Abstract
Vertebrate genomes are partitioned into chromatin domains or topologically associating domains (TADs), which are typically bound by head-to-head pairs of CTCF binding sites. Transcription at domain boundaries correlates with better insulation; however, it is not known whether the boundary transcripts themselves contribute to boundary function. Here we characterize boundary-associated RNAs genome-wide, focusing on the disease-relevant INK4a/ARF and MYC TAD. Using CTCF site deletions and boundary-associated RNA knockdowns, we observe that boundary-associated RNAs facilitate recruitment and clustering of CTCF at TAD borders. The resulting CTCF enrichment enhances TAD insulation, enhancer–promoter interactions, and TAD gene expression. Importantly, knockdown of boundary-associated RNAs results in loss of boundary insulation function. Using enhancer deletions and CRISPRi of promoters, we show that active TAD enhancers, but not promoters, induce boundary-associated RNA transcription, thus defining a novel class of regulatory enhancer RNAs.
The genomes of multicellular eukaryotes are organized into self-interacting units known as topologically associating domains (TADs) (Dixon et al. 2012). TADs are identified in Hi-C interaction matrices as contact domains, and contact domains that show corner dots (at the apex of a loop) are known as loop domains (Beagan and Phillips-Cremin 2020). Loop and contact domains are both proposed to be formed by loop extrusion (Dixon et al. 2012; Dekker and Mirny 2016; Fudenberg et al. 2016; Rao et al. 2017; Nuebler et al. 2018; Szabo et al. 2020) and are dependent on CTCF and cohesin (Nora et al. 2017; Rao et al. 2017; Schwarzer et al. 2017; Wutz et al. 2017; Szabo et al. 2020). The length of an extruded loop is determined by the presence and orientation of CTCF-bound regions that interact within the loop (Rao et al. 2014). By preventing enhancers and promoters within the loop from interacting outside of the loops, boundary CTCF pairs insulate genes within a TAD (Krivega and Dean 2017; Nora et al. 2017; Rowley et al. 2017; Szabo et al. 2020). However, the presence of CTCF consensus motifs or their orientation at contact domains does not accurately predict TAD boundaries, suggesting that additional factors must influence CTCF binding and, therefore, TAD formation and insulation.
Mutation of CTCF sites at TAD boundaries affects gene expression, ranging from no effect to severe effect in the same and neighboring TADs (Narendra et al. 2015; Flavahan et al. 2016; Hanssen et al. 2017; Despang et al. 2019; Sima et al. 2019; Williamson et al. 2019; Yang et al. 2019; Kloetgen et al. 2020). Mutation of all CTCF sites within a boundary region or depletion of CTCF may be required to display effects on transcription (Williamson et al. 2019; Yang et al. 2019; Khoury et al. 2020), suggesting redundancy among multiple CTCF sites at a TAD boundary. However, it is unclear how multiple CTCF sites within the boundary coordinate to achieve better insulation. Active transcription at TAD boundaries correlates with better insulation and CTCF occupancy (Bonev et al. 2017; Hsieh et al. 2020; Khoury et al. 2020) and has been speculated to increase TAD strength (Giorgetti et al. 2016; Zhang et al. 2019; Zhang et al. 2020a), consistent with observations that inhibition of transcription or Ribonuclease A (RNase A) treatment disrupts 3D chromatin architecture (Barutcu et al. 2019; Thakur et al. 2019; Swygert et al. 2021). Furthermore, CTCF dimerization, important for loop extrusion, is RNA dependent, and RNA binding of CTCF is required for TAD chromatin organization (Hansen et al. 2019; Saldaña-Meyer et al. 2019), but how CTCF:RNA interactions maintain TADs is not clear.
In some instances, TAD boundaries are found near sites of enhancers (Nandy Mazumdar et al. 2020). Also, TADs containing enhancer clusters show strong boundaries (Gong et al. 2018; Zhang et al. 2020a). However, it is not known whether these enhancers play a role in boundary insulation. Moreover, boundaries themselves display enhancer-like features (Sun et al. 2019; Swygert et al. 2021), although the levels of H3K27ac, RNA Polymerase II (Pol II), and H3K4me3 on TAD boundaries are far less than on active enhancers in general (Matthews and Waxman 2018). These observations suggest a relationship between certain TAD boundaries and enhancers, but the mechanistic basis for their putative interactions remains unknown.
Results
Transcribed TAD boundaries insulate better than nontranscribed boundaries
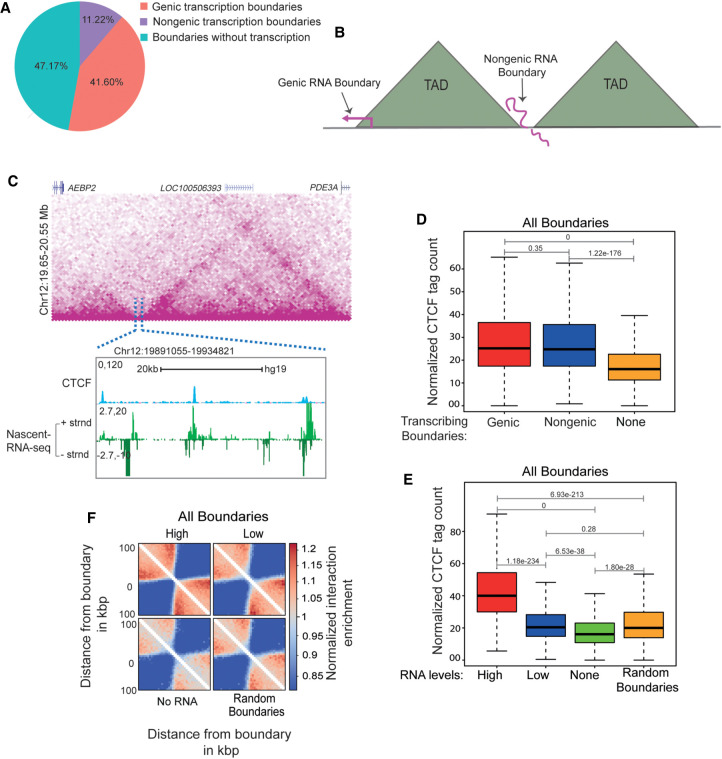
To determine the frequency of transcribed TAD boundaries, we measured run-on transcription with GRO-seq (Andersson et al. 2014) on TAD boundaries identified by HOMER's findTADsAndLoops.pl. This program identifies both contact and loop domain TADs based on intra-domain interaction frequency. Both TADs and loops are thought to be formed by loop extrusion and to be dependent on CTCF and cohesin (Nora et al. 2017; Rao et al. 2017; Schwarzer et al. 2017; Wutz et al. 2017). HOMER identified 15,397 TAD boundaries at 5-kb resolution using Hi-C data from HeLa cells (Rao et al. 2014). GRO-seq detected at least 10 normalized counts within about half of the TAD boundaries identified by HOMER (8134, 52.8%) (Fig. 1A). Almost 80% of transcribed boundaries are genic, and remaining 20% had no coding or noncoding gene but still showed transcription (Fig. 1A). Thus, domain boundaries are either nontranscribed, genic transcribed, or nongenic transcribed (Fig. 1B,C).
Figure 1.
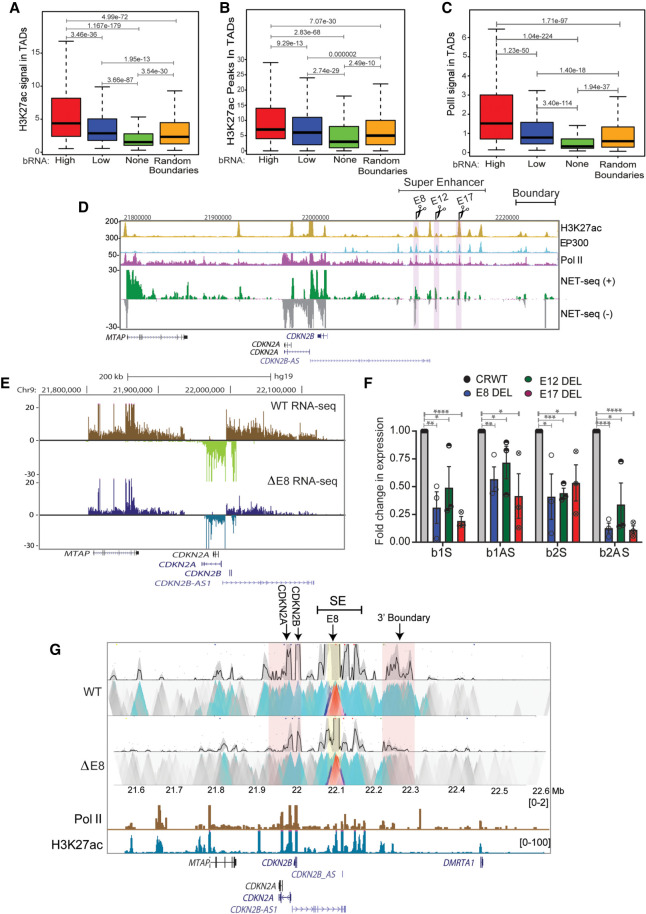
Transcribed TAD boundaries insulate better than nontranscribed boundaries. (A) Pie chart displaying the percentage of boundaries that show genic transcription, nongenic de novo transcription, and no transcription. (B) Schematic depicting genic and nongenic transcribed boundaries. (C) Browser shot displaying a TAD structure from Hi-C data. The zoomed-in box shows a de novo nongenic transcribed boundary region that is overlaid with CTCF and mNET-seq signal. (D) Boxplots showing CTCF enrichment on genic, nongenic, and nontranscribed boundaries. (E) Boxplots showing CTCF enrichment on all boundaries showing varying levels of RNAs versus nontranscribed and random boundaries. (F) Pile up (aggregated normalized Hi-C interactions) plot centered at high transcribed, low transcribed, nontranscribed, and random boundaries at a 5-kb resolution. The 100-kb distances are taken from the boundary region. The P-values in boxplots were calculated using the Wilcoxon rank-sum test. The boxplots depict the minimum (Q1 – 1.5 × IQR), first quartile, median, third quartile, and maximum (Q3 + 1.5 × IQR) without outliers.
TAD boundaries are enriched for CTCF binding. We determined if strong CTCF binding at TAD boundaries is associated with increased transcription. Using high-resolution CTCF CUT&RUN data (Skene and Henikoff 2017), we observed that both genic and nongenic transcribed TAD boundaries had significantly higher CTCF enrichment than nontranscribed boundaries (Fig. 1D). This relationship between transcription and CTCF enrichment at TAD boundaries was also observed for different RNA levels (Fig. 1E). Within nongenic transcribed boundaries, CTCF enrichment was also correlated with GRO-seq signal (Supplemental Fig. S1A,B). Similar results were seen using CTCF ChIP-seq (Supplemental Fig. S1C–E; Zhang et al. 2020b). We also tested this by plotting the ChIP-seq data in high, low, none and random boundaries as violin plots. We observed a clear shift in distributions displayed in the violin plot (Supplemental Fig. S1F). We conclude that transcribed TAD boundaries are more enriched for CTCF binding than nontranscribed boundaries.
We next asked if transcribed TAD boundaries that show high CTCF binding are stronger insulators. We noted a positive correlation between the levels of RNA at the transcribed boundaries and the TAD insulation (Fig. 1F). Even TADs with the least transcribed boundaries were better insulated compared with nontranscribed boundaries (Fig. 1F). We observed a similar positive correlation between boundary RNA levels and TAD insulation using two different bin sizes (5 kb and 20 kb) in our analyses, thereby ruling out resolution-related biases (Supplemental Fig. S2). A total of 69.2% (10,658 out of 15,397) of TADs boundaries were found to be transcribed (Supplemental Fig. S2A). The CTCF enrichment was stronger on transcribed genic and nongenic boundaries at both the 5-kb and 20-kb bin sizes (Supplemental Fig. S2B–E). Insulation scores on transcribed genic and nongenic boundaries were correlated with levels of RNA at the boundaries (Supplemental Fig. S2F–H).
We also examined the relationship between transcription of TAD boundaries and CTCF binding using HiCCUPS, which identifies loop anchors. In total, 3525 loop anchors were identified with 5-kb bin size, of which 43% (1519) were genic and 19% (675) were nongenic (Supplemental Fig. S3A). As was the case for TAD boundaries identified by HOMER, we found a positive correlation between transcription of loop anchors and CTCF enrichment using both the CUT&RUN (Supplemental Fig. S3B,C) and ChIP-seq data sets (Supplemental Fig. S3D,E). For subsequent analyses, we focused on TADs called by HOMER with 5-kb boundary regions.
We also observed positive associations between CTCF enrichment, insulation score, and transcription at TAD boundaries in IMR-90 fibroblasts (Supplemental Fig. S4A–D; Rao et al. 2014) and lymphoblastoid cells K562 (Supplemental Fig. S4E–H; Zirkel et al. 2018). Together, our results suggest that CTCF enrichment and enhanced insulation are general features of transcribed TAD boundaries.
RNA increases CTCF occupancy at TAD boundaries
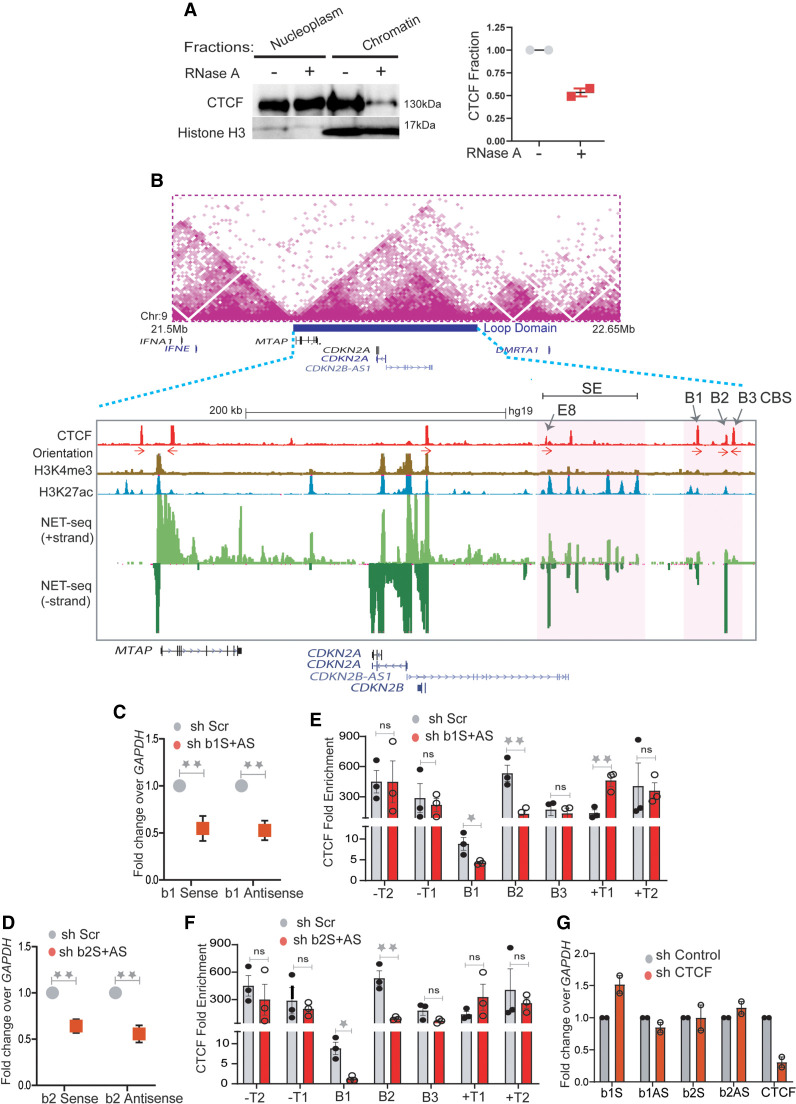
We wondered whether elevated levels of transcription and CTCF at TAD boundaries are causally related. To test this possibility, we treated HeLa cells with RNase A and performed western blot analysis on the nuclear and chromatin fractions. We observed a significant loss of CTCF in the chromatin fraction (Fig. 2A; Supplemental Fig. S5A), consistent with the reported dependence of CTCF on RNA for chromatin binding (Hansen et al. 2019; Saldaña-Meyer et al. 2019; Thakur et al. 2019). TAD boundaries in general show high binding of CTCF. Moreover, transcribing boundaries showed even higher CTCF enrichment compared with nontranscribing so we asked if the RNA at the transcribing boundaries has a functional role in CTCF enrichment.
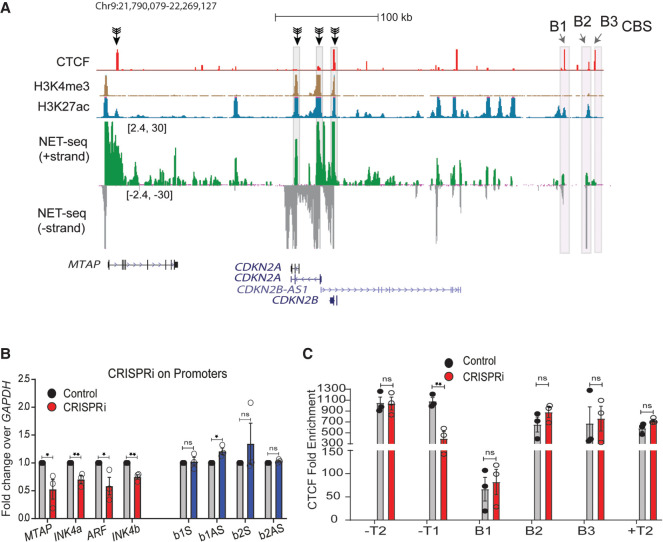
Figure 2.
RNA increases CTCF occupancy at TAD boundaries. (A) Immunoblotting with CTCF and histone H3 on soluble nucleoplasm and chromatin-bound fractions of the nucleus treated with or without RNase A (left). The right panel shows the quantified loss of CTCF from chromatin fractions from two replicates. (B) TAD structure at INK4a/ARF locus on 9p21 region (Rao et al. 2014). The loop domain (blue bar) is overlaid by the position of CTCF peaks, CTCF motif orientation, H3K4me3, H3K27ac (enhancers), mNET-seq tracks, and gene annotations. The highlighted region shows the B1, B2, and B3 CTCF sites on the 3′ boundary and super-enhancer with the position of enhancer E8 marked by an arrow. (C) qRT-PCRs showing the levels of sense and antisense noncoding RNA at B1 CTCF site upon B1 RNA knockdown. (D) qRT-PCRs showing the levels of sense and antisense RNA at the B2 CTCF site upon B2 RNA knockdown. (E) CTCF ChIP enrichment before and after shRNA-mediated knockdown of sense and antisense RNA at B1 CTCF sites. The bars show CTCF fold enrichment on three CTCF sites (B1, B2, and B3) at the 3′ boundary, on −T1 (5′ boundary) of the INK4a/ARF TAD, on -T2 on the adjacent TAD boundary upstream, +T1 (on the adjacent TAD boundary downstream), and +T2, the boundary of the following TAD downstream. (F) CTCF ChIP enrichment before and after shRNA-mediated knockdown of sense and antisense RNA at B2 CTCF sites. The bars show CTCF on three CTCF sites (B1, B2, and B3) at the 3′ boundary, on −T1 (5′ boundary) of the INK4a/ARF TAD, on −T2 of the adjacent TAD boundary upstream, on +T1 of the adjacent TAD boundary downstream, and on +T2 of the boundary of the following TAD downstream. (G) qRT-PCRs of sense and antisense RNAs from B1 and B2 CTCF sites upon CTCF knockdown. The drops in CTCF levels are shown in the last two bars. Error bars denote SEM from three biological replicates. P-values were calculated by the Student's two-tailed unpaired t-test in C–F. (**) P < 0.01, (*) P < 0.05, (ns) P > 0.05.
Toward this, we chose the ∼500-kb INK4a/ARF TAD that has nongenic transcribing boundary. The TAD spans the CDKN2A, CDKN2B, and MTAP cell cycle regulatory genes and a enhancer cluster (Fig. 2B, highlighted region; Harismendy et al. 2011; Farooq et al. 2021). The enhancer cluster within this TAD contains several active enhancers that are enriched for H3K27ac modification and enhancer RNAs (eRNAs) (Fig. 2B, mNET-seq panel; Farooq et al. 2021). We confirmed 5′ TAD boundary locations on the transcribed gene MTAP and 3′ boundary on the nongenic region containing three CTCF ChIP-seq peaks (B1, B2, and B3). The position of the boundary region is persistent in Hi-C data from other cell types. CTCF peaks B1 and B2 but not B3 show strong nascent transcription based on publicly available mNET-seq, GRO-seq, and PRO-seq data (Supplemental Fig. S6; Bouvy-Liivrand et al. 2017; Nilson et al. 2017; Schlackow et al. 2017) and so are classified as eRNAs. The GRO-seq signal at the boundary is moderate and is similar to that of most transcribing boundaries, whereas the CTCF signal at this boundary is toward the higher end of the distribution as shown by the GRO-seq and CTCF population density plots (Supplemental Fig. S5E,F). To test the role of RNA at B1 and B2 CTCF sites in CTCF binding, we depleted sense and antisense eRNAs at the B1 and B2 CTCF sites using shRNAs (Fig. 2C,D) and confirmed the drop of these eRNAs from chromatin fraction by qRT-PCR (Supplemental Fig. S5B). Knockdowns using shRNAs to B1 and B2 but not B3 and nearby TAD boundaries (T1 and T2) resulted in the loss of CTCF binding at these sites (Fig. 2E,F; Supplemental Fig. S5C). Notably, the loss of RNAs at B1 reduced CTCF occupancy not only at B1 but also at B2 and vice versa (Fig. 2E,F). This suggests that CTCF on transcribed boundaries is maintained in part by eRNAs, and different transcribed CTCF sites within a boundary are potentially interdependent. Notably, shRNA-mediated knockdown or overexpression of CTCF had minor effects on eRNA expression (Fig. 2G; Supplemental Fig. S5D), which suggests that CTCF enrichment at TAD boundaries depends on RNA but not vice versa.
Boundary RNAs enhance TAD transcription
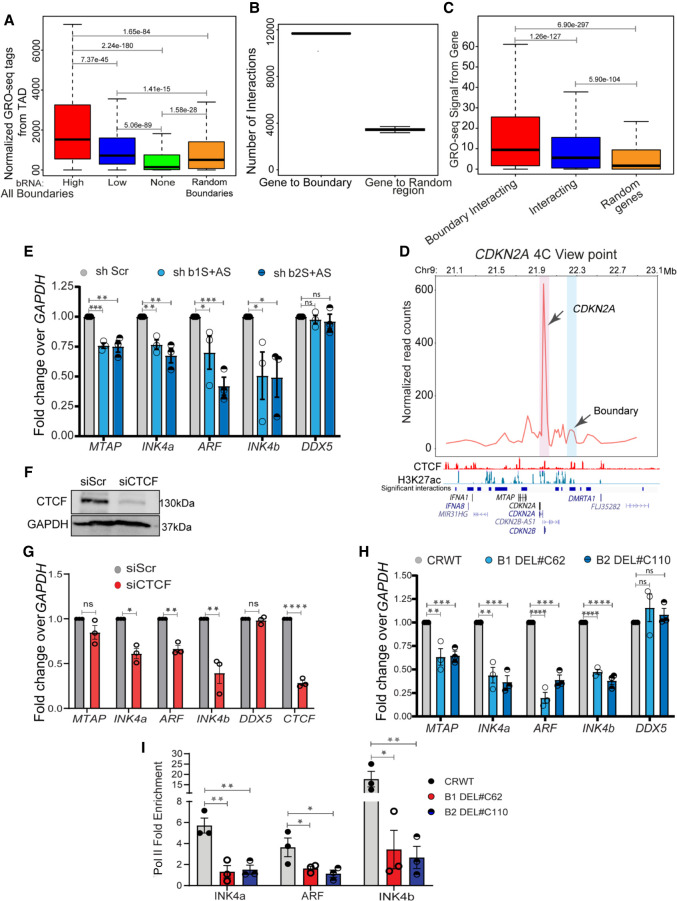
The enhanced insulation we observed at sites of boundary eRNAs (Supplemental Fig. S2F,H) suggested that genes within the TADs of transcribed boundaries might be expressed at higher levels than those in TADs of nontranscribed boundaries. Accordingly, we compared the transcriptional levels of TADs with high, low, and randomly selected transcribed or nontranscribed boundaries. We found that the transcriptional output of entire TADs (Fig. 3A) or of individual genes within TADs (Supplemental Fig. S7A) is correlated with the levels of RNAs at boundaries, and we obtained similar results for nongenic boundaries (Supplemental Fig. S7B). To determine whether this positive association resulted from direct interactions between promoters and boundaries, we analyzed available promoter capture-seq data in HeLa cells (Thiecke et al. 2020). Boundaries were captured by promoters far more than by random regions (Fig. 3B), and genes that interacted with boundaries were relatively more active than genes that interacted with other genomic regions (Fig. 3C). Together, these data suggest that transcribed boundaries tend to interact with promoters and are associated with higher rates of TAD transcription. We confirmed the interaction of the CDKN2A promoter with the nearby transcribed boundary within the INK4a/ARF TAD using 4C-seq assays (Fig. 3D).
Figure 3.
Boundary RNAs enhance TAD transcription. (A) Boxplots showing GRO-seq tags from the entire TAD with high, low, nontranscribed, and random boundaries. (B) Graph showing the number of interactions between genes and boundaries, and 100 randomizations of interactions between boundaries and random regions. (C) Boxplots showing the GRO-seq tag counts from genes when they interact with boundaries or any region versus random interactions. (D) 4C plot at the CDKN2A promoter viewpoint, where the interaction with the boundary is highlighted in blue. Below the plot, significant interactions, H3K27ac, CTCF, and gene annotations for HeLa cells are marked. (E) qRT-PCR plot shows the change in gene expression upon shRNA-mediated knockdowns of boundary RNAs arising from B1 and B2 CTCF sites at the INK4a/ARF TAD boundary. (F) Immunoblot showing the drop in CTCF protein levels upon its siRNA-mediated knockdown. (G) qRT-PCRs showing down-regulation of INK4a/ARF, DDX5, and CTCF upon CTCF knockdown. (H) qRT-PCR showing the changes in expression of different genes in the INK4a/ARF TAD and DDX5 upon Cas9-mediated independent deletion of B1 and B2 CTCF sites. (I) Pol II ChIP signal at INK4a, ARF, and INK4b promoters in WTs and in cells with B1 and B2 CTCF sites deletions. Error bars denote SEM from three biological replicates. P-values were calculated by the Student's two-tailed unpaired t-test in E and G–I. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, (ns) P > 0.05. P-values in boxplots were calculated by the Wilcoxon two-sided rank-sum test. The boxplots depict the minimum (Q1 – 1.5 × IQR), first quartile, median, third quartile, and maximum (Q3 + 1.5 × IQR) without outliers.
The CDKN2A and CDKN2B loci encode cell-cycle regulators INK4A, ARF, and INK4B, respectively. Next, we asked if boundary RNAs play a direct role in transcriptional activity of genes within this TAD. Using shRNAs to sense and antisense B1 and B2 CTCF sites, we observed down-regulation of INK4, ARF, INK4b, and MTAP, whereas the transcription of DDX5, a non-INK4a/ARF TAD gene, remained unchanged (Fig. 3E). We use INK4a/ARF and INK4b to describe mRNAs studied. These data suggest that boundary eRNAs positively regulate the transcription of the genes within the INK4a/ARF TAD by direct physical interaction between promoters and boundaries.
Because the loss of specific boundary RNAs also caused loss of CTCF binding at the TAD boundary, we asked if the role of boundary RNA in TAD transcription is CTCF dependent. Depletion of global CTCF using a pool of CTCF-specific siRNAs resulted in down-regulation of genes within INK4a/ARF TAD (Fig. 3F,G). To test the effects of the loss of CTCF binding only at the INK4a/ARF TAD boundary, we deleted the B1 and B2 CTCF sites individually in HeLa cells (Supplemental Fig. S8A,B) and observed down-regulation of CDKN2A and CDKN2B genes within the TAD (Fig. 3H) and the loss of Pol II at promoters (Fig. 3I). We also observed that boundary eRNA knockdown and boundary deletion reduced the levels of the INK4A protein encoded by CDKN2A gene (Supplemental Fig. S8C). These data suggest that boundary RNAs enrich the CTCF at the domain boundary.
Noncoding RNAs and CTCF interact at the INK4a/ARF TAD boundary
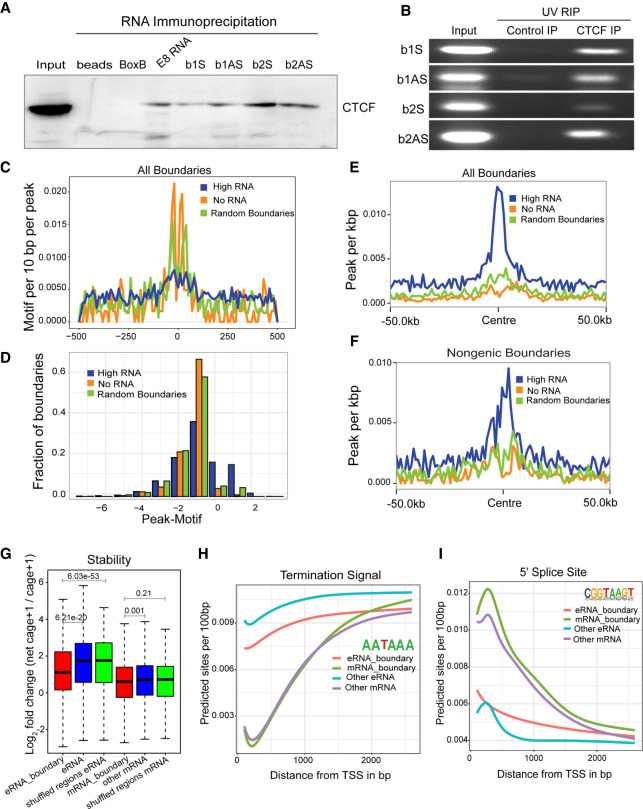
We next asked whether RNA and CTCF interact at functional boundaries. We first performed RNA pulldowns of sense and antisense RNAs transcribed from B1 and B2 CTCF sites in the INKa/ARF TAD followed by immunoblotting with a CTCF antibody (Fig. 4A). We observed CTCF interactions with B1 and B2 sense and antisense RNA and also with an eRNA from the CTCF-bound E8 enhancer within the enhancer cluster, but not with the BoxB negative control. CTCF binding to B1 and B2 sense and antisense RNAs was confirmed by UV-RIP, an RNA immunoprecipitation method (Fig. 4B). Notably, the CTCF motifs present at transcribed boundaries are much sparser than at nontranscribed or randomly selected boundaries (Fig. 4C,D). The top de novo motif identified in each of the categories is that of canonical CTCF and is similar across the categories with subtle differences (highlighted); for example, the weight of “T” (ninth base in the motif logo) was higher in boundaries with low to no transcription as opposed to high transcribing boundaries that show “G” as predominant base (Supplemental Fig. S9A). Further, upon looking at the canonical motif (Jasper), the frequency of CTCF motif was far lower over the background in transcribing boundaries (Supplemental Fig. S9A). However, we found that transcribed boundaries are more densely packed with CTCF peaks than are nontranscribed or randomly selected boundaries (Fig. 4E,F). These data suggest that transcription or the presence of eRNA can reinforce CTCF binding to multiple weak CTCF motifs.
Figure 4.
Noncoding RNAs and CTCF interact at the INK4a/ARF TAD boundary. (A) Immunoblot for CTCF on RNA pulldowns performed on various RNAs (n = 2). (B) Agarose gel image showing the UV-RIP RT-PCRs for sense and antisense RNA arising from B1 and B2 CTCF sites (n = 2). (C) Line plots show the CTCF motif per 10 bp per peak on various classes of boundaries. (D) Fraction of boundaries with differential occupancy of CTCF peaks over the presence of the CTCF motif. (E) CTCF peaks per kilobase on all transcribed boundaries with high RNAs, no RNAs, and random boundaries (50-kb flanks on both sides). (F) CTCF peaks per kilobase on nongenic transcribed boundaries with high RNAs, no RNAs, and random boundaries. (G) Log2FC of net-cage + 1/cage + 1, indicating stability of RNA at TSSs identified from NET-CAGE at boundaries and TSSs not at boundaries. eRNA_boundary denotes the eRNAs from nongenic boundaries; mRNA_boundary denotes the mRNAs from genic boundaries. (H) Average predicted TSS from the TSS identified in different classes of RNA by NET-CAGE. (I) Average predicted 5′ SS sites from the TSS identified by NET-CAGE. The P-values in boxplots were calculated by the Wilcoxon rank-sum test. The boxplots depict the minimum (Q1 – 1.5 × IQR), first quartile, median, third quartile, and maximum (Q3 + 1.5 × IQR) without outliers.
Next, we compared the stability, polyadenylation, and splicing features of genic RNAs and eRNAs at TAD boundaries with those that are not within boundaries. In general, eRNAs show fast turnover as seen by a higher ratio of nascent-to-mature eRNAs compared with genic RNAs (Andersson et al. 2014). By this measure, eRNAs on nongenic boundaries were significantly more stable than other eRNAs, and RNAs on genic boundaries were slightly more stable than other genic RNAs (Fig. 4G).
The AATAAA polyadenylation motif is rare around transcriptional start sites (TSSs) of genic transcripts, increasing in abundance downstream as expected (Fig. 4H). eRNAs on boundaries showed less enrichment of premature AATAAA than other eRNAs (Fig. 4H), consistent with their higher stability as seen in Figure 4G. Although the size of eRNAs and GC content at the boundary was not different from other eRNAs, mRNAs at the boundaries were longer in length and showed slightly higher GC content compared with the mRNAs elsewhere in the genome (Supplemental Fig. S9B,C). As expected, 5′ splice consensus sequences were strongly enriched in mRNAs relative to eRNAs (Fig. 4I), consistent with greater stability of mRNAs. Together, the enhanced stability, higher CTCF enrichment, and insulation-promoting activity of boundary eRNAs suggest that they have evolved to facilitate CTCF binding by direct physical interaction.
Active enhancers within the INK4a/ARF TAD directly regulate boundary eRNA transcription
What is responsible for INK4a/ARF boundary transcription? Because HeLa cells are addicted to high levels of proteins encoded within the TAD (McLaughlin-Drubin et al. 2013; Pauck et al. 2014), instead of deleting gene promoters, we used CRISPR inhibition (CRISPRi) using specific guide RNAs (gRNAs). We targeted dCas9-KRAB on promoters of CDKN2A, CDKN2B, and MTAP and assayed transcript levels at B1 and B2 CTCF sites (Fig. 5A). CRISPRi of these promoters resulted in transcriptional inhibition of these genes (Fig. 5B, red bars). However, boundary RNA levels were unaffected (Fig. 5B, blue bars). Consistent with unperturbed boundary RNA expression, there was no change in CTCF occupancy at B1 and B2 CTCF sites. However, we did observe loss of CTCF on the 5′ boundary that overlaps the MTAP gene, likely owing to down-regulation of MTAP (Fig. 5C). The boundary of flanking TADs also did not show any perturbations in CTCF occupancy (Fig. 5C). These data suggest that initiation of transcription from CDKN2A and CDKN2B promoters is not responsible for eRNAs at the boundary.
Figure 5.
Transcriptional activity of promoters does not regulate nongenic transcription at boundaries. (A) Browser shots displaying the promoters targeted by CRISPRi (black arrows); the track is overlaid with CTCF, H3K4me3, H3K27ac, and mNET-seq tracks. (B) qRT-PCR assessing the changes in INK4a/ARF mRNA, sense, and antisense boundary RNA arising from B1 and B2 CTCF sites upon CRISPRi on promoters. (C) CTCF enrichment on boundaries of INK4a/ARF and its flanking TADs upon CRISPRi on INK4a/ARF promoters. Error bars denote SEM from three biological replicates. P-values were calculated by the Student's two-tailed unpaired t-test in B and C. (**) P < 0.01, (*) P < 0.05, (ns) P > 0.05.
We next asked whether the INK4a/ARF TAD enhancers regulate genes within the TAD and boundary eRNAs. Genome-wide, transcribed boundary TADs are enriched in H3K27ac (Fig. 6A,B) and Pol II (Fig. 6C) relative to nontranscribed boundaries and randomly selected genic or nongenic boundaries (Fig. 6C; Supplemental Fig. S9D,E). We first tested the enhancer cluster using the E8, E12, and E17 enhancer deletion lines (Fig. 6D) in which CDKN2A and CDKN2B genes were seen to be down-regulated based on RNA-seq data (Fig. 6E; Farooq et al. 2021), confirming that these enhancers are functional enhancers. We then asked if these enhancers indeed regulate the boundaries as well. Toward that, we observed down-regulation of sense and antisense boundary RNAs at the B1 and B2 CTCF sites upon E8, E12, or E17 deletion (Fig. 6F), which suggests that enhancers activate boundary transcription. To determine whether down-regulation might be caused by direct contact, we also performed circular chromatin conformation capture 4C-seq on an enhancer beside E8 enhancer. We observed strong interactions with other enhancers in the enhancer cluster, with CDKN2A and CDKN2B promoters and with the boundary region (Fig. 6G, right highlighted pink). Importantly, deletion of the E8 enhancer conspicuously reduced 4C-seq signal, confirming that the interaction between the enhancer cluster and distant B1 and B2 CTCF sites regulates boundary transcription (Fig. 6G). Although interactions between boundaries and enhancers have been previously reported (Sun et al. 2019) no regulatory relationship had been established, but our data show that apart from activating their target promoters, functional enhancers positively regulate the transcription at the boundaries by direct interactions.
Figure 6.
Active enhancers within a TAD directly regulate boundary eRNA transcription. (A) Boxplots displaying enrichment of H3K27ac in TADs with high, low, nontranscribed, and random boundaries. (B) Boxplot showing number of H3K27ac peaks in TADs with various boundaries. (C) Levels of Pol II in TADs with different boundaries. (D) UCSC browser shot showing H3K27ac, EP300, Pol II ChIP-seq, and mNET-seq on the INK4a/ARF TAD. The tracks are overlaid by the gene annotations. The highlighted regions represent the enhancers (E8, E12, and E17) that were deleted. (E) Browser shot shows RNA-seq at INK4a/ARF TAD in WT and E8 enhancer delete lines. (F) qRT-PCR shows levels of sense and antisense boundary RNA arising from B1 and B2 CTCF sites upon E8, E12, and E17 enhancer deletions. (G) 4C-seq plot in WT and in E8 deletion lines on an enhancer beside E8 enhancer as viewpoint (yellow highlight), showing interactions with promoters and 3′ boundary region (pink highlight). Below are Pol II, H3K27ac ChIP-seq tracks, and gene annotations. Error bars denote SEM from three biological replicates. P-values were calculated by the Student's two-tailed unpaired t-test in F. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, (*) P < 0.05. P-values in boxplots were calculated by the Wilcoxon rank-sum test. The boxplots depict the minimum (Q1 – 1.5 × IQR), first quartile, median, third quartile, and maximum (Q3 + 1.5 × IQR) without outliers.
To ascertain whether high levels of boundary RNAs enhance insulation of the boundary in vivo, we interrogated the INK4a/ARF TAD upon senescence, where TAD transcription increases owing to its cell cycle regulatory role. Upon senescence, bidirectional eRNA expression increases several-fold at the enhancer cluster (Supplemental Fig. S10A, blue highlighted region), and INK4a/ARF levels and boundary RNA expression also increase (Supplemental Fig. S10A, pink highlighted region). We validated this increase in gene transcription, boundary RNA expression, and CTCF enrichment upon X-ray-induced senescence (nonreplicative senescence) (Supplemental Fig. S10B–D), and also observed elevated INK4A and CDKN1A (also known as P21) protein levels (Supplemental Fig. S10E). Using Hi-C data, we observed an increase in intra-TAD interactions at this TAD in senescent cells compared with proliferative cells (Supplemental Fig. S10F). Together these data suggest that increase in TAD enhancer activity accompanies increase in their interactions with boundaries to induce boundary RNA expression, which in turn recruits more CTCF to enhance insulation.
Boundary eRNAs strengthen TAD insulation
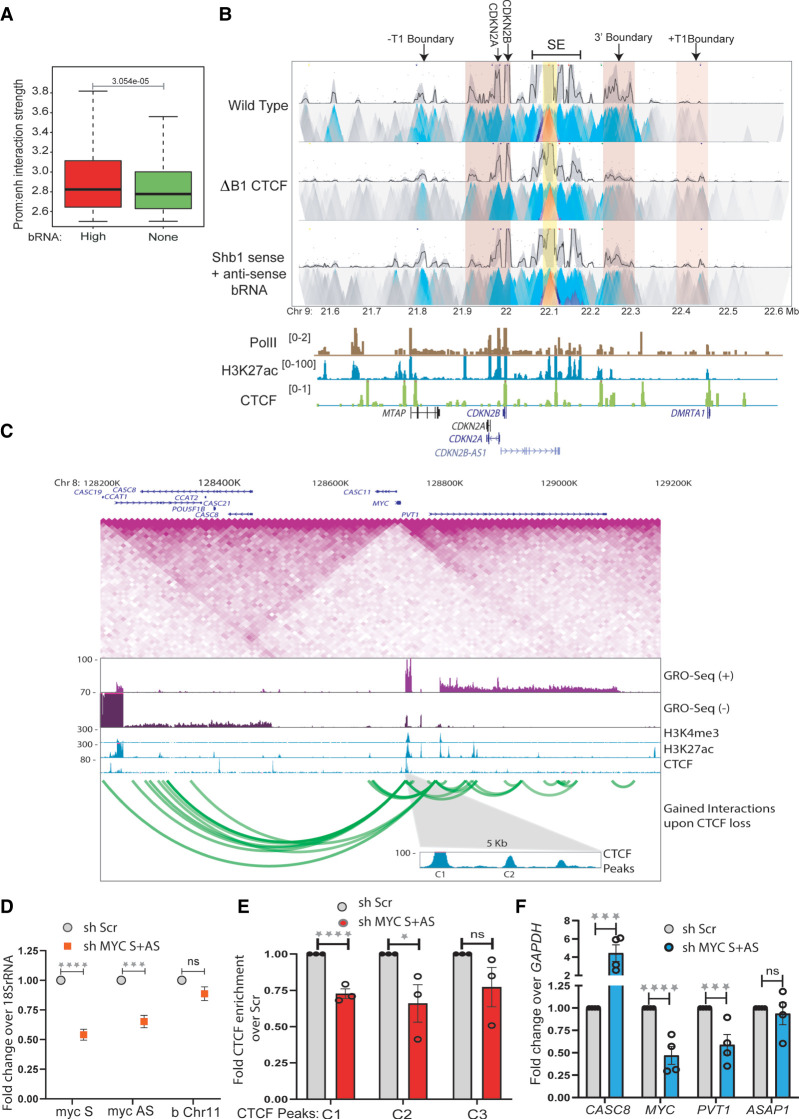
We noticed that TADs with transcribed boundaries showed higher strength of enhancer/promoter interactions than TADs with nontranscribed boundaries genome-wide (Fig. 7A). To test whether the boundary function of the INK4a/ARF TAD affects enhancer cluster/promoter interactions in a boundary RNA dependent manner, we performed 4C-seq assays on an enhancer beside E8 enhancer upon knockdown of sense and antisense boundary RNAs at the B1 CTCF site. As expected, we observed loss of boundary/enhancer interactions, but in addition, we observed weakening of the INK4a/ARF enhancer/promoter interactions (Fig. 7B). Knockdown of B1 RNAs also resulted in increased interaction with the +T1 boundary of the neighboring TAD (Fig. 7B), which also showed a gain in CTCF binding (Fig. 2E). To confirm that boundary RNA loss was accompanied by reduced CTCF binding, we performed 4C-seq at the E8 enhancer using cells with a B1 CTCF site deletion. We again observed a loss of enhancer/promoter interaction and a concomitant increase in the interaction with the +T1 boundary of the neighboring TAD (Fig. 7B).
Figure 7.
Boundary eRNAs strengthen TAD insulation. (A) Enhancer–promoter interaction strength within TADs with high and nontranscribed boundaries. (B) 4C-seq plot with the enhancer beside E8 enhancer as viewpoint (yellow highlight) in WT and upon B1 CTCF site deletion (ΔB1) and knockdown of B1 sense and antisense boundary RNAs. The anchored region shows interactions with promoters and the 3′ and +T1 boundaries (pink highlights). Below are Pol II, H3K27ac, and CTCF ChIP-seq tracks and gene annotations. (C) TAD structure at MYC locus on 8q24 region. The TAD diagram is overlaid by the position of CTCF, H3K4me3, H3K27ac, GRO-seq tracks, and gene annotations. The highlighted region shows the C1 and C2 CTCF sites on the 5′ boundary. The green looping tracks show the gain of interactions upon global CTCF knockdown. (D) qRT-PCRs showing the levels of sense and antisense RNA from MYC gene upon their shRNA-mediated knockdowns in a chromatin-associated RNA fraction. The level of RNA at Chr 11 boundary does not change. (E) CTCF ChIP enrichment before and after shRNA-mediated knockdown of sense and antisense MYC RNA at CTCF sites (C1, C2 at the 5′ TAD boundary, and C3 at upstream TAD boundary). (F) qRT-PCR showing the changes in expression of different genes in and around the MYC TAD upon shRNA-mediated knockdown of sense and antisense MYC RNA. Error bars denote SEM from three/four biological replicates. P-values were calculated by the Student's two-tailed unpaired t-test in D–F. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, (ns) P > 0.05.
To test if genic RNA transcribed at genic boundaries would also affect the CTCF binding at the boundary, we chose 8q24 locus, which has the MYC and PVT1 genes in the same TAD and several lncRNA genes in the adjacent TAD. The TAD harbors many enhancers that are distributed around the PVT1 and on its gene body in HeLa cells (Fig. 7C). However, the enhancers are fewer compared with the enhancer cluster at INK4a/ARF locus. The 5′ boundary of this TAD is around MYC gene (Fig. 7C). The promoter proximal regions and part of MYC gene overlap with three CTCF peaks that are conserved in several cell types and have been shown to act as a boundary (Fig. 7C; Supplemental Fig. S11A; Gombert and Krumm 2009; Schuijers et al. 2018). We depleted the sense and antisense RNA transcribing from MYC gene using pool of shRNAs and confirmed the knockdown from the chromatin and cellular fractions (Fig. 7D; Supplemental Fig. S11B). We observed, similar to INK4a/ARF TAD, reduced CTCF enrichment on C1 and C2 sites around MYC gene, whereas CTCF was unchanged at the 3′ boundary (C3) of downstream TAD (Fig. 7E). To rule out the effect of MYC RNA down-regulation on overall CTCF protein levels, we performed immunoblotting and found that CTCF levels do not change under these perturbations (Supplemental Fig. S11C).
Similar to INK4a/ARF expression, the sense and antisense RNA knockdown from MYC gene caused the down regulation of PVT1 in the same TAD, whereas the ASAP1 gene in downstream TAD was unaffected (Fig. 7F). Such dependencies of MYC and PVT1 expression on the affected CTCF binding have been reported previously (Gombert and Krumm 2009; Schuijers et al. 2018).
Further, we observed the increased interaction of enhancers in MYC TAD with regions in adjacent TAD (Fig. 7C, green looping tracks) upon CTCF knockdown, suggesting boundary functions were affected (Thiecke et al. 2020). We hypothesized, if CTCF binding was affected at the boundary owing to boundary RNA knockdown and if it had any effects on TAD boundary strength, the expression of genes in adjacent TAD should also be affected. Toward this, from GRO-seq, only the CASC8 and CASC19 genes transcribe in the corresponding TAD in HeLa cells (Fig. 7C); we tested the levels of CASC8 and found it to be up-regulated upon knockdown of MYC sense and antisense RNA, whereas the expression of ASAP1 gene in distal TAD remained unchanged (Fig. 7F).
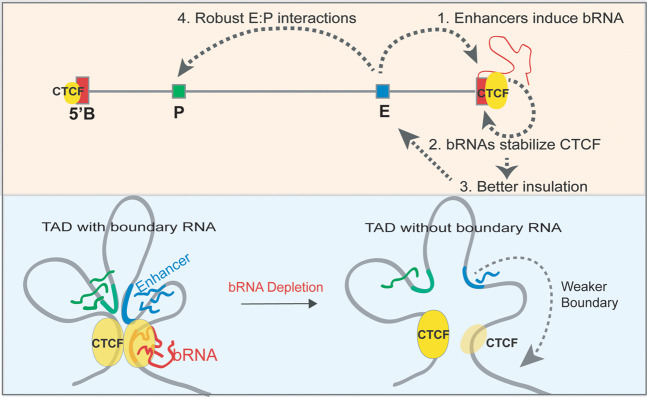
Together, these observations show that loss of a boundary RNA reduces CTCF binding and weakens insulation, resulting in aberrant enhancer interaction with an element outside of the TAD. We conclude that boundary RNAs are functional components of TAD boundaries (Fig. 8).
Figure 8.
Model. (1) An active enhancer physically interacts with a boundary and activates boundary RNA transcription. (2) The boundary RNA in return stabilizes the CTCF at these boundaries, thereby (3) strengthening insulation of these TADs. (4) This favors intra-TAD enhancer–promoter interactions to facilitate robust gene transcription. The loss of boundary RNA/enhancer deletion reduces the boundary RNA levels, which triggers the loss of CTCF and insulation of TADs. Enhancer–promoter interactions weaken in these scenarios, which causes concomitant loss of gene transcription.
Discussion
Interactions between CTCF and RNA have been implicated in genome organization (Hansen et al. 2019; Saldaña-Meyer et al. 2019), but the exact role of these interactions remains unknown. Here we show that eRNAs transcribed from TAD boundaries function in CTCF binding, TAD insulation, and gene regulation. Our data provide direct evidence that boundary eRNAs create an insulated neighborhood by associating with CTCF-bound sites and facilitating enhancer–promoter interactions for robust gene activation within a given TAD. We found that the transcriptional activity of promoters present within the TAD does not regulate nongenic boundary transcription. Using a series of enhancer and CTCF site deletions at the INK4a/ARF TAD, we have shown that enhancers interact with and positively regulate the expression of both genic and nongenic TAD boundary RNAs. Boundary RNAs, in turn, increase CTCF binding at TAD boundaries, strengthening weak CTCF motifs and increasing insulation. Transcription of boundary RNAs at the INK4a/ARF and MYC TAD is regulated by the enhancers within the TAD to facilitate CTCF binding and increased insulation at the boundary. This may explain why enhancer cluster–containing TADs show stronger boundaries (Gong et al. 2018; Matthews and Waxman 2018; Sun et al. 2019).
Boundary RNAs resemble eRNAs at active enhancers in being bidirectional (Li et al. 2013, 2016), and, as our study shows, making direct contact with distant regulatory elements. Popular chromosome conformation capture assays are strictly pairwise and cannot determine if enhancer elements activate promoters and boundary elements independently or together. However, more recently introduced 3D contact methods, including SPRITE (Quinodoz et al. 2018) and GAM (Beagrie et al. 2017), are able to capture multiple simultaneous contacts and, in the future, may be applied to understand how enhancers regulate both gene promoter activity and transcription at boundaries to strengthen TADs.
Furthermore, in the absence of the RNA-binding domain of CTCF, cells show a decrease in the total number of TADs and an increase in the size of individual TADs owing to reduced insulation between neighboring TADs (Hansen et al. 2019). A clear mechanistic basis for this observation comes from our findings in which TAD enhancer–mediated CTCF–RNA interactions maintain strong TAD boundaries by increasing insulation between neighboring TADs. Thus, the inability of CTCF mutants to interact with RNA will compromise CTCF enrichment and its insulating function at TAD boundaries, leading to the merging of adjacent TADs into larger TADs. In such cases, less CTCF at the boundary might allow the cohesin ring to slip to the next TAD, thereby compromising the insulation (Hansen 2020; Kentepozidou et al. 2020). Thus, a positive feedback loop is established whereby enhancer clusters maintain high specificity for promoters within the TAD by also activating boundary transcription and preventing interactions with neighboring promoters.
Further, gene expression phenotypes would depend largely on the local 3D chromatin topology, chromatin state, and the presence and location of enhancers within the TAD. Hence, upon boundary perturbations in the TAD of interest, the neighboring genes could display a whole range of dysregulation.
TAD boundaries are enriched for H3K27ac and Pol II, suggesting the presence of active regulatory elements and transcription (Gong et al. 2018; Matthews and Waxman 2018; Sun et al. 2019; Hsieh et al. 2020). However, activation of transcription at the boundary alone is not sufficient to maintain a TAD boundary (Bonev et al. 2017; Hug et al. 2017). Our study shows that the products of the TAD boundary transcription, that is, boundary RNAs, are required to strengthen TAD insulation by increasing CTCF occupancy. Transcription-assisted TAD boundary strengthening may also be a factor in zygotic activation–dependent boundary formation in which early transcribing genes serve as the nucleation sites for TAD boundaries (Hug et al. 2017). Similarly, transcription and the resulting RNA may be involved in forming distinct TADs boundaries around transcribed escape genes on the inactive chromosome during X Chromosome inactivation (Giorgetti et al. 2016). The study aims to understand the role of RNA at the chromatin domain boundaries and enhancers in CTCF recruitment and gene regulation.
Methods
Cell culture
HeLa cells were obtained from ATCC and were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The cells were maintained at 37°C in the presence of 5% CO2 in a humidified incubator.
Chromatin immunoprecipitation
Cells were cross-linked with 1% formaldehyde for 10 min at room temperature with constant shaking. Glycine was added to a final concentration of 125 mM to quench the formaldehyde. Cells were washed three times with 1× ice-cold PBS and then were scraped and pelleted-down in 1× PBS at 4°C. Cells were gently resuspended in nuclear lysis buffer (50 mM Tris-HCl at pH 7.4, 1% SDS, 10 mM EDTA at pH 8.0) supplemented with 1× PIC and incubated on ice for 10 min. The lysate was subjected to fragmentation using a Diagenode Bioruptor pico for 20 cycles (30 sec on and 30 sec off) to generate fragments of ∼500 bp. The cell lysate was cleared at 12,000 rpm for 12 min. For each IP, 100 µg of sheared chromatin was used. The lysates were diluted by adding dilution buffer (DB) (20 mM Tris-HCl at pH 7.4, 100 mM NaCl, 2 mM EDTA at pH 8.0, 0.5% Triton X-100) supplemented with 1× PIC in a 1:1.5 ratio (1 volume of sheared chromatin and 1.5 volumes of DB). One microgram of antibody was added to immunoprecipitate the DNA and was incubated on a rocking platform overnight at 4°C. Fifteen microliters of preblocked (with 1% BSA) Protein G dynabeads (Invitrogen 10004D) was added to the tubes and rotated for 4 h at 4°C. Beads were collected, flow-through was discarded, and the samples were washed with wash buffer I (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.1% SDS, 2 mM EDTA at pH 8.0, 1% Triton X-100) at 4°C on a rocking platform. Washes were sequentially repeated with wash buffer II (20 mM Tris-HCl at pH 7.4, 500 mM NaCl, 2 mM EDTA at pH 8.0, 1% Triton X-100), wash buffer III (10 mM Tris-HCl at pH 7.4, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA at pH 8.0) and 1× TE (10 mM Tris-HCL at pH 8.0, 1 mM EDTA at pH 8.0). Chromatin was eluted with 200 µL of elution buffer (100 mM NaHCO3, 1% SDS) for 30 min at 37°C in a thermomixer. The supernatant was collected in separate tubes, and 14 μL NaCl (5 M) was added to eluted samples and kept overnight at 65°C for de-cross-linking. Immunoprecipitated DNA was purified by phenol:chloroform:isoamyl alcohol (PCI), followed by ethanol precipitation. The final air-dried DNA pellet was dissolved in 100 µL of 1× TE. These samples were then used for qRT-PCRs using SYBR green. Mean values for all the regions analyzed in different conditions were expressed as fold-enrichment compared over beads. Statistically significant differences between different conditions were computed using a t-test (P-value < 0.05); ChIP primers are listed in Supplemental Table S1.
RNA isolation and cDNA synthesis
Cells were lysed in 1 mL of TRIzol (Thermo Fisher Scientific). To each sample, 200 µL chloroform was added; they were briefly vortexed and centrifuged at 12,000 rpm for 12 min. The aqueous phase was carefully collected and transferred to the fresh tube. One volume of isopropanol was added to the sample and incubated at room temperature for 10 min to precipitate the RNA. The samples were centrifuged at 12,000 rpm for 12 min, and supernatants were discarded without disturbing the pellet. The pellet was washed with 75% ethanol. The pellet was air-dried and dissolved in RNase-free water. The RNA was treated with ezDNase (Invitrogen) to remove the traces of contaminating DNA. One microgram of RNA was used for each cDNA synthesis reaction by SuperScript IV (Invitrogen) and random hexamers as per the manufacturer's recommendation. The CFX96 touch (Bio-Rad) real-time PCR instrument was used for qRT-PCRs. qRT-PCRs were performed using three technical replicates for each sample. Fold change was calculated by the ΔΔCt method, and individual expression data were normalized to GAPDH mRNA. The P-values were calculated by the Student's unpaired two-tailed t-test. qRT-PCR primers are listed in Supplemental Table S1.
CRISPR-Cas9-mediated deletion
gRNAs were designed with the crispr.mit.edu tool. gRNAs were selected based on the highest score and the least number of off-targets. gRNAs were cloned in a pgRNA humanized vector (Addgene 44248, a gift from the Stanley Qi laboratory) between BstX1 and Xho1 restriction sites. gRNAs were cotransduced with a lenti-Cas9 vector (Addgene 52962, a gift from the Zhang laboratory). Cells were placed under selection using puromycin (3 µg/mL) for 48 h. Single cells were seeded in a 96-well plate. Wells with single cells were marked and allowed to grow until they formed colonies. The cells were trypsinized and shifted to 48-well plates and allowed to grow to confluence, and then half of the cells were taken for the surveyor assay and the other half was plated again. Surveyor assays were performed using a PCR-based method using primers listed in the Supplemental Table S1.
CRISPRi gRNA design and cloning
We used GPP sgRNA Designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) to design the gRNAs against TSSs of MTAP, CDKN2A, and CDKN2B in the INK4a/ARF TAD. For each gene, two gRNAs were designed. The gRNAs were cloned in a customized PX459 vector (pSpCas9(BB)-2A-Puro V2.0, Addgene 62988, a gift from the Feng Zhang laboratory). The Cas9 enzyme cassette was removed from PX459 by digestion with XbaI and NotI, followed by blunt-end ligation. The cloning of the gRNAs in the customized PX459 plasmid was performed as per the Zhang laboratory's general cloning protocol. The dCas9 plasmid (Addgene 99372, a gift from Kristen Brennand), together with gRNA specific to targeted genes, was transfected in 70% confluent cells. Transfections were performed using Lipofectamine 2000 (Invitrogen 11668027). CRISPRi cells were harvested for ChIP and gene expression analysis.
shRNA design and transfection
We used an shRNA design tool (https://portals.broadinstitute.org/gpp/public/seq/search) to design the shRNAs against RNA at B1 and B2 CTCF sites within the INK4a/ARF TAD boundary. The shRNAs were cloned in a customized pLKO.1 puro, (Addgene 8453, a gift from Bob Weinberg). The cloning of the shRNAs in the pLKO.1 plasmid was performed as per the Addgene's pLKO.1 cloning protocol. shRNA-specific plasmid together with lentiviral packaging plasmids like VSVG (a gift from Bob Weinberg, Addgene 8454) and PAX2 (a gift from Didier Trono, Addgene 12260) was cotransfected using Lipofectamine 2000 (Invitrogen 11668027). Cells were selected by puromycin (3 µg/mL; Gibco A11138-03), and knockdowns were confirmed using qPCR oligos listed in Supplemental Table S1.
siRNA transfection
SMARTpools was used to design scramble (D-001810-10-05), and CTCF siRNAs (L-020165-00-0005) were purchased from GE Dharmacon. Transfections were performed using Lipofectamine 2000 (Invitrogen 11668027).
Lentiviral transduction
HEK293FT cells were grown in poly-D-lysine-coated culture dishes. These cells were cotransfected with lentiviral packaging plasmids like VSVG (a gift from Bob Weinberg, Addgene 8454) and PAX2 (a gift from Didier Trono, Addgene 12260) along with the plasmid of interest using Lipofectamine 2000 (Invitrogen 11668027). The medium was changed after 6 h. The viral supernatant was collected after 48 h and 72 h, pooled together, filtered with a 0.44-µm syringe filter, and then finally added to cells along with 8 µg/mL of polybrene. Transduction was stopped after 16 h.
RNA immunoprecipitation
For RNA, primers were designed to amplify desired genomic regions that correspond to peaks of boundary RNA (GRO-seq) at the B1 and B2 CTCF sites of the INK4a/ARF TAD boundary. The amplified products were cloned into the pcDNA3 BoxB plasmid (a gift from Howard Chang, Addgene 29729). All clones were confirmed by Sanger sequencing and subsequently used for RNA synthesis using T7 RNA polymerase (Promega P207e) and biotin RNA labeling mix (Roche 11685597910). Primer sequences are listed in Supplemental Table S1.
RNA immunoprecipitation was adopted from Jayani et al. (2017), with some modifications. HeLa cells were grown to 90%–95% confluency and were harvested by scraping and washed twice with ice-cold 1× PBS to gently resuspend in 2 mL of PBS. A volume of 2 mL of nuclear isolation buffer (NIB; 40 mM Tris–HCl at pH 7.5, 20 mM MgCl2, 1.28 M sucrose, 4% Triton X-100, 1 mM PMSF, protease inhibitors, and 20 U/mL SUPERase inhibitor; Thermo Fisher Scientific AM2694) was added, and the pellet was gently resuspended. Then, 6 mL of distilled water was added and kept on ice for 20 min with intermittent gentle shaking. Nuclei were pelleted by centrifugation at 2500g for 5 min at 4°C. The pellet was resuspended in 1 mL of RIP buffer (25 mM Tri–HCl at pH 7.4, 150 mM KCl, 0.5 mM DTT, 0.5% NP-40, 1 mM PMSF, protease inhibitors, and 20 U/mL SUPERase inhibitor) and incubated on ice for 5 min. Nuclei were sheared by 10 cycles of sonication (30 sec on and 30 sec off) in a Bioruptor (Diagenode) followed by centrifugation at 12,000 rpm for 10 min at 4°C. One microgram of biotinylated RNA was incubated with 20 μL of RNA structure buffer (RSB; 10 mM Tris–HCl at pH 7.0, 100 mM KCl, 10 mM MgCl2, 1 mM PMSF, protease inhibitors, and 20 U/mL SUPERase inhibitor) for 5 min at room temperature. Folded RNA was mixed with 1 mg of nuclear extract in 500 µL of RIP buffer and rotated for 1 h at 4°C. Fifteen microliters of Dynabeads MyOne streptavidin beads were added, and rotation continued for one more hour. Samples were washed three times with RIP buffer, and beads with proteins were boiled in 2×SDS for 10 min. Immunoblotting for CTCF was performed using CTCF antibody (Cell Signaling Technology 3418).
UV RNA immunoprecipitation
Ultraviolet RNA immunoprecipitation (RIP) was performed as previously described (Rahnamoun et al. 2018) with some modifications. HeLa cells at a confluency of 80%–90% were cross-linked by UV irradiation in a Stratalinker UV cross-linker. Cells were washed three times with cold 1 × PBS and scraped in PBS before subjecting to centrifugation at 2500 rpm. Pellets were lysed in RIP lysis buffer (25 mM HEPES-KOH at pH 7.5, 150 mM KCl, 0.5% NP-40, 1.5 mM MgCl2, 10% glycerol, 1 mM EDTA, 0.4 U RNase inhibitor; Invitrogen 18091050), and 1× protease inhibitor cocktail (PIC) on ice for 30 min. Cleared cell lysates were subjected to IP with CTCF antibody-bound Protein G dynabeads (Invitrogen 10004D) overnight. Samples were subsequently washed three times with RIP lysis buffer, and RNA samples were eluted using TRIzol reagent. Before cDNA synthesis, RNA was treated with RNase-free DNase (Qiagen 79256) to remove the traces of contaminating DNA. cDNA was prepared using random hexamers by SuperScript IV (Invitrogen 18091050) as per the manufacturer's recommendation and was analyzed by PCR primers listed in Supplemental Table S1 and were run on 1% agarose gel.
Cell fractionation
Cell fractionation and RNase A treatment of nuclei was adapted from the method of Gagnon et al. (2014). Briefly, cells were washed three times with cold 1 × PBS and scraped in 1 mL of PBS and pelleted-down at 2500 rpm for 5 min at 4°C. Pellets were then resuspended in hypotonic lysis buffer (HLB; 10 mM Tris HCl at pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.3% NP-40, 10% glycerol, 1 × PIC); the mix was incubated on ice for 10 min and then centrifuged at 800g for 8 min at 4°C. The supernatant was transferred to a new tube and marked as cytoplasmic fraction. The nuclei pellet after centrifugation was washed twice with HLB and centrifuged at 200g for 2 min at 4°C. The pellet was then resuspended in 700 µL of ice-cold modified Wuarin–Schiebler buffer (MWS; 10 mM Tris-HCl at pH 7.5, 300 mM NaCl, 4 mM EDTA, 1 M urea, 1% NP-40, 1% glycerol, 1 × PIC) with or without 0.1 µL of 100 mg/mL RNase A (Qiagen 19101) for 15 min on ice, briefly tapped few times during incubation time, and then centrifuged at 1000g for 5 min at 4°C. The supernatant was collected as a nucleoplasmic fraction. The chromatin pellet was washed twice with MWS buffer and centrifuged at 500g for 3 min at 4°C. The pellet was then dissolved in 500 µL of nuclear lysis buffer (20 mM Tris HCl at pH 7.5, 150 mM KCl, 3 mM MgCl2, 0.3% NP-40, 10% glycerol, 1 × PIC) and sonicated for 10 cycles (30 sec on, 30 sec off) in a Bioruptor (Diagenode). SDS loading dye was added, and the samples were boiled before immunoblotting.
Chromatin-associated RNA isolation
Chromatin-associated RNA isolation was adapted from the method of Conrad and Ørom (2017). Trypsinized cells were collected in 1.5-mL tubes and washed twice with 1 × PBS and pelleted-down at 200g for 2 min. To each pellet was added 400 µL of cell lysis buffer (10 mM Tris HCl at pH 7.4, 150 mM NaCl, 0.15% Igepal), gently pipetted three to five times, and then incubated for 5 min on ice. To the Lo-bind 1.5-mL tube was added 1 mL (2.5 volume) of cold sucrose buffer (10 mM Tris HCl at pH 7.4, 150 mM NaCl, 24% sucrose), which was gently overlaid with the cell lysate followed by centrifugation at 3500g for 5 min to collect the nuclear pellet. Isolated nuclei were rinsed with 1 mL of ice-cold 1 × PBS-EDTA solution followed by a short spin at 3500g. The nuclei were resuspended in 250 µL of glycerol buffer (20 mM Tris HCl at pH 7.4, 75 mM NaCl, 0.5 mM EDTA at pH 8.0, 50% glycerol), and then immediately 250 µL of urea buffer (10 mM Tris HCl at pH 7.4, 1 M Urea, 0.3 M NaCl, 7.5 mM MgCl2, 0.2 mM EDTA, 1% Igepal) was added; they were mixed by vortexing for 4 sec and then incubated on ice for 2 min. Chromatin was centrifuged at 13000g for 2 min to collect the chromatin–RNA complex. The chromatin–RNA pellet was washed with 1 × PBS-EDTA once, and RNA was isolated using TRIzol reagent. The RNA obtained was treated with ezDNase (Ambion) to remove the traces of contaminating DNA. One microgram of RNA was used for each cDNA synthesis reaction by SuperScript IV (Invitrogen) and random hexamers as per the manufacturer's recommendation. The CFX96 touch (Bio-Rad) real-time PCR was used for qRT-PCRs. qRT-PCRs were performed using three technical/biological replicates for each sample. Fold changes were calculated by the ΔΔCt method, and individual expression data were normalized to pre c-MYC. The P-values were calculated by the Student's unpaired two-tailed t-test. qRT-PCR primers are listed in Supplemental Table S1.
X-ray-induced senescence
BJ fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified incubator containing 5% CO2. Cells were first immortalized by expressing hTERT. Cells were then seeded at around 40% confluency. The day after seeding, a single dose of 10 Gy was delivered using a CIX3 cabinet x-ray irradiator (Xstrahl Life Sciences; 195 kV, 10 mA). Irradiated cells were not passaged, but the medium was changed on days 4, 8, 12, and 16, and cells were either harvested on day 10 or day 20 and used for RNA isolation or ChIP experiments, respectively. Senescence was confirmed by SA-β-gal staining (Cell Signaling Technology 9860) and CDKN1A (p21) expression (p21 Santa Cruz Biotechnology sc-397).
Antibodies
Antibodies used were CTCF (Cell Signaling Technologies, 3418 for immunoblotting and immunoprecipitation), DDX5 (Abcam ab-21696), CDKN2A/p16INK4a (Abcam ab-108349), p14ARF (Santa Cruz Biotechnology sc-53639), Pol II (Santa Cruz Biotechnology sc-55492), GAPDH (Santa Cruz Biotechnology sc-32233), and histone H3 (Sigma-Aldrich H0164).
ChIP-seq and CUT&RUN analysis
Sequenced reads were aligned to the hg19 assembly using default Bowtie 2 options (Langmead and Salzberg 2012). The aligned reads were filtered using SAMtools (Li et al. 2009). Tag directories were made from the aligned reads using the HOMER (Heinz et al. 2010; http://homer.ucsd.edu/homer/) makeTagDirectory command. A 200-bp sliding window was used to identify narrow peaks of transcription factors’ ChIP-seq and CTCF CUT&RUN. Artifacts from clonal amplification were neglected as only one tag from each unique genomic position was considered (−tbp 1). The threshold was set at a false discovery rate (FDR) of 0.001 determined by peak-finding using randomized tag positions in a genome with an effective size of 2 × 109 bp. For histone marks, seed regions were initially found using a peak size of 500 bp (FDR < 0.001) to identify enriched loci. Enriched regions separated by <1 kb were merged and considered as blocks of variable lengths. The HOMER (Heinz et al. 2010) makeUCSCfile command was used to generate bedGraph files of the read densities across the genome, and this track was uploaded to the UCSC Genome Browser (Kent et al. 2002). The tag directories generated were used to quantify signals at various regions of interest using annotatePeaks.pl. Heatmaps and profile plots were made using deepTools (Ramírez et al. 2016). The data sets used in the study are listed in Supplemental Table S2.
GRO-seq analysis
The sequenced reads were aligned to the hg19 assembly using default Bowtie 2 options (Langmead and Salzberg 2012). Tag directories were made from the aligned reads using the HOMER (Heinz et al. 2010) makeTagDirectory command. Tag directories were used to quantify signal and gene expression. The HOMER (Heinz et al. 2010) makeUCSCfile command was used to generate strand-specific bedGraph files of the read densities across the genome, and this track was uploaded to the UCSC Genome Browser (Kent et al. 2002).
TADs and loop domain calling
Raw paired-end reads were trimmed and individually mapped to the hg19 assembly using Bowtie 2 (Langmead and Salzberg 2012). The HOMER (Heinz et al. 2010) makeTagDirectory was first used to create tag directories with tbp 1 option. Data sets were further processed by HOMER (Heinz et al. 2010) in order to remove small fragment and self-ligations using makeTagDirectory with the following options: -removePEbg -removeSpikes 10000 5. Next, findTADsAndLoops.pl was used to obtain overlapping TADs (TADs and sub-TADs), produced at 5-kb resolution with 10-kb windows. This program was also used to generate two bedGraph files that describe the directionality index and the insulation score. tagDir2hicFile.pl was used to generate HIC files. The WashU (Li et al. 2019) epigenome browser was used to visualize the Hi-C data. Boundaries of the TADs were defined as 5-kb or 20-kb regions centered at the end of the TADs called. HiCCUPS (Rao et al. 2014; Durand et al. 2016) was used to obtain loop anchors from the HIC file at a 5-kb resolution. HOMER annotatePeaks.pl was used to quantify signal from tagDirectories, count peaks, quantify insulation, and annotate features at the TAD boundaries. Nongenic TAD boundaries were identified as those that did not intersect with an annotated gene body. GRO-seq signal was used to rank the boundaries and to then select 10% of unique boundaries in each category (high transcription, low transcription, no transcription, and random). The HIC file was converted to COOL. The file was then used to plot the averaged insulation score valleys at 5-kb resolution and 100 kb around the boundary using coolpup.py (Flyamer et al. 2020).
Out of the 6000 boundaries considered for the transcription-based classification analysis, only 19 (0.3%) regions were not mapped on hg38 when converted from hg19. Given the large number of regions spanning the genome and also a trend with varying levels, there would be negligible effects of a mapping to a different genome build.
Pile-up plots
The pile-up (aggregated normalized Hi-C interactions) plot cantered at high transcribed, low transcribed, nontranscribed, and random boundaries at a 5-kb resolution. The 100-kb distances are taken from the boundary region. The plot focuses on near-diagonal features. The normalized interaction frequency (from the Hi-C matrix), 100 kb around the boundary of interest, is averaged over the multiple boundary classes laid on top of each other to visualize insulation strength. The pile-ups are performed on the regions that lie on the main diagonal of the Hi-C map.
NET-CAGE and CAGE analysis
NET-CAGE and CAGE sequenced reads were aligned to hg19 using Bowtie 2 (Langmead and Salzberg 2012). The HOMER program makeTagDirectory was first used to create tag directories. findPeaks with “–style tss” was used to obtain the NET-CAGE TSSs. TSSs identified within 1 kb of each other on the same strand were merged. NET-CAGE identified TSSs intersecting the RefSeq-annotated TSSs were called mRNA TSSs, and the rest were defined as eRNA TSSs. Those TSSs that fell in boundaries were identified as bmRNAs or beRNAs. A span of 2 kb around each identified TSS was used to quantify NET-CAGE and CAGE signal and to calculate the log2 fold difference between the two.
5′ Splice site and transcription termination site sequence analysis
To identify the 5′ SS motif, HOMER de novo motif analysis was performed on the 500-bp region after the RefSeq-annotated TSSs with other H3K27ac 500-bp regions as background. The top hit was observed to be the splice acceptor site. The “AATAAA” motif was used as the 3′ poly(A) sequence. HOMER scanMotifGenomeWide was used to identify “predicted” splice sites and poly(A) terminator sites using previously identified motifs. The occurrence of predicted sites was plotted at the previously identified TSSs.
4C-seq
4C was performed as per the protocol described by van de Werken et al. (2012) with minor modifications. HeLa cells were fixed with fresh formaldehyde (1.5%) and quenched with glycine (125 mM) followed by washes with ice-cold PBS (2×) and were scraped, pelleted, and stored at −80°C. Lysis buffer (10 mM Tris-Cl pH 8.0, 10 mM NaCl, 0.2% NP-40, 1× PIC) was added to the pellets and homogenized with a Dounce homogenizer (20 stroked with pestle A followed by pestle B). The 3C digestion was performed with HindIII (400 units, NEB), and ligation was performed using the T4 DNA ligase and 7.61 mL ligation mix (745 μL 10% Triton X-100, 745 μL 10× ligation buffer [500 mM Tris-HCl at pH 7.5, 100 mM MgCl2, 100 mM DTT], 80 μL 10 mg/mL BSA, 80 μL 100 mM ATP, and 5.96 mL water). The ligated samples were then purified by PCI extraction and subjected to ethanol precipitation, and the pellet was eluted in TE (pH 8.0) to obtain the 3C library. The 4C digestion was performed using 50 units DpnII (NEB), and the samples were ligated, purified, and precipitated similar to the 3C library to obtain the 4C library. The 4C library was subjected to RNase A treatment and purified by the QIAquick PCR purification kit. The concentration of the library was then measured by NanoDrop and subjected to PCR using the oligos for the enhancer viewpoint. The samples were next PCR-purified using the same kit and subjected to Illumina HiSeq 2500 sequencing using 50-bp single-end reads.
4C method and analysis
The sequenced reads were aligned to the hg19 assembly as described in R.4Cker (Raviram et al. 2016; https://github.com/rr1859/R.4Cker) with Bowtie 2 (Langmead and Salzberg 2012). Data analysis was further performed using 4C-ker with default parameters. 4Cseqpipe (Van de Werken et al. 2012; http://compgenomics.weizmann.ac.il/tanay/?page_id=367) was also used to process the sequenced data. 4C-seq images were generated using truncated mean at a 10-kb resolution.
Motif analysis
annotatePeaks.pl was used with –hist option with the CTCF motif probability matrix to plot the motif occurrence in the peaks.
Promoter capture analysis
Processed CHiCAGO data were obtained fro m Thiecke et al. (2020) (https://osf.io/brzuc/). The BEDTools suite was used to obtain interactions of interest and perform further analysis (Quinlan and Hall 2010). The promoter interacting region of the BEDPE files of PCHi-C interactions was then intersected with boundaries to obtain promoters that interact with boundaries. The strength of the interactions obtained from the processed CHiCAGO data was plotted for interactions from boundary interacting promoters to enhancers (H3K27ac regions).
RNA-seq analysis
The raw reads were mapped to the hg19 assembly using HISAT2 (Kim et al. 2015) in a strand-specific manner. Tag directories were made from the aligned reads using the HOMER makeTagDirectory command. The HOMER makeUCSCfile command was used to generate strand-specific bedGraph files of the read densities across the genome, and this track was uploaded to the UCSC Genome Browser (Kent et al. 2002).
Supplementary Material
Acknowledgments
We thank D.N. laboratory members, Ranveer Jayani and Amanjot Singh, for the discussions. We acknowledge support of the Department of Atomic Energy, Government of India, under project no. 12-R&D-TFR-5.04-0800 and intramural funds from National Center for Biological Sciences–Tata Institute of Fundamental Research (NCBS-TIFR) (to D.N.). D.N. is an EMBO Global Investigator. We also acknowledge the funding support from Wellcome-IA (IA/1/14/2/501539) and Department of Science and Technology core grant (CRG/2019/005714) to D.N. U.F. is supported by the Council of Scientific and Industrial Research (CSIR, Government of India) senior research fellowship. K.W. and A.K.S. are supported by the TIFR-NCBS graduate program. K.W. is also supported by the CSIR-SPM (Government of India) fellowship. The genome sequence described/used in this research was derived from a HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We thank Henrietta Lacks, now deceased, and her surviving family members for their contributions to biomedical research. This study was reviewed by the National Institutes of Health HeLa Genome Data Access Working Group.
Author contributions: The study was conceptualized and designed by Z.I., B.S., and D.N. Most experiments were performed by Z.I. with help from U.F. and K.W. B.S. performed NGS data analysis. A.P. performed NGS library preparation and sequencing. A.K.S. and R.S. assisted in data analysis. D.N., Z.I., B.S., and S.H. wrote the manuscript with input from J.T. All authors read and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276643.122.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof, I, Bornholdt, J, Boyd, M, Chen, Y, Zhao, X, Schmidl, C, Suzuki, T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Blencowe BJ, Rinn JL. 2019. Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep 20: e48068. 10.15252/embr.201948068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan JA, Phillips-Cremin JE. 2020. On the existence and functionality of topologically associating domains. Nat Genet 52: 8–16. 10.1038/s41588-019-0561-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvy-Liivrand M, De Sande AH, Pölönen P, Mehtonen J, Vuorenmaa T, Niskanen H, Sinkkonen L, Kaikkonen MU, Heinäniemi M. 2017. Analysis of primary microRNA loci from nascent transcriptomes reveals regulatory domains governed by chromatin architecture. Nucleic Acids Res 45: 9837–9849. 10.1093/nar/gkx680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Ørom UA. 2017. Cellular fractionation and isolation of chromatin-associated RNA. Methods Mol Biol 1468: 1–9. 10.1007/978-1-4939-4035-6_1 [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M, et al. 2019. Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet 51: 1263–1271. 10.1038/s41588-019-0466-z [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq U, Saravanan B, Islam Z, Walavalkar K, Singh AK, Jayani RS, Meel S, Swaminathan S, Notani D. 2021. An interdependent network of functional enhancers regulates transcription and EZH2 loading at the INK4a/ARF locus. Cell Rep 34: 108898. 10.1016/j.celrep.2021.108898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Illingworth RS, Bickmore WA. 2020. Coolpup.py: versatile pile-up analysis of Hi-C data. Bioinformatics 36: 2980–2985. 10.1093/bioinformatics/btaa073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Janowski BA, Corey DR. 2014. Analysis of nuclear RNA interference in human cells by subcellular fractionation and argonaute loading. Nat Protoc 9: 2045–2060. 10.1038/nprot.2014.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombert WM, Krumm A. 2009. Targeted deletion of multiple CTCF-binding elements in the human C-MYC gene reveals a requirement for CTCF in C-MYC expression. PLoS One 4: e6109. 10.1371/journal.pone.0006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Lazaris C, Sakellaropoulos T, Lozano A, Kambadur P, Ntziachristos P, Aifantis I, Tsirigos A. 2018. Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nat Commun 5: 542. 10.1038/s41467-018-03017-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS. 2020. CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism. Nucleus 11: 132–148. 10.1080/19491034.2020.1782024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hsieh THS, Cattoglio C, Pustova I, Saldaña-Meyer R, Reinberg D, Darzacq X, Tjian R. 2019. Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol Cell 76: 395–411.e13. 10.1016/j.molcel.2019.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen LLP, Kassouf MT, Oudelaar AM, Biggs D, Preece C, Downes DJ, Gosden M, Sharpe JA, Sloane-Stanley JA, Hughes JR, et al. 2017. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol 19: 952–961. 10.1038/ncb3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, et al. 2011. 9p21 DNA variants associated with coronary artery disease impair interferon-γ 3 signalling response. Nature 470: 264–268. 10.1038/nature09753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin, YC, Laslo, P, Cheng, JX, Murre, C, Singh, H, Glass, CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol Cell 78: 539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Jayani RS, Singh A, Notani D. 2017. Isolation of nuclear RNA-associated protein complexes. Methods Mol Biol 1543: 187–193. 10.1007/978-1-4939-6716-2_9 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentepozidou E, Aitken SJ, Feig C, Stefflova K, Ibarra-Soria X, Odom DT, Roller M, Flicek P. 2020. Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol 21: 5. 10.1186/s13059-019-1894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury A, Achinger-Kawecka J, Bert SA, Smith GC, French HJ, Luu PL, Peters TJ, Du Q, Parry AJ, Valdes-Mora F, et al. 2020. Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nat Commun 11: 54. 10.1038/s41467-019-13753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetgen A, Thandapani P, Ntziachristos P, Ghebrechristos Y, Nomikou S, Lazaris C, Chen X, Hu H, Bakogianni S, Wang J, et al. 2020. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat Genet 52: 388–400. 10.1038/s41588-020-0602-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega I, Dean A. 2017. CTCF fences make good neighbours. Nat Cell Biol 19: 883–885. 10.1038/ncb3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498: 516–520. 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17: 207–223. 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- Li D, Hsu S, Purushotham D, Sears RL, Wang T. 2019. WashU Epigenome Browser update 2019. Nucleic Acids Res 47: W158–W165. 10.1093/nar/gkz348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Waxman DJ. 2018. Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. eLife 14: e34077. 10.7554/eLife.34077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Park D, Munger K. 2013. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc Natl Acad Sci 110: 16175–16180. 10.1073/pnas.1310432110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy Mazumdar M, Yin S, Paranjapye A, Kerschner JL, Swahn H, Ge A, Leir SH, Harris A. 2020. Looping of upstream cis-regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic Acids Res 48: 3513–3524. 10.1093/nar/gkaa089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347: 1017–1021. 10.1126/science.1262088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson KA, Lawson CK, Mullen NJ, Ball CB, Spector BM, Meier JL, Price DH. 2017. Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res 45: 11088–11105. 10.1093/nar/gkx724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115: E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauck A, Lener B, Hoell M, Kaiser A, Kaufmann AM, Zwerschke W, Jansen-Dürr P. 2014. Depletion of the cdk inhibitor p16INK4a differentially affects proliferation of established cervical carcinoma cells. J Virol 88: 5256–5262. 10.1128/JVI.03817-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools : a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, Lauberth SM. 2018. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol 25: 687–697. 10.1038/s41594-018-0102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz, JM, Perez, EM, Kieffer-Kwon, K-R, Sanborn, AL, Johnstone, SE, Bascom, GD, Bochkov, ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviram R, Rocha PP, Müller CL, Miraldi ER, Badri S, Fu, Y, Swanzey, E, Proudhon, C, Snetkova, V, Bonneau, R, et al. 2016. 4C-ker: a method to reproducibly identify genome-wide interactions captured by 4C-seq experiments. PLoS Comput Biol 12: e1004780. 10.1371/journal.pcbi.1004780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell 67: 837–852.e7. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jácome-López K, Nora EP, Bruneau BG, Tsirigos A, Furlan-Magaril M, Skok J, et al. 2019. RNA interactions are essential for CTCF-mediated genome organization. Mol Cell 76: 412–422.e5. 10.1016/j.molcel.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 65: 25–38. 10.1016/j.molcel.2016.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J, Manteiga JC, Weintraub AS, Day DS, Zamudio AV, Hnisz D, Lee TI, Young RA. 2018. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep 23: 349–360. 10.1016/j.celrep.2018.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, et al. 2019. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 176: 816–830.e18. 10.1016/j.cell.2018.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6: e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Chronis C, Kronenberg M, Chen XF, Su T, Lay FD, Plath K, Kurdistani SK, Carey MF. 2019. Promoter-enhancer communication occurs primarily within insulated neighborhoods. Mol Cell 73: 250–263.e5. 10.1016/j.molcel.2018.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swygert SG, Lin D, Portillo-Ledesma S, Lin P-Y, Hunt DR, Kao C-F, Schlick T, Noble WS, Tsukiyama T. 2021. Local chromatin fiber folding represses transcription and loop extrusion in quiescent cells. eLife 10: e72062. 10.7554/eLife.72062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Donjon A, Jerković I, Papadopoulos GL, Cheutin T, Bonev B, Nora EP, Bruneau BG, Bantignies F, Cavalli G. 2020. Regulation of single-cell genome organization into TADs and chromatin nanodomains. Nat Genet 52: 1151–1157. 10.1038/s41588-020-00716-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Fang H, Llagas T, Disteche CM, Henikoff S. 2019. Architectural RNA is required for heterochromatin organization. bioRxiv 10.1101/784835 [DOI]
- Thiecke MJ, Wutz G, Muhar M, Tang W, Bevan S, Malysheva V, Stocsits R, Neumann T, Zuber J, Fraser P, et al. 2020. Cohesin-dependent and -independent mechanisms mediate chromosomal contacts between promoters and enhancers. Cell Rep 32: 107929. 10.1016/j.celrep.2020.107929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter, E, Valdes-Quezada, C, Öz, Y, Bouwman, BAM, et al. 2012. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods 9: 969–972. 10.1038/nmeth.2173 [DOI] [PubMed] [Google Scholar]
- Williamson I, Kane L, Devenney PS, Flyamer IM, Anderson E, Kilanowski F, Hill RE, Bickmore WA, Lettice LA. 2019. Developmentally regulated Shh expression is robust to TAD perturbations. Development 146: dev179523. 10.1242/dev.179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros, DA, Stocsits, RR, Tang, W, Schoenfelder, S, Jessberger, G, Muhar, M, Hossain, MJ, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Vesterlund M, Siavelis I, Moura-Castro LH, Castor A, Fioretos T, Jafari R, Lilljebjörn H, Odom DT, Olsson L, et al. 2019. Proteogenomics and Hi-C reveal transcriptional dysregulation in high hyperdiploid childhood acute lymphoblastic leukemia. Nat Commun 10: 1519. 10.1038/s41467-019-09469-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY, et al. 2019. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet 51: 1380–1388. 10.1038/s41588-019-0479-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Huang P, Sharma M, Keller CA, Giardine B, Zhang H, Gilgenast TG, Phillips-Cremin JE, Hardison RC, Blobel GA. 2020a. Alteration of genome folding via contact domain boundary insertion. Nat Genet 52: 1076–1087. 10.1038/s41588-020-0680-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lee D, Dhiman V, Jiang P, Jie X, McGillivray P, Yang H, Liu J, Meyerson W, Clarke D, et al. 2020b. An integrative ENCODE resource for cancer genomics. Nat Commun 11: 3696. 10.1038/s41467-020-14743-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, et al. 2018. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70: 730–744.e6. 10.1016/j.molcel.2018.03.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.