Abstract
An expression system designed for cell surface display of hybrid proteins on Staphylococcus carnosus has been evaluated for the display of Staphylococcus aureus protein A (SpA) domains, normally binding to immunoglobulin G (IgG) Fc but here engineered by combinatorial protein chemistry to yield SpA domains, denoted affibodies, with new binding specificities. Such affibodies, with human IgA or IgE binding activity, have previously been selected from a phage library, based on an SpA domain. In this study, these affibodies have been genetically introduced in monomeric or dimeric forms into chimeric proteins expressed on the surface of S. carnosus by using translocation signals from a Staphylococcus hyicus lipase construct together with surface-anchoring regions of SpA. The recombinant surface proteins, containing the IgA- or IgE-specific affibodies, were demonstrated to be expressed as full-length proteins, localized and properly exposed at the cell surface of S. carnosus. Furthermore, these chimeric receptors were found to be functional, since recombinant S. carnosus cells were shown to have gained IgA and IgE binding capacity, respectively. In addition, a positive effect in terms of IgA and IgE reactivity was observed when dimeric versions of the affibodies were present. Potential applications for recombinant bacteria with redirected binding specificity in their surface proteins are discussed.
The display of heterologous proteins on the outer surface of bacteria has become an emerging topic in different fields of research within applied bacteriology, biotechnology, and vaccinology (7, 12, 45). The most-common application has aimed toward the development of live bacterial vaccine delivery systems by the exposure of foreign antigenic determinants at the outer cell surface of gram-negative or gram-positive bacteria. Escherichia coli and various Salmonella spp. have dominated among the gram-negative bacteria (12, 45), but various types of gram-positive bacteria have also been investigated, including attenuated mycobacteria (46), commensal streptococci (6, 37), and nonpathogenic food-grade lactococcal (35) and staphylococcal (22, 41, 45) species as well as sporulating Bacillus subtilis (1). Bacterial surface display has also been employed for surface expression of heterologous enzymes (9, 10, 47) and for the development of novel microbial biocatalysts. Polyhistidyl peptides have been surface exposed for capture of heavy metals, potentially with environmental applications (43). Single-chain scFv antibody fragments (i.e., the variable parts of the heavy and light chains genetically linked together into a single chain) have also been expressed in a surface-anchored functional form on both gram-negative (8, 11) and gram-positive (18) bacteria, and the potential use of such bacteria as whole-cell diagnostic devices has been discussed previously (18, 45). The gram-positive surface display systems have been reported to exhibit some advantages compared to gram-negative bacteria, since translocation through only one membrane is required and the gram-positive systems seem to allow surface display of larger proteins. Moreover, the gram-positive bacteria are considered to be more rigid, due to the thick cell wall surrounding the cells (7, 45). Such bacteria would be less likely to lyse through shear forces and would thus be more suitable in applications based on whole-cell reagents.
Two staphylococcal candidates which are being investigated extensively for various surface display applications are the nonpathogenic Staphylococcus xylosus and Staphylococcus carnosus (2, 22, 27, 28, 30, 31, 39), both of which traditionally have been used as starter cultures in meat fermentation applications (20, 26). Of the two staphylococcal species, the system based on the use of S. carnosus has been demonstrated to result generally in a more efficient display of heterologous surface proteins (39), on the order of 104 per bacterial cell (2). With S. carnosus as a host, the signal sequence and propeptide of a Staphylococcus hyicus lipase gene construct (13) have been used together with the staphylococcal protein A (SpA) cell surface-anchoring sequences (42) to achieve translocation and proper surface exposure.
In a previous study, we were able to demonstrate the expression of a murine anti-human-immunoglobulin E (IgE) scFv antibody fragment as surface exposed on S. xylosus and S. carnosus (18), and we could show that the recombinant bacteria, particularly S. carnosus, were capable of reacting in whole-cell assays to the antigen human IgE. Here we evaluated the possibility of exposing on the surface of S. carnosus tailor-made binding molecules, created by combinatorial protein engineering of an SpA domain, Z (32), which normally binds to IgG Fc (fragment crystallizable). An attempt to obtain such novel binding proteins with completely new specificities was recently initiated by using phage display in vitro selection technology. By using genetic engineering, libraries of the Z domain were created in which 13 surface residues (involved in the IgG Fc binding) of the domain were randomly and simultaneously substituted (34). This Z library was genetically fused to the coat protein III of filamentous phage M13, resulting in a phage library adapted for selection of novel specificities by biopanning (33). Novel Z variants, or “affibodies” (21, 33), have successfully been selected to diverse targets, such as Taq DNA polymerase, human insulin, a human apolipoprotein variant, and the G protein of human respiratory syncytial virus (21, 33). Recently, and analogous to the achievements of Nord and coworkers (33), such affibody ligands were selected against human IgA (38) and IgE (17), respectively.
Our overall objective in this study was to determine whether the IgA- and IgE-reactive affibodies could be expressed in an active form as parts of chimeric surface proteins on S. carnosus, thus creating a staphylococcal bacterium which has gained binding activity to human IgA or IgE, respectively. Gene fragments encoding either monomeric or dimeric forms of the affibodies were introduced into gene constructs encoding the chimeric surface proteins to evaluate the effects of ligand dimerization on IgA and IgE reactivity. The potential use of such recombinant staphylococci as whole-cell diagnostic devices or biosorbents or as alternatives to filamentous phages for the surface display of affibody libraries for selection is discussed.
MATERIALS AND METHODS
Strains and vectors.
The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain | Plasmid | Reference |
|---|---|---|
| E. coli RRIΔM15 | pKN1-dZIgA | 38 |
| pKN1-dZIgE | 17 | |
| pSPPmZIgAABPXM | This study | |
| pSPPdZIgAABPXM | This study | |
| pSPPmZIgEABPXM | This study | |
| pSPPdZIgEABPXM | This study | |
| S. carnosus TM300 | None | 13 |
| pSPPmABPXM | 41 | |
| pSPPmZIgAABPXM | This study | |
| pSPPdZIgAABPXM | This study | |
| pSPPmZIgEABPXM | This study | |
| pSPPdZIgEABPXM | This study |
Preparation and transformation of protoplasts.
The preparation and transformation of protoplasts were performed as described by Götz and collaborators (14, 15).
Antibodies.
Purified, myeloma-derived human IgA and IgE were obtained from Pharmacia and Upjohn Diagnostics (Uppsala, Sweden). Secondary antibodies used in this study were affinity-purified polyclonal rabbit anti-human IgA and three different mouse monoclonal IgG antibodies reactive with constant domains (Cɛ2, Cɛ3, or Cɛ4) of human IgE, all conjugated with a β-galactosidase enzyme (Pharmacia and Upjohn Diagnostics).
Selection of IgA- and IgE-reactive affibodies.
Affibodies to human IgA and human IgE, respectively, were selected by using the phage libraries Zlib-1 and Zlib-2, essentially as described by Nord and coworkers (33). Briefly, polyethylene glycol-precipitated phage stocks of libraries Zlib-1 and Zlib-2 were incubated with streptavidin-coated paramagnetic beads (M-280; Dynal AS, Oslo, Norway) with immobilized biotinylated target molecules, IgA, or a chimeric IgE (Pharmacia and Upjohn Diagnostics). After several washing steps, bound phages were eluted with low pH and used for reinfection of log-phase E. coli RRIΔM15 cells (40). The procedure was repeated, and potential binders were sequenced. The selected affibodies had the following amino acid substitutions relative to the original sequence of domain Z (33) at the randomized positions: Q9T, Q10I, N11Q, F13S, Y14Q, L17R, H18L, E24G, E25R, R27K, N28L, Q32H, and K35L for the IgA-specific affibody and Q9P, Q10T, N11A, F13S, Y14L, L17M, H18M, E24V, E25D, R27V, N28G, Q32G, and K35M for the IgE-specific affibody. A detailed description of the phage display-based panning procedures to obtain the two affibodies ZIgA and ZIgE as well as a detailed evaluation of their binding characteristics by real-time biospecific interaction analysis (BIA) with a model 2000 instrument (Biacore AB, Uppsala, Sweden) will be published elsewhere (17, 38). Separate injections of ZIgA and ZIgE proteins over sensor chip surfaces coated with human IgA, IgE, IgG showed that the selected affibodies bound only their respective targets and thus showed no detectable cross-reactivity (data not shown). The dissociation constants (Kd) for the monomeric ZIgA and ZIgE affibodies to their respective targets, as determined by BIA, were found to be 0.5 and 0.4 μM, respectively (17, 38).
DNA constructions.
Gene fragments encoding either monomeric or dimeric variants of the two affibodies, ZIgA and ZIgE could be amplified by PCR by using primers SAPA-27 (5′-GGGGGATCCTGTAGACAACAAATTCAACAAAG-3′) and SAPA-28 (5′-GGGGTCGACTTCGGCGCCTGAGCATC-3′) and with pKN1 phagemids (34), carrying inserts corresponding to dimeric versions of the two affibodies, as templates. Generated gene fragments were restricted with endonucleases SalI and BamHI and ligated to plasmid pSPPmABPXM (41), restricted by using the same enzymes. Solid-phase DNA sequencing (23) was performed by employing the indocarbocyanine dye ALFred (Cy5) phosphoramidite-labeled sequencing primers SAPA-25 (5′-TTACATCACAAGCGAGCGAC-3′) and SAPA-26 (5′-TGCTTTGGCTTTTGCTAGAG-3′) on a robotic workstation (Biomek 1000; Beckman Instruments, Fullerton, Calif.), and the obtained Sanger fragments were analyzed on an ALFexpress (Amersham Pharmacia Biotech, Uppsala, Sweden). The four verified expression vectors pSPPmZIgAABPXM, pSPPdZIgAABPXM, pSPPmZIgEABPXM, and pSPPdZIgEABPXM, designed for surface expression on S. carnosus, encode the surface-anchored fusion proteins PP-ZIgA-ABP-XM′, PP-ZIgA-ZIgA-ABP-XM′, PP-ZIgE-ABP-XM′, and PP-ZIgE-ZIgE-ABP-XM′, respectively. The expression vectors were used to transform S. carnosus protoplasts so as to generate the four recombinant S. carnosus strains, which for simplicity were denoted Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, and Sc:dZIgE. The m and d annotations indicate either monomeric or dimeric versions of the affibodies.
Extraction and purification of the chimeric surface proteins.
Extraction of the chimeric proteins from the cell wall of the recombinant S. carnosus was performed essentially as previously described (41). Briefly, cells harboring the parental vector pSSPmABPXM, denoted Sc:ABP, or the four new recombinant staphylococcal strains Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, and Sc:dZIgE were grown at 37°C in 50 ml of tryptone soy broth medium (TSB; Difco), supplemented with yeast extract (5 g/liter; Difco) and chloramphenicol (10 mg/liter) until the optical density at 578 nm (OD578) reached ≈1. Cells were washed twice in phosphate-buffered saline with 0.05% Tween 20 (PBST) and subjected to a cell wall-degrading treatment by using lysostaphin (Sigma). The cell pellets were dissolved in 6 ml of SMM solution (1 M sucrose, 40 mM maleic acid, and 40 mM MgCl2 [pH 6.5]), and 18 U of lysostaphin was added before incubation at 37°C for 1.5 h. The formed protoplasts were pelleted at 6,000 × g for 15 min, and the supernatants were diluted 10 times in Tris-buffered saline containing Tween (TST; 25 mM Trizma base-HCl [pH 8], 0.2 M NaCl, 1 mM EDTA, 0.05% Tween 20) and loaded onto a human serum albumin (HSA)-Sepharose column (36) for affinity purification. Eluted fractions were pooled and lyophilized prior to a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (4 to 20% polyacrylamide) analysis (Novex, San Diego, Calif.) under reducing conditions.
Enzymatic assay for detection of cell surface-exposed chimeric proteins.
Wild-type S. carnosus cells and the recombinant staphylococci Sc:ABP, Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, and Sc:dZIgE were grown overnight and diluted 1:200 in TSB medium supplemented with yeast extract (and chloramphenicol for the recombinant cells) and further grown at 37°C until the OD578 reached ≈1. The cells were harvested and washed twice in PBST. One milliliter of cell suspension, diluted in PBST to an OD578 of ≈1, was incubated with biotinylated HSA (biotinylated with d-biotinoyl-ɛ-aminocaproic acid N-hydroxysuccinimide ester [Boehringer] according to the supplier’s recommendations) at a final concentration of 62 nM for 30 min at room temperature. After the cells were washed three times in PBST, they were resuspended in 1 ml of PBST containing 0.5 U of streptavidin-alkaline phosphatase (Boehringer) and incubated for 30 min at room temperature. The mixtures were washed twice in PBST and once in substrate buffer (1 M diethanolamine-HCl [pH 9.8], 0.5 mM MgCl2) before the different cell types were resuspended in 5 ml of substrate buffer. Six 100-μl aliquots of each cell type were loaded in a microtiter plate before the addition of 100 μl of the substrate solution, p-nitrophenylphosphate (Sigma). The change in A405 was measured after 10 min in an enzyme-linked immunosorbent assay (ELISA) reader (SLT EAR 340AT; SLT-Labinstruments, Grödig, Austria).
Colorimetric assay to analyze the functionality of surface-displayed Ig binding surface proteins.
The different recombinant S. carnosus bacteria were grown and washed as described above, and 0.5 ml of cells corresponding to an OD578 of ≈1 were incubated with 2 μg of human IgA or human IgE for 45 min at room temperature. The samples were washed twice in PBST and incubated for 45 min at room temperature in 0.5 ml of PBST supplemented with β-galactosidase-conjugated polyclonal rabbit anti-IgA antibodies diluted 1:150 or a mixture of 1.5 μg of each of three different β-galactosidase-conjugated murine anti-IgE monoclonal antibodies (Pharmacia and Upjohn Diagnostics). The bacteria were washed twice in PBST and once in substrate buffer (0.2 M Na-phosphate buffer [pH 7.4], 2 mM MgCl2, 8% methanol, 0.25% Tween 20) before they were resuspended in 2.5 ml of substrate buffer. Six 100-μl aliquots were loaded in ELISA plate wells for each different cell type before the addition of 100 μl of the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) (9.2 mM in substrate buffer; Sigma). The change in A405 was determined after 60 min in an ELISA reader. As negative controls, similar experiments were performed, but the specific target protein human IgA or IgE was left out.
RESULTS
Background.
In this study, we investigated the possibility of displaying engineered SpA domains with novel binding specificities on the surface of S. carnosus in order to create recombinant staphylococci having binding capacities for IgA or IgE. Two such engineered SpA domains, termed affibodies, have been selected by phage display technology from protein libraries constructed by combinatorial strategies (17, 38) by using the 58-amino-acid Z domain (32) as a protein scaffold, analogous to work described by Nord and coworkers (33, 34). The two new SpA domains were successfully selected by biopanning by using human IgA and IgE antibodies, respectively, as target proteins. A detailed description of the panning procedures used to obtain the two affibodies ZIgA and ZIgE as well as a detailed evaluation of their binding characteristics by real-time biospecific interaction analysis will be published elsewhere (17, 38).
Expression vectors for surface display on S. carnosus.
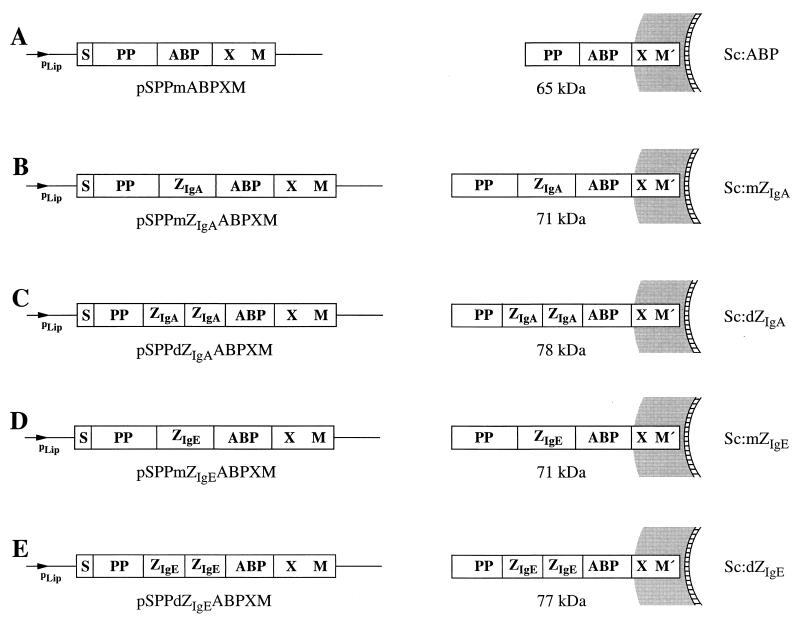
Four novel E. coli-staphylococcus shuttle vectors were constructed, designed for surface display of chimeric proteins containing the new ZIgA and ZIgE affibodies, on S. carnosus. The parental vector (41) as well as the four constructed expression vectors together with the encoded gene products PP-ABP-XM′, PP-ZIgA-ABP-XM′, PP-ZIgA-ZIgA-ABP-XM′, PP-ZIgE-ABP-XM′, and PP-ZIgE-ZIgE-ABP-XM′ are depicted in Fig. 1. M′ represents the processed and covalently anchored form (29, 42) of the M sequence of SpA. For simplicity, the five recombinant S. carnosus strains were denoted Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, Sc:dZIgE, and Sc:ABP. The S. carnosus expression vectors utilize the promoter, signal sequence, and propeptide sequence (PP) from an S. hyicus lipase gene construct, optimized for expression in S. carnosus (25). The vector system also contains gene fragments from the SpA gene (48) as follows: X, encoding a charged repetitive region postulated to interact with the peptidoglycan cell wall (19), and M, encoding a region common in gram-positive cell surface-bound receptors that is required for cell surface anchoring (29, 42). In addition, the gene encoding an albumin binding protein (ABP), derived from streptococcal protein G (36, 41), is also present in the expression vectors. The ABP region is expressed as the part of the chimeric surface proteins closest to the cell wall-anchoring motifs (41). It has been demonstrated to be useful as a reporter peptide in a colorimetric assay to analyze surface accessibility of the hybrid surface proteins (18, 39, 41) and as an affinity tag for recovery of the expressed recombinant surface proteins (18, 41) and has also been shown to act as a spacer protein to increase surface accessibility (44). Note that the PP from the S. hyicus lipase is not processed in S. carnosus (13), while it is processed in its homologous host, S. hyicus (3). This propeptide has been described as essential for the secretion of heterologous gene fusion products from S. carnosus (5) when the lipase signal peptide is used for secretion.
FIG. 1.
Expression vectors developed for surface display in S. carnosus shown with their encoded gene products illustrated as they are anchored to the cell surface. (A) Expression vector pSPPmABPXM (41), suitable for surface display in S. carnosus, with the processed gene fusion product PP-ABP-XM′ illustrated as anchored to the cell surface. (B and C) Expression cassettes of the surface expression vectors encoding the chimeric surface proteins containing the monomeric and dimeric versions of the ZIgA affibody, respectively, with their gene fusion products anchored to the S. carnosus cell surface. (D and E) Expression cassettes of the surface expression vectors encoding the chimeric surface proteins containing the monomeric and dimeric versions of the ZIgE affibody, respectively, with their cell surface-anchored gene products. The names of the constructed expression vectors are given below the expression cassettes, the molecular sizes of the encoded surface proteins are indicated in kilodaltons, and the abbreviated names of the recombinant staphylococci are given at the right.
Extraction, affinity purification, and characterization of the chimeric surface proteins.
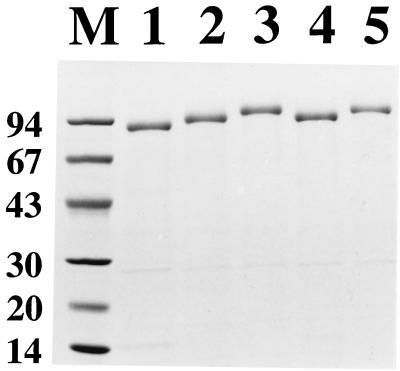
To investigate the expression of the different chimeric surface proteins, the recombinant staphylococci Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, and Sc:dZIgE (Fig. 1) and S. carnosus cells harboring the parental vector pSPPmABPXM (Sc:ABP was included as a reference) were cultivated to equal cell densities, harvested, and subjected to lysostaphin treatment to release cell wall-bound proteins. After centrifugation, the protein-containing supernatants were loaded onto HSA columns for ABP-mediated affinity purification of the recombinant surface proteins. Eluted proteins were analyzed by SDS-PAGE (Fig. 2). Extracted and affinity-purified material from cultures of Sc:ABP (lane 1) and Sc:mZIgA (lane 2), Sc:dZIgA (lane 3), Sc:mZIgE (lane 4), and Sc:dZIgE (lane 5) all showed major protein bands of essentially the expected sizes (65, 71, 78, 71, and 77 kDa, respectively). In accordance with earlier observations for proteins containing the lipase propeptide (13, 41), the recovered chimeric proteins migrated as slightly larger proteins. Furthermore, the purified chimeric proteins showed little or no proteolytic degradation (Fig. 2). The high stability to proteolytic degradation is perhaps not that surprising, considering that the ZIgA and ZIgE affibody domains are of staphylococcal origin, but it is of significant interest, since alternative protein domains, such as scFv antibody fragments, have been shown to be more susceptible to proteolysis when they are expressed as surface anchored in staphylococci (18). This assay thus indicates that the chimeric surface proteins were expressed as full-length gene products, which were correctly targeted to the cell wall of S. carnosus.
FIG. 2.
SDS-PAGE analysis on a 4 to 20% polyacrylamide gradient gel under reducing conditions. Eluted and pooled fractions from HSA-affinity-purified cell wall surface proteins from recombinant S. carnosus cells as follows: pSPPmABPXM transformed (lane 1), Sc:mZIgA (lane 2), Sc:dZIgA (lane 3), Sc:mZIgE (lane 4), and Sc:dZIgE (lane 5). Lane M, marker proteins with molecular masses (in kilodaltons).
Investigation of the surface accessibility and the Ig binding functions.
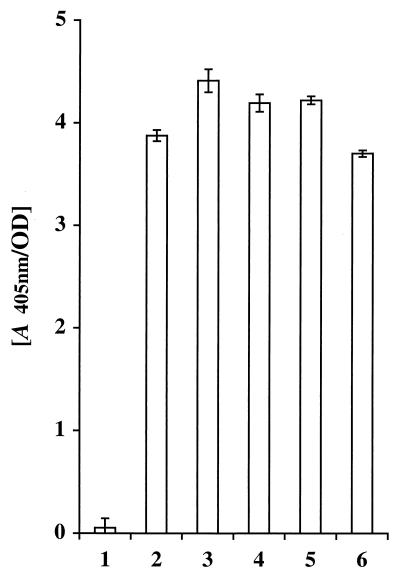
The surface accessibility of the expressed chimeric surface proteins was investigated by a colorimetric enzymatic assay (41), taking advantage of the albumin binding reporter moiety, ABP, present within the recombinant proteins. For wild-type and recombinant staphylococci Sc:ABP, Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, and Sc:dZIgE (Fig. 1), which were incubated with biotinylated HSA and a streptavidin-alkaline phosphatase conjugate, the presence of ABP-containing surface receptors was detected by using a chromogenic substrate. ABP was detectable on all the recombinant staphylococci (Fig. 3, bars 2 to 6), whereas wild-type S. carnosus cells, as expected, showed no albumin binding capacity (Fig. 3, bar 1). This demonstrates that the chimeric surface proteins with the capacity of binding to serum albumin were successfully targeted and anchored, in accessible forms, to the outer surface of the recombinant staphylococci. Interestingly, the levels of surface expression of chimeric proteins containing affibody domains (Fig. 3, bars 3 to 6), seemed to be of the same magnitude as those for the fusion protein construct encoded by the parental vector (Fig. 3, bar 2), suggesting that the surface density of chimeric proteins was not reduced by the introduction of monomeric and dimeric affibody domains. This contrasts with earlier results, in which the levels of surface expression of recombinant proteins appeared to be reduced when peptides and protein domains of various origins were introduced genetically into the surface proteins (18, 28, 39), and might be explained by the staphylococcal origin of the SpA-derived affibodies.
FIG. 3.
Histogram representation of the results from a colorimetric whole-cell assay for detection of surface-exposed proteins containing ABP. Biotinylated HSA is allowed to bind to the staphylococcal cells, and after the subsequent addition of a streptavidin-alkaline phosphatase conjugate and a chromogenic substrate, the change in A405 is monitored. S. carnosus wild type (bar 1) and recombinant S. carnosus cells as follows: pSPPmABPXM transformed (bar 2), Sc:mZIgA (bar 3), Sc:dZIgA (bar 4), Sc:mZIgE (bar 5), and Sc:dZIgE (bar 6).
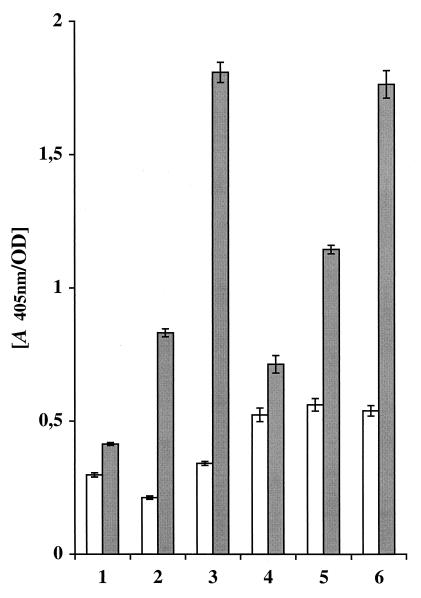
To evaluate if the affibody domains were functional when they were expressed as parts of the chimeric surface proteins on S. carnosus, the different recombinant staphylococcal cells expressing monomeric or dimeric IgA or IgE binding affibody domains were analyzed for their ability to bind their respective Ig targets. S. carnosus cells harboring the parental vector pSPPmABPXM were included as a negative control. Cultured cells were allowed to bind to human IgA or IgE, respectively, and the binding was analyzed by the addition of a β-galactosidase-conjugated polyclonal anti-IgA reagent or a mixture of three different murine β-galactosidase-conjugated IgE-specific monoclonal antibodies. When the staphylococcal cells expressing monomeric or dimeric ZIgA affibody domains were analyzed for their IgA binding capacity, a significant response was detected (Fig. 4, bars 2 and 3, shaded) compared to that in the cells harboring the parental vector (Fig. 4, bar 1, shaded), and a similar low background binding was observed for all constructs when IgA was omitted (Fig. 4, bars 1 to 3, white). When recombinant staphylococci carrying monomeric or dimeric ZIgE affibody variants were analyzed by a similar assay for their binding to IgE, a significant IgE reactivity was observed (Fig. 4, bars 5 and 6, shaded) compared to that for the control cells expressing the PP-ABP-XM′ protein (Fig. 4, bar 4, shaded). In this case, the negative control experiments, i.e., those with IgE omitted, resulted in a slightly higher background response (Fig. 4, bars 5 and 6, white), which was, however, of a magnitude similar to that of control cells (Fig. 4, bar 4, white).
FIG. 4.
Colorimetric assay showing whole-cell reactivity to human IgA or IgE. The different recombinant cells were cultured to the same cell density, harvested, and allowed to react with IgA or IgE. Thereafter, β-galactosidase-conjugated antibodies, either anti-IgA polyclonal serum or a mixture of three different monoclonal anti-IgE antibodies, were added, and after addition of the substrate, the shift in A405 was monitored. Shaded bars indicate the reactivity for recombinant S. carnosus cells to IgA (bar 1, pSPPmABPXM-transformed S. carnosus; bar 2, Sc:mZIgA; bar 3, Sc:dZIgA) or to IgE (bar 4, pSPPmABPXM-transformed S. carnosus; bar 5, Sc:mZIgE; bar 6, Sc:dZIgE. The white bars indicate the background reactivity when the target Ig, either IgA or IgE, was omitted.
Taken together, these results indicate a significant IgA and IgE binding capacity for the recombinant staphylococcal cells, carrying surface proteins containing ZIgA and ZIgE affibody domains, respectively. Six samples were compared each time, and the entire experiment was repeated, with highly reproducible results. Interestingly, significantly higher binding capacities for IgA and IgE were observed for the recombinant staphylococci with dimeric ZIgA and ZIgE affibody domains introduced into the chimeric surface proteins (Fig. 4, bars 3 and 6, shaded) compared with the corresponding constructs with monomeric affibody domains (Fig. 4, bars 2 and 5, shaded).
DISCUSSION
We have described how SpA domains, selected from phage-displayed combinatorial libraries, with binding specificity for human IgA and IgE, respectively, have been expressed as part of chimeric surface proteins in S. carnosus, thus generating staphylococcal cells capable of binding IgA and IgE, respectively. The different chimeric proteins were shown to be localized to the staphylococcal cell walls, from which they could be extracted and purified by affinity chromatography, by using an albumin-binding protein region, ABP, present in the chimeric surface proteins. The recovered surface proteins were shown to be proteolytically stable. In a previous study, it was demonstrated that S. carnosus cells transformed with the parental vector pSPPmABPXM carried approximately 104 surface-exposed chimeric proteins per cell (2). Since the recombinant staphylococci presented here, Sc:mZIgA, Sc:dZIgA, Sc:mZIgE, and Sc:dZIgE (Fig. 1), seem to express surface proteins at a level similar to that of the control cells (Fig. 3), it is likely that the numbers of surface proteins displayed by these recombinant staphylococci are in the same order of magnitude.
In an earlier study aimed to generate staphylococcal cells with IgE binding capacity, an anti-IgE scFv antibody construct was introduced into the S. carnosus surface expression system (18). In that study, a marked proteolytic degradation as well as a decreased surface expression of the chimeric surface proteins was observed (18). The staphylococcal origin of the SpA-derived affibodies used in this study might partially explain the improved expression and display characteristics observed.
Various applications of the strategy employed to create staphylococcal cells carrying protein A derivatives with redirected binding specificities can be envisioned. One practical application for this type of recombinant bacteria would be to use them as whole-cell monoclonal antibodies in different diagnostic tests. The described strategy could prove to be a straightforward and cost-effective way of producing monoclonal antibodies for diagnostic purposes. The resulting recombinant bacteria might also be suitable for immunoprecipitation experiments or, in an immobilized form (24), as inexpensive adsorbents for the recovery of IgE or IgA.
A second application for the described strategy would be to improve bacterial vaccine delivery systems by the selection and display of protein domains which would target the bacterial vaccine delivery vehicle to immunoreactive sites. Recombinant S. carnosus cells have been extensively investigated in the context of vaccine delivery (44), and attempts to improve immunogenicity include the surface display of the cholera toxin B subunit (CTB) (4, 28) or various fibronectin binding domains (27). The creation of purposely designed targeting domains would potentially improve such vaccine delivery systems.
Irrespective of the application, it is of the utmost importance to construct bacteria that bind efficiently to a desired target molecule. One way of improving the binding capacity has been demonstrated in the present study by dimerization of the affibody. However, it cannot be concluded from our results whether the obtained improved binding is due to sterical effects or an increased apparent affinity via avidity effects. Analogous to other affibodies selected from the naive Z domain libraries (21, 33), the affibodies used in this study have affinities (Kd) to their targets in the micromolar range. Gunneriusson and coworkers have shown that affibodies of significantly higher affinities can be isolated by using an affinity maturation strategy (16). Such affibodies might be useful for the creation of potent whole-cell diagnostic devices or efficient vaccine delivery vehicles.
Furthermore, it would be of obvious interest to investigate whether recombinant staphylococci could be used directly as an alternative to filamentous phages for the display of affibody libraries to achieve subsequent affinity selection of relevant affibodies by using fluorescence-activated cell sorting technology. This would thus circumvent the phage display selection procedure used in this study for the initial selection of the ZIgA and ZIgE affibodies.
ACKNOWLEDGMENTS
We are grateful to M. Uhlén and K. Nord for valuable discussions.
This work was financially supported by The Swedish National Board for Technical and Industrial Development (NUTEK).
REFERENCES
- 1.Acheson D W K, Sonenshein A L, Leong J M, Keusch G T. Heat-stable spore-based vaccines: surface expression of invasin-cell wall fusion proteins in Bacillus subtilis. In: Brown F, Burton D, Doherty P, Mekalanos J, Norrby E, editors. Vaccines 97. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 179–184. [Google Scholar]
- 2.Andréoni C, Goetsch L, Libon C, Samuelson P, Nguyen T N, Robert A, Uhlén M, Binz H, Ståhl S. Flow cytometric quantification of surface-displayed recombinant receptors on staphylococci. BioTechniques. 1997;23:696–704. [PubMed] [Google Scholar]
- 3.Ayora S, Lindgren P E, Götz F. Biochemical properties of a novel metalloprotease from Staphylococcus hyicus subsp. hyicus involved in extracellular lipase processing. J Bacteriol. 1994;176:3218–3223. doi: 10.1128/jb.176.11.3218-3223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cano, F., S. Liljeqvist, T. N. Nguyen, P. Samuelson, J.-Y. Bonnefoy, S. Ståhl, and A. Robert. A surface-displayed cholera toxin B peptide improves antibody responses using food-grade staphylococci for mucosal subunit vaccine delivery. FEMS Immunol. Med. Microbiol., in press. [DOI] [PubMed]
- 5.Demleitner G, Götz F. Evidence for importance of the Staphylococcus hyicus lipase pro-peptide in lipase secretion, stability and activity. FEMS Microbiol Lett. 1994;121:189–197. doi: 10.1111/j.1574-6968.1994.tb07098.x. [DOI] [PubMed] [Google Scholar]
- 6.Di Fabio S, Medaglini D, Rush C M, Corrias F, Panzini G L, Pace M, Verani P, Pozzi G, Titti F. Vaginal immunization of cynomolgus monkeys with Streptococcus gordonii expressing HIV-1 and HPV 16 antigens. Vaccine. 1998;16:485–492. doi: 10.1016/s0264-410x(97)80002-3. [DOI] [PubMed] [Google Scholar]
- 7.Fischetti V A, Medaglini D, Pozzi G. Gram-positive commensal bacteria for mucosal vaccine delivery. Curr Opin Biotechnol. 1996;7:659–666. doi: 10.1016/s0958-1669(96)80079-6. [DOI] [PubMed] [Google Scholar]
- 8.Francisco J A, Campbell R, Iverson B L, Georgiou G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc Natl Acad Sci USA. 1993;90:10444–10448. doi: 10.1073/pnas.90.22.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francisco J A, Earhart C F, Georgiou G. Transport and anchoring of β-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:2713–2717. doi: 10.1073/pnas.89.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francisco J A, Stathopoulos C, Warren R A, Kilburn D G, Georgiou G. Specific adhesion and hydrolysis of cellulose by intact Escherichia coli expressing surface anchored cellulase or cellulose binding domains. Bio/Technology. 1993;11:491–495. doi: 10.1038/nbt0493-491. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs P, Breitling F, Dubel S, Seehaus T, Little M. Targeting recombinant antibodies to the surface of Escherichia coli: fusion to a peptidoglycan associated lipoprotein. Bio/Technology. 1991;9:1369–1372. doi: 10.1038/nbt1291-1369. [DOI] [PubMed] [Google Scholar]
- 12.Georgiou G, Stathopoulos C, Daugherty P S, Nayak A R, Iverson B L, Curtiss R I. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol. 1997;15:29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- 13.Götz F. Staphylococcus carnosus: a new host organism for gene cloning and protein production. J Appl Bacteriol Symp Suppl. 1990;19:49S–53S. doi: 10.1111/j.1365-2672.1990.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 14.Götz F, Ahrne S, Lindberg M. Plasmid transfer and genetic recombination by protoplast fusion in staphylococci. J Bacteriol. 1981;145:74–81. doi: 10.1128/jb.145.1.74-81.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Götz F, Schumacher B. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol Lett. 1987;40:285–288. [Google Scholar]
- 16.Gunneriusson, E., K. Nord, M. Uhlén, and P.-Å. Nygren. Affinity maturation of a Taq DNA polymerase specific affibody by helix shuffling. Protein Eng., in press. [DOI] [PubMed]
- 17.Gunneriusson, E., J. Ringdahl, M. Högbom, H. Grönlund, and P.-Å. Nygren. Unpublished data.
- 18.Gunneriusson E, Samuelson P, Uhlén M, Nygren P-Å, Ståhl S. Surface display of a functional single-chain Fv antibody on staphylococci. J Bacteriol. 1996;178:1341–1346. doi: 10.1128/jb.178.5.1341-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guss B, Uhlén M, Nilsson B, Lindberg M, Sjöquist J, Sjödahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 20.Hammes W P, Bosch I, Wolf G. Contribution of Staphylococcus carnosus and Staphylococcus piscifermentans to the fermentation of protein foods. J Appl Bacteriol Symp Suppl. 1995;79:76S–83S. [Google Scholar]
- 21.Hansson M, Ringdahl J, Robert A, Power U, Goetsch L, Nguyen T N, Uhlén M, Ståhl S, Nygren P-Å. An in vitro selected binding protein (affibody) shows conformation-independent recognition of the respiratory syncytial virus (RSV) G protein. Immunotechnology. 1999;4:237–252. doi: 10.1016/s1380-2933(98)00026-8. [DOI] [PubMed] [Google Scholar]
- 22.Hansson M, Ståhl S, Nguyen T N, Bachi T, Robert A, Binz H, Sjölander A, Uhlén M. Expression of recombinant proteins on the surface of the coagulase-negative bacterium Staphylococcus xylosus. J Bacteriol. 1992;174:4239–4245. doi: 10.1128/jb.174.13.4239-4245.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hultman T, Ståhl S, Hornes E, Uhlén M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler S W. Use of protein A-bearing staphylococci for the immunoprecipitation and isolation of antigens from cells. Methods Enzymol. 1981;73:442–459. doi: 10.1016/0076-6879(81)73084-2. [DOI] [PubMed] [Google Scholar]
- 25.Liebl W, Götz F. Studies on lipase directed export of Escherichia coli beta-lactamase in Staphylococcus carnosus. Mol Gen Genet. 1986;204:166–173. doi: 10.1007/BF00330205. [DOI] [PubMed] [Google Scholar]
- 26.Liepe H-U. Bakterienkulturen und Rohwurst. Forum Mikrobiologie. 1982;5:10–15. [Google Scholar]
- 27.Liljeqvist S, Cano F, Nguyen T N, Uhlén M, Robert A, Ståhl S. Surface display of functional fibronectin-binding domains on Staphylococcus carnosus. FEBS Lett. 1999;446:299–304. doi: 10.1016/s0014-5793(99)00232-x. [DOI] [PubMed] [Google Scholar]
- 28.Liljeqvist S, Samuelson P, Hansson M, Nguyen T N, Binz H, Ståhl S. Surface display of the cholera toxin B subunit on Staphylococcus xylosus and Staphylococcus carnosus. Appl Environ Microbiol. 1997;63:2481–2488. doi: 10.1128/aem.63.7.2481-2488.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen T N, Gourdon M H, Hansson M, Robert A, Samuelson P, Libon C, Andréoni C, Nygren P-Å, Binz H, Uhlén M, Ståhl S. Hydrophobicity engineering to facilitate surface display of heterologous gene products on Staphylococcus xylosus. J Biotechnol. 1995;42:207–219. doi: 10.1016/0168-1656(95)00081-z. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen T N, Hansson M, Ståhl S, Bächi T, Robert A, Domzig W, Binz H, Uhlén M. Cell-surface display of heterologous epitopes on Staphylococcus xylosus as a potential delivery system for oral vaccination. Gene. 1993;128:89–94. doi: 10.1016/0378-1119(93)90158-y. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, Henrichson C, Jones T A, Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 33.Nord K, Gunneriusson E, Ringdahl J, Ståhl S, Uhlén M, Nygren P-Å. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 34.Nord K, Nilsson J, Nilsson B, Uhlén M, Nygren P-Å. A combinatorial library of an α-helical bacterial receptor domain. Protein Eng. 1995;8:601–608. doi: 10.1093/protein/8.6.601. [DOI] [PubMed] [Google Scholar]
- 35.Norton P M, Brown H W, Wells J M, Macpherson A M, Wilson P W, Le Page R W. Factors affecting the immunogenicity of tetanus toxin fragment C expressed in Lactococcus lactis. FEMS Immunol Med Microbiol. 1996;14:167–177. doi: 10.1111/j.1574-695X.1996.tb00284.x. [DOI] [PubMed] [Google Scholar]
- 36.Nygren P-Å, Eliasson M, Palmcrantz E, Abrahmsén L, Uhlén M. Analysis and use of the serum albumin binding domains of streptococcal protein G. J Mol Recognit. 1988;1:69–74. doi: 10.1002/jmr.300010204. [DOI] [PubMed] [Google Scholar]
- 37.Pozzi G, Contorni M, Oggioni M R, Manganelli R, Tommasino M, Cavalieri F, Fischetti V A. Delivery and expression of a heterologous antigen on the surface of streptococci. Infect Immun. 1992;60:1902–1907. doi: 10.1128/iai.60.5.1902-1907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ringdahl, J., E. Gunneriusson, H. Grönlund, and P.-Å. Nygren. Unpublished data.
- 39.Robert A, Samuelson P, Andréoni C, Bachi T, Uhlén M, Binz H, Nguyen T N, Ståhl S. Surface display on staphylococci: a comparative study. FEBS Lett. 1996;390:327–333. doi: 10.1016/0014-5793(96)00684-9. [DOI] [PubMed] [Google Scholar]
- 40.Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982;10:5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samuelson P, Hansson M, Ahlborg N, Andréoni C, Götz F, Bächi T, Nguyen T N, Binz H, Uhlén M, Ståhl S. Cell surface display of recombinant proteins on Staphylococcus carnosus. J Bacteriol. 1995;177:1470–1476. doi: 10.1128/jb.177.6.1470-1476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 43.Sousa C, Cebolla A, de Lorenzo V. Enhanced metalloadsorption of bacterial cells displaying poly-His peptides. Nat Biotechnol. 1996;14:1017–1020. doi: 10.1038/nbt0896-1017. [DOI] [PubMed] [Google Scholar]
- 44.Ståhl S, Samuelson P, Hansson M, Andréoni C, Goetsch L, Libon C, Liljeqvist S, Gunneriusson E, Binz H, Nguyen T N, Uhlén M. Development of non-pathogenic staphylococci as vaccine delivery vehicles. In: Pozzi G, Wells J M, editors. Gram-positive bacteria: vaccine vehicles for mucosal immunization. Georgetown, Tex: Landes Bioscience; 1997. pp. 62–81. [Google Scholar]
- 45.Ståhl S, Uhlén M. Bacterial surface display: trends and progress. Trends Biotechnol. 1997;15:185–192. doi: 10.1016/S0167-7799(97)01034-2. [DOI] [PubMed] [Google Scholar]
- 46.Stover C K, Bansal G P, Hanson M S, Burlein J E, Palaszynski S R, Young J F, Koenig S, Young D B, Sadziene A, Barbour A G. Protective immunity elicited by recombinant bacille Calmette-Guerin (BCG) expressing outer surface protein A (OspA) lipoprotein: a candidate Lyme disease vaccine. J Exp Med. 1993;178:197–209. doi: 10.1084/jem.178.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauss A, Götz F. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21:491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]
- 48.Uhlén M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]