Abstract
Cross-species comparative analyses of single-cell RNA sequencing (scRNA-seq) data allow us to explore, at single-cell resolution, the origins of the cellular diversity and evolutionary mechanisms that shape cellular form and function. Cell-type assignment is a crucial step to achieve that. However, the poorly annotated genome and limited known biomarkers hinder us from assigning cell identities for nonmodel species. Here, we design a heterogeneous graph neural network model, CAME, to learn aligned and interpretable cell and gene embeddings for cross-species cell-type assignment and gene module extraction from scRNA-seq data. CAME achieves significant improvements in cell-type characterization across distant species owing to the utilization of non-one-to-one homologous gene mapping ignored by early methods. Our large-scale benchmarking study shows that CAME significantly outperforms five classical methods in terms of cell-type assignment and model robustness to insufficiency and inconsistency of sequencing depths. CAME can transfer the major cell types and interneuron subtypes of human brains to mouse and discover shared cell-type-specific functions in homologous gene modules. CAME can align the trajectories of human and macaque spermatogenesis and reveal their conservative expression dynamics. In short, CAME can make accurate cross-species cell-type assignments even for nonmodel species and uncover shared and divergent characteristics between two species from scRNA-seq data.
Single-cell RNA sequencing (scRNA-seq) has rapidly emerged as a powerful tool to characterize a large number of single-cell transcriptomes in different tissues, organs, and species (Kolodziejczyk et al. 2015). It not only deepens our knowledge of cells but also provides novel insights into evolutionary and developmental biology (Marioni and Arendt 2017). Cross-species integration and comparison of scRNA-seq data sets allow us to explore, at single-cell resolution, the origins of cellular diversity and evolutionary mechanisms that shape cellular form and function (Marioni and Arendt 2017; Sebé-Pedrós et al. 2018; Tosches et al. 2018; Geirsdottir et al. 2019; Hodge et al. 2019; Shafer 2019; Drokhlyansky et al. 2020; Shami et al. 2020; Wang et al. 2021).
Cell-type assignment (or cell typing) and data integration are both vital steps involved in these analyses. For the cell-type assignment, a traditional approach includes three steps: clustering single cells, performing differentially expression analysis to find cluster-specific genes, and matching these genes with known markers. However, this strategy fails when clustering different cell types into one group and when analyzing many nonmodel species that lack prior knowledge of cell-type biomarkers. Several tools have been developed for this task recently (Abdelaal et al. 2019). Some existing approaches like CellAssign (Zhang et al. 2019a) and scCATCH (Shao et al. 2020) require prior knowledge of cell-type-specific markers. Some like SingleCellNet (Tan and Cahan 2019), SciBet (Li et al. 2020), SingleR (Aran et al. 2019), and Garnett (Pliner et al. 2019) were designed based on a reference data set and can achieve the cell-type assignment without providing marker information. In addition, several methods designed for data integration can also achieve cell-type assignment by transferring labels from the reference data set (Stuart et al. 2019; Gao et al. 2021). Seurat-v3 (Stuart et al. 2019) combines canonical correlation analysis and mutual nearest neighbors to perform data integration and label transfer based on “anchors.” Cell BLAST (Cao et al. 2020), ItClust (Hu et al. 2020), and scArches (Lotfollahi et al. 2022) make use of deep neural networks for both cell-type querying and cell embedding. LIGER (Welch et al. 2019) and CSMF (Zhang and Zhang 2019) extract the common and private features of two data sets, respectively, by joint nonnegative matrix factorization to achieve cell alignment across data sets and omics.
Despite all the progress, a tool for effective and robust cross-species integration and comparison is still immature and in demand. There are several computational challenges to be overcome. First, it is hard to determine cell identities for nonmodel species that lack prior knowledge of cell-type biomarkers, and most of the methods may fail when generalizing to cross-species label transfer. Second, many biological and technical factors, such as transcriptome variation between species, different experimental protocols, and inconsistent sequencing depths, can make cross-species data integration and comparison even more difficult. Third, homologous cell-type alignment requires quantifying the similarities of gene expression profiles, which usually vary across distinct normalizations and gene selections (Marioni and Arendt 2017). Fourth, cross-species cellular alignment is usually based on homologous genes, and current approaches are mostly restricted to one-to-one homologies shared by both organisms (Marioni and Arendt 2017; Sebé-Pedrós et al. 2018; Tosches et al. 2018; Geirsdottir et al. 2019; Hodge et al. 2019; Drokhlyansky et al. 2020; Shami et al. 2020; Wang et al. 2021), where non-one-to-one homologous genes characterizing cell-type conservative features could be lost. Lastly, evolutionary divergences are thought to be caused by transcriptional changes of groups of genes that evolve in a modular fashion and are controlled by transcription factors (Arendt et al. 2016). Extraction and comparison of gene modules between species will provide deep insights into evolutionary conservation and divergences (Oldham et al. 2006; Aibar et al. 2017; Wang et al. 2021). To this end, we develop a semisupervised heterogeneous graph neural network model, CAME, to achieve the aligned and interpretable cell and gene embeddings for cross-species cell-type assignment and gene module extraction.
Results
Overview of CAME
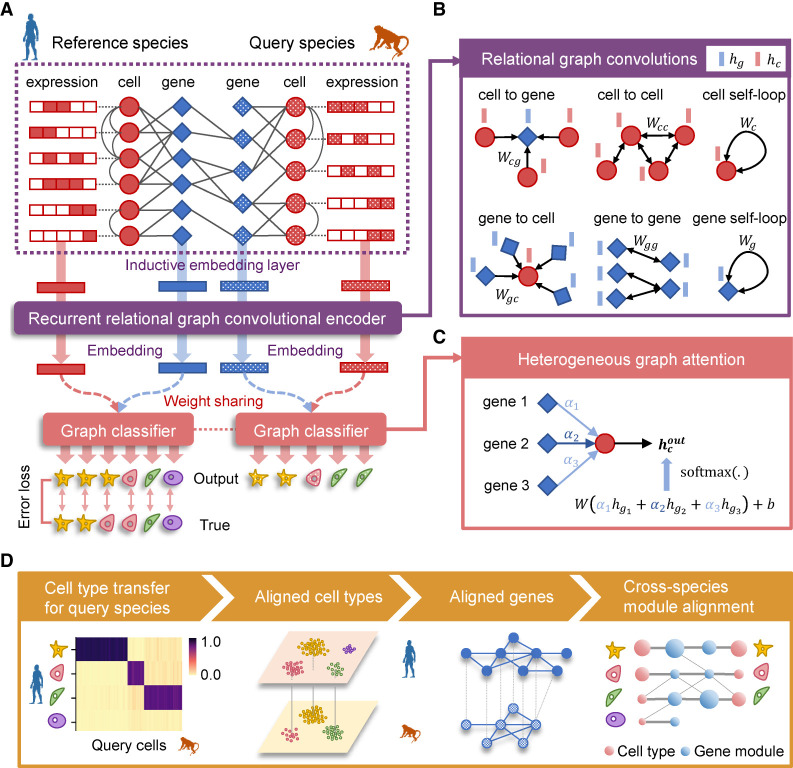
CAME takes two scRNA-seq data sets from different species, along with their homologous gene mappings as input. One data set with cell-type labels is taken as the reference, and the other whose cell types need to be assigned is the query (Fig. 1A). CAME encodes these two expression matrices and the mappings of homologous genes as a heterogeneous graph, where each node acts as either a cell or a gene, whereas a cell–gene edge indicates a non-zero expression of the gene in that cell, and an edge between a pair of genes indicates the homology between each other. Note that one-to-many and many-to-many homologies are allowed as well. In addition, CAME adopts single-cell networks precomputed from reference and query data sets using the k-nearest-neighbor (KNN) method, respectively, where a cell–cell edge indicates this pair of cells has similar transcriptomes with each other (Methods).
Figure 1.
Overview of CAME. (A) The architecture of the heterogeneous graph neural network in CAME. The scRNA-seq data of both reference and query species and their homology genes are encoded as a heterogeneous cell–gene graph. The cell–gene edge indicates that the cell has non-zero expression of the gene. The gene homologous mappings are represented by a gene–gene bipartite graph, with each edge indicating a gene homology. Note that the homologous gene mappings can be many-to-many homologies. To preserve the intrinsic data structure, the within-species cell–cell edges are adopted where an edge between a pair of cells indicates that one is the k nearest neighbor of the other (k = 5 by default). The heterogeneous graph and the gene expression profiles are input to CAME, passing through the inductive embedding layer, the recurrent relational graph neural network, and the graph classifier with attention mechanisms. The model is trained by minimizing the cross-entropy loss calculated between the model prediction and the given labels of the reference cells in an end-to-end manner. (B) Graph spatial convolutions for six different types of edges, including “a cell expresses a gene,” “a gene is expressed by a cell,” “cell–cell similarity,” “gene–gene homology,” “cell self-loop,” and “gene self-loop” with the edge type–specific convolution weights. (C) Heterogeneous graph attention classifier on the last layer, where each cell pays different attention to its neighbor genes. The output cell-type probabilities are calculated by the weighted sum of the neighbor-gene embeddings, followed by the softmax normalization. The attention weights are calculated from the concatenated cell and gene embeddings with a linear transformation, followed by activation and the softmax normalization among the neighbor genes of the cell. (D) The output of CAME includes the probabilistic cell-type assignment of the query species, as well as low-dimensional embeddings of the cells and genes from both species. The gene embeddings are used for joint module extraction that allows inter-species comparison of conservative or divergent characteristics.
CAME adopts a heterogeneous graph neural network to embed each node into a low-dimensional space (Methods) (Fig. 1B). For the initial cell embeddings, CAME takes the expression profiles followed by linear transformation with a nonlinear activation function, whereas for the initial gene-embeddings, CAME aggregates the expression profiles (called “messages”) from its neighbor cells that expressed it and then treats them with linear transformation and nonlinear activation, as performed for cells (Methods). Then the initial embeddings are input to two parameter-sharing graph convolution layers with heterogeneous edges and nodes. As a result, cells with more coexpressed genes are more likely to exchange the embedding message with each other and thus be encoded with similar embeddings; the same principle applies to genes. CAME further uses a heterogeneous graph attention mechanism (Veličković et al. 2017) to classify cells with embeddings of their neighbor genes as input, where each cell pays a distinct level of attention to each certain neighbor gene (Methods) (Fig. 1C). High attention paid by a cell to a gene implies that the gene is of relatively much importance for the cell to be characterized.
We note that a reference cell could be assigned with multiple labels in different hierarchies, and a cell type in query species might correspond to multiple ones in the reference. Thus, multilabel classification can be helpful to depict the state of a cell. CAME calculates the cross-entropy between the predicted cell-type probabilities and the true labels for the reference data to obtain both the multiclass and the multilabel loss, and sums them up as the training loss. Finally, CAME minimizes it by the backpropagation algorithm (Methods). The training process of CAME is semisupervised in an end-to-end manner. We found that the training process was quite stable, and the model tended to be well trained before 200–300 epochs (Supplemental Fig. S1A). In addition, CAME introduces the adjusted mutual information (AMI) between the predicted labels and preclustered ones of query cells to automatically determine the model checkpoint for downstream analysis (Methods) (Supplemental Fig. S1A).
CAME outputs the quantitative cell-type assignment for each query cell, that is, the probabilities of cell types that exist in the reference species, which enables the identification of the unresolved cell states in the query data. For most cells with homologous cell types in the reference, CAME assigns them with a maximal probability approximating one, whereas for those unobserved cell types or states, CAME would assign them to their analogs with relatively low confidences (Supplemental Fig. S2). In addition, CAME gives the aligned cell and gene embeddings across species, which facilitates low-dimensional visualization and joint gene module extraction (Methods) (Fig. 1D).
CAME shows superior accuracy and robustness for cell-type assignment compared with state-of-the-art methods
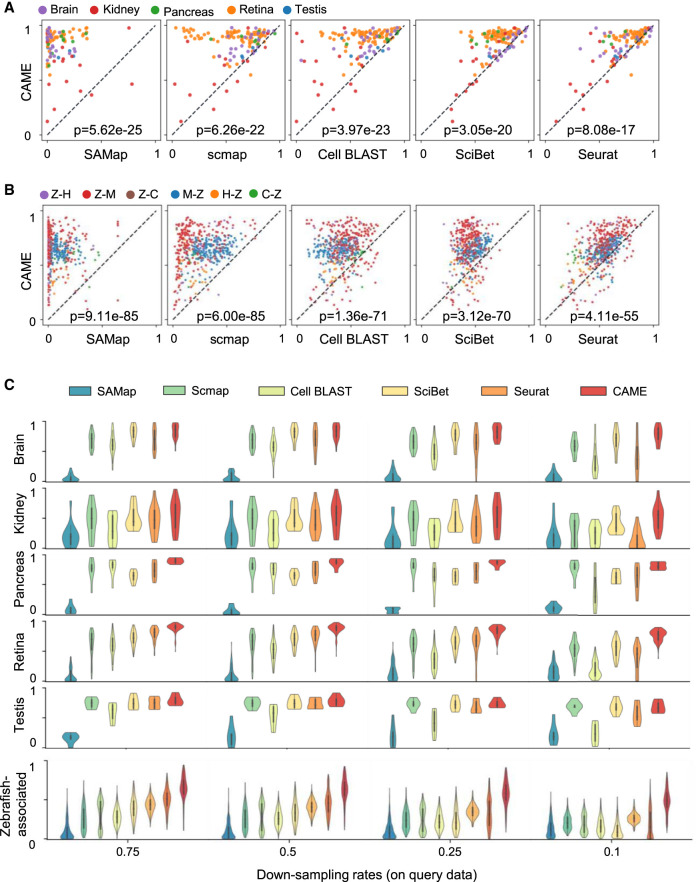
We collected 54 scRNA-seq data sets from five tissues across seven different species including human, macaque, mouse, chick, turtle, lizard, and zebrafish (Methods) (Supplemental Fig. S3A; Supplemental Table S1). More than a half of the homologous genes between zebrafish and other species are not one-to-one matched (Supplemental Fig. S3B). In addition, the proportion of non-one-to-one homologies between highly informative gene (HIG; Methods) sets with one associated with zebrafish (Hoang et al. 2020) is significantly higher than that of other cross-species data set pairs (60%–75% vs. 15%–40%) (Supplemental Fig. S3C). The ablation study shows that, when excluding non-one-to-one homologies, the cell typing accuracy of CAME suffers a significant drop (ranging from 1.5% to 8.7% for different species-pairs, 6.26% on average, with P-value = 7.8 × 10−23) on the zebrafish-associated data set pairs (Supplemental Figs. S3D, S4). Therefore, we divided these pairs into two scenarios: zebrafish-excluded (139 pairs) and zebrafish-associated (510 pairs; Methods).
We compared the cell typing performance of CAME with five current methods including one specifically designed for cross-species integration (SAMap) (Tarashansky et al. 2021), one deep-learning method (Cell BLAST) (Cao et al. 2020), and three other methods—SciBet (Li et al. 2020), scmap (Kiselev et al. 2018), and Seurat-v3 (Stuart et al. 2019)—in terms of accuracy, macro-F1 score, and weighted F1 score (Methods). CAME distinctly outperforms the others in most cases with statistical significance P-values < 10−16 and 10−54 using a Wilcoxon signed-rank test for both the zebrafish-excluded and zebrafish-associated scenarios, respectively (Fig. 2A,B; Supplemental Figs. S5, S6).
Figure 2.
Benchmarking cross-species cell-type assignment performance of CAME. (A,B) Performance comparison of CAME and five other approaches in terms of cell typing accuracy on 139 pairs of cross-species scRNA-seq data sets (A) and on 510 pairs of cross-species scRNA-seq data sets that associated with zebrafish (B), where each point represents a pair of cross-species data sets and is colored by tissue. The notation “X-Y” indicates that X is the reference and Y is the query. (H) Human, (M) mouse, (C) chick, (Z) zebrafish. (C) Performance comparison of the classification accuracies of CAME and the five other methods on different down-sampling rates (0.75, 0.5, 0.25, 0.1) for read counts.
To evaluate the robustness of CAME in the cases in which the reference and query data sets have inconsistent and insufficient sequencing depths, we performed down-sampling experiments (at various sampling rates 75%, 50%, 25%, 10%) for read counts on the reference, query, and both reference and query data sets. Again, CAME achieves superior performance compared with all five benchmarked methods (Fig. 2C; Supplemental Figs. S7, S8). In contrast, when the down-sampling rates are extremely unbalanced, some of these methods may fail. For example, at a down-sampling rate of 0.1 for query data sets, Seurat detects too few anchors to abort integration for label transfer, scmap fails to find enough genes because the median expression in the selected features is zero in each cell cluster, and SAMap fails because it cannot find cross-species edges to link the data sets. Also, we have tested the robustness of CAME under several hyperparameter settings including different hidden units, different hidden layers, and two gene selection strategies (highly variable gene [HVG] selection and the number of used genes) (Supplemental Fig. S9). Generally, the results of CAME are very robust under different settings. Although it may achieve a bit better performance under 512 hidden units, but the computational expense would be roughly four times as before, which may exceed the graphic memory limit of a typical graphic card on some data sets. When the graphic memory is enough, we suggest to use 512 hidden units to replace our default setting. All these results show that CAME is robust to the insufficient and inconsistent sequencing depths between reference and query pairs. The more comprehensive comparisons with other methods can be seen in Supplemental Figures S5 through S8.
CAME robustly aligns homologous cell types across species and multiple references
In addition to the accurate cross-species cell-type assignment, CAME is also capable of aligning homologous cell types from different species, even when crossing distant species. For example, when aligning cell types between the mouse (Tasic et al. 2018) and turtle (Tosches et al. 2018), CAME successfully distinguishes and aligns each major type, like inhibitory and excitatory neurons, whereas the alignments by Seurat are incapable. CAME also separates the neural progenitor cells from excitatory neurons, whereas LIGER merges these two groups. The visualization plots using a uniform manifold approximation and projection (UMAP) (McInnes et al. 2018) of cell embeddings of Cell BLAST tend to lose some relations between cell types. For instance, the inhibitory and excitatory neurons are not linearly separable on the two-dimensional (2D) UMAP plot (Fig. 3A; Supplemental Fig. S10).
Figure 3.
Alignment comparison of cell embeddings across data sets by CAME and four other methods. (A) The UMAP plots of the cell embeddings by CAME and four integration methods on the scRNA-seq data from turtle (reference) and mouse (query) brains. Cells are colored by their cell types (the first column) or data set identities (the second column). (B) Similar settings to A. Here the reference data sets are the human pancreatic scRNA-seq data from eight batches by three different platforms, and the query is from mouse pancreas cells. The UMAP plots of the third column show the reference cells, colored by batch identities. The scores under both the figures in A and B represent the graph connectivity score for each method.
When handling multiple references and batch information is unavailable, most integration methods will suffer from batch effects. In this situation, owing to the semisupervised manner, CAME can ignore the batch effects of reference data. In contrast, other integration tools may suffer from diverse sources of noises if the potential batch effects (such as noises from different individuals) are not considered. For instance, when aligning human and mouse pancreas cell types with a human reference composed of eight batches, cells of the same type (e.g., pancreatic A cell) but from different batches are still separated from each other. The query cells tend to be close to reference cells of the same protocol on the UMAP plot of Cell BLAST (Fig. 3B). Even when the batch labels are given, for some of the methods (e.g., LIGER [Welch et al. 2019] and Seurat-v3 [Stuart et al. 2019]), the reference batch effects still exist after cross-species integration (Supplemental Fig. S11). We also used the graph connectivity (GC) score (Luecken et al. 2022) to measure the quality of the integration across data sets derived from different species. CAME achieved the best performance compared with other methods in terms of this score (Fig. 3).
CAME accurately assigns cell types in mouse brains and reveals cell-type-specific gene modules
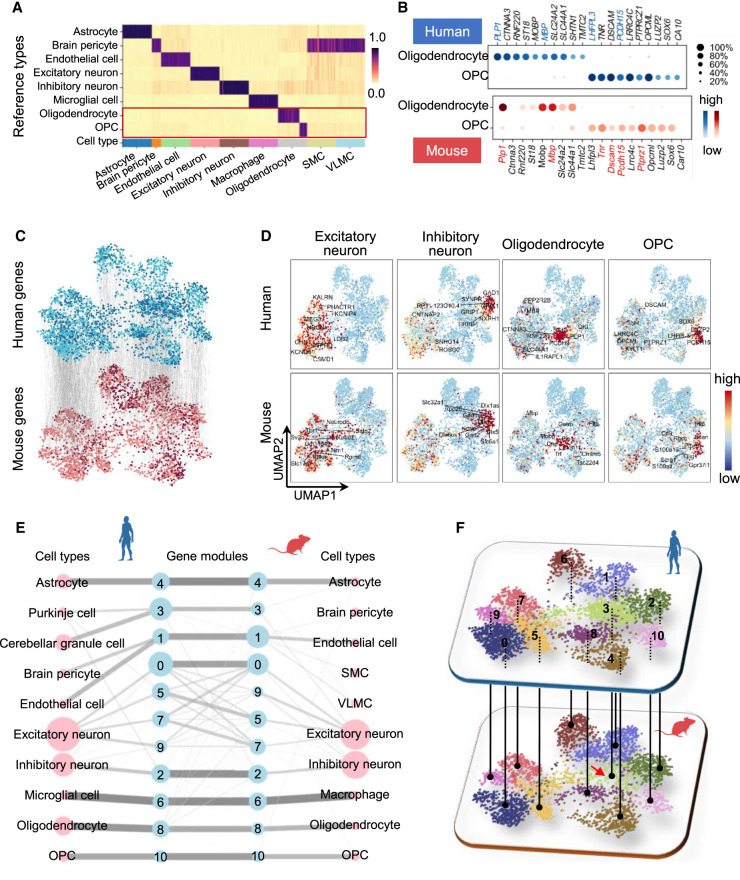
We applied CAME to assign the major types of single cells from the primary visual cortex and the anterior lateral motor cortex of mice (Tasic et al. 2018), and used human brain cells as the reference data set (Lake et al. 2018), which contains the cells from the hindbrain that are not included in the mouse data set. CAME achieves an accuracy of ∼98%, similar to that of Seurat and SciBet, superior to the other benchmarked methods (93% by Cell BLAST and only 55% by scmap). CAME also gets a higher macro-F1 score (0.55) than that of Seurat (0.44) and SciBet (0.46), indicating that CAME also accurately classifies the small groups. Specifically, those nonneuronal types accounting for a small proportion of mouse cells are accurately assigned, including endothelial cells (accounting for 0.6% of human cells and 0.85% of mouse cells) and its subclass, brain pericytes (0.61% of human cells and 0.14% of mouse cells). The macrophages (0.56% in mice) are classified as microglial cells (2.1% of human cells) that are biologically similar to this type (Diehl et al. 2016). Both oligodendrocyte precursor cells (OPCs) and oligodendrocytes in mice were originally assigned as oligodendrocytes (0.75% of mouse cells) by the investigators, but they are distinguished from each other in the reference of the human data (Fig. 4A). The identities of OPCs are also verified by examining the expression of typical marker genes (Zhang et al. 2019b) in each cell type (Fig. 4B). In addition, the genes with top attention from each cell type show high cell-type specificities, although these genes are quite different across species (Supplemental Fig. S12A). The UMAP plots of cell embeddings show that these major homologous cell types are well aligned with each other. This suggests that the major types of brain cells in humans and mice are well conserved (Supplemental Fig. S10). Meanwhile, the two cell types SMC and VLMC that did not appear in the reference data set were predicated by CAME as brain pericyte (Fig. 4A). It is reasonable because brain pericytes, VLMCs, and SMCs are all vascular cells (Saunders et al. 2018). Moreover, another study suggested that SMC and brain pericytes are both subtypes of mural cells (Hughes and Chan-Ling 2004). Compared with other cell types (e.g., neuron, endothelial, or oligo) in the reference, it is reasonable that SMCs and VLMCs in the query data set were predicted by CAME as brain pericytes with relatively low assignment scores. The weak connection from SMC and VLMC to gene modules indeed indicates that the assignment of these two cell types is of low confidence.
Figure 4.
Application of CAME to human and mouse scRNA-seq data of brain cells. (A) The predicted cell-type probabilities for each cell (each column) in the mouse brain scRNA-seq data. A maximum of 50 cells was subsampled from each type for visualization. The gene expressions of the human brain were taken as the reference. Each row indicates a cell type in human data. (OPC) Oligodendrocyte precursor cells, (SMC) smooth muscle cell, (VLMC) vascular and leptomeningeal cell. (B) The top homologous DEG expressions of oligodendrocytes and (predicted) OPCs in human and mouse data, including several marker genes reported by previous literature (collected from CellMarker; colored by red or blue). (C) Cross-species alignment of the gene embedding output by CAME, where each dot represents a gene, and each edge indicates the homology between a pair of genes. Genes shared between species are colored by light blue (human) or pink (mouse), and the other genes are colored by dark blue (human) or dark red (mouse). (D) The UMAP plots of gene embeddings showing the average expression patterns (z-scored across cell types for each gene) of four cell types (excitatory neurons, inhibitory neurons, oligodendrocytes, OPCs) of human and mouse brains, where the color of each dot is scaled by the expression level of that cell type in the gene. (E) Abstracted graph of the heterogenous cell–gene graph; each node represents a cell type (pink) or a gene module (light blue). The size of a node is scaled by the number of single cells in that type or the number of genes in that gene module. The width of an edge is scaled by either the normalized mean expression levels of a cell type in the connected gene module or the conservancy of inter-species gene modules based on the gene embeddings learned by CAME. (F) Gene modules detected by joint module extraction of genes from humans (above) and mice (below).
Similar results are observed when comparing four subtypes of the inhibitory neurons (VIP+, SST+, LAMP5+, PVALB+) between humans and mice. CAME still achieves a cell typing accuracy of 98.3% and 95.5% for human-to-mouse and mouse-to-human label transfers, respectively, which are consistently higher than that of the other methods (93.4% and 92.0% for SciBet, 98.0% and 78.9% for Cell BLAST, 69.5% and 78.9% for scmap, 94.2% and 87.2% for Seurat) (Supplemental Fig. S13A,B), although differentially expressed genes (DEGs) for each homologous subtype seem to not be transferrable across species (Supplemental Fig. S13C,D).
CAME can also give interpretable gene embeddings and enable us to explore both intra- and inter-species relationships between genes. The UMAP plots of gene embeddings show that the relative positions of human and mouse homologous genes are very consistent (Fig. 4C). We further showed the averaged gene expression profile on the UMAP plots of gene embeddings, where each point represents a gene (Fig. 4C; Supplemental Fig. S12B). It is worth noting that the neighbor genes tend to be coexpressed in the same cell types, such as those in excitatory or inhibitory neurons, oligodendrocytes, and OPCs (Fig. 4D). There are more cell-type-specific genes in human oligodendrocytes than in mice, indicating the evolutionary divergence between humans and mice. A population of genes is only detected in the human data set, and most of them are associated with Purkinje cells and cerebellum granule cells, which are not detected in the mouse data set owing to their sources from different brain regions. These genes are arranged where there are few mouse genes around (Fig. 4F; Supplemental Fig. S12B).
To further explore the influence of species-specific cell types on CAME and call species-specific features, we used the inhibitory cells from the mouse and human cortex as reference and query data, respectively (Tasic et al. 2018; Hodge et al. 2019). Using the inhibitory neuron homology provided by the investigators (Hodge et al. 2019), we combined the subclass of the data sets into five major cell types: LAMP5+, VIP+, SST+, PVALB+, and MEIS2+, where MEIS2+ is a mouse-specific cell type that was not detected in the human data. CAME still achieves a high cell typing accuracy of 95.3% for mouse-to-human label transfers, and few cell types in the human data set were predicted as MEIS2+ cells (Supplemental Fig. S14A). The embedding of MEIS2+ cells was consistent with our expectations, where it showed a distinct position and barely mixed with other human cells in the UMAP plot (Supplemental Fig. S14B). From the aspect of gene modules, the four shared cell types (LAMP5+, PVALB+, SST+, VIP+) are quite conservative, and we also found a MEIS2+-specific gene module 8 from mouse genes in the heterogeneous cell–gene graph (Supplemental Fig. S14C). The mouse genes from module 8 showed enrichment in forebrain development (Supplemental Fig. S14D). This is reasonable because MEIS2+ is mainly involved in the positive regulation of neuron differentiation and forebrain development in mice (Su et al. 2022). These results indicate that CAME preserves species-specific features, and it will not force the integration of species-specific cell types and related genes.
The aligned gene embeddings across species can facilitate us to jointly extract cell-type-specific gene modules with different degrees of conservancies between species, and each module corresponds to a cell type like OPCs or to related cell types like endothelial cells and their subtypes (Methods) (Fig. 4E). As expected, based on Gene Ontology (GO) (The Gene Ontology Consortium et al. 2000) enrichment analysis, we can see that the functions associated with most homologous gene modules are generally consistent with each other (Supplemental Table S2). For example, both the human and mouse genes in module 2 (which is associated with inhibitory neurons) tend to relate functions like “forebrain neuron differentiation” and “learning or memory.” Both the human and mouse genes in module 6 (corresponding to human microglia and mouse macrophage) are related to functions like “positive regulation of cytokine production” and “leukocyte migration.” In contrast, the function “ventral spinal cord development” is only enriched in human module 3 but not in mice, considering their gene members are quite different, although they are both associated with the function “cell differentiation in hindbrain” and “cerebellar cortex formation.”
CAME reveals conserved expression dynamics during spermatogenesis between human and macaque
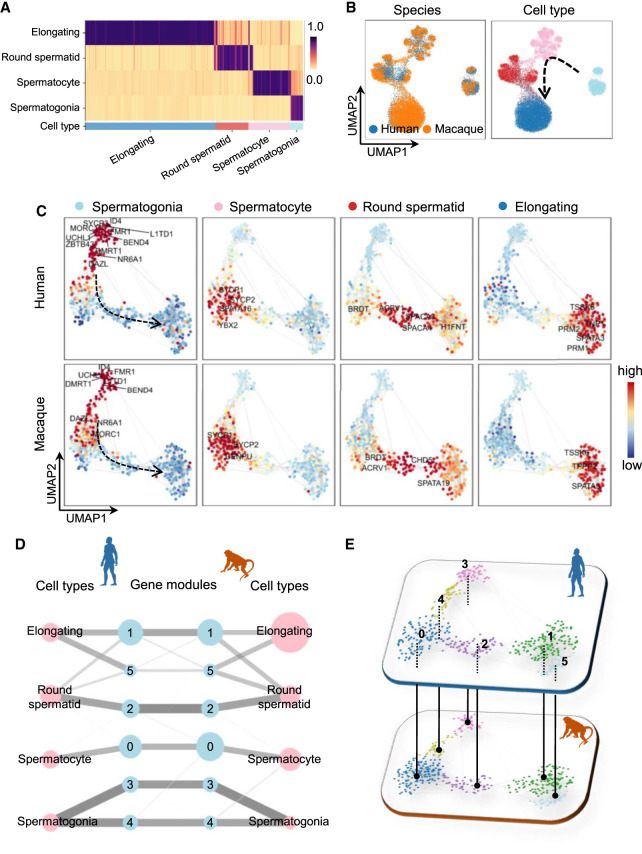
The comparison of continuous biological processes between two species is of much interest in evolutionary biology. We applied CAME to two scRNA-seq data sets from human and macaque testicular single cells (Shami et al. 2020) with the former as the reference. CAME achieves a very distinct cell typing accuracy of 95.0% (86.0% for SciBet, 76.1% for Cell BLAST, 87.3% for scmap, 89.1% for Seurat) and a precise alignment of the homologous cell types of human and macaque with each other (Fig. 5A,B). In addition, the labeled spermatogonia, spermatocyte, round spermatid, and elongating cells are correctly merged along the underlying differentiation trajectory. This suggests that CAME could well decipher the conserved four-stage spermatogenesis processes of humans and macaques.
Figure 5.
Application of CAME to human and macaque scRNA-seq data during spermatogenesis. (A) The predicted cell-type probabilities for each macaque testicular cell (each column). A maximum of 50 cells was subsampled from each type for visualization. Gene expression in human testis was taken as the reference. Each row indicates a cell type in the human data. (B) The UMAP plots of cell embeddings output by CAME, colored by data sets (left) or cell type (right). (C) 2D visualization of gene embeddings showing the average expression patterns (z-scored across cell types for each gene) of the four stages across spermatogenesis, where each point represents a gene, and the color of each scatter is scaled by the expression level of that cell type in the gene. (D) Abstracted graph of the heterogeneous cell–gene graph. Each node represents a cell type (pink) or a gene module (light blue). The size of a node is scaled by the number of single cells in that type or the number of genes in that gene module. The width of an edge is scaled by either the normalized mean expression levels of a cell type in the connected gene module or the conservancy of inter-species gene modules based on the gene embeddings learned by CAME. (E) Gene modules detected by joint module extraction of genes from humans (above) and macaques (below).
The continuously dynamic changing process of spermatogenesis can also be revealed by the UMAP plot of gene embeddings (Fig. 5C). As illustrated, the known stage-specific marker genes extracted by CAME (Shami et al. 2020) are highly coexpressed in the four main stages of spermatogenesis and form well-organized expression dynamics, suggesting the order of critical gene activations during spermatogenesis (Fig. 5C). By joint extraction of gene modules, we can see that the four stages of spermatogenesis are quite conservative from the aspect of gene modules (Fig. 5D,E), which cannot be revealed by separate module extraction (Supplemental Fig. S15). For example, modules 3, 4, and 0 are highly expressed in spermatogonia and spermatocyte, respectively, for both humans and macaques. And round spermatids and elongating spermatids share modules 2, 1, and 5 in different degrees. Typically, both human and macaque modules 4 are associated with functions like “RNA splicing,” and module 1 was associated with “sperm motility” and “spermatid development/differentiation,” which are typical characteristics of elongating spermatids (Supplemental Table S3).
CAME accurately assigns retinal cell types between distant species and shows superior power even for nonmodel species
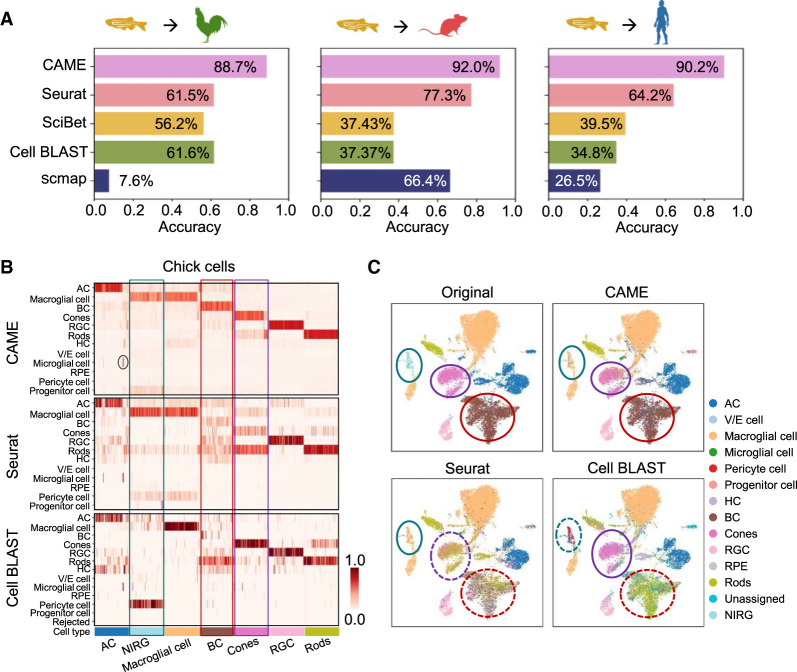
For distant species, the markers of homologous cell types can be quite diverse. For example, for the major cell types in the retina defined by scRNA-seq data, there are only a few shared DEGs for human, mouse, chicken, and zebrafish (Supplemental Fig. S16A), which limits the performance of many marker-based methods for cross-species cell-type assignment. Here we took the retinal scRNA-seq data of adult zebrafish (Hoang et al. 2020) as the reference, and applied CAME to assign the retinal cells for two distant species, human (Menon et al. 2019) and mouse (Macosko et al. 2015), and a nonmodel species, chick (Hoang et al. 2020). The prediction performance of CAME is significantly higher than the four other methods with distantly improved accuracy by 27.1%, 14.7%, and 26.0% compared with the second best one for the zebrafish–chick, zebrafish–mouse, and zebrafish–human pairs, respectively (Fig. 6A). As a distance-based method, scmap fails to make effective cell-type assignment between zebrafish and chick owing to the systematic differences in gene expression space cross-species. It is worth mentioning that even for chick, CAME achieves superior cell typing accuracy (with >27.1% improvement), showing its great application potential in diverse nonmodel species. Specifically, both Seurat and Cell BLAST fail to identify retinal bipolar cells in chick, probably because there are only two one-to-one-homologous genes that were differentially expressed in both the zebrafish and chick retina (Fig. 6B; Supplemental Fig. S16). Seurat mistakenly assigns retinal cone cells as rod cells for it found few anchors between reference and query cone cells. In addition, the nonastrocytic inner retinal glial (NIRG) cells in chick retina, which have no homologous type in the reference (zebrafish), are assigned as “macroglial cells” with the probabilities lower than the true macroglial cells (by 8% on average). The close location distributions of NIRG and macroglial cells in the UMAP plot indicate their high similarity (Fig. 6C), as described by previous studies (Fischer et al. 2010; Zelinka et al. 2012). Moreover, 53 cells originally annotated as “amacrine cell” are assigned as “microglial cell,” and canonical markers of “microglial cell” (C1QC, C1QA, CSF1R, and CD74) (Xu et al. 2022) are indeed highly expressed but rarely those of “amacrine cell” (Supplemental Fig. S16B), suggesting CAME could potentially correct prior annotations. Note that gene CD74 in chick and its two homologous genes cd74a and cd74b in zebrafish are more conserved than other DEGs, which indicates that non-one-to-one homologies can provide conservative features between distant species.
Figure 6.
Application of CAME to retina scRNA-seq data between distant species. (A) Performance comparison of CAME and four other approaches, including Seurat, scmap, SciBet, and Cell BLAST, in terms of cell typing accuracy on three pairs of cross-species scRNA-seq data sets about the retina with zebrafish as the reference and with three species (chick, mouse, and human) as the queries. (B) Heatmap comparison of the assignment probabilistic matrices of CAME and another top two methods (Seurat and Cell BLAST) in A for each query cell (column) about the retina with zebrafish as the reference and with chick as the query. For convenience, only 100 cells were subsampled from each cell type to visualize. Note that Cell BLAST only provides the P-value for each query cell to its k (k = 5 by default) nearest reference cells, so the probabilities were computed by averaging the labels of these reference cells with P-values lower than 0.05. (C) UMAP plots of the chick retinal cells colored by the original assignments and the predicted ones by CAME, Seurat, and Cell BLAST using zebrafish as the reference.
Discussion
Cross-species comparative and integrative analysis at single-cell resolution has deepened our understanding of the origin and evolutionary mechanisms of cellular states. Exploring the conservative and divergent characteristics of homologous cell states between human and other model and nonmodel species can help us to determine the animal model for studying human disease (Geirsdottir et al. 2019; Hodge et al. 2019; Drokhlyansky et al. 2020).
Existing approaches for cross-species integration were mainly based on one-to-one homologous genes, but when it is needed to align cell types across distant species, especially when a large number of gene duplications were involved during the evolution process (Glasauer and Neuhauss 2014; Ravi and Venkatesh 2018), considering only the one-to-one homologous genes will lead to significant loss of information. Even so, cells of homologous types are thought to have similar expression patterns; that is, they may coexpress a cell-type-specific combination of genes. These genes may not be easy to be identified as the marker genes with high expression levels but can act as “bridges” between cells that coexpress them. In addition, the gene-homology mappings can bridge the gene nodes of two species, where the non-one-to-one homologies can also be used. Based on this, we propose CAME to use a heterogeneous graph neural network to encode the cell–gene–gene–cell multipartite graph, boosting the “message-passing” from one species to the other. As a result, the utilization of non-one-to-one homologous gene mappings makes a significant improvement on cell-type assignment across zebrafish and other species. The heterogeneous graph is a good strategy to model the current task with cross-species scRNA-seq data and gene homology mapping. In the future, if one wants to adopt some prior knowledge about cells or genes (e.g., GO), the hypergraph strategy can potentially be adopted to expand the current model.
When handling multiple references, most integration approaches have to perform pairwise alignment for individual batches. However, the order of pairwise alignment can affect the results, and the computational complexity rises quadratically with the number of batches. Others like Harmony (Korsunsky et al. 2019) and Cell BLAST (Cao et al. 2020) can align multiple data sets simultaneously, once the batch labels are given. We showed that CAME can remove batch effects for multiple references even when batch labels are not provided. This is an important characteristic for integrating various data sets and constructing a unified cell typing reference.
By comparative analysis between human-versus-mouse brains and human-versus-macaque spermatogenesis process, we showed that CAME can not only achieve accurate and robust cell-type assignment but also give gene embeddings that facilitate the visualization of cell-type expression profiles on 2D UMAP plots. In addition, the extracted cell-type-specific gene modules can provide functional insights into the conservative and divergent characteristics between species. We note that CAME relies on the correct and reasonable annotation of reference data, which limits the use of CAME in unsupervised scenarios. We expect it will be solved in the future.
It should be noted that the inter-species gene homology mapping is still developing, and different methods like Ensembl_Compara (Vilella et al. 2009), Domainoid (Persson et al. 2019), and Hieranoid (Schreiber and Sonnhammer 2013; Kaduk and Sonnhammer 2017) could share different levels of inconsistency (Nevers et al. 2022). We compared the performance of CAME with the homologous gene relationships inferred by Ensembl_Compara and Domainoid, respectively. The homologous gene relationships inferred by them indeed showed different levels of inconsistency but still shared very larger proportions, ranging from 47.5% to 88.9% (Supplemental Fig. S17A). The accuracies of CAME with homologous genes provided by these two methods are very comparable and have no significant differences (i.e., P-value = 0.701 with the paired t-test, and P-value = 0.941 with the rank-sum test) (Supplemental Fig. S17B–D). These results show the stability and robustness of CAME under homologous gene relationships inferred by different methods.
Last but not least, the heterogeneous graph neural network structure of CAME can also be applied to the scenarios of within-species data integration or when we consider only the one-to-one homologous genes. The only adjustment is to replace each gene–gene edge with a single gene node. Moreover, this strategy can be applied for multi-omic label transfer and data integration. In summary, we believe that CAME will serve as a powerful tool for integrative and comparative analysis across species as well as multi-omic integration.
Methods
Building a heterogeneous cell–gene graph
Let us denote a gene expression matrix with N cells and M genes as X = (X1, X2, …, XN)T ∈ ℝN×M, where each row Xi = (xi1, xi2, …xiM) ∈ ℝM with an element xij representing the (normalized) expression value of a cell i in a gene j. We take and as the reference and query data sets, respectively; as the cell-type labels of the reference data set; and a set of gene pairs {(gi, gj)}ij to indicate the homology between two species. Note that MR is not necessarily equal to MQ.
The reference and query expression matrices and the homology together are represented as a heterogeneous cell–gene graph with binary edges, with each node acting as a cell or a gene (Fig. 1A). A cell–gene edge indicates that this cell has non-zero expression of the gene, a gene–gene edge indicates a homology between each other, and a cell–cell edge indicates the expression profiles of these two cells are similar to each other. In short, in this graph, there are two types of nodes, cell and gene, and six types of edges (relations) including “a cell expresses a gene,” “a gene is expressed by a cell,” “cell–cell similarity,” “gene–gene homology,” “cell self-loop,” and “gene self-loop.” Although the edge type “cell–cell similarity” seems redundant, we have observed an accuracy improvement of 1.29% on average when combining this edge type (Supplemental Fig. S1B).
Designing a heterogeneous graph neural network
CAME adopts a heterogeneous graph neural network, which was motivated by a relational graph convolutional network (Schlichtkrull et al. 2018), for a graph of homogeneous nodes but heterogeneous edges. We denote the convolution weights for these six edge types as Wcg, Wgc, Wcc, Wgg, Wc, and Wg, respectively (Fig. 1B). Note that the values of Wgg are assigned in the same way for one-to-one gene homologies and non-one-to-one homologies. For each cell i, its initial embedding (the 0-th layer) is calculated as
where σ is the leaky ReLU activation function with a negative slope of 0.05, is the gene expressions in the cell i (one-to-one homologous genes are taken as the common input features), and is the learnable bias vector. The genes, however, lack the initial embeddings in the 0-th layer and can be aggregated from their neighbor cells as follows:
where is the set of cells that have expressed the gene j, and is the normalization factor. This approach keeps the number of model parameters constant to the number of genes, which differs from the commonly used initialization that assigns a learnable embedding for those nodes without input features, where the increasing number of model parameters might lead to an overfitted model. It can also allow inductive learning for the genes not involved in the training process.
In each hidden layer l ≥ 1, the node features for the cell i and the gene j can be calculated as
and
respectively, and , , , and are the normalization factors. Note that we treat the edges between homologous genes and the self-loop on each gene identically; that is, . To boost the “message” flow between reference and query nodes, we adopt a recurrent convolution, where the parameters are shared across the hidden layers; that is, and for 1 ≤ l ≤ L, where L is the total number of the hidden layers. Ablation experiments showed that this recurrent manner gave an accuracy improvement of 2.09% on average. We recommend setting L as two or three in practice, and the default setting is two. We also adopt the layer normalization (Lei Ba et al. 2016) for all the hidden states to facilitate fast training convergence and high performance (Supplemental Fig. S1).
When it comes to the cell-type classifier, we adopt the attention mechanism for graph convolution (Veličković et al. 2017), where each cell pays distinct attention to its neighbor genes. Specifically, for each cell i, the output states for cell-type identification is only aggregated from their neighbor genes:
where αij is the attention that the cell i pays to the gene j, calculated as
with
In addition, we use multihead attention to enhance the model capacity and robustness, where there are several attention heads with their own parameters, and their outputs are merged by taking averages:
where K is the total number of attention-heads, set as eight by default. We found that the attention mechanism could give an accuracy improvement of 5.45% on average, and not accounting for the cell self-loop edges on classifier edges could give an improvement of 7.40% on average (Supplemental Fig. S1B).
Finally, the output layer states for cell-type classification were normalized in two different ways. First is the softmax function over cell types for multiclass classification:
where Y′ ∈ ℝN×T, and each row is the predicted probabilities over the T cell types for a cell. Second is the sigmoid function for multilabel classification:
where Y′′ ∈ ℝN×T, and each element is the predicted probability of the cell type t for the cell i. Note that Y′′ is the multilabel prediction that gives the probability independently for each cell type.
The classification loss and label smoothing
The classification loss for cells in reference data sets is calculated by the weighted cross-entropy loss between the ground-truth label Y and the predicted probabilities (Y′ and Y′′, respectively), combined with the L2 regularization as below:
where wt is the class-weight for cell-type t, satisfying . To avoid the model being dominated by the major populations and ignoring those rare types, we set , and Nt is the number of cells of cell type t in the reference data set. θ represents all the learnable parameters, λ is the penalization coefficient that controls the power of L2 regularization, and the default value of λ is 0.01.
To prevent the model from being overconfident and to improve the stability and generalization of the model, we use label smoothing (Szegedy et al. 2016). We minimize the cross-entropy between the modified targets and the model outputs Y′, where , and the final objective function is as below:
where ε controls the degree of smoothness, set as 0.1 by default. Finally, CAME adopts the Adam optimizer (Kingma and Ba 2015) with a learning rate of 0.001 for training.
Checkpoint selection
When training the heterogeneous graph neural network, we would like to choose the epoch where the classification result of query data sets achieves the highest accuracy. However, in practice, the exact type labels of the query cells are unknown, hindering us from choosing the best model. We put forward a metric to approximate the accuracy. Specifically, we first cluster the query cells to get the pseudolabel Ycluster for the query cells and introduce AMI (Vinh et al. 2010) to account for the chance between the model-predicted cell-type labels and the pseudolabels of the query cells to help decide when to stop. AMI is defined as
where H(X) is the entropy of X, and MI(X, Y) is the mutual information between variables X and Y. E[MI(Ycluster, Y′)] is the expected mutual information based on a “permutation model” (Ahrens 1971) in which cluster labels are generated randomly subject to having a fixed number of clusters and points in each cluster. We think that a well-trained model is expected to preserve the intrinsic data structure so that the predicted labels should be highly consistent with the pseudolabels to some extent. We run the model with 400 epochs and choose the checkpoint with the largest AMI. The clustering process is described in detail in the section Preclustering of the Query Cells.
Training using the mini-batches on subgraphs
When training CAME on the graphic processing unit (GPU), the size of a data set will be limited by the GPU memory. For example, training CAME on 100,000 cells could take about 13.75 GB of GPU memory, which exceeds the graphic memory of most GPUs. To handle this issue, we used a mini-batch training process by using the graph segmentation technique. Specifically, we first randomly divided all the cells (including cells in reference and query) into several groups, taken as mini-batches. For each mini-batch, we created a node-induced subgraph for a given group of cells, which contains all the cells in this group and all the genes expressed by these cells. Then, we iterated all subgraphs and fed the subgraphs to the graph neural network one by one. All the parameters were updated for each mini-batch training process. We performed extensive experiments by using the mini-batch training process and found it is suitable to choose the batch size as 8192 or more, for that achieved the comparable accuracy compared with whole graph training (Supplemental Fig. S18A), and the cost of GPU memory stays constant (2.4 GB) for data sets at different scales (Supplemental Fig. S18B). Such a low consumption of graphic memory means you can use CAME on almost all graphic cards. It is worth noting that the runtime of the batch-training process will be largely increased (Supplemental Fig. S18B) because we cannot feed forward the whole graph on a single epoch. To fully test whether CAME can be scaled to atlas-scale data with millions of cells, we synthesized a pair of data sets with about a million of cells in total by up-sampling from the human and mouse pancreas data sets, which is shown in Figure 3B. Here, the number of cells for both the reference and query data sets are 505,571 and 501,676, respectively. The peak GPU memory usage is still very low (5.78 GB) because we used a default mini-batch size of 8192 (which is relatively small compared with the number of cells); the peak RAM usage is 87 GB, and most servers can meet this memory requirement. The running time of CAME on such a large pair of data sets is 5.48 h, which is acceptable, and one can use a bigger batch size to reduce the running time if the GPU memory is large enough. All the tests were run on a 3.79-GHz AMD 3900X central processing unit with 128 GB of RAM and a 3090 graphic card with 24 GB of RAM.
Preprocessing of the single-cell data sets
For each scRNA-seq data set, we first normalized the counts of each cell by its library size (the total counts of that cell) with a scale factor multiplied (the median of library sizes by default) and log-transformed with a pseudocount added for the downstream analysis.
Gene selection
HVGs and DEGs are generally thought to be highly informative, and the latter is especially useful for cell-type characterization. Therefore, we used both HVGs and DEGs and extended them using homologous mappings to form the HIG sets for constructing the heterogeneous graph. We adopted the same approach as used in Seurat-v2 (Butler et al. 2018) with the SCANPY (Wolf et al. 2018) built-in function highly_variable_genes() to identify HVGs separately from both the reference and query data. Specifically speaking, it calculated the average expression and dispersion (variance/mean) for each gene and placed these genes into several bins based on the (log-transformed) average expression. The normalized dispersions were then obtained by scaling with the mean and standard deviation of the dispersions within each bin. We selected the top 2000 genes with the highest dispersions as HVGs of that data set. We computed the DEGs separately for reference and query data set by a Student's t-test, which is performed through the rank_genes_groups() function from the SCANPY package (Wolf et al. 2018). For reference data, cells are grouped by their cell-type labels, whereas for the query data, cells are grouped by their pseudolabels, that is, the preclustering labels.
Genes used as the cell-node features should be shared between species (or data sets). For both reference and query data sets, we first took the top 50 DEGs for each cell group and retained genes with one-to-one homology in the other species. We then took the union of the resulting two sets of genes for input. The resulting number of genes used for defining cell-node features ranges from 240 to 400 for distant species pairs (e.g., human to zebrafish) and from 400 to 900 for the others.
We combined both HVGs and DEGs from the reference and query data to decide the node genes used for training the graph neural network. Specifically, we first took the union of the HVGs and DEGs for each data set, denoted as and for reference and query, respectively. Then we extracted the genes that have homologies in from the query data, and the homologous genes for from the reference data, denoted as and respectively. Finally, we determined , the union of and , as the node genes for the reference species and as the node genes for the query species. The resulting number of node genes for each pair of data sets ranges from 5415 to 7050, with a median around 6787.
Orthology inference
We downloaded the gene homology information for each species pair from the BioMart web server (http://www.ensembl.org/biomart/martview) derived from the was derived from the Ensembl Compara pipeline (Kinsella et al. 2011). We also adopted Domainoid (Persson et al. 2019) to rerun our benchmarks. We downloaded protein sequence per gene (FASTA) for all the species used in this study from UniProt (Apweiler et al. 2004; https://www.uniprot.org/tool-dashboard) and then used Domainoid to infer orthologous protein pairs. After that, we converted the protein ID to the gene name (or Ensembl gene id) according to the gene format of each species involved in the data set by UniProt.
Construction of the single-cell graphs based on KNNs
The normalized expression matrices were centralized and scaled within each data set, followed by principal component analysis (PCA) to reduce the dimensionality. We searched approximate KNNs for each cell based on the top 30 PCs with the highest explained variances. We adopted k = 5 neighbors for each cell to make the graph sparse enough for computational efficiency. These neighbor connections provided “cell–cell” edges as a part of the heterogeneous graph.
Preclustering of the query cells
To facilitate model selection, we preclustered the query cells using a graph-based clustering method, that is, performing community detection using the Leiden algorithm (Traag et al. 2019) on the single-cell KNN graph. We constructed the KNN graph in almost the same way as described above, except that the number of neighbors k was set as 20, and the clustering resolution is set as 0.4 by default.
Unifying cell-type labels across data sets
For the data sets downloaded from the Cell BLAST web server (Cao et al. 2020), their cell-type labels were already unified by Cell Ontology (Diehl et al. 2016), a structured vocabulary for cell types, whereas for unifying annotations of the other data sets, we referred to Cell Ontology and manually adjusted the annotations. These annotations were used as ground truth.
Gene module extraction
To extract cell-type-specific gene modules shared between species, we took all the gene embeddings (of both species) on the last hidden layer and performed KNN searching for each gene. Like for clustering cells, we performed Leiden community detection on the KNN graph of genes. The clustering resolution was set as 0.8 by default. The expression information of HVGs and top DEGs for each cell type were used, and thus the gene modules mainly cover cell-type-specific ones. We can compute the z-scores of the gene-to-cell-type using the function “came.ana.module_enrichment_for_classes” in CAME to determine that. If some non-cell-type-specific genes are included in the scRNA-seq data, these genes will be assigned relatively low gene-to-cell-type z-scores. However, they will be assigned to a certain gene module anyway.
Calculating weights between gene modules
The weight Sij between homologous gene modules Modi and Modj on the abstracted graph was calculated as follows:
where hg is the embedding vector of gene g, and sim( · , · ) is the similarity function, cosine similarity by default. |Mod| represents the number of genes in this module.
Benchmarking cell-type assignment
For benchmarking cell-type assignment, we collected 54 scRNA-seq data sets from five tissues across seven different species (Supplemental Fig. 3A; Supplemental Table S1), paired data sets of different species within the same tissue, and filtered those pairs where >50% of query cells is unresolved in the reference cell types, resulting in 649 cross-species data set pairs. For each data set, we removed the cell types of fewer than 10 cells as in the method of Abdelaal et al. (2019). CAME was compared with five state-of-the-art methods including SAMap (Tarashansky et al. 2021), Seurat V3 (Stuart et al. 2019), scmap (Kiselev et al. 2018), SciBet (Li et al. 2020), and Cell BLAST (Cao et al. 2020). For SAMap, we preprocessed the raw data with the SAM() function and used the same homologous gene mappings as CAME. For Seurat V3, we input the raw data; used the default normalize process by NormalizeData() function; extracted the top 2000 HVGs by its FindVariableFeatures() function for reference and query, respectively; and performed further annotation process as described in its documentation. For scmap, we log-transformed the raw counts with pseudocount 1 added and used its inherited function selectFeaures() to select the top 2000 HVGs with a threshold = 0.1 in function scmapCluster() (which works better for the cross-species scenario than its default value). For SciBet, we used R to perform all the operations. We first input the library-size-normalized data calculated by the cpm() function of the package edgeR (Robinson et al. 2010), used the SelectGene_R() function from the SciBet package to select 2000 HVGs, and used SciBet_R() function to annotate the query data. For Cell BLAST, we used the raw data as input and used find_variable_genes() to select HVGs with default parameters and took the union of the HVGs between reference and query data sets. After that, the data sets were combined together to remove their batch effects by using function fit_DIRECTi() with lambad_reg = 0.001 as suggested by the original investigators to stabilize the training process. Cell BLAST also provides a supervised training process that leverages the cell-type labels of reference data sets to perform label transfer. However, it led to a 4% decrease in the average accuracy compared with their previous batch effect correction process. All hyperparameters not mentioned were set with default values in these four packages.
To evaluate the performance of the cell-type assignment, we adopted three metrics: Accuracy, MacroF1, and WeightedF1. Accuracy is the most common criterion, and it directly measures how many of the predictions are the same as the actual ones:
where is the sign of cardinality. Specifically, means the number of the total cells, and means the number of correctly predicted ones.
We also used MacroF1 and WeightedF1 which consider the F1-score for each cell type. For a binary classification task, precision and recall are calculated as
and
respectively, where TP, FP, and FN represent the number of true positives, false positives, and false negatives, respectively.
The F1-score is the harmonic mean of precision and recall:
and the MacroF1 is defined as the average of class-wise F1-scores,
where represents the F1-score for cell type t. The WeightedF1 considers the proportion of each class,
where Nt/N represents the proportion of type t in all cells.
We also used the graph connectivity (GC) metric (Luecken et al. 2022) to assess whether the kNN graph representation, G, of the integrated data directly connects all the cells with the same cell identity label. For each cell identity label c, we created the subset kNN graph G(NC;Ec) to contain only cells from a given label. The GC is calculated as
Here, C denotes the set of cell identity labels, |LCC()| means the number of nodes in the largest connected component of the graph, and |Nc| is the number of nodes with cell identity c. The GC has a range of (0, 1], where the bigger the GC score is, the better connection of the integration is. Here, k is set as 15.
Benchmarking data integration
Seurat-v3 (Stuart et al. 2019) was performed using the corresponding R package (R Core Team 2013) through SeuratWrapper, following the online documents with default settings. Seurat adopts normalization and the top 2000 HVGs by Seurat function NormalizeData and FindVariableFeatures, respectively. Harmony was performed on the PCA-reduced embeddings. The number of reduced dimensions for it was set as 50 for all pairs of data sets. LIGER (Welch et al. 2019) was input with the raw count data and run with the default pipeline (http://htmlpreview.github.io/?https://github.com/welch-lab/liger/blob/master/vignettes/Integrating_multi_scRNA_data.html). Cell BLAST (Cao et al. 2020) was performed using its Python package, following the standard pipeline with the default settings (https://cblast.gao-lab.org/doc-latest/start.html). SAMap (Tarashansky et al. 2021) was input with the results preprocessed by SAM (Tarashansky et al. 2019; https://github.com/atarashansky/self-assembling-manifold) and the same homologous gene mappings used by CAME. The cell-type annotations were transferred by using a diffusion process with the default pipeline of SAMap (https://github.com/atarashansky/SAMap/blob/main/SAMap_vignette.ipynb).
Software availability
The CAME algorithm is implemented in Python and is available on GitHub (https://github.com/zhanglabtools/CAME). CAME analysis scripts from this work are provided as Supplemental Code.
Supplementary Material
Acknowledgments
This work has been supported by the National Key Research and Development Program of China (nos. 2019YFA0709501 and 2021YFA1302500 to S.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (nos. XDA16021400 and XDPB17 to S.Z.), the Key-Area Research and Development of Guangdong Province (no. 2020B1111190001 to S.Z.), the National Natural Science Foundation of China (nos. 12126605 and 61621003 to S.Z.), and the CAS Project for Young Scientists in Basic Research (no. YSBR-034 to S.Z.).
Author contributions: S.Z. conceived and supervised the project. X.L. and Q.S. developed and implemented the CAME algorithm. X.L., Q.S., and S.Z. validated the methods and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276868.122.
Competing interest statement
The authors declare no competing interests.
References
- Abdelaal T, Michielsen L, Cats D, Hoogduin D, Mei H, Reinders MJT, Mahfouz A. 2019. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol 20: 194. 10.1186/s13059-019-1795-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens H. 1971. Lancaster, H. O.: the chi-squared distribution. Wiley & Sons, Inc., New York 1969. X, 366 S., 140 s. Biom Z 13: 363–364. 10.1002/bimj.19710130512 [DOI] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. 2017. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14: 1083–1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. 2004. UniProt: the universal protein knowledgebase. Nucleic Acids Res 32: D115–D119. 10.1093/nar/gkh131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20: 163–172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, et al. 2016. The origin and evolution of cell types. Nat Rev Genet 17: 744–757. 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z-J, Wei L, Lu S, Yang D-C, Gao G. 2020. Searching large-scale scRNA-seq databases via unbiased cell embedding with Cell BLAST. Nat Commun 11: 3458. 10.1038/s41467-020-17281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AD, Meehan TF, Bradford YM, Brush MH, Dahdul WM, Dougall DS, He Y, Osumi-Sutherland D, Ruttenberg A, Sarntivijai S, et al. 2016. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J Biomed Semantics 7: 44. 10.1186/s13326-016-0088-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drokhlyansky E, Smillie CS, Van Wittenberghe N, Ericsson M, Griffin GK, Eraslan G, Dionne D, Cuoco MS, Goder-Reiser MN, Sharova T, et al. 2020. The human and mouse enteric nervous system at single-cell resolution. Cell 182: 1606–1622.e23. 10.1016/j.cell.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Scott MA, Zelinka C, Sherwood P. 2010. A novel type of glial cell in the retina is stimulated by insulin-like growth factor 1 and may exacerbate damage to neurons and Müller glia. Glia 58: 633–649. 10.1002/glia.20950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Liu J, Kriebel AR, Preissl S, Luo C, Castanon R, Sandoval J, Rivkin A, Nery JR, Behrens MM, et al. 2021. Iterative single-cell multi-omic integration using online learning. Nat Biotechnol 39: 1000–1007. 10.1038/s41587-021-00867-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, Balic A, Giladi A, Sheban F, Dutertre C-A, et al. 2019. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 179: 1609–1622.e16. 10.1016/j.cell.2019.11.010 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium, Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer SM, Neuhauss SC. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics 289: 1045–1060. 10.1007/s00438-014-0889-2 [DOI] [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, et al. 2020. Gene regulatory networks controlling vertebrate retinal regeneration. Science 370: eabb8598. 10.1126/science.abb8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573: 61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Li X, Hu G, Lyu Y, Susztak K, Li M. 2020. Iterative transfer learning with neural network for clustering and cell type classification in single-cell RNA-seq analysis. Nat Mach Intelligence 2: 607–618. 10.1038/s42256-020-00233-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S, Chan-Ling T. 2004. Characterization of smooth muscle cell and pericyte differentiation in the rat retina in vivo. Investi Ophthalmol Vis Sci 45: 2795–2806. 10.1167/iovs.03-1312 [DOI] [PubMed] [Google Scholar]
- Kaduk M, Sonnhammer EJB. 2017. Improved orthology inference with Hieranoid 2. Bioinformatics 33: 1154–1159. 10.1093/bioinformatics/btw774 [DOI] [PubMed] [Google Scholar]
- Kingma DP, Ba J. 2015. Adam: a method for stochastic optimization. arXiv:1412.6980 [cs.LG]. 10.48550/arXiv.1412.6980 [DOI]
- Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011: bar030. 10.1093/database/bar030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Yiu A, Hemberg M. 2018. scmap: projection of single-cell RNA-seq data across data sets. Nat Methods 15: 359–362. 10.1038/nmeth.4644 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. 2015. The technology and biology of single-cell RNA sequencing. Mol Cell 58: 610–620. 10.1016/j.molcel.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S. 2019. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods 16: 1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, et al. 2018. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36: 70–80. 10.1038/nbt.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Ba J, Kiros JR, Hinton GE. 2016. Layer normalization. arXiv:1607.06450 [stat.ML]. 10.48550/arXiv.1607.06450 [DOI]
- Li C, Liu B, Kang B, Liu Z, Liu Y, Chen C, Ren X, Zhang Z. 2020. SciBet as a portable and fast single cell type identifier. Nat Commun 11: 1818. 10.1038/s41467-020-15523-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfollahi M, Naghipourfar M, Luecken MD, Khajavi M, Büttner M, Wagenstetter M, Avsec Ž, Gayoso A, Yosef N, Interlandi M, et al. 2022. Mapping single-cell data to reference atlases by transfer learning. Nat Biotechnol 40: 121–130. 10.1038/s41587-021-01001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken MD, Büttner M, Chaichoompu K, Danese A, Interlandi M, Müller MF, Strobl DC, Zappia L, Dugas M, Colomé-Tatché M, et al. 2022. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods 19: 41–50. 10.1038/s41592-021-01336-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Arendt D. 2017. How single-cell genomics is changing evolutionary and developmental biology. Annu Rev Cell Dev Biol 33: 537–553. 10.1146/annurev-cellbio-100616-060818 [DOI] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML]. 10.48550/arXiv.1802.03426 [DOI]
- Menon M, Mohammadi S, Davila-Velderrain J, Goods BA, Cadwell TD, Xing Y, Stemmer-Rachamimov A, Shalek AK, Love JC, Kellis M, et al. 2019. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun 10: 4902. 10.1038/s41467-019-12780-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers Y, Jones TE, Jyothi D, Yates B, Ferret M, Portell-Silva L, Codo L, Cosentino S, Marcet-Houben M, Vlasova AJ, et al. 2022. The Quest for Orthologs orthology benchmark service in 2022. Nucleic Acids Res 50: W623–W632. 10.1093/nar/gkac330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. 2006. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci 103: 17973–17978. 10.1073/pnas.0605938103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson E, Kaduk M, Forslund SK, Sonnhammer ELL. 2019. Domainoid: domain-oriented orthology inference. BMC Bioinformatics 20: 523. 10.1186/s12859-019-3137-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Shendure J, Trapnell C. 2019. Supervised classification enables rapid annotation of cell atlases. Nat Methods 16: 983–986. 10.1038/s41592-019-0535-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V, Venkatesh B. 2018. The divergent genomes of teleosts. Annu Rev Anim Biosci 6: 47–68. 10.1146/annurev-animal-030117-014821 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang SJC, et al. 2018. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174: 1015–1030.e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichtkrull M, Kipf TN, Bloem P, van den Berg R, Titov I, Welling M. 2018. Modeling relational data with graph convolutional networks. In The semantic web (ed. Gangemi A, et al.), pp. 593–607. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- Schreiber F, Sonnhammer ELL. 2013. Hieranoid: hierarchical orthology inference. J Mol Biol 425: 2072–2081. 10.1016/j.jmb.2013.02.018 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, Mukamel Z, Amit I, Hejnol A, Degnan BM, Tanay A. 2018. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol 2: 1176–1188. 10.1038/s41559-018-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer MER. 2019. Cross-species analysis of single-cell transcriptomic data. Front Cell Dev Biol 7: 175. 10.3389/fcell.2019.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shami AN, Zheng X, Munyoki SK, Ma Q, Manske GL, Green CD, Sukhwani M, Orwig KE, Li JZ, Hammoud SS. 2020. Single-cell RNA sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis. Dev Cell 54: 529–547.e12. 10.1016/j.devcel.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Liao J, Lu X, Xue R, Ai N, Fan X. 2020. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience 23: 100882. 10.1016/j.isci.2020.100882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wang Z, Lindtner S, Yang L, Shang Z, Tian Y, Guo R, You Y, Zhou W, Rubenstein JL, et al. 2022. Dlx1/2-dependent expression of Meis2 promotes neuronal fate determination in the mammalian striatum. Development 149: dev200035. 10.1242/dev.200035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. 2016. Rethinking the inception architecture for computer vision. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, pp. 2818–2826. [Google Scholar]
- Tan Y, Cahan P. 2019. SingleCellNet: a computational tool to classify single cell RNA-seq data across platforms and across species. Cell Syst 9: 207–213.e2. 10.1016/j.cels.2019.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarashansky AJ, Xue Y, Li P, Quake SR, Wang B. 2019. Self-assembling manifolds in single-cell RNA sequencing data. eLife 8: e48994. 10.7554/eLife.48994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarashansky AJ, Musser JM, Khariton M, Li P, Arendt D, Quake SR, Wang B. 2021. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10: e66747. 10.7554/eLife.66747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. 2018. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563: 72–78. 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. 2018. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science (New York, NY) 360: 881–888. 10.1126/science.aar4237 [DOI] [PubMed] [Google Scholar]
- Traag VA, Waltman L, van Eck NJ. 2019. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9: 5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veličković P, Cucurull G, Casanova A, Romero A, Liò P, Bengio Y. 2017. Graph attention networks. arXiv:1710.10903 [stat.ML]. 10.48550/arXiv.1710.10903 [DOI]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19: 327–335. 10.1101/gr.073585.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh NX, Epps J, Bailey J. 2010. Information theoretic measures for clusterings comparison: variants, properties, normalization and correction for chance. J Mach Learn Res 11: 2837–2854. 10.5555/1756006.1953024 [DOI] [Google Scholar]
- Wang J, Sun H, Jiang M, Li J, Zhang P, Chen H, Mei Y, Fei L, Lai S, Han X, et al. 2021. Tracing cell-type evolution by cross-species comparison of cell atlases. Cell Rep 34: 108803. 10.1016/j.celrep.2021.108803 [DOI] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. 2019. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177: 1873–1887.e17. 10.1016/j.cell.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19: 15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang C, Chen M, Yuan Y, Li L, Huang Z, Yuan Y, Yang H, Wang Q, Zhang X. 2022. Retina cell atlases of multiple species and an online platform for retina cell-type markers. J Genet Genomics 49: 262–265. 10.1016/j.jgg.2021.10.008 [DOI] [PubMed] [Google Scholar]
- Zelinka CP, Scott MA, Volkov L, Fischer AJ. 2012. The reactivity, distribution and abundance of non-astrocytic inner retinal glial (NIRG) cells are regulated by microglia, acute damage, and IGF1. PLoS One 7: e44477. 10.1371/journal.pone.0044477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang S. 2019. Learning common and specific patterns from data of multiple interrelated biological scenarios with matrix factorization. Nucleic Acids Res 47: 6606–6617. 10.1093/nar/gkz488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AW, O'Flanagan C, Chavez EA, Lim JLP, Ceglia N, McPherson A, Wiens M, Walters P, Chan T, Hewitson B, et al. 2019a. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat Methods 16: 1007–1015. 10.1038/s41592-019-0529-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, Luo T, Xu L, Liao G, Yan M, et al. 2019b. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res 47: D721–D728. 10.1093/nar/gky900 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.