Abstract
The seeds of Panax notoginseng (Burk.) F. H. Chen are typically characterized by their recalcitrance and after-ripening process and exhibit a high water content at harvest as well as a high susceptibility to dehydration. Storage difficulty and the low germination of recalcitrant seeds of P. notoginseng are known to cause an obstacle to agricultural production. In this study, the ratio of embryo to endosperm (Em/En) in abscisic acid (ABA) treatments (1 mg·l−1 and 10 mg·l−1, LA and HA) was 53.64% and 52.34%, respectively, which were lower than those in control check (CK) (61.98%) at 30 days of the after-ripening process (DAR). A total of 83.67% of seeds germinated in the CK, 49% of seeds germinated in the LA treatment, and 37.33% of seeds germinated in the HA treatment at 60 DAR. The ABA, gibberellin (GA), and auxin (IAA) levels were increased in the HA treatment at 0 DAR, while the jasmonic acid (JA) levels were decreased. ABA, IAA, and JA were increased, but GA was decreased with HA treatment at 30 DAR. A total of 4,742, 16,531, and 890 differentially expressed genes (DEGs) were identified between the HA-treated and CK groups, respectively, along with obvious enrichment in the ABA-regulated plant hormone pathway and the mitogen-activated protein kinase (MAPK) signaling pathway. The expression of pyracbactin resistance-like (PYL) and SNF1-related protein kinase subfamily 2 (SnRK2s) increased in the ABA-treated groups, whereas the expression of type 2C protein phosphatase (PP2C) decreased, both of which are related to the ABA signaling pathway. As a result of the changes in expression of these genes, increased ABA signaling and suppressed GA signaling could inhibit the growth of the embryo and the expansion of developmental space. Furthermore, our results demonstrated that MAPK signaling cascades might be involved in the amplification of hormone signaling. Meanwhile, our study uncovered that the exogenous hormone ABA could inhibit embryonic development, promote dormancy, and delay germination in recalcitrant seeds. These findings reveal the critical role of ABA in regulating the dormancy of recalcitrant seeds, and thereby provide a new insight into recalcitrant seeds in agricultural production and storage.
Keywords: recalcitrant seed, dormancy, abscisic acid, after-ripening process, Panax notoginseng
Introduction
The seed is divided into orthodox, intermediate, and recalcitrant types according to its storage behavior (Roberts, 1973). Orthodox seeds experience a dehydrating process at maturation and might be stored with a low water content (Matilla and Matilla-Vázquez, 2008). Correspondingly, recalcitrant seeds maintain a high-water content when they mature and fall off, thus making them sensitive to dehydration (Roberts, 1973; Smith and Berjak, 1995; Berjak and Pammenter, 2001). The viability is significantly reduced, and the germinability is obviously low, when the recalcitrant seeds of Acer saccharinum are dehydrated to be about 30% water content (Pukacka and Ratajczak, 2006; Kalemba and Ratajczak, 2018), and when the recalcitrant seeds of Castanea sativa are dehydrated to be below 17.4% water content (Costa et al., 2018). The recalcitrant Richilia dregeana seeds could maintain viability for several weeks at 16°C at the harvested water content, but they only exhibit 4% viability after 8 days of storage at the axis water content of 1.68 g·g−1 dry mass (Drew et al., 2000). The recalcitrant Araucaria angustifolia seeds show a complete loss of embryo viability after storage at 48% water content for 26 months (Gasparin et al., 2020). The dehydration sensitivity of recalcitrant seeds makes them impossible for long-term storage.
Seed dormancy is a strategy for plants to adapt to adverse environments during long-term phylogenies (Gubler et al., 2005; Song et al., 2008). Most recalcitrant seeds usually germinate immediately after they are shed from mother plants (Pritchard, 2004; Graeber et al., 2012), but some recalcitrant seeds show a markedly dormant characteristic (Liu et al., 2005). Morphophysiological dormancy (MPD) is one of the types of seed dormancy, and it is generally caused by incompletely developed embryos (Willis et al., 2014). The embryo of Paeonia lactiflora seeds with MPD dormancy is incompletely developed at harvest and thus has an after-ripening process of 75 days (Lu et al., 2022). After-ripening is a necessary process for MPD seeds before germination (Baskin and Baskin, 2004; Zhang and Shen, 2006; Chen et al., 2008; Yu et al., 2012). Abscisic acid (ABA) is continuously decreased and gibberellin (GA) is significantly increased in the MPD seeds of Musella lasiocarpa during the after-ripening process (Tang, 2014). The increase in endogenous GA and the changed rate of GA/ABA could promote the embryonic development of Paris polyphylla var. Yunnansensis seeds during the after-ripening process and consequently release dormancy (Pu et al., 2016). Panax quinquefolium seeds are characterized by MPD and a long period of dormancy, and exogenous GA treatment could promote them to complete the physiological after-ripening process (Zhao et al., 2000). However, it is not well investigated whether or not exogenous hormones play an important role in regulating the dormancy of recalcitrant seeds at the transcriptional level.
ABA is a critical positive regulator to maintain seed dormancy (Hilhorst, 1995; Kermode, 2005; Kucera et al., 2005). The germination of ABA-deficient mutant seeds is earlier than that of wild-type seeds (Frey et al., 2011), whereas dormancy is enhanced in transgenic lines of ABA-overaccumulated seeds (Qin and Zeevaart, 2002). ABA is highly accumulated in ABA catabolism mutants, which enhances the dormancy of Arabidopsis thaliana seeds (Matakiadis et al., 2009). The de novo synthesis of GA is necessary to release dormancy and is antagonistic to ABA in regulating seed dormancy (Shuai et al., 2017). GA-deficient mutants ga1 and ga2 exhibit enhanced dormancy and inhibited germination (Lee et al., 2002; Shu et al., 2013). The regulation of ABA/GA rate is associated with the metabolic transition required for dormancy release and germination (Yamaguchi, 2008; Nambara et al., 2010), and the maintenance of dormancy depends on a high rate of ABA/GA. The increased GA biosynthesis and the increased ABA degradation result in a low rate of ABA/GA, consequently releasing dormancy (Ali-Rachedi et al., 2004; Cadman et al., 2006). In addition, the application of exogenous auxin (IAA) prevents pre-harvest sprouting in wheat (Triticum aestivum) seeds (Ramaih et al., 2003). It has been demonstrated that the synergistic effect of jasmonic acid (JA) and ABA inhibits seed germination in several species, including Brassica napus, Linum usitatissimum, and A. thaliana (Staswick et al., 1992; Ellis and Turner, 2002). Nevertheless, relatively little effort has been made to investigate the effect of the exogenous hormone ABA on the regulation of recalcitrant seed dormancy.
The PYR/PYL/RCAR-PP2C-SnRKs signaling cascade is a major pathway for ABA to work (Cutler et al., 2010; Hubbard et al., 2010). ABA receptors pyracbactin resistance (PYR), pyracbactin resistance-like (PYL), and the regulatory component of ABA receptors (RCARs) could sense the ABA signal and inhibit the type 2C protein phosphatases (PP2Cs) in A. thaliana (Park et al., 2009; Tischer et al., 2017). The SNF1-related protein kinase subfamily 2 (SnRK2) positively regulates ABA signaling, as evidenced by the fact that the snrk2.2 snrk2.3 snrk2.6 triple mutant is insensitive to ABA (Fujii and Zhu, 2009; Nakashima et al., 2009). ABA signaling is not the only factor in regulating seed dormancy (Carrera et al., 2008). For example, the seeds of gid1a gid1b gid1c multiple mutants are dormant and unable to germinate in the absence of exogenous GA (Griffiths et al., 2006; Iuchi et al., 2007; Voegele et al., 2011), and it has been proven that MAPK signaling cascades are involved in ABA signal transduction and in regulating seed germination (Xing et al., 2008; Jammes et al., 2009). However, it is still not well understood about the role of endogenous ABA signaling in responding to exogenous hormone in recalcitrant seeds during the after-ripening process.
Panax notoginseng (Burk.) F. H. Chen belongs to the genus Panax in the Araliaceae family. The seeds are believed to be typically characterized by recalcitrance and the after-ripening process, having high water content at harvest and being sensitive to dehydration (Cui et al., 1993; Duan et al., 2014). The seed embryos of P. notoginseng belong to the type of MPD (Duan et al., 2010a; Duan et al., 2010b). The 30 days of after-ripening process (DAR) is a critical point for the release of the dormancy of P. notoginseng seeds, and it needs 45–60 days of lamination to complete the after-ripening of the embryos before germination (Yang et al., 2018; Yang et al., 2019; Ge et al., 2021). Our previous study has demonstrated that endogenous ABA shows a decreasing trend, while GA first increases and then decreases in P. notoginseng seeds during the after-ripening process (Yang et al., 2018), and some candidate genes related to seed dormancy have been obtained in P. notoginseng, including ABSCISIC ACID-INSENSITIVE 5 (ABI5), XANTHOXIN DEHYDROGENASE (ABA2), PROTEIN PHOSPHATASE 2C (PP2C), DELLA PROTEIN GA-INSENSITIVE (GAI), and GAINSENSITIVE DWARF 2 (GID2) (Yang et al., 2019). Our work has also shown that exogenous ABA treatment disturbs the balance of endogenous hormones, prolonging the after-ripening process in P. notoginseng seeds (Ge et al., 2021). However, it is still unclear whether exogenous ABA influences the dormant regulation of recalcitrant P. notoginseng seeds at the transcriptional level. In this study, the morphology of embryos, the endogenous hormones, and the differentially expressed genes (DEGs) were determined in control check (CK) and HA-treated P. notoginseng seeds. The results would provide a new insight into the mechanism of dormant regulation in recalcitrant seeds with after-ripening characteristics.
Materials and methods
Plant materials
In November, the mature and plump seeds were selected from the 3-year-old P. notoginseng that was planted at the experimental farm of Miaoxiang Sanqi Technology Company in Wenshan City, Yunnan Province, China. After harvesting, the red peels of seeds were removed, and then disinfected with 5% CuSO4 for 30 min, rinsed with double distilled water (ddH2O) twice, and dried indoors. The seeds were soaked in ABA hormone solution for 24 h, and their mass concentrations are 1 mg·l−1 and 10 mg· l−1 (LA and HA). The ratio is 1:3–4 between the seeds and the soaking solution, and the CK was treated with distilled water. Subsequently, the seeds were mixed with wet sand after high-temperature disinfection at a volume ratio of 1:5, and the humidity of the river sand is about 25%. The seeds were placed in well-ventilated, fully meshed baskets in a sand-laminated chamber at 15 ± 5°C and kept away from light for 50–60 days to accomplish the after-ripening process. Each treatment was designed with three replicates. At each sampling point (0, 30, and 50 DAR), the samples of the CK and ABA-treated groups were selected for further assays.
Measurement of morphological and physiological indicators
Seeds were selected to observe seed development at five time points: 0, 15, 30, 45, and 60 DAR. The seeds of P. notoginseng were cut along the embryo lengthwise and placed under a stereoscopic microscope (SteREO Discovery V.20, Zeiss, Germany, RRID : SCR_016980) equipped with a digital camera to measure the lengths of embryo (Em) and endosperm (En), and thus the rate of embryo and endosperm (Em/En) was calculated. The measurement was repeated 20 times independently, and three biological replicates were performed. At 45 DAR, the seeds were placed in seeding trays (100 seeds per tray) containing wet sand and incubated at 20°C in an incubator. The number of germinating seeds was recorded and counted until 60 DAR. The contents of endogenous hormones were determined at 0, 30, and 50 DAR by using HPLC-MS (high-performance liquid chromatography-tandem mass spectrometry), including ABA, GA, IAA, and JA in the CK and ABA-treated P. notoginseng seeds (Pan et al., 2010).
Total RNA extraction and transcriptome analysis
The TaKaRa MiniBEST Plant RNA Extraction Kit was used to extract total RNA from samples. The quality of RNA samples was tested by using 1% agarose electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA, RRID : SCR_013575). Sequencing libraries were obtained using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA, RRID : SCR_010233), and they were sequenced on an Illumina platform (NEB, USA, RRID : SCR_010233) by the Novegene Technology Company (Beijing, China). The clean reads were obtained by removing low-quality and adapter-containing reads and then compared to the P. notogensing reference genome (Fan et al., 2020) using HISAT2 v2.0.5 (RRID : SCR_015530). The transcript abundances of genes were estimated by fragments per kilobase of transcript per million fragments mapped (FPKM). The distribution of log2(FPKM + 1) showed relatively high gene expression ( Figure S1 ). Correlation analysis was performed to examine the repeatability of the experiment, and the reliability of pairwise correlation of all samples was assessed using the Pearson Correlation Coefficient (R) ( Figure S2 ). All R2 values were close to 1 between the three biological replicates of the six samples in this experiment, indicating that the sequencing quality of the transcriptome was satisfactory.
In this study, we used the DESeq2 R package (1.16.1, RRID : SCR_015687) to compare the treated group with the CK group. According to the method of Benjamini and Hochberg (1995), genes were designated as having differential expression at P-values < 0.05. The clusterProfiler R package (RRID : SCR_016884) was used for GO and KEGG analyses.
Validation of gene expression
The validation of gene expression was determined by quantitative real-time PCR (qRT-PCR). Primers for qRT-PCR were designed using the Primer3 website (https://primer3.ut.ee/ , RRID : SCR_003139) and synthesized by Tsingke Biotech Co., Ltd. (Yunnan, China). All primers are shown in Table S2 . The RNA was synthesized into single-strand cDNA by the Prime Script RT reagent kit after extracting from each sample (Takara Bio, Kyoto, Japan, RRID : SCR_021372). The QuantStudio12 K Flex System (Thermo Fisher Scientific, RRID : SCR_008452) was used to perform PCR with three technical replicates. The GLYCERALDEHYDE-3-PHOSPHATE DEHYDRO GENASE (GAPDH) was selected as the internal reference for P. notoginseng seeds. The relative expression of the candidate genes was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Statistical analysis of data
GraphPad Prism 8 (RRID : SCR_002798) and Adobe Photoshop CS6 (RRID : SCR_014199) were used for mapping, and Adobe Illustrator Artwork 21.0 was used for a combination. Statistics were analyzed using SPSS 26.0 (RRID : SCR_002865) by one-way ANOVA and Duncan’s multiple comparison method for a significant difference analysis (P < 0.05), and the variables are displayed as the mean ± SD (n = 3).
Results
Effects of ABA treatment on physiological indexes of P. notoginseng seeds
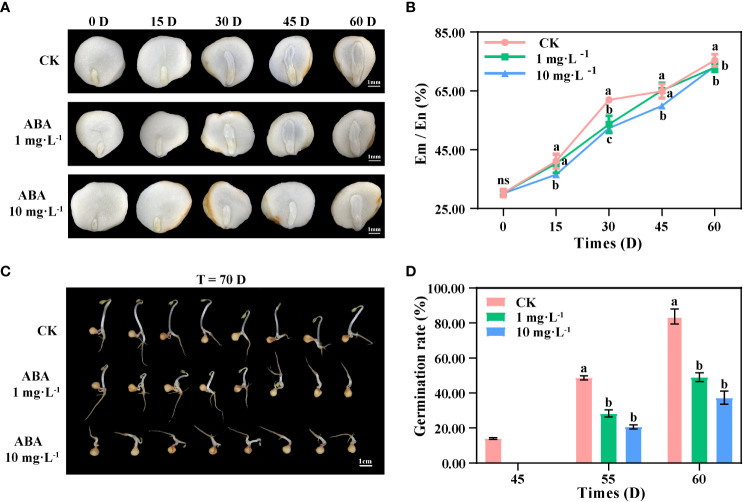
In this study, P. notoginseng seeds were cultivated in land stratification during the after-ripening process for observing the morphological development process. As shown in Figures 1A, B , the development of seed embryos was significantly inhibited during 0–30 DAR. In the LA-treated seeds and HA-treated seeds, the rates of Em/En at 30 DAR were 53.64% and 52.34%, being significantly lower than those in the CK (61.98%), whereas at 45 DAR, the rates of Em/En were 64.88% (CK), 65.16% (LA), and 59.92% (HA), respectively. It indicated that the development of seed embryos was delayed for at least 15 days under the HA treatment at the early stage of the after-ripening process. The development of seed embryos seemed to be accelerated with the ABA treatments after 30 DAR; the rates of Em/En in the CK, LA-treated seeds, and HA-treated seeds reached about 75% at 60 DAR ( Figures 1A, B ), indicating that the exogenous ABA had an inhibitory effect on the embryonic development of P. notoginseng seeds at the early after-ripening stage and that the inhibition of embryonic development was deepened by the increased ABA concentration ( Figures 1A, B ).
Figure 1.
Exogenous ABA inhibits the development of seed embryos and delays seed germination in Panax notoginseng during the after-ripening process. (A) The morphological development of P. notoginseng seed embryos. (B) Changes in the rate of Endosperm to Embryo (Em/En) of P. notoginseng seeds. (C) Appearance and morphology of P. notoginseng seeds after germination (T = 70 DAR (days during after-ripening)). (D) Changes in the rate of germination of P. notoginseng seeds. Values presented are the means ± SE (n = 3). Different letters indicate significant differences among treatments in the same period using Duncan’s test (P < 0.05).
At 45 DAR, 14% of the CK seeds germinated, while no seeds germinated in the LA-treated groups or the HA-treated groups ( Figure 1D ). At 55 DAR, the germination rate of LA-treated seeds and HA-treated seeds was 28.33% and 20.67%, being significantly lower than that of the CK (48.67%). At 60 DAR, the germination rates of the CK, LA-treated seeds, and HA-treated seeds were 83.67%, 49%, and 37.33%, respectively ( Figures 1C, D ). Our results showed that the germination of LA-treated seeds was delayed for about 5 days, while that of HA-treated seeds was delayed for more than 10 days.
Effects of ABA treatment on endogenous hormones of P. notoginseng seeds
As shown in Table 1 , the endogenous hormone ABA was extensively accumulated at 0 DAR and then decreased at 30 DAR and 50 DAR; however, the ABA content was increased under ABA treatment. Exogenous ABA increased GA content at 0 DAR, whereas endogenous GA accumulation was inhibited in the HA-treated seeds at 30 DAR and 50 DAR ( Table 1 ). During the after-ripening process, the IAA content was gradually decreased in the CK and HA-treated seeds, but the IAA content was higher in the HA-treated seeds than the ones in the CK at 0 and 30 DAR ( Table 1 ). The JA content was increased in both the CK and HA-treated seeds during the after-ripening process; however, in the HA-treated seeds, the JA content was higher than that in CK seeds at 30 and 50 DAR ( Table 1 ). Specifically, JA content was decreased in HA-treated seeds at 0 DAR ( Table 1 ). Furthermore, the reduction in GA and the increase in ABA resulted in a low rate of ABA/GA under the ABA treatment when compared with the CK ( Tables 1 , 2 ).
Table 1.
Changes of endogenous hormones in the control (CK) and10 mg·l−1 ABA-treated (HA) P. notoginseng seeds during the after-ripening process.
| Endogenous hormones | Time (DAR) | Treatments | |
|---|---|---|---|
| CK (ng·g−1) | 10 mg·l−1 ABA (ng·g−1) | ||
| ABA | 0 D | 0.25 ± 0.06 b | 2.83 ± 0.92 a |
| 30 D | 0.15 ± 0.02 a | 0.16 ± 0.16 a | |
| 50 D | 0.12 ± 0.03 a | 0.12 ± 0.01 a | |
| GA | 0 D | 0.43 ± 0.10 a | 0.77 ± 0.06 a |
| 30 D | 0.20 ± 0.20 a | 0.17 ± 0.17 a | |
| 50 D | 0.29 ± 0.10 a | 0.15 ± 0.03 a | |
| IAA | 0 D | 20.47 ± 1.90 a | 23.80 ± 3.52 a |
| 30 D | 12.70 ± 1.35 a | 13.91 ± 1.05 a | |
| 50 D | 7.11 ± 2.93 a | 6.70 ± 0.29 a | |
| JA | 0 D | 0.18 ± 0.03 a | 0.17 ± 0.01 a |
| 30 D | 0.23 ± 0.01 b | 0.64 ± 0.08 a | |
| 50 D | 0.96 ± 0.22 a | 1.25 ± 0.18 a | |
Values presented are the means ± SE (n = 3). Different letters indicate significant differences among treatments in the same period using Duncan’s test (P < 0.05).
Table 2.
Changes in the ratio of endogenous hormones ABA to GA (ABA/GA) in P. notoginseng seeds during the after-ripening process. .
| Time (DAR) | Treatments | ||
|---|---|---|---|
| CK | 1 mg·l−1 ABA | 10 mg·l−1 ABA | |
| 0 D | 0.58 ± 0.05 b | 0.09 ± 0.01 b | 3.67± 1.16 a |
| 30 D | 0.83 ± 0.18 a | 0.82 ± 0.02 a | 0.94 ± 0.20 a |
| 50 D | 0.62 ± 0.33 a | 0.82 ± 0.27 a | 0.81 ± 0.15 a |
Values presented are the means ± SE (n = 3). Different letters indicate significant differences among treatments in the same period using Duncan’s test (P < 0.05).
RNA-seq and gene annotation of P. notoginseng seeds
Based on the previous morphophysiological characteristics, samples of CK- and HA-treated seeds were collected from three time points (0, 30, and 50 DAR) and then were selected to investigate the regulatory pathways at the transcriptional level. Eighteen cDNA libraries (CK_0_1, CK_0_2, CK_0_3, CK_30_1, CK_30_2, CK_30_3, CK_50_1, CK_50_2, CK_50_3, HA_0_1, HA_0_2, HA_0_3, HA_30_1, HA_30_2, HA_30_3, and HA_50_1, HA_50_2, and HA_50_3) were sequenced to identify gene expression information of P. notoginseng seeds using Illumina HiSeq. The Q30 of eighteen samples was about 93%, and the base content was uniform ( Table S1 ). The clean reads were compared quickly and accurately with the genome of P. notoginseng (Fan et al., 2020). In all samples, mapped reads were above 90%, unique mapped reads were above 86%, and multiple mapped reads were below 5.2% ( Table 3 ).
Table 3.
Comparative genome statistics of samples.
| Sample | Total reads | Mapped reads | Unique mapped reads | Multiple mapped reads |
|---|---|---|---|---|
| CK_0_1 | 46815702 | 42910019(91.66%) | 40899591(87.36%) | 2010428(4.29%) |
| CK_0_2 | 46919124 | 42880938(91.39%) | 40775136(86.91%) | 2105802(4.49%) |
| CK_0_3 | 45916162 | 42104871(91.7%) | 40017119(87.15%) | 2087752(4.55%) |
| CK_30_1 | 46770276 | 43229185(92.43%) | 41469671(88.67%) | 1759514(3.76%) |
| CK_30_2 | 46087118 | 42530822(92.28%) | 40909769(88.77%) | 1621053(3.52%) |
| CK_30_3 | 50775950 | 46563721(91.7%) | 44146740(86.94%) | 2416981(4.76%) |
| CK_50_1 | 46450880 | 41971359(90.36%) | 39989166(86.09%) | 1982193(4.27%) |
| CK_50_2 | 44037090 | 40404986(91.75%) | 38490509(87.4%) | 1914477(4.35%) |
| CK_50_3 | 49358510 | 45429117(92.04%) | 43281381(87.69%) | 2147736(4.35%) |
| HA_0_1 | 46371594 | 42423231(91.49%) | 40335559(86.98%) | 2087672(4.5%) |
| HA_0_2 | 49958870 | 45764395(91.6%) | 43212512(86.5%) | 2551883(5.11%) |
| HA_0_3 | 46123694 | 42407335(91.94%) | 40478554(87.76%) | 1928781(4.18%) |
| HA_30_1 | 45367036 | 41508211(91.49%) | 39549766(87.18%) | 1958445(4.32%) |
| HA_30_2 | 44814232 | 41079675(91.67%) | 39150450(87.36%) | 1929225(4.3%) |
| HA_30_3 | 45870888 | 41875152(91.29%) | 39899776(86.98%) | 1975376(4.31%) |
| HA_50_1 | 44730048 | 40897348(91.43%) | 39038282(87.28%) | 1859066(4.16%) |
| HA_50_2 | 46684002 | 42801021(91.68%) | 40931415(87.68%) | 1869606(4.0%) |
| HA_50_3 | 45870028 | 41942451(91.44%) | 40168285(87.57%) | 1774166(3.87%) |
DEGs functional annotation and comparative analysis of P. notoginseng seeds
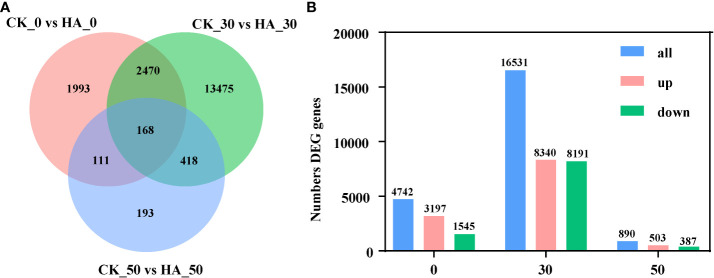
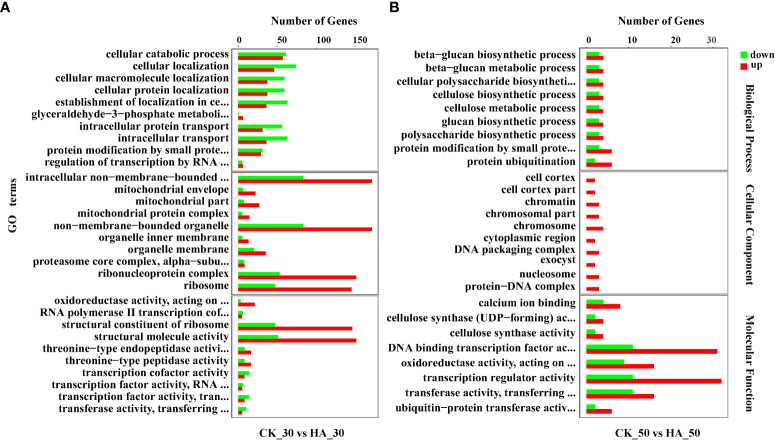
A total of 4,742, 16,531, and 890 DEGs were identified in pairwise comparisons of CK_0 vs HA_0, CK_30 vs HA_30, and CK_50 vs HA, respectively ( Figure 2A ). At 0 DAR, 3,197v DEGs were upregulated and 1,545 DEGs were downregulated in HA-treated seeds as compared with CK ( Figure 2B ). At 30 DAR, 8,340 DEGs were upregulated and 8,191 DEGs were downregulated in HA-treated seeds compared with CK ( Figure 2B ). At 50 DAR, 503 DEGs were upregulated and 387 DEGs were downregulated in HA-treated seeds ( Figure 2B ). The functions of DEGs were analyzed using GO annotation. Most of the DEGs were enriched in molecular function (MF) and biological process (BP) in the comparisons at CK_0 vs HA_0, CK_50 vs HA_50. The upregulated DEGs were mainly enriched in cellular component (CC), MF, and the downregulated DEGs were mainly enriched in BP and CC ( Figure 3 ; Figure S3 ). In CC classifications, it has been observed that the DEGs were enriched in organelles and organelle parts, and cellular processes, metabolic processes, and stress response cell parts were more active in BP classifications, while binding, catalysis, and transcription activity were more abundant in the MF classifications.
Figure 2.
Statistical analysis of differentially expressed genes (DEGs) in P. notoginseng seeds during the after-ripening process. (A) Venn diagram of DEGs in the control check (CK) and 10 mg·l−1 ABA-treated (HA-treated) P. notoginseng seeds. (B) Histogram of number DEG genes.
Figure 3.
GO functional annotation of differentially expressed genes (DEGs) in the CK and HA-treated P. notoginseng seeds. (A) Top 30 enriched GO terms of DEGs between CK_30 vs HA_30. (B) Top 30 enriched GO terms of DEGs between CK_50 vs HA_50. The Y-axis on the left represents GO terms, including biological process, cellular component, and molecular function, the X-axis indicates the gene number of each term. Upregulated genes are shown in a red bar, and downregulated genes are shown in a green bar.
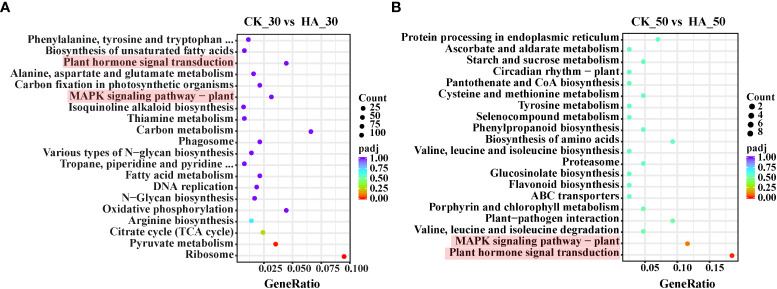
KEGG analysis revealed that the 442 upregulated differential genes were enriched in 97 pathways between the CK_0 and HA_0 groups, including plant–pathogen interaction, protein processing in the endoplasmic reticulum, the MAPK signaling pathway-plant, plant hormone signal transduction, and the biosynthesis of cofactors ( Figure S4A ). The 319 downregulated differential genes were enriched in 89 pathways, mainly the metabolism of amino acids and sugars, the citrate cycle (TCA cycle), the biosynthesis of amino acids and pantothenate, and CoA biosynthesis ( Figure S4B ). In the CK_30 and HA_30 groups, 1,732 upregulated differential genes were enriched in 115 pathways, including biosynthesis and metabolism of amino acids, biosynthesis of unsaturated fatty acids, plant hormone signal transduction, carbon fixation in photosynthetic organisms, MAPK signaling pathway–plant, carbon metabolism, various types of N-glycan biosynthesis, TCA cycle, and pyruvate metabolism ( Figure 4A ). In contrast, 1,483 downregulated differential genes were enriched in 114 pathways, mainly including some amino acid synthesis and metabolism, starch and sucrose metabolism, galactose metabolism, glycerolipid metabolism, and protein processing and transport ( Figure S4C ). The respiratory metabolism was enhanced, and the substance was synthesized and transported frequently in the seeds at 30 DAR. The 66 differentially upregulated genes were enriched in 38 pathways, mainly including protein processing in the endoplasmic reticulum, starch and sucrose metabolism, circadian rhythm–plant, plant–pathogen interaction, MAPK signaling pathway–plant, plant hormone signal transduction, and synthesis and metabolism of amino acids in the CK_50 and HA_50 groups ( Figure 4B ). The 62 differentially downregulated genes were enriched in 46 pathways, mainly including starch and sucrose metabolism, phenylpropanoid biosynthesis, galactose metabolism, glutathione metabolism, plant hormone signal transduction, and protein processing in the endoplasmic reticulum ( Figure S4D ). These results indicated that plant hormone signal transduction and the MAPK signaling pathway are active during the after-ripening process of P. notoginseng seeds.
Figure 4.
KEGG enrichment analysis of differentially expressed genes (DEGs) in the CK and HA-treated P. notoginseng seeds during the after-ripening process. (A) Top 20 enriched KEGG pathways of upregulated genes between CK_30 vs HA_30. (B) Top 20 enriched KEGG pathways of upregulated genes between CK_50 vs HA_50. The Y-axis on the left represents KEGG pathways, and the X-axis indicates the “gene ratio” represented by the ratio of upregulated gene numbers to total annotated gene numbers for each pathway. Low padj values are shown in the red circle, and high padj values are shown in the purple circle. The area of a circle represents DEG numbers.
Changes in signal transduction in response to exogenous ABA treatment of P. notoginseng seeds during after-ripening
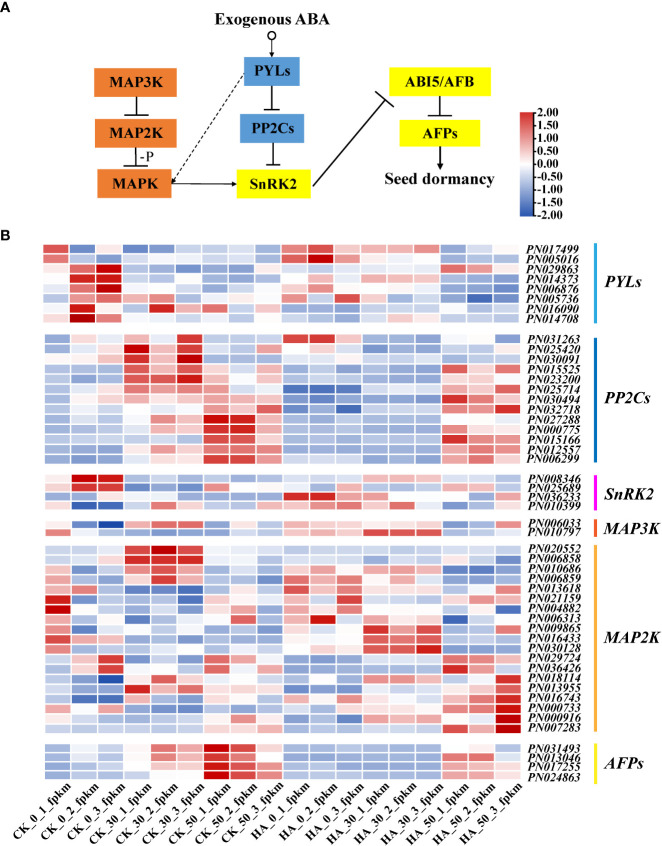
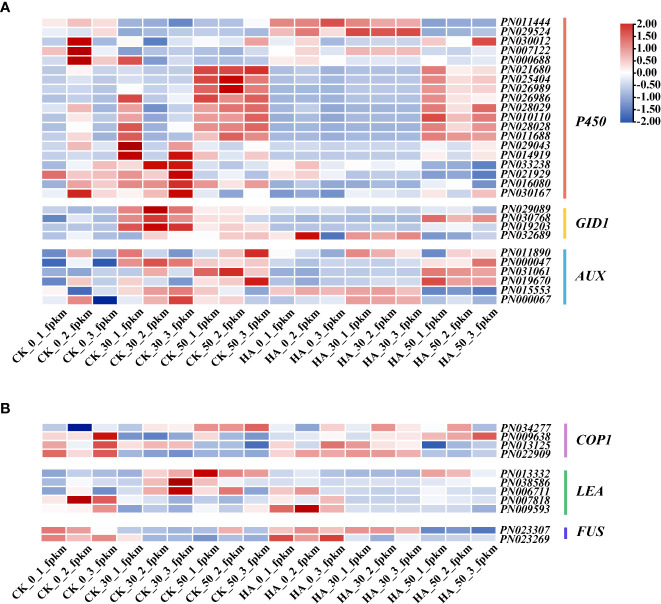
Some key DEGs of P. notoginseng seeds were analyzed, as shown in Figure 5 . The PYLs gradually decreased in the CK during the after-ripening process, and their expressions were downregulated in ABA-treated seeds at 0 DAR and upregulated at 30 DAR and 50 DAR ( Figure 5B ). The PP2Cs were gradually increased in the CK, especially at 50 DAR, whereas their expressions were downregulated in the ABA-treated groups ( Figure 5B ). The expression of SnRK2 was low in the CK but upregulated in the HA-treated groups ( Figure 5B ). The expression of ABRE-binding transcripts (AFPs) was downregulated in HA-treated seeds ( Figure 5B ). The expression of “AUX” (a receptor for the hormone IAA) was downregulated in HA-treated seeds ( Figure 6A ). Additionally, the expressions of cytochrome P-450 mono-oxygenases (P450) and the GA receptor (GID1) were significantly suppressed in HA-treated seeds ( Figure 6A ). The mitogen-activated protein kinase kinase (MAP2K) and mitogen-activated protein kinase kinase kinase (MAP3K) associated with MAPK signaling cascades were highly expressed in HA-treated seeds ( Figure 5B ). Among the DEGs associated with seed development, late embryogenesis abundant (LEA) was lowly expressed, while constitutively photomorphogenic1 (COP1) and FUSCA transcription factor (FUS) were relatively active in HA-treated seeds as compared with CK ( Figure 6B ). Therefore, we speculated that exogenous ABA treatment might prolong seed dormancy through the ABA signaling pathway and MAPK signaling cascades during the after-ripening process.
Figure 5.
Expression pattern analysis based on RNA-seq of the genes related to the ABA and MAPK signal transduction pathway. (A) The pathways of ABA signal transduction and MAPK signaling cascades. Arrows and dotted lines designate directly and indirectly interactions, respectively, and the dashed lines indicate a possible interaction. (B) Heat maps of genes associated with ABA signal transduction and MAPK signaling cascades in P. notoginseng seeds. In the heat map, different colors indicate the expression level changes.
Figure 6.
Effects of exogenous ABA on profiles of the transcriptome of P. notoginseng seeds during the after-ripening process. (A) A heat map showing the expression patterns of the candidate genes involved in GA and IAA signaling responses in P. notoginseng seeds. (B) A heat map showing the expression patterns of the candidate genes involved in embryonic development in P. notoginseng seeds. Different colors indicate the expression level changes.
qRT-PCR validation
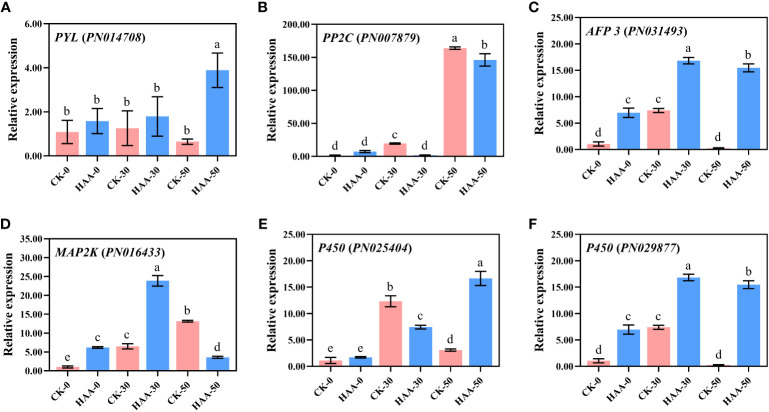
The candidate DEGs were selected for verification using qRT-PCR ( Figure 7 ), and it included genes related to ABA signal transduction (PYL, PP2C, AFP), MAPK signaling cascades (MAP3K), and GA Synthesis (P450).
Figure 7.
The DEG expression in response to ABA treatment of P. notoginseng seeds during the after-ripening process. Gene expressions were analyzed by RT-qPCR. (A) Pyrabactin resistance 1-like (PYL) (B) The protein phosphatase 2C family of genes (PP2C) (C) ABF-binding transcript 3 (AFP 3) (D) MAP kinase kinase (MAP2K) (E) Cytochrome P-450 mono-oxygenases (P450) (F) Cytochrome P-450 mono-oxygenases (P450). Values presented are the means ± SE (n = 3). Different letters indicate significant differences among treatments in the same period using Duncan’s test (P < 0.05).
Discussion
Exogenous ABA promote the dormancy of seed embryos and delay the germination of seeds
The hormone ABA is crucial in regulating seed development and dormancy (Shu et al., 2016b; Brookbank et al., 2021). ABA prevents a seed embryo from continuing to grow when the embryo is completely developed (Raz et al., 2001). Moreover, it has been demonstrated that GAs stimulate the growth potential and weaken the structures surrounding the embryos of A. thaliana and tomato (Solanum lycopersicum) seeds and thus accelerate the development of seed embryos (Yamaguchi and Kamiya, 2001). In our study, the development of embryos and the germination of seeds were delayed in the ABA-treated groups ( Figure 1 ). Similarly, it has been observed that the elongation growth of sprouts is evidently inhibited in the 10.0 mg·l−1 ABA-treated Oryza sativa seeds (Tang et al., 2003). This indicates that the exogenous hormone ABA might inhibit the elongation growth of the embryo and prevent the space for embryo development from expanding during the after-ripening process.
ABA positively regulates seed dormancy, while GA antagonizes ABA to promote seed germination (Chen et al., 2020; Abley et al., 2021; Zhao et al., 2022). Endogenous ABA is accumulated in A. thaliana seeds during the maturation process, which induces and maintains the dormancy of seeds, and then GA content is upregulated to release dormancy during the after-ripening process (Shu et al., 2016b). However, the endogenous ABA was increased and the endogenous GA was decreased in the HA-treated seeds ( Table 1 ), and the ratio of ABA to GA was elevated in the ABA-treated seeds of P. notoginseng ( Table 2 ). We presume that the levels of endogenous hormone and the rate of ABA/GA might be stimulated by exogenous ABA, thus influencing the dormancy of recalcitrant seeds. Further, it has been reported that ABA is accumulated in recalcitrant Dimocarpus longan and Litchi chinensis seeds during the after-ripening process (Peng and Fu, 1995), and the rate of ABA/GA is increased in dormant A. thaliana seeds (Cadman et al., 2006; Shu et al., 2016b; Yang et al., 2022). It has also been demonstrated that germination is inhibited in A. thaliana and T. aestivum seeds with the exogenous hormone IAA (Ramaih et al., 2003; Zhang et al., 2007). These results indicate that the levels of endogenous hormones are mainly increased by the increased synthesis of endogenous ABA in response to the application of exogenous ABA. Similarly, endogenous IAA was increased under exogenous ABA treatment ( Table 1 ), suggesting that increased IAA might enhance seed dormancy. These results are in line with the previous study that found that IAA stimulates ABA signaling to control seed dormancy in A. thaliana (Liu et al., 2013b). The JA precursor (12-oxo-phytodienoic acid (OPDA)) cooperates with ABA to inhibit the germination of A. thaliana seeds (Varshney and Majee, 2021; Nguyen et al., 2022). Consistently, JA was reduced in the HA-treated seeds than in the CK at 0 DAR ( Table 1 ), implying that ABA combined with JA precursor might induce seed dormancy at the early stage of the after-ripening process. Similarly, 80 mg·l−1 exogenous ABA treatment delays the germination process of Chenopodium quinoa seeds (Wang et al., 2022), as has also been observed in our study ( Figures 1C, D ). These results support the view that ABA treatment could inhibit the development of embryos and delay the germination of recalcitrant seeds.
The response of endogenous hormone ABA signaling to exogenous ABA in recalcitrant seeds
ABA receptors (PYR/PYL/RCAR family proteins), type 2C protein phosphatase (PP2C) proteins, SnRK2, and ABA-responsive element binding factors (ABF/AREB) respond to ABA and are involved in the regulation of seed dormancy (Fujii and Zhu, 2009; Nakashima et al., 2009; Ali et al., 2021). Similarly, a significant response has been found in the key gene expression of ABA signal transduction including PYL, PP2C, SnRK2, and ABFs in HA-treated P. notoginseng seeds ( Figure 5 ). In the HA-treated seeds, the PYLs were upregulated while they were gradually reduced during the after-ripening process ( Figure 5B ). This has also been evidenced by a previous study that found the ABA receptor PYL positively regulates the ABA signal transduction pathway in O. sativa seeds (Kim et al., 2012). The PYR/PYL/RCAR proteins directly bind to ABA and negatively regulate the protein phosphatase activity of the PP2C protein, which activates the ABA signaling (Wang et al., 2020). Our study showed that the expression of PP2Cs is lower at 0 DAR and 30 DAR has reversed expression at 50 DAR and is further suppressed under ABA treatment ( Figure 5B ). These findings support the view that members of the PP2C gene family negatively regulates the ABA signaling pathway (Miyazono et al., 2009; Park et al., 2009; Soon et al., 2012). A member of SnRK2 is also a key positive regulator of ABA signaling (Fujii and Zhu, 2009; Nakashima et al., 2009). SnRK2s are released from the inhibition of PP2Cs and become active when PP2C binds to ABA (Fujii and Zhu, 2009; Umezawa et al., 2009; Soon et al., 2012). Then ABA signaling is transmitted via the activated kinase SnRK2 phosphorylating various downstream targets, such as the transcription factors ABF2 and ABI5 (Miyazono et al., 2009; Kulik et al., 2011). Similarly, the expression of SnRK2 in CK was high at 0 DAR but almost absent at 30 and 50 DAR, whereas SnRK2 was consistently active in HA-treated seeds ( Figure 5B ). Unexpectedly, the expression of some AFPs was downregulated in HA-treated seeds ( Figure 5B ), suggesting that these transcription factors downstream of ABA might be differentially responsive to the ABA-mediated signaling regulation. In general, our results suggested that the genes (PYLs, PP2Cs, SnRK2s, and AFPs) involved in ABA signaling pathway might enhance seed dormancy and consequently delay the germination of P. notoginseng seeds with ABA treatment.
The cytochrome P-450 mono-oxygenases catalyze the formation of the GA precursor, GA12-aldehyde, thereby promoting GA synthesis (Pimenta Lange and Lange, 2006). The expression and activity of α-amylase are inhibited in mutant seeds of the receptor for GA signal (GID1) resulting in the inability of seeds to germinate (Griffiths et al., 2006). Similarly, it is necessary for seed germination that GA synthesis and GA signal transduction occur in soybean (Glycine max) (Shuai et al., 2017). Notably, the expression of P450 and GID1 was depressed in the HA-treated seeds, implying that the exogenous hormone ABA might inhibit GA synthesis and signaling transduction, thus suppressing seed germination ( Figure 6A ). The potential involvement of IAA in dormancy maintenance has been recorded, and likewise, IAA receptor mutants dramatically release seed dormancy (Liu et al., 2007). ABA could promote seed dormancy through enhancing IAA signaling in A. thaliana seeds (Griffiths et al., 2006). In particular, the expression of AUX (the response factor of IAA signaling) was downregulated in HA-treated seeds ( Figure 6A ), implying that IAA shows a diversified response in the regulation network of seed dormancy under ABA treatment. It also indicates that exogenous ABA could delay seed germination by inhibiting the synthesis of endogenous GA and the transduction of GA signals. Thus, we believe that exogenous ABA influences endogenous hormone signals, including the enhancement of the ABA signal and the reduction of the GA signal. This would contribute to promoting seed dormancy and delaying seed germination.
MAPK signaling cascades strengthen the regulation of embryonic development and seed dormancy with exogenous ABA
The MAPK signaling cascades could affect the phosphorylation of SnRK2 to activate ABA signaling and thus regulate seed germination (Vlad et al., 2009; Hasan et al., 2022). In O. sativa, it has been proved that OsMAPK5 is upregulated by ABA and is involved in ABA signaling transduction (Xiong and Yang, 2003; De Vleesschauwer et al., 2010), and ABA has been shown to regulate MAP3K gene expression, including OsDSM1 (O. sativa drought-hypersensitive mutant1) and OsEDR1 (O. sativa enhanced disease susceptibility1) (Kim et al., 2003; Ning et al., 2010). In our study, the expression of MAP3K and MAP2K associated with the MAPK signaling cascades was upregulated during the after-ripening process ( Figure 5B ). Similarly, MAPK11 upregulates ABA biosynthesis and phosphorylates SnRK2.2, which then negatively regulates tomato seed germination (Song et al., 2021). Hence, ABA signaling might be amplified through the MAPK signaling cascades to maintain seed dormancy and inhibit seed development. For example, AtFUS3 influences the development of seed embryos by inhibiting GA biosynthesis and promoting ABA accumulation (Gazzarrini et al., 2004). Consistently, FUS was increased in HA-treated seeds during the after-ripening process ( Figure 6B ). The expression of FUS3 is increased by exogenous ABA in A. Thaliana, supporting our results (Kagaya et al., 2005). Thus, we presume that MAPK signaling cascades could amplify the response of some endogenous signals to exogenous ABA and thus regulate the development of seed embryos and prolong seed dormancy. It provides a new method for prolonging the storage of recalcitrant seeds in production by regulating the dormant time.
Conclusion
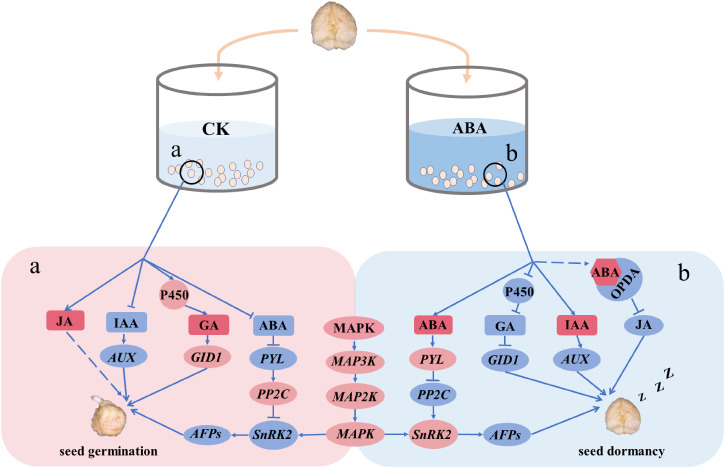
In conclusion, we found that exogenous ABA prolongs dormancy in recalcitrant P. notogensing seeds, upregulates ABA signal transduction, and downregulates endogenous GA-synthesis and GA signal transduction. Moreover, MAPK signaling cascades strengthen the regulation of embryonic development and seed dormancy when exogenous ABA is applied during the after-ripening process. In our study, a model is proposed to illustrate the dormant mechanism of recalcitrant seeds in response to exogenous ABA ( Figure 8 ). Exogenous hormone ABA could regulate embryonic development, prolong seed dormancy, and delay seed germination of recalcitrant seeds, and MAPK signaling cascades might be involved in amplifying the signaling modulation. These findings provide a novel insight into the dormant regulation of recalcitrant seeds in response to exogenous ABA during the after-ripening process and might contributed to the storage of recalcitrant seeds.
Figure 8.
A model explained the possible mechanism of exogenous ABA regulating the dormancy of P. notoginseng seeds during the after-ripening process at the transcriptional level. The increased genes are shown in a red circle, and the repressed genes are shown in a blue circle. A red square indicates that the content of hormone is enhanced, and a blue square indicates that the content of hormone is reduced. Arrows and dotted lines designate directly and indirectly interactions, respectively. The dashed lines indicate a possible interaction. (a) The hormonal regulatory network in the CK. (b) The hormonal regulatory network in ABA-treated seeds.
Data availability statement
The raw sequencing data in this study have been deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn/), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, accession number: CRA009677. Other data generated or analyzed during this study are included in this published article and its supplementary information files.
Author contributions
J-WC provided guidance on the whole process of the experiment and offered suggestions on the writing of the manuscript. Q-YW wrote the manuscript. Q-YW and LY participated most of the experiments. NG, J-SJ, R-MH, CC, Z-GM, and L-GL analyzed the relevant experimental data. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Glossary
| ABA | abscisic acid |
| ABF/AREB | ABA‐responsive element binding protein/ABRE‐binding factor |
| ABI5 | ABA insensitive 5 |
| AFPs | ABI5-binding proteins |
| BP | biological process |
| CC | cellular component |
| CK | the control check |
| COP1 | constitutively photomorphogenic1 |
| DAR | days of after-ripening process |
| ddH2O | double distilled water |
| DEGs | differentially expressed genes |
| Em | embryo |
| En | endosperm |
| Em/En | the rate of embryo to endosperm |
| FPKM | fragments per kilobase of transcript per million fragments mapped |
| FUS | FUSCA |
| GA | gibberellin |
| GID1 | GA insensitive dwarf1 |
| GO | Gene Ontology |
| HA | 10 mg l−1 ABA |
| HPLC-MS | high-performance liquid chromatography-tandem mass spectrometry |
| IAA | auxin |
| JA | jasmonic acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LA | 1 mg l−1 ABA |
| LEA | late embryogenesis abundant |
| MAP2K | mitogen-activated protein kinase kinase |
| MAP3K | mitogen-activated protein kinase kinase kinase |
| MAPK | mitogen-activated protein kinase |
| MF | molecular function |
| MPD | morphophysiological dormancy |
| OPDA | 12-oxo-phytodienoic acid |
| P450 | P-450 mono-oxygenases |
| PP2Cs | type 2C protein phosphatases |
| PYL | pyracbactin resistance-like |
| PYR | pyracbactin resistance |
| qRT-PCR | quantitative real-time PCR |
| RCARs | regulatory component of ABA receptors |
| SnRK2 | SNF1-related protein kinase subfamily 2 |
| TCA cycle | citrate cycle |
Funding Statement
This work is funded by the National Natural Science Foundation of China (32160248 and 81860676), the Major Special Science and Technology Project of Yunnan Province (202102AA310048), the National Key Research and Development Plan of China (2021YFD1601003), and the Innovative Research Team of Science and Technology in Yunnan Province (202105AE160016).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1054736/full#supplementary-material
References
- Abley K., Formosa-Jordan P., Tavares H., Chan E. Y., Afsharinafar M., Leyser O., et al. (2021). An ABA-GA bistable switch can account for natural variation in the variability of Arabidopsis seed germination time. eLife 10, e59485. doi: 10.7554/eLife.59485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F., Qanmber G., Li F., Wang Z. (2021). Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 35, 199–214. doi: 10.1016/j.jare.2021.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Rachedi S., Bouinot D., Wagner M. H., Bonnet M., Sotta B., Grappin P., et al. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the cape Verde islands ecotype, the dormant model of Arabidopsis thaliana . Planta 219, 479–488. doi: 10.1007/s00425-004-1251-4 [DOI] [PubMed] [Google Scholar]
- Baskin C. C., Baskin J. M. (2004). A classification system for plant dormancy. Seed Sci. Res. 14, 1–16. doi: 10.1079/SSR2003150 [DOI] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Start Soc B. 57, 289–300. doi: 10.2307/2346101 [DOI] [Google Scholar]
- Berjak P., Pammenter N. W. (2001). Seed recalcitrance-current perspectives. S. Afr. J. Bot. 67, 79–89. doi: 10.1016/S0254-6299(15)31111-X [DOI] [Google Scholar]
- Brookbank B. P., Patel J., Gazzarrini S., Nambara E. (2021). Role of basal ABA in plant growth and development. Genes 12 (12), 1936. doi: 10.3390/genes12121936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman C. S., Toorop P. E., Hilhorst H. W., Finsh-savage W. E. (2006). Gene expression profiles of Arabidopsis cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 46, 805–822. doi: 10.1111/j.1365-313X.2006.02738.x [DOI] [PubMed] [Google Scholar]
- Carrera E., Holman T., Medhurst A., Dietrich D., Footitt S., Theodoulou F. L., et al. (2008). Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis . Plant J. 53, 214–224. doi: 10.1111/J.1365-313X.2007.03331.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai X., Hu Z. H., Lei Y. N., Tan L. L. (2008). The development of embryo and endosperm and its influence on the germination of seeds of Bupleurum chinense . Bull. Entomol. Res. 28, 14–19. doi: 10.13313/j.issn.1673-4890.2008.10.001 [DOI] [Google Scholar]
- Chen H., Ruan J., Chu P., Fu W., Liang Z., Li Y., et al. (2020). AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J. 101 (2), 310–323. doi: 10.1111/tpj.14542 [DOI] [PubMed] [Google Scholar]
- Costa C. R., Lopes Pivetta K. F., de Souza G. R., Mazzini-Guedes R. B., Sajovic Pereira S. T., and Nogueira M. R., et al. (2018). Effects of temperature, light and seed moisture content on germination of Euterpe precatoria Palm. Am. J. Plant Sci. 9, 98–106. doi: 10.4236/ajps.2018.91009 [DOI] [Google Scholar]
- Cui X. M., Wang C. L., Li W., He C. F., Zhang Y., Wang Y. F. (1993). Study on the biological properties of Panax notoginseng seeds. Chin. J. Nat. Med. 12, 3–4. doi: 10.13863/j.issn1001-4454.1993.12.001 [DOI] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Yang Y., Cruz C. V., Hofte M. (2010). Abscisic acid-induced resistance against the brown spot pathogen cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152, 2036–2052. doi: 10.1104/pp.109.152702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew P., Pammenter N. W., Berjak P. (2000). ‘Sub-imbibed’ storage is not an option for extending longevity of recalcitrant seeds of the tropical species, Trichilia dregeana sond. Seed Sci. Res. 10, 355–363. doi: 10.1017/S0960258500000398 [DOI] [Google Scholar]
- Duan C. L., Li Z. T., Ding J. L., Xiao F. H. (2010. b). Physiologic characteristics of Panax notoginseng seeds during after-ripening process. China J. Chin. Mater. Med. 35, 2652–2656. doi: 10.4268/CJCMM20102002 [DOI] [PubMed] [Google Scholar]
- Duan Y. M., Yang L., Duan C. L. (2014). Desiccation sensitivity of Panax notoginseng seeds and membrane lipid peroxidation. Seed 33, 6–9. doi: 10.16590/j.cnki.1001-4705.2014.04.054 [DOI] [Google Scholar]
- Duan C. L., Yang L., Xiao F. H. (2010. a). A natomical observation on motphological development of Panax notoginseng embryo. Seed 29, 1–7. doi: 10.16590/j.cnki.1001-4705.2010.01.063 [DOI] [Google Scholar]
- Ellis C., Turner J. G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signaling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556. doi: 10.1007/s00425-002-0787-4 [DOI] [PubMed] [Google Scholar]
- Fan G. Y., Liu X. C., Sun S., Shi C. C., Du X., Han K., et al. (2020). The chromosome level genome and genome-wide association study for the agronomic traits of Panax notoginseng . iScience 23, 2589–0042. doi: 10.1016/j.isci.2020.101538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., et al. (2011). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. doi: 10.1111/j.1365-313X.2011.04887.x [DOI] [PubMed] [Google Scholar]
- Fujii H., Zhu J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparin E., Faria J. M. R., José A. C., Tonetti O. A. O., de Melo R. A., Hilhorst H. W. M. (2020). Viability of recalcitrant araucaria angustifolia seeds in storage and in a soil seed bank. J. For. Res. 31, 2413–2422. doi: 10.1007/s11676-019-01001-z [DOI] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell. 7, 373–385. doi: 10.1016/j.devcel.2004.06.017 [DOI] [PubMed] [Google Scholar]
- Ge N., Yang K., Yang L., Meng Z. G., Li L. G., Chen J. W. (2021). iTRAQ and RNA-seq analyses provide an insight into mechanisms of recalcitrance in a medicinal plant Panax notoginseng seeds during the after-ripening process. Funct. Plant Bio. 49 (1), 68–88. doi: 10.1071/FP21197 [DOI] [PubMed] [Google Scholar]
- Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., and Soppe W. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35(10), 1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x [DOI] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z. L., Powers S. J., et al. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18, 3399–3414. doi: 10.1105/tpc.106.047415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Millar A. A., Jacobsen J. V. (2005). Dormancy release, ABA and pre-harvest sprouting. Cur. Opin. Plant Bbiol. 8, 183–187. doi: 10.1016/j.pbi.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Hasan M. M., Liu X. D., Waseem M., Guang-Qian Y., Alabdallah N. M., Jahan M. S., et al. (2022). ABA activated SnRK2 kinases: an emerging role in plant growth and physiology. Plant Signal. 17 (1), 2071024. doi: 10.1080/15592324.2022.2071024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst H. W. M. (1995). A critical update on seed dormancy. i. primary dormancy. Seed Sci. Res. 5, 61–73. doi: 10.1017/S0960258500002634 [DOI] [Google Scholar]
- Hubbard K. E., Nishimura N., Hitomi K., Getzoff E. D., Schroeder J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Suzuki H., Kim Y. C., Iuchi A., Kuromori T., Ueguchi-Tanaka M., et al. (2007). Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 50, 958–966. doi: 10.1111/j.1365-313X.2007.03098.x [DOI] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20520–20525. doi: 10.1073/pnas.0907205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Okuda R., Ban A., Toyoshima R., Tsutsumida K., Usui H., et al. (2005). Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis . Plant Cell Physiol. 46, 300–311. doi: 10.1093/pcp/pci031 [DOI] [PubMed] [Google Scholar]
- Kalemba E. M., Ratajczak E. (2018). The effect of a doubled glutathione level on parameters affecting the germinability of recalcitrant Acer saccharinum seeds during drying. J. Plant Physiol. 223, 72–83. doi: 10.1016/j.jplph.2018.02.010 [DOI] [PubMed] [Google Scholar]
- Kermode A. R. (2005). Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24, 319–344. doi: 10.1007/s00344-005-0110-2 [DOI] [Google Scholar]
- Kim J. A., Agrawal G. K., Rakwal R., Han K. S., Kim K. N., Yun C. H., et al. (2003). Molecular cloning and mRNA expression analysis of a novel rice (Oryza sativa l.) MAPK kinase kinase, OsEDR1, an ortholog of Arabidopsis AtEDR1, reveal its role in defense/stress signalling pathways and development. Biochem. Biophys. Res. Commun. 300, 868–876. doi: 10.1016/s0006-291x(02)02944-3 [DOI] [PubMed] [Google Scholar]
- Kim H., Hwang H., Hong J. W., Lee Y. N., Ahn I. P., Yoon I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024. doi: 10.1093/jxb/err338 [DOI] [PubMed] [Google Scholar]
- Kucera B., Cohn M. A., Leubner-Metzger G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15, 281–307. doi: 10.1079/SSR2005218 [DOI] [Google Scholar]
- Kulik A., Wawer I., Krzywin´ska E., Bucholc M., Dobrowolska G. (2011). SnRK2 protein kinases–key regulators of plant response to abiotic stresses. OMICS 15, 859–872. doi: 10.1089/omi.2011.0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K. E., Wang W., He Y., Hussain A., et al. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. doi: 10.1101/gad.969002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. X., Koornneef M., Soppe W. J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 19, 433–444. doi: 10.1105/tpc.106.049221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Qiu Y. P., Zhang L., Chen J. (2005). Dormancy breaking and storage behavior of Garcinia cowa roxb. (Guttiferae) seeds: implications for ecological function and germplasm conservation. J. Integr. Plant Biol. 47, 38–49. doi: 10.1111/j.1744-7909.2005.00010.x [DOI] [Google Scholar]
- Liu X., Zhang H., Zhao Y., Feng Z., Li Q., Yang H. Q., et al. (2013. b). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis . Proc. Natl. Acad. Sci. U.S.A. 110 (38), 15485–15490. doi: 10.1073/pnas.1304651110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu Q. Y., Shen Y. B., Shi F. H. (2022). Study on embryo development and material metabolism of Paeonia lactiflora Pall. Mol. Breed. 1–9. Available at: http://kns.cnki.net/kcms/detail/46.1068.S.20220518.1139.006.html [Google Scholar]
- Matakiadis T., Alboresi A., Jikumaru Y., Tatematsu K., Pichon O., Renou J. P., et al. (2009). The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 149, 949–960. doi: 10.1104/pp.108.126938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla A. J., Matilla-Vázquez M. A. (2008). Involvement of ethylene in seed physiology. Plant Sci. 175, 87–97. doi: 10.1016/J.PLANTSCI.2008.01.014 [DOI] [Google Scholar]
- Miyazono K., Miyakawa T., Sawano Y., Kubota K., Kang H. J., Asano A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462 (7273), 609–614. doi: 10.1038/nature08583 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20, 55–67. doi: 10.1017/S0960258510000012 [DOI] [Google Scholar]
- Nguyen T. N., Tuan P. A., Ayele B. T. (2022). Jasmonate regulates seed dormancy in wheat via modulating the balance between gibberellin and abscisic acid. J. Exp. Bot. 73 (8), 2434–2453. doi: 10.1093/jxb/erac041 [DOI] [PubMed] [Google Scholar]
- Ning J., Li X., Hicks L. M., Xiong L. (2010). A raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890. doi: 10.1104/pp.109.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Welti R., Wang X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. F., Fu J. R. (1995). Changs of ABA content and the sensitivity to exogenous ABA during the development of litchi and longan seeds. J. Plant Physiol. 21, 159–165. [Google Scholar]
- Pimenta Lange M. J., Lange T. (2006). Gibberellin biosynthesis and the regulation of plant development. Plant Biol. (Stuttg) 8, 281–290. doi: 10.1055/s-2006-923882 [DOI] [PubMed] [Google Scholar]
- Pritchard H. W. (2004). “Classification of seed storage ‘types’ for ex situ conservation in relation to temperature and moisture,” in Exsitu plant conservation: supporting species survival in the wild. Eds. Guerrant E. O., Havens K., Maunder M. (Washington, America: Island Press; ), 139–161. [Google Scholar]
- Pukacka S., Ratajczak E. (2006). Antioxidative response of ascorbate-glutathione pathway enzymes and metabolites to desiccation of recalcitrant Acer saccharinum seeds. J. Plant Physiol. 163, 1259–1266. doi: 10.1016/j.jplph.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Pu M., Sun Y., Gao C., Li K. (2016). Relationship between endogenous hormone content and embryo growth and the seed germination of Paris polyphylla var. Yunnansensis . J. For. Res. 29, 268–273. doi: 10.13275/j.cnki.lykxyj.2016.02.019 [DOI] [Google Scholar]
- Qin X., Zeevaart J. A. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128, 544–551. doi: 10.1104/pp.010663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaih S., Guedira M., Paulsen G. M. (2003). Relationship of indoleacetic acid and tryptophan to dormancy and preharvest sprouting of wheat. Funct. Plant Biol. 30, 939–945. doi: 10.1071/FP03113 [DOI] [PubMed] [Google Scholar]
- Raz V., Bergervoet J. H., Koornneef M. (2001). Sequential steps for developmental arrest in Arabidopsis seeds. Development 128, 243–252. doi: 10.1242/dev.128.2.243 [DOI] [PubMed] [Google Scholar]
- Roberts E. H. (1973). Predicting the storage life of seeds. Seed Sci. Technol. 1, 499–514. [Google Scholar]
- Shuai H., Meng Y., Luo X., Chen F., Zhou W., Dai Y., et al. (2017). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci. Rep. 7, 12620. doi: 10.1038/s41598-017-13093-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Liu X. D., Xie Q., He Z. H. (2016. b). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis . PloS Genet. 9, e1003577. doi: 10.1371/journal.pgen.1003577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. T., Berjak P. (1995). “Deteriorative changes associated with the loss of viability of stored desiccation-tolerance and desiccation sensitive seeds,” in Seed development and germination. Ed. Kigel J. (New York, America: Routledge; ), 46–55. doi: 10.1201/9780203740071-26 [DOI] [Google Scholar]
- Song S. Q., Cheng H. Y., Jiang X. W. (2008). Seed biology (Beijing, China: Science Press; ). doi: 10.1007/978-981-15-4198-8_30 [DOI] [Google Scholar]
- Song J., Shang L., Wang X., Xing Y., Xu W., Zhang Y., et al. (2021). MAPK11 regulates seed germination and ABA signaling in tomato by phosphorylating SnRKs . J. Exp. Bot. 72, 1677–1690. doi: 10.1093/jxb/eraa56 [DOI] [PubMed] [Google Scholar]
- Soon F. F., Ng L. M., Zhou X. E., West G. M., Kovach A., Tan M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335 (6064), 85–88. doi: 10.1126/science.1215106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Su W., Howell S. H. (1992). Methyl jasmone inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89, 6837–6840. doi: 10.1073/pnas.89.15.6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A. J. (2014). Morphysiological dormancy and changes of endogenous ABA and GA4 contents in seeds of Musella lasiocarpa . J. Plant Physiol. 50, 419–425. doi: 10.13592/j.cnki.ppj.2013.0407 [DOI] [Google Scholar]
- Tang R. S., Wang J. P., Tong H. Y. (2003). Effect of ABA on the germination of rice seed and growth of its seeding. J. Agric. Sci. 19, 75–80. [Google Scholar]
- Tischer S. V., Wunschel C., Papacek M., Kleigrewe K., Hofmann T., Christmann A., et al. (2017). Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana . Proc. Natl. Acad. Sci. U.S.A. 114, 10280–10285. doi: 10.1073/pnas.1706593114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. doi: 10.1073/pnas.0907095106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney V., Majee M. (2021). JA shakes hands with ABA to delay seed germination. Trends Plant Sci. 26 (8), 764–766. doi: 10.1016/j.tplants.2021.05.00 [DOI] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis . Plant Cell. 21, 3170–3184. doi: 10.1105/tpc.109.069179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele A., Linkies A., Müller K., Leubner-Metzger G. (2011). Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J. Exp. Bot. 62, 5131–5147. doi: 10.1093/jxb/err214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ren Z., Cheng C., Wang T., Ji H., Zhao Y., et al. (2020). Counteraction of ABA-mediated inhibition of seed germination and seedling establishment by ABA signaling terminator in Arabidopsis . Mol. Plant 13 (9), 1284–1297. doi: 10.1016/j.molp.2020.06.011 [DOI] [PubMed] [Google Scholar]
- Wang D. L., Zhuang D. Y., Jiang R. Z., Fang D., Shi B. B., Fei L. Y., et al. (2022). Effect of exogenous ABA on seed germination of Chenopodium quinoa. China Seed Industry 2022, 93–96. doi: 10.19462/j.cnki.1671-895x.2022.02.026 [DOI] [Google Scholar]
- Willis C. G., Baskin C. C., Baskin J. M., Auld J. R., Venable D. L., Cavender-Bares J., et al. (2014). The evolution of seed dormancy: environmental cues, evolutionary hubs, and diversification of the seed plants. New Phytol. 203, 300–309. doi: 10.1111/nph.12782 [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis . Plant J. 54, 40–451. doi: 10.1111/j.1365-313X.2008.03433.x [DOI] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 15, 745–759. doi: 10.1105/tpc.008714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y. (2001). Gibberellins and light-stimulated seed germination. J. Plant Growth Regul. 20, 369–376. doi: 10.1007/s003440010035 [DOI] [PubMed] [Google Scholar]
- Yang B., Chen M., Zhan C., Liu K., Cheng Y., Xie T., et al. (2022). Identification of OsPK5 involved in rice glycolytic metabolism and GA/ABA balance for improving seed germination via genome-wide association study. J. Exp. Bot. 73 (11), 3446–3461. doi: 10.1093/jxb/erac071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Yang L., Fan W., Long G. Q., Xie S. Q., Meng Z. G., et al. (2019). Illumina-based transcriptomic analysis on recalcitrant seeds of Panax notoginseng for the dormancy release during the after-ripening process. Physiol. Plant 167, 597–612. doi: 10.1111/ppl.12904 [DOI] [PubMed] [Google Scholar]
- Yang K., Yang J. H., Liu S. W., Meng Z. G., Xie S. Q., Chen J. W. (2018). Embryo development and dynamic changes of six endogenous phytohormones during after-ripening process of Panax notoginseng seeds. J. Med. Chem. 41 (03), 519–523. doi: 10.13863/j.issn1001-4454.2018.03.004 [DOI] [Google Scholar]
- Yu M., Zhou S. B., Wu X. Y., Wang J., Chang L. L., Wang J. M. (2012). Dormancy break approaches and property of dormant seeds of wild Cryptotaenia japonica . Acta Ecol. Sin. 32, 1347–1354. doi: 10.5846/stxb201012221827 [DOI] [Google Scholar]
- Zhang Z., Li Q., Li Z., Staswick P. E., Wang M., Zhu Y., et al. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during arabidopsis-pseudomonas syringae interaction. Plant Physiol. 145, 450–464. doi: 10.1104/pp.107.106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Shen H. L. (2006). Regulation of seed dormancy and germination of Fraxinus species. J. Plant Physiol. 42, 354–360. doi: 10.1104/pp.107.106021 [DOI] [Google Scholar]
- Zhao Y. H., Liu H. Q., Liu T. C., Fu J. G., Liu W. Z., Zhang X. S. (2000). Affection of exogenous gibberellic acid (GA3) on endogenus hormones of Panax quinquefolium seed during its morphological afterripening period. J. Med. Chem. 23, 591–593. doi: 10.13863/j.issn1001-4454.2000.10.001 [DOI] [PubMed] [Google Scholar]
- Zhao H., Zhang Y., Zheng Y. (2022). Integration of ABA, GA, and light signaling in seed germination through the regulation of ABI5. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1000803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data in this study have been deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn/), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, accession number: CRA009677. Other data generated or analyzed during this study are included in this published article and its supplementary information files.