Abstract
The entomopathogenic bacterium Xenorhabdus bovienii exists in a mutualistic relationship with nematodes of the genus Steinernema. Free-living infective juveniles (IJs) of Steinernema prey on insect larvae and regurgitate X. bovienii within the hemocoel of a host larva. X. bovienii subsequently produces a complex array of specialized metabolites and effector proteins that kill the insect and regulate various aspects of the trilateral symbiosis. While Xenorhabdus species are rich producers of secondary metabolites, many of their biosynthetic gene clusters remain uncharacterized. Here, we describe a nonribosomal peptide synthetase (NRPS) identified through comparative genomics analysis that is widely conserved in Xenorhabdus species. Heterologous expression of this NRPS gene from X. bovienii in E. coli led to the discovery of a family of lipo-tripeptides that chromatographically appear as pairs, containing either a C-terminal carboxylic acid or carboxamide. Co-expression of the NRPS with the leupeptin protease inhibitor pathway enhanced production, facilitating isolation and characterization efforts. The new lipo-tripeptides were also detected in wildtype X. bovienii cultures. These metabolites termed bovienimides share an uncommon C-terminal D-citrulline residue. The NRPS lacked a dedicated chain termination domain resulting in product diversification and release from the assembly line through reactions with ammonia, water, or exogenous alcohols.
Introduction
The Xenorhabdus and Photorhabdus genera consist of entomopathogenic (insect pathogenic) Gammaproteobacteria that colonize the guts of nematodes in the genera Steinernema and Heterorhabditis, respectively.1, 2 These bacteria share a mutualistic symbiosis with the nematodes while engaging in parasitic relationships with insect hosts. The soil-dwelling infective juvenile (IJ) nematodes release the bacteria upon entering an insect larval prey, and the bacteria produce various bioactive secondary metabolites to facilitate colonization and killing of the insect prey, regulation of nematode development, and competition with other microbes feeding on the insect carcass.3–5 Genomic sequencing of Xenorhabdus and Photorhabdus isolates suggests that they dedicate as much as 6.5% of their genomes to natural product biosynthesis, which is comparable to the prolific Streptomyces natural product producers.4, 6
While many of the widely conserved biosynthetic gene clusters (BGCs) in both Xenorhabdus and Photorhabdus encode essential biological functions for their symbioses, specific metabolites produced by either Xenorhabdus or Photorhabdus have also been described, highlighting significant differences in BGCs and metabolic potential between the two genera.7 For example, Xenorhabdus produce xenocoumacin or amicoumacin antibacterials, whereas stilbenes are produced exclusively by all Photorhabdus species.8–14 Here, through comparative genomics analysis, we identified a nonribosomal peptide synthetase (NRPS) gene that is highly conserved in most Xenorhabdus species, but it is not present in Photorhabdus species. We characterized the metabolites of this pathway by heterologous expression in E. coli and compared their production levels with native metabolites in the representative X. bovienii strain SS-2004. These studies identified citrulline-functionalized lipo-tripeptides that appear to be diversified through a nucleophile-driven chemical offloading mechanism.
Results
Homologs of NRPS XBJ1_2367 are widely present in Xenorhabdus species.
A subset of polyketide synthase (PKS) and NRPS BGCs is conserved across the sequenced X. bovienii strains (Table S1).15 One such BGC, denoted as XBJ1_2367 in X. bovienii SS-2004, encodes a single 11.5 kb NRPS gene with three condensation-adenylation-thiolation (C-A-T) extension modules and an atypical C-T termination module (Figure 1A). The first starter condensation domain suggested an acylation as the first step of the biosynthesis, followed by three additional amide bond formation steps. The adenylation domain code prediction suggested the first amino acid residue to be leucine and the second to be alanine.16–18 However, the third adenylation domain sequence lacked candidate predictions. The second and fourth condensation domains were annotated as having dual condensation/epimerization (C/E) functionalities, suggesting two possible epimerization events. We confirmed this by sequence alignment with known C/E domains in the representative xenematide gene cluster (Figure S1).19 In X. bovienii strain CS03, the gene appears to be encoded on two open reading frames, with the last thiolation domain encoded in the second open reading frame (Table S2).
Figure 1.
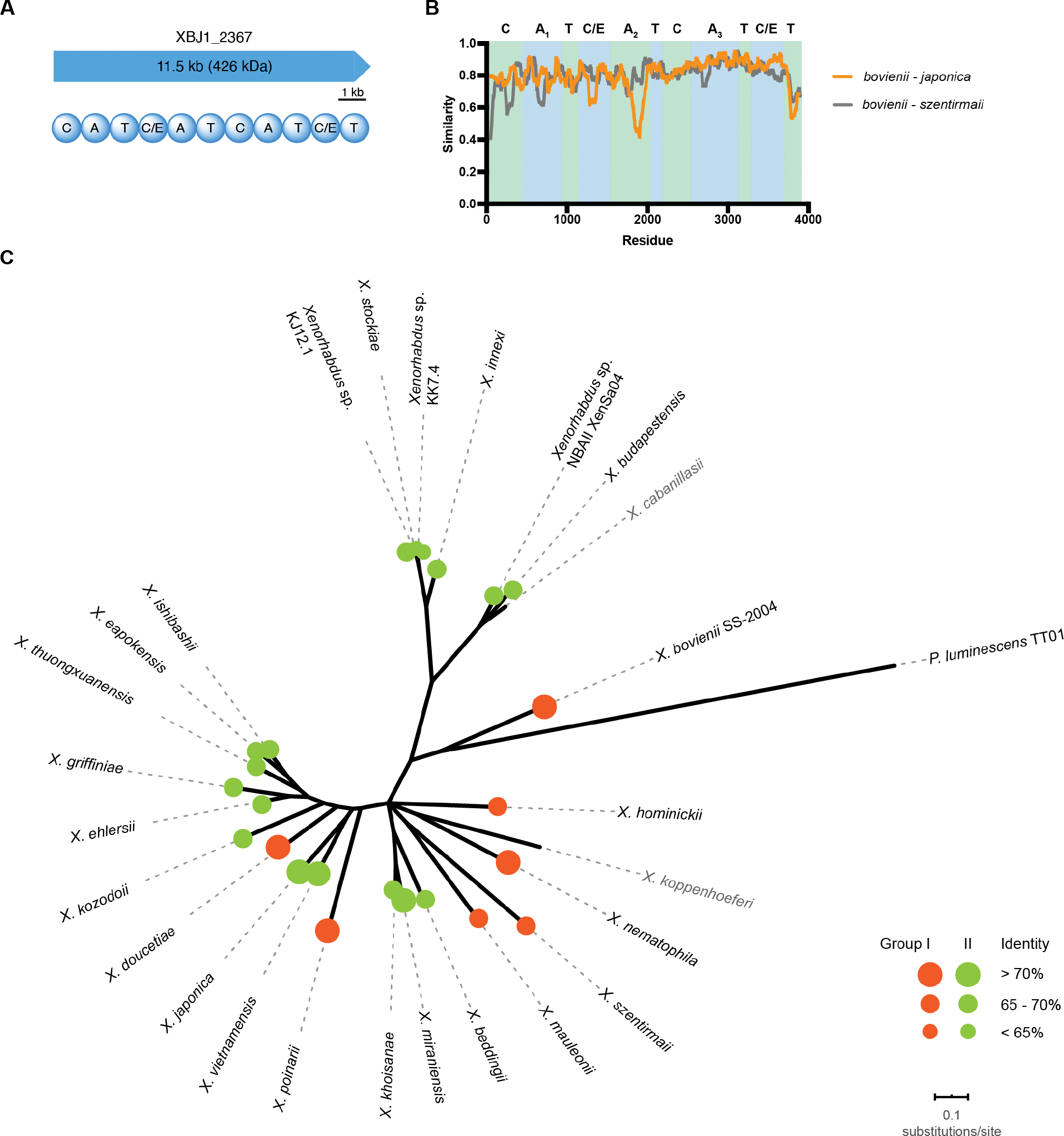
Domain architecture and distribution of XBJ1_2367 homologs in the genus Xenorhabdus. A, Domain architecture of NRPS XBJ1_2367. B, Sliding window analysis of amino acid sequences within group I (bovienii - szentirmaii) and between group I and II (bovienii - japonica). C, Unrooted phylogenetic tree of Xenorhabdus species and the distribution of XBJ1_2367 homologs. The maximum likelihood tree was based on the concatenated protein sequence alignments of five conserved housekeeping genes. Branches less than 50% of bootstrap frequencies were collapsed. Species without the gene homolog are shown in grey. Sizes of the circles represent protein sequence similarity to XBJ1_2367 (Orange circles: group I; Green circles: group II).
By searching the available Xenorhabdus genome assemblies in the National Center for Biotechnology Information (NCBI) Genome database, we found gene homologs of XBJ1_2367, with the same NRPS architecture, in most Xenorhabdus species (24 out of 26 analyzed, with the exception of X. cabanillasii and X. koppenhoeferi). In contrast, none of the Photorhabdus species contained this pathway, suggesting a function unique to Xenorhabdus (Figure 1). All identified homologs are more than 60% identical at the amino acid level compared to XBJ1_2367 in X. bovienii SS-2004 (Table S2). Additionally, we compared all three adenylation domains using an NRPS predictor algorithm16 and found that the A domain specificity codes are highly conserved across all strains, especially the unusual A3 domain (Figure S2, Table S2). The specificity codes of the A1 domains appeared to be highly conserved, whereas the specificity domain codes of the A2 domains were able to be classified into two groups (Figure S2). Group I, which includes XBJ1_2367, has the consensus sequence DLYNNALT, whereas the group II consensus sequence is DVW(H/Y)LSLI. Indeed, sliding window analyses between group I and group II proteins emphasizes a reduced conservation between group I and II A2 domains (Figure 1B).
NRPS XBJ1_2367 encodes a family of lipopeptides
The conserved nature of XBJ1_2367 prompted us to elucidate the molecules encoded by this pathway. The gene from X. bovienii SS-2004 was cloned into the pACYC-Duet expression vector and heterologously expressed in E. coli BAP1: an E. coli BL21(DE3) variant containing a promiscuous 4′-phosphopantetheinyl transferase (Sfp) from Bacillus subtilis20 that allows for diverse post-translational activation of NRPS and PKS systems. LC-HRMS-based analysis of ethyl acetate extracts from E. coli expressing the pathway versus an empty vector control revealed roughly 20 pathway-dependent metabolites with similar retention times and mass ranges (Figure 2, Table S4). Most of these metabolites were also detected in wildtype X. bovienii SS-2004 extracts in LB medium (X. bovienii SS-2004 was reisolated from an insect infection prior to analysis in LB), although the production ratios between the two systems were slightly different. For example, metabolites 5 and 6 were produced at similar levels in X. bovienii, whereas in E. coli cultured at 16°C, metabolite 6 was the major product. Tandem MS analysis of these metabolites supported a family of structurally related lipopeptides (Figure S5–15). As expected, none of the metabolites were detected in P. luminescens TT01 organic extracts.
Figure 2.
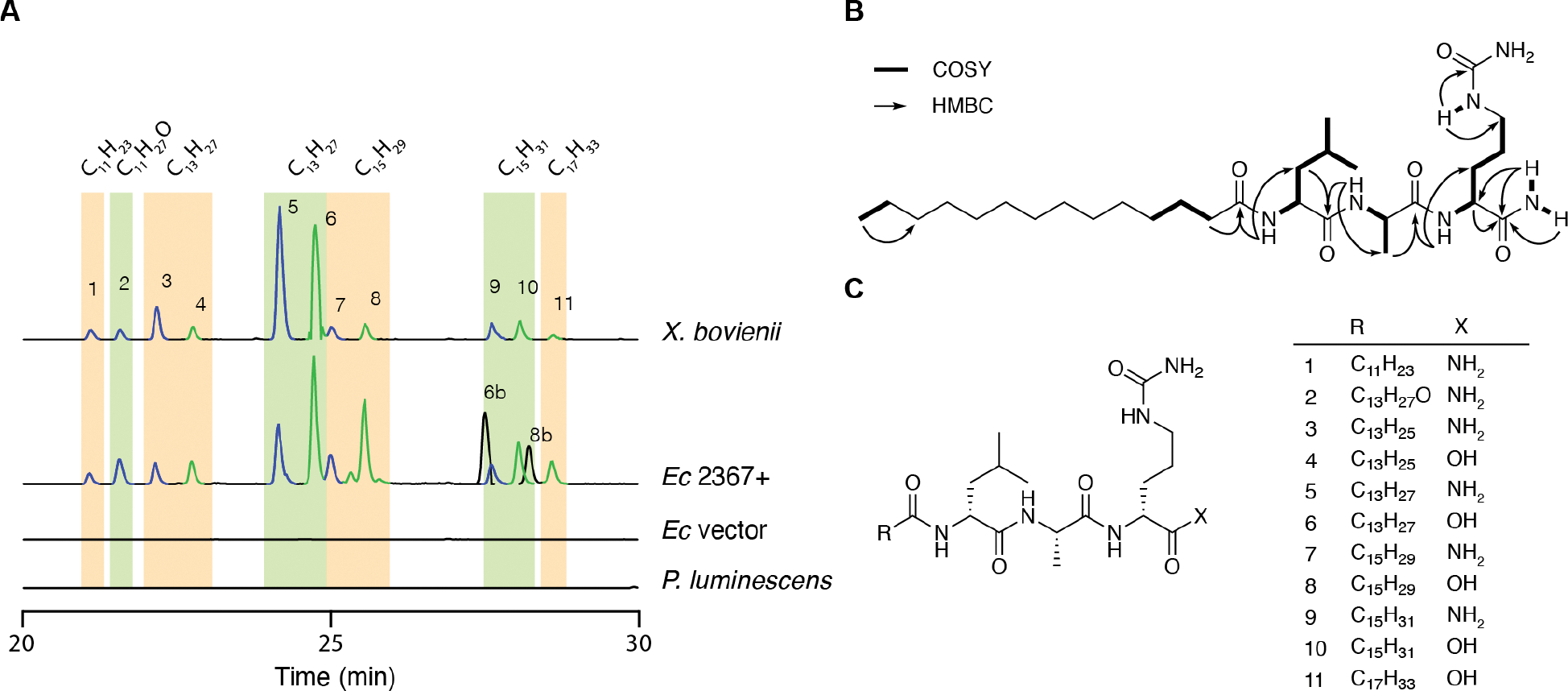
Characterization of XBJ1_2367 dependent metabolites. A, Extracted ion chromatograms of the 11 most abundant lipopeptides in X. bovienii. Carboxamide lipopeptides (X = NH2) are in blue LC traces, whereas free carboxylic acid lipopeptides are in green. Both X. bovienii and P. luminescens were cultured at 30°C, whereas E. coli BAP1 expressing XBJ1_2367 (Ec 2367+) or vector control were expressed at 16°C. B, Key NMR correlations of bovienimide A (5). C, Structures of 11 major bovienimides, which were predicted based on tandem MS and comparison with NMR characterized products 5 and 6.
To further characterize the structures of these small molecules, we purified the major pair of metabolites from E. coli BAP1 heterologously expressing XBJ1_2367. We found that production of these metabolites could be bolstered by co-expressing the leupeptin protease inhibitor pathway (pCDF-Leup)21 from X. bovienii with XBJ1_2367, presumably due to reduced cellular proteolytic degradation of the large multidomain NRPS protein. This workflow in E. coli enabled the ethyl acetate extraction and isolation of 5 and 6 (1.2 mg each) from a 6 L culture (Figure S3). Their molecular formulas were established as C29H56N6O5 and C29H55N5O6, respectively, using HR-MS (m/z 569.4397, Δ = 1.58 ppm; 570.4220, Δ = 1.86 ppm [M+H]+). 1H COSY and HSQC NMR spectra of 5 and 6 showed characteristic correlations of leucine, alanine, and a saturated fatty acid (Figure 2B, Figure S4), as anticipated from the bioinformatic analysis. By comparing tandem MS, HMBC cross-peaks and predicted molecular formulas, we determined that compounds 5 and 6 specifically contained C14 saturated fatty acid chains and unexpected citrulline (Cit) residues at their third amino acid positions. Interestingly, 5 contained an NH2 spin-system in which the N-H protons exhibited HMBC to the carbonyl carbon of the third Cit residue, suggesting a terminal carboxamide rather than an isomeric arginine residue. Only one of these N-H protons (δH 7.00) showed HMBC to the Cit α-carbon, likely due to the olefinic nature of the zwitterionic iminium resonance form.22 Furthermore, the neutral gas phase loss of isocyanic acid (43 Da) in 5 and 6 from the ureido group is also indicative of the presence of citrulline.23 The connectivity of the amino acids was established to be Leu1, Ala2, and Cit3 by tandem MS and HMBC. Given the high conservation of these two major lipopeptide metabolites in X. bovienii, we named them bovienimide A (5) and B (6).
We further established the stereocenters of the bovienimide amino acids using Marfey’s analysis.24 Briefly, we hydrolyzed purified bovienimide B (6) with 6 M HCl, neutralized the mixture, and reacted it with Marfey’s reagent. By comparing the LC-MS traces of the resulting Marfey’s derivatized products against derivatized standards of D/L leucine, alanine, and ornithine (the acid hydrolyzed product of citrulline), we confirmed the configuration to be D-Leu, L-Ala, and D-Cit (Figure S16). This is consistent with the dual C/E domain predictions of the C-2 and C-4 domains using antiSMASH.25
With these new structural insights, we were able to propose 11 pathway-dependent lipopeptides produced in both E. coli and X. bovienii by tandem MS (Figure 2, Figure S5–15). We found that the acyl chains varied from C12 to C18, with either a single unsaturation, one hydroxyl group, or full saturation. Most of the metabolites appeared as pairs, consistent with the free carboxylic acids versus carboxamides at the C-terminus. In addition, we also identified two metabolites, 6b and 8b, that were exclusively produced in E. coli; tandem MS predicted an extra C2H4 unit attached to the third residue (Figure S18–19). In our heterologous expression system in E. coli, chloramphenicol was used to maintain pACYC-XBJ1_2367. Given this, we suspected this C2H4 unit could be derived from the addition of ethanol, the chloramphenicol solvent vehicle. We then cultured our E. coli strain with chloramphenicol dissolved in methanol instead and indeed found a drastic reduction of 6b and 8b (Figure S17) with the concomitant detection of the expected methyl ester of 6 (6c, supported by tandem MS). Adding an equal volume of ethanol to the culture restored the intensity of 6b and 8b to the same level as cultures grown using a chloramphenicol-ethanol stock solution. Finally, addition of n-butanol gave rise to the corresponding butyl ester product (6d, supported by tandem MS) (Figure S20–21). Thus, we proposed the biosynthesis of bovienimides, where the Cstart domain utilizes acyl-CoA to catalyze amide bond formation with an activated L-Leu (Figure 3). The second C/E domain catalyzes the amide bond formation between L-Ala and also epimerizes L-Leu to D-Leu. The third C domain condenses activated L-citrulline. Finally, the last C/E domain catalyzes the epimerization reaction to form D-citrulline containing bovienimides, and it is conceivable that this domain participates in carboxamide offloading. The timing, assignment, and ordering of these proposed events would need to be supported in future protein biochemical studies. The lack of a dedicated thioesterase domain could alternatively suggest spontaneous hydrolysis by environmental nucleophiles such as water, alcohol, and ammonia, although we cannot rule out other enzymatic contributions in E. coli or X. bovienii. Interestingly, the accumulation of ammonia is thought to play a key role in triggering the emergence of infective juveniles from insect cadavers.26, 27
Figure 3.
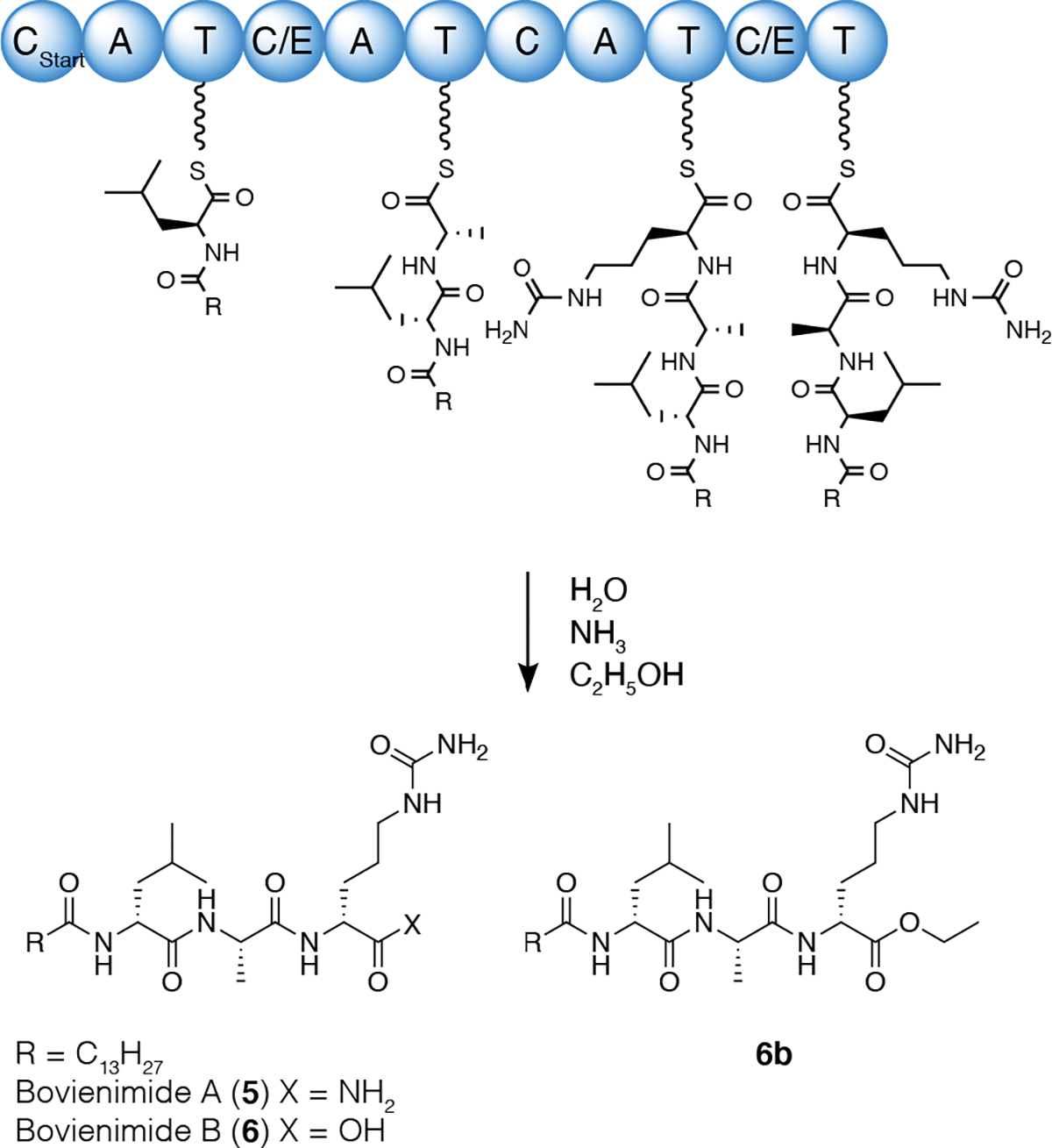
Proposed biosynthesis of bovienimides.
Phylogenetic analysis of XBJ1_2367 homologs in Xenorhabdus
We constructed the maximum likelihood phylogenetic tree of XBJ1_2367 using NGPhylogeny with all DNA sequences available on Magnifying Genomes (MaGe).28, 29 As this gene currently has no detectable homolog in species other than Xenorhabdus, we depicted it as an unrooted tree with bootstrap branch support (Figure 4). We found that the group I and group II NRPSs are separated into two distinct clades. In group I, branches between different species are relatively long compared to branches in group II. In group II clades, group IIA and group IIB have the same tree topology with the species phylogenetic tree, whereas a few species (X. japonica, X. vietnamensis, X. beddingii, X. miraniensis, X. khoisanae) are incongruent with their species tree topology. We also reconstructed the phylogenetic relationship between all the A domains in X. bovienii SS-2004 and these gene homologs (Figure S22). All A3 domains clustered in a single clade and were related to several A domains belonging to the PAX-peptide NRPS (paxABC).30 A1 domains also clustered into one clade, except for the A1 domain from X. hominickii. A2 domains were split into two clades, where group I and group II formed two separate clades. These adenylation domain analyses will aid in the refinement of adenylation domain predictions for other unknown NRPS systems.
Figure 4.
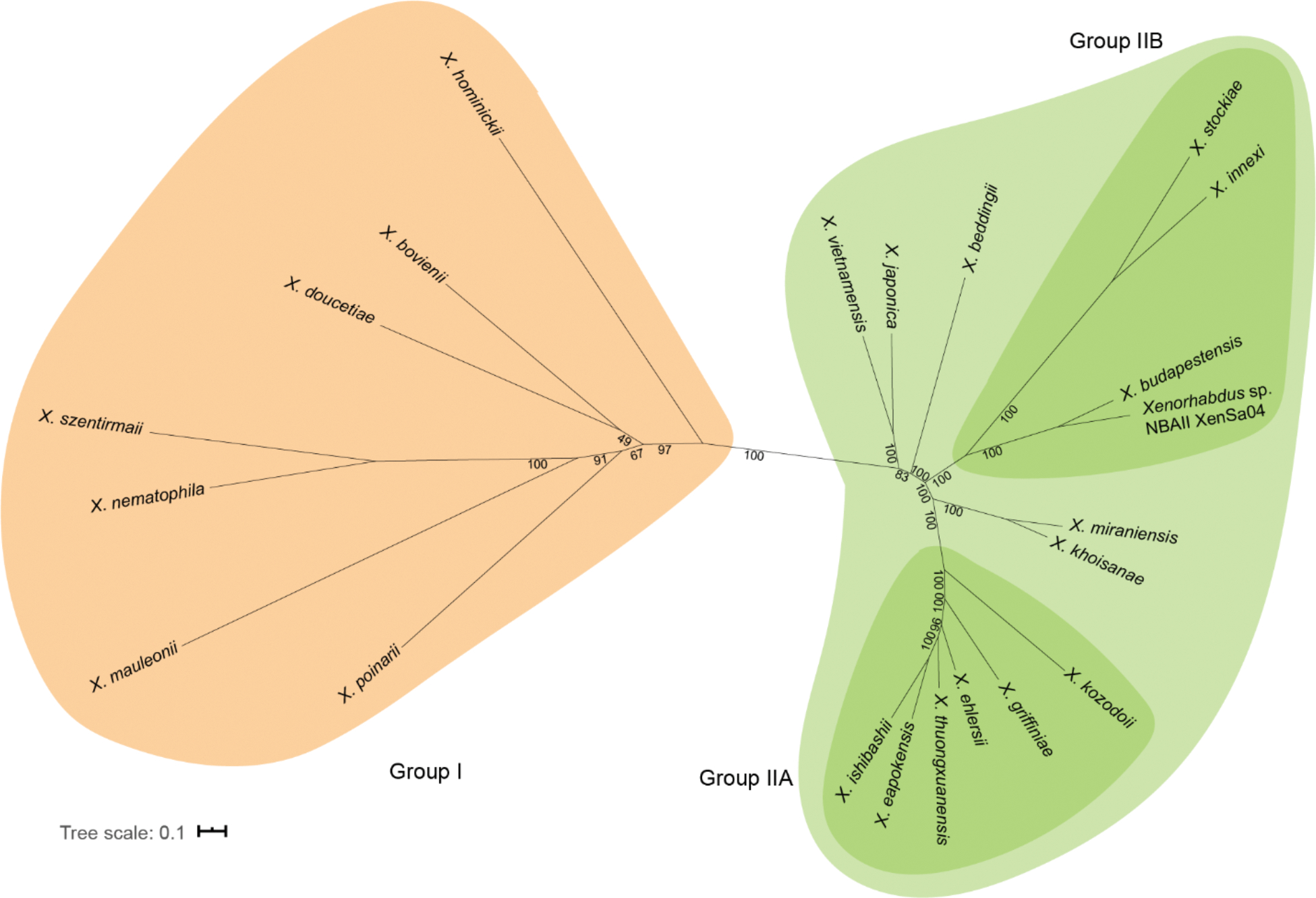
Phylogenetic relationship of XBJ1_2367 homologs. Maximum likelihood tree of XBJ1_2367. Group I members are in orange and group II members are in green. Numbers at the internal nodes indicate the percent bootstrap frequency (100 replicates).
Lipopeptides are a class of natural products with a wide range of bioactivity, including antibacterial, antifungal, cell signaling, and biosurfactant properties.31 We tested the antimicrobial activity of 5 and 6 against representative Gram-positive (Bacillus subtilis), Gram-negative (E. coli DH5α), and fungal species (Saccharomyces cerevisiae), but no growth inhibition was observed up to 500 μM. Additionally, no cellular toxicity was observed against a human cell line (THP-1) up to 100 μM. A Similarity Ensemble Approach (SEA) search with bovienimides A and B computationally predicted the human G-protein-coupled receptor (GPCR) C3a as a potential target;32 however, no activation of C3a was observed using the PRESTO-Tango assay system at concentrations up to 100 μM.33 Finally, we tested these compounds in human THP1-Dual™ reporter cells and no interferon stimulated gene (ISG) or NF-κB activation were observed at concentrations up to 100 μM. Establishing the biological function of these new metabolites remain an exciting future objective.
Discussion
Secondary metabolites provide reproductive fitness to their producer in its ecological niche. NRPS and PKS systems are major biosynthetic strategies for specialized metabolite biosynthesis, and their often modular organization provides a route to product diversification via gene duplication and rearrangement events of different modules/domains within these megasynth(et)ases. Examining the phylogenetic relationships of these biosynthetic pathways provides a snapshot of how these genes may have evolved and how to predict functionality in other uncharacterized biosynthetic systems. The highly conserved nature of XBJ1_2367 homologs in Xenorhabdus suggests the importance and possible vertical inheritance of this pathway. However, a closer look at the adenylation domains reveals major divergence in the A2 domain, resulting in two types of gene homologs that we designate as group I and group II. By reconstructing the phylogenetic tree of all the adenylation domains, we found that these two types of A2 domains are evolutionarily distant from each other despite the high overall sequence homology of the protein.
The A3 domain in XBJ1_2367 homologs are highly conserved in all the strains examined. Phylogenetic analysis of the A3 domain shows a close relationship to several A domains in the PAX peptide BGC (paxABC) in X. bovienii (Figure S22). PAX-peptides are lysine-rich, antifungal cyclic peptides originally isolated from X. nematophila.30, 34 These A domains in PaxABC have an acidic residue in their specificity code, which matches the lysine selective activation. The substitution of aspartate/glutamate to serine together with another hydrogen bond forming asparagine in the binding pocket of the A3 domain appear to have created an environment selective for citrulline rather than lysine. As citrulline is a less common NRPS substrate, this adenylation code assignment may improve the accuracy of NRPS bioinformatic prediction pipelines.
Lipopeptides are a diverse class of molecules containing a nonpolar fatty acid chain and amino acid residues of varying length and hydrophobicity. Many bacterial lipopeptides originate from NRPSs.35, 36 These amphiphilic molecules display a variety of bioactivities from antimicrobial, surfactant, to cell signaling.37 Most antimicrobial lipopeptides are macrocyclic; however, there are also several reports of linear lipopeptides with bioactivities such as paenipeptin (antibacterial) and holrhizin A (surfactant).38 Lipopeptides with less than 5 amino acids are not commonly reported.
It has been shown that lipo-tripeptides and lipo-pentapeptides from Mycobacterium avium have immunomodulatory properties in ruminants.39 Although bovienimide A and B were not stimulatory for the computationally predicted human GPCR C3a receptor or for interferon/NF-κB signaling in human THP-1 cell-based immunomodulatory assays under the conditions of our studies, it is possible that these metabolites mediate cell signaling programs to aid insect infection and/or nematode development with their Steinernema hosts. The XBJ1_2367 homolog in X. nematophila ATCC19061 (XNC1_2799) was shown to be negatively correlated with the leucine-responsive regulatory protein (lrp).40 Lrp homologs are LysR-type regulatory proteins that are conserved across different bacteria and are responsible for phenotypic lifestyle switching in Xenorhabdus. In X. nematophila, the phenotypic transition from pathogenic to mutualistic states is associated with the upregulation of Lrp.41, 42 Moreover, in IJs the Lrp expression level decreases prior to an infection cycle.43 Altogether, these observations suggest that bovienimides may serve as insect virulence factors or as IJ-nematode signaling molecules. Intriguingly, it has been shown that the hybrid PKS-NRPS in Caenorhabditis elegans produces lipopeptides (nemamides) to regulate larval development.44
In summary, we identified a conserved NRPS gene widely found in Xenorhabdus species. We characterized the structures encoded by this gene in X. bovienii and employed bioinformatic analysis to propose a biosynthesis for their construction. The A3 domain in this pathway, which exhibited an ambiguous bioinformatic specificity code, appears to activate citrulline. Finally, the citrulline-functionalized lipopeptide can be offloaded by various nucleophiles (i.e., alcohols, ammonia, and water). These studies add to our understanding of the structure, diversification, and biosynthesis of citrulline-containing natural products.
EXPERIMENTAL SECTION
Bioinformatics analyses
Orthologs of XBJ1_2367 were identified using National Center for Biotechnology Information (NCBI) blast search, and domain architectures were confirmed using PKS/NRPS Analysis Website 16. The unrooted trees for these sequences (nucleotide sequences) were inferred with NGPhylogeny.fr using the maximum likelihood method 28. The unrooted species tree was inferred from concatenated protein alignments of the following five conserved housekeeping proteins: AlaS, UvrC, RecN, RadA and PyrG. Pairwise sliding window analysis of the NRPS protein sequences was performed as follows: protein sequences of NRPS homologs were uploaded to The Scorecons Server (https://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/valdar/scorecons_server.pl) for similarity score calculation using default settings,45 and the similarity was calculated by averaging the scores with a 100-residue interval.
Cloning of XBJ1_2367 into E. coli BAP1
Xenorhabdus bovienii SS-2004 gDNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen) and used as a polymerase chain reaction (PCR) template. Primers ACYC_PacI and ACYC_HindIII were used to PCR amplify pACYC-Duet, and primers 2367_I_PacI, 2367_I_HindIII, 2367_II_HindIII, and 2367_II_XhoI were used to amplify XBJ1_2367 into 2 pieces with internal HindIII restriction sites (Table S3). NucleoSpin Gel and PCR Clean-up kit (Takara Bio) was used for purification of PCR products. Amplified pACYC-Duet and 2367_I were purified, restriction digested with PacI and HindIII at 37°C for 2 hours, purified, and ligated with T4 ligase (NEB) at 4°C overnight. The ligated product was transformed into E. coli DH5α through electroporation, recovered at 37°C for 1 hour, and plated on Luria-Bertani (LB) agar plates with 34 μg/mL chloramphenicol. Single colonies were inoculated for plasmid extraction using QIAprep Spin Miniprep Kit (Qiagen) to verify the correct incorporation of the first fragment (2367_I). Verified plasmid containing 2367_I and PCR amplified 2367_II were digested with HindIII and XhoI, and the plasmid was further dephosphorylated with calf intestinal alkaline phosphatase (NEB). Both products were purified and ligated with T4 ligase at 4°C overnight. The ligated product was transformed into DH5α through electroporation, recovered in SOC media at 37°C for 1 hour, and plated on LB agar plates with 34 μg/mL chloramphenicol. Colonies were inoculated for plasmid extraction and sequence verification (pACYC-XBJ1_2367). The sequence validated plasmids were transformed into E. coli BAP1 via heat shock, cells were recovered in LB at 37°C for 1 hour and plated on LB agar plates with 34 μg/mL chloramphenicol.
Metabolomics analysis
Both X. bovienii SS-2004 and P. luminescens TTO1 were inoculated from glycerol stock on LB agar plates at 30°C and grown for 2 days. Single colonies were inoculated in 5 mL of LB medium at 30°C for two days at 250 rpm. E. coli BAP1 cells (pACYC-XBJ1_2367 or pACYC-Duet control) were inoculated in LB supplemented with 34 μg/mL chloramphenicol at 37°C until around OD600 0.5, and the cultures were induced with 50 μM IPTG and cultivated overnight at 16°C. Cultures were extracted with 6 mL ethyl acetate, and 5 mL of the extract was dried under reduced pressure. All dried samples were dissolved in 200 μL methanol and subjected to LC-MS analysis. Multiple reaction monitoring (MRM) mode data were collected using an Agilent 6490 Triple-Quad (QQQ) MS system fitted with an electrospray ionization (ESI) source coupled to an Infinity 1290 high-performance liquid chromatography (HPLC) system and a Kinetex 1.7 μm C18 column (100 × 2.1 mm) using water and acetonitrile solvent systems containing 0.1% formic acid at 0.3 mL/min, 0–20 min, 5 to 100% acetonitrile. High-resolution electrospray ionization mass spectrometry (HRMS) data were obtained using an Agilent iFunnel 6550 quadrupole time-of-flight (QTOF) mass spectrometry instrument fitted with an ESI source coupled to an Agilent 1290 Infinity HPLC system and a Kinetex 5 μm C18 100 Å column (250 × 4.6 mm) with a water:acetonitrile gradient containing 0.1% formic acid at 0.7 mL/min: 0–30 min, 5 to 100% acetonitrile. The mass spectra were recorded in positive ionization mode with a mass range from m/z 100 to 1,700. Targeted MS/MS analysis was performed with Iso width set to “narrow width” (1.3 m/z) and fixed collision energies (CE 10, 20, 30, 40).
Isolation and structural elucidation of bovienimide A (5) and B (6)
E. coli BAP1 harboring pACYC-XBJ1_2367 and pCDF-Leup 21 was inoculated from an overnight LB culture into 6 ´ 1 L LB aliquots with addition of spectinomycin (50 μg/mL). The cultures were grown to OD 0.5 at 37°C and 250 rpm. The cultures were then cooled to 16°C, induced with 50 μM IPTG, and grown for 2 days at 16°C and 250 rpm. The cultures were centrifuged at 3,000 g for 30 min, and the supernatants were extracted twice with equal volumes of ethyl acetate. The combined ethyl acetate layer was dried under reduced pressure, yielding 460 mg of material. This crude extract was resuspended in a solution of 10% acetonitrile, 10% methanol, and 80% water. The extract was fractionated using an Agilent PrepStar HPLC system with an Agilent Polaris C18-A 5 μm (250 × 21.2 mm2) column with a gradient of 10% to 100% aqueous acetonitrile containing 0.01% TFA over 0 to 60 min at a flow rate of 8 mL/min. One fraction was collected per minute starting at 1 min, and fraction 51 contained compounds 5 and 6. Compounds 5 and 6 were further separated using the same mobile phase and gradient system with a Phenomenex Luna C18 (2) 100 Å (250 × 10 mm) column at a flow rate of 4 mL/min, yielding 1.2 mg of each compound. 1D- (1H and 13C) and 2D- (gCOSY, zTOCSY, gHSQCAD, and gHMBCAD) NMR spectral data were measured on Agilent 600 MHz NMR spectrometer equipped with a cold probe in a 3-mm tube, and the chemical shifts were recorded as δ values (ppm) referenced to solvent residual signals (See Table S5 and S6).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Sciences (1RM1GM141649-01 to J.M.C.), a Ford Foundation Pre-Doctoral Fellowship (to R.H.), and the National Institutes of Health Chemistry-Biology Interface Pre-Doctoral Training Grant program (5T32GM06754 3–12 to R.H.).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
1D and 2D NMR, assignments, HR-MS spectra, and bioinformatic analysis details. This material is available free of charge via the internet at http://pubs.acs.org
Contributor Information
Jhe-Hao Li, Department of Chemistry, Yale University, New Haven, CT 06511, USA; Institute of Biomolecular Design & Discovery, Yale University, West Haven, CT 06516, USA.
Wooyoung Cho, Department of Chemistry, Yale University, New Haven, CT 06511, USA; Institute of Biomolecular Design & Discovery, Yale University, West Haven, CT 06516, USA.
Randy Hamchand, Department of Chemistry, Yale University, New Haven, CT 06511, USA; Institute of Biomolecular Design & Discovery, Yale University, West Haven, CT 06516, USA.
Joonseok Oh, Department of Chemistry, Yale University, New Haven, CT 06511, USA; Institute of Biomolecular Design & Discovery, Yale University, West Haven, CT 06516, USA.
Jason M. Crawford, Department of Chemistry, Yale University, New Haven, CT 06511, USA; Institute of Biomolecular Design & Discovery, Yale University, West Haven, CT 06516, USA; Department of Microbial Pathogenesis, Yale University School of Medicine, New Haven, CT 06536 USA
REFERENCES
- 1.Herbert EE; Goodrich-Blair H, Friend and foe: the two faces of Xenorhabdus nematophila. Nat. Rev. Microbiol. 2007, 5 (8), 634–46. [DOI] [PubMed] [Google Scholar]
- 2.Forst S; Dowds B; Boemare N; Stackebrandt E, Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 1997, 51, 47–72. [DOI] [PubMed] [Google Scholar]
- 3.Han R; Ehlers RU, Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J. Invertebr. Pathol. 2000, 75 (1), 55–8. [DOI] [PubMed] [Google Scholar]
- 4.Shi YM; Bode HB, Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat. Prod. Rep. 2018, 35 (4), 309–335. [DOI] [PubMed] [Google Scholar]
- 5.Vizcaino MI; Guo X; Crawford JM, Merging chemical ecology with bacterial genome mining for secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 2014, 41 (2), 285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duchaud E; Rusniok C; Frangeul L; Buchrieser C; Givaudan A; Taourit S; Bocs S; Boursaux-Eude C; Chandler M; Charles JF; Dassa E; Derose R; Derzelle S; Freyssinet G; Gaudriault S; Medigue C; Lanois A; Powell K; Siguier P; Vincent R; Wingate V; Zouine M; Glaser P; Boemare N; Danchin A; Kunst F, The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 2003, 21 (11), 1307–13. [DOI] [PubMed] [Google Scholar]
- 7.Chaston JM; Suen G; Tucker SL; Andersen AW; Bhasin A; Bode E; Bode HB; Brachmann AO; Cowles CE; Cowles KN; Darby C; de Leon L; Drace K; Du Z; Givaudan A; Herbert Tran EE; Jewell KA; Knack JJ; Krasomil-Osterfeld KC; Kukor R; Lanois A; Latreille P; Leimgruber NK; Lipke CM; Liu R; Lu X; Martens EC; Marri PR; Medigue C; Menard ML; Miller NM; Morales-Soto N; Norton S; Ogier JC; Orchard SS; Park D; Park Y; Qurollo BA; Sugar DR; Richards GR; Rouy Z; Slominski B; Slominski K; Snyder H; Tjaden BC; van der Hoeven R; Welch RD; Wheeler C; Xiang B; Barbazuk B; Gaudriault S; Goodner B; Slater SC; Forst S; Goldman BS; Goodrich-Blair H, The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One 2011, 6 (11), e27909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McInerney BV; Taylor WC; Lacey MJ; Akhurst RJ; Gregson RP, Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 1991, 54 (3), 785–95. [DOI] [PubMed] [Google Scholar]
- 9.Li J; Chen G; Wu H; Webster JM, Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl. Environ. Microbiol. 1995, 61 (12), 4329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joyce SA; Brachmann AO; Glazer I; Lango L; Schwar G; Clarke DJ; Bode HB, Bacterial biosynthesis of a multipotent stilbene. Angew. Chem. Int. Ed. Engl. 2008, 47 (10), 1942–5. [DOI] [PubMed] [Google Scholar]
- 11.Park D; Ciezki K; van der Hoeven R; Singh S; Reimer D; Bode HB; Forst S, Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol. Microbiol. 2009, 73 (5), 938–49. [DOI] [PubMed] [Google Scholar]
- 12.Park HB; Perez CE; Perry EK; Crawford JM, Activating and attenuating the amicoumacin antibiotics. Molecules 2016, 21 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimer D; Pos KM; Thines M; Grun P; Bode HB, A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat. Chem. Biol. 2011, 7 (12), 888–90. [DOI] [PubMed] [Google Scholar]
- 14.Park HB; Sampathkumar P; Perez CE; Lee JH; Tran J; Bonanno JB; Hallem EA; Almo SC; Crawford JM, Stilbene epoxidation and detoxification in a Photorhabdus luminescens-nematode symbiosis. J. Biol. Chem. 2017, 292 (16), 6680–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murfin KE; Whooley AC; Klassen JL; Goodrich-Blair H, Comparison of Xenorhabdus bovienii bacterial strain genomes reveals diversity in symbiotic functions. BMC Genomics 2015, 16, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachmann BO; Ravel J, Chapter 8 Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. In Methods in Enzymology, Hopwood DA, Ed. 2009; Vol. 458, pp 181–217. [DOI] [PubMed] [Google Scholar]
- 17.Stachelhaus T; Mootz HD; Marahiel MA, The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999, 6 (8), 493–505. [DOI] [PubMed] [Google Scholar]
- 18.Challis GL; Ravel J; Townsend CA, Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000, 7 (3), 211–24. [DOI] [PubMed] [Google Scholar]
- 19.Crawford JM; Portmann C; Kontnik R; Walsh CT; Clardy J, NRPS substrate promiscuity diversifies the xenematides. Org. Lett. 2011, 13 (19), 5144–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer BA; Admiraal SJ; Gramajo H; Cane DE; Khosla C, Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 2001, 291 (5509), 1790–2. [DOI] [PubMed] [Google Scholar]
- 21.Li J-H; Oh J; Kienesberger S; Kim NY; Clarke DJ; Zechner EL; Crawford JM, Making and breaking leupeptin protease inhibitors in pathogenic gammaproteobacteria. Angew. Chem. Int. Ed. Engl. 2020. [DOI] [PubMed] [Google Scholar]
- 22.Stewart WE; Siddall TH, Nuclear magnetic resonance studies of amides. Chemical Reviews 1970, 70 (5), 517–551. [Google Scholar]
- 23.Hao G; Wang D; Gu J; Shen Q; Gross SS; Wang Y, Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J. Am. Soc. Mass Spectrom. 2009, 20 (4), 723–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii K; Ikai Y; Mayumi T; Oka H; Suzuki M; Harada K. i., A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: elucidation of limitations of Marfey’s method and of its separation mechanism. Anal. Chem. 1997, 69 (16), 3346–3352. [Google Scholar]
- 25.Blin K; Shaw S; Steinke K; Villebro R; Ziemert N; Lee SY; Medema MH; Weber T, antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47 (W1), W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.San-Blas E; Gowen SR; Pembroke B, Steinernema feltiae: ammonia triggers the emergence of their infective juveniles. Exp. Parasitol. 2008, 119 (1), 180–5. [DOI] [PubMed] [Google Scholar]
- 27.San-Blas E; Pirela D; Garcia D; Portillo E, Ammonia concentration at emergence and its effects on the recovery of different species of entomopathogenic nematodes. Exp. Parasitol. 2014, 144, 1–5. [DOI] [PubMed] [Google Scholar]
- 28.Lemoine F; Correia D; Lefort V; Doppelt-Azeroual O; Mareuil F; Cohen-Boulakia S; Gascuel O, NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47 (W1), W260–W265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallenet D; Calteau A; Dubois M; Amours P; Bazin A; Beuvin M; Burlot L; Bussell X; Fouteau S; Gautreau G; Lajus A; Langlois J; Planel R; Roche D; Rollin J; Rouy Z; Sabatet V; Medigue C, MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020, 48 (D1), D579–D589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs SW; Proschak A; Jaskolla TW; Karas M; Bode HB, Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 2011, 9 (9), 3130–2. [DOI] [PubMed] [Google Scholar]
- 31.Hamley IW, Lipopeptides: from self-assembly to bioactivity. Chem. Commun. 2015, 51 (41), 8574–83. [DOI] [PubMed] [Google Scholar]
- 32.Keiser MJ; Roth BL; Armbruster BN; Ernsberger P; Irwin JJ; Shoichet BK, Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25 (2), 197–206. [DOI] [PubMed] [Google Scholar]
- 33.Kroeze WK; Sassano MF; Huang XP; Lansu K; McCorvy JD; Giguere PM; Sciaky N; Roth BL, PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22 (5), 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gualtieri M; Aumelas A; Thaler JO, Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 2009, 62 (6), 295–302. [DOI] [PubMed] [Google Scholar]
- 35.Robbel L; Marahiel MA, Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 2010, 285 (36), 27501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L; Vo TD; Kaiser M; Bode HB, Phototemtide A, a Cyclic Lipopeptide Heterologously Expressed from Photorhabdus temperata Meg1, Shows Selective Antiprotozoal Activity. Chembiochem 2020, 21 (9), 1288–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peypoux F; Bonmatin JM; Wallach J, Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol 1999, 51 (5), 553–63. [DOI] [PubMed] [Google Scholar]
- 38.Niehs SP; Scherlach K; Hertweck C, Genomics-driven discovery of a linear lipopeptide promoting host colonization by endofungal bacteria. Org. Biomol. Chem. 2018, 16 (37), 8345–8352. [DOI] [PubMed] [Google Scholar]
- 39.Bannantine JP; Etienne G; Laval F; Stabel JR; Lemassu A; Daffe M; Bayles DO; Ganneau C; Bonhomme F; Branger M; Cochard T; Bay S; Biet F, Cell wall peptidolipids of Mycobacterium avium: from genetic prediction to exact structure of a nonribosomal peptide. Mol. Microbiol. 2017, 105 (4), 525–539. [DOI] [PubMed] [Google Scholar]
- 40.Engel Y; Windhorst C; Lu X; Goodrich-Blair H; Bode HB, The Global regulators Lrp, LeuO, and HexA control secondary metabolism in entomopathogenic bacteria. Front. Microbiol. 2017, 8, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao M; Patel T; Rickman T; Goodrich-Blair H; Hussa EA, High levels of the Xenorhabdus nematophila transcription factor Lrp promote mutualism with the Steinernema carpocapsae nematode host. Appl. Environ. Microbiol. 2017, 83 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casanova-Torres AM; Shokal U; Morag N; Eleftherianos I; Goodrich-Blair H, The global transcription factor Lrp Is both essential for and inhibitory to Xenorhabdus nematophila insecticidal activity. Appl. Environ. Microbiol. 2017, 83 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao M; Goodrich-Blair H, Xenorhabdus nematophila bacteria shift from mutualistic to virulent Lrp-dependent phenotypes within the receptacles of Steinernema carpocapsae insect-infective stage nematodes. Environ. Microbiol. 2020. [DOI] [PubMed] [Google Scholar]
- 44.Shou Q; Feng L; Long Y; Han J; Nunnery JK; Powell DH; Butcher RA, A hybrid polyketide-nonribosomal peptide in nematodes that promotes larval survival. Nat. Chem. Biol. 2016, 12 (10), 770–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valdar WS, Scoring residue conservation. Proteins 2002, 48 (2), 227–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


