Abstract
Single-nucleus RNA-sequencing technology has revolutionized understanding of nuanced changes in gene expression between cell types within tissues. Unfortunately, our understanding of regulatory RNAs, such as microRNAs (miRNAs), is limited through both single-cell and single-nucleus techniques due to the short length of miRNAs in the cytoplasm and the incomplete reference of longer primary miRNA (pri-miRNA) transcripts in the nucleus. We build a custom reference to align and count pri-miRNA sequences in single-nucleus data. Using young and aged subventricular zone (SVZ) nuclei, we show differential expression of pri-miRNAs targeting genes involved in neural stem cells (NSC) differentiation in the aged SVZ. Furthermore, using wild-type and 5XFAD mouse model cortex nuclei, to validate the use of primiReference, we find cell-type specific expression of pri-miRNAs known to be involved in Alzheimer’s Disease. pri-miRNAs likely contribute to NSC dysregulation with age and Alzheimer’s Disease pathology. primiReference is paramount in capturing a global profile of gene expression and regulation in single-nucleus data and can provide key insights into cell-type specific expression of pri-miRNAs, paving the way for future studies of regulation and pathway dysregulation. By looking at pri-miRNA abundance and transcriptional differences, regulation of gene expression by miRNAs in disease and aging can be further explored.
Keywords: primary-microRNA, Aging, subventricular zone, Alzheimer’s Disease, Single-Nucleus RNA-Seq, microRNA, Transcriptomics
Introduction
There are two neural stem cell (NSC) niches in the adult mammalian brain, the subventricular zone (SVZ) and the subgranular zone (SGZ) of the dentate gyrus in the hippocampus. NSCs in the SVZ are multipotent, giving rise to neurons, astrocytes, and oligodendrocytes while NSCs of the hippocampus are unipotent, giving rise to neurons. With age, NSCs decline in number, self-renewal capacity, and ability to produce new neurons (Conover and Shook, 2011; Daynac et al., 2016). The mechanisms leading to this decline are not well understood. Within the SVZ niche, there is close interaction among the cell-types (Lim and Alvarez-Buylla, 1999; Massirer et al., 2011; Matarredona et al., 2018). Ependymal cells line the ventricle wall and are in close contact with the cerebrospinal fluid, while astrocytes, oligodendrocytes, NSCs, and vasculature make up the majority of SVZ. Neuronal processes can protrude into the SVZ from the striatum nearby (Quinones-Hinojosa et al., 2006; Lim and Alvarez-Buylla, 2016). NSCs have been shown to interact with surrounding cells either directly through cell to cell contact or in a paracrine fashion through secreted factors (Lim and Alvarez-Buylla, 1999; Matarredona et al., 2018). Due to these interactions, age-associated NSC decline could be influenced by changes occurring in the surrounding cells within the niche in addition to changes within the NSCs directly. Therefore, it is important to understand changes occurring in both the NSCs and their adjacent, interacting cell-cell types in order to obtain a comprehensive assessment of niche decline.
Over the past decade, single-cell and single-nucleus sequencing techniques have revolutionized the transcriptomics and epigenomics fields. By performing RNA sequencing (RNA-seq) on a single-cell level, scientists can identify subtle changes in gene-expression on a cell-type specific basis as opposed to the overall changes in tissue observed with bulk-RNAseq. Using single-cell, microfluidics-based techniques such as Smart-Seq and 10X Genomics, thousands of cells per tissue can be profiled. On the downside, however, tissues such as brain have cells containing long processes that can be sheared resulting in insufficient capture of the cell types. To circumvent this issue, single-nucleus sequencing technologies arose. Single-nucleus and single-cell technologies have similar, overall sensitivity, however both technologies are missing key information on small regulatory RNAs such as microRNAs (miRNAs), due to their cytoplasmic location and small size (Ding et al., 2020; Thrupp et al., 2020). To fully understand how transcriptional changes with age can influence niche decline it is necessary to include miRNA expression, since miRNAs are potent regulators of gene expression and have been shown to be involved in intercellular communication, especially in regulating neurogenesis (Papagiannakopoulos and Kosik, 2009; Ji et al., 2013; Ma et al., 2019). To address this deficit, we built a reference that can be used for single-nucleus sequencing data to discern expression of both coding genes and primary-miRNAs (pri-miRNA) sequences.
miRNAs are generated in the nucleus as pri-miRNA transcripts by RNA Polymerase II (Kim and Kim, 2007). The pri-miRNA transcripts are subsequently processed into pre-miRNA sequences that are exported to the cytoplasm where they are further processed into the mature miRNAs (Ha and Kim, 2014; O’Brien et al., 2018). The mature miRNAs remain in the cytoplasm where they regulate the translation of their target mRNA transcripts. Pri-miRNAs can be monocistronic or polycistronic transcripts encoding miRNA gene sequences only, or contain protein coding and miRNA coding sequences simultaneously (Ha and Kim, 2014). Current single-cell sequencing technologies are unable to capture mature miRNA sequences due to their small size, which are in the range of 21–24 nucleotides (Lee et al., 2002; Ha and Kim, 2014). Most pri-miRNA transcripts are several kilobases long (Lee et al., 2002; Ha and Kim, 2014; Chang et al., 2015), allowing them to be detected in single-nucleus sequencing data. However, pri-miRNAs are not currently annotated in the reference genomes for single-nucleus sequencing data. To overcome this obstacle, we combined a publicly available dataset (Chang et al., 2015) of pri-miRNA sequences with the current mouse genome annotation (mm10) to create primiReference, which can be used to identify changes in the expression of both genes and pri-miRNA sequences in single-nucleus sequencing data. Here, we use primiReference to investigate cell-type specific changes in gene and pri-miRNA expression in the SVZ isolated from young and old mice. We expand upon this data by exploring cell-type specific changes in pri-miRNA and gene expression in the cortex of the 5XFAD Alzheimer’s Disease mouse model and wildtype (WT) mice (Zhou et al., 2020). By including the analysis of pri-miRNA expression, we are able to gain insight into possible mechanisms of age-associated decline and Alzheimer’s Disease pathology in a cell-type specific manner.
Results
primiReference creation
To identify cell-type specific changes in miRNA expression with age in the neurogenic niche, we performed single-nucleus sequencing of the SVZ isolated from young (4-month-old) and old (25–26-month-old) mice. Single-cell and single-nucleus technologies do not allow for the capture and identification of mature miRNAs due to their small size. To circumvent this issue, we identified overlaps between SVZ data aligned to the mm10 genome and a publicly available annotation of pri- miRNAs from six cell lines to identify pri-miRNA transcripts in the single-nucleus sequencing data (Chang et al., 2015). We subsequently added the pri-miRNA overlaps to the mm10 genome annotation to develop a reference to identify pri-miRNA transcripts and other coding sequences (primiReference). The pri-miRNA sequences identified by primiReference overlap with the annotated pre-miRNA sequence, which is usually around 70 bp, and additional sequence upstream and downstream (Fig. 1A). When viewing intragenic miRNAs, the pre-miRNA sequence is often within the intron, and the pri-miRNA transcript is the unspliced transcript of the entire gene (Fig. 1B). Accordingly, in primiReference intragenic pri-miRNAs are named as the gene name followed by the pri-miRNA name. By doing so, both gene and pri-miRNA transcript abundance is determined at once since their expression cannot be disentangled through single-nucleus sequencing. Consequently, the primiReference annotation leads to accurate mapping of pri-miRNA transcripts to the genome.
Fig. 1.
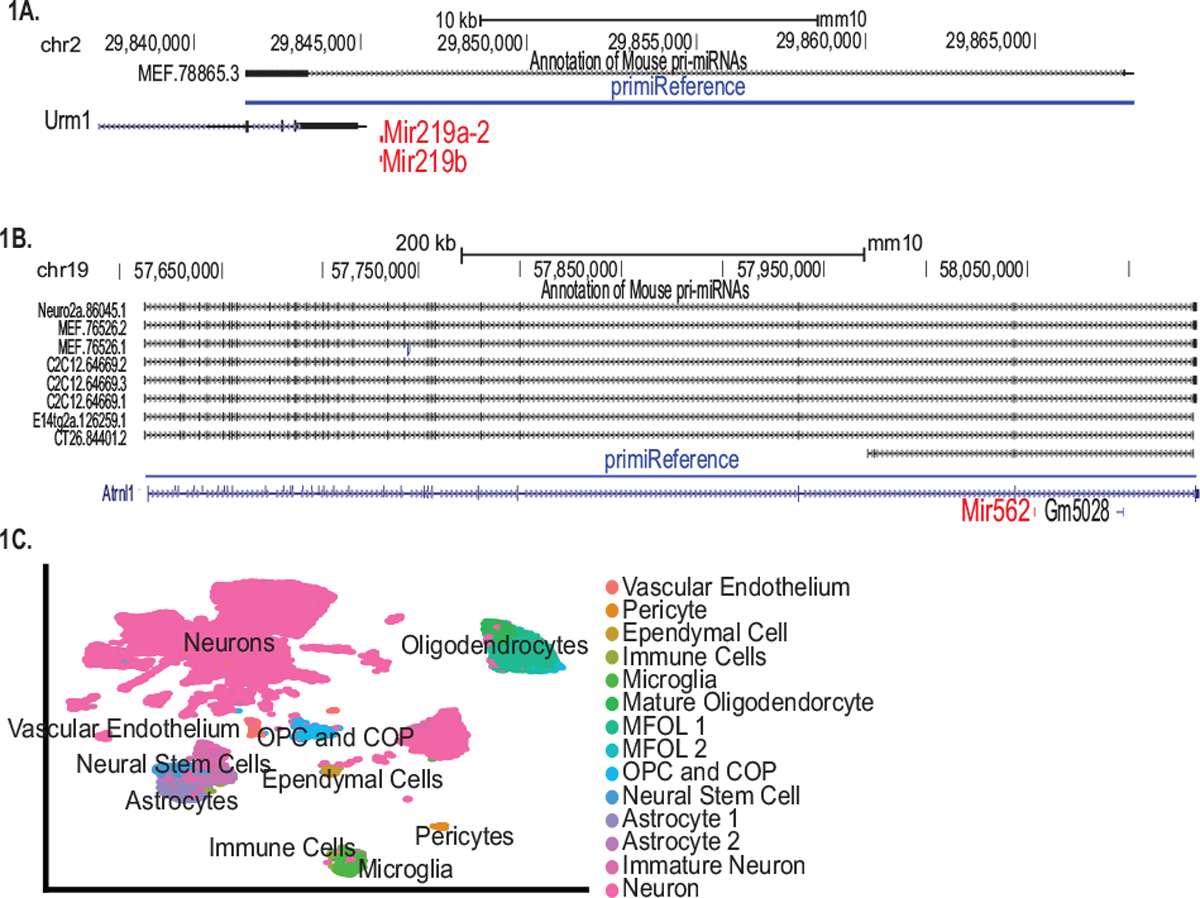
primiReference design and resulting clustering of cell types in the young and aged SVZ. A: Depicted using the UCSC genome browser, the mouse embryonic fibroblast sequence of the pri-miRNA for miR-219a-2 was shown. The primiReference generated alignment, shown in blue, overlaps this intergenic primary miRNA sequence. B: Several cell lines from a publicly available dataset were shown to have pri-miRNAs mapping to the Atrnl1 gene from which the primary-miR562 gene was transcribed intragenically from an intron. The primiReference alignment overlapped with the consensus of several cell lines. C: UMap dimensional reduction showing cell-type specific clusters in the SVZ.
Identification of cell clusters in the SVZ
Following alignment to primiReference, we filtered the data, conducted dimensional reduction and identification of clusters (Butler et al., 2018) (Figs. 1C, S1 and S2; Additional File 2). The clusters for the aging SVZ data show clear separation among the expected cell types found in the SVZ niche (Fig. 1C). The clusters were classified using the Allen Brain Atlas initially, and markers from literature were used to further define subtypes of mural cells (pericytes and other vessel-associated subtypes), oligodendrocytes, and astrocytes (Fig. 1C) (Herbert et al., 1986; Thiel, 1993; He et al., 2016; Marques et al., 2016; Tasic et al., 2016; Dulken et al., 2017; Saito et al., 2018; Shah et al., 2018; Zywitza et al., 2018; Stuart et al., 2019; Ximerakis et al., 2019). The neuronal subtypes were consolidated into a single neuronal class since they were likely striatal contamination due to the lack of defined borders of the SVZ. Furthermore, three clusters were unable to be fully defined based on the Allen Brain Atlas and literature, and these clusters, along with the choroid plexus, were excluded from the analysis.
The clusters defining the SVZ neurogenic niche - NSCs, oligodendrocyte progenitors (OPCs) and committed oligodendrocyte progenitors (COPs), mature oligodendrocytes, myelin forming oligodendrocytes (MFOLs), immature neurons, neurons, microglia, and mural cells – were strongly represented in samples (Figs. 1C and S2). Among the cell-types, there was no bias in pri-miRNA content within each cluster as a percentage of all genes expressed (Fig. S3A). Likewise, there was no bias in pri-miRNA content per condition as a percentage of all genes expressed (Fig. S3B). These results indicate that primiReference produces accurate mapping, since primiReference was shown to have comparable expression of pri-miRNAs to coding genes. Differential expression analysis was conducted between the young and aged male mice within the cell types of the SVZ niche to gain insights into the pri-miRNAs that may contribute to the overall decline of the niche with age (Additional File 3).
Differential expression of pri-miRNAs, within the cell-types of the subventricular zone niche
We examined changes in pri-miRNA expression with age in the major SVZ cell types and found that each cell type had a unique signature of age-associated changes in the pri-miRNAs (Fig. 2A; Table 1; Additional File 4). While there were several pri-miRNAs that changed in most or all the cell types examined, the majority of pri-miRNAs changed in only one or a few cell types (Fig. 2A). Some cell types have pri-miRNAs that change only in that cluster, indicating that changes in pri-miRNA expression with age can be cell-type specific (Fig. S4A). While immature neurons and pericytes had pri-miRNAs that changed with age (Additional File 4; Table 1), however, these will not be discussed further in this manuscript. We focused only on the pri-miRNAs that were differentially expressed in the major cell types that comprise the neurogenic niche in the SVZ.
Fig.2.
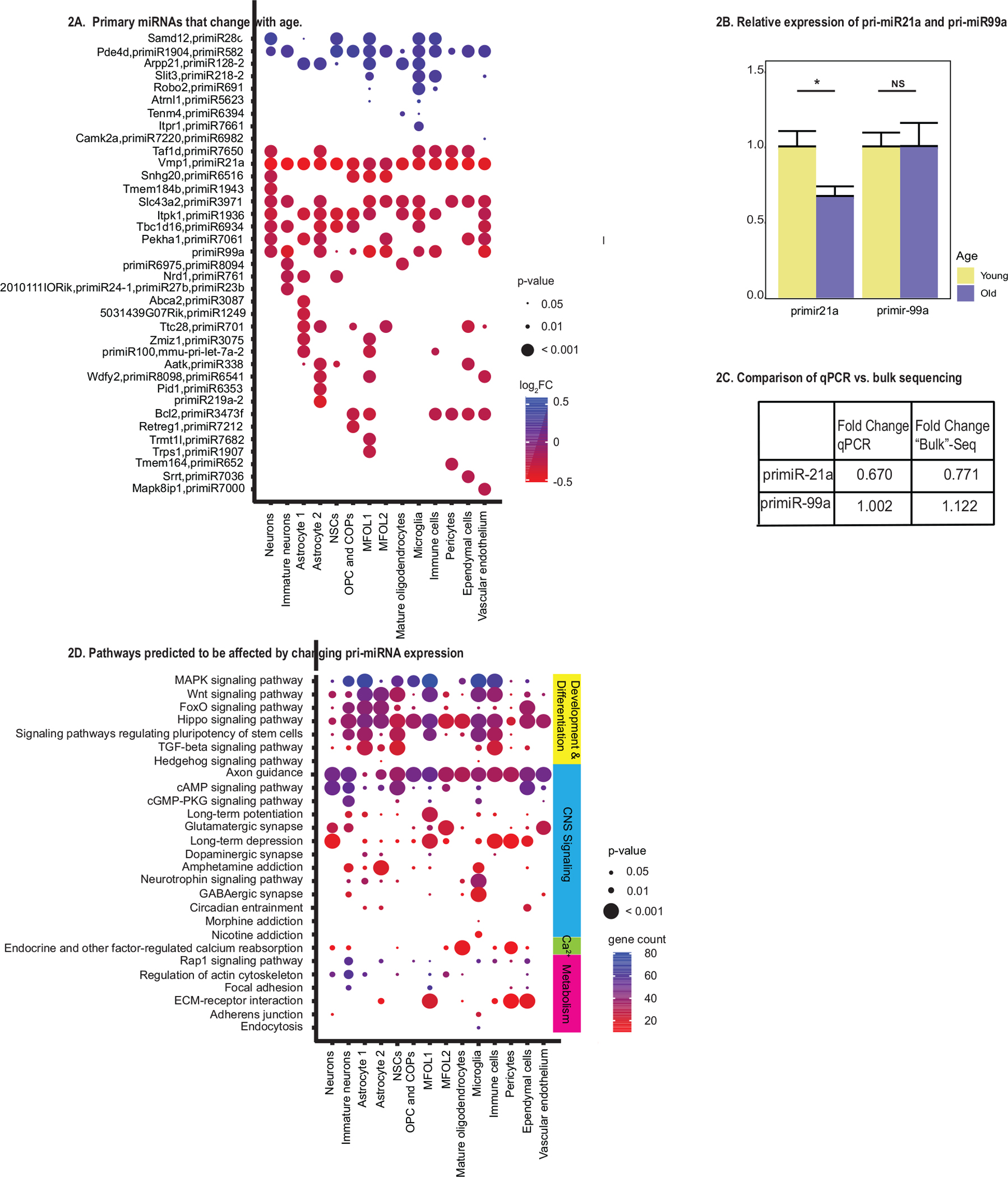
Primary miRNAs and pathways predicted to be affected in aged SVZ. A: Significantly changing primary miRNAs in the aged SVZ. Pri-miRNAs with significant differences in expression between young and old SVZ (P < 0.05). Per cell-type are shown by bubble plot. Those increasing in expression with age are shown in blue, with color intensity depicted by the log2 fold-change, while pri-miRNAs decreasing in expression with age are shown in red. B: The relative expression of primiR-21a and primiR-99a are shown from qPCR data. The yellow highlights the relative expression of whole young SVZ tissue, while the blue represents the relative expression of whole old SVZ tissue. For primiR-21a the fold change between old and young SVZ was significant (*, P < 0.05). C: Fold-change values between old and young SVZ for primiR-21a and primiR-99a in “bulk” sequencing data and qPCR. Note the change was similar using both methods. D: Bubble plot showing KEGG pathways enriched in predicted target genes of the mature miRNA sequences of pri-miRNAs that change in expression in the aged SVZ. The KEGG pathways depicted in this plot belonged to the general categories of development and differentiation, CNS signaling pathways, calcium signaling, and metabolism. All other pathways can be found in Additional File 6. Bubble color indicates the predicted number of genes affected by the changing pri-miRNAs in each pathway. Bubble size indicates the level of significance, with larger sizes indicating greater significance.
Table 1.
Function of Mature miRNAs of pri-miRNA Transcripts Differentially Expressed in Aging SVZ and 5XFAD Cortex.
| Tissue | miRNA | Cell types with DE of the pri-miRNA transcript in Aged SVZ compared to Young SVZ | Cellular Processes and Gene Targets |
|---|---|---|---|
|
| |||
| SVZ | miR-21a | ↓Expression: All cell types | Increases neurogenesis (Ma et al., 2019), found to be associated with the aging mouse (Kim et al., 2017) |
| miR-99a | ↓Expression: Neuron, Immature Neuron, Astrocyte2, NSC, OPC and COP, MFOL1, MFOL2, Microglia, Immune Cells, Vascular Endothelium | Decreases mTOR expression (Tsai et al., 2018). Likely involvement in commitment of progenitors to glial lineage (Stevanato and Sinden, 2014). | |
| miR-24–1 | ↓Expression: Immature Neuron | Knockout of this cluster leads to decreased NAD and other glycolytic metabolites (Jiang et al., 2021). Impaired glycolysis prevents neuronal differentiation and maturation (Agostini et al., 2016). Furthermore, this cluster is increased in terminally differentiated cells (Lal et al., 2009). | |
| miR-27b | ↓Expression: Immature Neuron | Mir27b targets AMPK (Wang et al., 2019b). Increases in AMPK are associated with aged hippocampal NSCs and their decline (Wang et al., 2019a). Knockout of this cluster leads to decreased NAD and other glycolytic metabolites (Jiang et al., 2021). Impaired glycolysis prevents neuronal differentiation and maturation (Agostini et al., 2016) Furthermore, this cluster is increased in terminally differentiated cells (Lal et al., 2009). | |
| miR-23b | ↓Expression: Immature Neuron | Knockout of this cluster leads to decreased NAD and other glycolytic metabolites (Jiang et al., 2021). Impaired glycolysis prevents neuronal differentiation and maturation (Agostini et al., 2016). Furthermore, this cluster is increased in terminally differentiated cells (Lal et al., 2009). | |
| miR-100 | ↓Expression: Astrocytel, MFOL1, Immune cells | Mir100 activates the innate immune response resulting in activated microglia and neurodegeneration (Wallach 2021, Chawla 2016). | |
| Let-7a | ↓Expression: Astrocyte1, MFOL1, Immune cells | Let7a activates the innate immune response by acting as a TLR7 ligand (Nazmi 2014). Involved in neuroinflammation through microglia polarization (Yang 2018). | |
| miR-219a-2 | ↓Expression: Astrocyte2, | Increases myelination and re-myelination as well as increasing OPC differentiation (Dugas et al., 2010; Wang et al., 2017; Osorio-Querejeta et al., 2020; Brandi et al., 2021). Decreased levels have been described in neurodegenerative diseases such as MS and AD (Bruinsma et al., 2017). In AD models, it has been shown that miR-219a can regulate tau expression (Arnes et al., 2019). | |
| miR-338 | ↓Expression: Astrocyte1, Astrocyte2, NSC, Ependymal Cell, | Knockout of miR-338 causes a worsened phenotype of dysregulated myelination of miR219 mutants (Wang et al., 2017). These miRNAs work together to induce proper myelination. | |
| miR-582 | ↑ Expression: Neuron, Immature Neuron, NSC, OPC and COP, MFOL1, MFOL2, Mature Oligodendrocyte, Microglia, Immune Cells, Pericyte, Ependymal Cell, Vascular Endothelium | Represses various target genes in the apoptosis pathway (Floyd et al., 2014). | |
| miR-128–2 | ↑ Expression: Astrocyte1, Astrocyte2, NSC, MFOL1, Mature Oligodendrocyte, Microglia | Directly targets stem cell factors such as KLF4, CSF1, NANOG, and SNAIL (Qian et al., 2012). Furthermore, it has been shown that this miRNA reduced NPC proliferation (Zhang et al., 2016). Also inhibits mTOR signaling (Chen et al., 2016). | |
| miR-218–2 | ↑ Expression: MFOL1, Microglia, Immune Cells, Vascular Endothelium | Increased expression in human Alzheimer’s Disease patients has been observed (Gugliandolo et al., 2020). Additionally, this miRNA can alter the Wnt signaling pathway leading to differentiation - an important part of neurogenesis (Hu et al., 2017). | |
| 5XFAD cortex | miR-124–1 | ↓Expression: L5 PT, NP, PValb, Lamp5, Immature Neuron, Astrocyte1, OPC1, MFOL1, MFOL2, MFOL3, VLMC | Targets BACE1 expression involved in Alzheimer’s Disease (An et al., 2017), and mir-124 was found to be decreased in the brains of AD patients (Smith et al., 2011). Additionally, the gene Drd2 is targeted by miR-124-1 and thus the decreased miR-124-1 expression decreases prefrontal cortex performance (Kozuka et al., 2019). |
| miR-3078 | ↓Expression: L5 PT, NP, PValb, Lamp5, Immature Neuron, Astrocyte1, OPC1, MFOL1, MFOL2, MFOL3, VLMC | Decreased in APP/PS1 mice compared to controls (Zhang et al., 2021). | |
Note: Column 1 indicates the mature miRNA name for pri-miRNAs that are differentially expressed with age in the SVZ or in the 5XFAD mouse cortex that have validated target genes in the literature. Column 2 indicates the direction of miRNA differential expression as well as which cell-types are differentially expressing these miRNAs in the SVZ with age. Column 3 indicates the direction and cell-type of differentially expressed miRNA in the 5XFAD cortex compared to wild-type. Column 4 describes relevant literature on the miRNA.
All cell types
Several of the mature miRNAs encoded by the pri-miRNAs that changed globally have documented functions in the central nervous system. pri-miR-21a and pri-miR-99a decreased in all or the majority of cell types, respectively (Fig. 2A). Interestingly, pri-miR-21a is one of the most significantly changed pri-miRNA in all cell types. Dysregulation of mature miR-21a and mir-99a have been linked to impaired neurogenesis and neurodegeneration (Stevanato and Sinden, 2014; Tsai et al., 2018; Ma et al., 2019). The polycistronic transcript encoding pri-miR-1904 and pri-miR-582 increased in the majority of cell types with age (Fig. 2A). While nothing is known about the targets of the mature miR-1904, the expression of mature miR-582 has been linked to neuronal injury and inflammation (Zhang and Zhang, 2020), and NSC maintenance (Zhang et al., 2022).
Astrocytes
Two clusters of astrocytes were defined in the SVZ (Fig. 1C), each with a distinct combination of pri-miRNAs that change with age (Fig. 2A). Overall, there were 19 pri-miRNAs that changed with age across the 2 clusters (Fig. 2A; Additional File 4), with 6 pri-miRNAs changing with age in both clusters (Fig. 2A; Additional File 4). In addition, there were six pri-miRNAs that changed in only in astrocyte cluster 1 and seven pri-miRNAs that changed only in astrocyte cluster 2. Moreover, four pri-miRNAs changed only in the astrocyte clusters and not in any other cell type (Fig. S4A). The astrocyte-specific pri-miRNAs changing with age include pri-miR-3087 and pri-miR-1249, which uniquely decrease in expression, with age, within the astrocyte cluster 1 while pri-miR-6353 and pri-mir-219a uniquely decreases in expression, with age, in the astrocyte cluster 2 (Fig. S4A).
NSCs
The changes occurring in NSC gene expression are paramount to our understanding of niche decline as a whole. Overall, nine pri-miRNAs significantly changed in expression with age in this population (Fig. 2A; Additional File 4). While none of the changing pri-miRNAs are unique to NSCs, four of the mature miRNAs are involved in NSC maintenance and differentiation. These include pri-miR-21a, pri-miR-582, pri-miR-128-2 and pri-miR-99a.
Oligodendrocytes
Within the SVZ, we identified several different subtypes of oligodendrocytes, including one cluster consisting of oligodendrocyte progenitor cells and committed oligodendrocyte progenitors (OPC and COPs, respectively), two myelin forming oligodendrocyte clusters (MFOL1 and MFOL2), and one cluster consisting of mature oligodendrocytes (Figs. 1C and S2A). Although all these cell-types are different subtypes of oligodendrocytes, changes in the pri-miRNA expression with age differs between the cell-types (Fig. 2A; Additional File 4). Overall, 20 pri-miRNA transcripts changed with age within these clusters. Only two pri-miRNA transcripts were found to change in all four clusters: pri-miR-21a, and the polycistronic pri-miR-1904, pri-miR-582 transcript (Fig. 2A; Additional File 4). The OPC and COP cluster contains one pri-miRNA that uniquely changes with age, pri-miR-7212 decreases with age (Figs. 2A and S4A). The two MFOL clusters show distinct patterns of differentially expressed pri-miRNAs clusters, (Fig. 2A; Additional File 4). There are 17 pri-miRNAs that change with age in MFOL1, whereas only 7 pri-miRNAs change with age in MFOL2. Of these, MFOL1 and MFOL2 clusters share five pri-miRNAs that change significantly with age (Fig. S4A). The mature oligodendrocyte cluster also has a unique pattern of differentially expressed pri-miRNAs. There are seven pri-miRNAs that are differentially expressed with age in this cluster (Fig. 2A). These data show that along the oligodendrocyte differentiation pathway, from OPCs to mature oligodendrocytes, each cell type has a unique pattern of differentially expressed pri-miRNAs with age, indicating that different physiological pathways may be affected throughout differentiation leading to cellular dysfunction.
Validation of primiReference
Changes in pri-miRNA expression should lead to changes in levels of mature miRNAs, which will result in changes in expression levels of their target genes. To determine if the changes in pri-miRNA expression in the nucleus lead to functional changes in mature miRNA expression in the cytoplasm, we used a publicly available single-cell RNA-seq data from young and aged SVZ containing expression levels of mRNAs located in the cytoplasm, where mature miRNAs are active, to determine if the expression levels of experimentally confirmed target mRNAs (TarBase (V8), [Karagkouni et al., 2018]) were changing in the predicted direction (Dulken et al., 2019). The single-cell data was analyzed in the same manner as single-nucleus data, and differential expression as conducted by age, per cell-type (Fig. S5). We determined the number of experimentally validated target genes that were differentially expressed in the single-cell dataset for each cell type, and then determined if the expression of the target genes was in the direction expected based on the change in pri-miRNA expression. For example, pri-miR21a decreased in expression in all cell-types, we determined if mmu-miR-21a target genes were differentially expressed in each cell type and if they showed an increase in expression as expected from a decrease in mmu-miR-21a expression. Interestingly, 60%–86% of the targets that were differentially expressed in the single-cell data set were changing in the predicted direction (increased in expression), suggesting that the change in pri-miRNA expression in the nucleus may lead to a corresponding change in the mature miRNA expression in the cytoplasm. For all pri-miRNAs that had experimentally confirmed targets, an average of 72% of the experimentally validated target genes changed in the direction predicted by pri-miRNA expression (Table 2). Given that miRNAs act on their targets by either leading to mRNA degradation or inhibition of translation, the actual effects on target mRNAs may be greater than we see in the single-cell dataset. These results indicate that changes in pri-miRNA expression reflect changes in mature miRNA expression and action in the cytoplasm.
Table 2.
Differential Expression of miRNA Target Genes in Single-cell Data.
| miRNA | Cell-type | Number of target genes present in single cell expression data | Percentage changing in predicted direction | P-value1 |
|---|---|---|---|---|
|
| ||||
| miR-21a-3p | Neuron | 20 | 65% | 0.180 |
| Astrocyte | 22 | 64% | 0.201 | |
| qNSC | 20 | 60% | 0.371 | |
| aNSC1 | 25 | 72% | 0.028 | |
| aNSC2 | 22 | 86% | 0.001 | |
| MFOL | 28 | 61% | 0.257 | |
| MOL | 24 | 75% | 0.014 | |
| Microglia | 25 | 80% | 0.003 | |
| Pericyte | 21 | 76% | 0.016 | |
| Vascular Endothelium | 16 | 81% | 0.012 | |
| miR-338 | Astrocyte | 20 | 70% | 0.074 |
| Vascular Endothelium | 14 | 71% | 0.109 | |
| miR-99a | aNSC1 | 4 | 25% | N.D. |
| aNSC2 | 2 | 100% | N.D. | |
| Astrocyte | 1 | 100% | N.D. | |
| MFOL | 2 | 50% | N.D. | |
| Microglia | 1 | 100% | N.D. | |
| Neuron | 2 | 50% | N.D. | |
| qNSC | 1 | 100% | N.D. | |
| Vascular Endothelium | 2 | 50% | N.D. | |
| miR-582 | aNSC1 | 1 | 100% | N.D. |
| Vascular Endothelium | 1 | 100% | N.D. | |
| Pericyte | 1 | 100% | N.D. | |
| miR-1943 | Neuron | 1 | 0% | N.D. |
|
| ||||
| Average | 72% | 2.9 × 10−12 | ||
Note: miRNAs that have experimentally validated target genes in TarBase are shown in column 1. Column 2 indicates the cell-types that show differential expression of the target genes in the single-cell sequencing dataset. Column 3 indicates the number of target genes that are differentially expressed in each cell type in the single-cell sequencing dataset. Column 4 indicates the percentage of target genes that change in the predicted direction given the change in pri-miRNA expression in the single nucleus dataset. Column 5 indicates the P-value for each cell type and each miRNA.
Chi-squared test. Significant values are bolded. Significance was not determined (N.D.) for cell types containing less than 5 predicted targets. P = 2.9 × 10−12, Chi-squared test, for the combination of all cell types and miRNAs.
To validate the changes in expression in pri-miRNAs found in our data, quantitative polymerase chain reaction (qPCR) was conducted on pri-miR-99a and pri-miR-21a since these pri-miRNAs were decreasing in most cell-types. To have a fair comparison of expression levels between whole isolated SVZ in qPCR data and our single-nucleus dataset, the single-nucleus data was combined to form “bulk” RNA-sequencing data, so that cell-type specific expression was eliminated and only expression of total reads in old SVZ compared to young SVZ was shown (Fig. 2B and 2C; Additional File 5). In bulk sequencing data, pri-miR-21a changed by 0.77 in old mice while in qPCR data it was shown to decrease 0.67 (p<0.05). Meanwhile, pri-miR-99a lost its differential expression when analyzed in bulk sequencing data and the same was shown in qPCR data (Fig. 2B and 2C). This underscores the validity of primiReference, as it highlighted the single-nucleus sequencing’s ability to pick up on subtle changes in gene-expression within cell-types that were not shown in bulk-sequencing.
Pathways of predicted target genes of pri-miRNAs differentially expressed in aged SVZ compared to young SVZ
To determine the possible physiological consequences of the changes in expression of the pri-miRNAs elucidated in this study, mirPath (Vlachos et al., 2015) was used to identify potential pathways affected by changes in predicted target genes of the differentially expressed pri-miRNA for each cell type (Figs. 2D and S4B; Additional File 6). As expected, each cell type had a unique set of pathways that may be affected by the changing pri-miRNAs. Overall, pathways involved in development and differentiation, central nervous system signaling, calcium signaling, and cell-cell interaction were predicted to be affected by the combination of changing pri-miRNAs with age in the SVZ (Figs. 2D; Additional File 6). Interestingly, the pathway that changed most significantly in most cell types was the axon guidance pathway (Fig. 2D).
MAPK signaling, Wnt signaling, FoxO signaling, and Hippo signaling were enriched for predicted target genes of pri-miRNAs differentially expressed with age in almost every cell-type. This is interesting, since all these pathways regulate development and differentiation. When compared to pathways affected by changing protein-coding gene expression in the same dataset, there were several additional pathways predicted to be affected by changing pri-miRNAs (Figs. 2D and S6). These included Wnt signaling, FoxO signaling, signaling pathways regulating pluripotency of stem cells, TGF-β signaling and Hedgehog signaling. Interestingly, these pathways are involved in regulating differentiation. These results suggest that changing pri-miRNA expression can affect pathways that are not affected by changes in gene expression, giving additional insight into possible mechanisms of the SVZ niche decline with age.
Several of the differentially expressed pri-miRNAs elucidated from astrocyte clusters have experimentally confirmed target genes (Table 1). However, most pri-miRNAs that change with age in the astrocyte clusters do not have experimentally validated target genes. Astrocyte clusters 1 and 2 have increased expression of pri-miR-128-2 and the mature sequence targets stem cell factors KLF4, CSF1, and SNAI1, which help maintain stemness (Qian et al., 2012). miRPath predicts that the mature miRNAs of changing pri-miRNAs will target mRNAs in pathways largely involved in development and differentiation, CNS pathways, and cell-cell interaction, with the most predicted target gene enrichment in MAPK signaling, Wnt signaling, FoxO signaling, and Hippo signaling (Fig. 2D; Additional File 6). This is interesting because these pathways, including Wnt signaling, in astrocytes have been shown to affect NSC differentiation (Lie et al., 2005).
In the NSC cluster, most predicted target genes belong to pathways involved in CNS pathways and pathways involved with differentiation and development. NSCs show differential expression of pri-miRNAs targeting genes involved in FoxO signaling, TGFβ signaling, signaling of pathways regulating pluripotency of stem cells, MAPK signaling, Wnt signaling, and Hippo signaling. Together, these data demonstrate that changing pri-miRNA expression in the aged NSCs may impact the pathways regulating their differentiation.
Oligodendrocyte progenitors have differentially expressed pri-miRNAs that are predicted to target genes that are largely associated with pathways involved in development and differentiation, as well as cell-cell interaction. MAPK signaling, Wnt signaling, FoxO signaling, Hippo signaling, are all enriched for predicted target genes of pri-miRNAs changing with age in the OPCs (Fig. 2D; Additional File 6). These pri-miRNAs may play into the dysfunction in oligodendrogenesis observed with age.
The two MFOL clusters have differentially expressed pri-miRNAs that are predicted to target genes involved in pathways associated with development and differentiation (Fig. 2D; Additional File 6). Interestingly, the MFOL_1 cluster has predicted target genes in the fatty acid biosynthesis pathway (Additional File 6). Fatty-acid biosynthesis is essential for myelin formation. Although the mature oligodendrocyte cluster does not contain any unique pri-miRNAs that change with age, the pathways belonging to predicted target genes of the distinct combination of altered pri-miRNA expression with age are enriched for genes involved in development and differentiation and CNS signaling (Fig. 2D; Additional File 6).
In microglia, pathways containing the predicted target genes of the differentially expressed pri-miRNAs are heavily enriched for pathways involved in development and differentiation, and CNS signaling (Fig. 2D; Additional File 6). As expected, microglia have the greatest number of predicted target genes involved in the immune response. Thirty-one predicted target genes are involved in mRNA surveillance pathways while 33 predicted target genes are involved in T-cell receptor signaling (Additional File 6). Changes in these pathways are significant since microglia play a crucial role in the immune response in the brain.
Analysis of wildtype and 5XFAD mouse cortex aligned to primiReference
To validate the utility of the reference in working with different single-nucleus datasets, we used primiReference to characterize the pri-miRNA and gene expression profiles in the cortex of wild-type and the 5XFAD Alzheimer’s Disease mouse model using a publicly available single-nucleus sequencing dataset (Zhou et al., 2020). As seen through the UMap, there were far more clusters of neurons and a greater diversity of neurons as expected from the cortex (Fig. 3A). Moreover, different layers of cortical interneurons were identified. Like the SVZ, there was no bias in pri-miRNA content within each cluster as a percentage of all genes expressed (Fig. S3C). Additionally, there was no significant difference in the pri-miRNA content per condition as a percentage of all genes expressed (Fig. S3D). Since primiReference did not bias clustering based on cell identity, nor did primiReference uncover bias in overall pri-miRNA expression between cell-types in conditions, the cortex was further analyzed to gain insights into 5XFAD associated pri-miRNA changes within each cell type (Figs.3B and S7; Additional File 7).
Fig.3.
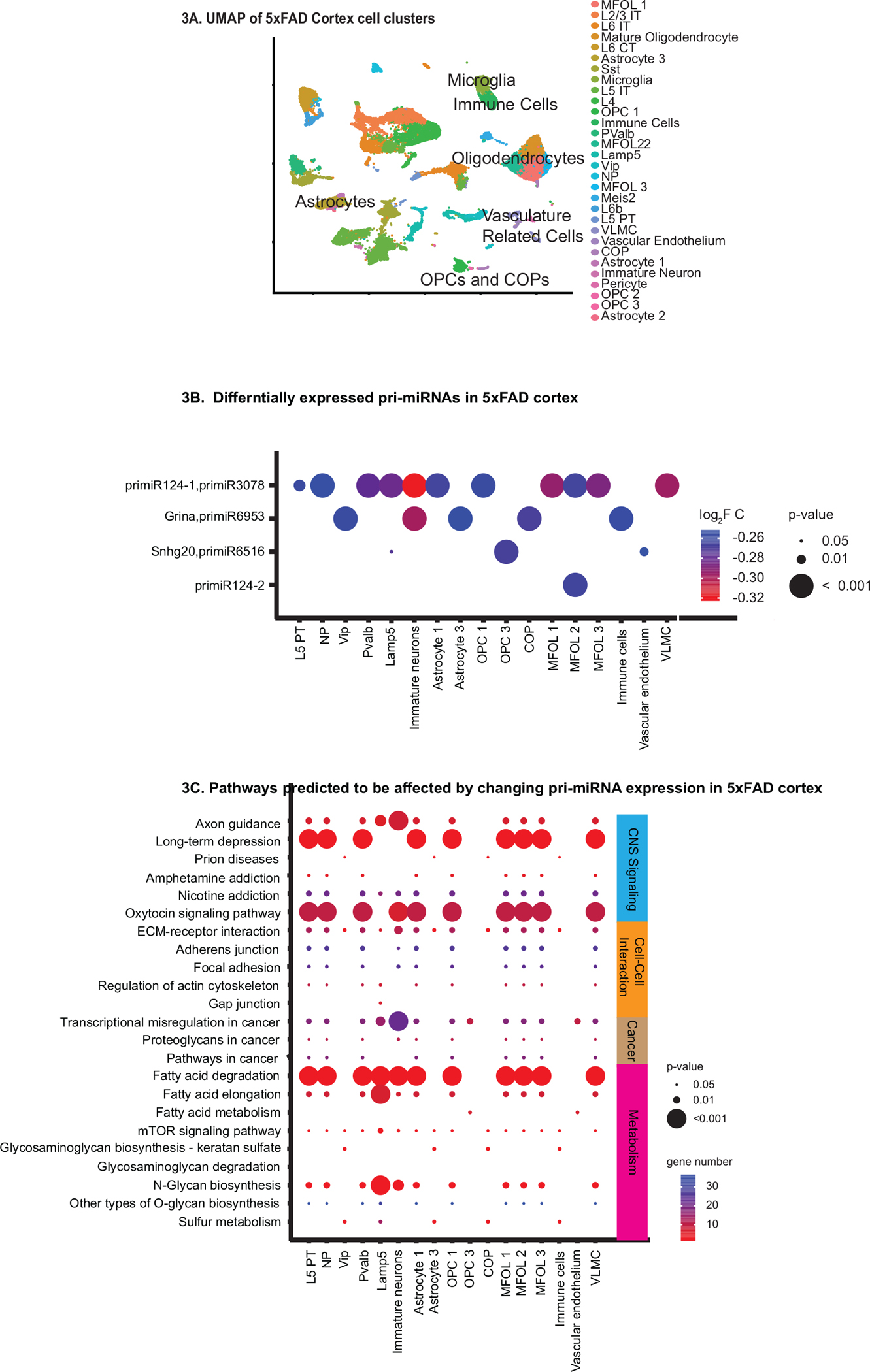
Clustering, pri-miRNA differential expression, and predicted target pathways of cell-types in WT and 5XFAD cortex. A: UMap dimensional reduction showing cell clusters in WT and 5XFAD cortex. B: Bubble plot showing pri-miRNAs with significant differences in expression between 5XFAD and wild-type cortex (P < 0.05) in each cell-type. Bubble color indicates the log2 fold change. Bubble size indicates the level of significance, with smaller size indicating greater significance. C: Bubble plot showing KEGG pathways enriched in predicted target genes of the mature miRNA sequences of pri-miRNAs that change in expression in the 5XFAD condition. The KEGG pathways belonged to CNS signaling pathways, cell-cell interaction, cancer, calcium signaling, and metabolism. All other pathways can be found in Additional file 6. Bubble color indicates the number of genes predicted to be affected by the changing pri-miRNAs in each pathway. Bubble size indicates significance, with larger size indicating greater significance.
Differential expression of pri-miRNAs within the cortex of 5XFAD mice compared to wildtype mice
All Cell-Types
There were far fewer pri-miRNAs changing in expression in the 5xFAD data than in the aged SVZ data, and not all cell types showed changes in pri-miRNA expression. Several cell types had unique profiles of differentially expressed pri-miRNAs (Fig. 3B). All of the differentially expressed pri-miRNAs decreased in expression in the 5XFAD mice compared to wild-type (Fig. 3B; Additional File 8). The primary transcript encoding primiR 124–1 and primiR-3078 was found to change in the majority of cell types. This is interesting since the mature miR-124-1 has been shown to have decreased expression in Alzheimer’s disease brains (Smith et al., 2011). These data indicate that changes in pri-miRNA expression in the 5XFAD model are cell-type specific and give insights into how changes in pri-miRNA expression can be associated with Alzheimer’s disease pathology.
Neurons
As expected from cortex data, there were more distinct populations of neurons identified in this dataset (Fig. 3A). These distinct populations were analyzed individually, instead of consolidating the clusters into one neuronal cell type as was done for the SVZ dataset, since it is important to understand the differences in the pri-miRNA expression in the different subtypes of neurons in the cortex in the context of Alzheimer’s Disease. There are fourteen subtypes of neurons identified in the cortex data, using the Allen Brain Atlas and the literature (Figs. 3A and S2B; Additional File 7).
The neuronal subtypes showed diversity in differential expression of pri-miRNA in the 5XFAD model. As expected, pri-miR-124 was found to decrease in expression within certain cell types in the 5XFAD condition, since this miRNA was shown to decrease in Alzheimer’s Disease (Santa-Maria et al., 2015; An et al., 2017; Arnes et al., 2019) (Fig. 3B; Table 1). pri-miR-124 deceases in expression in L5 PT, NP, PValb, Lamp5, immature neurons, astrocyte 1, OPC 1, all three MFOL clusters, and the VLMC cluster (Fig. 3B). These data showed that changes in pri-miRNA expression were cell-type specific in the 5XFAD model, suggesting that this may also be true in Alzheimer’s disease in humans. Although few pri-miRNAs found in this dataset have experimentally validated target genes, it is validated that the mature sequence of pri-miR-3078, which is co-transcribed with pri-miR-124-1, is found to decrease in APP/PS1 mice compared to wild-type mice thus showing consistency among Alzheimer’s models (Zhang et al., 2021) (Table 1).
Astrocytes
Three clusters of astrocytes were defined in this dataset (Fig. 3A). However, only astrocyte clusters 1 and 3 had differentially expressed pri-miRNAs. These astrocyte clusters only have one pri-miRNA transcript changing in the 5XFAD condition compared to wildtype, pri-miR-124-1 in astrocyte cluster 1 and pri-miR-6953 in astrocyte cluster 3. Overall, few of the mature miRNAs encoded by the pri-miRNAs elucidated in this study are a part of the scientific literature. Of those that are, miR-124-1, which is decreased in expression in the astrocyte 1 cluster, is known to decrease in expression in the brains of Alzheimer’s disease patients, and it is validated to target BACE1, which is heavily involved in Alzheimer’s disease pathology (Smith et al., 2011; An et al., 2017) (Table 1).
Oligodendrocytes
There were several additional distinct clusters of oligodendrocyte cell types defined in the cortex than in the aging SVZ (Fig. S2B). There are three OPC, one COP, three MFOL, and one mature oligodendrocyte clusters (Fig. 3A). Overall, four pri-miRNAs changed in the oligodendrocyte clusters. OPC clusters 1 and 3 showed decreases in a single pri-miRNA, the polycistronic pri-miR-124-1, pri-miR-3078 in cluster 1 and pri-miR-6516 in cluster 3. The COP cluster contained only one pri-miRNA that changed in the 5XFAD model, pri-miR-6953 decreased in expression in the 5xFAD model. Among the myelin forming oligodendrocytes (MFOLs), there were two pri-miRNAs changing in the 5XFAD condition (Fig. 3B). All 3 MFOL clusters had decreased expression of the polycistronic transcript primiR124-1, primiR3078. MFOL 2 also had decreased expression of pri-miR-124-2 in the 5XFAD model, which is encoded by an independent transcript from pri-miR-124-1 (Fig. 3B).
Pathways of predicted target genes of pri-miRNAs changing in the 5XFAD cortex compared to wild-type
The pathways affected by the predicted target genes of the mature sequences of the pri-miRNAs differentially expressed in the 5XFAD model fell into the CNS pathways, cell to cell interaction, cancer, and metabolism categories (Fig. 3C). There was no pathway that has enrichment of predicted target genes in all cell-types. This analysis showed the diversity of miRNA targeting in the cortex in the 5XFAD condition compared to wild-type.
Interestingly, there were a number of CNS pathways predicted to be affected by the changing pri-miRNAs in the 5XFAD model (Fig. 3C; Additional File 9). Several neuronal subtypes have predicted target genes within the axon guidance pathway. Alterations in this pathway could hinder CNS repair contributing to the neurodegeneration seen in Alzheimer’s Disease. Interestingly, the pri-miRNA differentially expressed in the Vip neurons in the 5XFAD model, pri-miR6953, has predicted target genes enriched for prion disease (Fig. 3C; Additional File 9). Prion protein has been implicated in Alzheimer’s Disease and serves as a potential therapeutic target (Um and Strittmatter, 2013; Salazar et al., 2017). L5 PT neurons have enrichment of predicted target genes for several pathways involved in cell-cell interaction (Fig. 3C; Additional File 9). And the pri-miRNAs that were differentially expressed in the 5XFAD condition in Lamp5 neurons largely targeted genes that fell into CNS signaling pathways and metabolic pathways (Fig. 3C; Additional File 9). There was diversity among enriched pathways among neuronal subtypes, supporting the idea that the 5XFAD mutations promoting the development of AD pathology affected pri-miRNA expression in a cell-type specific manner.
Astrocyte clusters 1 and 3 showed distinct pathways containing genes targeted by the differentially expressed pri-miRNAs (Fig. 3C; Additional File 9). The first astrocyte cluster has enrichment for pathways involved in CNS signaling, cell-cell interaction, cancer, and metabolism. The third cluster included unique pathways involved in prion disease, ECM-receptor interaction, and metabolism (Fig. 3C; Additional File 9). These data demonstrate that there is variability between the astrocyte clusters likely contributing to functional differences.
The oligodendrocyte clusters also show diversity in the pathways affected by changing pri-miRNAs in the 5XFAD model (Fig. 3C; Additional File 9). For the OPC clusters, only the first OPC cluster has predicted target genes belonging to pathways involved in CNS signaling and cell to cell interactions in the 5XFAD model compared to wild-type (Fig. 3C). The third OPC cluster had predicted target genes involved in fatty acid metabolism. In the COP cluster the mature sequences of the changing pri-miRNA were predicted to target genes involved in prion disease, ECM receptor interaction and metabolism (Fig. 3C; Additional File 9). The MFOL clusters had differential expression of the same pri-miRNAs. Pathways belonging to CNS signaling pathways, cell-cell interaction, cancer, and metabolism were predicted to be targeted by the mature forms of these miRNAs (Fig. 3C). These results indicate that changes induced by differential pri-miRNA expression in the oligodendrocyte clusters have different consequences in different cell-types.
Discussion
Currently, mature miRNAs are undetectable in single-cell data due to size exclusions used during sequencing library preparation, and pri-miRNAs are undetectable in single-nucleus data due to a lack of annotated reference. Since miRNAs serve as potent regulators of gene-expression, the inability for these transcripts to be detected on a single-nucleus/single-cell level is a great limitation to all fields utilizing this -omics technique. By creating an annotation of pri-miRNAs for single-nucleus sequencing data, this tool and strategy will now allow for novel insights into cell-type specific expression of pri-miRNA species.
Previously, only small-RNA-sequencing data was able to yield differential expression data of miRNAs via sequencing. This technique, however, was limited since information about gene expression was obtained from separate RNA-sequencing data. With use of primiReference, gene and pri-miRNA transcripts are analyzed within the same dataset. In addition, small RNA sequencing data had to be conducted in “bulk” – RNA sequencing libraries created from RNA isolated from whole tissues. Current single-cell and single-nucleus protocols do not capture small RNAs, meaning that there was no information about small RNA expression in individual cell types. With the development of single-nucleus sequencing, data can be derived about pri-miRNA expression on a cell-type specific basis. It also allows for discernment of changes in expression that may occur in only a small subset of cells that would have otherwise masked in bulk RNA-seq data.
To discover differential expression of pri-miRNAs with age in the brain, a neurogenic region of the brain, the SVZ, was selected. The SVZ contains several cell types, including NSCs, astrocytes, oligodendrocytes, neurons, ependymal cells, and mural cells. Since this region is neurogenic, there are also diverse subtypes of progenitors, since the NSCs are multipotent and can differentiate into astrocytes, neurons, and oligodendrocytes. Understanding cell-type specific differences in the SVZ gives insight into the functioning of the neurogenic niche, which relies heavily on cell-cell signaling (Song et al., 2002). Since miRNAs are highly involved in regulation of development and maturation of cell types, it was vital to explore differences in pri-miRNAs with age in this region. Additionally, the publicly available single-nucleus data set on wild-type and 5XFAD mice was analyzed using the same workflow to determine the differential pri-miRNA expression in an Alzheimer’s disease model in a cell-type specific manner.
Through our analyses using primiReference, we found several differentially expressed pri-miRNAs that may aid in our overall understanding of NSC decline within the subventricular niche and decline as a whole. For example, pri-miR-21a is decreased with age in NSCs, and in all the other cell-types. An increase in miR-21a is associated with increased neurogenesis and a decrease in this miRNA is associated with aging (Kim et al., 2017; Ma et al., 2019). Mature miR-21a has been shown to be packaged into extracellular vesicles, which are important mediators of intercellular communication and are produced by all cell types in the central nervous system (Ma et al., 2019; Schiera et al., 2019). Therefore, changes in miR-21a in other cell types of the SVZ can impact NSC activity. Moreover, miR-21a is predicted to target SIRT1. A reduction of miR-21a could lead to an increased SIRT1 expression, and increased SIRT1 has been shown to increase in NSC differentiation into astrocytes as opposed to neurons, which is a hallmark of the aging SVZ niche (Prozorovski et al., 2008; Conover and Shook, 2011). Therefore, the decreased expression of this pri-miRNA could contribute to the decrease in neurogenesis within the SVZ with age (Ma et al., 2019). Another pri-miRNA transcript that was found to be changed NSCs, and in most other SVZ cell types, is the polycistronic transcript encoding pri-miR1904 and pri-miR582, which increases in expression in all cell types except astrocytes. While the targets of miR-1904 have not been investigated, the mature form of pri-miR-582 is known to repress gene expression of various targets in the apoptosis pathway, and it has been associated with NSC maintenance and neuronal inflammation (Floyd et al., 2014; Zhang and Zhang, 2020; Zhang et al., 2022). In addition, pri-miR-128-2 increases in expression in NSCs and mature miR-128-2 has been shown to regulate neural differentiation (Zhang et al., 2016,) Taken together, these results show that age-associated changes in pri-miRNA expression can influence NSC’s ability to differentiate into neurons and may help explain the decline in the formation of new neurons with age.
The predicted mRNA targets of all pri-miRNAs differentially expressed with age in the SVZ fell into pathways associated with development and differentiation such as MAPK signaling, Hippo signaling, signaling pathways regulating pluripotency of stem cells, FoxO signaling, and Wnt signaling. There is likely dysregulation of these pathways, and this may contribute to NSC exhaustion with age (Conover and Shook, 2011; Daynac et al., 2016). It is interesting to note that only MAPK signaling and Hippo signaling are predicted to be affected by changes in gene expression in this dataset. Signaling pathways regulating pluripotency of stem cells, FoxO signaling, and Wnt signaling were identified solely from differences in pri-miRNA expression. Therefore, analyzing changes in pri-miRNA expression together with changes in gene expression provides greater insight into potential mechanisms of age-associated decline than looking at only changes in gene expression. Taken together, the validated and predicted targets of the pri-miRNAs changing in the SVZ could contribute to our understanding of the mechanisms behind the reduction in neurogenesis with age.
Astrocytes in the SVZ play a significant role in regulating NSC differentiation and proliferation through secretion of factors involved in Notch, Wnt, and Shh signaling (Lim and Alvarez-Buylla, 1999; Falk and Gotz, 2017). The two astrocyte clusters had pri-miRNAs differentially expressed with age that are predicted to target genes in pathways associated with NSC differentiation and proliferation. The predicted targets of all pri-miRNAs differentially expressed with age in the astrocyte clusters belong to pathways associated with development and differentiation such as Hippo signaling, signaling pathways regulating pluripotency of stem cells, FoxO signaling, and Wnt signaling. Dysregulated expression of some of the predicted target genes of these pathways resulting from altered miRNA expression may lead to differences in NSC differentiation or proliferation, since signaling proteins like Bmp2 and Wnt3 are predicted target genes.
Along with impaired neurogenesis, some pri-miRNAs may be involved in immune modification. pri-miRNA-6394 is increased in mature oligodendrocytes and microglia, and is predicted to target the enzyme Neu1, which degrades sialo-modifications, and St6galnac, which catalyzes the transfer of sialic acid residues (Puigdellivol et al., 2020). Interestingly, sialylation has implications in neurodegeneration and neuroinflammation since a reduction in sialyation leads to inappropriate phagocytosis of neurons by activated microglia in an inflammatory environment, contributing to neuronal loss (Puigdellivol et al., 2020). Additionally, miRNAs changing with age in the astrocyte clusters with age are predicted to target Ikbkb, which could impact NF-κB signaling. These results indicate that changes in pri-miRNA expression could contribute to neuroinflammation with age and in Alzheimer’s disease.
Together, our data indicate that several pri-miRNAs that change with age have either been demonstrated to target pathways involved in neurogenesis, oligodendrogenesis, and immature neuron maturation or are predicted to target genes in pathways which regulate these processes. Additionally, our data suggest that several pri-miRNAs that are differentially expressed in the aged SVZ are predicted to target pro-inflammatory cytokines and processes. Many miRNAs do not have validated target genes, and thus experimental studies on the predicted target genes and their respective pathways is needed to truly understand the breadth of miRNA impact on the aging subventricular niche.
There were significantly less pri-miRNAs differentially expressed in the 5xFAD cortex than seen in the aged SVZ. The miRNAs differentially expressed in the 5XFAD cortex have validated target genes that are highly implicated in the pathology of Alzheimer’s disease, thus demonstrating the utility of this tool to narrow down which cell types specifically play a role in the development of the disease. pri-miR-124-1 is decreased in expression in certain subpopulations of neurons, immature neurons, astrocytes, oligodendrocyte progenitors, and myelin forming oligodendrocytes in the 5XFAD mice. miR-124 is known to decrease in expression in the brains of Alzheimer’s patients, and this miRNA targets BACE1, which is directly involved in the development of the amyloid pathology of Alzheimer’s Disease (Smith et al., 2011; An et al., 2017). Furthermore, pri-miR-3078 decreases in expression in both 5XFAD models and APP/PS1 models of Alzheimer’s disease (Zhang et al., 2021). Interestingly, one of the predicted target genes, which would be increased in expression with the reduction of miR-3078, is Prnp. Prnp is implicated in behavioral insufficiencies seen in Alzheimer’s Disease pathology (Salazar et al., 2017). Therefore, by understanding which cell types have deficits in these pri-miRNAs, targeted therapeutic strategies can be developed to alleviate the pathology caused by their reduction, or targeted therapeutic strategies can be developed to artificially increase their expression.
Of importance, this study highlighted the specificity of each neuronal subtype in terms of differential expression of pri-miRNAs in the 5XFAD model compared to wild-type. Neurons are often treated as a group rather than by individual subtype in studies. Computational analysis of these different subpopulations highlights that the five familial AD mutations caused significantly different changes in differential expression of pri-miRNAs among the neuronal subtypes, providing insights for future studies to understand the impact of these changes on each subtype and their contributions to AD development.
Using the primiReference tool, cell-type specific changes in pri-miRNA expression within the aging SVZ and 5XFAD mouse cortex were elucidated. With these findings, new cell-type specific targets for neurogenesis are now available for study as well as new-cell type specific targets for Alzheimer’s Disease. By investigating the pri-miRNAs found through the primiReference tool, new avenues of research into manipulation of pri-miRNAs and their mature sequence can be conducted in a more targeted, cell-type specific manner, which may help mitigate neuropathology.
Materials and methods
Single-nucleus Preparation from the Male SVZ
Eight 3–4-month-old and eight 25–26-month-old C57BL/6 male mice were sacrificed in accordance with IACUC guidelines. Mice were obtained from the National Institute on Aging Aged Rodent Aged Rodent Colony (https://www.nia.nih.gov/research/dab/age-drodent-colonies-handbook). The mice were euthanized via CO2 at 2L/minute followed by cervical dislocation in accordance with Brown University’s IACUC guidelines (Protocol #19-01-0005). To minimize batch effects, all young mice were dissected in one batch and all old mice were dissected in another batch. The SVZ was dissected from two mice and combined to constitute a sample, so this experiment had four young and four old male samples each (Daynac et al., 2015). Upon dissection, we followed a modified protocol of the 10X Single-nucleus isolation protocol (10X Genomics, CG000124, Rev A).
Single-nucleus library preparation and sequencing
The nuclei suspension was used for library preparation using the 10X v3 3’ protocol (10X Genomics, CG000183, Rev A). Five thousand nuclei per sample were used to construct the libraries. The libraries were sequenced to a depth of 5,000 reads per nucleus or 250 million reads per sample using the HiSeq Sequencer via Genewiz. The accession number for these files is PRJNA890211.
primiReference construction
Fastq files from four young male and four old male samples were aligned to the mm10 genome using Cell Ranger (10X Genomics, Cell Ranger 3.0.0) (Zheng et al., 2017). Resulting files merged and overlapped with a publicly available reference for pri-miRNAs in six cell lines (Li et al., 2009; Chang et al., 2015). Using a mature miRNA gff3 from miRbase and bedops (Kozomara and Griffiths-Jones, 2014; Kozomara et al., 2019) sequences were relabeled with the mature miRNA name (Neph et al., 2012). The sequences were manually interrogated to ensure they properly overlapped with the location of pre-miRNAs already in the mm10 genome. The resulting GTF was appended to the mm10 GTF using 10X Cell Ranger (10X Genomics Cell Ranger 3.0.0). The mkref function was used to make a custom reference, and the Fastq files were aligned to this new ‘primiReference’.
primiReference analysis of young and aged SVZ
Once files were aligned and aggregated using 10X Cell Ranger, the resulting filtered feature matrices were input into Seurat (Stuart et al., 2019). Nuclei were filtered, so only nuclei with features between 200 and 4384 (2 standard deviations from the mean) as well as mitochondrial contamination less than 4.06% (2 standard deviations from the mean) were subset. All these nuclei were subsequently clustered to 50 principal components, based on Jackstraw and Elbow plots, and reduced using UMap. The cells were labeled via integration with the Allen Brain Atlas and using marker genes from published literature (Herbert et al., 1986; Mateo et al., 2015; Marques et al., 2016; Tasic et al., 2016; Boareto et al., 2017; Shah et al., 2018; Zywitza et al., 2018; Ximerakis et al., 2019). By using this method, pri-miRNA expression did not impact identification of cell types. Astrocytes, oligodendrocytes, NSCs, and neural progenitors were subset out and differential expression analysis was conducted between the two age groups using Seurat using default parameters. Briefly, we used the LogNormalize function for normalization of expression of each feature to the total expression for each cell type with a scale factor of 10,000. Adjusted P values, shown in supplemental tables, were calculated by Seurat using a Bonferroni correction. Following differential expression analysis, a heatmap was constructed using Morpheus (https://software.broadinstitute.org/morpheus) to demonstrate the statistically significant expressed (P < 0.05, non-parametric Wilcoxon rank sum test) pri-miRNAs within each cell type with age. These pri-miRNAs were input into miRPath using DIANATOOLS with FDR correction (Vlachos et al., 2015) using both the −3p and −5p mature sequences, since primiReference does not distinguish which form is expressed, and KEGG Pathways for targeted genes were determined.
Single-nucleus sequencing of wildtype and 5XFAD mouse cortex
Seven-month-old WT and 5XFAD samples were downloaded from GEO (GSE140511) as part of a study of single-nucleus sequencing between WT, 5XAD, and TREM2KO groups (Zhou et al., 2020). The original BAM files were downloaded then converted back to Fastq using 10X Genomics’ bamtofastq (10X Genomics, bamtofastq 1.3.2). These Fastq were aligned using ‘primiReference’ as described above. Upon alignment and aggregation, the data was input into Seurat (Stuart et al., 2019) and analyzed using the same standards as listed above for our own data. Therefore, permissible cells had between 200 and 4828.66 features and mitochondrial contamination less than 5.48% (both 2 standard deviations from the mean). The data was subsequently analyzed in the same manner as the SVZ data described above except neuronal identities, as identified by the Allen Brain Atlas (Tasic et al., 2016), were kept as defined rather than being consolidated into one neuronal class.
qPCR validation of pri-miRNA expression in the aged SVZ
To validate changing pri-miRNAs found using primiReference, single-nucleus data was re-analyzed as “bulk” RNA-sequencing data, so qPCR data of whole isolated SVZ can be compared to this dataset. To accomplish this, aligned primiReference BAM files of young and aged SVZ were analyzed using FeatureCounts (Liao et al., 2014) to determine the counts of each gene feature in a sample-specific rather than single-nucleus specific manner. Using FeatureCounts, the total reads of each cell within each sample are combined. DESeq2 (Love et al., 2014) was then used to compare gene expression of old SVZ to young SVZ using geometric mean for normalization. P-values were adjusted for multiple comparisons using the default Benjamini and Hochberg test default in DESeq2. Primers were made to pri-miR-21a and pri-miR-99a since these pri-miRNAs changed in most cell-types. The primers used were primiR21aF: TGTGAGAGTCGGTGTTGTGAG, primiR21aR: CCGCTGCACATACTGTTGGT; primiR99aF: TTCACAAGGACACGGTGAAA, primiR99aR: CTCCGGGCACGATTTAACTA. RNA was extracted from five young (4 months old) and five old (23 months old) SVZ using trizol and chloroform followed by the RNEasy kit (Qiagen, #74104). RNA concentration was measured and reverse transcribed into cDNA using Taqman reagents (Thermo Fisher Scientific, #4304134, Waltham, MA). qPCR reactions were performed in triplicate. pri-miRNA expression was normalized to GAPDH using GAPDHF: AGGTTGTCTCCTGCGACTTC and GAPDHR: TGTCATACCAGGAAATGAGCTTG. The fold change was compared to that of the pri-miRNAs in the “bulk” analysis of the single-nucleus data. The p-value of qPCR was determined using a two-sample T test. Ggplot2 was used to plot this data (Wickham, 2009).
Single-cell sequencing of young and aged SVZ
Using a publicly available single-cell sequencing dataset (Dulken et al., 2019) of the young and aged SVZ, we determined changes in cytoplasmic mRNA levels. Single-cell sequencing data was analyzed using Seurat (Stuart et al., 2019) in the same manner as our single-nucleus data. After clusters were established, differential expression was conducted between aged and young SVZ by cell-type.
Validation of target gene expression in single-cell data
TarBase V8 (Karagkouni et al., 2018) was used to determine the validated targets of pri-miRNAs changing in expression within the young and aged single-nucleus SVZ data set. Using the single-cell data set of young and aged SVZ, the validated TarBase target genes were overlapped with differential expression data per cell-type. Only cell-types where the pri-miRNA was changing with age were used. The number of those target genes found in both TarBase and DE data was given as a percentage. The expression of those intersecting target genes was viewed in the single-cell data set to determine if they were increasing or decreasing in expression, with age, within that given cell-type. If the direction was the opposite of the pri-miRNA, the target was considered validated. The number of genes that were changing in the expected direction, according to pri-miRNA expression, in the single-cell data was then given as a percentage of (number of genes in expected direction/total number of target genes intersected with TarBase). These were then referred to as validated targets (Table 2).
Statistical analyses
Wilcox rank sum test was used to determine significance of the differential expression data with a Bonferroni correction to the p value for multiple comparisons using Seurat (Seurat et al., 2019). Those pri-miRNAs with P > 0.05 are shown in supplementary tables, but they are excluded from in text figures. T-test was used to determine significance of qPCR data. Chi-squared test was used to determine significance of the miRNA validations tests (Table 2). No outliers were found or excluded in this dataset. All raw data information can be found in supplementary materials.
Supplementary Material
Acknowledgements
We thank the Computational Biology Core and Center for Computation and Visualization at Brown University for their help and support throughout this project. We acknowledge and thank the Sedivy laboratory for their generous provision of materials used to conduct selected experiments. We additionally would like to acknowledge the Brunet laboratory for providing additional materials from their single-cell dataset. Support for AE was provided by T32AG041688 and P20GM119943, funding for the research was provided by P20GM119943.
Footnotes
Conflict of interest
The authors declare they have no competing interests.
CRediT authorship contribution statement
JK: Conceptualization, Methodology, Data curation, Investigation, Validation, Writing - Original and revised draft. AE: Methodology, Data curation, Investigation, Validation, Writing - Original draft. BK: Investigation, Validation. TN: Investigation, Validation. All authors have read and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
Data is available, according to NIHGIMS practices. The datasets generated in this study can be found at the NCBI sequence read archive (BioProject PRJNA890211). All code associated with this manuscript can be found on GitHub (https://github.com/KreilingLab/primiReference; https://github.com/KreilingLab/SingleNucleusAnalysis). The genome file used is available from 10X Genomics as their mm10 fasta file. The 5XFAD and Wild-Type cortex dataset can be found at GEO (GSE140511) (Zhou et al., 2020). The single-cell dataset of young and aged SVZ can be found at BioProject PRJNA450425 (Dulken et al., 2019).
References
- Agostini M, Romeo F, Inoue S, Niklison-Chirou MV, Elia AJ, Dinsdale D, Morone N, Knight RA, Mak TW,Melino G, 2016. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 23, 1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Gong G, Wang Y, Bian M, Yu L,Wei C, 2017. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 8, 114065–114071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes M, Kim YA, Lannes J, Alaniz ME, Cho JD, McCabe BD,Santa-Maria I, 2019. MiR-219 deficiency in Alzheimer’s disease contributes to neurodegeneration and memory dysfunction through post-transcriptional regulation of tau-kinase network. bioRxiv, 607176. [Google Scholar]
- Boareto M, Iber D,Taylor V, 2017. Differential interactions between Notch and ID factors control neurogenesis by modulating Hes factor autoregulation. Development 144, 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi R, Fabiano M, Giorgi C, Arisi I, La Regina F, Malerba F, Turturro S, Storti AE, Ricevuti F, Amadio S, et al. , 2021. Nerve Growth Factor Neutralization Promotes Oligodendrogenesis by Increasing miR-219a-5p Levels. Cells 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma IB, van Dijk M, Bridel C, van de Lisdonk T, Haverkort SQ, Runia TF, Steinman L, Hintzen RQ, Killestein J, Verbeek MM, et al. , 2017. Regulator of oligodendrocyte maturation, miR-219, a potential biomarker for MS. J. Neuroinflammation 14, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E,Satija R, 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Pertea M, Lee S, Salzberg SL,Mendell JT, 2015. Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res. 25, 1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PH, Cheng CH, Shih CM, Ho KH, Lin CW, Lee CC, Liu AJ, Chang CK,Chen KC, 2016. The Inhibition of microRNA-128 on IGF-1-Activating mTOR Signaling Involves in Temozolomide-Induced Glioma Cell Apoptotic Death. PLoS One 11, e0167096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover JC,Shook BA, 2011. Aging of the Subventricular Zone Neural Stem Cell Niche. Aging Dis. 2, 149–163. [PMC free article] [PubMed] [Google Scholar]
- Daynac M, Morizur L, Chicheportiche A, Mouthon MA,Boussin FD, 2016. Age-related neurogenesis decline in the subventricular zone is associated with specific cell cycle regulation changes in activated neural stem cells. Sci. Rep. 6, 21505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynac M, Morizur L, Kortulewski T, Gauthier LR, Ruat M, Mouthon MA,Boussin FD, 2015. Cell Sorting of Neural Stem and Progenitor Cells from the Adult Mouse Subventricular Zone and Live-imaging of their Cell Cycle Dynamics. J. Vis. Exp. 103, e53247–e53255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, Hughes TK, Wadsworth MH, Burks T, Nguyen LT, et al. , 2020. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT,Barres BA, 2010. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken BW, Buckley MT, Navarro Negredo P, Saligrama N, Cayrol R, Leeman DS, George BM, Boutet SC, Hebestreit K, Pluvinage JV, et al. , 2019. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken BW, Leeman DS, Boutet SC, Hebestreit K,Brunet A, 2017. Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep. 18, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S,Gotz M, 2017. Glial control of neurogenesis. Curr Opin Neurobiol 47, 188–195. [DOI] [PubMed] [Google Scholar]
- Floyd DH, Zhang Y, Dey BK, Kefas B, Breit H, Marks K, Dutta A, Herold-Mende C, Synowitz M, Glass R, et al. , 2014. Novel anti-apoptotic microRNAs 582–5p and 363 promote human glioblastoma stem cell survival via direct inhibition of caspase 3, caspase 9, and Bim. PLoS One 9, e96239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliandolo A, Chiricosta L, Boccardi V, Mecocci P, Bramanti P,Mazzon E, 2020. MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain. Genes (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M,Kim VN, 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524. [DOI] [PubMed] [Google Scholar]
- He L, Vanlandewijck M, Raschperger E, Andaloussi Mae M, Jung B, Lebouvier T, Ando K, Hofmann J, Keller A,Betsholtz C, 2016. Analysis of the brain mural cell transcriptome. Sci. Rep. 6, 35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert J, Wilcox JN, Pham KT, Fremeau RT Jr., Zeviani M, Dwork A, Soprano DR, Makover A, Goodman DS, Zimmerman EA, et al. , 1986. Transthyretin: a choroid plexus-specific transport protein in human brain. The 1986 S. Weir Mitchell award. Neurology 36, 900–911. [DOI] [PubMed] [Google Scholar]
- Hu F, Sun B, Xu P, Zhu Y, Meng XH, Teng GJ,Xiao ZD, 2017. MiR-218 Induces Neuronal Differentiation of ASCs in a Temporally Sequential Manner with Fibroblast Growth Factor by Regulation of the Wnt Signaling Pathway. Sci. Rep. 7, 39427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji F, Lv X,Jiao J, 2013. The role of microRNAs in neural stem cells and neurogenesis. J. Genet. Genomics 40, 61–66. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Man YY, Liu Y, Yin CJ, Li JL, Shi HC, Zhao H,Zhao SG, 2021. Loss of miR-23b/27b/24–1 Cluster Impairs Glucose Tolerance via Glycolysis Pathway in Mice. Int. J. Mol. Sci. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, et al. , 2018. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 46, D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BR, Choi ES, Lee KM, Choi SK, Cho JH, Jeon WB,Kim E, 2017. Reverse Expression of Aging-Associated Molecules through Transfection of miRNAs to Aged Mice. Mol. Ther. Nucleic Acids 6, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK,Kim VN, 2007. Processing of intronic microRNAs. EMBO J 26, 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M,Griffiths-Jones S, 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A,Griffiths-Jones S, 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Omori Y, Watanabe S, Tarusawa E, Yamamoto H, Chaya T, Furuhashi M, Morita M, Sato T, Hirose S, et al. , 2019. miR-124 dosage regulates prefrontal cortex function by dopaminergic modulation. Sci. Rep. 9, 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. , 2009. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol. Cell 35, 610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S,Kim VN, 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21, 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R,Genome Project Data Processing S, 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK,Shi W, 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Desire L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, et al. , 2005. Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375. [DOI] [PubMed] [Google Scholar]
- Lim DA,Alvarez-Buylla A, 1999. Interaction between astrocytes and adult subventricular zone precursors stimulates neurogenesis. Proc. Natl. Acad. Sci. U S A 96, 7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA,Alvarez-Buylla A, 2016. The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb Perspect. Biol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W,Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li C, Huang Y, Wang Y, Xia X,Zheng JC, 2019. Exosomes released from neural progenitor cells and induced neural progenitor cells regulate neurogenesis through miR-21a. Cell. Commun. Signal. 17, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Haring M, Hochgerner H, Romanov RA, et al. , 2016. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massirer KB, Carromeu C, Griesi-Oliveira K,Muotri AR, 2011. Maintenance and differentiation of neural stem cells. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 107–114. [DOI] [PubMed] [Google Scholar]
- Matarredona ER, Talaveron R,Pastor AM, 2018. Interactions Between Neural Progenitor Cells and Microglia in the Subventricular Zone: Physiological Implications in the Neurogenic Niche and After Implantation in the Injured Brain. Front. Cell. Neurosci. 12, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo JL, van den Berg DL, Haeussler M, Drechsel D, Gaber ZB, Castro DS, Robson P, Lu QR, Crawford GE, Flicek P, et al. , 2015. Characterization of the neural stem cell gene regulatory network identifies OLIG2 as a multifunctional regulator of self-renewal. Genome Res. 25, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. , 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, Hayder H, Zayed Y,Peng C, 2018. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 9, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio-Querejeta I, Carregal-Romero S, Ayerdi-Izquierdo A, Mager I, A NL, Wood M, Egimendia A, Betanzos M, Alberro A, Iparraguirre L, et al. , 2020. MiR-219a-5p Enriched Extracellular Vesicles Induce OPC Differentiation and EAE Improvement More Efficiently Than Liposomes and Polymeric Nanoparticles. Pharmaceutics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T,Kosik KS, 2009. MicroRNA-124: micromanager of neurogenesis. Cell. Stem Cell 4, 375–376. [DOI] [PubMed] [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, et al. , 2008. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell. Biol. 10, 385–394. [DOI] [PubMed] [Google Scholar]
- Puigdellivol M, Allendorf DH,Brown GC, 2020. Sialylation and Galectin-3 in Microglia-Mediated Neuroinflammation and Neurodegeneration. Front. Cell. Neurosci. 14, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, Banerjee A, Wu ZS, Zhang X, Wang H, Pandey V, Zhang WJ, Lv XF, Tan S, Lobie PE, et al. , 2012. Loss of SNAIL regulated miR-128–2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 72, 6036–6050. [DOI] [PubMed] [Google Scholar]
- Quinones-Hinojosa A, Sanai N, Soriano-Navarro M, Gonzalez-Perez O, Mirzadeh Z, Gil-Perotin S, Romero-Rodriguez R, Berger MS, Garcia-Verdugo JM,Alvarez-Buylla A, 2006. Cellular composition and cytoarchitecture of the adult human subventricular zone: a niche of neural stem cells. J. Comp. Neurol. 494, 415–434. [DOI] [PubMed] [Google Scholar]
- Saito K, Koike T, Kawashima F, Kurata H, Shibuya T, Satoh T, Hata Y, Yamada H,Mori T, 2018. Identification of NeuN immunopositive cells in the adult mouse subventricular zone. J. Comp. Neurol. 526, 1927–1942. [DOI] [PubMed] [Google Scholar]
- Salazar SV, Gallardo C, Kaufman AC, Herber CS, Haas LT, Robinson S, Manson JC, Lee MK,Strittmatter SM, 2017. Conditional Deletion of Prnp Rescues Behavioral and Synaptic Deficits after Disease Onset in Transgenic Alzheimer’s Disease. J. Neurosci. 37, 9207–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Maria I, Alaniz ME, Renwick N, Cela C, Fulga TA, Van Vactor D, Tuschl T, Clark LN, Shelanski ML, McCabe BD, et al. , 2015. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J. Clin. Invest. 125, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiera G, Di Liegro CM,Di Liegro I, 2019. Cell-to-Cell Communication in Learning and Memory: From Neuro- and Glio-Transmission to Information Exchange Mediated by Extracellular Vesicles. Int. J. Mol. Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PT, Stratton JA, Stykel MG, Abbasi S, Sharma S, Mayr KA, Koblinger K, Whelan PJ,Biernaskie J, 2018. Single-Cell Transcriptomics and Fate Mapping of Ependymal Cells Reveals an Absence of Neural Stem Cell Function. Cell 173, 1045–1057 e1049. [DOI] [PubMed] [Google Scholar]
- Smith P, Al Hashimi A, Girard J, Delay C,Hebert SS, 2011. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J. Neurochem. 116, 240–247. [DOI] [PubMed] [Google Scholar]
- Song H, Stevens CF,Gage FH, 2002. Astroglia induce neurogenesis from adult neural stem cells. Nature 417, 39–44. [DOI] [PubMed] [Google Scholar]
- Stevanato L,Sinden JD, 2014. The effects of microRNAs on human neural stem cell differentiation in two- and three-dimensional cultures. Stem Cell Res. Ther. 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P,Satija R, 2019. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. , 2016. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, 1993. Synapsin I, synapsin II, and synaptophysin: marker proteins of synaptic vesicles. Brain Pathol. 3, 87–95. [DOI] [PubMed] [Google Scholar]
- Thrupp N, Sala Frigerio C, Wolfs L, Skene NG, Fattorelli N, Poovathingal S, Fourne Y, Matthews PM, Theys T, Mancuso R, et al. , 2020. Single-Nucleus RNA-Seq Is Not Suitable for Detection of Microglial Activation Genes in Humans. Cell Rep. 32, 108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TF, Lin JF, Chou KY, Lin YC, Chen HE,Hwang TI, 2018. miR-99a-5p acts as tumor suppressor via targeting to mTOR and enhances RAD001-induced apoptosis in human urinary bladder urothelial carcinoma cells. Onco. Targets Ther. 11, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW,Strittmatter SM, 2013. Amyloid-beta induced signaling by cellular prion protein and Fyn kinase in Alzheimer disease. Prion 7, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T,Hatzigeorgiou AG, 2015. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 43, W460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BZ, Yang JJ, Zhang H, Smith CA,Jin K, 2019a. AMPK Signaling Regulates the Age-Related Decline of Hippocampal Neurogenesis. Aging Dis. 10, 1058–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Moyano AL, Ma Z, Deng Y, Lin Y, Zhao C, Zhang L, Jiang M, He X, Ma Z, et al. , 2017. miR-219 Cooperates with miR-338 in Myelination and Promotes Myelin Repair in the CNS. Dev. Cell 40, 566–582 e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yuan Y, Zhang Z,Ding K, 2019b. Inhibition of miRNA-27b enhances neurogenesis via AMPK activation in a mouse ischemic stroke model. FEBS Open Bio. 9, 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, 2009. Ggplot2 : elegant graphics for data analysis. Springer, New York. [Google Scholar]
- Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C, et al. , 2019. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708. [DOI] [PubMed] [Google Scholar]
- Zhang W, Kim PJ, Chen Z, Lokman H, Qiu L, Zhang K, Rozen SG, Tan EK, Je HS,Zeng L, 2016. MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qian L, Liu Y, Liu Y, Yu W,Zhao Y, 2021. CircRNA-ceRNA Network Revealing the Potential Regulatory Roles of CircRNA in Alzheimer’s Disease Involved the cGMP-PKG Signal Pathway. Front. Mol. Neurosci. 14, 665788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y,Zhang Y, 2020. lncRNA ZFAS1 Improves Neuronal Injury and Inhibits Inflammation, Oxidative Stress, and Apoptosis by Sponging miR-582 and Upregulating NOS3 Expression in Cerebral Ischemia/Reperfusion Injury. Inflammation 43, 1337–1350. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Li XX, Cao XL, Ji CC, Gao XY, Gao D, Han H, Yu F,Zheng MH, 2022. MicroRNA-582–5p Contributes to the Maintenance of Neural Stem Cells Through Inhibiting Secretory Protein FAM19A1. Front. Cell. Neurosci. 16, 866020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. , 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. , 2020. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zywitza V, Misios A, Bunatyan L, Willnow TE,Rajewsky N, 2018. Single-Cell Transcriptomics Characterizes Cell Types in the Subventricular Zone and Uncovers Molecular Defects Impairing Adult Neurogenesis. Cell Rep. 25, 2457–2469 e2458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available, according to NIHGIMS practices. The datasets generated in this study can be found at the NCBI sequence read archive (BioProject PRJNA890211). All code associated with this manuscript can be found on GitHub (https://github.com/KreilingLab/primiReference; https://github.com/KreilingLab/SingleNucleusAnalysis). The genome file used is available from 10X Genomics as their mm10 fasta file. The 5XFAD and Wild-Type cortex dataset can be found at GEO (GSE140511) (Zhou et al., 2020). The single-cell dataset of young and aged SVZ can be found at BioProject PRJNA450425 (Dulken et al., 2019).


