Abstract
Pepper is a fluorogenic RNA aptamer tag that binds to a variety of benzylidene-cyanophenyl (HBC) derivatives with tight affinity and activates their fluorescence. To investigate how Pepper RNA folds to create a binding site for HBC, we used antibody-assisted crystallography to determine the structures of Pepper bound to HBC530 and HBC599 to 2.3 and 2.7 Å resolutions, respectively. The structural data show that Pepper folds into an elongated structure and organizes nucleotides within an internal bulge to create the ligand binding site, assisted by an out- of-plane platform created by tertiary interactions with an adjacent bulge. As predicted from a lack of K+ dependence, Pepper does not use a G-quadruplex to form a binding pocket for HBC. Instead, Pepper uses a unique base-quadruple•base-triple stack to sandwich the ligand with a U•G wobble pair. Site-bound Mg2+ ions support ligand binding structurally and energetically. This research provides insight into the structural features that allow the Pepper aptamer to bind HBC and show how Pepper’s function may expand to allow the in vivo detection of other small molecules and metals.
Graphical Abstract
Introduction
Fluorogenic RNAs1 were first developed to mimic the function of the well-understood and naturally derived green-fluorescent protein (GFP), which allows visualization of the proteome through fluorescence microscopy. Using GFP’s chromophore as a starting point, Jaffrey et al. engineered a synthetic RNA—Spinach—via SELEX to bind the small fluorophore DFHBI,2 a derivative of 4-hydroxybenzylidene imidazolinone (HBI) in GFP, which allows RNA transcripts to be visualized in real time without accessory proteins, as long as the DFHBI molecule is available for binding.2 Further research was conducted to improve upon Spinach, leading to next- generation aptamers, such as “Broccoli” RNA, which requires lower Mg2+ concentrations and exhibits better folding properties than Spinach, making it more versatile.3 Mango,4,5 Corn,6 and Chili7 have also been developed to fulfill a wide range of needs, from even tighter binding affinities and photo- bleaching resistance to optimization for FRET applications. These fluorogenic aptamers all function by binding and restricting bond rotation of a small-molecule chromophore to inhibit non-radiative pathways of decay and thereby promote fluorescence.8−10 Continued development and improvement of these tools require understanding their structure and biophysical properties.
Pepper is the latest addition to an existing suite of RNA aptamers that can bind small molecules to form fluorescent complexes. Developed by Chen et al. via SELEX,11 Pepper is able to bind variants of the HBC fluorophore, chosen as the fluorogenic ligand for its capacity to accommodate election- withdrawing groups that can promote red-shifted spectral properties (Figure 1). Accordingly, variants of HBC allow visualization of Pepper-containing RNA transcripts with many choices of excitation wavelengths.11 The predicted secondary structure of Pepper’s minimal motif showed three helical regions (P1−P3) with an asymmetric bulge (J1/2) separating P1 and P2 and a symmetrical bulge separating P2 and P3 (Figure S1). Mutational analysis of the Pepper construct11 indicated that the asymmetric bulge in the secondary structure contributes an essential function to fluorogenic activity (Figure 1). Additionally, mutations to the base of P2 abolished activity, whereas exchange of the terminal loop of P3 had no effect. Pepper fluorescence activation has a requirement for Mg2+ ions but not K+, indicating the absence of a K+-dependent G quadruplex in Pepper’s structure, in contrast to Spinach, Mango, Corn, and Chili.
Figure 1.
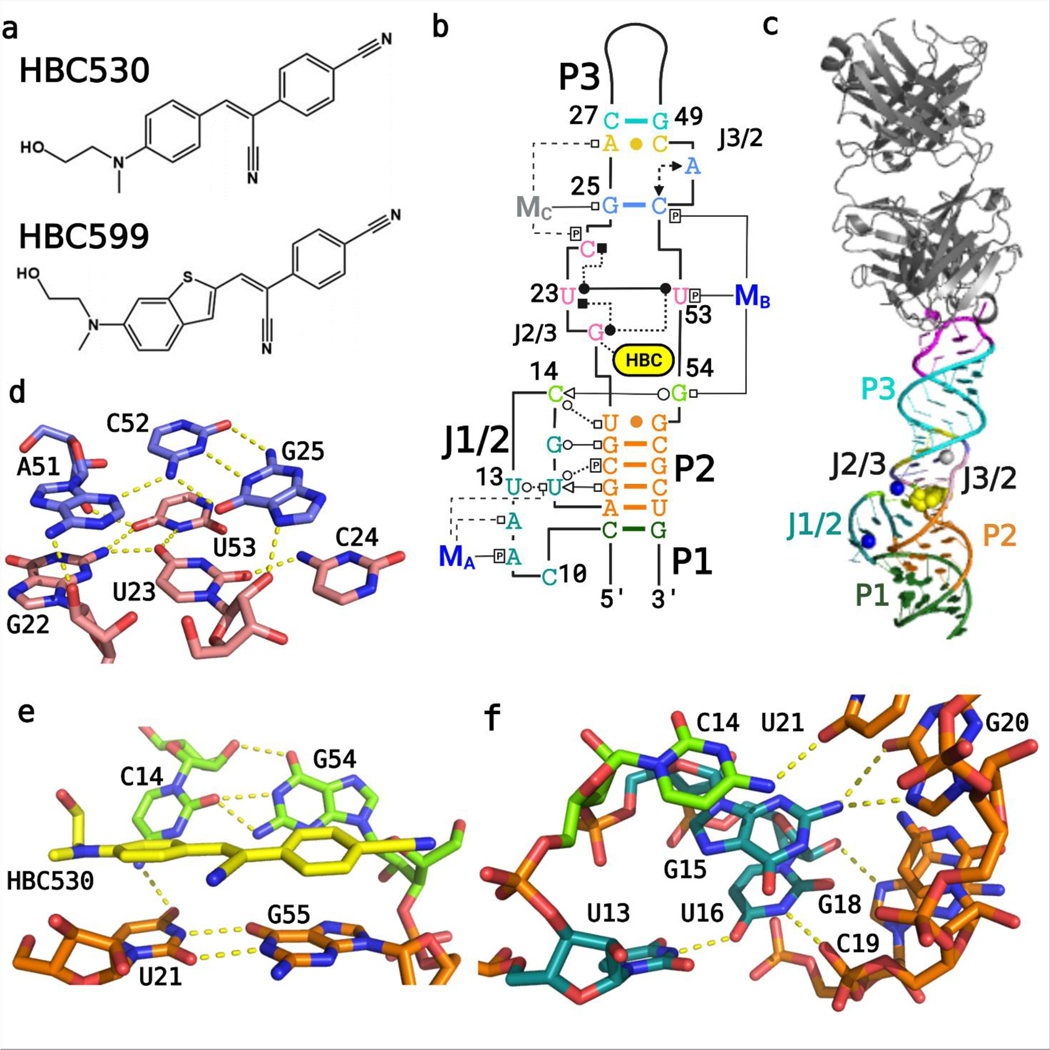
Overall structure of the Pepper-BL3–6 aptamer. (A) Chemical structures of the HBC ligands, HBC530 and HBC599 activated by Pepper RNA. (B) Tertiary structure of the Pepper aptamer bound to HBC530 utilizing the Leontis−Westhof nomenclature for RNA-interaction classification,12 with color-coded regions, and the two Mg2+ cations that are visible in both structures (MA, MB; blue) and the Na+ ion visible in the HBC530 complex only (MC; gray). (C) Fab BL3–6 bound to Pepper with the HBC530 ligand. (D) Tetrad (pink) and triplex (blue) structures above the ligand from the HBC599 complex, without MC. (E) Tertiary interactions between C14 and G54, adjacent to the bound ligand HBC530. (F) Tertiary interactions between J1/2 and P2.
To help understand this new member of the fluorogenic aptamer family, we deployed chaperone-assisted RNA crystallography using Fab BL3–6 to obtain two crystal structures of Pepper in complex with Fab chaperone bound to the ligands HBC530 and HBC599 to 2.3 and 2.7 Å resolutions, respectively. Using these structural data as a guide, we constructed and tested site-directed and atomic mutations to assess the importance of implicated RNA, ligand, and metal ion interactions, informing our understanding of how the RNA folds to create a binding site for the HBC ligand. Our findings were corroborated by the recent publication of a circularly permuted (cp) Pepper structure,12 which appeared during the preparation of our article.
Results
Construct and Crystallization
We started from the minimal T1 motif11 and made modifications that would assist crystallization without significantly changing the fluorogenic activity. We replaced the original stem loop of P3 with a cognate pentaloop hairpin (GAAACAC) to make a binding site for the Fab BL3–6 crystallization chaperone (Figure S1). The other main changes included extension of P1, addition of two G residues to the 5′ end to assist transcription, and extension of P3 to increase the distance between the Fab and the aptamer core. Fab binding to this construct was confirmed via electrophoretic-mobility-shift assays. To ensure that the resultant crystal structure would have relevance for the mechanism of fluorescence activation, we confirmed via fluorescence spectroscopy that the construct activates the fluorescence of HBC ligands in the presence of Fab.
In the search to identify a suitable construct-chaperone complex for crystallography, four Pepper constructs were designed that had variable P1 and P3 lengths; they were able to bind Fabs BL3–6,13 HAVx,14,15 BRG,16 and BL3–6-S97N17 and activate HBC to produce a strong fluorescent signal, and one construct, referred to hereafter as Pepper-BL3–6, formed diffracting crystals in complex with Fab BL3–6 in the presence of the saturating ligand (Table 1). The Pepper-BL3–6-HBC530 crystals were solved to 2.3 Å resolution via molecular replacement with Fab BL3–6 variable and constant regions (4kzd),18 followed by rounds of building and refinement of the RNA (Figure S2). Due to the high quality of the data, the ligand density became unambiguous as the RNA was built (Figure S3). Ligand orientations were decided by best fit with the electron density difference maps. Metal ions were assigned by density, coordination, occupancy, and temperature factors relative to adjacent RNA.19,20 The Pepper-BL3–6-HBC599 crystals were solved to 2.7 Å resolution via molecular replacement to place the RNA and Fab and then inserting the HBC599 ligand into the revealed density.
Table 1.
Data Collection and Refinement Statistics for the Pepper Aptamer in Complex with HBC530 (7SZU) and HBC599 (7U0Y).
| HBC530 | HBC599 | |
|---|---|---|
| data collection | ||
| space group | P212121 | P212121 |
| resolution | (2.32–2.24) | (2.76–2.66) |
| cell dimensions | ||
| a, b, c, (Å) | 61.61, 96.99, 148.26 | 61.25, 96.07, 148.17 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 |
| Rmerge (%) | 0.06286 (1.283) | 0.04627 (0.5635) |
| I/σ | 18.17 (1.22) | 15.61 (1.27) |
| completeness (%) | 99.9 (99.6) | 96.8 (83.0) |
| redundancy | 6.8 (7.0) | 3.3 (2.5) |
| CC(1/2) | 0.998 (0.56) | 0.997 (0.723) |
| refinement | ||
| no. reflections | 43498 (4281) | 24995 (2083) |
| R work /R free | 0.2198/0.2634 | 0.2182/0.2548 |
| R.M.S. deviations | ||
| bond angles (deg) | 0.58 | 0.83 |
| bond length (Å) | 0.002 | 0.005 |
| Ramachandran plot of protein residues | ||
| preferred regions (%) | 96.77 | 93.5 |
| allowed regions (%) | 2.77 | 5.34 |
| disallowed regions (%) | 0.46 | 1.16 |
| B-factors | 74.8 | 89.5 |
| RNA | 78.5 | 84.4 |
| protein | 73.8 | 91.5 |
| ligand | 72.8 | 90.5 |
| MA, MB, (MC) | 74.0, 48.7, (61.3) | 90.1, 86.8, (n/a) |
Overall Structure
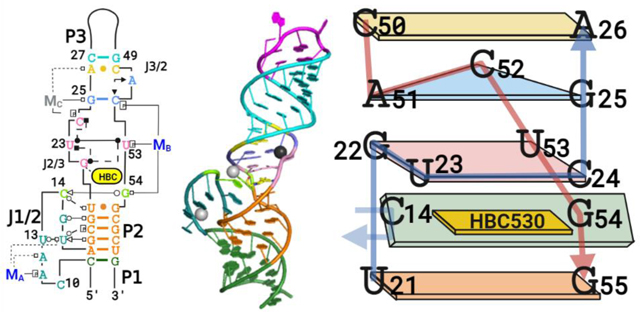
The Pepper-BL3–6 structures with bound ligand HBC derivatives (Figure 1A–C) mostly conform to secondary structure predictions from programs such as mfold,21 containing three A-form helical stems and two bulges (Figure 1B,C). P1 and P2 stack directly together; a joining region (J1/2) connects them on the 5′-side, but they share a continuous 3′ strand. The bulge between P2 and P3 harbors the ligand binding pocket and causes P3 to tilt into the ligand pocket and sit at a roughly 30° angle from the orientation of P1 and P2. Both structures contain site-bound Mg2+ cations, one located in J1/2 distal to the ligand binding site referred to herein as MA and one located in J3/2 proximal to the ligand binding site referred to herein as MB (Figure 1B). The HBC530 structure contains an additional site-bound Na+ also distal to the ligand binding site located in J2/3, referred to herein as MC.
J1/2 contains seven nucleotides, C10 to U16; this loop, which would be predicted to be structurally flexible based on minimum-free energy calculations (Figure S1), adopts a well- defined structure in the crystal, with J1/2 calculated to have average temperature factors similar to the paired regions of the aptamer after structure solving (Figure S4). The J1/2 bulge adopts a conformation that turns back on itself twice, with the backbone forming a tight turn at C14 and U16, with A17 re- joining the P2 helical stack above C9. This elaborate fold allows the J1/2 bulge to reach the ligand binding site and cooperatively interact with J3/2 to form essential interactions for ligand binding. In the crystal lattice, the first nucleotide of J1/2, C10, forms an intermolecular crystal contact with the 5′- most nucleotide, G1—an unintentional but beneficial inter- action that assists crystallization (Figure S5). At the apex of J1/ 2, C14 forms a tertiary interaction with G54 adjacent to the ligand binding pocket, and its amino group (C14−N4) lies within the range to form a tertiary interaction with U21’s keto group (U21−O4) (Figure 1E). In addition, J1/2 forms tertiary interactions with the minor groove of P2 through G15 and U16 without perturbing P2’s A-form helical geometry (Figure 1F).
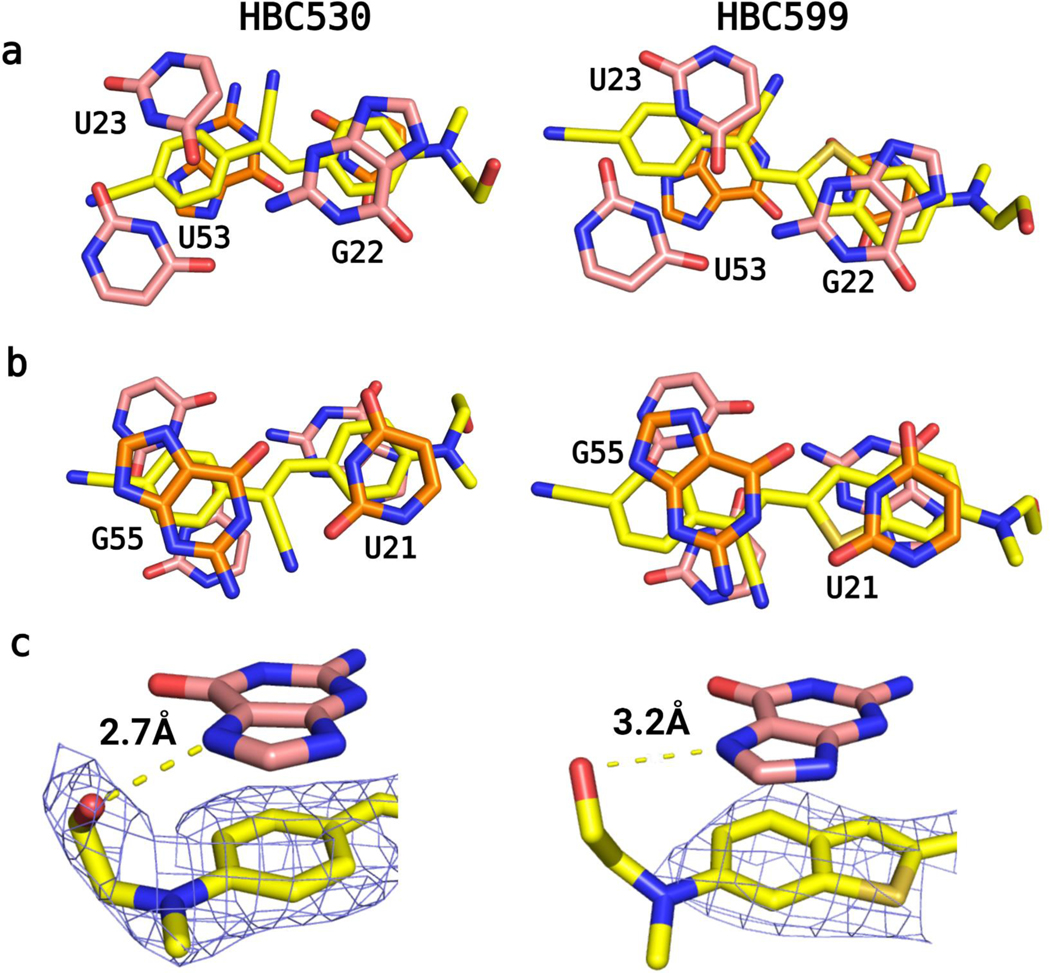
J1/2 leads into P2, a helix composed of four canonical Watson−Crick−Franklin (WCF) base pairs, followed by a U21•G55 wobble pair. This pair forms the base of the symmetric bulge that creates the ligand binding site. On the 5′ side of this J2/3 bulge starting with G22, the backbone of the next two nucleotides U23 and C24 runs perpendicular to the helical stack to allow G22, U23, and C24 to all lie in the same plane and form a quadruple base interaction that includes U53 on the 3′-side of the bulge (Figure 1C,E,F). In this quadruple layer, G22, U23, and U53 interact with each other, but the position of the fourth base, C24, differs slightly between the two structures. In the Pepper-HBC599 complex structure, the exocyclic amine of C24 resides within the hydrogen bonding distance of U23’s O2 keto group, but the two groups are 4 Å away in the Pepper-HBC530 complex (Figure S6). The MC present in the HBC530 structure but not in the HBC599 structure may contribute to this difference.
The base quadruple layer sits on top of the HBC ligand, sandwiching it with the U•G wobble pair below (Figures 2 and 3). With the U21−G22 backbone forming one side of the binding pocket, the aforementioned C14−G54 tertiary interaction closes the other side via a WCF/sugar-edge (SE) base pair. In this tertiary interaction, the SE of C14 from J1/2 interacts with the WCF face of G54, which adopts a C2′-endo pucker that facilitates its orientation toward C14.
Figure 2.
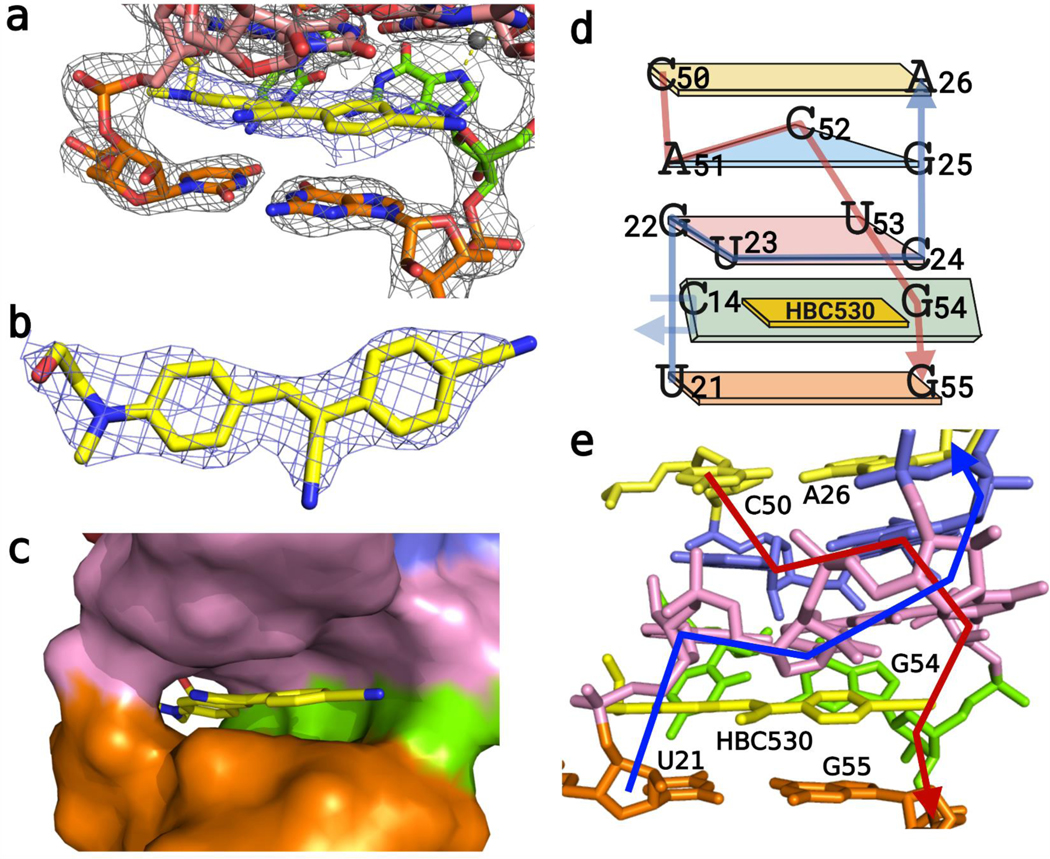
Structure of the HBC ligand pocket of Pepper-BL3–6. (A) Electron density of the ligand HBC appears within the pocket, carved to 2.0 rmsd. (B) Top-down view of the electron density of the HBC530 ligand to 2.0 rmsd. (C) Color-surface cartoon of the binding pocket. (D) Schematic of the J2/3 and J3/2 stacking. (E) Cartoon view of stacking with 5’ (blue) and 3′(red) backbones traced.
Figure 3.
HBC ligand orientation within the pocket. (A) View from above shows that one ring sits sandwiched between U21 and G22—possibly allowing the tail of the HBC ligands to form a H-bond with G22−N7. The tetrad (C24 not shown) has U23 and U53 making a wobble pair over the other ring of HBC. (B) U21−G55 wobble pair below the ligand. (C) Distances between G22−N7 and the tail of the HBC ligands, with the ligand density carved to 2.0 rmsd.
A base triple involving nucleotides G25, A51, and C52 is stacked upon the quadruple base layer (Figure 1C). G25 and C52 form a WCF base pair, and A51 completes the triple using its N3 endocyclic nitrogen to accept a hydrogen bond from the N4 exocyclic amino group of C52. The ribose of A51 adopts a C2′-endo pucker, which allows A51 and C52 to remain in the same plane. In the Pepper-HBC530 structure, G25 uses its N7 nitrogen to coordinate directly to MC, whereas in the Pepper- HBC599 structure, which lacks MC, N7 accepts a hydrogen bond from the U23’s2′-OH below (Figure 1D). Above the base triple, an A26−C50 wobble base pair closes the symmetric bulge with a distance of 2.8 Å between A26−N1 and C50−O2. This distance implies that A26−N1 is protonated, consistent with the crystal growth occurring at pH 6.5 or lower. Nevertheless, protonation of A26 appears unnecessary for Pepper’s fluorogenic activity as HBC530 fluorescence exhibited little change between pH 5 and 9 when bound to the RNA construct (Figure S7). P3 follows this symmetric bulge and contains the AAACA BL3–6 pentaloop, which binds Fab BL3–6 in a manner similar to previous Fab- BL3–6-RNA complexes.13,22−24
Ligand Binding Pocket
The HBC ligands sit in the tunnel-shaped pocket (Figure 2A–C) formed by the symmetric bulge between P2 and P3. Both ligands stack with one ring sandwiched between U21 below and G22 above, with the G22 ribose adopting a C2′-endo pucker, possibly to accommodate the increased phosphate linker distance from U21 (Figures 2D,E and 3A). The other ring of the ligand stacks between G55 below and the U23/U53 of the quadruple- base layer (Figure 3B). The π−π distances between the bases and ligand are about 3.4 Å as expected for such stacking interactions. Finally, both HBC ligands orient the central α,β unsaturated nitrile away from the C14−G54 pair, probably due to steric reasons (Figure 2).
Like other fluorogenic RNA aptamers, Pepper binds its ligand via a platform generated by a co-planar arrangement of multiple bases, although not using G-quadruplexes as observed in several other aptamers6,18,25−27 (Tables S1 and S2). The base quadruple in Pepper appears to be sequence-specific as mutations in the tetrad lead to loss of activity (see below), although the base triple above it is more forgiving. Chen et al. reported that changing the U21•G55 wobble pair on the underside of the ligand to a GC pair results in weaker fluorescence;11 possibly, a more stable G•C pair attenuates dynamics needed for ligand binding and activation or alters the base pair geometry in a way that affects the structure or stacking preferences of the binding pocket. Another possibility is that U21C disrupts the hydrogen bond with C14, reducing the fluorescence signal (Figure 1F).
Metal Binding Pockets
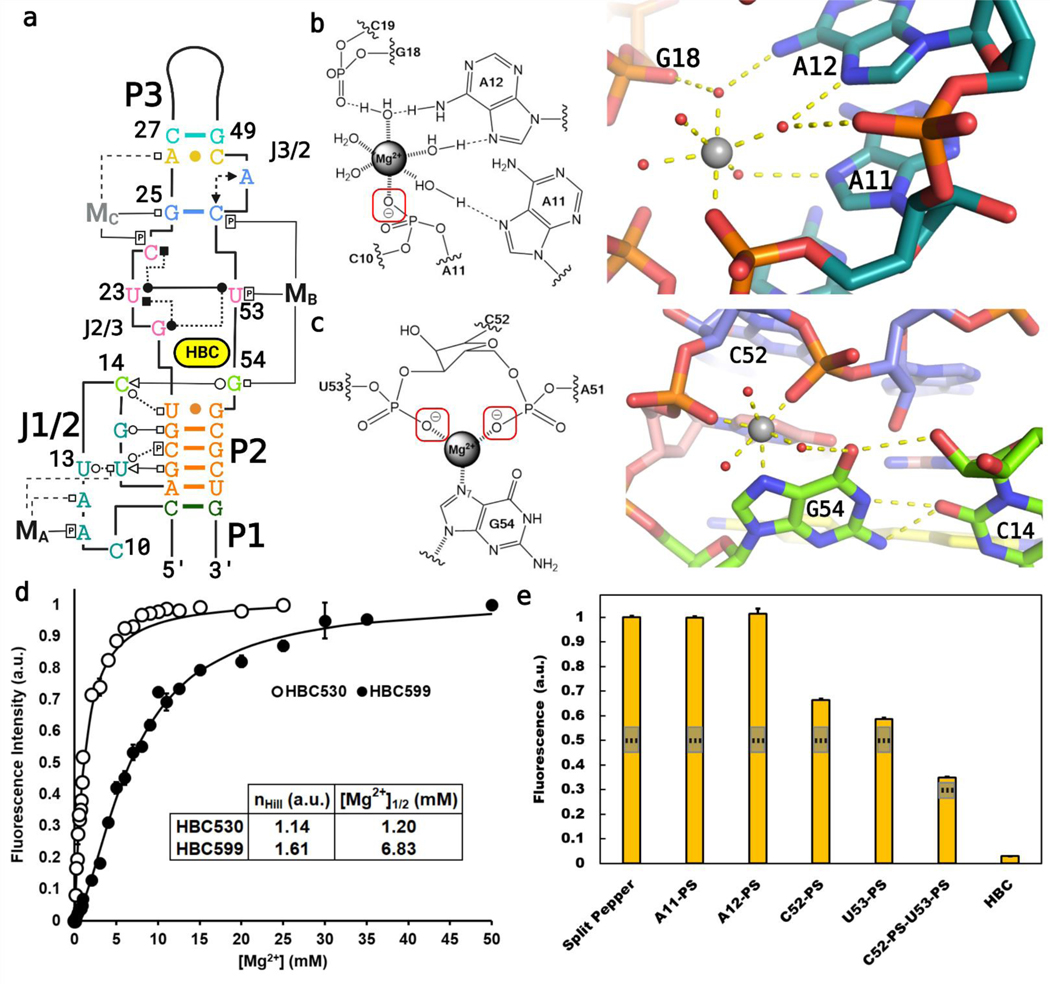
Pepper’s fluorogenic activity exhibits a strict dependence on divalent cations,11 consistent with the site-bound Mg2+ cations observed in our structures (Figure 4). The MA in J1/2 has an inner-sphere contact to the pro-RP phosphate oxygen of A11, supported by three outer- sphere contacts that coordinate to N7 of A11, N7 of A12, and N6 of A12 (Figure 4B). A second metal ion, MB, can also be assigned in both structures due to substantial electron densities above G54 and adjacent to the binding pocket. G54 forms an inner-sphere interaction with MB through N7, in addition to forming the tertiary interaction to C14 (Figure 4C). Additionally, MB forms two inner-sphere interactions with the pro-RP-phosphate oxygens of C52 and U53 and may make the A51 C2′-endo pucker more favorable. MB’s coordination to a purine N7 required scrutiny in assigning the cation identity as inner-sphere purine N7 contacts have often been misassigned as Mg2+;20 here, the 2.0 Å bond distance between the purine and Mg2+ and the two phosphate oxygen coordinations make this unlikely to be another cation. Only in the structure of Pepper bound to HBC530 is a third metal ion visible, MC, assigned as a sodium ion. MC coordinates directly to N7 of G25 (2.4 Å) and the pro-RP-oxygen of C24 (2.5 Å), causing C24 to shift relative to the HBC structure (Figure S6). As these coordination distances are substantially longer than expected for Mg2+ (2.4 Å vs the expected 2.0 Å), we ascribe MC to a sodium ion.20 In the pdb, the phosphate pro-RP oxygen represents the atom in RNA most likely to form inner-sphere interactions with Mg2+,11 and interactions between Na+ and purine N7s are ubiquitous.19,20 Pepper conforms to these trends. In contrast, MB’s inner-sphere coordination with a purine N7 is less common but not unprecedented.28−31
Figure 4.
Mg2+ sites identified in the crystallized Pepper-BL3–6 aptamers. (A) Tertiary structure diagram of Pepper bound to HBC530, with the magnesium ions (MA, MB) labeled. (B) Diagram and cartoon of the J1/2 Mg2+ (MA) with one inner-sphere contact to A11-PO. (C) Diagram and cartoon of the binding site Mg2+ (MB) with three inner-sphere contacts to G54−N7 and the C52 and U53 phosphates. The G54−C14 interaction is also highlighted. (D) 2 M NaCl-background fluorescence isotherm showing how fluorescence of Pepper-BL3–6 changes over [Mg2+], with nHill =1.14 ± 0.10 for binding HBC530 and nHill = 1.61 ± 0.09 for binding HBC599. With the saturated ion atmosphere from 2 M NaCl, Mg2+ binding is limited to non-monovalent binding sites on the aptamer. (E) Fluorescence of the split-Pepper system with phosphorothioate modifications to probe Mg−O inner-sphere interactions. 50 and 75% signals marked with dotted lines to gauge significance.
The dependence of Pepper on Mg2+ for folding and functions can reflect contributions to the folded RNA relative to the unfolded RNA from both site-bound metal ions and diffusely associated metal ions that form the ion atmosphere.32 To isolate the contribution of site-bound Mg2+ to Pepper functions, we saturated the ion atmosphere with high concentrations of monovalent cations (2 M NaCl) and conducted fluorescence measurements of samples with varying Mg2+ concentrations [conditions: see Methods 33 (Figure 4D)]. Fits of fluorescence versus Mg2+ yielded Hill coefficients (n) of 1.14 and 1.61 for the HBC530 and HBC599 ligands, respectively, indicating that at least one site-bound metal ion is required for each ligand. The different apparent Mg1/2 values for HBC599 ([Mg2+]1/2 = 6.83 mM) compared to HBC530 ([Mg2+]1/2 = 1.2 mM) suggest energetic coupling between Mg2+ and ligand binding. Below, we address this coupling further.
To further investigate how the structurally assigned Mg2+ contribute to Pepper functions, we incorporated phosphorothioate modifications into the aptamer to perturb inner- sphere Mg−O phosphate interactions (Figure 4E). To facilitate access to atomically perturbed Pepper constructs, we inferred a minimal motif from the crystal structure and designed a split RNA system that could be reconstituted from two oligoribonucleotides made by solid-phase synthesis (Figure S8). Incorporation of sulfur during solid-phase synthesis occurs stereorandomly, yielding RP and SP phosphorothioate diastereomers in roughly equal proportions. We tested the effect of sulfur substitution at each of the structurally inferred inner-sphere phosphate ligands to MA and MB. Phosphorothioate substitutions at positions C52 or U53 each diminished fluorescence by 35−45%, presumably reflecting a strong deleterious effect from the RP diastereomers, respectively. Substitution at C52 and U53 simultaneously, which yields four stereochemical configurations (RPRP, SPSP, RPSP, and SPRP), three of which would perturb an MB ligand, diminishes fluorescence even further (65%). Consistent with these results revealing the functional importance of the C52 and U53 phosphate oxygens, MB in our structures plays a critical role in organizing the J3/2 backbone and interacts directly with the out-of-plane nucleotide, G54. In contrast, phosphorothioate substitution at positions A11 had no effect on fluorescence, indicating that the aptamer can tolerate disruption of the MA phosphate interaction. Unlike MB, which makes three inner-sphere interactions with the RNA to directly organize the ligand binding site, MA makes only one inner- sphere interaction and resides more distal to the ligand binding site. Perhaps MA supports the backbone curvature of J1/2 through charge neutralization and does not require precise positioning. We also used our split system to probe MB and MC interactions with G54 and G25, respectively, using substitution with 7-deazaguanosine (n7cG). Incorporation of n7cG at G25 had no effect on fluorescence, but incorporation at G54 reduced fluorescence by about 60% (Figure S9), consistent with fluorescence activation being facilitated by the inferred MB metal interaction with G54. We assessed this interaction further by saturating the ion atmosphere (2 M NaCl) and monitoring the Mg2+ dependence of HBC530 fluorescence activation in the unmodified split system and the n7cG54- modified split system. We found that the modification shifted the fluorescence activation profile to higher Mg2+ concentrations, consistent with disruption of site-bound Mg2+ by n7cG54. We also tested the influence of the n7cG54 modification on HBC530 binding and observed a 10-fold reduction in affinity (HBC530 Kd = 44 ± 1.3 nM, vs 429 ± 101 nM; Figure S9), consistent with the structural coupling between MB and the ligand binding pocket via G54.
Mutational Analysis
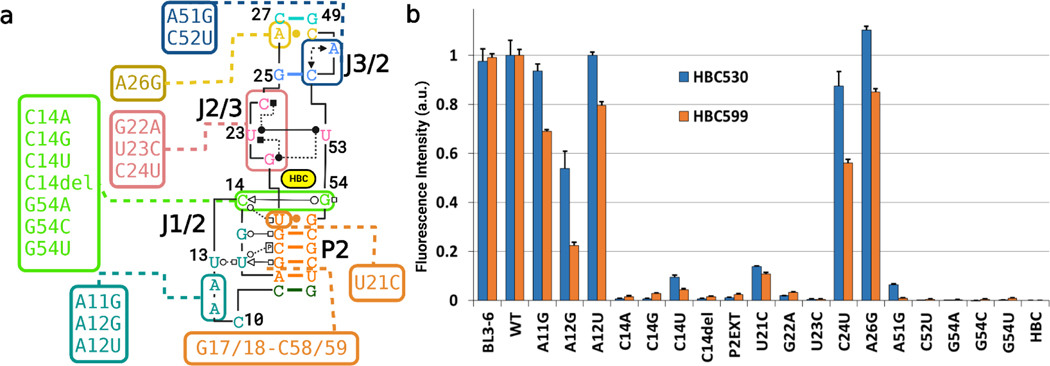
To probe the functional relevance of our structure for fluorogenic activity, mutational analysis was conducted (Figure 5). First, we tested mutations in several layers stacked above or below the ligand. The quadruple base layer was probed by separate mutations at G22 and U23. A G22A mutation reduced fluorescence to barely above back- ground with both HBC530 and HBC599, consistent with the importance of the hydrogen bonds mediated by the G22- exocyclic amine (Figure 1D). U23 has proximity to all other members of the tetrad and may serve as the keystone. A U23C mutation reduced fluorescence to almost background levels, potentially also due to disrupting the quadruple base platform for ligand binding. A C24U mutation mostly retained fluorescence, consistent with the structures implicating C24 to be more dynamic than the rest of the tetrad. In the G25− A51−C52 base triple above the tetrad, a C52U mutation, which breaks the canonical G25−C52 base pair at the heart of the triplex, abolishes fluorescence, supporting the importance of the WCF base pair in this triplex. A51G, which loses fluorescence, indicates that the interaction between A51 and C52 is also critical. The non-canonical A−C base pair at the transition between the bulge and P3 tolerates conversion to a canonical base pair through an A26G mutation with no change in signal. In the layer below the ligand, U21C causes a significant drop in signal. Presumably, this mutation converts the U21−G55 wobble pair to a CG WCF base pair, which could disrupt the hydrogen bond between the C14’s 2′-OH and U21’s O4 (Figure 1F).
Figure 5.
Assessment of base contributions to Pepper’s fluorogenic activity via mutational analysis with HBC530 (blue) and HBC599 (orange). Pepper-BL3–6 was incubated with an HBC ligand for 30 min at 25 °C. Measurements were made with 100 nM RNA, 1 μM HBC, 40 mM HEPES pH 7.5, 100 mM KCl, and 5 mM MgCl2.
Next, we tested the key nucleotides that sit out of plane to the ligand and form the back of the binding site—C14, and G54 (Figure 1E). G54 mutations (G54A, G54C, and G54U) all caused a complete loss of fluorescent signal, presumably by disrupting the interactions with C14 or MB, indicating that this residue makes a vital contribution to Pepper’s fluorogenic activity. Although G54A should be able to maintain the inner- sphere contact with the MB through N7 and a favorable interaction with C14’s 2′-OH via N6, the interactions with C14’s O2 keto group would be disrupted. Deletion of C14 (C14del) and transversion mutations (C14A, C14G) abolished the signal. However, the transition mutation, C14U, maintained around 10% WT fluorescence from HBC530 but abolished the signal from HBC599. P2ext—where an extra base pair was inserted into P2—increasing the distance helical turn between J1/2 and the binding pocket, which also reduced the signal to background.
Finally, we constructed mutations to further probe the Mg2+ binding site in J1/2. As noted above, MA forms one inner- sphere contact through the pro-RP oxygen of A11 and several outer sphere interactions to the Hoogsteen faces of A11 and A12. The first mutation—A11G—maintained fluorescence with HBC but led to a 30% loss with HBC599. A12U also maintained a high fluorescent signal, despite most likely changing the outer-sphere arrangements of MA significantly. A12G maintained a weaker signal, indicating that the structure of J1/2 was affected but not entirely disrupted.
Discussion
Using chaperone-assisted RNA crystallography, we successfully crystallized Pepper in complex with Fab BL3–6 and HBC530 or HBC599. Pepper binds the HBC ligand similar to other fluorogenic aptamers; the ligand is sandwiched between a base quadruple and a U•G wobble base pair with a supporting out- of-plane base system that completes the pocket (Scheme 1). A symmetric bulge including J2/3 and J3/2 harbors the stacking nucleobases, and the out-of-plane base system comprises a tertiary interaction between G54 from the J3/2 side of the symmetric bulge and C14 from the upstream asymmetric bulge, J1/2. The Mg2+ dependence of Pepper fluorescence in the context of a saturating ion atmosphere suggests a minimal requirement for two site-bound metal ions. MA and MB are present in both our crystal structures; MB coordinates directly to the out-of-plane base, G54, via N7, and MA indirectly positions the other out-of-plane base, C14, by supporting the conformation of J1/2. However, only MB exhibited sensitivity to phosphorothioate substitution. Mutations to other features of the motif including the base-quadruple, wobble pair, out-of-plane bases severely impaired activity, consistent with the functional relevance of the structure. Although the resolution of our structures clearly defines the location of the ligand, the electron density and surrounding binding site can accommodate multiple orientations so long as the alkene-nitrile points away from the C14•G54 face of the pocket for steric reasons.
Scheme 1. Synthesis of HBC Ligandsa.
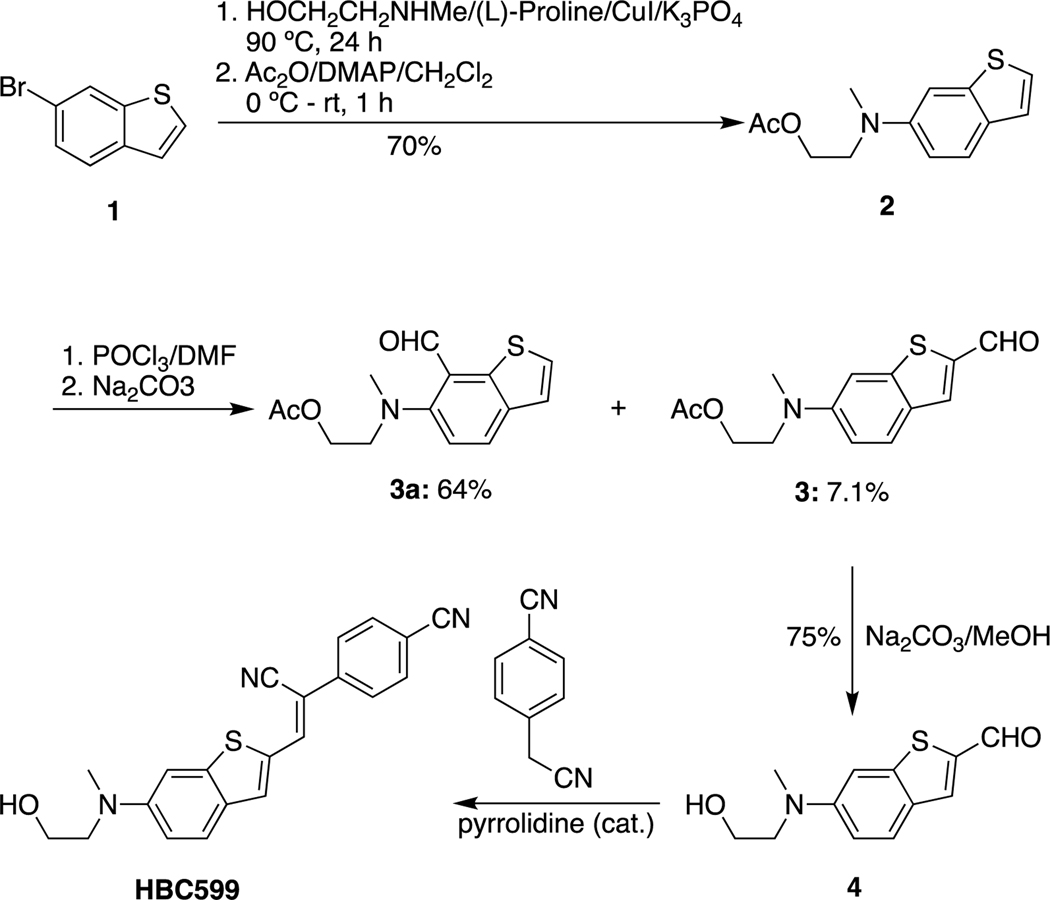
aHBC530 was prepared according to the reported procedure.11 HBC599 was also synthesized according to the reported procedure.1 However, in contrast to the literature report, the reaction of 2 produced 3 as the minor product rather than the major product (Scheme 1). The details for the step from 2 to 3 and the NMRs of the intermediates are provided in the methods section.
After we solved the structures of Pepper bound to ligands HBC530 and HBC599, respectively, Huang et al. reported the X-ray crystal structure of a circularly permuted (cp) Pepper construct bound to HBC530 and several other ligands,12 although not with HBC599. The overall structure reported is highly similar to our structures (Figures S10 and S11), despite different space groups and lattice packing interactions, supporting the functional relevance of the observed 3D architecture. The main differences include the angle subtended by P2 and P3, which is a few degrees more acute in the cp structure, and the number of metal ions visible. The cp structure contains five Mg2+ ions (M1−M5). M1 and M3 correspond directly to MA and MB in our structures. Huang et al. further confirmed the assignment of M3 as a divalent metal ion using the collected anomalous signal from Mn2+. M2, M4, and M5 have no counterpart in our structures, and MC (Na+) has no counterpart in the cp structure. M4 and M5 in the cp structure reside distal to the ligand binding site within the P1 helix, but the corresponding helix in our structure (P3) contains an entirely different sequence identical to the original aptamer.11 M2 binds in J1/2 along with M1/MA and similarly makes one inner-sphere interaction with the U13 pro-RP phosphate. Like M1/MA, M2 tolerates phosphorothioate substitution at U13 (see Figure 4E).
Most likely the modest differences between the structures reflect distinct lattice interactions (Figure S12). The cp RNA crystal lattice buries significantly more of the total RNA surface area compared to the Fab-RNA crystal lattice (8.6% vs 2.6%). In the cpRNA crystals, all lattice contacts involve RNA−RNA interactions, burying 732.9 Å2 of RNA surface area, whereas in the Fab-RNA crystals, only 18% of the lattice contacts involve RNA−RNA interactions, burying 286.6 Å2 of RNA surface area. These distinct lattices will strongly influence the electrostatic environment due to RNA’s negative charge, possibly accounting for the differences in metal ion occupancy between the crystallized Pepper constructs.
All known fluorogenic aptamers function by folding to create a binding pocket for organic chromophores bearing planar aromatic π-systems. The binding pockets consist of planar nucleobase layers that sandwich the ligand through stacking interactions and in most cases one or more out-of-plane nucleobases that wall off one side of the binding pocket (Table S1). The out-of-plane system can involve up to three nucleotides, as seen in the malachite green aptamer34 and Spinach.18,25 Various combinations of nucleobases comprise the two stacking layers and fluorogenic aptamers fall broadly into two categories. Those that use a G-quadruplex fold to form one of the stacking layers include Spinach,18,25 Mango,26,35,36 Chili,27 and Corn,27 and those that do not, include the malachite green aptamer,34 DIR2,34 Squash,34 and Pepper.34 The latter category of aptamers uses a base triple or base quadruple. The crystal structures of the G-quadruplex class all contain bound K+ to support the quadruplex fold. Within the non-G-quadruplex class, the crystal structures of Squash and Pepper contain site-bound Mg2+. This leaves DIR2 and malachite green as the only aptamers with crystal structures lacking a bound metal ion (Table S2).
Salt dependencies of fluorogenic aptamers are important as this determines the range of environments in which an aptamer can function. For example, the average Mg2+ concentrations across eukaryotic cells are approximately 0.5 mM,37 but mitochondrial environments tend to be much higher (above 10 mM).38 Pepper’s reliance on Mg2+ is not unique among fluorogenic RNAs; Squash also requires Mg2+,39 and even solely K+-reliant G-quadruplex-containing aptamers perform significantly better with divalent ions present. Of the four non- G-quadruplex aptamers, the malachite green and DIR2 aptamers are able to function without divalent ions,34 which correlates with their respective crystal structures lacking divalent ions in sites close to the aptamer core. Our work has made clear why Pepper has requirements for M2+ cations and cannot fluoresce with high monovalent concentrations alone.
Fluorogenic aptamers have served as sensors for small molecules through a rational engineering approach that couples the ligand binding capability of riboswitches to a fluorescence activation by the aptamer.40,41 Analogously, the structural coupling between the ligand binding pocket and the conformation of J1/2 observed here suggests how Pepper might serve as a scaffold for development of fluorogenic aptamers that can sense specific substrates. In vitro selection starting from a Pepper library containing the J1/2 sequence and length variations could give rise to aptamers that couple small-molecule-induced structuring of J1/2 to the formation of the chromophore binding pocket and fluorescence. Current examples of aptazymes and synthetic allosteric sensors all rely on communication modules—dsRNA regions that hybridize upon a riboswitch binding its target.42 However, as J1/2 is a short ssRNA region that binds M2+ and is integral to Pepper’s function, it could serve as a new type of communication module where the trigger is with J1/2 forming the interaction with the HBC binding site rather than simple hybridization.
Fab chaperones have been used previously to assist with crystal packing of RNAs and have successfully facilitated the crystallization of Pepper here. The lattice is composed of two sets of RNA-Fab polymers bonding through an intermolecular base pair (G1•C10; Figure S5). Interactions with Fab accounted for about 80% of the surface buried through lattice interactions; Fab−Fab and Fab−RNA contacts accounted for 2.7% (35.8 Å2) and 77% (1020.1 Å2), respectively. The RNA− RNA interface constituted 20.3% (268.6 Å2) of the crystal lattice interface, entirely though the G1•C10 intermolecular interaction (Figure S5). Compared to previous examples of RNA-Fab complexes, Pepper is very similar to the structure of BL3 Fab bound to the class I ligase ribozyme,43 where 65% of crystal contacts were Fab-RNA, and more generally, Fab-BL3–6 has tended to interface mostly with the RNA, mitigating the need for RNA−RNA contacts.
Methods
Pepper RNA Synthesis and Purification.
The dsDNA template used to transcribe the RNA was prepared via PCR of a single-stranded DNA template purchased from integrated DNA technologies (www.idtdna.com). The first two nucleotides of the reverse primer were modified with 2′-OMe to reduce transcriptional heterogeneity at the 3′ end.44 RNA was prepared via in vitro transcription for 3.5 h at 37 °C in a buffer composed of 40 mM Tris−HCl pH 7.9, 10 mM NaCl, 25 mM MgCl2, 2 mM Spermidine, 10 mM DTT, 30 U/mL RNase inhibitor, 2.5 U/mL TIPPase, 4 mM of each nucleotide tri-phosphate, 8 ng/uL of the DNA template, and 40 μg/mL of in-house-prepared T7 RNA polymerase. To remove the DNA template from the reaction, 10 U/mL RNase-free DNase I (www.promega.com) was added and the reaction incubated for a further 30 min at 37 °C. After the phenol/chloroform pH 4.3 extraction to remove proteins, the RNA was purified via denaturing polyacrylamide gel electrophoresis. The RNA band of interest was quickly visualized via UV shadowing and excised from the gel. Using the mash-and-soak method, the RNA was collected, aliquoted into small fractions, and stored at −80 °C until further use.
For split-Pepper experiments incorporating non-natural nucleotides (phosphorothioates, 7-deazaguanosines), oligonucleotides were synthesized in-house by solid-phase synthesis on a 1 μmol scale using an Expedite nucleic acid synthesis system (8900) by following standard RNA synthesis protocols. The oligonucleotides were released from the solid support with 3:1 NH4OH/EtOH at 55 °C for 8 h, desilylated with 300 μL 6:3:4 N-methylpyrrolidinone/triethylamine/ triethylamine-3HF at 65 °C for 2 h, and precipitated by n-BuOH. The oligomers were further purified by dPAGE, collected in pure water, and stored at −80 °C until further use.
Crystallization of the Pepper-BL3–6-Fab Complex.
The RNA sample was refolded in a buffer containing 10 mM Tris pH 7.5, 100 mM KCl, and 5 mM MgCl2 (supplied as 10×). For refolding, RNA was heated at 90 °C for 1 min in deionized water and then cooled on ice for 2 min before folding buffer and fluorogen dye was added. This was then followed by incubation at room temperature for 30 min. The refolded RNA was then introduced to 1.1 equiv of the BL3–6 Fab [expressed as soluble protein in phagemid as an expression vector and purified by affinity and ion-exchange chromatography using protein A, G, and heparin columns (GE Healthcare), respectively43 at room temperature for 30 min and concentrated to 3 mg/mL using a 10 kDa cutoff, Amicon Ultra-15 column. The formation of the Fab-RNA complex was confirmed by native poly-acrylamide gel electrophoresis. To prevent excess nucleation events, RNA was then passed through 0.2 μm cutoff, Millipore centrifugal filter units. A mosquito liquid handling robot (STP Labtech) was used to set up high-throughput hanging drop vapor diffusion crystallization screens at room temperature using commercially available screening kits from Hampton Research and Jena Bioscience. After additive screening with diffracting conditions, the best-diffracting crystals of the Pepper- BL3–6-Fab-HBC530 complex were obtained in a condition from the Natrix screen: 0.02 M magnesium sulfate hydrate, 0.002 M cobalt (II) chloride hexahydrate, 0.05 M sodium cacodylate trihydrate pH 6.0, 0.0005 M spermine, and 4% v/v 2,5-hexanediol. For the complex with the HBC599 fluorophore, the optimal condition was from the index screen: 0.2 M ammonium acetate, 0.1 M BIS-TRIS pH 6.5, and 45% v/v (+/−)-2-methyl-2,4-pentanediol. Crystals appeared and grew to full size within 2 days in 100 + 100 nL hanging drops. The crystals were looped without a cryo-protectant before being flash-frozen in liquid nitrogen. Diffraction data sets were collected at 80 K at the Advanced Photon Source NE-CAT section beamline 24-ID-C and 24- ID-E at Argonne National Lab.
Mg Stoichiometry Measurements.
Hill isotherm measurements were conducted using a Neo2 Synergy BioTek multi-mode plate reader. Ion-atmosphere saturation was achieved by using a 2 M NaCl, 40 mM HEPES (pH 7.5) buffer, and the Hill coefficient was calculated as previously described.29 RNA was heated to 95 °C for 1 min in water and cooled on ice for 2 min before the buffer [1×; 40 mM HEPES (pH 7.5), 2 M NaCl] was added as a 2× stock and various concentrations of Mg2+ (starting from 0.1 to 25 mM for HBC530 and starting from 0.1 to 50 mM for HBC599) were added as a 10× stocks. Then, the RNA in the buffer was incubated for 15 min at 50 °C before adding the ligand chromophore (1×; 5 μM for HBC530, 25 μM for HBC599) as a 10× stock. Finally, diluted RNA (1×; 100 nM) was then incubated in the buffer with the ligand chromophore for 30 min at 25 °C before measurements were taken (Ex/Em 485/530 for HBC530, 515/599 for HBC599, gain 100 ± 20) at 25 °C, and the reported results were the average of three measurements. The intensity data were fit initially to a four-parameter Hill equation as described in the Supporting Information (eq S1) and were subsequently normalized based on these fits. We then took the average of all the protection data at each divalent metal-ion concentration and fit the resulting points to the Hill equation to produce the final curves shown in Figures 4 and S9. For plots of fluorescence versus ligand or Mg2+ concentration in the split system, we used eq S1 to fit the data.
Split System Fluorescence Assay.
Fluorescence measurements were conducted using a Neo2 Synergy BioTek multi-mode plate reader. RNA was heated to 95 °C for 1 min in water and cooled on ice for 2 min before the buffer [1×; 40 mM HEPES (pH 7.5), 100 mM KCl, 5 mM MgCl2] and ligand chromophore (1×; 1 μM) were added as a 10× stocks. Then, the RNA (1×; 100 nM) was incubated in the buffer with the ligand chromophore for 15 min at 50 °C and for additional 30 min at 25 °C before measurements were taken (Ex/Em 485/530 for HBC530, gain 100 ± 20) at 25 °C, and the reported results were the average of three measurements.
Split System Binding Isotherm Assay.
Binding isotherm measurements were conducted using a Neo2 Synergy BioTek multi-mode plate reader. RNA was heated to 95 °C for 1 min in water and cooled on ice for 2 min before the buffer [1×; 40 mM HEPES (pH 7.5), 100 mM KCl, 1 mM MgCl2] and ligand chromophore (1×; 40 nM) were added as a 10× stocks. Then, the various concentrations of RNA (starting from 0.001 to 1 μM) in the buffer with the ligand chromophore were incubated for 15 min at 50 °C and for additional 30 min at 25 °C before measurements were taken (Ex/Em 485/530 for HBC530, gain 100 ± 20) at 25 °C, and the reported results were the average of three measurements.
HBC derivative synthesis.
Compounds 3 and 3a: Phosphorous oxychloride (0.11 mL, 1.16 mmol) was added dropwise to a stirred 0 °C solution of compound 4 (240 g, 0.96 mmol) in dry CH2Cl2 (10 mL) and dry DMF (1 mL) under the protection of Ar. The resulting mixture was warmed to room temperature and stirred for an additional 5 h. The mixture was quenched with a saturated solution of sodium carbonate and extracted with CH2Cl2. The organic layer was dried over Na2SO4; then the solvent was removed under reduced pressure to give the crude product which was purified by silica gel column chromatography, eluting with 15−25% ethyl acetate in hexane to yield compound 3a (0.171 g, 64%) [top spot on thin-layer chromatography (TLC)] and compound 3 (19 mg, 7.1%) (low spot on TLC). Compound 3 is more visible on TLC with long-wave UV than with short-wave UV.
Compound 3: 1H NMR (400 MHz, CDCl3): δ 9.88 (s, 1H), 7.78 (s, 1H), 7.67 (d, J = 9.0 Hz, 1H), 7.19 (s, 1H), 6.98 (d, J = 2.4 Hz,1H), 6.87 (dd, J = 9.0, 2.4 Hz, 1H), 4.23 (t, J = 6.0 Hz, 2H), 3.64 (t, J = 6.0 Hz, 2H), 3.03 (s, 3H), 1.94 (s, 3H).
Compound 3a: 1H NMR (400 MHz, CDCl3): δ 10.50 (s, 1H), 7.83 (d, J = 8.6 Hz, 1H), 7.35 (d, J = 5.5 Hz, 1H), 7.17 (d, J = 5.5 Hz, 1H), 7.14 (d, J = 8.6 Hz, 1H), 4.15 (t, J = 5.7 Hz, 2H), 3.37 (t, J = 5.7 Hz, 2H), 2.90 (s, 3H), 1.85 (s, 3H); 13C NMR (101 MHz, CDCl3): δ 190.25, 170.77, 155.36, 139.06, 136.28, 129.89, 128.69, 123.05, 122.45, 117.68, 61.44, 57.58, 43.70, 20.79.
Supplementary Material
Acknowledgements
We thank the staff of the Advanced Photon Source (APS) at Argonne National Laboratory for providing advice and assistance on X-ray data collection. We thank Anna Lewicka for expression and purification of Fab BL3-6. We thank Christina Roman and Phoebe Rice for useful discussions and guidance in solving the structures and members of the Piccirilli laboratory and the reviewers for constructive comments on the manuscript. Figures were made using BioRender, and figures that show 3D models of structures were made with Pymol (The PyMOL Molecular Graphics System, Version 4.3 Schrödinger, LLC.). This work was supported by a grant from the US National Institute of Health to J.A.P. (GM102489).
Footnotes
Supplementary Information
Complete secondary structure diagrams and data from supporting assays; crystal lattice packing, effect of MC on C24, comparisons between our Pepper structure and the recently published structure of the circularly permuted Pepper construct, and Pepper in the context of the currently known fluorogenic aptamers.
References
- (1).Grate D; Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 6131–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Paige JS; Wu KY; Jaffrey SR RNA Mimics of Green Fluorescent Protein. Science 2011, 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Filonov GS; Moon JD; Svensen N; Jaffrey SR Broccoli Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014, 136, 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dolgosheina EV; Jeng SCY; Panchapakesan SSS; Cojocaru R; Chen PSK; Wilson PD; Hawkins N; Wiggins PA; Unrau PJ RNA Mango aptamer-fluorophore: A bright, high- affinity complex for RNA labeling and tracking. ACS Chem. Biol 2014, 9, 2412–2420. [DOI] [PubMed] [Google Scholar]
- (5).Jeng SCY; Trachman RJ; Weissenboeck F; Truong L; Link KA; Jepsen MDE; Knutson JR; Andersen ES; Ferré- D’Amaré AR; Unrau PJ Fluorogenic aptamers resolve the flexibility of RNA junctions using orientation-dependent FRET. RNA 2021, 27, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Warner KD; Sjeklocá L; Song W; Filonov GS; Jaffrey SR; Ferré-D’Amaré AR. A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat. Chem. Biol. 2017, 13, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Steinmetzger C; Palanisamy N; Gore KR; Höbartner C. A Multicolor Large Stokes Shift Fluorogen-Activating RNA Aptamer with Cationic Chromophores. Chem.—Eur. J. 2019, 25, 1931–1935. [DOI] [PubMed] [Google Scholar]
- (8).Dao NT; Haselsberger R; Khuc MT; Phan AT; Voityuk AA; Michel-Beyerle ME Photophysics of DFHBI bound to RNA aptamer Baby Spinach. Sci. Rep. 2021, 11, 7356–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Santra K; Geraskin I; Nilsen-Hamilton M; Kraus GA; Petrich JW Characterization of the Photophysical Behavior of DFHBI Derivatives: Fluorogenic Molecules that Illuminate the Spinach RNA Aptamer. J. Phys. Chem. B 2019, 123, 2536–2545. [DOI] [PubMed] [Google Scholar]
- (10).Truong L; Ferré-D’Amaré AR From fluorescent proteins to fluorogenic RNAs: Tools for imaging cellular macromolecules. Protein Sci. 2019, 28, 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chen X; Zhang D; Su N; Bao B; Xie X; Zuo F; Yang L; Wang H; Jiang L; Lin Q; et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 2019, 37, 1287–1293. [DOI] [PubMed] [Google Scholar]
- (12).Huang K; Chen X; Li C; Song Q; Li H; Zhu L; Yang Y; Ren A. Structure-based investigation of fluorogenic Pepper aptamer. Nat. Chem. Biol. 2021, 17, 1289–1295. [DOI] [PubMed] [Google Scholar]
- (13).Koldobskaya Y; Duguid EM; Shechner DM; Suslov NB; Ye J; Sidhu SS; Bartel DP; Koide S; Kossiakoff AA; Piccirilli JA A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 2011, 18, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Koirala D; Lewicka A; Koldobskaya Y; Huang H; Piccirilli JA Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation. ACS Chem. Biol. 2020, 15, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Krochmal D; Shao Y; Li N-S; DasGupta S; Shelke SA; Koirala D; Piccirilli JA Structural basis for substrate binding and catalysis by a self-alkylating ribozyme. Nat. Chem. Biol. 2022, 18, 376. [DOI] [PubMed] [Google Scholar]
- (16).Shao Y; Huang H; Qin D; Li N-S; Koide A; Staley JP; Koide S; Kossiakoff AA; Piccirilli JA Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment. J. Mol. Biol. 2016, 428, 4100–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Koirala D; Shelke SA; Dupont M; Ruiz S; DasGupta S; Bailey LJ; Benner SA; Piccirilli JA Affinity maturation of a portable Fab−RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res. 2018, 46, 2624–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Huang H; Suslov NB; Li N-S; Shelke SA; Evans ME; Koldobskaya Y; Rice PA; Piccirilli JA A G-quadruplex- containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Auffinger P; Grover N; Westhof E. Metal ion binding to RNA. Met. Ions Life Sci. 2011, 9, 1. [PubMed] [Google Scholar]
- (20).Leonarski F; D’Ascenzo L; Auffinger P. Mg2+ ions: Do they bind to nucleobase nitrogens? Nucleic Acids Res. 2017, 45, 987–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shelke SA; Shao Y; Laski A; Koirala D; Weissman BP; Fuller JR; Tan X; Constantin TP; Waggoner AS; Bruchez MP; et al. Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat. Commun. 2018, 9, 4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Koirala D; Shelke SA; Dupont M; Ruiz S; DasGupta S; Bailey LJ; Benner SA; Piccirilli JA Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res. 2018, 46, 2624–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Roman C; Lewicka A; Koirala D; Li N-S; Piccirilli JA The SARS-CoV-2 Programmed −1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 Å Using Chaperone-Assisted RNA Crystallography. ACS Chem. Biol. 2021, 16, 1469–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Warner KD; Chen MC; Song W; Strack RL; Thorn A; Jaffrey SR; Ferré-D’Amaré AR Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014, 21, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Trachman RJ III; Demeshkina NA; Lau MWL; Panchapakesan SSS; Unrau PJ; Ferré-D’Amaré AR Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Physiol. Behav. 2017, 13, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mieczkowski M; Steinmetzger C; Bessi I; Lenz AK; Schmiedel A; Holzapfel M; Lambert C; Pena V; Höbartner C. Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine. Nat. Commun. 2021, 12, 3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mir A; Golden BL Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry 2016, 55, 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lee T-S; López CS; Giambasu̧ GM; Martick M; Scott WG; York DM Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J. Am. Chem. Soc. 2008, 130, 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ganguly A; Weissman BP; Giese TJ; Li N-S; Hoshika S; Rao S; Benner SA; Piccirilli JA; York DM Confluence of theory and experiment reveals the catalytic mechanism of the Varkud satellite ribozyme. Nat. Chem. 2020, 12, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ren A; Vusǔrovic N; Gebetsberger J; Gao P; Juen M; Kreutz C; Micura R; Patel DJ. Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage. Nat. Chem. Biol. 2016, 12, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Frederiksen JK; Li N-S; Das R; Herschlag D; Piccirilli JA Metal-ion rescue revisited: Biochemical detection of site-bound metal ions important for RNA folding. RNA 2012, 18, 1123–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Das R; Travers KJ; Bai Y; Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J. Am. Chem. Soc. 2005, 127, 8272–8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Baugh C; Grate D; Wilson C. 2.8 Å Crystal structure of the malachite green aptamer. J. Mol. Biol. 2000, 301, 117–128. [DOI] [PubMed] [Google Scholar]
- (35).Trachman RJ; Cojocaru R; Wu D; Piszczek G; Ryckelynck M; Unrau PJ; Ferré-D’Amaré AR Structure-Guided Engineering of the Homodimeric Mango-IV Fluorescence Turn-on Aptamer Yields an RNA FRET Pair. Structure 2020, 28, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Trachman RJ; Autour A; Jeng SCY; Abdolahzadeh A; Andreoni A; Cojocaru R; Garipov R; Dolgosheina EV; Knutson JR; Ryckelynck M; et al. Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nat. Chem. Biol. 2019, 15, 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Goytain A; Quamme GA Identification and characterization of a novel family of membrane magnesium transporters, MMgT1 and MMgT2. Am. J. Physiol. 2008, 294, 495–502. [DOI] [PubMed] [Google Scholar]
- (38).Pilchova I; Klacanova K; Tatarkova Z; Kaplan P; Racay P. The Involvement of Mg2+ in Regulation of Cellular and Mitochondrial Functions. Oxid. Med. Cell. Longevity 2017, 2017, 6797460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Dey SK; Filonov GS; Olarerin-George AO; Jackson BT; Finley LWS; Jaffrey SR Repurposing an adenine riboswitch into a fluorogenic imaging and sensing tag. Nat. Chem. Biol. 2022, 18, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Su Y; Hammond MC RNA-based fluorescent biosensors for live cell imaging of small molecules and RNAs. Curr. Opin. Biotechnol. 2020, 63, 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Truong J; Hsieh Y-F; Truong L; Jia G; Hammond MC Designing fluorescent biosensors using circular permutations of riboswitches. Methods 2018, 143, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhong G; Wang H; Bailey CC; Gao G; Farzan M. Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. eLife 2016, 5, No. e18858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Koldobskaya Y; Duguid EM; Shechner DM; Suslov NB; Ye J; Sidhu SS; Bartel DP; Koide S; Kossiakoff AA; Piccirilli JA A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 2011, 18, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kao C; Rüdisser S; Zheng M. A simple and efficient method to transcribe RNAs with reduced 3′ heterogeneity. Methods 2001, 23, 201–205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.