Abstract
DNA polymerases catalyze DNA synthesis with high efficiency, which is essential for all life. Extensive kinetic and structural efforts have been executed in exploring mechanisms of DNA polymerases, surrounding their kinetic pathway, catalytic mechanisms, and factors that dictate polymerase fidelity. Recent time-resolved crystallography studies on DNA polymerase η (Pol η) and β have revealed essential transient events during the DNA synthesis reaction, such as mechanisms of primer deprotonation, separated roles of the three metal ions, and conformational changes that disfavor incorporation of the incorrect substrate. DNA-embedded ribonucleotides (rNs) are the most common lesion on DNA and a major threat to genome integrity. While kinetics of rN incorporation has been explored and structural studies have revealed that DNA polymerases have a steric gate that destabilizes ribonucleotide triphosphate binding, the mechanism of extension upon rN addition remains poorly characterized. Using steady-state kinetics, static and time-resolved X-ray crystallography with Pol η as a model system, we showed that the extra hydroxyl group on the primer terminus does alter the dynamics of the polymerase active site as well as the catalysis and fidelity of DNA synthesis. During rN extension, Pol η error incorporation efficiency increases significantly across different sequence contexts. Finally, our systematic structural studies suggest that the rN at the primer end improves primer alignment and reduces barriers in C2′-endo to C3′-endo sugar conformational change. Overall, our work provides further mechanistic insights into the effects of rN incorporation on DNA synthesis.
Keywords: Ribonucleotide extension, Primer alignment, Sugar pucker, DNA polymerase fidelity
Abbreviations: dA, deoxyribose adenine; dC, deoxyribose cytosine; dGTP, deoxyribose guanine triphosphate; dN, deoxynucleotide; dNTP, deoxynucleotide triphosphate; dT, deoxyribose thymine; DTT, dithiothreitol; MES, 2-(N-morpholino) ethanesulfonic acid; Pol η, DNA polymerase η; rA, ribose adenine; rG, ribose guanine; rN, ribonucleotide; rNTP, ribonucleotide triphosphate; rU, ribose uridine
DNA polymerases catalyze the essential process of DNA synthesis during DNA replication and DNA repair. Although distinct in their sequences and structures and categorized into seven enzymatic families, DNA polymerases contain similar active sites, follow similar kinetic pathways, and employ similar catalytic mechanisms (1, 2). They adopt a right-hand architecture with the active site in the palm domain, the thumb domain that interacts with primer:template DNA duplex, and the finger domain that interacts with the incoming nucleotide (Fig. 1A). Conserved acidic residues line the active site, and once DNA and the incoming nucleotide bind, replicative polymerases (A, B, C, D, and RT-family) and some X-family polymerases (β and λ) exhibit large-scale finger domain conformational changes, while Y-family polymerases and some X-family polymerases do not. Following finger domain movement, multiple metal ions are recruited in promoting DNA synthesis (Fig. 1C) (1, 3, 4, 5, 6). The B-site metal ion (Me2+B) is associated with the triphosphate motif of the incoming deoxynucleotide triphosphate (dNTP) and stabilizes its binding; the A-site metal ion (Me2+A) lies between the primer end and the incoming dNTP and aligns the primer 3′-OH with the substrate α-phosphate to promote primer 3′-OH deprotonation and nucleophilic attack; following the binding of dNTP and the Me2+A and Me2+B, the C-site metal ion (Me2+C) binds between the α- and β-phosphates on the other side of the active site and drives α-β-phosphate bond breakage. Following product formation in A-, B- and some X-family DNA polymerases (7, 8, 9, 10, 11, 12, 13, 14), the primer terminus sugar pucker remains in a C3′-endo conformation to avoid steric clashes. For Y-family polymerases like DNA polymerase η (Pol η) (15), the primer terminus sugar pucker changes from a C3′-endo conformation to a C2′-endo conformation to avoid steric clashes with the nonbridging oxygen on the incoming nucleotide. Concurrently, the metal ions and pyrophosphate dissociate from the polymerase active site, and the newly synthesized primer end translocates out of the nucleotide insertion site for the next round of dNTP incorporation.
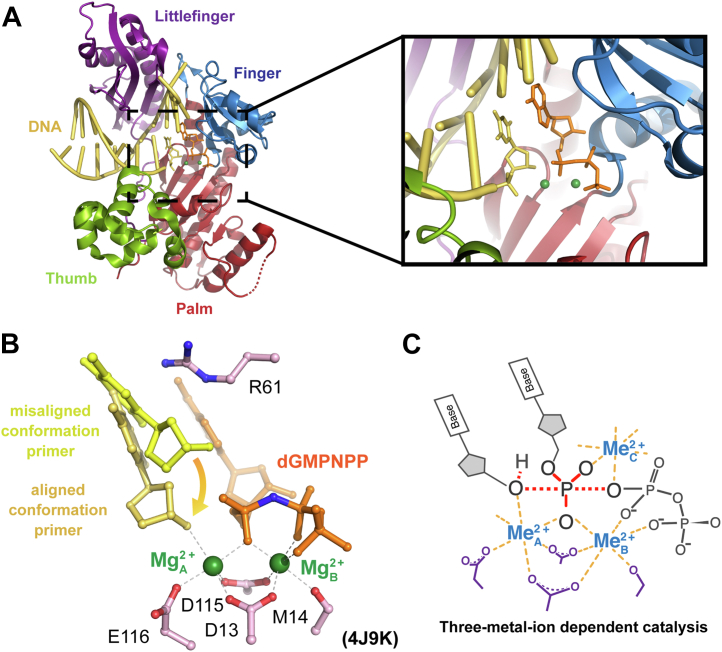
Figure 1.
DNA polymerase with bound DNA and substrate nucleotide. A, polymerase η with the DNA colored in yellow and the substrate nucleotide colored in orange. The Me2+A and Me2+B are colored in green in the zoom-in panel. The palm, finger, thumb, and little finger domains are colored in red, blue, green, and purple, respectively. B, stick representation of the primer terminus in yellow and yellow-orange, nucleotide in orange, the Me2+A and Me2+B in green, and protein residues in pink. C, scheme of the three-metal-ion dependent enzyme catalysis and transition state stabilization. Divalent metal ions optimally bind to their ligands with bond lengths of 2.0 to 2.2 Å at 90 ˚.
Genomic DNA constantly faces endogenous and environmental assaults. Although there exist proficient repair pathways, polymerases inevitably face DNA lesions while traveling along DNA. Such lesions may create bulky obstacles or alter base-pairing, pi-stacking, and the DNA backbone, promoting polymerase stalling and replication stress (16). Eukaryotic cells contain repair and translesion polymerases that can bypass various lesions (2). Different translesion mechanisms are used to bypass different lesions with varying chemical properties (2). DNA-embedded ribonucleotides (rNs) are the most common lesion and pose major threats to genome integrity (17, 18). A vast majority of rNs are incorporated during DNA replication by DNA polymerases δ and ε (19, 20). In addition, rNs are incorporated by Pol α as primers during Okazaki fragment formation (21). During nonhomologous end joining or base excision repair, rN incorporation by X-family polymerases facilitates downstream ligation (22, 23). Having a OH group on the 2′ carbon (Fig. 2A), DNA embedded rNs can lead to nicks on the phosphate backbone and the accumulation of nonsynonymous mutations (20, 24). Although rN incorporation does not significantly affect the processivity of Pol η, Pol α, Pol δ, and Pol ε (25, 26), rNs, if left incorporated on the nuclear or mitochondria template strand, can lead to error-prone impairment and stalling of the replication and transcription machinery (26, 27, 28, 29). Because of their lethal nature, the cell has evolved two specific pathways to remove DNA-embedded rNs. The first involves ribonuclease H2 (RNaseH2), which nicks at the rN site for error-free ribonucleotide excision repair (30, 31). The second involves topoisomerase I, which can lead to small DNA deletions (24, 32, 33, 34). In addition, mismatch repair has been implicated as an alternative pathway in rN removal (35), but the mechanism of rN recognition is unclear.
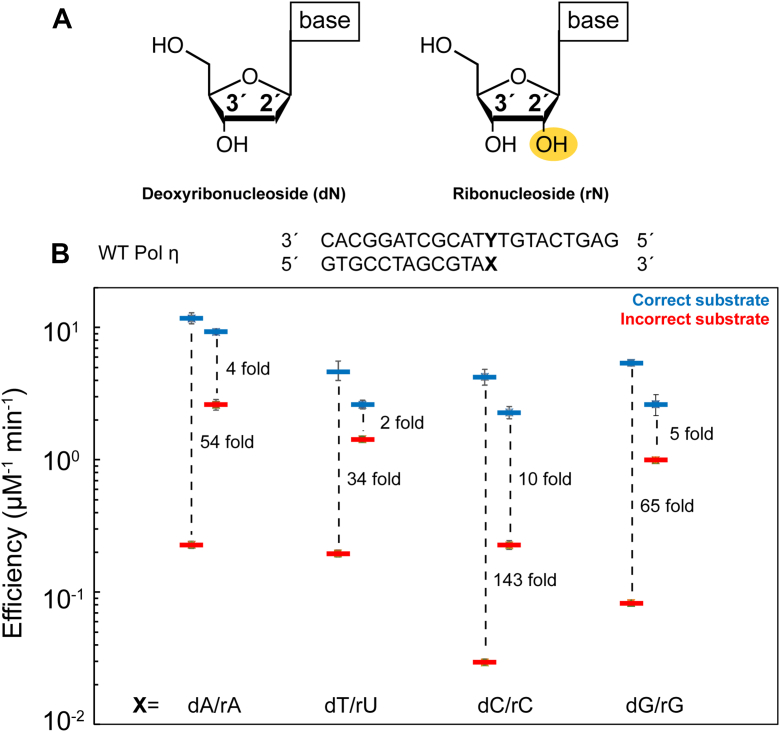
Figure 2.
The ratio of correct versus incorrect nucleotide incorporation efficiency decreases during ribonucleotide extension by DNA polymerase η. A, chemical representation of deoxyribonucleoside (dN) and ribonucleoside (rN). Hydrogens on the sugar moieties have been removed for clarity. B, WT Pol η DNA polymerase correct versus incorrect nucleotide incorporation efficiencies during dN or rN extension in the presence of Mg2+ based on steady-state kinetics. The bars represent the mean of triplicate measurements for the catalytic efficiencies (kcat/KM) for incorporation of dATP (blue) and dGTP (red) opposite dT for WT Pol η. The errors bars represent the standard deviation for the measurements. The distance between the respective catalytic efficiencies represents a ratio of the efficiencies and is a measure of discrimination. Data are generated from Table S1. dGTP, deoxyribose guanine triphosphate; Pol η, DNA polymerase η.
The structural basis of rN incorporation by DNA polymerases is well understood. A steric gate for discriminating the 2′-OH group of rN was first proposed in 1995 and 1997 (36, 37). In A-family (38), B-family (39, 40), and Y-family polymerases (41), a steric gate formed by tryptophan and phenylalanine residues sterically clashes with the hydroxyl group on the 2′ carbon of ribonucleotide triphosphates (rNTPs) and prevents rNTP binding. As a consequence, the A-family Klenow fragment polymerase, B-family RB69, and Y-family pol η discriminate rNTP incorporation by 3400- (42), 6400- (39), and 770- to 3400-folds (25), respectively. X-family polymerases such as DNA polymerase μ, β, and λ contain only a peptide backbone in place of a steric gate. With a tyrosine backbone in place of this steric gate, Pol β and Pol λ discriminate rNTPs by 2000- to 8200-fold (43) and 4000-fold (44), respectively. On the other hand, Pol μ which contains a glycine backbone instead, prefers to incorporate dNTP over rNTP by only 2-fold (45). During DNA replication, if a rN does get incorporated, DNA synthesis extending from the rN can still occur because the rN contains a functional 3′-OH group. Recently, it was revealed that the catalytic rate of DNA polymerase ε drops 3300-fold during rN extension compared to deoxynucleotide (dN) extension (46), hinting the involvement of translesion polymerases in rN extension. Kinetic analysis of Pol β revealed insignificant changes in catalytic efficiency (Kcat/KM) between dN and rN extension (43). Structural studies of DNA polymerase λ showed minimal differences when the 3′-end of primer was a dN versus a rN during correct nucleotide incorporation (47). Despite such efforts, whether and how the 2′-OH at the primer end affects the structure and dynamics of the active site as well as polymerase catalysis and fidelity are not fully explored.
Pol η is a Y-family translesion polymerase responsible for bypassing cyclobutane pyrimidine dimers (48, 49). Additionally, Pol η has been implicated in translesion DNA synthesis against a variety of lesions (2). People with mutations on the Pol η gene develop a predisposition for skin cancer and xeroderma pigmentosum (50, 51). Furthermore, Pol η widely participates during lagging strand synthesis (52) and has the ability to incorporate rNs and exhibits reverse transcriptase activity on RNA:DNA duplexes, although the biological role of the latter remains unclear (53). Pol η has been used as a model system for investigating mechanisms of polymerase catalysis. Kinetic studies have revealed that Pol η follows a similar kinetic pathway to other polymerases (54). The reaction process of Pol η was captured at atomic resolution with recent time-resolved crystallography (3, 4, 5, 55, 56, 57). By tracking the DNA synthesis by Pol η in crystallo, we have showed that the Me2+C binds between the α- and β-phosphates on the opposite side of Me2+A and Me2+B and plays an essential role in driving α-β-phosphate bond breakage. For Pol η, the primer terminus sugar pucker changes from a C3′-endo conformation to a C2′-endo conformation to avoid steric clashes with the nonbridging oxygen on the incoming nucleotide (15). In addition, primer alignment by Me2+A is perturbated during misincorporation and contributes to intrinsic polymerase fidelity (5). Moreover, structural snapshots of Pol η bypassing various lesions such as cyclobutene pyrimidine dimers, 8,5′-cyclo-2′-deoxyadenosine, phenanthriplatin, and cisplatin were captured for illustrating mechanisms of translesion synthesis (49, 58, 59, 60). Bypassing a cyclobutene pyrimidine dimer and cisplatin was promoted by finger domain movement that helped minimize DNA changes in pi-stacking and alignment. On the other hand, perturbations in primer-substrate alignment prevented Pol η-mediated bypass of 8,5′-cyclo-2′-deoxyadenosine and phenanthriplatin on the DNA template. Thus, we sought to use the Pol η system to investigate the consequences of rN extension at atomic resolution.
Here, we present biochemical and structural studies of Pol η extending rNs. Steady-state kinetic data on correct and incorrect single-rN extension suggest that Pol η can extend rNs with high efficiency. Interestingly, the rN primer end significantly decreases substrate discrimination. Corresponding crystal structures of Pol η complexed with both Mg2+ and Mn2+ and single rN-primed DNA substrate suggest that having a rN at the primer terminus stabilizes it in a productive conformation for nucleophilic attack. Furthermore, we compare the misincorporation process extending from ribose uridine (rU) and deoxyribose thymine (dT) primed DNA substrate with time-resolved crystallography. The results further confirm that the decreased Pol η fidelity during rN extension is due to the stabilization of the active aligned conformation during nucleophilic attack.
Results
Kinetics and misincorporation efficiencies of rN extension by pol η
During the DNA synthesis reaction, the primer 3′-OH aligns with the α-phosphate to initiate the nucleophilic attack. It was recently revealed that primer 3′-OH alignment promoted by Me2+A is the key step in substrate discrimination (5, 61). Interestingly, Gregory et al. captured the rN primer terminus in the aligned conformation in the absence of the Me2+A (57), suggesting the 2′-OH group alters conformation of the sugar ring and possibly facilitates the nucleophilic attack. We thus hypothesized that a rN at the 3′-primer end may promote primer 3′-OH alignment and stimulate incorrect nucleotide incorporation. Steady-state kinetic assays of Pol η with native and single-rN primed DNA substrate were conducted to detect for changes in misincorporation efficiencies. The correct nucleotide incorporation efficiency during rN extension was 1.3 to 2-fold lower than during dN extension (Fig. 2B and Table S1). However, misincorporation efficiency from a rN primer was enhanced by over 10-fold compared to dN extension. Previous studies suggested that the misincorporation efficiencies of Pol η is sequence dependent and decreases when extending from deoxyribose adenine (dA) and dT, also known as the WA (W, A or T) motif (62). We thus also examined DNA substrates with different terminal nucleotides. Similar as reported, Pol η catalyzed DNA synthesis with different misincorporation efficiencies on substrates with different primer termini, with deoxyribose cytosine (dC) as the highest and dA/dT as the lowest. For all sequence context, substrate discrimination dropped over 10-fold with a rN primer end. Notably for rU extension, correct incorporation was only twice more efficient than misincorporation during rN extension.
Misincorporation efficiencies of S113 A pol η
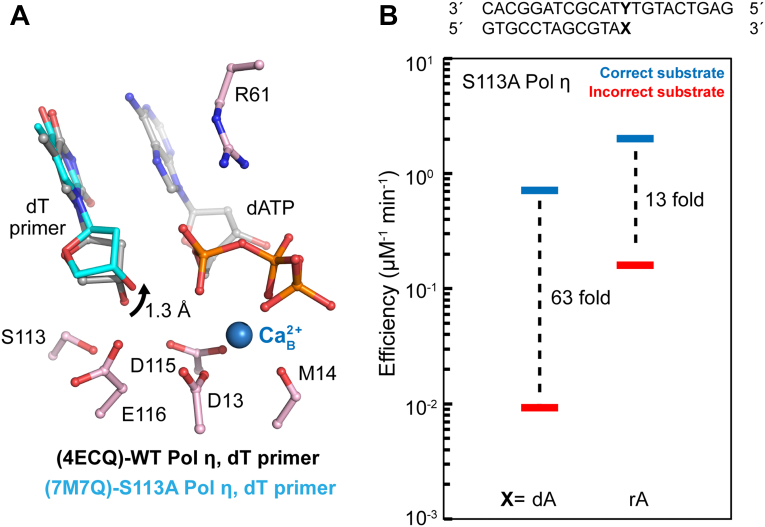
Previous crystal structures of Pol η showed that the primer is coordinated in the down aligned conformation by serine 113 in the active site before Me2+A binding (Fig. 3A) (3, 57). This widely conserved serine among Y-family polymerases is important for primer alignment. We investigated whether a S113 A Pol η mutant would perturb primer alignment and alter polymerase extension. With a dN primer end, the S113 A mutant catalyzed DNA synthesis with 6-fold lower KM and 3-fold lower kcat compared to the WT (Table S2), consistent with previous studies (3). Misincorporation of deoxyribose guanine triphosphate (dGTP) over dT template affected both KM and Kcat, resulting in a 63-fold drop in catalytic efficiency compared to correct dATP incorporation over dT (Table S2). A ribose adenine (rA)-ended primer increased the catalytic efficiency of correct dATP incorporation around 3-fold but stimulated incorrect dGTP incorporation around 16-fold, resulting in a 5-fold change in misincorporation efficiency. The stimulation of the misincorporation by rA-ended primer is mainly due to the elevated kcat (Fig. 3B and Table S2). Our kinetic studies were consistent with previous studies, where rA at the primer termini overrides the effect of S113 to help in aligning the primer for nucleophilic attack. The results further confirmed the significant role of primer alignment in fidelity control and the conformation selection of the rN primer end.
Figure 3.
Mechanisms of S113 A polymerase η catalysis and misincorporation. A, WT Pol η ground state with the dT primer terminus close to S113 (PDB ID 4ECQ). The primer 3′-OH is slightly too low or close to S113 to be in an aligned conformation. The angle of the primer 3′ oxygen, substrate phosphorus, and its bridging oxygen are at 160˚. dT primer terminus close to S113 (light blue) in the S113 A Pol η ground state (PDB ID 7M7Q). The dT primer terminus moves 1.3 A toward the misaligned conformation when an alanine is substituted in place of S113. B, S113 A Pol η DNA polymerase correct and incorrect substrate incorporation efficiency during dN or rN extension in the presence of Mg2+ based on steady-state kinetics. The fold-change between correct and incorrect substrate incorporation efficiencies represents a measurement of substrate discrimination. The bars represent the mean of triplicate measurements for the catalytic efficiencies (kcat/KM) for incorporation of dATP (blue) and dGTP (red) opposite dT. Data are generated from Table S2. dGTP, deoxyribose guanine triphosphate; dN, deoxynucleotide; Pol η, DNA polymerase η; rN, ribonucleotide; dT, deoxyribose thymine.
Binary complex of pol η with dN and rN ended primer
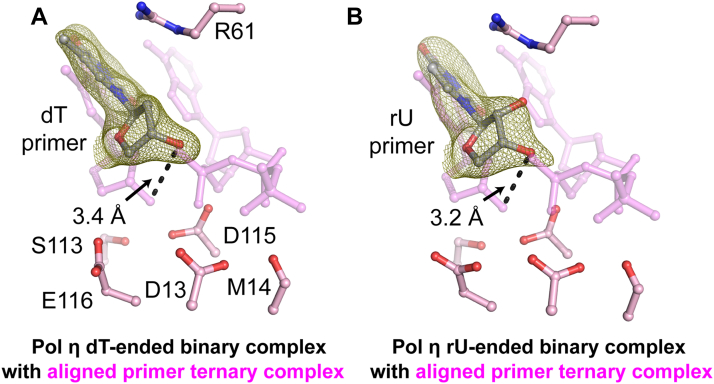
To test how the rN primer end would affect primer alignment, we first obtained crystals of Pol η in the absence of bound incoming nucleotide. We captured the binary state of Pol η complexed with dT-ended DNA at 1.75 Å resolution and rU-ended DNA at 2.1 Å resolution (Fig. 4). Both structures were similar to the previous ternary complex (RMSD 0.2 and 0.3, respectively) and binary complex (RMSD 0.3 and 0.4, respectively), confirming that there are no significant conformational changes during incoming nucleotide binding (63). The dT and rU structures are almost identical. In addition, the sugar ring conformations were the same. Compared to the ternary complex, the sugar ring for both primers termini in the binary complex is in a C2′ endo geometry. The 3′-OH group exists in the misaligned up conformation with the substrate phosphate 4.4 Å and 3.5 Å relative to the aligned 3′-OH group and stabilized by R61 through hydrogen bonds within the dT and rU structures, respectively. These binary structures suggested that the primer 3′-end with either dN or rN is not aligned in the absence of the incoming dNTP and Me2+.
Figure 4.
Pol η primer terminus alignment upon favorable substrate binding. A, B, binary structure of Pol η complexed with dT-primed in A or rU-primed DNA in B. Superimposition of binary (DNA only) and ternary (DNA with substrate nucleotide 4ECR) structures (pink) during dT extension during correct nucleotide incorporation in A, or rU extension during incorrect incorporation in B indicates primer movement upon substrate binding. The Fo-Fc omit map for the primer (green) was contoured at 3 σ (σ values represent r.m.s. density values). dT, deoxyribose thymine; rU, ribose uridine; Pol η, DNA polymerase η.
Structures of pol η misincorporation complex with rN at the primer terminus
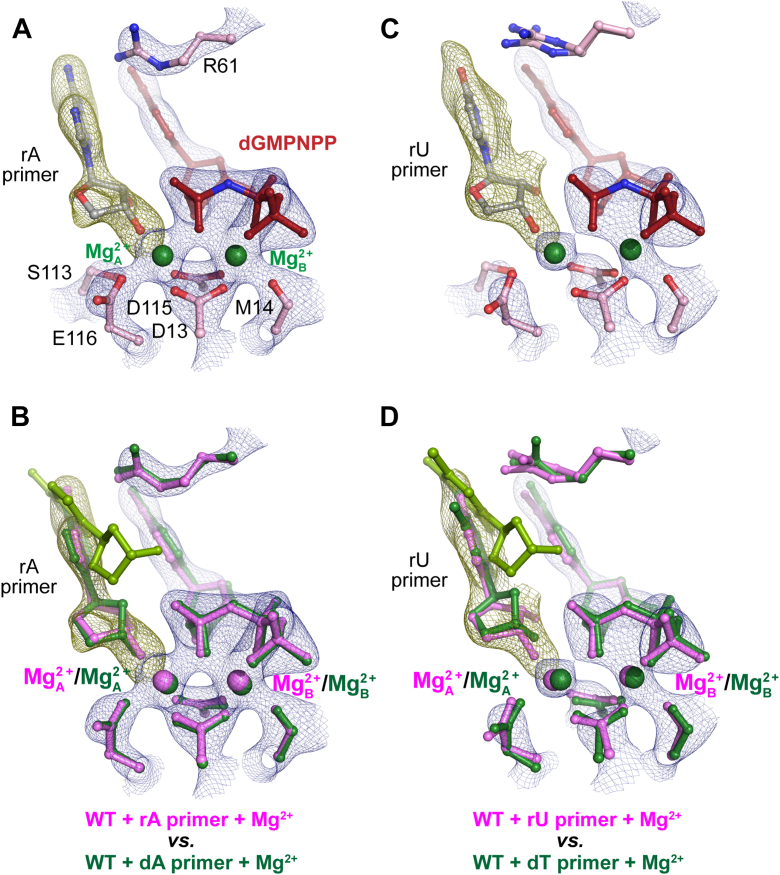
We further investigated how a rN primer end affects primer alignment by capturing the ternary misincorporation complex. To prevent catalysis, we prepared ternary Pol η structures with rN-ended primers complexed with 2′-deoxyguanosine-5′-[(α,β)-imido]triphosphate (dGMPNPP). We determined structures of Pol η with rA, rU, rC, ribose guanine (rG) at the primer terminus. In the rA structure, the primer is 100% aligned even with the incorrect substrate, in contrast to only 25% in the dA structure (Fig. 5, A and B) (5). It is interesting to note that for the rA primed structure, the sugar pucker of the rA base was already in a C3′endo conformation, as opposed to that of the dA structure, which was in a C2′ endo conformation (Figure 5, Figure 6). Similarly, in the rU structure, 100% of the rU primer existed in a C3′endo aligned down conformation (Fig. 5, C and D). In contrast, the primer termini in the rC and rG structures existed in the misaligned up conformation, similar to the structures with dC and deoxyribose guanine (dG) (Fig. 7, A and B and Fig. S1) (62). The decreased efficiency of bypass mediated by perturbations in primer termini alignment has also been observed during 8,5′-cyclo-2′-deoxyadenosine and phenanthriplatin bypass (49, 58, 59). Interestingly, all of our rN primed structures showed minimal changes in base-pairing and planar geometries similar to what has been observed during Pol η-mediated cyclobutene pyrimidine dimer and cisplatin bypass (Figs. S2 and S3) (49, 60). In contrast, during phenanthriplatin bypass weaker pi-stacking interactions between a template phenanthriplatin and the incoming nucleotide were suggested to explain the 6-fold drop in nucleotide binding.
Figure 5.
Structural comparison between reactant states of incorrect nucleotide incorporation during dN or rN extension show increased inclination of primer terminus alignment during rA or rU extension misincorporation. A,C, structure of rA extension (A) and rU extension (C) WT Pol η mismatch reactant state complexed with Mg2+. B, D, structural overlay of dN (green) (PDB ID 4J9M and 4J9K) or rN (pink) extension structures with dGMPNPP:dT base-pair indicating differences in 3′-OH alignment. The misaligned primer terminus in the dN structure (green) is colored with yellow green. All electron density maps apply to the molecule colored in pink. The 2Fo-Fc map for everything including Me2+A and Me2+B, dGMPNPP, and catalytic residues and S113 (blue) was contoured at 2 σ. The Fo-Fc omit map for the primer terminus (green) was contoured at 3.5 σ in A, B, and 2.7 σ in C, D. rU, ribose uridine; dN, deoxynucleotide; Pol η, DNA polymerase η; rN, ribonucleotide; dT, deoxyribose thymine.
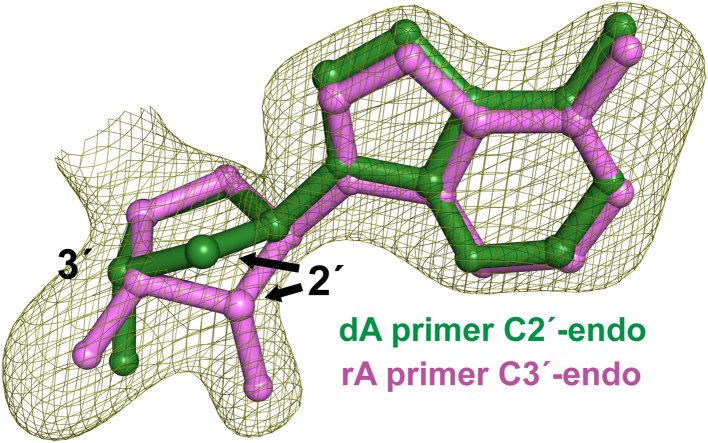
Figure 6.
Sugar pucker geometry comparison between primer terminus of dN versus rN extension. The Fo-Fc omit map for the pink rA primer terminus structure was contoured at 3.5 σ. dN, deoxynucleotide; rN, ribonucleotide.
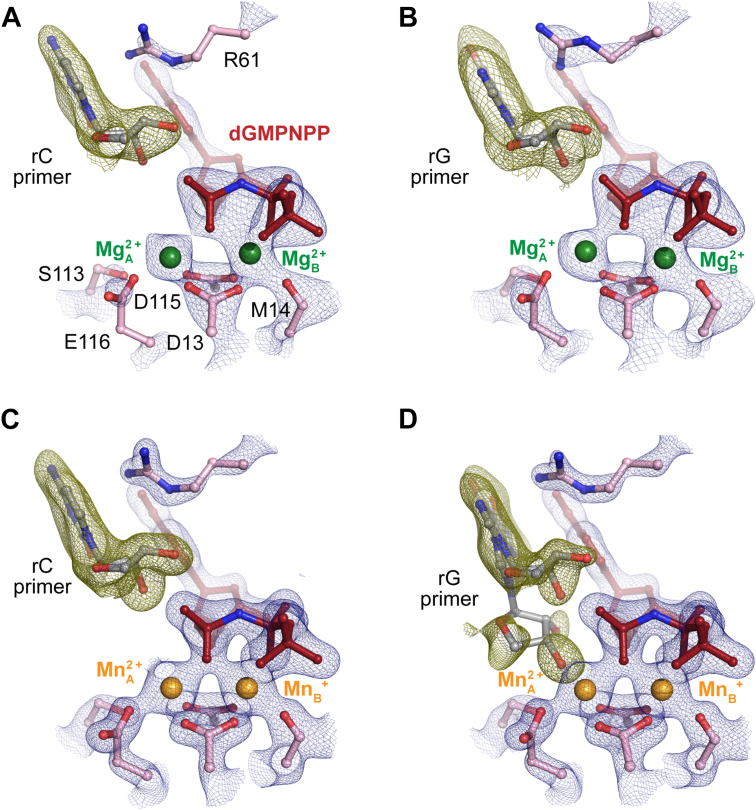
Figure 7.
Structural comparison between reactant states of incorrect nucleotide incorporation during rC and rG extension.A,B, structure of rC extension (A) and rG extension (B) misincorporation with Mg2+ by WT Pol η. C, D, structure of rC extension (C) and rG extension (D) misincorporation with Mn2+ by Pol η. The 2Fo-Fc map for everything including Me2+A and Me2+B, dGMPNPP, and catalytic residues and S113 (blue) was contoured at 2 σ. The Fo-Fc omit map for the primer terminus (green) was contoured at 3.8 σ in A and C and 2.0 σ in B and D. Pol η, DNA polymerase η; rG, ribose guanine.
We suspect that the primer end is in dynamic equilibrium between the misaligned and aligned conformations as observed in previous in crystallo studies (5, 61). The occupancy might be too low for the aligned conformation of rC and rG structures to be refined at the current resolution. Because Mn2+ has been shown to improve primer alignment, we captured the same rC and rG-ended Pol η mismatch structures with Mn2+. The rG primed structure with Mn2+ showed increased presence of the primer terminus in the down aligned conformation (from 0% to 30%) (Fig. 7C and Fig. S1). The sugar pucker of the down conformation was also in C3′endo configuration. In contrast, for the structures with rC at the primer terminus, the primer remained in the up misaligned conformation, consistent with the kinetic studies, showing dC/rC-primed DNA with the highest incorrect nucleotide discrimination (Fig. 7D and Fig. S1).
In crystallo rN extension of pol η
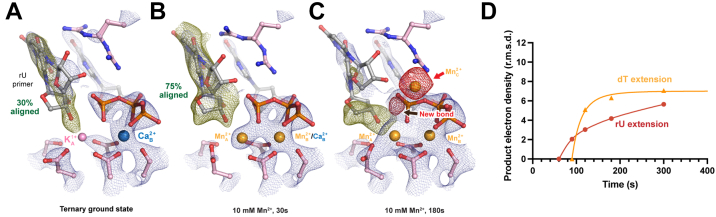
Previous studies have shown that nonhydrolyzable nucleotide analogs such as dNMPNPPs may impede primer alignment (5). Thus, we visualized the misincorporation process of dGTP across dT in the presence of Mn2+ from a single uridine primed DNA substrate. We crystalized Pol η complexed with dATP, Ca2+, K1+ to prevent the reaction. In this ground state, 30% of the rU primer termini existed in the aligned down conformation and 3.6 Å away from the target α-phosphate (Fig. 8A). dGTP formed a wobble base-pair with the template dT. Similar to dT extension (5), 70% of R61 in the ground state was already flipped away from the triphosphate motif of the incoming nucleotide to stabilize the wobble dG-dT base pair.
Figure 8.
In crystallo visualization of WT Pol η rU extension misincorporation with Mn2+.A–C, structures of WT Pol η during in crystallo catalysis after 10 mM Mn2+ soaking for 30s (B) and 180s (C). The 2Fo-Fc map for the primer terminus up conformation (blue) was contoured at 1.2 σ. The 2Fo-Fc map for the Me2+A, Me2+B, dGTP, and catalytic residues (blue) was contoured at 2.0 in A, 2.5 in B, and 2.0 σ in C. The Fo-Fc omit map for the down conformation of the primer terminus (green) was contoured at 3 σ in A–C. The Fo-Fc omit map for all the Mn2+C, and newly formed bond (red) was contoured at 3.5 σ. D, timescale of reaction product of Pol η rU extension misincorporation in crystallo with Mn2+. Pol η, DNA polymerase η; rU, ribose uridine; dGTP, deoxyribose guanine triphosphate.
To track the reaction process, we have determined six structures of Pol η soaking in 10 mM Mn2+ for 30 to 300s (Table S3). Mn2+ was chosen over Mg2+ due to the higher occupancies of the incoming nucleotide and its better signal in X-ray diffraction. The resolution of these structures are similar with different soaking time, ranging from 2.05 to 2.2 Å. After 30s soaking in 10 mM Mn2+, 70% of the Me2+A was already saturated with Mn2+, and 75% of the rU primer had moved down in the aligned conformation, residing 3.6 Å away from the target α-phosphate (Fig. 8B). In comparison, after 30s soaking with a dT-ended primer, 65% of the Me2+A was saturated with Mn2+, and 40% of the thymine primer was aligned for misincorporation. The Me2+A during rU extension was assigned with 70% occupancy and was in an optimal octahedral geometry. Despite the improvement in alignment, the 3′-OH of rU resided 2.6 Å away from the Me2+A, 0.5 Å further compared to that during correct incorporation. After 180s of soaking, clear electron density for newly formed bond was visible and assigned to 50% between the rU 3′-OH and target α-phosphate (Fig. 8, C and D). Unlike during dT extension in which the Me2+C appeared before product formation, the Me2+C here appeared simultaneously with product formation.
Discussion
Many efforts have investigated the kinetic and structural mechanism surrounding polymerase fidelity (64, 65, 66, 67, 68, 69, 70, 71, 72, 73). It was proposed that the exonuclease proofreading and the finger domains' conformational changes (64, 65) play significant roles in substrate discrimination. However, polymerases that lack proofreading exonuclease and finger domain conformational changes still incorporate correct bases versus incorrect bases more efficiently than what can be provided by Watson–Crick base pairing melting energies, which has been estimated to be only ∼0.3 kcal/mol (2, 66, 69, 74, 75, 76). More recently, studies of Pol β and Pol η have shown that primer alignment contributes to the intrinsic polymerase incorrect nucleotide discrimination and is perturbed during misincorporation (5, 61). Here, we show that during rN extension, misincorporation efficiency increases over 10-fold (Fig. 2B). Systematic structural studies confirmed that this rise in misincorporation is likely due to improved primer alignment in the presence of the incorrect incoming nucleotide (Figure 5, Figure 7, Figure 8). Pol η misincorporation is sequence dependent, with dA/dT at the primer end showing higher misincorporation and dC with lower misincorporation (62). The wobble base pair during rN extension looked identical to the previous reported dN extension mismatch structures, suggesting similar pi-stacking interactions (62). Consistent with the trend of misincorporation, the rA and rU primer ends are better aligned compared to rC (Figure 5, Figure 7). These multiple levels of correlation of primer end alignment and misincorporation highlighted the critical role of primer alignment in polymerase substrate discrimination control. All of our binary, ternary, and in crystallo reaction structures were determined in the same space group with similar parameters, and the observed difference in primer alignment was not likely influenced by the crystal lattice.
During Pol η-promoted DNA synthesis, the primer terminus overcomes a C2′-endo to C3′-endo barrier (3) to avoid steric clashes with the nonbridging oxygen of the incoming nucleotide. Our structures suggested that an extra 2′-OH on the sugar motif like on a rN affects sugar pucker conformation. The rN primer termini are more readily in the C3′-endo form in the aligned conformation before product formation (Fig. 6). In addition, this suggests weaker barriers for product formation compared to DNA extension. Minimal steric clashes of rNs at the penultimate primer (P-2) position in A-family Pol I, B-family RB69, X-family Pol β, and Y-family Pol η may suggest that the C3′-endo conformation is preferred as the product form during rN incorporation (Fig. S4). The combination of improved primer alignment and weaker C2′-endo to C3′-endo sugar pucker conversion might explain the changes in substrate discrimination by Pol η. Many nucleoside analog drugs have modifications on the 3′ and 2′ carbon of the sugar ring and inhibit DNA synthesis at the extension step (77). Cytarabine, which is similar to dC but has a 2′-OH in the β direction, is an effective chain terminator and a drug for leukemia treatment (77, 78, 79, 80). Elevated Pol η expression has been found to help bypass cytarabine (81, 82). Structural studies show that cytarabine exists as a C2′-endo at the primer terminus even during correct nucleotide incorporation (83). The altered sugar ring conformation might increase barriers in C2′-endo to C3′-endo conversion and thus also inhibit polymerase extension (83, 84). Similarly, nucleoside analog drugs such as entecavir and galidesivir contain C2′-modifications and possibly exert their inhibitory effect through similar mechanisms.
Our study on rN extension suggests that the 2′-OH significantly affects polymerase misincorporation. As Me2+A-mediated primer alignment is a required step in DNA synthesis, we hypothesize that the observed elevated incorporation error rate might be a universal property for all polymerases. However, polymerases from different families may have evolved specific structural features to discriminate the 2′-OH at the primer end. Although Pol η and X-family polymerases can extend past rN primers with high efficiency, Pol ε efficiency decreases over 3300-fold during rN extension (46). Further biochemical and structural studies of different polymerases may be needed to clarify the effect of the 2′-OH group on catalysis, misincorporation, and primer extension. The reduced efficiency of Pol ε in extending rN primer might indicate involvement of translesion polymerases in extending rN primers (5, 61).
Experimental procedures
Protein expression and purification
Wildtype human polymerase η (Pol η) (residues 1–432) was cloned into a modified pET28p vector with a N-terminal 6-histidine tag and a PreScission Protease cleavage site as described (56). For protein expression, this Pol η plasmid was transformed into BL21 DE3 Escherichia coli cells. When the absorbance of the E. coli cells reached 0.8, isopropyl ß-D-1-thiogalactopyranoside was added to a final concentration of 1 μM isopropyl ß-D-1-thiogalactopyranoside. After 20 h (16 ˚C) of induction, the cell paste was collected via centrifugation and re-suspended in a buffer that contained 20 mM Tris (pH 7.5), 1 M NaCl, 20 mM imidazole, and 5 mM ß-mercaptoethanol. After sonification, Pol η was loaded onto a HisTrap HP column (GE Healthcare), which was pre-equilibrated with a buffer that contained 20 mM Tris (pH 7.5), 1 M NaCl, 20 mM imidazole, and 5 mM ß-mercaptoethanol. The column was washed with 300 ml of buffer to remove nonspecific–bound proteins and was eluted with buffer that contained 20 mM Tris (pH 7.5), 1 M NaCl, 300 mM imidazole, and 3 mM dithiothreitol (DTT). The eluted Pol η was incubated with PreScission Protease to cleave the N-terminal 6-histidine-tag. Afterwards, Pol η was buffer-exchanged and desalted to 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.0), 250 mM KCl, 10% glycerol, 0.1 mM ethylenediaminetetraacetic acid, and 3 mM DTT and was loaded onto a MonoS 10/100 column (GE Healthcare). The protein was eluted with an increasing salt (KCl) gradient. Finally, Pol η was cleaned with a Superdex 200 10/300 Gl column (GE Healthcare) with a buffer that contained 20 mM Tris (pH 7.5), 450 mM KCl, and 3 mM DTT.
DNA synthesis assay
The nucleotide incorporation activity was assayed by the following: The reaction mixture contained 1.3 to 180 nM Pol η (WT or S113 A), 5 μM DNA, 0 to 400 μM dNTP (either dATP or dGTP), 100 mM KCl, 50 mM Tris (pH 7.5), 5 mM MgCl2, 3 mM DTT, 0.1 mg/ml bovine serum albumin, and 4% glycerol. The incorporation assays were executed using DNA template and 5′-fluorescein–labeled primer listed in Table 1. Reactions were conducted at 37 °C for 5 min and were stopped by adding formamide quench buffer to the final concentrations of 40% formamide, 50 mM ethylenediaminetetraacetic acid (pH 8.0), 0.1 mg/ml xylene cyanol, and 0.1 mg/ml bromophenol. After heating to 97 °C for 5 min and immediately placing on ice, reaction products were resolved on 22.5% polyacrylamide urea gels. The gels were visualized by a Sapphire Biomolecular Imager and quantified using the built-in software. Quantification of Kcat, KM, VMax and fitting and graphic representation were executed by Graph Prism. Source data of urea gels are provided as a Source Data file.
Table 1.
Kinetic assay DNA sequences
| DNA sequence N = DNA N = RNA | Name |
|---|---|
| 3′CACGGATCGCATTTGTACTGAG5′ 5′ GTGCCTAGCGTAA 3′ |
dA primed |
| 3′CACGGATCGCATATGTACTGAG5′ 5′ GTGCCTAGCGTAT 3′ |
dT primed |
| 3′CACGGATCGCATGTGTACTGAG5′ 5′ GTGCCTAGCGTAC 3′ |
dC primed |
| 3′CACGGATCGCATCTGTACTGAG5′ 5′ GTGCCTAGCGTAG 3′ |
dG primed |
| 3′CACGGATCGCATTTGTACTGAG5′ 5′ GTGCCTAGCGTAA 3′ |
rA primed |
| 3′CACGGATCGCATATGTACTGAG5′ 5′ GTGCCTAGCGTAU 3′ |
rU primed |
| 3′CACGGATCGCATGTGTACTGAG 5′ 5′ GTGCCTAGCGTAC 3′ |
rC primed |
| 3′CACGGATCGCATCTGTACTGAG5′ 5′ GTGCCTAGCGTAG 3′ |
rG primed |
Crystallization
Pol η was concentrated to 300 μM in buffer that contained 20 mM Tris (pH 7.5), 0.45 M KCl, and 3 mM DTT. Then DNA, dGTP or dGMPNPP, and Ca2+ and low salt buffer [20 mM Tris (pH 7.5), and 3 mM DTT] were added to this polymerase solution at the molar ratio of 1 : 1.2: 1: 1 for Pol η, DNA, dGTP or dGMPNPP, and Ca2+, bringing Pol η′s concentration to 100 μM. Then after this solution was kept on ice for 10 min, more dGTP or dGMPNPP was added to a final concentration of 0.5 mM. DNA template and primer used for crystallization are listed in Table 2. All crystals were obtained using the hanging-drop vapor-diffusion method against a reservoir solution containing 0.1 M MES (pH 6.0) and 9 to 15% (w/v) PEG2K-MME at room temperature within 4 days.
Table 2.
Crystallization DNA sequences
| DNA sequence N = DNA N = RNA | Name |
|---|---|
| 3′ TCGCAGTTTTAC 5′ 5′ AGCGTCAA 3′ |
rA primed |
| 3′ TCGCAGTATTAC 5′ 5′ AGCGTCAU 3′ |
rU primed |
| 3′ TCGCAGTGTTAC 5′ 5′ AGCGTCAC 3′ |
rC primed |
| 3′ TCGCAGTCTTAC 5′ 5′ AGCGTCAG 3′ |
rG primed |
| 3′ TCGCAGTGTTAC 5′ 5′ AGCGTCAC 3′ |
dC primed |
| 3′ TCGCAGTCTTAC 5′ 5′ AGCGTCAG 3′ |
dG primed |
Chemical reaction in crystallo
The crystals were first transferred and incubated in a prereaction buffer containing 0.1 M MES (pH 7.0, titrated by KOH), 100 μM dGTP, and 20% (w/v) PEG2K-MME for 30 min. The chemical reaction was initiated by transferring the crystals into a reaction buffer containing 0.1 M MES (pH 7.0), 20% (w/v) PEG2K-MME, and 10 mM MnCl2. After incubation for a desired time period, the crystals were quickly dipped in a cryo-solution supplemented with 20% (w/v) glycerol and flash-cooled in liquid nitrogen.
Data collection and refinement
Diffraction data were collected at 100 K on LS-CAT beam lines 21-D-D, 21-ID-F, and 21-ID-G at the Advanced Photon Source (Argonne National Laboratory). Data were indexed in space group P61, scaled, and reduced using XDS (85). Isomorphous Pol η structures with Mg2+ PDB: was used as initial models for refinement using PHENIX (86) and COOT (87). Initial occupancies were assigned for the substrate, reaction product, PPi, Me2+A, Me2+B, and Me2+C, for the ternary ground state, following the previous protocol (4). After there were no significant Fo-Fc peaks and each atom’s B value had roughly similar values to its ligand, we assigned occupancies for the same regions for the timepoints in between (Figs. S5–S7). Occupancies were assigned to the misaligned and aligned conformations of the primer termini until there were no more significant Fo-Fc peaks. For the structures in which both primer conformations were at an equilibrium (not 100% in either misaligned or aligned), occupancies were assigned until the Fo-Fc peaks for both conformations (while they still remained) did not increase. In addition, for the structures in which some positive Fo-Fc peaks were present around the Me2+ binding sites or primer termini, no change in the assigned occupancy was executed when a 10% change in occupancy (e.g., 100% to 90%) failed to significantly change the intensity of the Fo-Fc peaks. Source data of the electron densities in r.m.s. density are provided as a Source Data file. Each structure was refined to the highest resolution data collected, which ranged between 1.75 and 2.2 Å. Software applications used in this project were compiled and configured by SBGrid (88). Source data of data collection and refinement statistics are summarized in Table S3, A–C. All structural figures were drawn using PyMOL (http://www.pymol.org).
Data availability
The coordinates, density maps, and structure factors for all the structures have been deposited in Protein Data Bank (PDB) under accession codes: 8E85, 8E86, 8E87, 8E88, 8E89, 8E8A, 8E8B, 8E8C, 8E8D, 8E8E, 8E8F, 8E8G, 8E8H, 8E8I, 8E8J, and 8E8K.
Supporting information
This article contains supporting information.
Conflict of interests
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Our sincere appreciation to the members of the Gao lab, and Drs. Phillips, Nikonowicz, and Lu who serve on CC’s thesis committee. We thank the APS LS-CAT beam technicians and research scientists Drs. Anderson, Wawrzak, Kondrashkina, and Focia. We thank Aaron Lin and Grace Zhou for critical reading of the manuscript. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Author contribution
Y. G. conceptualization; C. C. S. E., G. Z., C. L. L., and A. L. investigation; C. C. data curation; C. C. formal analysis; C. C. and Y. G. writing–original draft.
Funding and additional information
This work is supported by CPRIT (RR190046) and the Welch Foundation (C-2033–20200401) to Y.G. and a predoctoral fellowship from the Houston Area Molecular Biophysics Program (NIH Grant No. T32 GM008280, Program Director Dr. Theodore Wensel) to C.C.
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Supplementary information
References
- 1.Wu W.-J., Yang W., Tsai M.-D. How DNA polymerases catalyse replication and repair with contrasting fidelity. Nat. Rev. Chem. 2017;1:68. [Google Scholar]
- 2.Yang W., Gao Y. Translesion and repair DNA polymerases: diverse structure and mechanism. Annu. Rev. Biochem. 2018;87:239–261. doi: 10.1146/annurev-biochem-062917-012405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura T., Zhao Y., Yamagata Y., Hua Y.-j., Yang W. Watching DNA polymerase η make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y., Yang W. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016;352:1334–1337. doi: 10.1126/science.aad9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C., Lee Luo C., Gao Y. In crystallo observation of three metal ion promoted DNA polymerase misincorporation. Nat. Commun. 2022;13:2346. doi: 10.1038/s41467-022-30005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W., Weng P.J., Gao Y. A new paradigm of DNA synthesis: Three-metal-ion catalysis. Cell Biosci. 2016;6:51. doi: 10.1186/s13578-016-0118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doublié S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Korolev S., Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 10.Berman A.J., Kamtekar S., Goodman J.L., Lázaro J.M., de Vega M., Blanco L., et al. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson S.J., Taylor J.S., Beese L.S. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F., Yang W. Structural insight into translesion synthesis by DNA pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Diaz M., Bebenek K., Krahn J.M., Pedersen L.C., Kunkel T.A. Role of the catalytic metal during polymerization by DNA polymerase lambda. DNA Repair. 2007;6:1333–1340. doi: 10.1016/j.dnarep.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batra V.K., Beard W.A., Shock D.D., Krahn J.M., Pedersen L.C., Wilson S.H. Magnesium-Induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W., Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marians K.J. Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 2018;87:217–238. doi: 10.1146/annurev-biochem-062917-011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z.-X., Williams J.S., Lujan S.A., Kunkel T.A. Ribonucleotide incorporation into DNA during DNA replication and its consequences. Crit. Rev. Biochem. Mol. Biol. 2021;56:109–124. doi: 10.1080/10409238.2020.1869175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldecott K.W. Ribose—an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- 19.Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundström E.B., Burgers P.M., et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams J.S., Lujan S.A., Kunkel T.A. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 2016;17:350–363. doi: 10.1038/nrm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clausen A.R., Lujan S.A., Burkholder A.B., Orebaugh C.D., Williams J.S., Clausen M.F., et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pryor J.M., Conlin M.P., Carvajal-Garcia J., Luedeman M.E., Luthman A.J., Small G.W., et al. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science. 2018;361:1126–1129. doi: 10.1126/science.aat2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McElhinny S.A.N., Ramsden D.A. Polymerase Mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McElhinny S.A.N., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundström E.-B., et al. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Y., Egli M., Guengerich F.P. Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem. 2016;291:3747–3756. doi: 10.1074/jbc.M115.706226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausen A.R., Murray M.S., Passer A.R., Pedersen L.C., Kunkel T.A. Structure–function analysis of ribonucleotide bypass by B family DNA replicases. Proc. Natl. Acad. Sci. 2013;110:16802–16807. doi: 10.1073/pnas.1309119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh M., Posse V., Peter B., Falkenberg M., Gustafsson Claes M. Ribonucleotides embedded in template DNA impair mitochondrial RNA polymerase progression. Nucleic Acids Res. 2022;50:989–999. doi: 10.1093/nar/gkab1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clausen A.R., Zhang S., Burgers P.M., Lee M.Y., Kunkel T.A. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair. 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt D.L., Johansson E., Burgers P.M., Kunkel T.A. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair. 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparks J.L., Chon H., Cerritelli S.M., Kunkel T.A., Johansson E., Crouch R.J., et al. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Figiel M., Chon H., Cerritelli S.M., Cybulska M., Crouch R.J., Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutières syndrome defects. J. Biol. Chem. 2011;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams Jessica S., Smith Dana J., Marjavaara L., Lujan Scott A., Chabes A., Kunkel Thomas A. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekiguchi J., Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 34.Kim N., Huang S.N., Williams J.S., Li Y.C., Clark A.B., Cho J.E., et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghodgaonkar M.M., Lazzaro F., Olivera-Pimentel M., Artola-Borán M., Cejka P., Reijns M.A., et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao G., Orlova M., Georgiadis M.M., Hendrickson W.A., Goff S.P. Conferring RNA polymerase activity to a DNA polymerase: A single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. U. S. A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgiadis M.M., Jessen S.M., Ogata C.M., Telesnitsky A., Goff S.P., Hendrickson W.A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 38.Wang W., Wu E.Y., Hellinga H.W., Beese L.S. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J. Biol. Chem. 2012;287:28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang G., Franklin M., Li J., Lin T.C., Konigsberg W. A conserved tyr residue is required for sugar selectivity in a pol α DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 40.Bonnin A., Lázaro J.M., Blanco L., Salas M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase11Edited by A. R. Fersht. J. Mol. Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- 41.DeLucia A.M., Grindley N.D.F., Joyce C.M. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Astatke M., Ng K., Grindley N.D., Joyce C.M. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavanaugh N.A., Beard W.A., Wilson S.H. DNA polymerase beta ribonucleotide discrimination: Insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown J.A., Fiala K.A., Fowler J.D., Sherrer S.M., Newmister S.A., Duym W.W., et al. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon A.F., Pryor J.M., Ramsden D.A., Kunkel T.A., Bebenek K., Pedersen L.C. Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res. 2017;45:9138–9148. doi: 10.1093/nar/gkx527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lisova A.E., Baranovskiy A.G., Morstadt L.M., Babayeva N.D., Tahirov T.H. Efficient discrimination against RNA-containing primers by human DNA polymerase ε. Scientific Rep. 2022;12:10163. doi: 10.1038/s41598-022-14602-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gosavi R.A., Moon A.F., Kunkel T.A., Pedersen L.C., Bebenek K. The catalytic cycle for ribonucleotide incorporation by human DNA Pol λ. Nucleic Acids Res. 2012;40:7518–7527. doi: 10.1093/nar/gks413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masutani C., Kusumoto R., Iwai S., Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biertümpfel C., Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J.Y., et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Limoli C.L., Giedzinski E., Morgan W.F., Cleaver J.E. Polymerase η deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc. Natl. Acad. Sci. 2000;97:7939. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 52.Kreisel K., Engqvist M.K.M., Kalm J., Thompson L.J., Boström M., Navarrete C., et al. DNA polymerase η contributes to genome-wide lagging strand synthesis. Nucleic Acids Res. 2019;47:2425–2435. doi: 10.1093/nar/gky1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su Y., Egli M., Guengerich F.P. Human DNA polymerase η accommodates RNA for strand extension. J. Biol. Chem. 2017;292:18044–18051. doi: 10.1074/jbc.M117.809723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Washington M.T., Prakash L., Prakash S. Yeast DNA polymerase eta utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107:917–927. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 55.Yang W., Weng P.J., Gao Y. Erratum to: a new paradigm of DNA synthesis: Three-metal-ion catalysis. Cell Biosci. 2017;7:32. doi: 10.1186/s13578-017-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samara N.L., Gao Y., Wu J., Yang W. Detection of reaction intermediates in Mg2+-dependent DNA synthesis and RNA degradation by time-resolved X-ray crystallography. Methods Enzymol. 2017;592:283–327. doi: 10.1016/bs.mie.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory M.T., Gao Y., Cui Q., Yang W. Multiple deprotonation paths of the nucleophile 3′-OH in the DNA synthesis reaction. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2103990118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weng P.J., Gao Y., Gregory M.T., Wang P., Wang Y., Yang W. Bypassing a 8,5′-cyclo-2′-deoxyadenosine lesion by human DNA polymerase η at atomic resolution. Proc. Natl. Acad. Sci. 2018;115:10660. doi: 10.1073/pnas.1812856115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gregory M.T., Park G.Y., Johnstone T.C., Lee Y.-S., Yang W., Lippard S.J. Structural and mechanistic studies of polymerase η bypass of phenanthriplatin DNA damage. Proc. Natl. Acad. Sci. 2014;111:9133. doi: 10.1073/pnas.1405739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y., Biertümpfel C., Gregory M.T., Hua Y.J., Hanaoka F., Yang W. Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7269–7274. doi: 10.1073/pnas.1202681109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freudenthal Bret D., Beard William A., Shock David D., Wilson Samuel H. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y., Gregory M.T., Biertümpfel C., Hua Y.-J., Hanaoka F., Yang W. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase η. Proc. Natl. Acad. Sci. 2013;110:8146. doi: 10.1073/pnas.1303126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ummat A., Silverstein T.D., Jain R., Buku A., Johnson R.E., Prakash L., et al. Human DNA polymerase η is pre-aligned for dNTP binding and catalysis. J. Mol. Biol. 2012;415:627–634. doi: 10.1016/j.jmb.2011.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong I., Patel S.S., Johnson K.A. An induced-fit kinetic mechanism for DNA replication fidelity: Direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 65.Patel S.S., Wong I., Johnson K.A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 66.Tsai Y.-C., Johnson K.A. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson S.J., Beese L.S. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 68.Wu E.Y., Beese L.S. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an "ajar" intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rothwell P.J., Mitaksov V., Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 70.Sucato C.A., Upton T.G., Kashemirov B.A., Batra V.K., Martínek V., Xiang Y., et al. Modifying the beta,gamma leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase beta. Biochemistry. 2007;46:461–471. doi: 10.1021/bi061517b. [DOI] [PubMed] [Google Scholar]
- 71.Showalter A.K., Tsai M.-D. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 72.Arndt J.W., Gong W., Zhong X., Showalter A.K., Liu J., Dunlap C.A., et al. Insight into the catalytic mechanism of DNA polymerase β: structures of intermediate complexes. Biochemistry. 2001;40:5368–5375. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 73.Bakhtina M., Lee S., Wang Y., Dunlap C., Lamarche B., Tsai M.-D. Use of viscogens, dNTPαS, and rhodium(III) as probes in stopped-flow experiments to obtain new evidence for the mechanism of catalysis by DNA polymerase β. Biochemistry. 2005;44:5177–5187. doi: 10.1021/bi047664w. [DOI] [PubMed] [Google Scholar]
- 74.Oertell K., Harcourt E.M., Mohsen M.G., Petruska J., Kool E.T., Goodman M.F. Kinetic selection vs. free energy of DNA base pairing in control of polymerase fidelity. Proc. Natl. Acad. Sci. 2016;113:E2277. doi: 10.1073/pnas.1600279113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bebenek K., Kunkel T.A. Functions of DNA polymerases. Adv. Protein. Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 76.Dunlap C.A., Tsai M.-D. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase β †. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 77.Hewish M., Martin S.A., Elliott R., Cunningham D., Lord C.J., Ashworth A. Cytosine-based nucleoside analogs are selectively lethal to DNA mismatch repair-deficient tumour cells by enhancing levels of intracellular oxidative stress. Br. J. Cancer. 2013;108:983–992. doi: 10.1038/bjc.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lichtman M.A. A historical perspective on the development of the cytarabine (7days) and daunorubicin (3days) treatment regimen for acute myelogenous leukemia: 2013 the 40th anniversary of 7+3. Blood Cell Mol. Dis. 2013;50:119–130. doi: 10.1016/j.bcmd.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 79.Kadia T.M., Ravandi F., O'Brien S., Cortes J., Kantarjian H.M. Progress in acute Myeloid leukemia. Clin. Lymphoma Myeloma Leuk. 2015;15:139–151. doi: 10.1016/j.clml.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Magina K.N., Pregartner G., Zebisch A., Wölfler A., Neumeister P., Greinix H.T., et al. Cytarabine dose in the consolidation treatment of AML: A systematic review and meta-analysis. Blood. 2017;130:946–948. doi: 10.1182/blood-2017-04-777722. [DOI] [PubMed] [Google Scholar]
- 81.Srivastava A.K., Han C., Zhao R., Cui T., Dai Y., Mao C., et al. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. 2015;112:4411–4416. doi: 10.1073/pnas.1421365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y.W., Cleaver J.E., Hanaoka F., Chang C.F., Chou K.M. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol. Cancer Res. 2006;4:257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- 83.Rechkoblit O., Choudhury J.R., Buku A., Prakash L., Prakash S., Aggarwal A.K. Structural basis for polymerase η–promoted resistance to the anticancer nucleoside analog cytarabine. Scientific Rep. 2018;8:12702. doi: 10.1038/s41598-018-30796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Konerding D., James T.L., Trump E., Soto A.M., Marky L.A., Gmeiner W.H. NMR structure of a gemcitabine-substituted model Okazaki fragment. Biochemistry. 2002;41:839–846. doi: 10.1021/bi015678l. [DOI] [PubMed] [Google Scholar]
- 85.Kabsch W. Xds. Acta Crystallographica. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M., et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates, density maps, and structure factors for all the structures have been deposited in Protein Data Bank (PDB) under accession codes: 8E85, 8E86, 8E87, 8E88, 8E89, 8E8A, 8E8B, 8E8C, 8E8D, 8E8E, 8E8F, 8E8G, 8E8H, 8E8I, 8E8J, and 8E8K.