ABSTRACT
Allosteric HIV-1 integrase (IN) inhibitors (ALLINIs) are an emerging class of small molecules that disrupt viral maturation by inducing the aberrant multimerization of IN. Here, we present cocrystal structures of HIV-1 IN with two potent ALLINIs, namely, BI-D and the drug candidate Pirmitegravir. The structures reveal atomistic details of the ALLINI-induced interface between the HIV-1 IN catalytic core and carboxyl-terminal domains (CCD and CTD). Projecting from their principal binding pocket on the IN CCD dimer, the compounds act as molecular glue by engaging a triad of invariant HIV-1 IN CTD residues, namely, Tyr226, Trp235, and Lys266, to nucleate the CTD-CCD interaction. The drug-induced interface involves the CTD SH3-like fold and extends to the beginning of the IN carboxyl-terminal tail region. We show that mutations of HIV-1 IN CTD residues that participate in the interface with the CCD greatly reduce the IN-aggregation properties of Pirmitegravir. Our results explain the mechanism of the ALLINI-induced condensation of HIV-1 IN and provide a reliable template for the rational development of this series of antiretrovirals through the optimization of their key contacts with the viral target.
KEYWORDS: antiretroviral drugs, HIV-1, integrase, allosteric inhibitor, ALLINI, LEDGIN, Pirmitegravir, STP0404, BI-D
INTRODUCTION
Highly active antiretroviral therapy has transformed HIV-1 infection from a terminal illness to a manageable condition with a near normal life expectancy. The combinatorial treatment approach relies on multiple drugs with independent modes of action, typically targeting two or three viral enzymes. A lack of drug supply and/or insufficient adherence leads to the emergence of drug resistance and virologic failure. Infection with drug-resistant HIV-1 strains may then greatly limit treatment options. Therefore, to stay ahead of viral resistance, it is important to continue developing established as well as novel classes of antiretrovirals.
Integrase (IN) is the retroviral enzyme that is responsible for the insertion of the DNA copy of the viral RNA genome into a host cell chromosome (reviewed in [1]). As such, it is a target for strand transfer inhibitors, which are a class of small molecules that bind in the IN active site and arrest HIV-1 replication (2, 3). Five strand transfer inhibitors, including the long-acting second-generation compound cabotegravir, have been approved for the treatment of HIV-1 infection. Due to their shared mode of action, strand transfer inhibitors display overlapping resistance profiles (4–7). Allosteric HIV-1 IN inhibitors (ALLINIs) are an emerging class of antiretrovirals with an orthogonal mode of action. These small molecules were discovered and optimized by independent groups that used different approaches. Consequently, they are also known as LEDGF-IN site inhibitors (LEDGINs) (8), IN-LEDGF allosteric inhibitors (INLAIs) (9, 10), multimerization IN inhibitors (MINIs) (11), or noncatalytic IN inhibitors (NCINIs) (12, 13).
ALLINIs bind with high affinity in a small pocket at the HIV-1 IN catalytic core domain (CCD) dimer interface (8, 9, 11, 14–18). This pocket is also involved in the interaction with the host protein LEDGF/p75 that directs lentiviral DNA integration to bodies of active transcription units (19–22). Consequently, ALLINIs compete with LEDGF/p75 for binding to HIV-1 IN (8, 10, 23). Although gene-tropic HIV-1 integration can accordingly be suppressed in the presence of ALLINIs (23–25), these compounds more potently disrupt the viral life cycle at the stage of particle morphogenesis (10, 15, 26, 27). When HIV-1 virions are produced in the presence of ALLINIs, the genomic RNA is mislocalized outside the viral core (10–12, 15, 26–28), and, upon their entry into target cells, these virions are unable to complete reverse transcription (11, 12, 15, 26, 28, 29). Strikingly similar phenotypes are displayed by the class II HIV-1 IN mutant viruses (30, 31). Unlike class I HIV-1 IN mutants, which are typified by amino acid substitutions within the IN active site and are specifically blocked for integration, class II mutants are characterized by pleiotropic defects and are not confined to a specific IN region or domain. Recent evidence has indicated that HIV-1 IN in the presence of ALLINIs, as well as class II IN mutant proteins, are compromised for RNA binding, which is a noncatalytic function of HIV-1 IN that is essential for viral morphogenesis (31–33). Plausibly, the disruption of IN-RNA interactions may be responsible for the mislocalization of the viral genomic material within affected virions.
ALLINIs trigger aberrant HIV-1 IN multimerization within virions as well as the rapid aggregation of the recombinant protein in vitro (10, 15, 26). This property relies on an unnatural, inhibitor-induced intermolecular interaction that involves the dimeric CCD of one IN oligomer as well as the carboxyl-terminal domain (CTD) of a separate IN multimer (16, 34). Under physiological conditions, lentiviral INs have the propensity to form tetramers and larger order functional oligomers in the presence of viral DNA and are stabilized by the homomeric dimerization of the CCD and the CTD (35–40). However, by inducing the additional CTD-CCD interface, the compounds promote the uncontrolled propagation of linear and branched IN polymers (41). The principal ALLINI binding pocket at the HIV-1 IN CCD dimer interface has been well-characterized via X-ray crystallography, and the crystal structures were instrumental in the development of these compounds (8, 10, 11, 15). However, the ALLINI-induced CTD-CCD interface has only been visualized at 4.4 Å resolution (16), which was insufficient to explain the role of the small molecules in its formation. The shared ALLINI functionality, which is composed of carboxyl, tert-butoxyl, and bulky hydrophobic groups (Fig. 1A), has been visualized at a comparatively high resolution in HIV-1 IN CCD dimer cocrystal structures. However, the apparent lack of similarity outside these CCD-binding moieties may appear inconsistent with their mechanism of action, which requires the least conserved portion of the chemical structure to serve a shared role. Here, we describe the high-resolution cocrystal structures of a two-domain HIV-1 IN construct with two highly potent ALLINIs, namely, BI-D (10, 15, 42, 43) and the current drug candidate Pirmitegravir (PIR; originally designated STP0404) (17). The structures reveal atomistic details of the ALLINI-induced CTD-CCD interface and provide reliable means for the further optimization of this promising class of antiretrovirals.
FIG 1.
Cocrystallization of HIV-1 IN with ALLINIs. (A) Chemical structures of BI-D (upper) and PIR (lower). The functional groups in the ALLINI pharmacophore are color-coded: carboxyl groups (red); tert-butoxyl (green); bulky hydrophobic side chains, chromanyl of BI-D, and chlorophenyl of PIR (blue). Central aromatic scaffolds (quinoline in BI-D and pyrrolopyridine in PIR) are black. (B) Structure of the HIV-1 IN-PIR complex. The protein backbone is shown in a cartoon representation, with CTDs in yellow and CCD dimer chains in cyan and green. The ALLINI molecules are shown as sticks, with carbon, nitrogen, and oxygen atoms in pink, blue, and red, respectively.
RESULTS
HIV-1 IN-ALLINI cocrystal structures.
The rapid aggregation of HIV-1 IN in the presence of ALLINIs greatly impedes the cocrystallization of the viral protein with these small molecules. To overcome this obstacle, we sought to design novel HIV-1 IN constructs with a reduced propensity for aggregation, while preserving the key components for ALLINI interactions. HIV-1 IN is comprised of three flexibly linked globular domains, the N-terminal domain (NTD), CCD, and CTD, each of which is involved in the functional oligomerization of the full-length protein (31). In particular, CTD dimerization (36, 39) is crucial for the formation of higher-order lentiviral IN oligomers (40, 44). Moreover, the CTD dimer interface contributes to the propagation of ALLINI-induced aberrant polymers of HIV-1 IN but not to the interaction with ALLINIs (41). Therefore, we evaluated the oligomerization properties of HIV-1 CTD variants harboring single amino acid substitutions within the dimerization interface. Multiangle laser light scattering coupled with size exclusion chromatography (SEC-MALLS) revealed that whereas the isolated HIV-1 IN CTD existed in a monomer-dimer equilibrium, the mutants L242E and W243E behaved as monomeric species (Fig. S1A).
Development, validation, and X-ray crystallography of the HIV-1 IN construct in complex with ALLINIs. (A) Disruption of HIV-1 IN CTD dimerization by a single amino acid substitution. Wild type (left), L242E (middle), or W243E (right) HIV-1 IN CTD constructs were analyzed using SEC-MALLS. Solid lines report the refractive index for each elution, and the scatter points are weight-averaged molar masses that were determined at 1 s intervals throughout the elution of the observed peaks. The colors correspond to loading protein concentrations of 8 (red), 4 (magenta), 2 (green), 1 (blue), or 0.5 (puce) mg/mL. Grey dashes indicate the molecular masses corresponding to the CTD monomer (6.0 kDa) and dimer (12 kDa) species. (B) The CTD enhances the affinity of HIV-1 IN for PIR. SPR sensorgrams of the association and dissociation of PIR from HIV-1 IN CCD (F185H) (left) and CTD-CCD (F185K/W243E) (right). Compound was injected at concentrations of 0.015 to 1 μM, as indicated. The lower panel shows the association and dissociation rate constants (kon and koff), the equilibrium dissociation constants (KD) that were determined from the SPR data, and the fold change between the parameters between the two IN constructs. (C) Electron density maps of the BI-D (left) and PIR (right) cocrystal structures. Shown are wall-eyed stereo views of the ALLINI-induced CTD-CCD interface with final weighted 2Fo-Fc maps that are contoured at a 1.0 rms deviation as a blue mesh. Protein residues and ligands are depicted as sticks, and water molecules are depicted as small red spheres. The carbon atoms of the amino acid residues are colored by chain (CTD, yellow; subunits 1 and 2 of the CCD dimer, cyan and green, respectively). The carbon atoms of the ALLINIs are colored pink. The carbon atoms of PEG and ethylene glycol are colored grey. The remaining atoms are colored according to conventions: oxygen, red; nitrogen, blue. Download FIG S1, PDF file, 3.1 MB (3.2MB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Having tested a range of recombinant HIV-1 IN constructs, we focused on a reverse-oriented CTD-CCD variant that spanned the CTD with the dimer-disrupting substitution W243E, which was in turn linked via a flexible peptide to the CCD harboring the solubilization mutation F185K (35, 44). The linker was comprised of the HIV-1 IN 18-residue C-terminal tail region, which is unstructured (31), fused to 6 residues of the flexible NTD-CCD linker. The resulting construct lacked the NTD, which is involved in lentiviral IN tetramerization (38, 45) but does not contribute to ALLINI binding (41). Our design was inspired by the idea that linking the domains via a flexible sequence could promote intramolecular CTD-ALLINI-CCD contacts and thus limit the aggregation of the IN-drug assemblies.
Surface plasmon resonance (SPR) measurements revealed that the CTD-CCD fusion construct bound PIR, a clinical candidate ALLINI (17), with an approximately 30-fold higher affinity and an approximately 10-fold slower dissociation rate than did the isolated HIV-1 IN CCD (Fig. S1B). Moreover, the affinity of PIR for the CTD-CCD fusion (KD ≈ 5 nM) closely correlated with the antiviral activity of the ALLINI, which displayed a 50% effective concentration (EC50) of approximately 12 nM to inhibit HIV-1 infection (see below) (17). Collectively, these results are concordant with the key involvement of the CTD in the formation of HIV-1 IN-ALLINI complexes (16, 34) and validate the ability of our engineered CTD-CCD protein to meaningfully interact with the small molecules.
Following extensive screening, we were able to cocrystallize the CTD-CCD fusion with two highly potent ALLINIs: BI-D (10, 15, 42, 43) and PIR (17) (Fig. 1A). The BI-D and PIR containing crystals diffracted X-rays to the resolution of 1.8 and 2.1 Å, respectively (Table S1). The asymmetric units contained a canonical CCD dimer with two associated CTDs, each of which sandwiched an ALLINI molecule (Fig. 1B; Fig. S1C and S2A). 22 and 15 residues of the interdomain linker region were disordered in the BI-D and PIR cocrystals, respectively, suggesting that the linker was free to adopt variable conformations throughout the crystal lattices. Therefore, it is unlikely that the artificial linkage affected the well-ordered portions of the protein, including the critical CTD-CCD interface. The four independently observed CTD-CCD interfaces in the crystal structures were highly similar (Fig. S2A). Moreover, the observed relative positioning of the CTDs and CCD dimers in our structures correspond to those in the 4.4 Å resolution cocrystal structure of full-length HIV-1 IN with ALLINI GSK1264 (PDB entry 5HOT) (Fig. S2B and C) (16), further validating our crystallography strategy. Pairwise alignments of the CCD dimers in our crystal structures resulted in rms deviations between CTD Cα atom positions of 2.2 ± 0.7 Å, whereas the same metric for the CTD β1 and β5 strands that are directly involved in the interface was 1.2 ± 0.5 Å (Fig. S2C). Thus, the ALLINI-induced interface allows for minor relative adjustments of the CTD and the CCD dimer.
Conservation of the ALLINI-induced CTD-CCD interface in the crystal structures. (A) Superposition of the two crystal structures determined in this study. Protein backbones are shown as cartoons, with CTDs in yellow and CCD dimer chains in cyan and green. The ALLINI molecules (indicated with arrowheads) are shown as sticks, with carbon, nitrogen, and oxygen atoms in pink, blue, and red, respectively. (B) Left panel: a linear polymer of HIV-1 IN dimers observed in a cocrystal structure with GSK1264 (PDB entry 5HOT) is stabilized by ALLINI-induced association of the CCD dimers with CTDs provided by neighboring dimers. An example of this substructure is boxed. Right panel: the CCD dimer with its associated CTD chains is shown in the same orientation as the crystal structures in panel A. (C) Left panel: rhe four independent CTD-ALLINI-CCD interfaces that were observed in the crystal structures that are reported here as well as the related substructure from 5HOT were superposed, based on the CCD dimers. The CTDs from cocrystal structures with BI-D, PIR, and GSK1264 are shown as ribbons and are colored orange, yellow, and brown, respectively. Right panel: the superposition of the CTDs from the cocrystal structures with BI-D (orange) and PIR (yellow) with a prior drug-free HIV-1 IN CTD structure (PDB entry 6T6E). Individual β strands of the five-bladed SH3 fold are indicated (β1 to β5). Note that the CTD structure does not undergo substantial conformation changes as a result of ALLINI binding. Download FIG S2, PDF file, 0.6 MB (626.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallography statistics and free energy perturbation data. Download Table S1, XLSX file, 0.02 MB (21.3KB, xlsx) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ALLINI-induced CTD-CCD interface.
ALLINIs share three chemical features that are attached to an aromatic scaffold: a bulky hydrophobic group (chromanyl and chlorophenyl in BI-D and PIR, respectively), a tert-butoxyl side chain, and an invariant carboxylate (shown in blue, green, and red, respectively, in Fig. 1A). In agreement with prior observations (8, 10, 11, 15, 17), the compounds insert into the principal ALLINI binding pocket at the CCD dimer interface. Within it, the carboxylate group forms a bidentate hydrogen bond with the main chain amides of Glu170 and His171, whereas the tert-butoxyl and the bulky aromatic groups make hydrophobic interactions with side chains of both CCD dimer subunits (Gln95, Tyr99, Leu102, Tr125, Trp132, Thr174, and Met178) (Fig. 2 and 3).
FIG 2.
Details of the IN-ALLINI interface. The CTD-CCD interface observed in cocrystals of BI-D (upper) and PIR (lower) IN complexes. The left panels show stereo wall-eyed views of the interfaces, and the right panels show the key CTD residues involved in the stacking interactions with the ALLINIs. Selected amino acid residues, along with the inhibitor molecules, are labeled and shown as sticks. Black dashes on the left panels indicate hydrogen bonds. In the right panels, the aromatic and lysine residue side chains that make delocalized interactions with inhibitor molecules are shown.
FIG 3.
ALLINI-induced CTD-CCD contacts outside the immediate vicinity of the bound inhibitors. The last residues that originate from the HIV-1 IN C-terminal tail region in the refined models are indicated with arrowheads (Asp272 and Asp279 in the BI-D and PIR complexes, respectively). Other labels and colorings are the same as in Figure 2.
In addition to these well-characterized ALLINI-CCD interactions, our crystal structures now reveal details of the ALLINI-induced CTD-CCD interface (Fig. 2 and 3). The aromatic scaffolds of the compounds (quinoline in BI-D and pyrrolopyridine in PIR [shown in black in Fig. 1A]) protrude sufficiently from the CCD dimer to make T-shaped π-π stacking interactions with the side chains of the invariant CTD residues Tyr226 and Trp235. Remarkably, this configuration allows the side chain of the invariant CTD residue Lys266 to pack along the corner formed by the aromatic planes of the inhibitor scaffold and Trp235. This orients Lys266 for ionic interactions with the side chain of CCD Glu170 and carboxylate groups of the inhibitors. Additionally, the methylpyrazole side chain in PIR forms a parallel π-π stacking interaction with Trp235. Within the buried CCD dimer interface, the chromanyl of BI-D and the chlorophenyl of PIR engage in hydrophobic interactions with the side chain of CTD Ile268. This residue, although not invariant among circulating HIV-1 strains, is nevertheless highly conserved, with the occasional occurrence of Leu at this position in a minority of HIV-1 isolates (46). Thus, the compounds harness four invariant (Glu170, Tyr226, Lys266, and Trp235) and one highly conserved (Ile268) HIV-1 IN residues to facilitate the CTD-CCD interaction (Fig. 4).
FIG 4.
Conservation of HIV-1 IN amino acid residues involved in the ALLINI-induced CTD-CCD interface. Open-book view of the interface with the CTD (upper) and the CCD dimer (lower) shown in surface representations and colored according to amino acid conservation, as shown on the scale: from dark blue (invariant and most conserved) to red (most variable). Selected residues are indicated, and PIR is shown as sticks. The conservation scores were calculated using the ConSurf server (https://consurf.tau.ac.il/) (98) from an alignment of IN amino acid sequences of representative HIV-1 strains, including the subtypes M, N, and O as well as recombinant viruses and simian immunodeficiency viruses from chimpanzees (46).
The extent of the ALLINI-induced CTD-CCD interface occludes a total of approximately 1,200 Å2 of molecular surface, and it is stabilized by interactions of highly conserved or invariant HIV-1 IN residues outside the direct vicinity of the ALLINIs (Fig. 2 to 4). Overall, the CTD-CCD contacts are devoid of water molecules and are primarily mediated by hydrophobic and π stacking interactions. This includes the side chain of CTD residue Arg224, which forms a cation-π stacking interaction with the invariant CCD residue Trp131 (Fig. 2 and 3). In addition, the invariant CTD residue Tyr271, which is located at the beginning of the C-terminal tail immediately abutting the CTD, makes hydrophobic interactions with CCD residues Val165, Ile182 (both highly conserved), and Trp132 (invariant). The superposition of the CTD chains in our refined models with a crystal structure of the isolated CTD (47) revealed a rms deviation of Cα atom positions of only 0.4 Å (Fig. S2C). Therefore, the formation of the ALLINI-induced interface does not require substantial conformational adaptation of the IN CTD.
Energetics and dynamics of the ALLINI-induced CTD-CCD interface.
We next applied computational chemistry to quantitatively assess the interactions observed in the cocrystal structures. To this end, we extended the experimental structures by modeling polypeptide loops that were disordered in the crystals (Fig. S3A), placed them in an appropriate box of solvent (Fig. S3B–D), and conducted 1 μs all-atom molecular dynamics (MD) simulations in the absence and presence of BI-D or PIR. Initially, we computed heavy atom rms deviations for the CCD and CTD chains with respect to the first frame of the corresponding simulation (Fig. S4A to C). When the MD was performed in the absence of ALLINIs, the CCD and CTD chains diverged from the initial states by 3.23 ± 0.38 Å and 6.62 ± 1.16 Å, respectively (Fig. S4A). As expected, the ALLINIs strongly stabilized the interface, considerably diminishing the divergence of both domains during the simulations. In particular, the rms deviations of the CTD in the presence of BI-D or PIR reduced to 4.39 ± 0.32 or 3.03 ± 0.30 Å, respectively (Fig. S4B and C). Snapshots of the simulation in the absence of the ALLINIs revealed that each CTD chain underwent partial unfolding and drifted from its initial position (Fig. S4A). In contrast, in the presence of BI-D or PIR, the CTD remained stably engaged with the CCD dimer throughout the MD simulations (Fig. S4B and C). The examination of the rms fluctuations per protein residue further confirmed that the presence of either compound greatly rigidified the CTD-CCD interface (Fig. S4D). To identify the events that were responsible for the comparatively dramatic structural fluctuations observed in the absence of ALLINIs, we performed rms deviation matrix analyses between pairwise structures across the MD trajectory. Figure S4E demonstrates the extents to which the CTDs of the apo form of our construct transitioned through various conformational meta states, including its dislodgement from the CCD dimer.
MD setup. (A) Modeled loops. BI-D (left) and PIR (right) cocrystal structures completed by modeling the missing polypeptide loops. For clarity, ALLINI molecules are not shown. The protein mainchains are shown as cartoons, with the CCDs in orange, CTDs in red, and modeled loops in blue. (B–D) MD simulation domains. Simulation domains for the systems without ALLINI (B), with BI-D (C), or with PIR (D). Protein chains are shown as cartoons, with CCD in orange and CTD in red. Inhibitors are shown in space-filling representations. The purple, green, and cyan spheres represent sodium, chloride, and Mg2+ ions, respectively. The grey boxes are solvent water. (E) An example of the thermodynamic cycle of BI-D binding. The horizontal legs represent the alchemical transformation of an amino acid residue (Lys266) into Ala either in the bound state (complex, upper panel) or in the unbound state (free, bottom panel). The vertical legs represent binding. Download FIG S3, PDF file, 0.3 MB (339.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MD simulation details. (A–C) The rms deviation time traces of all of the protein heavy atoms are shown for the CCDs (green) and CTDs (yellow) throughout the MD simulations that were conducted either in the absence of ALLINIs (A) or in the presence of BI-D (B) or PIR (C). Corresponding snapshots of the protein backbone and bound compounds are shown for each system, with the CCD and the CTD in green and yellow, respectively. BI-D and PIR are presented in space-filling representations. (D) The rms fluctuation per residue trace in the absence of ALLINI (red), in the presence of BI-D (blue), or in the presence of PIR (purple). (E) All-to-all rms deviation matrix analysis. Shown is the rms deviation matrix (81) between the pairwise structures in the MD trajectory of the protein in the absence of ALLINIs. Representative snapshots are shown to the right (I–VI, indicated with arrowheads on the matrix plot) of the protein, highlighting various transitions observed along the MD trajectory. (F) Dynamics of ALLINIs binding to IN. Snapshots are shown from the MD simulations of the CTD-CCD interface in the presence of BI-D (upper) or PIR (lower) at 0, 250, 500, 750, and 1,000 ns of the MD simulations. The ALLINIs and selected side chains are shown in ball-and-stick mode in separate colors and are labelled on the left diagrams that correspond to the initial states (0 ns). Download FIG S4, PDF file, 0.4 MB (466.9KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Importantly, the results of our MD simulations are in accord with the experimental evidence that ALLINIs stabilize the HIV-1 IN CTD-CCD interface. To quantify the contributions of individual amino acid residues to the ALLINI-induced CTD-CCD interface, we next performed free energy perturbation (FEP) calculations. IN residues that make direct contacts with ALLINIs were alchemically transformed to alanine, and the final free energy differences (ΔΔG) were computed using the thermodynamic cycle (Fig. S3E). Additionally, we computed ΔΔG values for selected IN amino acid residues (Arg224, Trp131, and Lys264) which, while not directly interacting with the ALLINIs, were located within the specified force field cutoff for nonbonded interactions (12 Å). The resulting ΔΔG values are reported in Fig. 5 and Table S1. A positive ΔΔG value indicates that the side chain of the truncated residue has a stabilizing role within the IN-ALLINI interface. Throughout the MD simulations in the presence of BI-D or PIR, Lys266 interacted with the aromatic ALLINI scaffold via cation-π stacking and formed a salt bridge to the inhibitor carboxylate group (Fig. S4F). FEP analyses revealed that the Lys266 side chain had a strong contribution to ALLINI binding, with corresponding ΔΔG values of 3.72 ± 0.54 and 3.63 ± 0.90 kcal/mol for the BI-D and PIR-induced interfaces, respectively. Likewise, the MD simulations confirmed that both BI-D and PIR formed stable T-shaped π-π stacking interactions with the side chain of Tyr226 (Fig. S4F), which accounted for 2.22 ± 1.35 kcal/mol and 3.58 ± 1.34 kcal/mol of binding energy, respectively. Throughout the simulations, the side chain of Ile268 was oriented to form hydrophobic interactions with the chromanyl and chlorophenyl groups of BI-D and PIR, contributing ΔΔG of 1.69 ± 0.64 kcal/mol and 3.35 ± 0.93 kcal/mol, respectively (Fig. 5). Our computations also suggested that the energetic contributions of some of the IN residues may depend on the specific ALLINI. Thus, the side chain of Trp235 engaged in stable T-shaped π-π stacking with BI-D (Fig. S4F), contributing as much as 3.43 ± 1.1 kcal/mol of binding energy. In contrast, the stacking of Trp235 with PIR was less extensive (Fig. S4F), and the substitution of Trp235 with Ala was associated with a ΔΔG of 0.49 ± 0.87 kcal/mol. Likewise, the side chain of the CCD residue Glu170 contributed more binding energy to BI-D (3.29 ± 1.06 kcal/mol) than to PIR (−0.28 ± 0.70 kcal/mol) due to stability of the salt bridge to Lys266 during the MD simulations. Moreover, according to the FEP analysis, we observed that the CCD residues Gln168, Thr174, and Glu170 were favorable for the recognition of BI-D, whereas the CCD residues Met178, Trp132, and Thr124 were favorable for the recognition of PIR (Fig. 5; Table S1). These subtle differences between individual compounds notwithstanding, computational chemistry confirmed the importance of the conserved HIV-1 IN CTD residues Lys266, Trp235, Tyr226, and Ile268 for the interactions with the ALLINIs.
FIG 5.
Energetics of ALLINI binding to HIV-1 IN. The calculated relative free energy differences (ΔΔG) of Ala substitutions within the CTD-CCD interface induced by BI-D (upper) or PIR (lower). The error bars represent the standard deviations of the mean values from four independent simulations (Table S1).
Site directed mutagenesis of the residues at the CTD-CCD interface.
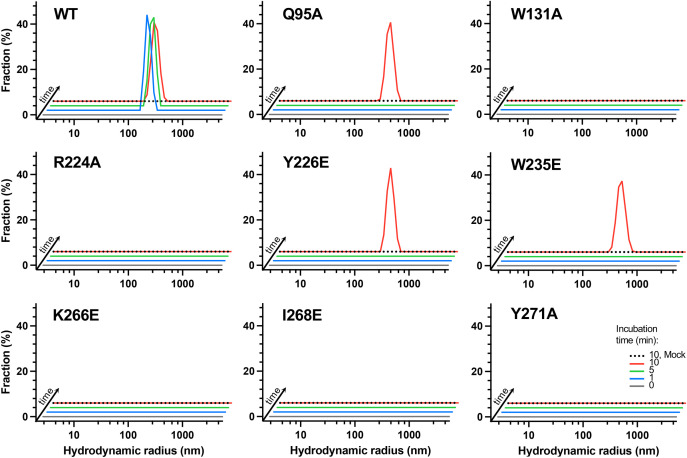
ALLINIs are known to cause the rapid aggregation of HIV-1 IN (10, 15, 16, 26, 41). Concordantly, the incubation of 4 μM recombinant wild type IN with near stoichiometric amounts (2.5 or 5 μM) of PIR resulted in the near quantitative precipitation of the protein (Fig. S5). Furthermore, dynamic light scattering (DLS) revealed the formation of protein aggregates within minutes of supplementing 12.5 nM HIV-1 IN with 62.5 nM PIR (Fig. 6). To probe the contributions of individual residues positioned at the ALLINI-induced CTD-CCD interface to the inhibitor activity, we prepared full-length HIV-1 IN proteins with the following amino acid substitutions: Q95A, W131A, R224A, Y226A, W235A, K266A, I268A, and Y171A. Because some of these proteins displayed aberrant aggregation (for example, the Y226A and K266A INs precipitated in the absence of an ALLINI [Fig. S5]), we additionally produced Y226E, W235E, K266E, and I268E IN proteins.
FIG 6.
Amino acid substitutions within the ALLINI-induced CTD-CCD interface reduce IN aggregation by PIR. Full-length wild type and mutant IN proteins were incubated in the absence (black dotted lines) or presence of PIR for 0, 1, 5, or 10 min (gray, blue, green, and red lines, respectively). The plots show the distribution of the observed hydrodynamic radii (nm) that were determined via dynamic light scattering.
Effects of amino acid substitutions within the CTD-CCD interface on PIR-induced IN aggregation. Full-length wild type and mutant HIV-1 IN proteins (indicated, right) were incubated in the absence or presence of 2.5 or 5 μM PIR. Centrifugally separated precipitated (P) and soluble (S) fractions were analyzed via SDS polyacrylamide electrophoresis, and IN was detected by staining with Coomassie brilliant blue. The migration positions of the protein molecular mass standards are indicated to the left (kDa). Download FIG S5, PDF file, 0.3 MB (330.7KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DLS and analytical size exclusion chromatography analyses confirmed that the Q95A, W131A, R224A, Y226E, W235E, K266E, I268E, and Y271A INs behaved as soluble oligomeric species, although relative amounts of tetramers, dimers, and monomers varied among the variants (Fig. 6; Fig. S5, S6). As revealed by DLS, and in agreement with the cocrystal structures, the mutations suppressed HIV-1 IN aggregation in the presence of PIR (Fig. 6). Although DLS provides a time course for ALLINI-induced IN aggregation, this technique does not delineate what fraction of the initial, soluble protein is converted into higher order aggregates. Therefore, we additionally tested our panel of HIV-1 IN mutants in the ALLINI-induced precipitation assay to monitor both the soluble and the aggregated fractions. Substitutions of IN residues that play key roles in the interface with the inhibitors, namely, Y226E, W235E, K266A, K266E, I268A, and I268E, conferred resistance to precipitation in the presence of PIR (Fig. S5; note that the compound solubilized K266A IN). In contrast, the substitution of Gln95, which plays a relatively minor role at the periphery of the ALLINI-induced CTD-CCD interface (Fig. 2; Table S1), did not reduce ALLINI-mediated precipitation (Fig. S5). The majority of the remaining mutants displayed intermediate phenotypes. Thus, whereas W131A IN was less susceptible to precipitation in the presence of 2.5 μM PIR and was effectively precipitated by 5 μM PIR, the R224A and Y271A INs were comparatively more resistant to ALLINI-induced precipitation (Fig. S5).
Analytical size exclusion chromatography of HIV-1 IN variants. Elution chromatograms (A280) of full-length wild type or mutant recombinant HIV-1 IN (as indicated) from a Superdex-200 10/30 column. The expected elution volumes of the tetramer (T), dimer (D), and/or monomer (M) forms of the protein are indicated above each chromatogram. Download FIG S6, PDF file, 0.1 MB (66.9KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also tested our panel of mutants for their effects on viral infectivity. The key CTD residues involved in the ALLINI-induced interface with the CCD are either invariant or highly conserved (Fig. 4). Unsurprisingly, the majority of mutations within the interface abrogated HIV-1 infectivity (Fig. S7). In particular, the IN mutations Y226E, W235E, K266E, I268E, and Y271A, each of which substantially compromised the ability of PIR to induce IN precipitation (Fig. S5), resulted in a loss of infectivity. These results are in concordance with earlier observations that mutations of conserved residues within HIV-1 IN CTD can cause severe replication defects (30, 48). Of the viruses we tested, only those with Q95A, W131A, and R224A mutations retained infectivity, with W131A showing partial (approximately fourfold) resistance to PIR (Fig. S7).
Effects of amino acid substitutions within the ALLINI-induced CTD-CCD interface on HIV-1 infectivity and PIR antiviral activity. Left panel: relative infectivity of HIV-1 variants carrying the indicated mutations within IN. The infectivity of wild type HIV-1NL4-3 was considered to be 100%. The bars indicate the mean values. The error bars are the standard deviations from n = 3 biological replicates for each condition. The open circles correspond to individual measurements. Right panel: PIR 50% effective inhibitory concentrations (EC50) on the wild type and HIV-1 IN mutants. Download FIG S7, PDF file, 0.02 MB (19.9KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
ALLINIs fill a small and relatively rigid pocket at the HIV-1 IN CCD dimer interface (8, 9, 11, 14–17) and act as molecular glue to paste a CTD from one IN protomer to the CCD dimer of another. The compounds bind the CCD dimer using common chemical functionalities (Fig. 1A), elements of which mimic the key amino acid residues at the tip of the LEDGF/p75 integrase-binding domain (Asp366 and Ile365) (8, 19). However, the apparent lack of similarity outside their CCD-binding function may appear inconsistent with the mode of action of these compounds. Herein, we asked how these diverse molecules bind the same face of the CTD. Our cocrystal structures with two potent ALLINIs and accompanying computational chemistry reveal that a planar aromatic scaffold is the key feature that allows these small molecules to recruit the CTD. The scaffold protrudes from the CCD pocket sufficiently to nucleate the CTD-CCD interface by engaging a triad of invariant HIV-1 IN CTD residues in a network of π-π stacking, cation-π, and hydrophobic interactions. Remarkably, while the CTD does not undergo substantial conformational changes to achieve a geometric fit to the CCD dimer (Fig. S2C), the CTD-CCD interface has not been observed within characterized functional IN complexes. Hence, this likely entirely unnatural interface can be considered to be a viral Achilles heel that is templated and leveraged by ALLINIs.
The surface of the CTD recruited by the ALLINIs shows remarkable conservation among circulating HIV-1 strains (Fig. 4). Likely due to variations within the IN CCD regions, ALLINIs specifically inhibit HIV-1 lineage viruses without any evidence for affecting closely related lentiviruses, including HIV-2 (9). Yet, it is noteworthy that the triad of the CTD residues recruited by the ALLINI aromatic scaffolds is highly conserved across diverse lentiviral IN proteins (Fig. S8A). The remarkable conservation is explained by the multiple functions played by the IN CTD in the viral life cycle and during integration, in particular. Lentiviral intasomes are thought to contain as many as 12 to 16 IN subunits (6, 39, 40, 49), and the CTDs belonging to different IN chains play diverse roles, depending on their locations within the complex. For example, the CTD residues involved in binding ALLINIs pack against the extended IN NTD-CCD linker in one type of IN chain and interact with the alpha-helical IN CCD-CTD linker region in another (Fig. S8B). Although both types of CTDs interact with viral DNA, they approach it in different ways (Fig. S8B). Therefore, mutations within this patch of the CTD surface would necessitate simultaneous adaptive changes in several positions of the IN amino acid sequence. Moreover, this compact domain has been implicated in multiple HIV-1 IN functions outside the process of integration, which may additionally limit the mutational rates at some of its key positions (50–53). Amino acid substitutions within the HIV-1 IN CTD can cause class I or class II lethal phenotypes, and the replication-defective HIV-1 K266E and W235E mutant viruses (Fig. S7) were previously characterized as class II and class I, respectively (30, 48).
Variable roles of IN CTDs in lentiviral integration. (A) Residues corresponding to HIV-1 IN Tyr226, Trp235, and Lys266 are highly conserved among lentiviral INs. The IN CTD regions from HIV-1, HIV-2, maedi-visna virus (MVV), equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV), and bovine immunodeficiency virus (BIV) are aligned. Red and yellow boxes highlight the amino acid residues in positions that are invariant across the alignment or are conserved through a similarity of physicochemical properties, respectively. The sequence numbering and secondary structure elements (extracted from PDB entry 6T6E) that are shown above the alignment correspond to HIV-1 IN. The key HIV-1 IN residues involved in ALLINI binding are indicated with red arrowheads (Tyr226, Trp235, and Lys266). The figure was prepared using the ESPript server (https://espript.ibcp.fr) (67). (B) The IN CTD residues involved in the ALLINI-induced interface play multiple roles within the lentiviral intasome assembly. (Top panel) HIV-1 intasome core assembly (PDB entry 6PUY [6]), with protein and viral DNA chains shown as cartoons. Catalytic IN chains are colored green and cyan. The remaining IN chains are colored according to their functions within the twofold symmetric assembly. Individual IN domains (NTD, CCD, and CTD) that belong to different IN chains and are resolved in the structure are indicated. (Lower panels) Detailed view of the CTDs belonging to IN chains that are colored orange and magenta in the top panel. DNA and selected amino acid side chains are shown as sticks, with the amino acid residues labelled. Also indicated are the alpha helical CCD-CTD and an extended NTD-CCD linker of the catalytic IN subunits. Note that the triad of IN CTD residues (Tyr226, Trp235, and Lys266) have different functions within these two types of intasomal subunits. Download FIG S8, PDF file, 0.5 MB (479.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The strict conservation of the HIV-1 IN CTD residues involved in interactions with ALLINIs explains why the majority of the mutations associated with resistance to this class of small molecules map in and around the binding pocket on the CCD dimer, which is considerably more variable (Fig. 4) (9, 13, 14, 16, 17, 54, 55). In most instances where resistance has occurred, it has arisen from either the exclusion of the inhibitor from its principal binding pocket at the CCD dimer interface or the loss of interaction within the pocket (54). However, at least two mutations resulting in partial resistance to ALLINIs, namely, W131C and N222K (14, 16), could not be explained by the previous cocrystal structures that lacked the CTD. Indeed, neither Trp131 nor Asn222 make direct contact with ALLINIs (Fig. 2 and 3). In our structures, the CCD residue Trp131 is involved in hydrophobic and cation-π stacking interactions with CTD residue Arg224 and makes Van der Waals contact with three additional CTD residues (Asn222, Ile268, and Asp270) (Fig. 2 and 3). The extents of these interactions would be considerably reduced upon the substitution of the bulky aromatic Trp sidechain for a Cys sulfhydryl. Although our FEP analyses suggested that the contribution of the Trp131 side chain to the CTD-CCD interface may be ALLINI-dependent (Fig. 5), we observed the partial resistance of the recombinant W131A IN to aggregation in the presence of PIR (Fig. 6; Fig. S5). Concordantly, the W131A IN virus displayed reduced sensitivity to the compound (Fig. S7). Although pinpointing the mechanism of resistance associated with N222K IN will require additional work, given the Van der Waals interaction of Asn222 with Trp131, we speculate that this amino acid substitution reduces the geometric fit between the domains and thereby compromises the ability of the drug to induce the CTD-CCD interface.
Our structures reveal atomistic details of what is likely the complete HIV-1 IN-ALLINI interface, and, as such, they will be invaluable for the further development of this promising class of antiretrovirals. Most importantly, the structures will allow for the optimization of the aromatic scaffold for improved interactions with the key HIV-1 IN CTD residues uncovered here (Tyr226, Trp235, and Lys266). By improving the ALLINI-CTD interactions, it might be possible to gain activity against HIV-1 variants that are resistant to current ALLINIs due to polymorphisms or mutations within the CCD binding pocket.
MATERIALS AND METHODS
Recombinant proteins.
The protein construct used for the X-ray crystallography was based on HIV-1 isolate NL4-3 and contained the IN residues 220 to 288 (CTD with the amino acid substitution W243E, including the complete C-terminal tail), which were directly followed by residues 50 to 212 (CCD, with the solubilizing mutation F185K). In this inverted design, the unstructured HIV-1 IN C-terminal tail region (residues 270 to 288), together with a portion of the NTD-CCD linker (residues 50 to 55), form a flexible linker to join the globular IN domains. The amino acid sequence of the composite linker is YGKQMAGDDCVASRQDEDMHGQVD (the underlined residues are those derived from the C-terminal tail). Isolated CTD (wild type and mutants) were used in MALLS experiments (Fig. S1A) and spanned HIV-1 IN residues 220 to 270 (corresponding to the CTD and lacking the C-terminal tail). Both proteins were produced in bacteria with N-terminal hexa-histidine (His6)-Sumo tags. Escherichia coli BL21-CodonPlus (DE3) cells (Agilent) that were transformed with the desired recombinant expression plasmid were grown in Luria broth in shaker flasks at 30°C. Protein expression was induced via the addition of 0.01% (wt/vol) isopropyl β-D-1-thiogalactopyranoside (IPTG) to log phase cultures for 4 h. To isolate the recombinant proteins, the cells were lysed via sonication in core buffer (0.5 M NaCl and 20 mM Tris-HCl, pH 7.5) supplemented with 1 mM phenylmethylsulfonyl fluoride and cOmplete EDTA-free protease inhibitor cocktail (Roche). The supernatant, precleared by centrifugation, was incubated with Ni-NTA agarose (Qiagen) at 4°C in the presence of 15 mM imidazole for 1 h. The resin was extensively washed with core buffer supplemented with 15 mM imidazole, and the recombinant protein was eluted with 200 mM imidazole in core buffer. The eluted protein, supplemented with 2 mM dithiothreitol (DTT), was incubated with Ulp1 sumo protease (using 0.01 mg of protease per mg of recombinant protein) at 4°C for 16 h to release the His6-Sumo tag. The next day, the cleaved protein was diluted to adjust the NaCl concentration to 150 mM, injected into a 5-mL HiTrap SP HP column (Cytiva), and eluted with a linear 0.15 to 1.0 M NaCl gradient in 20 mM Tris-HCl, pH 7.5. The peak fractions were separated via size exclusion chromatography (SEC) through a HiLoad 16/600 Superdex 200 pg column (Cytiva) that was equilibrated in core buffer. For the SPR experiments, the removal of the His6-Sumo tag was omitted.
Full-length wild type and mutant HIV-1 IN proteins were expressed in E. coli BL21(DE3) cells and purified via sequential chromatography on nickel and heparin affinity columns as described (56, 57). Isolated CCD constructs spanning HIV-1 IN residues 50 to 212 with the solubilizing mutation F185H and an N-terminal His6 tag were produced in E. coli as described (57). The protein was purified via chromatography on a nickel affinity column, and this was followed by SEC using a HiLoad 16/600 Superdex column (Cytiva), with the elution buffer containing 0.5 M NaCl, 20 mM HEPES (pH 7.5), 10% glycerol, 0.5 mM EDTA, and 2 mM DTT.
SEC-MALLS.
Size exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS) was used to determine the molar mass distribution of the wild type and mutant HIV-1 IN CTDs. Samples (100 μL) were applied to a Superdex-75 Increase 10/300 column (Cytiva) that was equilibrated in 0.5 M NaCl, 25 mM Tris-HCl (pH 7.5), 3 mM NaN3 and 0.5 mM TCEP and was mounted on a JASCO-4000 semimicro HPLC system. Chromatography was performed at 25°C, at a flow rate of 1 mL/min. The scattered light intensities and protein concentrations of the eluted peaks were recorded using a DAWN-HELEOS II laser photometer and an OPTILAB-TrEX differential refractometer (Wyatt Technology), respectively. The weight-averaged molar mass of the material contained in the chromatographic peaks was determined using the combined data from both detectors and dn/dc = 0.186 mL/g in the ASTRA software version 7.3.2 (Wyatt Technology).
ALLINIs.
BI-D was obtained from MedChemExpress (product number HY-18601). PIR was synthesized as previously described (58) and verified via mass spectrometry (Mr = 495.1 Da).
Surface plasmon resonance.
SPR studies were performed using a four-channel Reichert4 SPR instrument (AMETEK, Reichert Technologies). His6-tagged CCD (F185H) and (His6)-Sumo-CTD-CCD (F185K/W243E) were captured onto an NTA chip to about 4,500 response units, using HBS-P+ buffer containing 10 mM HEPES, 150 mM NaCl, and 0.05% (vol/vol) surfactant P20 (Cytiva) supplemented with 5% dimethyl sulfoxide (DMSO). A method development capture assay was executed to establish the best association and dissociation times for both the ligand and the analyte. After the determination of the parameters, experiments were carried out in triplicate for each ligand. The data were analyzed using the Reichert4SPR and Tracedrawer software packages to calculate the kon, koff, and KD values via a 1:1 binding model.
Analytical SEC.
The self-association of recombinant wild type and mutant INs was analyzed on a Superdex 200 10/300 GL column (GE Healthcare). The column was equilibrated in a running buffer that contained 20 mM HEPES (pH 7.5), 1 M NaCl, 10% (vol/vol) glycerol, 7.5 mM 3-([3-cholamidopropyl] dimethylammonio)-1-propanesulfonate, and 5 mM β-mercaptoethanol at a flow rate of 0.45 mL/min. The proteins were diluted to 20 μM with the running buffer and incubated for 1 h at 4°C. This was followed by centrifugation at 10,000 × g for 20 min. The retention volumes for the IN tetramers, dimers, and monomers were approximately 12.3 mL, 14.0 mL, and 15.0 mL, respectively.
Protein crystallization.
Stock solutions of BI-D and PIR were prepared for the cocrystallization experiments in DMSO at concentrations of 50 mM and 60 mM, respectively. To prepare the IN-ALLINI complexes, W243E/F185K CTD-CCD (0.6 mg/mL in core buffer) was supplemented with 25 μM ALLINI (diluted directly from the stocks in DMSO) in the absence (BI-D) or presence (PIR) of 5% (vol/vol) glycerol. Following incubation for 10 min on ice, the IN-ALLINI complexes were concentrated to 5 mg/mL using a 10-kDa cutoff VivaSpin device (Sigma-Aldrich). Crystals were grown at 18°C in hanging drops that were formed by combining 1 μL of protein with 1 μL of a reservoir solution, which contained either 10% polyethylene glycol (PEG) 4,000, 17.5% glycerol, 30 mM MgCl2, 30 mM CaCl2, and 0.1 M Tris-Bicine (pH 8.5) (BI-D cocrystals) or 10% (wt/vol) PEG 8,000, 20% ethylene glycol, 30 mM MgCl2, 30 mM CaCl2, and 0.1 M imidazole-MES (pH 6.5) (PIR cocrystals). The crystals, which were cryoprotected in mother liquor supplemented with 30% glycerol, were flash-frozen via plunging into liquid nitrogen.
X-ray data collection and structure refinement.
The diffraction data were collected at beamline I04 of the Diamond Light Source (Oxford, UK) at 100 K, using a wavelength of 0.9795 Å, a transmission of 100%, and a 43 × 30 μm beam with either 0.1-s exposure and 0.2° rotation per image (BI-D cocrystals) or 0.3-s exposure and 0.5° rotation per image (PIR cocrystals). The single-crystal diffraction data were integrated, scaled, and merged with DIALS (59) within the Xia2 automatic data processing pipeline (60). The structures of the HIV-1 IN CTD (PDB entry 6T6E [47]) and CCD (PDB entries 4O55 [11] and 2B4J [19]) were used as the search models for molecular replacement in PHASER (61) within the Phenix software suite (62). The resulting models were adjusted with multiple rounds of interactive fitting in Coot (63) and refinement using phenix.refine (version 1.20.1-4487-000) (62, 64). Protein chains were initially extended where possible, and then water, PEG, glycerol, and/or ethylene glycol molecules were added, as is visualized in the electron density maps. ALLINI molecules were then fitted into prominent positive difference densities. The ligand geometry definition files were generated using eLBOW (65). The final refinements included 5 translation/libration/screw (TLS) anisotropic B factor groups per protein chain, guided by the TLSMD server (66). The refined models fit well into the electron density and had good geometry, as assessed by the MolProbity server (http://molprobity.biochem.duke.edu) (66) (Table S1; Fig. S1C). The crystal structure illustrations were prepared using PyMol software (https://pymol.org), and the IN amino acid sequence alignment (Fig. S8A) was formatted using ESPript (https://espript.ibcp.fr) (67).
Parameterization of ALLINI molecules.
The calculation of the atomic interaction potentials for molecular dynamics requires empirical parameters for every bond, angle, dihedral, and Van der Waals interaction. We obtained the initial parameters for BI-D from the CHARMM General Force Field (CGenFF) (68). Further, initial parameters with an assigned penalty higher than 10 (Fig. S9A) were optimized using the VMD Force Field Toolkit (ffTK) (69) and Gaussian-16 (70). For this process, to reduce the computational cost of the calculations, a divide and conquer approach was applied. First the BI-D molecule was fragmented into three moieties (Fig. S9C and D). Then, the fragments were chosen to span the dihedral angles to be parameterized and to facilitate the reassembly of the full molecule. The geometry of each fragment was first optimized using Gaussian at a MP2/6-31* level of theory under water solvent to ensure that no hydrogen bonds were generated between parts of the same moiety. Then, the partial charge of each water-accessible atom was optimized to reproduce its role as a hydrogen bond acceptor or donor, following the CHARMM conventions. A water molecule was placed 2 Å apart from the atom of interest with either a hydrogen atom (for the hydrogen bond acceptors) or the oxygen atom (for the hydrogen bond donors) of water oriented toward the atom. The distance and orientation of the water-molecule complex was optimized quantum mechanically in Gaussian at the b3lyp/6-31g(d,p) level of theory. These were then transferred to ffTK to fit the atomic partial charges. The bond and angle parameters were optimized in ffTK by comparing the molecular mechanics (MM) potential energy distribution (PES) with the quantum mechanics PES that was derived from the Hessian matrix calculation for each fragment calculated in Gaussian at the MP2/6-31G* theory level. Finally, the dihedral parameters were optimized by calculating the torsion angle scans (MP2/6-31G*) and fitting the MM PES to reproduce the maxima and minima. The resulting MM PES and QM PES from our parameterization (Fig. S9B) yielded a root mean square difference between the energy profiles of 1.44 kcal/mol. Once all of the high penalty parameters were optimized from the fragments, the optimized partial charges that were assigned to the original molecule by moving the charges of overlapping atoms to their closest nonoverlapping neighbors were used to conserve the total charge of the molecule. The force field parameters for PIR were obtained from CGenFF and used as is, since we observed that they reproduce the stability of this molecule in the binding pocket.
Parameterization scheme and molecular fragmentation of BI-D. (A) Dihedral angle parameters with CGenFF penalties above 10. (B) Parameterized molecular mechanics potential (solid blue line) fitted to the quantum chemistry calculated potential (dotted black line). The rms difference between both potentials was 1.44 kcal/mol. (C) 3D model of BI-D. (D) To perform dihedral scans in the parameterization scheme, the molecule was fragmented into three moieties, and these are shown as stick diagrams (left) and 3D models (right). Download FIG S9, PDF file, 0.2 MB (197.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Molecular dynamics setup.
We initiated all of the molecular dynamics (MD) and free energy perturbation (FEP) simulations of the HIV-1 IN structures with bound BI-D or PIR from the crystal structures that are presented in the manuscript. Additionally, we conducted MD simulations of an apo conformation of the HIV-1 IN, which was obtained by removing the BI-D molecules from the binding pockets. Before conducting the simulations, we modeled and added the following loop residues, which were missing from the crystal structure of the HIV-1 IN in the presence of BI-D: residues 140 to 148 and 189 to 192 (chain A), residues 141 to 148 and 189 to 192 (chain B), residues 230 to 231 (chain C), and residues 230 to 233 (chain D). The following missing loop residues were modeled and added to the crystal structure of the HIV-1 IN in the presence of PIR: residues 145 to 148 (chain A) and residues 141 to 147 (chain B). The modeling of the missing residues was performed using the Modeller software (Fig. S3A) (71). We retained two Mg2+ ions from the crystal structures. Hydrogen atoms were added to the HIV-1 IN, BI-D, and PIR inhibitors while retaining the (S) form chirality of the BI-D and PIR inhibitors, as the (S) form was determined to be more predominant and bioactive than was the (R) form for these compounds (17, 72). Further, we solvated each system in a periodic box of TIP3P water molecules and ionized them with Na+ and Cl− ions to a salt concentration of 150 mM (Fig. S3B and D) using VMD software (73). The final simulation domains contained approximately between 132,000 and 142,000 atoms, with an overall system size of approximately 116 Å × 109 Å × 120 Å.
Each simulation domain was equilibrated in the presence and absence of inhibitors using NAMD2.14 simulation software (74), according to the following procedure. In the first step, we applied restraints to the protein and inhibitors while the solvent molecules were energy minimized using the conjugate gradient scheme until the gradient converged to values below 10 kcal mol−1 Å−1. In the next step, the restraints on the protein and inhibitors were released, and the system was energy minimized using the conjugate gradient scheme until the gradient converged to values below 10 kcal mol−1 Å−1. Each minimization was followed by the tempering of the simulation domain from 50 K to 310 K in increments of 5 K over 0.5 ns. During the second tempering step, we applied positional restraints to the heavy backbone atoms of the HIV-1 IN and to the heavy atoms of the inhibitors with a force constant of 10 kcal mol−1 Å−1. Subsequently, we performed the equilibration simulations of each system while the positional restraints on the heavy backbone atoms of the HIV-1 IN and on the heavy atoms of the inhibitors were gradually released at a rate of 1 kcal mol−1 Å−1 per 0.5 ns from 10 kcal mol−1 Å−1 to 0 kcal mol−1 Å−1 over 5 ns. During the equilibration simulations, we maintained the temperature at 310 K with a coupling constant of 1 ps−1, using a Langevin thermostat. The pressure was maintained at 1 atm, with the period and decay parameters set to 100 ps and 50 ps, using a Nose-Hoover barostat.
After performing the equilibration simulations, we conducted production MD and FEP simulations in the NVT ensemble with the temperature set to 310 K and the coupling constant set to 1 ps−1. These were maintained using a Langevin thermostat. We used periodic boundary conditions in all of the MD and FEP simulations with a 2 fs timestep. The electrostatic interactions were calculated using the particle mesh Ewald method with the cutoff for short-range electrostatics interactions being set to 12 Å. The coordination number between two Mg2+ ions and protein atoms which were located within a cutoff of 5 Å from the Mg2+ ions was constrained in all of the simulations by using the coordNum function in the Colvar module (75) of NAMD. We conducted all of the simulations using the CHARMM36m force field for proteins (76), the Roux force field parameters for ions (77), and the TIP3P water model (78). We derived the force field parameters for the BI-D inhibitor by using quantum mechanical calculations in conjunction with the CGenFF2.5 software package (79). The force field parameters for PIR were determined solely using the CGenFF2.5 software package with the CGenFF force field version 4.5 (68, 80). For each system (without or with ALLINI), we generated a 1 μs MD simulation, which resulted in an overall 3 μs data set and saved frames every 10 ps. An all-to-all rms deviation matrix analysis was performed as described (81).
Free energy perturbation (FEP) protocol.
We determined the relative free energy differences (ΔΔG) associated with the mutations of selected amino acid residues that comprise the binding pockets of BI-D and PIR inhibitors via the in silico transformation of wild type residues into alanine using the FEP method (82) in conjunction with a thermodynamic cycle (Fig. S3E). FEP is an old but commonly used technique (83) to compute free energy differences, which has previously been successfully applied to determine the energetics of RNA/protein interactions (84, 85), protein/protein interactions (86, 87), ligand/protein interactions (88, 89), and ligand/DNA interactions (90), as well as to address other scientific problems (91). In this work, we conducted alchemical transformations that correspond to the horizontal arms in the thermodynamic cycle (Fig. S3E), both in the unbound HIV-1 IN (free state) and in the bound HIV-1 IN with BI-D or PIR molecules (complex state). The vertical arms in the thermodynamic cycle represent the binding of BI-D or PIR to IN (Fig. S3E). We calculated the relative free energy differences according to , in which and correspond to free energy changes along the horizontal arms of the thermodynamic cycle, and and correspond to the free energy changes along the vertical arms of the thermodynamic cycle and characterize the binding of the ALLINIs to the wild type HIV-1 IN and mutant HIV-1 IN (Fig. S3E).
We applied the dual-topology paradigm (92), in which a hybrid energy function (U) is utilized to represent a mixture of the initial and final states of one of the horizontal arms of the thermodynamic cycle. The initial and final states were connected by a coupling parameter (λ) that corresponds to a series of individual simulations of the intermediate states (93). The coupling parameter spans values between λ = 0 and λ = 1, which correspond to the physical initial and final states, respectively, and the intermediate values correspond to mixed unphysical states. Each window of the alchemical transformation was prefaced by 0.2 ns of equilibration of the simulation domain, and this was followed by 0.8 ns of data generation for the free energy calculations.
We used the bidirectional approach (94) in each alchemical transformation by propagating the simulations in both the forward and backward directions to improve the accuracy of the free energy calculations. We determined the free energy changes ( and ) with their associated statistical errors over the forward and backward simulations with 20 equally spaced λ windows by using the Bennet acceptance ratio estimator (95) which is implemented in the ParseFEP plugin (96) in VMD1.9.3. Each transformation was repeated in quadruplicate, and the resulting free energy changes were averaged to yield the final free energy difference. All of the free energy differences, along with the final free energy difference for each alchemical transformation, are reported in Table S1. We conducted all of the FEP simulations using NAMD3.0 simulation software (74).
We computed the heavy atom (C, N, O, and S) rms deviation for the CCD and CTD with respect to the initial frame of the corresponding simulation in each system to establish the effects of the ALLINIs on the protein domain structure. The calculated rms deviation values indicate changes in the CCD or CTD with respect to a reference state. We also computed the per-residue rms fluctuations based on the alpha carbons in the protein backbone to further investigate the flexibility of each amino acid and each protein domain.
HIV-1 IN aggregation assays.
For DLS, wild type and mutant IN proteins were diluted from 200 nM stocks to 12.5 nM in buffer containing 1 M NaCl, 2 mM MgCl2, 2 mM DTT, and 50 mM HEPES-NaOH (pH 7.5) that were prepared with Milli-Q grade water and filtered twice with a 0.2 μm filter. PIR (62.5 nM) was added, and particle size distributions for the wild type and for each protein were recorded over a course of 10 min at 1 min intervals using a Zetasizer Nano ZS90 instrument (Malvern Panalytical). Any signal below a 2 nm threshold was considered to be background.
To observe ALLINI-induced precipitation, 4 μM wild type or mutant IN was incubated with increasing concentrations of PIR in 20 μL of assay buffer that contained 1 M NaCl, 5 mM β-mercaptoethanol, and 20 mM HEPES-NaOH (pH 7.5) at 4°C overnight. Following centrifugation at 10,000 × g for 10 min, the supernatant was mixed with 100 μL of 1.25× Novex NuPAGE LDS sample buffer. The remaining pellet was washed 3 times with 40 μL of assay buffer before resuspension in 120 μL of Novex NuPAGE LDS sample buffer to match the overall volume of the corresponding supernatant fraction. The supernatant and pellet fractions were analyzed via SDS-PAGE, and the IN was visualized via staining with Coomassie blue.
Virus infectivity assay.
No cell lines from the list of known misidentified cell lines that is maintained by the International Cell Line Authentication Committee were used in this study. HEK293T (American Type Culture Collection) and HeLa TZM-bl (NIH AIDS Reference and Reagent Program) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) that was supplemented with 10% fetal bovine serum (Sigma-Aldrich), 100 U/mL penicillin, and 100 U/mL streptomycin (Thermo Fisher Scientific). The cell lines were maintained at 37°C in a humidified 5% CO2 atmosphere, and they were tested monthly for Mycoplasma contamination. For the virus infectivity assays, HEK293T cells were seeded at concentrations of 2 to 4 × 105 cells/mL in 6-well plates 1 d prior to transfection with 2 μg of replication-competent pNL4-3 (wild type or IN mutants), using the HilyMax transfection reagent (Dojindo Molecular Technologies) in a 1:3 ratio, following the manufacturer’s protocol. The medium was replaced with fresh medium at 12 to 16 h posttransfection and incubated at 37°C. 48 h posttransfection, viruses containing clarified supernatant were filtered through a 0.45 μm filter, and the level of p24 (CA) was quantified via Western blotting. The level of CA was similar in all viruses, so 50 μL of filtered viral supernatant from the HEK293T cells were used to infect the TZM-bl cells (seeded at 50,000 cells/well) and incubated at 37°C for 3 to 4 h. Then, the medium was removed and replaced with fresh medium. The cells were collected at 48 h postinfection, and the virus infectivity was measured via a luciferase assay (Promega).
To test the STP-0404 antiviral activity, full replication cycle experiments (wild type or IN mutants) were performed as described previously (11, 97). Briefly, viruses were prepared from HEK293T cells in the presence of various concentrations of STP-0404 or with DMSO alone as a control. Target HeLa TZM-bl cells were similarly preincubated with STP-0404 or with DMSO. The cells were then infected with viruses for 3 to 4 h at 37°C, the medium was replaced, and fresh inhibitors were added. 48 h postinfection, the cells were harvested, and the infectivity was measured via a luciferase assay. The STP-0404 EC50 values were calculated using Origin software (OriginLab). All of the virus infections were performed in the presence of 8 μg/mL Polybrene, and the values are expressed as the mean ± the standard deviation (SD).
Data availability.
The data that support this study are available from the corresponding authors upon reasonable request. The refined models and the associated X-ray diffraction data were deposited into the Protein Data Bank under accession codes 8A1P (BI-D cocrystal structure) and 8A1Q (PIR).
ACKNOWLEDGMENTS
We thank P. Walker and A. Purkiss for the computer support and the help with X-ray data collection arrangements as well as the staff of Diamond Light Source beamline I04 for the assistance during data collection. Synchrotron access was provided via block allocation group MX13775. This work was funded by the US National Institutes of Health grants P50AI150481 (P.C. and A.N.E.) and U54AI170791 (P.C., J.R.P., and A.N.E.), R01AI143649 and U54AI150472 (M.K.), R01AI141327 (B.K.), R37AI039394 (A.N.E.), and P20GM104316 (J.R.P.). The laboratories of P.C. and I.A.T. are supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2058 and CC2029), the UK Medical Research Council (CC2058 and CC2029), and the Wellcome Trust (CC2058 and CC2029). We acknowledge computational support through the Delaware Advanced Research Workforce and Innovation Network (DARWIN) as well as the Caviness cluster.
Kyungjin Kim is the chief executive officer of ST Pharm Co. Ltd. No other authors declare a potential conflict of interest.
Footnotes
This article is a direct contribution from Alan N. Engelman, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Henry Levin, NIH/NICHD, and Rafael Bernardi, Auburn University.
Contributor Information
Juan R. Perilla, Email: JPerilla@udel.edu.
Mamuka Kvaratskhelia, Email: Mamuka.Kvaratskhelia@cuanschutz.edu.
Peter Cherepanov, Email: Peter.Cherepanov@crick.ac.uk.
Kellie Jurado, University of Pennsylvania.
REFERENCES
- 1.Maertens GN, Engelman AN, Cherepanov P. 2022. Structure and function of retroviral integrase. Nat Rev Microbiol 20:20–34. doi: 10.1038/s41579-021-00586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci USA 99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith SJ, Zhao XZ, Burke TR, Jr, Hughes SH. 2018. Efficacies of Cabotegravir and Bictegravir against drug-resistant HIV-1 integrase mutants. Retrovirology 15:37. doi: 10.1186/s12977-018-0420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook NJ, Li W, Berta D, Badaoui M, Ballandras-Colas A, Nans A, Kotecha A, Rosta E, Engelman AN, Cherepanov P. 2020. Structural basis of second-generation HIV integrase inhibitor action and viral resistance. Science 367:806–810. doi: 10.1126/science.aay4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passos DO, Li M, Jóźwik IK, Zhao XZ, Santos-Martins D, Yang R, Smith SJ, Jeon Y, Forli S, Hughes SH, Burke TR, Craigie R, Lyumkis D. 2020. Structural basis for strand-transfer inhibitor binding to HIV intasomes. Science 367:810–814. doi: 10.1126/science.aay8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SJ, Zhao XZ, Passos DO, Pye VE, Cherepanov P, Lyumkis D, Burke TR, Jr, Hughes SH. 2021. HIV-1 integrase inhibitors with modifications that affect their potencies against drug resistant integrase mutants. ACS Infect Dis 7:1469–1482. doi: 10.1021/acsinfecdis.0c00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. 2010. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 9.Amadori C, van der Velden YU, Bonnard D, Orlov I, van Bel N, Le Rouzic E, Miralles L, Brias J, Chevreuil F, Spehner D, Chasset S, Ledoussal B, Mayr L, Moreau F, Garcia F, Gatell J, Zamborlini A, Emiliani S, Ruff M, Klaholz BP, Moog C, Berkhout B, Plana M, Benarous R. 2017. The HIV-1 integrase-LEDGF allosteric inhibitor MUT-A: resistance profile, impairment of virus maturation and infectivity but without influence on RNA packaging or virus immunoreactivity. Retrovirology 14:50. doi: 10.1186/s12977-017-0373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. 2013. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. 2014. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog 10:e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan M, Yant SR, Tsai L, O'Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. 2013. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS One 8:e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohata Y, Tomonaga M, Watanabe Y, Tomura K, Kimura K, Akaki T, Adachi K, Kodama EN, Matsuzaki Y, Hayashi H. 2022. Antiviral activity and resistance profile of the novel HIV-1 non-catalytic site integrase inhibitor JTP-0157602. J Virol 96:e0184321. doi: 10.1128/JVI.01843-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. 2012. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J Biol Chem 287:21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. 2013. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci USA 110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta K, Turkki V, Sherrill-Mix S, Hwang Y, Eilers G, Taylor L, McDanal C, Wang P, Temelkoff D, Nolte RT, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. 2016. Structural basis for inhibitor-induced aggregation of HIV integrase. PLoS Biol 14:e1002584. doi: 10.1371/journal.pbio.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maehigashi T, Ahn S, Kim UI, Lindenberger J, Oo A, Koneru PC, Mahboubi B, Engelman AN, Kvaratskhelia M, Kim K, Kim B. 2021. A highly potent and safe pyrrolopyridine-based allosteric HIV-1 integrase inhibitor targeting host LEDGF/p75-integrase interaction site. PLoS Pathog 17:e1009671. doi: 10.1371/journal.ppat.1009671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parcella K, Wang T, Eastman K, Zhang Z, Yin Z, Patel M, Tu Y, Zheng BZ, Walker MA, Saulnier MG, Frennesson D, Bowsher M, Gillis E, Peese K, Belema M, Cianci C, Dicker IB, McAuliffe B, Ding B, Falk P, Simmermacher J, Parker DD, Sivaprakasam P, Kish K, Lewis H, Hanumegowda U, Jenkins S, Kadow JF, Krystal M, Meanwell NA, Naidu BN. 2022. Discovery and preclinical profiling of GSK3839919, a potent HIV-1 allosteric integrase inhibitor. ACS Med Chem Lett 13:972–980. doi: 10.1021/acsmedchemlett.2c00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA 102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 21.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng L, Dharmarajan V, Serrao E, Hoyte A, Larue RC, Slaughter A, Sharma A, Plumb MR, Kessl JJ, Fuchs JR, Bushman FD, Engelman AN, Griffin PR, Kvaratskhelia M. 2016. The Competitive interplay between allosteric HIV-1 integrase inhibitor BI/D and LEDGF/p75 during the early stage of HIV-1 replication adversely affects inhibitor potency. ACS Chem Biol 11:1313–1321. doi: 10.1021/acschembio.6b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vranckx LS, Demeulemeester J, Saleh S, Boll A, Vansant G, Schrijvers R, Weydert C, Battivelli E, Verdin E, Cereseto A, Christ F, Gijsbers R, Debyser Z. 2016. LEDGIN-mediated inhibition of integrase-LEDGF/p75 interaction reduces reactivation of residual latent HIV. EBioMedicine 8:248–264. doi: 10.1016/j.ebiom.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh PK, Li W, Bedwell GJ, Fadel HJ, Poeschla EM, Engelman AN. 2022. Allosteric integrase inhibitor influences on HIV-1 integration and roles of LEDGF/p75 and HDGFL2 host factors. Viruses 14:1883. doi: 10.3390/v14091883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. 2013. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Bel N, van der Velden Y, Bonnard D, Le Rouzic E, Das AT, Benarous R, Berkhout B. 2014. The allosteric HIV-1 integrase inhibitor BI-D affects virion maturation but does not influence packaging of a functional RNA genome. PLoS One 9:e103552. doi: 10.1371/journal.pone.0103552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. 2015. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol 89:9765–9780. doi: 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madison MK, Lawson DQ, Elliott J, Ozanturk AN, Koneru PC, Townsend D, Errando M, Kvaratskhelia M, Kutluay SB. 2017. Allosteric HIV-1 integrase inhibitors lead to premature degradation of the viral RNA genome and integrase in target cells. J Virol 91:e01048-19. doi: 10.1128/JVI.00821-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv Virus Res 52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 31.Engelman AN, Kvaratskhelia M. 2022. Multimodal functionalities of HIV-1 Integrase. Viruses 14:926. doi: 10.3390/v14050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessl JJ, Kutluay SB, Townsend D, Rebensburg S, Slaughter A, Larue RC, Shkriabai N, Bakouche N, Fuchs JR, Bieniasz PD, Kvaratskhelia M. 2016. HIV-1 integrase binds the viral RNA genome and is essential during virion morphogenesis. Cell 166:1257–1268. doi: 10.1016/j.cell.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elliott JL, Eschbach JE, Koneru PC, Li W, Puray-Chavez M, Townsend D, Lawson DQ, Engelman AN, Kvaratskhelia M, Kutluay SB. 2020. Integrase-RNA interactions underscore the critical role of integrase in HIV-1 virion morphogenesis. Elife 9:e54311. doi: 10.7554/eLife.54311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng N, Hoyte A, Mansour YE, Mohamed MS, Fuchs JR, Engelman AN, Kvaratskhelia M, Levy R. 2016. Allosteric HIV-1 integrase inhibitors promote aberrant protein multimerization by directly mediating inter-subunit interactions: structural and thermodynamic modeling studies. Protein Sci 25:1911–1917. doi: 10.1002/pro.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 36.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. 1995. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 37.McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. 2008. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem 283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog 5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballandras-Colas A, Maskell DP, Serrao E, Locke J, Swuec P, Jonsson SR, Kotecha A, Cook NJ, Pye VE, Taylor IA, Andresdottir V, Engelman AN, Costa A, Cherepanov P. 2017. A supramolecular assembly mediates lentiviral DNA integration. Science 355:93–95. doi: 10.1126/science.aah7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballandras-Colas A, Chivukula V, Gruszka DT, Shan Z, Singh PK, Pye VE, McLean RK, Bedwell GJ, Li W, Nans A, Cook NJ, Fadel HJ, Poeschla EM, Griffiths DJ, Vargas J, Taylor IA, Lyumkis D, Yardimci H, Engelman AN, Cherepanov P. 2022. Multivalent interactions essential for lentiviral integrase function. Nat Commun 13:2416. doi: 10.1038/s41467-022-29928-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta K, Allen A, Giraldo C, Eilers G, Sharp R, Hwang Y, Murali H, Cruz K, Janmey P, Bushman F, Van Duyne GD. 2021. Allosteric HIV integrase inhibitors promote formation of inactive branched polymers via homomeric carboxy-terminal domain interactions. Structure 29:213–225. doi: 10.1016/j.str.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsantrizos YS, Bailey MD, Bilodeau F, Carson RJ, Coulombe R, Fader L, Halmos T, Kawai S, Landry S, Laplante S, Morin S, Parisien M, Poupart M-A, Simoneau B. 2009. Inhibitors of human immunodeficiency virus replication. Germany patent WO 2009/062285 A1WO 2009/062285 A1.
- 43.Wang H, Jurado KA, Wu X, Shun MC, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P, Kvaratskhelia M, Hughes SH, Engelman A. 2012. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res 40:11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins TM, Engelman A, Ghirlando R, Craigie R. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem 271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 45.Zheng R, Jenkins TM, Craigie R. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA 93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korbe B. 2018. HIV Sequence Compendium. Los Alamos National Laboratory, Los Alamos, New Mexico, USA. [Google Scholar]
- 47.Kanja M, Cappy P, Levy N, Oladosu O, Schmidt S, Rossolillo P, Winter F, Gasser R, Moog C, Ruff M, Negroni M, Lener D. 2020. NKNK: a new essential motif in the C-terminal domain of HIV-1 group M integrases. J Virol 94:e01035-20. doi: 10.1128/JVI.01035-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu R, Ghory HZ, Engelman A. 2005. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J Virol 79:10356–10368. doi: 10.1128/JVI.79.16.10356-10368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. 2017. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson TA, Januszyk K, Phillips ML, Tekeste SS, Zhang M, Miller JT, Le Grice SF, Clubb RT, Chow SA. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J Biol Chem 284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winans S, Goff SP. 2020. Mutations altering acetylated residues in the CTD of HIV-1 integrase cause defects in proviral transcription at early times after integration of viral DNA. PLoS Pathog 16:e1009147. doi: 10.1371/journal.ppat.1009147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu S, Phan AT, Mao D, Wang X, Gao G, Goff SP, Zhu Y. 2022. HIV-1 exploits the Fanconi anemia pathway for viral DNA integration. Cell Rep 39:110840. doi: 10.1016/j.celrep.2022.110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mugisha CS, Dinh T, Kumar A, Tenneti K, Eschbach JE, Davis K, Gifford R, Kvaratskhelia M, Kutluay SB. 2022. Emergence of compensatory mutations reveal the importance of electrostatic interactions between HIV-1 integrase and genomic RNA. mBio 13:e0043122. doi: 10.1128/mbio.00431-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slaughter A, Jurado KA, Deng N, Feng L, Kessl JJ, Shkriabai N, Larue RC, Fadel HJ, Patel PA, Jena N, Fuchs JR, Poeschla E, Levy RM, Engelman A, Kvaratskhelia M. 2014. The mechanism of H171T resistance reveals the importance of Ndelta-protonated His171 for the binding of allosteric inhibitor BI-D to HIV-1 integrase. Retrovirology 11:100. doi: 10.1186/s12977-014-0100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura T, Nakamura T, Amano M, Miyakawa T, Yamagata Y, Matsuoka M, Nakata H. 2020. A conformational escape reaction of HIV-1 against an allosteric integrase inhibitor. J Virol 94:e00486-20. doi: 10.1128/JVI.00486-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. 2012. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koneru PC, Francis AC, Deng N, Rebensburg SV, Hoyte AC, Lindenberger J, Adu-Ampratwum D, Larue RC, Wempe MF, Engelman AN, Lyumkis D, Fuchs JR, Levy RM, Melikyan GB, Kvaratskhelia M. 2019. HIV-1 integrase tetramers are the antiviral target of pyridine-based allosteric integrase inhibitors. Elife 8:e46344. doi: 10.7554/eLife.46344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim BJ, Lee IY, Kim JH, Shin HS, Son JC, Lee C-K, Kim K, Kim U-I, Nam HJ. 2022. Novel pyrrolopyridine derivative, method for producing same, and use thereof. USA patent US11214570B2.
- 59.Winter G, Waterman DG, Parkhurst JM, Brewster AS, Gildea RJ, Gerstel M, Fuentes-Montero L, Vollmar M, Michels-Clark T, Young ID, Sauter NK, Evans G. 2018. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr D Struct Biol 74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winter G, Lobley CM, Prince SM. 2013. Decision making in xia2. Acta Crystallogr D Biol Crystallogr 69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moriarty NW, Grosse-Kunstleve RW, Adams PD. 2009. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zucker F, Champ PC, Merritt EA. 2010. Validation of crystallographic models containing TLS or other descriptions of anisotropy. Acta Crystallogr D Biol Crystallogr 66:889–900. doi: 10.1107/S0907444910020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD, Jr. 2010. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. 2013. Rapid parameterization of small molecules using the force field toolkit. J Comput Chem 34:2757–2770. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA, Jr, Peralta JE, Ogliaro F. 2016. Gaussian 16 Rev. C.01, Wallingford, CT. [Google Scholar]
- 71.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 72.Fader LD, Malenfant E, Parisien M, Carson R, Bilodeau F, Landry S, Pesant M, Brochu C, Morin S, Chabot C, Halmos T, Bousquet Y, Bailey MD, Kawai SH, Coulombe R, LaPlante S, Jakalian A, Bhardwaj PK, Wernic D, Schroeder P, Amad M, Edwards P, Garneau M, Duan J, Cordingley M, Bethell R, Mason SW, Bos M, Bonneau P, Poupart MA, Faucher AM, Simoneau B, Fenwick C, Yoakim C, Tsantrizos Y. 2014. Discovery of BI 224436, a noncatalytic site integrase inhibitor (NCINI) of HIV-1. ACS Med Chem Lett 5:422–427. doi: 10.1021/ml500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 74.Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Henin J, Jiang W, McGreevy R, Melo MCR, Radak BK, Skeel RD, Singharoy A, Wang Y, Roux B, Aksimentiev A, Luthey-Schulten Z, Kale LV, Schulten K, Chipot C, Tajkhorshid E. 2020. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys 153:e044130. doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fiorin G, Klein ML, Henin J. 2013. Using collective variables to drive molecular dynamics simulations. Molecular Physics 111:3345–3362. doi: 10.1080/00268976.2013.813594. [DOI] [Google Scholar]
- 76.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD, Jr. 2017. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beglov D, Roux B. 1994. Finite representation of an infinite bulk system - solvent boundary potential for computer-simulations. J Chemical Physics 100:9050–9063. doi: 10.1063/1.466711. [DOI] [Google Scholar]
- 78.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chemical Physics 79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 79.Vanommeslaeghe K, MacKerell AD, Jr. 2012. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J Chem Inf Model 52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu W, He X, Vanommeslaeghe K, MacKerell AD, Jr. 2012. Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J Comput Chem 33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stone JE, Perilla JR, Cassidy CK, Schulten K. 2017. Chapter 11 - GPU-accelerated molecular dynamics clustering analysis with OpenACC, p 215–240. In Farber R (ed), Parallel Programming with OpenACC. Morgan Kaufmann, Boston. doi: 10.1016/B978-0-12-410397-9.00011-1. [DOI] [Google Scholar]
- 82.Zwanzig RW. 1954. High-temperature equation of state by a perturbation method. I. nonpolar gases. J Chem Phys 22:1420–1426. doi: 10.1063/1.1740409. [DOI] [Google Scholar]
- 83.Chen H, Maia JDC, Radak BK, Hardy DJ, Cai W, Chipot C, Tajkhorshid E. 2020. Boosting Free-energy perturbation calculations with GPU-accelerated NAMD. J Chem Inf Model 60:5301–5307. doi: 10.1021/acs.jcim.0c00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar A, Vashisth H. 2021. Conformational dynamics and energetics of viral RNA recognition by lab-evolved proteins. Phys Chem Chem Phys 23:24773–24779. doi: 10.1039/d1cp03822b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar A, Aqvist J, Satpati P. 2019. Principles of tRNA(Ala) selection by alanyl-tRNA synthetase based on the critical G3.U70 base pair. ACS Omega 4:15539–15548. doi: 10.1021/acsomega.9b01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pavlova A, Zhang Z, Acharya A, Lynch DL, Pang YT, Mou Z, Parks JM, Chipot C, Gumbart JC. 2021. Machine learning reveals the critical interactions for SARS-CoV-2 spike protein binding to ACE2. J Phys Chem Lett 12:5494–5502. doi: 10.1021/acs.jpclett.1c01494. [DOI] [PubMed] [Google Scholar]
- 87.Panel N, Villa F, Fuentes EJ, Simonson T. 2018. Accurate PDZ/peptide binding specificity with additive and polarizable free energy simulations. Biophys J 114:1091–1102. doi: 10.1016/j.bpj.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L, Wu Y, Deng Y, Kim B, Pierce L, Krilov G, Lupyan D, Robinson S, Dahlgren MK, Greenwood J, Romero DL, Masse C, Knight JL, Steinbrecher T, Beuming T, Damm W, Harder E, Sherman W, Brewer M, Wester R, Murcko M, Frye L, Farid R, Lin T, Mobley DL, Jorgensen WL, Berne BJ, Friesner RA, Abel R. 2015. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J Am Chem Soc 137:2695–2703. doi: 10.1021/ja512751q. [DOI] [PubMed] [Google Scholar]
- 89.Sun Q, Ramaswamy VSK, Levy R, Deng N. 2021. Computational design of small molecular modulators of protein-protein interactions with a novel thermodynamic cycle: allosteric inhibitors of HIV-1 integrase. Protein Sci 30:438–447. doi: 10.1002/pro.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cruz J, Wickstrom L, Yang D, Gallicchio E, Deng N. 2020. Combining alchemical transformation with a physical pathway to accelerate absolute binding free energy calculations of charged ligands to enclosed binding sites. J Chem Theory Comput 16:2803–2813. doi: 10.1021/acs.jctc.9b01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. 2010. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 92.Gao J, Kuczera K, Tidor B, Karplus M. 1989. Hidden thermodynamics of mutant proteins: a molecular dynamics analysis. Science 244:1069–1072. doi: 10.1126/science.2727695. [DOI] [PubMed] [Google Scholar]
- 93.Jorgensen WL, Thomas LL. 2008. Perspective on free-energy perturbation calculations for chemical equilibria. J Chem Theory Comput 4:869–876. doi: 10.1021/ct800011m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pohorille A, Jarzynski C, Chipot C. 2010. Good practices in free-energy calculations. J Phys Chem B 114:10235–10253. doi: 10.1021/jp102971x. [DOI] [PubMed] [Google Scholar]
- 95.Bennett CH. 1976. Efficient estimation of free-energy differences from Monte-Carlo data. J Computational Physics 22:245–268. doi: 10.1016/0021-9991(76)90078-4. [DOI] [Google Scholar]
- 96.Liu P, Dehez F, Cai W, Chipot C. 2012. A toolkit for the analysis of free-energy perturbation calculations. J Chem Theory Comput 8:2606–2616. doi: 10.1021/ct300242f. [DOI] [PubMed] [Google Scholar]
- 97.Bester SM, Wei G, Zhao H, Adu-Ampratwum D, Iqbal N, Courouble VV, Francis AC, Annamalai AS, Singh PK, Shkriabai N, Van Blerkom P, Morrison J, Poeschla EM, Engelman AN, Melikyan GB, Griffin PR, Fuchs JR, Asturias FJ, Kvaratskhelia M. 2020. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 370:360–364. doi: 10.1126/science.abb4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Armon A, Graur D, Ben-Tal N. 2001. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol 307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Development, validation, and X-ray crystallography of the HIV-1 IN construct in complex with ALLINIs. (A) Disruption of HIV-1 IN CTD dimerization by a single amino acid substitution. Wild type (left), L242E (middle), or W243E (right) HIV-1 IN CTD constructs were analyzed using SEC-MALLS. Solid lines report the refractive index for each elution, and the scatter points are weight-averaged molar masses that were determined at 1 s intervals throughout the elution of the observed peaks. The colors correspond to loading protein concentrations of 8 (red), 4 (magenta), 2 (green), 1 (blue), or 0.5 (puce) mg/mL. Grey dashes indicate the molecular masses corresponding to the CTD monomer (6.0 kDa) and dimer (12 kDa) species. (B) The CTD enhances the affinity of HIV-1 IN for PIR. SPR sensorgrams of the association and dissociation of PIR from HIV-1 IN CCD (F185H) (left) and CTD-CCD (F185K/W243E) (right). Compound was injected at concentrations of 0.015 to 1 μM, as indicated. The lower panel shows the association and dissociation rate constants (kon and koff), the equilibrium dissociation constants (KD) that were determined from the SPR data, and the fold change between the parameters between the two IN constructs. (C) Electron density maps of the BI-D (left) and PIR (right) cocrystal structures. Shown are wall-eyed stereo views of the ALLINI-induced CTD-CCD interface with final weighted 2Fo-Fc maps that are contoured at a 1.0 rms deviation as a blue mesh. Protein residues and ligands are depicted as sticks, and water molecules are depicted as small red spheres. The carbon atoms of the amino acid residues are colored by chain (CTD, yellow; subunits 1 and 2 of the CCD dimer, cyan and green, respectively). The carbon atoms of the ALLINIs are colored pink. The carbon atoms of PEG and ethylene glycol are colored grey. The remaining atoms are colored according to conventions: oxygen, red; nitrogen, blue. Download FIG S1, PDF file, 3.1 MB (3.2MB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conservation of the ALLINI-induced CTD-CCD interface in the crystal structures. (A) Superposition of the two crystal structures determined in this study. Protein backbones are shown as cartoons, with CTDs in yellow and CCD dimer chains in cyan and green. The ALLINI molecules (indicated with arrowheads) are shown as sticks, with carbon, nitrogen, and oxygen atoms in pink, blue, and red, respectively. (B) Left panel: a linear polymer of HIV-1 IN dimers observed in a cocrystal structure with GSK1264 (PDB entry 5HOT) is stabilized by ALLINI-induced association of the CCD dimers with CTDs provided by neighboring dimers. An example of this substructure is boxed. Right panel: the CCD dimer with its associated CTD chains is shown in the same orientation as the crystal structures in panel A. (C) Left panel: rhe four independent CTD-ALLINI-CCD interfaces that were observed in the crystal structures that are reported here as well as the related substructure from 5HOT were superposed, based on the CCD dimers. The CTDs from cocrystal structures with BI-D, PIR, and GSK1264 are shown as ribbons and are colored orange, yellow, and brown, respectively. Right panel: the superposition of the CTDs from the cocrystal structures with BI-D (orange) and PIR (yellow) with a prior drug-free HIV-1 IN CTD structure (PDB entry 6T6E). Individual β strands of the five-bladed SH3 fold are indicated (β1 to β5). Note that the CTD structure does not undergo substantial conformation changes as a result of ALLINI binding. Download FIG S2, PDF file, 0.6 MB (626.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallography statistics and free energy perturbation data. Download Table S1, XLSX file, 0.02 MB (21.3KB, xlsx) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MD setup. (A) Modeled loops. BI-D (left) and PIR (right) cocrystal structures completed by modeling the missing polypeptide loops. For clarity, ALLINI molecules are not shown. The protein mainchains are shown as cartoons, with the CCDs in orange, CTDs in red, and modeled loops in blue. (B–D) MD simulation domains. Simulation domains for the systems without ALLINI (B), with BI-D (C), or with PIR (D). Protein chains are shown as cartoons, with CCD in orange and CTD in red. Inhibitors are shown in space-filling representations. The purple, green, and cyan spheres represent sodium, chloride, and Mg2+ ions, respectively. The grey boxes are solvent water. (E) An example of the thermodynamic cycle of BI-D binding. The horizontal legs represent the alchemical transformation of an amino acid residue (Lys266) into Ala either in the bound state (complex, upper panel) or in the unbound state (free, bottom panel). The vertical legs represent binding. Download FIG S3, PDF file, 0.3 MB (339.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MD simulation details. (A–C) The rms deviation time traces of all of the protein heavy atoms are shown for the CCDs (green) and CTDs (yellow) throughout the MD simulations that were conducted either in the absence of ALLINIs (A) or in the presence of BI-D (B) or PIR (C). Corresponding snapshots of the protein backbone and bound compounds are shown for each system, with the CCD and the CTD in green and yellow, respectively. BI-D and PIR are presented in space-filling representations. (D) The rms fluctuation per residue trace in the absence of ALLINI (red), in the presence of BI-D (blue), or in the presence of PIR (purple). (E) All-to-all rms deviation matrix analysis. Shown is the rms deviation matrix (81) between the pairwise structures in the MD trajectory of the protein in the absence of ALLINIs. Representative snapshots are shown to the right (I–VI, indicated with arrowheads on the matrix plot) of the protein, highlighting various transitions observed along the MD trajectory. (F) Dynamics of ALLINIs binding to IN. Snapshots are shown from the MD simulations of the CTD-CCD interface in the presence of BI-D (upper) or PIR (lower) at 0, 250, 500, 750, and 1,000 ns of the MD simulations. The ALLINIs and selected side chains are shown in ball-and-stick mode in separate colors and are labelled on the left diagrams that correspond to the initial states (0 ns). Download FIG S4, PDF file, 0.4 MB (466.9KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effects of amino acid substitutions within the CTD-CCD interface on PIR-induced IN aggregation. Full-length wild type and mutant HIV-1 IN proteins (indicated, right) were incubated in the absence or presence of 2.5 or 5 μM PIR. Centrifugally separated precipitated (P) and soluble (S) fractions were analyzed via SDS polyacrylamide electrophoresis, and IN was detected by staining with Coomassie brilliant blue. The migration positions of the protein molecular mass standards are indicated to the left (kDa). Download FIG S5, PDF file, 0.3 MB (330.7KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analytical size exclusion chromatography of HIV-1 IN variants. Elution chromatograms (A280) of full-length wild type or mutant recombinant HIV-1 IN (as indicated) from a Superdex-200 10/30 column. The expected elution volumes of the tetramer (T), dimer (D), and/or monomer (M) forms of the protein are indicated above each chromatogram. Download FIG S6, PDF file, 0.1 MB (66.9KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effects of amino acid substitutions within the ALLINI-induced CTD-CCD interface on HIV-1 infectivity and PIR antiviral activity. Left panel: relative infectivity of HIV-1 variants carrying the indicated mutations within IN. The infectivity of wild type HIV-1NL4-3 was considered to be 100%. The bars indicate the mean values. The error bars are the standard deviations from n = 3 biological replicates for each condition. The open circles correspond to individual measurements. Right panel: PIR 50% effective inhibitory concentrations (EC50) on the wild type and HIV-1 IN mutants. Download FIG S7, PDF file, 0.02 MB (19.9KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variable roles of IN CTDs in lentiviral integration. (A) Residues corresponding to HIV-1 IN Tyr226, Trp235, and Lys266 are highly conserved among lentiviral INs. The IN CTD regions from HIV-1, HIV-2, maedi-visna virus (MVV), equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV), and bovine immunodeficiency virus (BIV) are aligned. Red and yellow boxes highlight the amino acid residues in positions that are invariant across the alignment or are conserved through a similarity of physicochemical properties, respectively. The sequence numbering and secondary structure elements (extracted from PDB entry 6T6E) that are shown above the alignment correspond to HIV-1 IN. The key HIV-1 IN residues involved in ALLINI binding are indicated with red arrowheads (Tyr226, Trp235, and Lys266). The figure was prepared using the ESPript server (https://espript.ibcp.fr) (67). (B) The IN CTD residues involved in the ALLINI-induced interface play multiple roles within the lentiviral intasome assembly. (Top panel) HIV-1 intasome core assembly (PDB entry 6PUY [6]), with protein and viral DNA chains shown as cartoons. Catalytic IN chains are colored green and cyan. The remaining IN chains are colored according to their functions within the twofold symmetric assembly. Individual IN domains (NTD, CCD, and CTD) that belong to different IN chains and are resolved in the structure are indicated. (Lower panels) Detailed view of the CTDs belonging to IN chains that are colored orange and magenta in the top panel. DNA and selected amino acid side chains are shown as sticks, with the amino acid residues labelled. Also indicated are the alpha helical CCD-CTD and an extended NTD-CCD linker of the catalytic IN subunits. Note that the triad of IN CTD residues (Tyr226, Trp235, and Lys266) have different functions within these two types of intasomal subunits. Download FIG S8, PDF file, 0.5 MB (479.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Parameterization scheme and molecular fragmentation of BI-D. (A) Dihedral angle parameters with CGenFF penalties above 10. (B) Parameterized molecular mechanics potential (solid blue line) fitted to the quantum chemistry calculated potential (dotted black line). The rms difference between both potentials was 1.44 kcal/mol. (C) 3D model of BI-D. (D) To perform dihedral scans in the parameterization scheme, the molecule was fragmented into three moieties, and these are shown as stick diagrams (left) and 3D models (right). Download FIG S9, PDF file, 0.2 MB (197.5KB, pdf) .
Copyright © 2023 Singer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data that support this study are available from the corresponding authors upon reasonable request. The refined models and the associated X-ray diffraction data were deposited into the Protein Data Bank under accession codes 8A1P (BI-D cocrystal structure) and 8A1Q (PIR).