Abstract
Activation of scramblases is one of the mechanisms that regulate the exposure of phosphatidylserine to the cell surface, a process that plays an important role in tumor immunosuppression. Here we show that chemotherapeutic agents induce overexpression of Xkr8, a scramblase activated during apoptosis, at the transcriptional level in cancer cells, both in vitro and in vivo. Given this finding, we developed a nanocarrier for co-delivery of Xkr8 siRNA and the FuOXP pro-drug to tumors. Intravenous injection of our nanocarrier led to significant inhibition of tumor growth in colon and pancreatic cancer models along with increased antitumor immune response. Targeting Xkr8 in combination with chemotherapy may represent a novel strategy for the treatment of various types of cancers.
Editor Summary:
Downregulation of specific proteins named scramblases might enhance tumour immunosuppression. In this paper the authors first show that the scramblase Xrk8 is overexpressed in tumour cells upon treatment with chemotherapeutics, and then developed a nanomedicine platform for co-delivery of a cancer pro-drug and of a siRNA directed against the Xrk8 gene, showing therapeutic effect and enhanced immune response in animal tumour models.
Phosphatidylserine (PS) has long been implicated in cancer immunosuppression1,2. As an important component of membrane structure, PS is confined to the inner leaflet and this asymmetric distribution is maintained by the action of P4-ATPases such as ATP11A and 11C, which function as flippases in the plasma membrane to actively translocate PS from the outer leaflet to the inner leaflet3. Scramblases collapse membrane asymmetry thereby randomizing all phospholipid species between leaflets, which effectively increases the accumulation of PS on the external side of the membrane4,5. As an important scramblase, Xkr8 carries a caspase 3 recognition site in its C-terminal region3,5. It is generally believed that Xkr8 is regulated post-transcriptionally and is activated by caspases during apoptosis6,7. Meanwhile flippases are inactivated by caspases, resulting in irreversible exposure of PS on the cell surface8.
Macrophages engulf apoptotic cells but not living cells and PS serves as an important “eat me” signal9. The engagement of PS on apoptotic cells with its receptors on immune cells leads to profound immunosuppression and this evolutionally conserved mechanism is critical for the silent clearance of apoptotic cells in normal animal development10. PS also functions as an upstream immune checkpoint that contributes to the immunosuppressed tumor microenvironment by preventing immune reactions11.
The discovery of PS receptors and the underlying signaling suggests a new strategy for cancer immunotherapy through blocking the immunosuppressive pathway12. Blockade of PS via annexin A5 (Annexin V) or PS-specific antibody (Ab) is another attractive strategy1. Thorpe’s group has developed a family of PS-targeting antibodies13. Unfortunately, despite the demonstrated efficacy and safety in preclinical and phase I/II clinical studies, a larger phase III trial failed to demonstrate benefit over chemotherapy alone14. Another concern is targeting of the antibody to other PS-expressing cells under physiological conditions and/or resulting from comorbid diseases such as cardiovascular diseases, especially in combination with systemic chemotherapy. Strategies that specifically target PS on tumor cells but do not affect PS on normal cells15, and those that prevent or decrease the quantity of PS exposure when in combination with other anticancer drugs, may represent a more attractive approach.
In this study we report that treatment of tumors with chemotherapeutic drugs led to significant upregulation of Xkr8 expression in vitro and in vivo, suggesting that Xkr8 may serve as a novel therapeutic target for cancer treatment. We have further shown that codelivery of Xkr8 siRNA (siXkr8) and a chemotherapeutic agent using a new nanocarrier led to significant inhibition of tumor growth and enhanced antitumor immunity in animal tumor models,
Xkr8 was induced by chemotherapeutic agents in vitro and in vivo
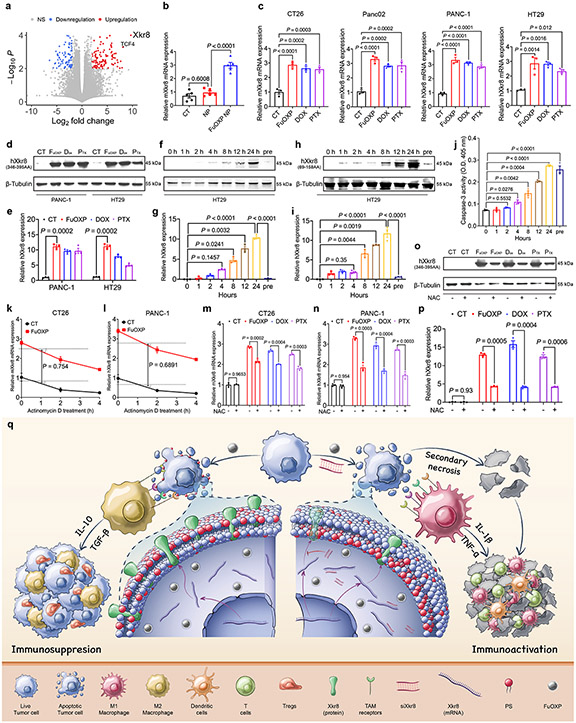
In an independent study to define the mechanism of chemoresistance in colorectal cancer (CRC), RNA-seq was conducted to examine changes in gene expression profile after treatment of CT26 tumor-bearing mice with FuOXP, a prodrug conjugate of 5-FU and oxoplatin16. As shown in Fig. 1a, murine Xkr8 (mXkr8) was one of the top genes whose mRNA expression was significantly upregulated. Induction of Xkr8 mRNA expression in vivo was further confirmed by qRT-PCR although the fold of increase was much lower in qRT-PCR (Fig. 1b). Induction of Xkr8 mRNA expression by FuOXP as well as two other drugs (doxorubicin – DOX, and paclitaxel – PTX) was also demonstrated in cultured murine and human CRC (CT26 and HT-29) and pancreatic cancer (PCa) (Panc02 and PANC-1) cell lines, respectively (Fig. 1c & Suppl Fig. 1). We further confirmed induction of the human Xkr8 (hXkr8) at the protein level in human cancer cell lines (Fig. 1d-e & Suppl Fig. 2a). Xkr8 induction appeared to occur earlier than the activation of caspase 3 after FuOXP treatment followed by the caspase-mediated cleavage Fig. 1f-j & Suppl Fig. 2b-c).
Fig. 1: Xkr8 was induced by chemotherapeutic agents in vitro and in vivo.
a: Volcano plot for the RNA-seq analysis of CT26 tumors treated with FuOXP NPs versus empty NPs. Statistical analysis of RNA-seq was performed by two-tailed Student’s t-test for comparison using Cuffdiff of Cufflinks. b: qRT-PCR analysis of mXkr8 mRNA expression in CT26 tumors treated with FuOXP NPs or empty NPs (normalized against GAPDH). N = 5 tumors per group. c: qRT-PCR analysis of Xkr8 mRNA expression in various types of tumor cells at 24 h following various treatments. N = 3 replicates. d: Western analysis of hXkr8 protein expression in PANC-1 or HT29 cells using anti-hXkr8 (346-395AA) antibody that detects only full-length hXkr8 at 24 h following various treatments. hXkr8 MW: 45 kDa, β-Tubulin MW: 55 kDa. e: Densitometry analysis of protein bands in d. f & h: Western blot analysis of hXkr8 protein expression in HT29 cells using anti-hXkr8 (346-395AA) antibody (f) or anti-hXkr8 (69-158AA) antibody that detects both the full-length and truncated (caspase 3-cleaved) hXkr8 (h). Cells were treated with FuOXP for various times or pre-treated with siXkr8 for 72 h followed by 24 h of FuOXP treatment. g & i: Densitometry analysis of protein bands in f (g)) or h (i). j: Caspase-3 activity in HT29 cells following various treatments as described in f. N= 3 replicates. k & l: Effect of actinomycin D on the FuOXP-induced Xkr8 mRNA expression levels over time in CT26 (k) and PANC-1 (l) cells. N= 3 replicates. m~p: Effect of NAC on drug-induced changes in the expression levels of Xkr8 mRNA in CT26 (m) and PANC-1(n), and Xkr8 protein in PANC-1 cells (o & p). p: Densitometry analysis of protein bands in Western blot (o). q: Proposed strategy of reversing chemotherapy drug-induced Xkr8 induction and immunosuppression through in situ codelivery with siXkr8. Data are presented as mean ± SEM and statistical analysis was performed by two-tailed Student’s t-test for comparison in k and l and one-way analysis of variance (ANOVA) with Tukey post hoc test for comparison in b, c, e, g, i, j, m, n, and p. Data are representative of 2 independent experiments in b, j, k, l, m and n, and 3 independent experiments in c-i, o and p.
Fig. 1k shows the kinetics of mXkr8 mRNA levels in CT26 cells following 12 h of FuOXP treatment followed by another 2 h of treatment with actinomycin D (ActD), a transcription inhibitor. Cells with or without ActD co-treatment showed similar kinetics of decline in the level of mXkr8 mRNA, implying that FuOXP did not affect the rate of mXkr8 mRNA degradation. Similar results were observed in PANC-1 cells (Fig. 1l), suggesting that FuOXP caused Xkr8 induction through enhanced gene transcription. Furthermore, pretreatment with the antioxidant N-acetylcysteine (NAC) partially attenuated the Xkr8 induction by the 3 drugs in CT26 (Fig. 1m) and PANC-1 cells (Fig. 1n-p) at both mRNA (Fig. 1m-n) and protein (Fig. 1o-p & Suppl Fig. 2d) levels, suggesting a role of oxidative stress in Xkr8 induction.
Characterization of PMBOP-CP nanocarrier for codelivery of siXkr8 and FuOXP
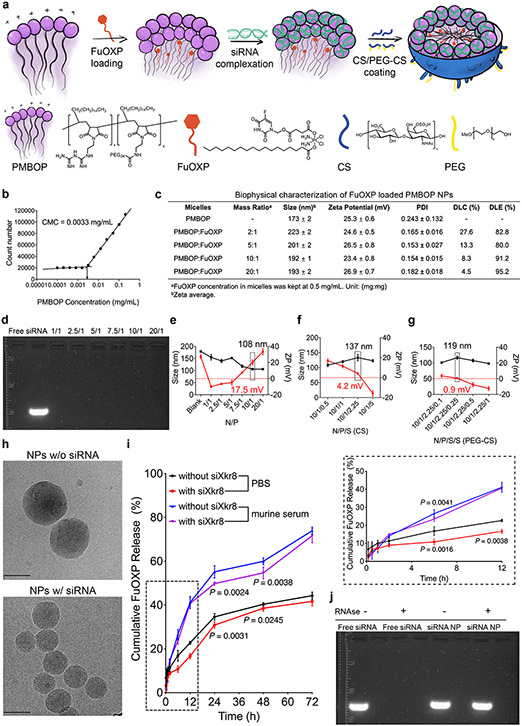
Since no small molecule inhibitors of Xkr8 are available, we developed a new nanocarrier, PMBOP-CP, for codelivery of murine siXkr8 and FuOXP (Fig. 1q). Fig. 2a shows the major components and steps in the development of the PMBOP-CP nanocarrier. The 1-octadecene lipid motif in the amphiphilic PMBOP polymer is expected to facilitate the interaction with cell membrane and improve transfection17, while also helping to improve the loading of FuOXP into the hydrophobic/lipophilic core. The biguanidine motif was designed to enhance the interaction with siRNA due to its highly cationic nature. The synthesis route of PMBOP is shown in Scheme 1 (Suppl Fig. 3) and its structure was confirmed by nuclear magnetic resonance (1H NMR) spectroscopy (Suppl Fig. 4).
Fig. 2: Development and biophysical characterization of PMBOP-CP-based nanocarrier for codelivery of siXkr8 and FuOXP.
a: A schematic diagram of the protocol for the preparation of FuOXP/siRNA-coloaded PMBOP-CP NPs. b: CMC of PMBOP polymer. c: Biophysical characterization of PMBOP/FuOXP mixed micelles at various carrier/drug ratios (w/w). N=3 replicates. d: Gel retardation assay of PMBOP/FuOXP/siRNA complexes at various N/P ratios. e: Sizes and zeta potentials of PMBOP/FuOXP/siRNA complexes at various N/P ratios. N=3 replicates. f: Sizes and zeta potentials of FuOXP/siRNA-coloaded PMBOP-C NPs (coated with CS alone) at various N/P/S ratios. N=3 replicates. g: Sizes and zeta potentials of FuOXP/siRNA-coloaded PMBOP-CP NPs (coated with a mixture of CS and PEG-CS) at various N/P/S(CS)/S(PEG-CS) ratios. N=3 replicates. h: Spherical morphology of PMBOP-CP NPs with and without coloaded siRNA by cryo-EM. Scale bar, 100 nm. i: Cumulative FuOXP release from PMBOP-CP NPs with or without siRNA complexation in PBS or murine serum. N=3 replicates. j: Protection of siRNA in PMBOP-CP NPs against the digestion by RNAse. Data are presented as mean ± SEM and statistical analysis was performed by one-way analysis of variance (ANOVA) with Tukey post hoc test for comparison in i. Data are representative of 2 independent experiments in h and 3 independent experiments in b-g, i and j.
PMBOP polymer had a low CMC of 0.0033 mg/mL (Fig. 2b) and readily formed micelles in PBS with a size of 173 nm. FuOXP could be loaded into PMBOP micelles at a carrier/drug weight ratio as low as 2/1 (Fig. 2c). Both drug-free and FuOXP-loaded PMBOP micelles readily formed complexes with siRNA. Gel retardation assay shows that siRNA was effectively incorporated into micelles at nitrogen/phosphate (N/P) ratios of 1 and above (Fig. 2d), resulting in formation of compact PMBOP/siRNA complexes (Suppl Fig. 5a-b). At an N/P ratio of 10/1, the resulting PMBOP/FuOXP/siRNA complexes were positively charged (+17.5 mV) and 108 nm in size, smaller than PMBOP micelles loaded with FuOXP alone (192 nm) (Fig. 2e), suggesting that siRNA wrapped around and stabilized the micelles. Similar results were shown when siRNA formed complexes with drug-free PMBOP micelles (Suppl Fig. 6).
It is well known that drug carriers with cationic surface are not suitable for systemic delivery to distant solid tumors18. Therefore, the PMBOP/FuOXP/siRNA complexes formed at an N/P ratio of 10/1 were subjected to surface coating with a mixture of chondroitin sulfate (CS) and PEG-CS to form PMBOP-CP nanoparticles (NPs). CS is a highly negatively charged molecule and can be used to decrease the surface positive charge of the resulting NPs19. CS, like hyaluronic acid (HA) is also a natural ligand for CD44, which is overexpressed in various types of cancer cells and tumor endothelial cells (ECs). CS- and HA-based NPs have been studied extensively as carriers for tumor targeting20,21. One barrier that limits the effectiveness of HA- or CS-mediated tumor targeting is the expression of CD44 on liver sinusoidal endothelial cells (LSECs) that, due to their abundance, rapidly remove most circulating NPs22,23. Therefore, small amount of PEG-CS was included with the expectation of improving the EPR effect by minimizing the “nonspecific” uptake by liver. Fig. 2f-2g show the sizes and surface charges of the NPs at various N/P/S (sulphate of CS)/S (PEG-CS) ratios. The morphology of PMBOP-CP NPs with and without loaded siRNA at a N/P/S/S ratio of 10/1/2.25/0.25 was examined by cryo-electron microscopy (cryoEM) revealing spherical particles of relatively uniform diameter in each case (Fig. 2h). Fig. 2i shows the kinetics of FuOXP release from PMBOP-CP NPs in PBS or in mouse serum (Fig. 2i). Moreover, siRNA loaded into PMBOP-CP NPs was well protected from degradation by RNAse (Fig. 2j).
CS/CD44-mediated tumor targeting of PMBOP-CP nanocarrier in vivo
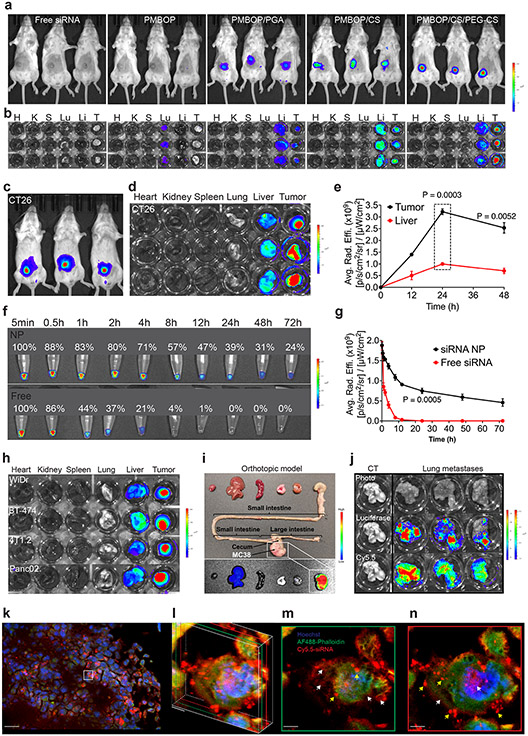
The in vivo distribution of the Cy5.5-siRNA in tumors (s.c. CT26) and liver was examined by IVIS imaging at 24 h following i.v. injection of different NPs. Cy5.5 signals were barely detectable in tumors following injection of free Cy5.5-siRNA (Fig. 3a-b & Suppl Fig. 7). Positively charged PMBOP/Cy5.5-siRNA complexes were mainly entrapped in lung. Surface coating with polyglutamic acid (PGA) led to improved accumulation at tumor site (Fig. 3a-b). CS coating was more effective than PGA in improving the tumor uptake of NPs. Surface coating with both CS and PEG-CS led to further improvement in tumor accumulation with a decrease in liver uptake (Fig. 3a-b).
Fig. 3: Optimizing PMBOP-CP NPs for effective tumor targeting in vivo.
a: NIRF whole body imaging of CT26 tumor-bearing mice at 24 h following i.v. administration of free Cy5.5-siXkr8 or various Cy5.5-siXkr8 NPs. N=3 mice per group. b: Ex vivo imaging of tumors (CT26) and major organs at 24 h following i.v. administration of free Cy5-siXkr8 or various Cy5.5-siXkr8 NPs. N=3 mice per group. c: NIRF whole body imaging of CT26 tumor-bearing mice at 24 h following i.v. administration of Cy5.5-siXkr8-loaded PMBOP-CP NPs. d: Ex vivo imaging of tumors (CT26) and major organs at 24 h following i.v. administration of Cy5.5-siXkr8-loaded PMBOP-CP NPs. e: Fluorescence (Cy5.5-siXkr8) intensity at tumors and liver at different times following i.v. administration of Cy5.5-siXkr8-loaded PMBOP-CP NPs. N= 3 mice per timepoint. f & g: Changes in fluorescence (Cy5.5-siXkr8) intensity in blood at different times following i.v. administration of free Cy5.5-siXkr8 or Cy5.5-siXkr8-loaded PMBOP-CP NPs. N= 3 mouse blood samples per timepoint. h & i: Ex vivo imaging of tumors and major organs at 24 h following i.v. administration of Cy5.5-siXkr8-loaded PMBOP-CP NPs in various types of s.c. tumor models (h), a MC38 orthotopic model (i), and a CT-26 lung metastasis model (j), respectively. N=3 mice. k-n: Confocal laser scanning microscopic images of tumor (s.c. CT26) sections at 24 h following i.v. administration of Cy5.5 siXkr8-loaded PMBOP-CP NPs. k: A merged image of cell nuclei (Hoechst), F-actin (Alexa Fluor® 488 Phalloidin) and siRNA (Cy5.5) at a 20x magnification. Scale bar, 30 μm. l: The 3D Z-stacking of two layers of 2D scanning images from different depths. Green: 0.1 μm layer from the first scan (m). Red: 0.5 μm layer from the first scan (n). Magnification, 600x. Scale bar, 1 μm. Yellow arrow: punctuated pattern. White arrow: diffused pattern. Data are presented as mean ± SEM and statistical analysis was performed by two-tailed Student’s t-test for comparison in e and g. Data are representative of 2 independent experiments in a-h and j, and 3 tumor tissues in k-n.
We then further evaluated the in vivo distribution of the Cy5.5-labeled siRNA in tumors and liver by fluorescence microscopy at 24 h following i.v. injection of various PMBOP-CP NPs that were prepared at an N/P ratio of 10/1 and coated with various amounts of CS/PEG-CS, respectively. Increasing the N/P/S (CS) ratio from 10/1/1 to 10/1/2.25 was associated with a gradual increase of Cy5.5 signal in tumors and a decrease of signal in liver. Further increase in the amount of CS resulted in decrease of the signal in tumor and increased signal in liver (Suppl Fig. 8a). We then kept the N/P/S ratio at 10/1/2.25 while gradually increasing the amount of PEG-CS. As shown in Suppl Fig. 8b, increasing the ratio of PEG-CS/CS from 0.1/2.25 to 0.25/2.25 led to further increase of Cy5.5 signal in tumors while the signal in liver was decreased. Further increase in the amount of PEG-CS resulted in decreased signal in tumors with increased signal in liver (Suppl Fig. 8b). All subsequent studies were conducted with NPs prepared at an N/P/S(CS)/S(PEG-CS) ratio of 10/1/2.25/0.25.
Fig. 3c-e show the NIRF images at different times following i.v. injection of Cy5.5-siRNA NPs. The Cy5.5 signal was largely concentrated in the tumor at 24 h (Fig. 3c-d & Suppl Fig. 9). The siRNA signal in tumors increased over time, peaked at 24 h, and slowly declined thereafter (Fig. 3e). The siRNA NPs stayed in the blood significantly longer than free siRNA (Fig. 3f & 3g). We also demonstrated effective tumor targeting in several other s.c. tumor models including human CRC (WiDr), human breast cancer (BCa) (BT-474), murine BCa (4T1.2) and murine PCa (Panc02) (Fig. 3h & Suppl Fig. 10). A similar result was also observed in an orthotopic murine CRC model (Fig. 3i). One potential limitation of NPs is their low efficiency in targeting disseminated tumors and metastases due to the limited EPR in these small tumor lesions24. Interestingly, a preliminary study showed that PMBOP-CP NPs effectively accumulated in metastatic tumors in the lung established by tail vein injection of CT26 tumor cells while minimal signal was seen in normal lung (Fig. 3j & Suppl Fig. 11). Fig. 3k-n and Suppl Fig. 12-13 show widespread distribution of Cy5.5 signal in tumor sections as well as intracellular delivery.
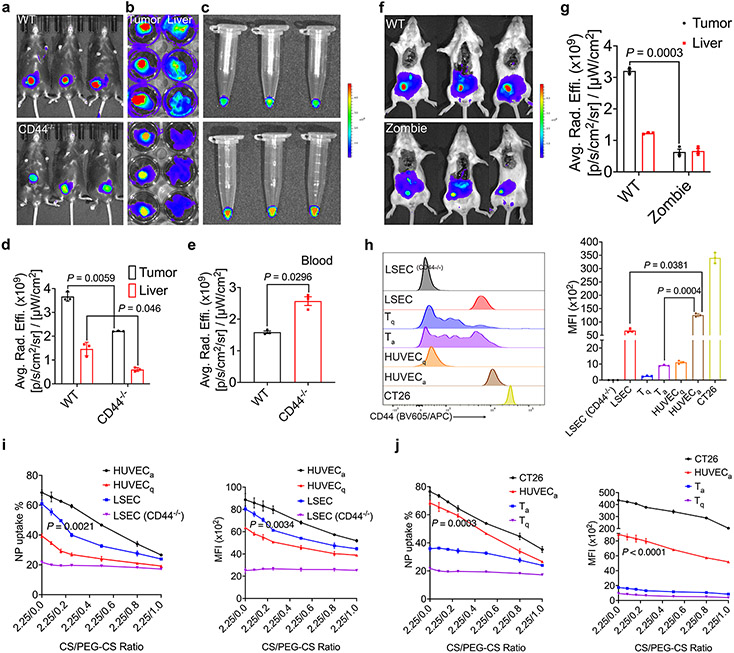
The decrease in tumor uptake that was associated with increased PEG shielding (PEG-CS/CS > 0.5/2.25) suggests that CS-mediated active targeting likely plays a role in the overall tumor targeting. To examine whether the CD44-mediated ECs targeting plays a role, NIRF imaging was performed similarly in CD44−/− mice. As shown in Fig. 4a-b, the Cy5.5 signal in tumor was decreased significantly in CD44−/− mice. The uptake of Cy5.5 siRNA NPs was also decreased in the liver of CD44−/− mice (Fig. 4b & 4d). Interestingly, the Cy5.5 siRNA signal in blood was increased (Fig. 4c & 4e), suggesting that Cy5.5 siRNA NPs were stable in the blood for a significant period of time and that the CD44-mediated targeting of tumor ECs contributed to the overall tumor targeting. Meanwhile, CD44 in the LSECs and possibly other cell types may also contribute to the uptake of the NPs in the liver.
Fig. 4. CD44-mediated vascular targeting plays a role in tumor-targeting.
a & b: NIRF whole body (a) and ex vivo (b) imaging of CT26 tumor targeting in WT and CD44−/− mice at 24 h following i.v. administration of Cy5.5-siXkr8-loaded PMBOP-CP NPs. c: Ex vivo imaging of blood collected from WT and CD44−/− mice bearing CT26 tumors. d & e: Fluorescence (Cy5.5-siXkr8) intensity in tumors and liver (d), and in blood (e) at 24 h following i.v. administration of the NPs. N= 3 mice per group. f: NIRF whole body imaging of CT26 tumor-bearing Zombie mice. Mice were fixed prior to circulation of Cy5.5-siXkr8-loaded PMBOP-CP NPs for 6 h using trans-cardiac perfusion. N= 3 mice per group. g: Fluorescence (Cy5.5-siXkr8) intensity in tumors and livers at 6 h following trans-cardiac perfusion of the NPs. N= 3 mice per group. h: Flow analysis of CD44 expression in mouse LSECs (WT and CD44−/−), Tq, Ta, HUVECq, HUVECa and CT26 cells. N= 3 replicates. i: Quantitative analysis of the % of Cy5.5+ cells and the MFI at 4 h following incubation of Cy5.5-siXkr8-loaded PMBOP-CP NPs with LSECs (WT and CD44−/−), HUVECq, and HUVECa cells, respectively. N= 3 replicates. j: Quantitative analysis of cellular uptake of Cy5.5-siXkr8-loaded PMBOP-CP NPs by CT26, HUVECa, Tq, and Ta cells. N= 3 replicates. Data are presented as mean ± SEM and the statistical analysis was performed by two-tailed Student’s t-test for comparison in d, e and g, and one-way analysis of variance (ANOVA) with Tukey post hoc test for comparison in h, i and j. Data are representative of 2 independent experiments in all panels.
The tumor-targeting efficiency of PMBOP-CP NPs was also significantly decreased in the Zombie mouse model25 (Fig. 4f-g) in which EPR remains active while the active trans-endothelial transport is inhibited. These data, together with the data from CD44−/− mice (Fig. 4c & 4e) suggest that both EPR and tumor EC active targeting contribute to the overall tumor targeting by PMBOP-CP NPs.
To further investigate the respective roles of CD44 in tumor ECs and LSECs in interacting with PMBOP-CP NPs, we examined the uptake of Cy5.5 siRNA NPs by primary mouse LSECs and human umbilical vein endothelial cells (HUVECs). HUVECs cultured in the absence of growth factors are quiescent (HUVECq) and express a low level of CD44 while HUVECs cultured with growth factors become activated (HUVECa) and express a higher level of CD4426, which are often used to model tumor ECs27. Fig. 4h shows that the expression level of CD44 on HUVECa was about ~1.9- and ~13.5-fold higher than that on LSECs and HUVECq, respectively. NPs coated with CS only were effectively taken up by both HUVECa and LSECs with more NPs being taken up by HUVECa (69.3% vs 61.5%, P < 0.01) (Fig. 4i). It is also apparent that HUVECq took up significantly less amount of the NPs compared to HUVECa (Fig. 4i). CD44−/− LSECs showed the lowest level of uptake of NPs among all groups, suggesting that CD44-mediated endocytosis likely plays a role in the cellular uptake of the CS-coated NPs by both tumor ECs and LSECs.
Incorporation of PEG-CS led to decreased cellular uptake of the NPs in a PEG dose-dependent manner for both HUVECa and LSECs. However, PEG-CS clearly showed more impact on the uptake by LSECs compared to HUVECa (Fig. 4i). The above data strongly indicated that CD44 plays a role in the active interaction of PMBOP-CP NPs with both tumor ECs (mimicked by HUVECa) and LSECs and possibly other cells as well, and incorporation of PEG (PEG-CS) differentially inhibited the respective interaction. Cells with a higher level of CD44 such as activated HUVECs (mimic of tumor ECs) were less affected than cells with a lower level of CD44 such as LSECs. Our in vitro and in vivo data support the notion that incorporation of “optimal” amount of PEG (PEG-CS) could indirectly improve the overall tumor targeting through more effective inhibition of uptake by the abundant LSECs, rendering more NPs available in blood circulation for effective tumor accumulation through both EPR and CD44-mediated tumor ECs targeting.
One concern is the expression of CD44 on immune cells, such as the activated T cells (Ta). However, their CD44 levels were significantly lower than those of tumor cells and “tumor ECs” (HUVECa) (Fig. 4h). Accordingly, the uptake of NPs by Ta was significantly less than that by tumor cells or tumor ECs (Fig. 4j).
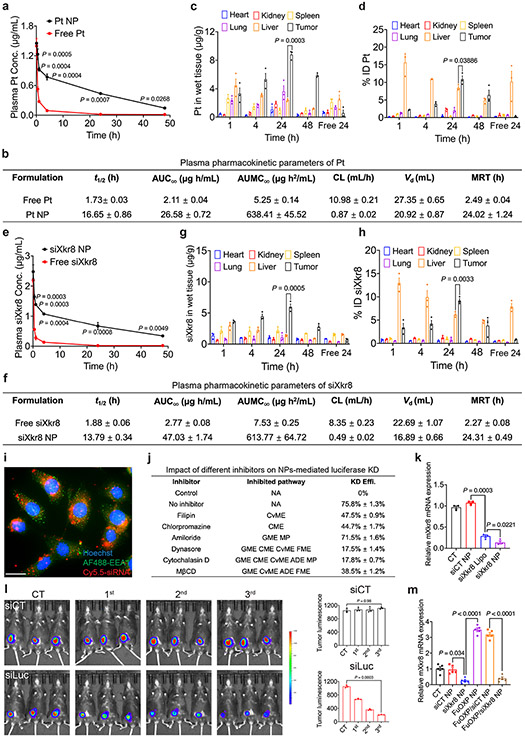
We further quantitatively examined the PK and tissue distributions of FuOXP and Cy5.5-siXkr8 via ICP-MS (for FuOXP)28 and qRT-PCR or fluorometer (for Cy5.5-siXkr8)29, respectively. Fig. 5a shows the concentrations of Pt over time in blood following injection of free FuOXP/siXkr8 or FuOXP/siXkr8 NPs into CT26 tumor-bearing mice. FuOXP formulated in the NPs showed substantially greater t1/2, AUC, and Cmax while its Vd and CL were significantly lower than those of free FuOXP (Fig. 5b). The concentration of Pt in tumors increased over time following i.v. administration. The Pt concentration in the tumors was about ~ 3.7-fold higher than in liver (Fig. 5c) and the total amount of Pt in the tumors was 10.9% of injected does (ID) at 24 h (Fig. 5d). Pt was found largely in the liver and was barely detectable in tumors after injection of free FuOXP (Fig. 5c-d). Similar results were observed for the PK and tissue distribution of siXkr8 by either qRT-PCR (Fig. 5e-h or fluorescence measurement (Suppl Fig. 15a-c). We acknowledge some discrepancy between imaging and qRT-PCR data due to the limitation of imaging in fluorescence quantification as reported in literature30.
Fig. 5. In vivo PK and tissue distribution of siXkr8 and FuOXP following i.v. administration of FuOXP/siXkr8 NPs, and the efficiency of gene knockdown.
a: ICP-MS analysis of plasma concentrations of Pt after i.v. injection of free FuOXP/siXkr8 (in Cremophor EL) or FuOXP/siXkr8 NPs in BALB/c mice bearing CT26 tumors at a dose of 1 and 5 mg/kg for siRNA and FuOXP, respectively. N=3 mice per group. b: Pharmacokinetic parameters of Pt were analyzed by a non-compartmental model. N=3 mice per group. c & d: Biodistribution of Pt in different organs following same treatments in a. N= 3 mice per group. e: qRT-PCR analysis of plasma concentrations of siXkr8 following same treatments in a. N=3 mice per group. f: Pharmacokinetic parameters of siXkr8 were analyzed by a non-compartmental model. N=3 mice per group. g & h: Biodistribution of siXkr8 in different organs with same treatments in a. N= 3 mice per group. i: Fluorescence microscopic images of cultured CT26 tumors cells at 2 h following treatment with Cy5.5-siXkr8-loaded NPs. Scale bar, 10 μm. j: Cultured MC38-Luc cells with or without pretreatment of different inhibitors (1 h) were treated with siLuc NPs for 24 h followed by luciferase assay. N=3 replicates. k: qRT-PCR analysis of Xkr8 mRNA expression levels in CT26 cells 24 h following various treatments. N= 3 replicates. l: C57BL/6 mice bearing MC38-Luc tumors received i.v. administration of siLuc NPs once every 5 days and the mice were subjected to whole body bioluminescence imaging one day after each treatment. N= 3 mice per group. m: CT26 tumor-bearing mice received various treatments for 3 times once every 5 days at 1 and 5 mg/kg for siRNA and FuOXP, respectively. The mRNA expression levels of Xkr8 in tumors were examined by qRT-PCR one day after the last treatment. N= 5 mice per group. Data are presented as mean ± SEM and the statistical analysis was performed by two-tailed Student’s t-test for comparison in a and e, and one-way analysis of variance (ANOVA) with Tukey post hoc test for comparison in c, d, g, h, k, l and m. Data are representative of 2 independent experiments in all panels.
Fig. 5i and Suppl Fig. 16 show that Cy5.5-siRNA NPs were effectively taken up by cultured CT26 cells. Delivery of luciferase siRNA (siLuc) to MC38-Luc cells led to significantly decreased luciferase activity (Fig. 5j). The efficiency of silence was significantly attenuated by pretreatment with various endocytosis inhibitors (Fig. 5j), suggesting the involvement of multiple pathways in the cellular uptake of our NPs31. Fig. 5k shows that the mRNA levels of Xkr8 were significantly decreased in cultured CT26 cells at 20 h following treatment with siXkr8 NPs.
In vivo delivery of siLuc NPs also led to a gradual decrease in luciferase activity in MC38-Luc tumor-bearing mice upon repeated injections as assessed by whole-body bioluminescence imaging (Fig. 5l). In a side-by-side comparison with a liver-specific formulation for Onpattro (a siRNA-based clinical drug)32, PMOBP-CP NPs selectively inhibited the expression of a target gene in tumors with minimal impact on liver (Suppl Fig. 17a-b). Fig. 5m shows that siXkr8 NPs effectively inhibited both basal and FuOXP-induced Xkr8 mRNA levels in vivo.
Biological consequences of Xkr8 knockdown in vitro and in vivo
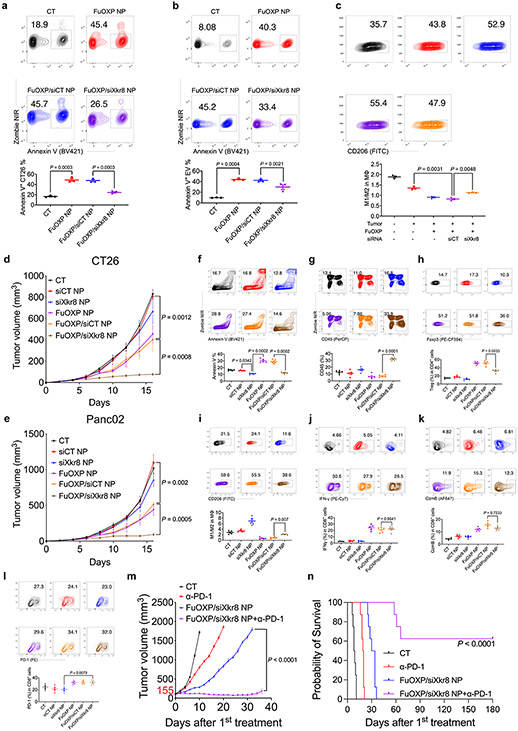
Consistent with previous studies33, treatment of CT26 tumor cells with FuOXP NPs led to a significant increase in the level of surface PS (Fig. 6a). This increase was almost abolished when the cancer cells were pre- and then co-treated with siXkr8. Pretreatment with siXkr8 NPs also partially abolished the FuOXP-induced increase in the amount of the PS-positive extracellular vesicles (EVs) (Fig. 6b) that have also been reported to be highly immunosuppressive34,35.
Fig. 6. Biological consequences of Xkr8 knockdown in vitro and in vivo.
a: CT26 tumor cells were treated with control or siXkr8 NPs for 72 h followed by treatment with FuOXP/siRNA-coloaded NPs. The numbers of Annexin V+ cells were examined by flow 24 h later. N= 3 replicates. b: CT26 tumor cells received similar treatments as described in a and the amount of secreted Annexin V+ EVs was examined by flow. N= 3 replicates. c: CT26 tumor cells received similar treatment as described in a and the tumor cells were then co-cultured with resident peritoneal macrophages. The percentages of M1 and M2 cells were then quantified. N= 3 replicates. d & e: Mice bearing CT26 (d) or Panc02 (e) tumors received various treatments once every 5 days for 3 times at 1 and 5 mg/kg for siRNA and FuOXP, respectively. Tumor volumes were followed once every 2 days. N= 5 mice per group. f-k: Single cell suspensions were prepared at the completion of therapy study described in d and subjected to flow analysis including Annexin V+ cells (f), CD45+ cells (g), Treg cells (h), M1/M2-like ratios (i), IFNγ+ CD8+ cells (j), and GzmB+ CD8+ cells (k), respectively. N= 5 mice per group. l: Single cell suspensions were prepared at the completion of therapy study described in e and subjected to flow analysis of PD-1+ CD8+ cells. N= 5 mice per group. m~n: Mice bearing Panc02 tumors received various treatments when the tumors reached ~155 mm3 in sizes once every 5 days for 3 times at a dose of 1, 5 and 10 mg/kg for siXkr8, FuOXP, and anti-PD-1, respectively. Tumor growth (m) and survival (n) were followed. N= 8 mice per group. Data are presented as mean ± SEM and statistical analysis was performed by one-way analysis of variance (ANOVA) with Tukey post hoc test for comparison in all panels. All related gating strategies are shown in Suppl Fig. 17. Data are representative of 2 independent experiments in panels a-l.
Fig. 6c shows that co-culture of mouse resident peritoneal macrophages with CT26 cells pretreated with FuOXP/control siRNA (siCT) NPs resulted in a decrease in M1/M2-like ratio. These changes were significantly reversed when CT26 cells were first treated with siXkr8 NPs followed by FuOXP/siXkr8 cotreatment. The engagement of PS+ tumor cells with macrophages has been reported to induce IL10 production by macrophages36 and that IL-10 directs macrophage polarization to an immunosuppressive phenotype37. Indeed, co-culture of macrophages with FuOXP-treated CT26 cells caused increased production of IL-10 (Suppl Fig. 18). This increase was significantly attenuated by the siXkr8 co-treatment, suggesting that siXkr8 may block the M2 polarization in part through inhibiting IL-10 production by macrophages (Suppl Fig. 18).
Fig. 6d & Suppl. Fig. 19 show that SiXkr8 NPs alone slightly inhibited the growth of CT26 tumors. This is likely due to the specific Xkr8 knockdown as a CT26 subline with stable Xkr8 knockdown showed comparable proliferation rate in vitro but significant delay of tumor growth in immunocompetent mice (Suppl. Fig. 20a-b). FuOXP NPs alone showed modest effect in controlling tumor growth (Fig. 6d & Suppl. Fig. 19). However, the combination of both led to a significant improvement in antitumor activity (Fig. 6d & Suppl. Fig. 19). Similar results were observed in Panc02 model (Fig. 6e & Suppl. Fig. 21).
The frequency of PS+ cells was reduced following treatment with siXkr8 NPs alone (Fig. 6f). Treatment with FuOXP NPs led to a drastic increase in the frequency of PS+ cells. This increase was almost abolished when FuOXP and siXkr8 were co-delivered to tumors via PMBOP-CP NPs (Fig. 6f).
Treatment with FuOXP/siXkr8-coloaded NPs led to significant increases in the percentages of CD45+ cells, IFNγ+ CD8+ T cells, and GzmB+ CD8+ T cells as well as a decrease in the percentage of CD4+ Treg cells compared to control or the group treated with FuOXP NPs alone (Fig. 6g-6k. Treatment with PMBOP-CP NPs loaded with FuOXP alone or co-loaded with FuOXP and siCT led to a significant decrease in the M1/M2-like ratio (Fig. 6i). These changes were significantly reversed in the group treated with FuOXP/siXkr8-coloaded NPs (Fig. 6i). Similar flow data were obtained in Panc02 tumor model (Suppl Fig. 22).
Fig. 6l shows that FuOXP NPs with or without co-loaded siXkr8 also caused significant upregulation of PD-1 expression in CD8+ T cells in the Panc02 model, suggesting potential for a combination therapy with anti-PD-1 antibody. Indeed, combination of FuOXP/siXkr8 NPs with anti-PD-1 led to a drastic improvement in therapeutic efficacy as evident from significant inhibition of tumor growth (Fig. 6m) and prolongation of survival time in a relatively advanced tumor model (Fig. 6n).
The above data suggest that Xkr8 knockdown in combination with chemotherapy may represent a new and effective treatment for cancer. The significant slowdown of tumor growth of Xkr8 knockdown tumor cells (Suppl. Fig. 20b) may be attributed to inhibition of engulfment of apoptotic cells by macrophages (Suppl. Fig. 20c), which promotes antitumor immune response following secondary necrosis and inflammatory response1,12. This is in consistent with previous reports that inhibition of engulfment of apoptotic tumor cells by anti-PS antibody38 or direct injection of necrotic tumor cells39 induced antitumor immune response. The much less inhibitory effect with siXkr8 NPs (Fig. 6d & e) is likely due to the fact that the treatment was started at a time when the tumors were already well established. The drastically improved antitumor activity with the combined treatment is likely due to a combination of tumor debulking by chemotherapy and an amplified immune response as a result of increased numbers of non-engulfed apoptotic cells due to the in situ Xkr8 knockdown in FuOXP-induced apoptotic cells.
FuOXP/siRNA-coloaded NPs were well tolerated at the doses used as shown by normal body weight gains (Suppl Fig. 24a), minimal changes in the serum levels of ALT and AST (Suppl Fig. 24b), and normal histology of several major organs examined (Suppl Fig. 24c). In addition, unlike siRNA complexed with cationic DOTAP liposomes that induced significant increases in serum levels of TNF-α and IL-640, siRNA PMBOP-CP NPs did not affect the serum levels of the two proinflammatory cytokines (Suppl Fig. 24d). Interestingly, FuOXP NPs also showed minimal impact on the level of mXkr8 mRNA (Suppl Fig. 24e) as well as the number of PS+ cells (Suppl Fig. 24f) in liver at the dose used in therapy study. In addition, there were no obvious change in the number of CD45+ cells following treatment with FuOXP/siXkr8 NPs (Suppl Fig. 24g).
Conclusions
We have shown that Xkr8 expression in tumor cells was significantly induced by chemotherapeutic agents. In addition, our hypothesis-driven study led to the development of a PMBOP-CP-based nanocarrier that is highly effective in tumor targeting through effective tumor ECs-mediated active targeting while minimizing LSECs-mediated liver uptake. In addition to enhanced delivery of both types of therapeutics to tumors, this strategy has the advantage of selectively delivering siXkr8 to those tumor cells that are exposed to chemodrugs. Consequently, our codelivery approach is particularly effective in antagonizing the Xkr8 mRNA that is induced in situ by co-delivered chemotherapeutic drug. Codelivery of siXkr8 and FuOXP led to significant improvement in the tumor immune microenvironment and enhanced antitumor activity. It is interesting to note that Xkr8−/− mice are developmentally normal, do not develop autoimmunity in C57BL/6 mice41, suggesting that targeting Xkr8 in tumor cells is a safe approach. Safety concerns with Xkr8 targeting can be further minimized through tumor-targeted delivery using NPs. Targeting Xkr8 in combination with chemotherapy may represent a novel and effective immunochemotherapy for the treatment of various types of cancers.
Methods
Mice.
Female C57BL/6, BALB/c, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) and B6.129(Cg)-Cd44tm1Hbg/J (CD44−/−) mice aged between 4–6 weeks were purchased from The Jackson Laboratories (CT, USA). Mice were housed under pathogen-free conditions according to AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) guidelines. The mouse-related experiments were performed in full compliance with institutional guidelines and approved by the Animal Use and Care Administrative Advisory Committee at the University of Pittsburgh under Protocol #: 21099779. Mice were housed at an ambient temperature of 22 °C (22–24 °C) and humidity of 45%, with a 14/10 day/night cycle (on at 6:00, off at 20:00), and allowed access to food ad libitum.
Tumor cell lines.
CT26 (CRL-2638) CRC cell lines, HT29 (HTB-38) and WiDr (CCL-218) human CRC cell lines, Panc02 (CRL-2553) murine PCa cell line, PANC-1 (CRL-1469) human PCa cell line, 4T1.2 (CRL-3406) murine BCa cell line, and BT-474 (HTB-20) human BCa cell line were obtained from ATCC (VA, USA). MC38 CRC (ENH204-FP) cell lines were originally obtained from Kerafast (MA, USA). CT26 cells were cultured in RPMI-1640 medium supplemented with 10% FBS and penicillin/streptomycin (100 U/mL). MC38, Panc02, PANC-1, 4T1.2, and BT-474 cells were cultured in DMEM medium supplemented with 10% FBS and penicillin/streptomycin (100 U/mL). HT29 cells were cultured in McCoy’s 5A medium supplemented with 10% FBS and penicillin/streptomycin (100 U/mL). WiDr cells were cultured in EMEM medium supplemented with 10% FBS and penicillin/streptomycin (100 U/mL). The cells were all cultured at 37 °C in a humidified atmosphere with 5% CO2. For establishment of mouse tumor models, tumor cells between passages 3–10 in 100 μL of saline were subcutaneously (s.c.) inoculated into the right lower abdomen using a 271/2 G needle.
Xkr8 siRNA and other oligonucleotides.
Murine Xkr8 siRNA (siXkr8) and control siRNA (siCT) were designed and synthesized by Ambion (TX, USA). Cy5.5-siXkr8 was synthesized by Sigma-Aldrich (MO, USA). Cy5.5 was introduced to siXkr8 via phosphate linkage using phosphoramidite chemistry. Primers for RT-PCR amplification of mXkr8 and hXkr8 mRNAs, and siXkr8 were provided by IDT (IA, USA). The sequences of siXkr8, siCT, and other primers are shown in Suppl Table. 1.
Analysis of Xkr8 mRNA level by qRT-PCR.
Groups of 5 BALB/c mice bearing s.c. CT26 tumors (~200 mm3) received FuOXP NPs, empty NPs or PBS as described above and tumors were collected one day after the last treatment and subjected to qRT-PCR analysis as detailed below.
To examine the effect of different chemotherapeutic agents on the expression level of Xkr8 mRNA, CT26, Panc02, PANC-1 or HT29 cells at a confluency of 80~90% were treated with various concentrations of FuOXP, DOX or PTX. Twenty-four h later, cells were collected and subjected to qRT-PCR of mXkr8 or hXkr8 as detailed below.
To examine the impact of ActD treatment on the expression level of Xkr8 mRNA, CT26 or PANC-1 tumor cells were treated with FuOXP (10 μM) for 12 h followed by addition of ActD (2 μM). At 2 or 4 h following ActD treatment, cells were collected and subjected to qRT-PCR of Xkr8 mRNA as described below. In another study, cells were treated with NAC (2.5 mM) for 12 h followed by treatment with FuOXP, DOX or PTX for another 12 h. qRT-PCR of Xkr8 mRNA was then similarly performed.
Tumor tissues or cells collected from the above experiments were subjected to RNA extraction by TRIzol™. cDNA was generated from the purified RNA using QuantiTect Reverse Transcription Kit (Qiagen, MD, USA) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using SYBR Green Mix on a 7900 HT PCR instrument (Applied Biosystems, MA, USA). Relative target mRNA levels were analyzed using delta-delta-Ct calculations and normalized to GAPDH42. The primer sequences are shown in Suppl Table 1.
Analysis of hXkr8 protein level by Western blot.
Cultured HT29 or PANC-1 cells received similar treatments as described above. Cells were then lysed with RIPA lysis buffer (Thermo Fisher Scientific, MA, USA) by gently shaking on ice for 30 min. After centrifugation at 14,500 g for 10 min, the supernatants were collected, and the concentrations of proteins were measured using Pierce BCA Protein Assay Kit (ThermoFisher Scientific, MA, USA). The protein samples were denatured by boiling for 5 min and loaded onto 10% SDS-PAGE gel for electrophoresis. The proteins in the gels were subsequently transferred onto PVDF membranes (Bio-Rad, CA, USA). The membranes were then incubated in blocking buffer (5% non-fat dry milk in TBST) for 1 h at RT, followed by incubation with either anti-hXkr8 (346-395AA, dilution: 1/2000) or anti-hXkr8 (69-158AA, dilution: 1/1500) polyclonal antibody (ThermoFisher Scientific, MA, USA) in antibody dilution buffer (5% BSA in TBST, 1/2000 dilution) with gentle agitation overnight at 4 °C. Anti-hXkr8 (346-395AA) antibody only detects full-length hXkr8 but not truncated (caspase 3-cleaved) hXkr8 in which the C-terminal motif is removed. On the other hand, anti-hXkr8 (69-158AA) antibody detects both full-length and truncated hXkr8. After washing with TBST for three times, the membranes were subsequently incubated with the secondary HRP-linked goat anti-rabbit IgG antibody (Cell Signaling Technology, MA, USA, dilution: 1/5000) for 1 h at RT. After another three washes with TBST, the membranes were incubated with Pierce™ ECL Western Blotting Substrate (ThermoFisher Scientific, MA, USA) for 1 min. Protein expression was normalized against β-Tubulin expression (Cell Signaling Technology, MA, USA, dilution: 1/2000). In a separate experiment, HT29 cells were treated with FuOXP (10 μM) for 0, 1, 2, 4, 8, 12 and 24 h, respectively and the protein level of hXkr8 was similarly examined as described above. The protein expression of murine Xkr8 could not be examined due to lack of antibody.
In vitro caspase-3 activity assay.
Caspase-3 activity was determined using a colorimetric assay kit (Abcam, MA, USA) following the manufacturer’s instruction. Briefly, HT29 cells were treated with FuOXP (10 μM) for 0, 1, 2, 4, 8, 12 and 24 h, respectively. Cells were collected at various timepoints, and lysed in the supplied lysis buffer for 30 min at 4 °C. Supernatants were collected and incubated with the supplied reaction buffer containing dithiothreitol and DEAD-pNA as substrates at 37 °C. One and a half h later, the caspase-3 activity was determined by following changes in the absorbance at 405 nm using a microplate reader.
Preparation of FuOXP/siRNA-coloaded PMBOP NPs.
Synthesis of PMBOP polymer was detailed in Supplementary Information. PMBOP polymer (5 mg) and FuOXP (0.5 mg) of 10:1 ratio (w/w) were dissolved in DMSO and added to water with 15 times of the initial volume. The mixture was then transferred to a Amicon® Ultra 3K (3,000 MWCO) centrifugal filter device (Sigma-Aldrich, MO, USA). The device was centrifuged at 17,000 g for 15 min followed by the addition of 1 mL water. This step was repeated 3 times to remove any residual DMSO and concentrate the FuOXP-loaded micelles to a desired volume. Greater than 99% of PMBOP monomer was found to be incorporated into micelles based on the quantification of free FITC-labeled PMBOP following the filtration of FITC-labeled PMBOP micelles through a membrane of 100 nm pore size. SiXkr8 or siCT (0.1 mg/mL) was then mixed with FuOXP-loaded micelles at a 10:1 N:P ratio at RT for 20 min to form PMBOP/FuOXP/siXkr8 or PMBOP/FuOXP/siCT complexes. Formation of stable PMBOP/siXkr8 complexes was confirmed by gel-retardation assay. Controls include the complexes of siRNA with DOTAP liposome, Lipofectamine RNAiMAX (ThermoFisher Scientific, MA, USA) or PEG-derivatized chitosan polymer. Subsequent incubation with a mixture of CS/PEG-CS of various ratios at RT for 20 min led to the formation of CS/PEG-CS-decorated, FuOXP/siXkr8 co-loaded PMBOP-CP NPs. CS was found to be quantitatively (99.12%) incorporated into PMBOP-CP NPs under the “optimal” condition using FITC-labeled CS. The particle size (zeta average), zeta potential and polydispersity index (PDI) were measured by a Zetasizer from three batches of formulation. Drug loading capacity (DLC) and drug loading efficiency (DLE) of FuOXP were determined by high-performance liquid chromatography (HPLC)43. The siRNA concentration in the NPs was determined by Ribogreen assay after the NPs were disrupted by adding SDS (0.05%)44 and greater than 99% of siRNA was also found to be incorporated into the PMBOP-CP NPs. To examine the resistance of FuOXP/siXkr8 co-loaded NPs against nuclease-mediated degradation, the siRNA NPs (0.1 mg/mL siRNA, 0.35 mg/mL PMBOP) were incubated with RNase (50 U/mL) (NEB, MA, U.S.A) at 37 °C. One h later, the NPs were disrupted by 0.05% SDS and the integrity of siRNA was examined by electrophoresis. Free siRNA was used as a control. The abbreviated empty NPs in this communication refer to PMBOP micelles coated with CS/PEG-CS while FuOXP NPs refer to FuOXP-loaded PMBOP micelles coated with CS/PEG-CS. FuXOP/siRNA NPs refer to PMBOP/FuOXP/siRNA complexes coated with CS/PEG-CS.
Whole-body near-infrared fluorescence (NIRF) imaging and ex vivo imaging.
Groups of 3 BALB/c mice were each inoculated with 5 ×105 CT26 cells s.c. at the right lower abdomen. When the tumors grew to ~300 mm3, the mice were i.v. administered with Cy5.5-siXkr8-loaded PMBOP-CP NPs at a siRNA dose of 1 mg/kg. Controls include free Cy5.5-siXkr8, PMBOP/Cy5.5-siRNA complexes without surface coating of anionic polymer, PMBOP/Cy5.5-siRNA complexes coated with PGA, and PMBOP/Cy5.5-siRNA complexes coated with CS alone at the same siRNA dose. At 12 h, 24 h and 48 h time points, the mice were imaged by IVIS 200 system (Perkin Elmer, MA, USA) at a constant 1s exposure time with excitation at 679 nm and emission at 702 nm for all the groups. After whole-body imaging, mice were euthanized, tumor and various organs were excised for ex vivo imaging following our previous protocol45. For the study of PK in blood, blood was collected in Li-Heparin-containing tubes at 5 min, 0.5 h, 1 h, 2 h, 4 h, 8 h, 12 h, 48 h and 72 h time points and plasma samples were prepared by centrifugation at 14,500 g for 10 min and imaged by IVIS 200 system. Similar studies were performed in several other s.c. tumor models including human colon cancer (WiDr), human breast cancer (BT-474), murine pancreatic cancer (Panc02), and murine breast cancer (4T1.2, inoculated into the mammary fat pad), and an orthotopic murine colon cancer model (MC38)46. For evaluation of targeting efficiency in a lung metastasis model, BALB/c mice were injected with CT26-luc cells (2 × 105 in 100 μL DPBS) through the tail vein. Fifteen days after tumor cell injection, whole-body and ex vivo imaging was conducted as described above. Tumor-free mice injected with Cy5.5-siRNA-loaded NPs were used as control. In addition, imaging study was conducted in CD44−/− mice bearing MC38 tumors (s.c.) and compared to that in WT mice.
Blood pharmacokinetics of siXkr8.
Groups of 3 BALB/c mice bearing s.c. CT26 tumors (~300 mm3) received tail vein injection of PMBOP-CP NPs loaded with FuOXP/Cy5.5-mXkr8 siRNA or free FuOXP/Cy5.5-mXkr8 siRNA at a dose of 5 and 1 mg/kg for FuOXP and siRNA, respectively. For the free drug combination, FuOXP was dissolved in Cremophor EL and mixed with siRNA prior to injection. At 5 min, 30 min, 1 h, 4 h, 24 h and 48 h post injection, blood was collected, and plasma was prepared. The amount of siRNA in the samples was quantified by both qRT-PCR and fluorescence measurement47 as detailed below.
SiXkr8 quantification by qRT-PCR.
An aliquot of the plasma (100 μL) was diluted with PBS to make the final volume 500 μL. Then, methanol (1.0 mL) and chloroform (0.5 mL) were added, and the samples were vortexed for 2 min until a clear, single-phase solution was obtained. The mixture was then centrifuged at 14,500 g for 30 min, and the aqueous phase containing Cy5.5-mXkr8 siRNA was collected. SuperScript III reverse transcription kit (InVitrogen, MA, USA) was used to convert siRNA into cDNA. For reverse transcription, 6 μL of RT mastermix (2 μL of water, 2 μL 5x buffer, 0.5 μL of 0.1 M DTT, 0.5 μL of 10 mM dNTPs (InVitrogen, MA, USA), 0.5 μL of RNAse OUT (InVitrogen, MA, USA), and 0.5 μL of SuperScript III enzyme) were combined with 2 μL of 0.5 μM GS primer and 2 μL of template in a 96-well plate. GS primer, and template were premixed, heated at 85°C for 2 min, snap-chilled on ice, and RT premix was added. The 10 μL RT reaction was incubated at 50°C for 30 min, 85°C for 5 min, cooled to room temperature, and diluted 10-fold with 90 μL of water. Following reverse transcription, quadruplicate measurements of 2 μL of cDNA were made in 10 μL final reaction volumes by qPCR in a 384-well optical PCR plate using a 7900 HT PCR instrument (Applied Biosystems, MA, USA). SYBR green PCR mix contained 5 μL of 2x SYBR green PCR master mix (Applied Biosystems, MA, USA), 1.4 μL of water, 0.8 μL of 10 μM universal primer, 0.8 μL of 10 μM LNA-R primer, and 2 μL of sample. The primer sequences are shown in Suppl Table. 1. A standard curve was generated by spiking 100 μL of plasma aliquots from untreated animals with the NPs containing Cy5.5-mXkr8 siRNA at concentrations ranging from 0 to 20 μg/mL.
Fluorescence measurement.
Extraction of Cy5.5-mXkr8 siRNA from blood was performed as described above. The fluorescence was measured using a Spectramax M5 multiplate reader (Molecular Devices, CA) at an excitation wavelength of 679 nm and an emission wavelength of 702 nm. A standard curve was similarly generated as described above. The PK parameters were obtained by fitting the blood siRNA concentration versus time using a noncompartment model43.
Biodistribution of siXkr8.
Groups of 3 BALB/c mice bearing s.c. CT26 tumors (~300 mm3) received tail vein injection of PMBOP-CP NPs loaded with FuOXP/Cy5.5-mXkr8 siRNA or free FuOXP/Cy5.5-mXkr8 siRNA at a dose of 5 and 1 mg/kg for FuOXP and siRNA, respectively. Hearts, livers, spleens, lungs, kidneys, and tumors were collected at 1 h, 4 h, 24 h and 48 h and homogenized in 1,000 μL TRIzol™. Two-hundreds μL of chloroform was added to the homogenized tissues. After 5 min of incubation, the samples were centrifuged at 14,500 g for 15 min. The upper aqueous phase was similarly used for qRT-PCR and florescence quantification as described above. A standard curve was generated by spiking known amount of Cy5.5-mXkr8 siRNA (0-20 μg/mL) in the tissues obtained from non-treated control animals and used to calculate the amount of siRNA in the samples.
Quantification of platinum (Pt) by ICP-MS28.
Plasma and tissue samples were collected as described above. Samples were placed into a pre-weighed Purillex PFA bottle (Savillex, MN, USA) and the net weights were recorded. Plasma and tissue samples were freezed at −80 °C and lyophilized. Four (4) mL HNO3 (69.0% w/w) and 2 mL HCl (37% w/w) were added into each PFA bottle, which was then immersed into 90 °C water bath for sample digestion to obtain free Pt ion in the lysates. The lysates were then dried down for 12 h at 50 °C to get rid of residual acid. HCl (5%) was added and the samples were transferred into a 15 mL centrifuge tube and subsequently Pt concentrations in the samples were measured using a PerkinElmer Nexion 300x Inductively Coupled Plasma-Mass Spectrometer (ICP-MS). A standard curve for Pt on the ICP-MS was created by diluting a Pt single element standard (1,000 μg/mL, for AA and ICP, Spex CertiPrep, NJ, USA).
Zombie fixation and nanoparticle circulation.
NIRF imaging in a zombie mouse model was conducted according to a previously published protocol45. Mice were fixed using trans-cardiac perfusion with a 4% formaldehyde and 0.5% glutaraldehyde PBS solution for 60 min. Perfusion was then performed with Cy5.5-siXkr8-loaded PMBOP-CP NPs at a siRNA concentration of 22.22 μg/mL at a physiologically relevant flow rate (6 mL min−1) for 6 h using a peristaltic pump. The mice were then imaged by IVIS 200 system for Cy5.5 detection.
Cellular uptake.
Mouse liver sinusoidal endothelial cells (LSECs) were isolated according to a previously published protocol48 from both WT C57BL/6 and B6.129(Cg)-Cd44tm1Hbg/J (CD44−/−) mice. Briefly, perfused mouse liver was cut out from the mice and grinded to release the cells. Cell suspension was then centrifuged several times at different speed and the suspended pellet was loaded on top of Percoll gradient. Non-parenchymal cells (NPC) were collected from the interface between the two density cushions of 25% with 50% Percoll and Kupffer cells were removed by selective adherence. LSECs were harvested by seeding the cells on collagen-coated cell-culture plastic dish. T cells were isolated from naïve mouse spleen and activated T cells were obtained by adding IL-2 (50 IU/mL) every two days for 1 week. For cellular uptake study, LSECs from both WT and CD44−/− mice, mouse T cells (quiescent and activated) as well as sub-confluent HUVECs with or without treatment of 1% endothelial cell growth supplement (containing growth factors, hormones, and proteins for the culture of human microvascular endothelial cells) (ECGS, ScienCell, CA, USA) were incubated with Cy5.5-siRNA-loaded PMBOP-CP NPs coated with various amounts of CS/PEG-CS. Cellular uptake was examined by flow cytometry after 4 h. The expression of CD44 in LSECs, mouse T cells, and HUVECs with or without treatment of growth factors was examined by flow cytometry using both APC rat anti-mouse CD44 antibody (BD Biosciences, NJ, USA, dilution: 1/200) and BV605 mouse anti-human CD44 antibody (BD Biosciences, NJ, USA, dilution: 1/200).
In vivo mXkr8 knockdown.
CT26 tumor cells were s.c. inoculated into the right lower abdomen of BALB/c mice. When the tumor volume reached ~50 mm3, mice were randomly grouped (n = 5) and intravenously administered with DPBS (CT), siCT NPs, siXkr8 NPs, FuOXP NPs, FuOXP/siCT NPs or FuOXP/siXkr8 NPs three times at an interval of 5 days (day 5, 10 and 15). The doses for FuOXP and siRNA were 5 and 1 mg/kg, respectively. Twenty-four h after final injection, tumors were collected and subjected to qRT-PCR of mXkr8 expression as detailed above.
Effect of Xkr8 knockdown on the PS expression levels of tumor cells and EVs following FuOXP treatment.
CT26 cancer cells were first treated with siXkr8 NPs for 72 h and then treated with FuOXP/siXkr8-coloaded NPs for another 24 h. EVs were isolated from culture medium using gradient ultra-centrifugation according to a previously established protocol49. Briefly, culture medium was collected and centrifuged for 20 min at 300 g at 4°C to clear dead cells and cell debris. The supernatant was then transferred to a polycarbonate ultracentrifuge tube and centrifuged at 10,000 g for 35 min at 4°C to collect larger EVs. The supernatant was centrifuged again at 100,000 g for 80 min at 4°C and the pellet was resuspended in 100 μL of PBS for smaller EVs collection. The levels of surface PS on tumor cells and EVs were analyzed by flow using BV421-labeled Annexin V (Biolegend, CA, USA, dilution: 1/200). For detection of EVs by flow cytometry, electronic “Height” (-H) parameter rather than the “Area” (-A) parameter was used to allow optimal signal detection.
Tumor cells/macrophages co-culture study.
Mouse macrophages were isolated from peritoneal cavity according to a previously established protocol50. CT26 cancer cells were first treated with siXkr8 NPs for 72 h and then treated with FuOXP/siXkr8-coloaded NPs for another 24 h in a 24-well plate. The treated cancer cells were then transferred and co-cultured with macrophages at a 10:1 ratio for 24 h. Macrophages were subjected to flow analysis of M1/M2 markers including F4/80 (macrophage marker) (Biolegend, CA, USA, dilution: 1/200) and CD206 (M2-like macrophage marker) (Biolegend, CA, USA, dilution: 1/200)51. Culture supernatants were also collected, and IL-10 concentrations were measured using a mouse IL-10 ELISA kit (R&D Systems, MN, USA) according to the instructions of the manufacturer. Controls include macrophages without cancer cell co-culture, macrophages co-cultured with cancer cells without FuOXP treatment, and macrophages co-cultured with cancer cells treated with FuOXP NPs with or without coloaded siCT.
In vivo therapeutic efficacy of FuOXP/siXkr8-coloaded NPs.
CT26 or Panc02 tumor cells were s.c. inoculated into the right lower abdomen of BALB/c or C57/BL6 mice. When the tumor volume reached ~50 mm3, mice were randomly grouped (n = 5) and intravenously administered with DPBS (CT), siCT NPs, siXkr8 NPs, FuOXP NPs, FuOXP/siCT NPs or FuOXP/siXkr8 NPs three times at an interval of 5 days (day 5, 10 and 15). The doses for FuOXP and siRNA were 5 and 1 mg/kg, respectively. Mice were followed once every 2 days for tumor sizes and body weights. To test the therapeutic effect of combinational therapy of FuOXP/siXkr8 NPs with anti-PD-1, the treatment was started when the tumors reached ~155 mm3 in sizes. Mice were treated with anti–PD-1 (clone RMP1-14, Bio X Cell, NH, USA) alone, FuOXP/siXkr8 NPs alone or the combination once every 5 days for 3 times. Anti-PD-1 was administered at 200 μg per dose intraperitoneally (i.p.) while FuOXP/siXkr8 NPs were given i.v. at a dose of 5 mg/kg for FuOXP and 1 mg/kg for siXkr8. Mice were followed until death or sacrificed if tumor size reached 2000 mm3, the maximal tumor size permitted by the Animal Use and Care Administrative Advisory Committee at the University of Pittsburgh.
Analysis of tumor-infiltrating lymphocytes and monocytes.
Flow cytometry was performed with the instrument LSRII (BD Biosciences, NJ, USA) and Aurora (Cytek Biosciences, CA, USA) and analyzed by FlowJo (BD Biosciences, NJ, USA). Spleens and tumors were harvested one day after the last treatment (Day 16 post initial tumor inoculation). Single-cell suspensions were prepared from mouse spleens or tumors as previously described52. Briefly, tumors were dissected and transferred into RPMI 1640. Tumors were disrupted mechanically using scissors, digested with a mixture of deoxyribonuclease I (0.3 mg/mL, Sigma-Aldrich, MO, USA) and TL Liberase (0.25 mg/mL, Roche, Basal, Switzerland) in serum-free RPMI 1640 at 37 °C for 30 min, and dispersed through a 40-μm cell strainer (BD Biosciences, NJ, USA). After red blood cell lysis, live/dead cell discrimination was performed using Zombie NIR Fixable Viability Kit (BioLegend, CA, USA, dilution: 1/1000) at 4°C for 30 min in DPBS. Surface staining was performed at 4°C for 30 min in FACS staining buffer (1× phosphate-buffered saline/5% FBS/0.5% sodium azide) containing designated antibody cocktails (Brilliant Violet 421 Annexin V antibody, PerCP anti-mouse CD45 antibody, Brilliant Violet 785 anti-mouse CD4 antibody, FITC anti-mouse CD8 antibody, APC anti-mouse CD11b antibody, Brilliant Violet 711 anti-mouse Gr-1 antibody, APC/Cyanine7 anti-mouse F4/80 antibody, and Pacific Blue anti-mouse MHC II antibody. Dilution: 1/200 for all antibodies). For intracellular proteins staining (PE-CF594 anti-mouse Foxp3 antibody and FITC anti-mouse CD206 antibody. Dilution: 1/200 for both antibodies), cells were fixed and permeabilized using the BD Cytofix/Cytoperm kit, following the manufacturer’s instructions. For intracellular cytokine staining (PE-Cy7 anti-mouse IFN-γ antibody and Alexa Fluor 647 anti0human/mouse Granzyme B antibody. Dilution: 1/200 for both antibodies), cells were stimulated with phorbol 12-myristate 13-acetate (100 ng/mL) and ionomycin (500 ng/mL) for 6 h in the presence of Monensin. Cells were fixed/permeabilized using the BD Cytofix/Cytoperm kit before cell staining. More detailed information and gating strategies can be found in Fig. S17.
Statistical analysis.
All values were presented as mean ± standard error of mean (SEM). Statistical analysis was performed with two-tailed Student’s t-test for comparison between two groups and one-way analysis of variance (ANOVA) with Tukey post hoc test for comparison between multiple groups. Results were considered statistically significant if p <0.05. Prism 9.4.0 (GraphPad Software) was used for the data analysis and graph plotting.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health grants R01CA219399, R01CA223788 (to SL), R01CA219716 (to BL and SL), and a grant from Shear Family Foundation. We thank Dr. Robert Gibbs for his advice on statistical analysis.
Footnotes
Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The bulk messenger RNA-seq data mapped to the mouse genome (GRCm38: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/) are available in the NCBI Gene Expression Omnibus (GEO) under accession no. GSE214881 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214881). Source data are provided with this paper. All data generated or analyzed during this study are included in this published article and its Supplementary information files.
References
- 1.Birge RB et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death and Differentiation 23, 962–978, doi: 10.1038/cdd.2016.11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar S, Calianese D & Birge RB Efferocytosis of dying cells differentially modulate immunological outcomes in tumor microenvironment. Immunol Rev 280, 149–164, doi: 10.1111/imr.12587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagata S, Suzuki J, Segawa K & Fujii T Exposure of phosphatidylserine on the cell surface. Cell Death Differ 23, 952–961, doi: 10.1038/cdd.2016.7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hankins HM, Baldridge RD, Xu P & Graham TR Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16, 35–47, doi: 10.1111/tra.12233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki J, Denning DP, Imanishi E, Horvitz HR & Nagata S Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406, doi: 10.1126/science.1236758 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Suzuki J, Imanishi E & Nagata S Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc Natl Acad Sci U S A 113, 9509–9514, doi: 10.1073/pnas.1610403113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Q et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med 17, 860–866, doi: 10.1038/nm.2385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakuragi T, Kosako H & Nagata S Phosphorylation-mediated activation of mouse Xkr8 scramblase for phosphatidylserine exposure. Proc Natl Acad Sci U S A 116, 2907–2912, doi: 10.1073/pnas.1820499116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravichandran KS Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J Exp Med 207, 1807–1817, doi: 10.1084/jem.20101157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochreiter-Hufford A & Ravichandran KS Clearing the Dead: Apoptotic Cell Sensing, Recognition, Engulfment, and Digestion. Cold Spring Harbor Perspectives in Biology 5, doi:ARTN a008748 10.1101/cshperspect.a008748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang TH et al. Annexin A5 as an immune checkpoint inhibitor and tumor-homing molecule for cancer treatment. Nature Communications 11, doi:ARTN 1137 10.1038/s41467-020-14821-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang W, Fa H, Xiao D & Wang J Targeting phosphatidylserine for Cancer therapy: prospects and challenges. Theranostics 10, 9214–9229, doi: 10.7150/thno.45125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorpe PE Targeting anionic phospholipids on tumor blood vessels and tumor cells. Thromb Res 125 Suppl 2, S134–137, doi: 10.1016/S0049-3848(10)70031-1 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Sun A & Benet LZ Late-Stage Failures of Monoclonal Antibody Drugs: A Retrospective Case Study Analysis. Pharmacology 105, 145–163, doi: 10.1159/000505379 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Shin SA, Moon SY, Park D, Park JB & Lee CS Apoptotic cell clearance in the tumor microenvironment: a potential cancer therapeutic target. Arch Pharm Res 42, 658–671, doi: 10.1007/s12272-019-01169-2 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Zhang R, Song X-Q, Liu R-P, Ma Z-Y & Xu J-Y Fuplatin: An Efficient and Low-Toxic Dual-Prodrug. Journal of medicinal chemistry 62, 4543–4554 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Li M, Schlesiger S, Knauer SK & Schmuck C A tailor-made specific anion-binding motif in the side chain transforms a tetrapeptide into an efficient vector for gene delivery. Angewandte Chemie 127, 2984–2987 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Mitchell MJ et al. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov 20, 101–124, doi: 10.1038/s41573-020-0090-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M et al. Drug delivery systems based on CD44-targeted glycosaminoglycans for cancer therapy. Carbohydr Polym 251, 117103, doi: 10.1016/j.carbpol.2020.117103 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Mattheolabakis G, Milane L, Singh A & Amiji MM Hyaluronic acid targeting of CD44 for cancer therapy: from receptor biology to nanomedicine. J Drug Target 23, 605–618, doi: 10.3109/1061186X.2015.1052072 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Luo Z, Dai Y & Gao H Development and application of hyaluronic acid in tumor targeting drug delivery. Acta Pharm Sin B 9, 1099–1112, doi: 10.1016/j.apsb.2019.06.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qhattal HS, Hye T, Alali A & Liu X Hyaluronan polymer length, grafting density, and surface poly(ethylene glycol) coating influence in vivo circulation and tumor targeting of hyaluronan-grafted liposomes. ACS Nano 8, 5423–5440, doi: 10.1021/nn405839n (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng C et al. Desirable PEGylation for improving tumor selectivity of hyaluronic acid-based nanoparticles via low hepatic captured, long circulation times and CD44 receptor-mediated tumor targeting. Nanomedicine 24, 102105, doi: 10.1016/j.nano.2019.102105 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Subhan MA, Yalamarty SSK, Filipczak N, Parveen F & Torchilin VP Recent Advances in Tumor Targeting via EPR Effect for Cancer Treatment. J Pers Med 11, doi: 10.3390/jpm11060571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sindhwani S et al. The entry of nanoparticles into solid tumours. Nature Materials 19, 566-+, doi: 10.1038/s41563-019-0566-2 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Griffioen AW et al. CD44 is involved in tumor angiogenesis; an activation antigen on human endothelial cells. Blood 90, 1150–1159 (1997). [PubMed] [Google Scholar]
- 27.Auerbach R, Akhtar N, Lewis RL & Shinners BL Angiogenesis assays: problems and pitfalls. Cancer Metastasis Rev 19, 167–172, doi: 10.1023/a:1026574416001 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Vojtek M et al. Fast and reliable ICP-MS quantification of palladium and platinum-based drugs in animal pharmacokinetic and biodistribution studies. Anal Methods 12, 4806–4812, doi: 10.1039/d0ay01328e (2020). [DOI] [PubMed] [Google Scholar]
- 29.Kumar V et al. Pharmacokinetics and biodistribution of polymeric micelles containing miRNA and small-molecule drug in orthotopic pancreatic tumor-bearing mice. Theranostics 8, 4033–4049, doi: 10.7150/thno.24945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H & Guo P Radiolabeled RNA Nanoparticles for Highly Specific Targeting and Efficient Tumor Accumulation with Favorable In Vivo Biodistribution. Mol Pharm 18, 2924–2934, doi: 10.1021/acs.molpharmaceut.1c00035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vocelle D, Chan C & Walton SP Endocytosis Controls siRNA Efficiency: Implications for siRNA Delivery Vehicle Design and Cell-Specific Targeting. Nucleic Acid Ther 30, 22–32, doi: 10.1089/nat.2019.0804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Y, Siegwart DJ & Anderson DG Strategies, design, and chemistry in siRNA delivery systems. Adv Drug Deliv Rev 144, 133–147, doi: 10.1016/j.addr.2019.05.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song W et al. Synergistic and low adverse effect cancer immunotherapy by immunogenic chemotherapy and locally expressed PD-L1 trap. Nat Commun 9, 2237, doi: 10.1038/s41467-018-04605-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lima LG, Chammas R, Monteiro RQ, Moreira ME & Barcinski MA Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett 283, 168–175, doi: 10.1016/j.canlet.2009.03.041 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Sharma R, Huang X, Brekken RA & Schroit AJ Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br J Cancer 117, 545–552, doi: 10.1038/bjc.2017.183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proto JD et al. Regulatory T Cells Promote Macrophage Efferocytosis during Inflammation Resolution. Immunity 49, 666–677 e666, doi: 10.1016/j.immuni.2018.07.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi L et al. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget 7, 71673–71685, doi: 10.18632/oncotarget.12317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray MJ et al. Phosphatidylserine-targeting antibodies augment the anti-tumorigenic activity of anti-PD-1 therapy by enhancing immune activation and downregulating pro-oncogenic factors induced by T-cell checkpoint inhibition in murine triple-negative breast cancers. Breast Cancer Res 18, 50, doi: 10.1186/s13058-016-0708-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder AG et al. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci Immunol 4, doi: 10.1126/sciimmunol.aaw2004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Hardie J, Zhang X & Rotello VM Effects of engineered nanoparticles on the innate immune system. Semin Immunol 34, 25–32, doi: 10.1016/j.smim.2017.09.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawano M & Nagata S Lupus-like autoimmune disease caused by a lack of Xkr8, a caspase-dependent phospholipid scramblase. Proceedings of the National Academy of Sciences of the United States of America 115, 2132–2137, doi: 10.1073/pnas.1720732115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S et al. Effect of immune response on gene transfer to the lung via systemic administration of cationic lipidic vectors. The American Journal of Physiology 276, 796–804 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Chen Y et al. An immunostimulatory dual-functional nanocarrier that improves cancer immunochemotherapy. Nature Communications 7, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shum K & Rossi JJ SiRNA delivery methods : methods and protocols. (Humana Press, 2016). [Google Scholar]
- 45.Sun J et al. A prodrug micellar carrier assembled from polymers with pendant farnesyl thiosalicylic acid moieties for improved delivery of paclitaxel. Acta Biomaterialia 43, 282–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tseng W, Leong X & Engleman E Orthotopic mouse model of colorectal cancer. J Vis Exp, 484, doi: 10.3791/484 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP & Johnson JM Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11, 1737–1744, doi: 10.1261/rna.2148705 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynch RW et al. An efficient method to isolate Kupffer cells eliminating endothelial cell contamination and selective bias. J Leukoc Biol 104, 579–586, doi: 10.1002/JLB.1TA0517-169R (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorgun C et al. Isolation and Flow Cytometry Characterization of Extracellular-Vesicle Subpopulations Derived from Human Mesenchymal Stromal Cells. Curr Protoc Stem Cell Biol 48, e76, doi: 10.1002/cpsc.76 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Ray A & Dittel BN Isolation of mouse peritoneal cavity cells. J Vis Exp, doi: 10.3791/1488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horuluoglu BH, Kayraklioglu N, Tross D & Klinman D PAM3 protects against DSS-induced colitis by altering the M2:M1 ratio. Sci Rep 10, 6078, doi: 10.1038/s41598-020-63143-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turnis ME et al. Interleukin-35 Limits Anti-Tumor Immunity. Immunity 44, 316–329, doi: 10.1016/j.immuni.2016.01.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk messenger RNA-seq data mapped to the mouse genome (GRCm38: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/) are available in the NCBI Gene Expression Omnibus (GEO) under accession no. GSE214881 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214881). Source data are provided with this paper. All data generated or analyzed during this study are included in this published article and its Supplementary information files.