Abstract
There are currently no non-invasive imaging methods available for astrogliosis assessment or mapping in the central nervous system despite its essential role in the response to many disease states, such as infarcts, neurodegenerative conditions, traumatic brain injury and infection. Multidimensional MRI is an increasingly employed imaging modality that maximizes the amount of encoded chemical and microstructural information by probing relaxation (T1 and T2) and diffusion mechanisms simultaneously. Here, we harness the exquisite sensitivity of this imagining modality to derive a signature of astrogliosis and disentangle it from normative brain at the individual level using machine learning. We investigated ex vivo cerebral cortical tissue specimens derived from seven subjects who sustained blast-induced injuries, which resulted in scar-border forming astrogliosis without being accompanied by other types of neuropathological abnormality, and from seven control brain donors. By performing a combined post-mortem radiology and histopathology correlation study we found that astrogliosis induces microstructural and chemical changes that are robustly detected with multidimensional MRI, and which can be attributed to astrogliosis because no axonal damage, demyelination or tauopathy were histologically observed in any of the cases in the study. Importantly, we showed that no one-dimensional T1, T2 or diffusion MRI measurement can disentangle the microscopic alterations caused by this neuropathology. Based on these findings, we developed a within-subject anomaly detection procedure that generates MRI-based astrogliosis biomarker maps ex vivo, which were significantly and strongly correlated with co-registered histological images of increased glial fibrillary acidic protein deposition (r = 0.856, P < 0.0001; r = 0.789, P < 0.0001; r = 0.793, P < 0.0001, for diffusion-T2, diffusion-T1 and T1–T2 multidimensional data sets, respectively). Our findings elucidate the underpinning of MRI signal response from astrogliosis, and the demonstrated high spatial sensitivity and specificity in detecting reactive astrocytes at the individual level, and if reproduced in vivo, will significantly impact neuroimaging studies of injury, disease, repair and aging, in which astrogliosis has so far been an invisible process radiologically.
Keywords: astrogliosis, traumatic brain injury, blast, multidimensional MRI, diffusion, relaxation, machine learning, radiological–pathological correlations, GFAP
Can astrogliosis be viewed non-invasively? Benjamini et al. employ machine learning with multidimensional MRI and produce maps of blast-induced astrogliosis at the individual subject level. The method could be used to study injury, disease, repair and ageing, in which astrogliosis has previously been an invisible process radiologically.
Introduction
Astrocytes are glial cells that are spread throughout the mammalian CNS where they represent the most abundant cell population in the brain.1 As part of their many functions in the healthy CNS, astrocytes respond to CNS damage and disease through a process called astrogliosis, which occurs in multiple CNS disorders including traumatic brain injury (TBI),2,3 autoimmune disease,4 stroke,5 neoplasia6 and neurodegenerative diseases,7 and which plays an essential role in regulating CNS inflammation. The phenotypic cellular changes in astrocytes that are associated with astrogliosis can range from mild, with variable degrees of hypertrophy of cell body and stem processes, to that seen in scar-border forming astrogliosis, where cell processes overlap and intertwine to form compact borders.8 The degree of glial fibrillary acidic protein (GFAP) deposition in reactive astrocytes often parallels the severity of the neuropathology9 and is therefore the most widely used marker of astrogliosis.
Although reactive astrocytes are integral and essential components of CNS innate immunity and have numerous beneficial functions,10,11 they can also cause harmful effects2,8,12 that are regarded as detrimental to clinical outcomes. Regardless of the role astrogliosis plays in different conditions, it is a dominant feature and common component of almost all CNS disorders. However, the successful development of non-invasive imaging techniques, primarily ones based upon MRI, to make astrogliosis visible has been elusive, mainly because of the failure of conventional MRI methods to detect it, but also, and perhaps more importantly, due to the experimental difficulty of disentangling astrogliosis from comorbid pathologies. The latter is especially true in MRI and diffusion tensor imaging (DTI) studies involving TBI animal models that result in axonal injury, demyelination, neurodegeneration, oedema or neuroinflammatory processes that are concurrent with astrogliosis.13–17 Studying astrogliosis is particularly difficult because of the challenges of decoupling the response to cellular alterations it generates from the response to the other microstructural and chemical processes that take place due to comorbidities.18,19
In addition to the experimental difficulty of isolating astrogliosis in brain tissue, a basic limitation of MRI is its low spatial resolution—on the order of 2 mm3 on clinical scanners. Although relaxation and diffusion contrast mechanisms carry information about components at the micron length scale, coarse, voxel-averaged images ‘flatten’ any intra-voxel heterogeneity, leading to loss of sensitivity and specificity in detecting microstructural and compositional changes induced by astrogliosis. There has been a recent push within the neuroimaging community to maximize the amount of information in an image by using a combination of magnetic field profiles to probe relaxation and diffusion mechanisms simultaneously, i.e. multidimensional MRI.20,21 That, combined with theoretical21–24 and technological innovations,25,26 has allowed the acquisition of MR images with effectively subvoxel resolution, resulting in the identification of multiple biological components within a given voxel.27–31
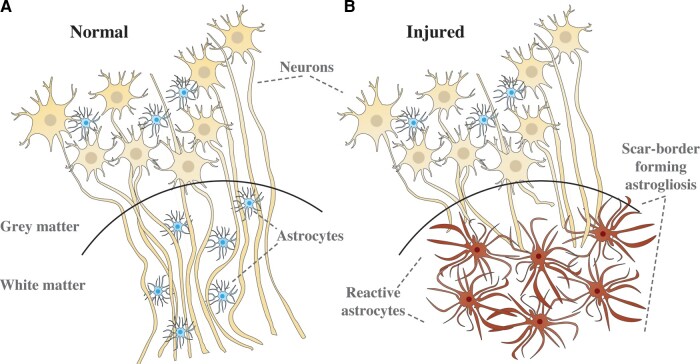
In this study we developed a multidimensional MRI machine learning framework to map astrogliosis in individual ex vivo brains by focusing on blast-induced TBI. Blast TBI is prevalent in the military cohort,32 and our understanding of the neuropathology following blast exposure is still in its infancy, particularly concerning its chronic sequelae. Recent studies suggest that both blast or mild impact neurotrauma are characterized by scar-border forming astrogliosis at brain interfaces including the subpial glial plate, around penetrating cortical blood vessels, at the grey–white matter interface (as illustrated in Fig. 1) and structures lining the ventricles.33–35 Importantly, such interface astrogliosis in the samples we examined here was not accompanied by axonal damage, demyelination or phosphorylated tau (pTau) pathology, making these blast TBI cases ideally suited for studying whether and how astrogliosis changes the multidimensional MRI signature.
Figure 1.
Illustration of microstructural changes occurring in the grey–white matter junction when scar-border forming astrogliosis is present. In A, axons are tightly aligned, forming a densely packed cellular environment. In B, scar-border forming reactive astrocytes have overlapping processes that sequester damaged tissue and inflammation while preventing injured axons from growing through the border. These changes are hypothesized to be reducing the overall cellular density in the white matter.
Here, we performed a combined post-mortem multidimensional MRI and histopathology study to investigate the ways in which astrogliosis affects MRI relaxation and diffusion, and to establish whether multidimensional MRI can be used to map the presence of scar-border forming astrogliosis in brain tissue. We compared brain sections with histologically confirmed astrogliosis and from control subjects using robust quantitative radiological–pathological correlations and developed a multidimensional MRI machine learning framework to map astrogliosis. We showed the spatial accuracy and sensitivity of the proposed framework and its ability to produce results at the individual subject level. We stress that our findings do not shed light on the underlying cause of astrogliosis in these cases, but rather provide the ability to non-invasively map astrogliosis, which we hope would become an important new tool for investigating and diagnosing a wide array of CNS disorders.
Materials and methods
Donor specimens
We evaluated 14 autopsy-derived brain specimens from two different human brain collections. Formalin-fixed portions of approximately 20 × 20 × 10 mm3 from the frontal lobe were obtained from two civilian subjects enrolled in the Transforming Research and Clinical Knowledge in Traumatic Brain Injury study (TRACK-TBI; https://tracktbi.ucsf.edu/transforming-research-and-clinical-knowledge-tbi; Cases 2 and 3) and 12 military subjects from the Department of Defense/Uniformed Services University Brain Tissue Repository (DoD/USU BTR, https://www.researchbraininjury.org, Uniformed Services University of the Health Sciences, Bethesda, MD; Cases 1, 4–14). For each case, the next-of-kin or legal representative provided written consent for donation of the brain for use in research. The brain tissues used have undergone procedures for donation of the tissue, its storage and use of available clinical information that have been approved by the USU Institutional Review Board (IRB) prior to the initiation of the study. All experiments were performed in accordance with current federal, state, DoD and NIH guidelines and regulations for post-mortem analysis. A detailed description of demographics for the subjects from whom brain tissue samples were obtained is listed in Table 1 and Supplementary Table 1.
Table 1.
Main demographic and histopathological findings in patients with history of TBI and healthy controls
| Casea | Age | Manner of death | PMI (h) | Blast exposure | Impact TBI | Astrogliosisb |
|---|---|---|---|---|---|---|
| 1 | 63 | Natural (cardiovascular) | 12 | None reported | None reported | None |
| 2 | 70 | Accident (MVA) | <12 | None reported | MVA | Mild |
| 3 | 65 | Undetermined | N/A | None reported | Fall | Moderate |
| 4 | 48 | Natural (cardiovascular) | 22 | None reported | None reported | None |
| 5 | 52 | Suicide | N/A | None reported | Multiple concussions; MVA (4 months prior to death) | None |
| 6 | 32 | Suicide | N/A | None reported | None reported | Mild |
| 7 | 59 | Suicide | 21 | None reported | Multiple MVAs (2015, 2020) | None |
| 8 | 38 | Suicide | N/A | IED exposure with LOC | As secondary injury | Scar-forming |
| 9 | 46 | Suicide | N/A | Multiple | Multiple concussions; MVA (at age 14) | Scar-forming |
| 10 | 29 | Suicide | <48 | Multiple | Fall with LOC (at age 27) | Scar-forming |
| 11 | 35 | Undetermined | <56 | Multiple | None reported | Scar-forming |
| 12 | 40 | Suicide | N/A | At least one exposure | Multiple concussions | Scar-forming |
| 13 | 64 | Natural (cardiovascular) | <48 | None reported | None reported | Scar-forming |
| 14 | 44 | Suicide | N/A | Multiple | Multiple MVAs (at age 5 and at early twenties) | Scar-forming |
IED = improvised explosive device; LOC = loss of consciousness; MVA = motor vehicle accident; N/A = not applicable; PMI = post-mortem interval.
All subjects in this study were males.
Microscopically confirmed GFAP-positive.
Of the total 14 cases evaluated, there were 7 cases with known interface astrogliosis and 7 control cases negative for interface astrogliosis, based on prior neuropathological examination at the DoD/USU BTR. Interface astrogliosis pathology was diagnosed in these cases from microscopic examination of cerebral sections immunostained for GFAP, based on the presence of prominent scar-border forming astrogliosis involving subpial glial plate, penetrating cortical blood vessels, grey–white matter junctions and structures lining the ventricles, as has been described and published by authors in this study (D.P.P.).33 However, initially, tissues from all 14 of these cases were received from the DoD/USU BTR for blinded MRI examination without access to the corresponding histopathology findings, TBI history or other medical history.
MRI acquisition
Prior to MRI scanning, each formalin-fixed brain specimen was transferred to a phosphate-buffered saline-filled container for 12 days to ensure that any residual fixative was removed from the tissue. The specimen was then placed in a 25-mm tube and immersed in perfluoropolyether (Fomblin LC/8, Solvay Solexis, Italy), a proton-free fluid void of a proton-MRI signal. Specimens were imaged using a 7 T Bruker vertical bore MRI scanner equipped with a microimaging probe and a 25-mm quadrupole RF coil.
Multidimensional data were acquired using a 3D inversion recovery diffusion-weighted sequence with a repetition time of 1000 ms, in-plane resolution of 200 × 200 μm2 and slice thickness of 300 μm. To encode the multidimensional MR space spanned by T1 and T2 (i.e. T1–T2), by T1 and mean diffusivity (i.e. T1-MD) and by T2 and mean diffusivity (i.e. T2-MD), 56, 302 and 302 images were acquired, respectively, according to a previously published sampling scheme.31,36 Additional parameters of the MRI pulse sequence can be found in the Supplementary material.
A standard DTI imaging protocol was applied with the same imaging parameters as the multidimensional data and using 21 diffusion gradient directions and four b-values ranging from 0 to 1400 s/mm2.
Lastly, a high-resolution MRI scan with an isotropic voxel dimension of 100 µm was acquired using a fast low angle shot (FLASH) sequence37 with a flip angle of 49.6° to serve as a high-resolution reference image and facilitate co-registration of histopathological and MR images.
Histology and immunohistochemistry
After MRI scanning, each tissue block was transferred for histopathological processing. Tissue blocks from each brain specimen were processed using an automated tissue processor (ASP 6025, Leica Biosystems). After tissue processing, each tissue block was embedded in paraffin and cut in a series of 5-μm thick consecutive sections. The first section was selected for haematoxylin and eosin (H&E) stains, while the remaining sections were selected for immunohistochemistry procedures using a Leica Bond III automated immunostainer with a diaminobenzidine chromogen detection system (DS9800, Leica Biosystems). Immunohistochemistry was performed for GFAP to evaluate presence of astrogliosis, for amyloid precursor protein (APP) for the detection of axonal injury, for abnormally phosphorylated tau (AT8) protein, and myelin basic protein (MBP) to evaluate possible myelin loss. Two sections per antibody were stained at 300 μm apart from each other, in accordance with the MRI slice thickness. More details regarding immunohistochemistry can be found in the Supplementary material.
All stained sections were digitally scanned using an Aperio scanner system (Aperio AT2—High Volume, Digital whole-slide scanning scanner, Leica Biosystems, Inc.) for further assessment and analyses. A Zeiss Imager A2 (ImagerA2 microscope, Zeiss) bright-field microscope with ×40 and ×63 magnification lenses was used to identify and photograph histologic and pathologic details, as needed.
Quantification of astrogliosis
Images of GFAP-stained sections were digitized using an Aperio whole-slide scanning scanner system (Leica Biosystems) at ×20 magnification. The following steps, all implemented using MATLAB (The Mathworks, Natick, MA), were taken to allow for a quantitative analysis of the GFAP images. First, the images were transformed into a common, normalized space to enable improved quantitative analysis.38 Then, the normalized images were deconvolved to unmix the primary (GFAP) and secondary (H&E) stains, and background to three separate channels.39 Once an GFAP-only image has been obtained, a final thresholding step individualized for each slice was taken to exclude non-specific staining and to allow for a subsequent % area calculation.
Histology–MRI co-registration
The high-resolution MR images were used as anatomical references to which the histological images were registered. Areas in the histological images that grossly diverged from the wet tissue state (i.e. the MR images) due to deformation were manually removed, while maintaining the image aspect ratio. Following convergence of 2D affine co-registration of histology and MR images (Image Processing Toolbox, MATLAB, The Mathworks, Natick, MA), we performed a 2D diffeomorphic registration refinement between the GFAP image slices and MRI volumes. This was done to recover true in-plane tissue shape and bridge over residual differences between the modalities. The diffeomorphic registration procedure in this study was performed using an efficient implementation of the greedy diffeomorphic algorithm,40 provided as an open-source software package (greedy, https://github.com/pyushkevich/greedy). The greedy software was initialized and used as previously described.41 The transformed histology images were overlaid on MR images to assess the quality of the co-registration and the Jaccard index42 was computed to quantify the overlap scores between the co-registered modalities (Supplementary Fig. 1).
T1 and T2 maps and diffusion tensor MRI processing
Diffusion tensor imaging parameters,43 axial diffusivity (AD), radial diffusivity (RD) and fractional anisotropy (FA), were calculated using in-house MATLAB (The Mathworks, Natick, MA) code based on previous work.44
Conventional quantitative relaxation maps were first computed by fitting the signal decay to mono-exponential functions. The T1 value was computed by fitting a subset of the multidimensional data that included 20 images with inversion times in the range of 12 ms and 980 ms. The T2 value was computed by fitting a subset of the multidimensional data that included 20 images with echo times in the range of 10.5 ms and 125 ms.
We also applied a commonly used strategy45,46 to correct for possible between-subject differences arising from post-mortem effects; we adjusted each voxel-averaged MRI parameter by dividing them by the mean for that parameter across all the normal-appearing white matter (WM) voxels in each brain sample.
Image domain masks
The FA maps were used to derive WM and grey matter (GM) image masks (defined using a threshold of 0.2). The co-registered GFAP density image and its inverse image were used as ‘injured’ and ‘normal’ image domain masks, respectively. Normal-appearing WM and GM image domain masks were obtained by multiplying the WM and GM masks with the ‘normal’ tissue mask. These image masks were eroded using a disk-shaped structuring element with a radius of 1 voxel to avoid partial volume bias from adjacent structures or from the edges of the brain tissue block. Grey–white matter interface image masks were obtained by adding the following products: (i) five voxels dilation of the GM image mask multiplied by the WM image mask; and (ii) two voxels dilation of the WM image mask multiplied by the GM image mask; their addition resulted in grey–white matter interface masks that include 1 mm of WM and 0.4 mm of GM.
Multidimensional MRI processing
Prior to processing, multidimensional MRI data were denoised using an adaptive non-local multispectral filter (i.e. NESMA47), which was shown to reduce noise and improve the accuracy of the resulting injury MRI biomarker maps.48 The filtered data were then processed using a marginally constrained, ℓ2-regularized, non-negative least-square optimization to compute the multidimensional distribution in each voxel, as previously described.27,31,36 It is a well-tested approach that has been proved robust and reliable,49–53 which in this study resulted in three types of distributions in each voxel: T1–T2, T2-MD, and T1-MD.
We implemented the following procedure to correct for possible between-subject differences arising from post-mortem effects. First, the normal-appearing WM mask was applied, and the maximal peak location in the spectral domain (e.g. T1–T2) was automatically found (this step was repeated for each subject). A control subject was then selected to be used as a reference (Case 1 in this study), to which all the remaining cases were aligned to in the spectral domain. This procedure ensures standardization across subjects, equivalent to the well-established strategy employed for voxel-average images, in which image values are divided by the mean across all the normal-appearing WM voxels in each brain sample.45,46 Despite the known effect of fixation, the variability of T1, T2 and MD values in WM across all subjects was limited. We report here the median and IQR of these values: T1 (from T1–T2) = 281 (94) ms, T2 (from T1–T2) = 27.0 (7) ms, T1 (from T1-MD) = 281 (91) ms, MD (from T1-MD) = 0.35 (0.14) µm2/ms, T2 (from T2-MD) = 27.0 (2.4) ms and MD (from T2-MD) = 0.22 (0.09) µm2/ms.
Statistical analysis
A linear mixed-effects model framework was used to study correlations between MRI-derived maps and GFAP density images. Random effects were added to model the within-subject correlation among histological samples. A whole-image approach (as opposed to regions of interest), in which all regions from the MRI maps and histological images were included, was used to achieve the most objective measures of correlations. Potential spatial correlation and co-dependencies within subjects were accounted for in two ways: (i) both MRI and histological maps were downsampled by a factor of 12 to reduce spatial dependencies54; and (ii) a Gaussian structured noise covariance matrix was included in the linear mixed-effects regression process to account for the spatial correlations among voxels (further details in the Supplementary material). Subject age was included as covariate. A two-sample t-test was performed on all pairs of regions of interest (i.e. normal-appearing and astrogliosis) to determine whether they are significantly different from one another.
False discovery rate (FDR) correction was carried out to take the overall number of pairwise contrasts into account.55 A P-value of 0.05 was considered statistically significant. RStudio was used for all computations.
Data availability
The data sets generated and analysed during the current study are available from the corresponding author on reasonable request.
Results
Neuropathological findings
Table 1 and Supplementary Table 1 summarize the main demographic data and known medical history for each examined subject and histopathological findings observed in each studied tissue block. Of the total 14 cases evaluated, there were seven cases with known interface astrogliosis and seven control cases negative for interface astrogliosis.
Scar-border forming astrogliosis pathology is demonstrated in immunostained sections for GFAP from four representative cases in Fig. 2. The astrogliosis pathology in our cohort was notably present at the grey–white matter junction in WM, without associated accumulation of pTau in involved cortical regions, and otherwise not seen in the pattern of a known tauopathy (e.g. chronic traumatic encephalopathy, CTE). Astrogliosis did not coexist with axonal injury or with demyelination that would be indicated by APP and MBP immunohistochemistry, respectively (APP, pTau and MBP sections from representative cases are shown in Supplementary Fig. 2). Additionally, H&E-based staining did not reveal evidence of ischaemic–necrotic lesions, presence of vascular lesions, microhaemorrhages or tissue rarefaction.
Figure 2.
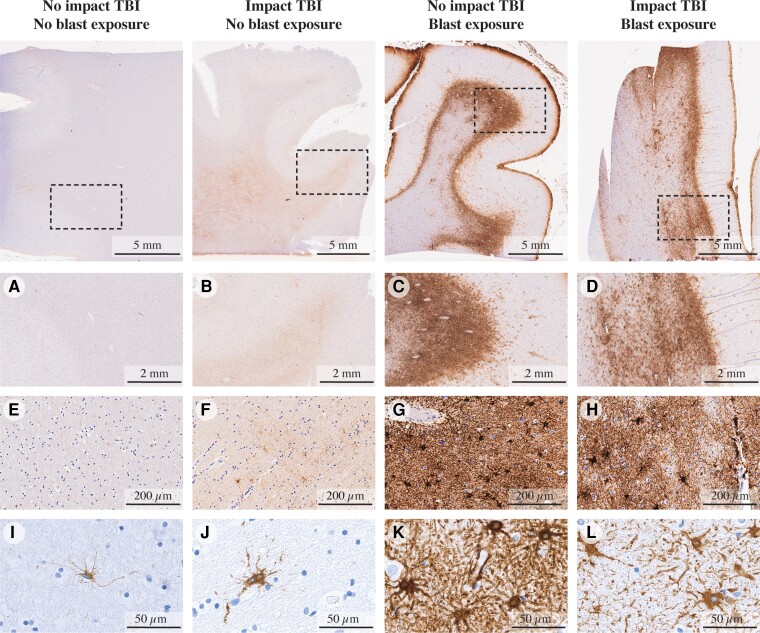
GFAP immunoreactivity in specimens. Figure shows specimens without impact or blast exposure TBI, with impact TBI but without blast exposure, without impact TBI but with blast exposure and with both impact and blast exposure TBI cases, at different magnification levels (×2, ×20 and ×80, from top to bottom). A, E and I show minimal GFAP immunoreactivity (Case 1). B, F and J show limited GFAP immunoreactivity with mild reactive astrocytes (Case 3). C, G and K show dense scar-border forming astrogliosis at the grey–white matter junction (Case 11). D, H and L show a similar pattern of dense scar-border forming astrogliosis at the grey–white matter junction (Case 8).
Of the seven cases with reported blast neurotrauma, five have reported a history of impact TBI at various degrees and time points. Given that increased GFAP expression at the cortical grey–white matter interface can indicate mild blast or impact neurotrauma,35 the specificity of scar-forming astrogliosis for the diagnosis of blast TBI cannot be demonstrated here. Additionally, there was a single interface astrogliosis-positive case in which there is no reported history of blast or impact TBI exposure (Case 13); the cause of the pathology in this case is uncertain and may relate to underreported TBI history. Despite that, the goal of this study was to map astrogliosis regardless of its underlying cause and to advance the field towards a non-invasive diagnostic tool.
Astrogliosis has a multidimensional MRI signature
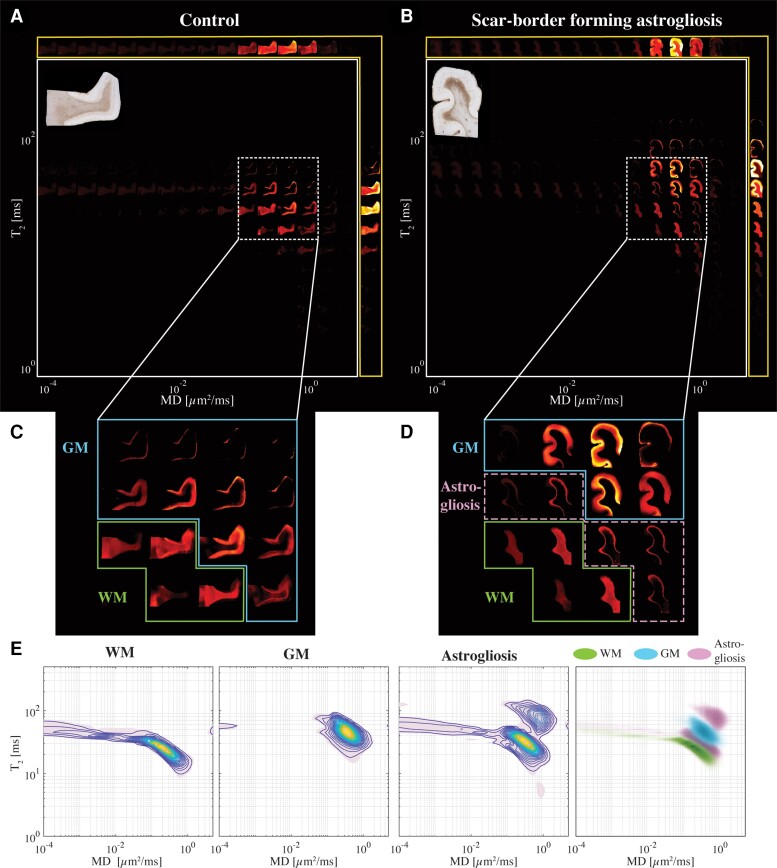
Our multidimensional MRI data reveal that a distinct signature exists for scar-border forming astrogliosis, which cannot be seen using one-dimensional MRI measurements. Investigation of the spatially resolved subvoxel multidimensional spectral components illustrates these findings, and to this end, it is useful to summarize the rich data set in a visually accessible manner. Each image slice of these data contains 4D information consisting of spatially resolved spectra with 50 × 50 elements in each voxel. We can visualize these data as arrays of maps with varying subvoxel T1, T2 and MD values. To make them more readable, the 50 × 50 spectra were subsampled on a 16 × 16 grid. Such summarized data of the T2-MD contrast from representative control (Case 7) and injured (Case 10) subjects are shown in Fig. 3A and B, respectively. In addition, the marginal distributions (i.e. 1D spectra) of subvoxel MD values (top row) and subvoxel T2 values (right column) are shown to illustrate the information content of any 1D approach (yellow frames in Fig. 3A and B). The GFAP histological image of each case is also shown on the upper-left corner of each panel, for reference.
Figure 3.
Changes in the T2-MD multidimensional MR signature induced by confirmed astrogliosis. Maps of 2D spectra of subvoxel T2-MD values reconstructed on a 16 × 16 grid of a representative (A) control (Case 7) and (B) injured (Case 10) subjects, along with their respective GFAP histological image (top left of each panel). (C) Magnified spectral region from the control case shows the clear separation of white (yellow frame) and grey (teal frame) matter according to their diffusion and T2 values. (D) The same magnified spectral region as in C from the injured case shows that while the WM and GM spectral information content is still clearly separable (yellow and teal frames, respectively), distinct spectral components can be seen on the grey–white matter interface (purple frame), which is qualitatively similar to the GFAP staining pattern of the sample. (E) T2-MD spectra averaged across all subjects in WM, GM and GFAP-positive spatial regions of interest (left to right) and a superposition of the average spectra from the three regions of interest. It should be noted that the peak in normal-appearing WM was forced to align between subjects, but not in GM or in injured tissue.
The scar-border forming astrogliosis multidimensional MRI signature can be seen by examining the T2-MD range that contains most of the spectral information, highlighted as white rectangles. Magnifications of this range in the spectra are shown below in Fig. 3C and D for the control and injured subjects, respectively. A clear separation of grey (blue frame) and white matter (green frame) can be seen in both control and injury states. However, we identified a distinct diagonal T2-MD spectral region (pink frame, Fig. 3D) in which intensities are concentrated at the grey–white matter junction, primarily on the WM side; these intensities follow closely the GFAP histological pattern (see inset image in Fig. 3B), while this newly found spectral information is absent in the control subject (Fig. 3C). Furthermore, the diagonal pattern in T2-MD points directly at a joint dependency with respect to T2 and MD, making it clear that this unique injury-related information cannot be seen by looking at T2 or MD separately (i.e. conventional 1D relaxation or diffusion MRI).
Generalizing these findings, we move from representative cases to averaged normal-appearing WM, GM and astrogliosis T2-MD spectra across the entire study (Fig. 3E left to right, respectively). A visualization of these three spectral clusters together is shown in the outermost right panel in Fig. 3E. The clear GM–WM separation can be easily seen, and in addition, the astrogliosis-related spectral information can be seen with most intensities lying in between the GM and WM peaks (grey–white matter interface) and some residual intensities towards higher values of T2 and MD (GM and meningeal border). A quantitative assessment of the difference between the different spectral signatures was performed by using the Jensen Distance (dJ),56 a well-established method of measuring distance between two probability distributions and which is bounded by 0 and 1. The Jensen Distances between regions with astrogliosis and normal-appearing WM, and between regions with astrogliosis and normal-appearing GM T2-MD spectral signatures, were 0.35 and 0.32, respectively. For reference, the Jensen Distances between normal-appearing WM and GM was 0.53.
We also examined the T1-MD and T1–T2 datasets. We found that while they contain astrogliosis-related information, it is significantly reduced and harder to distinguish as compared with the T2-MD contrast, thus the latter provides the most potentially useful information. Nevertheless, summarized T1-MD and T1–T2 data are shown in Supplementary Figs 3 and 4, respectively.
Anomaly detection in individuals
Although we demonstrated in the previous section that a multidimensional MRI signature associated with astrogliosis exists, being able to detect and refine it in an unsupervised and objective manner presents a significant challenge, especially when the information is hard to discern (e.g. T1-MD and T1–T2 in Supplementary Figs 3 and 4). In this work we build on previous frameworks31,36 and propose a within-subject anomaly detection procedure that results in MRI neuropathology biomarker maps. Conceptually, the principle is to first define what the ‘normal’ (i.e. uninjured in the desired context) multidimensional MRI signature is for a given individual and then look for deviations from that ‘normal’. Here we implemented this framework by integrating co-registered histological images with multidimensional MRI data and using the GFAP density to define what is normative. Alternately, in cases with little to no GFAP density half of the grey–white matter interface voxels were artificially excluded from what is considered normative to avoid bias towards detecting the interface as anomalous, thus averting circular reasoning of the machine learning algorithm. In the future, the normative brain multidimensional signature could be defined by collecting baseline multidimensional MRI data from healthy participants in vivo.
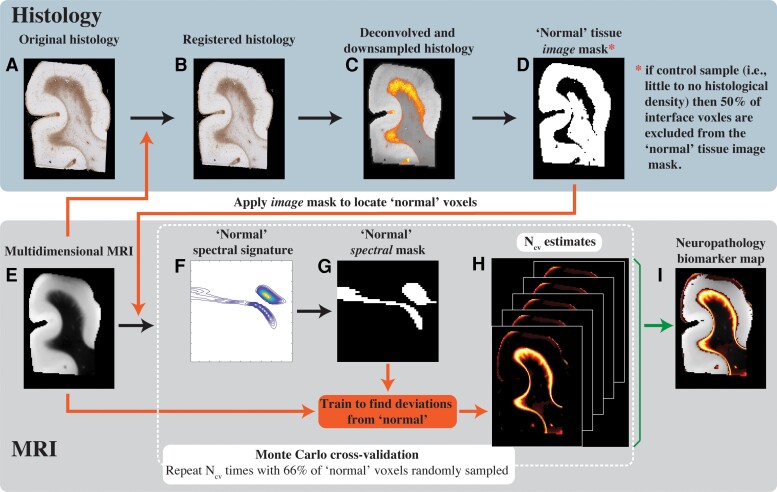
A schematic representation of the pipeline is shown in Fig. 4, where after the co-registration and deconvolution steps (Fig. 4A–C), the GFAP density image is inverted to obtain a ‘normal’ mask in the image domain (Fig. 4D). An additional step was taken in control cases (Cases 1–7), in which a grey–white matter interface image mask was used to artificially exclude from the analysis 50% of the interface voxels (randomly sampled). This additional step was taken to ensure that all samples contain a significant amount of grey–white matter boundary voxels. The resulting mask is then applied, after binary erosion (with a conservative radius of 4 voxels), on the multidimensional MRI data set to isolate voxels outside of the injury regions. Once all normal-appearing voxels are identified within a subject, a Monte Carlo cross-validation procedure57 is used to create multiple random splits (Ncv = 1000 in this study) of 66% and 34% of the normal-appearing voxels into training and validation data, respectively. For each such split, the average multidimensional signature is computed using the training data (Fig. 4F), then thresholded (at 1% of the maximal intensity) to obtain a binary spectrum (i.e. spectral mask) of the normal-appearing tissue (Fig. 4G). In the next step, the inverse of this spectral mask is assumed to contain abnormal spectral information and is used voxelwise on the full multidimensional data to result in a map of abnormal signal components. This process is repeated Ncv = 1000 times to allow the assessment of uncertainty and predictive accuracy, resulting in a set of neuropathology-related maps (Fig. 4H). The results are then averaged over the splits, yielding the final neuropathology MRI biomarker map (Fig. 4I).
Figure 4.
Schematic representation of the proposed anomaly detection framework. (A) The original GFAP histological image is processed in two steps: (B) co-registration to the MRI data set and (C) subsequent deconvolution and downsampling to match the MRI resolution. (D) This GFAP density image is then thresholded, inverted and used as an image domain mask for normative brain voxels on the (E) multidimensional MRI data. A Monte Carlo cross-validation procedure is used to create Ncv = 1000 multiple random splits of 66% and 34% of the normal-appearing voxels into training and validation data, respectively, resulting in a 1000 (F) normative spectral signatures, each of which is binarized to obtain (G) spectral masks of normative brain. To detect anomalies, the normative spectral mask is inverted and is used on the full multidimensional data to directly obtain (H) Ncv = 1000 versions of abnormal signal component maps, which are then averaged to yield the final (I) neuropathology MRI biomarker map.
Special attention was paid to the design of the machine learning algorithm, and while our approach provides reasonable mitigation of circular reasoning, it does not completely avert it. The inherent difference between the control and pathological cases, and the way in which they are treated by the learning algorithm (i.e. masking out 50% of random voxels in the grey–white matter boundary or applying a contiguous GFAP mask, for control or pathological cases, respectively) remains a general limitation of the radiological–pathological integration approach.
This strategy to detect anomaly in individuals was used separately on each subject and with each of the T2-MD, T1-MD and T1–T2 data sets. It should be noted that it can be applied to any multidimensional data, with any number of dimensions, as long as the data are processed to produce distributions and not scalar values. In addition, we amended this strategy to be able to handle scalar value maps to allow a fair comparison between multidimensional MRI and conventional voxel-averaged MRI (please refer to the Supplementary material).
Multidimensional MRI maps of astrogliosis
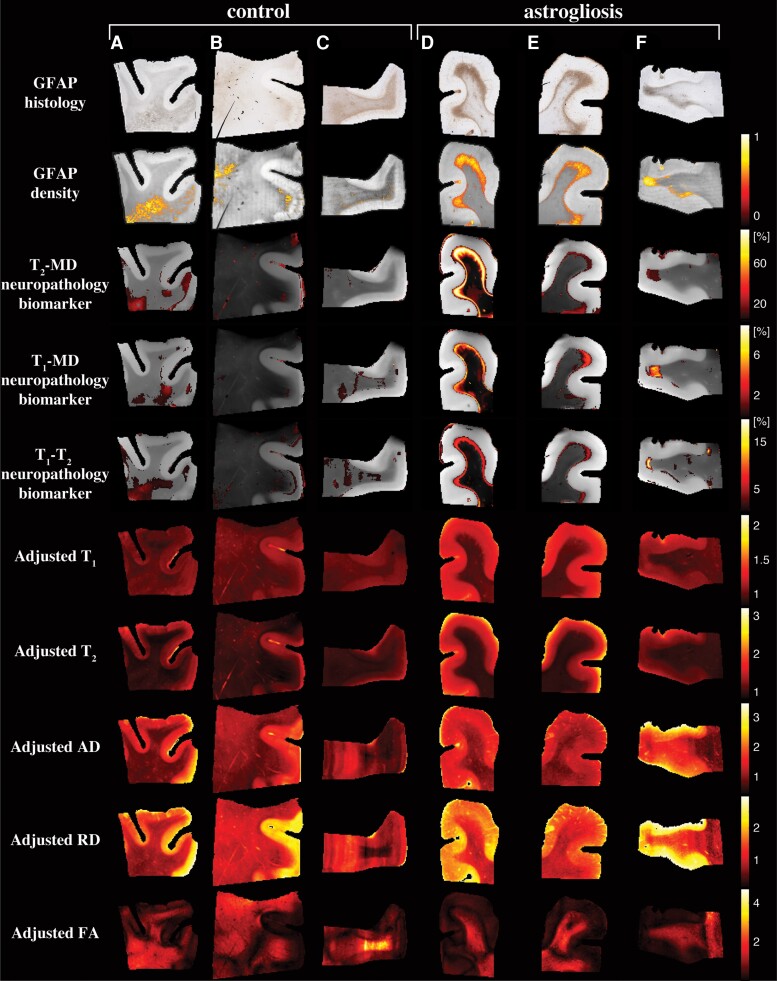
First, we examined the performance of our machine learning framework in visualizing astrogliosis by assessing their spatial sensitivity and specificity. Fig. 5 shows conventional MRI and DTI maps, multidimensional MRI maps and histological GFAP density images of six representative control and injured cases (Cases 3, 4, 7, 10, 11, 12 shown in Fig. 5A–F, respectively).
Figure 5.
Multidimensional and voxel-averaged MRI maps. A–C are subjects without severe astrogliosis (Cases 3, 4 and 7), while D–F had substantial GFAP overexpression (Cases 10–12). The different rows correspond to the different MRI contrasts, including all the conventional relaxation and DTI parameters, and the proposed multidimensional astrogliosis maps. In addition, the co-registered histological GFAP images and density maps are shown. Multidimensional neuropathology maps overlaid onto proton density images show substantial injury along the grey–white matter interface, while conventional MRI maps of T1, T2, AD, RD and FA do not show visible abnormalities. Note that to facilitate visualization, the multidimensional neuropathology MRI biomarker maps were thresholded at 10% of the maximal intensity and overlaid on greyscale proton density images.
Qualitatively, the multidimensional MRI neuropathology maps follow closely the GFAP histology and density images, with the T2-MD maps having a significantly larger dynamic range of intensities as compared with the T1-MD and T1–T2 maps, pointing to increased sensitivity. The standard deviations of the MRI neuropathology biomarkers could be computed from the multidimensional processing framework, thus providing a quantified measure of the biomarker uncertainty. Maps of the standard deviations of the cases in Fig. 5 are shown in Supplementary Fig. 5. The standard deviations are under an order of magnitude smaller than the corresponding means, pointing to relative stability and low uncertainty.
Conventional MRI and DTI maps provided useful anatomical macroscopic information, especially grey–white matter separation; however, they fail to capture the microstructural or compositional changes that are induced by astrogliosis. To allow for a direct comparison between multidimensional and conventional MRI and DTI, we further processed the voxel-averaged relaxation and diffusion maps using an adjusted anomaly detection machine learning algorithm (Supplementary material and Supplementary Fig. 6). The resulting unimodal neuropathology maps lacked similarity and agreement with GFAP histology (Supplementary Figs 7 and 8), which indicates that the microstructural and chemical composition changes induced by astrogliosis are averaged-out when scalar value MRI maps are used.
Strong correlation between MRI measures and GFAP density
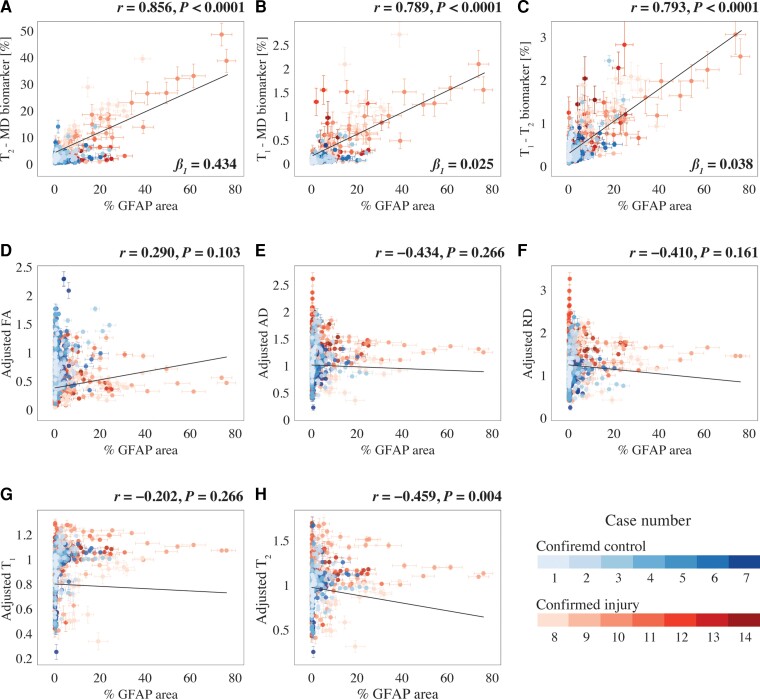
The multimodal data set in this study allows one to investigate the strength of the relationships between the multidimensional MRI-derived biomarkers we discovered and histologically based astrogliosis and their spatial agreement. A whole-image approach (as opposed to regions of interest), in which all regions from the MRI maps and histological images were included, was selected to achieve the most objective measures of correlations. After matching the GFAP density images resolution to their MRI counterparts, both MRI and histological maps were downsampled by a factor of 12 to account for co-registration errors and to reduce spatial dependencies (Supplementary Fig. 9), resulting in a total of 556 pairs of MR image volumes and GFAP densities from all 14 subjects. Fig. 6 summarizes the association between the investigated MR metrics—multidimensional MRI neuropathology biomarker maps and conventional voxel-averaged images—and the pathological findings across the entire images, grouped according to the Case number. This allows for inferences about both within and between subject correlations (see the Statistical Analysis section for more details).
Figure 6.
Radiological-pathological correlations between MRI metrics and GFAP density. GFAP density (% area) from 556 tissue regions from 14 subjects (colour-coded, see legend) and the corresponding MR parameter correlations. Individual data points represent the mean value from each post-mortem tissue sample. Scatterplots of the mean (with 95% CI error bars) % area GFAP and (A) T2-MD, (B) T1-MD and (C) T1–T2 injury MRI biomarkers show strong positive and significant correlation with GFAP density. The conventional MRI metrics in D–G did not result in strong and significant correlations with % area GFAP, apart from weak yet significant correlation of (H) voxel-averaged T2.
We found that GFAP density was strongly and significantly correlated with the T2-MD neuropathology biomarker (r = 0.856, P < 0.0001), the T1-MD neuropathology biomarker (r = 0.789, P < 0.0001) and the T1–T2 neuropathology biomarker (r = 0.793, P < 0.0001). Importantly, these results indicated that higher intensity of the multidimensional neuropathology MRI biomarkers is associated with increased astrogliosis severity, regardless of the tissue type studied. Notably, while the slopes (β1 in Fig. 6) from the T1-MD and T1–T2 regression analyses indicated potentially low sensitivity (0.025 and 0.038, respectively), the slope from the T2-MD neuropathology MRI biomarker was an order of magnitude larger (0.434). From the conventional voxel-averaged images, the only measure that had a significant yet weak correlation with GFAP density was the adjusted T2 (r = −0.459, P = 0.004). The effect of the subject’s age was insignificant for all MRI contrasts.
Astrogliosis is detectable in individuals
After showing that the multidimensional signature of interface astrogliosis is distinct from normal-appearing WM and GM, we now turn to establishing whether it is distinguishable from normal-appearing grey–white matter boundary. We then compare the multidimensional MRI astrogliosis maps intensities from normal-appearing and injured regions to test whether our approach can be used to image astrogliosis in a single subject.
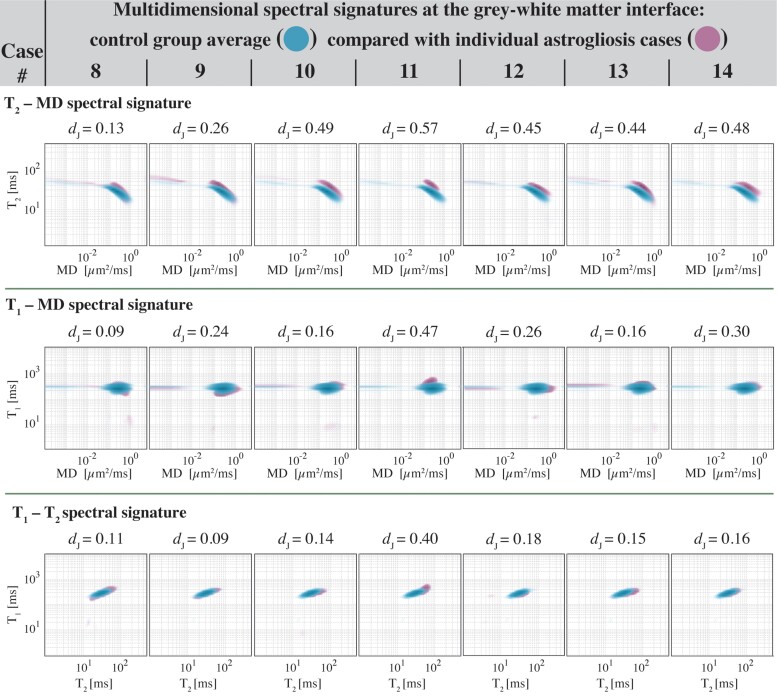
Investigating the uniqueness of the interface astrogliosis multidimensional signature, interface regions of interest were manually drawn on proton density MR images (regions of interest shown in Supplementary Fig. 10), and T2-MD, T1-MD and T1–T2 spectral signatures from all control cases (Cases 1–7) were averaged, yielding a grey–white matter interface spectral signature control group average for each MR data set (Fig. 7, blue). Then, grey–white matter interface spectral signatures from each GFAP-positive case (Cases 8–14) were individually compared with the control group average (Fig. 7, purple). The signatures were overlaid to allow for a qualitative assessment of the differences at the individual subject level. A quantitative assessment was performed by computing the Jensen Distances between control group average and individual signatures and is shown in Fig. 7. These results indicate that in addition to being distinct from normal-appearing WM and GM (Fig. 3E and Supplementary Figs 2 and 3), multidimensional signature of astrogliosis is also qualitatively and quantitatively distinct from normal-appearing grey–white matter interface. The results demonstrate that the microstructural and chemical composition changes caused by astrogliosis are being detected most substantially with T2-MD encoding (consistent with our findings, e.g. β1 in Fig. 6).
Figure 7.
Comparisons of normal-appearing grey–white matter interface and interface astrogliosis multidimensional signatures. Spectral signatures from all control cases were averaged, yielding a grey–white matter interface spectral signature control group average for each MR dataset (blue). Interface astrogliosis spectral signatures from each GFAP-positive case (purple) are individually compared with the control group average and the Jensen Distance between control group average and individual signatures are shown for each case. As a reference, the Jensen Distances between normal-appearing WM and GM across the entire study was 0.53, 0.13 and 0.21 for the T2-MD, T1-MD and T1–T2 spectral signatures, respectively.
Moving from the spectral domain to image domain analysis, normal-appearing and injured regions of interest were defined automatically based on the GFAP density image (e.g. Fig. 4D) and were used as binary masks on the multidimensional MRI neuropathology maps to obtain average and 95% CIs of the intensity values. Supplementary Fig. 11 shows these comparisons for the neuropathology MRI biomarkers for all subjects. Apart from Case 9, the astrogliosis maps were shown to be capable of detecting astrogliosis in individuals, illustrated by significant differences between the regions of interest (P < 0.0001 for all cases, except for P < 0.05 for Case 14’s T1-MD and T1–T2 biomarkers).
Discussion
This is the first report of a non-invasive MRI framework to directly map astrogliosis in individual brains ex vivo. In this study we showed that astrogliosis induces microstructural and compositional changes that result in a distinct multidimensional MRI spectral signature. Further, we developed a novel approach to utilize this information and obtain MRI maps of astroglial neuropathology in individual human brains. We found that the multidimensional MRI astrogliosis biomarker maps are significantly and strongly correlated with co-registered histological images of increased GFAP expression. We showed that our approach has the spatial sensitivity to detect altered tissue states at the individual level by establishing the distinction of interface astrogliosis spectral signature from normal grey–white matter interface, and by comparing normal-appearing and histologically confirmed regions within the same brain.
Mounting evidence points to increased GFAP expression at the cortical grey–white matter interface as a neuropathological hallmark of blast neurotrauma.33,34 However, interface astrogliosis cannot be considered diagnostic or specific to blast TBI given its presence in mild impact neurotrauma cases as well.35,58 Our goal in this study was to investigate whether astrogliosis produces a multidimensional MRI signature, and if so to map the injury, regardless of its underlying cause. This study was not designed and does not provide evidence to support the specificity of interface astrogliosis towards blast neurotrauma. All the examined chronic blast TBI cases, five of which have had in addition a history of impact TBI, were characterized by scar-border forming astrogliosis but importantly, absent additional cellular changes, most notably without evidence of tau pathology, axonal injury or demyelination. This neuropathological uniqueness towards astrogliosis presented an opportunity for a targeted study that provided inferences with a good degree of specificity. Conversely, very few studies have directly investigated the effect astrogliosis has on MRI diffusion and relaxation properties, and those who have did so by using animal injury models that always resulted in substantial axonal damage and other major microscopic changes in addition to gliosis.13–16
Our results show that scar-border forming astrogliosis was mainly present in superficial WM and involved the grey–white matter junction in all the cases we examined in this study. Therefore, focusing on changes to the multidimensional spectral signature between regions with astrogliosis and normal-appearing WM (Fig. 3E), and normal grey–white matter interface (Fig. 7), could elucidate the chemical and microstructural alterations induced by this type of neuropathology. In terms of T2 relaxation, severe astrogliosis causes an increase in T2 compared with normal-appearing WM and grey–white matter interface, and similarly, regions with astrogliosis are characterized by faster MD compared with normal-appearing WM (pink highlight in Fig. 3D and E) and with grey–white matter interface (Fig. 7). Increases in both relaxation times and diffusivities point to a reduction in the degree of axonal packing and density that is expected to result from the presence of highly reactive astrocytes14,59 (see Fig. 1). These measurable microscopic changes can be attributed to astrogliosis because no axonal damage or demyelination were histologically observed in any of the cases in the study. In addition, we recently reported that axonal injury has a distinct multidimensional MRI signature characterized by shortening of T1 and T2.31 These findings along with the current results indicate that decoupling of axonal injury and astrogliosis using a single framework should be possible because of the opposing effects these neuropathologies have on their respective multidimensional MRI spectra (i.e. reduction or increase in T1 and T2 for axonal injury and astrogliosis, respectively).
The novel framework we propose here helped to elucidate the underpinning of MRI signal response from astrogliosis and, importantly, showed that no one-dimensional T1, T2 or diffusion MRI measurement can disentangle the microscopic alterations caused by this neuropathology. It is therefore not surprising that very little progress has been made thus far towards the radiological assessment and mapping of astrogliosis, as this study appears to be the first one to use multidimensional MRI to address this problem. In principle, our framework can be readily extended to include more MR dimensions (e.g. diffusion orientation24 and magnetization transfer60) to improve sensitivity and more histological stains (e.g. axonal damage, myelin) to improve specificity.
DTI metrics are well-studied in the context of microstructural alterations due to TBI45,61,62; however, it is becoming increasingly evident that they are inconsistent in their observed response.63–65 Moreover, one of the only comprehensive MRI studies that examined the contribution of astrogliosis to DTI metrics showed that while significant increase in FA was associated with astrogliosis in cortical GM, significant changes in WM were related to demyelination and not astrogliosis.14 These findings are consistent with our results that showed correlations between DTI metrics and astrogliosis in WM were not significant, reflecting the heterogeneity and variability of real life TBI cases, as opposed to animal models.
Although we used histological data to infer the spatial distribution of astrogliosis as an integrated part of our machine learning framework, our approach is not limited to ex vivo studies. Using histology to locate normal-appearing regions of the brain can be replaced by collecting baseline multidimensional MRI spectra from healthy participants that will define a normative brain, and then apply our approach to detect abnormalities in the rich data. Acquiring such data in a clinical setting has become feasible following recent developments of multidimensional MRI clinical protocols by multiple groups.28,66,67 As is the case with any other single-patient analysis methods, substantial amounts of normative data will be required to establish a reference ‘atlas’.68,69 Additional limitations and confounds specific to our study include the effects of postmortem decay, fixation and resulting dehydration. The fixation process by itself and the delay in fixation from the time of death (i.e. post-mortem interval, PMI) cause changes to tissue properties and affect measured MRI parameters,70 which prevents direct comparison with in vivo data. Furthermore, information regarding the PMI was available from only about half of the subjects and cannot be controlled for in our study. The variability in T1, T2 and diffusion parameters in the current study was quantified by recording the distribution of the WM spectral peak locations across the different subjects (see Multidimensional MRI processing Section). Although intersubject variability was present as expected, it was not adverse, as seen by the relatively low average coefficients of variation (computed as the ratio between the median absolute deviation and the median) of 0.15, 0.11 and 0.19 in T1, T2 and MD dimensions, respectively. Although showing that the GFAP deposition corresponds to changes in the multidimensional signature was only possible using combined ex vivo MRI and immunostaining, it remains to be demonstrated in vivo. This study was designed to mostly investigate scar-border forming astrogliosis; however, future studies that examine the full range of astrogliosis severity are warranted to establish the sensitivity of our approach towards its detection and mapping.
In summary, being able to selectively focus on sub-voxel relaxation and diffusion components combined with a simple yet effective machine learning approach to detect anomalies in individual subjects provides a framework for mapping of astrogliosis with high precision. While MRI may offer promise to detect subtle microscopic differences at the group level, the goal of clinical neuroimaging is to be applicable at the individual level, potentially facilitating individualized diagnosis and subsequent therapy. This work emphasizes the importance and the potential of combining relaxation and diffusion MRI with artificial intelligence for studying human brain astroglial reactivity noninvasively.
Supplementary Material
Acknowledgements
We thank the subjects’ families that consented for brain donations for the better understanding of TBI consequences. The authors thank Dr Murat Bilgel for insights into the statistical analysis. We also thank Patricia Lee, Nichelle Gray and Paul Gegbeh for their valuable technical work. We are grateful to Stacey Gentile, Deona Cooper and Harold Kramer Anderson for their administrative assistance. We thank the TRACK-TBI Investigators.
Contributor Information
Dan Benjamini, Section on Quantitative Imaging and Tissue Sciences, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, Bethesda, MD 20891, USA; Multiscale Imaging and Integrative Biophysics Unit, Laboratory of Behavioral Neuroscience, National Institute on Aging, NIH, Baltimore, MD 21224, USA; Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
David S Priemer, Department of Pathology, F. Edward Hébert School of Medicine, Uniformed Services University, Bethesda, MD 20814, USA; The Department of Defense/Uniformed Services, University Brain Tissue Repository, Bethesda, MD 20814, USA; The Henry M. Jackson Foundation for the Advancement of Military Medicine (HJF), Bethesda, MD 20817, USA.
Daniel P Perl, Department of Pathology, F. Edward Hébert School of Medicine, Uniformed Services University, Bethesda, MD 20814, USA; The Department of Defense/Uniformed Services, University Brain Tissue Repository, Bethesda, MD 20814, USA.
David L Brody, Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA; Department of Neurology, F. Edward Hébert School of Medicine, Uniformed Services University, Bethesda, MD 20814, USA; Laboratory of Functional and Molecular Imaging, National Institute of Neurological Disorders and Stroke, NIH, Bethesda, MD 20892, USA.
Peter J Basser, Section on Quantitative Imaging and Tissue Sciences, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, Bethesda, MD 20891, USA; Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Funding
This research was partially supported by a grant from the U.S. Department of Defense, Program Project 308430 USUHS. Support for this work also included funding from the U.S. Department of Defense to the Brain Tissue Repository and Neuropathology Core, Center for Neuroscience and Regenerative Medicine (CNRM). D.B. was supported by the CNRM Neuroradiology-Neuropathology Correlation Core. D.P., D.P.P. and D.L.B. were supported by the CNRM and USUHS. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Aging and the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The opinions expressed herein are those of the authors and are not necessarily representative of those of the Uniformed Services University of the Health Sciences (USUSH), the Department of Defense (DOD), the NIH or any other US government agency.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Sofroniew MV, Vinters HV. Astrocytes: Biology and pathology. Acta Neuropathol. 2010;119:7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bush TG, Puvanachandra N, Horner CH, et al. Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar-forming, reactive astrocytes in adult transgenic mice. Neuron. 1999;23:297–308. [DOI] [PubMed] [Google Scholar]
- 3. Wanner IB, Anderson MA, Song B, et al. Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via STAT3-dependent mechanisms after spinal cord injury. J Neurosci. 2013;33:12870–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Voskuhl RR, Peterson RS, Song B, et al. Reactive astrocytes form scar-like perivascular barriers to leukocytes during adaptive immune inflammation of the CNS. J Neurosci. 2009;29:11511–11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pekny M, Pekna M. Astrocyte reactivity and reactive astrogliosis: Costs and benefits. Physiol Rev. 2014;94:1077–1098. [DOI] [PubMed] [Google Scholar]
- 6. Heiland D H, Ravi VM, Behringer SP, et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun. 2019;10:2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shao W, zhen Zhang S, Tang M, et al. Suppression of neuroinflammation by astrocytic dopamine D2 receptors via αB-crystallin. Nature. 2013;494:90–94. [DOI] [PubMed] [Google Scholar]
- 8. Sofroniew MV. Astrocyte barriers to neurotoxic inflammation. Nat Rev Neurosci. 2015;16:249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Escartin C, Guillemaud O, Carrillo-de Sauvage M. Questions and (some) answers on reactive astrocytes. Glia. 2019;67:2221–2247. [DOI] [PubMed] [Google Scholar]
- 10. Anderson MA, Burda JE, Ren Y, et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature. 2016;532:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herrmann JE, Imura T, Song B, et al. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci. 2008;28:7231–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wheeler MA, Clark IC, Tjon EC, et al. MAFG-driven astrocytes promote CNS inflammation. Nature. 2020;578:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwartz ED, Duda J, Shumsky JS, Cooper ET, Gee J. Spinal cord diffusion tensor imaging and fiber tracking can identify white matter tract disruption and glial scar orientation following lateral funiculotomy. J Neurotrauma. 2005;22:1388–1398. [DOI] [PubMed] [Google Scholar]
- 14. Budde MD, Janes L, Gold E, Turtzo LC, Frank JA. The contribution of gliosis to diffusion tensor anisotropy and tractography following traumatic brain injury: Validation in the rat using Fourier analysis of stained tissue sections. Brain. 2011;134:2248–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhuo J, Xu S, Proctor JL, et al. Diffusion kurtosis as an in vivo imaging marker for reactive astrogliosis in traumatic brain injury. Neuroimage. 2012;59:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chary K, Nissi MJ, Nykänen O, et al. Quantitative susceptibility mapping of the rat brain after traumatic brain injury. NMR Biomed. 2021;34:e4438. [DOI] [PubMed] [Google Scholar]
- 17. Benjamini D, Hutchinson EB, Komlosh ME, et al. Direct and specific assessment of axonal injury and spinal cord microenvironments using diffusion correlation imaging. Neuroimage. 2020;221:117195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishiki A, Harada R, Kai H, et al. Neuroimaging-pathological correlations of [18F]THK5351 PET in progressive supranuclear palsy. Acta Neuropathol Commun. 2018;6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hatakeyama T, Kawakita K, Kawai N, et al. Temporal and spatial changes in reactive astrogliosis examined by 18F-THK5351 positron emission tomography in a patient with severe traumatic brain injury. Eur J Hybrid Imaging. 2021;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benjamini D, Basser PJ. Multidimensional correlation MRI. NMR Biomed. 2020;33:e4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Slator PJ, Palombo M, Miller KL, et al. Combined diffusion-relaxometry microstructure imaging: Current status and future prospects. Magn Reson Med. 2021;86:2987–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benjamini D, Basser P. Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. J Magn Reson. 2016;271:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim D, Doyle EK, Wisnowski JL, Kim JH, Haldar JP. Diffusion–relaxation correlation spectroscopic imaging: A multidimensional approach for probing microstructure. Magn Reson Med. 2017;78:2236–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Topgaard D. Multidimensional diffusion MRI. J Magn Reson. 2017;275:98–113. [DOI] [PubMed] [Google Scholar]
- 25. Hutter J, Slator PJ, Christiaens D, et al. Integrated and efficient diffusion–relaxometry using ZEBRA. Sci Rep. 2018;8:15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manhard MK, Stockmann J, Liao C, et al. A multi-inversion multi-echo spin and gradient echo echo planar imaging sequence with low image distortion for rapid quantitative parameter mapping and synthetic image contrasts. Magn Reson Med. 2021;86:866–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini D, Basser PJ. Magnetic resonance microdynamic imaging reveals distinct tissue microenvironments. Neuroimage. 2017;163:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slator PJ, Hutter J, Palombo M, et al. Combined diffusion–relaxometry MRI to identify dysfunction in the human placenta. Magn Reson Med. 2019;82:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martins JP dA, Tax CMW, Reymbaut A, et al. Computing and visualising intra-voxel orientation-specific relaxation–diffusion features in the human brain. Hum Brain Mapp. 2021;42:310–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reymbaut A, Critchley J, Durighel G, et al. Toward nonparametric diffusion—Characterization of crossing fibers in the human brain. Magn Reson Med. 2021;85:2815–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benjamini D, Iacono D, Komlosh ME, Perl DP, Brody DL, Basser PJ. Diffuse axonal injury has a characteristic multidimensional MRI signature in the human brain. Brain. 2021;144:800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phipps H, Mondello S, Wilson A, et al. Characteristics and impact of U.S. military blast-related mild traumatic brain injury: A systematic review. Front Neurol. 2020;11:559318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shively SB, Horkayne-Szakaly I, Jones RV, Kelly JP, Armstrong RC, Perl DP. Characterisation of interface astroglial scarring in the human brain after blast exposure: A post-mortem case series. Lancet Neurol. 2016;15:944–953. [DOI] [PubMed] [Google Scholar]
- 34. Schwerin SC, Chatterjee M, Hutchinson EB, et al. Expression of GFAP and tau following blast exposure in the cerebral cortex of ferrets. J Neuropathol Exp Neurol. 2021;80:112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Babcock KJ, Abdolmohammadi B, Kiernan PT, et al. Interface astrogliosis in contact sport head impacts and military blast exposure. Acta Neuropathol Commun. 2022;10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pas K, Komlosh ME, Perl DP, Basser PJ, Benjamini D. Retaining information from multidimensional correlation MRI using a spectral regions of interest generator. Sci Rep. 2020;10:3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matthaei D, Frahm J, Haase A, Hanicke W. Regional physiological functions depicted by sequences of rapid magnetic resonance images. Lancet. 1985;326:893. [DOI] [PubMed] [Google Scholar]
- 38. Macenko M, Niethammer M, Marron JS, et al. A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE international symposium on biomedical imaging: From nano to macro. IEEE; 2009:1107–1110. [Google Scholar]
- 39. Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- 40. Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23:S151–S160. [DOI] [PubMed] [Google Scholar]
- 41. Adler DH, Wisse LEM, Ittyerah R, et al. Characterizing the human hippocampus in aging and Alzheimer’s disease using a computational atlas derived from ex vivo MRI and histology. Proc Natl Acad Sci. 2018;115:4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jaccard P. The distribution of the flora in the alpine zone. New Phytol. 1912;11:37–50. [Google Scholar]
- 43. Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barmpoutis A, Vemuri BC. A unified framework for estimating diffusion tensors of any order with symmetric positive-definite constraints. In: 2010 IEEE international symposium on biomedical imaging: From nano to macro. IEEE;2010:1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holleran L, Kim JH, Gangolli M, et al. Axonal disruption in white matter underlying cortical sulcus tau pathology in chronic traumatic encephalopathy. Acta Neuropathol. 2017;133:367–380. [DOI] [PubMed] [Google Scholar]
- 46. Cooper G, Finke C, Chien C, et al. Standardization of T1w/T2w ratio improves detection of tissue damage in multiple sclerosis. Front Neurol. 2019;10:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bouhrara M, Bonny JM, Ashinsky BG, Maring MC, Spencer RG. Noise estimation and reduction in magnetic resonance imaging using a new multispectral nonlocal maximum-likelihood filter. IEEE Trans Med Imaging. 2017;36:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benjamini D, Bouhrara M, Komlosh ME, et al. Multidimensional MRI for characterization of subtle axonal injury accelerated using an adaptive nonlocal multispectral filter. Front Phys. 2021;9:737374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Provencher SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput Phys Commun. 1982;27:213–227. [Google Scholar]
- 50. Kroeker RM, Henkelman MR. Analysis of biological NMR relaxation data with continuous distributions of relaxation times. J Magn Reson. 1986;69:218–235. [Google Scholar]
- 51. Mitchell J, Chandrasekera TC, Gladden LF. Numerical estimation of relaxation and diffusion distributions in two dimensions. Prog Nucl Magn Reson Spectrosc. 2012;62:34–50. [DOI] [PubMed] [Google Scholar]
- 52. Celik H, Bouhrara M, Reiter DA, Fishbein KW, Spencer RG. Stabilization of the inverse Laplace transform of multiexponential decay through introduction of a second dimension. J Magn Reson. 2013;236:134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benjamini D, Basser PJ. Water mobility spectral imaging of the spinal cord: Parametrization of model-free Laplace MRI. Magn Reson Imaging. 2019;56:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hyun D, Crowley ALC, Dahl JJ. Efficient strategies for estimating the spatial coherence of backscatter. IEEE Trans Ultrason Ferroelectr Freq Control. 2017;64:500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 56. Endres DM, Schindelin JE. A new metric for probability distributions. IEEE Trans Inf Theory. 2003;49:1858–1860. [Google Scholar]
- 57. Xu QS, Liang YZ. Monte Carlo cross validation. Chemom Intell Lab Syst. 2001;56:1–11. [Google Scholar]
- 58. Hsu ET, Gangolli M, Su S, et al. Astrocytic degeneration in chronic traumatic encephalopathy. Acta Neuropathol. 2018;136:955–972. [DOI] [PubMed] [Google Scholar]
- 59. Donald CL M, Barber J, Andre J, Panks C, Zalewski K, Temkin N. Longitudinal neuroimaging following combat concussion: Sub-acute, 1 year and 5 years post-injury. Brain Commun. 2019;1:fcz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sled JG, Pike GB. Quantitative imaging of magnetization transfer exchange and relaxation properties in vivo using MRI. Magn Reson Med. 2001;46:923–931. [DOI] [PubMed] [Google Scholar]
- 61. Donald CL M, Dikranian K, Bayly P, Holtzman D, Brody D. Diffusion tensor imaging reliably detects experimental traumatic axonal injury and indicates approximate time of injury. J Neurosci. 2007;27:11869–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bourke NJ, Yanez Lopez M, Jenkins PO, et al. Traumatic brain injury: A comparison of diffusion and volumetric magnetic resonance imaging measures. Brain Commun. 2021;3:fcab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Budde MD, Annese J. Quantification of anisotropy and fiber orientation in human brain histological sections. Front Integr Neurosci. 2013;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kamnaksh A, Budde MD, Kovesdi E, Long JB, Frank JA, Agoston DV. Diffusion tensor imaging reveals acute subcortical changes after mild blast-induced traumatic brain injury. Sci Rep. 2015;4:4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dennis EL, Wilde EA, Newsome MR, et al. ENIGMA military brain injury: A coordinated meta-analysis of diffusion MRI from multiple cohorts. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018). IEEE; 2018:1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Avram A V, Sarlls JE, Basser PJ. Whole-brain imaging of subvoxel T1-diffusion correlation spectra in human subjects. Front Neurosci. 2021;15:671465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Martin J, Reymbaut A, Schmidt M, et al. Nonparametric D-R1-R2 distribution MRI of the living human brain. Neuroimage. 2021;245:118753. [DOI] [PubMed] [Google Scholar]
- 68. Chamberland M, Genc S, Tax CMW, et al. Detecting microstructural deviations in individuals with deep diffusion MRI tractometry. Nat Comput Sci. 2021;1:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jolly AE, Bălăeţ M, Azor A, et al. Detecting axonal injury in individual patients after traumatic brain injury. Brain. 2021;144:92–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shepherd TM, Flint JJ, Thelwall PE, et al. Postmortem interval alters the water relaxation and diffusion properties of rat nervous tissue—Implications for MRI studies of human autopsy samples. Neuroimage. 2009;44:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and analysed during the current study are available from the corresponding author on reasonable request.