ABSTRACT
Norovirus is a major human pathogen that can cause severe gastroenteritis in vulnerable populations. The extensive viral diversity presented by human noroviruses constitutes a major roadblock for the development of effective vaccines. In addition to the large number of genotypes, antigenically distinct variants of GII.4 noroviruses have chronologically emerged over the last 3 decades. The last variant to emerge, Sydney_2012, has been circulating at high incidence worldwide for over a decade. We analyzed 1449 capsid sequences from GII.4 Sydney_2012 viruses to determine genetic changes indicative of antigenic diversification. Phylogenetic analyses show that Sydney_2012 viruses scattered within the tree topology with no single cluster dominating during a given year or geographical location. Fourteen residues presented high variability, 7 of which mapped to 4 antigenic sites. Notably, ~52% of viruses presented mutations at 2 or more antigenic sites. Mutational patterns showed that residues 297 and 372, which map to antigenic site A, changed over time. Virus-like particles (VLPs) developed from wild-type Sydney_2012 viruses and engineered to display all mutations detected at antigenic sites were tested against polyclonal sera and monoclonal antibodies raised against Sydney_2012 and Farmington_Hills_2002 VLPs. Minimal changes in reactivity were detected with polyclonal sera and only 4 MAbs lost binding, with all mapping to antigenic site A. Notably, reversion of residues from Sydney_2012 reconstituted epitopes from ancestral GII.4 variants. Overall, this study demonstrates that, despite circulating for over a decade, Sydney_2012 viruses present minimal antigenic diversification and provides novel insights on the diversification of GII.4 noroviruses that could inform vaccine design.
IMPORTANCE GII.4 noroviruses are the major cause of acute gastroenteritis in all age groups. This predominance has been attributed to the continued emergence of phylogenetically discrete variants that escape immune responses to previous infections. The last GII.4 variant to emerge, Sydney_2012, has been circulating at high incidence for over a decade, raising the question of whether this variant is undergoing antigenic diversification without presenting a major distinction at the phylogenetic level. Sequence analyses that include >1400 capsid sequences from GII.4 Sydney_2012 showed changes in 4 out of the 6 major antigenic sites. Notably, while changes were detected in one of the most immunodominant sites over time, these resulted in minimal changes in the antigenic profile of these viruses. This study provides new insights on the mechanism governing the antigenic diversification of GII.4 norovirus that could help in the development of cross-protective vaccines to human noroviruses.
KEYWORDS: antigenic variation, calicivirus, gastroenteritis, norovirus, phylogenetic analysis
INTRODUCTION
Norovirus is a major cause of acute gastroenteritis affecting people from all age groups. The human norovirus RNA genome is organized in 3 open reading frames (ORF1-3). These ORFs encode for the nonstructural proteins (e.g., VPg, protease, and polymerase), the major capsid protein (VP1), and the minor capsid protein (VP2), respectively. The expression of recombinant VP1 results in virus-like particles (VLPs) that resemble the native virion. In the absence of traditional cell culture systems for human noroviruses, VLPs have been used to determine the structure and study the antigenic properties of VP1. The structural model of norovirus VP1 shows 2 domains: the shell (S) and protruding (P). The former makes the scaffold of the inner capsid, and the latter is the most exposed region of the viral capsid. The P domain is further subdivided into P1 and P2 (1–3). Further demonstrating their relevance, previous studies have utilized norovirus VLPs to determine that the capsid binds to histo-blood group antigen (HBGA) carbohydrates, and this interaction facilitates human norovirus infection (4, 5). Moreover, elicitation of antibodies capable of blocking this VP1:HBGA interaction is strongly correlated with disease protection and virus neutralization (1, 2, 6–10).
Human noroviruses present a great diversity (>40 genotypes), with one genotype, GII.4, accounting for ~50% of all norovirus infections globally (11). The predominance of GII.4 norovirus has been associated with the chronological emergence of phylogenetically discrete variants that are antigenically different, allowing escape from previous infections. The major differences among these GII.4 variants map to variable antigenic sites/motifs (A-I) located at the outermost surface of the VP1 (7, 12, 13). Six major global epidemics have been associated with the emergence of GII.4 variants (11, 14, 15). The first pandemic was linked to the emergence and circulation of the Grimsby_1995 virus, the second to the Farmington_Hills_2002 virus, and the third to the Hunter_2004 virus. In 2006, the Yerseke_2006a and Den_Haag_2006b variants emerged, with the latter causing outbreaks worldwide (16). These variants were both superseded by the New_Orleans_2009 virus in 2009. In 2012, the Sydney_2012 variant emerged with a completely different set of nonstructural proteins ([P31], named based on the polymerase type). This variant replaced all others and has been circulating for over a decade in the human population (11, 13). In 2015, this variant was detected with a new polymerase, Sydney_2012[P16], which quickly predominated alongside Sydney_2012 [P31] viruses worldwide (17). This polymerase could have been acquired by recombination with the GII.2[P16] viruses that were first detected in 2014 and that caused large outbreaks in different countries during 2016 to 2017 (18). Other GII.4 variants have been detected (e.g., Sakai_2003, Yerseke_2006a, Osaka_2007, Apeldoorn_2007, and HongKong_2019); however, their impact on gastroenteritis outbreaks was limited to specific geographical regions (14).
While most predominant GII.4 variants only circulated for 2 to 4 years, Grimsby_1995 and Sydney_2012 predominated for more than 8 years (Fig. 1A) (11, 19). The rapid turnover of variants during the mid-2000s seems to be associated with the single immunodominance of antigenic site A, while the Grimsby_1995 and Sydney_2012 variants present co-immunodominance of antigenic sites A and G (7). Advances in genome sequencing technologies over the last decade resulted in a larger data set of norovirus genome information (11, 13, 20), which facilitates the study of intra-variant diversification. Here, we harnessed over 1400 GII.4 norovirus capsid sequences to study the intra-variant evolution of the Sydney_2012 variant, and interrogate whether this variant has undergone antigenic diversification after more than a decade of circulation in the human population. While genomics analyses suggest antigenic variation in 4 out of the 6 major antigenic sites, the overall antigenicity of Sydney_2012 remained very similar for over a decade. These data contradict the notion that the predominance of GII.4 noroviruses is only linked to antigenic variation of the capsid protein.
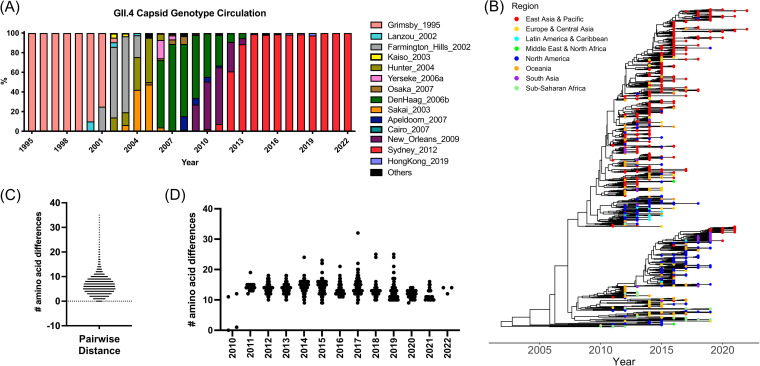
FIG 1.
Evolution of the major capsid protein (VP1) from GII.4 Sydney_2012 noroviruses. (A) GII.4 variant yearly distribution tabulated using all sequences available in public databases (11). The last variant to emerge, GII.4 Sydney_2012 variant, has been circulating for over a decade. (B) Time-scaled phylogenetic tree of GII.4 Sydney_2012 noroviruses showing the circulation of different clusters every given year. The phylogenetic tree was calculated using 1,449 nucleotide sequences encoding the VP1 of Sydney_2012 noroviruses and the Markov chain Monte Carlo (MCMC) method, as implemented in BEAST v1.10.4 (41). (C) Amino acid pairwise differences among all Sydney_2012 viruses calculated using the same data set used for phylogenetic tree reconstruction. (D) Amino acid pairwise differences of all Sydney_2012 viruses in each year compared to the earliest virus detected in 2010. Intra-variant diversification reveals an overall variation of 2 to 11 (10th to 90th percentile; range 1 to 35) residues.
RESULTS
Genetic diversification of the Sydney_2012 variant.
We retrieved 3,145 capsid sequences from GII.4 noroviruses from GenBank and selected 1,449 sequences that cluster within the Sydney_2012 variant. The phylogenetic tree of the Sydney_2012 variant shows that viruses are scattered within the tree topology, with no single cluster dominating during a given year (Fig. 1B). The sequences were collected from 34 countries from 5 continents (Table S1). While differences in the number of sequences are observed for each given geographical region, viruses from different countries were observed throughout the multiple clusters of the phylogenetic tree (Fig. 1). Of note, the co-dominance of 2 viruses, Sydney_2012[P31] and Sydney_2012[P16], from 2 major clusters, has occurred since 2015 (Fig. S1). The capsid of Sydney_2012 viruses presented clock-like linear evolution on its genome (Fig. S2), with multiple differences at the amino acid level (range, 1 to 35; mean, 6.5; 10th percentile, 2; 90th percentile, 11) (Fig. 1C). Notably, these amino acid differences did not accumulate over time and a similar number of differences were detected for viruses circulating in any given year (Fig. 1D).
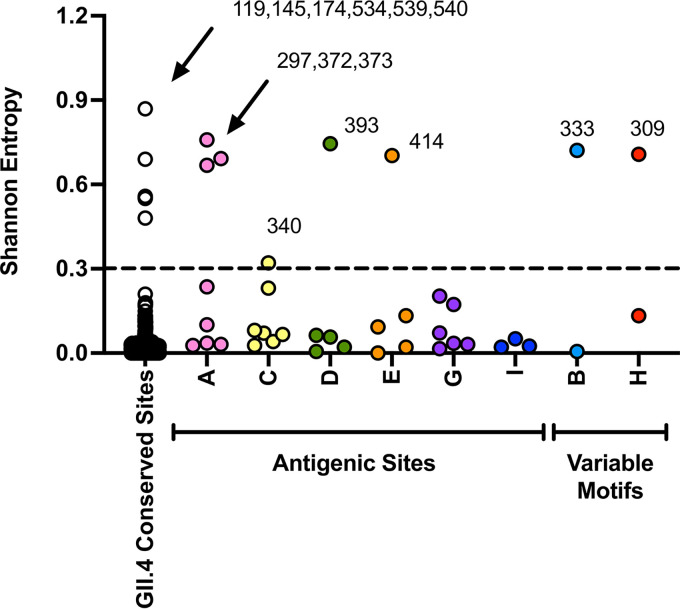
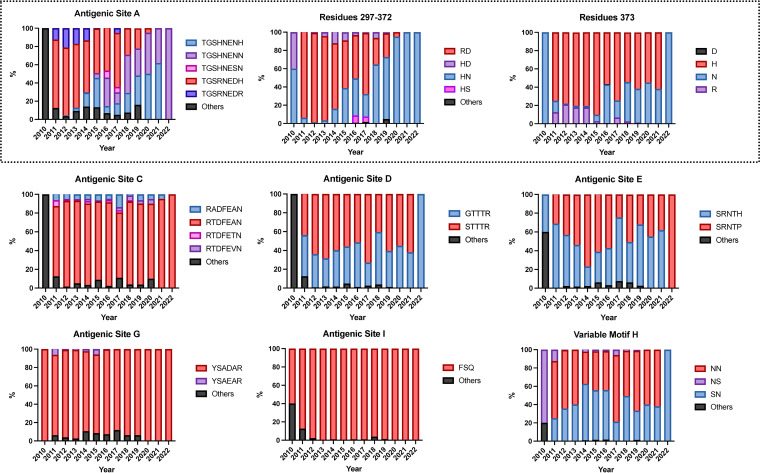
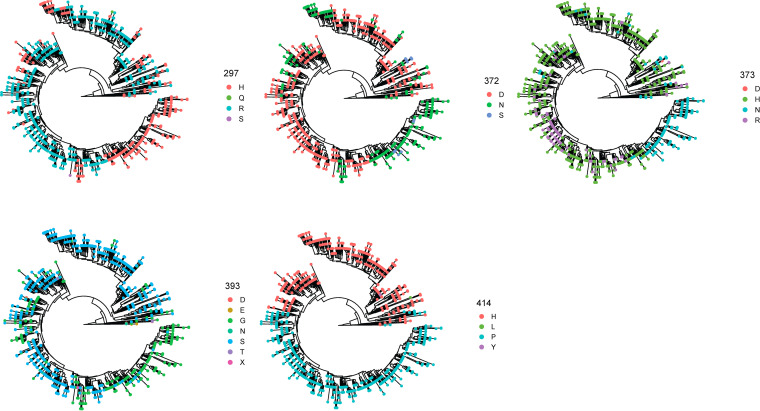
We then determined where these mutations mapped on the norovirus capsid protein. We found that 14 residues presented Shannon entropy values above 0.3, which is indicative of high variability (13). Seven residues mapped on major antigenic sites or variable motifs: 297, 372, and 373 (antigenic site A); 333 (variable motif B); 340 (antigenic site C); 393 (antigenic site D); 414 (antigenic site E); and 309 (variable motif H). No variable residues were detected on antigenic sites G or I (7, 13). The other variable residues mapped on the S domain (119, 145, and 174) and at the 3′-end of the P1 domain (534, 539, 540), and their role on the norovirus phenotype is unknown (Fig. 2). Profiling of the temporal frequency of the amino acid sequence patterns (mutational patterns) for each of the antigenic sites shows that only antigenic site A changed its amino acid sequence over time. This change was attributed to mutations occurring at residues 297 and 372 (Fig. 3). The profiling of antigenic sites D, E, and H suggests the co-circulation of 2 different virus populations.
FIG 2.
Conservation analysis of the major capsid protein (VP1) from GII.4 Sydney_2012 noroviruses. Shannon entropy was used to calculate the amino acid variation for each site in the VP1. Analysis was carried out using 1,449 complete (or nearly complete) sequences of Sydney_2012 noroviruses and the Shannon Entropy-One tool, as implemented in Los Alamos National Laboratory. Residues were grouped on those mapping outside antigenic sites (non-antigenic sites) and those mapping on antigenic sites (columns A to E, G to I). Residues presenting entropy values ≥0.3 are considered as variables (13). Variable motifs are predicted to be relevant to virus diversification, but not yet experimentally demonstrated to be an antigenic site.
FIG 3.
Temporal amino acid patterns on the major antigenic sites from GII.4 Sydney_2012 noroviruses. Colors of the bars correspond to the most frequent sequence patterns presented at any given antigenic site. Amino acid changes occurring on the 3 (297, 372, and 373) variable residues from antigenic site A are shown separately. Analysis was carried out using 1,449 complete (or nearly complete) sequences of Sydney_2012 noroviruses and scripts implemented in R (13, 38).
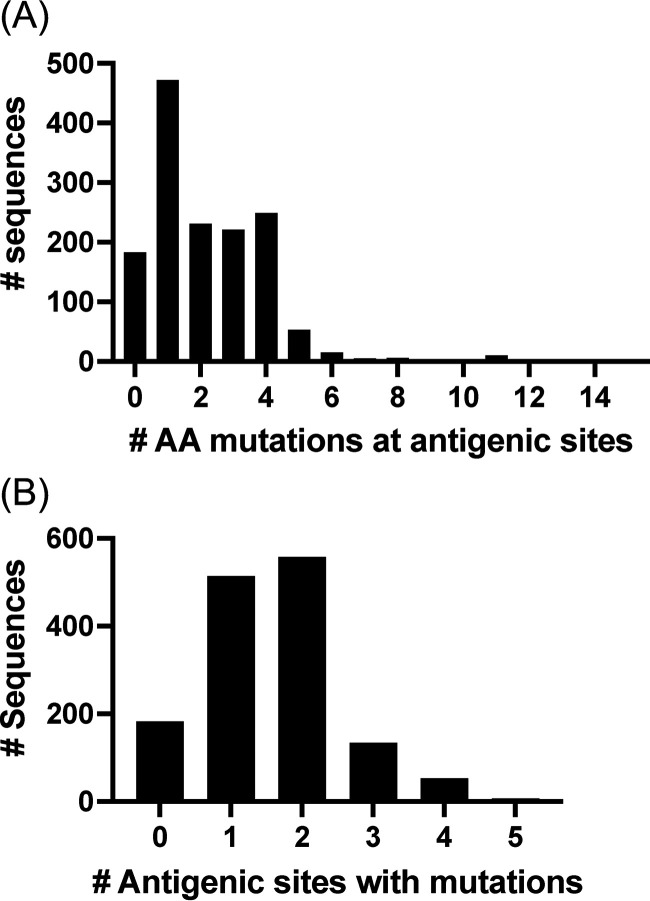
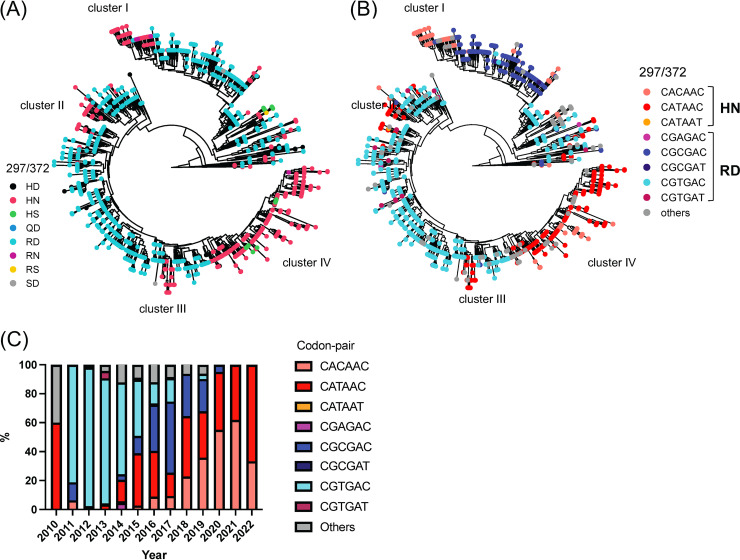
Next, we investigated how many viruses present simultaneous mutations in multiple antigenic sites. To quantify this information, we determined the consensus sequence of the Sydney_2012 variant and calculated the number of amino acid mutations on the antigenic sites for any given virus compared to the consensus sequence. Notably, most viruses (88%, 1268/1449) presented ≥1 mutation at antigenic sites (Fig. 4A) and ~52% (754/1449) of the viruses presented mutations at 2 or more antigenic sites (Fig. 4B). We next investigated whether these were amino acid mutations that followed similar evolutionary histories. To visualize this, we plotted the amino acids present in each strain from the phylogenetic tree (Fig. 5). Patterns of residue composition confirm that 2 Sydney_2012 virus populations co-circulated globally (Fig. 5), and that residues 297 and 372 from antigenic site A co-evolved (Fig. 6A). Notably, 4 independent clusters (clusters I-IV) on the phylogenetic tree show genetic linkage of these 2 residues (297H, 372N), suggesting that convergence is the mechanism for this mutational profile. Indeed, we note that different codon usage supported convergence of 2 different evolutionary trajectories to display the same residues at these 2 sites (Fig. 6B). Thus, the viruses from cluster I use the CACAAC codon pair, while those grouping together in clusters II to IV mostly use the CATAAC codon pair. The mutational pattern of these 2 codon pairs shows that these 2 populations co-circulated in the human population (Fig. 6C). Notably, the convergence for cluster I required 2 steps of mutations from the ancestral codon pair (CGTGAC to CGCGAC to CACAAC) (Fig. 6B). Mapping residues 297 and 372 on the major capsid protein shows that they are very close to each other (Fig. S3). Overall, the genomic analyses suggest that changes on one of the most immunodominant antigenic sites (site A) could account for intra-variant antigenic diversification of the Sydney_2012 variant.
FIG 4.
Quantification of amino acid mutations on the major antigenic sites from GII.4 Sydney_2012 noroviruses. (A) Distribution of viruses by the number of mutations presented at antigenic sites as compared with the consensus sequence of Sydney_2012 variant. (B) Distribution of viruses by the number of antigenic sites presenting any one mutation as compared with the consensus sequence of Sydney_2012 variant. The consensus sequence for the Sydney_2012 variant was calculated from 1,449 complete (or nearly complete) sequences of Sydney_2012 noroviruses and the Entropy tool as described in Materials and Methods.
FIG 5.
Mutations on antigenic sites are associated to specific phylogenetic clusters. The tips of the time-scaled phylogenetic tree were color-coded based on the residue presented by every viral sequence. The phylogenetic trees were calculated as indicated in Fig. 1 and the Materials and Methods section.
FIG 6.
Amino acid mutational pattern from antigenic site A from GII.4 Sydney_2012 variant is associated to differential genetic codon usage. (A) Amino acid distribution in the phylogenetic trees shows evolutionary convergence in residues 297 and 372 from major capsid protein (VP1) of GII.4 Sydney_2012 noroviruses. (B) Phylogenetic tree color-coded based on the codon presented for the codon pair encoding residues 297 and 372. (C) Temporal genetic diversification of the codon pair encoding amino acids 297 and 372. Colors of the bars correspond to the most frequent sequence patterns presented at those 2 codon positions. The analyses were done using 1,449 complete (or nearly complete) sequences of Sydney_2012 noroviruses. The phylogenetic trees were calculated as indicated in Fig. 1 and the Materials and Methods section. Clusters were arbitrarily assigned, in order to facilitate description, to monophyletic branching of strains with similar codon pair usage.
Antigenic diversification of the Sydney_2012 variant.
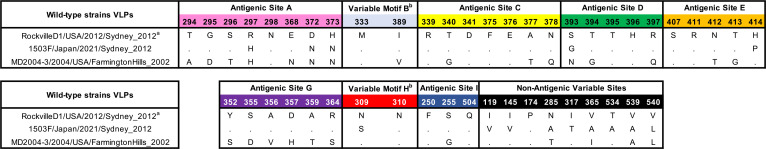
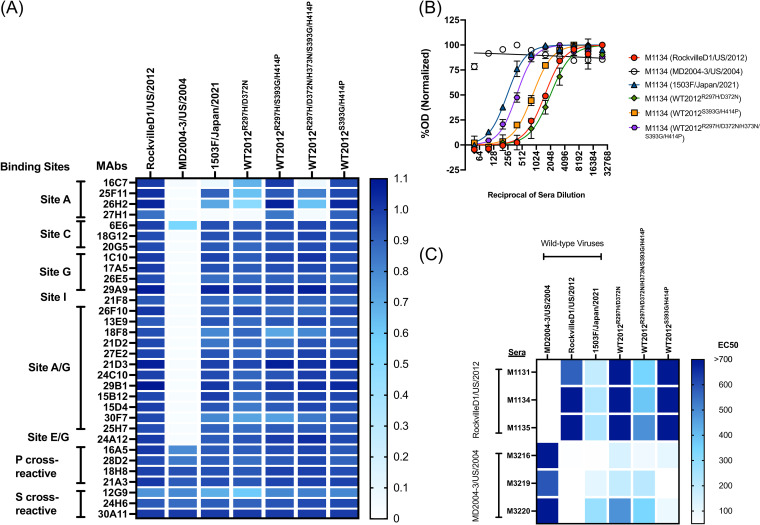
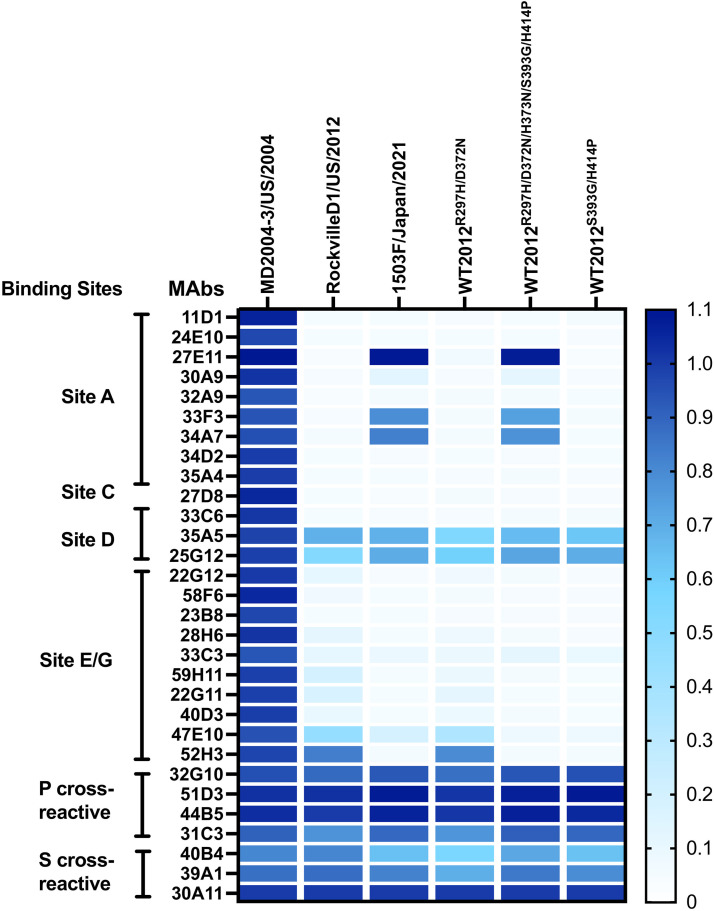
To determine if these mutations play a role in the antigenic characteristics of the Sydney_2012 variant, we developed VLPs of a wild-type virus (1503F/Japan/2021) that was detected in 2021, which presented multiple mutations on the antigenic sites (Table 1). We also developed mutant VLPs presenting the major intra-variant mutations in the backbone of the ancestral Sydney_2012 variant virus: RockvilleD1/US/2012. These VLPs were used to test for binding changes of mouse monoclonal antibodies (MAbs) developed against the ancestral Sydney_2012 variant (RockvilleD1/US/2012) (13, 21). We examined a total of 32 MAbs, 25 of them mapping to variable antigenic sites, 4 mapping to cross-reactive epitopes in the P domain, and 3 mapping to cross-reactive epitopes in the S domain (Fig. 7A) (7). Twenty-four MAbs mapping to variable antigenic sites were negative and 1 MAb (6E6, antigenic site C) was positive for MD2004-3/US/2004 VLPs, a variant of the Farmington_Hills_2002 virus. Regarding the Sydney_2012 VLPs, only 4 MAbs lost ≥50% of binding (16C7, 25F11, 26H2, and 27H1), with all mapping to antigenic site A. The MAb 16C7 completely lost binding to VLPs presenting 3 simultaneous changes at residues 297, 372, and 373, and lost partial binding with the VLPs with mutations in residues 297 and 372. The MAb 25F11 also lost partial (~30%) binding with the VLPs with mutations in residues 297 and 372. The MAb 26H2 lost >50% binding to VLPs with mutations at residues 297 and 372, but lost only ~30% binding with the VLP presenting the 5 mutations (297, 372, 373, 393, and 414). The latter suggests that additional mutations compensate for the binding of 26H2. The MAb 27H1 lost >75% binding with all VLPs with mutations in residues 297 and 372. Binding profiles with mutant VLPs presenting 3 mutations (297, 393, and 414) show that a single mutation (R297H) in antigenic site A did not alter binding of any of the MAbs tested here. None of the MAbs mapping to the A/G epitope lost binding with any of the mutants. We did not have MAbs specific to antigenic sites D and E, and therefore could not test the role of mutations 393 and 414 in the binding of MAbs. Since there are some antigenic differences related to the mutations that emerged recently, particularly the pair 297H and 372N, and because we do not have MAbs mapping to antigenic sites D and E, we tested the mutant VLPs using polyclonal sera raised against the ancestral wild-type Sydney_2012 and Farmington_Hills_2002 variants (Fig. 7B and Fig. S4). Mouse sera against the homologous VLPs presented a strong carbohydrate blocking titer (EC50 ranging: 793 to 1502), but no (or minimal) blocking to the heterologous VLPs (EC50 ranging: <50 to 53; P = 0.004 in Dunnett multiple comparison test). Mice immunized with RockvilleD1/US/2012 showed strong blockade titer to double mutant (R297H/D372N and S393G/H414P) VLPs, and an intermediate blockade titer to the 1503F/Japan/2021 VLPs (EC50: 189 to 233, with statistical difference of P = 0.015) and penta mutant (R297H/D372N/H373N/S393G/H414P; EC50: 350 to 488, P = 0.064) (Fig. 7C). The differences in reactivity for each of the sera against the 1503F/Japan/2021 or mutant VLPs ranged from 0.6 to 6.6-fold, which is below the 16-fold difference calculated as indicative of antigenic differences between the GII.4 noroviruses (11). Notably, sera from mice immunized with the Farmington_Hills_2002 variant, MD2004-3/US/2004, showed distinct patterns of reactivity. Overall, there was no statistical difference against RockvilleD1/US/2012 VLPs due to variations among individual mice, but Mice 3219 and 3220 showed intermediate blockade titers to 1503F/Japan/2021 VLPs (EC50: 113 to 280), the double mutant R297H/D372N (EC50: 187 to 487), and penta mutant VLPs (EC50: 211 to 341) (Fig. 7B), while mouse 3216 showed minimal blocking activity against Sydney VLPs (EC50: 51 to 140). This pattern of reactivity detected for the mice immunized with the Farmington_Hills_2002 variant can be explained by the similar amino acids presented by these viruses at residues 297, 372, 373, and 414 (Table 1).
TABLE 1.
Mutations presented at the major antigenic sites by the wild-type viruses selected in this study
aViruses name: Strain/Country of isolation/Date of isolation/Variant designation.
bSites predicted to be relevant to virus diversification, but not yet experimentally demonstrated to be an antigenic site.
FIG 7.
GII.4 Sydney_2012 noroviruses present minimal intra-variant antigenic variation. (A) A monoclonal antibody (MAb) library developed against a wild-type GII.4 Sydney_2012 virus (RockvilleD1/US/2012) indicates binding lost only by a subset of MAbs targeting antigenic site A. The heatmap indicates MAb-VLP binding as detected by ELISA, and represents the normalized OD405nm values. The columns denote wild-type and mutant VLPs. The OD405 nm were normalized to the cross-reactive MAb 30A11 (1.0) and the negative control (0). (B) Polyclonal sera responses from animals immunized with RockvilleD1/US/2012 VLPs. Normalized curves are shown from 1 mouse polyclonal serum as example. (C) The heatmap indicates the EC50 value from HBGA-blocking titers from the polyclonal sera from animals immunized with RoclvilleD1/US/2012 and MD2004-3/US/2004 (a wild-type Farmington_Hills_2002 virus) VLPs. The columns denote wild-type and mutant VLPs, and the rows show individual polyclonal sera.
Sydney_2012 viruses show reversion to ancestral GII.4 epitopes.
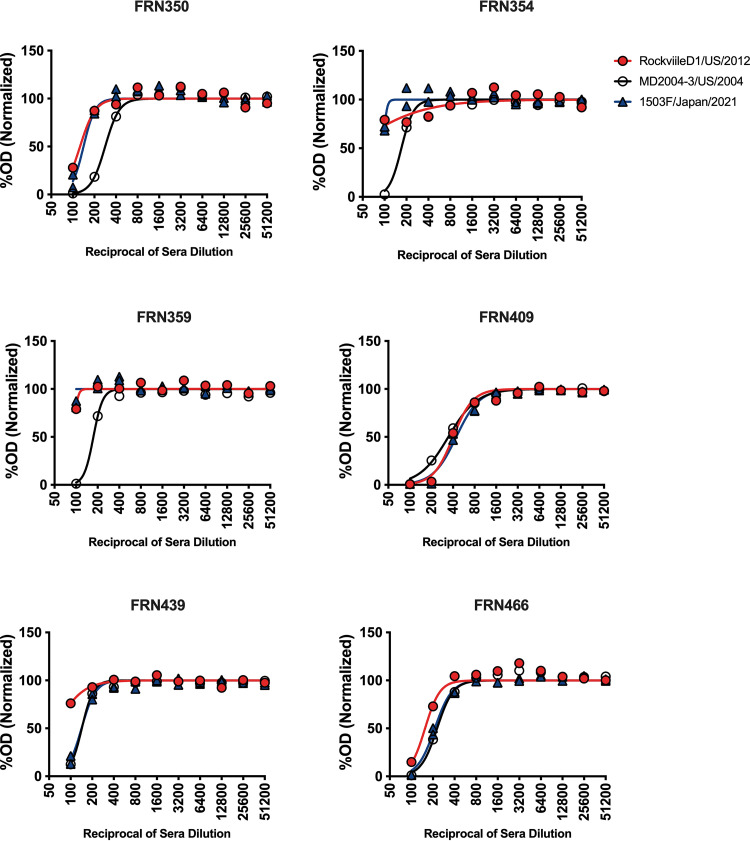
To confirm the findings described above, we used a panel of 29 MAbs raised against the MD2004-3/US/2004 VLPs. All P and S cross-reactive MAbs showed reactivity with all VLPs tested (Fig. 8). Four MAbs mapping to variable antigenic sites, 2 mapping to site D, and 2 mapping to site E/G were positive for RockvilleD1/US/2012 VLPs. Three (27E11, 33F3, and 34A7) out of the 9 MAbs mapping to antigenic site A gained reactivity to the 1503F/Japan/2021 virus and penta mutant VLPs. None of the Farmington_Hills_2002 MAbs gained reactivity to the double mutant R297H/D372N VLPs; however, 1 weak, cross-reactive MAb (52H3) that maps to E/G antigenic sites lost binding with those VLPs presenting the mutation at position 414 (Fig. 8). Finally, we tested the carbohydrate blocking activity of sera from 6 adult individuals challenged with a Farmington_Hills_2002 variant virus, 031693/US/2003 (22). The 031693/US/2003 virus does not have mutations on major antigenic sites except D295G on antigenic site A, as compared with MD2004-3/US/2004 virus. There was no overall significant difference of blocking titers among VLPs (P = 0.55 in One-way ANOVA). However, as 6 challenged individuals might have different infection histories, reactivity patterns were very different among them. Thus, the sera from the challenged individuals showed strong blocking titers (mean, 223; range, 143 to 344) against the Farmington_Hills_2002 VLPs (MD2004-3/US/2004), and varying blocking titers against the Sydney_2012 VLPs, RockvilleD1/US/2012 (mean, 147; range, <50 to 402) and 1503F/Japan/2021 (mean, 180; range, <50 to 444). Two individuals, FRN439 and FRN466, presented blocking titers that indicate closer antigenicity between the 1503F/Japan/2021 and MD2004-3/US/2004 viruses. The sera from 3 individuals (FRN350, FRN354, and FRN359) reacted similarly to the Sydney_2012 VLPs, and the sera from individual FRN40 reacted strongly to all 3 VLPs (Fig. 9).
FIG 8.
GII.4 Sydney_2012 noroviruses present epitopes from ancestral GII.4 variants. A monoclonal antibody (MAb) library developed against a wild-type GII.4 Farmington_Hills_2002 virus (MD2004-3/US/2004) indicates reactivity of MAbs, mapping to antigenic site A, against newer Sydney_2012 viruses. The columns denote wild-type and mutant VLPs. The background of the mutant VLPs is that from the wild-type RockvilleD1/US/2012 virus. The OD405 nm were normalized to the cross-reactive MAb 30A11 (1.0) and the negative control (0).
FIG 9.
Sera from individuals challenged with a Farmington_Hills_2002 virus present HBGA-blocking activity against modern Sydney_2012 VLPs. Normalized curves are shown from sera from 6 individuals challenged with the 031693/US/2003 virus (22). Two individuals, FRN439 and FRN466, presented blocking titers that indicate closer antigenicity between the modern Sydney_2012 virus (1503F/Japan/2021) and the Farmington_Hills_2002 virus (MD2004-3/US/2004). The 1503F/Japan/2021 virus presents an antigenic site A that resembles those from Farmington_Hills_2002 virus (Table 1).
DISCUSSION
Norovirus genotypes present 2 distinct patterns of evolution: GII.4 noroviruses present a chronological emergence of variants and accumulation of mutations at the major capsid protein that alter antigenicity (7, 12, 13, 23), while non-GII.4 noroviruses remain very similar over several decades of circulation in humans (19). Overall, GII.4 variants differ by ~25 mutations (~5%) on the whole capsid protein (19), with most of these mutations occurring at the major antigenic sites, A to I (7, 12, 13). Notably, while Sydney_2012 viruses could present up to 35 mutations, these viruses did not show a marked accumulation of mutations over time. The mutational profile shows that GII.4 noroviruses acquired a considerable number of mutations very early in the circulation of this virus (circa 2010), and then presented a large period of great genetic stability, as shown for most non-GII.4 noroviruses (19). A very similar set of events was reported for the emerging and transiently predominant GII.17 viruses (19).
Tracking the evolutionary history and genetic diversification of viruses provides fundamental knowledge about the natural history of disease for any given virus (24–30). This is best exemplified by the unprecedented effort to sequence the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus and to link amino acid changes to their effect on immunity, transmission, and pathobiology of COVID-19 (27–30). Using large-scale genomics, we identified patterns of genetic diversification that could be associated with antigenic changes in Sydney_2012 viruses. Mutational patterns of 2 residues, 297 and 372, which map to antigenic site A, showed a very tight link in that mutating one resulted in mutations on the other residue. Notably, these 2 residues presented strong signal for coevolution throughout the diversification of GII.4 noroviruses (23), further supporting evolutionary constraints and the reversion to ancestral epitopes. These 2 residues are very close to each other on the surface of the viral capsid, and map on a critical region that not only plays an important role in defining the antigenicity of the virus but also in binding to HBGA carbohydrates (Fig. S4). Of note, we recently showed that mutations in antigenic site A could abrogate binding to HBGA carbohydrates (7). Mutant VLPs presenting mutations in these 2 residues, 297 and 372, still showed binding to HBGA carbohydrates. The antigenic site A is composed of 8 residues, so there are >109 possible combinations of amino acids for that antigenic site, yet the intra-variant evolution of Sydney_2012 resulted in its predominance with epitopes that resemble those from Farmington_Hills_2002 viruses. This suggest that there are evolutionary constraints and a limited number of possible combinations that can result in viable viruses. A better understanding of the determinants of the genetic robustness of the immunodominant antigenic site A in terms of antigenic diversification and HBGA carbohydrate binding could help in the design of cross-protective vaccines for GII.4 norovirus.
Even in the presence of mutations mapping on epitopes involved in viral neutralization and protection, antigenic stability has been described for other norovirus genotypes (31), measles (32), and respiratory syncytial viruses (33). The stability of measles has been attributed to the co-immunodominance of multiple antigenic sites, which restricts the virus to change multiple sites simultaneously to escape immune responses (32, 34). Noroviruses circulating during 2002 to 2006 presented a strong immunodominance on antigenic site A, which gradually changed to antigenic site G, and ultimately resulted in the immunodominance of epitope(s) shared by antigenic sites A and G in Sydney_2012 viruses (7). While antigenic site A was shown to be variable, antigenic site G remained highly stable in the Sydney_2012 variant. The major effect on Sydney_2012 variant antigenicity was attributed to changes occurring in synchrony at residues 297 and 372. Antibodies mapping to these residues do not map to the immunodominant epitope(s) shared by antigenic sites A and G (7), thus explaining the antigenic stability of Sydney_2012 at the polyclonal antibody level. Why the residues from antigenic site G are conserved for over a decade is unknown. A similar pattern of antibody immunodominance and long circulation was observed with the Grimsby_1995 viruses (7). Notably, the subsequent variant, Farmington_Hills_2002, presented a unique insertion on the antigenic site D and a drastic change in the immunodominance toward single antigenic site A (7, 23, 35, 36). Thus, it is tempting to speculate that the next pandemic GII.4 variant might present unique mutations (e.g., insertions and deletions) or drastic changes in the chemical properties of the residues mapping to the antigenic sites. If this is the case, it would remain to be seen if a new cycle of rapid turnover of variants (2 to 3 years) occurs, such as the turnover that occurred during 2002 to 2012.
The current dogma is that the predominance of GII.4 is linked to the chronological emergence of antigenically distinct variants that can escape from immunity acquired from previous infections. On the contrary, the predominance of Sydney_2012 has been recently attributed to a lack in the establishment of strong Sydney 2012 immunity in adults, which facilitates the continued circulation and predominance of this variant over the last decade (37). Notably, the same study showed (i) strong blocking (i.e., neutralizing) titers to Sydney_2012 viruses in children infected with Sydney_2012 viruses, and (ii) strong blocking titers to Farmington_Hills_2002 viruses in adults infected with early Sydney_2012 viruses; suggesting that there is strong herd immunity to limit Sydney_2012 infections and that antigenic variation of Sydney_2012 in residues 297 and 372 would not facilitate immune escape in individuals that were exposed to Farmington_Hills_2002 viruses (37).
Overall, this study demonstrates that, despite circulating for over a decade, Sydney_2012 viruses present minimal antigenic changes in their capsid protein and provides novel insights on the intra-variant diversification of GII.4 noroviruses that could inform vaccine design. Moreover, since the Sydney_2012 variant has been recorded as the predominant strain globally, this study also raises new questions about the determinants for the predominance of GII.4 variants in the human population.
MATERIALS AND METHODS
Genetic diversification analyses of GII.4_Sydney_2012 norovirus using bioinformatics.
A total of 3,145 full-length (1,623 nucleotides [nt]) and nearly full-length (≥1,500 nt) VP1 sequences of the GII.4 genotype were downloaded from GenBank (GB). Sequences obtained from immunocompromised patients or environmental samples were omitted from the data set. A total of 1,452 sequences were typed as Sydney_2012 using Norovirus Typing Tool (https://www.rivm.nl/mpf/typingtool/norovirus/), and sequence analyses were performed in MEGA v7. After examination of the diversity and mutational profile, 3 sequences (MW045405/1065/Botswana/2017, MT221660/CUHK-NS-2467/China/2020, and KX354139/2016-SP-0151_020316_CA/US/2016) were removed from the data set as they presented a very large number of substitutions, and their origin was not fully determined. The final Sydney_2012 data set included 1,449 sequences that included viruses detected from 2010 to 2022. Shannon entropy values and the Sydney_2012 consensus sequence were calculated using the Shannon Entropy-One tool as implemented in Los Alamos National Laboratory (https://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy.html). Profiling of mutational patterns of antigenic sites over time and comparison with the Sydney_2012 consensus sequence were performed using R (38), as described elsewhere (13). Three-dimensional structure of norovirus capsid P domain (PDB: 2OBS) and mapping of residues were rendered using UCSF Chimera v1.16 (https://www.cgl.ucsf.edu/chimera/). Temporal phylogenetic analysis was performed using TempEst v1.5.3 (39), and a maximum-likelihood tree was built with PhyML 3.3 (40). A time-scaled tree was estimated using Bayesian Markov chain Monte Carlo (MCMC) methodology, as implemented in BEAST v1.10.4 (41). The 4 MCMC runs were independently performed using the Shapiro-Rambaut-Drummond-2006 (SRD06) substitution model and strict clock model, and combined after removing burn-in from each run to obtain convergence of all the parameters. The maximum clade credibility (MCC) tree was generated from combined posterior trees using TreeAnnotator v1.10.4, and visualized using ggtree package in R.
If available, polymerase types from GII.4 Sydney_2012 sequences were determined using Norovirus Typing Tool (https://www.rivm.nl/mpf/typingtool/norovirus/) and annotated on the phylogenetic tree using R. Yearly distribution of the polymerase types from all GII.4 Sydney_2012 viruses (n = 1,237, including dual-typed sequences with ≥100 nt length collected from 1995 to 2019) were obtained from a previous study (11).
VLP production, carbohydrate binding, and immunoassays.
The VP1-encoding sequences of wild-type GII.4 viruses (RockvilleD1/US/2012 [GB: KY424328] and 1503F/Japan/2021 [GB: LC644993]) were synthesized and cloned into pFastBac1 vectors (GenScript). Baculoviruses expressing the norovirus VP1s were obtained by using the Bac-to-Bac Baculovirus Expression System (Gibco). The produced VLPs were purified using 25% sucrose cushion, followed by cesium chloride gradient as described previously (7, 23, 35). Expression of VP1 protein was confirmed by Western blotting with norovirus VP1-specific cross-reactive MAb (7, 42), and VLP integrity was confirmed by electron microscopy. Mutant VLPs were designed by introducing the different mutations into the backbone of RockvilleD1/US/2012. The ORF2s with these mutations were synthesized, cloned, and subjected to the Bac-to-Bac Baculovirus Expression System as described above. Binding of VLPs to the HBGA carbohydrate was performed using PGM III as described elsewhere (38). The reactivity of a large panel of mouse MAbs and polyclonal sera (7) developed against the RockvilleD1/US/2012 and MD2004-3/US/2004 was tested using ELISA and HBGA-blocking assays, as described elsewhere (7, 23). Animal protocol was approved by the IACUC (protocol number 2018-41). Moreover, human sera collected from 6 individuals challenged with a Farmington_Hills virus (031693/US/2003 [GB: JQ965810]) (22) were tested for HBGA-blocking assays. The history of norovirus infection in these individuals is unknown, but the challenges were done prior to the circulation of the Sydney_2012 variant. The use of human samples was approved by the institutional review board ([IRB], protocol number CBER IRB 16-069B). The concentration of VLPs and MAbs used for the ELISA was 0.5 μg/mL and 10 μg/mL, respectively. The concentration of VLPs used for HBGA-blocking assays ranged from 0.25 to 0.5 μg/mL. Those concentrations were chosen based on the binding to PGM III, i.e., lowest dilution that provides an OD405 nm value of 1.5 to 2. Binding signals were measured as OD405 nm using a SPECTROstar Nano plate reader and values were plotted using GraphPad Prism v9. Statistical analyses were performed using One-way ANOVA and post hoc Dunnett multiple comparison test as implemented in GraphPad Prism v9.
Data availability.
The sequences used in this study were downloaded from the GenBank public database. The list of sequences and their GenBank accession numbers is available in Table S2. The VLPs and MAbs used in this study are available upon request.
ACKNOWLEDGMENTS
We thank all colleagues that submit their sequences into the public repositories so this type of analyses can be conducted. We thank the animal technicians and veterinarians at the Division of Veterinary Services, Food and Drug Administration for their support with the animal experiments. Financial support for this work was provided by the Food and Drug Administration intramural funds (program number Z01 BK 04012 LHV to G.I.P.).
G.I.P. conceived the project; K.T. and G.I.P. conducted bioinformatics analyses and designed the VLPs panel; K.T., L.A.F.-S., J.A.K., P.E., and G.I.P. generated VLPs; K.T., L.A.F.-S., P.E., K.A.P., and G.I.P. conducted immunoassays; Y.G. acquired electron microscopies; G.I.P. supervised the research activity and wrote the original draft; and all authors reviewed and edited the paper.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Gabriel I. Parra, Email: gabriel.parra@fda.hhs.gov.
Anice C. Lowen, Emory University School of Medicine
REFERENCES
- 1.Ford-Siltz LA, Tohma K, Parra GI. 2021. Understanding the relationship between norovirus diversity and immunity. Gut Microbes 13:1–13. 10.1080/19490976.2021.1900994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ford-Siltz LA, Wales S, Tohma K, Gao Y, Parra GI. 2020. Genotype-specific neutralization of norovirus is mediated by antibodies against the protruding domain of the major capsid protein. J Infect Dis 225:1205–1214. 10.1093/infdis/jiaa116. [DOI] [PubMed] [Google Scholar]
- 3.Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290. 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 4.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977. 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, Stewart P, LePendu J, Baric R. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat Med 9:548–553. 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 6.Atmar RL, Ettayebi K, Ayyar BV, Neill FH, Braun RP, Ramani S, Estes MK. 2020. Comparison of microneutralization and histo-blood group antigen-blocking assays for functional norovirus antibody detection. J Infect Dis 221:739–743. 10.1093/infdis/jiz526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tohma K, Ford-Siltz LA, Kendra JA, Parra GI. 2022. Dynamic immunodominance hierarchy of neutralizing antibody responses to evolving GII.4 noroviruses. Cell Rep 39:110689. 10.1016/j.celrep.2022.110689. [DOI] [PubMed] [Google Scholar]
- 8.Alvarado G, Ettayebi K, Atmar RL, Bombardi RG, Kose N, Estes MK, Crowe JE, Jr. 2018. Human monoclonal antibodies that neutralize pandemic GII.4 noroviruses. Gastroenterology 155:1898–1907. 10.1053/j.gastro.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atmar RL, Bernstein DI, Lyon GM, Treanor JJ, Al-Ibrahim MS, Graham DY, Vinje J, Jiang X, Gregoricus N, Frenck RW, Moe CL, Chen WH, Ferreira J, Barrett J, Opekun AR, Estes MK, Borkowski A, Baehner F, Goodwin R, Edmonds A, Mendelman PM. 2015. Serological correlates of protection against a GII.4 norovirus. Clin Vaccine Immunol 22:923–929. 10.1128/CVI.00196-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, Graham DY, Atmar RL. 2010. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis 202:1212–1218. 10.1086/656364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kendra JA, Tohma K, Parra GI. 2022. Global and regional circulation trends of norovirus genotypes and recombinants, 1995–2019: a comprehensive review of sequences from public databases. Rev Med Virol 32:e2354. 10.1002/rmv.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debbink K, Lindesmith LC, Donaldson EF, Baric RS. 2012. Norovirus immunity and the great escape. PLoS Pathog 8:e1002921. 10.1371/journal.ppat.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tohma K, Lepore CJ, Gao Y, Ford-Siltz LA, Parra GI. 2019. Population genomics of GII.4 noroviruses reveal complex diversification and new antigenic sites involved in the emergence of pandemic strains. mBio 10:e02202-19. 10.1128/mBio.02202-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra GI. 2019. Emergence of norovirus strains: a tale of two genes. Virus Evol 5:vez048. 10.1093/ve/vez048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J Virol 81:9932–9941. 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siebenga JJ, Vennema H, Zheng DP, Vinje J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O'Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis 200:802–812. 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 17.Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magana LC, Gregoricus N, Marine RL, Chhabra P, Vinje J. 2017. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol 55:2208–2221. 10.1128/JCM.00455-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barclay L, Cannon JL, Wikswo ME, Phillips AR, Browne H, Montmayeur AM, Tatusov RL, Burke RM, Hall AJ, Vinje J. 2019. Emerging novel GII.P16 noroviruses associated with multiple capsid genotypes. Viruses 11. 10.3390/v11060535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra GI, Squires RB, Karangwa CK, Johnson JA, Lepore CJ, Sosnovtsev SV, Green KY. 2017. Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog 13:e1006136. 10.1371/journal.ppat.1006136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tohma K, Lepore CJ, Martinez M, Degiuseppe JI, Khamrin P, Saito M, Mayta H, Nwaba AUA, Ford-Siltz LA, Green KY, Galeano ME, Zimic M, Stupka JA, Gilman RH, Maneekarn N, Ushijima H, Parra GI. 2021. Genome-wide analyses of human noroviruses provide insights on evolutionary dynamics and evidence of coexisting viral populations evolving under recombination constraints. PLoS Pathog 17:e1009744. 10.1371/journal.ppat.1009744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parra GI, Green KY. 2014. Sequential gastroenteritis episodes caused by 2 norovirus genotypes. Emerg Infect Dis 20:1016–1018. 10.3201/eid2006.131627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frenck R, Bernstein DI, Xia M, Huang P, Zhong W, Parker S, Dickey M, McNeal M, Jiang X. 2012. Predicting susceptibility to norovirus GII.4 by use of a challenge model involving humans. J Infect Dis 206:1386–1393. 10.1093/infdis/jis514. [DOI] [PubMed] [Google Scholar]
- 23.Kendra JA, Tohma K, Ford-Siltz LA, Lepore CJ, Parra GI. 2021. Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses. Proc Natl Acad Sci USA 118:e2015874118. 10.1073/pnas.2015874118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, Holmes EC. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327–332. 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 25.Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. 2016. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci USA 113:E1701–E1709. 10.1073/pnas.1525578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudas G, Carvalho LM, Bedford T, Tatem AJ, Baele G, Faria NR, Park DJ, Ladner JT, Arias A, Asogun D, Bielejec F, Caddy SL, Cotten M, D'Ambrozio J, Dellicour S, Di Caro A, Diclaro JW, Duraffour S, Elmore MJ, Fakoli LS, Faye O, Gilbert ML, Gevao SM, Gire S, Gladden-Young A, Gnirke A, Goba A, Grant DS, Haagmans BL, Hiscox JA, Jah U, Kugelman JR, Liu D, Lu J, Malboeuf CM, Mate S, Matthews DA, Matranga CB, Meredith LW, Qu J, Quick J, Pas SD, Phan MVT, Pollakis G, Reusken CB, Sanchez-Lockhart M, Schaffner SF, Schieffelin JS, Sealfon RS, Simon-Loriere E, et al. 2017. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature 544:309–315. 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Lai S, Gao GF, Shi W. 2021. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 600:408–418. 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 28.Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva Filipe A, Wojcechowskyj JA, Davis C, Piccoli L, Pascall DJ, Dillen J, Lytras S, Czudnochowski N, Shah R, Meury M, Jesudason N, De Marco A, Li K, Bassi J, O'Toole A, Pinto D, Colquhoun RM, Culap K, Jackson B, Zatta F, Rambaut A, Jaconi S, Sreenu VB, Nix J, Zhang I, Jarrett RF, Glass WG, Beltramello M, Nomikou K, Pizzuto M, Tong L, Cameroni E, Croll TI, Johnson N, Di Iulio J, Wickenhagen A, Ceschi A, Harbison AM, Mair D, Ferrari P, Smollett K, Sallusto F, Carmichael S, Garzoni C, Nichols J, Galli M, COVID-19 Genomics UK (COG-UK) Consortium, et al. 2021. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184:1171–1187. 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Consortium C-GU, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attwood SW, Hill SC, Aanensen DM, Connor TR, Pybus OG. 2022. Phylogenetic and phylodynamic approaches to understanding and combating the early SARS-CoV-2 pandemic. Nat Rev Genet 23:547–562. 10.1038/s41576-022-00483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanstrom J, Lindesmith LC, Donaldson EF, Yount B, Baric RS. 2014. Characterization of blockade antibody responses in GII.2.1976 Snow Mountain virus-infected subjects. J Virol 88:829–837. 10.1128/JVI.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Alia MA, Nace RA, Zhang L, Russell SJ. 2021. Serotypic evolution of measles virus is constrained by multiple co-dominant B cell epitopes on its surface glycoproteins. Cell Rep Med 2:100225. 10.1016/j.xcrm.2021.100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trento A, Abrego L, Rodriguez-Fernandez R, Gonzalez-Sanchez MI, Gonzalez-Martinez F, Delfraro A, Pascale JM, Arbiza J, Melero JA. 2015. Conservation of G-protein epitopes in respiratory syncytial virus (Group A) despite broad genetic diversity: is antibody selection involved in virus evolution? J Virol 89:7776–7785. 10.1128/JVI.00467-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greaney AJ, Welsh FC, Bloom JD. 2021. Co-dominant neutralizing epitopes make anti-measles immunity resistant to viral evolution. Cell Rep Med 2:100257. 10.1016/j.xcrm.2021.100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parra GI, Abente EJ, Sandoval-Jaime C, Sosnovtsev SV, Bok K, Green KY. 2012. Multiple antigenic sites are involved in blocking the interaction of GII.4 norovirus capsid with ABH histo-blood group antigens. J Virol 86:7414–7426. 10.1128/JVI.06729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dingle KE, Norovirus Infection Control in Oxfordshire Communities H . 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J Clin Microbiol 42:3950–3957. 10.1128/JCM.42.9.3950-3957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindesmith LC, Boshier FAT, Brewer-Jensen PD, Roy S, Costantini V, Mallory ML, Zweigart M, May SR, Conrad H, O'Reilly KM, Kelly D, Celma CC, Beard S, Williams R, Tutill HJ, Becker Dreps S, Bucardo F, Allen DJ, Vinje J, Goldstein RA, Breuer J, Baric RS. 2022. Immune imprinting drives human norovirus potential for global spread. mBio 13:e0186122. 10.1128/mbio.01861-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 39.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 41.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parra GI, Azure J, Fischer R, Bok K, Sandoval-Jaime C, Sosnovtsev SV, Sander P, Green KY. 2013. Identification of a broadly cross-reactive epitope in the inner shell of the norovirus capsid. PLoS One 8:e67592. 10.1371/journal.pone.0067592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Tables S1 and S2. Download jvi.01716-22-s0001.pdf, PDF file, 1.2 MB (1.3MB, pdf)
Data Availability Statement
The sequences used in this study were downloaded from the GenBank public database. The list of sequences and their GenBank accession numbers is available in Table S2. The VLPs and MAbs used in this study are available upon request.