Abstract
Purpose
The global protein profile of the aqueous humor has been found to correlate with the severity of retinal vascular disease. Studying the aqueous humor in central retinal vein occlusion (CRVO) with proteomic techniques may bring insights to the molecular mechanisms underlying the condition.
Methods
Aqueous humor samples from treatment naïve patients with CRVO complicated by macular edema (n = 28) and age-matched controls (n = 20) were analyzed by label-free quantification liquid chromatography - tandem mass spectrometry. Best corrected visual acuity (BCVA) was measured as logMAR, and the severity of macular edema was evaluated as central retinal thickness (CRT) with optical coherence tomography. Control samples were obtained prior to cataract surgery. Significantly changed proteins were identified by a permutation-based calculation with a false discovery rate of 0.05.
Results
A total of 177 proteins were differentially expressed in CRVO. Regulated proteins were involved in complement activation, innate immune response, blood coagulation, and cell adhesion. Upregulated proteins that correlated with BCVA and CRT included fibrinogen alpha, beta, and gamma chains, fibronectin, Ig lambda-6 chain C region, Ig alpha-1 chain C region, and complement C7. Downregulated proteins that correlated negatively with BCVA, and CRT, included procollagen C-endopeptidase enhancer 1, clusterin, opticin, reelin, fibrillin-1, and cadherin-2. Monocyte differentiation antigen CD14 and lipopolysaccharide-binding protein were increased in CRVO.
Conclusions
Fibrinogen chains, fibronectin, and immunoglobulin components correlated with BCVA and CRT, suggesting a multifactorial response. Protective anti-angiogenic proteins, including procollagen C-endopeptidase enhancer 1, clusterin, and opticin, were downregulated in CRVO and correlated negatively with BCVA and CRT.
Keywords: retina, retinal vasculature, mass spectrometry, proteomics, aqueous humor
Central retinal vein occlusion (CRVO) is a visually disabling condition caused by a thrombus of the central retinal vein, which is the major outflow vessel of the eye.1,2 Macular edema is the most common cause of vision loss in CRVO3 and visual acuity following CRVO generally remains below 20/40, unless treatment is initiated.4 CRVO results in increased resistance to blood flow in retinal arterioles leading to closure of retinal capillaries and small arterioles. Retinal hypoxia resulting from vascular occlusion drives increased production of vascular endothelial growth factor A (VEGF-A), and an inflammatory response mediated by interleukin (IL)-6, IL-8, and monocyte chemotactic protein-1. VEGF-A and the inflammatory response increase vascular permeability thereby giving rise to macular edema.3,5
Intravitreal VEGF-neutralizing agents are the first-line therapy for patients with macular edema secondary to CRVO. Dexamethasone intravitreal implants, which are used as second-line treatment, effectively downregulate the inflammatory driving force in macular edema.3,6–8 Despite advances in the treatment of CRVO, management of the condition has several challenges. Approximately 45% of patients with macular edema due to CRVO need anti-VEGF therapy for more than 4 years.9 Reports on real-world data indicate that 28.1% of eyes do not achieve resolution of macular edema.10 A suboptimal response to anti-VEGF neutralization may be observed, because several permeability factors other than VEGF-A contribute to the formation of macular edema.5,11,12
The objective of a proteome analysis is to identify and quantify the entire set of proteins in a given body fluid or tissue.12,13 We previously showed that the aqueous humor proteome reflects the severity of retinal vascular disease.14 To the best of our knowledge, the aqueous humor proteome in CRVO has never been studied.12 Studying the aqueous humor from patients with CRVO may generate important knowledge about mechanisms that contribute to visual loss, formation of macular edema, and resistance to anti-VEGF therapy. Optical coherence tomography (OCT) continues to improve the diagnostic workup and management of retinal diseases.15,16 Correlating the proteome of CRVO to OCT features has the potential to bring novel insights to the pathogenesis of macular edema in retinal vascular disease. Here, we report on a proteomic analysis of aqueous humor samples from 28 treatment-naïve patients with CRVO complicated by macular edema, which were compared to samples from an age-matched control group.
Methods
Samples
The study was conducted in compliance with the Institutional Review Board of Kyoto Prefectural University of Medicine which approved the study (permission RBMR-C-864-6). The study adhered to the tenets of the Helsinki Declaration. Aqueous humor samples from treatment-naïve patients with CRVO complicated by macular edema with onset within 3 months (n = 28) and age-matched controls (n = 20) were donated from the biobank of Kyoto Prefectural University of Medicine, Kyoto, Japan (Table 1). Informed consent to use samples from the biobank was obtained from all patients after explaining the nature and possible consequences of the study. There were no statistically significant differences in age between the two groups as verified by Student's t-test (see Table 1). In the CRVO group, the inclusion criteria were ≥20 years of age, symptom onset of visual disturbance within 3 months, and macular edema >300 µm by OCT. Exclusion criteria in the CRVO group were iris rubeosis, hyphema, neovascular glaucoma, vitreous hemorrhage, retinal neovascularization, previous retinal photocoagulation, other retinal disease, or use of topical treatments within the last 3 months. Control samples were from age-matched patients from whom aqueous humor samples were obtained prior to cataract surgery. Patients in the control group had no ocular disease except for cataract. The data, including best corrected visual acuity (BCVA) were collected from the electronic charts of patients at Kyoto Prefectural University of Medicine. BCVA was measured using the Japanese standard Landolt visual acuity chart, and then converted to the logarithm of the minimum angle of resolution (logMAR). Swept source OCT was used (DRI-OCT Triton; Topcon, Tokyo, Japan). The severity of macular edema was measured as central retinal thickness (CRT), which was defined as the distance between the outer border of the hyper-reflective retinal pigment epithelium and the inner border of the internal limiting membrane at the center of the fovea measured using the caliper tool of the Topcon OCT software. The grader (author K.K.) was masked to the proteomics data and ELISA data. CRT was measured two times and the mean value was calculated. The intraclass correlation coefficient of the grader was 0.95. Fluorescein angiography (FA) was performed using a confocal scanning laser ophthalmoscope (Heidelberg Retina Angiograph 2; Heidelberg Engineering, Heidelberg, Germany) and the area of retinal non-perfusion was measured in optic disc areas using the “draw lesion” tool in Heidelberg Retinal Angiography 2.
Table 1.
Samples for Proteomic Analysis
| CRVO | Control | P Value | |
|---|---|---|---|
| Number of samples | 28 | 20 | |
| Age, y | 71.3 ± 15.9 | 75.3 ± 11.4 | 0.35 |
| Sex (M/F) | 17/11 | 13/7 | |
| Size of macular edema (µm) | 725 ± 281 | ||
| BCVA (logMAR) | 0.76 ± 0.53 | ||
| Patients with retinal area of non-perfusion ≤10 disc areas | 22 | ||
| Patients with retinal area of non-perfusion >10 disc areas | 6 |
Data are expressed as n or mean ± standard deviation.
Additional aqueous humor samples from patients with CRVO (n = 15) and control samples (n = 5) were obtained from the biobank for validation by enzyme-linked immunosorbent assay (ELISA; Table 2). CRVO samples and control samples were age-matched and selected according to the inclusion and exclusion criteria specified above. For samples obtained for ELISA, the Mann-Whitney U test was used to verify that there was no significant difference in age between the groups (see Table 2).
Table 2.
Samples for ELISA Validation
| CRVO | Control | P Value | |
|---|---|---|---|
| Number of samples | 15* | 5 | |
| Age, y | 70.9 ± 15.7 | 75.6 ± 11.3 | 0.46 |
| Size of macular edema (µm) | 766 ± 249 | ||
| BCVA (logMAR) | 0.87 ± 0.47 | ||
Fibrinogen alpha chain was quantified in all samples. Nine of the samples had sufficient material for quantification of VEGF.
Data are expressed as mean ± standard deviation unless otherwise noted.
Sample Preparation for Mass Spectrometry
Samples were stored at −80°C until preparation was initiated. Measurement of protein concentration and sample preparation according to the S-Trap Micro spin column digestion protocol from ProtiFi (Huntington, NY, USA) were performed as described previously,14 including the reduction of disulfide bonds, alkylation of cysteines, and tryptic digestion.14 The peptide concentration was measured as described previously.17 The samples were dried in a vacuum centrifuge and stored at −80°C until further use.
Quantitative Mass Spectrometry by Label-Free Quantification Nano Liquid Chromatography - Tandem Mass Spectrometry
Samples were re-suspended in 0.1% formic acid and analyzed by label-free quantification nano liquid chromatography – tandem mass spectrometry (LFQ nLC-MS/MS). For each sample, 1 µg was analyzed in replicates, except for one sample that was analyzed only once due to technical reasons. Mass spectrometry was performed on an Orbitrap Fusion Tribrid mass spectrometer equipped with an EasySpray ion source coupled to a Dionex UltiMate 3000 RSLC nano system (Thermo Fisher Scientific Instruments, Waltham, MA, USA). Liquid chromatography and label-free quantification (LFQ) were conducted as described previously.14 The samples were generally analyzed as technical duplicates run with several days of intermission. The sequence of samples run in the analysis was mixed, distributing the samples from each group throughout the whole sequence. Using MaxQuant software version 1.6.6.0 for LFQ analysis,18 raw data files were searched against the UniProt Homo sapiens database as described previously.19 Unfiltered results of the database search are provided in Supplementary File S1.
Mass spectrometry data were further processed with Perseus software20 (version 1.6.2.3). Removal of poorly identified proteins was performed in Perseus as described previously.21 The LFQ values were log2 transformed and mean LFQ values were calculated. For successful protein identification, at least two unique peptides were required. Proteins were required to be successfully identified and quantified in at least 70% of the samples in each of the 2 groups. For each technical duplicate sample analyzed, we calculated the median coefficient of variation of the analyzed proteins. The average of the median coefficient of variation among the analyzed samples was below 12%.
Statistics
Statistical analysis was performed using Student's t-test in Perseus to compare CRVO to controls. A subgroup analysis was performed with the Student's t-test to compare ischemic CRVO to non-ischemic CRVO. Correction for multiple hypothesis testing was performed using the permutation-based method in Perseus22 with the number of randomizations set to 250 and an S0 parameter of 0.1. The false-discovery rate (FDR) was set to 0.05.
Correlations were calculated in STATA version 16.0 (StataCorp, College Station, TX, USA) using Pearson's correlation coefficient (r). Correlations were considered significant if P < 0.05. Scatter plots with prediction from a linear regression were generated in STATA version 16.0.
Gene Ontology analysis of biological processes was performed in GeneCodis 4.023 software as described previously.24 Cluster analysis of significantly regulated proteins was performed with STRING 11.5 (string-db.org),25–27 as described previously,14 and the minimum required interaction score set to 0.90. Principal component analysis was performed in Perseus using default settings with imputation of missing values from the normal distribution.
Enzyme-Linked Immunosorbent Assay
Aqueous concentrations of fibrinogen alpha chain and VEGF were measured by ELISA using the SEB154Hu ELISA kit for Fibrinogen Alpha Chain (Cloud-Clone Corp., Wuhan, China) and the ab222510 Human VEGF SimpleStep ELISA Kit (Abcam, UK), respectively.
For quantification of fibrinogen alpha chain, the samples were diluted 1:8. Assay preparation was performed according to the manufacturer's instructions. A volume of 100 µL of standard or sample was added to the wells and incubated for 1 hours at 37°C. Wells were aspirated and 100 µL of Detection Reagent A added, followed by 1 hour incubation at 37°C. Each well was washed three times with wash buffer (1:20 Wash Buffer from ELISA kit, Cloud-Clone Corp. Wuhan, China, in deionized water). A volume of 100 µL of Detection Reagent B was added to each well, followed by incubation for 30 minutes at 37°C. Each well was washed five times with the wash buffer. A volume of 90 µL of Substrate Solution (provided in kit) was added to each well and the plate incubated for 20 minutes at 37°C. A volume of 50 µL of Stop Solution (provided in kit) was added to each well and read at an optical density of 450 nm. For quantification of VEGF, the samples were diluted 1:2, and quantification of VEGF was performed as described in a previous report.14 The Mann-Whitney U test performed in STATA version 16.0 was used to calculate differences in fibrinogen alpha chain and VEGF between CRVO and controls. Correlations were calculated as the Pearson's correlation coefficient (r) in STATA version 16.0.
Results
A total of 891 proteins were successfully identified in the combined set of aqueous samples (Supplementary File S2). In total, 255 aqueous humor proteins were successfully identified and quantified in at least 70% of the samples in each group (Supplementary File S3) and statistical analysis was performed on these proteins.
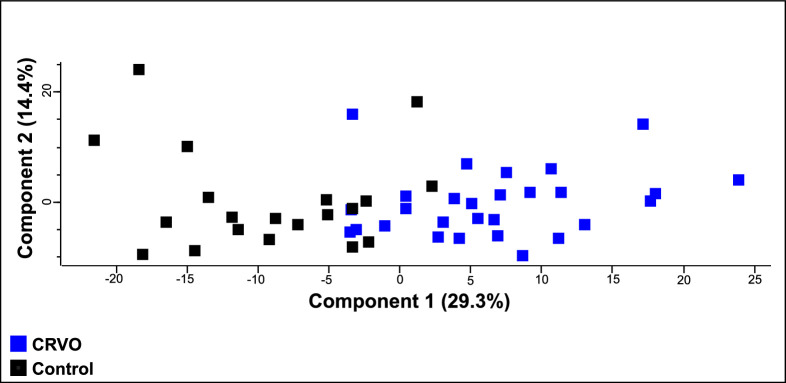
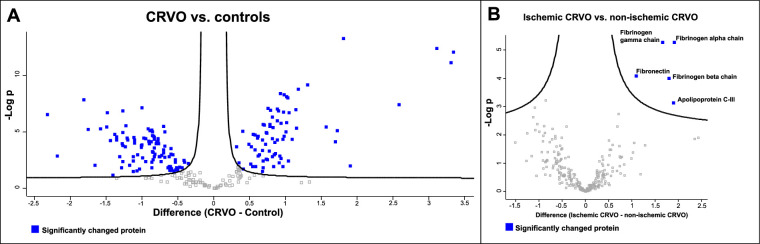
Samples from patients with CRVO could nearly be separated from control samples based on their proteomes (Fig. 1). After correction for multiple hypothesis testing, a total of 177 proteins were significantly regulated in CRVO compared to controls (Table 3, Fig. 2A). Among the significantly regulated proteins, 75 proteins were increased in CRVO, whereas 102 proteins were decreased in content (see Table 3). Five proteins were increased in ischemic CRVO versus non-ischemic CRVO (Fig. 2B), including apolipoprotein C-III (P = 0.00074), fibrinogen alpha chain (P = 5.45 × 10−6), fibrinogen beta chain (P = 0.0001), fibrinogen gamma chain (P = 5.32 × 10−6), and fibronectin (P = 8.35 × 10−5).
Figure 1.
Principal component analysis (PCA). The PCA suggested that samples from patients with CRVO could nearly be separated from control samples based on their proteomes.
Table 3.
Significantly Regulated Proteins in CRVO versus Controls
| Protein ID | Protein Names | Gene Names | P Value | Fold Change CRVO/Control |
|---|---|---|---|---|
| P02675 | Fibrinogen beta chain | FGB | 8.28 × 10−13 | 10.20 |
| P02671 | Fibrinogen alpha chain | FGA | 6.56 × 10−12 | 9.96 |
| P02679-2 | Fibrinogen gamma chain | FGG | 3.74 × 10−13 | 8.68 |
| P02656 | Apolipoprotein C-III | APOC3 | 3.55 × 10−8 | 6.00 |
| P01871 | Ig mu chain C region | IGHM | 0.010 | 3.74 |
| P02751-1 | Fibronectin | FN1 | 4.96 × 10−14 | 3.52 |
| P06312 | Ig kappa chain V-IV region | IGKV4-1 | 7.36 × 10−6 | 3.31 |
| P04432 | Ig kappa chain V-I region Daudi | IGKV1-12 | 7.01 × 10−5 | 3.25 |
| P01876 | Ig alpha-1 chain C region | IGHA1 | 3.60 × 10−6 | 2.97 |
| P01031 | Complement C5 | C5 | 6.59 × 10−10 | 2.48 |
| Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 1.5 × 10−9 | 2.28 |
| P18428 | Lipopolysaccharide-binding protein | LBP | 4.43 × 10−6 | 2.23 |
| P06727 | Apolipoprotein A-IV | APOA4 | 9.91 × 10−8 | 2.22 |
| P27169 | Serum paraoxonase/arylesterase 1 | PON1 | 4.4 × 10−5 | 2.14 |
| P07360 | Complement component C8 gamma chain | C8G | 1.92 × 10−7 | 2.08 |
| P00734 | Prothrombin | F2 | 0.0035 | 2.06 |
| P08603 | Complement factor H | CFH | 9.47 × 10−9 | 2.05 |
| P08697 | Alpha-2-antiplasmin | SERPINF2 | 7.57 × 10−9 | 2.03 |
| P02647 | Apolipoprotein A-I | APOA1 | 1.5 × 10−6 | 2.02 |
| P07358 | Complement component C8 beta chain | C8B | 1.02 × 10−5 | 2.01 |
| P05543 | Thyroxine-binding globulin | SERPINA7 | 2.33 × 10−5 | 1.98 |
| P05546 | Heparin cofactor 2 | SERPIND1 | 5.19 × 10−9 | 1.97 |
| P04004 | Vitronectin | VTN | 1.67 × 10−7 | 1.97 |
| P02792 | Ferritin light chain | FTL | 0.012 | 1.96 |
| P13671 | Complement component C6 | C6 | 4.68 × 10−5 | 1.95 |
| P02750 | Leucine-rich alpha-2-glycoprotein | LRG1 | 0.0021 | 1.92 |
| P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 1.59 × 10−6 | 1.92 |
| P01008 | Antithrombin-III | SERPINC1 | 3.78 × 10−9 | 1.91 |
| P13796 | Plastin-2 | LCP1 | 0.0014 | 1.90 |
| A0A0C4DH38 | Immunoglobulin heavy variable 5–51 | IGHV5-51 | 0.0053 | 1.89 |
| P01042 | Kininogen-1 | KNG1 | 1.28 × 10−7 | 1.87 |
| P03952 | Plasma kallikrein | KLKB1 | 5.27 × 10−5 | 1.85 |
| P08185 | Corticosteroid-binding globulin | SERPINA6 | 9.34 × 10−8 | 1.85 |
| P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1 | 9.65 × 10−7 | 1.85 |
| A0A075B6S5 | Immunoglobulin kappa variable 1-27 | IGKV1-27 | 0.00078 | 1.82 |
| P10643 | Complement component C7 | C7 | 8.22 × 10−5 | 1.81 |
| P02760 | Protein AMBP | AMBP | 0.0011 | 1.80 |
| A0A075B6J9 | Immunoglobulin lambda variable 2-18 | IGLV2-18 | 0.012 | 1.79 |
| Q96IY4 | Carboxypeptidase B2 | CPB2 | 3.96 × 10−6 | 1.78 |
| Q96PD5-2 | N-acetylmuramoyl-L-alanine amidase | PGLYRP2 | 4.11 × 10−5 | 1.77 |
| P04217 | Alpha-1B-glycoprotein | A1BG | 4.88 × 10−7 | 1.76 |
| P0DOX2 | Immunoglobulin alpha-2 heavy chain | n/a | 0.0014 | 1.76 |
| P01024 | Complement C3 | C3 | 1.22 × 10−7 | 1.73 |
| P06681 | Complement C2 | C2 | 2.15 × 10−6 | 1.72 |
| P43652 | Afamin | AFM | 3.01 × 10−6 | 1.72 |
| P20396 | Pro-thyrotropin-releasing hormone | TRH | 0.00051 | 1.71 |
| P29622 | Kallistatin | SERPINA4 | 7.82 × 10−8 | 1.69 |
| P35858 | Insulin-like growth factor-binding protein complex acid labile subunit | IGFALS | 0.0028 | 1.69 |
| P01009 | Alpha-1-antitrypsin | SERPINA1 | 1.17 × 10−5 | 1.67 |
| P01011 | Alpha-1-antichymotrypsin | SERPINA3 | 1.15 × 10−5 | 1.66 |
| P02748 | Complement component C9 | C9 | 0.0024 | 1.66 |
| P04180 | Phosphatidylcholine-sterol acyltransferase | LCAT | 0.00012 | 1.66 |
| P02746 | Complement C1q subcomponent subunit B | C1QB | 2.55 × 10−6 | 1.63 |
| P01619 | Ig kappa chain V-III region B6 | IGKV3-20 | 0.015 | 1.62 |
| P08571 | Monocyte differentiation antigen CD14 | CD14 | 2.1 × 10−6 | 1.61 |
| Q14520-2 | Hyaluronan-binding protein 2 | HABP2 | 0.0038 | 1.61 |
| P01023 | Alpha-2-macroglobulin | A2M | 4.69 × 10−5 | 1.60 |
| P19652 | Alpha-1-acid glycoprotein 2 | ORM2 | 0.00041 | 1.60 |
| P04278-5 | Sex hormone-binding globulin | SHBG | 0.032 | 1.60 |
| P26927 | Hepatocyte growth factor-like protein | MST1 | 0.0010 | 1.56 |
| P25311 | Zinc-alpha-2-glycoprotein | AZGP1 | 2.43 × 10−5 | 1.54 |
| P0DOY3 | Ig lambda-6 chain C region | IGLC6 | 0.0013 | 1.54 |
| P04196 | Histidine-rich glycoprotein | HRG | 3.27 × 10−5 | 1.53 |
| Q9UGM5 | Fetuin-B | FETUB | 0.012 | 1.50 |
| P02765 | Alpha-2-HS-glycoprotein | AHSG | 0.00040 | 1.49 |
| P02766 | Transthyretin | TTR | 1.78 × 10−5 | 1.49 |
| P01019 | Angiotensinogen; angiotensin 1–9 | AGT | 1.62 × 10−5 | 1.47 |
| P00748 | Coagulation factor XII | F12 | 0.012 | 1.46 |
| P01859 | Ig gamma-2 chain C region | IGHG2 | 0.00020 | 1.44 |
| P00747 | Plasminogen | PLG | 0.0066 | 1.43 |
| P51884 | Lumican | LUM | 0.00012 | 1.41 |
| P07357 | Complement component C8 alpha chain | C8A | 0.020 | 1.33 |
| P00450 | Ceruloplasmin | CP | 8.38 × 10−6 | 1.32 |
| P00751 | Complement factor B | CFB | 0.0053 | 1.28 |
| P05156 | Complement factor I | CFI | 0.00020 | 1.25 |
| P10909 | Clusterin | CLU | 0.0060 | 0.78 |
| Q9UBP4 | Dickkopf-related protein 3 | DKK3 | 0.0046 | 0.75 |
| O00391-2 | Sulfhydryl oxidase 1 | QSOX1 | 0.0033 | 0.75 |
| P18065 | Insulin-like growth factor-binding protein 2 | IGFBP2 | 0.015 | 0.74 |
| P15291-2 | Beta-1,4-galactosyltransferase 1 | B4GALT1 | 0.013 | 0.72 |
| P24592 | Insulin-like growth factor-binding protein 6 | IGFBP6 | 0.025 | 0.71 |
| P10745 | Retinol-binding protein 3 | RBP3 | 0.015 | 0.70 |
| P39060-2 | Collagen alpha-1(XVIII) chain | COL18A1 | 0.021 | 0.70 |
| P16035 | Metalloproteinase inhibitor 2 | TIMP2 | 0.0029 | 0.69 |
| Q6EMK4 | Vasorin | VASN | 0.0046 | 0.69 |
| P12109 | Collagen alpha-1(VI) chain | COL6A1 | 0.029 | 0.68 |
| P61812 | Transforming growth factor beta-2 | TGFB2 | 0.035 | 0.68 |
| P23471-3 | Receptor-type tyrosine-protein phosphatase zeta | PTPRZ1 | 0.0011 | 0.67 |
| Q99435-4 | Protein kinase C-binding protein NELL2 | NELL2 | 0.020 | 0.66 |
| Q99972 | Myocilin | MYOC | 0.020 | 0.66 |
| Q8N475 | Follistatin-related protein 5 | FSTL5 | 0.024 | 0.66 |
| P10645 | Chromogranin-A | CHGA | 0.018 | 0.66 |
| P35443 | Thrombospondin-4 | THBS4 | 0.00030 | 0.66 |
| Q7Z3B1 | Neuronal growth regulator 1 | NEGR1 | 0.00091 | 0.66 |
| Q9BSG5 | Retbindin | RTBDN | 0.012 | 0.66 |
| P07195 | L-lactate dehydrogenase B chain | LDHB | 0.024 | 0.65 |
| Q9BY67-2 | Cell adhesion molecule 1 | CADM1 | 0.0015 | 0.65 |
| P27797 | Calreticulin | CALR | 0.011 | 0.65 |
| P15586 | N-acetylglucosamine-6-sulfatase | GNS | 0.014 | 0.64 |
| P01034 | Cystatin-C | CST3 | 1.00 × 10−5 | 0.62 |
| P41222 | Prostaglandin-H2 D-isomerase | PTGDS | 0.00018 | 0.62 |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 1.00 × 10−5 | 0.62 |
| P51693 | Amyloid-like protein 1 | APLP1 | 0.0016 | 0.62 |
| Q14118 | Dystroglycan | DAG1 | 0.00073 | 0.62 |
| P23142 | Fibulin-1 | FBLN1 | 3.17 × 10−6 | 0.62 |
| Q9UBM4 | Opticin | OPTC | 0.00037 | 0.61 |
| P13591-5 | Neural cell adhesion molecule 1 | NCAM1 | 0.0013 | 0.61 |
| Q92765 | Secreted frizzled-related protein 3 | FRZB | 0.0042 | 0.60 |
| Q15113 | Procollagen C-endopeptidase enhancer 1 | PCOLCE | 0.0015 | 0.59 |
| P10599 | Thioredoxin | TXN | 0.0015 | 0.59 |
| Q9HCB6 | Spondin-1 | SPON1 | 0.00011 | 0.59 |
| Q16270 | Insulin-like growth factor-binding protein 7 | IGFBP7 | 3.82 × 10−5 | 0.58 |
| Q99969 | Retinoic acid receptor responder protein 2 | RARRES2 | 5.53 × 10−5 | 0.58 |
| P09486 | SPARC | SPARC | 7.51 × 10−5 | 0.58 |
| Q14055 | Collagen alpha-2(IX) chain | COL9A2 | 0.00010 | 0.57 |
| P08123 | Collagen alpha-2(I) chain | COL1A2 | 0.00092 | 0.57 |
| O43505 | Beta-1,4-glucuronyltransferase 1 | B4GAT1 | 0.00025 | 0.56 |
| Q16769-2 | Glutaminyl-peptide cyclotransferase | QPCT | 3.19 × 10−5 | 0.56 |
| O75326 | Semaphorin-7A | SEMA7A | 0.0014 | 0.56 |
| Q8IZJ3-2 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8 | CPAMD8 | 4.51 × 10−5 | 0.56 |
| Q14515-2 | SPARC-like protein 1 | SPARCL1 | 2.48 × 10−5 | 0.56 |
| P61916 | Epididymal secretory protein E1 | NPC2 | 0.00065 | 0.56 |
| P80188 | Neutrophil gelatinase-associated lipocalin | LCN2 | 0.0030 | 0.56 |
| O94985-2 | Calsyntenin-1 | CLSTN1 | 9.62 × 10−6 | 0.55 |
| P19022 | Cadherin-2 | CDH2 | 1.95 × 10−5 | 0.55 |
| Q96KN2 | Beta-Ala-His dipeptidase | CNDP1 | 0.00053 | 0.55 |
| Q92520 | Protein FAM3C | FAM3C | 6.77 × 10−6 | 0.55 |
| P09972 | Fructose-bisphosphate aldolase C | ALDOC | 0.0048 | 0.55 |
| P12259 | Coagulation factor V | F5 | 0.014 | 0.55 |
| Q9UHL4 | Dipeptidyl peptidase 2 | DPP7 | 6.89 × 10−5 | 0.55 |
| Q12805-2 | EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 7.20 × 10−6 | 0.54 |
| Q12860 | Contactin-1 | CNTN1 | 0.00010 | 0.54 |
| P05067 | Amyloid-beta precursor protein | APP | 9.57 × 10−5 | 0.54 |
| Q02818 | Nucleobindin-1 | NUCB1 | 7.89 × 10−5 | 0.54 |
| Q9NQ79-3 | Cartilage acidic protein 1 | CRTAC1 | 6.22 × 10−5 | 0.53 |
| Q9BRK5-6 | 45 kDa calcium-binding protein | SDF4 | 4.82 × 10−6 | 0.53 |
| Q12841 | Follistatin-related protein 1 | FSTL1 | 0.0039 | 0.52 |
| Q9P121-3 | Neurotrimin | NTM | 4.1 × 10−5 | 0.52 |
| Q92823-3 | Neuronal cell adhesion molecule | NRCAM | 3.63 × 10−5 | 0.51 |
| P06733 | Alpha-enolase | ENO1 | 0.0020 | 0.50 |
| P30086 | Phosphatidylethanolamine-binding protein 1 | PEBP1 | 7.09 × 10−8 | 0.50 |
| P07686 | Beta-hexosaminidase subunit beta | HEXB | 0.00014 | 0.50 |
| P62987 | Ubiquitin-60S ribosomal protein L40 | UBA52 | 0.00022 | 0.50 |
| Q14767 | Latent-transforming growth factor beta-binding protein 2 | LTBP2 | 0.00016 | 0.50 |
| O15537 | Retinoschisin | RS1 | 0.019 | 0.49 |
| P51888 | Prolargin | PRELP | 0.0010 | 0.48 |
| P11021 | 78 kDa glucose-regulated protein | HSPA5 | 0.029 | 0.48 |
| Q9BU40 | Chordin-like protein 1 | CHRDL1 | 5 × 10−5 | 0.47 |
| Q9Y5W5 | Wnt inhibitory factor 1 | WIF1 | 8.91 × 10−5 | 0.46 |
| P08294 | Extracellular superoxide dismutase [Cu-Zn] | SOD3 | 3.12 × 10−5 | 0.46 |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | PPIA | 0.030 | 0.46 |
| Q92563 | Testican-2 | SPOCK2 | 0.0016 | 0.46 |
| O14773 | Tripeptidyl-peptidase 1 | TPP1 | 0.0039 | 0.46 |
| P04075 | Fructose-bisphosphate aldolase A | ALDOA | 0.0084 | 0.44 |
| P16870-2 | Carboxypeptidase E | CPE | 4.76 × 10−5 | 0.44 |
| O95428-6 | Papilin | PAPLN | 3.36 × 10−5 | 0.43 |
| Q13822 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | ENPP2 | 2.86 × 10−6 | 0.43 |
| P14618 | Pyruvate kinase PKM | PKM | 0.00020 | 0.42 |
| Q08380 | Galectin-3-binding protein | LGALS3BP | 1.16 × 10−5 | 0.42 |
| Q99574 | Neuroserpin | SERPINI1 | 1.36 × 10−7 | 0.42 |
| P78509 | Reelin | RELN | 0.0044 | 0.41 |
| P31025 | Lipocalin-1 | LCN1 | 0.00016 | 0.41 |
| P98164 | Low-density lipoprotein receptor-related protein 2 | LRP2 | 0.00011 | 0.41 |
| P63104 | 14-3-3 protein zeta/delta | YWHAZ | 0.015 | 0.41 |
| P09211 | Glutathione S-transferase P | GSTP1 | 0.0016 | 0.40 |
| Q7Z7G0 | Target of Nesh-SH3 | ABI3BP | 2.71 × 10−5 | 0.39 |
| Q02413 | Desmoglein-1 | DSG1 | 5.25 × 10−5 | 0.39 |
| Q06481 | Amyloid-like protein 2 | APLP2 | 1.68 × 10−5 | 0.38 |
| Q08554-2 | Desmocollin-1 | DSC1 | 5.81 × 10−5 | 0.37 |
| Q08629 | Testican-1 | SPOCK1 | 2.00 × 10−7 | 0.36 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | HSPG2 | 3.48 × 10−6 | 0.36 |
| O00468-6 | Agrin | AGRN | 5.27 × 10−6 | 0.33 |
| Q9NZT1 | Calmodulin-like protein 5 | CALML5 | 0.0086 | 0.32 |
| P00558 | Phosphoglycerate kinase 1 | PGK1 | 5.74 × 10−6 | 0.30 |
| P35555 | Fibrillin-1 | FBN1 | 1.32 × 10−8 | 0.28 |
| P15924 | Desmoplakin | DSP | 0.0014 | 0.22 |
| P22914 | Beta-crystallin S | CRYGS | 2.65 × 10−7 | 0.20 |
Figure 2.
Volcano plots. Log2 transformed abundance ratios for each protein are plotted on the x-axis. Negative log10 transformed P values are plotted on the y-axis. A false discovery rate (FDR) of 0.05 was applied. Significantly regulated proteins are localized above the full curves. (A) CRVO versus control samples. A total of 177 significantly changed proteins (blue squares) were identified. (B) Five proteins were increased in ischemic CRVO compared to non-ischemic CRVO, including fibrinogen chains alpha, beta and gamma, apolipoprotein C-III, and fibronectin.
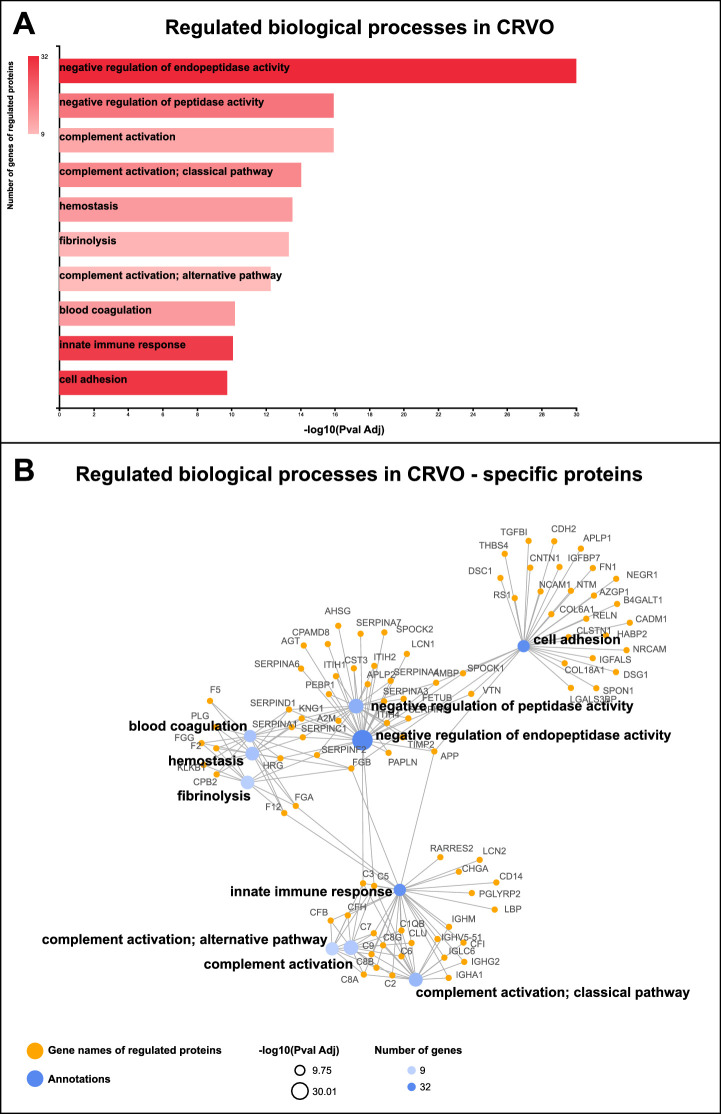
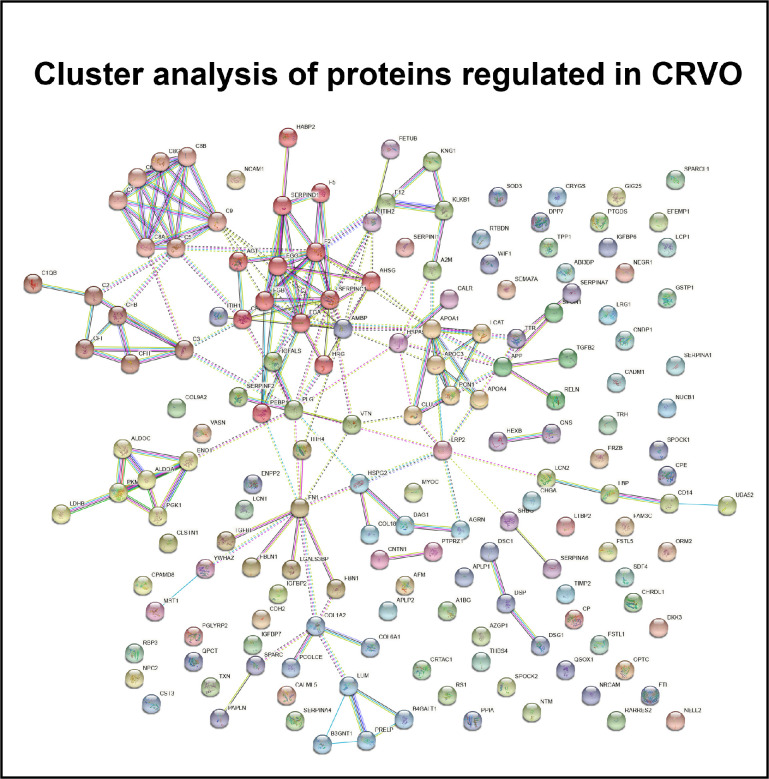
CRVO was associated with the regulation of endopeptidase activity, complement activation, innate immune response, blood coagulation, and cell adhesion (Figs. 3A, 3B). Proteins involved in the innate immune response and complement activation included complement factors, immunoglobulin chains, lipopolysaccharide-binding protein (LBP), monocyte differentiation antigen CD14 (CD14), neutrophil gelatinase-associated lipocalin, retinoic acid receptor protein 2, and chromogranin-A (see Fig. 3B). Similarly, STRING cluster analysis revealed regulation of a large cluster of interacting complement factors (Fig. 4). A large group of proteins involved in blood coagulation, hemostasis, and fibrinolysis were upregulated in CRVO, including fibrinogen chains, prothrombin, coagulation factor 12, histidine-rich glycoprotein, plasminogen, coagulation factor V, coagulation factor XIII, alpha-2-macroglobulin, kininogen-1, plasma kallikrein, carboxypeptidase B2, antithrombin-III, heparin cofactor 2, and alpha-1-antitrypsin (see Fig. 3B). STRING cluster analysis also identified a major cluster of proteins consisting of fibrinogen chains, prothrombin, coagulation factor V, histidine-rich glycoprotein, angiotensinogen, antithrombin-III, and heparin cofactor 2 (see Fig. 4). Another major group of regulated proteins in CRVO were proteins involved in cell adhesion, including cell adhesion molecule 1, neuronal cell adhesion molecule, fibronectin, neural cell adhesion molecule, retinoschisin, neurotrimin, spondin-1, contactin-1, reelin, desmoglein-1, hyaluronan-binding protein, cadherin-2, zinc-alpha-2-glycoprotein, neuronal growth regulator, calsyntenin-1, galectin-3 binding protein, desmocollin-1, and thrombospondin-4 (see Fig. 3B).
Figure 3.
Bioinformatic analyses of significantly regulated proteins. (A) CRVO resulted in the regulation of proteins involved in negative regulation of endopeptidase activity, complement activation, hemostasis, fibrinolysis, blood coagulation, innate immune response, and cell adhesion. (B) CRVO was associated with increased levels of proteins involved in the innate immune response and complement activation, including complement components (CFB, CFH, CFI, C1QB, C2, C3, C5, C6, C7, C8A, C8B, and C8G), lipopolysaccharide-binding protein (LBP), monocyte differentiation antigen CD14 (CD14), neutrophil gelatinase-associated lipocalin (LCN2), retinoic acid receptor responder protein 2 (RARRES2), and chromogranin-A (CHGA). Proteins involved in blood coagulation included fibrinogen chains alpha, beta and gamma (FGA, FGB, and FGG), prothrombin (F2), and coagulation factors (F5 and F12). CRVO was also associated with changes in proteins involved in cell adhesion, including fibronectin (FN1), collagen chains (COL18A1 and COL6A1), spondin-1 (SPON1), reelin (RELN), calsyntenin-1 (CLSTN1), neural cell adhesion molecule (NCAM1), neuronal cell adhesion molecule (NRCAM), contactin-1 (CNTN1), and cadherin-2 (CDH2).
Figure 4.
STRING cluster analysis of regulated proteins in CRVO. A major cluster (light brown nodes) was formed by complement factors (C1QB, C2, C3, C5, C6, C7, C8A, C8B, C8G, C9, CFB, CFH, and CFI). Another major cluster (red nodes) consisted of fibrinogen chains (FGA, FGB, and FGG), prothrombin (F2), coagulation factor V (F5), angiotensinogen (AGT), antithrombin-III (SERPINC1), and heparin factor 2 (SERPIND1). CRVO was also associated with the regulation of a cluster of proteins consisting of apolipoproteins (APO1A, APOA4, and APOC3), serum paraoxonase/arylesterase 1 (PON1), and clusterin (CLU) whereas another cluster (yellow nodes) was comprised of fructose-bisphosphate aldolases (ALDOA and ALDOC), alpha-enolase (ENO1), and phosphoglycerate kinase 1 (PGK1).
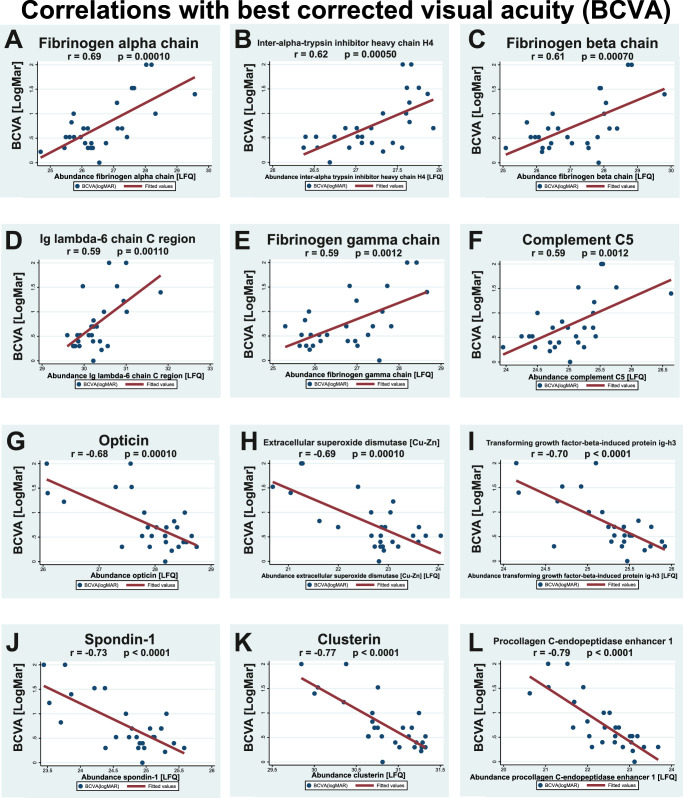
Among the 177 significantly regulated proteins, 78 proteins correlated significantly with BCVA (Table 4, examples are shown in Fig. 5) and 42 proteins correlated significantly with the severity of macular edema (see Table 5, examples are shown in Fig. 6). Strong correlations with BCVA were observed for fibrinogen chains alpha, beta and gamma, inter-alpha-trypsin inhibitor heavy chain H4, Ig lambda-6 chain C region, and complement factors C5, H, and C9 (see Table 4, examples are shown in Fig. 5). Strong negative correlations with BCVA were observed for procollagen C-endopeptidase enhancer 1, clusterin, spondin-1, transforming growth factor-beta-induced protein ig-h3, extracellular superoxide dismutase [Cu-Zn] and opticin (see Table 4, examples are shown in Fig. 5).
Table 4.
Correlations Between Proteins and Best Corrected Visual Acuity
| Protein ID | Protein Names | Correlation, r | P Value |
|---|---|---|---|
| P02671 | Fibrinogen alpha chain | 0.69 | 0.00010 |
| Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | 0.62 | 0.00050 |
| P02675 | Fibrinogen beta chain | 0.61 | 0.00070 |
| P0DOY3 | Ig lambda-6 chain C region | 0.59 | 0.00110 |
| P02679-2 | Fibrinogen gamma chain | 0.59 | 0.0012 |
| P01031 | Complement C5 | 0.59 | 0.0012 |
| P01876 | Ig alpha-1 chain C region | 0.59 | 0.0013 |
| P08603 | Complement factor H | 0.58 | 0.0014 |
| P02748 | Complement component C9 | 0.58 | 0.0016 |
| P02750 | Leucine-rich alpha-2-glycoprotein | 0.57 | 0.0018 |
| P02656 | Apolipoprotein C-III | 0.57 | 0.0022 |
| P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | 0.56 | 0.0027 |
| P02760 | Protein AMBP | 0.55 | 0.0029 |
| P02751-1 | Fibronectin | 0.54 | 0.0036 |
| P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | 0.54 | 0.0038 |
| P13671 | Complement component C6 | 0.52 | 0.0060 |
| P10643 | Complement component C7 | 0.48 | 0.012 |
| P02647 | Apolipoprotein A-I | 0.44 | 0.022 |
| P04278-5 | Sex hormone-binding globulin | 0.42 | 0.033 |
| P26927 | Hepatocyte growth factor-like protein | 0.41 | 0.038 |
| P25311 | Zinc-alpha-2-glycoprotein | 0.40 | 0.036 |
| P01011 | Alpha-1-antichymotrypsin | 0.40 | 0.041 |
| P00751 | Complement factor B | 0.39 | 0.042 |
| P06727 | Apolipoprotein A-IV | 0.39 | 0.045 |
| O00391-2 | Sulfhydryl oxidase 1 | −0.40 | 0.038 |
| Q08629 | Testican-1 | −0.42 | 0.033 |
| P31025 | Lipocalin-1 | −0.42 | 0.028 |
| P61916; | Epididymal secretory protein E1 | −0.43 | 0.026 |
| O43505 | Beta-1,4-glucuronyltransferase 1 | −0.43 | 0.025 |
| Q9BU40 | Chordin-like protein 1 | −0.43 | 0.032 |
| P30086 | Phosphatidylethanolamine-binding protein 1 | −0.44 | 0.023 |
| Q99972 | Myocilin | −0.44 | 0.021 |
| Q14515-2 | SPARC-like protein 1 | −0.44 | 0.020 |
| Q12805-2 | EGF-containing fibulin-like extracellular matrix protein 1 | −0.45 | 0.038 |
| O00468-6 | Agrin | −0.45 | 0.019 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | −0.45 | 0.019 |
| Q9BSG5 | Retbindin | −0.45 | 0.018 |
| P35555 | Fibrillin-1 | −0.45 | 0.020 |
| Q99574 | Neuroserpin | −0.45 | 0.020 |
| Q96KN2 | Beta-Ala-His dipeptidase | −0.46 | 0.021 |
| P12259 | Coagulation factor V | −0.46 | 0.017 |
| P16035 | Metalloproteinase inhibitor 2 | −0.48 | 0.012 |
| Q02818 | Nucleobindin-1 | −0.48 | 0.011 |
| P10745 | Retinol-binding protein 3 | −0.48 | 0.011 |
| P13591-5 | Neural cell adhesion molecule 1 | −0.51 | 0.0075 |
| P23142 | Fibulin-1 | −0.52 | 0.0051 |
| P12109 | Collagen alpha-1(VI) chain | −0.54 | 0.0046 |
| Q92520 | Protein FAM3C | −0.54 | 0.0035 |
| Q14118 | Dystroglycan | −0.54 | 0.0042 |
| O75326 | Semaphorin-7A | −0.54 | 0.0042 |
| Q9Y5W5 | Wnt inhibitory factor 1 | −0.55 | 0.0037 |
| Q9NQ79-3 | Cartilage acidic protein 1 | −0.55 | 0.0029 |
| Q6EMK4 | Vasorin | −0.55 | 0.0028 |
| P19022 | Cadherin-2 | −0.56 | 0.0025 |
| P16870-2 | Carboxypeptidase E | −0.56 | 0.0024 |
| Q08380 | Galectin-3-binding protein | −0.56 | 0.0029 |
| P51888 | Prolargin | −0.57 | 0.0019 |
| Q9BY67-2 | Cell adhesion molecule 1 | −0.57 | 0.0029 |
| Q12841 | Follistatin-related protein 1 | −0.58 | 0.0078 |
| Q99969 | Retinoic acid receptor responder protein 2 | −0.59 | 0.0012 |
| Q92823-3 | Neuronal cell adhesion molecule | −0.59 | 0.0024 |
| Q16769-2 | Glutaminyl-peptide cyclotransferase | −0.60 | 0.0010 |
| Q9UBP4 | Dickkopf-related protein 3 | −0.60 | 0.00090 |
| Q8IZJ3-2 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8 | −0.61 | 0.00080 |
| Q14055 | Collagen alpha-2(IX) chain | −0.61 | 0.00090 |
| O94985-2 | Calsyntenin-1 | −0.61 | 0.00070 |
| P01034 | Cystatin-C | −0.63 | 0.00050 |
| Q13822 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | −0.64 | 0.00030 |
| P78509 | Reelin | −0.66 | 0.00040 |
| P41222 | Prostaglandin-H2 D-isomerase | −0.67 | 0.00010 |
| Q16270 | Insulin-like growth factor-binding protein 7 | −0.68 | 0.00010 |
| Q9UBM4 | Opticin | −0.68 | 0.00010 |
| P08294 | Extracellular superoxide dismutase [Cu-Zn] | −0.69 | 0.00010 |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | −0.70 | P < 0.0001 |
| Q9HCB6 | Spondin-1 | −0.73 | P < 0.0001 |
| P10909 | Clusterin | −0.77 | P < 0.0001 |
| Q15113 | Procollagen C-endopeptidase enhancer 1 | −0.79 | P < 0.0001 |
Figure 5.
Correlations with best corrected visual acuity (BCVA). A total of 78 proteins correlated with BCVA. Correlations are shown for the six proteins with the strongest positive correlations with BCVA and for the six proteins with strongest negative correlations with BCVA. Correlations were calculated as Pearson's correlation coefficient, r. Label-free quantification (LFQ) values denote the protein content measured in the proteomic analysis. (A-F) The proteins with the strongest positive correlations with BCVA (LogMAR) were fibrinogen alpha chain, inter-alpha-trypsin inhibitor heavy chain H4, fibrinogen beta chain, Ig lambda-6 chain C region, fibrinogen gamma chain and complement C5. (G-L) The strongest negative correlations with BCVA were observed for opticin, extracellular superoxide dismutase, transforming growth factor-beta-induced protein ig-h3, spondin-1, clusterin, and procollagen C-endopeptidase enhancer 1.
Table 5.
Correlations Between Proteins and Severity of Macular Edema
| Protein ID | Protein Names | Correlation, r | P Value |
|---|---|---|---|
| P01876 | Ig alpha-1 chain C region | 0.53 | 0.0036 |
| P0DOY3 | Ig lambda-6 chain C region | 0.52 | 0.0046 |
| P01871 | Ig mu chain C region | 0.50 | 0.0075 |
| P02671 | Fibrinogen alpha chain | 0.49 | 0.0078 |
| P02675 | Fibrinogen beta chain | 0.48 | 0.0097 |
| P04432 | Ig kappa chain V-I region Daudi | 0.47 | 0.015 |
| Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | 0.46 | 0.014 |
| P06727 | Apolipoprotein A-IV | 0.45 | 0.016 |
| P02751-1 | Fibronectin | 0.45 | 0.017 |
| P10643 | Complement component C7 | 0.44 | 0.020 |
| P0DOX2 | Immunoglobulin alpha-2 heavy chain | 0.43 | 0.022 |
| P02679-2 | Fibrinogen gamma chain | 0.42 | 0.025 |
| P02760 | Protein AMBP | 0.39 | 0.040 |
| P03952 | Plasma kallikrein | 0.39 | 0.049 |
| P02656 | Apolipoprotein C-III | 0.39 | 0.041 |
| Q9HCB6 | Spondin-1 | −0.38 | 0.049 |
| P08294 | Extracellular superoxide dismutase [Cu-Zn] | −0.38 | 0.045 |
| Q14055 | Collagen alpha-2(IX) chain | −0.39 | 0.044 |
| Q08380 | Galectin-3-binding protein | −0.39 | 0.043 |
| Q13822 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | −0.40 | 0.035 |
| Q02818 | Nucleobindin-1 | −0.40 | 0.034 |
| P41222 | Prostaglandin-H2 D-isomerase | −0.40 | 0.034 |
| P51888 | Prolargin | −0.40 | 0.034 |
| O00391-2 | Sulfhydryl oxidase 1 | −0.41 | 0.031 |
| Q9NQ79-3 | Cartilage acidic protein 1 | −0.42 | 0.028 |
| Q14118 | Dystroglycan | −0.42 | 0.029 |
| P16870-2 | Carboxypeptidase E | −0.43 | 0.024 |
| O00468-6 | Agrin | −0.43 | 0.021 |
| P01034 | Cystatin-C | −0.44 | 0.020 |
| Q16270 | Insulin-like growth factor-binding protein 7 | −0.44 | 0.019 |
| Q16769-2 | Glutaminyl-peptide cyclotransferase | −0.45 | 0.016 |
| Q9BY67-2 | Cell adhesion molecule 1 | −0.45 | 0.020 |
| Q99969 | Retinoic acid receptor responder protein 2 | −0.46 | 0.014 |
| P10909 | Clusterin | −0.47 | 0.011 |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | −0.52 | 0.0050 |
| Q8IZJ3-2 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8 | −0.53 | 0.0036 |
| P35555 | Fibrillin-1 | −0.54 | 0.0037 |
| P19022 | Cadherin-2 | −0.54 | 0.0030 |
| Q9UBM4 | Opticin | −0.55 | 0.0025 |
| Q15113 | Procollagen C-endopeptidase enhancer 1 | −0.58 | 0.0013 |
| P78509 | Reelin | −0.59 | 0.0021 |
| Q9UHL4 | Dipeptidyl peptidase 2 | −0.59 | 0.0025 |
Figure 6.
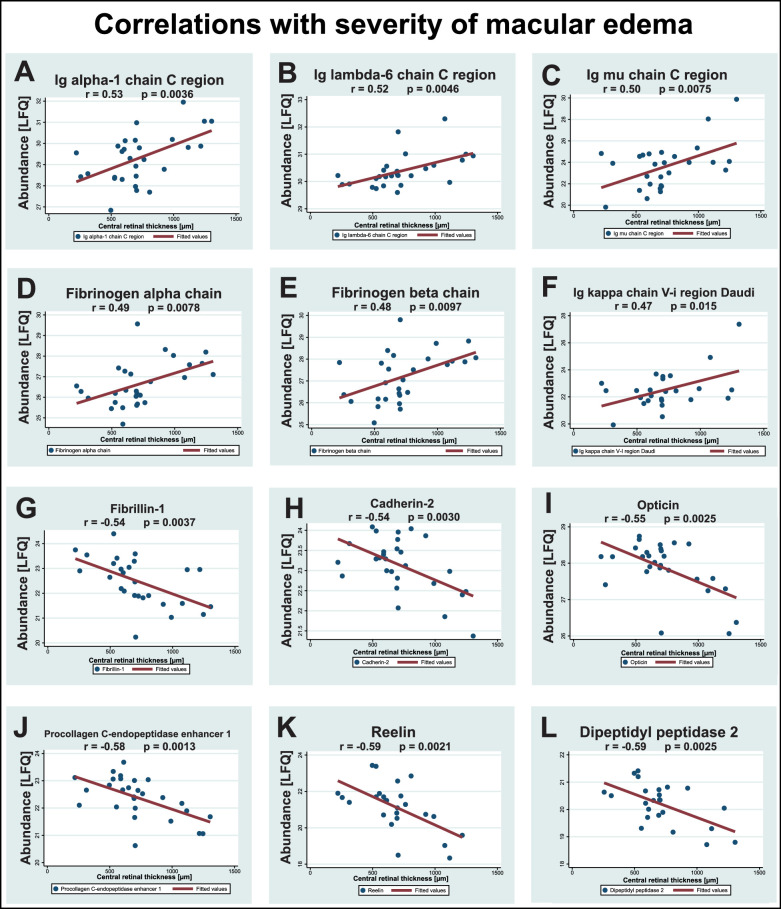
Correlations with severity of macular edema. A total of 42 proteins correlated with severity of macular edema. Correlations are shown for the six proteins with the strongest positive correlations and the six proteins with the strongest negative correlations with the severity of macular edema. Correlations were calculated as Pearson's correlation coefficient, r. (A-F) The strongest positive correlations with severity of macular edema were observed for Ig alpha-1 chain C region, Ig lambda-6 chain C region, Ig mu chain C region, fibrinogen alpha chain, fibrinogen beta chain, and Ig kappa chain V-I region Daudi. (G-L) The strongest negative correlations with severity of macular edema were observed for fibrillin-1, cadherin-2, opticin, procollagen endopeptidase enhancer 1, reelin, and dipeptidyl peptidase 2.
The strongest correlations between the proteome and severity of macular edema were observed for Ig alpha-1 chain C region, Ig lambda-6 chain C region, Ig mu chain C region, fibrinogen alpha and beta chains, and Ig kappa chain V-i region Daudi (see Table 5, examples are shown in Fig. 6). The strongest negative correlations between the proteome and severity of macular edema were observed for fibrillin-1, cadherin-2, opticin, procollagen C-endopeptidase enhancer 1, reelin, and dipeptidyl peptidase 2 (see Table 5, examples are shown in Fig. 6).
A number of proteins correlated significantly with both BCVA and severity of macular edema, including fibrinogen chains alpha and beta, fibronectin, Ig lambda-6 chain C region, Ig alpha-1 chain C region, inter-alpha-trypsin inhibitor heavy chain H4, and complement component C7 (see Tables 4, 5; Figs. 5, 6). Proteins that correlated negatively with BCVA and severity of macular edema, included reelin, procollagen C-endopeptidase enhancer 1, opticin, fibrillin-1, cadherin-2, C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8, transforming growth factor-beta-induced protein ig-h3, clusterin, glutaminyl-peptide cyclotransferase, retinoic acid receptor responder protein 2, agrin, and sulfhydryl oxidase 1 (see Tables 4, 5; Figs. 5, 6).
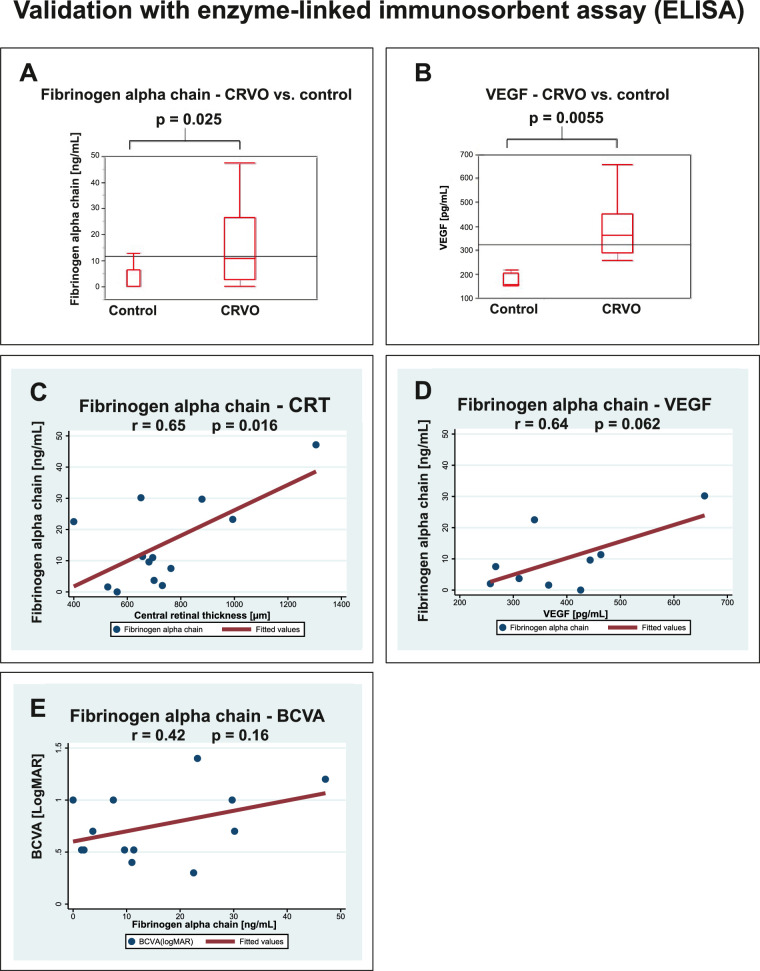
ELISA confirmed the increased level of fibrinogen alpha chain in CRVO (P = 0.025; Fig. 7A). Aqueous VEGF was elevated in CRVO (P = 0.0055; Fig. 7B). ELISA confirmed a significant correlation between fibrinogen alpha chain and severity of macular edema (r = 0.65, P = 0.016; Fig. 7C). ELISA also indicated a correlation between fibrinogen alpha chain and VEGF, without reaching significance (r = 0.64, P = 0.062; Fig. 7D). The correlation between fibrinogen alpha chain and BCVA was not confirmed with ELISA (r = 0.42, p = 0.16; Fig. 7E).
Figure 7.
Validation by ELISA. Correlations were calculated as Pearson's correlation coefficient, r. (A) ELISA confirmed an increased level of aqueous fibrinogen alpha chain in CRVO. (B) CRVO was associated with an increased level of VEGF. (C) ELISA confirmed a significant correlation between fibrinogen alpha chain and the severity of macular edema. (D) ELISA indicated a correlation between fibrinogen alpha chain and VEGF, but the correlation was not statistically significant. (E) The correlation between fibrinogen alpha chain and BCVA observed by proteomics was not confirmed by ELISA.
Discussion
This study aimed to elucidate intraocular molecular changes in CRVO through proteomic analysis of the aqueous humor. A multitude of proteins were regulated, supporting a multifactorial pathogenesis in macular edema secondary to CRVO. A total of 177 proteins were regulated in CRVO compared to controls; 78 proteins correlated with BCVA and 42 proteins correlated with the severity of macular edema. In our previous study of aqueous humor from patients with branch retinal vein occlusion (BRVO), we identified 52 significantly regulated proteins, including 13 proteins that correlated with the severity of macular edema, and one protein that correlated with BCVA. Overall, aqueous proteome changes were stronger in CRVO than in BRVO.14
Important clinical implications can be derived by comparing the proteomes in CRVO and BRVO. The pronounced protein changes in CRVO compared to BRVO support urgent and aggressive management of CRVO. Furthermore, the strong protein changes in CRVO indicate a potential need for shorter injection intervals and frequent follow-up visits. The number of significantly regulated proteins was higher in CRVO than BRVO, suggesting a multifactorial response in which additional proteins and pathways are activated when the entire neuroretina is affected by retinal vein occlusion. The inflammatory driving force was particularly severe in CRVO, with higher levels of pro-inflammatory proteins, including CD14, LBP, and complement factors. Iglicki and co-workers8 previously demonstrated the efficacy of dexamethasone intravitreal implants in cases that are resistant to anti-VEGF agents. The multifaceted nature and inflammatory profile of macular edema observed in our study supports a prompt switch to second-line therapy with dexamethasone intravitreal implants in eyes refractory to anti-VEGF therapy.
Fibrinogen chains alpha, beta and gamma, fibronectin and apolipoprotein C-III were more abundant in ischemic CRVO than non-ischemic CRVO, linking these proteins to ischemic processes. The strong correlations with BCVA observed in our study for fibrinogen chains, fibronectin and apolipoproteins may be related to retinal ischemia. At the retinal level, we previously observed that fibrinogen and fibronectin increase with the degree of retinal ischemia in experimental CRVO in porcine eyes.21,28
The role of VEGF in the formation of macular edema secondary to CRVO is well-established.29 Using two fundamentally different quantitative techniques, we show that the fibrinogen alpha chain was closely associated with the severity of macular edema, highlighting the importance of additional proteins. In addition, the aqueous level of fibrinogen was higher in ischemic CRVO compared to non-ischemic CRVO. Despite complete resolution of macular edema, visual impairment may persist due to macular ischemia.3 As the fibrinogen alpha chain was associated with ischemia in CRVO, the protein may be a potential target in therapies directed at reducing macular ischemia. When the coagulation cascade is activated, fibrinogen is converted to insoluble fibrin by thrombin, leading to clot formation.30 Our study suggests an interplay between VEGF and fibrinogen alpha chain. ELISA did not confirm a correlation between BCVA and fibrinogen alpha chain, but the sample size for proteomic analysis was larger than the sample size used for ELISA.
We previously showed in BRVO that aqueous fibronectin correlates with BCVA and the severity of macular edema.14 Interestingly, the same observation was made for CRVO. The soluble form of fibronectin, which is present in the aqueous humor, regulates thrombosis and accelerates wound healing.31,32 In laser induced CRVO in porcine eyes, fibronectin is deposited in the endothelium of retinal vessels,28 indicating that the upregulation of fibronectin may be caused by local changes and not merely be the result of a disrupted blood-retinal barrier.
The levels of several complement factors were increased in CRVO. Complement factors are likely to contribute to the inflammatory response. Increased levels of complement C3 have also been observed in vitreous samples from patients with retinal vein occlusion33 and in porcine retinas with laser-induced CRVO.28 Increased aqueous levels of the inflammatory proteins CD14 and LBP were observed in CRVO. CD14 and LBP are involved in the recognition of lipopolysaccharide, a major component of the outer membrane of Gram-negative bacteria and have regulatory functions in the innate immune system.34,35 Although CD14 and LBP are likely to be inflammatory driving forces in CRVO, the proteins did not correlate with BCVA and severity of macular edema. Features discovered by OCT continue to improve the diagnostic work-up and management of retinal diseases.16 Future studies may investigate the correlation between inflammatory proteins and specific OCT biomarkers of inflammation established in previous studies.15
A number of proteins correlated negatively with BCVA and the severity of macular edema, including cadherin-2, agrin, opticin, procollagen C-proteinase enhancer 1, clusterin, fibrillin-1, and reelin. The negative correlations with clinical parameters suggest a downregulation of protective proteins in CRVO. Cadherins contribute to a number of functions at the retinal level, including tissue morphogenesis, neuronal survival, and photoreceptor development and survival.36 Agrin is a basement membrane proteoglycan known to be abundant in retinal blood vessels,37 but its function needs to be further elucidated. Opticin belongs to the family of small-leucine rich repeat proteoglycans37 and was previously found to be downregulated in vitreous humor samples from patients with CRVO.33 Opticin exerts an anti-angiogenic effect in hypoxia-induced retinopathy in zebrafish38 and is downregulated in retinopathy of prematurity.39 Procollagen C-proteinase enhancer 1 is a glycoprotein with anti-angiogenic features involved in assembly of the extracellular matrix.40,41 Clusterin has anti-inflammatory features and was previously found to inhibit vascular permeability induced by VEGF through restoration of tight junction proteins.42,43 Loss of the anti-angiogenic response in CRVO due to downregulation of opticin, procollagen C-proteinase enhancer 1, and clusterin needs to be further elucidated. Fibrillin-1 and reelin were previously found to be downregulated in aqueous humor from patients with BRVO,14 but the roles of these proteins in retinal vascular disease are poorly understood.
Detection of low abundance proteins remains a challenge in the proteomic analysis of aqueous humor. Our proteomic analysis did not detect low-abundant proteins such as VEGF, IL-6, and IL-8.12,13 A number of factors, including sample complexity, technical variation, and fragmentation efficiency, are known to limit the detection of low-abundant proteins.44,45 In our study, ELISA was necessary for successful quantification of VEGF-A. The sample material was another limitation. Due to the low volumes and low protein concentrations of aqueous humor samples, there was only sufficient material to validate fibrinogen alpha chain and VEGF in our study.
Conclusions
Multiple proteins were regulated in CRVO complicated by macular edema, supporting a multifactorial pathogenesis. Positive correlations with BCVA and severity of macular edema were observed for fibrinogen chains and fibronectin. The aqueous content of fibrinogen chains and fibronectin were higher in ischemic CRVO versus non-ischemic CRVO, suggesting that the proteins were involved in ischemic processes. Complement factors C5, C6, C7, C9, B, and H were upregulated in CRVO and correlated with BCVA. Procollagen C-endopeptidase enhancer 1, opticin, and clusterin were downregulated in CRVO and correlated negatively with BCVA and severity of macular edema, indicating decreased levels of anti-angiogenic and anti-inflammatory proteins. The pro-inflammatory proteins LBP and CD14 were upregulated in CRVO and may be driving forces in the inflammatory response in CRVO.
Supplementary Material
Acknowledgments
The authors thank Mona Britt Hansen, Aarhus University, Aarhus, Denmark, for her expert technical assistance. The authors thank Fight for Sight Denmark, Helene og Viggo Bruuns Fond, the Svend Andersen Foundation, Synoptik-Fonden, the Herta Christensen Foundation, the North Denmark Region (2013-0076797), Speciallæge Heinrich Kopps Legat, the Danish Society of Ophthalmology, and Overlægerådets Forskningsfond, Odense University Hospital, Odense, Denmark, for their generous support. The mass spectrometers used for this study were funded by A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til almene Formaal.
Disclosure: L.J. Cehofski, None; K. Kojima, None; N. Kusada, None; M. Rasmussen, None; D.V. Muttuvelu, None; J. Grauslund, None; H. Vorum, None; B. Honoré, None
References
- 1. Green WR, Chan CC, Hutchins GM, Terry JM.. Central retinal vein occlusion: a prospective histopathologic study of 29 eyes in 28 cases. Trans Am Ophthalmol Soc. 1981; 79: 371–422. [PMC free article] [PubMed] [Google Scholar]
- 2. Hayreh SS, Podhajsky PA, Zimmerman MB.. Natural history of visual outcome in central retinal vein occlusion. Ophthalmology. 2011; 118: 119–133.e111-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campochiaro PA, Akhlaq A.. Sustained suppression of VEGF for treatment of retinal/choroidal vascular diseases. Prog Retin Eye Res. 2020; 83: 100921. [DOI] [PubMed] [Google Scholar]
- 4. McIntosh RL, Rogers SL, Lim L, et al.. Natural history of central retinal vein occlusion: an evidence-based systematic review. Ophthalmology. 2010; 117: 1113–1123.e1115. [DOI] [PubMed] [Google Scholar]
- 5. Noma H, Mimura T, Yasuda K, Shimura M.. Role of soluble vascular endothelial growth factor receptor signaling and other factors or cytokines in central retinal vein occlusion with macular edema. Invest Ophthalmol Vis Sci. 2015; 56: 1122–1128. [DOI] [PubMed] [Google Scholar]
- 6. Brown DM, Campochiaro PA, Singh RP, et al.. Ranibizumab for macular edema following central retinal vein occlusion: six-month primary end point results of a phase III study. Ophthalmology. 2010; 117: 1124–1133.e1121. [DOI] [PubMed] [Google Scholar]
- 7. Haller JA, Bandello F, Belfort R Jr., et al.. Dexamethasone intravitreal implant in patients with macular edema related to branch or central retinal vein occlusion twelve-month study results. Ophthalmology. 2011; 118: 2453–2460. [DOI] [PubMed] [Google Scholar]
- 8. Iglicki M, Busch C, Zur D, et al.. Dexamethasone Implant For Diabetic Macular Edema In Naive Compared With Refractory Eyes: The International Retina Group Real-Life 24-Month Multicenter Study. The IRGREL-DEX Study. Retina. 2019; 39: 44–51. [DOI] [PubMed] [Google Scholar]
- 9. Campochiaro PA, Sophie R, Pearlman J, et al.. Long-term outcomes in patients with retinal vein occlusion treated with ranibizumab: the RETAIN study. Ophthalmology. 2014; 121: 209–219. [DOI] [PubMed] [Google Scholar]
- 10. Hogg HDJ, Talks SJ, Pearce M, Di Simplicio S.. Real-World Visual and Neovascularisation Outcomes from anti-VEGF in Central Retinal Vein Occlusion. Ophthalmic Epidemiol. 2021; 28: 70–76. [DOI] [PubMed] [Google Scholar]
- 11. Campochiaro PA, Hafiz G, Mir TA, et al.. Pro-Permeability Factors After Dexamethasone Implant in Retinal Vein Occlusion; the Ozurdex for Retinal Vein Occlusion (ORVO) Study. Am J Ophthalmol. 2015; 160: 313–321.e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cehofski LJ, Honore B, Vorum H.. A Review: Proteomics in Retinal Artery Occlusion, Retinal Vein Occlusion, Diabetic Retinopathy and Acquired Macular Disorders. Int J Mol Sci. 2017; 18(5): 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cehofski LJ, Mandal N, Honore B, Vorum H.. Analytical platforms in vitreoretinal proteomics. Bioanalysis. 2014; 6: 3051–3066. [DOI] [PubMed] [Google Scholar]
- 14. Cehofski LJ, Kojima K, Terao N, et al.. Aqueous Fibronectin Correlates With Severity of Macular Edema and Visual Acuity in Patients With Branch Retinal Vein Occlusion: A Proteome Study. Invest Ophthalmol Vis Sci. 2020; 61: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iglicki M, Loewenstein A, Barak A, Schwartz S, Zur D.. Outer retinal hyperreflective deposits (ORYD): a new OCT feature in naïve diabetic macular oedema after PPV with ILM peeling. Br J Ophthalmol. 2020; 104: 666–671. [DOI] [PubMed] [Google Scholar]
- 16. Iglicki M, Busch C, Loewenstein A, et al.. Underdiagnosed Optic Disk Pit Maculopathy: Spectral Domain Optical Coherence Tomography Features For Accurate Diagnosis. Retina. 2019; 39: 2161–2166. [DOI] [PubMed] [Google Scholar]
- 17. Honore B. Proteomic Protocols for Differential Protein Expression Analyses. Methods Mol Biol. 2020; 2110: 47–58. [DOI] [PubMed] [Google Scholar]
- 18. Tyanova S, Temu T, Cox J.. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016; 11: 2301–2319. [DOI] [PubMed] [Google Scholar]
- 19. Christakopoulos C, Cehofski LJ, Christensen SR, Vorum H, Honore B.. Proteomics reveals a set of highly enriched proteins in epiretinal membrane compared with inner limiting membrane. Exp Eye Res. 2019; 186: 107722. [DOI] [PubMed] [Google Scholar]
- 20. Tyanova S, Temu T, Sinitcyn P, et al.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016; 13: 731–740. [DOI] [PubMed] [Google Scholar]
- 21. Cehofski LJ, Kruse A, Kirkeby S, et al.. IL-18 and S100A12 Are Upregulated in Experimental Central Retinal Vein Occlusion. Int J Mol Sci. 2018; 19(11): 3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tusher VG, Tibshirani R, Chu G.. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001; 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia-Moreno A, López-Domínguez R, Villatoro-García JA, et al.. Functional Enrichment Analysis of Regulatory Elements. Biomedicines. 2022; 10(3): 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cehofski LJ, Kruse A, Kjaergaard B, Stensballe A, Honore B, Vorum H.. Proteins involved in focal adhesion signaling pathways are differentially regulated in experimental branch retinal vein occlusion. Exp Eye Res. 2015; 138: 87–95. [DOI] [PubMed] [Google Scholar]
- 25. Szklarczyk D, Franceschini A, Wyder S, et al.. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D, Gable AL, Lyon D, et al.. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47: D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szklarczyk D, Morris JH, Cook H, et al.. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45: D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cehofski LJ, Kruse A, Alsing AN, et al.. Proteome Analysis of Aflibercept Intervention in Experimental Central Retinal Vein Occlusion. Molecules. 2022; 27(11): 3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petri AS, Boysen K, Cehofski LJ, et al.. Intravitreal Injections with Vascular Endothelial Growth Factor Inhibitors: A Practical Approach. Ophthalmol Ther. 2020; 9: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petersen MA, Ryu JK, Akassoglou K.. Fibrinogen in neurological diseases: mechanisms, imaging and therapeutics. Nat Rev Neurosci. 2018; 19: 283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faralli JA, Filla MS, Peters DM.. Role of Fibronectin in Primary Open Angle Glaucoma. Cells. 2019; 8(12): 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lemanska-Perek A, Adamik B.. Fibronectin and its soluble EDA-FN isoform as biomarkers for inflammation and sepsis. Adv Clin Exp Med. 2019; 28: 1561–1567. [DOI] [PubMed] [Google Scholar]
- 33. Reich M, Dacheva I, Nobl M, et al.. Proteomic Analysis of Vitreous Humor in Retinal Vein Occlusion. PLoS One. 2016; 11: e0158001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zanoni I, Granucci F.. Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol. 2013; 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stasi A, Intini A, Divella C, et al.. Emerging role of Lipopolysaccharide binding protein in sepsis-induced acute kidney injury. Nephrol Dial Transplant. 2017; 32: 24–31. [DOI] [PubMed] [Google Scholar]
- 36. Yusuf IH, Garrett AM, MacLaren RE, Charbel Issa P. Retinal cadherins and the retinal cadherinopathies: Current concepts and future directions. Prog Retin Eye Res. 2022; 90: 101038. [DOI] [PubMed] [Google Scholar]
- 37. Keenan TD, Clark SJ, Unwin RD, Ridge LA, Day AJ, Bishop PN.. Mapping the differential distribution of proteoglycan core proteins in the adult human retina, choroid, and sclera. Invest Ophthalmol Vis Sci. 2012; 53: 7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu X, Xing Y, Liu X, Zeng L, Ma J.. Opticin Ameliorates Hypoxia-Induced Retinal Angiogenesis by Suppression of Integrin α2-I Domain-Collagen Complex Formation and RhoA/ROCK1 Signaling. Invest Ophthalmol Vis Sci. 2022; 63: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patnaik S, Rai M, Jalali S, et al.. An interplay of microglia and matrix metalloproteinase MMP9 under hypoxic stress regulates the opticin expression in retina. Sci Rep. 2021; 11: 7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Massoudi D, Germer CJ, Glisch JM, Greenspan DS.. Procollagen C-proteinase enhancer 1 (PCPE-1) functions as an anti-angiogenic factor and enhances epithelial recovery in injured cornea. Cell Tissue Res. 2017; 370: 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Potthoff J, Bojarski KK, Kohut G, et al.. Analysis of Procollagen C-Proteinase Enhancer-1/Glycosaminoglycan Binding Sites and of the Potential Role of Calcium Ions in the Interaction. Int J Mol Sci. 2019; 20(20): 5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilson MR, Satapathy S, Jeong S, Fini ME.. Clusterin, other extracellular chaperones, and eye disease. Prog Retin Eye Res. 2022; 89: 101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim JH, Kim JH, Yu YS, Min BH, Kim KW.. Protective effect of clusterin on blood-retinal barrier breakdown in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2010; 51: 1659–1665. [DOI] [PubMed] [Google Scholar]
- 44. Harney DJ, Hutchison AT, Su Z, et al.. Small-protein Enrichment Assay Enables the Rapid, Unbiased Analysis of Over 100 Low Abundance Factors from Human Plasma. Mol Cell Proteomics. 2019; 18: 1899–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee HY, Kim EG, Jung HR, et al.. Refinements of LC-MS/MS Spectral Counting Statistics Improve Quantification of Low Abundance Proteins. Sci Rep. 2019; 9: 13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.