Abstract
This review reports recent findings on the specification and patterning of neurons that establish the larval nervous system of the sea urchin embryo. Neurons originate in three regions of the embryo. Perturbation analyses enabled construction of gene regulatory networks controlling the several neural cell types. Many of the mechanisms described reflect shared features of all metazoans and others are conserved among deuterostomes. This nervous system with a very small number of neurons supports the feeding and swimming behaviors of the larva until metamorphosis when an adult nervous system replaces that system.
Keywords: sea urchin, neural, larval nervous system, neural specification, neural patterning
Two nervous systems are produced during the life of a sea urchin, one for the larval period until metamorphosis, and a completely new nervous system that serves adult life. The adult nervous system is little explored and not used as a model system for neural function. By contrast, the larva is bilaterally symmetric and, as an early branching deuterostome, has a nervous system that has attracted the curiosity of several laboratories interested in the development and evolution of this system relative to vertebrates and other deuterostomes (Angerer et al., 2011; Bisgrove and Burke, 1986; Bisgrove and Burke, 1987; Burke et al., 2006a; Burke et al., 2006b; Cheatle Jarvela et al., 2016; Hinman and Burke, 2018; Yaguchi and Katow, 2003; Yankura et al., 2013). Embryonic development of the sea urchin has long been used as a model system for learning how specification and patterning contribute to basic cell and molecular mechanisms of development. Consequently, most recent work on neural development has been aimed toward identification of patterning inputs and molecular information on specification of neurons. This review focuses on those aspects of the embryonic period during which the larval nervous system is assembled. After a brief introduction to orient the reader to the sea urchin larva and to the anatomy of the nervous system, the three parts of the nervous system are reviewed first to provide known patterning information that establishes a tissue niche for neural specification. Then, the focus switches to what is known about the actual specification of the neurons in that territory. Included for each cell type will be a gene regulatory model that reflects pertinent known molecular information both for the patterning and for the neural specification. A final section briefly reviews recent data on the function of the larval nervous system. A remarkable feature of this system is that it begins functioning in many species within two days of fertilization and at that time the entire nervous system consists of only about 50 neurons. 35–40 of those neurons are in the ciliary band or in the postoral territory and 5–10 serotonergic neurons are in a small anterior domain. Additional neurons are added as larval development proceeds, but even with only 50 neurons the larva can control swimming, feeding, swallowing, peristaltic movement of food, a response to light, and probably additional behaviors yet to be identified.
Several species of sea urchin have been used for studies on neural development. Based on the anatomy, and on the molecular experiments conducted with multiple species, there appears to be a high degree of conservation in the patterning and specification of neurons. For that reason, this review will consider all studies simply to be on the sea urchin. This may be a flawed assumption since some species differences may be discovered when examinations are made in greater detail. However, those differences, if they are uncovered, are likely to be minor. As a convention we will use embryonic stages to describe temporal events. This is because embryos of different species develop at different rates depending upon temperature, yet seminal events of neural development occur at similar developmental stages when different species are compared.
Development and anatomy of the sea urchin larval nervous system
The sea urchin nervous system is specified beginning at the mesenchyme blastula stage and is established in three regions of the embryo. The animal pole is considered anterior because it is proximal to the eventual location of the mouth, and the vegetal pole of the embryo where invagination of the archenteron occurs, is designated as posterior because the anus develops there. Zygotic specification begins quite early to establish the endomesoderm in the posterior half of the embryo and ectoderm in the anterior half. The three locations of neural patterning and specification are the anterior neurectoderm (ANE) located at the anterior end of the embryo, the ciliary band that separates the dorsal and ventral ectoderm, and in the endoderm where an enteric nervous system will control movement of food through the larva (Range et al., 2013; Wei et al., 2016; Wei et al., 2011; Yaguchi et al., 2010; McClay et al., 2018). Several Serotonergic neurons are specified in the ANE as seen using either antibodies to serotonin (Bisgrove and Burke, 1986), or tryptophan 5-hydroxylase (Lv-tph), an enzyme required for serotonin synthesis (Yaguchi and Katow, 2003; Yaguchi and Katow, 2003; Slota and McClay, 2018). Later as the larval stage is reached neurons populate the ciliary band. The serotonergic neurons establish connections to form a network when axons are extended early in the pluteus larval stage. Two kinds of neurons and perhaps more, differentiate in or next to the ciliary band, cholinergic and dopaminergic.
Patterning the Anterior neurectoderm (ANE)
The ANE (Range et al., 2013; Wei et al., 2009) (also called APD based on slightly different criteria (Wei et al., 2009)) is a region at the anterior end of the embryo that is characterized by an apical tuft of long cilia and a group of 4–10 serotonergic neurons (depending on species) in a small domain just dorsal to the ciliary band. The ciliary band separates the ventral from the dorsal ectoderm and develops between late gastrulation and the prism stage. At the anterior end of the embryo the ciliary band bisects the ANE and contains neurons distinct from the serotonergic neurons in neurotransmitter release. The ANE neurons are specified between cleavage and the prism stage. They differentiate and begin functioning by the time the larva begins to feed. The patterning phase of ANE organization involves signaling throughout the entire embryo and it begins with maternal information. That patterning provides the appropriate environment for neurogenesis and it contributes to processes that set aside the correct number of cells that will be specified as neurons. Neurogenesis overlaps in time with the patterning, but to simplify the descriptions these two phases are separated with the patterning presented first, followed by current information on the specification sequence of the serotonergic neurons.
Patterning the ANE as a neurogenic territory is a restrictive process. Canonical Wnt signaling has an enormous impact on establishing the Anterior-Posterior axis. Experimental inhibition of nuclear accumulation of β-catenin results in a massive expansion of neural specification over the entire embryo (Yaguchi et al., 2006). Conversely, ectopic overexpression of β-catenin results in absence of neurons (Yaguchi et al., 2006). Thus, the Wnt pathway is involved in restricting neurogenesis to the ANE as well as in other domains. Several anterior markers were identified and used to further determine how the ANE becomes restricted. The transcription factor Six3 was found to be crucial. Six3 is expressed broadly in the anterior half of the embryo during early cleavage but gradually becomes restricted to, and is necessary for retention of neurogenic potential in the ANE. Posterior Wnt signaling is responsible for this restriction because absence of cWnt signaling results in expansion of six3 expression posteriorly, and over-expression of six3 causes an expansion of the ANE (Wei et al., 2009). Microarray analysis of control embryos vs embryos perturbed by a Six3 knockdown indicate that Six3 is responsible for expression of a number of ANE markers including the Wnt inhibitors sfrp1/5 and dkk3 (Wei et al., 2009). Other Wnt inhibitors include dkk1 which, oddly enough, appears to be expressed downstream of a Wnt-Fzl5/8 expression sequence indicating that DKK1 is a feedback inhibitor of that pathway (Range et al., 2013). Dkk1 expression is important for expression of the dkk3 and sfrp1/5 as revealed by perturbation analysis, indicating that dkk1 could be one of the primary Wnt inhibitors allowing the ANE to specify neural (Range et al., 2013). Later studies indicate that Wnt signaling through Fzl5/8 also activates sfrp1/5 expression suggesting again that neurogenesis at the anterior end of the embryo requires inhibition of Wnt signaling through several pathways (Khadka et al., 2018), a theme that is in common with other deuterostomes. Thus, Wnt signaling starts at the posterior and sequentially is relayed toward the anterior end where it progressively restricts its own signaling (Range et al., 2013). Expression of six3 and Wnt inhibitors at the anterior end of the embryo provides a region that is competent for neurogenesis and establishes the ANE, a disc-like domain that later is subdivided into dorsal and ventral subdomains.
Orthogonal to the Wnt A-P signaling, the dorsal-ventral axis of the embryo is established through Nodal/BMP2/4 signaling (Duboc et al., 2004). The ANE, in response to these signals, establishes a subregion where neurogenesis occurs. After identifying BMP2/4 as a signal in early sea urchin development (Angerer et al., 2000), Nodal was identified, and the function of these TGFβ signals revealed how the dorsal-ventral axis is established (Duboc et al., 2004). Nodal is expressed in the ventral ectoderm, and in addition to patterning the ventral ectoderm, the nodal expressing cells also express bmp2/4. BMP diffuses to the dorsal half of the embryo where it signals to activate dorsal ectoderm specification (Duboc et al., 2004). Nodal signaling is also responsible for establishing left-right asymmetries later in development (Duboc et al., 2005), and for patterning dorsal-ventral mesoderm and endoderm (Duboc et al., 2010). That extensive involvement of Nodal/BMP2/4 in establishing axial identities was soon explored in detail to determine the degree to which these signals impact patterning of neural territories.
Two genes examined during Nodal/BMP2/4 patterning of the ANE provide important insights into ANE patterning. FoxQ2 is a zygotically expressed transcription factor that has been used in a number of studies to delineate the ANE (Yaguchi et al., 2016). At the gastrula stage another transcription factor, homeobrain (hbn), is also expressed in the ANE. The limits of foxq2 expression are established by the above-mentioned Wnt-six3 sequential restriction mechanism. Foxq2 expression establishes the ANE within the ring of six3 expression. As development advances toward prism and the pluteus stage, foxq2 expression is retained in the ventral ANE and its expression gradually disappears from the dorsal ANE, while at the same time hbn expression diminishes from the ventral ANE and is retained in the dorsal half, both responses to the Nodal/BMP2/4 D-V patterning (Fig.2). Serotonergic neurons are specified along the margin of the dorsal patch of hbn expression (Yaguchi et al., 2016). Although expression of hbn does not overlap with cells expressing serotonin, knockdown of hbn inhibits development of serotonergic neurons, indicating that the cells of the ANE that express hbn are necessary to somehow influence serotonergic neuron specification (Yaguchi et al., 2016). Both Nodal and BMP2/4 continue to participate in the dynamic sub-patterning of the ANE. Nodal signaling maintains foxQ2 expression in the ventral ANE. Knockdown of Nodal with a morpholino leads to a reduction in foxQ2 expression while knockdown of Lefty, the antagonist of Nodal, causes an expansion of foxQ2 expression. The opposite occurs with hbn expression. Knockdown of Nodal expands hbn expression throughout much of the ANE and increases the number of serotonergic neurons produced. Knockdown of Lefty greatly reduces hbn expression and eliminates serotonergic neurons (Yaguchi et al., 2016). Expression of bmp2/4 is necessary for dorsal expression of hbn (Yaguchi et al., 2016), and the level of BMP2/4 input into the dorsal ANE is limited by a circuit involving the transcription factor Fez (Yaguchi et al., 2011). Later studies investigating neural specification in the ANE confirmed these necessary patterning inputs from Nodal, BMP2/4, FoxQ2, and Hbn (McClay et al., 2018).
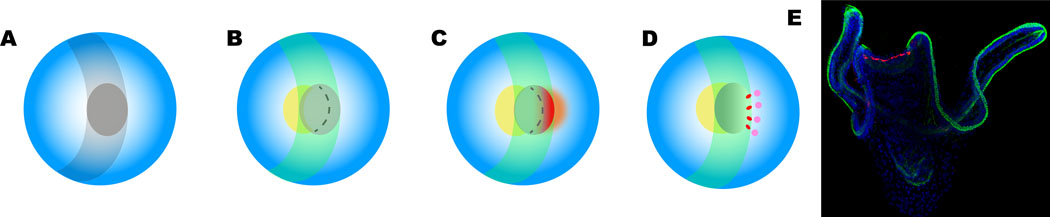
Fig. 2. Development of serotonergic neurons in the anterior neurectoderm (ANE) domain.
The diagrams look down onto the anterior end of the blue embryo with ventral to the left. A. The ANE (dark gray) is formed by repressing Wnt signaling through Wnt inhibitors and expression of Six3 (white). B. When dorsal-ventral signaling is activated, Nodal forces an expansion of the ventral ANE (yellow), followed by C, BMP2/4 establishing a crescent dorsal domain (red). D. A group of cells in the dorsal ANE express hbn (red) and immediately adjacent to these cells serotonergic neurons (pink) are specified. At the gastrula stage the ciliary band appears (green in B-D) and bisects the ANE just ventral to the serotonergic neurons. E. The image of the pluteus larva shows the location of serotonergic neurons (red)and the ciliary band (green). Nuclei (blue).
The dynamics of patterning the ANE through both Wnt and TGFβ-pathways also indicates several other relationships. For example, Wnt signaling reduces foxQ2 expression in the ANE. If Wnt is knocked down and foxq2 expression is expanded, or if foxq2 is over-expressed, nodal expression is reduced or eliminated, indicating that the Wnt-directed restriction of foxq2 expression to the ANE is necessary for the normal expression of nodal (and bmp2/4) (Yaguchi et al., 2008).
Neurogenesis in the ANE
Neurons are specified in two locations in the ANE. Cholinergic neurons are specified throughout the ciliary band, part of which passes through the ANE. Specification of those neurons will be covered in a later section since ciliary neurons, regardless of location, appear to be similarly specified. Here, the accumulated evidence on specification of serotonergic neurons is reviewed since their specification is unique to their origins in the ANE (Bisgrove and Burke, 1986; McClay et al., 2018; Yaguchi and Katow, 2003). While a number of transcription factors have been identified and localized to the ANE, and if knocked down block expression of neural markers, that criterion alone is not sufficient to know whether the transcription factor is expressed directly in the serotonergic neural progenitors. Therefore, the few transcription factors known to be in serotonergic neurons were identified initially in double in situs that included either expression of tph, an enzyme in the serotonergic pathway, or serotonin itself. With those criteria in mind several contributing transcription factors were identified and perturbed to provide a gene regulatory network model (GRN) of the ANE using BioTapestry, a program for building graphic models of GRNs based on results of perturbation analyses (Longabaugh et al., 2009) (Fig. 3).
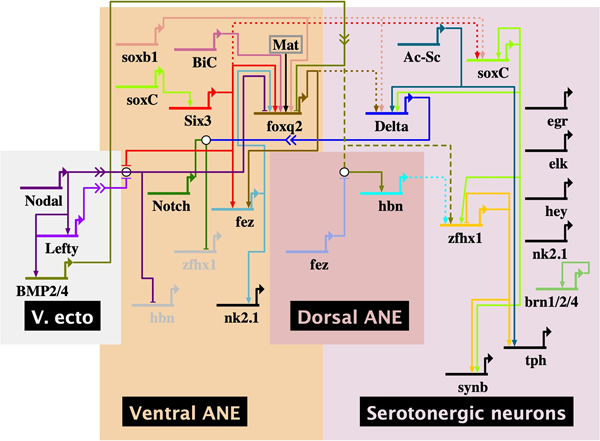
Fig. 3. Serotonergic neuron specification in the animal neurectoderm domain.
Prior to initiation of neurogenesis the ANE is patterned through Wnt signaling (not shown) and Nodal/BMP2/4 signaling from the ventral ectoderm (Nodal) and dorsal ectoderm through BMP2/4. The serotonergic neurons are specified at the boundary between the dorsal and ventral ANE. At that location cells expressing hbn somehow activate the adjacent serotonergic neurons. The BioTapestry model shown is based on perturbation data from a number of laboratories (solid lines). Dotted lines are based on perturbations of a transcription factor in one cell affecting expression of a gene in another cell where the signal between cells is not known. The dashed lines represent proposed signal transductions. Mat = maternal input.
Partial gene regulatory networks directing specification of ANE serotonergic neurons have been published (McClay et al., 2018; Yaguchi et al., 2012). These include perturbation studies on Six3, SoxB1 and FoxQ2 as ANE territorial drivers and Delta, SoxC, Sip1 and Insm as transcription factors involved in specification of the neurons (McClay et al., 2018). FoxQ2, Six3, Delta, Fez, Zfhx1, Tph, SynB, and Serotonin were studied in a perturbation analysis to identify the role of Zfhx1 (Yaguchi et al., 2012). Also, Ac/Sc was shown to be upstream of Tph (Slota et al., 2019). A number of additional transcription factors have been identified to play a role in the serotonergic neuron specification pathway including hbn (Yaguchi et al., 2016), and an aristaless-related homeodomain gene (ars in humans (Lim et al., 2019)). Loss of function experiments with bicaudalC (bicC) indicate that this gene is also involved in neurogenesis since serotonergic neurons are absent in bicC knockdowns, yet expression of foxq2 is unaffected (Yaguchi et al., 2014). Additionally, Brn1/2/4 and z167 have been reported as downstream of six3 (Garner et al., 2016). Rx and several other transcription factors were reported to be in or near the neurons in the ANE as part of the gene survey of the Strongylocentrotus purpuratus genome (Burke et al., 2006a). Recently, egr (Early growth factor response), a zinc finger transcription factor, hey (hairy/enhancer of split related), and elk (ets related transcription factor) were identified and localized to single cells in the ANE (Slota et al., 2019). The BioTapestry model (Fig. 3) provides an update of known molecular relationships. The ANE domain includes connections known to be involved in establishing that domain based on perturbation analyses. The model includes those transcription factors that have been shown to be co-expressed with the serotonergic markers. In each case the perturbation is based on morpholino knockdowns. Connections may be indirect since cis-regulatory relationships have yet to be established for any of the genes included in the model. Relative time runs from top to bottom in the model though the position of a gene may not accurately reflect its timing of first expression relative to nearby genes.
Patterning the ciliary band domain.
Ciliary band neurons are thought to become the peripheral nervous system of the larva (assuming the equivalent of the central nervous system is located in the ANE)(Hinman and Burke, 2018). Ciliary band neurons begin their specification sequence as early as the mesenchyme blastula stage, prior to the appearance of the ciliary band. When the ciliary band appears, coincident with the expression of onecut (Barsi et al., 2015; Otim et al., 2004), ciliary band neurons either are enfolded in place by the ciliary band or actually move a short distance to enter that domain (McClay et al., 2018; Slota, 2019; Slota et al., 2020). In addition, a group of neurons called the postoral neurons are specified below the mouth and later differentiate and reside just outside the ciliary band (Fig. 1). The neurons in the ciliary band, at least those that have been characterized by neurotransmitter release, are cholinergic, while the neurons located in the postoral position are both cholinergic and dopaminergic (Slota and McClay, 2018). Additional neurons in the ciliary band (and in the ANE and endoderm) release at least 38 identified neuropeptides (Wood et al., 2018), and some of those neuropeptides may be released by the neurons that release acetylcholine, dopamine, serotonin, or GABA (Bisgrove and Burke, 1987; Wood et al., 2018).
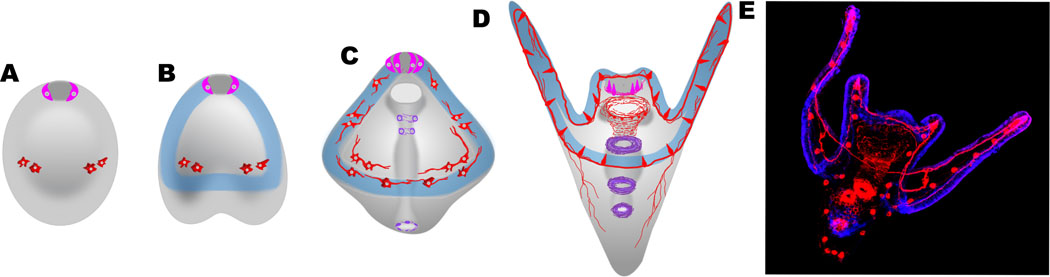
Fig. 1. Neural development during embryogenesis.
A. Proneural cells first appear in two locations, at the animal pole domain (ANE) (pink), and on either side of the ventral ectoderm (red). B. The ciliary band (blue) appears during gastrulation and shortly thereafter additional proneural cells appear. C. The cells in the ANE that become serotonergic neurons localize just dorsal to the ciliary band. The proneural cells in the ventral ectoderm become the postoral neurons and are both cholinergic and dopaminergic (red inside ciliary band), and ciliary band neurons appear in the ventral ectoderm and later differentiate in the ciliary band (red). In the gut, a set of enteric neurons (violet) are specified and later in the larval stage (D), these surround the mouth and the sphincters separating the chambers of the gut. E. A fluorescent image of a pluteus larva is stained with an antibody to L1, a pan-neural marker showing the entire nervous system plus the cell bodies of the skeletogenic cells (red). The nervous system is largely associated with the ciliary band (blue). Diagram after Garner et al., (2016).
The ciliary band is patterned by the Nodal-BMP2/4 signaling system during the patterning of the dorsal-ventral ectoderm territories (Duboc et al., 2004). Briefly, an early asymmetric event that is only partially characterized leads to the expression of Nodal in the ventral ectoderm early in cleavage (Bradham and McClay, 2006; Coffman et al., 2009; Coffman and Davidson, 2001; Coffman and Denegre, 2007; Coffman et al., 2004; Coffman et al., 2014; Duboc et al., 2004; Modell and Bradham, 2011; Molina et al., 2017). Nodal establishes a signaling center that regulates its own expression using a community effect signaling mechanism (Bolouri and Davidson, 2009; Gurdon, 1988). Nodal-producing cells also express the Nodal regulator lefty which restricts the area influenced by Nodal signaling (Duboc et al., 2008). The nodal-expressing cells also express bmp2/4 (Duboc et al., 2004) which will establish the dorsal ectoderm. The movement of BMP2/4 to the dorsal ectoderm is facilitated by Chordin which is also expressed in the ventral ectoderm. Mechanistically, Chordin binds to BMP2/4 inhibiting BMP function in the ventral ectoderm. Chordin is then modeled as a chaperone that diffuses to the dorsal ectoderm while bound to BMP2/4. There Chordin is degraded, and BMP able to signal (Ashe and Levine, 1999; Shimmi et al., 2005; Yu et al., 2000). This allows BMP2/4 to function in specification of dorsal ectoderm (Bradham et al., 2009; van Heijster et al., 2014). A number of additional molecules are used in this system (Haillot et al., 2015; Lapraz, 2009; Lapraz et al., 2015; Molina and Lepage, 2020; Range et al., 2007; Saudemont et al., 2010; Yaguchi et al., 2007; Zito et al., 2003) and these have been summarized recently and the combined data used to model the Nodal/BMP2/4 patterning system (Floc’hlay et al., 2021).
The ciliary band appears at the border between the ventral and the dorsal ectoderm. It is established by the Nodal-BMP signaling system and their antagonists Lefty and Chordin. The ciliary band is delineated by expression of onecut, and is a band of columnar ciliated cells tightly packed, 4–6 cells wide and it also harbors the cell bodies and many neurites of the larval nervous system (Fig. 1) (Bisgrove and Burke, 1987; Burke et al., 2006b; Garner et al., 2016). The boundaries of the ciliary band are reinforced at a molecular level by a series of repressors acting in adjacent tissues to prevent expansion of the ciliary band cell GRN into neighboring cells (Barsi et al., 2015). The Barsi, et al., study found that the ciliary band can be subdivided into four regions based on genes expressed in each region. The repressors prevent ciliary band specification in dorsal and ventral ectoderm tissues adjacent to the ciliary band. Repressors performing this function differ in cells bordering the four regions. Within the ciliary band itself, Barsi et al. identified 23 transcription factors and signals that operate, including four transcription factors that are expressed in all four regions of the CB (onecut, foxg, otxβ1/2, and z166). The cells in the ciliary band appear to be stabilized by a feedback circuit involving expression of onecut. Perturbation experiments established that Soxb1 activates onecut expression (Saudemont et al., 2010), and onecut activates expression of otxβ1/2, which then activates expression of foxg, and FoxG feeds back to activate otxβ1/2 (Barsi et al., 2015). Additional nodes for the ciliary band GRN are presented in Barsi et al., (2015). Thus, as a consequence of Nodal and BMP2/4 signaling, the ciliary band is patterned and provides a niche for neural differentiation.
Neural specification in and near the ciliary band.
Specification of ciliary band and postoral neurons begins before a ciliary band is present, i.e. prior to increased expression of onecut (Barsi and Davidson, 2016), and the cell shape change of the ciliated cells that will become the ciliary band. The earliest transcription factors known to contribute to neural specification are SoxB1 and SoxC (Garner et al., 2016). Fig. 1 shows the time course of appearance of proneural and neural cells in the ciliary band. The first proneural cells to express soxC become the postoral dopaminergic-cholinergic cells that align themselves just outside the ciliary band at the pluteus stage (Slota and McClay, 2018). SoxC is expressed in these cells throughout specification until its expression is lost when the cells differentiate (Garner et al., 2016; Slota et al., 2019). The appearance of proneural cells reflects several canonical molecular events commonly seen during neurogenesis in other organisms throughout the animal kingdom. Early patterning establishes a cellular environment conducive for neurogenesis. Details of that environment are not clear, but a number of experiments provide some information on the signaling that impacts the neurogenic field. As discussed earlier, blockage of Wnt signaling at the outset of development results in a vast excess of neurogenesis (Yaguchi et al., 2006). A number of experiments have detailed both the Wnts and Frizzleds involved and the inhibitors that block their function in the ectoderm (Range, 2014; Range, 2018; Range et al., 2013; Range and Wei, 2016; Wei et al., 2016). In vertebrates that environment also must exclude BMP signaling, and in the sea urchin neurogenesis takes place in the ventral ectodermal region, away from the dorsal ectoderm where BMP2/4 signals. Nodal signaling is necessary in the ventral ectoderm for early specification of postoral and ciliary band neurogenesis since expression of soxC, soxb1, sip1, insm, brn1/2/4, and ngn are inhibited by knockdown of nodal expression (McClay et al., 2018). The second canonical requirement for neural development is that differentiation of neurons occurs only when the proneural cells become postmitotic. That criterion occurs in the sea urchin at some time soon after the proneural cells express soxC (Garner et al., 2016). In that study, mitotic events were observed after soxC expression was first observed, so the early SoxC-dependent specification occurs prior to the mitotic arrest that signals upcoming neural differentiation. A third requirement for proneural development originally discovered in Drosophila is Notch-Delta signaling that limits the number of ectodermal cells that become neural (Artavanis-Tsakonas et al., 1990; Artavanis-Tsakonas et al., 1983). Knockdown of delta expression results in an excess numbers of proneural cells (McClay et al., 2018; Mellott et al., 2017). Thus, a checkpoint in neural specification is Delta-Notch signaling which limits the number of competent ectoderm cells that proceed forward toward neural differentiation.
In vertebrates, a number of inhibitors are produced by the embryonic organizer to inhibit BMP signaling, thereby allowing ectoderm to become neural, a process that has been called the “default” mechanism of neurogenesis (Hemmati-Brivanlou and Melton, 1997; Hemmati-Brivanlou and Melton, 1994). Further studies in vertebrates suggested that all TGFβ signaling was inhibitory for neural specification (Chang and Harland, 2007). However, experiments in sea urchin development suggest that Nodal and BMP, while inhibitory in some locations, actually are necessary in others for neurogenesis (McClay et al., 2018; Yaguchi et al., 2016). In the ANE, for example, BMP signaling is necessary for expression of hbn, and Hbn is necessary for specification of serotonergic neurons (Yaguchi et al., 2016). Neurogenesis in the ventral ectoderm occurs in a location where Nodal signaling is active and knockdown of nodal or overexpression of lefty actually blocks early neurogenesis (McClay et al., 2018). The explanation for this apparent difference compared to vertebrates could be that Nodal doesn’t inhibit specification of the neural fate trajectory, but instead blocks neural differentiation as has been seen in development of embryonic stem cells toward neurons (Smith et al., 2008; Vallier et al., 2004). In fact, Nodal promotes neural specification of vertebrate embryonic stem cells but appears to maintain pleuripotency (Smith et al., 2008; Vallier et al., 2009). Thus, in the sea urchin the perturbation results might best be explained that an inhibition of early Wnt signaling in the anterior half of the embryo provides the necessary signaling that later allows Delta-Notch proneural signaling to restrict the number of proneural cells. Nodal signaling in the ventral ectoderm, necessary for ventral-dorsal axial patterning, does not inhibit neural specification in the ventral ectoderm prior to the appearance of the ciliary band and in fact appears to support that early specification. However, neural differentiation may require the environment in and around the ciliary band to be a TGFβ-free zone. This may well be facilitated by the known loss of nodal expression in the ventral ectoderm before neurons differentiate there (Duboc et al., 2004; Duboc et al., 2005).
Fig. 4 illustrates in graphic form the GRN governing neural specification in the ciliary band. It models genes known to be expressed in the proneural specification state, through Delta signaling, up to and including activation of neurotransmitter genes. That model has several deficiencies in addition to being incomplete. A major deficiency is timing. Neurons are not specified synchronously and as a consequence the timing of appearance of transcription factors by in situ or by other measurements doesn’t help in temporally sequencing specification events. Neurons also differentiate asynchronously so the duration of the specification sequence is unclear. Perturbation studies also suffer from some ambiguity; that is, since most perturbation studies involve introduction of morpholinos at fertilization, or introduction of inhibitors long before the reaction targeted is functional, it is difficult to accurately deduce the timing of specific GRN events of specification, and potential off target effects are a concern. Thus, it is difficult to deduce which events are direct and which are indirect. Some of the deficiencies can be resolved by knowing cis-regulatory information, but there too, timing is a difficult parameter to discern. New technologies such as single cell sequencing, ATAC-seq, and optogenetic methods for activating or inactivating genes provide promise for overcoming many of these deficiencies. In the meantime, with those significant caveats in mind, Fig. 4 is a model that reflects current information on the status of ciliary band neuron specification. As with the serotonergic neurons in the ANE, the signaling inputs provide an environment for neural specification. Thus, the ventral ectoderm is the environment in which initial neural specification occurs. The organization of the ciliary band environment is best known from a nanostring perturbation study using a probe set of 181 transcription factors and signals known to be expressed during early embryogenesis (Barsi et al., 2015). The Barsi, et al. (2015) study summarizes how the ciliary band environment is established and sets the stage for neurogenesis of the ciliary band neurons (Fig. 4). Genes activated in the postoral region are modeled separately from ciliary band neurons. There is good reason to believe that these neurons are specified differently. In situ analysis shows postoral neurons being specified first. Then later, in a different location near the ciliary band, the first ciliary band neurons initiate specification. Perturbation studies also indicate a distinct specification sequence. A growing number of neural genes have been identified that contribute to neural specification of these two cell types (Wei et al., 2016; Wei et al., 2009; McClay et al., 2018; Slota and McClay, 2018; Slota et al., 2019). Perturbation studies have begun to establish the GRN relationships between these transcription factors (Fig. 4). Two of these studies also asked whether neural cells are specified within the ciliary band or initially outside the ciliary band. The conclusion of the first study is that cells, if they migrate, do not migrate very far from their position at the end of cleavage (McClay et al., 2018). A later study followed individual fluorescently labeled cells and showed that some proneural cells move but only one to two cell diameters (Slota et al., 2020). Differentiation of neurons is marked by expression of synaptotagminB (Burke et al., 2006b), by expression of chat (choline acetyltransferase) an enzyme necessary for synthesis of acetylcholine, th (tyrosine hydroxylase), an enzyme necessary for synthesis of dopamine, and tph (tryptophan 5-hydroxylase), an enzyme in the serotonin synthesis pathway (Slota and McClay, 2018). Of the roughly 50 neurons that are present when larval behavior is initiated, 35–40 of those neurons are ciliary band and postoral neurons.
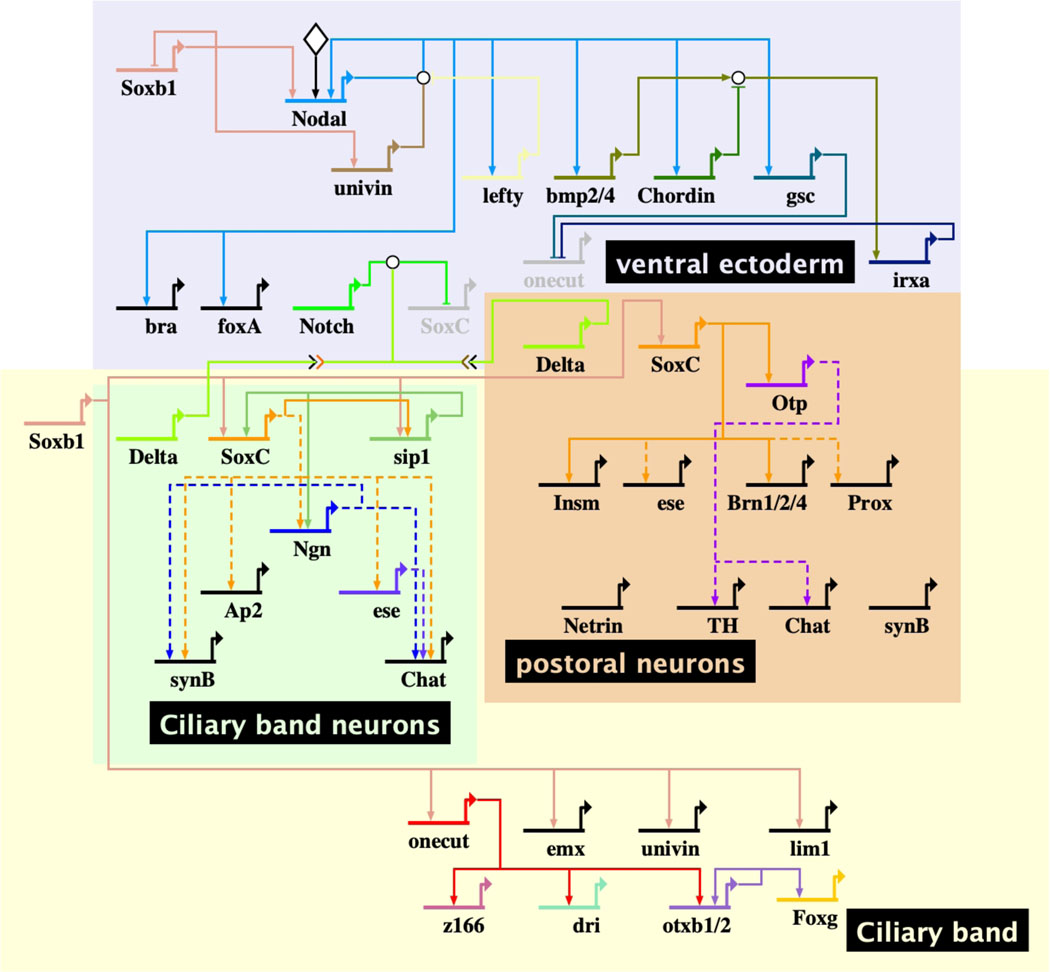
Fig. 4. Ciliary band neurons.
BioTapestry model of neurogenesis of postoral and ciliary band neurons. Specification occurs following the early patterning of the ventral ectoderm. First the postoral neurons begin to be specified. soxC expression is activated by Soxb1. Delta is activated shortly thereafter to signal through Notch to neighboring cells resulting in repression of neurogenesis in the Notch-activated cells, thereby restricting the number of neurons specified. When the ciliary band appears, ciliary band neurons complete their specification and differentiate. Solid lines represent perturbations reported in the literature. The connections may or may not be direct since cis-regulatory confirmation has not been conducted. Dashed lines represent likely connections that are only preliminarily established. Diamond input into Nodal represents an early asymmetry of uncertain origin.
Patterning the endomesoderm
The larva contains enteric neurons and ganglia at the site of each sphincter along the gut (Yaguchi and Yaguchi, 2019) (Fig. 1). In a surprising discovery the endodermal neurons originate from the endomesoderm (Wei et al., 2011). The Wei et al. paper referred to the embryonic tissue as endoderm based on the observation that expression of six3 was first detected at 9th cleavage, a cleavage after the first definitive endoderm cells are detected. That detection is at or near the boundary of endoderm and mesoderm, and recent scRNA-seq analyses indicate there is an extended intermediate state in a number of endomesoderm cells (Massri, 2021; McClay et al., 2018), so endomesoderm will be used here as a conservative estimation of the tissue of origin. The Wei et al., paper focused only on neurons in the pharyngeal region but staining with L1 (NgCAM), a neural differentiation marker, shows a ring of neurons around the pharyngeal, pyloric and anal sphincters as well (McClay et al., 2018). Yaguchi and Yaguchi (2019), showed enteric neurons in these locations (Fig. 1). Experiments with a photoconvertible dye showed there was not a significant migration of proneural cells into the gut region suggesting that the neurons are specified there from endomesoderm cells and not imported from elsewhere (Wei et al., 2011). Independent lineage analyses confirmed these results by showing that progeny of early ectodermal blastomeres formed neurons in the ANE and ciliary band but not in the endomesoderm, whereas progeny of endomesoderm formed SoxC-positive cells in locations that later formed neural ganglia (McClay et al., 2018). The reason the Wei et al., finding was so surprising is that it defied conventional wisdom about neural specification. Elsewhere in the embryo neural development is inhibited by Wnt signaling while endomesodermal specification requires Wnt signaling (Logan et al., 1999). In demonstrating an endomesodermal origin of neurons Wei et al., (2011) showed that soxb1, six3, and nkx3–2 expression are necessary for specification of endodermal neurons using SynaptotagminB as a marker for neural cells. Recent work showed SynB-expressing neurons in the pyloric sphincter (between the midgut and hindgut) also express nitric oxide synthase. Release of nitric oxide by these enteric neurons relaxes the sphincter muscles, a property shared with vertebrates (Yaguchi and Yaguchi, 2019). Thus, in patterning the endoderm, both endodermally derived muscle (Sherwood and McClay, 1999), and endodermally derived neurons are produced to control movement of food through the gut.
Specification of neurons originating along the gut.
A band of soxb1 expression precedes expression of a band of six3 expression in the foregut and both are necessary for the later appearance of a band of nkx3.2 expression (Wei et al., 2011). That band of endomesoderm includes more cells than later differentiate as neurons so it is likely that these three transcription factors help establish competence for neural specification but may not be sufficient for that specification. When differentiation occurs nkx3.2 expression is retained only in cells that co-express synaptotagminB (Wei et al., 2011).
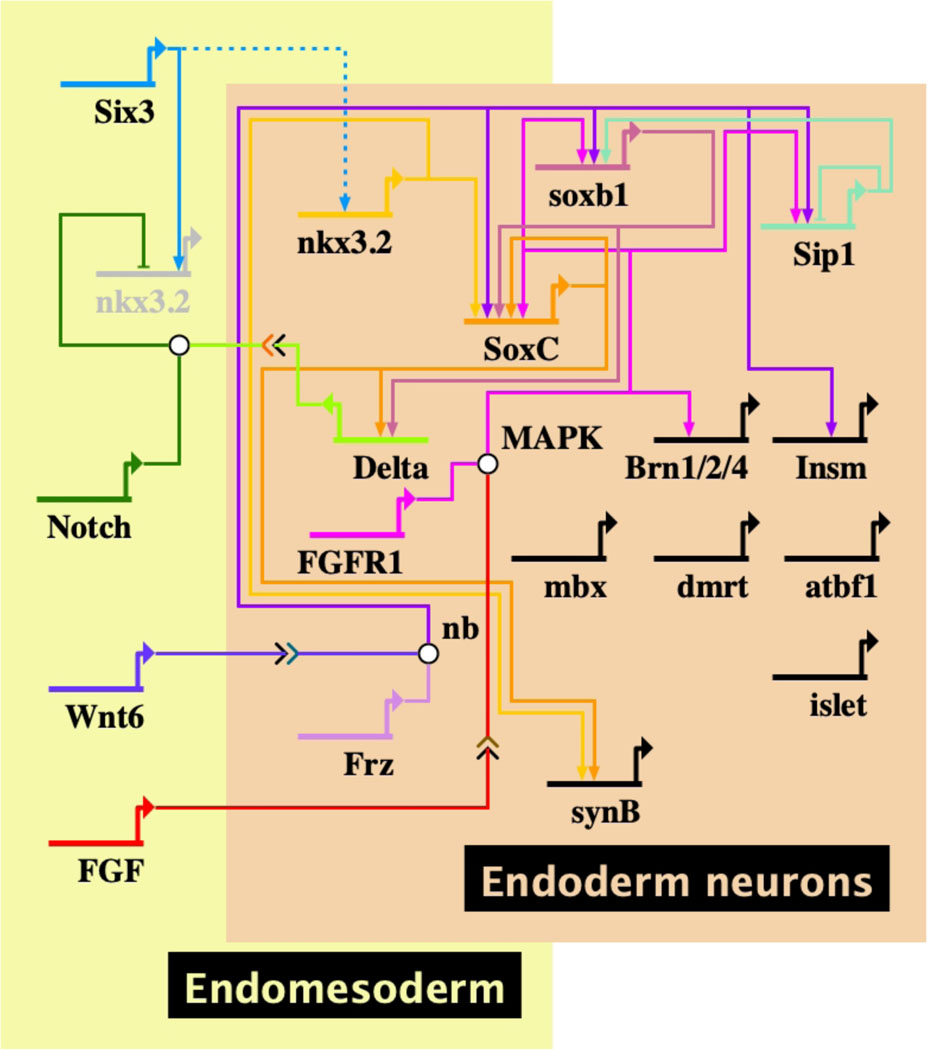
Nodal and BMP signaling perturbations have little effect on specification of the endomesodermal neurons while Wnt6 perturbations have a strong effect as does perturbation of FGF signaling (McClay et al., 2018). A gene regulatory network of the gut neurons is modeled in Fig. 5 based on perturbation experiments by (Wei et al., 2011; McClay et al., 2018; Yaguchi and Yaguchi, 2019), and expression studies of probable neural specification genes (Slota et al., 2019).
Fig. 5. Endoderm neurons.
The BioTapestry model includes the genes that have been studied with the connections based on published perturbations. A number of additional genes are included in the neurons but their position in the network has yet to be established. The dotted connection is a transcription factor in one cell affecting the proneural cell and must require a signal between the cells.
A functional nervous system in the larva.
As indicated above, when the larval stage is reached the embryo has produced about 50 neurons and evidence suggests that circuits established by these 50 neurons immediately begin to function. Little is known about the circuits but given the growing body of information on development of the neurons, the circuitry and their role in behaviors offers a rich frontier for future research, especially since so few neurons are involved. Some information is beginning to emerge on neural behavior. As described by Strathmann (Strathmann, 1975, 2007; Strathmann et al., 1972), cilia in the ciliary band beat coordinately to move the larva forward. Encountering obstacles or the air-water interface causes a rapid coordinated ciliary reversal. As observed by microscopy, a larva repeatedly moves up in the water column in a slow spiral with the pluteus arms pointing toward the direction of movement. When the larva encounters a particle, the cilia rapidly reverse causing the larva to fall in the water column. At the same time another ciliary-directed current directs small particles into the mouth (Strathmann, 1975, 2007). The extent to which neural behavior controls a particle selection process isn’t known. Particles that accumulate in the foregut are periodically moved into the midgut using muscle fibers that surround the foregut (Sherwood and McClay, 1999; Wessel et al., 1990). That movement is a peristaltic movement presumably coordinated by neural activity. About eight muscle bands surround the foregut, and these contract while at the same time endodermally-derived pharyngeal sphincter muscles relax to open the gut tube and allow the food particles to enter the midgut. Those movements are coordinated by the larval nervous system though very little is known about the neural circuit involved. While in the gut much of the food is digested and continuously moved by long cilia in the gut. The remaining debris is then moved to the hindgut and ultimately released at the anus. A recent paper offers a glimpse of the kinds of studies that can connect knowledge of neural specification and differentiation with behavior. Yaguchi and Yaguchi (2021) show in a series of experiments that light stimulates opsin at the anterior end of the larva. This activates serotonergic neurons. An action potential is relayed to stimulate the release of NO from enteric neurons in the pyloric sphincter. The NO, in turn, causes a neural response triggering the muscle-directed opening of the pyloric sphincter allowing movement of the food from the midgut to the hindgut.
The Yaguchi and Yaguchi study demonstrates that light is used for a feeding behavior while other studies suggest that light is also detected by the larva and used for movement in the water column (Lowe et al., 2018; Valencia et al., 2019). Additional behaviors are likely to be discovered as behavior of larva is examined in greater detail. The fact that sea urchins express a number of neuropeptides suggest that many behaviors may exist to support larval physiology and behavior prior to metamorphosis (Elphick and Thorndyke, 2005; Wood et al., 2018). The processes and behaviors regulated by these peptides are unknown but it will be most interesting to discover how these work, and given the fact that knowledge of specification, patterning, and differentiation at a molecular level can now be coupled with an idea of how different behaviors operate, the field has reached a systems level of analysis.
Conclusion.
Molecular studies have advanced an understanding of the specification and patterning required for competent neurogenic territories, and perturbation studies have provided an early glimpse into the specification sequence of the several neuronal cell types. Sea urchin larvae begin to feed within days of fertilization using a nervous system containing only about 50 neurons. Those feeding behaviors involve neurons produced in three locations in the embryo: serotonergic neurons at the anterior end, cholinergic and dopaminergic neurons within and next to the ciliary band, and enteric neurons at several locations in the gut.
Acknowledgements:
The author appreciates the input provided by members of the McClay lab. Also I thank Dr. Shunsuke Yaguchi and two anonymous reviewers who provided valuable input that measurably improved the accuracy of the information reviewed. The graphics were produced by Esther Miranda who has been essential to research in the McClay lab for more than 20 years. Her work is much appreciated. Thanks also to a former graduate student, Dr. Leslie Slota, who initiated research in the lab on neural development. Funding for this work was provided by NIH RO1 HD-14483 and a grant from the Feidelson family.
References
- Angerer LM, Oleksyn DW, Logan CY, McClay DR, Dale L, Angerer RC, 2000. A BMP pathway regulates cell fate allocation along the sea urchin animal-vegetal embryonic axis. Development 127, 1105–1114. [DOI] [PubMed] [Google Scholar]
- Angerer LM, Yaguchi S, Angerer RC, Burke RD, 2011. The evolution of nervous system patterning: insights from sea urchin development. Development 138, 3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Delidakis C, Fehon R, Hartley D, Herndon V, Johansen K, Markopoulou K, Preiss A, Rebay I, Scottgale N, et al. , 1990. Notch and the molecular genetics of neuroblast segregation in Drosophila. Molecular Reproduction and Development 27, 23–27. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch MA, Yedvobnick B, 1983. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A 80, 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Levine M, 1999. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature 398, 427–431. [DOI] [PubMed] [Google Scholar]
- Barsi JC, Davidson EH, 2016. cis-Regulatory control of the initial neurogenic pattern of onecut gene expression in the sea urchin embryo. Dev Biol 409, 310–318. [DOI] [PubMed] [Google Scholar]
- Barsi JC, Li E, Davidson EH, 2015. Geometric control of ciliated band regulatory states in the sea urchin embryo. Development 142, 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Burke RD, 1986. Development of serotonergic neurons in embryos of the sea urchin, Strongylocentrotus purpuratus. Devel. Growth & Differ. 28, 569–574. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Burke RD, 1987. Development of the nervous system of the pluteus larva of Strongylocentrotus droebachiensis. Cell and Tissue Research 248, 335–343. [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH, 2009. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Dev Biol 340, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradham CA, McClay DR, 2006. p38 MAPK is essential for secondary axis specification and patterning in sea urchin embryos. Development 133, 21–32. [DOI] [PubMed] [Google Scholar]
- Bradham CA, Oikonomou C, Kuhn A, Core AB, Modell JW, McClay DR, Poustka AJ, 2009. Chordin is required for neural but not axial development in sea urchin embryos. Dev Biol 328, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RD, Angerer LM, Elphick MR, Humphrey GW, Yaguchi S, Kiyama T, Liang S, Mu X, Agca C, Klein WH, Brandhorst BP, Rowe M, Wilson K, Churcher AM, Taylor JS, Chen N, Murray G, Wang D, Mellott D, Olinski R, Hallbook F, Thorndyke MC, 2006a. A genomic view of the sea urchin nervous system. Dev Biol 300, 434–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RD, Osborne L, Wang D, Murabe N, Yaguchi S, Nakajima Y, 2006b. Neuron-specific expression of a synaptotagmin gene in the sea urchin Strongylocentrotus purpuratus. J Comp Neurol 496, 244–251. [DOI] [PubMed] [Google Scholar]
- Chang C, Harland RM, 2007. Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development 134, 3861–3872. [DOI] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, Yankura KA, Hinman VF, 2016. A gene regulatory network for apical organ neurogenesis and its spatial control in sea star embryos. Development 143, 4214–4223. [DOI] [PubMed] [Google Scholar]
- Coffman JA, Coluccio A, Planchart A, Robertson AJ, 2009. Oral-aboral axis specification in the sea urchin embryo III. Role of mitochondrial redox signaling via H2O2. Dev Biol 330, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA, Davidson EH, 2001. Oral-aboral axis specification in the sea urchin embryo. I. Axis entrainment by respiratory asymmetry. Dev Biol 230, 18–28. [DOI] [PubMed] [Google Scholar]
- Coffman JA, Denegre JM, 2007. Mitochondria, redox signaling and axis specification in metazoan embryos. Dev Biol 308, 266–280. [DOI] [PubMed] [Google Scholar]
- Coffman JA, McCarthy JJ, Dickey-Sims C, Robertson AJ, 2004. Oral-aboral axis specification in the sea urchin embryo II. Mitochondrial distribution and redox state contribute to establishing polarity in Strongylocentrotus purpuratus. Dev Biol 273, 160–171. [DOI] [PubMed] [Google Scholar]
- Coffman JA, Wessels A, Deschiffart C, Rydlizky K, 2014. Oral-aboral axis specification in the sea urchin embryo, IV: Hypoxia radializes embryos by preventing the initial spatialization of nodal activity. Dev Biol 386, 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Besnardeau L, Lepage T, 2008. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev Biol 320, 49–59. [DOI] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Saudemont A, Bessodes N, Mekpoh F, Haillot E, Quirin M, Lepage T, 2010. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development 137, 223–235. [DOI] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Besnardeau L, Lepage T, 2004. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell 6, 397–410. [DOI] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Lapraz F, Besnardeau L, Lepage T, 2005. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell 9, 147–158. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Thorndyke MC, 2005. Molecular characterisation of SALMFamide neuropeptides in sea urchins. J Exp Biol 208, 4273–4282. [DOI] [PubMed] [Google Scholar]
- Floc’hlay S, Molina MD, Hernandez C, Haillot E, Thomas-Chollier M, Lepage T, Thieffry D, 2021. Deciphering and modelling the TGF-beta signalling interplays specifying the dorsal-ventral axis of the sea urchin embryo. Development 148. [DOI] [PubMed] [Google Scholar]
- Garner S, Zysk I, Byrne G, Kramer M, Moller D, Taylor V, Burke RD, 2016. Neurogenesis in sea urchin embryos and the diversity of deuterostome neurogenic mechanisms. Development 143, 286–297. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, 1988. A community effect in animal development. Nature 336, 772–774. [DOI] [PubMed] [Google Scholar]
- Haillot E, Molina MD, Lapraz F, Lepage T, 2015. The Maternal Maverick/GDF15-like TGF-beta Ligand Panda Directs Dorsal-Ventral Axis Formation by Restricting Nodal Expression in the Sea Urchin Embryo. PLoS Biol 13, e1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton D, 1997. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell 88, 13–17. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA, 1994. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell 77, 273–281. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Burke RD, 2018. Embryonic neurogenesis in echinoderms. Wiley Interdiscip Rev Dev Biol 7, e316. [DOI] [PubMed] [Google Scholar]
- Khadka A, Martinez-Bartolome M, Burr SD, Range RC, 2018. A novel gene’s role in an ancient mechanism: secreted Frizzled-related protein 1 is a critical component in the anterior-posterior Wnt signaling network that governs the establishment of the anterior neuroectoderm in sea urchin embryos. Evodevo 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapraz F, Besnardeau L, Lepage T, 2009. Patterning of the dorsal-ventral axis in echinoderms: insights into the evolution of the BMP-chordin signaling network. . PLoS Biol. 7, e1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapraz F, Haillot E, Lepage T, 2015. A deuterostome origin of the Spemann organiser suggested by Nodal and ADMPs functions in Echinoderms. Nat Commun 6, 8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y, Cho IT, Shi X, Grinspan JB, Cho G, Golden JA, 2019. Arx Expression Suppresses Ventralization of the Developing Dorsal Forebrain. Sci Rep 9, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR, 1999. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development 126, 345–357. [DOI] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, and Bolouri H 2009. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta 1789, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe EK, Garm AL, Ullrich-Luter E, Cuomo C, Arnone MI, 2018. The crowns have eyes: multiple opsins found in the eyes of the crown-of-thorns starfish Acanthaster planci. BMC Evol Biol 18, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massri AJ, Greenstreet L, Afanassiev A, Berrio A, Wray GA, Schiebinger G, and McClay DR, 2021. Developmental single-cell transcriptomics in the Lytechinus variagatus sea urchin embryo. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay DR, Miranda E, Feinberg SL, 2018. Neurogenesis in the sea urchin embryo is initiated uniquely in three domains. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellott DO, Thisdelle J, Burke RD, 2017. Notch signaling patterns neurogenic ectoderm and regulates the asymmetric division of neural progenitors in sea urchin embryos. Development 144, 3602–3611. [DOI] [PubMed] [Google Scholar]
- Modell JW, Bradham CA, 2011. Mitochondrial gradients and p38 activity in early sea urchin embryos. Molecular Reproduction and Development 78, 225. [DOI] [PubMed] [Google Scholar]
- Molina MD, Lepage T, 2020. Maternal factors regulating symmetry breaking and dorsal-ventral axis formation in the sea urchin embryo. Curr Top Dev Biol 140, 283–316. [DOI] [PubMed] [Google Scholar]
- Molina MD, Quirin M, Haillot E, Jimenez F, Chessel A, Lepage T, 2017. p38 MAPK as an essential regulator of dorsal-ventral axis specification and skeletogenesis during sea urchin development: a re-evaluation. Development 144, 2270–2281. [DOI] [PubMed] [Google Scholar]
- Otim O, Amore G, Minokawa T, McClay DR, Davidson EH, 2004. SpHnf6, a transcription factor that executes multiple functions in sea urchin embryogenesis. Dev Biol 273, 226–243. [DOI] [PubMed] [Google Scholar]
- Range R, 2014. Specification and positioning of the anterior neuroectoderm in deuterostome embryos. Genesis 52, 222–234. [DOI] [PubMed] [Google Scholar]
- Range R, Lapraz F, Quirin M, Marro S, Besnardeau L, Lepage T, 2007. Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-beta related to Vg1. Development 134, 3649–3664. [DOI] [PubMed] [Google Scholar]
- Range RC, 2018. Canonical and non-canonical Wnt signaling pathways define the expression domains of Frizzled 5/8 and Frizzled 1/2/7 along the early anterior-posterior axis in sea urchin embryos. Dev Biol 444, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Range RC, Angerer RC, Angerer LM, 2013. Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol 11, e1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Range RC, Wei Z, 2016. An anterior signaling center patterns and sizes the anterior neuroectoderm of the sea urchin embryo. Development 143, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudemont A, Haillot E, Mekpoh F, Bessodes N, Quirin M, Lapraz F, Duboc V, Rottinger E, Range R, Oisel A, Besnardeau L, Wincker P, Lepage T, 2010. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet 6, e1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood DR, McClay DR, 1999. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 126, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O’Connor MB, 2005. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120, 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota L, 2019. Cell type specification and evolution of the developing sea urchin nervous and digestive systems. 1–180. [Google Scholar]
- Slota LA, McClay DR, 2018. Identification of neural transcription factors required for the differentiation of three neuronal subtypes in the sea urchin embryo. Dev Biol 435, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota LA, Miranda E, Peskin B, McClay DR, 2020. Developmental origin of peripheral ciliary band neurons in the sea urchin embryo. Dev Biol 459, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota LA, Miranda EM, McClay DR, 2019. Spatial and temporal patterns of gene expression during neurogenesis in the sea urchin Lytechinus variegatus. Evodevo 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Vallier L, Lupo G, Alexander M, Harris WA, Pedersen RA, 2008. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol 313, 107–117. [DOI] [PubMed] [Google Scholar]
- Strathmann RR, 1975. Larval Feeding in Echinoderms. Am Zool 15, 717–730. [Google Scholar]
- Strathmann RR, 2007. Time and extent of ciliary response to particles in a non-filtering feeding mechanism. Biol Bull 212, 93–103. [DOI] [PubMed] [Google Scholar]
- Strathmann RR, Fonseca JRC, Jahn TL, 1972. Suspension Feeding by Marine Invertebrate Larvae - Clearance of Particles by Ciliated Bands of a Rotifer, Pluteus, and Trochophore. Biological Bulletin 142, 505-+. [Google Scholar]
- Valencia JE, Feuda R, Mellott DO, Burke RD, Peter IS, 2019. Ciliary photoreceptors in sea urchin larvae indicate pan-deuterostome cell type conservation. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA, 2009. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136, 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Rugg-Gunn PJ, Bouhon IA, Andersson FK, Sadler AJ, Pedersen RA, 2004. Enhancing and diminishing gene function in human embryonic stem cells. Stem Cells 22, 2–11. [DOI] [PubMed] [Google Scholar]
- van Heijster P, Hardway H, Kaper TJ, Bradham CA, 2014. A computational model for BMP movement in sea urchin embryos. J Theor Biol 363, 277–289. [DOI] [PubMed] [Google Scholar]
- Wei Z, Angerer LM, Angerer RC, 2016. Neurogenic gene regulatory pathways in the sea urchin embryo. Development 143, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM, 2011. Direct development of neurons within foregut endoderm of sea urchin embryos. Proc Natl Acad Sci U S A 108, 9143–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yaguchi J, Yaguchi S, Angerer RC, Angerer LM, 2009. The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development 136, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Zhang W, Klein WH, 1990. Myosin heavy chain accumulates in dissimilar cell types of the macromere lineage in the sea urchin embryo. Dev Biol 140, 447–454. [DOI] [PubMed] [Google Scholar]
- Wood NJ, Mattiello T, Rowe ML, Ward L, Perillo M, Arnone MI, Elphick MR, Oliveri P, 2018. Neuropeptidergic Systems in Pluteus Larvae of the Sea Urchin Strongylocentrotus purpuratus: Neurochemical Complexity in a “Simple” Nervous System. Frontiers in endocrinology 9, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi J, Angerer LM, Inaba K, Yaguchi S, 2012. Zinc finger homeobox is required for the differentiation of serotonergic neurons in the sea urchin embryo. Dev Biol 363, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi J, Takeda N, Inaba K, Yaguchi S, 2016. Cooperative Wnt-Nodal Signals Regulate the Patterning of Anterior Neuroectoderm. PLoS Genet 12, e1006001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi J, Yaguchi S, 2019. Evolution of nitric oxide regulation of gut function. Proc Natl Acad Sci U S A 116, 5607–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi J, Yaguchi S, 2021. Sea urchin larvae utilize light for regulating the pyloric opening. BMC Biol 19, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi S, Katow H, 2003. Expression of tryptophan 5-hydroxylase gene during sea urchin neurogenesis and role of serotonergic nervous system in larval behavior. J Comp Neurol 466, 219–229. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, 2008. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell 14, 97–107. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, Burke RD, 2010. TGFbeta signaling positions the ciliary band and patterns neurons in the sea urchin embryo. Dev Biol 347, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD, 2006. Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development 133, 2337–2346. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD, 2007. Sp-Smad2/3 mediates patterning of neurogenic ectoderm by nodal in the sea urchin embryo. Dev Biol 302, 494–503. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Inaba K, 2014. bicaudal-C is required for the formation of anterior neurogenic ectoderm in the sea urchin embryo. Sci Rep 4, 6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Wei Z, Jin Y, Angerer LM, Inaba K, 2011. Fez function is required to maintain the size of the animal plate in the sea urchin embryo. Development 138, 4233–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankura KA, Koechlein CS, Cryan AF, Cheatle A, Hinman VF, 2013. Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc Natl Acad Sci U S A 110, 8591–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Srinivasan S, Shimmi O, Biehs B, Rashka KE, Kimelman D, O’Connor MB, Bier E, 2000. Processing of the Drosophila Sog protein creates a novel BMP inhibitory activity. Development 127, 2143–2154. [DOI] [PubMed] [Google Scholar]
- Zito F, Costa C, Sciarrino S, Poma V, Russo R, Angerer LM, Matranga V, 2003. Expression of univin, a TGF-beta growth factor, requires ectoderm-ECM interaction and promotes skeletal growth in the sea urchin embryo. Dev Biol 264, 217–227. [DOI] [PubMed] [Google Scholar]