Abstract
Freshwater Unionid bivalves have recently faced ecological upheaval through pollution, barriers to dispersal, harvesting, and changes in fish–host prevalence. Currently, over 70% of species in North America are threatened, endangered or extinct. To characterize the genetic response to recent selective pressures, we collected population genetic data for one successful bivalve species, Megalonaias nervosa. We identify megabase-sized regions that are nearly monomorphic across the population, signals of strong, recent selection reshaping diversity across 73 Mb total. These signatures of selection are greater than is commonly seen in population genetic models. We observe 102 duplicate genes with high dN/dS on terminal branches among regions with sweeps, suggesting that gene duplication is a causative mechanism of recent adaptation in M. nervosa. Genes in sweeps reflect functional classes important for Unionid survival, including anticoagulation genes important for fish host parasitization, detox genes, mitochondria management, and shell formation. We identify sweeps in regions with no known functional impacts, suggesting mechanisms of adaptation that deserve greater attention in future work on species survival. In contrast, polymorphic transposable elements (TEs) appear to be detrimental and underrepresented among regions with sweeps. TE site frequency spectra are skewed toward singleton variants, and TEs among regions with sweeps are present at low frequency. Our work suggests that duplicate genes are an essential source of genetic novelty that has helped this species succeed in environments where others have struggled. These results suggest that gene duplications deserve greater attention in non-model population genomics, especially in species that have recently faced sudden environmental challenges.
Keywords: Unionidae, Megalonaias nervosa, population genomics, gene family expansion, transposable element evolution, detox genes, environmental change
Introduction
The origins of genetic innovation during ecological change remain among the most challenging questions in evolutionary theory. The ways that genetic variation appears in populations and then responds to strong shifts in selective pressures is fundamental to understanding how organisms evolve in nature (Ranz and Parsch 2012). Among classes of mutations that can contribute to adaptation, duplicate genes and related chimeric constructs are held as a key source of innovation (Ohno 1970; Conant and Wolfe 2008; Rogers et al. 2017; Rogers and Hartl 2011).
Theory suggests that gene duplications can create redundancy that frees sequences from selective constraint, allowing neofunctionalization, and adaptive subfunctionalization (Ohno 1970; Conant and Wolfe 2008; Des Marais and Rausher 2008). These mutations can produce novel proteins and changes in expression that are difficult to mimic via point mutations (Rogers and Hartl 2011; Rogers et al. 2017; Stewart and Rogers 2019). Duplications may be less likely to be neutral than other classes of mutations, and may serve as mutations of large effect when large effects are needed (Emerson et al. 2008; Schrider and Hahn 2010; Rogers et al. 2015). Transposable elements (TEs) and duplications show an interplay, where TE content and activity is expected to produce higher duplication rates (Bennetzen 2000; Yang et al. 2008). How these mutations contribute to evolutionary outcomes as selective pressures are altered is key to understanding the role that genetic novelty plays in adaptation. Characterizing the genetic response to strong selection will clarify where the limits may lie in survival under fundamental shifts in environment.
The final outcomes of strong selection during habitat changes can be observed in the spectrum of diversity after selection (Tajima 1989; Nielsen 2005; Hartl 2020). Regions of the genome that have contributed to adaptation will show reductions in nucleotide diversity across linked regions and highly skewed site frequency spectra (Tajima 1989). Genome scans with population genetics can clarify what genetic variation has contributed to adaptation in a way that is agnostic to function and free from human-centric biases regarding animal survival (Nielsen 2005; Ellegren 2014). Examining genetic diversity among regions targeted by selection, we can discern the types of mutations and functional classes that are most important for survival and reproduction (Nielsen 2005; Ellegren 2014). Such approaches are commonly implemented in population genetic model systems, where sequencing and functional analysis are straightforward (Sella et al. 2009; Rogers et al. 2010; Rogers and Hartl 2011; Langley et al. 2012; Mackay et al. 2012).
As genome sequencing has advanced, similar analysis can identify outcomes of selection in new organisms (Ellegren 2014) where extreme environmental shifts have resulted in challenges to species survival. These technological advances allow us to finally explore questions of genetic novelty in alternative evolutionary systems with species that have faced sudden shifts in selective pressures. These goals will help clarify whether the genetic response during adaptation is fundamentally different under ecological upheaval compared with species that experience ecological stability. The evolutionary response to ecological changes is often studied under deep time using phylogenetics. However, in the modern anthropogenic era, we can observe challenges to species under threat happening in a single human lifetime (Lake et al. 2000). This unfortunate opportunity allows us to study how species respond over shorter timescales than before, and determine genetic factors that allow some organisms to adapt while other species go extinct.
Unionidae as a Population Genetic Model
Unionidae (and their relatives in Margaritaferidae) are notable for their unusual life cycle. Adults are benthic filter feeders that settle into the substrate (Williams et al. 2008; Patterson et al. 2018). Most species are dioecious with separate male and female sexes (Williams et al. 2008; Haag 2012). Females are fertilized internally and brood offspring in their gills until releasing them into the water column where they parasitize fish hosts (Williams et al. 2008; Haag 2012; Patterson et al. 2018). As larvae mature, they drop off fish hosts to develop into mature adults (Williams et al. 2008), a key developmental timepoint for species survival (Haag and Williams 2014; Modesto et al. 2018). Phenotypic studies reveal arms races with fish hosts. Unionidae are also unusual for their dual uniparental inheritance of mitochondria, where males inherit mitochondria from the paternal lineage and females inherit mitochondria from the maternal line (Liu et al. 1996; Breton et al. 2007; Wen et al. 2017). Karyotype data for Unionida show no evidence of heterogametic sex chromosomes (Kongim et al. 2015). These unusual biological processes offer a unique setting to study parasite–host coevolution, parental care, a transition from filter feeding to blood feeding, and unusual mitochondrial function.
Freshwater Unionidae represent one clade that has recently experienced ecological upheaval. Over 70% of species in North America are threatened, endangered, or extinct (Williams et al. 1993; Strayer et al. 2004; Régnier et al. 2009; Haag and Williams 2014). Other species have thrived even in the face of these same ecological pressures, some even experiencing recent population expansion. SNP chips (Pfeiffer et al. 2019), single locus studies (Pfeiffer et al. 2018), mtDNA (Campbell et al. 2005), and reference genomes for the clade are just now being initiated (Renaut et al. 2018; Rogers et al. 2021; Smith 2021), opening doors for evolutionary analysis in Unionid bivalves. These resources can clarify phylogenetic relationships, especially in cases where morphology, mtDNA and nuclear loci might differ (Campbell et al. 2005; Pfeiffer et al. 2019). However, to gather a complete portrait of genetic novelty and the adaptive response to strong shifts in selection, whole-genome population samples are required.
Muscle Shoals offers a focal location, with high species diversity for freshwater bivalves. Historically over 80 species have been identified in the Tennessee River and surrounding tributaries (Williams et al. 2008). Rivers in the area have been dammed, preventing dispersal of aquatic fauna with impacts on species diversity (Ortmann 1924; Cahn 1936). Water quality has been affected with pesticide and fertilizer runoff and industrial pollution, threatening avian and aquatic wildlife (Woodside 2004). Bivalves experienced additional pressures, as they were harvested for shells in the freshwater pearl and button industries, with up to 6,700 tons produced per year during peak historical demand (Williams et al. 2008). Many freshwater bivalves in the Southeast now compete with invasive species such as Corbicula fluminea (Asian clams) (Strayer 1999) and Dreissena polymorpha (Zebra mussels) that have just recently been observed in the Tennessee River. Some 32 species have not been seen at Muscle Shoals since the 1930s, with another 10 species listed as federally endangered (Garner and McGregor 2001). Current estimates suggest roughly 40 species remain (Garner and McGregor 2001).
During these modifications, other species that were able to thrive in flooded overbank habitats experienced intense population expansion (Ahlstedt and McDonough 1992; Garner and McGregor 2001; Williams et al. 2008). Among these, Megalonaias nervosa is a success story where populations have expanded instead of contracting. Surveys suggest that M. nervosa is among the most common species with tens of millions of individuals in one reservoir (Ahlstedt and McDonough 1992). As a benchmark for adaptation in this clade where so many species are under threat, we have assembled and scaffolded a reference genome (Rogers et al. 2021) and produced a whole-genome population sequencing panel for M. nervosa. Using scans of selection, we can identify adaptive genetic variation that has recently spread through populations due to natural selection (Nielsen 2005; Ellegren 2014).
In the data presented here, we observe signals of strong, recent selective sweeps reshaping genetic diversity in freshwater bivalves that have recently experienced ecological upheaval. We use a genes-up approach that is agnostic to function, yet still observe genetic variation that recapitulates the biology of the organism. In addition, we identify targets of selection that have no clear functional annotation. These loci reveal as yet poorly understood mechanisms of adaptation in bivalves that deserve future study to discover the full genetic response to strong, recent selection. We observe key contributions from duplicate genes, which serve as causative agents of adaptive changes in M. nervosa, including functional classes known to influence the biology of the organism. Together, these results indicate that duplicate genes are an essential part of the evolutionary response to strong selection that deserve greater attention in conservation genomics and non-model evolutionary genetics.
Results
Across the genome of M. nervosa Tajima’s D is negative on average with a mean of −1.16 and a median of −1.2 suggesting population expansion in the past. Mean diversity π is θ = 0.0054 (supplementary table S2, Supplementary Material online), slightly lower from single-genome estimates of heterozygosity in a deeply sequenced reference specimen (Rogers et al. 2021). Simulations of a 10-fold population expansion roughly 5,000 generations ago recapitulate background diversity measures (supplementary fig. S1, Supplementary Material online). Inference of ancient population expansion is consistent with a species range expansion at the end of the last glaciation as greater habitat became available after glaciers receded (Elderkin et al. 2007; Keogh et al. 2021). Diversity π and Tajima’s D varies across scaffolds, with a roughly normal distribution genomewide (fig. 1). We used the heterozygosity at 4-fold nonsynonymous sites where any of the three possible nucleotide substitutions alters amino acid sequence compared with 4-fold synonymous sites where none of the nucleotide alters amino acid sequence to measure constraint in the M. nervosa genome. We find that πN/πS = 0.47 and SN/SS = 0.48. These numbers are consistent with estimates from the reference genome suggesting HN/HS = 0.46.
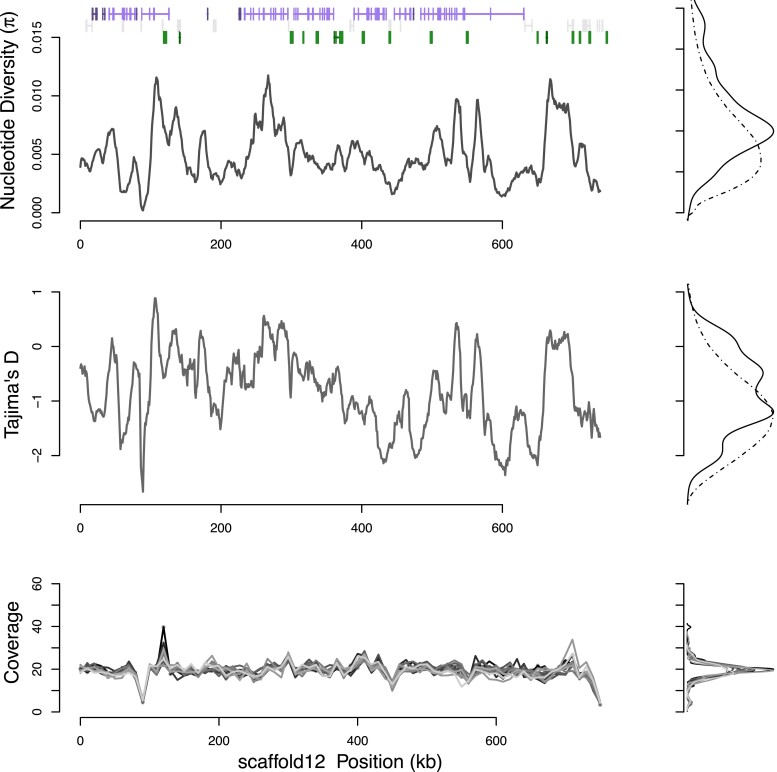
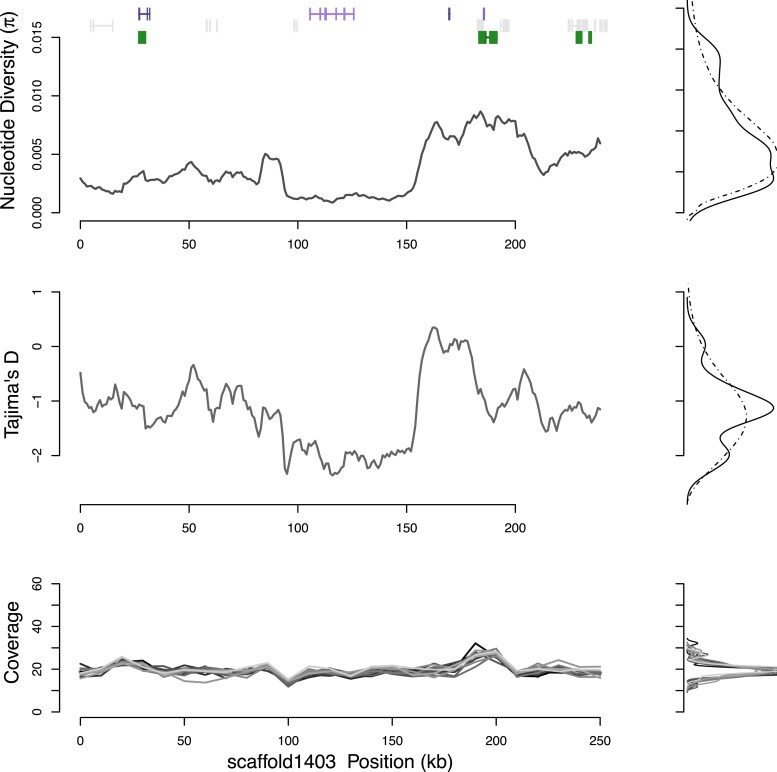
Fig. 1.
Population genetic diversity π, Tajima’s D, and normalized sample coverage for a typical scaffold in M. nervosa. Density plots of each metric are shown to the side for the scaffold (solid line) and genomewide background (dashed line). Genetic diversity, π is centered about 0.0054 and varies across the genetic region. Normalized coverage for the 13 samples (shaded gray lines) shows a single polymorphic CNV/repeat around 470 kb. Gene models confirmed with RNAseq data are shown in purple, unconfirmed in RNAseq data shown in gray, and repetitive elements in green.
We identified the Site Frequency Spectrum (SFS) for putatively neutral 4-fold synonymous sites and for 4-fold nonsynonymous sites. The SFS suggests a modest difference in the impacts of amino acid changing SNPs compared with synonymous SNPs in its slightly more U-shaped spectra with greater numbers of highest and/or lowest frequency alleles, but differences are not significant (Wilcoxon Rank Sum Test W = 1331540915, P = 0.1086; Kolmogorov–Smirnov D = 0.0066259, P = 0.2524; fig. 2). Such results indicate mild constraint on polymorphism across the genome.
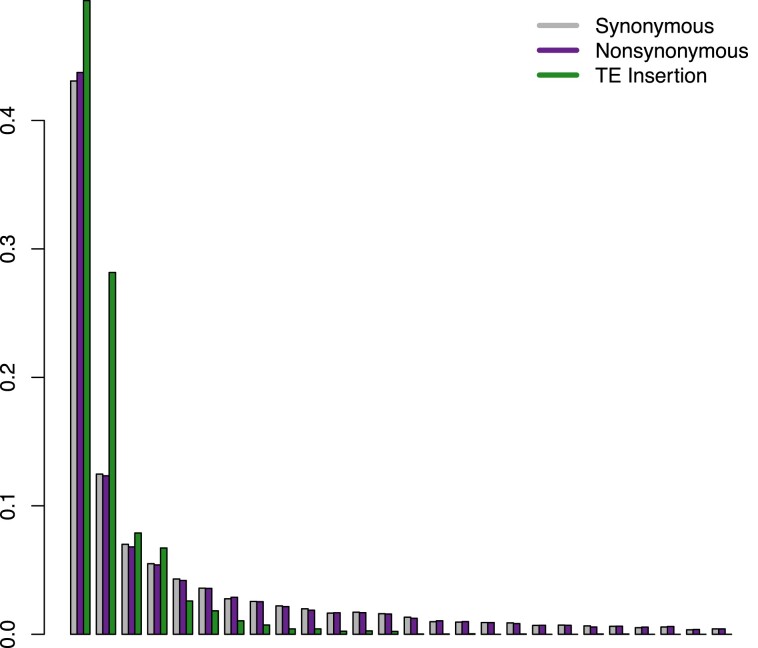
Fig. 2.
SFS for synonymous SNPs, nonsynonymous SNPs, and TE insertions in M. nervosa. The SFS for TE insertions is skewed toward extreme allele frequencies, suggesting non-neutral impacts compared with synonymous and nonsynonymous SNPs. Few polymorphic TE insertions reach moderate frequency and none are at high frequency in M. nervosa.
Strong, Recent Selective Sweeps
We used nucleotide diversity π, θW, and Tajima’s D to identify hard selective sweeps with signatures of natural selection altering genetic diversity in the population of M. nervosa. We identify signatures of very strong, very recent selective sweeps in M. nervosa that have reshaped diversity in M. nervosa (fig. 3). Across the M. nervosa genome 73 Mb on 851 contigs is contained in regions that are borderline monomorphic for extended tracts (where π < 0.0015). These regions constitute 2.7% of total genome (2.6 Gb) and 6.2% of assayable sequence in scaffolds 10 kb or longer (1.16 Gb). Such signals of genetic diversity show strong linkage at swept regions, and are in addition to putative ancient selective sweeps with classic v-like signatures in diversity data (fig. 1). Regions with sweeps include 31.5% coding sequence compared with background rates of 35.7%, demonstrating that swept regions are not gene poor. Results are not explained by deletions, low coverage or unidentified repetitive sequence (fig. 4). The presence of C. gigas orthologs and typical genomic coverage confirms that these sequences are in fact of mollusk origin, not driven by contaminants. Specimens are different ages (supplementary table S1, Supplementary Material online) precluding the possibility that they might be derived from the same brood. Fish host dispersal (Williams et al. 2008) or even avian transport (Darwin 1878) minimizes the likelihood of incidental relatedness from microgeographic effects. The genus shows little geographic differentiation (Pfeiffer et al. 2018). Simulations suggest that such signals cannot be recapitulated through extreme bottlenecks with small numbers of potentially related individuals (supplementary text, Supplementary Material online). These extended tracts of low genetic diversity are unusual in comparison with scans of selection in standard population genetic models (Sella et al. 2009; Rogers et al. 2010; Rogers and Hartl 2011; Langley et al. 2012; Mackay et al. 2012).
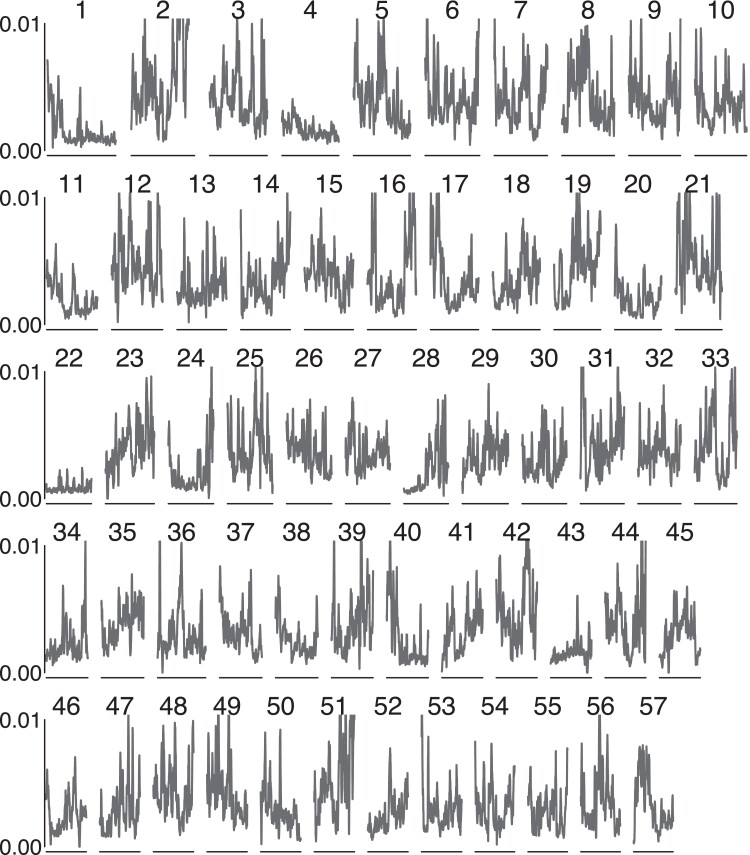
Fig. 3.
Nucleotide diversity π on the 57 largest genome scaffolds in order of scaffold size. Scaffold 4, Scaffold 22, Scaffold 43, and most of Scaffold 20 are nearly monomorphic across most of the sequence. Scaffold 28 begins with reduced diversity and then returns to normal levels of polymorphism. Diversity varies between 0.00 and 0.01, with mean genetic diversity of 0.0054. The largest scaffold shown is 980 kb, and the shortest is 579 kb.
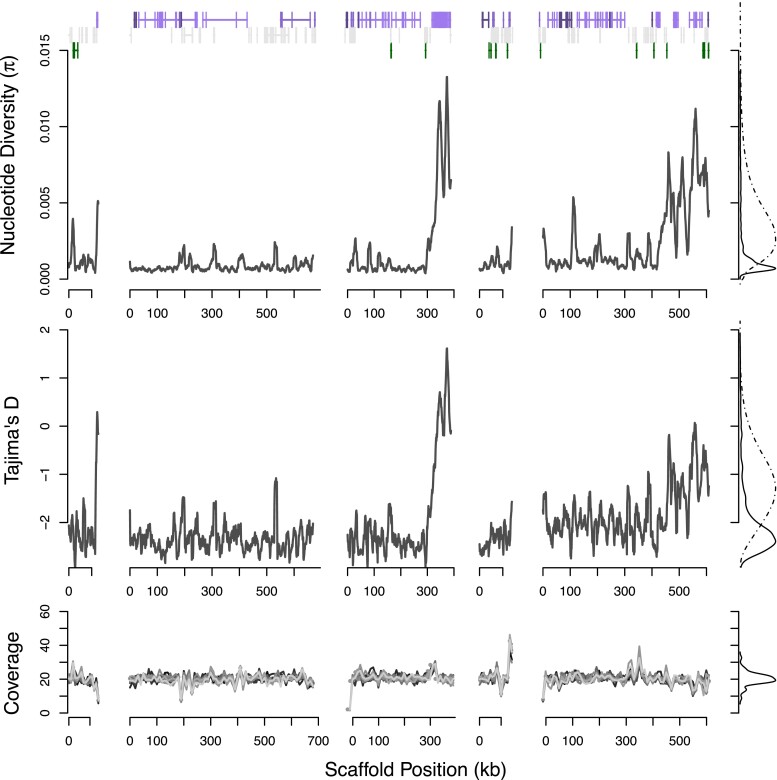
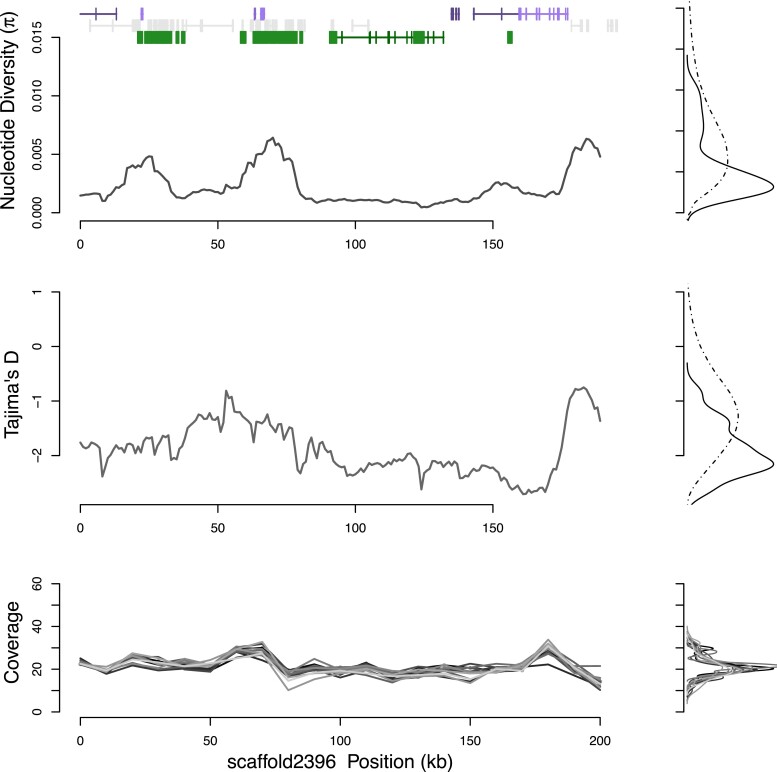
Fig. 4.
Diversity on five scaffolds encompassing a 1.9 Mb region syntenic with Potamilus streckersonii is nearly monomorphic (scaffold4763, scaffold22, scaffold372 - reversed, scaffold4254, scaffold40-reversed). Nucleotide diversity π shows a marked departure compared with background genomic levels, as supported by changepoint statistics. Tajima’s D is below −2.2 throughout the region. Annotated genes match with marine bivalve C. gigas, eliminating possibility that results are driven by contaminating sequences. The population genetic parameters suggest a strong, recent selective sweep that has driven out diversity from the population. Genes in the region include anticoagulation gene Furin on scaffold22.
Historical data are well documented, and are not consistent with population bottlenecks (Isom et al. 1973; Garner and McGregor 2001). However, we performed simulations that suggest even an unobserved 5-fold reduction in population sizes cannot explain these sweep-like signals in the population (supplementary fig. S5, Supplementary Material online). Ancient bottlenecks do not reduce genetic diversity to the levels close to zero, as observed in sweep-like signals (supplementary fig. S5, Supplementary Material online). Recent bottlenecks are expected to reduce population diversity according to Ht = H0 (1 − 1/2N)t. Assuming 20 generations since the formation of the dams along the Tennessee River (Ortmann 1924) even a 10-fold reduction in population size would not reduce diversity below 99.96% of normal levels in the past century. This effect is less than the rounding error diversity estimates. Coalescent times in M. nervosa are expected to be 777,000 generations (Rogers et al. 2021), suggesting that neutral genetic diversity captures demographic effects far in the past prior to formation of dams. Hence, we suggest that neutral forces of genetic drift act too slowly to reshape genetic diversity in this species on historical timescales.
In contrast, very strong selective sweeps can produce 1–2 Mb reductions in diversity even in less than 20 generations, though sweeps further in the past show similar patterns (supplementary text, Supplementary Material online). Hence we suggest that these reductions in diversity indicate a response to strong selective pressures, in recent evolutionary time. Although we do not have sufficient resolution to differentiate between sweeps on 10 generations compared with 1,000 generations, the results are compatible with genetic changes occurring on historical timescales or in the recent evolutionary past. We would suggest that these genomic regions contain alleles that were essential to survival or reproduction in the recent past.
The M. nervosa reference genome has improved with additional long-read sequencing (supplementary text, Supplementary Material online), but still has an N50 of 125 kb. A total of 96% of the genome is in scaffolds 10 kb or larger, sufficient to estimate population genetic statistics and 80% is now in scaffolds 50 kb or larger. To help anchor sweep regions, we used syntenic comparisons with the more contiguous genome of Potamilus streckersonii (N50 = 2 Mb) (Smith 2021) to align scaffolds likely to belong in the same genomic regions. One selective sweep identified spans the majority of five scaffolds, with 1.6 out of 1.9 Mb nearly monomorphic across the region (fig. 4). This scaffold (Scaffold 22) contains Furin, upstream in a pathway from von Willebrand factors that are known to have rampant duplication and elevated amino acid substitutions across paralogs (dN/dS > 1.0) (Rogers et al. 2021). Karyotpe data for Unionidae suggests little rearrangement across species (Kongim et al. 2015). However, if synteny were broken in these regions, these regions would constitute multiple shorter selective sweeps rather than one large selective sweep.
We identify 851 regions in extreme selective sweeps. Only 350 of these have a gene annotation in them with functional info in C. gigas (Zhang et al. 2012) or from Interproscan. Sweeps span 0-9 genes. Some 185 regions have only 1 functional annotation based on comparison with C. gigas (Zhang et al. 2012) or Interproscan annotations. Homology-mediated annotation may be more limited for this species, as well annotated marine relatives are between 200 million and 500 million years divergent (Bolotov et al. 2017; Kumar et al. 2017; Crouch et al. 2021). For these regions containing only a single gene, we would suggest that it is most likely the causative agent of the selective sweeps. Examples of two such single-gene sweeps include a chitin synthtase (fig. 5) and a cytochrome P450 gene (fig. 6). Both represent categories that have experienced adaptive gene family amplification (Rogers et al. 2021). Functional categories in regions with strong, recent selection include functions involved in shell formation, toxin resistance, mitochondria, and parasite–host co-interactions like molecular mimicry or anticoagulation, and stress tolerance (supplementary table S5, Supplementary Material online). These classifications are consistent with adaptive gene duplications in the reference genome (Rogers et al. 2021). Curiously, ABC transporters do not appear multiple times in sweeps, in spite of rampant duplication and their known interactions with cytochromes.
Fig. 5.
Population genetic diversity π, Tajima’s D, and normalized sample coverage for a chitin synthetase gene. Density plots of each metric are shown to the side for the scaffold (solid line) and genomewide background (dashed line). Normalized coverage for the 13 samples (shaded gray lines) shows a small polymorphic deletion at 100 kb and a small polymorphic duplication at 190 kb. The chitin synthetase is the only functional annotation within this region.
Fig. 6.
Population genetic diversity π, Tajima’s D, and normalized sample coverage for a selective sweep at the locus of a Cytochrome P450 gene in M. nervosa. Density plots of each metric are shown to the side for the scaffold (solid line) and genomewide background (dashed line). Such signals would require strong, recent selection to substantially alter genetic diversity for an extended region. The cytochrome gene is the only functional annotation within this selective sweep. Normalized coverage for the 13 samples (shaded gray lines) shows small polymorphic CNV/repeats at the locus of a TE around 60 kb that drives a brief spike in diversity.
Genes in sweeps also include categories of DNA repair, development, apoptosis inhibitors (supplementary table S5, Supplementary Material online), as well as less well-characterized functional categories like Zinc fingers or Zinc knuckles, cell adhesion, cytoskeleton proteins, and WD-40 repeats. Although these conserved domains have less clear functional impacts based on the current information, but point to putative coevolution of large protein complexes, the exact nature of which remains unknown. Other regions have no known conserved domains or functional annotation, suggesting they contribute to bivalve survival, even when we cannot explain why. Duplicate gene analysis and selective sweeps were performed in a high throughput, whole-genome setting without preconceived bias regarding which functional classes should be represented. Yet, in these scans of selection we identify functional classes that reflect the biology and environmental history of the organism. Here, these computational approaches can offer a more complete account of factors that are important for organism survival and reproduction. As Unionid genomics improves and greater functional information is provided, we may be able to resolve what other biological functions have contributed to adaptation in Unionidae.
Adaptive Gene Duplication
We identified gene families in reannotated sequences in version 2 of the M. nervosa reference genome and estimated dN/dS across paralogs (supplementary text, Supplementary Material online), a signature of selection for adaptive bursts of amino acid substitutions (Goldman and Yang 1994). This metric of selection should offer a gene-specific analysis. We observe no correlation between dN/dS on terminal branches and nucleotide diversity π (P = 0.89, R2 = −0.00047), indicating that these two tests of selection are independent from selective sweep analysis (supplementary fig. S21, Supplementary Material online). These genes include Cytochrome P450 genes, von Willebrand proteins, and shell formation genes (supplementary table S5, Supplementary Material online, figs. 5 and 6). We observe duplicate genes with high dN/dS across paralogs that are also located in strong recent selective sweeps. Hence, such genes are strong candidates of causative agents during adaptation as they have two independent signals of recent selection.
We identify a total of 102 genes with signatures of selective sweeps and high dN/dS on their own terminal branch. Terminal branches represent timescales most compatible with recent selective sweeps that can be assayed with population genetics. Under a null expectation, we would expect only 14 (6% of the 241 duplicate pairs with dN/dS > 1.0) to be found among regions with strong selection. Hence, we suggest that duplicate genes with high amino acid divergence across paralogs are 10-fold more likely to be associated with selective sweeps than we would expect based on background rates. Among these are 2 cytochromes, 1 von Willebrand factor, 1 chitin sythetase, and A-macroglobulin TED domain important for anticoagulation. Others of unknown function include DUFs, WD40s, and Zinc Knuckles. In addition, some sweeps contain adjacent copies of duplicate genes with the same functional annotation with no other causative factors, even when dN/dS < 1.0. These likely represent adaptation without the burst of amino acid substitutions. A total of 245 have dN/dS > 1.1 on any past branch. These include two additional von Willebrand, two more cytochromes, one Thioredoxin, and some DUFS, more Zinc Fingers, inhibitors of apoptosis, and mitochondrial genes.
We identify these two independent measures of selection in elevated dN/dS across paralogs and independently identified sweep signals from population diversity. Such concordance represents a rare case of multiple measures of selection pointing to gene duplication as a causative source of recent adaptation in M. nervosa.
TE Insertions are Detrimental
TEs are selfish constructs that proliferate in genomes even at the expense of their hosts. These repetitive sequences are intimately related to gene duplication rates, as they can facilitate ectopic recombination, form retrogenes, and translocate copies of neighboring DNA. Signals of TE proliferation, especially of Gypsy and Polinton elements, were observed in the reference genome of M. nervosa (Rogers et al. 2021). To determine whether these TE insertions may be adaptive or detrimental, we surveyed frequencies of TE insertions in the population of M. nervosa. We identified genome rearrangements with abnormally mapping read pairs and used BLAST to identify those with transposon sequences at one of the two genomic locations. We assume that the TE-associated region is the donor and the region without TE sequences is the acceptor region where the new TE copy lands. We identify 4,971 TE insertions that can be characterized in this reference genome. TE insertions appear to be highly detrimental in M. nervosa, with a skewed SFS showing an excess of singletons that is significantly different from SNPs (Wilcoxon Rank Sum Test W = 150176864, P < 2.2e × 10−16; Kolmogorov–Smirnov Test D = 0.30443, P < 2.2e × 10−16fig. 2).
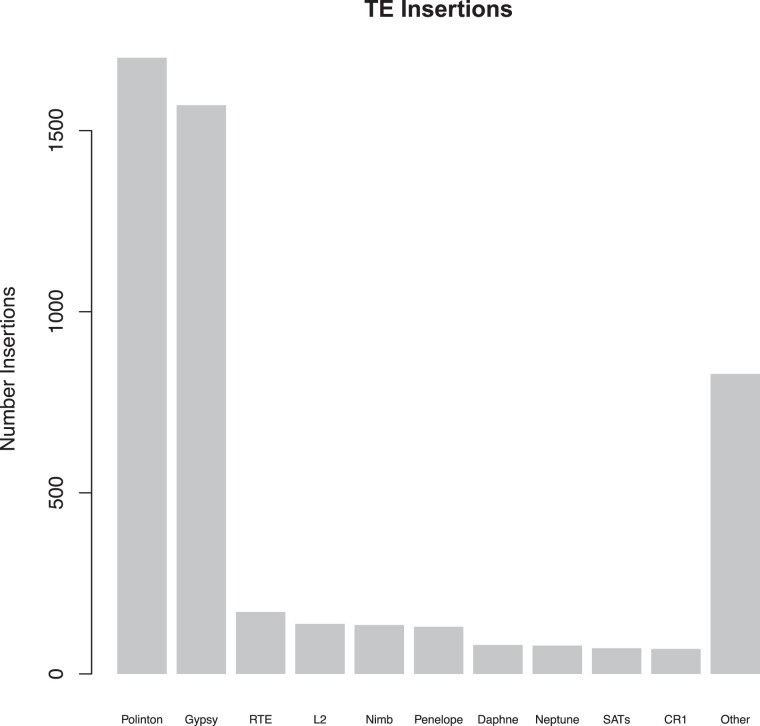
TE movement is dominated by Polinton DNA transposon activation and Gypsy retroelements (supplementary table S6, Supplementary Material online, fig. 7). We also observe Neptune element activity, consistent with the presence of these elements in the M. nervosa reference but not in other species surveyed like Venustaconcha ellipsiformis or Elliptio hopetonensis (Rogers et al. 2021). TEs are underrepresented among reference genome sequence in regions with selective sweeps (P < 10−5). We find only 8.9 Mb (1.5%) of TEs identified via RepDeNovo compared against an expectation of 34 Mb if TE content were allocated proportionally to the amount of the genome captured by sweeps. We identify 18 Mb of RepeatScout TEs, against an expectation of 55 Mb TEs. Only low-frequency insertions at a frequency of 1/26-3/26 are identified in swept regions, precluding the possibility that these mutations are causative agents of selection in strong, recent selective sweeps. Combined with the SFS, we conclude that these mutations are unlikely to be adaptive in M. nervosa, and are rather forming detrimental insertions throughout the genome.
Fig. 7.
Polymorphic TE insertions identified in populations of M. nervosa. Gypsy elements are most common, followed by Polinton elements, consistent with recent TE proliferation identified in the reference genome of M. nervosa.
Discussion
Selection under Environmental Upheaval
Scans of selection can clarify genetic variation that has contributed to survival and reproduction, with fewer a priori biases about what functions should be represented (Nielsen 2005; Ellegren 2014). This reverse ecological genetics can identify the most likely candidates driving selective sweeps, offering more complete information about how adaptation occurs in nature (Ellegren 2014). Historical records suggest population expansion, excluding the possibility of a bottleneck event (Ahlstedt and McDonough 1992; Garner and McGregor 2001). Working in such a species with detailed census records is an advantage for population genetics over model organisms that typically lack extensive historical or fossil records. Moreover, simulations of bottleneck events do not show similar genetic signals compared with strong selective sweeps (supplementary text, Supplementary Material online). These selective sweeps occurred in recent evolutionary time, though resolution on historical time is difficult. If historic DNA could be acquired for the same species at the same location, or through comparisons at other locations, it might resolve how much of these adaptive changes are the product of anthropogenic influence over the past 100 years.
These results recapitulate and confirm the functional categories represented among adaptive gene family amplification in a single genome (Rogers et al. 2021). These selection scans offer greater detail and more information about adaptation than analyses that can be done with single reference genomes. We also identify strong selection in regions with less clear functional implications, such as WD-40 domains, Zinc Fingers, Zinc Knuckles, and apoptosis genes, or even regions with no known functional impacts. These signatures of selection on regions with unknown functions that open questions remain that will need to be explored in future work to fully understand the drivers of genetic adaptation in Unionidae. If future functional analysis can determine what lies in the regions with unannotated genes, we can better understand factors that contribute to success of M. nervosa and how to help endangered populations.
Gene Duplications and Adaptation
Gene duplications have long been held as a source of evolutionary innovation that can contribute new genes with novel functions (Ohno 1970; Conant and Wolfe 2008). Theory suggests that duplicate genes can produce new copies of genes that are functionally redundant. Under reduced constraint copies may accumulate divergence and thereby explore novel functions. Alternatively, duplicate genes may specialize in ancestral functions and offer adaptive subfunctionalization in escape from adaptive conflict (Des Marais and Rausher 2008). Our results showing duplications followed by a burst of amino acid substitutions and strong signals of selective sweeps are consistent with these evolutionary models. It has long been proposed that mutations of large effect appear first, that later are fine-tuned through mutations with narrower functional impacts to reach evolutionary optima (Orr 2006). Empirical evidence suggests that such outcomes are a regular product of gene duplication in other systems (Long and Langley 1993; Jones and Begun 2005; Conant and Wolfe 2008; Des Marais and Rausher 2008). Such models would be consistent with duplications serving as such mutations of large effect that later are fine-tuned through amino acid substitutions.
It is striking to observe such independent signals of selection on multiple duplicate genes: high dN/dS across paralogs and the presence in strong, recent selective sweeps. Over 100 duplications contribute to recent adaptive changes with elevated dN/dS on the terminal branch, and 245 duplicate genes in recent selective sweeps are members of gene families with adaptive signals further in the past. The genomic patterns are overwhelming that these mutations are key contributors to innovation in this species that has experienced recent environmental threat. Duplication in gene families important for the biology of M. nervosa appear to be key for survival and reproduction in this robust species. Fish–host interactions, detox pathways and shell formation, suggest strong recent selective pressures consistent with known ecological challenges are reshaping genetic variation in M. nervosa. Parallel analysis on marine bivalves has revealed adaptive duplications in adaptation to environmental changes (Sun et al. 2017; Hu et al. 2022). Hence, we expect that these principles hold true outside this single species.
We observe an association between detox genes and related stress resistance in a species that has recently been exposed to high levels of pesticides, herbicides, and other pollutants. These chemicals gained widespread use at Muscle Shoals after the 1940s through mosquito and malaria control efforts, farming, and other industrial activities along the river (Woodside 2004). The high pollution load has had extreme effects on wildlife throughout the region (Woodside 2004). Pesticide and herbicide use is known to induce very strong selective pressures in other evolutionary systems. Previous studies have observed parallel cases of detox gene duplication or rearrangements of lesser magnitude in Drosophila (Aminetzach et al. 2005; Schmidt et al. 2010), Morning Glories (Van Etten et al. 2020), and rodents (Nelson et al. 2004). It is likely that the gene duplications observed in M. nervosa are an important part of the genetic response to these selective pressures.
TE Bursts
TEs are selfish genes that amplify themselves even at the expense of host genomes. They can break gene sequences, remodel expression for neighboring genes, and create chimeric TE-gene products (Feschotte 2008; Schaack et al. 2010; Dubin et al. 2018). It is hypothesized that TEs may offer a source of innovation, especially when species experience environmental stress. Recent TE amplification for Gypsy and Polinton elements was previously observed in the reference genome of M. nervosa (Rogers et al. 2021), but it remained unclear whether such population expansion was adaptive or a detrimental byproduct of TE escape from silencing. In this new population genetic data for M. nervosa, we observe no support for adaptive TE insertions.
TE insertions appear to be detrimental with a skewed SFS and underrepresentation in selective sweeps. A species with very large population sizes may be able to weed out detrimental TE insertions while retaining adaptive variation, especially if unlinked from beneficial variation. In light of these results, it seems most likely that the recent proliferation of Gypsy and Polinton elements may have escaped conflict in the short term, but are prevented from spreading through populations under strong selective constraint.
These two different classes of elements represent both LTR retroelement expansion and DNA transposon activity, rather than expansion only within a single class. Gypsy elements carry chromodomains, that can modify heterochromatin organization and modify gene expression for neighboring genes, with more widespread impacts than local dynamics of gene damage (Chen and Corces 2001; Gause et al. 2001; Gao et al. 2008). Under expectations of genetics arms races, we might expect strong selection to favor suppressors of TE activity in the future (Cosby et al. 2019).
There may be interplay between TE content and duplication rates, as repetitive elements can facilitate gene family amplification (Bennetzen 2000; Yang et al. 2008), but these are indirect effects not tied to individual TE copies. The cost of genetic innovation and TE activity may be different in small populations where evolutionary dynamics may be more permissive to TE proliferation (Lynch 2007). Future analysis of the spectrum of TE variation in other species of Unionidae with different population dynamics will help answer questions about the interplay of TEs and adaptive duplication during habitat shifts. Regardless, individual TE insertions in M. nervosa largely appear to be maladaptive.
Implications for Imperiled Species
In scans of selection in Megalonaias nervosa, we identify many genes that point to fish–host interactions in glochidia, a known point of attrition for many species (Modesto et al. 2018). Strong selection at glochidia stages, may favor larger brood sizes to overcome attrition in some reproductive strategies (Haag 2013). M. nervosa females can produce 1 million offspring per reproductive cycle (Haggerty et al. 2005), a factor that may influence their success. Genomics also points to detox genes, shell formation genes, stress response genes, and mitochondria genes, all consistent with factors important for Unionida.
Not all species may have sufficient genetic variation to solve these challenges under shifting selective pressures, especially when population sizes are small (Maynard Smith 1971; Hermisson and Pennings 2005). M. nervosa with its large population expansion represents a successful species that can be used as a genetic benchmark for adaptive changes. In threatened or endangered species with smaller population sizes, it is possible that genetic variation may be limited, and genetic drift may impede adaptive walks. In M. nervosa, some gene duplication is likely to have been adaptation prior to the modern era as dS > 0. The likelihood of adaptation from standing variation for gene duplications was high in M. nervosa (Rogers et al. 2021).
As duplicate genes appear to be important sources of innovation, we suggest that they deserve better characterization across Unionidae and in other species experiencing ecological threat. Species that lack gene duplications may struggle to survive in the face of these same environmental pressures. Conversely, current results would suggest that new TE insertions are primarily rare and non-adaptive. They may are unlikely, based on current evidence, to represent those genetic changes that are most essential for species survival. Highly variable low-frequency TEs like Gypsy and Polinton elements in M. nervosa may serve as poor markers for species tracking, and may require masking for the design of species markers. These insights and genetic resources can help conservation biologists working in mussel management as they design analyses to monitor and aid threatened or endangered species of Unionidae. Similar work in population genetics for endangered Unionids may also offer and empirical setting to explore alternative population genetic models relevant to high variance in reproductive success (Matuszewski et al. 2018).
Future cross species comparisons may help address the genetic basis of adaptation in this clade that has experienced recent environmental upheaval. How do species with smaller population sizes respond under these environmental pressures? What is missing from the genomes of species that are most threatened? If we can begin to approach these genetic questions for Unionidae, we can better understand how evolutionary processes differ in species that are under threat and determine how genomes influence species survival.
Methods
Specimen Collection
JT Garner collected specimens of Megalonaias nervosa at Pickwick Reservoir (supplementary table S1, Supplementary Material online). The largest (Specimen #3) was selected as the reference genome specimen, sequenced and annotated as previously described (Rogers et al. 2021). Some 14 specimens were dissected and sequenced using Illumina short read sequences for population genomic data. The reference specimen N50 in previous sequencing was roughly 50 kb, adequate for many applications in evolutionary genomics, but with limited information regarding linkage across distance. Scaffolding attempts with HiC and OmniC have not been successful, with zero cross-linked read pairs (Rogers et al. 2021). To improve the reference genome for population genetic applications, we generated additional long-read sequence data to scaffold the assembly into longer contigs with an N50 of 120 kb. We reannotated according to previously used methods (Rogers et al. 2021) similar to those used to reannotate non-model Drosophila (Rogers et al. 2014) (supplementary text, Supplementary Material online).
Identification of Regions with Extended Selective Sweeps
Sequences were aligned to the reference genome, and used to estimate genetic diversity statistics commonly used population genetic inference π, Wattersons θ, and Tajima’s D. We required that windows have full coverage for at least 75% of sites across all strains. Population genetic statistics were corrected for the number of sites in 10 kb windows with full coverage. We used msprime v1.1.1 (Baumdicker et al. 2022) and SLiM v3.7 (Messer 2013; Haller and Messer 2019; Haller et al. 2019) to model expectations of diversity under demographic scenarios and with natural selection for these populations. Additional detail is available in supplementary text, Supplementary Material online. We identified SNPs from 4-fold synonymous sites where none of the three possible nucleotide substitutions alter amino acid sequence, and 4-fold nonsynonymous sites where any of the three possible substitution alters amino acid sequence and estimated site frequency spectra (SFS) for each and tested for significant differences using both Kolmogorov–Smirnov tests and Wilcoxon rank sum tests.
Extended regions with reduced genetic diversity display stronger signals than the classic V-shape most often described for typical selective sweeps (Sella et al. 2009; Hartl 2020). They do not fit with theoretical models that could be used to place boundaries on the timing and selection coefficient of selective sweeps using diversity and recombination rates (Kaplan et al. 1989; Sella et al. 2009). To objectively place boundaries on recent selective sweeps, we use Bayesian changepoint statistics. Changepoint statistics are agnostic to the direction, magnitude, and duration of effects. They identify regions of the data that depart from the background patterns in the remainder of the data, where shorter signals with greater magnitude may be significant as can longer signals of lesser magnitude. We identified changepoints and posterior means of π, and Tajima’s D for regions between changepoints in the R package bcp (Erdman and Emerson 2007) (supplementary text, Supplementary Material online). Genome assembly for freshwater molluscs remains challenging because of repeats and difficulty of long molecule extraction, with limited N50s even using long-read data (Rogers et al. 2021; Smith 2021; Renaut et al. 2018). The best assembly to date is for Potamilus streckersonii, with an N50 of 2 Mb after 100X coverage of PacBio and 48X coverage of 10X sequencing (Smith 2021). To anchor scaffolds identified in the longest sweeps, we used syntenic mapping against this more contiguous bivalve assembly to identify sections of M. nervosa likely to be from similar genetic locations (supplementary text, Supplementary Material online).
Adaptive Gene Duplications
We identified 4,758 gene families (with two or more paralogs) using a First-Order Fuzzy Reciprocal Best Hit Blast (Han et al. 2009) on the re-annotated the scaffolded genome as per previous methods (Rogers et al. 2021) (supplementary text, Supplementary Material online). Protein sequences were aligned in clustalw (Thompson et al. 1994) then back-translated to the original nucleotide sequence. Synonymous and nonsynonymous substitutions were analyzed with the codeml package of PAML (Yang 1997) with the F1x4 codon model using the clustalw generated guide tree. We excluded 44 out of 4,758 gene families that proved computationally intractable, with failed alignments or failed PAML runs. We then identified duplication events with elevated amino acid substitutions across at least one branch for paralogs, suggesting selection for amino acid replacements (high dN/dS ≫ 1.0), a gene-specific measure of selection (Goldman and Yang 1994). The locations of these adaptive gene duplications were matched with locations of strong, recent selective sweeps to identify cases where they therefore likely contribute as causative agents of selective sweeps.
TE Insertions
We identified polymorphic TE insertions using a paired-end read approach (Cridland et al. 2013). We identified polymorphic read pairs that map to different scaffolds, indicative of DNA moving from one location to the other. We required at least five abnormally mapping read pairs support each mutation, clustering read-pairs within 325 bp, based on the Illumina sequencing insert size. Mutations with insertion sites within 325 bp were clustered across samples. We took 1000 bp on either side of each breakpoint and matched these in a tblastx against the RepBase database (Bao et al. 2015) at an E-value of 10−20, requiring hits at least 100 bp long to a repetitive element sequence on one side, but not both sides of rearrangements. These mutations were considered to be novel TE insertions compared with the ancestral state. We used coverage >1.75X and <5X whole-genome background levels to identify homozygous mutations.
Supplementary Material
Acknowledgements
We thank Cathy Moore for advice on molecular assays. We thank Karen Lopez for support with local Nanopore sequencing at UNCC. Jon Halter, Michael Moseley, Chad DeWitt, Chuck Price, and Chris Maher provided help with software installation and functionality on the UNCC HPC system. We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Shared Resource, which provided Illumina sequencing services for M. nervosa genomes and transcriptomes. De Novo Genomics generated a subset of the Oxford Nanopore sequences for this study. All analyses were run on the UNCC High Performance Computing cluster, supported by UNC Charlotte and the Department of Bioinformatics.
Contributor Information
Rebekah L Rogers, Department of Bioinformatics and Genomics, University of North Carolina, Charlotte, NC 28223, USA.
Stephanie L Grizzard, Department of Biological Sciences, Old Dominion University, Norfolk, VA, USA.
Jeffrey T Garner, Division of Wildlife and Freshwater Fisheries, Alabama Department of Conservation and Natural Resources, Florence, AL, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
R.L.R. designed experiments and analyses; J.G. collected specimens from wild populations; R.L.R. and S.L.G. performed experiments; R.L.R. performed analyses with assistance from S.L.G.; R.L.R. and J.G. wrote and edited the manuscript with input from S.L.G..
Data Availability
Megalonaias nervosa genomic sequence data are available in PRJNA646917 and PRJNA929048 and transcriptome data are available at SRA PRJNA646778. The Megalonaias nervosa genome assembly is available at PRJNA681519. Supplementary data files are described in a README file. These include annotations (MergedScaffold2.isos.gff MergedScaffold2.isos.pt.fa.gz MergedScaffold2.isos.pt.interpro.tsv), Population Genetic Data Files (ChangePtMeans.pi.fmt ChangePtMeans.tajD.fmt, ThetaSlide.combined.fmt.gz, DnDs.out), and a syntenic map (Synteny.fmt.gz).
Funding
This work was supported by startup funding from the Department of Bioinformatics and Genomics at the University of North Carolina, Charlotte. R.L.R. is funded in part by NIH NIGMS R35 GM133376. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- Ahlstedt SA, McDonough TA. 1992. Quantitative evaluation of commercial mussel populations in the Tennessee river portion of Wheeler Reservoir, Alabama. In Conservation and Management of Freshwater Mussels. Proceedings of a UMRCC Symposium. p. 12–14. Norris and Knoxville, Tennessee: Tennessee Valley Authority Water Resources Aquatic Biology Department.
- Aminetzach YT, Macpherson JM, Petrov DA. 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309(5735):764–767. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumdicker F, Bisschop G, Goldstein D, Gower G, Ragsdale AP, Tsambos G, Zhu S, Eldon B, Ellerman EC, Galloway JG, et al. 2022. Efficient ancestry and mutation simulation with msprime 1.0. Genetics 220(3):iyab229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. 2000. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 42(1):251–269. [PubMed] [Google Scholar]
- Bolotov IN, Kondakov AV, Vikhrev IV, Aksenova OV, Bespalaya YV, Gofarov MY, Kolosova YS, Konopleva ES, Spitsyn VM, Tanmuangpak K, et al. 2017. Ancient river inference explains exceptional oriental freshwater mussel radiations. Sci Rep. 7(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Beaupre HD, Stewart DT, Hoeh WR, Blier PU. 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough? Trends Genet. 23(9):465–474. [DOI] [PubMed] [Google Scholar]
- Cahn AR. 1936. The molluscan fauna of the Clinch River below Norris Dam upon the completion of that structure. Norris, Tennessee: Tennessee Valley Authority. [Google Scholar]
- Campbell DC, Serb JM, Buhay JE, Roe KJ, Minton RL, Lydeard C. 2005. Phylogeny of North American amblemines (Bivalvia, Unionoida): prodigious polyphyly proves pervasive across genera. Invertebr Biol. 124(2):131–164. [Google Scholar]
- Chen S, Corces VG. 2001. The gypsy insulator of Drosophila affects chromatin structure in a directional manner. Genetics 159(4):1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 9(12):938–950. [DOI] [PubMed] [Google Scholar]
- Cosby RL, Chang N -C, Feschotte C. 2019. Host–transposon interactions: conflict, cooperation, and cooption. Genes Dev. 33(17-18):1098–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Macdonald SJ, Long AD, Thornton KR. 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol Biol Evol. 30(10):2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch NM, Edie SM, Collins KS, Bieler R, Jablonski D. 2021. Calibrating phylogenies assuming bifurcation or budding alters inferred macroevolutionary dynamics in a densely sampled phylogeny of bivalve families. Proc R Soc B. 288(1964):20212178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1878. Transplantation of shells. Nature 18:120–121. [Google Scholar]
- Des Marais DL, Rausher MD. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454(7205):762–765. [DOI] [PubMed] [Google Scholar]
- Dubin MJ, Scheid OM, Becker C. 2018. Transposons: a blessing curse. Curr Opin Plant Biol. 42:23–29. [DOI] [PubMed] [Google Scholar]
- Elderkin C, Christian A, Vaughn C, Metcalfe-Smith J, Berg D. 2007. Population genetics of the freshwater mussel, Amblema plicata (say 1817) (Bivalvia: Unionidae): evidence of high dispersal and post-glacial colonization. Conserv Genet. 8(2):355–372. [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1):51–63. [DOI] [PubMed] [Google Scholar]
- Emerson J, Cardoso-Moreira M, Borevitz JO, Long M. 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320(5883):1629–1631. [DOI] [PubMed] [Google Scholar]
- Erdman C, Emerson JW. 2007. bcp: an R package for performing a Bayesian analysis of change point problems. J Stat Softw. 23(3):1–13. [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 9(5):397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. 2008. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 18(3):359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner J, McGregor S. 2001. Current status of freshwater mussels (Unionidae, Margaritiferidae) in the Muscle Shoals area of Tennessee River in Alabama (Muscle Shoals revisited again). Am Malacol Bull. 16(1-2):155–170. [Google Scholar]
- Gause M, Morcillo P, Dorsett D. 2001. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 21(14):4807–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11(5):725–736. [DOI] [PubMed] [Google Scholar]
- Haag WR. 2012. North American freshwater mussels: natural history, ecology, and conservation. New York: Cambridge University Press. [Google Scholar]
- Haag WR. 2013. The role of fecundity and reproductive effort in defining life-history strategies of North American freshwater mussels. Biol Rev. 88(3):745–766. [DOI] [PubMed] [Google Scholar]
- Haag WR, Williams JD. 2014. Biodiversity on the brink: an assessment of conservation strategies for North American freshwater mussels. Hydrobiologia 735(1):45–60. [Google Scholar]
- Haggerty TM, Garner JT, Rogers RL. 2005. Reproductive phenology in Megalonaias nervosa (Bivalvia: Unionidae) in Wheeler Reservoir, Tennessee River, Alabama, USA. Hydrobiologia 539(1):131–136. [Google Scholar]
- Haller BC, Galloway J, Kelleher J, Messer PW, Ralph PL. 2019. Tree-sequence recording in SLiM opens new horizons for forward-time simulation of whole genomes. Mol Ecol Resour. 19(2):552–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, Messer PW. 2019. SLiM 3: forward genetic simulations beyond the Wright-Fisher model. Mol Biol Evol. 36(3):632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. 2009. Adaptive evolution of young gene duplicates in mammals. Genome Res. 19(5):859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. 2020. A primer of population genetics and genomics. New York: Oxford University Press. [Google Scholar]
- Hermisson J, Pennings PS. 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169(4):2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Song H, Feng J, Zhou C, Yang M-J, Shi P, Yu Z-L, Li Y-R, Guo Y-J, Li H-Z, et al. 2022. Massive heat shock protein 70 genes expansion and transcriptional signatures uncover hard clam adaptations to heat and hypoxia. Front Marine Sci. 9:898669. [Google Scholar]
- Isom BG, Yokley P, Gooch CH. 1973. Mussels of the Elk River Basin in Alabama and Tennessee-1965–1967. Am Midl Nat. 89:437–442. [Google Scholar]
- Jones CD, Begun DJ. 2005. Parallel evolution of chimeric fusion genes. Proc Natl Acad Sci U S A. 102(32):11373–11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. 1989. The “hitchhiking effect” revisited. Genetics 123(4):887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh SM, Johnson NA, Williams JD, Randklev CR, Simons AM. 2021. Gulf coast vicariance shapes phylogeographic history of a north American freshwater mussel species complex. J Biogeogr. 48(5):1138–1152. [Google Scholar]
- Kongim B, Sutcharit C, Panha S. 2015. Cytotaxonomy of unionid freshwater mussels (unionoida, unionidae) from northeastern thailand with description of a new species. ZooKeys. 27(514):93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. Timetree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lake PS, Palmer MA, Biro P, Cole J, Covich AP, Dahm C, Gibert J, Goedkoop W, Martens K, Verhoeven J. 2000. Global change and the biodiversity of freshwater ecosystems: impacts on linkages between above-sediment and sediment biota: all forms of anthropogenic disturbance—changes in land use, biogeochemical processes, or biotic addition or loss—not only damage the biota of freshwater sediments but also disrupt the linkages between above-sediment and sediment-dwelling biota. BioScience. 50(12):1099–1107. [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192(2):533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-P, Mitton JB, Wu S-K. 1996. Paternal mitochondrial DNA differentiation far exceeds maternal mitochondrial DNA and allozyme differentiation in the freshwater mussel, Anodonta grandis grandis. Evolution 50(2):952–957. [DOI] [PubMed] [Google Scholar]
- Long M, Langley CH. 1993. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260(5104):91–95. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland (Mass): Sinauer Associates. p. 494. [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszewski S, Hildebrandt ME, Achaz G, Jensen JD. 2018. Coalescent processes with skewed offspring distributions and nonequilibrium demography. Genetics 208(1):323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. 1971. What use is sex? J Theor Biol. 30(2):319–335. [DOI] [PubMed] [Google Scholar]
- Messer PW. 2013. SLiM: simulating evolution with selection and linkage. Genetics 194(4):1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesto V, Ilarri M, Souza AT, Lopes-Lima M, Douda K, Clavero M, Sousa R. 2018. Fish and mussels: importance of fish for freshwater mussel conservation. Fish Fisheries. 19(2):244–259. [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. 2004. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenet Genomics. 14(1):1–18. [DOI] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu Rev Genet. 39:197–218. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin, New York: Springer-Verlag. [Google Scholar]
- Orr HA. 2006. The distribution of fitness effects among beneficial mutations in Fisher’s geometric model of adaptation. J Theor Biol. 238(2):279–285. [DOI] [PubMed] [Google Scholar]
- Ortmann A. 1924. Mussel shoals. Science 60(1564):565–566. [DOI] [PubMed] [Google Scholar]
- Patterson MA, Mair RA, Eckert NL, Gatenby CM, Brady T, Jones JW, Simmons BR, Devers JL. 2018. Freshwater mussel propagation for restoration. Cambridge University Press. [Google Scholar]
- Pfeiffer JM, Breinholt JW, Page LM. 2019. Unioverse: a phylogenomic resource for reconstructing the evolution of freshwater mussels (Bivalvia, Unionoida). Mol Phylogenet Evol. 137:114–126. [DOI] [PubMed] [Google Scholar]
- Pfeiffer JM, Sharpe AE, Johnson NA, Emery KF, Page LM. 2018. Molecular phylogeny of the Nearctic and Mesoamerican freshwater mussel genus Megalonaias. Hydrobiologia 811(1):139–151. [Google Scholar]
- Ranz JM, Parsch J. 2012. Newly evolved genes: moving from comparative genomics to functional studies in model systems: how important is genetic novelty for species adaptation and diversification? Bioessays. 34(6):477–483. [DOI] [PubMed] [Google Scholar]
- Régnier C, Fontaine B, Bouchet P. 2009. Not knowing, not recording, not listing: numerous unnoticed mollusk extinctions. Conserv Biol. 23(5):1214–1221. [DOI] [PubMed] [Google Scholar]
- Renaut S, Guerra D, Hoeh WR, Stewart DT, Bogan AE, Ghiselli F, Milani L, Passamonti M, Breton S. 2018. Genome survey of the freshwater mussel Venustaconcha ellipsiformis (Bivalvia: Unionida) using a hybrid de novo assembly approach. Genome Biol Evol. 10(7):1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Bedford T, Lyons AM, Hartl DL. 2010. Adaptive impact of the chimeric gene Quetzalcoatl in Drosophila melanogaster. Proc Natl Acad Sci U S A. 107(24):10943–10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, Thornton KR. 2015. Tandem duplications and the limits of natural selection in Drosophila yakuba and Drosophila simulans. PLoS ONE. 10(7):e0132184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Grizzard SL, Titus-McQuillan JE, Bockrath K, Patel S, Wares JP, Garner JT, Moore CC. 2021. Gene family amplification facilitates adaptation in freshwater unionid bivalve Megalonaias nervosa. Mol Ecol. 30(5):1155–1173. [DOI] [PubMed] [Google Scholar]
- Rogers RL, Hartl DL. 2011. Chimeric genes as a source of rapid evolution in Drosophila melanogaster. Mol Biol Evol. 29(2):517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Shao L, Sanjak JS, Andolfatto P, Thornton KR. 2014. Revised annotations, sex-biased expression, and lineage-specific genes in the Drosophila melanogaster group. G3 4(12):2345–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Shao L, Thornton KR. 2017. Tandem duplications lead to novel expression patterns through exon shuffling in Drosophila yakuba. PLoS Genet. 13(5):e1006795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25(9):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, Martin J, Daborn PJ, Goddard ME, Batterham P, et al. 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6(6):e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hahn MW. 2010. Gene copy-number polymorphism in nature. Proc R Soc B: Biol Sci. 277(1698):3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5(6):e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CH. 2021. A high-quality reference genome for a parasitic bivalve with doubly uniparental inheritance (Bivalvia: unionida). Genome Biol Evol. 13(3):evab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart NB, Rogers RL. 2019. Chromosomal rearrangements as a source of new gene formation in Drosophila yakuba. PLoS Genet. 15(9):e1008314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer DL. 1999. Effects of alien species on freshwater mollusks in North America. J N Am Benthol Soc. 18(1):74–98. [Google Scholar]
- Strayer DL, Downing JA, Haag WR, King TL, Layzer JB, Newton TJ, Nichols JS. 2004. Changing perspectives on pearly mussels, North America’s most imperiled animals. BioScience. 54(5):429–439. [Google Scholar]
- Sun J, Zhang Y, Xu T, Zhang Y, Mu H, Zhang Y, Lan Y, Fields CJ, Hui JHL, Zhang W, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 1(5):1–7. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten M, Lee KM, Chang S-M, Baucom RS. 2020. Parallel and nonparallel genomic responses contribute to herbicide resistance in Ipomoea purpurea, a common agricultural weed. PLoS Genet. 16(2):e1008593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen HB, Cao ZM, Hua D, Xu P, Ma XY, Jin W, Yuan XH, Gu RB. 2017. The complete maternally and paternally inherited mitochondrial genomes of a freshwater mussel Potamilus alatus (Bivalvia: Unionidae). PLoS ONE. 12(1):e0169749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JD, Bogan AE, Garner JT. 2008. Freshwater mussels of Alabama and the Mobile basin in Georgia, Mississippi, and Tennessee. Tuscaloosa (AL): University of Alabama Press. [Google Scholar]
- Williams JD, Warren ML Jr, Cummings KS, Harris JL, Neves RJ. 1993. Conservation status of freshwater mussels of the United States and Canada. Fisheries 18(9):6–22. [Google Scholar]
- Woodside MD. 2004. Water quality in the lower Tennessee River Basin, Tennessee, Alabama, Kentucky, Mississippi, and Georgia, 1999–2001. Vol. 1233. Reston (VA): US Geological Survey. [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13(5):555–556. [DOI] [PubMed] [Google Scholar]
- Yang S, Arguello JR, Li X, Ding Y, Zhou Q, Chen Y, Zhang Y, Zhao R, Brunet F, Peng L, et al. 2008. Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet. 4(1):e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418):49–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Megalonaias nervosa genomic sequence data are available in PRJNA646917 and PRJNA929048 and transcriptome data are available at SRA PRJNA646778. The Megalonaias nervosa genome assembly is available at PRJNA681519. Supplementary data files are described in a README file. These include annotations (MergedScaffold2.isos.gff MergedScaffold2.isos.pt.fa.gz MergedScaffold2.isos.pt.interpro.tsv), Population Genetic Data Files (ChangePtMeans.pi.fmt ChangePtMeans.tajD.fmt, ThetaSlide.combined.fmt.gz, DnDs.out), and a syntenic map (Synteny.fmt.gz).