Abstract
Castor (Ricinus communis L.) is an important industrial multipurpose non-edible oilseed C3 crop belongs to spurge family popularly known as Euphorbiaceae. Its oil has exceptional properties which provides an industrial importance to this crop. The present investigation is aimed to judge the stability and performance of yield and yield assigning traits and selection of suitable genotype for varied locality of western rainfed regions of India. During the study with 90 genotypes, the genotype × environment interaction was found to be significant for seed yield per plant as well as for plant height up to primary raceme, total length of primary raceme, effective length of primary raceme, capsules on main raceme and effective number of racemes per plant. E1 is the least interactive and highly representative site for seed yield. Which won where and what biplot decipher ANDCI 10-01 as vertex genotype for E3 while ANDCI 10-03 and P3141 for E1 and E2. Average Environment co-ordinate identify ANDCI 10-01, P3141, P3161, JI 357 and JI 418 as tremendously stable and high seed yielding genotypes. The study outlined the pertinency of Multi Trait Stability Index, that calculated based on the genotype-ideotype distance as the multiple interacting variables. MTSI evaluated all genotypes and sort ANDCI 12-01, JI 413, JI 434, JI 380, P3141, ANDCI 10-03, SKI 215, ANDCI 09, SI 04, JI 437, JI 440, RG 3570, JI 417 and GAC 11 with maximum stability and high mean performance of analyzed interacting traits.
Keywords: Castor, AMMI, GGE, MTSI, Oilseed
1. Introduction
Castor (Ricinus communis L.) is an annual or perennial flowering non-edible versatile oilseed species of Euphorbiaceae family. It is a fast-growing monotypic C3 plant having 10 diploid set of chromosomes. Castor bean is domesticated 3200 years ago from Ethiopian center of origin as a non-edible vegetable oil seed crop [1,2]. Mirza [3] reported 50–55% oil and 26–30% protein in seed. It makes uniform solution with alcohol at standard temperature and pressure which enable several chemical reactions for study [4]. Castor oil has unique fatty acid named “Ricinoleic acid” which has anti-inflammatory effects [5]. Oil has more than 700 uses ranges from medicine to cosmetics in daily life while used as a biofuel in today's energy option, used as manure and organic termite controller in agriculture [6].
Castor is mostly dispersed in tropical, subtropical and warm-temperate areas of the globe. It is particularly widespread on fellow land, curbside and residences in rural and urban zones, as well as alongside drought - prone rivers at elevation ranges from 400 to 2700 m. Castor is noticeably cultivated in (sub) tropical and temperate zones. In India, it is agriculturally most imperative crop having the 900 (‘000hm2) acreage with 1198 (‘000 t) output [7]. In Asian continent, India is the major producer and exporter of castor seed. It is cultivated largely in Gujarat, Andhra Pradesh and Rajasthan in Indian mainland.
Seed yield is an ultimate product for farmers but it is a polygenic dependent variable influenced by another self-governing variable as well as prevailing environment. Environment plays an important role in production of seed yield in castor. As a result, knowledge of these yield components is required in order to have a meaningful influence on yield studies aimed at improvement [8]. Use of stable genotypes could be a source of yield and oil enhancement in castor across varied environments. However, genotype environment interaction (GEI) is an important feature of plant breeding programs for introduction of different crop types in vast region [8]. Baker [9] defined GEI as a change in genotypic performance under the influence of different environmental conditions. Different crop varieties have different buffering capacity and they have different response towards changing their micro climate like soil type, nutrient availability, biotic and abiotic influences [10,11]. It is therefore domineering that the investigation of GEI takes focal point when assessing varieties for adaptation. GEI analysis is prerequisite in breeding programme to recommend specifically or generally adopted cultivar which gives high and stable seed yield. As a result, it is critical that the GEI being addressed, properly understood and studied using diverse locations and seasons before release of variety for commercial cultivation.
Various methodologies have been proposed for crop stability evaluation, such as Roemer [12], who initially proposed stability analysis utilizing variation across environments as a yield stability indicator. Wricke [13] proposed the notion of ecovalence to get access to the stability of each genotype. Shukla [14] recommended to use the variance component of each genotype across a variety of environments to assess phenotypic stability. The most common sort of stability analysis is joint regression analysis, which aids in estimating genotype characteristics in a liner response to environmental fluctuations [15]. Yates and Cochran [16] developed the joint regression analysis strategy which was later improved by Finlay and Wilkinson [17] and Eberhart and Russell [18] and is now a frequently used method but it is not able to delineate the environments into few mega environments.
Among all discussed stability parameters, Additive Main Effect and Multiplicative Interaction (AMMI) model and Genotype × Genotype × Environment (GGE) biplot technique is more efficient and frequently used to process multi environmental data. AMMI model gives information regarding main and interaction effects including biplot [19]. It is specifically efficient for illustrating adaptive response and buffering capacity. But AMMI is incapable to identify close relation between high mean performance and stability. This problem is overcome through GGE biplot method [20]. It includes both genotype main effects and GEI effects for the study [21]. As a result, the GGE biplot model is used to determine the optimal genotypes and test locations [22].
Nowadays, there is a great demand for stable cultivars that perform well in a wide range of environments. This requirement was full filled through Smith and Hazel selection index [23,24]. But this index is suffering from multicollinearity effects when applied to MET data. This constrain is solved by Multi trait selection index [25]. It evolved as unique techniques for choosing superior genotypes with high yield stability and desirable features that might perform better across varied environmental circumstances [26]. Castor is most important crop of world especially Brazil, India and China but its improvement is slower compared to other oilseed crops though around 50 varieties and hybrids have been developed in India. But to give a pace to castor breeding around the globe, identification of high yielding genotypes is prerequisite. Studies are there in this direction but multi-location experiments are very limited moreover with very limited genotypes. To generate the information and to fill the gap in this direction, current experiment was conducted to evaluate the performance of castor germplasm at multiple locations for stability analysis of genotypes. On the other side, no information on biplot, AMMI and MTSI is reported in castor. On the light of above discussion, MET data of castor genotypes were assessed to identify highly stable and high performing genotype for cultivation in wide range of locations and seasons.
2. Materials and methods
2.1. Planting material and locations for evaluation
A total of ninety diverse castor genotypes were used in this study (Suppl. Table 1). The field experiment was directed at three different locations in 2020–21 in Kharif (an Indian term for monsoon season cropping). These locations are come under agroclimatic zone III of middle Gujarat, India but these all location have different soil profile like soil of E1 (Agricultural Research Station, AAU, Sansoli) environment is acidic in nature with 6.7 pH value and sandy in texture with poor water holding capacity. E2 (Pulse Research Station, AAU, Vadodara) has loamy sand texture with good water holding capacity. Soil reaction was slightly towards alkaline having pH 7.7. E3 (Agricultural research Station, AAU, Derol) has neutral soil reaction with 7.1 pH. The soil was sandy loam in texture, deprived in organic carbon, average in available phosphorous and rich in available potash.
2.2. Experimental design and package of practices
The experiment used a Randomized Complete Block Design (RCBD) with two replications in each site. Each genotype was grown in a single row of 6 m length with 120 cm of between row distance and 60 cm of plant-to-plant distance. To avoid damage and border effect, the experiment was ringed by a guard row. The crop was successfully raised by following the necessary agronomical and plant protection methods. Pendimethalin 30% EC was sprayed as a pre-emergence herbicide shortly after planting to address the initial weed issue.
2.3. Trait phenotyping
Thirteen yield and yield attributing traits like days to 50% flowering, days to maturity, node up to primary raceme, plant height up to primary raceme (cm), total length of primary raceme (cm), effective length of primary raceme (cm), capsules on main raceme, effective racemes per plant, seed yield per plant (g), 100 seed weight (g), oil content (%), shelling out turn (%) and L:B ratio of seed were observed from five competitive plants per genotype per replication. For 100-seed weight, the harvested capsules were hand cleaned and threshed using a manual decorticator. The weight of 100 seeds of each entry was recorded in grams and used in statistical analysis. Shelling out turn (%) was estimated as per Sapovadiya [27]. Days to 50% flowering and days to maturity was recorded on plot basis as per Akhila et al. [28].
2.4. Analysis of variance
The data of seed yield and its related traits under three test environments were analyzed through pooled analysis of variance where genotypes assumed as fixed and test environments as random factors [29].
2.5. GEI analysis
2.5.1. AMMI analysis
The data of seed yield per plant was subjected to AMMI analysis [30,31]. Regular ANOVA explained the additive main effects of genotype and environments while PCA revealed non-additive portion. The location wise stable genotypes identified by AMMI analysis assessed for significance using the Gollob [32] F-test approach [33]. The main effect of means vs the first principal component axis (PCA I) and between the first two principal component axes were used to create AMMI biplots. The AMMI stability values (ASV) and yield stability index (YSI) were also used to rank genotypes [34,35].
2.5.2. GGE biplots
The site regression genotype-genotype environment interaction (GGEI) biplot models are regarded as a potent tool for successful analysis and interpretation of multi-environment data structures in plant breeding [20,36]. The GGE study revealed that the first two components which are best match for creating GGE biplots that explain most of the variance. All of the biplots in the research were constructed using environment-centered data and the symmetrical technique of singular value partitioning (SVP) method. Mean versus stability biplot created with SVP's row metric preservation approach.
2.6. Multi-trait stability index (MTSI)
To measure the stability of each genotype, SVD (Singular Value Decomposition) of the matrix of Basic Linear Unbiased Predictions (BLUP) for the GE interaction effects was created using a linear mixed model (LMM). The stability of each genotype was determined by calculating the Weighted Average of Absolute Scores (WAAS) from the singular value decomposition of the matrix of best linear unbiased predictions for the GEI effects derived by a linear mixed-effect model and concurrent selection for mean performance and stability were accomplished by employing the WAASBY index [26].
The genotype with the lowest MTSI score is more similar to the ideotype and hence has a high mean performance and stability over the environments for all traits under study. The most desired genotypes with the best production and stability were chosen with a 15% selection intensity. MTSI scores were plotted to demonstrate these selected and non-selected genotypes.
By generating a Y × WAAS biplot, the genotypes were categorized into four distinct groups, allowing for the combined interpretation of stability and mean performance in varied contexts. This four-quadrant biplot was created with seed yield on the x-axis and WAASB values on the y-axis.
2.7. Statistical packages
Pooled analysis of variance, AMMI analysis, ASV, YSI, GGE biplots and MTSI were calculated using RStudio, R version 4.0.3 by using ‘agricolae’ and ‘metan’ R packages.
3. Results and discussion
3.1. ANOVA and mean performance
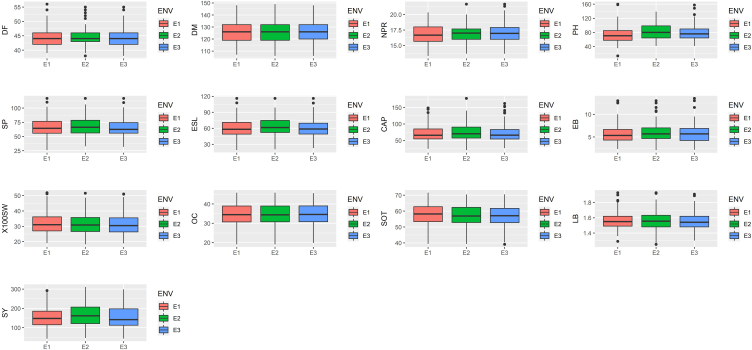
A joint analysis of variance revealed a difference among all genotypes for all attributes under evaluation. Total length of primary raceme exhibited the most variance towards overall (G + E + GEI) variability (19.48%) followed by effective length of primary raceme (17.31%), effective number of racemes per plant (15.86%) and seed yield per plant (11.09%) (Table 1). While genotypes shared the highest variation for all characteristics tested across all locations, environment shared the least variation for all traits yet it was found significant in all features except Oil content, implying that all environments evaluated were different. The highest variance for environment among all variables was 3.51% for plant height up to main raceme and 1.07% for effective number of racemes per plant. In the current study, seed yield showed considerable GEI in pooled analysis of variance, indicating that genotype stability should be investigated further. Patel et al. [37] and Patel et al. [38] found similar result for pooled analysis of variance for all the traits and suggested to go for stability analysis of all characters through Eberhart and Russell model [18]. Mean seed yield was 159.63 g/plant over the locations with the range of 154.93 (E1) to 167.69 g/plant (E3). Likewise, Table 2 showed mean over environments of the other characters studied which were 44.38 (DF), 126.63 (DM), 16.92 (NPR), 78.05 (PH), 66.58 (SP), 60.74 (ESL), 72.25 (CAP), 5.77 (EB), 31.69 (100SW), 34.39 (OC), 57.16 (SOT) and 1.56 (LB) and Boxplot (Fig. 1) depict mean of characters of individual environments. Movaliya et al. [39] and Akhila et al. [28] reported similar kind of mean and range of different characters. Chaudhari et al. [40] also reported comparable results for all traits over the environments. Authors advised that the appearance of diversity across all genotypes for all attributes might be a result of differential ability of castor genotype and variable interaction of genes with changing environment [40].
Table 1.
Combined analysis of variance for yield and its attributing traits towards total variation among 90 castor genotypes tested at three locations in Kharif 2020-21.
| Source of variation |
Environment (df: 2) |
Genotype (df: 89) |
GEI (df: 178) |
Residual (df: 267) |
CV (%) | |||
|---|---|---|---|---|---|---|---|---|
| Trait | Mean Squares | % (G + E + GEI) | Mean Squares | % (G + E + GEI) | Mean Squares | % (G + E + GEI) | Mean Squares | |
| DF | 12.91** | 0.54 | 51.38** | 96.31 | 0.84 | 3.15 | 2.58 | 3.626 |
| DM | 6.11 | 0.03 | 498.23** | 99.44 | 1.33 | 0.53 | 2.84 | 1.332 |
| NPR | 2.60* | 0.56 | 9.53** | 91.56 | 0.41 | 7.88 | 0.85 | 5.461 |
| PH | 4267.94** | 3.51 | 2280.41** | 83.58 | 176.12** | 12.91 | 68.54 | 10.607 |
| SP | 322.73** | 0.57 | 1026.12** | 79.95 | 125.04** | 19.48 | 36.21 | 9.039 |
| ESL | 617.43** | 0.87 | 1311.06** | 81.82 | 138.72** | 17.31 | 49.26 | 11.556 |
| CAP | 805.37** | 0.49 | 3328.78** | 89.85 | 178.87** | 9.66 | 98.89 | 13.764 |
| EB | 9.16** | 1.07 | 16.03** | 83.07 | 1.53** | 15.86 | 0.83 | 15.855 |
| 100SW | 9.96** | 0.08 | 294.62** | 99.80 | 0.18 | 0.12 | 0.19 | 1.400 |
| OC | 0.15 | 0.00 | 191.66** | 99.94 | 0.06 | 0.06 | 0.23 | 1.406 |
| SOT | 29.63** | 0.27 | 241.69** | 99.15 | 0.70 | 0.57 | 2.31 | 2.660 |
| LB | 0.005* | 0.16 | 0.07** | 97.07 | 0.001 | 2.77 | 0.001 | 2.388 |
| SY | 8851.78** | 0.97 | 17997.54** | 87.94 | 1135.11** | 11.09 | 321.16 | 11.226 |
*,** Significant at p < 0.05 and p < 0.01, respectively, df: degrees of freedom, DF: Days to 50% flowering, DM: Days to maturity, NPR: Nodes up to primary raceme, PH: Plant height up to primary raceme, SP: Total length of primary raceme, ESL: Effective length of primary raceme, CAP: Capsules on main raceme, EB: Effective number of racemes per plant, 100 SW: 100 seed weight, OC: Oil content, SOT: Shelling out turn, LB: Length/Breadth ratio of seed and SY: Seed yield per plant.
Table 2.
Mean and range of quantitative traits of castor genotypes tested at three locations in Kharif 2020-21.
| Traits | Sansoli |
Vadodara |
Derol |
Mean over environments | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | Mean | Min | Max | Mean | Min | Max | ||
| DF | 44.07 | 39.00 | 55.00 | 44.49 | 39.00 | 54.50 | 44.57 | 39.50 | 54.50 | 44.38 |
| DM | 126.59 | 107.50 | 146.50 | 126.83 | 107.00 | 146.50 | 126.47 | 106.50 | 148.50 | 126.63 |
| NPR | 16.80 | 13.67 | 20.00 | 17.04 | 13.84 | 20.61 | 16.91 | 14.17 | 20.17 | 16.92 |
| PH | 72.93 | 26.81 | 159.17 | 78.60 | 42.33 | 153.00 | 82.62 | 44.95 | 137.10 | 78.05 |
| SP | 66.06 | 27.00 | 106.30 | 65.57 | 33.95 | 113.34 | 68.10 | 32.99 | 107.83 | 66.58 |
| ESL | 59.66 | 19.50 | 103.08 | 59.68 | 24.15 | 111.65 | 62.88 | 21.61 | 103.83 | 60.74 |
| CAP | 70.93 | 25.33 | 146.56 | 71.13 | 26.78 | 158.17 | 74.69 | 21.47 | 158.96 | 72.25 |
| EB | 5.63 | 2.50 | 12.75 | 5.65 | 2.35 | 13.25 | 6.03 | 2.32 | 12.75 | 5.77 |
| 100SW | 31.91 | 19.07 | 51.56 | 31.44 | 19.16 | 51.00 | 31.72 | 18.52 | 51.58 | 31.69 |
| OC | 34.37 | 19.79 | 45.32 | 34.37 | 20.15 | 45.23 | 34.42 | 19.61 | 45.58 | 34.39 |
| SOT | 57.55 | 39.69 | 70.44 | 56.74 | 39.09 | 70.24 | 57.19 | 39.54 | 69.42 | 57.16 |
| LB | 1.56 | 1.29 | 1.92 | 1.55 | 1.31 | 1.90 | 1.57 | 1.26 | 1.93 | 1.56 |
| SY | 154.93 | 46.42 | 283.10 | 156.28 | 45.65 | 291.97 | 167.69 | 56.19 | 290.65 | 159.63 |
DF: Days to 50% flowering, DM: Days to maturity, NPR: Nodes up to primary raceme, PH: Plant height up to primary raceme, SP: Total length of primary raceme, ESL: Effective length of primary raceme, CAP: Capsules on main raceme, EB: Effective number of racemes per plant, 100 SW: 100 seed weight, OC: Oil content, SOT: Shelling out turn, LB: Length/Breadth ratio of seed and SY: Seed yield per plant.
Fig. 1.
Box plots showing mean performance of the studied traits across all three environments during Kharif 2020-21. Box plots of mean performance for various characters. (A)DF: Days to 50% flowering, (B) DM: Days to maturity, (C) NPR: Nodes up to primary raceme, (D) PH: Plant height up to primary raceme, (E) SP: Total length of primary raceme, (F) ESL: Effective length of primary raceme, (G) CAP: Capsules on main raceme, (H) EB: Effective number of racemes per plant, (I) 100 SW: 100 seed weight, (J) OC: Oil content, (K) SOT: Shelling out turn, (L) LB: Length/Breadth ratio of seed (M) SY: Seed yield per plant, ENV: Environments, E1: Sansoli, E2: Vadodara, E3: Derol.
3.2. GEI analysis
AMMI analysis for seed yield revealed a significant difference for the additive component of the total sum of squares provided by genotypic influence (87.94%), GEI interaction effect (11.09%) and environment (0.97%) (Table 3). This finding revealed that altering environmental conditions had a significant impact on seed yield. Dave et al. [41] found parallel results for oil content and Chaudhari et al. [40] for seed yield in castor. The AMMI model separates the GEI effect into two interaction components known as multiplicative effect which was revealed using principal component analysis [42]. According to Yan et al. [43], strong GEI impacts diminish the gain for quantitative characteristics such as seed yield, but in the current study, its proportion was found to be medium to low which provides potential for seed yield improvement across a wide range of environments.
Table 3.
Analysis of variance based on AMMI model for seed yield per plant across three locations (Kharif-2020-21).
| Source | df | MS | Total Explained variation (%) | GEI Contributed (%) |
|---|---|---|---|---|
| Environment (E) | 2 | 8851.78** | 0.97 | – |
| Replication (E) | 3 | 190.77 | – | – |
| Genotype (G) | 89 | 17997.54** | 87.94 | – |
| GEI | 178 | 1135.11** | 11.09 | – |
| PC1 | 90 | 1815.66** | – | 80.88 |
| PC2 | 88 | 439.10* | – | 19.12 |
| Residuals | 267 | 321.16 | – | – |
| Total | 717 | 2942.69 | – | – |
*,** Significant at p < 0.05 and p < 0.01, respectively, df: degrees of freedom, MS: Mean sum of square, PC: Principal component, GEI: Genotype environment interaction.
3.3. AMMI biplot
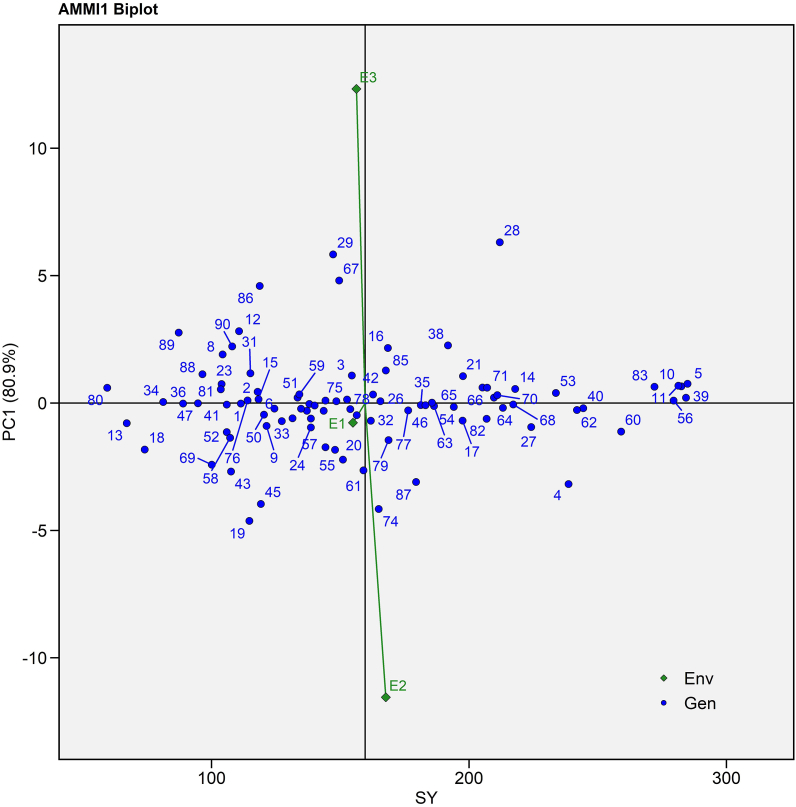
Different biplots were used to depict the potential of genotypes for seed yield, correlation with the environment under investigation and genotype stability. The AMMI I biplot was created by presenting mean seed yield across environment on the X-axis, known as main effects, and IPCA I score on the Y-axis, which represents multiplicative effects (Fig. 2) [42].
Fig. 2.
AMMI I biplot (seed yield per plant vs IPCA I) of 90 castor genotypes (Blue text) and three environments (Green text) for seed yield evaluated during Kharif 2020-21. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3 showed a significant GEI with 11.09% variance, which was further subdivided into two interactive principal components that explained 80.88 and 19.12% interactive variation, respectively. As a result, the IPCA I axis accounted the majority of the variance (Fig. 2). E1 placed to near the origin with the shortest vector suggested the least changeable environment, whilst E2 and E3 had longer vector from the origin indicated highly interactive environment. In the biplot, E2 placed on the right side of the mean seed yield line indicated the most favourable environment for high seed yield while E1 and E3 were in left side indicated poor environments for seed yield (Fig. 2). Aina et al. [44] validated the current findings by revealing 87.97% deviation for IPCA I axis in cassava. P3 141 (G39), JI 380 (G56), ANDCI 12-01(G10), and P3163(G40) had near-zero IPCA I value with high mean; therefore, they were positioned on the right side of the overall mean line (Fig. 2). They were also portrayed the least interaction with the environment because they positioned on the IPCA I line in AMMI I biplot (Suppl. Table 2 and Fig. 2). According to Ebdon and Gauch [42], SH 08 (G78), MI 16 (G26), RB01 (G42), and JI 446 (G75) placed near the origin were widely accepted to all environments with near-average seed yield performance (Fig. 2).
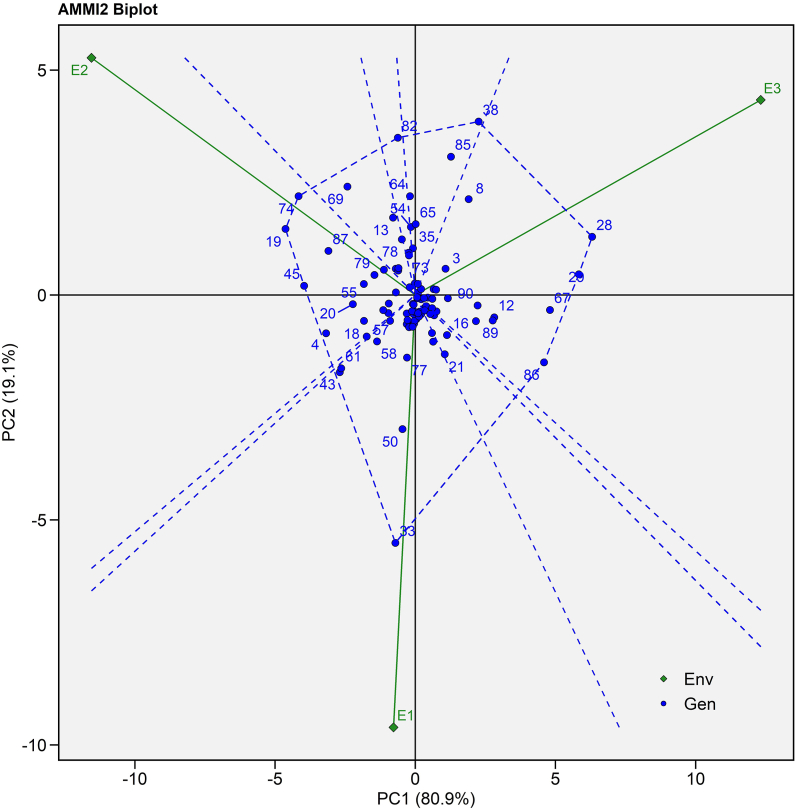
In the current study, IPCA II was also found to be significant in nature, which successfully explained the interaction of genotype with environment. To investigate the IPCA II, AMMI II biplot was created (Fig. 3). The dotted line connecting the vertex genotypes in this plot polygon view indicated maximum seed yield in a certain environment. Vertical projection from genotype to environment vector demonstrated the degree of interaction with the specific environment. The biplot showed E1 is highly interactive for DCS 103 (G33) which contribute mainly to GEI. Genotype MI 27 (G28), SI 14 (G20), SKI 316 (G86) exceptionally performed well in E3 while SI 09 (G19), RG 2561 (G45) and JI 444 (G74) performed well in E2 (Fig. 3).
Fig. 3.
AMMI II biplot (IPCA I vs IPCA II) of 90 castor genotypes (Blue text) and three environments (Green text) for seed yield evaluated during Kharif 2020-21. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
AMMI Stability Value (ASV) claimed that ANDCI 07-B (G1), MI 16 (G26), SKI 264 (G84), JI 434 (G68) and JI 35 (G47) had the lowest ASV value and ranked first to fifth in terms of environmental stability (Suppl. Table 4). However, according to the yield stability index, ANDCI 10-03 (G5), P3141 (G39), ANDCI 12-01 (G10), ANDCI 14 (G11) and JI 380 (G56) ranked first to fifth in the environment (Suppl. Table 4). ASV solely assessed genotypic stability, but YSI takes into account genotypic stability as well as genotype mean performance. These findings are in accordance with Singamsetti et al. [45]. They obtained a similar finding in maize hybrid during evaluating them for different moisture regimes.
3.4. GGE biplots
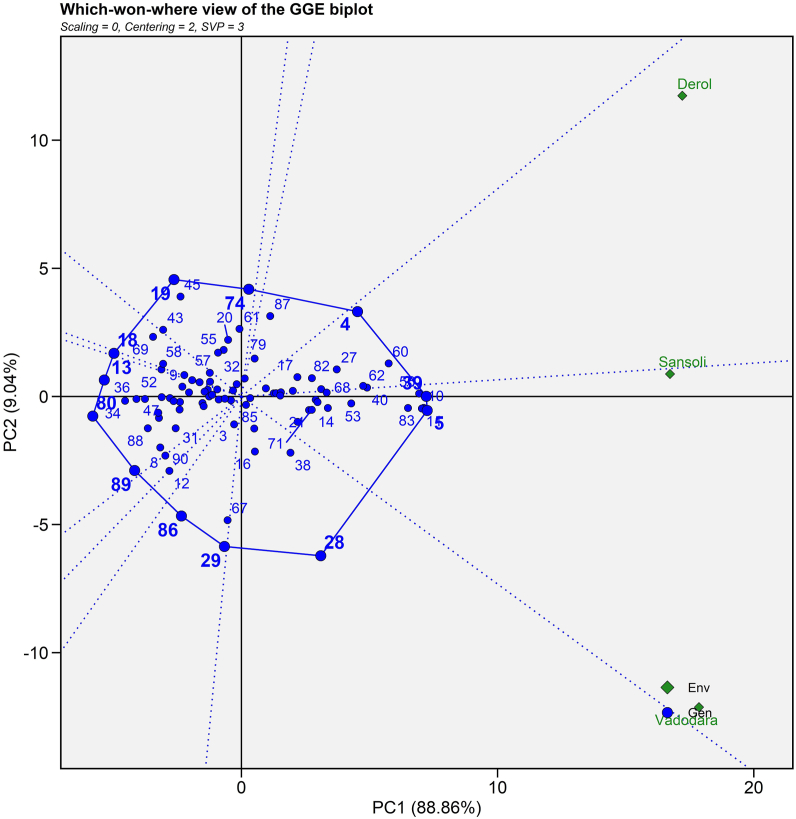
3.4.1. Which-won-where and what?
Which-won-where and what? pattern analysis assists in determining the best genotype for a particular environment. In this biplot, the vertex genotypes are combined to form a polygon (Fig. 4). The polygon is further subdivided into several separate sectors by rays (dotted lines) that originate from the plot's origin and proceed perpendicular to the polygon's sides. Gauch and Zobel [46] proposed that this division aids in genotype recommendation for a specific environment. The biplot depicted two mega environments. Sansoli (E1) and Vadodara (E2) are in a single mega environment in this biplot but Derol (E3) is in a distinct mega environment. This result was compared with Sakhare et al. [47] in castor by getting two seasons in a single section. Biplot revealed that ANDCI 10-01 (G4) is a high yielding vertex genotype for the E3 environment, whereas ANDCI 10-03 (G5) and P3141 (G39) are high yielding vertex genotypes for the E1 and E2 environments, respectively (Fig. 4). GP 640 (G13), SI 06 (G18), SI 09 (G19), MI 27 (G28), MI 33 (G29), SKI 003 (G80), SKI 316 (G86) and SKI 346 (G89) genotypes were also vertex genotypes, however no environment falls in their sector suggested that these genotypes were low yielding genotypes at some or all sites. Genotypes around the biplot's origin were best suited to low yielding conditions because they are stable in nature. These findings are consistent with those made by Nzuve et al. [48] for maize hybrids. They discovered three mega-environments through which won where biplot and selected the X16, X24 and X40 as the top performers for the mega-environments of Kabate, Kiboko and Kakamega, respectively. Similarly, Sakhare et al. [47] carried out GGE biplot analysis for castor hybrids and extracted Aruna, 48-1, AKC 1, DCH 177 and DCS 9 as a poor yielder for seed yield.
Fig. 4.
Which-won-where view of 45 genotypes of 90 castor genotypes (Blue text) and three environments (Green text) for seed yield evaluated during Kharif 2020-21. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
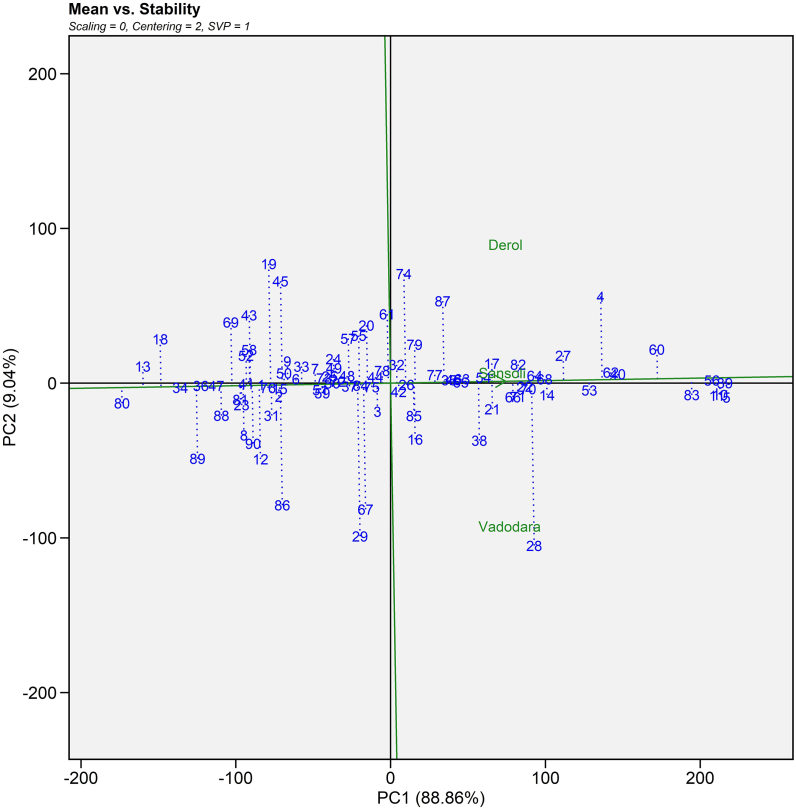
3.4.2. Mean seed yield vs stability
2-D visualization of seed yield vs stability constructed on PC I and PC II scores and was developed according to reports of Laurie and Booyse [49] and Samyuktha et al. [50]. This plot is ideal for genotype assessment because it was created using the row metric preservation approach (Supplemental Fig. S1). The genotypes' stability was shown using an average environment coordination (AEC) view with an arrow on the AEC line indicating the increasing mean direction. A short perpendicular line to the AEC axis denoted very stable genotypes. ANDCI 12-01 (G10), P3141 (G39), P3 163 (G40), JI 357 (G53), JI 380 (G56) and JI 418 (G62) genotypes were extremely stable and with good yielding ability. Sakhare et al. [47] used this biplot (mean seed yield versus stability plot) to decipher high seed yield generating hybrids GCH-7 and DCH 519 with good and poor stability, respectively.
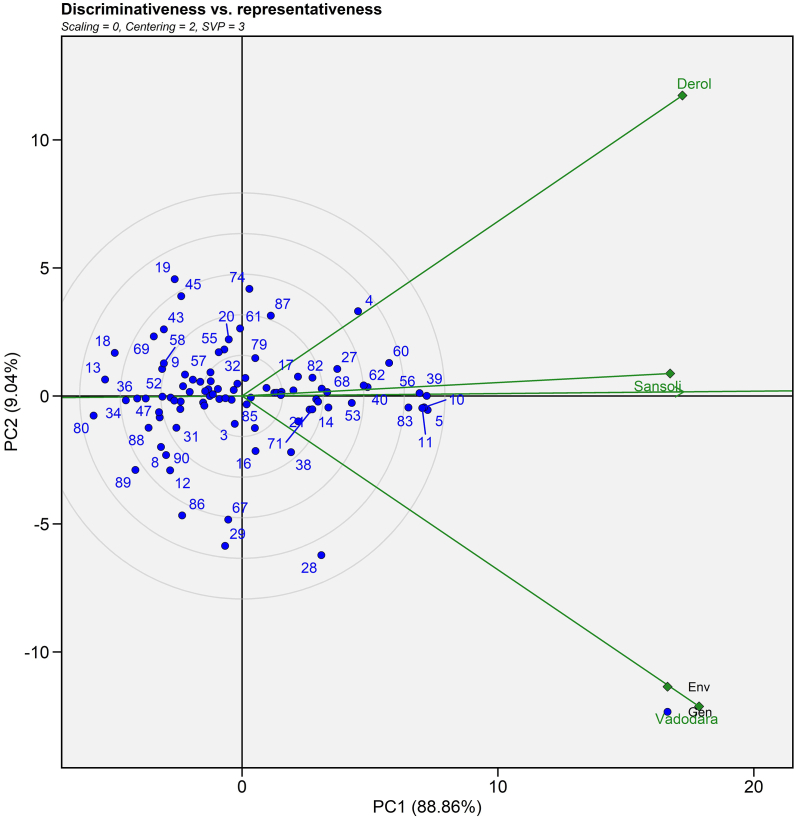
3.4.3. Discriminativeness vs representativeness
The relationship between two environments is explained by the cosine angle between environmental vectors [51]. A perfect 90° angle between two environments implies no association, an acute angle indicates positive correlation and an obtuse angle indicates negative correlation [52]. Supplemental Fig. 2 revealed a positive association among all settings. In contrast, the longest vector with E2 (Vadodara) and E3 (Derol) environments revealed the most discriminative environment. On the biplot, the length of the environment's vector is proportional to the standard deviation within the same environment and provides information about the environment's discriminating capacity [50]. To determine the representativeness of the environment, the average environment axis (AEA) must be used. The greater the angle between the AEA and the environmental vector, the lower the representativeness and vice versa. E1 (Sansoli) is a highly representative environment. This study demonstrated that generally adopted genotypes were chosen from the E1 (Sansoli) environment, whereas specially adapted genotypes were chosen from the E2 (Vadodara) and E3 (Derol) environments. The results for selecting an optimum environment for a general and specific genotype were consistent as per the results of Sserumaga et al. [53], Samyuktha et al. [50] and Sholihin [54].
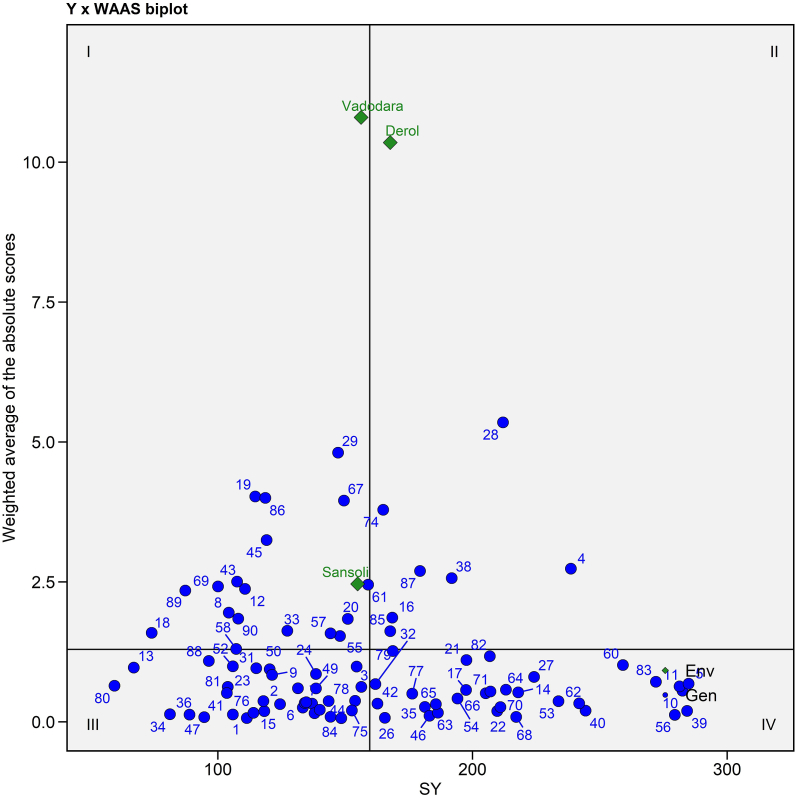
3.4.4. Y × WAAS biplot
Y × WAAS biplot built using seed yield on the X-axes and WAAS value on the Y-axes (Fig. 5 and Supplementary Table S3). This plot separates genotypes into four quadrants, allowing for simultaneous genotype selection with high seed yield and stability of included variables. According to the biplot (Figs. 5), 17 genotypes including E1 (Sansoli) and E3 (Vadodara) are in the first quadrant demonstrated greater instability of genotypes with low productivity and a strong discriminating ability, whereas 7 genotypes including E2 (Derol) are in the second quadrant featured with poor stability and higher seed yield which advised for paying extra attention to the environment in order to increase seed yield [55]. The third quadrant included 35 genotypes having more stability (due to the low WAAS value) but lesser productivity while the remaining 31 genotypes were found in the fourth quadrant had greater stability and fantastic performance for seed yield production.
Fig. 5.
Seed yield per plant × WAASB biplot based on combined interpretation of productivity (SY) and stability (WAASB) for 90 castor genotypes evaluated across three environments during Kharif 2020-21.
3.5. Multi-trait stability index (MTSI)
Traditional stability indicators based on first degree statistics were utilized by the majority of plant breeders. The assortment of a stable genotype based on mean, regression and deviation from regression parameters may not be sufficient to provide a simple interpretation of mean performance and trait stability. As a result, the MTSI technique has evolved into a sophisticated quantitative genetic tool for exploiting suitable variants in all crop species [26].
PH, SP, ESL, CAP, EB and SY were found significant for GEI (p < 0.05) in pooled ANOVA. MTSI included all GEI significant traits. The Weighted Average of Absolute Scores of Stability with Yield (WAASBY) values generated using a Pearson's correlation matrix and the retrieved high magnitude relationships were combined as a common factor. Exploratory factor analysis using six characters, resulted two PCs with cumulatively variation of 70.65% (Table 4). Communality, an indication of shared variance among traits, ranged from 0.133 (PH) to 0.936 (ESL) with an average value of 0.706 after varimax rotation. Six traits were grouped in the two factors by extracting WAASBY value from each character given in Table 5. PH, SP and ESL were grouped in the FA1 CAP, EB and SY were in FA2 (Table 5). The selection differential for mean performance as well as WAASBY index were positive. The range of mean performance was found between 1.84 (EB) to 67.54 (SY) while range of WAASBY index was found between 5.73 (SP) to 22.73 (SY) (Table 5). The selection performed in Fig. 6 was used as a basis to calculate mean of the selected genotypes (XS) which was higher than the mean of the original population for mean performance of particular trait. Based on the WAASB index, Abdelghany et al. [56] selected ZDD12828 and ZDD12832 genotype for protein content, WDD01583 and WDD03025 for oil content, ZDD23040 for palmitic acid, WDD00033 for stearic acid, ZDD23822 for oleic acid, ZDD11183 for linoleic acid, and ZDD08489 for linolenic acid. A similar approach adopted to evaluate the relative effects of abiotic stress in sunflower by Kaya et al. [57].
Table 4.
Eigen values, explained variance, factorial loadings after varimax rotation, communalities and uniqueness obtained in the factor analysis of the six GEI significant variables studied in 90 castor genotypes across three environments during Kharif 2020-21.
| Traits | FA1 | FA2 | Communality | Uniqueness |
|---|---|---|---|---|
| PH | −0.231 | 0.283 | 0.133 | 0.867 |
| SP | −0.956 | 0.009 | 0.913 | 0.087 |
| ESL | −0.965 | 0.068 | 0.936 | 0.064 |
| CAP | −0.861 | 0.237 | 0.798 | 0.202 |
| EB | 0.082 | 0.835 | 0.703 | 0.297 |
| SY | −0.179 | 0.850 | 0.755 | 0.245 |
| Eigen value | 2.824 | 1.415 | 0.706a | |
| Variance (%) | 47.065 | 23.590 | ||
| Accumulated (%) | 47.065 | 70.655 |
FA: Factor Analysis, PH: Plant height up to primary raceme, SP: Total length of primary raceme, ESL: Effective length of primary raceme, CAP: Capsules on main raceme, EB: Effective number of racemes per plant and SY: Seed yield per plant.
Average of the communality.
Table 5.
Selection differential for mean of the traits and WAASBY index for 6 traits of 90castor genotypes across three environments during Kharif-2020-21.
| Traits | Factor | Mean performance |
WAASBY |
||||
|---|---|---|---|---|---|---|---|
| Overall (X0) | Selected genotype (Xs) | SD (%) | Overall (X0) | Selected genotype (Xs) | SD (%) | ||
| SP | FA 1 | 66.57 | 70.24 | 3.67 | 56.053 | 61.783 | 5.730 |
| ESL | FA 1 | 60.73 | 66.83 | 6.09 | 58.444 | 65.714 | 7.270 |
| CAP | FA 1 | 72.24 | 93.40 | 21.16 | 51.888 | 65.055 | 13.167 |
| PH | FA 2 | 78.05 | 86.70 | 8.65 | 45.978 | 55.132 | 9.154 |
| EB | FA 2 | 5.76 | 7.61 | 1.84 | 48.162 | 61.346 | 13.184 |
| SY | FA 2 | 159.63 | 227.17 | 67.54 | 57.180 | 79.911 | 22.731 |
FA: Factor Analysis, SP: Total length of primary raceme, ESL: Effective length of primary raceme, CAP: Capsules on main raceme, PH: Plant height up to primary raceme, EB: Effective number of racemes per plant, SY: Seed yield per plant, SD: Selection differential and WAASBY: Weighted average of absolute scores of stability with yield.
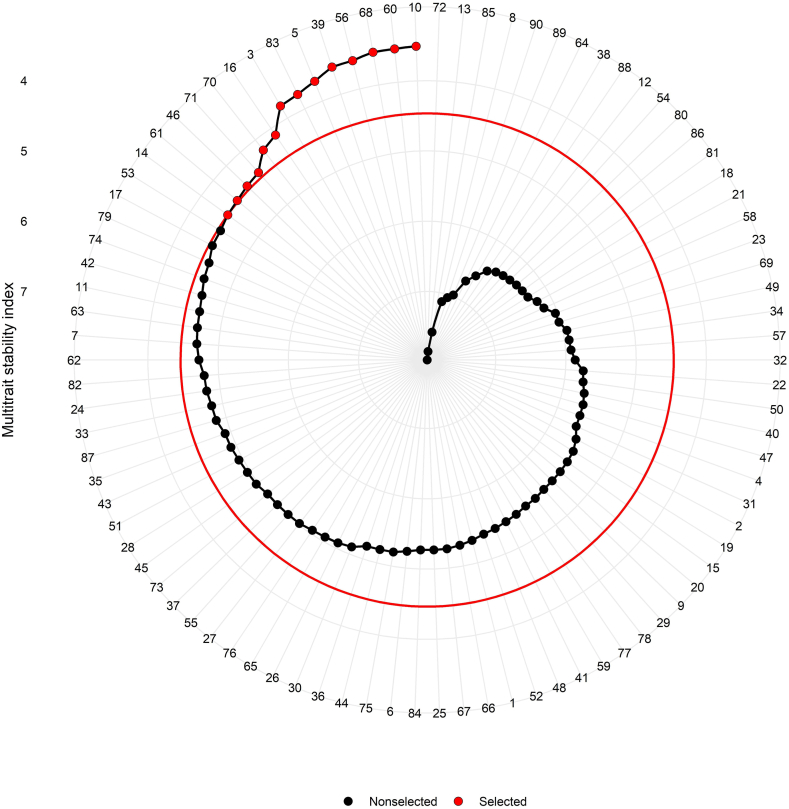
Fig. 6.
Genotype ranking and selected genotypes among 90 castor genotypes for multi trait stability index (MTSI) considering 15% selection intensity.
The most significant aspect of stability analysis is the selection of stable genotypes with higher mean performance for various characteristics [58]. It was carried out with the use of genotype-ideotype Euclidian distance-based scores. Exploratory factor analysis yielded scores for 90 genotypes as well as ideotype estimates in the first two factors (Supplementary Table S4).
MTSI help to select genotype having higher stability with a higher mean performance of all significant interacting variables. Likewise, Benakanahalli et al. [59] selected guar genotypes by the MTSI index presented desired values for 13 out of 15 productive and biochemical traits. Lower MTSI valued genotypes were selected with 15% selection intensity. According to Fig. 6, ANDCI 12-01 [G10 (MTSI = 3.508)], JI 413 [G60 (MTSI = 3.526)], JI 434 [G68 (MTSI = 3.529)], JI 380 [G56 (MTSI = 3.586)], P3141 [G39 (MTSI = 3.593)], ANDCI 10-03 [G5 (MTSI = 3.700)], SKI 215 [G83 (MTSI = 3.772)], ANDCI 09 [G3 (MTSI = 3.801)], SI 04 [G16 (MTSI = 4.114)], JI 437 [G70 (MTSI = 4.187)], JI 440 [G71 (MTSI = 4.387)], RG 3570 [G46 (MTSI = 4.413)], JI 417 [G61 (MTSI = 4.447)] and GAC 11 [G14 (MTSI = 4.466)] were selected with maximum stability and high mean performance of analyzed traits. Red circle in Fig. 6 indicated the cutoff point with MTSI value of 4.466 for GAC 11 (G14). Genotype JI 442 (G72) had a higher MTSI value (MTSI = 7.976) followed by GP 640 [G13 (MTSI = 7.854)], SKI 315 [G85 (MTSI = 7.574)] and ANDCI 10-08 [G8 (MTSI = 7.120)], these genotypes were recognized as an unstable genotype with the poor performance for traits under study. Similarly, Koundinya et al. [60] selected highly stable twenty-five cassava genotypes for increasing performance of leaf area index, yield per plant, harvest index, dry matter and starch yield per plant.
MTSI aids in the selection of genotypes with a high mean for multiple characteristics that are more stable. ANDCI 10-03 (G5), SKI 215 (G83), JI 380 (G56), P3141 (G39), JI 413 (G60) and ANDCI 12-01 (G10) were detected in the fourth quadrant of the Y × WAAS biplot, which were stable with low MTSI value indicating high seed yield generating genotypes. SKI 346 (G89), SKI 316 (G86), SKI 003 (G80), GP 640 (G13), SI 06 (G18), SI 09 (G19) and JI 444 (G74) were also vertex genotypes for Which won where and what biplot indicating unstable nature for seed yield. High MTSI values also validated this conclusion. While genotypes ANDCI 10-03 (G5), ANDCI 12-01 (G10) and P3141 (G39) contribute to mega environments, they also had lower MTSI values and were more competitive. These selected genotypes can contribute to a more stable genetic basis for future breeding programmes for wide tracts. Similar genotype selection was adopted by Abdelghany et al. [56] in soyabean. They selected 14 genotypes for their average performance for all traits with greater stability through MTSI with 10% selection intensity.
4. Conclusions
Seed yield is an important character as it is highly influenced by the altering environment. In the present study, selected material showed considerable variability over wide range of environment for different characters. Due to the course of evolution castor became stable over different environments so, most of the characters show continuous stability over environment. Only yield attributing traits are responsive to light period, irrigation, fertilizer and micro environment of particular locations. These attributes are unstable in nature. AMMI model helps to select stable and high yielding genotype which can be used for commercial cultivation as well as breeding purpose in wide range of environment. According to which won where and what biplot, ANDCI 10-01 (G4) as a vertex genotype for mega environment E3 while ANDCI 10-03 (G5) and P3141 (G39) recommended for the E1 and E2, respectively. Moreover, ANDCI 10-03 (G5) and P3141 (G39) were selected through MTSI for all interacting traits and they had high enough exploratory factor analysis score give the strong support to its ideotype. Ideotype of this selected genotype performed constant in fluctuating environment. Constant seed yielding ability of ANDCI 10-03 (G5) and P3141 (G39) was also effectively proven by Y × WAAS biplot. These were fallen in the fourth quadrant which indicated perfect stability and high seed yield across the environment regimes. So, selection through MTSI is as effective as other parameters. As per the MTSI, ANDCI 12-01 (G10), JI 413 (G60), JI 434 (G68), JI 380 (G56), P3141 (G39), ANDCI 10-03 (G5), SKI 215 (G83), ANDCI 09 (G3), SI 04 (G16), JI 437 (G70), JI 440 (G71), RG 3570 (G46), ANDCI 10-04 (G6) and GAC 11 (G14) were the best suited genotypes for increased performance for interacting traits under wide range of environment.
Author contribution statement
Juned Memon, Rumit Patel: Performed the experiments; Wrote the paper.
Sushil Kumar, Juned Memon, Dipak A. Patel, Bharat N. Patel: Conceived and designed the experiments.
Juned Memon, Dinesh J. Parmar, Rumit Patel: Analyzed and interpreted the data.
Néel A. Patel, Pankaj Katba: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
The data that has been used is confidential.
Declaration of interest's statement
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13515.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Supplemental Fig. S1.
Average environment coordination (AEC) view of the GGE-biplot based on environment-focused scaling, genotype focused singular value partitioning for the mean performance and stability of 90 castor genotypes (Blue text) and three environments (Green text) for seed yield evaluated during Kharif 2020-21.
Supplemental Fig. S2.
GGE biplot view of the discriminativeness and representativeness developed through environment focused centering and symmetrical method of singular value partitioning of 90 castor genotypes (Blue text) and three environments (Green text) for seed yield evaluated during Kharif 2020-21.
References
- 1.Rukhsar, Patel M.P., Parmar D.J., Kalola A.D., Kumar S. Morphological and molecular diversity patterns in castor germplasm accessions. Ind. Crop. Prod. 2017;97:316–323. [Google Scholar]
- 2.Xu W., Wu D., Yang T., Sun C., Wang Z., Han B., Wu S., Yu A., Chapman M.A., Maruguri S., Tan Q., Wang W., Bao Z., Liu A., Li D.Z. Genomic insights into the origin, domestication and genetic basis of agronomic traits of castor bean. Genome Biol. 2021;22(1):1–27. doi: 10.1186/s13059-021-02333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirza M.Y. NARC; Islamabad: 2009. Oil Seed Program. [Google Scholar]
- 4.Rukhsar, Patel M.P., Parmar D.J., Kumar S. Genetic variability, character association and genetic divergence studies in castor (Ricinus communis L.) Ann. Agrar. Sci. 2018;16(2):143–148. [Google Scholar]
- 5.Vieira C., Evangelista S., Cirillo R., Lippi A., Maggi C.A., Manzini S. Effect of ricinoleic acid in acute and subchronic experimental models of inflammation. Mediat. Inflamm. 2000;9(5):223–228. doi: 10.1080/09629350020025737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiti S., Hegde M.R., Chattopadhyay S.B. Oxford and IBH Publishing Co. (PVt). Ltd.; New Delhi: 1988. Handbook of Oil Seed Crops; p. 317. [Google Scholar]
- 7.Undata . 2018. Castor Oil Seed. http://data.un.org/Data.aspx?d¼FAO& f¼itemCode %3A265 #FAO. [Google Scholar]
- 8.Asungre P.A., Akromah R., Kena A.W., Gangashetty P. Genotype by environment interaction on grain yield stability and iron and zinc content in OPV of pearl millet in Ghana using the AMMI method. Int. J. Agron. 2021;2021 doi: 10.1155/2021/9656653. [DOI] [Google Scholar]
- 9.Baker R.J. 1988. Differential Response to Environmental Stress.https://agris.fao.org/agris-search/search.do?recordID=US8863668 [Google Scholar]
- 10.Joshi H.J., Maheta D.R., Jadon B.S. Phenotypic stability and adaptability of castor hybrids. Indian J. Agric. Res. 2002;36(4):269–273. http://arccarticles.s3.amazonaws.com/webArticle/articles/ijar2364008.pdf [Google Scholar]
- 11.Dixon A., Nukenine E. Statistical analysis of cassava yield trials with the additive main effects and multiplicative interaction (AMMI) model. Afr. J. Root Tuber Crops. 1997;3:46–50. https://hdl.handle.net/10568/103846 [Google Scholar]
- 12.Roemer J. Sinde die ertagdreichen Sorten ertagissicherer. Mitt DLG. 1917;32(1):87–89. [Google Scholar]
- 13.Wricke G. Zur berechning der okovalenz bei sommerweizen und hafer. Zeitschrift fur Pflanzenzuchtung. 1965;52:127–138. doi: 10.4236/ojbm.2021.96148. [DOI] [Google Scholar]
- 14.Shukla G.K. Some statistical aspects of partitioning genotype-environmental components of variability. Heredity. 1972;29(2):237–245. doi: 10.1038/hdy.1972.87. [DOI] [PubMed] [Google Scholar]
- 15.Freeman G.H. Statistical methods for the analysis of genotype-environment interactions. Heredity. 1973;31(3):339–354. doi: 10.1038/hdy.1973.90. [DOI] [PubMed] [Google Scholar]
- 16.Yates F., Cochran W.G. The analysis of groups of experiments. J. Agric. Sci. 1938;28:556–580. doi: 10.1017/S0021859600050978. [DOI] [Google Scholar]
- 17.Finlay K.W., Wilkinson G.N. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963;14:742–754. doi: 10.1071/AR9630742. [DOI] [Google Scholar]
- 18.Eberhart S.A., Russell W.A. Stability parameters for comparing varieties. Crop Sci. 1966;6(1):36–40. doi: 10.2135/cropsci1966.0011183X000600010011x. [DOI] [Google Scholar]
- 19.Annicchiaricom P. Additive main effects and multiplicative interaction (AMMI) analysis of genotype–location interaction in variety trials repeated over years. Theor. Appl. Genet. 1997;94(8):1072–1077. doi: 10.1007/s001220050517. [DOI] [Google Scholar]
- 20.Yan W., Hunt L.A., Sheng Q., Szlavnics Z. Cultivar evaluation and mega‐environment investigation based on the GGE biplot. Crop Sci. 2000;40(3):597–605. doi: 10.2135/cropsci2000.403597x. [DOI] [Google Scholar]
- 21.Miranda G.V., Souza L.V.D., Guimarães L.J.M., Namorato H., Oliveira L.R., Soares M.O. Multivariate analyses of genotype x environment interaction of popcorn. Pesqui. Agropecuária Bras. 2009;44:45–50. doi: 10.1590/S0100-204X2009000100007. [DOI] [Google Scholar]
- 22.Ding M., Tier B., Yan W.K. Australasian Forest Genetics Conference. 2007. Application of GGE biplot analysis to evaluate genotype (G), environment (E) and G×E interaction on P. radiata: case study; pp. 11–14. April 2007 (Hobart, TAS: The Old Woolstore) [Google Scholar]
- 23.Smith H.F. A discriminant function for plant selection. Ann. Eugen. 1936;7:240–250. doi: 10.1111/j.1469-1809.1936.tb02143.x. [DOI] [Google Scholar]
- 24.Hazel L.N. The genetic basis for constructing selection indexes. Genetics. 1943;28:476–490. doi: 10.1093/genetics/28.6.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocha, J.R.D.A.S.D.C. Machado J.C., Carneiro P.C.S. Multi-trait index based on factor analysis and ideotype-design: proposal and application on elephant grass breeding for bioenergy. GCB Bio-Energy. 2018;10:52–60. doi: 10.1111/gcbb.12443. [DOI] [Google Scholar]
- 26.Olivoto T., Lúcio A.D., da Silva J.A., Sari B.G., Diel M.I. Mean performance and stability in multienvironment trials II: selection based on multiple traits. J. Agron. 2019;111(6):2961–2969. doi: 10.2134/agronj2019.03.0221. [DOI] [Google Scholar]
- 27.Sapovadiya M.H., Dobariya K.L., Babariya C.A., Mungra K.S., Vavdiya P.A. Heterosis for seed yield and its components over environments in castor (Ricinus communis L.) Electron. J. Plant Breed. 2015;6(4):1118–1123. [Google Scholar]
- 28.Akhila S.R., Kumar S., Sakure A.A., Patel D.A., Patel M.P. Integration of morpho-physico-biochemical traits with SSR and SRAP markers for characterization of castor genotypes of Indian origin. Oil Crop Sci. 2022;7(1):22–30. [Google Scholar]
- 29.Peterson D.D. McGraw-Hill Book Company, Inc; New York and London: 1939. Statistical Techniques in Agricultural Research: A Simple Exposition of Practice and Procedure in Biometry. [Google Scholar]
- 30.Bradu D., Gabriel K.R. The biplot as a diagnostic tool for models of two-way tables. Technometrics. 1978;20(1):47–68. doi: 10.1080/00401706.1978.10489617. [DOI] [Google Scholar]
- 31.Gauch H.G., Jr. Biometrics; 1988. Model Selection and Validation for Yield Trials with Interaction; pp. 705–715. [DOI] [Google Scholar]
- 32.Gollob H.F. A statistical model which combines features of factor analytic and analysis of variance techniques. Psychometrika. 1968;33(1):73–115. doi: 10.1007/BF02289676. [DOI] [PubMed] [Google Scholar]
- 33.Vargas M., Crossa J. International Maize and Wheat Improvement Center, Edo Mex; Mexico: 2000. The AMMI Analysis and Graphing the Biplot. Biometrics & Statistics Unit, Genetic Resources Program. [Google Scholar]
- 34.Farshadfar E. Incorporation of AMMI stability value and grain yield in a single non-parametric index (GSI) in bread wheat. Pakistan J. Biol. Sci. 2008;11(14):1791. doi: 10.3923/pjbs.2008.1791.1796. [DOI] [PubMed] [Google Scholar]
- 35.Atta B.M., Shah T.M., Abbas G., Haq M.A. Genotype x environment interaction for seed yield in kabuli chickpea (Cicer arietinum L.) genotypes developed through mutation breeding. Pakistan J. Bot. 2009;41(4):1883–1890. [Google Scholar]
- 36.Zobel R.W., Wright M.J., Gauch H.G., Jr. Statistical analysis of a yield trial. J. Agron. 1988;80(3):388–393. doi: 10.2134/agronj1988.00021962008000030002x. [DOI] [Google Scholar]
- 37.Patel C.M., Patel J.M., Patel C.J. Gene-Environment interaction and stability analysis for yield and yield determinant traits in Castor (Ricinus Communis L) J. Agr. Veter. Sci. 2015;8:68–72. [Google Scholar]
- 38.Patel V.R., Dumancas G.G., Vishwanath L.C., Maples R., Subong B.J. Castor oil: properties, uses and optimization of processing parameters in commercial production. Lipid Insights. 2016;9 doi: 10.4137/LPI.S40233. LPI–S40233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Movaliya H.M., Chovatia V.P., Madariya R.B., Mungala R.A., Pipaliya H.R., Bhuva S.K. Study of variability and correlation for seed yield and its attributes in castor (Ricinus communis L.) J. Pharmacogn. Phytochem. 2018;7(2):1474–1477. [Google Scholar]
- 40.Chaudhari B.A., Patel M.P., Soni N.V., Patel A.M., Makwana R.R., Patel A.B. Genotype× Environment interactions and stability analysis for seed yield and yield attributing characters in Castor (Ricinus communis L.) Int. J. Curr. Microb. Appl. Sci. 2019;8(5):2475–2481. [Google Scholar]
- 41.Dave P.B., Patel B.N., Parmar D.J., Patel N.A. Interpretation of genotype× environment effect on oil content in castor. IJTA (Int. J. Trop. Agric.) 2017;35(3):517–523. https://www.cabdirect.org/cabdirect/abstract/20183105631 [Google Scholar]
- 42.Ebdon J.S., Gauch H.G., Jr. Additive main effect and multiplicative interaction analysis of national turfgrass performance trials: I. Interpretation of genotype × environment interaction. Crop Sci. 2002;42(2):489–496. doi: 10.2135/cropsci2002.4890. [DOI] [Google Scholar]
- 43.Yan W., Cornelius P.L., Crossa J., Hunt L.A. Two types of GGE biplots for analyzing multienvironment trial data. Crop Sci. 2001;41(3):656–663. doi: 10.2135/cropsci2001.413656x. [DOI] [Google Scholar]
- 44.Aina O.O., Dixon A.G., Akinrinde E.A. Effect of soil moisture stress on growth and yield of cassava in Nigeria. Pakistan J. Biol. Sci.: PJBS. 2007;10(18):3085–3090. doi: 10.3923/pjbs.2007.3085.3090. [DOI] [PubMed] [Google Scholar]
- 45.Singamsetti A., Shahi J.P., Zaidi P.H., Seetharam K., Vinayan M.T., Kumar M., Singla S., Shikha K., Madankar K. Genotype× environment interaction and selection of maize (Zea mays L.) hybrids across moisture regimes. Field Crop. Res. 2021;270 [Google Scholar]
- 46.Gauch H.G., Jr., Zobel R.W. Identifying mega‐environments and targeting genotypes. Crop Sci. 1997;37(2):311–326. doi: 10.2135/cropsci1997.0011183X003700020002x. [DOI] [Google Scholar]
- 47.Sakhare S.B., Nagdeve S.S.M., Deshmukh D.T. GGE Bi-plot analysis in castor (Riccinus communis L.) for vidarbha region of Maharashtra state. Electron. J. Plant Breed. 2018;9(2):768–772. [Google Scholar]
- 48.Nzuve F., Githiri S., Mukunya D.M., Gethi J. Analysis of genotype x environment interaction for grain yield in maize hybrids. J. Agril. Sci. 2013;5(11):75–85. [Google Scholar]
- 49.Laurie S.M., Booyse M. Employing the GGE SREG model plus Elston index values for multiple trait selection in sweet potato. Euphytica. 2015;204(2):433–442. [Google Scholar]
- 50.Samyuktha S.M., Malarvizhi D., Karthikeyan A., Dhasarathan M., Hemavathy A.T., Vanniarajan C., Sheela V., Hepziba S.J., Pandiyan M., Senthil N. Delineation of genotype× environment interaction for identification of stable genotypes to grain yield in mungbean. Front. Agron. 2020;2:17. doi: 10.3389/fagro.2020.577911. [DOI] [Google Scholar]
- 51.Yan W. Singular-value partitioning in biplot analysis of multi environment trial data. Agron. J. 2002;94:990–996. doi: 10.1016/j.jssas.2016.10.001. [DOI] [Google Scholar]
- 52.Yousaf M.I., Akhtar N., Mumtaz A., Shehzad A., Arshad M., Shoaib M., Mehboob A. Yield stability studies in indigenous and exotic maize hybrids under genotype by environment interaction. Pakistan J. Bot. 2021;53(3):941–948. [Google Scholar]
- 53.Sserumaga J.P., Oikeh S.O., Mugo S., Asea G., Otim M., Beyene Y., Kikafunda J. Genotype by environment interactions and agronomic performance of doubled haploids testcross maize (Zea mays L.) hybrids. Euphytica. 2016;207(2):353–365. doi: 10.1007/s10681-015-1549-2. [erratum: 214:204] [DOI] [Google Scholar]
- 54.Sholihin . Vol. 2331. AIP Publishing LLC; 2021. GGE and AMMI biplot for interpreting interaction of genotype X environments of cassava promising genotypes. (AIP Conference Proceedings). No. 1. [Google Scholar]
- 55.Huang X., Jang S., Kim B., Piao Z., Redona E., Koh H.J. Evaluating Genotype× environment interactions of yield traits and adaptability in rice cultivars grown under temperate, subtropical and tropical environments. Agriculture. 2021;11(6):558. doi: 10.3390/agriculture11060558. [DOI] [Google Scholar]
- 56.Abdelghany A.M., Zhang S., Azam M., Shaibu A.S., Feng Y., Qi J., Li J., Li Y., Tian Y., Hong H., Lamlom S.F., Li B., Sun J. Exploring the phenotypic stability of soybean seed compositions using multi-trait stability index approach. Agronomy. 2021;11(11):2200. doi: 10.3390/agronomy11112200. [DOI] [Google Scholar]
- 57.Kaya M.D., Okcub G., Ataka M., Cikilic Y., Kolsaricia O. Seed treatments to overcome salt and drought stress during germination in sunflower (Helianthus annuus L.) Eur. J. Agron. 2006;24:291–295. doi: 10.1016/j.eja.2005.08.001. [DOI] [Google Scholar]
- 58.Yue H., Jiang X., Wei J., Xie J., Chen S., Peng H., Bu J. A study on genotype× environment interactions for the multiple traits of maize hybrids in China. Agron. J. 2021;113(6):4889–4899. doi: 10.1002/agj2.20907. [DOI] [Google Scholar]
- 59.Benakanahalli N.K., Sridhara S., Ramesh N., Olivoto T., Sreekantappa G., Tamam N., Abdelmohsen S.A. A framework for identification of stable genotypes Basedon MTSI and MGDII indexes: an example in guar (Cymopsis tetragonoloba L.) Agronomy. 2021;11(6):1221. doi: 10.3390/agronomy11061221. [DOI] [Google Scholar]
- 60.Koundinya A.V.V., Ajeesh B.R., Hegde V., Sheela M.N., Mohan C., Asha K.I. Genetic parameters, stability and selection of cassava genotypes between rainy and water stress conditions using AMMI, WAAS, BLUP and MTSI. Sci. Hortic. 2021;281 doi: 10.1016/j.scienta.2021.109949. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.