V transformants had begun to grow hyphally. Unlike that of the wild type, where new hyphal growth is identical in cell width to the yeast cells from which it has extended, the hyphal growth of the cflAD120A and cflAG14V transformants was substantially thinner. After 3 days, the extension of new hyphal growth was more apparent (Fig. 6B). The cultures were a mixture of morphologically normal hyphae and a small number of swollen yeast cells. After 5 days the cultures were predominantly hyphal.
DISCUSSION
The P. marneffei cflA gene encodes a highly conserved Cdc42p-like protein containing all of the predicted domains previously shown to be necessary for GTPase function. cflA is highly conserved not only with CDC42 homologs in other closely related fungal species but also with genes in more distantly related organisms such as mammals. The high level of structural conservation indicates that the general function of cflA is also likely to be conserved. For example, one of the human Cdc42 isoforms has been shown to complement the S. cerevisiae dominant-negative CDC42 mutation (CDC42D118A ) (34). Cdc42p homologs from other organisms play diverse roles during development, but all characteristically regulate changes in morphology by initiating signal transduction pathways and by mediating changes in the organization of the actin cytoskeleton (reviewed in references 18 and 25).
The cflA gene produces two transcripts, differing in polyadenylation sites, which are expressed at all stages of growth. It is possible that the two transcripts differ in stability or result in different cellular localizations, but their role remains to be determined. A transitory increase in the level of the cflA transcripts was observed when cells were switched from 37 to 25°C, indicating a possible requirement for higher levels of CflA during the onset of vegetative hyphal growth.
The dominant-negative cflAD120A and dominant-activated cflAG14V transformants resulted in morphologically defective phenotypes. This indicates that CflA cycles from the inactive GDP-bound state to the active GTP-bound state. In addition, overexpression of wild-type cflA has no morphological consequences. This result is in concordance with the proposed mechanism in which CDC42-like proteins act as molecular switches (8). Similar results were generated by overexpression of the wild-type homologs in S. cerevisiae and S. pombe (27).
Attempts to obtain a cflA deletion strain were not successful, possibly because deletion of cflA confers a lethal phenotype. A CDC42 deletion results in a lethal phenotype in both S. cerevisiae and S. pombe (1, 27). The null mutation in S. pombe gives rise to small cells in which the cell cycle has been blocked (27). A CDC42 deletion in S. cerevisiae results in large, multinucleate cells which are unable to bud (1). However, unlike CDC42 in S. cerevisiae and S. pombe, P. marneffei cflA does not appear to be associated with the cell cycle, as nuclear division proceeds in the dominant-negative cflA transformants. It is likely that P. marneffei cflA deletion transformants cannot be recovered due to a loss of vegetative polarized growth, which was seen in the dominant-negative transformants but which would be expected to be more severe in a deletion strain.
The generation of the dominant-negative (cflAD120A) and dominant-activated (cflAG14V) cflA mutant transformants allowed us to examine the function of CflA in P. marneffei. These mutant transformants showed pleiotropic effects in several processes involving polarized growth. The cflAD120A and cflAG14V transformants showed a decrease and increase in the rate of germination, respectively. Germination requires a switch from isotropic spore growth to polarized growth and the emergence of the primary germ tube. In A. nidulans rasA functions to switch isotropic growth to polarized growth in a nutrient-dependent manner (33, 38). It is as yet unknown how the A. nidulans CDC42 homolog, modA, functions during germination. However, it may be activated by rasA after the switch from isotropic growth to polarized growth in order to extend the primary germ tube. The RasA homolog in S. cerevisiae, Ras2p, has been shown to activate Cdc42p during pseudohyphal growth, a process which requires the elongation of cells, similar to hyphal extension. The dominant-negative CDC42D118A mutation blocks Ras2G19Vp activation, placing Cdc42p downstream of Ras2p (31). This model is supported by the proposed function of P. marneffei cflA, which is involved in the extension of the germ tube, possibly after activation by a RAS2 homolog in P. marneffei.
Hyphal extension requires the transport of secretory vesicles along the polarized cytoskeleton to the hyphal tip and the deposition of new cell wall material in a narrow zone at the hyphal apex. Cylindrical yeast cells also require polarized growth, directing cell wall material toward one or both ends of the cell, in addition to the cell center, during division. In other organisms Cdc42 functions during polarized growth to localize actin and other proteins required for polarized growth to the correct location (3); therefore it was proposed that, in addition to germination, CflA would be involved in other processes in P. marneffei requiring changes in polarization, such as vegetative hyphal growth at 25°C and yeast-like growth at 37°C. The P. marneffei cflAD120A and cflAG14V transformants exhibit cell growth which extends in all directions, not just at the hyphal tip, and this results in swollen and misshapen hyphal cells which appear reduced in length. The P. marneffei cflAD120A and cflAG14V transformants also exhibit abnormal hyphal tips, consistent with a loss of polarized growth. The role of CflA during polarized hyphal growth is also supported by preliminary localization experiments which have shown that a CflA::GFP fusion protein was localized to hyphal tips (data not shown). The nucleus-to-septum ratio remains the same, indicating that, in P. marneffei, septation may be controlled by a mechanism similar to that of A. nidulans, by nuclear division and positioning, in addition to cell volume (30, 39). This also supports the proposal that the cells appear reduced in size due to isotropic growth rather than a septation defect. The cell remains the same size but has an altered shape.
The cflA gene is most closely related to the homologous gene from E. dermatitidis. The equivalent CDC42G14V mutation in E. dermatitidis results in a phenotype similar to that of P. marneffei, resulting in nonpolarized growth that leads to cell enlargement, multiple nucleation, and repressed hyphal growth. However, unlike what was found for P. marneffei, the dominant-negative mutation (cdc42D120A) was not found to affect fungal cell polarization (40). The phenotypes of the P. marneffei dominant-activated and -negative mutations are also similar to those produced by the equivalent mutations in S. pombe, which both give rise to enlarged, round, or misshapen cells which, in addition, can be abnormally elongated (27). The equivalent CDC42G12V and CDC42D118A mutations in S. cerevisiae CDC42 also lead to a loss of polarized growth and to growth defects during the pheromone response and mitotic budding. The lethal S. cerevisiae CDC42G12V mutation generates large, multibudded cells, and the temperature-sensitive CDC42D118A mutation generates small, unbudded cells at 23°C but not at 30°C (14). The equivalent mutations in higher eukaryotes are also associated with a loss of actin localization and polarized growth (18, 25).
In S. cerevisiae and S. pombe, Cdc42p is linked to the cell cycle. In S. cerevisiae, Cdc42p activates Clb2p-Cdc28p and Cla4p, two cell cycle proteins. However, in S. cerevisiae loss of correct Cdc42p function does not lead to a block in the cell cycle unlike in S. pombe (27). P. marneffei cflA does not appear to be linked to the cell cycle since, in the cflAD120A mutant, nuclear division can proceed. Both cflAD120A and cflAG14V mutations result in similar phenotypes, suggesting that correct hyphal growth requires the continual cycling between the inactive GDP-bound and active GTP-bound forms of CflA. This may not be the case in other fungi. The dominant-negative and -activated mutations in S. cerevisiae and E. dermatitidis result in different phenotypes, whereas in S. pombe the mutants display identical phenotypes (14, 27, 40). Our results suggest that P. marneffei CflA plays a role during the initiation of polarized growth, the extension of germ tubes, and the maintenance of this polarized growth in the vegetative hyphae at 25°C.
CflA is also involved in yeast-like growth at 37°C and seems to function to control cell shape and morphology. The cflAD120A transformants produce branched yeast cells which are septate, and true yeast cells dividing by fission could not be identified, indicating that active GTP-bound CflA is required for correct separation of yeast cells. P. marneffei cflA may play a role during the separation of cells, as it has been previously shown to be necessary for cytokinesis in S. cerevisiae (35). It is expected that the polarization of P. marneffei yeast cells would be similar to that of S. pombe, where growth is specifically directed to both ends of the cylindrical yeast cell (27). Both the cflAG14V and cflAD120A transformants produce yeast cells which are swollen and misshapen. This suggests that CflA acts to polarize the yeast cell and hence determines correct cell shape. Both mutants have similar phenotypes, and therefore, yeast-like growth may be similar to hyphal growth in requiring continual cycling of GTP- and GDP-bound CflA.
Asexual development in fungi is a conserved developmental program which requires a change in polarized growth and morphology. Conidiophores are produced from the differentiation of hyphal cells. A stalk extends from a hyphal foot cell, and from this structure uninucleate cells are produced in a mechanism similar to S. cerevisiae budding. The stalk produces metulae, which subsequently produce phialides, which sequentially bud off multiple conidia. If CflA plays a global role in polarization, then it could be expected that CflA would be involved during asexual development. In addition, S. cerevisiae Cdc42p is essential for budding (14). Surprisingly, the cflA mutations did not affect either the timing of conidiation or the structure of conidiophores during asexual development, despite CflA playing a role during polarized growth in the basal mycelium. It is likely therefore that the process of polarized growth in asexual development may be controlled by another related GTPase.
However, when the P. marneffei cflA mutant constructs were introduced into A. nidulans, conidiation was completely blocked. This suggests that, unlike what was found for P. marneffei, correct Cdc42 function is required for conidiation in A. nidulans, indicating that the mechanisms and GTPases controlling development may differ among fungal species. Other members of the Rho GTPase family, such as Rac and Rho, also play roles in organizing the actin cytoskeleton and consequently in polarized growth and cell morphology. For example, in mammalian fibroblast cells, Cdc42 induces actin-rich surface protrusions called filopodia, whereas Rac induces formation of actin-rich lamellipodia and Rho activation leads to the assembly of actin stress fibers (reviewed in reference 18). Likewise, in Drosophila melanogaster, the five Rho GTPases, Rho1, Rac1, Rac2, Cdc42, and RhoL, all play unique and specific roles during polarized growth in oogenesis, gastrulation, dorsal closure, and muscle, eye, and neuronal development (reviewed in reference 25). Therefore differences in specialized functions of Rho type GTPases among fungal species are suggested by our results.
In addition to a lack of effect of the cflAG14V and cflAD120A mutations during conidiation, CflA does not appear to be involved in determining dimorphic switching in response to temperature shift. The role of CflA is to regulate the changes in morphology which occur throughout the switch. Both kinds of mutant transformants were able to undergo the growth transition from hyphal to yeast cells and vice versa, with the resultant cell types displaying abnormal morphology.
It will be of considerable interest to isolate other Rho type GTPases and determine their role in specific developmental processes in P. marneffei.
ACKNOWLEDGMENTS
We thank Steven Harris for providing the A. nidulans CDC42 homolog modA.
This work was supported by a grant from the Australian Research Council. K.J.B was supported by an Australian Postgraduate Award (APA) scholarship.
REFERENCES
- 1.Adams A E, Johnson D I, Longnecker R M, Sloat B F, Pringle J R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benton B K, Tinkelenberg A, Gonzalez I, Cross F R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman A R, Hynes M J, Andrianopoulos A. The abaA homologue of Penicillium marneffei participates in two developmental programs: conidiation and dimorphic growth. Mol Microbiol. 2000;38:1034–1047. doi: 10.1046/j.1365-2958.2000.02202.x. [DOI] [PubMed] [Google Scholar]
- 7.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. A STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 8.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 9.Chang F, Woollard A, Nurse P. Isolation and characterisation of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- 10.Chant J, Stowers L. GTPase cascades choreographing cellular behaviour: movement, morphogenesis, and more. Cell. 1995;81:1–4. doi: 10.1016/0092-8674(95)90363-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Lim H H, Lim L. The CDC42 homologue from Caenorhabditis elegans. J Biol Chem. 1993;18:13280–13285. [PubMed] [Google Scholar]
- 12.Cooper C R J. From bamboo rats to humans: the odyssey of Penicillium marneffei. ASM News. 1998;64:390–397. [Google Scholar]
- 13.Cove D. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 14.Davis C R, Richman T J, Deliduka S B, Blaisdell J O, Collins C C, Johnson D I. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J Biol Chem. 1998;273:849–858. doi: 10.1074/jbc.273.2.849. [DOI] [PubMed] [Google Scholar]
- 15.d'Enfert C. Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet Biol. 1997;21:163–172. [Google Scholar]
- 16.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarised morphogenesis. Science. 1997;276:118–121. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 17.Gow N A R. Growth and guidance of the fungal hypha. Microbiology. 1994;140:3193–3205. doi: 10.1099/13500872-140-12-3193. [DOI] [PubMed] [Google Scholar]
- 18.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 19.Harris S D, Morrell J L, Hamer J E. Identification and characterisation of Aspergillus nidulans mutants defective in cytokinesis. Genetics. 1994;136:517–532. doi: 10.1093/genetics/136.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris S D, Hamer J E. sepB: an Aspergillus nidulans gene involved in chromosome segregation and the initiation of cytokinesis. EMBO J. 1995;14:5244–5257. doi: 10.1002/j.1460-2075.1995.tb00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris S D, Hamer L, Sharpless K E, Hamer J E. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 1997;16:3474–3483. doi: 10.1093/emboj/16.12.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris S D, Hofman A F, Tedford H W, Lee M P. Identification and characterisation of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics. 1999;151:1015–1035. doi: 10.1093/genetics/151.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imwidthaya P. Update of penicillosis in Thailand. Mycopathologia. 1994;127:135–137. doi: 10.1007/BF01102912. [DOI] [PubMed] [Google Scholar]
- 24.Kozminski K G, Chen A J, Rodal A A, Drubin D G. Functions and functional domains of the GTPase Cdc42p. Mol Biol Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Settleman J. The role of rho family GTPases in development: lessons from Drosophila melanogaster. Mol Cell Biol Res Commun. 1999;1:87–94. doi: 10.1006/mcbr.1999.0119. [DOI] [PubMed] [Google Scholar]
- 26.May G S. Cloning, mapping and molecular analysis of the pyrG (orotidine-5′-phosphate decarboxylase) gene of Aspergillus nidulans. Gene. 1987;61:385–399. doi: 10.1016/0378-1119(87)90201-0. [DOI] [PubMed] [Google Scholar]
- 27.Miller P J, Johnson D I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirabito P M, Adams T H, Timberlake W E. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell. 1989;57:859–868. doi: 10.1016/0092-8674(89)90800-3. [DOI] [PubMed] [Google Scholar]
- 29.Momany M, Westfall P J, Abramowsky G. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics. 1999;151:557–567. doi: 10.1093/genetics/151.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris R N. Mitotic mutants of Aspergillus nidulans. Genet Res. 1976;26:237–254. doi: 10.1017/s0016672300016049. [DOI] [PubMed] [Google Scholar]
- 31.Mosch H U, Roberts R L, Fink G R. Ras2 signals via the cdc42/ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohya Y, Qadota H, Anraku Y, Pringle J R, Bolstein D. Suppression of yeast geranylgeranyl transferase I defect by alternative prenylation of two target GTPases, Rho1p and Cdc42p. Mol Biol Cell. 1993;4:1017–1025. doi: 10.1091/mbc.4.10.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signalling and protein synthesis. Genetics. 2000;155:647–656. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peter M, Neiman A M, Park H, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 35.Richman T J, Sawyer M M, Johnson D I. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J Biol Chem. 1999;274:16861–16870. doi: 10.1074/jbc.274.24.16861. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning, a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schmidt A, Hall M N. Signalling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 38.Som T, Kolaparthi V S R. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol Cell Biol. 1994;14:5333–5348. doi: 10.1128/mcb.14.8.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolkow T D, Harris S D, Hamer J E. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci. 1996;109:2179–2188. doi: 10.1242/jcs.109.8.2179. [DOI] [PubMed] [Google Scholar]
- 40.Ye X, Szaniszlo P J. Expression of a constitutively active Cdc42 homologue promotes development of sclerotic bodies but represses hyphal growth in the zoopathogenic fungus Wangiella (Exophiala) dermatitidis. J Bacteriol. 2000;182:4941–4950. doi: 10.1128/jb.182.17.4941-4950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]