Introduction
Physicians are increasingly using clinical sequencing tests to establish diagnoses of patients who might have genetic disorders, so that accuracy of sequencing and interpretation are important elements in ensuring the benefits of genetic testing. In the past, clinical sequencing tests were designed to detect specific, prespecified variants or unknown variants that were in limited regions of an individual’s genome. The raw data for each detected variant was then manually reviewed for errors in sequencing and for its potential clinical importance. Newer technology allows for assessment of exomes or entire genomes and can identify millions of genetic variants in each sequenced individual. The shift from limited, targeted sequencing to genome sequencing requires automated algorithms to parse through raw data, to help distinguish true variants from those caused by systematic errors. Errors can result from incorrectly read bases in particular sequence contexts and from mapping short sequences incorrectly to the human reference genome. New developments in sequencing and analysis as well as standard quality measures will be critical to ensure the accuracy of sequencing results intended for medical use.
How It Works
The output of high-throughput sequencing (HTS) instruments is not the complete genome sequence of the individual being analyzed. HTS yields billions of short sequences, known as “reads”. Each individual read is only 100-100,000s of basepairs while the complete human genome sequence is approximately 3.2 billion base pairs long. Mapping is used to align the short sequences to known human genome reference sequences. Comparisons are made between the newly mapped individual’s sequences and reference sequences to find differences which are called variants. These variants can be very small, such as single nucleotide variants (SNVs), or much larger “structural variants” up to the size of a chromosome.
The accuracy of what gets identified as a variant differs by the type of variant, how repetitive the genome sequence is, and sequencing technology. The easiest 80% to 90% of the genome can be accessed by the most commonly used “short-read” HTS technologies that read sequences of 100s of basepairs with low per-base error rates ~0.1%. However, 10% to 20% of the genome contains large repetitive structures that make it difficult or impossible to map short sequences accurately. Similarly, many structural variants occur in repetitive sequences or introduce new sequence much larger than short reads, so that these variants are difficult to detect with short reads. Newer technologies sequence single molecules, enabling much longer sequences of 10,000s to 100,000s basepairs. Techniques for generating longer sequences are currently more expensive per sequenced basepair and the raw reads have an error in the sequence every 5 to 20 basepairs, but new methods can read the same 10,000s basepair molecule multiple times, yielding fewer than one error per 100 basepairs. For HTS, each location in the genome is sequenced many (typically 10s to 1000s) times, depending on sample type (tumor typically require more than normal). Statistical models and heuristics use all the sequences at a given position to distinguish real variants from errors, including systematic errors in particular DNA sequences and mis-alignments of these sequences to the reference [see Figure for sources of errors]. More recently deep learning techniques enable faster adoption of new technologies with complicated error processes by taking advantage of the very large volumes of data to minimize errors in variant detection.
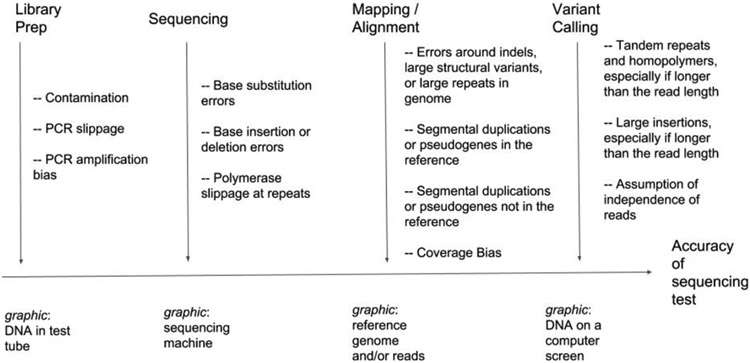
Figure. Analytical sources of error in a clinical sequencing assay.
Examples of sources of error and bias introduced in the stages of the sequencing process from DNA to variant calls: (1) Library prep – preparation of the DNA for sequencing, (2) Sequencing – measuring the sequence of the DNA molecules, (3) Mapping/alignment – comparing the DNA sequences to the Reference Genome, and (4) Variant Calling – determining differences (variants) between the individual being sequenced and the Reference Genome.
Analytic validity is defined as how well a sequencing instrument coupled with automated algorithms can accurately and reliably detect genetic variants. That is, when a genome is sequenced and analyzed, are some true variants missed or false variants detected? A high degree of analytical validity is critical for making accurate diagnoses. Although analytical validity of HTS has steadily improved over the years, it is still imperfect, creating the potential for errors when using genetic sequencing for diagnostic purposes. For older targeted clinical sequencing tests, analytical validity of a testing method can be established for a particular set of variants of interest. For HTS, analytical validity cannot be established for every possible variant, so laboratories establish sensitivity and specificity for examples of different types and sizes of variants in different types of repetitive and non-repetitive regions. The performance in these example areas then serve as proxies for the analytic performance of the sequencing method for similar variants of clinical interest occurring in other areas. To help develop standards for analytic validity, the National Institute of Standards and Technology formed the Genome in a Bottle Consortium (GIAB), an open-science endeavor that has extensively characterized 7 genomes as reference materials. GIAB integrated sequencing data from many technologies on the same genomes to provide high-confidence sequences that can be used as reference standards for benchmarking any sequencing method.1 The Global Alliance for Genomics and Health Benchmarking Team developed sophisticated, standardized benchmarking tools that enable laboratories to use reference standards to help establish analytical validity for different types of variants and repetitive and non-repetitive regions.2
Important Care Considerations
Because HTS methods are generally highly accurate in detecting small variants in non-repetitive regions of the genome, a recent article proposed a systematic approach to separate the harder variants that need confirmation by another method from those that are unlikely to be errors. 3 However, larger variants and variants in repetitive regions can be challenging to detect with standard NGS methods. 4 5 Missing these variants (false negatives) or calling inaccurate variants (false positives) can result in misdiagnosis. For example, variants in tandem repeats longer than short sequences can cause Muscular Dystrophy; large structural variants can cause intellectual disability disorders;6 and variants in the gene PMS2, which has a closely related pseudogene that makes mapping of short reads challenging, can cause Lynch Syndrome. Each of these disease entities may be missed or misdiagnosed by some diagnostic tests based on HTS. A variety of technical advancements currently under development could enable genome sequencing to be applied clinically to diagnose diseases associated with challenging variants and regions.
Value and Evidence Base
Sequencing the first human genome from the Human Genome Project was a massive effort, costing ~$2.7 billion. Now, it only costs ~$1,000 to sequence a person’s genome using HTS technologies. There is an increasing use of HTS data in the clinical medicine; for example, a HTS assay of 29 genes associated with hereditary risk of cancer had accuracy for 750 variants comparable to accuracy of older tests designed for single genes.5 To aid the growing number of clinical laboratories using HTS, the Association for Molecular Pathology and the College of American Pathologists developed guidelines to aid in validating HTS algorithms.7 The FDA has recognized the need for innovation in regulatory science for HTS and has launched a series of PrecisionFDA community challenges using the GIAB data to benchmark algorithms.2 These challenges showed, for a blinded sample, the best methods had SNV calling accuracy around 99.92% recall at 99.97% precision, and small insertion and deletion mutations were approximately an order of magnitude worse with 99.3% recall at 99.5% precision. While even the best methods still had thousands of errors in the GIAB high-confidence regions, many more errors exist outside the (large, but easier) benchmark regions, so GIAB is developing expanded benchmarks for difficult regions. The PrecisionFDA Truth Challenge V2 held in 2020 showed error rates are higher in the difficult regions covered by the new benchmarks, but new HTS technologies and algorithms are improving characterization of these difficult regions.
Bottom Line
The analytic validity of HTS is high for selected regions of the human genome. It is important for clinical decision-makers to understand both the strengths and limitations of any particular clinical sequencing test. Robust sequencing technologies with long, high-accuracy reads as well as reference materials are needed for difficult variants and difficult genomic regions to reach the full potential of clinical sequencing assays to detect all clinically important variants.
Footnotes
Statement of Conflict of Interest
Authors RP is an employee of Google, Inc., which has products that perform variant calling, and MD is an employee of BigHat Biosciences.
References
- 1.Zook JM, McDaniel J, Olson ND, et al. An open resource for accurately benchmarking small variant and reference calls. Nat Biotechnol. 2019;37(5):561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krusche P, Trigg L, Boutros PC, et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat Biotechnol. 2019;37(5):555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lincoln SE, Truty R, Lin C-F, et al. A Rigorous Interlaboratory Examination of the Need to Confirm Next-Generation Sequencing–Detected Variants with an Orthogonal Method in Clinical Genetic Testing. J Mol Diagn. 2019;21(2). doi: 10.1016/j.jmoldx.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wenger AM, Peluso P, Rowell WJ, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. August 2019. doi: 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lincoln SE, Kobayashi Y, Anderson MJ, et al. A Systematic Comparison of Traditional and Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Genes in More Than 1000 Patients. J Mol Diagn. 2015;17(5):533–544. [DOI] [PubMed] [Google Scholar]
- 6.Sanders SJ, Ercan-Sencicek AG, Hus V, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy S, Coldren C, Karunamurthy A, et al. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines: A Joint Recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn. 2018;20(1):4–27. [DOI] [PubMed] [Google Scholar]