ABSTRACT
Coronaviruses (CoVs) of genera α, β, γ, and δ encode proteins that have a PDZ-binding motif (PBM) consisting of the last four residues of the envelope (E) protein (PBM core). PBMs may bind over 400 cellular proteins containing PDZ domains (an acronym formed by the combination of the first letter of the names of the three first proteins where this domain was identified), making them relevant for the control of cell function. Three highly pathogenic human CoVs have been identified to date: severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2. The PBMs of the three CoVs were virulence factors. SARS-CoV mutants in which the E protein PBM core was replaced by the E protein PBM core from virulent or attenuated CoVs were constructed. These mutants showed a gradient of virulence, depending on whether the alternative PBM core introduced was derived from a virulent or an attenuated CoV. Gene expression patterns in the lungs of mice infected with SARS-CoVs encoding each of the different PBMs were analyzed by RNA sequencing of infected lung tissues. E protein PBM of SARS-CoV and SARS-CoV-2 dysregulated gene expression related to ion transport and cell homeostasis. Decreased expression of cystic fibrosis transmembrane conductance regulator (CFTR) mRNA, essential for alveolar edema resolution, was shown. Reduced CFTR mRNA levels were associated with edema accumulation in the alveoli of mice infected with SARS-CoV and SARS-CoV-2. Compounds that increased CFTR expression and activity, significantly reduced SARS-CoV-2 growth in cultured cells and protected against mouse infection, suggesting that E protein virulence is mediated by a decreased CFTR expression.
KEYWORDS: antivirals, CFTR, coronavirus, envelope (E) protein, lung edema resolution
INTRODUCTION
Coronaviruses (CoVs) are cytoplasmic, positive-sense RNA viruses with a large genome (around 30 kb (kb)) (1). CoVs infect a wide variety of animals including humans. Seven human CoVs (hCoVs) have been identified. Four (229E, OC43, HKU1, and NL63) are responsible for approximately 15 to 30% of common colds (2, 3). In contrast, the other three zoonotic CoVs severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and SARS-CoV-2 are highly pathogenic.
SARS-CoV was first identified in China in 2002 and spread worldwide to infect more than 8,000 people with a mortality rate of around 10% (4, 5). In 2012, MERS-CoV was identified in Saudi Arabia (6, 7). Since then, 2,562 laboratory-confirmed cases and 890 deaths have been reported by WHO as of May 2022, corresponding to a fatality rate of 35% (https://www.who.int/emergencies/mers-cov/en/). SARS-CoV-2 was discovered in December 2019 and is the causative agent of COVID-19, the most important pandemic of the 21st century (8). Since then, the European Center for Disease Control (ECDC) has reported 600 million cases worldwide and more than 6,000,000 deaths with a mortality rate of 1.2% (https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases). Studies of CoV-host interactions are essential to developing safe and effective antiviral therapies and vaccines, which are required to resolve the ongoing international health emergency and to prevent and treat future pandemics caused by emerging zoonotic highly pathogenic CoVs (9–11).
Our group has described that the envelope (E) protein of SARS-CoV and MERS-CoV is implicated in virus replication and virulence (12–14). Two domains of the SARS-CoV E protein play a major role in virulence, its ion channel activity and the PDZ-binding motif (PBM) (15, 16). CoV E protein is also essential for intracellular virus trafficking (17).
CoVs have several proteins that include PBMs that may modify cell behavior to benefit the virus. PBMs are specific sequences usually located at the C terminus of proteins, normally within the last four residues and interact with PDZ domains (an acronym formed by the combination of the first letter of the names of the three first proteins where this domain was identified: PSD-95, Discs large 1, and Zonula occludens 1, respectively), consisting of around 80 to 90 amino acids (aa). There are 266 PDZ domains present within more than 400 protein isoforms in the human genome (18). Three different classes of PBMs are recognized based on the nature of their core sequence motifs: class I PBMs consist of X-S/T-X-Φ-COOH, where X can be any amino acid and Φ is a hydrophobic amino acid (normally V, I, or L), class II PBMs are X-Φ-X-Φ-COOH, and class III PBMs are X-D/E-X-Φ-COOH. Due to the functional versatility of proteins containing PDZ domains, viruses can modify a wide range of cellular functions through their PBMs to enhance viral replication and dissemination, and influence disease pathogenesis (19). Although the E proteins of all hCoVs have a PBM in their C terminus, the PBM of SARS-CoV E protein is the only one that has been studied in detail. The role of E protein PBMs from other hCoVs in virulence remains unknown (15).
SARS-CoV E protein is a small integral membrane protein of 76 aa, including the PBM formed by the last four aa of the carboxy terminus (-DLLV-COOH). An identical PBM sequence is present in the SARS-CoV-2 E protein. The SARS-CoV E protein PBM is a known virulence factor (15), inducing an exaggerated proinflammatory response by binding to the host PDZ-containing protein syntenin-1 and possibly to additional host proteins. The interaction of SARS-CoV E protein PBM and syntenin-1 leads to acute respiratory distress syndrome, pulmonary edema, and death at least through the phosphorylation and activation of p38-MAPK, a protein regulating the expression of proinflammatory cytokines (15).
Here, we use reverse genetics to generate variants of SARS-CoV, MERS-CoV, and SARS-CoV-2 that lack the E protein PBM to investigate their pathogenicity in mice. We found that the PBMs of these three hCoVs are virulence factors required for optimal replication and the generation of alveolar edema. Furthermore, we constructed a collection of SARS-CoV mutants in which the E protein PBM was replaced with one derived from virulent or attenuated hCoVs and studied their virulence. A gradient of virulence was observed, depending on whether the E protein was derived from an attenuated or virulent hCoV. Following infection of mice with SARS-CoV, we performed transcript profiling in lung tissues using RNA sequencing (RNA-seq). These studies showed that the E protein PBMs of SARS-CoV or SARS-CoV-2 altered the expression of genes involved in ion transport and cell homeostasis. Of note, we observed decreased expression of cystic fibrosis transmembrane conductance regulator (CFTR) mRNA, encoding a protein involved in liquid absorption in the alveolus. The reduction in CFTR transcript abundance was associated with increased alveolar edema in the lungs of mice infected with SARS-CoV or SARS-CoV-2. We further investigated the effects of small molecule CFTR modulators on the replication of SARS-CoV-2 in cultured cells and observed that these compounds significantly reduced SARS-CoV-2 replication and protected against severe disease in a mouse model. These results demonstrate the importance of the E protein PBMs in CoV replication and virulence and identify novel cellular targets for the selection of antivirals.
RESULTS
Relevance of E protein PBM from virulent hCoVs on viral replication in vitro and in vivo.
Using an infectious cDNA clone engineered in our laboratory, we constructed a collection of mutant viruses by replacing the 4 residues of the PBM core with glycines (Fig. 1A). The growth kinetics of mouse-adapted SARS-CoV and SARS-CoV-2 wild-type and deletion mutants were assessed in Vero E6 cells. Similar studies of modified MERS-CoV were performed in Huh-7 cells. While the growth of rSARS-CoV with or without E PBM was similar, the E PBM deletion mutants of SARS-CoV-2 and MERS-CoV exhibited between 10 and 1,000-fold lower viral titers, respectively, than the parental (wt) viruses (Fig. 1B). This suggests that SARS-CoV-2 and MERS-CoV required an E PBM for optimal virus replication in cultured cells.
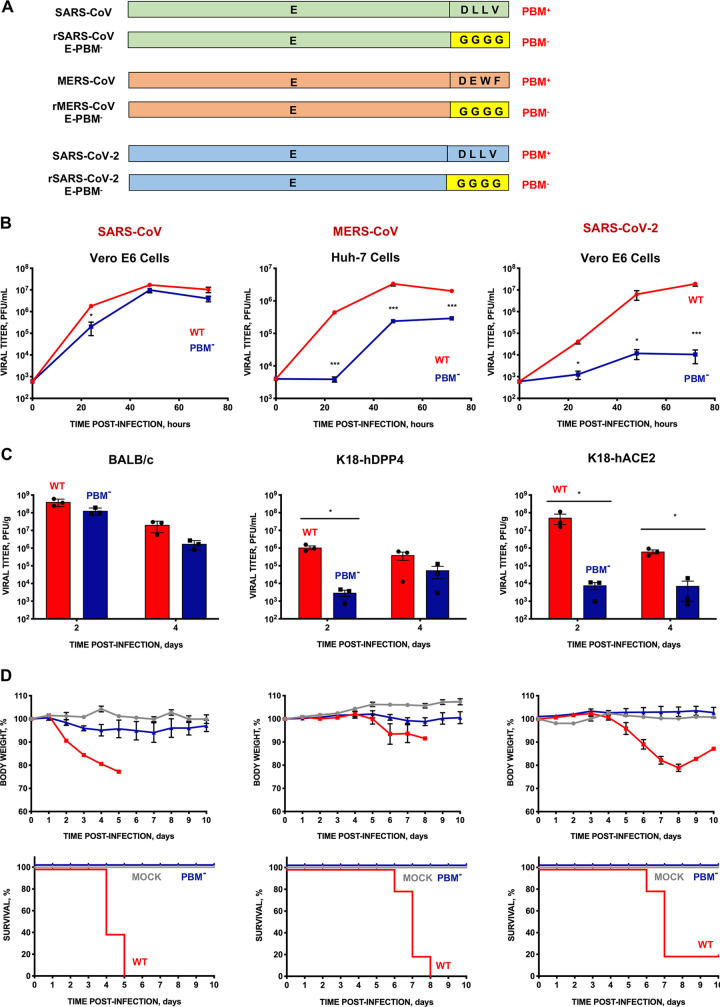
FIG 1.
Growth and virulence of SARS-CoV, MERS-CoV, and SARS-CoV-2 mutants lacking E protein PBM. (A) Diagram mutant pairs, with (PBM+) and without the PBM (PBM−), were generated for each of the three viruses. In the rSARS-CoV-E-PBM−, rMERS-CoV-E-PBM−, and rSARS-CoV-2-E-PBM− mutants, the PBM of E protein was replaced by four glycines. (B) Subconfluent Vero E6 or Huh-7 cells were infected with an MOI of 0.001 with each of the three viruses. Wild- type (WT) viruses are shown with red lines and symbols. Mutants without E protein PBM (PBM−), with blue lines and symbols. Supernatants were collected at 24, 48, and 72 hpi and titrated by the lysis plaque formation method. (C) Groups of six 16-week-old BALB/c, K18-hDPP4, or K18-hACE2 mice were intranasally inoculated with 10,000 PFU of the WT viruses (red columns) or each of the mutants lacking E protein PBM (blue columns). Three mice from each group were euthanized at 2 and 4 dpi to analyze virus production in lung. Vertical bars represent the standard error of the mean. Statistically significant data are represented according to the P value obtained in Student’s t test analysis: *, P < 0.05; ***, P < 0.001. (D) Groups of five 16-week-old BALB/c, K18-hDPP4, or K18-hACE2 mice were mock-infected (gray lines) or intranasally inoculated with 10,000 PFU with each of the parental (WT) virus (red lines), or with their corresponding mutants without PBM of E protein (PBM−) (blue lines). Weight loss (top) and survival (bottom) of mice were monitored for 10 days postinfection. Vertical bars represent the standard error of the mouse weight mean.
To investigate the role of E protein PBM in virus replication in vivo, BALB/c, K18-hDPP4, or K18-hACE2 mice were infected with either wild-type (wt) SARS-CoV, MERS-CoV, or SARS-CoV-2, respectively, or their corresponding mutants lacking E PBMs. Lung tissue titers were determined at days 2 and 4 postinfection (dpi) (Fig. 1C). The MERS-CoV and SARS-CoV-2 E protein deletion mutants grew to 1,000- to 10,000-fold lower titers that the corresponding parental viruses, indicating that E protein PBMs contributed to efficient virus replication in vitro and in vivo. We then monitored weight loss and survival through 10 dpi. All mice infected with parental viruses lost weight and exhibited mortality ranging from 80% to 100%, as expected (Fig. 1D). In contrast, all mice infected with virus lacking E protein PBM expression survived, further supporting the conclusion that the E protein PBM is a CoV virulence factor.
Human CoVs cause mild to severe respiratory disease. To assess whether disease severity is influenced by the source of the PBM, we engineered a set of recombinant SARS-CoVs expressing the E protein PBM sequences of virulent or attenuated β-hCoV, including SARS-CoV-2, MERS-CoV, HCoV-OC43, and HCoV-HKU1 (Fig. 2A). As a control, a rSARS-CoV without a PBM core sequence was generated (rSARS-PBM−). We inoculated BALB/c mice intranasally with the recombinant viruses and monitored weight loss (Fig. 2B) and survival (Fig. 2C) for 10 dpi. While all mock-infected mice and mice infected with SARS-CoV lacking E protein PBM survived, mice infected with recombinant viruses containing the parental virus PBM from SARS-CoV-2 (PBM-SARS2) or MERS-CoV (PBM-MERS) lost weight and died between 4 and 6 dpi, respectively. Interestingly, we observed a gradient of lethality among these highly virulent viruses. All mice infected with PBM-SARS2 died with less than 20% weight loss. In contrast, all mice infected with PBM-MERS exhibited more than 25% weight loss and required euthanasia. Mice infected with viruses containing the E protein PBM core sequences from the attenuated hCoVs HKU1 and OC43 demonstrated intermediate survival phenotypes with 40% and 80% survival, respectively. Thus, substituting the SARS-CoV E protein PBM core sequence with those from other hCoVs altered virulence. Lung tissue virus titers from SARS-CoV with native or modified PBMs were similar, indicating that the observed differences in virulence were not due to changes in virus titers (Fig. S1 in the supplemental material).
FIG 2.
Virulence of SARS-CoV mutants with the PBM of E protein from attenuated or virulent β-hCoVs. (A) E protein sequence of parental and the corresponding mutants in which the PBM of E protein was replaced by that of an alternative virulent or attenuated β-hCoV (in red). Groups of five 16-week-old BALB/c mice were inoculated intranasally with DMEM (uninfected, gray), or with 100,000 PFU of parental SARS-CoV (PBM-SARS2), or with the generated mutants: PBM-MERS (orange line), PBM-OC43 (green line), and PBM-HKU1 (dark blue line), and PBM− (pink line). (B and C) Weight loss (B) and survival (C) were monitored for 10 dpi. Vertical bars represent the standard error of the mean weight of the mice. TMD, transmembrane domain.
Growth kinetics of SARS-CoV E protein PBM mutants. (A) Subconfluent Vero E6 were infected with SARS-CoV E protein PBM mutants at a MOI of 0.001. The cells infected with different virus mutants are indicated with lines of different color: native viruses (wt) are shown with red lines and the mutants without E protein PBM (PBM−) with pink lines and symbols. PBM-MERS-CoV (orange lines), PBM-hCoV-OC43 (green lines), and PBM-hCoV-HKU1 (blue lines). Supernatants were collected at 24, 48, and 72 hpi and titrated by the lysis plate formation method. (B) Groups of six 16-week-old BALB/c mice were intranasally inoculated with 100,000 pfu of the native viruses (red columns), or PBM− (pink column), PBM-MERS-CoV (orange column), PBM-OC43 (green column), and PBM-HKU1 (blue column). Three mice from each group were euthanized at 2 and 4 dpi to analyze virus production in lungs. Vertical bars represent the standard error of the mean titers obtained by repeating the experiment three times. Statistically significant data are represented according to the P value obtained in Student’s t analysis: **, P < 0.01. Download FIG S1, TIF file, 0.3 MB (325.1KB, tif) .
Copyright © 2023 Honrubia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E protein PBM conditions β-hCoV pathogenesis.
At 2 dpi, SARS-CoV-2-infected K18-hACE2 mice displayed mild alveolar septal thickening and mild perivascular and peribronchiolar mononuclear infiltrates. These lesions increased by 4 dpi, and mice also developed alveolar edema (Fig. 3A and B) with a histopathology score of 14.0. However, mice infected with the mutant SARS-CoV-2-PBM− exhibited no significant inflammatory or vascular changes having a score of 4.7. K18-hDPP4 mice infected with MERS-CoV showed moderate lung inflammatory lesions at 2 dpi that increased in severity by day 4 pi with a score of 11.4, including the presence of diffuse alveolar septal thickening, and moderate perivascular and peribronchiolar mononuclear infiltrates. In contrast, mice infected with the mutant MERS-CoV-PBM− showed a score of around 6.0 with no significant inflammatory lesions at either time point (Fig. 3C and D). In BALB/c mice infected with SARS-CoV, lung tissue pathology was more severe than in BALB/c mice infected with the SARS-CoV-PBM− mutant with histopathology scores of 17.7 and 13.5 at 2 dpi, respectively. These differences included severe interstitial pneumonia, perivascular and peribronchiolar mononuclear infiltrates, and alveolar edema (Fig. 3E and F).
FIG 3.
Lung pathology associated with E protein PBM of SARS-CoV, MERS-CoV, and SARS-CoV-2. Lung histopathology scores were examined in lung samples taken from K18-hACE2 (A), K18-hDPP2 (C), and BALB/c mice (E) infected with SARS-CoV-2, MERS-CoV, and SARS-CoV parental and mutant viruses, respectively, as indicated in Materials and Methods (n = 3/group), and euthanized at day 2 and 4 postinfection (dpi). Representative lung histopathological sections (H&E staining) from infected K18-hACE2 (B), K18-hDPP4 (D), and BALB/c (F) mice are shown (magnification: 10×). Mean and SD of cumulative histopathological lesion scores are represented. Unpaired t test: *, P < 0.05; ***, P < 0.001.
We performed additional histologic analysis of lung tissue sections from mice infected with rSARS-CoVs with PBMs from attenuated or virulent CoVs at 2 or 4 dpi to assess immune cell infiltration and alveolar edema. Mice infected with virus lacking the E protein PBM core sequence (PBM−) showed minimal inflammatory cell infiltrates and edema only at 4 dpi (Fig. 4). Mice infected with rSARS-CoV with an E protein PBM from HCoV-HKU1 (PBM-HKU1) or HCoV-OC43 (PBM-OC43) developed significant cellular infiltrates at 2 dpi that persisted at 4 dpi, while mice infected with virulent SARS-CoVs showed progressive infiltrates at the same time points (Fig. 4). Mice infected with viruses encoding the E protein PBM core from SARS-CoV-2 showed moderate interstitial and peribronchiolar cell infiltration at 2 dpi and more alveolar edema at 4 dpi coincident with the greater lethality of this virus. Interestingly, mice infected with recombinant viruses encoding an E protein PBM core from MERS-CoV showed a phenotype similar to that observed for PBM-SARS2 at 2 dpi but with a significant increase in cell infiltration and alveolar and bronchiolar wall thickening at 4 dpi, compared to mice infected with viruses encoding E PBMs from attenuated CoVs (Fig. 4). These data indicated that virus virulence correlated with lung tissue pathology and the E protein PBM core sequence contributed to these findings.
FIG 4.
Lung pathology in SARS-CoV-infected mice due to the presence of E protein PBM from attenuated or virulent β-hCoVs. Groups of six 16-week-old mice were mock infected or inoculated with 100,000 PFU of a SARS-CoV including E proteins with a PBM derived either from the SARS/SARS-2-CoV, or from MERS-CoV, hCoV-OC43, hCoV-HKU1, or with the PBM replaced by four glycines (PBM−). Three mice from each group were euthanized, and lungs were collected at 2 and 4 dpi. Lungs were fixed in formalin with zinc, embedded in kerosene, and cut and stained with hematoxylin and eosin. Cellular infiltration (A) and edema (B) scores were determined in lung samples as indicated in Materials and Methods (n = 3/group). The vertical bars represent the mean and the standard deviation of the data obtained in the lung of each of the three mice used for each experimental setting. Unpaired t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Effect of E protein PBM virus origin on host gene expression.
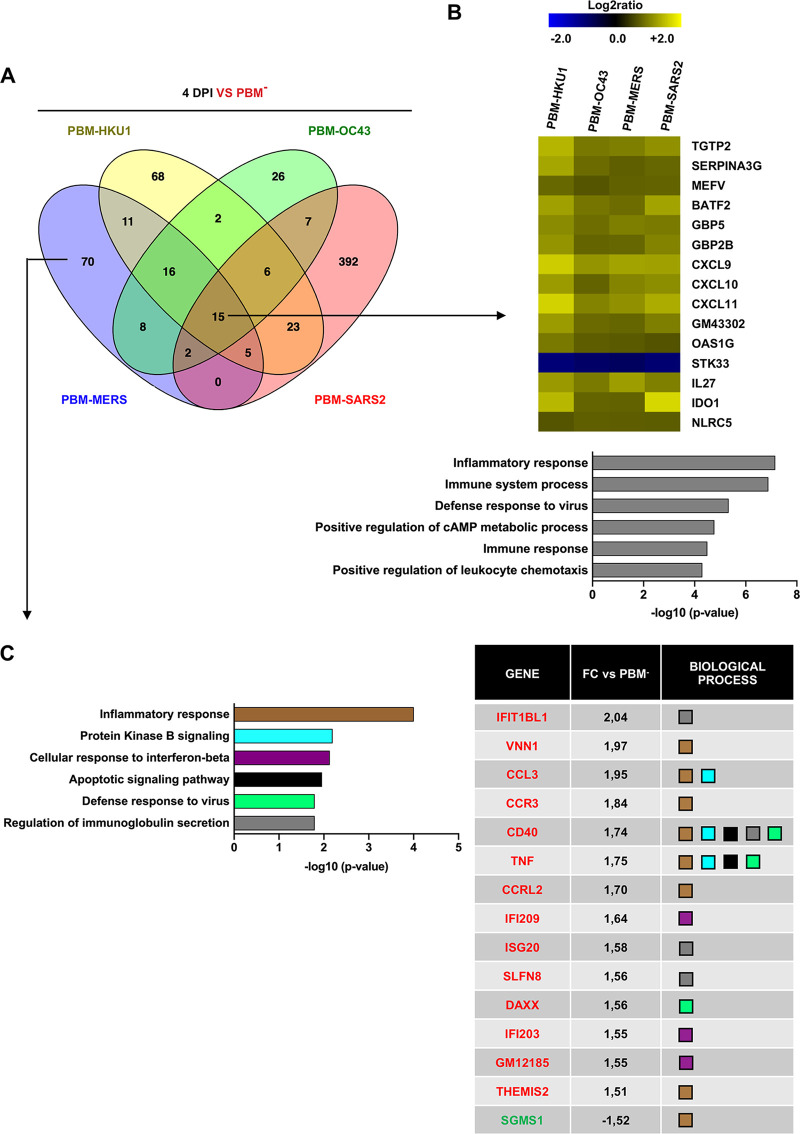
To investigate the virulence mechanisms of rSARS-CoV with PBMs from the different hCoVs, we performed RNA-seq analysis on lung tissue from infected mice. Because the observed association of virulence and lung histopathology scores was greater at 4 dpi, we focused on this time point. A Venn diagram of differentially expressed genes (DEGs) with a fold change (FC) higher than 1.5 and a false discovery rate (FDR) lower than 0.05 (|FC|>1.5; FDR <0.05, respectively) of mice infected with rSARS-CoVs with PBMs from hCoVs causing a range of pathology compared to the PBM− mutant was determined (Fig. 5A). We sought to identify transcripts with expression driven by the presence of an E protein PBM from attenuated or virulent β-hCoVs. The presence of an E protein PBM from attenuated β-hCoVs reduced SARS-CoV mortality (Fig. 2B and C). A set of 14 transcripts that increased their abundance after infection with all four viruses, including PBM-HKU1, PBM-OC43, PBM-MERS, or PBM-SARS2, was upregulated in all cases, whereas 1 (STK33) was downregulated in comparison with the infection by a SARS-CoV including a nonfunctional PBM that was replaced by four glycines (Fig. 5B). These transcripts were classified using the Gene Ontology analysis (DAVID software) as mainly related to immune and inflammatory responses (Fig. 5B), suggesting that the presence of a functional E protein PBM from either an attenuated or virulent β-hCoV triggered a host innate immune response.
FIG 5.
Differential and common gene expression patterns among the different rSARS-CoV with E protein PBM from virulent or attenuated β-hCoVs. (A) Venn diagram of genes differentially expressed at least 1.5-fold with an FDR <0.05 in the lungs of mice infected with the different rSARS-CoVs with the PBMs from the indicated hCoVs, compared to rSARS-CoV without the E protein PBM (PBM−), at 4 dpi. (B) Expression profile of the most frequently expressed genes due to the presence of a PBM from an attenuated or virulent hCoV. Analysis of the 15 genes with highest variation, due to the presence of E protein specific PBMs. The clustering of genes based on the biological process terms defined in Gene Ontology are shown. The six most relevant biological processes at 4 dpi into which the 15 commonly expressed genes are classified, have been listed. The values on the x axis indicate the −log10 (P value) values obtained with the DAVID software. (C) Biological activity types into which the 70 genes that are differentially expressed in the lungs of rSARS-CoV-PBM-MERS-infected mice are grouped, compared to rSARS-CoVs without PBM or with PBMs from other hCoVs. The grouping of these genes into distinct sets is based on the biological processes defined in Gene Ontology. The six most relevant biological processes are described. A table with the fold change (FC) and the biological processes in which genes of the six most relevant biological processes are involved is provided. The names of the genes in red and green correspond to those that increase or decrease their expression, respectively.
Replacing the SARS-CoV E protein PBM core with the homologous sequence of a virulent hCoV (PBM-MERS or PBM-SARS2) increased lung injury and virus pathogenicity compared to replacement by the PBM core from an attenuated hCoV (PBM-HKU1 and PBM-OC43). We next focused on the 70 and 392 DEGs that were solely changed in expression by viruses encoding the E protein PBM from MERS-CoV or SARS-CoV-2 at 4 dpi, respectively (Fig. 5A). The 70 DEGs enriched in tissues from mice infected with CoVs encoding a PBM-MERS were associated with host immune and inflammatory responses (Fig. 5C).
Among the 392 DEGs enriched during infection with viruses containing the E PBM from SARS-CoV or SARS-CoV-2 (Fig. 6A), 52 were present within biological activity processes related to ion transport and homeostasis (Fig. 6A). At least 34 of these 52 transcripts were identified in a highly interactive network (Fig. 6B). The genes with increased expression were mainly associated with the regulation of cytosolic Ca2+ concentration while those with decreased expression were associated with ion transport and cell homeostasis (Fig. 6C). These transcriptional changes were associated with greater pulmonary edema scores and mortality. Among these genes we noted that CFTR transcript abundance was reduced only in the presence of E protein PBM from SARS-CoV and SARS-CoV-2. The reduction in CFTR mRNA expression in the lungs of mice infected with SARS-CoV, containing a PBM identical to that of SARS-CoV-2, was confirmed by quantitative PCR at 4 dpi (Fig. 6D). SARS-CoV and SARS-CoV-2 infection reduced CFTR mRNA transcript abundance in Calu-3 2b4 cells and lung tissue of infected mice (Fig. 7A and B).
FIG 6.
Biological activity groups of genes differentially expressed exclusively due to the presence of E protein PBM from either SARS-CoV and SARS-CoV-2. (A) Clustering of 392 genes that are differentially expressed in the lungs of mice infected with the native virus compared to rSARS-CoVs without a PBM or with a PBM from another hCoV. The clustering of these genes is based on the biological process terms defined in Gene Ontology. The six more relevant biological processes are represented. (B) Network of interactions between genes that belong to the most relevant biological processes according to the STRING database. The thickness of the lines indicates the robustness of the data supporting each interaction and all those interactions with a score >0.4 were considered statistically significant. (C) Table with information on the rate of change (FC) and the biological processes to which the genes of the most relevant interaction networks belong. The names of the genes are shown in red or in green depending on whether correspond to those that increased or decreased their expression. (D) RT-qPCR of CFTR mRNA expression in lungs from mice infected with SARS-CoV E protein PBM mutants. RNA from lungs of mock-infected mice (gray column)or infected with PBM-SARS2 (red column) or with the E protein PBM mutants PBM-MERS (orange column), PBM-HKU1 (blue column), PBM-OC43 (green column), and PBM− (pink column) were quantified by RT-qPCR at 4 dpi. Mean values and their standard deviations are presented. Statistically significant data are indicated according to the P value obtained in Student’s t analysis: **, P < 0.01.
FIG 7.
CFTR mRNA expression decreased during SARS-CoV and SARS-CoV-2 infection in the presence of E protein PBM. (A and B) CFTR mRNA expression in Calu-3 cells (A) and lungs (B) from mice infected with SARS-CoV or SARS-CoV-2 with and without E protein PBM was analyzed by qPCR at 48 hpi and 4 dpi, respectively. (C and D) Edema scores examined in lung samples taken from K18-hACE2 or BALB/c mice infected with SARS-CoV-2 or SARS-CoV parental or mutant viruses, respectively, as indicated in Materials and Methods (n = 3/group), and euthanized at 2 and 4 days postinfection (dpi). Mean and SD of cumulative histopathological lesion scores. Unpaired t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CFTR modulators modify virus replication in infected cells.
In the alveolus, CFTR plays an important role in fluid absorption and thus loss of CFTR function could contribute to alveolar edema (20–23). Consistent with this, mice infected with SARS-CoV-2 or SARS-CoV had more alveolar edema compared to the PBM-variants (Fig. 7C and D). Recently, small molecule CFTR modulators were discovered that improve the function of some mutant forms of CFTR protein associated with cystic fibrosis. The corrector compound VX809 (lumacaftor) acts as a chemical chaperone and partially rescues CFTR function in cells expressing the misfolded F508del CFTR protein (24, 25). The potentiator VX770 (ivacaftor) increases the probability of the CFTR anion channel associated with G551D and related conductance mutations being in an open conformation (26–30). Of note, these CFTR modulators also enhance the function of wt CFTR (31–33).
We hypothesized that CFTR modulator treatment would partially rescue CFTR function and modify the disease phenotype. To test this hypothesis, CFTR modulators VX809 and VX770 were administered to Calu-3-2B4 cells and to K18-hACE2 mice infected with SARS-CoV-2. SARS-CoV-2 titers were measured at 48 h postinfection in cell culture, the time point at which the highest titer is reached (Fig. S2). CFTR modulator treatment resulted in a 10- and 7-fold reduction in virus titers in vitro and in vivo, respectively, in relation to nontreated cells or mice (Fig. 8A and B). At these concentrations, the modulators were nontoxic to Calu-3-2B4 cells (Fig. S3). Remarkably, the combined treatment of mice infected with SARS-CoV-2 with the VX809 and VX770 was associated with 80% survival, while mock-treated animals had 100% mortality. Still, a minimal effect on weight loss was observed, which is in line with previous observations that weight loss is a more sensitive measure of virus pathogenesis than survival (reference 15). Mice receiving VX770 or VX809 alone exhibited 40% and 20% survival, respectively (Fig. 8C and D). Treatment with the combination of both CFTR modulators delayed the appearance of clinical signs due to SARS-CoV-2 infection. SARS-CoV-2-infected and nontreated mice showed the highest clinical score (3.6) at both 7 and 8 dpi. In contrast, infected mice treated with both CFTR modulators showed a clinical score of 2.6 and 0.5 at the same times, respectively (Fig. 8E). Moreover, infected mice treated with CFTR modulators individually showed less edema score (4.9 for both cases) at 6 dpi in comparison with untreated mice (7.9). The combination of both CFTR modulators also reduced the edema score (4.0) (Fig. 8F).
FIG 8.
Protection of mice infected with SARS-CoV-2 by the modulation of CFTR expression. (A) Subconfluent Calu-3 2B4 cells were infected (MOI of 0.1) with SARS-CoV-2 wt virus. Infected cells were nontreated (red lines) or incubated with lumacaftor (VX809; 50 μM) (orange column), ivacaftor (VX770; 5 μM) (blue column) or both (green column). Culture supernatants were collected at 48 hpi and extracellular virus was titrated by the lysis plaque formation method. (B) Groups of five 20-week-old K18-hACE2 mice were intranasally inoculated with 10,000 PFU and daily intraperitoneally inoculated until 8 dpi with lumacaftor (12 mg/kg; orange column), ivacaftor (6 mg/kg; blue column), or both (green column). Three mice from each group were euthanized at 3 and 6 dpi to analyze virus production in lung. Vertical bars represent the standard error of the mean titers obtained by repeating the experiment three times. Statistically significant data are represented according to the P value obtained in Student’s t test: *, P < 0.05. Weight loss (C) and survival (D) of mice were monitored for 10 days postinfection. Vertical bars represent the standard error of the mouse mean weight. (E) Clinical score of mice infected with SARS-CoV-2 in the absence (red circle) or presence of VX809 (orange circle), VX770 (blue circle), or both CFTR modulators (green circle). Five mice per group were monitored daily for 10 dpi and their clinical signs of disease were classified in five grades of severity. Vertical bars represent the standard deviation of the mean derived from clinical scores obtained from five mice. Statistically significant data are represented according to the P value obtained in Student’s t analysis: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (F) Edema scores were determined in lung samples as indicated in Materials and Methods (n = 3/group). The vertical bars represent the mean and the standard deviation of the data obtained in the lung of each of the three mice used for each experimental setting. Unpaired t test: *, P < 0.05; ****, P < 0.0001. (G) mRNA quantification by RT-qPCR of inflammation mediators at 3 and 6 dpi in the lungs of infected mice treated with lumacaftor and ivacaftor. mRNA levels were compared to those in mock-infected mice, using the 2−ΔΔCT method and the 18S rRNA as the normalization endogenous control. Error bars indicate the standard error of the mean. Statistical significance was calculated by two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ***,P < 0.001.
Growth kinetics of SARS-CoV-2 in Calu-3 2B4. Subconfluent Calu-3 2B4 cells were infected with SARS-CoV-2 parental virus (red line) at a MOI of 0.1. Supernatants were collected at 24, 48 and 72 hpi and titrated. Vertical bars represent the standard error of the mean. Download FIG S2, TIF file, 0.1 MB (120.1KB, tif) .
Copyright © 2023 Honrubia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Viability of Calu-3 2B4 cells in the presence of different CFTR protein modulators. Subconfluent and uninfected Calu-3 2B4 cells were incubated with different concentrations (1 to 50 μM) of ivacaftor (A) and lumacaftor (B) and with the combination of ivacaftor (5 μM) and lumacaftor (50 μM) (green column) (C). Supernatants were collected at 48 hpi and cell viability was determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazole bromide] reduction assay (88). Mean values of cell viability and their standard deviations are shown. Download FIG S3, TIF file, 0.2 MB (205.2KB, tif) .
Copyright © 2023 Honrubia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of CFTR modulators on inflammatory cytokines induced by SARS-CoV-2 in the lungs of infected mice.
SARS-CoV-2 E protein modulates the NLRP3 inflammasome (34). CFTR modulators reduced NLRP3 inflammasome-mediated activation in monocytes derived from CF patients (35). We investigated the effects of CFTR modulators on NLRP3 mRNA transcript abundance and NLRP3-dependent cytokines (TNF-α, IL-6, and IL-1β) and chemokines (CXCL10 and CXCL2) in the lung tissues of SARS-CoV-2-infected K18-hACE2 mice by qRT-PCR (Fig. 8G) (36–38). SARS-CoV-2 infection increased the expression of NLRP3 cytokines and chemokines at 3 dpi. In contrast, the lungs of CFTR modulator-treated mice infected with SARS-CoV-2 showed lower levels of NLRP3 and NLRP3-dependent cytokines and chemokines (Fig. 8G) with more obvious effects when both modulators were used. These results suggest that CFTR modulators might also have an anti-inflammatory effect during SARS-CoV-2 infection (39–41).
DISCUSSION
Relevance of E protein PBM core motifs as virulence determinants.
In this article, the role of the E protein PBMs in human CoV replication and virulence is investigated, leading to the discovery that E protein PBM from SARS-CoV and SARS-CoV-2 reduces the transcript abundance for several genes involved in ion transport and cellular homeostasis (Fig. 6A), including CFTR (Fig. 6B and C). It is also shown that the combination of two drugs previously used to treat cystic fibrosis increased by 80% the survival of humanized mice after their infection by SARS-CoV-2.
Using reverse genetics, we generated a collection of SARS-CoV mutants by replacing its E protein PBM with homologous PBMs from attenuated or virulent β-hCoVs (SARS-CoV-2, MERS-CoV, hCoV-OC43, and hCoV-HKU1). The engineered mutants showed significant differences in virulence in mice. The PBMs from common cold hCoVs partially attenuated SARS-CoV and showed less cellular infiltrates and alveolar edema, compared to mice infected with the parental virus (Fig. 4). In contrast, rSARS-CoVs with PBM cores from virulent hCoVs caused the death of all infected mice and were associated with diffuse cellular infiltrates and alveolar edema. While only a small number of viruses were analyzed, these results support the notion that the PBMs play a critical role in virus virulence. Analysis of additional CoVs should be performed to confirm (or not) this interesting association.
SARS-CoV and SARS-CoV-2 share identical E protein PBMs. Infection of mice with these two viruses caused the greatest alveolar edema and mortality. In contrast, the introduction of the MERS-CoV PBM core was associated with increased cellular infiltrates and alveolar wall thickening. These results suggest that the E protein PBMs from all three virulent human CoVs are drivers of virus pathogenicity. PBMs encoded by viral proteins have identified roles in the virulence of other viruses. For example, The NS1 protein of the influenza virus also contains a PBM in its terminal carboxyl that is involved in virulence (42, 43). Rabies virus (RABV) strains with different G protein PBMs showed a spectrum of virulence regulated by the activation of different signaling pathways (44, 45).
Effects of E protein PBMs on host gene expression.
We identified host mRNA transcripts with expression significantly altered by infection with recombinant SARS-CoVs in which only the E protein PBM was modified. The introduction of the MERS-CoV PBM into SARS-CoV maintained expression of inflammatory response-related genes, particularly late in the infection, unlike the PBMs from other virulent viruses (Fig. 5C). For example, high levels of SLFN8, CCL3, CCR3, and CCRL2 were observed in addition to factors related to T-cell proliferation, leukocyte recruitment, eosinophil accumulation, and recruitment of effector immune cells (46–49). Increases in these factors could contribute to the cellular infiltrates observed in the lungs of mice infected with SARS-CoV mutants encoding the MERS-CoV PBM core domain.
We have shown that E protein PBM from SARS-CoV and SARS-CoV-2 reduces the transcript abundance for several genes involved in ion transport and cellular homeostasis (Fig. 6A), including CFTR (Fig. 6B and C).
Influence of CFTR on the lung pathology produced by SARS-CoV and SARS-CoV-2.
CFTR dysfunction in alveolar epithelial cells impairs alveolar fluid absorption by reducing Cl- permeability across the alveolar epithelium (23). CFTR is required to establish the osmotic gradient for fluid absorption from the alveolar space via aquaporin channels and paracellular pathways (20). Interactions between the SARS-CoV and SARS-CoV-2 E protein PBMs and CFTR may increase alveolar capillary and endothelium permeability, favoring edema formation. CFTR may function in an anti-inflammatory capacity (50–55). The membrane protein (M2) of influenza A has intrinsic ion channel activity and also inhibits CFTR during infection (56). Therefore, it is possible that CFTR inhibition could be a mechanism to induce respiratory virus pathogenesis. In fact, previous studies identified an important role for CFTR in ion and fluid transport across the alveolus. Adenovirus-mediated CFTR expression in rat lungs increased fluid absorption (57), whereas CFTR-deficient animals exhibit impaired alveolar fluid absorption in epithelial cells (58). We found that the CFTR modulators ivacaftor and lumacaftor reduced SARS-CoV-2 replication. Furthermore, infection with native SARS-CoV-2 virus reduced CFTR mRNA expression, while a deletion mutant without an E protein PBM did not (Fig. 7A and B) (59). Similar observations were reported using in vitro and in vivo models of influenza A virus infection (60). Influenza A virus infection reduced CFTR protein abundance and ion channel activity, and these changes were restored by CFTR modulators (60). Ivacaftor also improved alveolar liquid clearance in isolated pig lung lobes ex vivo and reduced edema in a volume overload in vivo pig model of hydrostatic pulmonary edema (61).
Loss of CFTR function also alters Ca2+ homeostasis and contributes to increased inflammatory responses in CF (62). Viruses can also disrupt Ca2+ homeostasis by forming membrane structures used for viral particle formation (63). Ivacaftor and lumacaftor normalize mitochondrial and intracellular Ca2+ levels and reduce oxidative stress and proinflammatory cytokines levels in CF lungs (64–66). The E protein PBM of SARS-CoV-2 may affect virus replication by altering Ca2+ homeostasis, leading to the reduction in SARS-CoV-2 titers observed in cells infected and treated with lumacaftor or ivacaftor (Fig. 8) (67–69). In fact, genes responsible for Ca2+ homeostasis, such as ATP2A3 and ATP2B2, also are underregulated due to the presence of E protein PBM from SARS-CoV-2 (Fig. 6C). Complementary studies have demonstrated that the use of Ca2+ channel blockers may reduce the mortality from SARS-CoV-2 in humans (70, 71).
In summary, loss of CFTR function in the setting of severe CoV infection may contribute to lung injury through multiple mechanisms including impaired fluid and electrolyte transport, altered Ca2+ homeostasis, and NLRP3 inflammasome activation. Ivacaftor and lumacaftor treatment partially reversed this pathology and improved survival in a mouse model of severe disease. FDA-approved CFTR modulators, now widely used for patients with cystic fibrosis, may also help to reduce morbidity and mortality due to high-pathogenicity human coronaviruses.
MATERIALS AND METHODS
Ethics statement.
All animal experimental protocols were approved by the NIH-CDC in accordance with US legislation, the Environmental Council of Madrid (permit number: PROEX 146.6/20), and the Ethical Committee of the Center for Animal Health Research (CISA-CSIC) (permit numbers: CBS 2014/005 and CEEA 2014/004), in strict accordance with Spanish National Royal Decree (RD 53/2013) and international European Union guidelines 2010/63/UE on the protection of animals used for experimentation and other scientific purposes and Spanish National law 32/2007 about animal welfare. All work with infected animals was performed in a biosafety level 3+ (BSL3+) laboratory of the Center for Animal Health Research (CISA-CSIC, Madrid, Spain). Infected mice were housed in a self-contained ventilated rack (Allentown, NJ).
Viruses.
Mouse-adapted SARS-CoV (MA15) (72) parental wild-type (wt) and recombinant viruses were rescued from infectious cDNA clones generated in a bacterial artificial chromosome (BAC). rSARS-CoV-PBM− was generated in our lab previously (15).
Generation of recombinant virus infectious clones.
The cDNA of SARS-CoV-MA15 assembled in a pBAC (pBAC-SARS-CoV-MA15) (73) was used to introduce the corresponding mutations affecting the E protein of SARS-CoV. To generate viruses with mutations in the SARS-CoV E protein carboxy terminus, DNA fragments including nucleotides 26017 to 26884 of the SARS-CoV-MA15 genome flanked by BamHI and XcmI restriction sites were generated using the oligonucleotides indicated in Table 1. These fragments carried different mutations located on the E gene generating amino acid changes at the E protein carboxy terminus: PBM-MERS, PBM-HKU1, and PBM-OC43. The fragments were digested with enzymes BamHI and XcmI and introduced into an intermediate plasmid pBAC-BamHI-RsrII-SARS-CoV containing nucleotides 26044 to 29783 of the SARS-CoV infectious clone. Next, these plasmids were digested with enzymes BamHI and RsrII, and the fragments containing the different mutations were inserted into plasmid pBAC-SARS-CoV-MA15 digested with the same enzymes, obtaining the respective infectious clones. The integrity of cDNA was verified by restriction analysis with the enzyme HindIII and by Sanger sequencing.
TABLE 1.
PCR primers used to engineer SARS-CoV-E-PBM mutants
| Virus/primer | Sequence (5′→3′) |
|---|---|
| PBM-MERS | |
| SARS-E-VS | CTCTTCAGGAGTTGCTAATCCAGCAATGG |
| SARS-E-PBM-MERS-RS | TTAGAACCATTCATCAGGAACTCCTTCAGAAGAGTTCAG |
| SARS-E-PBM-MERS-VS | TCTGAAGGAGTTCCTGATGAATGGTTCTAAACGAACTAACTATTA |
| SARS-26885-RS | GGTCCTTAATGTCACAGCGCCC |
| PBM-HKU1 | |
| SARS-E-VS | CTCTTCAGGAGTTGCTAATCCAGCAATGG |
| SARS-E-PBM-HKU1-RS | CGTTTAGATCAGATCATCAGGAACTCCTTCAGAAGAGTT |
| SARS-E-PBM-HKU1-VS | GAAGGAGTTCCTGATGATCTGATCTAAACGAACTAACTATTA |
| SARS-26885-RS | GGTCCTTAATGTCACAGCGCCC |
| PBM-OC43 | |
| SARS-E-VS | CTCTTCAGGAGTTGCTAATCCAGCAATGG |
| SARS-E-PBM-OC43-RS | TTAGACGTCATCAACAGGAACTCCTTCAGAAGAG |
| SARS-E-PBM-OC43-VS | TCTGAAGGAGTTCCTGTTGATGACGTCTAAACGAACTAACTATTA |
| SARS-26885-RS | GGTCCTTAATGTCACAGCGCCC |
| MERS-CoV PBM− | |
| SA27502VS | GCTTATCGTTTAAGCAGCTC |
| Rs mutPBM | CATATTAGACATTATGAAGGAGTTCGTTATCCTCCTCCTCCAGGTGGT |
| VS mutPBM | AACCCCCTCTACCACCTGGAGGAGGAGGATAACGAACTCCTTCATAAT |
| SA28319RS | TCTGTCGTAGTCACAAGCAC |
SARS-CoV-2 cDNA was assembled in a pBAC (pBAC-SARS-CoV-2) following previously described procedures (13, 74, 75). This infectious cDNA clone was used to introduce the corresponding mutations affecting the PBM of the SARS-CoV-2 E protein. First, a DNA fragment was obtained by chemical synthesis (GeneScript) that included nucleotides 26386 to 26751 of the SARS-CoV-2 genome, flanked by HpaI and AgeI restriction sites. This fragment carried the carboxy terminus mutation eliminating the PBM (DLLV → GGGG). The fragment digested with HpaI and AgeI was introduced into the corresponding sites of plasmid psL-F6-SARS-CoV-2, which contains nucleotides 25314 to 28610 of the SARS-CoV-2 genome, generating plasmid psL-F6-SARS-CoV-2-E-PBM−. Plasmid psL-F6-SARS-CoV-2-E-PBM− was then digested with restriction enzymes BamHI and AvrII and the resulting fragment was cloned into an intermediate plasmid pBAC-F6-SARS-CoV-2. Subsequently, this plasmid with was digested with BamHI and RsrII and the fragment containing the E gene mutation was inserted into the unique BamHI and RsrII cloning sites of the pBAC-SARS-CoV-2 plasmid to generate the corresponding cDNA infectious clone. The integrity of the cloned DNA was verified by restriction analysis with the enzyme EcoRI and by Sanger sequencing.
The cDNA infectious clone of MERS-CoV strain EMC/2012, assembled in pBAC (pBAC-SA-FL) (13), was used to generate the mutants with a modified MERS-CoV E protein PBM. First, DNA fragments including nucleotides 27502 to 28319 of the MERS-CoV genome flanked by KflI and Pfl23II restriction sites were generated by PCR using the oligonucleotides indicated in Table 1. The fragment obtained was introduced into an intermediate plasmid pBAC-SA-F6 including nucleotides 25841 to 30162 of the MERS-CoV genome using the restriction enzymes KflI and Pfl23II. The 25,841 to 30,162 region of the MERS-CoV genome is flanked by the PacI and RsrII unique restriction sites in plasmids pBAC-SA-FL and pBAC-SA-F6. The PacI-RsrII region of intermediate plasmid pBAC-SA-F6, with the desired E gene mutation, was cloned into pBAC-SA-FL to generate the complete infectious cDNA clone of MERS-CoV including E protein PBM mutations. All cloning steps were verified by Sanger sequencing.
Recovery of recombinant virus variants from cDNA clones.
Vero E6, Huh-7, and Vero E6-TMPRSS2 cells were grown to 95% confluence in 12.5 cm2 flasks and transfected with 6 μg of each infectious SARS-CoV, MERS-CoV, and SARS-CoV cDNA clones, and 18 μL of Lipofectamine 2000 (Invitrogen), according to the manufacturer’s specifications. At 6 h posttransfection the transfection medium was replaced by standard Dulbecco’s medium with 10% fetal calf serum and incubated at 37°C for 72 h. Cell supernatants were harvested and passaged once on fresh cells, and the recovered viruses were cloned by three rounds of plaque purification, following standard procedures for SARS-CoV. In the case of MERS-CoV and SARS-CoV-2, the supernatants were collected at 72 h posttransfection and titrated by plaque lysis formation in semisolid medium. Viruses were then amplified in 75 cm2 flasks, by infecting monolayers of the cells indicated for each virus at a multiplicity of infection (MOI) of 0.001. The supernatants were collected at 72 hpi and titrated. The 3′-end genome of different viruses, from the end of the replicase to the untranslated region, was sequenced. All sequences were compared to that of the parental wt virus sequence using SeqMan software (Lasergene, Madison, WI).
Growth kinetics.
Subconfluent monolayers (around 90% confluence) of Vero E6 and Huh-7 cells were infected at an MOI of 0.001 with parental viruses or the respective mutant viruses without E protein PBM. Culture supernatants were collected at 24, 48, and 72 hpi, and virus titers were determined as previously described (12).
Mice.
To determine mutant virus pathogenesis, SARS-CoV, MERS-CoV, and SARS-CoV-2 viruses were inoculated into the corresponding mice strains: 16-week-old BALB/c OlaHsd (Harlan), K18-hDPP4 (76), and K18-hACE2 (77) (Jackson Laboratories), respectively. Mice were anesthetized with isoflurane and inoculated intranasally using 100,000 or 10,000 PFU of the corresponding virus in a 50 μL volume of DMEM supplemented with 2% FBS. Manipulation of infected mice was carried out in a level 3+ biological containment laboratory at CISA-CSIC (Algete, Madrid) equipped with the required containment infrastructure for animal and cell cultures work. Scores for clinical signs of disease were determined based on the animal physical appearance and behavior. They were classified in five grades of severity (0, normal strength; 1, less active; 2, dull or sluggish movements, ruffled fur, squinting eyes; 3, ataxia, tremors; and 4, motionless, lying down, or hunched posture). Virus titrations were performed as previously described (12). Viral titers were expressed as PFU counts per gram of tissue.
Lung viral titers of infected mice.
Lungs from infected mice were harvested on different days postinfection. Lungs were homogenized in 2 mL of PBS containing 100 IU/mL penicillin, 0.1 mg/mL streptomycin, 50 μg/mL gentamicin, and 0.5 μg/mL amphotericin B (Fungizone), using gentleMACS Dissociator (Miltenyibiotec). Virus titers were determined as previously described (12).
Lung histopathology.
The entire left lung lobe was removed from each mouse and immersion-fixed in zinc formalin (Sigma-Aldrich) for 48 h. After the fixation period, samples were routinely processed and embedded in paraffin blocks that were then sectioned at a 4-μm thickness on a microtome, mounted onto glass slides, and routinely stained with hematoxylin and eosin (H&E). Lung sections were microscopically evaluated using an Olympus BX43 microscope by a single veterinary pathologist who was blinded to the identity of mice (Veterinary Pathology Department, Animal Health Research Center-CISA-CSIC, Valdeolmos, Spain). To assess the presence and severe histopathological lesions, lung inflammation parameters based on previous reports on SARS-CoV-2 infection in mouse models were used (78). The histopathological parameters evaluated were as follows: alveolar hemorrhage, alveolar edema, perivascular edema, alveolar septal thickening (interstitial pneumonia), inflammatory cell infiltration in alveoli, bronchi/bronchioles with epithelial necrosis, detached epithelium or inflammatory cells in the lumen (bronchitis/bronchiolitis), peribronchial/peribronchiolar and perivascular mononuclear infiltrates, and cytopathic effect in pneumocytes or syncytium formation. The histopathological parameters were graded following a semiquantitative scoring system as follows: 0, no lesion;1, minimal lesion; 2, mild lesion; 3, moderate lesion; and 4, severe lesion. The cumulative scores of histopathological lesions provided the total score per animal. In each experimental group, the individual scores were used to calculate the group average.
Extraction and analysis of RNA.
Cell culture or lung RNA samples were collected and incubated in RNAlater reagent (Ambion). To extract total RNA, lungs were homogenized in 2 mL of RLT lysis buffer (Qiagen) supplemented with 1% β-mercaptoethanol using the GentleMACS Dissociator system (Miltenyi Biotec) following the protocol proposed by the manufacturer. RNA was purified from supernatants using the rNeasy minikit reagent (Qiagen), as previously described. To generate cDNAs from the purified RNAs, a reverse transcription (RT) reaction was carried out using the High Capacity cDNA RT kit reagent (Applied Biosystems). To verify the sequence of the mutations introduced into the viral genome, 4 μL of the cDNA generated in the RT reaction was used as a template in a PCR using the enzyme Taq polymerase (Invitrogen). Cellular mRNA expression was analyzed by quantitative reverse transcription-quantitative PCR (RT-qPCR), using TaqMan technology with commercial probes (ThermoFisher Scientific) (Table 2). In all cases, the reaction was performed with the FastStart Universal Probe Master reagent (Rox) (Roche). qPCRs were performed in a 7500 Real Time PCR System (ThermoFisher Scientific). qPCR data were analyzed using the 7500 software v2.0.6 (ThermoFisher Scientific). All experiments met the recommendations for analysis of gene expression by RT-qPCR (MIQE) (79). Each result is the average of three independent experiments in which each sample was tested in triplicate. The relative quantification of gene expression was performed from the mean values of CT (cycle in which the amplification curve cuts at the threshold level using the 2−ΔΔCT method) (80). The level of 18S rRNA was used as an endogenous control to normalize cellular mRNA levels in mouse lungs, while hydroxymethylbilane synthase (HMBS) was used to normalize cellular mRNA levels in cell cultures.
TABLE 2.
TaqMan probes used to quantify cellular mRNAs by RT-qPCR
| Gene | TaqMan assay ID |
|---|---|
| mCFTR | Mm00445197_m1 |
| hCFTR | Hs00357011_m1 |
| mNLRP3 | Mm00840904_m1 |
| mTNF-α | Mm00443258_m1 |
| mIL-6 | Mm00446190_m1 |
| mIL-1β | Mm01336189_m1 |
| mCXCL10 | Mm00445235_m1 |
| mCXCL2 | Mm00436450_m1 |
| 18S rRNA | Mm03928990_g1 |
| HMBS | Hs00609297_m1 |
Deep sequencing of mRNAs.
Total RNA extracted from the lungs of infected mice was used for rNaseq sequencing. The samples were digested with the DNAsaI (Roche) to eliminate possible DNA contamination. RNA was processed to remove rRNA, a major component in large RNA samples, and enrich the resulting sequences for less abundant expressed RNAs such as mRNAs. RNA libraries were constructed (Illumina method) and 300 nt paired-end reads were obtained in which the complementary strand was marked to distinguish the original sense of the sequences. Three replicates per variable and per day were sequenced. From each sample, 80 million sequences were obtained.
Genomic alignment of Illumina paired-end reads.
Paired-end reads in FASTQ format were quality checked with FASTQC (www.bioinformatics.babraham.ac.uk/projects/fastqc; 2021). As no limitations were detected for each sample, all unprocessed reads were aligned to the mouse genome (GRCm38, primary assembly) using RNA-STAR (81) (–alignIntronMax 1000000 –alignMatesGapMax 1000000). Duplicated reads (i.e., those aligned at identical genomic positions) were marked using MarkDuplicates function of picard-tools (https://broadinstitute.github.io/picard/; 2021). Alignment files in SAM format were compressed, sorted, and indexed with corresponding commands of Samtools package (82). The IGV browser (83) was used to visualize alignment results.
Quantification of mouse genes and differential expression determination.
All mouse genes (ENSEMBL release 101) were quantified using featureCounts function of bioconductor package Rsubread (84) (ignoreDup=TRUE, requiredBothEndsMapped=TRUE, primaryOnly=TRUE). To calculate differential expression between pairs of replicated samples, DESeq and lfcShrink functions of bioconductor package DESeq2 (85) were used (cooksCutoff=FALSE, independentFiltering=TRUE, type=”normal”). Differential expression results, including adjusted P values by FDR (86), were stored as text-tabulated files and converted to FIESTA format (https://bioinfogp.cnb.csic.es/tools/FIESTA; 2021) to facilitate the visualization of overexpressed and underexpressed genes in form of MA plots. For each comparison, genes with |logRatio| ≥ 1.5 and FDR ≤ 0.05 were considered differentially expressed.
Functional analysis of relevant genes.
The DAVID tool (87) was used to determine overrepresentation of Gene Ontology terms (Biological Process) in differentially expressed genes (both overexpressed and underexpressed). The six most representative biological processes were selected based on the P value.
Treatment of mice with CFTR modulators.
Mice (n = 11) were treated by intraperitoneal injection administration with an appropriate concentration of compounds 24 h before infection with 10,000 PFU of SARS-CoV-2. Mock-infected or infected animals were treated once daily with 12 or 6 mg/kg of Lumacaftor or Ivacaftor, respectively. All treatments ceased after day 8 postinfection. Three mice from each group were euthanized at 3 and 6 dpi to analyze virus production in the lungs.
Cell viability assay.
To determine the cytotoxicity of commercial compounds that modulate CFTR protein activity, cells were incubated with different concentrations (1 to 50 μM) of ivacaftor (VX-770, Merck) or lumacaftor (VX-809; Selleck Chemicals) and cell viability was determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazol bromide] reduction assay (88).
Statistical analysis.
Two-tailed, unpaired Student’s t tests were used to analyze the differences in mean values between groups. The statistical significances are indicated in the figures as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ACKNOWLEDGMENTS
This work was supported by grants from the Government of Spain (BIO2016-75549-R; PID2019-107001RB-I00 AEI/FEDER, UE; SEV 2017-0712 and PIE_INTRAMURAL_LINEA 1-202020E079), CSIC (PIE_INTRAMURAL-202020E043), the European Zoonotic Anticipation and Preparedness Initiative (ZAPI) (IMI_JU_115760), the European Commission (H2020-SC1-2019, ISOLDA Project No. 848166-2), and the U.S. National Institutes of Health (NIH) (2P01AI060699). J.M.H. received a contract from Comunidad de Madrid (Y2020/BIO-6576, COVID-PREclinical-MODels-CM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. In vivo experiments were performed at INIA-CISA (Madrid, Spain).
We thank Marga Gonzalez (CNB-CSIC) for technical assistance.
Footnotes
This article is a direct contribution from Luis Enjuanes, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Shinji Makino, The University of Texas Medical Branch, and Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai.
Contributor Information
Luis Enjuanes, Email: l.enjuanes@cnb.csic.es.
Kanta Subbarao, NIAID, NIH.
REFERENCES
- 1.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garbino J, Crespo S, Aubert JD, Rochat T, Ninet B, Deffernez C, Wunderli W, Pache JC, Soccal PM, Kaiser L. 2006. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (Non-SARS)-related human coronavirus infection. Clin Infect Dis 43:1009–1015. doi: 10.1086/507898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh K, Kapikian AZ, Hardison KA, Hartley JW, Chanock RM. 1969. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J Immunol 102:1109–1118. [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen M-H, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Günther S, Osterhaus ADME, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Bermingham A, Chand M, Brown C, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody R, Green H, Boddington N, Gopal R, Price N, Newsholme W, Drosten C, Fouchier R, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 17:20290. [PubMed] [Google Scholar]
- 7.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z, Lian X, Su X, Wu W, Marraro GA, Zeng Y. 2020. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res 21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA 102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menachery VD, Yount BL, Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menachery VD, Yount BL, Jr, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci USA 113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeDiego ML, Alvarez E, Almazan F, Rejas MT, Lamirande E, Roberts A, Shieh WJ, Zaki SR, Subbarao K, Enjuanes L. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almazan F, DeDiego ML, Sola I, Zuñiga S, Nieto-Torres JL, Marquez-Jurado S, Andres G, Enjuanes L. 2013. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4:e00650-13. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez-Alvarez J, Honrubia JM, Fernandez-Delgado R, Wang L, Castano-Rodriguez C, Zuniga S, Sola I, Enjuanes L. 2021. Genetically engineered live-attenuated Middle East Respiratory Syndrome Coronavirus viruses confer full protection against lethal infection. mBio 12:e00103-21. doi: 10.1128/mBio.00103-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, Enjuanes L. 2014. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein Is a determinant of viral pathogenesis. PLoS Pathog 10:e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieto-Torres JL, Dediego ML, Verdia-Baguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, Alcaraz A, Torres J, Aguilella VM, Enjuanes L. 2014. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog 10:e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruch TR, Machamer CE. 2012. The coronavirus E protein: assembly and beyond. Viruses 4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luck K, Charbonnier S, Trave G. 2012. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett 586:2648–2661. doi: 10.1016/j.febslet.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Javier RT, Rice AP. 2011. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J Virol 85:11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthay MA. 2014. Resolution of pulmonary edema. Thirty years of progress. Am J Respir Crit Care Med 189:1301–1308. doi: 10.1164/rccm.201403-0535OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthay MA, Folkesson HG, Clerici C. 2002. Lung epithelial fluid transport and the resolution of pulmonary edema. Physiol Rev 82:569–600. doi: 10.1152/physrev.00003.2002. [DOI] [PubMed] [Google Scholar]
- 22.Matthay MA, Ware LB, Zimmerman GA. 2012. The acute respiratory distress syndrome. J Clin Invest 122:2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saint-Criq V, Gray MA. 2017. Role of CFTR in epithelial physiology. Cell Mol Life Sci 74:93–115. doi: 10.1007/s00018-016-2391-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiedorczuk K, Chen J. 2022. Mechanism of CFTR correction by type I folding correctors. Cell 185:158–168.e11. doi: 10.1016/j.cell.2021.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J, Olson ER, Wine JJ, Frizzell RA, Ashlock M, Negulescu PA. 2011. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA 108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anonymous . 2018. Clinical Review Report: lumacaftor/Ivacaftor (Orkambi): (Vertex Pharmaceuticals (Canada) Incorporated): indication: for the treatment of cystic fibrosis in patients aged six years and older who are homozygous for the F508del mutation in the cystic fibrosis transmembrane conductance regulator gene. Canadian Agency for Drugs and Technologies in Health, Ottawa, Canada. [PubMed] [Google Scholar]
- 27.Accurso FJ, Rowe SM, Clancy JP, Boyle MP, Dunitz JM, Durie PR, Sagel SD, Hornick DB, Konstan MW, Donaldson SH, Moss RB, Pilewski JM, Rubenstein RC, Uluer AZ, Aitken ML, Freedman SD, Rose LM, Mayer-Hamblett N, Dong Q, Zha J, Stone AJ, Olson ER, Ordonez CL, Campbell PW, Ashlock MA, Ramsey BW. 2010. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N Engl J Med 363:1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones AM, Helm JM. 2009. Emerging treatments in cystic fibrosis. Drugs 69:1903–1910. doi: 10.2165/11318500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 29.Jih KY, Hwang TC. 2013. Vx-770 potentiates CFTR function by promoting decoupling between the gating cycle and ATP hydrolysis cycle. Proc Natl Acad Sci USA 110:4404–4409. doi: 10.1073/pnas.1215982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckford PD, Li C, Ramjeesingh M, Bear CE. 2012. Cystic fibrosis transmembrane conductance regulator (CFTR) potentiator VX-770 (ivacaftor) opens the defective channel gate of mutant CFTR in a phosphorylation-dependent but ATP-independent manner. J Biol Chem 287:36639–36649. doi: 10.1074/jbc.M112.393637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csanady L, Torocsik B. 2019. Cystic fibrosis drug ivacaftor stimulates CFTR channels at picomolar concentrations. Elife 8:e46450. doi: 10.7554/eLife.46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Cai Z, Gosling M, Sheppard DN. 2018. Potentiation of the cystic fibrosis transmembrane conductance regulator Cl− channel by ivacaftor is temperature independent. Am J Physiol Lung Cell Mol Physiol 315:L846–L857. doi: 10.1152/ajplung.00235.2018. [DOI] [PubMed] [Google Scholar]
- 33.Xue X, Mutyam V, Thakerar A, Mobley J, Bridges RJ, Rowe SM, Keeling KM, Bedwell DM. 2017. Identification of the amino acids inserted during suppression of CFTR nonsense mutations and determination of their functional consequences. Hum Mol Genet 26:3116–3129. doi: 10.1093/hmg/ddx196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yalcinkaya M, Liu W, Islam MN, Kotini AG, Gusarova GA, Fidler TP, Papapetrou EP, Bhattacharya J, Wang N, Tall AR. 2021. Modulation of the NLRP3 inflammasome by Sars-CoV-2 Envelope protein. Sci Rep 11:24432. doi: 10.1038/s41598-021-04133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarosz-Griffiths HH, Scambler T, Wong CH, Lara-Reyna S, Holbrook J, Martinon F, Savic S, Whitaker P, Etherington C, Spoletini G, Clifton I, Mehta A, McDermott MF, Peckham D. 2020. Different CFTR modulator combinations downregulate inflammation differently in cystic fibrosis. Elife 9:e54556. doi: 10.7554/eLife.54556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boro M, Balaji KN. 2017. CXCL1 and CXCL2 regulate NLRP3 Inflammasome activation via G-protein-coupled receptor CXCR2. J Immunol 199:1660–1671. doi: 10.4049/jimmunol.1700129. [DOI] [PubMed] [Google Scholar]
- 37.Freeman TL, Swartz TH. 2020. Targeting the NLRP3 inflammasome in severe COVID-19. Front Immunol 11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng J, Xie X, Feng XL, Xu L, Han JB, Yu D, Zou QC, Liu Q, Li X, Ma G, Li MH, Yao YG. 2022. Specific inhibition of the NLRP3 inflammasome suppresses immune overactivation and alleviates COVID-19 like pathology in mice. EBioMedicine 75:103803. doi: 10.1016/j.ebiom.2021.103803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopp BT, Fitch J, Jaramillo L, Shrestha CL, Robledo-Avila F, Zhang S, Palacios S, Woodley F, Hayes D, Jr, Partida-Sanchez S, Ramilo O, White P, Mejias A. 2020. Whole-blood transcriptomic responses to lumacaftor/ivacaftor therapy in cystic fibrosis. J Cyst Fibros 19:245–254. doi: 10.1016/j.jcf.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanda MK, Cebotaru L. 2021. VX-809 mitigates disease in a mouse model of autosomal dominant polycystic kidney disease bearing the R3277C human mutation. FASEB J 35:e21987. doi: 10.1096/fj.202101315r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruffin M, Roussel L, Maille E, Rousseau S, Brochiero E. 2018. Vx-809/Vx-770 treatment reduces inflammatory response to Pseudomonas aeruginosa in primary differentiated cystic fibrosis bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 314:L635–L641. doi: 10.1152/ajplung.00198.2017. [DOI] [PubMed] [Google Scholar]
- 42.Golebiewski L, Liu H, Javier RT, Rice AP. 2011. The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions. J Virol 85:10639–10648. doi: 10.1128/JVI.05070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 44.Caillet-Saguy C, Maisonneuve P, Delhommel F, Terrien E, Babault N, Lafon M, Cordier F, Wolff N. 2015. Strategies to interfere with PDZ-mediated interactions in neurons: what we can learn from the rabies virus. Prog Biophys Mol Biol 119:53–59. doi: 10.1016/j.pbiomolbio.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Prehaud C, Wolff N, Terrien E, Lafage M, Megret F, Babault N, Cordier F, Tan GS, Maitrepierre E, Menager P, Chopy D, Hoos S, England P, Delepierre M, Schnell MJ, Buc H, Lafon M. 2010. Attenuation of rabies virulence: takeover by the cytoplasmic domain of its envelope protein. Sci Signal 3:ra5. doi: 10.1126/scisignal.2000510. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa K, Narayanan K, Wada M, Makino S. 2018. Inhibition of stress granule formation by middle east respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol 92:e00902-18. doi: 10.1128/JVI.00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagome K, Nagata M. 2020. Possible mechanisms of eosinophil accumulation in eosinophilic pneumonia. Biomolecules 10:638. doi: 10.3390/biom10040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Prete A, Sozio F, Schioppa T, Ponzetta A, Vermi W, Calza S, Bugatti M, Salvi V, Bernardini G, Benvenuti F, Vecchi A, Bottazzi B, Mantovani A, Sozzani S. 2019. The Atypical Receptor CCRL2 Is Essential for Lung Cancer Immune Surveillance. Cancer Immunol Res 7:1775–1788. doi: 10.1158/2326-6066.CIR-19-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin W, Li Y, Song Y, Zhang J, Wu C, Chen Y, Miao Y, Lin C, Lin Y, Yan D, Chen J, He R. 2021. CCRL2 promotes antitumor T-cell immunity via amplifying TLR4-mediated immunostimulatory macrophage activation. Proc Natl Acad Sci USA 118:e2024171118. doi: 10.1073/pnas.2024171118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng HP, Jennings S, Wellems D, Sun F, Xu J, Nauseef WM, Wang G. 2020. Myeloid CFTR loss-of-function causes persistent neutrophilic inflammation in cystic fibrosis. J Leukoc Biol 108:1777–1785. doi: 10.1002/JLB.3A0520-193RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adib-Conquy M, Pedron T, Petit-Bertron AF, Tabary O, Corvol H, Jacquot J, Clément A, Cavaillon JM. 2008. Neutrophils in cystic fibrosis display a distinct gene expression pattern. Mol Med 14:36–44. doi: 10.2119/2007-00081.Adib-Conquy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lara-Reyna S, Holbrook J, Jarosz-Griffiths HH, Peckham D, McDermott MF. 2020. Dysregulated signalling pathways in innate immune cells with cystic fibrosis mutations. Cell Mol Life Sci 77:4485–4503. doi: 10.1007/s00018-020-03540-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Shrestha CL, Kopp BT. 2018. Cystic fibrosis transmembrane conductance regulator (CFTR) modulators have differential effects on cystic fibrosis macrophage function. Sci Rep 8:17066. doi: 10.1038/s41598-018-35151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonfield TL, Hodges CA, Cotton CU, Drumm ML. 2012. Absence of the cystic fibrosis transmembrane regulator (Cftr) from myeloid-derived cells slows resolution of inflammation and infection. J Leukoc Biol 92:1111–1122. doi: 10.1189/jlb.0412188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Song K, Painter RG, Aiken M, Reiser J, Stanton BA, Nauseef WM, Wang G. 2013. Cystic fibrosis transmembrane conductance regulator recruitment to phagosomes in neutrophils. J Innate Immun 5:219–230. doi: 10.1159/000346568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Londino JD, Lazrak A, Noah JW, Aggarwal S, Bali V, Woodworth BA, Bebok Z, Matalon S. 2015. Influenza virus M2 targets cystic fibrosis transmembrane conductance regulator for lysosomal degradation during viral infection. FASEB J 29:2712–2725. doi: 10.1096/fj.14-268755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mutlu GM, Adir Y, Jameel M, Akhmedov AT, Welch L, Dumasius V, Meng FJ, Zabner J, Koenig C, Lewis ER, Balagani R, Traver G, Sznajder JI, Factor P. 2005. Interdependency of beta-adrenergic receptors and CFTR in regulation of alveolar active Na+ transport. Circ Res 96:999–1005. doi: 10.1161/01.RES.0000164554.21993.AC. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Comellas AP, Karp PH, Ernst SE, Moninger TO, Gansemer ND, Taft PJ, Pezzulo AA, Rector MV, Rossen N, Stoltz DA, McCray PB, Jr, Welsh MJ, Zabner J. 2012. CFTR is required for maximal transepithelial liquid transport in pig alveolar epithelia. Am J Physiol Lung Cell Mol Physiol 303:L152–L160. doi: 10.1152/ajplung.00116.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morales L, Oliveros JC, Enjuanes L, Sola I. 2022. Contribution of host miRNA-223-3p to SARS-CoV-induced lung inflammatory pathology. mBio 13:e0313521. doi: 10.1128/mbio.03135-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brand JD, Lazrak A, Trombley JE, Shei RJ, Adewale AT, Tipper JL, Yu Z, Ashtekar AR, Rowe SM, Matalon S, Harrod KS. 2018. Influenza-mediated reduction of lung epithelial ion channel activity leads to dysregulated pulmonary fluid homeostasis. JCI Insight 3:e123467. doi: 10.1172/jci.insight.123467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X, Vargas Buonfiglio LG, Adam RJ, Stoltz DA, Zabner J, Comellas AP. 2017. Cystic fibrosis transmembrane conductance regulator potentiation as a therapeutic strategy for pulmonary edema: a proof-of-concept study in pigs. Crit Care Med 45:e1240–e1246. doi: 10.1097/CCM.0000000000002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rimessi A, Vitto VAM, Patergnani S, Pinton P. 2021. Update on calcium signaling in cystic fibrosis lung disease. Front Pharmacol 12:581645. doi: 10.3389/fphar.2021.581645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Frey TK, Yang JJ. 2009. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium 46:1–17. doi: 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Philippe R, Antigny F, Buscaglia P, Norez C, Becq F, Frieden M, Mignen O. 2015. SERCA and PMCA pumps contribute to the deregulation of Ca2+ homeostasis in human CF epithelial cells. Biochim Biophys Acta 1853:892–903. doi: 10.1016/j.bbamcr.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Rimessi A, Bezzerri V, Patergnani S, Marchi S, Cabrini G, Pinton P. 2015. Mitochondrial Ca2+-dependent NLRP3 activation exacerbates the Pseudomonas aeruginosa-driven inflammatory response in cystic fibrosis. Nat Commun 6:6201. doi: 10.1038/ncomms7201. [DOI] [PubMed] [Google Scholar]
- 66.Vachel L, Norez C, Becq F, Vandebrouck C. 2013. Effect of VX-770 (ivacaftor) and OAG on Ca2+ influx and CFTR activity in G551D and F508del-CFTR expressing cells. J Cyst Fibros 12:584–591. doi: 10.1016/j.jcf.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 67.Straus MR, Tang T, Lai AL, Flegel A, Bidon M, Freed JH, Daniel S, Whittaker GR. 2020. Ca(2+) ions promote fusion of middle east respiratory syndrome coronavirus with host cells and increase infectivity. J Virol 94:e00426-20. doi: 10.1128/JVI.00426-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai AL, Millet JK, Daniel S, Freed JH, Whittaker GR. 2017. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J Mol Biol 429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, Takvorian PM, Bleck C, Hsu VW, Fehr AR, Perlman S, Achar SR, Straus MR, Whittaker GR, de Haan CAM, Kehrl J, Altan-Bonnet G, Altan-Bonnet N. 2020. beta-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang LK, Sun Y, Zeng H, Wang Q, Jiang X, Shang WJ, Wu Y, Li S, Zhang YL, Hao ZN, Chen H, Jin R, Liu W, Li H, Peng K, Xiao G. 2021. Author Correction: calcium channel blocker amlodipine besylate therapy is associated with reduced case fatality rate of COVID-19 patients with hypertension. Cell Discov 7:29. doi: 10.1038/s41421-021-00267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Solaimanzadeh I. 2020. Nifedipine and amlodipine are associated with improved mortality and decreased risk for intubation and mechanical ventilation in elderly patients hospitalized for COVID-19. Cureus 12:e8069. doi: 10.7759/cureus.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts A, Deming D, Paddock CD, Cheng A, Yount B, Vogel L, Herman BD, Sheahan T, Heise M, Genrich GL, Zaki SR, Baric R, Subbarao K. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fett C, DeDiego ML, Regla-Nava JA, Enjuanes L, Perlman S. 2013. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J Virol 87:6551–6559. doi: 10.1128/JVI.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gutiérrez-Álvarez J, Honrubia JM, Sanz-Bravo A, González-Miranda E, Fernández-Delgado R, Rejas MT, Zúñiga S, Sola I, Enjuanes L. 2021. Middle East respiratory syndrome coronavirus vaccine based on a propagation-defective RNA replicon elicited sterilizing immunity in mice. Proc Natl Acad Sci USA 118:e2111075118. doi: 10.1073/pnas.2111075118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong LR, Zheng J, Wilhelmsen K, Li K, Ortiz ME, Schnicker NJ, Thurman A, Pezzulo AA, Szachowicz PJ, Li P, Pan R, Klumpp K, Aswad F, Rebo J, Narumiya S, Murakami M, Zuniga S, Sola I, Enjuanes L, Meyerholz DK, Fortney K, McCray PB, Jr, Perlman S. 2022. Eicosanoid signaling blockade protects middle-aged mice from severe COVID-19. Nature 605:146–151. doi: 10.1038/s41586-022-04630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, Jr, Baric RS, Enjuanes L, Gallagher T, McCray PB, Jr, Perlman S. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA 111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCray PB, Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun J, Zhuang Z, Zheng J, Li K, Wong RL, Liu D, Huang J, He J, Zhu A, Zhao J, Li X, Xi Y, Chen R, Alshukairi AN, Chen Z, Zhang Z, Chen C, Huang X, Li F, Lai X, Chen D, Wen L, Zhuo J, Zhang Y, Wang Y, Huang S, Dai J, Shi Y, Zheng K, Leidinger MR, Chen J, Li Y, Zhong N, Meyerholz DK, McCray PB, Jr, Perlman S, Zhao J. 2020. Generation of a broadly useful model for covid-19 pathogenesis, vaccination, and treatment. Cell 182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 80.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 81.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 87.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. 2022. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth kinetics of SARS-CoV E protein PBM mutants. (A) Subconfluent Vero E6 were infected with SARS-CoV E protein PBM mutants at a MOI of 0.001. The cells infected with different virus mutants are indicated with lines of different color: native viruses (wt) are shown with red lines and the mutants without E protein PBM (PBM−) with pink lines and symbols. PBM-MERS-CoV (orange lines), PBM-hCoV-OC43 (green lines), and PBM-hCoV-HKU1 (blue lines). Supernatants were collected at 24, 48, and 72 hpi and titrated by the lysis plate formation method. (B) Groups of six 16-week-old BALB/c mice were intranasally inoculated with 100,000 pfu of the native viruses (red columns), or PBM− (pink column), PBM-MERS-CoV (orange column), PBM-OC43 (green column), and PBM-HKU1 (blue column). Three mice from each group were euthanized at 2 and 4 dpi to analyze virus production in lungs. Vertical bars represent the standard error of the mean titers obtained by repeating the experiment three times. Statistically significant data are represented according to the P value obtained in Student’s t analysis: **, P < 0.01. Download FIG S1, TIF file, 0.3 MB (325.1KB, tif) .
Copyright © 2023 Honrubia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth kinetics of SARS-CoV-2 in Calu-3 2B4. Subconfluent Calu-3 2B4 cells were infected with SARS-CoV-2 parental virus (red line) at a MOI of 0.1. Supernatants were collected at 24, 48 and 72 hpi and titrated. Vertical bars represent the standard error of the mean. Download FIG S2, TIF file, 0.1 MB (120.1KB, tif) .
Copyright © 2023 Honrubia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Viability of Calu-3 2B4 cells in the presence of different CFTR protein modulators. Subconfluent and uninfected Calu-3 2B4 cells were incubated with different concentrations (1 to 50 μM) of ivacaftor (A) and lumacaftor (B) and with the combination of ivacaftor (5 μM) and lumacaftor (50 μM) (green column) (C). Supernatants were collected at 48 hpi and cell viability was determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazole bromide] reduction assay (88). Mean values of cell viability and their standard deviations are shown. Download FIG S3, TIF file, 0.2 MB (205.2KB, tif) .
Copyright © 2023 Honrubia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.