Abstract
ZNF410 is a highly-conserved transcription factor, remarkable in that it recognizes a 15-base pair DNA element but has just a single responsive target gene in mammalian erythroid cells. ZNF410 includes a tandem array of five zinc-fingers (ZFs), surrounded by uncharacterized N- and C-terminal regions. Unexpectedly, full-length ZNF410 has reduced DNA binding affinity, compared to that of the isolated DNA binding ZF array, both in vitro and in cells. AlphaFold predicts a partially-folded N-terminal subdomain that includes a 30-residue long helix, preceded by a hairpin loop rich in acidic (aspartate/glutamate) and serine/threonine residues. This hairpin loop is predicted by AlphaFold to lie against the DNA binding interface of the ZF array. In solution, ZNF410 is a monomer and binds to DNA with 1:1 stoichiometry. Surprisingly, the single best-fit model for the experimental small angle X-ray scattering profile, in the absence of DNA, is the original AlphaFold model with the N-terminal long-helix and the hairpin loop occupying the ZF DNA binding surface. For DNA binding, the hairpin loop presumably must be displaced. After combining biophysical, biochemical, bioinformatic and artificial intelligence-based AlphaFold analyses, we suggest that the hairpin loop mimics the structure and electrostatics of DNA, and provides an additional mechanism, supplementary to sequence specificity, of regulating ZNF410 DNA binding.
INTRODUCTION
DNA recognition by transcription factors is essential for appropriate expression of genes in any living organism (1). These DNA binding proteins exert their effects by binding at appropriate genomic locations on the double helix. Zinc finger (ZF) proteins, containing Cys2-His2 (C2H2)-coordinated zinc ligands, probably comprise the largest family of regulatory proteins in mammals (2,3). Two recent studies identified ZNF410, also known as another partner for tumor suppressor ARF (APA-1) (4), as the DNA binding protein that activates a single gene, CHD4, via binding to its promoter and regulatory enhancer regions (5,6). CHD4, in turn, is a component of the nucleosome remodeling and deacetylase complex (NuRD (7)), that is required for the silencing of the fetal type globin genes.
ZNF410 isoform b is a 478-amino-acid protein containing five C2H2 zinc fingers, flanked by N-terminal (NT) and C-terminal (CT) regions of unknown function (Figure 1A). ZNF410 is made in multiple isoforms (Figure S1), adding to the apparent complexity of its functioning. [In this study, we used only isoform b or its truncations.] ZNF410 is highly conserved among mammals (Figure S2), and more generally among vertebrates (Figure S3). Its DNA sequence specificity also appears to be highly conserved, as the characteristic densely clustered ZNF410 binding ‘boxes’ (5,6) occur upstream of the genes for CHD4 orthologs across the vertebrates (interestingly, always in the same orientation; Figure S3C).
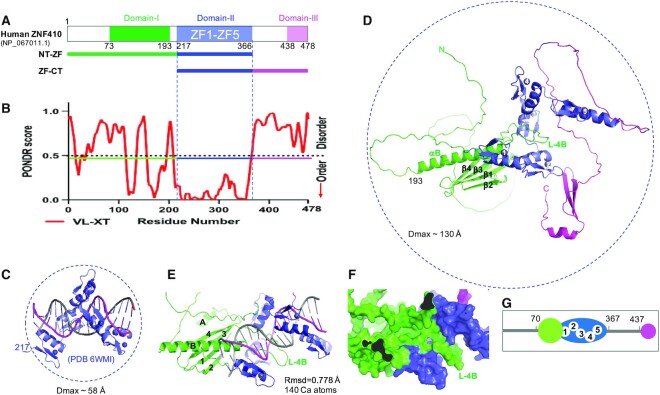
Figure 1.
Structure prediction of full length human ZNF410. (A) Schematic of ZNF410 comprising three regions: N-terminal (NT; green), a tandem of five zinc fingers (ZF; blue) and C-terminal (CT; magenta). (B) PONDR prediction shows two ordered regions of residues 100–200 and the central ZF domain. (C) The crystal structure of five ZFs (residues 217–366) in complex with DNA (PDB 6WMI). (D) AlphaFold prediction of ZNF410 structure in the absence of DNA. NT, ZF and CT are represented in green, blue and magenta respectively. The secondary structural elements of the NT domain are labeled (helices αA and αB and strands β1–β4). (E) Superposition of AlphaFold model and experimentally determined structure of the ZF-DNA complex (PDB 6WMI). (F) Surface representation of the 30-residue-long helix (αB) and its preceding loop (L4B), occupying the DNA binding surface of the ZF domain. (G) Cartoon representation of ordered regions in ZNF410, represented as in (A, D, E and F) in green, blue and magenta. Five ordered ZFs are numbered. The green colored N-terminal domain occupied the DNA binding surface in the absence of DNA.
We previously characterized the crystal structure of ZNF410’s five tandem ZFs, in complex with a 17-base pair oligonucleotide taken from the consensus sequence of the CHD4 promoter and enhancer (5). Besides the DNA binding domain, many transcription factors contain an activation domain, an oligomerization (dimerization) domain, and/or a ligand binding domain, e.g. NF-κB (8–10), p53 (11,12) and nuclear receptors (13–15). Usually, these domains are connected by disordered linkers.
Here, we study the solution behavior of full-length ZNF410, in the presence and absence of DNA, using small angle X-ray scattering (SAXS). SAXS is emerging as a powerful tool for structural characterization of biological macromolecules in solution, particularly for disordered linkers and dynamic complexes involving protein-protein and protein-nucleic acid interactions (16–19). We observed that ZNF410 is primarily a monomeric protein, which adopts a range of conformations in solution. Upon binding DNA, ZNF410 undergoes significant conformational changes in solution, thus modulating its ability to form a stable ZNF410/DNA complex. The wide range of conformational states adopted by ZNF410 can be attributed to the extended regions of intrinsic disorder that connect functional domains. We report here that one of these AlphaFold-predicted domains contains an apparently DNA-mimicking hairpin loop, enriched in acidic (aspartate/glutamate) and hydroxyl (serine/threonine) residues, that appears to regulate the ability of the ZF array to bind DNA via a cis-acting allosteric mechanism.
MATERIALS AND METHODS
Recombinant proteins expression and purification
The DNA fragments coding for the full length (FL) human ZNF410 (NP_001229855.1; residues 1–478; pXC2179), and four different subsections: NT (residues 1–216; pXC2288), ZF (residues 217–366; pXC2180), NT-ZF (residues 1–366; pXC2218), ZF-CT (residues 217–478; pXC2217), were ligated into pGEX-6P-1 vector with a GST fusion tag. The plasmids were transformed into Escherichia coli strain BL21-Codon-plus (DE3)-RIL (Stratagene). Bacteria were grown in Lysogeny broth (LB) in a shaker at 37°C until reaching the log phase (A600 nm between 0.4 and 0.5), at which time the shaker temperature was set to 16°C and 25 μM ZnCl2 (final concentration) was added to the cell culture to ensure Zn incorporation into the ZF domains. When the A600 nm reached ∼0.8, protein expression was induced by the addition of 0.2 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG), with subsequent growth for 20 h at 16°C. The ZF, NT-ZT, ZF-CT and FL proteins were purified as described, via a three-column chromatography protocol (5).
To purify NT, cells were collected by centrifugation and the pellet was suspended in lysis buffer [20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 5% glycerol, 0.5 mM tris(2-carboxyethl)phosphine (TCEP)]. Cells were lysed by sonication, and debris was removed by centrifugation for 30 min at 47 000 × g. The supernatant was loaded onto a 5 ml GSTrap column (GE Healthcare). The resin was washed with lysis buffer, and bound protein was eluted with elution buffer (100 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 0.5 mM TCEP and 20 mM reduced form glutathione). The GST fusions were digested with PreScission protease (produced in-house) to remove the GST fusion tag. The cleaved protein was loaded onto a 5 ml HiTrap-Q-HP column (GE Healthcare). The protein was eluted by an NaCl gradient from 0.1 to 1 M in 20 mM Tris–HCl, pH 8.0, 5% glycerol and 0.5 mM TCEP. The peak fractions were pooled, and loaded onto the GSTrap column again to remove free GST. The flowthrough sample was concentrated, and loaded onto a HiLoad 16/60 Superdex S200 column (GE Healthcare) equilibrated with 20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5% glycerol and 0.5 mM TCEP. The protein was frozen and stored at −80°C prior to use.
Mutagenesis
We generated seven substitutions of hydroxyl residues (S148D, S154D, S155D, S157D, T158E, S160D and S161D) within the hairpin-loop in the context of the full length (FL, pXC2319) or NT-ZF fragment (pXC2312) of ZNF410. The first substitution, S148D (pXC2330), was introduced by one-step PCR-based mutagenesis (Table S1). We introduced the other six mutations (S154D, S155D, S157D, T158E, S160D and S161D) on top of the S148D mutant, using a two-step PCR amplification procedure. In the first step, two fragments overlapping at the region to be altered were generated using a pair of primers (Table S1). In the second step, the two fragments containing the S/T-to-D/E substitutions were used as the templates with the 5’ forward and the 3’ reverse primer pair to generate the final mutant constructs. For the D/E-to-A substitutions at acidic residues D143, E144, E146 and D147, the same two-step PCR amplification procedure was adopted, via two fragments overlapping at the targeted region using a pair of primers (Table S1). All mutants were confirmed by sequencing, and cloned into pGEX-6P-1 vector with an N-terminal GST tag.
The expression and purification procedures for ZNF410 from the mutants were similar to those of WT protein, as described (5), with two modifications. First, the cell lysate was treated with 0.1% (w/v) polyethylenimine instead of 0.3% (w/v). Second, the mutant protein was eluted from a 5-mL HiTrap-SP-HP (GE Healthcare) cation exchange chromatography column, instead of an anion exchange column, by a NaCl gradient from 0.1 to 1 M in 20 mM Tris–HCl, pH 7.0, 5% glycerol and 0.5 mM TCEP. From the sizing column, the majority of mutant protein eluted early, as aggregates, but a smaller monomer peak eluted in later fractions and was collected for DNA binding assays.
DNA binding assays
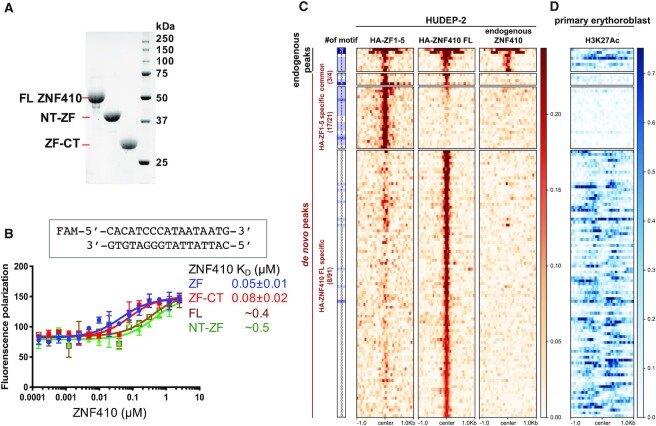
Fluorescence polarization (FP) was used to measure the binding affinity, using a Synergy 4 Microplate Reader (BioTek). Aliquots (5 nM) of 6-carboxy-fluorescein (FAM)-labeled DNA duplex (FAM-5′-CAC ATC CCA TAA TAA TG-3′ and 3′-GTG TAG GGT ATT ATT AC-5′) were incubated with varied amounts of proteins (0 to 2.5 μM) in 20 mM Tris–HCl, pH 7.5, 300 mM NaCl, 5% glycerol and 0.5 mM TCEP for 10 min at room temperature. The data were processed using Graphpad Prism (version 9.0) with equation [mP] = [maximum mP] × [C] / (KD +[C]) + [baseline mP], in which mP is millipolarization and [C] is protein concentration. The KD value for each protein–DNA interaction was derived from two replicated experiments.
Electrophoretic mobility shift assay (EMSA) was performed by incubating 0.1 μM ZF and varied amount of NT (serial 2× dilution of 2.5 μM) and 5 nM (FAM)-labeled DNA in 20 mM Tris–HCl, pH 7.5, 300 mM NaCl, 5% glycerol and 0.5 mM TCEP for 10 min at room temperature. Aliquots of 10 μl of reactions were loaded onto an 8% native 1× TBE polyacrylamide gel and run at 200 V for 30 min in 0.5× TBE buffer. The gel was imaged using a ChemiDoc Imaging System (BIO-RAD).
PONDR analysis
The amino acid sequence of human ZNF410 was retrieved from Uniprot (Q86VK4) and given as input in FASTA format for analysis at the PONDR-VLXT (20) webserver, to predict the propensity for intrinsic disorder in ZNF410.
SEC-SAXS data collection and processing
The protein samples of ZNF410 FL, NT-ZF and ZF-CT were concentrated respectively to 4, 5 and 13 mg/ml. The complex samples were prepared by incubating protein with DNA at a 1:1 molar ratio. SEC-SAXS data were collected at the SIBYLS beamline 12.3.1 at the Advanced Light Source (21). X-ray wavelength was set to λ = 1.127 Å, and the sample to detector distance was 2100 mm, resulting in scattering vectors (q) ranging from 0.01 to 0.4 Å−1. A total of 60 μl of purified proteins, with and without DNA respectively, were prepared in running buffer (20 mM Tris pH 7.5, 250 mM NaCl, 0.1% β-mercaptoethanol). The Shodex KW803 column was equilibrated with running buffer, with a flow rate of 0.5 ml/min. A 55 μl volume of each sample was injected into the SEC column, and 3 s X-ray exposures were recorded continuously for 30 min. SCÅTTER (https://bl1231.als.lbl.gov/scatter/) was used for buffer subtraction. The subtracted curves were merged and used for Guinier analysis, Dimensionless Kratky plots, and computing P(r) plots. The P(r) plots were normalized based to unity at their maxima (22). MW was calculated using SAXSMoW 2.0 (built-in ATSAS package), which applies a correction factor to the Porod volume (23). This method has a ∼10–12% error rate.
Solution structure modeling
The AlphaFold protein structure database (24) was used to predict the model of the full length ZNF410. The crystal structure of the zinc fingers ZF1–ZF5 in complex with 17-bp DNA (PDB ID: 6WMI) was superposed and compared to the predicted model. The AlphaFold model was used as a template to perform rigid body modeling. Minimal molecular dynamic simulations were performed on the flexible regions within the structure so as to explore the conformational space adopted by FL and individual constructs of ZNF140 using BILBOMD (25). In BILBOMD every ∼100 ps one conformational state is stored for further SAXS fitting. We record 10 000 conformers in total, varying in Rg and Dmax values for consequent SAXS fitting and multistate validation (26). Because of the dynamic and flexible character of ZNF410, a minimal ensemble search method was used to identify the multi-state model (27) required to best fit the experimental data. The scattering profile from all 10 000 models was first computed and subsequent genetic algorithm-selection operators were performed to shortlist the best models. The experimental scattering profiles from each construct were then compared with the theoretical scattering profile of the shortlisted best atomistic models generated by BILBOMD, using FOXS followed by multistate model selection by MULTIFOXS (27).
SEC-MALS
Eluent was subsequently split 3 to 1 between the SAXS line and a series of UV absorbance determinations at 280 and 260 nm, MALS, quasi-elastic light scattering (QELS), and refractometer detector. MALS experiments were performed using an 18-angle DAWN HELEOS II light scattering detector connected in tandem to an Optilab refractive index concentration detector (Wyatt Technology). System normalization and calibration were performed with bovine serum albumin using a 45 μl sample at 10 mg/ml in the same SEC running buffer, and a dn/dc value of 0.19. The light scattering experiments were used to perform analytical scale chromatographic separations for MW determination of the principal peaks in the SEC analysis. UV, MALS, and differential refractive index data were analyzed using Wyatt Astra 7 software to monitor the homogeneity of the sample across the elution peak.
ChIP-peak calling and motif analysis
We analyzed the available ChIP-seq data (GSE154960) published previously for HA-ZF1-5 and HA-ZNF410-FL, each expressed in HUDEP-2 cells (5). Files in FASTQ format were trimmed by trim_galore (v0.4.4_dev), and mapped to the human reference genome (hg38) using Bowtie (v1.2.2) (28). Peaks were identified by MACS2 (v2.1.2) (29), and heatmaps were generated by deeptools (v3.3.0) (30) and pheatmap (v1.0.12). Motifs were scanned, allowing up to two mismatches to ‘CATCCCATAATA’ by scanMotifGenomeWide.pl (homer2 v4.10.1) (31).
RESULTS
ZNF410 is predicted to contain three ordered regions
To investigate the solution features of ZNF410, we first performed analysis using predictor of natural disordered regions (PONDR) (20), which predicts that two short stretches within residues 100–200 and in the central ZF domain are ordered (PONDR score close to 0; Figure 1B). The boundaries of the central ZF domain correspond well with the fragment we previously used for crystallographic study (Figure 1C) (5). During the course of our current study, AlphaFold (24) released protein structure predictions for the human proteome, including human ZNF410 (Figure 1D), which is predicted to contain three folded domain structures: N-terminal residues 73–193, the central ZF domain, and C-terminal residues 438–478. The AlphaFold model has a very low model confidence (pLDDT < 50 in a scale of 100) for both N- and C-terminal domains, but higher confidence (between 70 and 90) for the ZF domain. Interestingly, superimposition of the AlphaFold ZF model onto that of our experimentally-determined ZF-DNA complex resulted in root-mean-square (rms) deviations of <0.8 Å over 140 residues (Figure 1E); though we do not think that the ZF array would have the same conformation in the absence of DNA. An earlier solution NMR study revealed relative orientation and mobilities of each finger within a three-finger array (32), and comparison of X-ray structure of DNA-bound zinc fingers and solution NMR structure of DNA-free zinc fingers revealed that the flexibility of the linker between the two fingers in the absence of DNA may allow tandem ZF arrays to sample a great number of conformations (see figure 4H of reference (33)).
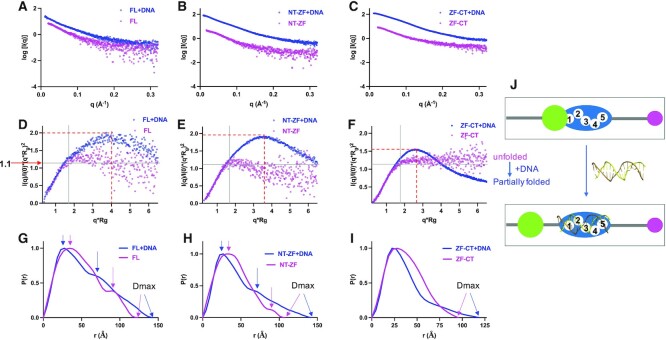
Figure 4.
SAXS data analysis from three ZNF410 protein samples (FL, NT-ZF, ZF-CT) in the absence and presence of a 17-bp DNA duplex. (A–C) Experimental scattering intensity profiles. Magenta, samples without DNA; blue, with DNA. (D–F) Dimensionless Kratky plots. Note the shifts of profiles from unfolded to partially folded sates induced by DNA. For a reference point, a bell-shaped peak is commonly observed for a globular system at Y axis of ∼1.1 (or q*Rg ∼√3). (G–I) Normalized pair-wise distribution function. P(r) plots computed from the indirect Fourier transformation of measured SAXS data in the absence (magenta) and presence of DNA (blue). The P(r) plots are normalized to unity at their maxima. Arrows indicate Dmax values or possible multi-domain linked by disordered loops. (J) A cartoon showing the transition from unfolded state to partially folded state upon DNA binding.
We note that in the AlphaFold model the N-terminal 30-residue-long helix (residues 164–193, named as αB) and its preceding loop (named as Loop-4B between strand β4 and αB) occupy the DNA binding surface of the ZF domain (Figure 1F). This intramolecular interaction could stabilize the ZF domain, but would have to be removed to allow DNA binding. Similarly, the corresponding long αB helix predicted for Bos taurus and mouse ZNF410 orthologs would block DNA binding (Figure S4). Taken together, the predictions of PONDR and AlphaFold suggest that ZNF410 has two disordered regions: the extreme N-terminal residues 1-70, and residues 367–437 connecting the ZF and C-terminal domains (Figure 1G). We note that the respective maximum dimensions, measured from the existing ZF-DNA complex structure (PDB 6WMI) and the AlphaFold model of ZNF410 full-length protein, are ∼58 Å and ∼130 Å (Figure 1C and D).
The N-terminal region of ZNF410 reduces the ZF DNA binding affinity
To further investigate the ZNF410 features outside of the ZF domain in solution, we purified ZNF410 full-length (FL, 1–478) and the two fragments containing the N-terminal residues 1–366 (NT-ZF) or C-terminal residues 217–478 (ZF-CT) (Figure 2A). All three recombinant proteins contain the DNA binding ZF domain (residues 217–366). Next, we quantified and compared the DNA binding affinities of the isolated DNA binding ZF domain with that of three longer fragments, containing either N- or C-terminal addition or both (FL), by fluorescence polarization (34). The DNA binding ZF domain and ZF-CT gave a similar dissociation constant (KD) of ∼50–80 nM, whereas the NT-ZF and FL exhibited significantly weaker DNA binding (∼5–8×; KD ∼0.4–0.5 μM) under the same conditions (Figure 2B). These binding data suggest that the CT region is largely dispensable and does not contribute to the DNA binding, at least in vitro. On the other hand, the presence of the NT region restricts DNA binding, possibly by competing for the DNA binding surface of the ZFs, as hinted by the AlphaFold prediction (Figure 1E).
Figure 2.
ZF1-5 has stronger DNA binding in the absence of the N-terminal domain. (A) An 18% SDS-PAGE gel of Bio-RAD stain-free imaging showing the recombinant protein samples used in this study. (B) Fluorescence polarization assay of recombinant ZNF410 protein segments with and without N- or C-terminal additions. Data represent the mean ± SD of three independent determinations (N = 3). (C) Unique binding sites of HA-ZF1-5 and HA-ZNF410 FL in HUDEP-2 cells. Heatmaps of the signal intensities of the HA-ZF1-5 (left column), HA-ZNF410 FL (middle) and endogenous ZNF410 (right) ChIP-seq peaks in HUDEP-2 cells (GSE154960). At the far left is an indication of binding motif occurrence. (D) The corresponding H3K27ac mark in primary human erythroblasts. Note that HUDEP-2 cells exhibit similar gene expression patterns to human HSPC-derived erythroid cells (primary erythroblasts) (59).
At the CHD4 regulatory region, forcibly expressed ZF domain (HA-ZF1-5) displayed a binding pattern similar to that of overexpressed HA-ZNF410 FL (5). In addition, overexpressed HA-ZF1–5 was capable of competing with endogenous ZNF410 for chromatin binding at the CHD4 regulatory regions. To explore whether the isolated ZF domain has altered chromatin binding properties in intact cells, we interrogated genome-wide binding profiles of overexpressed HA-tagged ZNF410 FL and HA-ZF1-5 in HUDEP-2 human erythroid precursor cells (5) (GSE154960). We thus detected 25 de novo peaks in cells overexpressing HA-ZF1-5, and 95 de novo peaks in cells overexpressing HA-ZNF410 FL (Figure 2C). Unexpectedly, there were only four shared de novo peaks between the two constructs, three of which contained the ZNF410 binding motif. Further motif analysis showed that 17 out of the 21 uniquely gained HA-ZF1-5 peaks (81%) harbor at least one binding motif, while only 8 out of 91 (9%) uniquely gained HA-ZNF410 FL peaks contained the motif (Figure 2C). This suggests that HA-ZF-associated peaks were mainly dependent on the ZF–DNA interaction. The new sites occupied by ZNF410 FL might in part be due to overexpression and/or facilitated by portions of ZNF410 outside the ZF domain and, by inference, not always involve direct DNA binding. However, significantly, even for the four shared de novo peaks, the signal densities from HA-ZF1–5 cells were visually larger than those from HA-ZNF410 FL cells (Figure 2C), suggesting stronger binding affinity of the isolated ZF domain in the absence of the NT and CT regions.
Next, we examined whether these de novo peaks are associated with open chromatin, as indicated by histone H3 lysine 27 acetylation (H3K27ac). Notably, the 21 gained peaks upon HA-ZF1-5 expression were only marginally associated with H3K27ac signals, whereas almost all of the 91 gained peaks during HA-ZNF410 FL expression were located in H3K27ac-marked regions (Figure 2D). This result implies that HA-ZNF410 FL was likely recruited to these open chromatin regions by interactions outside of the ZF DNA binding domain. Together, these observations suggest that the ZNF410 domains outside of ZF (i) have negative effects on binding DNA properly in the genome, and (ii) they are involved in additional interactions for chromatin targeting.
ZNF410 is monomer in solution
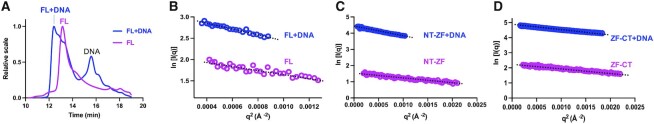
To further characterize ZNF410 in solution, we performed synchrotron-based small-angle X-ray scattering (SAXS) experiments (35) on the three protein segments in the absence and presence of DNA (total of six samples) (Table 1). We used the same 17-bp double-stranded oligonucleotide as was used in the co-crystallization (5). All samples were subjected to size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS) (36). The same fractions eluted from the size exclusion chromatography were simultaneously examined by SAXS. The shift in the maxima of the peak fraction of the protein and DNA mixtures relative to the elution volume of individual components suggests the formation of protein–DNA complexes (Figure 3A). Standard analysis tools (see Materials and Methods) were used to obtain model-independent parameters (Table 1), including the absolute masses of the ZNF410 proteins and their complexes with DNA. As a positive control, the unbound DNA has the exact molecular weight, maximum dimension (Dmax) and radius of gyration (Rg) as expected for a 17-bp oligo (Table 1 and Figure S5).
Table 1.
Biophysical parameters derived from SAXS and MALS data
| Instrument = ALS beamline 12.3.1; q range = 0.01–0.35 (Å−1); exposure time = 3 s; temperature = 20°C | ||||||||
|---|---|---|---|---|---|---|---|---|
| Parameters | Full length (FL) | NT + ZF | ZF + CT | DNA | ||||
| No DNA | + DNA | No DNA | + DNA | No DNA | + DNA | |||
| Concentration | (mg/ml) | 4 | 4 | 5 | 5 | 13 | 13 | 5 |
| (μM) | ∼77 | ∼124 | ∼448 | ∼454 | ||||
| MW (kDa) | Calculated (monomer) | 52 | 63 | 40 | 51 | 29 | 40 | 11.1 |
| SEC-MALS | 56 | 63 | 46 | 52 | 33 | 40 | 11.2 | |
| SAXS MoW | 55.9 | 64 | 43.7 | 53.8 | 31.7 | 38.1 | 10.8 | |
| Porod volume (Å3) | 105 839 | 117 525 | 78 713 | 94 146 | 55 180 | 70 985 | 15 604 | |
| D max (Å) | 122 | 142 | 107 | 141 | 95 | 119 | 58 | |
| Low-q range (Å−1) used for Guinier analysis | 0.02–0.04 | 0.02–0.03 | 0.01–0.05 | 0.01–0.03 | 0.02–0.05 | 0.01–0.04 | 0.02–0.07 | |
| I(0) (Å) | from Guinier | 8.41 | 21.81 | 4.72 | 30.28 | 9.79 | 130.50 | 25.90 |
| from P(r) | 8.41 | 21.81 | 4.72 | 30.28 | 9.78 | 130.50 | 25.91 | |
| R g (Å) | from Guinier | 38.54 | 43.80 | 31.95 | 40.60 | 29.71 | 30.72 | 18.33 |
| from P(r) | 38.69 | 43.85 | 31.11 | 40.62 | 29.71 | 30.89 | 18.38 | |
Values of Rg and I(0) parameters were estimated from Guinier plot and P(r) plot analyses.
Figure 3.
ZNF410 is monomer in solution in the presence and absence of DNA. (A) Overlay of profiles from size exclusion chromatography (SEC) of ZNF410 FL in the presence (blue line) and absence of DNA (magenta line). (B–D) Guinier plots from the measured scattering intensity I(q) as a function of the scattering vector q in the low-q regions.
The molecular weights obtained from SEC-MALS and SAXS analyses for each construct (FL, NT-ZF and ZF-CT) are in close agreement with the expected molecular weight of monomers, and show that all the constructs bind DNA in 1:1 stoichiometry (Table 1). We note that the measured MWs of protein–DNA complexes are close to the calculated (expected) values, whereas the MW of protein alone (for all three fragments) are larger than the calculated numbers by a few Da. This could result from the larger conformational variation of proteins in the absence of DNA (see next section), and the methods used for estimation might not fully account for the dynamics. Furthermore, the Guinier plot analysis for all constructs (with and without DNA) shows nearly a linear relationship of measured scattering intensity as a function of the scattering vector (q) in the low-q regions (Figure 3B–D), suggesting that each sample was essentially monodisperse (free from aggregation, dissociation or interparticle interactions). This also indicates that neither the N- nor C-terminal domains mediate multimerization or aggregation of ZNF410.
ZNF410 has high conformational flexibility in the absence of DNA
The SAXS curves from the absorption peak region from the SEC column were buffer subtracted and averaged, to obtain a scattering curve for each sample (Figure 4A-C). To check the flexibility of each sample, we generated a dimensionless Kratky plot (Figure 4D–F), and made the following observations. First, the Kratky plots do not exhibit a ‘bell-shape’ peak at low-q, as is typical for samples having well-ordered globular shapes, generally having a characteristic peak at Y-axis ∼1.1 and X-axis √3 (35,37). All six samples have pronounced flexibility, as indicated by the Kratky plots failing to converge to the X-axis in the high-q region. Second, the FL and NT-ZF samples (magenta colors in Figure 4D, E) have similar Kratky profiles with diffuse scattering at high-q, suggesting a mixture of high conformational flexibility and long disordered regions (38). Third, the ZF-CT profile shows a classic hyperbolic plateau (magenta in Figure 4F), suggesting an unfolded state for ZF-CT in the absence of DNA. Fourth, we note that all three protein samples reached a plateau with the Y-axis of ∼1.1 at low-q, suggesting that the samples are not in a fully extended conformation. The AlphaFold model predicts inter-domain interactions between the NT (helix αB and the loop-L4B) and the ZF domain (Figure 1F), potentially stabilizing part of the protein. However, such intra-molecular interaction will not occur for the ZF-CT fragment (which lacks the NT region).
Binding to DNA induces conformational changes in ZNF410
Binding DNA by all three protein samples resulted in the appearance of the bell-like features in the Kratky plot (blue lines in Figure 4D–F), indicating increase of a folded portion as described in the preceding section. There were also bell-like peaks shifting right into the higher-q region and upwards at Y-axis between 1.5 and 2.0 (red dashed lines in Figure 4D–F), indicating the existence of flexible linkers or extended loops between the compacted domains. It is evident from the pair distribution functions [P(r)] that FL and NT-ZF are each two-domain proteins connected by flexible linkers (magenta lines in Figure 4G, H). In the presence of DNA, extended tails on the P(r) plot show a slower approach to the X-axis, suggesting an increase in the Dmax values (blue lines in Figure 4G, H). Coincidentally, the maximum dimension predicted by the AlphaFold model of ZNF410 (∼130 Å in Figures 1D and S4) is between the DNA-free (122 Å) and DNA-bound forms (142 Å in Figure 4G). This is consistent with DNA binding by the ZF presumably displacing the NT domain from the DNA binding ZF surface (Figure 1F), and making the DNA-bound molecule slightly larger (Figure 4J).
For the NT-ZF fragment, the maximum dimension in the absence of DNA is shorter than the FL protein (107 Å versus 123 Å), as expected since it is missing the CT portion. However, upon binding to DNA, the maximum dimension becomes the same for the NT-ZF and FL proteins (141 Å versus 142 Å) (Figure 4H), suggesting that the carboxyl region might become ordered and fold back onto the rest of the protein, and thus does not contribute to the overall arrangement. The binding of DNA to unfolded ZF-CT results in a conformational change of five individual ZF units, which bind to a single piece of DNA, thus helping to attain substantial structure by folding the protein (Figure 4F). Like the two other tested proteins, the observed Dmax increases to 119 Å in the DNA-bound form, from 95 Å in the DNA-free form (Figure 4I).
An auto-inhibitory model of the N-terminal helix of ZNF410 in the absence of DNA
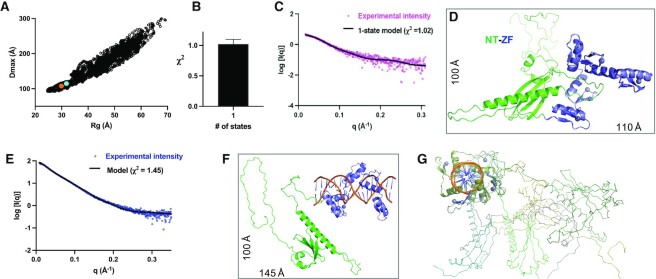
Using the AlphaFold model (after virtually incorporating a zinc ion into each finger), we used a molecular dynamics-based conformational sampling method, BILBOMD (25), to model the population of different conformations adopted by ZNF410. To reduce the number of variables, we kept the three structured domains constant, and allowed the long loops to be flexible. BILBOMD generated 10 000 conformational models for each construct (FL, NT-ZF and ZF-CT) with varying values of Rg and Dmax (Figure 5A, Supplementary Figures S5A and S6A). Multi-state modeling with SAXS profiles (MultiFoXS) (27) suggests that FL exists in two states (Supplementary Figure S6B), NT-ZF in one state (Figure 5B) and ZF-CT in at least two states (Supplementary Figure S7B). Surprisingly, the single best-fit model for NT-ZF, as compared to the experimental scattering profile with χ2 = 1.02 (Figure 5C), is the original AlphaFold model with the NT long-helix αB and loop-L4B occupying the ZF DNA binding interface (Figure 5D) with a calculated Rg = 32 Å and Dmax = 107 Å. Significantly, these calculated numbers from the model are in nearly perfect agreement with the experimental parameters (Rg = 32 Å and Dmax = 108 Å) (Table 1).
Figure 5.
Rigid body modeling of NT-ZF. (A) Plot of Rg versus Dmax values for all 10 000 models obtained in MD conformational sampling. The starting AlphaFold model is indicated by an orange dot and the best fit model is in a cyan dot. (B) MULTIFOXS analysis suggests that NT-ZF exists in a single state. (C, D) Fitting of experimental X-ray scattering profile (magenta dots) to the calculated scattering (black line) of the best-fit model (panel D) in the absence of DNA. (E) The fitting of experimental X-ray scattering profile (blue dots) to the calculated scattering (black line) of the fitting model in the presence of DNA. (F) One example of DNA bound NT-ZF model. (G) A sampling of DNA bound NT-ZF models with varied conformations of NT; the DNA helical axis is perpendicular to the page.
Addition of the CT residues, in the context of FL or ZF-CT, introduced flexibility, due to the CT loop adopting a large range of conformational spaces (from compact to extended). Fitting the scattering profile of the two-state model (FL and ZF-CT) with the experimental scattering profile yields a fit with χ2 = 2.38 and 3.07 respectively, which is considered a good fit given the high signal-to-noise ratio for these data (Supplementary Figures S6C, D and S7C, D).
Release of auto-inhibitory loop upon DNA binding
To evaluate the DNA-bound conformations of ZNF410, we manually modeled the DNA into the AlphaFold model (Figure 1E), and removed the NT helix αB and its associated loop-L4B from a steric clash with the DNA. Analysis of the best fit model for the FL suggests that DNA binding results in the transition of FL from two states to a single state, with the calculated profile of the model fitting the experimental data with χ2 = 1.43 (Supplementary Figure S6E, F). The same model of the displaced N-terminal helix applies to the NT-ZF fragment, with the model Rg value (39.5 Å) matching the experimental Rg (40.6 Å) with a χ2 = 1.45 (Figure 5E, F). The fragment ZF-CT still exists in multiple states (Supplementary Figure S7E, F).
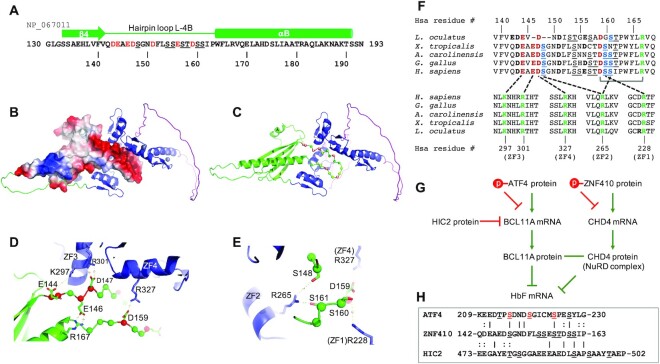
The acidic asp/glu-rich hairpin loop mimics DNA
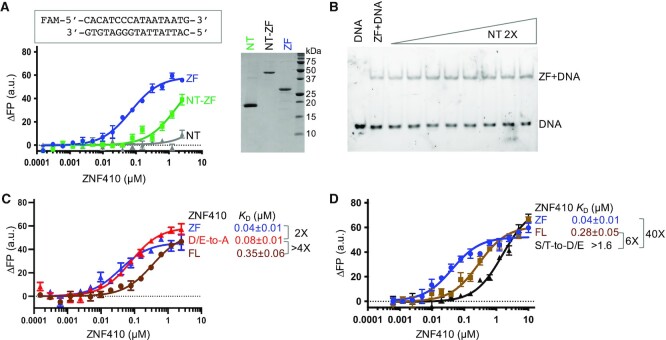
We note that the loop-L4B contains seven acidic residues (four aspartates and three glutamates), comprising a third of its 21 residues (Figure 6A). The loop appears to adopt a hairpin-like structure, and can fold onto the DNA binding surface of the ZFs, with the loop's negatively-charged surface complementing the positively-charged surface of the ZF array (Figure 6B, C). We suspect that the hairpin loop mimics DNA structurally and electrostatically, as known DNA-mimicry proteins do (39–41). These DNA-mimicking proteins, using electrostatic mimicry of DNA phosphates by protein sidechain carboxyl groups (39), modulate the activities of proteins as diverse as CRISPR-Cas9 (42), BRCA2 (43) and NF-κB (44), but usually the mimicry proteins act in trans. In the case of ZNF410, the isolated NT region (residues 1–216), as expected, does not bind DNA (Figure 7A). However, when added in trans it also does not prevent DNA from binding to the ZF array (Figure 7B). Interestingly in this regard, the hairpin loop-containing N-terminal fragment (BAG62922) and ZF-containing C-terminal fragments (NP_001229857.1 or EAW81139.1) are also expressed as different isoforms of human ZNF410 (Supplementary Figure S1A). It is possible that these two ZNF410 isoforms containing ZFs but not the hairpin loop can bind DNA unconstrained by the allosteric control described here.
Figure 6.
A model of DNA-mimic hairpin loop. (A) Sequence of ZNF410 in the secondary structural element of β4-hairpin loop-αB. (B) The hairpin loop displays an electronegative surface (blue for positive, red for negative and white for neutral). (C) The hairpin loop forms potential interactions with four out of five ZFs. (D) The hairpin loop forms four pairs of electrostatic interactions. (E) Three serine residues link two arginine residues of ZF1 and ZF2. (F) Pairwise interactions between hairpin loop (top) and ZFs (bottom). (G) ATF4 and HIC2 control BCL11A expression, and ZNF410 controls CHD4 expression, which is a component of NuRD. Together, BCL11A and NuRD silence fetal hemoglobin expression. Phosphorylation of either or both of ATF4 and ZNF410 might activate fetal hemoglobin expression. (H) Sequence alignment of known ATF4 phosphorylation sites (red S/T), ZNF410 hairpin loop and the HIC2 loop inserted between two ZF units.
Figure 7.
Opposite effects of mutagenesis on DNA binding by removing or introducing acidic residues within the hairpin loop. (A) The isolated NT fragment (residues 1–216) does not bind to DNA but the NT-ZF fragment (residues 1–366) has reduced DNA binding potential (left) (N = 7). An 18% SDS-PAGE stain-free gel (right panel) showing the recombinant protein samples used in DNA binding assays. (B) The isolated NT fragment added in trans does not prevent the DNA from binding to the ZF array. (C) The mutant with four D/E-to-A substitutions (red line) within the hairpin loop increased DNA binding in the context of full-length ZNF410. (D) The mutant with seven S/T-to-D/E substitutions (black line) within the hairpin loop further reduced DNA binding in the context of full-length ZNF410 (N = 6). ZF, blue lines and FL, brown lines.
Among the seven acidic residues in the human ortholog loop, four of them (E144, E146, D147 and D159) are predicted to form salt-bridge electrostatic pairs respectively with K297 and R301 of ZF3, R327 of ZF4 and R167 of the helix αB (Figure 6D). In addition, three serine residues (S148, S160 and S161) are located near R228 of ZF1 and R265 of ZF2 (Figure 6E). Thus, the hairpin loop appears capable of interacting with four out of the five ZF units, and in particular with basic ZF residues important for DNA phosphate and base-specific interactions (5). This apparent loop-based allosteric control of human ZNF410 is conserved among vertebrates as distant as fish—of the seven D/E residues most likely to provide the DNA-mimicking negative charges in the loop, seven are present in the corresponding but shorter loop of the L. oculatus ortholog (Figure 6F and Supplementary Figure S3D). Significantly, all of these interacting residues (seven in the loop, one in αB, and five among the ZFs) are fully conserved from primate to amphibian, while the fish ortholog has 10/13 conserved (Figure 6F and Supplementary Figure S3D). In addition, the tip of the hairpin loop, S154SEST158, could likely reach ZF5, meaning that the cis-acting regulatory region would contact all five ZFs.
Mutagenesis of D/E to A, and S/T to D/E, within the hairpin loop
A key test of the model that the N-terminal domain's hairpin loop acts as a DNA mimic, limiting DNA binding by the ZF array, is mutationally altering the charges on the hairpin loop. We did this in two ways: first replacing Asp/Glu with Ala, which per the model should increase DNA binding by the full-length ZNF410, and second by replacing Ser/Thr with Asp/Glu, which should decrease DNA binding (by increasing hairpin loop affinity for the ZF domain). Indeed, replacing the acidic residues by alanine (abbreviated as DEAED-to-AAAAA) at residues D143, E144, E146 and D147, we observed an increased binding affinity by >4× to that of wild-type and 2× difference from the isolated ZF DNA binding domain (Figure 7C).
In addition to the negatively charged amino acids, the hairpin loop also harbors six serine residues and one threonine. Per the DNA-mimicry model, altering these residues to acidic ones would be expected to increase affinity of the loop for the positively-charged DNA-binding surface of the ZFs. Accordingly, we substituted the six serine residues within the hairpin loop with negatively charged aspartate and the threonine with glutamate. As expected, introducing these new negative charges resulted in further-reduced DNA binding of >6× from that of the wild-type or >40× from the isolated ZF DNA binding domain (Figure 7D). This result is consistent with the model that the hairpin loop attenuates ZNF410 DNA binding activity by acting as an inhibitory DNA mimic.
DISCUSSION
Structure and apparent auto-inhibition of ZNF410
Here we used SAXS data to model different conformations of ZNF410, which harbors a DNA binding domain consisting of five tandem ZF units (5) along with, as predicted by AlphaFold, an N-terminal four-stranded and two-helix domain containing a long helix αB and less-ordered CT region outside the DNA binding domain. These folded units are linked by highly flexible and intrinsically disordered loops. The existence of extended regions of intrinsic disorder is a common feature for transcription factors (45), needed for flexibility-associated binding and downstream signaling. In the case of an array of ZF units (five in ZNF410), the flexibility of the linker between any two fingers may allow the multi-finger array to sample a variety of conformations in the absence of DNA (for example, see Figure 4 of (33)). Upon binding to DNA, it is possible that the specific contacts with the ZF array are formed sequentially, or even uni-directionally along the DNA, until all five fingers sit along the major groove of DNA. These substrate DNA-induced conformational changes could explain the behavior of the ZF-CT fragment, as observed in many DNA-binding proteins, particularly for sequence-specific recognition (46).
What is somewhat unique to ZNF410 is that the NT domain appears to function as a negative auto-regulator of DNA binding, reducing the ZF DNA binding affinity by occluding the DNA binding surface (Figure 2B). Transcription factors typically bind to thousands of sites genome-wide and regulate large cohorts of genes. ZNF410 is remarkable in that it recognizes a 15-bp DNA sequence element but has only a single responsive target in the mammalian genome in erythroid cells (5,6). Clustering of these elements is part of the answer, but additional mechanism(s), supplementary to the sequence specificity, must control ZNF410 DNA binding. Indeed, our in vitro and in-cell data indicate that the ZF DNA-binding domain exhibits higher binding affinity for DNA, compared to the full-length ZNF410 protein. As noted earlier, ZNF410 isoform e lacks the hairpin loop region (Supplementary Figure S1A), so the ratio of isoforms b and e being produced may affect overall ZNF410 activity.
Possible role of hairpin loop phosphorylation
The Ser/Thr residues in the hairpin loop are, in theory, sites of potential protein phosphorylation. Such phosphorylation would introduce additional negative charges to the hairpin loop. The phosphorylation of transcription factors is a common mechanism used by the cell to link signaling pathways to the control of gene expression (47). Protein phosphorylation can directly regulate distinct aspects of transcription factor function, including cellular localization, protein stability, protein-protein interactions and DNA binding (reviewed in (48,49) and references therein). Phosphorylation within or near a DNA binding domain introduces a negative charge, which is incompatible with efficient binding of the polyanionic DNA, though in this case it would perhaps act by increasing binding of the DNA-mimicking loop to the positively-charged ZF DNA-binding surface. We thus suggest that phosphorylation of ZNF410, at the hairpin loop (by yet-to-be determined protein kinases) could attenuate ZNF410 DNA binding activity. Significantly, ZNF410 inhibition represents a new strategy for the treatment of hemoglobinopathies (50,51). Indeed, a recent report that combines shRNA mediated BCL11A depletion with ZNF410 depletion is more potent that either factor alone (52). Pharmacologic modulation of ZNF410 e.g. by altering its phosphorylation state might be a way to raise fetal hemoglobin levels in patients with sickle cell disease or beta-thalassemia (Figure 6G).
At present, there is no evidence that any of the seven S/T residues in the hairpin loop is actually phosphorylated. Our substitutions of S/T with D/E were made to test the model that the ZNF410 hairpin loop acts as an inhibitory DNA mimic, but the substitutions mimic the effects of protein phosphorylation and they did decrease DNA binding by ZNF410 (Figure 7D). For comparison, the phosphorylation of c-Jun on S/T residues located N-terminal to the basic leucine-zipper (bZIP) DNA binding domain inhibits the DNA binding of c-Jun (53). Like c-Jun, the DNA binding activity of the zinc-finger-containing Wilms’ tumor protein WT1 is inhibited by phosphorylation of two serine residues within the zinc-finger region (54). ATF4, another known transcription factor indirectly involved in regulating expression of fetal hemoglobin (55), contains phosphorylated S/T sites N-terminal to the bZIP DNA binding domain (56,57). We note that, like the ZNF410 hairpin loop, the ATF4 phosphorylation sites are located in an acidic environment and the corresponding regions of the two proteins share sequence similarity (Figure 6H). HIC2, another ZF transcription factor, controls developmental hemoglobin switching by repressing BCL11A transcription, and contains a 5-ZF DNA binding domain, with an insertion that is rich in negatively charged glutamate and aspartate between the first two fingers, which reduced DNA binding (58).
DNA mimicry by proteins
DNA mimicry by proteins has been studied, for a number of proteins, in the regulation of enzymatic/binding activity on DNA (reviewed by Dryden (39)). Here, we summarize the similarities and differences between the Dryden model of DNA mimicry and the hairpin-loop we propose for ZNF410. First, both use electrostatic mimicry of DNA phosphates by protein sidechain carboxyl groups. Second, ZNF410 may be subject to posttranslational phosphorylation by one or more protein kinases, which represents a potential mechanism of regulation. We have no direct evidence for phosphorylation of the hairpin loop, though our S/T substitutions with D/E did inhibit DNA binding (Figure 7D). Third, the Dryden model (exemplified by the bacteriophage T7 Ocr protein) uses a rigid protein structure functioning in trans, whereas ZNF410 uses a flexible hairpin loop functioning only in cis. Fourth, the trans interaction works against several proteins (type I restriction enzymes in the case of Ocr) that recognize different DNA sequences, whereas the cis interaction in ZNF410 affects one protein recognizing one DNA sequence.
Summary
We have used a combination of artificial intelligence-based AlphaFold analyses, small-angle X-ray scattering, biochemistry and bioinformatics to characterize the human transcription factor ZNF410. The results strongly suggest that this highly conserved protein, which plays a critical role in erythroid cell development, uses its NT region as a DNA mimic in order to attenuate accessibility of its DNA binding region. Unlike most cases of DNA mimicry by proteins (39), in this case the inhibitory polypeptide is part of the protein being allosterically regulated. The roles of alternative ZNF410 isoforms, and of possible modification by protein kinase(s), remain to be determined. The cis competition with DNA for DNA binding could tie DNA binding to specific physiological states (if phosphorylation is involved, for example), simply reduce ZNF410 binding to all but the highest-affinity sites, or some combination of these two possibilities. Our study also demonstrates that a combination of AlphaFold and SAXS advances studies of protein dynamics, particularly for the functional domains connected by extended regions of intrinsic disorder.
DATA AVAILABILITY
The SAXS experimental data that support the findings of this study are deposited in SASBDB with accession codes: SASDQF5 (ZNF410), SASDQG5 (DNA), SASDQH5 (ZNF410 + DNA), SASDQJ5 (NT-ZF), SASDQK5 (NT-ZF + DNA), SASDQL5 (ZF-CT), SASDQM5 (ZF-CT + DNA).
https://www.sasbdb.org/data/SASDQF5/j64atksk7r
https://www.sasbdb.org/data/SASDQG5/aa5tmxufck
https://www.sasbdb.org/data/SASDQH5/yz4s0v6ce4
https://www.sasbdb.org/data/SASDQJ5/mci6edmtp3
https://www.sasbdb.org/data/SASDQK5/54zx4gmvnl
https://www.sasbdb.org/data/SASDQL5/ttlzbup5uo
https://www.sasbdb.org/data/SASDQM5/o6m9jsp4wy
A project summary can be found here: https://www.sasbdb.org/project/1731/bjh4u9o653
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Chris Bosey, Dr Chi-Lin Tsi and Dr John Tainer for discussions on SAXS data. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authors contributions: G.K. is responsible for coordinating and performed SAXS data analysis, performed writing of original draft. R.R. performed cloning, protein purifications, mutagenesis and DNA binding assays. M.H. performed SAXS data collection and helped to generate SAXS models using BILBOMD. J.R.H. helped with computer modeling. J.Y. and Y.C. performed mutagenesis, mutant protein purification and mutant-DNA binding assay. C.H., F.L. and X.L. analyzed the ChIP-seq data. X.L and G.A.B. provided the initial ZNF410 construct; R.M.B. carried out bioinformatic analyses and participated in writing the manuscript. X.Z performed supervision, conceptualization and project administration. X.C performed writing, reviewing and editing of the manuscript, conceptualization and funding acquisition.
Contributor Information
Gundeep Kaur, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Ren Ren, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Michal Hammel, Molecular Biophysics and Integrated Bioimaging, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
John R Horton, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Jie Yang, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Yu Cao, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Chenxi He, Shanghai Key Laboratory of Medical Epigenetics, International Laboratory of Medical Epigenetics and Metabolism, Ministry of Science and Technology, Institutes of Biomedical Sciences, Fudan University and Key Laboratory of Carcinogenesis and Cancer Invasion, Ministry of Education, Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai 200032, China.
Fei Lan, Shanghai Key Laboratory of Medical Epigenetics, International Laboratory of Medical Epigenetics and Metabolism, Ministry of Science and Technology, Institutes of Biomedical Sciences, Fudan University and Key Laboratory of Carcinogenesis and Cancer Invasion, Ministry of Education, Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai 200032, China.
Xianjiang Lan, Department of Systems Biology for Medicine, School of Basic Medical Sciences; Department of Liver Surgery and Transplantation, Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai 200032, China; Division of Hematology, the Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Gerd A Blobel, Division of Hematology, the Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA; Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Robert M Blumenthal, Department of Medical Microbiology and Immunology, and Program in Bioinformatics, The University of Toledo College of Medicine and Life Sciences, Toledo, OH 43614, USA.
Xing Zhang, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Xiaodong Cheng, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (NIH) [R35GM134744 to X.C., R01HL119479 to G.A.B.]; Cancer Prevention and Research Institute of Texas (CPRIT) [RR160029 to X.C. who is a CPRIT Scholar in Cancer Research]; National Cancer Institute grants for Structural Biology of DNA Repair (SBDR) [CA092584]; SAXS data collection at SIBYLS is funded through NIGMS [P30 GM124169-01, ALS-ENABLE]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interests statement
None declared.
REFERENCES
- 1. Latchman D.S. Transcription factors: an overview. Int. J. Exp. Pathol. 1993; 74:417–422. [PMC free article] [PubMed] [Google Scholar]
- 2. Iuchi S. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 2001; 58:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fedotova A.A., Bonchuk A.N., Mogila V.A., Georgiev P.G.. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae. 2017; 9:47–58. [PMC free article] [PubMed] [Google Scholar]
- 4. Benanti J.A., Williams D.K., Robinson K.L., Ozer H.L., Galloway D.A.. Induction of extracellular matrix-remodeling genes by the senescence-associated protein APA-1. Mol. Cell. Biol. 2002; 22:7385–7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lan X., Ren R., Feng R., Ly L.C., Lan Y., Zhang Z., Aboreden N., Qin K., Horton J.R., Grevet J.D.et al.. ZNF410 uniquely activates the NuRD component CHD4 to silence fetal hemoglobin expression. Mol. Cell. 2021; 81:239–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vinjamur D.S., Yao Q., Cole M.A., McGuckin C., Ren C., Zeng J., Hossain M., Luk K., Wolfe S.A., Pinello L.et al.. ZNF410 represses fetal globin by singular control of CHD4. Nat. Genet. 2021; 53:719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allen H.F., Wade P.A., Kutateladze T.G.. The NuRD architecture. Cell. Mol. Life Sci. 2013; 70:3513–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siebenlist U., Franzoso G., Brown K.. Structure, regulation and function of NF-kappa B. Annu. Rev. Cell Biol. 1994; 10:405–455. [DOI] [PubMed] [Google Scholar]
- 9. Zheng C., Yin Q., Wu H.. Structural studies of NF-kappaB signaling. Cell Res. 2011; 21:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghosh G., Wang V.Y., Huang D.B., Fusco A.. NF-kappaB regulation: lessons from structures. Immunol. Rev. 2012; 246:36–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arrowsmith C.H. Structure and function in the p53 family. Cell Death Differ. 1999; 6:1169–1173. [DOI] [PubMed] [Google Scholar]
- 12. Joerger A.C., Fersht A.R.. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 2016; 85:375–404. [DOI] [PubMed] [Google Scholar]
- 13. Mueller-Fahrnow A., Egner U.. Ligand-binding domain of estrogen receptors. Curr. Opin. Biotechnol. 1999; 10:550–556. [DOI] [PubMed] [Google Scholar]
- 14. Davey R.A., Grossmann M.. Androgen receptor structure, function and biology: from bench to bedside. Clin Biochem Rev. 2016; 37:3–15. [PMC free article] [PubMed] [Google Scholar]
- 15. Frank F., Ortlund E.A., Liu X.. Structural insights into glucocorticoid receptor function. Biochem. Soc. Trans. 2021; 49:2333–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Putnam C.D., Hammel M., Hura G.L., Tainer J.A.. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007; 40:191–285. [DOI] [PubMed] [Google Scholar]
- 17. Rambo R.P. Resolving individual components in protein-RNA complexes using small-angle X-ray scattering experiments. Methods Enzymol. 2015; 558:363–390. [DOI] [PubMed] [Google Scholar]
- 18. Hammel M., Tainer J.A.. X-ray scattering reveals disordered linkers and dynamic interfaces in complexes and mechanisms for DNA double-strand break repair impacting cell and cancer biology. Protein Sci. 2021; 30:1735–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trewhella J. Recent advances in small-angle scattering and its expanding impact in structural biology. Structure. 2022; 30:15–23. [DOI] [PubMed] [Google Scholar]
- 20. Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z.. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005; 3:35–60. [DOI] [PubMed] [Google Scholar]
- 21. Dyer K.N., Hammel M., Rambo R.P., Tsutakawa S.E., Rodic I., Classen S., Tainer J.A., Hura G.L.. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol. Biol. 2014; 1091:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rambo R.P., Tainer J.A.. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013; 496:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piiadov V., Ares de Araujo E., Oliveira Neto M., Craievich A.F., Polikarpov I.. SAXSMoW 2.0: online calculator of the molecular weight of proteins in dilute solution from experimental SAXS data measured on a relative scale. Protein Sci. 2019; 28:454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pelikan M., Hura G.L., Hammel M.. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009; 28:174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schneidman-Duhovny D., Hammel M.. Modeling structure and dynamics of protein complexes with SAXS profiles. Methods Mol. Biol. 2018; 1764:449–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A.. FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016; 44:W424–W429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng J., Liu T., Qin B., Zhang Y., Liu X.S.. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bruschweiler R., Liao X., Wright P.E.. Long-range motional restrictions in a multidomain zinc-finger protein from anisotropic tumbling. Science. 1995; 268:886–889. [DOI] [PubMed] [Google Scholar]
- 33. Hashimoto H., Wang D., Horton J.R., Zhang X., Corces V.G., Cheng X.. Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol. Cell. 2017; 66:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patel A., Hashimoto H., Zhang X., Cheng X.. Characterization of how DNA modifications affect DNA binding by C2H2 zinc finger proteins. Methods Enzymol. 2016; 573:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kikhney A.G., Svergun D.I.. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015; 589:2570–2577. [DOI] [PubMed] [Google Scholar]
- 36. Some D., Amartely H., Tsadok A., Lebendiker M.. Characterization of proteins by size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). J. Vis. Exp. 2019; 148:e59615. [DOI] [PubMed] [Google Scholar]
- 37. Receveur-Brechot V., Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 2012; 13:55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rambo R.P., Tainer J.A.. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011; 95:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dryden D.T. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotechnol. 2006; 24:378–382. [DOI] [PubMed] [Google Scholar]
- 40. Wang H.C., Chou C.C., Hsu K.C., Lee C.H., Wang A.H.. New paradigm of functional regulation by DNA mimic proteins: recent updates. IUBMB Life. 2019; 71:539–548. [DOI] [PubMed] [Google Scholar]
- 41. Wang Z., Wang H., Mulvenna N., Sanz-Hernandez M., Zhang P., Li Y., Ma J., Wang Y., Matthews S., Wigneshweraraj S.et al.. A bacteriophage DNA mimic protein employs a non-specific strategy to inhibit the bacterial RNA polymerase. Front. Microbiol. 2021; 12:692512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shin J., Jiang F., Liu J.J., Bray N.L., Rauch B.J., Baik S.H., Nogales E., Bondy-Denomy J., Corn J.E., Doudna J.A.. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017; 3:e1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao W., Vaithiyalingam S., San Filippo J., Maranon D.G., Jimenez-Sainz J., Fontenay G.V., Kwon Y., Leung S.G., Lu L., Jensen R.B.et al.. Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA mimicry. Mol. Cell. 2015; 59:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jennings E., Esposito D., Rittinger K., Thurston T.L.M.. Structure-function analyses of the bacterial zinc metalloprotease effector protein GtgA uncover key residues required for deactivating NF-kappaB. J. Biol. Chem. 2018; 293:15316–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J., Perumal N.B., Oldfield C.J., Su E.W., Uversky V.N., Dunker A.K.. Intrinsic disorder in transcription factors. Biochemistry. 2006; 45:6873–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andrabi M., Mizuguchi K., Ahmad S.. Conformational changes in DNA-binding proteins: relationships with precomplex features and contributions to specificity and stability. Proteins. 2014; 82:841–857. [DOI] [PubMed] [Google Scholar]
- 47. Hunter T., Karin M.. The regulation of transcription by phosphorylation. Cell. 1992; 70:375–387. [DOI] [PubMed] [Google Scholar]
- 48. Whitmarsh A.J., Davis R.J.. Regulation of transcription factor function by phosphorylation. Cell. Mol. Life Sci. 2000; 57:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Puertollano R., Ferguson S.M., Brugarolas J., Ballabio A.. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018; 37:e98804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim J., Dean A.. A zinc finger transcription factor faithfully dedicated to only a single target gene in erythroid cells. Mol. Cell. 2021; 81:218–219. [DOI] [PubMed] [Google Scholar]
- 51. Tumburu L., Thein S.L.. Targeting ZNF410 as a potential beta-hemoglobinopathy therapy. Nat. Genet. 2021; 53:589–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu B., Brendel C., Vinjamur D.S., Zhou Y., Harris C., McGuinness M., Manis J.P., Bauer D.E., Xu H., Williams D.A.. Development of a double shmiR lentivirus effectively targeting both BCL11A and ZNF410 for enhanced induction of fetal hemoglobin to treat beta-hemoglobinopathies. Mol. Ther. 2022; 30:S1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin A., Frost J., Deng T., Smeal T., al-Alawi N., Kikkawa U., Hunter T., Brenner D., Karin M.. Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell. 1992; 70:777–789. [DOI] [PubMed] [Google Scholar]
- 54. Sakamoto Y., Yoshida M., Semba K., Hunter T.. Inhibition of the DNA-binding and transcriptional repression activity of the Wilms' tumor gene product, WT1, by cAMP-dependent protein kinase-mediated phosphorylation of Ser-365 and Ser-393 in the zinc finger domain. Oncogene. 1997; 15:2001–2012. [DOI] [PubMed] [Google Scholar]
- 55. Huang P., Peslak S.A., Lan X., Khandros E., Yano J.A., Sharma M., Keller C.A., Giardine B., Qin K., Abdulmalik O.et al.. The HRI-regulated transcription factor ATF4 activates BCL11A transcription to silence fetal hemoglobin expression. Blood. 2020; 135:2121–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M.et al.. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004; 117:387–398. [DOI] [PubMed] [Google Scholar]
- 57. Elefteriou F., Benson M.D., Sowa H., Starbuck M., Liu X., Ron D., Parada L.F., Karsenty G.. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006; 4:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang P., Peslak S.A., Ren R., Khandros E., Qin K., Keller C.A., Giardine B., Bell H.W., Lan X., Sharma M.et al.. HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription. Nat. Genet. 2022; 54:1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Masuda T., Wang X., Maeda M., Canver M.C., Sher F., Funnell A.P., Fisher C., Suciu M., Martyn G.E., Norton L.J.et al.. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016; 351:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SAXS experimental data that support the findings of this study are deposited in SASBDB with accession codes: SASDQF5 (ZNF410), SASDQG5 (DNA), SASDQH5 (ZNF410 + DNA), SASDQJ5 (NT-ZF), SASDQK5 (NT-ZF + DNA), SASDQL5 (ZF-CT), SASDQM5 (ZF-CT + DNA).
https://www.sasbdb.org/data/SASDQF5/j64atksk7r
https://www.sasbdb.org/data/SASDQG5/aa5tmxufck
https://www.sasbdb.org/data/SASDQH5/yz4s0v6ce4
https://www.sasbdb.org/data/SASDQJ5/mci6edmtp3
https://www.sasbdb.org/data/SASDQK5/54zx4gmvnl
https://www.sasbdb.org/data/SASDQL5/ttlzbup5uo
https://www.sasbdb.org/data/SASDQM5/o6m9jsp4wy
A project summary can be found here: https://www.sasbdb.org/project/1731/bjh4u9o653