Abstract
Purpose:
Preclinical data suggest that antiprogestins inhibit the growth of luminal breast carcinomas that express higher levels of progesterone receptor isoform A (PRA) than isoform B (PRB). Thus, we designed a presurgical window of opportunity trial to determine the therapeutic effects of mifepristone in patients with breast cancer, based on their high PRA/PRB isoform ratio (MIPRA; NCT02651844).
Patients and Methods:
Twenty patients with luminal breast carcinomas with PRA/PRB > 1.5 (determined by Western blots), and PR ≥ 50%, naïve from previous treatment, were included for mifepristone treatment (200 mg/day orally; 14 days). Core needle biopsies and surgical samples were formalin fixed for IHC studies, while others were snap-frozen to perform RNA sequencing (RNA-seq), proteomics, and/or Western blot studies. Plasma mifepristone levels were determined using mass spectrometry. The primary endpoint was the comparison of Ki67 expression pretreatment and posttreatment.
Results:
A 49.62% decrease in Ki67 staining was observed in all surgical specimens compared with baseline (P = 0.0003). Using the prespecified response parameter (30% relative reduction), we identified 14 of 20 responders. Mifepristone induced an increase in tumor-infiltrating lymphocytes; a decrease in hormone receptor and pSer118ER expression; and an increase in calregulin, p21, p15, and activated caspase 3 expression. RNA-seq and proteomic studies identified downregulated pathways related to cell proliferation and upregulated pathways related to immune bioprocesses and extracellular matrix remodeling.
Conclusions:
Our results support the use of mifepristone in patients with luminal breast cancer with high PRA/PRB ratios. The combined effects of mifepristone and estrogen receptor modulators warrant clinical evaluation to improve endocrine treatment responsiveness in these patients.
Translational Relevance.
We propose that progesterone receptor (PR) ligands may serve as therapeutic tools for luminal breast cancer. Our preclinical data suggest that antiprogestins inhibit tumor growth and metastasis in breast cancer models with higher levels of PR isoform A (PRA) than isoform B (PRA-H tumors), while they stimulate the metastatic burden in those with the opposite ratio. The MIPRA study showed that mifepristone neoadjuvant treatment benefits patients with PRA-H tumors and underscores the relevance of testing the PR isoform ratio before administering antiprogestins to patients with breast cancer. Proteomics coupled with RNA sequencing profiling revealed mifepristone-modulated biological processes that explain and strengthen the Ki67 data. The fact that lymph node metastases retain the PRA/PRB ratio as the primary tumor posits this subgroup of patients as recipients of antiprogestin treatment, even in adjuvant settings. Our findings suggest that mifepristone may be included in the armamentarium against breast cancer in the future.
Introduction
A total of 70% of breast carcinomas express estrogen receptor alpha (ER) and progesterone receptors (PR) and are currently treated with therapies that target ER (reviewed in refs. 1, 2). Although there is clear evidence that PR participates in tumor growth (3–5), to date, PR has only been evaluated as a prognostic factor.
Antiprogestins, such as mifepristone (RU486; refs. 6–8), onapristone (9), and lonaprisan (10), have been used in the past to treat patients who failed other treatments, and positive responses were obtained in selected patients treated with mifepristone [9% overall response rate (ORR; ref. 6); 11% ORR, 95% confidence interval (CI): 2–28 (7)] or onapristone [ORR: 56% partial response (9); reviewed in refs. 3, 11]. A recent window of opportunity trial (WOT) using antiprogestin telapristone acetate (TLP) in patients naïve to other treatments also showed a mild response (12), suggesting the necessity of identifying PR+ patients who will benefit from antiprogestin treatment.
PR mainly comprises two isoforms encoded by the same gene: isoform A (PRA) and isoform B (PRB). There is consensus that PRA is the prevailing isoform in human breast carcinomas (reviewed in ref. 3). Because commercial PR antibodies recognize both isoforms (13), the relative abundance of one isoform over the other is usually unknown (reviewed in ref. 3). Preclinical studies (3, 14, 15) and human breast cancer samples cultured ex vivo (16) showed that tumors with a high PRA/PRB ratio (PRA-H) are antiprogestin-responsive tumors and that those with the opposite ratio (PRB-H) may show mild responses, no response, or may even be stimulated to proliferate or metastasize under this treatment (15).
Initially, mifepristone was developed as an antiglucocorticoid, and its antiprogestin effects were subsequently discovered (17). Currently, it is used for the early termination of pregnancy and it exerts beneficial effects in the treatment of uterine fibroids, endometriosis, meningiomas, and Cushing disease (reviewed in ref. 18).
In this scenario, the Mifepristone for Breast Cancer Patients with Higher Levels of Progesterone Receptor Isoform A than Isoform B (MIPRA) clinical trial was designed as a WOT, being the first to select patients based on their prevailing PR isoforms, including only patients with PRA-H naïve breast cancer, to be treated with mifepristone for 14 days. Mifepristone was selected among other antiprogestins because it is freely available and its side effects are well documented. As our hypothesis suggests that PRB-H patients might be stimulated by mifepristone, only PRA-H patients were included for ethical reasons. We show herein that mifepristone treatment decreased Ki67 expression in 70% of PRA-H patients, providing a powerful personalized therapy for selected patients with (ER+, PR+) breast cancer.
Patients and Methods
Study type
This study was an open-label, interventional, prospective, single-arm clinical trial (ClinicalTrials.gov identifier: NCT02651844).
Patient population
The trial included patients who spontaneously attended the Magdalena V de Martinez Hospital at General Pacheco, Buenos Aires (Hospital PMVM). Postmenopausal women with untreated breast cancer whose curative treatment plan included surgical resection were screened. If they met the inclusion criteria, they were derived for possible accrual. Eligibility criteria included: (i) postmenopausal status more than 1 year after the last menses, (ii) tumors larger than 15 mm, (iii) PRA/PRB ratios higher than 1.5 determined by Western blot (WB), (iv) PR total levels ≥ 50% evaluated by IHC, (v) World Health Organization condition of 1 with the adequate function of organs and systems: absolute neutrophil count 1,500/mL; platelets ≥100,000/mL; hemoglobin ≥10 g/dL; CD4 count ≥400; creatinine <1.5 mg/dL; total bilirubin, aspartate aminotransferase, and alanine aminotransferase <1.5 U/L × upper limit of institutional normal. Patients were excluded if they (i) received any other treatment for cancer, (ii) had hepatitis or human immunodeficiency virus infection, (iii) had cognitive alterations that limited their understanding of the protocol, (iv) experienced a prolonged QT/QTc basal interval, or (v) had asthma or other autoimmune diseases. The representativeness of the 20 study participants is presented in Supplementary Table S1.
Study endpoints
The primary endpoint was the comparison of the Ki67 index before and after treatment. We prespecified that a 30% of relative reduction in Ki67 would be considered a positive result. The secondary outcomes included the evaluation of apoptotic and PR-regulated proteins, changes in cellular pathways (transcriptomics/proteomics), plasma mifepristone levels, tumor size, and ultrasound measurements, although the latter were not expected to change. Only changes greater than 20% would be considered.
Study design
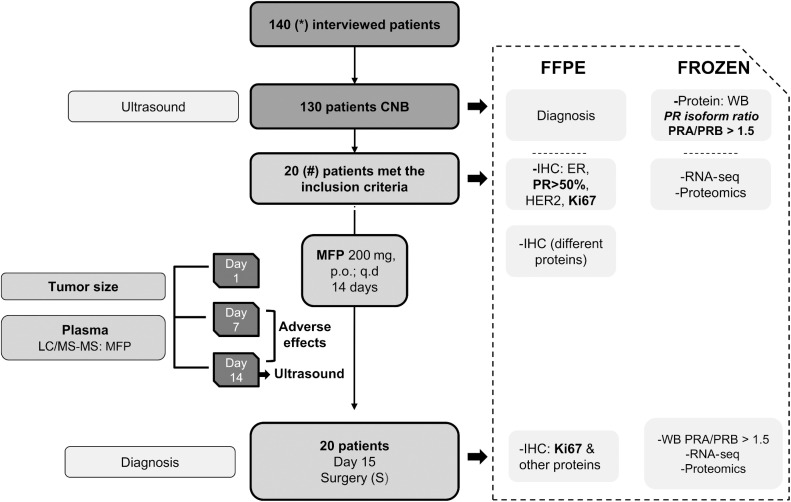
The protocol design is illustrated in Fig. 1. Ultrasound-guided core needle biopsies (CNB) were performed using 14 G, 0.8 Promag Short throw needles; two samples were immediately snap-frozen, and the other two were formalin fixed and paraffin embedded. Plasma was collected on days 0, 7, and 14 and stored at −80°C for further studies. During surgery, normal tissue and tumor samples were collected and processed as described for CNB. We set a priori that a sample size of 20 patients would be necessary to test our hypothesis with a power of 87%, a type I error of 0.05, and a type II error of 0.1.
Figure 1.
CONSORT diagram. MIPRA trial was a single-arm, open-label trial designed to explore the biological and clinical activity of mifepristone (MFP) treatment for patients with breast cancer with higher levels of PRA than PRB. A summary of the trial workflow is shown. *From the 140 patients interviewed, 1 patient did not sign the informed consent, 4 declined after signing it, 5 patients could not be biopsied for different reasons, 19 were considered non-neoplastic, 2 had metastatic disease and chemo-neoadjuvant treatment was recommended. # 21 tumors (1 patient had bilateral breast cancer). FFPE, formalin-fixed paraffin-embedded; IHC, immunohistochemistry; ER, estrogen receptors alpha; WB, Western blot; p.o., oral administration; q.d, once a day; CNB, core needle biopsy.
Frozen samples
Frozen CNB were immediately transported on dry ice to the Instituto de Biologia y Medicina Experimental. Frozen samples were pulverized and separated using TRIzol reagent (Life Technologies) for RNA extraction or using NE-PER Nuc and Cyt extraction reagents (Pierce; Thermo Fisher Scientific) for protein extraction [WB and mass spectrometry (MS) studies]. The surgically frozen samples were processed similarly.
Formalin-fixed paraffin-embedded samples used for diagnosis
After confirming the diagnosis of breast cancer and the PRA/PRB ratio, blank slides of CNB samples were sent to a private certified pathology laboratory [GAH, San Isidro Laboratory], to determine ER, PR, HER2, and Ki67 expression using American Society of Clinical Oncology recommended guidelines (19–21). Antibodies against ER (NCL-ER-6F11/2, RRID:AB_876939), PR (RTU-PGR-312, RRID:AB_563966), and Ki67 (RTU-Ki67-MM1, RRID:AB_563840) were purchased from Leica Biosystems, and HER2 expression was determined using the Benchmark XT Ventana Pathway (790-2991, RRID:AB_2335975) following the approved guidelines (20). p53 staining was evaluated in surgical samples using the P53 (NCL-L-p53-DO7, RRID:AB_563936) Novocastra antibody. The same methods and controls were used to stain Ki67 in the CNB and surgical samples.
Mifepristone administration
Mifepristone tablets (200 mg) were purchased from PharmaWeb. Each patient received one pill of 200 mg/day for 14 days by trained volunteers from La Liga Argentina de Lucha contra el Cáncer (LALCEC, Tigre) on the patient's home-signing forms to corroborate compliance. Surgery was performed on day 15.
Clinical control and adverse events
Laboratory analyses were performed before and 7 or 14 days after treatment initiation. At the same time, three plasma tubes from each patient were frozen. At each visit, the patients were examined to detect possible adverse effects that were graded according to the NCI Common Toxicity Criteria for Adverse Events (v 4.0). To determine changes in tumor size, the initial ultrasound measurement obtained on the day of CNB was considered as the initial size. The second ultrasound measurement was performed the day before surgery.
IHC
Assays were performed as described previously (16). Because limited slides from CNBs were available, only the selected pairs were evaluated. Antibodies against HER-2/ErbB2 (2165, RRID:AB_10692490), PR (8757, RRID:AB_2797144), and pser167ER (64508, RRID:AB_2799660) were purchased from Cell Signaling Technology; against ER (ab108398, RRID:AB_10863604), pser118ER (ab32396, RRID:AB_732252), active caspase 3 (ab2302, RRID:AB_302962), and p15 (ab53034, RRID:AB_2078578) from Abcam; against calregulin (sc-373863, RRID:AB_10915425) and MUC-1 (sc-7313, RRID:AB_626983) from Santa Cruz Biotechnology, and against p21 (556430, RRID:AB_396414) from BD Biosciences Pharmingen.
Ki67 quantitation
Ki67 changes were quantified independently by two pathologists following the accepted guidelines (21). Pathologists were blinded to the patients’ data. All cancer cells were counted in CNB and at least 10 40× fields/slide in the surgical samples. Software counting was performed using the AT2 Aperio-Scanscope and quantification was performed using the open-source software platform for whole-slide image analysis, QuPath (Alfredo Molinolo, Moores Cancer Center, San Diego; ref. 22). In the few cases in which there were discrepancies that compromised the 30% cut-off limit owing to the use of different technologies, both pathologists revisited the case together and reached a consensus value.
WB studies
Cytosol (Cyt) and nuclear (Nuc) protein extracts and WB were performed as described previously (16).
Morphologic evaluation of CNBs and matched surgical samples
Hematoxylin and eosin (H&E)-stained tumor sections from CNB and surgical samples were evaluated by a staff hospital pathologist and a second pathologist (S.I. Vanzulli), blinded to the Ki67 data. The details are included in the Supplementary Materials and Methods section.
Mifepristone plasma levels
Mifepristone was measured in 200 μL of plasma from each patient using the supported liquid extraction method and LC/MS-MS (23). The LC system was a Water I class, Mobile phase A of 50 mmol/L Amm Fluroide and Mobile phase B of 50 mmol/L AmF in MeOH: Column Kinetex C18 (150×2.1 mm; 2.6 μmol/L): Mass Spect: Qtrap 6500+. The study was performed at the mass spectrometry core of the Edinburgh Clinical Research Facility, University of Edinburgh (Edinburgh, Scotland).
RNA sequencing analysis
RNA extracted (EtOH precipitated; total RNA > 0.01 μg) from CNB and surgical samples from 13 patients was sent to Macrogen, Inc to be processed. Only paired samples from 8 patients passed the initial quality control. Library construction was performed using SMART-Seq v4 Ultra Low Input RNA for sequencing (data output, 40 M reads; Macrogen). The details are provided in the Supplementary Materials and Methods section.
Proteomics
Paired Nuc and Cyt extracts from ten patients were used for proteomic analysis. Only extracts with 50–100 μg, and concentrations greater than 3 μg/μL were selected. Extracts were solubilized in SDS 5% and 50 mmol/L triethyl ammonium bicarbonate pH 7.5 and 100 μg of protein was digested using the Protifi S-Trap Micro Spin Column Digestion Protocol, as described previously (24). LC/MS-MS was performed by coupling an UltiMate 3000 LC system with a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). The details are provided in the Supplementary Materials and Methods section.
Statistical analysis
The Wilcoxon matched-pair signed-rank test was used to compare pretreatment and posttreatment changes (n = 20). To compare laboratory measurements before and after 7 or 14 days of treatment, ANOVA for matched values and the Friedman test were used. The statistics involved in the proteomic and RNA sequencing (RNA-seq) analyses are explained in the respective sections.
Statement of ethics
Written informed consent was obtained from all patients. This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Boards of the hospital and IBYME (2012-026).
Data availability
The RNA-seq data generated in this study are publicly available in the Gene Expression Omnibus at GSE212690 and proteomic data at ProteomeXchange at PXD036515.
Results
The study was conducted from April 2016 to October 2019. A summary of the trial workflow is shown in Fig. 1. The clinical features of the patients are presented in Table 1. The median (range) time elapsed from CNB to surgery was 49 (25–119) days. The individual time points for each patient are listed in Supplementary Table S2. In all the cases, treatment was initiated once the surgical day was established.
Table 1.
Clinicopathologic features of ER+PR+ mifepristone-treated patients.
| Clinicopathologic parameters | n/total | % | |
|---|---|---|---|
| Age (years; median, range) | 66 (54–84) | 20 | |
| Tumor size (T)a | |||
| T1 | 7/21b | 33.3 | |
| T2 | 11/21 | 52.4 | |
| T3 | 3/21 | 14.3 | |
| Lymph node status (N) | |||
| N0 | 8/21 | 38.1 | |
| N1 | 6/21 | 28.6 | |
| N2 | 4/21 | 19.0 | |
| N3 | 3/21 | 14.3 | |
| Tumor stage | |||
| I | 5/21 | 23.8 | |
| II | 13/21 | 61.9 | |
| III | 3/21 | 14.3 | |
| Histologic type | |||
| IC-NST | 14/21 | 66.7 | |
| ILC | 4/21 | 19.0 | |
| PAP | 2/21 | 9.5 | |
| MUC | 1/21 | 4.8 | |
| Histologic gradea | |||
| 1 | 0/21 | 0 | |
| 2 | 3/21 | 14.3 | |
| 3 | 18/21 | 85.7 | |
| HER2a | |||
| Positive | 1/21 | 4.8 | |
| Negative | 20/21 | 95.2 | |
| ER (%; median, range)a | |||
| 100 (70–100) | 21/21 | 100 | |
| PR (%; median, range)a | |||
| 90 (50–100) | 21/21 | 100 | |
Abbreviations: ER, estrogen receptors alpha; IC-NST, invasive carcinoma of no special type; ILC, invasive lobular carcinoma; MUC, mucinous carcinoma; PAP, papillary carcinoma; PR, progesterone receptors.
aPretreatment.
b20 patients and 21 tumors.
Changes in Ki67 expression, tumor size, and clinical features pretreatment and posttreatment
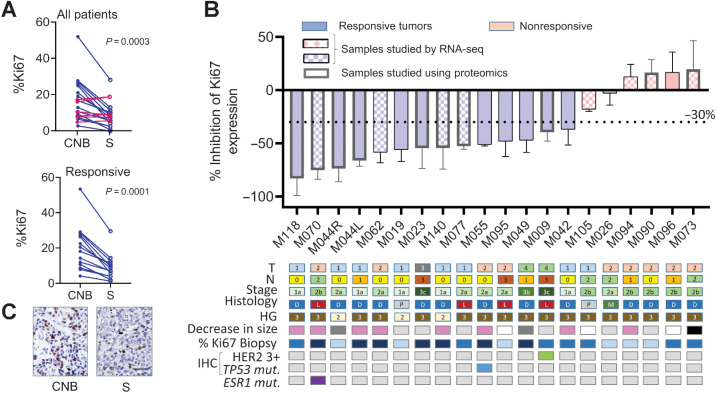
Using the prespecified response parameter (30% relative reduction in Ki67), we identified 14 of 20 (70%) responders (Fig. 2A). The median (range) Ki67 value of CNB was 15.1% (4.1–53.4) and for the surgical sample 8.6% (1.2–29.5). The individual values are listed in Supplementary Table S2. A decrease (median: 49.6%; CI: 55.9–17.9) in the percentage of Ki67 was registered in surgery compared with baseline (P = 0.0003). A 54.4% (median) inhibition was observed when only the responsive tumors were considered (P = 0.0001). A waterfall plot depicting the changes in Ki67 expression in individual participants, highlighting their clinical and histologic characteristics, is shown in Fig. 2B. A representative image of Ki67 staining is shown in Fig. 2C. Tumor size was recorded as an exploratory measure and in 7 of 16 evaluable patients, a decrease greater than 20% was observed. No differences in basal PR expression were found between responsive and unresponsive tumors (responsive: 83.6% ± 16.5% vs. unresponsive: 93.3% ± 12.1%; P = 0.15).
Figure 2.
Primary endpoint: Ki67 expression. A, Ki67 expression before (CNB) and after (S) mifepristone treatment. Top, all samples were included (n = 20), except sample M124. The differences in Ki67 values were studied using the Wilcoxon matched pairs signed-rank test. The pairing was significant (P = 0.0004); rs Spearman: 0.6932. Bottom, only responsive cases were included (n = 14). The pairing was significant (P = 0.0001); rs Spearman: 0.8637. B, Top, waterfall plot showing the percentage of inhibition in each individual patient. Negative values below 30% indicate inhibitory effects. The samples that were chosen for RNA-seq studies are denoted in the graph using a different fill texture, and those used in proteomics with a thicker column frame. Bottom: squares represent different parameters, such as tumor size (T1-4), lymphatic nodes (N0-3), tumor stage (1–3), histologic type (D: IC-NST, L: ILC, P: papillary, and M: mucinous), histologic grade (2 and 3), HER2 expression (green box), p53 mutation (blue box), and ESR1 missense mutation (RNA-seq data; violet box). Decreases in tumor size greater than 20% in ultrasound studies are shown in pink squares; in dark gray, those not determined; in white, those not considered because different operators performed the studies; light gray, no change; and black, increase in size. The Ki67 values in CNB < 10% are shown in light blue, 10%–20% in blue, and >20% in dark blue. C, Representative IHC of Ki67 staining (patient M055). Bar: 100 μm.
WB assays
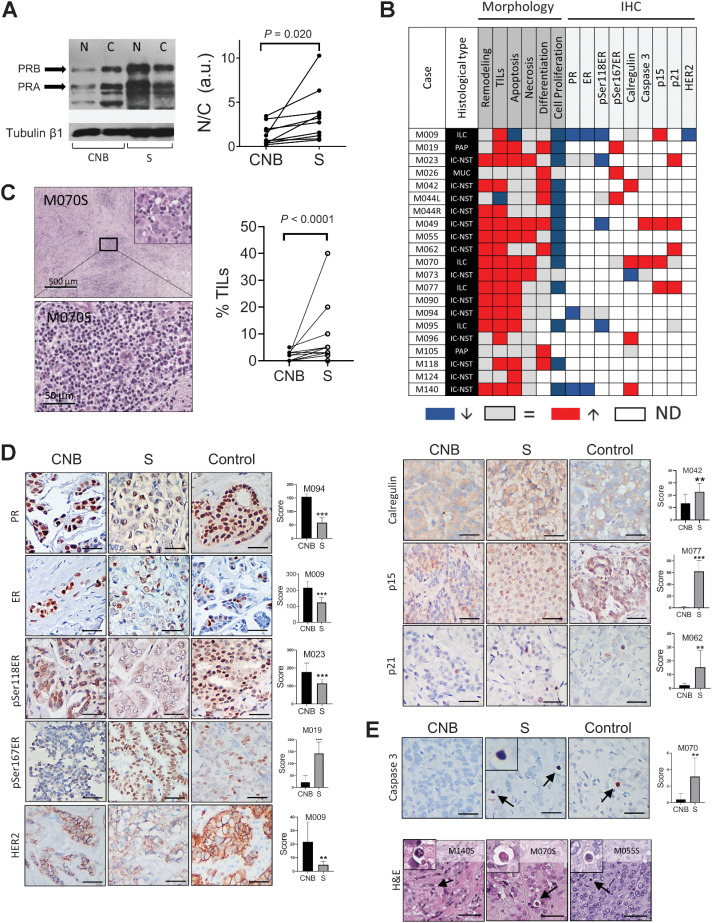
All Nuc fractions from the CNB were PRA-H (inclusion criteria). However, in three cases, the Cyt fraction was PRB-H (Supplementary Fig. S1A). After treatment, the most significant change was the increase in PR in the Nuc fraction. Upshifted PR bands were observed in most of the surgical samples, suggesting protein activation. Figure 3A (left) illustrates a WB of the M009 patient, and WBs from all patients are shown in Supplementary Fig. S1B. Quantification of Nuc and Cyt PR is shown in Fig. 3A (right).
Figure 3.
Western blots, morphologic, and IHC studies. A, Left, Western blot analysis of PR in the nuclear (N) and cytosolic (C) extracts of CNB and surgical samples of patient M009. Right, Total PR was quantified relative to loading control in both fractions, and the relation between N/C was plotted to illustrate that after treatment, PR is mainly located in the nuclei (n = 10). B, Chart summarizing morphologic changes (left columns) observed between CNB and surgical (S) samples, and summary of protein expression evaluated by IHC (right columns). Red means that higher levels were found in surgical samples compared with CNB; blue, lower levels; and gray, no changes. ND, not done (white boxes); ILC, invasive lobular carcinoma; PAP, papillary invasive carcinoma; MUC, mucinous invasive carcinoma; IC-NST, invasive carcinoma of no special type. C, Left, images of TILs after mifepristone treatment. Intratumor TILs (Inset) or stromal TILs present in the surgical sample of a patient in whom almost no TILs were observed in the CNB. Right, quantification of TILs; Wilcoxon test (left). D, Representative images of IHC staining of different proteins in the CNB, in their matched surgical samples (S), and in samples from luminal breast carcinomas used as positive controls (control); n = 3–6 pairs/protein. Right, quantification of the staining in the surgical samples versus the respective CNB in each case. The score of protein expression was calculated by evaluating the staining intensity (low: 1, intermediate: 2, high: 3) and the percentage of positive cells. Bar: 50 μm. E, Top, activated caspase 3 (Cas3) in the CNB (left), in the matched surgical sample (middle), and in a positive control (right). Bottom, H&E of apoptotic cells in three different surgical samples. Bar: 50 μm.
Morphologic changes
Individual histologic characteristics before and after treatment are shown in Supplementary Table S3. A chart summarizing these changes is shown in Fig. 3B. An increase in stroma formed by loose connective tissue with a reduction in dense fibrosis and greater stromal matrix was frequently observed (Supplementary Fig. S2A and S2B). Areas of tissue remodeling (Supplementary Fig. S2C) were observed in 62% of patients. In the trial design, the quantification of tumor-infiltrating lymphocytes (TIL) was not originally planned. However, because (i) TILs quantification has been introduced as a parameter to be considered in standard histopathologic practice (25), and (ii) preclinical studies have shown that mifepristone-primed PRA-H experimental mammary carcinomas respond to an immune checkpoint inhibitor (26), we included TILs quantification in the morphologic evaluation. As expected, low number of TILs was observed in the CNB (<5%). A total of 81% of patients showed an increase after treatment (Fig. 3C). A minimal increase in the amount of isolated apoptosis was observed in 62% of cases analyzed. The presence of differentiation areas (Supplementary Fig. S2D and S2E) and/or secretory vacuoles (Supplementary Fig. S2F) was observed in 43% of cases analyzed. Although we cannot discard the possibility that tumors would have had previous to treatment differentiated areas not observed in the CNB, the differences observed in several of these parameters point toward a treatment effect.
Protein expression by IHC
A significant decrease in the expression of ER and PR was observed after treatment when comparing values from clinical records (ER, P < 0.001; PR, P < 0.05). In our laboratory, IHC assays were simultaneously performed on CNB and surgical samples under nonsaturated conditions in selected cases. A significant decrease in the expression levels of PR, ER, and pSer118ER was observed. Increased nuclear expression of pSer167ER, p21, and p15, and increased membrane localization of calregulin were observed in surgical samples. HER2 expression was downregulated in the HER2+ patient. Only patient M055 was positive for P53 expression. Representative images of these stainings are shown in Fig. 3D, and the evaluated pairs are shown in Fig. 3B (right). The modest increase in apoptosis observed morphologically was validated by measuring activated caspase 3 expression (Fig. 3E). The fact that PR was downregulated by IHC, but not WB, suggests that protein folding in mifepristone-bound PR may hide reactive epitopes.
Taken together, these data suggest that mifepristone treatment decreases pSer118ER expression without affecting pSer167ER and may increase p15, p21, caspase 3 expression related to cytostasis/apoptosis, and membrane calregulin expression related to immunogenic cell death.
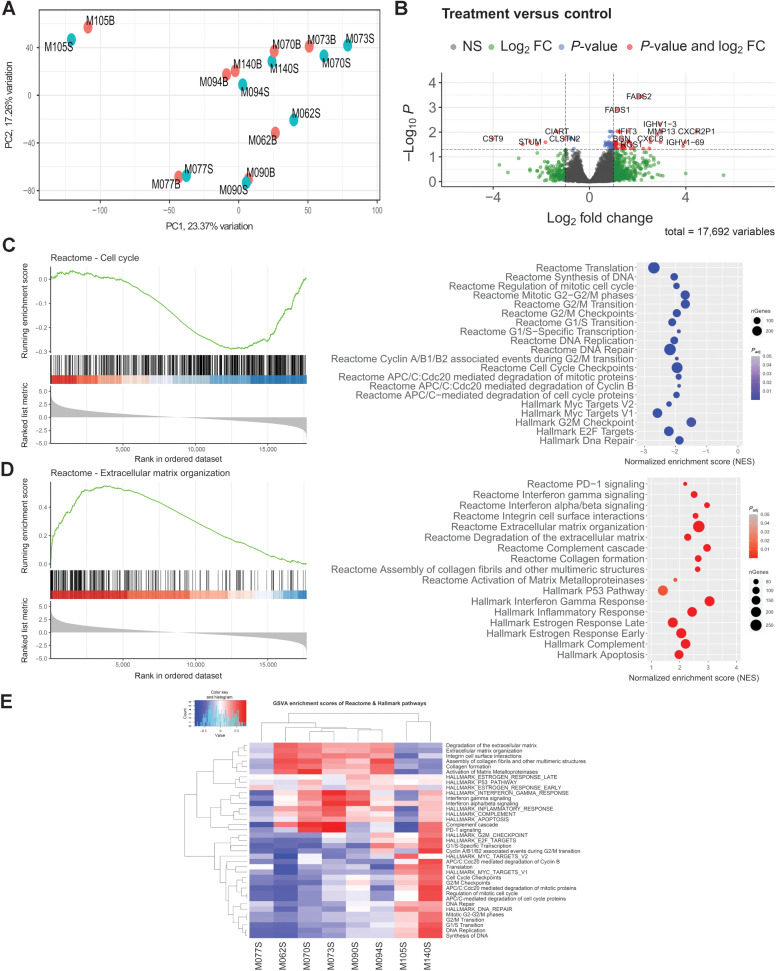
Transcriptome analysis of paired samples
Unsupervised analysis showed that the paired samples clustered together, but neither principal component analysis nor hierarchical clustering showed any relevant clusters (Fig. 4A). Supplementary Figure S3 shows the contribution of the different principal components and the percentage of variance explained for each dimension. When the eight CNBs and their respective surgical samples were paired, differential expression analysis identified 11 and 76 downregulated and upregulated genes [log fold change (LFC) > 1 and FDR < 0.05; Fig. 4B]. Gene set enrichment analysis based on the Hallmark and Reactome databases showed downregulation of cell proliferation pathways (Fig. 4C), and upregulation of tissue remodeling, apoptosis, early and late estrogen-related genes, and immune bioprocesses (Fig. 4D). When we separated mifepristone-responsive (n = 4) and mifepristone-unresponsive (n = 4) tumors, we found that the nonresponsive group shared some modulated pathways with the responsive group (Supplementary Fig. S4A and S4B). Remarkably, in Gene Set Variation Analysis (GSVA), tumor M073 (unresponsive by Ki67 criteria), clustered together with tumor M070, the most responsive tumor in most of the pathways shown in Fig. 4E. The individual values of the RNA-seq data are presented in Supplementary Table S4.
Figure 4.
RNA-seq analysis. A, MDS plot. B, Volcano plot of deregulated genes; 48 genes were upregulated and 11 genes downregulated (log2FC >1; Padj < 0.05). C and D, Representative GSEA plots (left) and dot plot of relevant enriched pathways from GSEA results (Reactome and Hallmark database; right). nGenes corresponds to the number of genes involved in the pathway analyzed. E, GSVA enrichment scores of Reactome and Hallmark pathways. Data were regularized log transformed before analysis.
Using the xCell deconvolution algorithm based on RNA-seq profiles, we estimated immune cell fractions. A significant increase in the immune score, macrophages, M1 macrophages, activated myeloid dendritic cells, and CD4 memory effector T cells was observed after treatment (Supplementary Fig. S4C). A trend was observed for CD8+ cells (P = 0.3525). The tumors with the most prominent increases were M070 and M073, in which the levels of TILs increased the most.
Using RNA-seq data, we identified possible mutations in the 99 most relevant breast cancer driver genes (https://www.intogen.org/download; Supplementary Table S5; Supplementary Fig. S5). Tumor M070, one of the most responsive tumors, shows a missense mutation in ESR1 (p.E380Q; COSM3829320) associated with endocrine resistance (27). Others included missense variants in ERBB2 and FOXA1 in 44% and 56% of the samples tested, respectively, with no reported pathologic effects, and a conservative in-frame deletion in KAT6B of the M105 patient, a resistant tumor to Ki67 criteria.
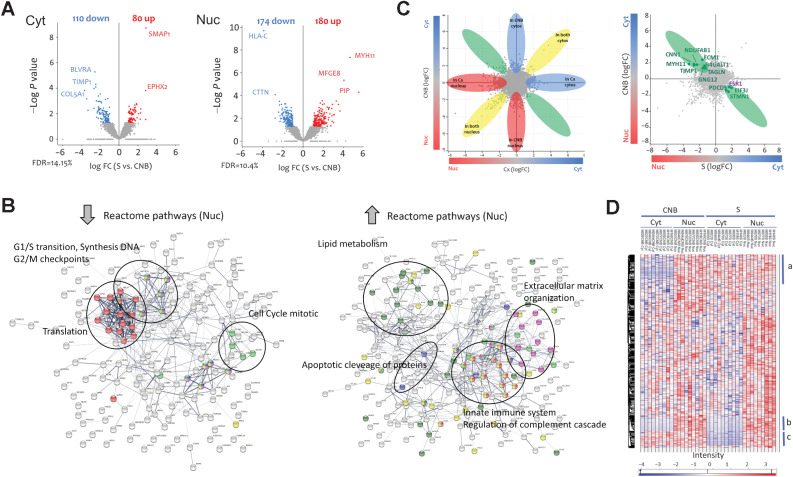
Proteomics
Nuc and Cyt extracts from CNB and surgical samples from 10 patients were analyzed using LC/MS-MS. Differential expression analysis identified 544 of 4,852 deregulated proteins (190 in Cyt and 354 in Nuc; LFC > 1, FDR < 0.05). Figure 5A shows volcano plots of the most deregulated proteins. As depicted in Fig. 5B, in the nucleus, pathways related to cell proliferation that were downregulated in RNA-seq studies were also downregulated in the proteome, together with other pathways, such as the Wnt signaling pathway. Pathways related to extracellular matrix organization, innate immune system, and apoptotic cleavage proteins were upregulated in the Nuc fractions (FDR < 0.05; Supplementary Table S6). An increase in proteins related to cell differentiation such as MUC1, CALML3, and B4GAL1 was also observed. When the Cyt fractions were evaluated, the pathways of lipid metabolism and caspase-mediated cleavage of cytoskeletal proteins (ADD1, MAPT, and VIM) were upregulated, while peptidyl-proline modification, extracellular matrix organization, and collagen biosynthesis were downregulated (Supplementary Table S7). We explored the differential location of the proteins, considering that all samples were processed using the same experimental procedures; hence, the background related to fraction purity would be similar (Fig. 5C and D). Analysis of these proteins is presented in Supplementary Table S8. Among the proteins that may have shuttled into the cytoplasm were HLA-DRB1, HLA-DQB1, HLA-DRA, HLA-DPA1, and HLA-DPB1, all of which are related to PD1 signaling and CTSS related to MHC class II antigen presentation.
Figure 5.
Proteomic analysis. A, Volcano plot showing deregulated proteins in the Cyt fractions (left) and in the Nuc fraction (right). B, Protein–protein association scheme (String) illustrating downregulated (left) and upregulated (right) Reactome pathways after analyzing Nuc fractions from tumors, before and after mifepristone treatment, which were also observed deregulated in RNA-seq studies. C, Flower scheme illustrating the distribution of proteins between compartments and those that shuttled between the Nuc and Cyt compartments after treatment. D, Heatmap illustrating proteins that were normally in the Nuc compartment and that after treatment were found in both compartments (a); Nuc proteins that remained unchanged after treatment (b) and proteins that were in both compartments and after treatment were only observed in the Nuc (c). The complete protein lists and pathways are available in Supplementary Tables S6–S8.
Supplementary Figure S4D shows a cell-cycle KEGG graph illustrating deregulated genes and/or proteins by RNA-seq and MS. Although the individual role of each protein or gene is beyond the scope of this study, as a whole it is clear that Ki67 data, morphologic analysis, RNA-seq, and MS datapoint out that mifepristone exerts an antiproliferative effect in this cohort of patients and disclose relevant bioprocesses that explain its therapeutic effect.
Determination of mifepristone levels in plasma and adverse events
No significant differences were observed between mifepristone levels on day 7 or 14 after treatment [day 7 (mean ± SD):300.3 ± 31.7 ng/mL (690 nmol/L); day 14:320 ± 54.3 ng/mL (745 nmol/L; n = 11); Supplementary Fig. S6A].
Adverse effects are shown in Supplementary Fig. S6B. On day 7, 45% of patients declared no adverse effects, 30% only one, 15% two of them, and 10% more than two. On day 14, 65% had no adverse effects and 35% only one adverse effect. Only 1 patient (M019) had fatigue graded as type 2 on day 7, which turned to grade 1 on day 14. All laboratory values were within reference ranges (Supplementary Fig. S6C).
Discussion
MIPRA was the first clinical trial in which patients were categorized according to their PR isoform ratio. In this single-arm study, we demonstrated that mifepristone inhibited the proliferation of breast carcinomas with higher levels of PRA than of PRB. The study met the primary endpoint of a 30% reduction in Ki67 between CNB and surgery. Morphologic evaluation, transcriptomic, and proteomic studies support the therapeutic effect of mifepristone, and suggest that responses may be underestimated with the unique measurement of Ki67. Although transcriptomic studies have been performed to reinforce Ki67 data in different breast cancer WOT studies (28), to the best of our knowledge, this is the first study in which paired samples were characterized by proteomics.
Originally, we estimated that we had to evaluate 100 patients to reach 20 who met the inclusion criteria. However, we recruited 140 patients because, in some cases, the biopsy failed to have sufficient cancer cells, and the sensitivity of WB was not sufficient to obtain a reliable result.
Following the design of a similar study (29), we did not include a placebo group to compare the intrinsic variation in Ki67 expression between CNB and surgical samples. In trials in which control patients have been included, some authors reported a slight increase in Ki67 expression (30–33), while others reported a decrease between the surgical sample and the CNB that never exceeded 20% (12, 34), which is below the decrease obtained in this study (49%). Ki67 evaluation, as a primary endpoint, has become the gold standard in WOT studies (35–37). Our results are similar to those originally reported for tamoxifen treatment in patients with ER+ breast cancer (30, 38). In line with other studies, such as the IMPACT (39), POETIC (40), and neoMONARCH (28) trials, MIPRA patients received mifepristone for only 2 weeks to avoid surgery delays.
Although a decrease in tumor size determined by ultrasound was not expected, it was achieved in several cases, even considering that the first ultrasound measurement was registered 49 days (median) before the surgical sample at the time of CNB. Considering that luminal tumors may increase in size by 0.17%–0.21% per day (41), the real decrease in size might be even higher than the one recorded.
Six of the 20 tumors did not meet the prespecified criteria for treatment response. As Ki67 evaluation was the primary endpoint, we named these tumors as unresponsive. Three of these patients had very low Ki67 levels on CNB, suggesting that a possible therapeutic effect might have been masked. Moreover, RNA-seq analysis performed using four of six unresponsive tumors showed the activation of pathways similar to those in responsive tumors. GSVA showed a clear inhibitory effect for patients M090 and M073 and less sharp suppression for M094, whereas M105 had a different response. Several morphologic signs of drug response were also observed in three of six unresponsive cases, and the analysis was performed blinded to the Ki67 data.
RNA-seq and MS studies identified downregulated pathways related to cell proliferation, and although the individual MKI67 mRNA (Ki67 gene) or protein did not appear to be one of the deregulated candidates, proliferating cell nuclear antigen and MCM2–7 proteins, which are also considered surrogate markers of cell proliferation, were downregulated. This highlights the importance of simultaneously evaluating several biomarkers to improve the data accuracy. As Ki67 and RNA-seq/proteomic data originated from different CNB or surgical samples, inclusion of both analyses increased the robustness of our data.
Common enriched pathways found in both proteomic and RNA-seq studies were those related to the innate immune system and those related to cell-matrix organization. This agrees with the increase in TILs observed in most mifepristone-treated tumors and with recent preclinical findings showing that mifepristone may prime PRA-H tumors for a second treatment with an immune checkpoint inhibitor (26), and with those from Werner and colleagues, who suggested the use of antiprogestins to increase immune infiltrates in tumors by targeting PR (42). Proteins related to PD1 signaling were found in the Cyt extracts of mifepristone-treated tumors. Calreticulin/calregulin, a protein related to immunogenic cell death (43), was highly expressed in the cell membranes of several mifepristone-treated samples (IHC assays). Similar immune signatures were observed in trials using CDK4/6 inhibitors (44). However, it may be argued that the CNB procedure elicits an inflammatory effect. In the NeoMONARCH study, CNB and surgical samples were collected after 14 days of single or combined treatment, and an increase in immune-related pathways was only observed in the combined treatment group, ruling out the possible assumption (45).
Apoptosis has not been identified as a hierarchical pathway involved in the success of endocrine therapies, showing only mild increases after tamoxifen or fulvestrant treatment (46). However, in preclinical studies in which mifepristone induced almost complete tumor regression involving differentiation and/or tissue remodeling, a significant increase in apoptosis was observed after 24 or 48 hours of mifepristone administration (47). Thus, we expected to find an increase in apoptotic cells in mifepristone-treated tumors. In the MIPRA trial, a modest increase in apoptosis was observed morphologically, as confirmed by IHC and proteomic studies, in which an increase in several proapoptotic proteins was observed.
In our study, we also included patients with advanced stages that were naïve to any other treatment for this cancer, which makes this study unique compared with other trials using mifepristone, in which patients who failed to respond to other treatments were included. Romieu and colleagues (8) and Klijn and colleagues (6) enrolled patients with tamoxifen resistance. Perrault and colleagues identified PR+ patients who received no other treatment for recurrence but were previously treated for their primary tumors (7). In preclinical models, most endocrine-resistant tumors are PRB-H (3), which may partially explain the poor responses observed in the aforementioned clinical trials.
Different reasons may explain why some patients in our cohort were unresponsive to mifepristone, and they should be considered in future studies. Two of the unresponsive tumors, M026 and M105, and the responsive M140 tumor were classified as PRA-H based on the WB of the Nuc extract; however, in the three tumors, the Cyt fraction was PRB-H. It may be suggested that tumors in which both the Nuc and Cyt fractions are PRA-H may represent the best candidates for mifepristone treatment, as they truly represent the total protein ratio. Tumor M140 showed good responsiveness in the Ki67 assay, a decrease in tumor size, an increase in the immune bioprocess pathways and apoptosis, but unexpectedly, an increase in proliferative pathways. This may be because the frozen CNB sample also contained adjacent nontumor mammary cells, which may have masked the mifepristone-induced effects.
Among Ki67 responsive patients, a HER2+ patient (CNB evaluation) was cataloged as negative in the surgical sample by the Hospital Pathology Department. We also found a decrease in membrane staining for HER2 (IHC), and HER2/ERBB2 was one of the downregulated proteins reported in the Cyt extracts (proteomics). Similar observations were made by Lee and colleagues, who used TLP to show a decrease in HER2-related genes in TLP-treated tumors. Moreover, they proposed that HER2+ luminal tumors might respond best to antiprogestin therapy (12).
IHC for p53 indirectly detects p53 mutations (48). The only tumor that was positive for p53 was sensitive to mifepristone, as was the case with T47D xenografts that have a pathogenic p53 mutation (49) and respond to mifepristone treatment (50). RNA-seq analysis allowed us to investigate possible mutations in the eight tumors studied. It is worth mentioning the missense mutation in ESR1, which, as reported previously, is associated with endocrine resistance (27). As this patient was responsive to mifepristone, the effect of mifepristone treatment in patients with activated ESR1 mutations deserves further investigation.
The decrease in ER observed by IHC was consistent with the possible displacement of ER from the nuclear compartment to the cytoplasm, as observed in proteomic studies. ER is usually phosphorylated at Ser118 in response to MAPK activation or estradiol binding, whereas Ser167 is phosphorylated by Akt, RSK, and casein kinase II in addition to MAPK (51, 52). As we only observed a decrease in pSer118ER expression following mifepristone treatment, it can be speculated that mifepristone treatment compromises MAPK-mediated ER signaling.
As mentioned previously, mifepristone, in addition to its antiprogestin action, exerts antiglucocorticoid effects (53). Taking advantage of the immunomodulatory effects of mifepristone, an ongoing clinical trial intends to exploit this property therapeutically using high mifepristone doses in patients with breast cancer (NCT03225547). Using experimental PRA-H tumor models, we showed that mifepristone inhibits tumor growth even when tumors are transplanted into immunosuppressed mice (26). Moreover, in vitro, mifepristone exerts antiproliferative effects only in PRA-H+ cells (16), suggesting that the direct antiprogestin effect of mifepristone is the prevailing effect, without ruling out a contribution from its antiglucocorticoid or its antiandrogenic effects (reviewed in ref. 15).
In summary, our clinical study showed that preclinical findings are valid in patients with breast cancer, suggesting that mifepristone can be used for the treatment of luminal PRA-H breast cancer. Further studies are needed to evaluate the effects of combined treatment with antiprogestins and tamoxifen in PRA-H patients. Preclinical studies have suggested that a stronger response can be obtained with this combination (54). Aromatase inhibitors or ER degraders may not work together with mifepristone, because they inhibit PR expression. Because we have already shown that lymph node metastases maintain the same PR isoform ratio as the primary tumor (15), it may be speculated that this treatment might prove suitable in an adjuvant setting, or alternatively, in a neoadjuvant setting, if further immunotherapy is suggested. Ongoing studies are currently evaluating the combination of mifepristone and CDK4/6 inhibitors in preclinical PRA-H models. Finally, because WB does not seem to be an ideal method for discriminating PRA-H patients in hospital facilities, companion diagnostic tools to improve screening should be developed.
Strengths
Originality, proof of concept after years of preclinical research, primary endpoint met, combination of approaches: standard Ki67 supported by exhaustive morphologic evaluation, IHC validation, WB, transcriptomics, and proteomics. Most secondary outcomes were met, including studies based on frozen samples and studies using different formalin-fixed samples. The evaluation of mifepristone in plasma guarantees treatment compliance.
Limitations
This study had a small number of patients, no placebo group, limited material from CNB for further analysis, and a short treatment period.
Supplementary Material
Morphological evaluation of CNBs and matched surgical samples; RNA-Seq analysis; Proteomics
A, Prevailing PR isoforms before and after mifepristone treatment and their prevailing nuclear (Nuc) or cytosolic (Cyt) localization in western blot studies. *: Prevailing PR isoforms in Western blot studies; #: Distribution of PR in the Nuc or Cyt compartments; CNB: Core needle biopsy, S: Surgical sample. In grey font: Mifepristone unresponsive tumors by Ki67 criteria. B, Representative western blots of CNB and surgical samples from all patients. The upper band is PRB and the lower band PRA, as labeled in the first set.
Morphological features after mifepristone treatment. A-D, Images illustrating increased stromal tissue (A), thin and scarce collagen fibers, and increased stromal matrix (arrow; B), an area of tissue remodeling (C), and an area of increased differentiation (D) observed in the surgical samples (S) compared to the respective core needle biopsies (CNB). E, Image illustrates an area of necrosis (dotted arrow) together with a differentiated structure (black arrow) in a surgical sample. F, Cells with Alcian blue+ (left), PAS+ (middle) and MUC-1+ (immunohistochemistry; right) vacuoles observed in a mifepristone-treated tumor. Inset: High power field of a differentiated gland/duct.
A, Library size before and after low count filtering. B, P-value histogram distribution observed in the differential expression analysis. C, Pairwise scatter plots of the first five principal components from the principal component analysis of RNA-Seq data.
RNA-Seq and Proteomics analysis. A and B, Dot plot of relevant enriched pathways from GSEA results (Reactome and Hallmark databases) separating responsive (A) and unresponsive tumors (B). C, Immune cell deconvolution (xcell) in the different tumors analyzed by RNA-Seq. Plots with significant p values analyzed by Wilcoxon test and the T cell CD8+ plot are shown. D, Kegg diagram of the Cell cycle pathway, that may explain mifepristone therapeutics effects. A colored code was used: the first half of the box is colored according to the RNA-Seq data (8 tumors) and the last half of the box is colored according to the proteomic data (n=10 tumors; nuclear fraction).
Mutations identified in RNA-Seq analysis. The data obtained in RNA-Seq studies were graphed using the Oviz-Bio tool (https://academic.oup.com/nar/article/48/W1/W415/5835823). Only driver breast cancer genes (99) were selected from the Intogene data base (https://www.intogen.org/download) and we found mutations in 19 driver genes after read depth filtering (DP >50). Of note is the missense mutation found in ESR1 in the M070 patient. Left, Percentage of samples with mutations. Color defines the type of mutations.
Mifepristone plasma levels and mifepristone-related side effects. A, Plasma levels of mifepristone 7 or 14 days after mifepristone treatment. The data from 9-10 patients are shown. Data were analyzed with ANOVA for matched samples (Friedman test). B, Clinical adverse effects grade 1 recorded in mifepristone (MFP)-treated patients. C, Laboratory values. Patients M049, M095 and M140 were treated for diabetes. Patients M009, M019, M023, M042, M049, M062, M070, M077, M090, M095, M105 and M140 were treated for hypertension and patients M009, M019 and M124 were treated for hypothyroidism. Dotted areas represent normal levels. Patients under mifepristone treatment did not interrupt standard medicine care.
Representativeness of Study Participants
Evaluation of Ki67 expression in paired samples before and after mifepristone treatment by two independent pathologists, and time elapsed between core needle biopsy and surgery.
Morphological features of tumors before and after mifepristone treatment
RNA-Seq DEG and GSEA
RNA-Seq Variant calling
Proteomics: DE and Reactome of Cyt proteins
Proteomics: DE and Reactome of Nuc proteins
Proteomics: Enrichment analysis from heatmap
Acknowledgments
The clinical study was funded by MINCyT, Agencia Nacional de Promoción Científica y Tecnológica - Argentina, ANAPCYT (PIDC 2012-084) to C. Lanari, with participation of CONICET and Ministerio de Salud de la Provincia de Buenos Aires. RNA-seq and plasma studies were funded by Fundación Sales, grant 2020 to C. Lanari. Proteomic studies were funded by the Horizon 2020, Program of the European Union, project EPIC-XS 823839, and grants from Fundação para a CiÊncia e Technologia (FCT), projects UIDB/04501/2020 and UIDP/04501/2020, and Comissão de Coordenação e Desenvolvimento Regional do Centro (CCDRC), Project MEDISIS (CENTRO-01-0246-FEDER-000018) to Luisa Helguero.
We are grateful to Marisa Tiratel (Pharmacist) for keeping the mifepristone tablets at the hospital's pharmacy; Mrs. Analia Presta and Mrs. Elidia Acosta for taking the medication to the patient's home every day; and Mrs. Elsa Arias, Mrs. Marcela Lino, Mrs. Marcela Chiachiarelli, and Mr. Ariel Quinteros for their excellent technical assistance. We also wish to thank LALCEC Tigre, Estudio Mendez y Asociados, Rotary Club from General Pacheco and Cooperadora del HospitalPMVM for paying the insurance, Dr. Fernando Abramzon and Dr. Leticia Borrino from Centro Diagnóstico Municipal, Tigre and Dr. Gustavo San Martín, HospitalPMVM, for their help in mammographic and ultrasound studies respectively, to Natalie Homer and Ruth Andrew of the University of Edinburgh for their help in the measurement of mifepristone in plasma, to Dr. Veronica Solernou and Dr. Fabiana Lubienieck from Garrahan Hospital for kindly sharing the scanscope during Covid pandemic, and to Dr. Alfredo Molinolo, Moores Cancer Center, UCSD, San Diego, for his help in the digital evaluation of Ki67 in scanned samples.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 831
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
P. Rojas reports a patent for WO2013086379A2 issued. C. Lanari reports a patent for WO2013086379A2 issued. No disclosures were reported by the other authors.
Authors' Contributions
A. Elía: Data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. L. Saldain: Validation, methodology. S.I. Vanzulli: Formal analysis, supervision, writing–original draft. L.A. Helguero: Data curation, funding acquisition, methodology, writing–review and editing. C.A. Lamb: Formal analysis, writing–review and editing. V. Fabris: Formal analysis. G. Pataccini: Methodology. P. Martínez-Vazquez: Methodology. J. Burruchaga: Methodology. I. Caillet-Bois: Methodology. E. Spengler: Methodology. G. Acosta Haab: Methodology. M. Liguori: Methodology. A. Castets: Supervision. S. Lovisi: Supervision. M.F. Abascal: Methodology. V. Novaro: Supervision. J. Sánchez: Data curation, formal analysis. J. Muñoz: Formal analysis, supervision. J.M. Belizán: Writing–review and editing. M.C. Abba: Data curation, formal analysis, supervision. H. Gass: Conceptualization, resources, supervision, methodology, project administration, deceased 2019. P. Rojas: Conceptualization, supervision, methodology, writing–review and editing. C. Lanari: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Burstein HJ, Somerfield MR, Barton DL, Dorris A, Fallowfield LJ, Jain D, et al. Endocrine treatment and targeted therapy for hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer: ASCO guideline update. J Clin Oncol 2021;39:3959–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fanning SW, Greene GL. Next-generation ERalpha inhibitors for endocrine-resistant ER+ breast cancer. Endocrinology 2019;160:759–69. [DOI] [PubMed] [Google Scholar]
- 3. Giulianelli S, Lamb CA, Lanari C. Progesterone receptors in normal breast development and breast cancer. Essays Biochem 2021;65:951–69. [DOI] [PubMed] [Google Scholar]
- 4. Dwyer AR, Truong TH, Ostrander JH, Lange CA. 90 YEARS OF PROGESTERONE: Steroid receptors as MAPK signaling sensors in breast cancer: let the fates decide. J Mol Endocrinol 2020;65:T35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scabia V, Ayyanan A, De Martino F, Agnoletto A, Battista L, Laszlo C, et al. Estrogen receptor positive breast cancers have patient specific hormone sensitivities and rely on progesterone receptor. Nat Commun 2022;13:3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klijn JG, de Jong FH, Bakker GH, Lamberts SW, Rodenburg CJ, Alexieva-Figusch J. Antiprogestins, a new form of endocrine therapy for human breast cancer. Cancer Res 1989;49:2851–6. [PubMed] [Google Scholar]
- 7. Perrault D, Eisenhauer EA, Pritchard KI, Panasci L, Norris B, Vandenberg T, et al. Phase II study of the progesterone antagonist mifepristone in patients with untreated metastatic breast carcinoma: a national cancer institute of Canada clinical trials group study. J Clin Oncol 1996;14:2709–12. [DOI] [PubMed] [Google Scholar]
- 8. Romieu G, Maudelonde T, Ulmann A, Pujol H, Grenier J, Cavalie G, et al. The antiprogestin RU486 in advanced breast cancer: preliminary clinical trial. Bull Cancer 1987;74:455–61. [PubMed] [Google Scholar]
- 9. Robertson JF, Willsher PC, Winterbottom L, Blamey RW, Thorpe S. Onapristone, a progesterone receptor antagonist, as first-line therapy in primary breast cancer. Eur J Cancer 1999;35:214–8. [DOI] [PubMed] [Google Scholar]
- 10. Jonat W, Bachelot T, Ruhstaller T, Kuss I, Reimann U, Robertson JF. Randomized phase II study of lonaprisan as second-line therapy for progesterone receptor-positive breast cancer. Ann Oncol 2013;10:2543–8. [DOI] [PubMed] [Google Scholar]
- 11. Goyeneche AA, Telleria CM. Antiprogestins in gynecological diseases. Reproduction 2015;149:R15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee O, Sullivan ME, Xu Y, Rodgers C, Muzzio M, Helenowski I, et al. Selective progesterone receptor modulators in early stage breast cancer: a randomized, placebo-controlled phase II window of opportunity trial using telapristone acetate. Clin Cancer Res 2020;26:25–34. [DOI] [PubMed] [Google Scholar]
- 13. Fabris V, Abascal MF, Giulianelli S, May M, Sequeira GR, Jacobsen B, et al. Isoform specificity of progesterone receptor antibodies. J Pathol Clin Res 2017;3:227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wargon V, Helguero LA, Bolado J, Rojas P, Novaro V, Molinolo A, et al. Reversal of antiprogestin resistance and progesterone receptor isoform ratio in acquired resistant mammary carcinomas. Breast Cancer Res Treat 2009;116:449–60. [DOI] [PubMed] [Google Scholar]
- 15. Abascal MF, Elia A, Alvarez M, Pataccini G, Sequeira G, Riggio M, et al. Progesterone receptor isoform ratio dictates antiprogestin/progestin effects on breast cancer growth and metastases: a role for NDRG1. Int J Cancer 2022;150:1481–96. [DOI] [PubMed] [Google Scholar]
- 16. Rojas PA, May M, Sequeira GR, Elia A, Alvarez M, Martinez P, et al. Progesterone receptor isoform ratio: a breast cancer prognostic and predictive factor for antiprogestin responsiveness. J Natl Cancer Inst 2017;109:djw317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baulieu EE. Contragestion and other clinical applications of RU 486, an antiprogesterone at the receptor. Science 1989;245:1351–7. [DOI] [PubMed] [Google Scholar]
- 18. Islam S, Afrin S, Jones SI, Segars J. Selective progesterone receptor modulators-mechanisms and therapeutic utility. Endocr Rev 2020;41:bnaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American society of clinical oncology/college of American pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med 2010;134:907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997–4013. [DOI] [PubMed] [Google Scholar]
- 21. Dowsett M, Nielsen TO, A'Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst 2011;103:1656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Denham SG, Just G, Kyle CJ, Richardson J, Lee P, Simpson JP, et al. Automated supported liquid extraction for the analysis of a panel of 12 endogenous steroids in human plasma by LC-MS/MS. Preprints 2020, 2020110551. [Google Scholar]
- 24. Juste YR, Kaushik S, Bourdenx M, Aflakpui R, Bandyopadhyay S, Garcia F, et al. Reciprocal regulation of chaperone-mediated autophagy and the circadian clock. Nat Cell Biol 2021;23:1255–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs working group 2014. Ann Oncol 2015;26:259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sequeira GR, Sahores A, Dalotto-Moreno T, Perrotta RM, Pataccini G, Vanzulli SI, et al. Enhanced antitumor immunity via endocrine therapy prevents mammary tumor relapse and increases immune checkpoint blockade sensitivity. Cancer Res 2021;81:1375–87. [DOI] [PubMed] [Google Scholar]
- 27. Lei JT, Gou X, Seker S, Ellis MJ. ESR1 alterations and metastasis in estrogen receptor positive breast cancer. J Cancer Metastasis Treat 2019;5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hurvitz SA, Martin M, Press MF, Chan D, Fernandez-Abad M, Petru E, et al. Potent cell-cycle inhibition and upregulation of immune response with abemaciclib and anastrozole in neoMONARCH, phase II neoadjuvant study in HR(+)/HER2(-) breast cancer. Clin Cancer Res 2020;26:566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Day TA, Shirai K, O'Brien PE, Matheus MG, Godwin K, Sood AJ, et al. Inhibition of mTOR signaling and clinical activity of rapamycin in head and neck cancer in a window of opportunity trial. Clin Cancer Res 2019;25:1156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Decensi A, Robertson C, Viale G, Pigatto F, Johansson H, Kisanga ER, et al. A randomized trial of low-dose tamoxifen on breast cancer proliferation and blood estrogenic biomarkers. J Natl Cancer Inst 2003;95:779–90. [DOI] [PubMed] [Google Scholar]
- 31. Dowsett M, Dixon JM, Horgan K, Salter J, Hills M, Harvey E. Antiproliferative effects of idoxifene in a placebo-controlled trial in primary human breast cancer. Clin Cancer Res 2000;6:2260–7. [PubMed] [Google Scholar]
- 32. Dowsett M, Bundred NJ, Decensi A, Sainsbury RC, Lu Y, Hills MJ, et al. Effect of raloxifene on breast cancer cell Ki67 and apoptosis: a double-blind, placebo-controlled, randomized clinical trial in postmenopausal patients. Cancer Epidemiol Biomarkers Prev 2001;10:961–6. [PubMed] [Google Scholar]
- 33. Robertson JF, Nicholson RI, Bundred NJ, Anderson E, Rayter Z, Dowsett M, et al. Comparison of the short-term biological effects of 7alpha-[9-(4,4,5,5,5-pentafluoropentylsulfinyl)-nonyl]estra-1,3,5, (10)-triene-3,17beta-diol (Faslodex) versus tamoxifen in postmenopausal women with primary breast cancer. Cancer Res 2001;61:6739–46. [PubMed] [Google Scholar]
- 34. Shike M, Doane AS, Russo L, Cabal R, Reis-Filho JS, Gerald W, et al. The effects of soy supplementation on gene expression in breast cancer: a randomized placebo-controlled study. J Natl Cancer Inst 2014;106:dju189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones EF, Hathi DK, Freimanis R, Mukhtar RA, Chien AJ, Esserman LJ, et al. Current landscape of breast cancer imaging and potential quantitative imaging markers of response in ER-positive breast cancers treated with neoadjuvant therapy. Cancers 2020;12:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arnedos M, Roulleaux Dugage M, Perez-Garcia J, Cortes J. Window of Opportunity trials for biomarker discovery in breast cancer. Current Opin Oncol 2019;31:486–92. [DOI] [PubMed] [Google Scholar]
- 37. Nielsen TO, Leung SCY, Rimm DL, Dodson A, Acs B, Badve S, et al. Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in breast cancer working group. J Natl Cancer Inst 2021;113:808–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clarke RB, Laidlaw IJ, Jones LJ, Howell A, Anderson E. Effect of tamoxifen on Ki67 labelling index in human breast tumours and its relationship to oestrogen and progesterone receptor status. Br J Cancer 1993;67:606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, Griffith C, et al. Short-term changes in Ki-67 during neoadjuvant treatment of primary breast cancer with anastrozole or tamoxifen alone or combined correlate with recurrence-free survival. Clin Cancer Res 2005;11:951s–8s. [PubMed] [Google Scholar]
- 40. Smith I, Robertson J, Kilburn L, Wilcox M, Evans A, Holcombe C, et al. Long-term outcome and prognostic value of Ki67 after perioperative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): an open-label, multicentre, parallel-group, randomised, phase 3 trial. Lancet Oncol 2020;21:1443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee SH, Kim YS, Han W, Ryu HS, Chang JM, Cho N, et al. Tumor growth rate of invasive breast cancers during wait times for surgery assessed by ultrasonography. Medicine 2016;95:e4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Werner LR, Gibson KA, Goodman ML, Helm DE, Walter KR, Holloran SM, et al. Progesterone promotes immunomodulation and tumor development in the murine mammary gland. J Immunother Cancer 2021;9:e001710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 2012;12:860–75. [DOI] [PubMed] [Google Scholar]
- 44. Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hadad SM, Jordan LB, Roy PG, Purdie CA, Iwamoto T, Pusztai L, et al. A prospective comparison of ER, PR, Ki67 and gene expression in paired sequential core biopsies of primary, untreated breast cancer. BMC Cancer 2016;16:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ellis PA, Saccani-Jotti G, Clarke R, Johnston SR, Anderson E, Howell A, et al. Induction of apoptosis by tamoxifen and ICI 182780 in primary breast cancer. Int J Cancer 1997;72:608–13. [DOI] [PubMed] [Google Scholar]
- 47. Vanzulli S, Efeyan A, Benavides F, Helguero L, Peters G, Shen J, et al. p21, p27 and p53 in estrogen and antiprogestin-induced tumor regression of experimental mouse mammary ductal carcinomas. Carcinogenesis 2002;23:749–58. [DOI] [PubMed] [Google Scholar]
- 48. Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol 2008;28:4365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wargon V, Riggio M, Giulianelli S, Sequeira GR, Rojas P, May M, et al. Progestin and antiprogestin responsiveness in breast cancer is driven by the PRA/PRB ratio via AIB1 or SMRT recruitment to the CCND1 and MYC promoters. Int J Cancer 2015;136:2680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 1995;270:1491–4. [DOI] [PubMed] [Google Scholar]
- 52. Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem 2001;276:9817–24. [DOI] [PubMed] [Google Scholar]
- 53. Gaillard RC, Riondel A, Muller AF, Herrmann W, Baulieu EE. RU 486: a steroid with antiglucocorticosteroid activity that only disinhibits the human pituitary-adrenal system at a specific time of day. Proc Natl Acad Sci U S A 1984;81:3879–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. El Etreby MF, Liang Y. Effect of antiprogestins and tamoxifen on growth inhibition of MCF-7 human breast cancer cells in nude mice. Breast Cancer Res Treat 1998;49:109–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological evaluation of CNBs and matched surgical samples; RNA-Seq analysis; Proteomics
A, Prevailing PR isoforms before and after mifepristone treatment and their prevailing nuclear (Nuc) or cytosolic (Cyt) localization in western blot studies. *: Prevailing PR isoforms in Western blot studies; #: Distribution of PR in the Nuc or Cyt compartments; CNB: Core needle biopsy, S: Surgical sample. In grey font: Mifepristone unresponsive tumors by Ki67 criteria. B, Representative western blots of CNB and surgical samples from all patients. The upper band is PRB and the lower band PRA, as labeled in the first set.
Morphological features after mifepristone treatment. A-D, Images illustrating increased stromal tissue (A), thin and scarce collagen fibers, and increased stromal matrix (arrow; B), an area of tissue remodeling (C), and an area of increased differentiation (D) observed in the surgical samples (S) compared to the respective core needle biopsies (CNB). E, Image illustrates an area of necrosis (dotted arrow) together with a differentiated structure (black arrow) in a surgical sample. F, Cells with Alcian blue+ (left), PAS+ (middle) and MUC-1+ (immunohistochemistry; right) vacuoles observed in a mifepristone-treated tumor. Inset: High power field of a differentiated gland/duct.
A, Library size before and after low count filtering. B, P-value histogram distribution observed in the differential expression analysis. C, Pairwise scatter plots of the first five principal components from the principal component analysis of RNA-Seq data.
RNA-Seq and Proteomics analysis. A and B, Dot plot of relevant enriched pathways from GSEA results (Reactome and Hallmark databases) separating responsive (A) and unresponsive tumors (B). C, Immune cell deconvolution (xcell) in the different tumors analyzed by RNA-Seq. Plots with significant p values analyzed by Wilcoxon test and the T cell CD8+ plot are shown. D, Kegg diagram of the Cell cycle pathway, that may explain mifepristone therapeutics effects. A colored code was used: the first half of the box is colored according to the RNA-Seq data (8 tumors) and the last half of the box is colored according to the proteomic data (n=10 tumors; nuclear fraction).
Mutations identified in RNA-Seq analysis. The data obtained in RNA-Seq studies were graphed using the Oviz-Bio tool (https://academic.oup.com/nar/article/48/W1/W415/5835823). Only driver breast cancer genes (99) were selected from the Intogene data base (https://www.intogen.org/download) and we found mutations in 19 driver genes after read depth filtering (DP >50). Of note is the missense mutation found in ESR1 in the M070 patient. Left, Percentage of samples with mutations. Color defines the type of mutations.
Mifepristone plasma levels and mifepristone-related side effects. A, Plasma levels of mifepristone 7 or 14 days after mifepristone treatment. The data from 9-10 patients are shown. Data were analyzed with ANOVA for matched samples (Friedman test). B, Clinical adverse effects grade 1 recorded in mifepristone (MFP)-treated patients. C, Laboratory values. Patients M049, M095 and M140 were treated for diabetes. Patients M009, M019, M023, M042, M049, M062, M070, M077, M090, M095, M105 and M140 were treated for hypertension and patients M009, M019 and M124 were treated for hypothyroidism. Dotted areas represent normal levels. Patients under mifepristone treatment did not interrupt standard medicine care.
Representativeness of Study Participants
Evaluation of Ki67 expression in paired samples before and after mifepristone treatment by two independent pathologists, and time elapsed between core needle biopsy and surgery.
Morphological features of tumors before and after mifepristone treatment
RNA-Seq DEG and GSEA
RNA-Seq Variant calling
Proteomics: DE and Reactome of Cyt proteins
Proteomics: DE and Reactome of Nuc proteins
Proteomics: Enrichment analysis from heatmap
Data Availability Statement
The RNA-seq data generated in this study are publicly available in the Gene Expression Omnibus at GSE212690 and proteomic data at ProteomeXchange at PXD036515.