Abstract
Coccidioidomycosis (“Valley fever”) is caused by Coccidioides immitis and C. posadasii. These fungi are thermally dimorphic, cycling between mycelia and arthroconidia in the environment and converting into spherules and endospores within a host. Coccidioides can cause a broad spectrum of disease that can be difficult to treat. There has been a steady increase in disease, with an estimated 350,000 new infections per year in the United States. With the increase in disease and difficulty in treatment, there is an unmet need to increase research in basic biology and identify new treatments, diagnostics, and vaccine candidates. Here, we describe protocols required in any Coccidioides laboratory, such as growing, harvesting, and storing the different stages of this dimorphic fungal pathogen.
Basic Protocol 1:
Growth and harvest of liquid mycelia cultures for extractions
Alternate Protocol 1:
Large-volume growth and harvest of liquid mycelia cultures
Basic Protocol 2:
Mycelial growth on solid medium
Alternate Protocol 2:
Maintaining mycelial growth on solid medium
Basic Protocol 3:
Harvesting and quantification of arthroconidia
Alternate Protocol 3:
Long-term storage of arthroconidia
Basic Protocol 4:
Parasitic spherule growth and harvest
Alternate Protocol 4:
Obtaining endospores from spherules
Basic Protocol 5:
Intranasal infection of murine models
Keywords: arthroconidia, BSL-3, Coccidioides, dimorphic fungi, endospores, mycelia, spherules
INTRODUCTION
Coccidioides immitis and C. posadasii are soil-dwelling dimorphic fungi endemic to arid and semi-arid regions in the Americas (Nguyen et al., 2013). These neglected fungal pathogens are the etiological agents of coccidioidomycosis, which is commonly referred to as “Valley fever.” Recent reports indicate that the regions recognized to be endemic for these fungi have increased and are predicted to expand further in response to climate change and rising average temperature (Baptista-Rosas, Hinojosa, & Riquelme, 2007; Gorris, Cat, Zender, Treseder, & Randerson, 2018; Hamm, Hutchison, Leonard, Melman, & Natvig, 2019; Litvintseva et al., 2015; Tong, Wang, Gill, Lei, & Wang, 2017). Although most cases of Valley fever are asymptomatic (60%), disease in humans typically ranges from acute to chronic respiratory infection. In rare cases, coccidioidomycosis can result in disseminated lethal disease with diverse patterns of extrapulmonary dissemination (Fisher, Koenig, White, & Taylor, 2002; Nguyen et al., 2013; Odio, Marciano, Galgiani, & Holland, 2017; Van Dyke, Thompson, Galgiani, & Barker, 2019). Disease can occur in both immunocompetent and immunocompromised individuals, and new estimates suggest that about 350,000 new infections occur each year in the United States (Chiller, 2019) many of which are misdiagnosed (Tsang et al., 2010; Valdivia et al., 2006). Current antifungal drugs against Coccidioides spp. have side effects and are generally fungistatic, and there is no vaccine available to prevent infection (see current reviews on efforts against Coccidioides spp.; Thompson, Lewis, Nix, & Patterson, 2019; Van Dyke et al., 2019). Given the potential for endemic range expansion and the current increase in cases, greater research effort is needed to understand the basic biology of these dimorphic fungal pathogens in order to develop improved antifungal treatment strategies, rapid and accurate diagnostics, and potential vaccines.
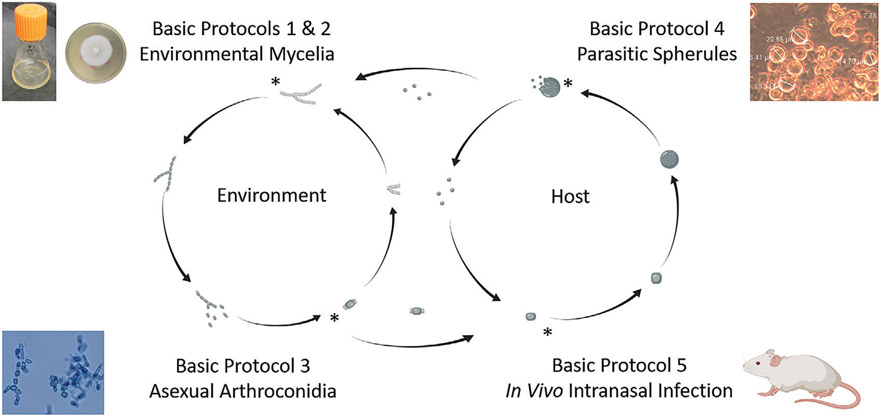
Herein, we provide detailed protocols for growing and maintaining Coccidioides spp. for both stages of the dimorphic lifecycle. An overview of these protocols and how they relate to the lifecycle is shown in Figure 1. In the environment, these fungi grow as vegetative filamentous mycelia, reproducing asexually via arthroconidia. This is achieved in vitro in liquid medium or on agar plates at ambient or low temperatures (about 22° to 30°C). Within the lungs, inhaled arthroconidia swell into spherules that mature, septate, and become an encasement for multiple haploid endospores. The spherules rupture and release endospores, thus leading to a significant increase in infection (Lewis, Bowers, & Barker, 2015). In vitro, morphogenesis to the parasitic lifecycle is triggered by exposing arthroconidia to high temperatures (about 37° 39°C) and to 5% to 20% CO2 in liquid medium.
Figure 1.
Graphical protocol overview across the Coccidioides lifecycle. Illustration created with BioRender.
The protocols described herein provide details on obtaining mycelia in liquid, with information on harvesting these cultures for nucleic acid extractions (Basic Protocol 1) and in large batches (Alternate Protocol 1). Next, we demonstrate the use of long-term mycelia cultures (Basic Protocol 2 and Alternate Protocol 2) to obtain infectious arthroconidia (Basic Protocol 3) and how to maintain these structures in long-term storage (Alternate Protocol 3). Then, we transition to protocols specific to the parasitic lifecycle. We discuss growing spherules in vitro (Basic Protocol 4), encouraging endosporulation (Alternate Protocol 4), and a basic method to infect mice using nasal instillation of arthroconidia (Basic Protocol 5). For each of the protocols, we provide detailed instructions, including time considerations and potential troubleshooting for each experiment in an attempt to encourage reproducibility and increase research on this important fungal pathogen.
STRATEGIC PLANNING
Obtaining Coccidioides Cultures
Coccidioides isolates can be obtained through BEI Resources (https://www.beiresources.org/). Commonly used and cited laboratory strains are C. posadasii C735 (NR-48945) and Silveira (NR-48944) and C. immitis RS (NR-48942) and 2006 (NR-48934). We also recommend the attenuated strain C. posadasii Δcts2/Δard1/Δcts3 (NR-166), which can be handled in a biosafety level 2 (BSL-2) environment and has been extensively characterized in vitro and in vivo (Hung, del Pilar Jimenez-Alzate, et al., 2014; Hung, Zhang, et al., 2018; Hung, Castro-Lopez, & Cole, 2014; Hung, Gonzalez, Wuthrich, Klein, & Cole, 2011; Mead, Roe, et al., 2020; Mead, Teixeira, Galgiani, & Barker, 2019; Xue et al., 2009). Isolates arrive as a 1-ml glycerol stock that should be stored at −80°C until use.
Biosafety Considerations
Wild-type Coccidioides is designated as a risk group level 3 organism. Researchers should follow the Centers for Disease Control and Prevention recommendations outlined in the “Biosafety in Microbiological and Biomedical Laboratories” resource for biosafety level 3 (BSL-3) or animal BSL-3 (ABSL-3) research (Centers for Disease Control and Prevention, 2009). We recommend a thorough risk assessment and consultation with biosafety experts prior to working with this organism.
Alternatively, there is a risk group level 2 strain of Coccidioides that was originally developed as a live attenuated vaccine that can be handled safely in a BSL-2 space (Xue et al., 2009). This strain behaves similarly to wild-type strains and can be used for personnel training, standard operating procedure (SOP) development, and obtaining fungal material or derivatives. The strain can grow as mycelia or arthroconidia and creates non-rupturing spherules. It has been shown to infect mice but not cause disease. Attenuation was achieved by deletion of chitinase 2 (cts2), d-arabinotol-2-dehydrogenase (ard1), and chitinase 3 (cts3). For more detail on this strain, we recommend several publications (Hung, Gonzalez, et al., 2011; Mead, Blackmon, Vogler, & Barker, 2019; Xue et al., 2009).
Engineering controls
All manipulations of Coccidioides should occur in a class II or class III biosafety cabinet (BSC). Centrifugation should be performed using an aerosol-resistant rotor, which should be opened inside of the BSC only.
Personal protective equipment (PPE)
Our personnel wear coverall suits (Tyvek, DuPont) over designated laboratory scrubs, long boot covers (Tyvek) over designated laboratory shoes, primary extended-cuff nitrile gloves (Kimberly-Clark), secondary nitrile or latex gloves (Kimberly-Clark), and a powered air-purifying respirator (Versaflo, 3M) to work within the BSL-3 and ABSL-3 facilities. We acknowledge that there are many acceptable options for PPE and recommend that this be determined by the institutional biosafety committee prior to beginning work.
Coccidioides-specific considerations
Arthroconidia are the infectious particles that cause disease and can be easily aerosolized. Consequently, working with arthroconidia produces the greatest risk of accidental infection to laboratorians. We recommend wearing Tyvek sleeves (Kimberly-Clark) when harvesting arthroconidia to further reduce this risk. We have previously shown that Coccidioides can be inactivated using 70% ethanol for 2 min or fresh 10% bleach for 2 min (Vogler, Nottingham, Parise, Keim, & Barker, 2015). Other researchers report effective use of Virex and report issues with 70% ethanol (Mitchell, Magee, Grys, & Lake, 2019).
NOTE: Please see the biosafety considerations above, including regarding BSC use, before starting a protocol below.
BASIC PROTOCOL 1 GROWTH AND HARVEST OF LIQUID MYCELIA CULTURES FOR EXTRACTIONS
Mycelia are the saprobic phase of Coccidioides and are useful for studying physiology related to the environmental phase of the lifecycle (Fisher, Koenig, et al., 2001; Kollath, Miller, & Barker, 2019). These fungi are often associated with rodents/rodent burrows, armadillos, or archeological sites and are more likely to degrade animal proteins than those associated with plants (Baptista-Rosas, Catalán-Dibene, et al., 2012; Del Rocio Reyes-Montes et al., 2016; Emmons, 1942; Kollath, Teixeira, Funke, Miller, & Barker, 2020; Sharpton et al., 2009). Therefore, it is hypothesized that the environmental niche of the fungi is connected to the presence of mammalian tissue (Taylor & Barker, 2019). This protocol describes how to grow and harvest a liquid culture of Coccidioides mycelia, which is useful for extracting nucleic acids, proteins, cell wall components, or other derivatives. If you require large volumes, we recommend Alternate Protocol 1.
Materials
2× glucose yeast extract (2× GYE) liquid medium (see recipe)
Fungal glycerol stocks (see Alternate Protocol 3)
1× phosphate-buffered saline (PBS)
DNA extraction kit (Quick-DNA Fungal/Bacterial Miniprep Kit Zymo Research, cat. no. D6005, or DNeasy UltraClean Microbial Kit, Qiagen, cat. no. 12224)
RNA extraction kit (Qiagen RNeasy Mini Kit, cat. no. 74124)
Vented 50-ml conical tubes (Corning, cat. no. 431720) and appropriate rack
30°C shaking incubator or standard incubator with shaking platform (e.g., Benchmark Orbi-Shaker with MAGic Clamp)
70-μm cell strainers (Corning, cat. no. 431751)
50-ml conical tubes
10-μl loops
Collection tubes (1.5-ml snap-cap tubes or collection tubes from DNA or RNA extraction kit) and appropriate rack
Additional reagents and equipment for DNA and RNA extraction
NOTE: Experiments involving PCR and RNA require extremely careful technique to prevent contamination and RNA degradation.
Start mycelia culture
-
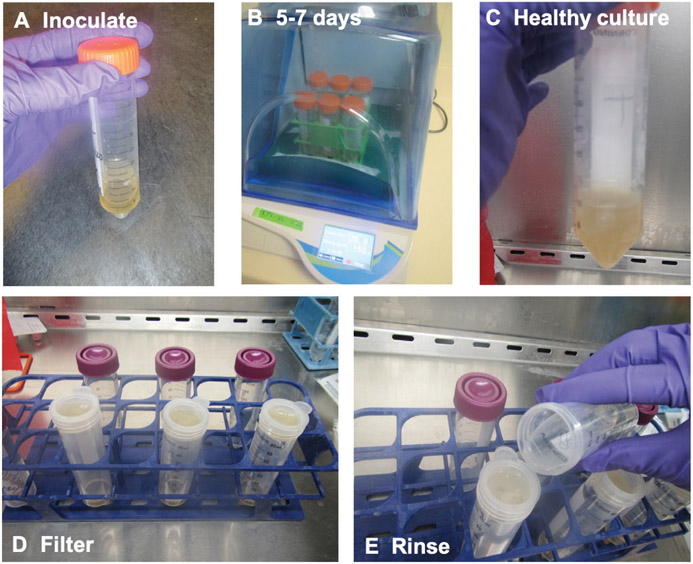
Add 30 ml of 2× GYE liquid medium to a vented 50-ml conical tube in an appropriate rack in a BSC (Fig. 2A).
We cover the vent with laboratory tape prior to placing it in the BSC to protect the filter from disinfectant. Once the tube is removed from the BSC, remove the tape to ensure gas exchange.
We suggest including a blank medium control to ensure pure cultures.
-
Pipet 30 μl fungal glycerol stock into tube (Fig. 2A).
Any Coccidioides material can be used to start liquid cultures. We frequently use arthroconidia in PBS (see Basic Protocol 3) or a fungal plug from a 1- to 2-week-old 2× GYE plate (see Alternate Protocol 2). Mycelia on solid medium (see Basic Protocol 2) can also be used. New users will likely start from slants obtained from collaborators or from glycerol stocks. The more material you add, the faster the nutrition will be used up, so plan accordingly.
Remove culture from the BSC and secure it in a 30°C shaking incubator or a standard incubator with a shaking platform.
-
Shake at 150 rpm at 30°C for ≤1 week (Fig. 2B).
Reducing the temperature will slow the growth. If you need to buy yourself time, such as over the weekend, lower it to 25° to 28°C.
Collect desired material (see step 6) while the culture is still fluffy(Fig. 2C). Older liquid cultures become stringy and hard to manipulate.
Figure 2.
Small mycelia cultures are ideal for nucleic acid extractions. (A) Inoculate 2× GYE in a vented 50-ml conical tube. (B) Incubate at 30°C for 5 to 7 days. (C) Healthy cultures are fluffy, rather than stringy. (D) Pour mycelia through a nested filter. (E) Rinse fungal material with PBS until white.
Harvest mycelia
-
5.
Pour culture into a nested filter (two 70-μm cell strainers) placed on a 50-ml conical tube (Fig. 2D).
If pouring makes you uncomfortable, transfer using a serological pipet will work, too.
-
6.
Rinse pellet with 1× PBS (Fig. 2E).
You will notice the fungal mass begin to lose color as you rinse. Keep going until it looks like all the medium has been removed.
-
7.
Use a 10-μl loop to transfer material into a collection tube in appropriate rack.
-
8.
Perform DNA extraction using a DNA extraction kit according to the manufacturer’s instructions.
Kit extractions are acceptable for PCR. We have used Zymo Research and Qiagen kits without issue, although alternative kits exist. These kits also work using fungi grown on agar (see Basic Protocol 2).
Effective bead beating with a homogenizer (based on your kit’s recommendation; e.g., Fisherbrand Bead Mill 24 Homogenizer) is crucial for success. We recommend using 4 to 5 cycles of 1 min each. Our homogenizer setting is 5 m/s with a 2-min rest between cycles. When in doubt, check for cell lysis using a microscope (e.g., Leica DMI3000 B). The Zymo Research kit includes beads, and we use pre-filled glass beads (BeadBug prefilled tubes, 2.0-ml capacity, containing 1.0-mm acid-washed silica glass beads, Millipore Sigma, cat. no. Z763756) with the Qiagen kit.
The centrifuge used should be based on your kit’s recommendation (e.g., Eppendorf 5810R).
If you are extracting from multiple isolates for whole-genome sequencing, we recommend heat-killing mycelia and performing a phenol chloroform extraction in lower containment. See our publication for details (Mead et al., 2019).
-
9.
Perform RNA extraction using a RNA extraction kit according to the manufacturer’s instructions.
We have used Qiagen RNeasy kits without issue, although many alternatives exist.
Effective bead beating is crucial for success; we recommend using 3 to 5 cycles of 30 s each. Our homogenizer setting is 6 m/s with a 1-min rest between cycles, and we cool the tubes on ice between rounds. When in doubt, check for cell lysis using a microscope (e.g., Leica DMI3000 B).
For RNA sequencing, we recommend using organic solvents. Methods are outlined in prior publications (Mead, Roe, et al., 2020; Whiston et al., 2012).
ALTERNATE PROTOCOL 1 LARGE-VOLUME GROWTH AND HARVEST OF LIQUID MYCELIA CULTURES
Similar to Basic Protocol 1, this protocol describes how to grow and harvest a liquid culture of Coccidioides mycelia, but in large volumes.
Additional Materials (also see Basic Protocol 1)
Vented 250-ml Erlenmeyer flasks with filter protection (Corning, cat. no. 431407)
Centrifuge (e.g., Eppendorf 5810R) with 50-ml-capacity fixed-angle rotor
Start mycelia culture
-
Add 100 ml of 2× GYE liquid medium to a vented 250-ml Erlenmeyer flask with filter protection in a BSC.
We purchase flasks with filter protection to prevent damage by disinfectant. You can also cover the vent with laboratory tape prior to placing it in the BSC. Once the flasks are removed from the BSC, remove the protection to ensure gas exchange.
We suggest including a blank medium control to ensure pure cultures.
-
Pipet 100 μl fungal glycerol stock into flask.
Any Coccidioides material can be used to start liquid cultures. We frequently use arthroconidia in PBS (see Basic Protocol 3) or 3 to 4 fungal plugs from a 2× GYE plate (see Alternate Protocol 2). Mycelia on solid medium (see Basic Protocol 2) can also be used. New users will likely start from slants obtained from collaborators or from glycerol stocks. The more material you add, the faster the nutrition will be used up, so plan accordingly.
Remove culture from the BSC, remove filter tape, and secure it in a 30°C shaking incubator or standard incubator with a shaking platform.
-
Shake at 150 rpm at 30°C for ≤1 week.
Reducing the temperature will slow fungal growth. If you need to buy yourself time, such as over the weekend, lower it to 25° to 28°C.
Collect desired material while the culture is still fluffy. Older liquid cultures become stringy and hard to manipulate.
Harvest mycelia
-
5.
Pour culture into a 50-ml conical tube.
-
6.
Using a centrifuge with a 50-ml-capacity fixed-angle rotor, centrifuge conical tubes for 8 min at 12,000 × g to pellet cells.
Ensure that the conical tubes are rated for this speed, which is often designated on the tube itself.
These cultures can be difficult to pellet. Use a low volume (~25 ml) for best results.
Mycelia can also be collected in a nested filter and then rinsed. This is suitable for small cultures but not for large ones. This is detailed in Basic Protocol 1.
-
7.
Pour off medium into a waste container.
Expect a robust white-to-tan pellet.
If your culture is fluffy and hard to pellet, this is still acceptable. Instead of pouring off the medium, use a serological pipet to remove it.
-
8.
Resuspend cell pellet in 5 ml of 1× PBS.
Inverting or vortexing the tube (e.g., with a Scientific Industries Vortex-Genie 2) can help rinse the material.
-
9.
Using the same centrifuge and rotor, centrifuge conical tubes for 8 min at 12,000 × g to pellet cells.
Again, if your culture is fluffy and hard to pellet, this is fine. Instead of pouring off the medium, use a serological pipet to remove it.
-
10.
Pour off medium into a waste container.
-
11.
Store mycelia at −80°C until further use.
The storage duration depends on downstream experimental goals. In general, we recommend use within 6 months of collection.
BASIC PROTOCOL 2 MYCELIAL GROWTH ON SOLID MEDIUM
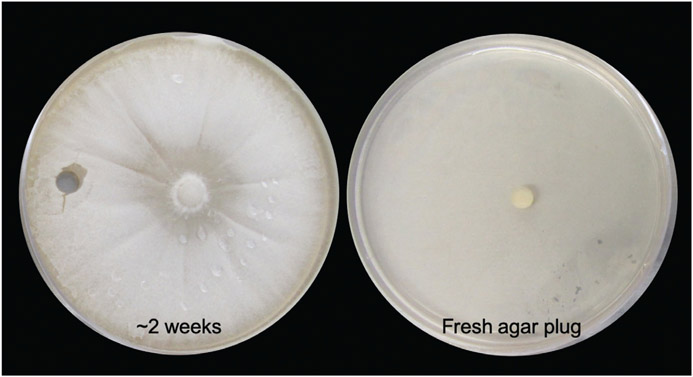
As mentioned above in Basic Protocol 1, mycelia are the environmental phase of Coccidioides. When the culture is grown on solid medium, the filamentous colony will cover the surface of the plate within ~2 weeks (Fig. 3). These plates are necessary to obtain arthroconidia (Basic Protocol 3), the asexual structures that develop into endosporulating spherules (Basic Protocol 4), or material used in infection models (Basic Protocol 5). For any experiment using arthroconidia, you will want to start these cultures 4 to 6 weeks prior to planned use. Additionally, use of these plates is a common method of maintaining a Coccidioides culture with minimal effort for future use. In this context, a fungal plug can be placed on the center of a plate and left for several weeks before another subculture is necessary; this is outlined in Alternate Protocol 2.
Figure 3.
Maintenance of active culture on solid medium. Fungal growth covers the plate in ~2 weeks (left). A fungal transfer tube can be used to subculture a fresh agar plug onto a new plate (right).
Materials
Fungal glycerol stocks (see Alternate Protocol 3)
2× GYE solid agar plates (see recipe; 20 total)
10-μl loops or hockey sticks
Laboratory tape
Unicorn bags (type 10T; Unicorn Bags) or plate seals (Scientific Device, cat. no. 261)
30°C standard incubator
-
Pipet 100 μl fungal glycerol stock onto each 2× GYE solid agar plate in a BSC.
This procedure can also be started with existing mycelia on solid medium (see Alternate Protocol 2) or arthroconidia (see Basic Protocol 3). In this case, we particularly recommend that you use a 10-μl loop or hockey stick to spread the fungal matter across the plate to ensure even growth (see step 2).
As an alternative to solid agar plates, vented suspension flasks containing 2× GYE solid agar (see annotation to recipe for plates; 10 total) can be used.
-
Use a 10-μl loop or hockey stick to ensure fungal material is adequately spread across the surface.
If you chose to use vented flasks, simply spread the material by tilting the flask from side to side or stick a loop into the neck of flask.
-
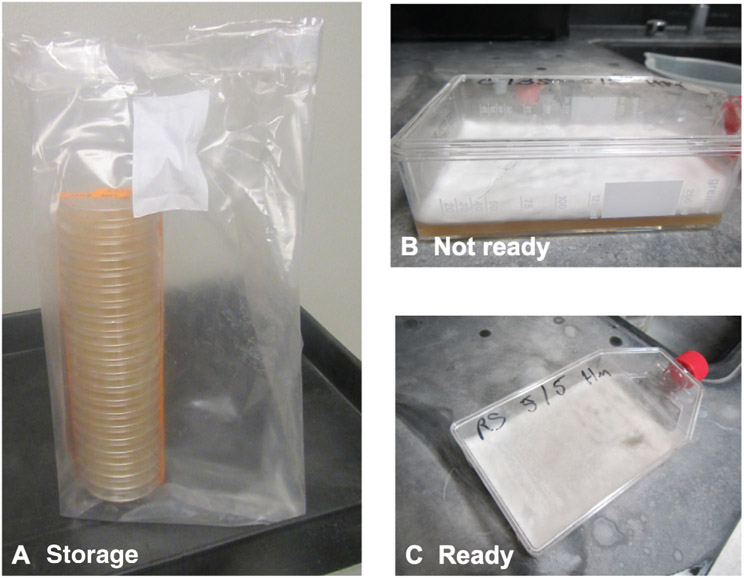
Use laboratory tape to tape plates together in a stack. After they have been appropriately decontaminated and removed from the BSC, place them in a unicorn bag and seal top with tape or wrap them in plate seals (Fig. 4A).
If you chose to use vented flasks, ensure that each lid is tightened. Avoid getting the vent wet with culture from the inside or with disinfectant from the outside. We cover the vent with laboratory tape prior to placing it in the BSC to protect the filter from disinfectant. Once the flasks are removed from the BSC, remove the tape to ensure gas exchange (Fig. 4B and 4C).
-
Store plates for 4 to 6 weeks in a 30°C standard incubator to develop arthroconidia.
Monitor the plates for moisture. Wet plates will slow the development of arthroconidia.
The plates will also show growth at room temperature, but the progress will be slower.
Figure 4.
Long-term storage of solid mycelia cultures promotes development of arthroconidia. (A) Stack plates, tape them tightly together, and store them in Unicorn bags for 4 to 6 weeks. (B-C) Vented flasks are an alternative storage option. Mycelia that are fluffy or moist are not ready for harvest (B). Notably, dry cultures demonstrate small aerial structures. This indicates the presence of arthroconidia (C).
ALTERNATE PROTOCOL 2 MAINTAINING MYCELIAL GROWTH ON SOLID MEDIUM
Basic Protocol 2 describes growing mycelia on solid agar in large quantities with the goal of producing arthroconidia. If your desire is to maintain an active Coccidioides culture for future use, a fungal plug in the center of a plate is more than sufficient and is described below.
Materials
2× GYE solid agar plates with fungal growth (see Basic Protocol 2)
2× GYE solid agar plates (see recipe; one plate per isolate)
Fungal transfer tubes (Spectrum Laboratories, cat. no. 190195)
Plate seals (Scientific Device, cat. no. 261)
30°C standard incubator
-
Use a fungal transfer tube to remove a plug from an existing 2× GYE solid agar plate with fungal growth in a BSC (Fig. 3).
This procedure should be started with mycelia on solid medium (see Basic Protocol 2). However, if you do not have an existing plate, then medium can be inoculated with arthroconidia (see Basic Protocol 3), or glycerol stocks (see Alternate Protocol 3).
-
Place plug on a 2× GYE solid agar plate. Seal plate with a plate seal. Allow seal to fully dry prior to removing the plate from the BSC.
The plug can be fungus side up or down.
-
Incubate for ≤2 weeks in a 30°C standard incubator.
Store upright for ~24 hr until the fungal plug has adhered to the agar. After this, you can invert the plate if desired.
After 2 weeks, the culture margins will likely be at the edges of the plate. We recommend subculturing approximately every 2 to 3 weeks. Plates can be subcultured beyond 3 weeks. However, arthroconidia begin to form between 2 and 3 weeks, so additional care should be taken to reduce aerosols (see Strategic Planning: Biosafety Considerations).
Storing plates at room temperature will slow growth substantially.
BASIC PROTOCOL 3 HARVESTING AND QUANTIFICATION OF ARTHROCONIDIA
In the environment, as conditions become less favorable, filamentous mycelia segment into small barrel-shaped arthroconidia that are easily aerosolized. These structures are the infectious propagules that, upon inhalation, can cause the disease Valley fever. In vitro, these asexual structures develop on solid medium after 4 to 6 weeks of growth, as outlined in Basic Protocol 2. In Basic Protocol 2, the fungal growth usually covers the plate after ~2 weeks. At this point, the mycelia on the plate are fluffy and cottony and can appear moist (Fig. 4B); arthroconidia are not yet present in large quantities. Once the colony appears dry and often powdery, you can see aerial hyphae (Fig. 4C), rather than cottony growth. This is a good indication that the plates are ready to be harvested. The time needed can vary by laboratory and strain, but in general, 6 weeks is a good rule. Harvesting too soon can reduce overall yield, and harvesting too late can reduce viability. For long-term storage, see Alternate Protocol 3.
Materials
Sterile 1× PBS
4- to 6-week-old cultures of Coccidioides (see Basic Protocol 2)
Sterile 1% (w/v) formalin [dilute 10% formalin with 1× PBS and filter-sterilize (0.2 μm)]
2× GYE solid agar plates (see recipe)
70-μm cell strainers (Corning, cat. no. 431751)
50-ml conical tubes and appropriate rack
Cell scrapers
Vortex (e.g., Vortex-Genie 2, Scientific Industries)
Centrifuge (e.g., Eppendorf 5810R) with 50-ml-capacity fixed-angle rotor
2-ml screw-cap tubes or 1.5-ml snap-cap tubes and appropriate rack
Standard hemocytometer
Microscope (e.g., Leica DMI3000 B)
Sterile 1.5-ml tubes
10-μl loops or hockey sticks
Laboratory tape or plate seals (Scientific Device, cat. no. 261)
30°C standard incubator
Harvest dry cells from plate
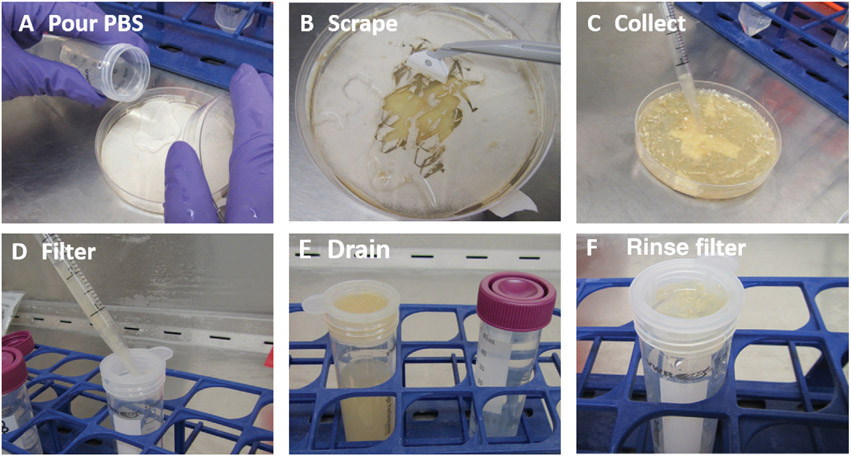
Place a 70-μm cell strainer into mouth of a 50-ml conical tube in an appropriate rack in a BSC.
-
Pour sterile 1× PBS into 4- to 6-week-old cultures of Coccidioides (Fig. 5A).
Use ~5 ml per petri dish or 25 ml per flask.
This procedure requires mycelia growing on solid medium for 4 to 6 weeks to obtain arthroconidia (see Basic Protocol 2). Time considerations and volumes are based on 20 plates or 10 flasks.
Harvest one strain at a time to prevent cross-contamination.
-
Use a cell scraper to dislodge culture from the agar (Fig. 5B).
Cells will be hydrophobic and in clumps; this is normal.
-
Use a serological pipet to transfer PBS and fungal culture to the cell strainer (Fig. 5C and 5D).
Add small volumes of liquid to the filter and allow the liquid to drain into the conical tube (Fig. 5E). Using multiple filters and conical tubes at once can expedite the process.
Periodically rinse the filter with ~0.5 ml PBS to maximize collection (Fig. 5F).
Each conical tube should contain ≤30 ml. Higher volumes take additional time to pellet.
Change filters and/or use additional conical tubes as needed.
-
Repeat until all plates have been scraped and their contents passed through a filter.
Some fungal material will remain on the filter. A loop can be used to gently “stir” this material while rinsing to maximize collection. After the final filter rinse, discard the filter and remaining material.
Dispose of scraped plates and filters.
-
Cap conical tube and vortex for 30 s at max speed.
Vortexing between washes ensures that you obtain single arthroconidia rather than chains.
Figure 5.
Arthroconidia harvest process. (A) Pour PBS onto dry cultures. (B) Scrape the fungal material into solution. (C) Use a serological pipet to transfer the fungal material and PBS solution. (D) Gently release the material into the filter, allowing time for the solution to pass into the conical tube. (E) Arthroconidia in solution are often yellow/brown in color. (F) After the solution passes through the filter, rinse the filter with small volumes of PBS to maximize collection.
Clean and concentrate arthroconidia
-
8.
Using a centrifuge with a 50-ml-capacity fixed-angle rotor, centrifuge conical tubes for 8 min at 12,000 × g to pellet cells.
Ensure that the conical tubes are rated for this speed, which is often designated on the tube itself.
The next steps (steps 9 to 15) remove cell debris and break the chains of arthroconidia into individual cells. We recommend two vortex/wash steps.
Reduce the volume of PBS with each wash step to concentrate the arthroconidia.
-
9.
Pour off PBS into a waste container.
Expect a tan-to-brown pellet. Light coloration of the supernatant is normal. We have observed significant color variation between strains.
The arthroconidia will pellet and stick to the tube wall. Any debris that is in solution is unneeded.
-
10.
Resuspend each cell pellet in 5 ml sterile 1× PBS.
This will help concentrate the arthroconidia.
This is a good time to combine tubes. For example, if you started with six tubes holding ~30 ml PBS each, in this step, combine the pellets into three tubes holding 10 ml each.
-
11.
Vortex conical tube for 1 min at max speed.
We have tried mixing with a serological pipet or vigorous manual shaking. Although this may work, the vortex method produces the best results.
-
12.
Using the same centrifuge and rotor, centrifuge conical tubes for 8 min at 12,000 × g to pellet cells.
-
13.
Pour off PBS into a waste container.
The arthroconidia will pellet and stick to the tube wall. Any debris that is in solution is unneeded.
-
14.
Combine all cell pellets into a final volume of 5 ml sterile 1× PBS.
The easiest method is to use a serological pipet to resuspend one pellet and to then transfer the entire contents (pellet + PBS) to the next pellet. Repeat until all pellets have been resuspended and combined.
When in doubt, keep final volumes low. We recommend having approximately 5 to 7 ml PBS in the final tube after harvesting 20 plates, with 106 to 108 total conidia. You can dilute after quantification (see step 23) if desired.
-
15.
Vortex conical tube for 1 min at max speed.
-
16.
Store ≤6 months at 4°C or proceed immediately to quantification and viability assessment.
Cultures remain viable beyond 6 months, but we have observed reduced viability over time. We have observed that concentrated cultures (>106) maintain viability better than more dilute cultures. If possible, maintain a stock culture of arthroconidia (>106). Perform any needed dilutions just before use (within 1 week).
Counting of arthroconidia does not indicate viability, and viability stains have not been optimized to date. Arthroconidia counting is performed using a standard hemocytometer (see step 23). In our facility, this is done outside of containment; therefore, arthroconidia must be inactivated (see steps 17 to 22). If this is not needed for your laboratory, proceed to step 23. Depending on the estimated concentration based on pellet size, it is likely that you will need to perform counts using a 10−3 or 10−4 dilution factor.
Not all arthroconidia are viable, with viability depending on the strain used and the age of the plates used for harvest. To accurately determine the quantity of viable conidia, plate colony-forming units (CFUs; steps 24 to 30).
Quantification
-
17.
Vortex each arthroconidia suspension (see step 16) to be quantified to mix well.
-
18.
Transfer 500 to 1000 μl of each arthroconidia suspension to be quantified into a 2-ml screw-cap tube or 1.5-ml snap-cap tube in an appropriate rack.
Volumes can be adjusted based on harvest volume.
-
19.
Centrifuge tubes for 4 min at 9000 × g.
-
20.
Carefully remove PBS without disturbing the pellet.
-
21.
Resuspend pellet in an equal volume (see step 18) of sterile 1% formalin and vortex well.
This will fix (i.e., inactivate) the arthroconidia. We recommend that you verify inactivation procedures in house, as variation between laboratories/solutions can occur.
-
22.
Fix cells for 24 hr to inactivate.
We recommend that you use 10% of the total volume to verify inactivation prior to removing the tube from the BSL-3 space. This can be done by spinning down 10%, removing the formalin, resuspending in PBS, plating, and then incubating at 30°C for 48 hr along with a positive control to ensure that there is no evidence of growth before working with the cells outside of the BSL-3 space.
-
23.
Quantify cells using a standard hemocytometer and a microscope.
Expect to dilute the cells (factor of 10−3 or 10−4) in order to have a reasonable number to count.
Viability assessment
-
24.
Add 450 μl sterile 1× PBS to seven sterile 1.5-ml tubes.
-
25.
Vortex each arthroconidia suspension (see step 16) to be quantified to mix well.
Arthroconidia settle quickly, so vortexing just before use will increase overall accuracy.
-
26.
Transfer 50 μl stock conidia to the first tube of PBS.
This is a 1:10 dilution. If you plate 50 μl, then it becomes a 1:100 dilution.
-
27.
Vortex tube and continue dilution series, taking care to vortex each tube prior to making the next dilution.
-
28.
Plate 50 μl (10%) of at least the last three tubes on 2× GYE solid agar plates.
If plating 10%, then your tubes should represent 10−2, 10−3, 10−4, 10−5, 10−6, 10−7, and 10−8 dilutions.
-
29.
Spread well with a 10-μl loop or hockey stick. Seal plates with laboratory tape or plate seals.
We seal all plates that will be kept for >1 week using shrink seals. If you plan to count the cells on the plates and then dispose of them before 1 week, tape is sufficient.
-
30.
Incubate for 3 to 4 days in a 30°C standard incubator and inspect for initial colony formation (Fig. 6).
Colonies can be hard to see in the beginning. Hold the plates up to the light to look for small white star-like shapes.
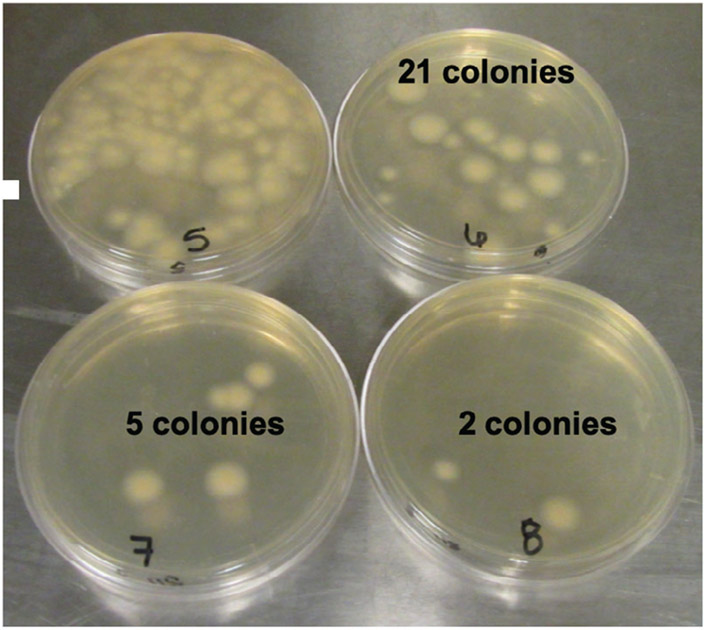
Colonies become more obvious with time but can grow into one another. Plating multiple dilutions and replicates can help with accuracy. Determine concentration using the following calculation:For example, in Figure 6, there are 21 colonies on the 10−6 plate, 5 colonies on the 10−7 plate, and 2 colonies on the 10−8 plate (the 10−5 plate has too many to count accurately). The calculation is as follows: -
31.
Store arthroconidia suspensions for ≤6 months at 4°C.
Overall viability remains for ~6 months, after which we harvest fresh cells for best results. If you have old conidia (>6 months), these can be used to start new solid medium plates (see Basic Protocol 2). Before you throw away old stocks of arthroconidia that are <6 months old, use them to make glycerol stocks, as outlined in Alternate Protocol 3.
Figure 6.
Quantification of viable arthroconidia. Plate multiple dilutions to obtain accurate numbers. The 10−5 plate has too many colonies to count accurately. There are 21 colonies on the 10−6 plate, 5 colonies on the 10−7 plate, and 2 colonies on the 10−8 plate.
ALTERNATE PROTOCOL 3 LONG-TERM STORAGE OF ARTHROCONIDIA
Arthroconidia (Basic Protocol 3) are viable with long-term storage at −80°C. We recommend making at least two stocks of new isolates and checking the viability ~30 days later before discarding active growth plates/slants.
Materials
Harvested arthroconidia suspended in PBS (see Basic Protocol 3)
Fungal freezing medium (see recipe)
2-ml screw-cap cryovial tubes and appropriate rack
-
Place 500 μl harvested arthroconidia suspended in PBS in a 2-ml screw-cap cryovial tube in an appropriate rack in a BSC.
Volumes are flexible; however, we recommend including at least ~106 cell/ml in one glycerol stock.
Alternatively, cells can be scraped directly from a 3- to 6-week-old plate (see Basic Protocol 3, step 3). For long-term storage, straining and rinsing is not necessary.
We have recently experimented with taking 8 to 10 fungal plugs directly from a plate (see Alternate Protocol 2). So far, this method has been successful, with viability confirmed up to 1 year later.
Add 500 μl fungal freezing medium, using a pipet to mix tube contents.
-
Store at ~80°C.
We recommend that you maintain an active culture until you have confirmed the viability of the glycerol stocks. This can be either active growth on a plate (see Alternate Protocol 2) or arthroconidia (see Basic Protocol 3).
We have observed viability for up to 7 to 10 years.
BASIC PROTOCOL 4 PARASITIC SPHERULE GROWTH AND HARVEST
Spherules are the parasitic structure of the Coccidioides lifecycle that develops when environmental arthroconidia are inhaled into the respiratory system of a susceptible host. Arthroconidia undergo isotropic growth and develop into multinucleated spherules, which then segment internally to create endospores. This process occurs cyclically approximately every 5 days (Huppert, Sun, & Harrison, 1982). Endospores are premature spherules that individually swell and develop into mature spherules that rupture and release endospores. For in-depth investigations of this critical stage in disease establishment, see several prior publications (Cole & Hung, 2001; Cole, Seshan, et al., 1988; Drutz & Huppert, 1983; Hector & Pappagianis, 1983; Mandel, Galgiani, Kroken, & Orbach, 2006; Mead, Roe, et al., 2020; Viriyakosol, Singhania, et al., 2013; Whiston et al., 2012; Wise, Hung, Whiston, Taylor, & Cole, 2013). Morphogenesis is triggered by exposure to high temperatures (>37°C), gas concentrations (5% to 15% CO2), and other factors that have not been precisely defined (Breslau & Kubota, 1964; Klotz, Drutz, Huppert, Sun, & DeMarsh, 1984). In vitro, spherules can be obtained using either chemically defined Converse medium (Converse, 1955) or RPMI1640 supplemented with 10% fetal bovine serum (FBS; RPMI-sph medium) (Petkus, Baum, Ellis, Stern, & Danley, 1985). Most literature reports the use of Converse medium; therefore, it may be necessary to use this medium to replicate some previously published results. However, in our investigations, RPMI-sph medium produces very similar spherule morphology and yield, is simple to make, and uses a formulation that represents a more host-like nutrition source (Mead et al., 2019).
Materials
Converse medium (see Table 1) or RPMI-sph medium [RPMI-1640 (Gibco, cat. no. 11835055) with 10% (v/v) FBS (Gibco, cat. no. 16000044)], room temperature or 39°C
Quantified arthroconidia suspension (see Basic Protocol 3)
1× PBS
Vented 125-ml Erlenmeyer flasks with filter protection (Corning, cat. no. 431405)
Vortex (e.g., Vortex-Genie 2, Scientific Industries)
39°C, 10% CO2 shaking incubator or standard incubator (with gas exchange at 5% to 20% CO2; e.g., Caron Oasis 6400) with shaking platform (e.g., Benchmark Orbi-Shaker with MAGic Clamp)
70-μm cell strainers (Corning, cat. no. 431751)
50-ml conical tubes and appropriate rack
Centrifuge (e.g., Eppendorf 5810R) with 50-ml-capacity fixed-angle rotor
Microscope (e.g., Leica DMI3000 B)
Hemocytometer
Table 1.
Stock Solutions for 1 L Modified Converse Medium
| Reagenta | MW | g/L | Molarity or concentration |
Volume/1 Lb |
|---|---|---|---|---|
| Ammonium acetate | 77.08 | 123.0 | 1.596 M | 10 ml |
| KH2PO4 anhydrousc | 136.09 | 51.0 | 0.37 M | 10 ml |
| K2HPO4 anhydrousc | 174.18 | 52.3 | 0.3 M | 10 ml |
| MgSO47H2Oc | 246.5 | 39.5 | 0.16 M | 10 ml |
| ZnSO47H2Oc | 287.56 | 0.72 | 2.5 mM | 5 ml |
| NaCl | 58.44 | 1.4 | 24 mM | 10 ml |
| CaCl22H2Oc | 147.02 | 0.6 | 4.08 mM | 5 ml |
| NaHCO3 | 84.01 | 1.2 | 14.3 mM | 10 ml |
| Tamold | NA | 50.0 | 50 g/L | 10 ml |
| Glucosee | 180.86 | 40.0 | 40 g/L | 100 ml |
| N-Z aminee | NA | 5.0 | 5 g/L | 10 ml |
Prepare each stock solution in 1 L bottled water (distilled deionized water in clean glassware) and number as shown above. The stocks may be stored ≤1 year at 4°C.
Allocate 820 ml distilled deionized water into a container for solid medium and 810 ml for liquid medium. Pipet individual reagents into the flask, swirling to mix after each addition. pH to 6.5. Autoclave for 30 min. For solid medium, add 18 g/L purified agar. When cool, add sterile-filtered glucose and N-Z amine if making liquid medium and only sterile-filtered glucose if making solid Converse plates.
The hydrated form of these particular reagents can be substituted, but the weight will need to be adjusted to obtain the correct molarity.
Omit from the medium for agar plates.
Filter-sterilize and add after autoclaving.
Start spherule culture
-
Place 50 ml Converse medium or RPMI-sph medium into a vented 125-ml Erlenmeyer flask with filter protection in a BSC.
Volumes can be scaled up or down depending on experimental goals. Similar to what is done with mycelia in Basic Protocol 1, we have used vented 50-ml conical tubes when small cultures are needed.
Pre-warming the medium to 39°C is ideal but not necessary.
Protect the filter from disinfectant before placing it into the BSC.
-
Vortex quantified arthroconidia suspension.
Arthroconidia settle quickly, so vortex just before use to ensure accuracy.
-
Inoculate medium to a final concentration of 106 arthroconidia/ml.
Depending on your goals, inoculation concentrations may vary, or a specific concentration may not be necessary. The concentration of 106 arthroconidia/ml consistently produces good results.
Remove cultures from BSC and remove filter tape.
-
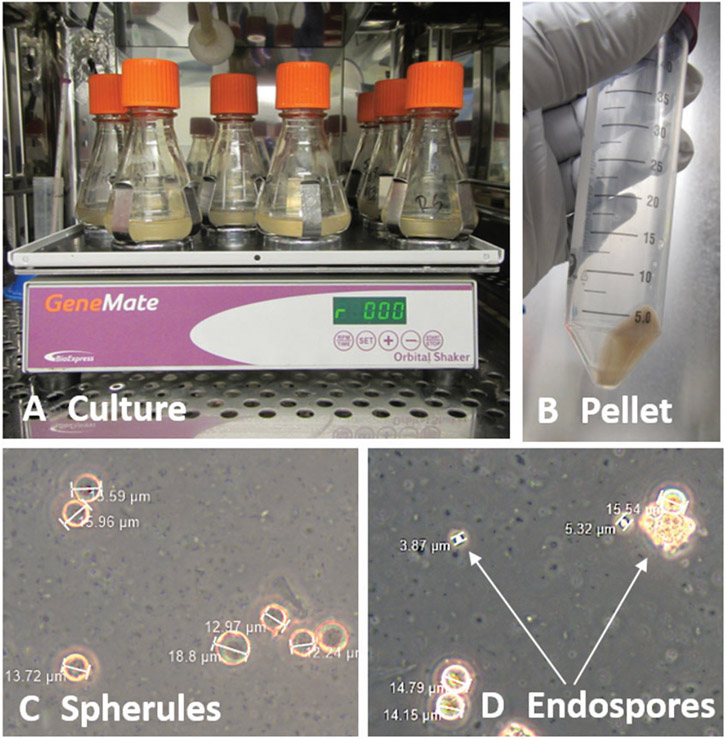
Place cultures on a 39°C, 10% CO2 shaking incubator or standard incubator with a shaking platform at 150 rpm for ≥3 days (Fig. 7A).
A CO2 level of 5% to 20% is standard.
Spherules can be harvested at any point after 3 days. Day 5 is often when endosporulation will occur. Endosporulation is more robust with a medium change (see Alternate Protocol 4). Change the medium if you intend to keep the culture beyond 6 to 7 days.
Figure 7.
Spherule growth. (A) Culture arthroconidia at ~1 × 106 cells/ml in RPMI-sph medium at 39°C, 10% CO2, for ≥3 days. (B) Pour cultures into a 50-ml conical tube and pellet cells by centrifugation. (C) After resuspension of the pellet, confirm the presence of spherules. Shown is C. posadasii Silveira on day 4, after growth in RPMI-sph medium. (D) Packets of endospores are present after medium change. Shown is C. posadasii Silveira on day 7 after changing the RPMI-sph medium on day 4.
Harvest spherules
-
6.
Place nested filter (two 70-μm cell strainers) in a 50-ml conical tube in an appropriate rack.
-
7.
Pour spherule culture through the filter to remove any hyphal fragments.
Minor fragments will be present and do not necessarily need to be removed. Skip this step if desired.
-
8.
Remove filter and cap conical tube.
-
9.
Using a centrifuge with a 50-ml-capacity fixed-angle rotor, centrifuge conical tubes for 8 min at 9000 × g to pellet cells.
Cell pellets can be rinsed with PBS if desired. Expect a white pellet (Fig. 7B).
-
10.
Pour off medium into a waste container.
-
11.
Resuspend in 50 ml of 1× PBS to maintain concentration.
Adjust the volume as needed.
-
12.
Confirm presence of spherules using a microscope (Fig. 7C).
-
13.
Count spherules using a hemocytometer and microscope.
ALTERNATE PROTOCOL 4 OBTAINING ENDOSPORES FROM SPHERULES
Basic Protocol 4 describes how to grow and harvest spherules. Although 5- to 7-day-old cultures of spherules will contain endospores, we have observed that spherule cultures require fresh medium in order to encourage synchronized endosporulation. In the protocol below, we describe how to incite this behavior.
Additional Materials (also see Basic Protocol 4)
4- to 5-day-old active spherule cultures (see Basic Protocol 4, steps 1 to 5)
-
Place 50 ml fresh Converse medium or RPMI-sph medium into a vented 125-ml Erlenmeyer flask with filter protection in a BSC.
Pre-warming the medium to 39°C is ideal but not necessary.
Protect the filter from disinfectant before placing it into the BSC.
-
Inoculate fresh medium with 2 to 4 ml of 4- to 5-day-old active spherule culture.
From our experience, an exact volume is not important. You just want to provide fresh nutrition and reduce the cell density to encourage endosporulation.
Remove cultures from the BSC and remove filter tape.
-
Place cultures on a 39°C, 10% CO2 shaking incubator or standard incubator with a shaking platform at 150 rpm for 24 hr (Fig. 7A).
A CO2 level of 5% to 20% is standard.
-
Confirm presence of endospores using a microscope (Fig. 7D).
Cultures will be a mix of spherules and endospores.
Small (1- to 5-μm) filters can be used for endospore isolation.
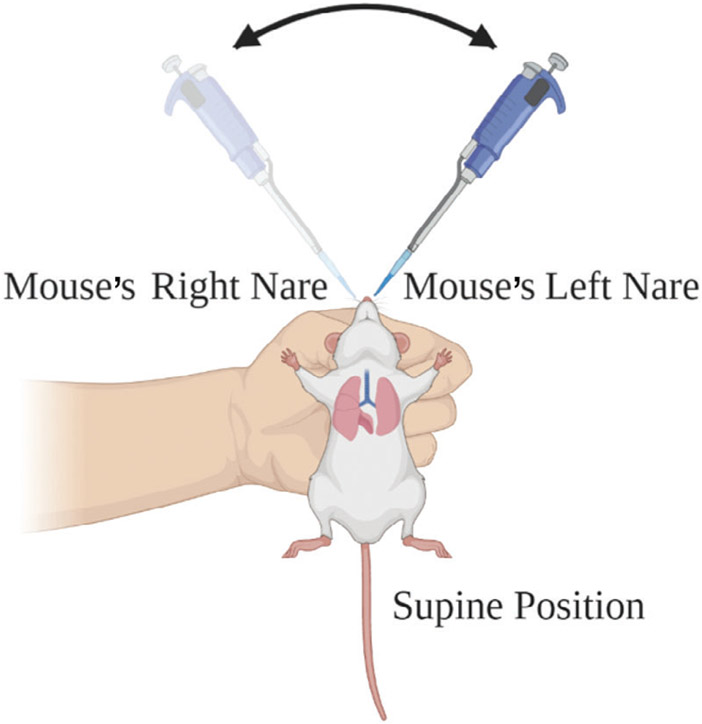
BASIC PROTOCOL 5 INTRANASAL INFECTION OF MURINE MODELS
Arthroconidia are the infectious propagules of Coccidioides. Infection of murine models through intranasal delivery is the preferred and widely used method to understand Coccidioides pathology (Campuzano et al., 2020; Castro-Lopez & Hung, 2017; Hayden et al., 2019; Mead, Roe, et al., 2020; Viriyakosol, Kapoor, et al., 2019). Susceptibility to Coccidioides varies among mouse strains, with studies having shown that C57BL/6 and BALB/c mice are highly susceptible and that DBA/2 mice are more resistant (Kirkland & Fierer, 1983). Therefore, you must consider which model is most appropriate when designing a new mouse experiment. To infect mice, start with Basic Protocol 3. Inoculum should be made with freshly quantified (within 2 weeks) arthroconidia.
Materials
<6-month-old harvested arthroconidia (see Basic Protocol 3)
Sterile 1× PBS
Mice
70% (v/v) ethanol
Appropriate tube (large enough to hold inoculum) and appropriate rack
Vortex (e.g., Vortex-Genie 2, Scientific Industries)
Sharpie marker or ear punch
Gauze pad
Additional reagents and equipment for ketamine/xylazine or isoflurane anesthesia and viability assessment (see Basic Protocol 3, steps 21 to 30)
-
Make inoculum of <6-month-old harvested arthroconidia in sterile 1× PBS in an appropriate tube in an appropriate rack in a BSC based on the following formulas:
where x = μl of stock needed. Then, subtract total μl of final inoculum from μl of stock needed to calculate the volume of PBS needed.For example, if your stock is 1 × 106 CFU/ml, you want to infect mice with 1000 CFU in 30 μl, and you want a final stock volume of 5 ml or 5000 μl.We recommend quantifying arthroconidia (see Basic Protocol 3) just before infection to ensure accurate dosage.
-
Lightly anesthetize mouse either by intraperitoneal injection of ketamine/xylazine or by isoflurane inhalation.
We recommend ketamine/xylazine, as determined by our institutional veterinarian. A dose of ketamine/xylazine of 80/8 mg/kg is commonly used in C57BL/6 mice. Mouse strains may differ in required dosage. Weigh mice to determine the appropriate dosage.
-
Ensure that mouse is sufficiently anesthetized.
Mice should be responsive to toe pinch but not mobile.
-
Vortex inoculum and draw up appropriate volume of inoculum into a pipet.
We recommend 30 μl total using a 200-μl pipet tip.
-
Gently scruff back of the neck and cup mouse in the less dominant hand (Fig. 8). To ensure that the mouse is sufficiently anesthetized, lightly tap nose with the pipet tip. If the mouse flinches, allow sedation to progress further. With the dominant hand, pipet inoculum into the mouse nares ~5 μl at a time, switching between the left and right nares (Fig. 8).
Allow the natural breathing rhythm of the mouse to bring the inoculum into the nares.
Sneezing may occur and prevents delivering the full inoculum. Ensure that the mouse is sufficiently anesthetized to avoid sneezing or swallowing of inoculum.
-
Label mouse as appropriate for the laboratory (i.e., with a Sharpie marker or ear punch) and dab nose with a gauze pad dipped in 70% ethanol. Then, return mouse to its cage.
Be gentle with the ethanol; this removes exterior arthroconidia.
Repeat steps 2 to 6 with remaining mice.
Ensure that all mice are awake and alert before leaving facility.
Plate inoculum using a dilution series as described in Basic Protocol 3, steps 21 to 30.
Read CFUs of the inoculum plates in 3 to 4 days.
Figure 8.
Graphical overview of an intranasal murine Coccidioides infection. Using thumb and index finger, with the nondominant hand, hold the mouse skin behind the neck and cradle the mouse in hand. Using the dominant hand, pipet arthroconidia slowly, alternating between the mouse’s left and right nares. Illustration created with BioRender.
REAGENTS AND SOLUTIONS
Fungal freezing medium
1% (w/v) glucose (VWR)
1% (w/v) yeast extract (Difco)
10% (v/v) glycerol
Autoclave at 121°C for 15 min
Add sterilized 0.01% (w/v) Tween-20
Store ≤1 year at 4°C
Glucose yeast extract (GYE) liquid medium, 2x
2% (w/v) glucose (VWR)
1% (w/v) yeast extract (Difco)
Autoclave at 121°C for 15 min
Store ≤6 months at 4°C
Glucose yeast extract (GYE) solid agar plates, 2x
2% (w/v) glucose (VWR)
1% (w/v) yeast extract (Difco)
1.5% (w/v) bacteriological agar (Difco)
Autoclave at 121°C for 15 min
Pour 20 ml into 100 × 15–mm petri dishes (VWR)
Store ≤6 months at 4°C
As an alternative to plates, pour 35 ml into vented 125-ml Erlenmeyer flasks (VWR).
COMMENTARY
Background Information
There are a limited number of publications detailing laboratory methodology for Coccidioides species. Historically, knowledge has been only included in the methods section of published experimental findings or shared between research laboratories rather than published. The methods included herein to culture the environmental phase of the lifecycle, namely filamentous mycelia or arthroconidia, are based on methods widely used by the Coccidioides research community, with small variations. For example, some laboratories use less glucose (1% rather than 2%) and yeast (0.5% rather than 1%) in solid or liquid medium (Hung, Wise, & Cole, 2012). We have used these formulations as well and did not observe differences in fungal growth or experimental outcomes. Similarly, when harvesting arthroconidia, some laboratories use syringes containing nylon fiber rather than the nested filters that we suggest (Hung, Wise, et al., 2012). This is simply a matter of preference and should not impact the overall outcome. The parasitic spherule structure has routinely been grown in Converse medium since 1955. This medium formulation can be tedious to make because of the 11 different components but has the benefit of being widely used and cited (Cole & Hung, 2001; Cole, Seshan, et al., 1988; Hung, Yu, Seshan, Reichard, & Cole, 2002; Viriyakosol, Singhania, et al., 2013; Whiston et al., 2012; Wise et al., 2013). We routinely use RPMI-sph medium, which was originally described in 1985 and compared to Converse medium (Mead et al., 2019). This medium has the advantage of being simple to make and compatible with cell lines. Endospores are discussed in the literature; however, methods for encouraging endosporulation are limited. One study suggested that removing hyphal fragments, replacing the medium, and maintaining 10% CO2 produces a continuous culture of spherules, and therefore endospores (Breslau & Kubota, 1964). We based our laboratory methodology and Alternate Protocol 4 on this publication. For techniques that are beyond the scope of this publication, we recommend Hung, Wise, et al. (2012) for genetic modification strategies and Mitchell, Grys, & Lake (2020) for protein extraction methodology.
Critical Parameters and Troubleshooting
Basic Protocol 1 and Alternate Protocol 1: Coccidioides can take 3 to 5 days to show visible growth at 30°C and up to 1 week at room temperature. If you do not observe growth after 1 week, restart the cultures. If the cultures are stringy and clumpy, they are overgrown. These observations are based on common laboratory strains, and variation has been observed. If working with a novel strain, we recommend including a blank medium control to rule out contamination issues. If the extraction yield is low, it is most likely that the fungal cells were not lysed. We recommend using a microscope to see whether intact cells are present. Simply compare the non-homogenized cells to homogenized ones.
Basic Protocol 2: Coccidioides can take 3 to 5 days to show visible growth at 30°C and up to 1 week at room temperature. If you do not observe growth after 1 week, restart the cultures. Again, variation among strains exists, so if using a novel strain, microscopic assessment is advised. Keep in mind that moisture buildup on the plates will slow the development of arthroconidia. We have found that opening the incubator weekly can reduce overall moisture buildup. Alternatively, plates can be moved to the bench for 1 to 2 days to allow moisture to dry and then returned to the incubator. If you cannot prevent moisture buildup in the incubator, we recommend growing the strain at room temperature, which should relieve the issue.
Alternate Protocol 2: Coccidioides can take 3 to 5 days to show visible growth at 30°C, and up to 1 week at room temperature. If you do not observe growth after 1 week, restart the cultures.
Basic Protocol 3: If your harvest produces a significantly lower yield than expected, it is possible that the arthroconidia are not fully developed or are overly dry/old. In our laboratory, 6 weeks produces optimal cultures; however, variation can occur. This could be due to elevation, humidity, laboratory-specific, and/or strain-specific factors. See Figures 4 and 6 for phenotypic evidence of arthroconidia. If you see a significant drop-off in viability, concentrate your cells into a lower volume. We have observed a rapid decline in viability when arthroconidia are stored at concentrations <105 cells/ml.
Alternate Protocol 3: If glycerol stocks are not viable, increase the concentration of cells per tube.
Basic Protocol 4: There can be several reasons that arthroconidia do not develop into spherules. Keep in mind that spherule development varies among strains. We have observed this in our laboratory, and it has been reported in the literature (Berman, Friedman, Roessler, & Smith, 1956; Friedman, Papagianis, Berman, & Smith, 1953). Successful growth of these structures can be complex and may require one or two attempts before success. First, ensure that the arthroconidia are viable by plating on 2× GYE. Second, be sure to calibrate the temperature and CO2 of your incubator. Finally, we have found that RPMI-sph medium is more reliable than Converse medium. If your experimental goals require the use of Converse medium, make sure it is fresh. See our publication for in-depth discussion of these medium options (Mead et al., 2019).
Alternate Protocol 4: Endospores are, by definition, premature spherules. Discerning endospores (2 to 4 μm) from small spherules (4 to 8 μm) can be difficult. Unfortunately, there are very few publications with methods that focus on obtaining endospores.
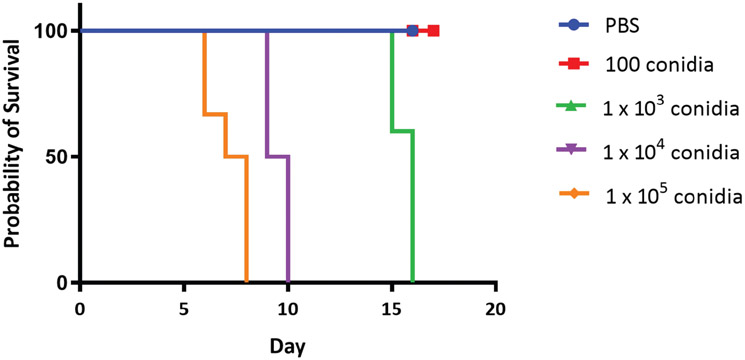
Basic Protocol 5: Symptoms and survival are based on the inoculum (Fig. 9). Upon completion of fungal burden analysis of tissue at the study endpoint, if it appears that one of the mice did not get infected (compared to control mice) or did not receive a full dose based on the fungal burden of the experimental groups, this could be for several reasons. Mice that are not fully anesthetized can either swallow or sneeze out inoculum, preventing full infection. Ensure that the mice are completely anesthetized before intranasal infection. Ensure also that you have used the desired inoculum by plating after infection. Additionally, be sure to protect the pipet tips and inoculum from disinfectant spray, as this could inactivate the inoculum.
Figure 9.
Survival of CD1 mice infected with a range of dosages of the C. posadasii strain Silveira. CD1 mice received an intranasal infection with 100, 1 × 103, 1 × 104, or 1 × 105 arthroconidia of strain Silveira (n = 5 to 7) or were given control PBS (n = 3). Mice were observed for up to 17 days post-infection for survival analysis.
Understanding Results
Basic Protocol 1 and Alternate Protocol 1: You should expect a healthy, fluffy fungal culture that can be easily caught and rinsed in the nested filter after 3 to 5 days of growth. There should be enough material for several nucleic acid extractions per sample.
Basic Protocol 2: When the culture is grown on solid medium, the filamentous colony will cover the surface of the plate within ~2 weeks. Arthroconidia will develop as nutrition depletes and the colony dries. Most Coccidioides cultures are white, with even margins (C. posadasii Silveira, C735, or C. immitis 2006, RS). However, slight pigmentation and irregular margins have been observed in other strains.
Alternate Protocol 2: When a fungal plug is grown on solid medium at 30°C, the filamentous colony will cover the surface of the plate within ~2 weeks. Most Coccidioides cultures are white, with even margins (C. posadasii Silveira, C735, or C. immitis 2006, RS). However, slight pigmentation and irregular margins have been observed.
Basic Protocol 3: If using the common laboratory strains C. posadasii Silveira or C. immitis RS, you would expect that 20 plates or 10 vented suspension flasks containing 2× GYE solid agar should result in ~108 cells/ml in 5 to 7 ml PBS. Concentrate or dilute as necessary.
Alternate Protocol 3: When stored at −80°C, isolates should be viable for years (we have observed viability for up to 7 to 10 years). As a precaution, confirm viability before disposing of active growth.
Basic Protocol 4: Macroscopically, cultures appear slightly cloudy compared to an uninoculated medium control. Microscopically, structures should be evident after 48 to 72 hr of growth, with minimal hyphae present. In vitro, spherules are distinctly round and range from 8 to 25 μm in diameter (Figs. 1 and 7). Expect a small number of nonviable arthroconidia to remain in the culture; these structures are barrel shaped and 2 to 4 μm in diameter (Fig. 1).
Alternate Protocol 4: Macroscopically, cultures appear slightly cloudy compared to an uninoculated medium control. Microscopically, endospores should be evident 24 hr after medium change. Endospores will be small (2 to 5 μm) and often clumped together (Fig. 7D).
Basic Protocol 5: Infection studies vary depending on dosage and the strain of mice used. Endosporulation occurs in vivo between 96 and 120 hr, increasing fungal burden and disease symptoms. Figure 9 demonstrates the results for outbred CD1 mice given varying doses of the C. posadasii Silveira strain. CD1 mice infected with 100 conidia or given control PBS demonstrated a 100% survival rate at day 17 post-infection (Fig. 9), whereas mice infected with 1 × 103, 1 × 104, or 1 × 105 conidia demonstrated a median survival rate of 16, 9.5, and 7.5 days post-infection, respectively. These results demonstrate that CD1 mice are more resistant to infection with Coccidioides compared to other mouse strains, such as BALB/c or C57BL/6 (Cox, Kennell, Boncyk, & Murphy, 1988; Hung, Zhang, et al., 2018; Magee & Cox, 1995).
Time Considerations
Basic Protocol 1: Starting liquid cultures takes ~15 min per isolate, and harvesting takes ~15 min per isolate.
Alternate Protocol 1: Starting liquid cultures takes ~15 min per isolate, and harvesting takes ~45 min per isolate.
Basic Protocol 2: Starting 20 plates or 10 flasks takes ~1 hr.
Alternate Protocol 2: Subculture of one isolate takes ~15 min.
Basic Protocol 3: Harvesting 20 plates or 10 flasks takes ~3 hr.
Alternate Protocol 3: Creating long-term storage stocks for one isolate takes ~10 min.
Basic Protocol 4: Starting spherule cultures takes ~15 min per isolate, and harvesting takes ~45 min.
Alternate Protocol 4: Passaging spherules takes ~30 min per isolate.
Basic Protocol 5: Infecting 20 mice takes ~3 hr with two personnel.
Acknowledgments
Table 1, detailing the stock solutions needed to prepare 1 L Converse medium, was developed by Rosemary Hayden. Funding to support this work was provided to BMB by ABRC 14-082975, ABRC 16-162415, NIH/NIAID K22-A1104801, NIH/NIAID R21 AI128536, State of Arizona TRIF funding, and the Flinn Foundation.
Footnotes
Internet Resources
https://www.beiresources.org/BEIHighlights1.aspx?ItemId=71&ModuleId=14004
BEI Resources, for ordering Coccidioides cultures
https://unicornbags.com/product/type-10tbox-of-1000/
Unicorn Bags, for long-term storage of stacks of plates
http://www.scientificdevice.com/shrink-seals/
Shrink seals, for sealing individual plates
https://us.vwr.com/store/product/4637081/transfertube-disposable-harvesters-spectrum-laboratories
Transfertube® Disposable Harvesters (Spectrum® Laboratories), for subculturing fungal plates
The Coccidioidomycosis Study Group, which group oversees conferences, annual meetings, and research studies; much of the documented knowledge of the pathogenesis, mycology, and clinical aspects of Coccidioidomycosis originated from studies performed by this research group
https://pubmed.ncbi.nlm.nih.gov/22328372/
Resource for genetic manipulation of Coccidioides
https://pubmed.ncbi.nlm.nih.gov/31691790/
Resource for spherulin
Literature Cited
- Baptista-Rosas RC, Catalán-Dibene J, Romero-Olivares AL, Hinojosa A, Cavazos T, & Riquelme M (2012). Molecular detection of Coccidioides spp. from environmental samples in Baja California: Linking Valley Fever to soil and climate conditions. Fungal Ecology, 5(2), 177–190. doi: 10.1016/j.funeco.2011.08.004. [DOI] [Google Scholar]
- Baptista-Rosas RC, Hinojosa A, & Riquelme M (2007). Ecological niche modeling of Coccidioides spp. in western North American deserts. Annals of the New York Academy of Sciences, 1111, 35–46. doi: 10.1196/annals.1406.003. [DOI] [PubMed] [Google Scholar]
- Berman RJ, Friedman L, Roessler WG, & Smith CE (1956). The virulence and infectivity of twenty-seven strains of Coccidioides immitis. American Journal of Hygiene, 64(2), 198–210. doi: 10.1093/oxfordjournals.aje.a119834. [DOI] [PubMed] [Google Scholar]
- Breslau AM, & Kubota MY (1964). Continuous in vitro cultivation of spherules of Coccidioides immitis. Journal of Bacteriology, 87, 468–472. doi: 10.1128/JB.87.2.468-472.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano A, Zhang H, Ostroff GR, Dos Santos Dias L, Wuthrich M, Klein BS, … Hung CY (2020). CARD9-associated Dectin-1 and Dectin-2 are required for protective immunity of a multivalent vaccine against Coccidioides posadasii infection. Journal of Immunology, 204(12), 3296–3306. doi: 10.4049/jimmunol.1900793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Lopez N, & Hung CY (2017). Immune response to coccidioidomycosis and the development of a vaccine. Microorganisms, 5(1), 13. doi: 10.3390/microorganisms5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2009). Biosafety in microbiological and biomedical laboratories (5th ed.). Washington, D.C.: Centers for Disease Control and Prevention. Retrieved from https://www.cdc.gov/labs/BMBL.html. [Google Scholar]
- Chiller T (2019). Overview of endemic mycoses. Paper presented at the Vaccine Strategies for Endemic Fungal Pathogens meeting, Rockville, MD. [Google Scholar]
- Cole GT, & Hung CY (2001). The parasitic cell wall of Coccidioides immitis. Medical Mycology, 39(Suppl 1), 31–40. doi: 10.1080/mmy.39.1.31.40. [DOI] [PubMed] [Google Scholar]
- Cole GT, Seshan KR, Franco M, Bukownik E, Sun SH, & Hearn VM (1988). Isolation and morphology of an immunoreactive outer wall fraction produced by spherules of Coccid ioides immitis. Infection and Immunity, 56(10), 2686–2694. doi: 10.1128/IAI.56.10.2686-2694.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse JL (1955). Growth of spherules of Coccidioides immitis in a chemically defined liquid medium. Proceedings of the Society for Experimental Biology and Medicine, 90(3), 709–711. doi: 10.3181/00379727-90-22144. [DOI] [PubMed] [Google Scholar]
- Cox RA, Kennell W, Boncyk L, & Murphy JW (1988). Induction and expression of cell-mediated immune responses in inbred mice infected with Coccidioides immitis. Infection and Immunity, 56(1), 13–17. doi: 10.1128/IAI.56.1.13-17.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rocio Reyes-Montes M, Perez-Huitron MA, Ocana-Monroy JL, Frias-De-Leon MG, Martinez-Herrera E, Arenas R, & Duarte-Escalante E (2016). The habitat of Coccidioides spp. and the role of animals as reservoirs and disseminators in nature. BMC Infectious Diseases, 16(1), 550. doi: 10.1186/s12879-016-1902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drutz DJ, & Huppert M (1983). Coccidioidomycosis: Factors affecting the host-parasite interaction. Journal of Infectious Diseases, 147(3), 372–390. doi: 10.1093/infdis/147.3.372. [DOI] [PubMed] [Google Scholar]
- Emmons C (1942). Isolation of Coccidioides from soil and rodents. Public Health Reports, 57(4), 109–111. doi: 10.2307/4583988.19315853 [DOI] [Google Scholar]
- Fisher MC, Koenig GL, White TJ, & Taylor JW (2002). Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia, 94(1), 73–84. doi: 10.1080/15572536.2003.11833250. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Koenig GL, White TJ, San-Blas G, Negroni R, Alvarez IG, … Taylor JW (2001). Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proceedings of the National Academy of Sciences, 98(8), 4558–4562. doi: 10.1073/pnas.071406098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Papagianis D, Berman RJ, & Smith CE (1953). Studies on Coccidioides immitis: Morphology and sporulation capacity of forty-seven strains. Journal of Laboratory and Clinical Medicine, 42(3), 438–444. doi: 10.5555/uri:pii:0022214353902571. [DOI] [PubMed] [Google Scholar]
- Gorris ME, Cat LA, Zender CS, Treseder KK, & Randerson JT (2018). Coccidioidomycosis dynamics in relation to climate in the Southwestern United States. Geohealth, 2(1), 6–24. doi: 10.1002/2017GH000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm PS, Hutchison MI, Leonard P, Melman S, & Natvig DO (2019). First analysis of human Coccidioides isolates from New Mexico and the Southwest Four Corners Region: Implications for the distributions of C. posadasii and C. immitis and human groups at risk. Journal of Fungi, 5(3), 74. doi: 10.3390/jof5030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden CA, Hung CY, Zhang H, Negron A, Esquerra R, Ostroff G, … Howard JA (2019). Maize-produced Ag2 as a subunit vaccine for Valley fever. The Journal of Infectious Diseases, 220(4), 615–623. doi: 10.1093/infdis/jiz196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RF, & Pappagianis D (1983). Inhibition of chitin synthesis in the cell wall of Coccidioides immitis by polyoxin D. Journal of Bacteriology, 154(1), 488–498. doi: 10.1128/JB.154.1.488-498.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Castro-Lopez N, & Cole GT (2014). Vaccinated C57BL/6 mice develop protective and memory T cell responses to Coccidioides posadasii infection in the absence of interleukin-10. Infection and Immunity, 82(2), 903–913. doi: 10.1128/IAI.01148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Gonzalez A, Wuthrich M, Klein BS, & Cole GT (2011). Vaccine immunity to coccidioidomycosis occurs by early activation of three signal pathways of T helper cell response (Th1, Th2, and Th17). Infection and Immunity, 79(11), 4511–4522. doi: 10.1128/IAI.05726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, del Pilar Jimenez-Alzate MP, Gonzalez A, Wuthrich M, Klein BS, & Cole GT (2014). Interleukin-1 receptor but not Toll-like receptor 2 is essential for MyD88-dependent Th17 immunity to Coccidioides infection. Infection and Immunity, 82(5), 2106–2114. doi: 10.1128/IAI.01579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Wise HZ, & Cole GT (2012). Gene disruption in Coccidioides using hygromycin or phleomycin resistance markers. Methods in Molecular Biology, 845, 131–147. doi: 10.1007/978-1-61779-539-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Yu JJ, Seshan KR, Reichard U, & Cole GT (2002). A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory Fungal pathogen. Infection and Immunity, 70(7), 3443–3456. doi: 10.1128/iai.70.7.3443-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Zhang H, Castro-Lopez N, Ostroff GR, Khoshlenar P, Abraham A, … Yu JJ (2018). Glucan-Chitin particles enhance Th17 Response and improve protective efficacy of a multivalent antigen (rCpa1) against pulmonary Coccidioides posadasii infection. Infection and Immunity, 86(11), e00070–18. doi: 10.1128/iai.00070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert M, Sun SH, & Harrison JL (1982). Morphogenesis throughout saprobic and parasitic cycles of Coccidioides immitis. Mycopathologia, 78(2), 107–122. doi: 10.1007/bf00442634. [DOI] [PubMed] [Google Scholar]
- Kirkland TN, & Fierer J (1983). Inbred mouse strains differ in resistance to lethal Coccidioides immitis infection. Infection and Immunity, 40(3), 912–916. doi: 10.1128/IAI.40.3.912-916.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz SA, Drutz DJ, Huppert M, Sun SH, & DeMarsh PL (1984). The critical role of CO2 in the morphogenesis of Coccidioides immitis in cell-free subcutaneous chambers. The Journal of Infectious Diseases, 150(1), 127–134. doi: 10.1093/infdis/150.1.127. [DOI] [PubMed] [Google Scholar]
- Kollath DR, Miller KJ, & Barker BM (2019). The mysterious desert dwellers: Coccidioides immitis and Coccidioides posadasii, causative fungal agents of coccidioidomycosis. Virulence, 10(1), 222–233. doi: 10.1080/21505594.2019.1589363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollath DR, Teixeira MM, Funke A, Miller KJ, & Barker BM (2020). Investigating the role of animal burrows on the ecology and distribution of Coccidioides spp. in Arizona soils. Mycopathologia, 185(1), 145–159. doi: 10.1007/s11046-019-00391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ER, Bowers JR, & Barker BM (2015). Dust devil: The life and times of the fungus that causes valley Fever. PLOS Pathogens, 11(5), e1004762. doi: 10.1371/journal.ppat.1004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marsden-Haug N, Hurst S, Hill H, Gade L, Driebe EM, … Chiller T (2015). Valley fever: Finding new places for an old disease: Coccidioides immitis found in Washington State soil associated with recent human infection. Clinical Infectious Diseases, 60(1), e1–3. doi: 10.1093/cid/ciu681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee DM, & Cox RA (1995). Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infection and Immunity, 63(9), 3514–3519. doi: 10.1128/IAI.63.9.3514-3519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Galgiani JN, Kroken S, & Orbach MJ (2006). Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genetics and Biology, 43(11), 775–788. doi: 10.1016/j.fgb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Mead HL, Blackmon VA, Vogler JA, & Barker MB (2019). Heat inactivation of Coccidioides posadasii and Coccidioides immitis for use in lower biosafety containment. Applied Biosafety, 24, 123–128. doi: 10.1177/1535676019856525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead HL, Roe CC, Higgins Keppler EA, Van Dyke MCC, Laux KL, Funke AL, … Barker BM (2020). Defining critical genes during spherule remodeling and endospore development in the fungal pathogen, Coccidioides posadasii. Frontiers in Genetics, 11, 483. doi: 10.3389/fgene.2020.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead HL, Teixeira MM, Galgiani JN, & Barker BM (2019). Characterizing in vitro spherule morphogenesis of multiple strains of both species of Coccidioides. Medical Mycology, 57(4), 478–488. doi: 10.1093/mmy/myy049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell NM, Grys TE, & Lake DF (2020). Carbo-loading in Coccidioides spp.: A quantitative analysis of CAZyme abundance and resulting glycan populations. Glycobiology, 30(3), 186–197. doi: 10.1093/glycob/cwz092. [DOI] [PubMed] [Google Scholar]
- Mitchell NM, Magee DM, Grys TE, & Lake DF (2019). Evaluation of Virex(R) II 256 and Virex(R) Tb as disinfectants of the dimorphic fungi Coccidioides immitis and Coccidioides posadasii. Applied Biosafety, 24(1), 30–33. doi: 10.1177/1535676018818560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C, Barker BM, Hoover S, Nix DE, Ampel NM, Frelinger JA, … Galgiani JN (2013). Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clinical Microbiology Reviews, 26(3), 505–525. doi: 10.1128/CMR.00005- 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odio CD, Marciano BE, Galgiani JN, & Holland SM (2017). Risk factors for disseminated Coccidioidomycosis, United States. Emerging Infectious Diseases, 23(2), 308–311. doi: 10.3201/eid2302.160505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkus AF, Baum LL, Ellis RB, Stern M, & Danley DL (1985). Pure spherules of Coccidioides immitis in continuous culture. Journal of Clinical Microbiology, 22(2), 165–167. doi: 10.1128/JCM.22.2.165-167.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, … Taylor JW (2009). Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Research, 19(10), 1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, & Barker BM (2019). The endozoan, small-mammal reservoir hypothesis and the life cycle of Coccidioides species. Medical Mycology, 57(Supplement_1), S16–S20. doi: 10.1093/mmy/myy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GR 3rd, Lewis JS 2nd, Nix DE, & Patterson TF (2019). Current concepts and future directions in the pharmacology and treatment of Coccidioidomycosis. Medical Mycology, 57(Supplement_1), S76–S84. doi: 10.1093/mmy/myy029. [DOI] [PubMed] [Google Scholar]
- Tong DQ, Wang JXL, Gill TE, Lei H, & Wang B (2017). Intensified dust storm activity and Valley fever infection in the southwestern United States. Geophysical Research Letters, 44(9), 4304–4312. doi: 10.1002/2017GL073524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CA, Anderson SM, Imholte SB, Erhart LM, Chen S, Park BJ, … Sunenshine RH (2010). Enhanced surveillance of coccidioidomycosis, Arizona, USA, 2007-2008. Emerging Infectious Diseases, 16(11), 1738–1744. doi: 10.3201/eid1611.100475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia L, Nix D, Wright M, Lindberg E, Fagan T, Lieberman D, … Galgiani JN (2006). Coccidioidomycosis as a common cause of community-acquired pneumonia. Emerging Infectious Diseases, 12(6), 958–962. doi: 10.3201/eid1206.060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke MCC, Thompson GR, Galgiani JN, & Barker BM (2019). The rise of Coccidioides: Forces against the dust devil unleashed. Frontiers in Immunology, 10,2188. doi: 10.3389/fimmu.2019.02188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viriyakosol S, Kapoor M, Okamoto S, Covel J, Soltow QA, Trzoss M, … Fierer J (2019). APX001 and other Gwt1 inhibitor pro-drugs are effective in experimental Coccidioides immitis pneumonia. Antimicrobial Agents and Chemotherapy, 63(2), e01715–18. doi: 10.1128/AAC.01715-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viriyakosol S, Singhania A, Fierer J, Goldberg J, Kirkland TN, & Woelk CH (2013). Gene expression in human fungal pathogen Coccidioides immitis changes as arthroconidia differentiate into spherules and mature. BMC Microbiology, 13, 121. doi: 10.1186/1471-2180-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AJ, Nottingham R, Parise KL, Keim P, & Barker BM (2015). Effective disinfectants for Coccidioides immitis and C. posadasii. Applied Biosafety, 20(3), 154–158. doi: 10.1177/153567601502000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiston E, Zhang Wise H, Sharpton TJ, Jui G, Cole GT, & Taylor JW (2012). Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLOS One, 7(7), e41034. doi: 10.1371/journal.pone.0041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HZ, Hung CY, Whiston E, Taylor JW, & Cole GT (2013). Extracellular ammonia at sites of pulmonary infection with Coccidioides posadasii contributes to severity of the respiratory disease. Microbial Pathogenesis, 59-60, 19–28. doi: 10.1016/j.micpath.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Chen X, Selby D, Hung CY, Yu JJ, & Cole GT (2009). A genetically engineered live attenuated vaccine of Coccidioides posadasii protects BALB/c mice against coccidioidomycosis. Infection and Immunity, 77(8), 3196–3208. doi: 10.1128/IAI.00459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]