Abstract
DcrB is an 18 kilodalton lipoprotein that contains a single domain of unknown function. DcrB is found within Enterobacteriaceae, a family of Gram-negative bacteria which includes pathogens that can cause food-borne illness and hospital-acquired infections. In Salmonella enterica serovar Typhimurium, DcrB is up-regulated by conditions that promote the production of known virulence factors. We determined the structure of a truncated form of DcrB from Salmonella to 1.92 Å resolution by X-ray crystallography. This truncated form, DcrBΔ37, contains the entire domain of unknown function but lacks the lipoprotein signal sequence (residues 1–20) as well as residues 21–37. The DcrBΔ37 monomer contains the Mog1p/PsbP-like fold, which is found in functionally diverse proteins in mammals, yeast, plants, and cyanobacteria. Interestingly, DcrBΔ37 crystallized as a domain-swapped homodimer in which the N-terminal β-hairpin extends from one protomer to interact with the core of the second protomer. This domain-swapping indicates that the N-terminal portion of the Mog1p/PsbP-like fold likely has conformational flexibility. Overall, our results provide the first example of an enterobacterial protein that contains the Mog1p/PsbP-like fold and expands knowledge of the structural and phylogenetic diversity of Mog1p/PsbP-like proteins.
Keywords: Mog1p/PsbP-like fold, Salmonella pathogenesis, lipoprotein, domain of unknown function
1. Introduction
Antibiotic resistance in bacterial pathogens poses an extreme risk to public health worldwide (Frieden, 2013). One strategy to confront antibiotic-resistant pathogens is to target the ability of bacteria to adapt to hostile conditions encountered during infection of a host, (Clatworthy et al., 2007; Lee et al., 2003), a process that is largely controlled by virulence regulatory systems (Beier and Gross, 2006; Calva and Oropeza, 2006). Nevertheless, many of the proteins known to be activated by virulence regulatory systems contain domains of unknown structure and/or function, which prevents understanding of their role in bacterial virulence.
Salmonella enterica is a facultative intracellular pathogen that is a major cause of food-borne gastroenteritis and typhoid fever worldwide (Dougan and Baker, 2014; Galán, 2016; Ohl and Miller, 2001). A key regulator of Salmonella virulence is the PhoP/PhoQ two-component system (Fields et al., 1989; Groisman, 2001). The PhoP/PhoQ system is required for the virulence of Salmonella enterica and other pathogenic enterobacteria (Fields et al., 1989; Galán and Curtiss, 1989; Garcia Véscovi et al., 1994; Groisman, 2001; Moss et al., 2000; Oyston et al., 2000). PhoP/PhoQ-activated gene products confer resistance to host innate immune defenses (Dalebroux and Miller, 2014; Ernst et al., 2006).
Although certain PhoP-activated gene products have known functions, PhoP also up-regulates hypothetical proteins of unknown function (Charles et al., 2009; Yu and Guo, 2011). One of these PhoP-activated proteins is encoded by the gene STM3580. This 185 amino acid protein is an ortholog of the Escherichia coli DcrB protein, a cytoplasmic membrane lipoprotein that is required for phage C1 infection (Likhacheva et al., 1996; Samsonov et al., 2002) but has no reported biochemical function. The STM3580 protein, which we will refer to as DcrB, consists of an N-terminal lipoprotein signal sequence (Met1–Cys20), a short linker region (residues 21–50), and a single predicted domain of unknown function (DUF), termed DUF1795 (residues 51–181). Orthologs of DcrB are found in several enterobacterial pathogens and contain a highly conserved amino acid sequence (Supplementary Fig. 1).
Here, we report the first three-dimensional structure of the DcrB protein from Salmonella enterica, determined by X-ray crystallography to 1.92 Å resolution. The structure reveals that DcrB contains the Mog1p/PsbP-like fold. Interestingly, the structure of DcrB reveals a domain-swapped dimer. These results provide the foundation for understanding the function of DcrB proteins in enterobacterial pathogens.
2. Materials and Methods
2.1. Materials
Oligonucleotide primers used in this study are listed in Supplementary Table 1 and were obtained from Integrated DNA Technologies (Coralville, IA). All chemicals were molecular biology grade. LB (Luria-Bertani) broth and LB agar were obtained as premixed powder from BD Difco (Franklin Lakes, NJ). Terrific broth and 1× M9 minimal salts media were prepared as described (Green and Sambrook, 2012). l-selenomethionine and imidazole were obtained from Acros Organics (Fair Lawn, NJ). HisPur™ Ni-NTA resin was obtained from Thermo Scientific (Rockford, IL). Ammonium sulfate was obtained from Fisher Scientific (Fair Lawn, NJ). Isopropyl β-D-1-thiogalactopyranoside (IPTG) was obtained from GoldBio (St. Louis, MO). All other chemicals were obtained from Sigma Aldrich (St. Louis, MO). Phusion® High-Fidelity DNA polymerase, T4 DNA ligase, and restriction endonucleases were obtained from New England Biolabs (Ipswich, MA). Taq DNA Polymerase was obtained from GenScript (Piscataway, NJ). Ampicillin was used at a final concentration of 50 μg/mL in media. Strain JM273, a ΔphoA748::kan derivative of C41(DE3) Escherichia coli, was prepared by P1 transduction using strain JW0374–1 as donor as previously described (Miller, 1972). Plasmid pBR322-dcrB was generously provided by Eduardo Groisman (Yale School of Medicine). Bacterial strains and plasmids used in this study are listed in Supplementary Table 2.
2.2. Construction of a plasmid for production of His6-DcrBΔ20
The sequence coding for amino acids 21–185 of DcrB (referred to as DcrBΔ20) was amplified using pBR322-dcrB as the template, primers JM012 and JM013, and Taq DNA polymerase. Primer JM012 was designed to incorporate a His6-tag at the N-terminus of DcrBΔ20. The resulting PCR product was digested with NdeI and HindIII-HF and ligated to the pET-22b(+) vector that had been digested with NdeI and HindIII-HF. The sequence of the resulting plasmid, pET-22b-His6dcrBΔ20, was verified by DNA sequence analysis using primers T7-Promoter and T7-Terminator. A separate plasmid, pET-22b-His6dcrBΔ37, encoding His6-DcrBΔ37 (corresponding to amino acid residues 38–185) was obtained from GenScript USA (Piscataway, NJ).
2.3. Production and purification of His6-DcrBΔ20 and His6-DcrBΔ37
LB supplemented with ampicillin (50 μg/mL) was inoculated with strain JM280 (for His6-DcrBΔ20) or strain JM296 (for His6-DcrBΔ37) and incubated at 37 °C with aeration (240 rpm) overnight. Terrific Broth supplemented with ampicillin (50 μg/mL) was inoculated 1/250 with the overnight culture and incubated at 37 °C with aeration (140 rpm) until an OD600 of 0.50 was reached. IPTG was added to the culture to a final concentration of 1 mM. After addition of IPTG, the culture was incubated at 37 °C with aeration (140 rpm) for 3 hours, and cells were harvested from the culture by centrifugation at 7,000 × g for 10 minutes.
The cell pellet from 250 mL of culture was resuspended in buffer A (20 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol (Tris) pH 8.0, 300 mM NaCl). The cell suspension was sonicated on ice with a Fisher Scientific 550 sonic dismembrator. The sonicated extract was centrifuged at 21,130 × g for 15 minutes. The supernatant was loaded onto HisPur™ Ni-NTA resin that had been equilibrated with buffer A using a Biologic low-pressure chromatography system (Bio-Rad) at a flow rate of 3.0 mL/minute. His6-DcrB was eluted using a linear gradient to 100% buffer B (20 mM Tris pH 8.0, 300 mM NaCl, 400 mM imidazole). Fractions were analyzed for purity by SDS-PAGE with Coomassie blue staining (Laemmli, 1970). Fractions containing purified protein were combined, and ethylenediaminetetraacetic acid (EDTA) was added to 1 mM final concentration. The sample was dialyzed against 4 L of 20 mM Tris pH 8.0, 1 mM EDTA and then against 4 L of 20 mM Tris pH 8.0 using dialysis tubing with 6,000–8,000 nominal molecular weight cutoff. The dialyzed sample was concentrated to 15 mg/mL with Amicon Ultra-15 regenerated cellulose centrifugal filters (10,000 nominal molecular weight cutoff). Protein concentrations were determined by measuring UV absorbance at 280 nm, using the calculated extinction coefficient of 2,980 M−1cm−1 (Gasteiger et al., 2005).
2.4. Production of selenomethionine-incorporated His6-DcrBΔ20
Selenomethionine was incorporated into His6-DcrBΔ20 using a previously reported procedure (Van Duyne et al., 1993). SeMet His6-DcrBΔ20 was purified using the same procedure as described above for His6-DcrBΔ20.
2.5. Analysis of secondary structure by circular dichroism
Circular dichroism (CD) wavelength scans were carried out using an AVIV Biomedical Inc. Circular Dichroism Spectrometer, Model 420. CD signal of 20 μM DcrB in 20 mM Tris pH 8.0 was analyzed from 190–300 nm with a step length of 1 nm, an averaging time of 2 seconds, and a bandwidth of 2 nm at 25 °C.
2.6. Determination of the structure of DcrB using X-ray crystallography
Crystallization conditions were identified for native His6-DcrBΔ20 using a sparse matrix screen (Crystal Screen 2, HR2–112, Hampton Research, Aliso Viejo, California) and subsequently refined. Crystals used for diffraction studies of SeMet-incorporated His6-DcrBΔ20 were grown in 2.2 M ammonium sulfate and 100 mM Tris pH 8.5. Crystals used for diffraction studies of His6-DcrBΔ37 were grown in 2.0 M ammonium sulfate and 100 mM Tris pH 8.6. Protein (12 mg/mL) was mixed with reservoir solution at a 1:1 ratio and crystallized by hanging drop vapour diffusion at 295 K. Ten microliters of reservoir solution containing 25% glycerol were added directly to the drops with crystals, which were subsequently flash frozen in liquid nitrogen. X-ray diffraction data was collected at 100 K at the Advanced Photon Source on beamline LS-CAT 21-ID-D at a wavelength of 0.97895 Å for SeMet-incorporated His6-DcrBΔ20 or at a wavelength of 0.97851 Å for native His6-DcrBΔ37.
Diffraction data were indexed and scaled using XDS (Kabsch, 2010) and AIMLESS (Evans, 2011) using the autoPROC software (Vonrhein et al., 2011). The structure of SeMet-incorporated His6-DcrBΔ20 was solved using single-wavelength anomalous dispersion (SAD) phasing to a resolution of 2.30 Å. Selenium positions were identified and an initial model was built using the CRANK2 experimental phasing pipeline within CCP4i2 (Skubák and Pannu, 2013; Winn et al., 2011). Statistics for data collection and phasing of SeMet DcrBΔ20 are shown in Supplementary Table 3. A monomer from the SeMet DcrBΔ20 model was used as the search model for solving the structure of His6-DcrBΔ37 by molecular replacement using Phaser within PHENIX (Adams et al., 2010). The final model of native His6-DcrBΔ37 was obtained through successive manual model building using Coot (Emsley et al., 2010) and refinement using REFMAC (Murshudov et al., 2011). Final stages of refinement used 5 translation-libration-screw (TLS) groups per chain, which were defined using the TLSMD server (Painter and Merritt, 2006). Model validity was checked using the MolProbity web server (Chen et al., 2010). Coordinates and structure factors for native His6-DcrBΔ37 were deposited in the RCSB Protein Data Bank, under the accession ID 6E8A. Final statistics for data collection and refinement for DcrBΔ37 are shown in Table 1.
Table 1.
X-ray diffraction data collection and refinement statistics
| DcrBΔ37 | |
|---|---|
|
| |
| Data Collection | |
|
| |
| Beamline | 21-ID-D, LS-CAT, APS |
| Wavelength, Å | 0.97851 |
| Resolution range (high resolution bin), Å | 44.45–1.92 (1.95–1.92) |
| Space Group | P 65 2 2 |
| Unit Cell (a, b, c (Å)) | 41.715, 41.715, 533.438 |
| (α, β, γ (°)) | 90.0, 90.0, 120.0 |
| Completeness, % | 99.2 (100.0) |
| Total Reflections | 837,305 (35,493) |
| Unique Reflections | 22,978 (1,086) |
| Redundancy | 36.4 (32.7) |
| <I/σI> | 21.2 (2.2) |
| Rmerge†, % | 9.7 (179) |
|
| |
| Refinement | |
|
| |
| Resolution, Å | 1.92 |
| Rwork/Rfree,‡ % | 22.2 / 23.5 |
| rms deviations | |
| Bonds, Å | 0.008 |
| Angles, ° | 1.218 |
| Ramachandran statistics, % | |
| Most favored | 99.3 |
| Allowed | 0.7 |
| Generously allowed | 0 |
| Disallowed | 0 |
| # atoms | |
| Protein | 2,199 |
| Solvent | 27 |
| <B factor>, Å2 | |
| Protein | 71.3 |
| Solvent | 50.4 |
Rmerge = ΣΣj|Ij −<I>|ΣIj, where Ij is the intensity measurement for reflection j and <I> is the mean intensity for multiply recorded reflections.
Rwork / Rfree = Σ||Fobs| − |Fcalc||/|Fobs|, where the working and free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections (5.17% of the total) were held aside throughout refinement.
2.7. Multiple sequence alignment and structural homology search.
A multiple sequence alignment of amino acid sequences of DcrB proteins was generated using CLUSTAL Omega (Sievers et al., 2011), using DcrB homologs from the following enterobacteria: Salmonella enterica (UniProt accession number Q7CPJ3); Shigella dysenteria (UniProt accession number E7SI71); Escherichia coli (UniProt accession number P0AEE1); Klebsiella pneumoniae (UniProt accession number A0A170ZDP4); and Yersinia pestis (UniProt accession number Q7CKX4). Analysis of quaternary structure was performed using Proteins, Interfaces, Structures and Assemblies (PISA) (Krissinel and Henrick, 2007). A search for structures with similarity to DcrB was performed using the Dali server (Holm and Laakso, 2016). Structural superpositions of DcrB and similar structures were generated using the PyMOL molecular graphics system, version 2.0.1 (Schrodinger LLC, 2015).
3. Results and Discussion
3.1. DcrB consists of an N-terminal segment connected to a three-layered α-β-α sandwich
We purified His6-DcrBΔ20, which lacks the lipoprotein signal sequence, using Ni2+-NTA affinity chromatography. We obtained protein at a level of purity amenable to crystallization (Supplementary Fig. 2). We identified and refined crystallization conditions using a sparse matrix screen followed by refinement experiments.
An initial structural model of DcrB was obtained by experimental SAD phasing of SeMet His6-DcrBΔ20 diffraction data, but electron density was not observed for DcrB amino acid residues 21–37 (Fig. 1A). Therefore, we purified His6-DcrBΔ37, which lacks these residues, to a similar level of purity as His6-DcrBΔ20 (Supplementary Fig. 2), and collected X-ray diffraction data on crystals of His6-DcrBΔ37. We used a monomer of the initial SeMet His6-DcrBΔ20 model as the search model for solving the structure of His6-DcrBΔ37 by molecular replacement. In the structure of native His6-DcrBΔ37, there are two DcrB molecules per asymmetric unit, giving a solvent content of 37.70% and a Matthews coefficient of 1.97 Å3/Da. The native DcrB model was refined to a final Rwork of 22.3% and Rfree of 23.5% (Table 1). Because electron density for the N-terminal purification tag (MetHis6Ala) of chains A and B as well as for residues 38–39, 66–67 and 185 of chain B was poor, these residues were excluded from the final structural model of DcrB.
Figure 1.
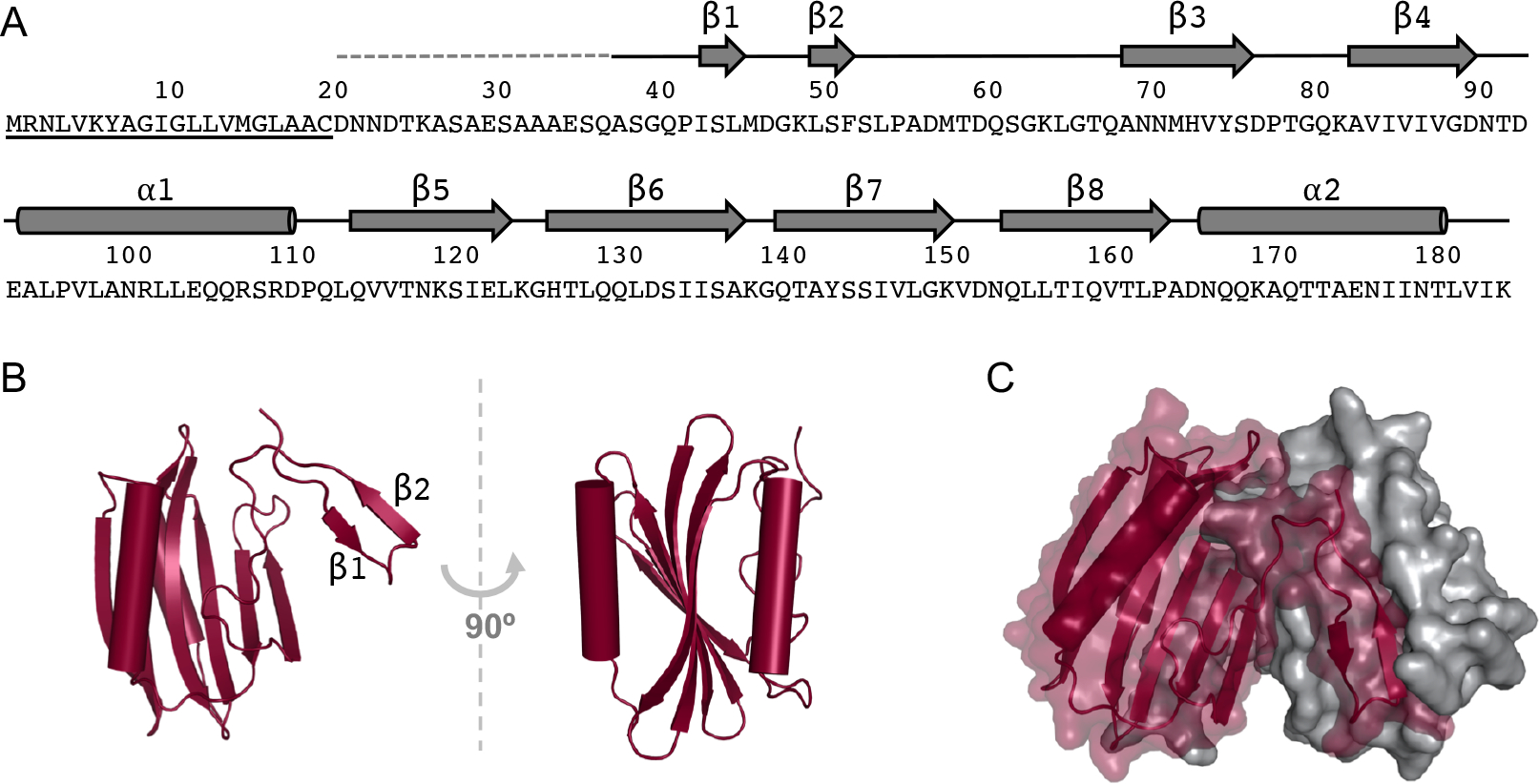
Sequence and structure of Salmonella DcrB. A) The secondary structure that was observed in the structure of DcrB is indicated over the amino acid sequence of DcrB. The lipoprotein signal sequence is underlined. The dashed gray line indicates the portion of purified SeMet DcrBΔ20 protein that was not observed in electron density maps. Solid black lines correspond to regions of DcrBΔ37 that were observed in electron density maps but were not part of α-helices or β-strands. Residues 50–181 comprise the predicted domain of unknown function 1795 (DUF1795). B) The DcrBΔ37 monomer, with α-helices indicated by cylinders and β-strands indicated by arrows. The N-terminal β-hairpin is labeled with the corresponding β-strands. C) The domain-swapped DcrBΔ37 dimer, with α-helices indicated by cylinders and β-strands indicated by arrows. The first protomer is maroon, rendered as a transparent surface with secondary structure, and the second protomer is in gray, rendered as a solid surface.
Each DcrB monomer consists of the following secondary structure, listed from N-terminus to C-terminus: β1-β2-β3-β4-α1-β5-β6-β7-β8-α2 (Fig. 1A). The β1 and β2 strands form a β-hairpin at the N-terminus. The remaining secondary structural elements form a 3-layered α-β-α sandwich in which the α1- and α2-helices flank a 6-stranded antiparallel β-sheet (β3-β4-β8-β7-β6-β5) (Fig. 1B). Analysis of the His6-DcrBΔ37 protein by circular dichroism (CD) showed one peak at 215 nm (Supplementary Fig. 3), indicating that the predominant secondary structure of DcrB in solution is β-sheet, consistent with the observed crystallographic structure.
3.2. DcrBΔ37 forms domain-swapped crystallographic dimers
Each DcrBΔ37 monomer from the asymmetric unit forms a homodimer with a crystallographic symmetry mate: chain A•chain A and chain B•chain B (Fig. 1C). Analysis of the DcrBΔ37 model by PISA showed a complex significance score (CSS) of 1.000 for the A•A dimer and of 1.000 for the B•B dimer, indicating the highest probability of dimer formation. The A•A interface buries a total of 5,114 Å2 of surface area, which corresponds to 27.4% of each monomer’s surface area and a change in solvent free energy of −42.3 kcal/mol. The B•B interface buries a total of 4,790 Å2 of surface area, which corresponds to 26.2% of each monomer’s total surface area and a change in solvent free energy of −39.6 kcal/mol.
Interestingly, domain-swapping contributes to the crystallographic dimer interface: the N-terminal β-hairpin (β1–β2) of one protomer extends outward via a 16-amino acid linker region (residues 53–68) to interact with the α–β–α sandwich of the second protomer (Fig. 1C). The domain-swapping of one protomer is stabilized by salt bridges formed by the side-chains of Lys49 (chain A1) and Asp153 (chain A2) and by the side-chains of Asp57 (chain A1) and Lys82 (chain A2) (Supplementary Fig. 4). Additionally, each protomer forms 13 hydrogen-bonding interactions with the other protomer (for a total of 26 hydrogen bonds in the dimer interface); 12 of these 13 hydrogen bonds occur between the swapped domain and the core of the other protomer.
Analysis of His6-DcrBΔ20 by size exclusion chromatography with multi-angle light scattering (carried out at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University) indicated that His6-DcrBΔ20 exists as a monomer in solution (Supplementary Table 4). This result suggests that DcrB forms dimers at the higher protein concentrations used for crystallization and/or in the specific crystallization conditions. A defining feature of proteins that form domain-swapped oligomers is a conformationally flexible hinge loop region that connects the swapped domain with the remainder of the protein (Gronenborn, 2009; Liu and Eisenberg, 2002). In general, this hinge loop adopts a closed conformation in the monomer but an extended conformation in the domain-swapped oligomer. Therefore, the domain-swapped dimeric structure of DcrB indicates that the N-terminal linker region (residues 53–68) is a conformationally flexible hinge loop that enables formation of the domain-swapped dimer from monomeric protein in solution.
3.3. DcrB is a member of the Mog1p/PsbP-like superfamily
Comparison of the DcrBΔ37 monomer with known protein structures using the Dali server revealed that the DcrBΔ37 monomer structurally aligns with proteins that contain the Mog1p/PsbP-like fold (Fig. 2). DcrB is the first example of an enterobacterial protein with the Mog1p/PsbP-like fold, which occurs in phylogenetically and functionally diverse proteins. The yeast Mog1p protein and human MOG1 protein regulate nuclear transport (Marfatia et al., 2001; Oki and Nishimoto, 1998; Stewart and Baker, 2000). Mog1p has strong structural similarity with PsbP proteins, which are membrane-extrinsic proteins that are a subunit of photosystem II in plants and cyanobacteria (Bricker et al., 2013; Cao et al., 2015; Ifuku et al., 2004; Jackson et al., 2012; Michoux et al., 2010). A structural alignment of DcrBΔ37 with Mog1p (PDB ID 1EQ6) by the Dali sever gave a root mean square deviation (rmsd) of 3.1 Å, and a structural alignment of DcrBΔ37 with a PsbP protein (PDB ID 2XB3) gave an rmsd of 2.8 Å.
Figure 2.
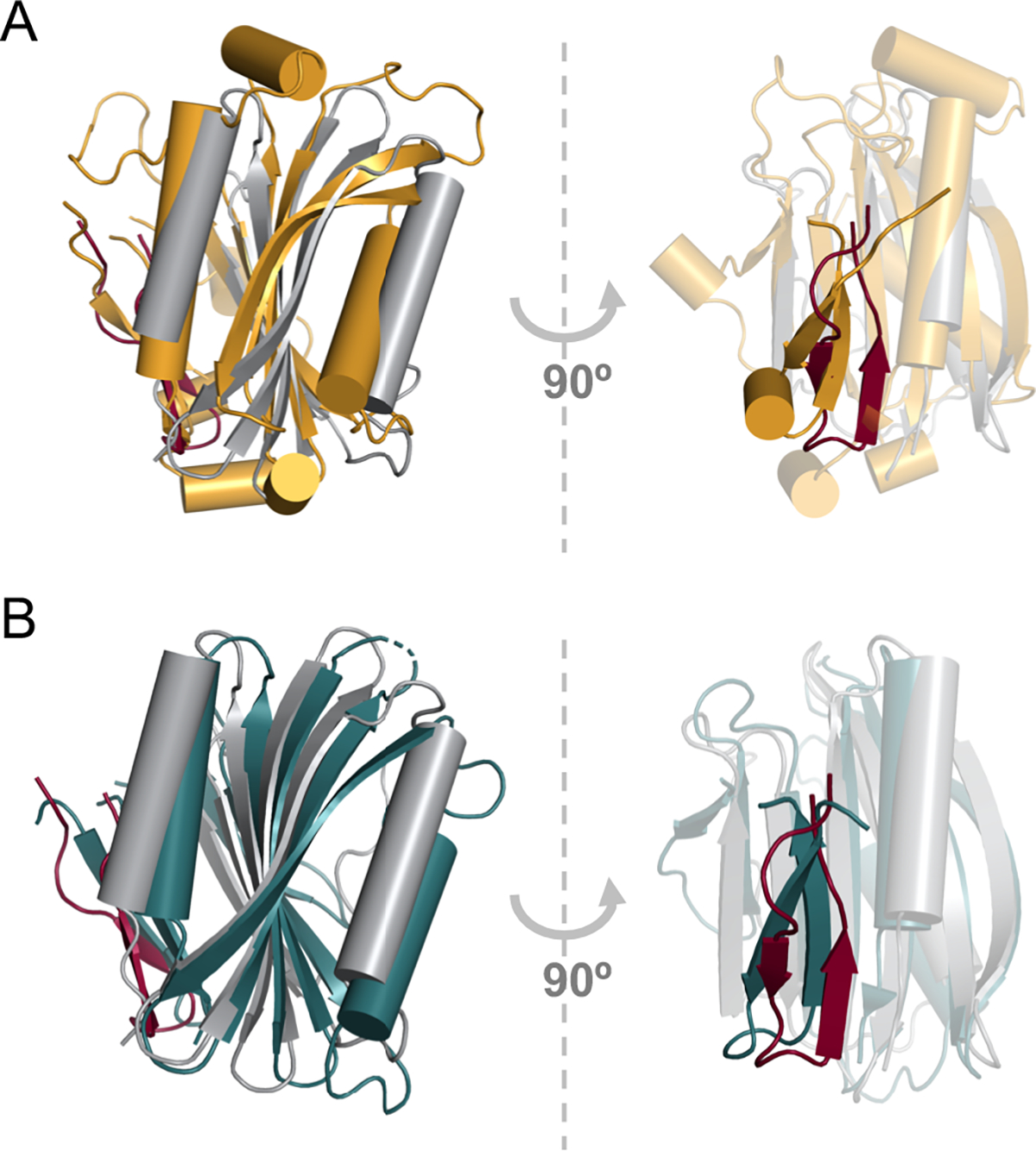
DcrB contains the Mog1p/PsbP-like fold. Structural superposition of the DcrB monomer (gray) with A) yeast Mog1p (amber, PDB ID 1EQ6) and B) cyanobacterial PsbP (teal, PDB ID 2XB3). The domain-swapped region of DcrB from the second protomer is indicated in maroon. α-helices are indicated by cylinders and β-strands are indicated by arrows.
To explore regions of structural similarity and structural divergence, we superimposed the DcrBΔ37 monomer with Mog1p (PDB ID 1EQ6) and cyanobacterial PsbP (PDB ID 2XB3) (Fig. 2). The major region of structural similarity is the 3-layered α-β-α sandwich, in which two α-helices flank a 6-stranded antiparallel β-sheet. The high conservation of the 3-layered sandwich underscores that it is the defining feature of the Mog1p/PsbP-like fold. The small loops between β5 and β6, β6 and β7, and β7 and β8 (DcrB numbering) are very similar between DcrB and the other Mog1p/PsbP structures. Additionally, the length of the C-terminal α2-helix and its orientation relative to the β-sheet are highly conserved, suggesting that these properties of α2-helix are critical for stabilizing the Mog1p/PsbP fold.
DcrB and Mog1p/PsbP proteins also have regions of structural divergence (Fig. 2). Notably, the N-terminal β-hairpin that undergoes domain-swapping in the DcrB dimer does not undergo domain swapping in Mog1p or PsbP proteins. In Mog1p or PsbP, the N-terminal β-hairpin interacts with the same location on the α-β-α sandwich as the swapped β-hairpin does in DcrB (Fig. 2). Overall, the domain-swapping in DcrBΔ37 indicates that the N-terminal portion of the Mog1p/PsbP can extend away from the core α-β-α sandwich to participate in intermolecular interactions.
3.4. Implications for the orientation of DcrB relative to the cytoplasmic membrane
Because DcrB proteins contain a consensus lipoprotein signal sequence (Fig. 1A), DcrB is predicted to be trafficked to the periplasm, followed by proteolytic cleavage of the signal sequence and covalent lipid-modification of the strictly conserved Cys20 that immediately follows the cleavage site (Okuda and Tokuda, 2011). In the DcrBΔ37 structure, the membrane-anchored Cys20 is predicted to connect to Ala38 via the unstructured tether Cys20-Gln37 (Fig. 3). The predicted membrane-proximal face of the DcrB monomer has regions of positive charge on its surface (Supplementary Fig. 5). Conversely, the predicted membrane-distal face of the DcrB monomer has regions of negative charge on its surface (Supplementary Fig. 5). This predicted orientation would position the negatively charged surface of DcrB away from the cytoplasmic membrane, which carries a net negative charge due to its phospholipid composition (Matsuzaki et al., 1997; Raetz and Dowhan, 1990).
Figure 3.
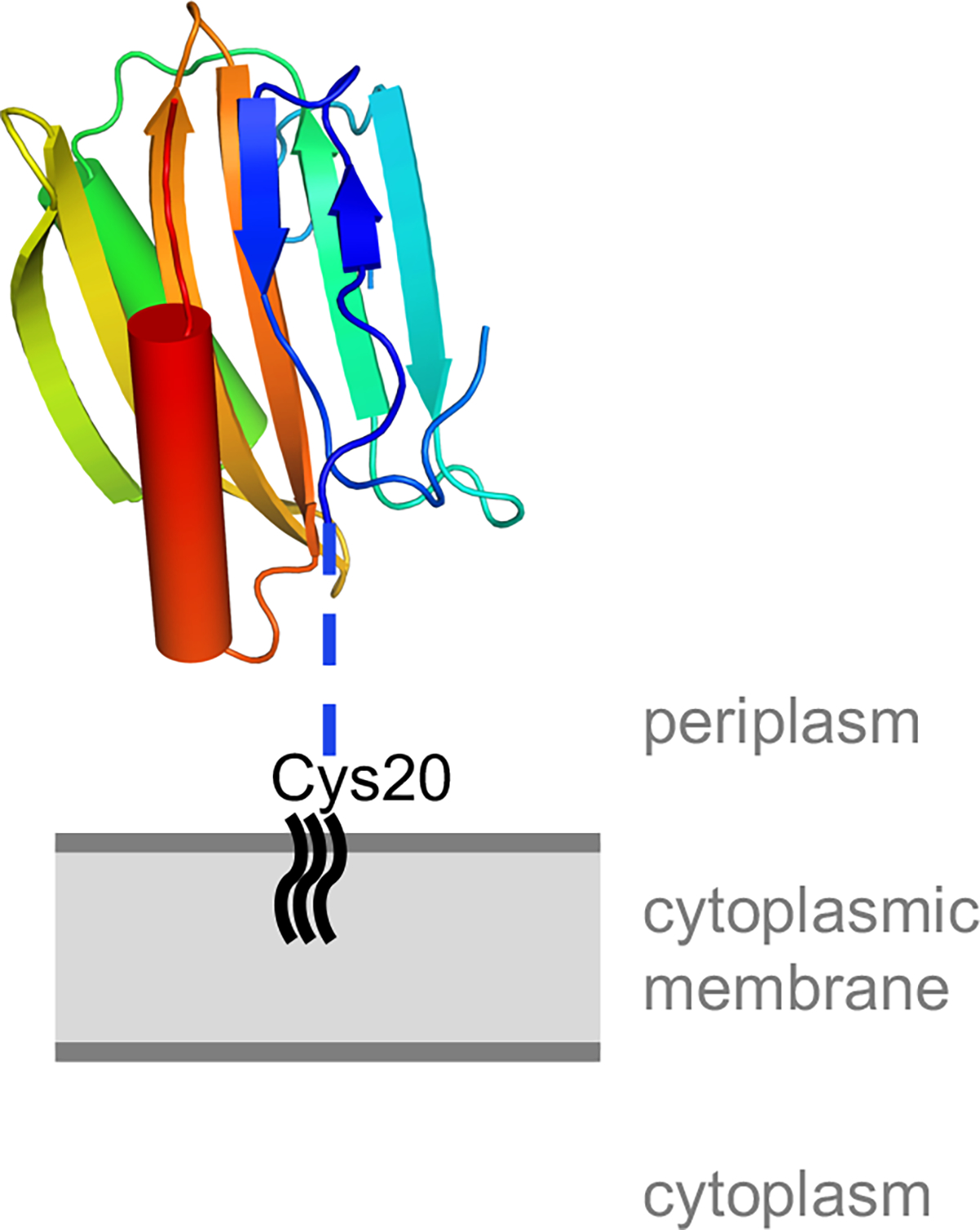
Predicted orientation of DcrB relative to the cytoplasmic membrane. The DcrB monomer is shown with the N-terminal β–hairpin in the predicted closed conformation, and is colored using the chainbows setting from PyMOL from N-terminus (blue) to C-terminus (red). The blue dotted line represents the unstructured region, corresponding to amino acids 21–37, that connects the lipid-modified Cys20 to the structured domain of DcrB.
4. Conclusions
The first three-dimensional structure of Salmonella enterica DcrB establishes that the domain of unknown function in DcrB (DUF1795) consists of the Mog1p/PsbP-like fold, which is found in a functionally diverse group of proteins in cyanobacteria, mammals, yeast, and plants. Interestingly, DcrB forms domain-swapped crystallographic homodimers in which the N-terminal β-hairpin of one protomer interacts with the core α-β-α sandwich of the other protomer. These finding reveals a flexibility in the N-terminal segment of the Mog1p/PsbP-like fold and expands knowledge of the phylogenetic diversity of this fold. The structure of an enterobacterial DcrB protein provides the foundation for understanding the biochemical function of this lipoprotein and its role in the physiology of enterobacteria.
Supplementary Material
Acknowledgements
The authors would like to thank James Keck (University of Wisconsin-Madison) for providing initial assistance with X-ray diffraction data collection and helpful discussions, Todd Weaver (University of Wisconsin-La Crosse) for helpful discussions, and Joseph Brunzelle (LS-CAT) for assistance with collecting and processing the DcrBΔ20 SeMet X-ray diffraction data. This research was supported by UW-La Crosse Faculty Research Grants (JFM), UW-La Crosse Undergraduate Research Grants (DMR, RWS, and TJD), the UW-La Crosse Eagle Apprentice Program (RWS), a UW-La Crosse College of Science and Health Dean’s Distinguished Fellowship (RWS), and the UW-La Crosse McNair Scholars Program (DMR). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). The authors would like to thank the Biophysics Resource of the Keck Facility at Yale Univeristy for SEC-MALS analysis. The SEC-LS/UV/RI instrumentation was supported by NIH Award Number 1S10RR023748-01. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Gross R, 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9, 143–152. 10.1016/j.mib.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Bricker TM, Roose JL, Zhang P, Frankel LK, 2013. The PsbP family of proteins. Photosynth. Res. 116, 235–250. 10.1007/s11120-013-9820-7 [DOI] [PubMed] [Google Scholar]
- Calva E, Oropeza R, 2006. Two-Component Signal Transduction Systems, Environmental Signals, and Virulence. Microb. Ecol. 51, 166–176. 10.1007/s00248-005-0087-1 [DOI] [PubMed] [Google Scholar]
- Cao P, Xie Y, Li M, Pan X, Zhang H, Zhao X, Su X, Cheng T, Chang W, 2015. Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change. Mol. Plant 8, 664–6. 10.1016/j.molp.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Charles RC, Harris JB, Chase MR, Lebrun LM, Sheikh A, LaRocque RC, Logvinenko T, Rollins SM, Tarique A, Hohmann EL, Rosenberg I, Krastins B, Sarracino DA, Qadri F, Calderwood SB, Ryan ET, 2009. Comparative proteomic analysis of the PhoP regulon in Salmonella enterica serovar Typhi versus Typhimurium. PLoS One 4, e6994. 10.1371/journal.pone.0006994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT, 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548. 10.1038/nchembio.2007.24 [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Miller SI, 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol. 17, 106–113. 10.1016/j.mib.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G, Baker S, 2014. Salmonella enterica Serovar Typhi and the Pathogenesis of Typhoid Fever. Annu. Rev. Microbiol. 68, 317–336. 10.1146/annurev-micro-091313-103739 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K, 2010. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Guina T, Miller SI, 2006. Outer membrane remodeling of Salmonella typhimurium and host innate immunity. Microbes Infect. Inst. Pasteur 3, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Evans PR, 2011. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 282–292. 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Groisman EA, Heffron F, 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243, 1059–62. [DOI] [PubMed] [Google Scholar]
- Frieden T, 2013. Antibiotic resistance threats in the United States, 2013, Cdc. Centers for Disease Control and Prevention (US), Atlanta, GA. https://doi.org/CS239559-B [Google Scholar]
- Galán JE, 2016. Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc. Natl. Acad. Sci. 113, 6338–6344. 10.1073/pnas.1606335113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, Curtiss R, 1989. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog. 6, 433–443. 10.1016/0882-4010(89)90085-5 [DOI] [PubMed] [Google Scholar]
- Garcia Véscovi E, Soncini FC, Groisman EA, 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res. Microbiol. 145, 473–480. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A, 2005. Protein Identification and Analysis Tools on the ExPASy Server, The Proteomics Protocols Handbook. Humana Press, Totowa, NJ. 10.1385/1-59259-890-0:571 [DOI] [Google Scholar]
- Green MR, Sambrook J, 2012. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- Groisman EA, 2001. The Pleiotropic Two-Component Regulatory System PhoP-PhoQ. J. Bacteriol. 183, 1835–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn AM, 2009. Protein acrobatics in pairs—dimerization via domain swapping. Curr. Opin. Struct. Biol. 19, 39–49. 10.1016/j.sbi.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Laakso LM, 2016. Dali server update. Nucleic Acids Res. 44, W351–5. 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Nakatsu T, Kato H, Sato F, 2004. Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep. 5, 362–367. 10.1038/sj.embor.7400113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, Hinds MG, Eaton-Rye JJ, 2012. Solution structure of CyanoP from Synechocystis sp. PCC 6803: new insights on the structural basis for functional specialization amongst PsbP family proteins. Biochim. Biophys. Acta 1817, 1331–8. 10.1016/j.bbabio.2012.02.032 [DOI] [PubMed] [Google Scholar]
- Kabsch W, 2010. XDS. Acta Cryst (2010). D66, 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K, 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–97. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Laemmli UK, 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lee YM, Almqvist F, Hultgren SJ, 2003. Targeting virulence for antimicrobial chemotherapy. Curr. Opin. Pharmacol. 3, 513–519. 10.1016/j.coph.2003.04.001 [DOI] [PubMed] [Google Scholar]
- Likhacheva NA, Samsonov VV, Sineoky SP, 1996. Genetic control of the resistance to phage C1 of Escherichia coli K-12. J. Bacteriol. 178, 5309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D, 2002. 3D domain swapping: As domains continue to swap. Protein Sci. 11, 1285–1299. 10.1110/ps.0201402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfatia KA, Harreman MT, Fanara P, Vertino PM, Corbett AH, 2001. Identification and characterization of the human MOG1 gene. Gene 266, 45–56. 10.1016/S0378-1119(01)00364-X [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Sugishita K, Harada M, Fujii N, Miyajima K, 1997. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim. Biophys. Acta - Biomembr. 1327, 119–130. 10.1016/S0005-2736(97)00051-5 [DOI] [PubMed] [Google Scholar]
- Michoux F, Takasaka K, Boehm M, Nixon PJ, Murray JW, 2010. Structure of CyanoP at 2.8 Å: Implications for the evolution and function of the PsbP subunit of photosystem II. Biochemistry 49, 7411–7413. 10.1021/bi1011145 [DOI] [PubMed] [Google Scholar]
- Miller JH, 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Moss JE, Fisher PE, Vick B, Groisman EA, Zychlinsky A, 2000. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell. Microbiol. 2, 443–452. 10.1046/j.1462-5822.2000.00065.x [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, 2011. REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 355–367. 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl ME, Miller SI, 2001. Salmonella: A Model for Bacterial Pathogenesis. Annu. Rev. Med. 52, 259–274. 10.1146/annurev.med.52.1.259 [DOI] [PubMed] [Google Scholar]
- Oki M, Nishimoto T, 1998. A protein required for nuclear-protein import, Mog1p, directly interacts with GTP-Gsp1p, the Saccharomyces cerevisiae ran homologue. Proc. Natl. Acad. Sci. U. S. A. 95, 15388–15393. 10.1073/pnas.95.26.15388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Tokuda H, 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–59. 10.1146/annurev-micro-090110-102859 [DOI] [PubMed] [Google Scholar]
- Oyston PCF, Dorrell N, Williams K, Li SR, Green M, Titball RW, Wren BW, 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68, 3419–3425. 10.1128/IAI.68.6.3419-3425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA, 2006. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111. 10.1107/S0021889805038987 [DOI] [Google Scholar]
- Raetz CR, Dowhan W, 1990. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 265, 1235–1238. [PubMed] [Google Scholar]
- Samsonov VV, Samsonov VV, Sineoky SP, 2002. DcrA and dcrB Escherichia coli genes can control DNA injection by phages specific for BtuB and FhuA receptors. Res. Microbiol. 153, 639–646. 10.1016/S0923-2508(02)01375-X [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC, 2015. The PyMOL Molecular Graphics System, Version 1.8.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG, 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubák P, Pannu NS, 2013. Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777. 10.1038/ncomms3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Baker RP, 2000. 1.9 A resolution crystal structure of the Saccharomyces cerevisiae Ran- binding protein Mog1p. J Mol Biol 299, 213–223. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J, 1993. Atomic Structures of the Human Immunophilin FKBP-12 Complexes with FK506 and Rapamycin. J. Mol. Biol. 229, 105–124. 10.1006/jmbi.1993.1012 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G, D. F, M. MS, A. LG, 2011. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 293–302. 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JL, Guo L, 2011. Quantitative proteomic analysis of Salmonella enterica serovar Typhimurium under PhoP/PhoQ activation conditions. J. Proteome Res. 10, 2992–3002. 10.1021/pr101177g [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


