Abstract
Many animals perceive odorant molecules by collecting information from ensembles of olfactory neurons, where each neuron uses receptors that are tuned to recognize certain odorant molecules with different binding affinity. Olfactory systems are able, in principle, to detect and discriminate diverse odorants using combinatorial coding strategies. We have combined microfluidics and multineuronal imaging to study the ensemble-level olfactory representations at the sensory periphery of the nematode Caenorhabditis elegans. The collective activity of C. elegans chemosensory neurons reveals high-dimensional representations of olfactory information across a broad space of odorant molecules. We reveal diverse tuning properties and dose-response curves across chemosensory neurons and across odorants. We describe the unique contribution of each sensory neuron to an ensemble-level code for volatile odorants. We show that a natural stimuli, a set of nematode pheromones, are also encoded by the sensory ensemble. The integrated activity of the C. elegans chemosensory neurons contains sufficient information to robustly encode the intensity and identity of diverse chemical stimuli.
The responses of C. elegans neurons to many odors were found to contain sufficient information to encode odor identity.
INTRODUCTION
Many animals exhibit diverse behaviors—navigating the world, finding food, avoiding dangers—in response to olfactory cues. To do this, their olfactory systems distinguish the identity and intensity of numerous odorant molecules.
Insect and mammalian olfactory systems use large ensembles of olfactory sensory neurons to detect odorants and pheromones (1–6). Each olfactory sensory neuron usually expresses a specific olfactory receptor tuned to recognize odorant molecules by ligand-receptor binding affinity (7). A given receptor is typically activated by many different odorant molecules, and each odorant can activate multiple receptors (1, 8). These olfactory systems can potentially use combinatorial coding strategies to distinguish and identify large numbers of odorant molecules.
Caenorhabditis elegans senses many odorants across wide concentration ranges (9–11). However, its olfactory circuits have a compact cellular and molecular organization that differs from insects and mammals (Fig. 1A). The C. elegans genome encodes >1000 putative chemosensory G protein–coupled receptors (GPCRs) (12, 13); at least 200 GPCRs are expressed by its 11 pairs of amphid chemosensory neurons. This suggests both a substantial capacity for olfactory detection and a coding strategy where the properties of each neuron are shaped by many receptors (12–14).
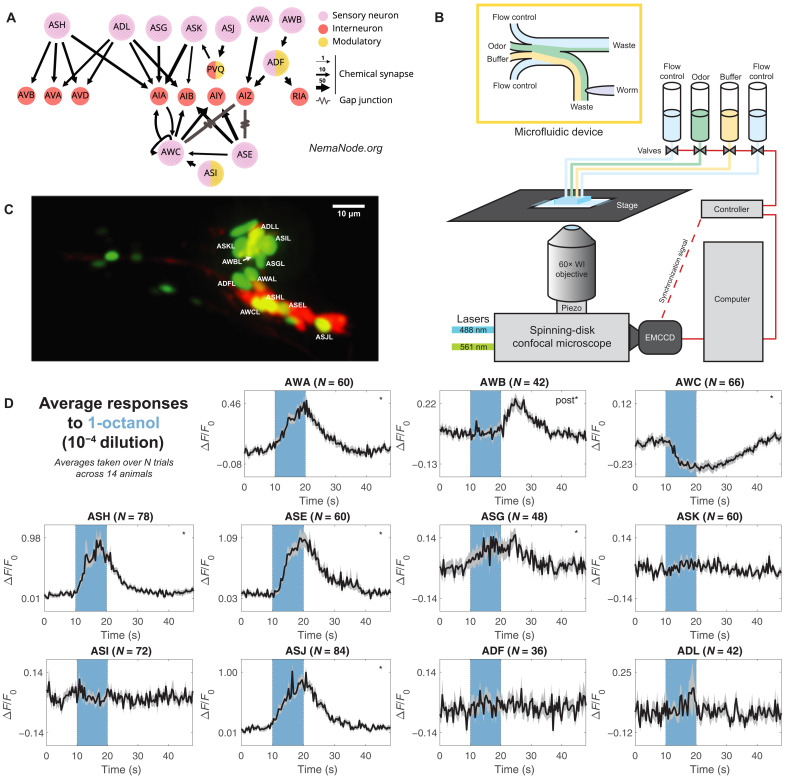
Fig. 1. Labeling and recording from chemosensory neurons.
(A) Downstream partners of the 11 chemosensory neurons in the C. elegans connectome (55, 56). Panel generated at nemanode.org. (B) Adult C. elegans were immobilized inside a microfluidic device and controllably presented with odorant solutions. Each animal was volumetrically imaged at 2.5 Hz with a spinning-disk confocal microscope during stimulus presentations. EMCCD, electron multiplying charge-coupled device; WI, water-immersion. (C) Animals expressed nuclear-localized GCaMP6s in all ciliated sensory neurons. A sparse wCherry landmark distinguished the 11 chemosensory neurons. Here, a dual-color maximum projection image shows the head of the worm. The 11 chemosensory neurons on the near (L) side are labeled. For clarity, the chemosensory neurons on the far side and other ciliated neurons are not labeled. (D) Neuronal activity traces of the 11 chemosensory neurons in response to a single odorant presentation (1-octanol, 10−4 dilution), averaged across multiple trials across 14 animals. The number of trials varies across neurons because neurons that were occluded or improperly tracked were excluded from the dataset (see Materials and Methods). The 10-s odorant delivery period is shown by the colored bar. Significant responses (q ≤ 0.01) are marked with stars, with “post” indicating a significant response to stimulus removal (OFF response). Error bars (gray) are SEM.
C. elegans chemosensory neurons have been characterized as individual sensors for specific modalities from volatile odorants (AWA, AWB, and AWC) (15–21), soluble chemicals (ASE) (22, 23), and ascaroside pheromones (ADL, ASK, and ADF) (24–29) to nociception (ASH) (10, 30–33). A few chemosensory neurons detect gases (CO2 and O2) or temperature changes, in addition to odorants (12, 34).
Behavioral studies first established AWA, AWB, and AWC to be the primary detectors of odorants. Laser ablation of AWA or AWC severely degrades chemotaxis toward selected attractive odorants. However, when both AWA and AWC are ablated, animals could move toward odorant sources (35). In similar experiments with selected organic compounds and salts, ablation of other chemosensory neurons—ASE, ADF, ASG, ASI, ASJ, and ASK—degrades chemotaxis to a lesser extent (10). Therefore, although some neurons are more important for chemotaxis toward some odorants than others, chemosensation does not rely on single neurons.
The stimulus-evoked properties of C. elegans chemosensory neurons have also been described through the detection of selected odorants by selected neurons (15–21). For example, isoamyl alcohol is detected by AWC, AWB, and ASH (15), and benzaldehyde is detected by AWA, AWB, AWC, and ASE (17). AWA responds to a wide range of volatile odorants (16). Diacetyl is detected by AWA at low concentrations and by ASH at high concentrations, as well as by AWC, ASK, and ASE. In some cases, the left and right pairs of a chemosensory neuron type detect different odorant molecules. For example, the left and right AWC neurons, AWCL and AWCR, are stochastically asymmetric, where one detects butanone and the other detects 2,3-pentanedione (36, 37). The ASE neurons, primarily gustatory, respond asymmetrically to different ions, where ASEL detects sodium ions and ASER detects chloride and potassium ions (22, 23).
We know less about the response properties of odorant receptors. ODR-10 remains its most thoroughly characterized odorant receptor, expressed in AWA and sensitive to diacetyl, an attractive stimulus. Ectopic expression of ODR-10 in AWB leads to diacetyl repulsion. This suggests that the attractive and aversive behaviors are encoded by the neuron, instead of specific odorants (38). Consistently, AWB and AWC are also needed for aversive olfactory learning (39, 40) and activating different subsets of chemosensory neurons changes the activity of different downstream interneurons (41).
Complex properties of individual neurons and their relationships to behaviors have been extensively examined. AWC, ASH, and ASE exhibit adaptation: When a chemical stimulus is prolonged from minutes to hours, both neuronal activity and behavioral responses diminish (16, 42–45). AWA and AWC change their response properties in a context-dependent manner (46, 47). AWA neurons fire action potentials that may encode stimulus-specific features (48). Complex activity patterns of AWA have been directly mapped to behavioral patterns (16, 17, 38, 48, 49).
Although odorant-evoked responses in many individual C. elegans chemosensory neurons are well characterized, how their collective dynamics might represent odorant information as an ensemble remains unexamined. We set out to characterize how this chemosensory ensemble responds to a chemically diverse space of odorants at different concentrations and how the tuning properties of each chemosensory neuron might relate to an ensemble-level code. We assembled a panel of olfactory stimuli spanning a diverse molecular chemistry and used microfluidics to deliver these odorants at multiple concentrations (Fig. 1B). To efficiently record neuronal responses at the sensory periphery, we used a transgenic animal that allowed simultaneous measurement of intracellular calcium dynamics in all amphid chemosensory neurons (Fig. 1, A and C).
We found that most odorant-evoked responses are widespread across the chemosensory ensemble. Dose-response curves are different for different odorant molecules, whether comparing the responses of the same neuron to different odorants or comparing the responses of different neurons to the same odorant. Odorant identity and intensity information can be reliably decoded by the collective activity of the chemosensory ensemble. A set of pheromones also evokes ensemble-level responses but with a distinct pattern from volatile odorants. We conclude that the ensemble-level representations of different odorants in the small sensory system of C. elegans contain sufficient information to accurately distinguish the identity and intensity of odorant molecules across olfactory stimulus space.
RESULTS
Calcium imaging of chemosensory neurons with representative odorant stimuli
We developed a GCaMP6s calcium reporter line to simultaneously record calcium dynamics in all ciliated sensory neurons (Supplementary Methods and fig. S1). We focused on the 11 pairs of amphid chemosensory neurons: AWA, AWB, AWC, ASE, ASG, ASH, ASI, ASJ, ASK, ADL, and ADF (Fig. 1A). We immobilized and positioned young adult C. elegans in a microfluidic device that allows odorants to flow past its nose (Fig. 1B) (50). We adapted a multichannel microfluidic device (4) to control the delivery of pulses of single and mixed odorant solutions. Volumetric imaging was performed at 2.5 Hz with a spinning-disk confocal microscope (Fig. 1, B to D, and fig. S2).
We assembled a stimulus panel of 23 odorants, selected to span the chemical diversity of 122 previously studied molecules in C. elegans olfaction (35, 51). We included exemplars of six chemical classes: alcohols (1-pentanol, 1-hexanol, 1-heptanol, 1-octanol, 1-nonanol, isoamyl alcohol, and geraniol), aromatics (benzaldehyde and methyl salicylate), esters (ethyl acetate, ethyl butyrate, pentyl acetate, ethyl heptanoate, and butyl butyrate), ketones (2-butanone, diacetyl, 2-heptanone, 2-nonanone, and 2,3-pentanedione), pyrazines (2,5-dimethyl pyrazine and 2-methyl pyrazine), and thiazoles (2-isobutylthiazole and 2,4,5-trimethylthiazole). To assess chemical diversity, we constructed a geometrical odor space using physical and chemical descriptors of molecular structure (52). Our 23 odorants broadly sample this geometrical space (fig. S3, A and B) (52).
We recorded the responses of all amphid chemosensory neurons to >70 stimulus conditions, testing each of the 23 odorants at multiple concentrations. Individual animals were repeatedly presented with series of 10-s odorant pulses separated by 30-s buffer blanks (fig. S2, A and B). For each stimulus condition, we recorded the responses to ∼80 odor presentations across multiple animals (Fig. 2, A to C, and fig. S3, C and D). The highest concentrations we tested were 10−4 dilutions. The lowest concentrations we tested (10−8 dilutions) did not elicit significant responses from any neuron.
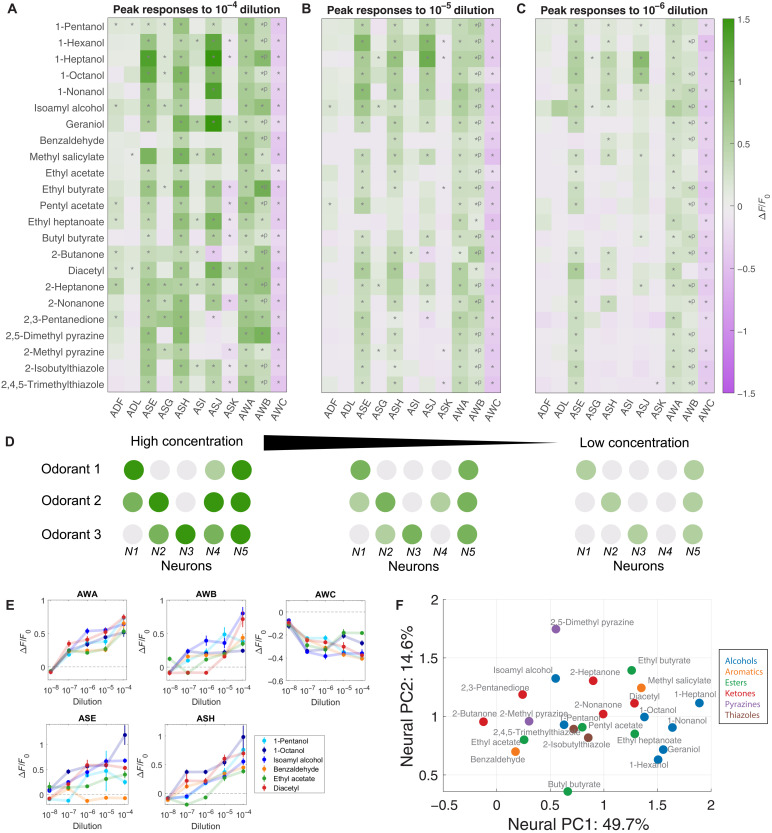
Fig. 2. Ensemble responses to a broad odorant panel.
Average peak responses of the 11 chemosensory neurons to odorants at (A) high concentration (10−4 dilution), (B) medium concentration (10−5 dilution), and (C) low concentration (10−6 dilution). Peaks were computed from a time window from onset of odor delivery to 10 s after odor removal. Responses are reported as ∆F/F0. Significant responses (q ≤ 0.01, two-tailed, paired t tests) are indicated with stars. Significant OFF responses are indicated with *P. Most odorants elicit significant responses from unique neuron combinations. (D) Schematic of coding strategy observed in (A) to (C). Different odorants evoke responses in distinct subsets of sensory neurons. Responses are generally stronger at high concentrations. Additional neurons are activated as concentration increases. (E) Dose responses of the peak responses of AWA, AWB, AWC, ASE, and ASH are diverse, with distinct concentration-dependent curves in response to different odorants. See fig. S3F for dose responses of the other six sensory neurons. Error bars are SEM. (F) PC space built from standardized peak average neural responses. Chemical class is indicated by color. Some odorant classes, such as alcohols and ketones, have more similar neural representations, while other odorant classes, such as esters, have more diverse representations. See fig. S3I for PC loadings.
Odorants elicit ensemble responses
Across our odorant panel, calcium imaging captured many sensory neuron responses, some previously characterized and some unknown. Nearly every odorant reliably activated more sensory neurons than previously described. For example, diacetyl, attractive at low concentrations (53), reliably activated AWA upon odor onset at all concentrations (Fig. 2, A to C). 1-Octanol, a repellent (54), reliably activated ASH and inhibited AWC across concentrations (Fig. 2, A to C). However, additional reliable responses were also uncovered. For example, AWC was inhibited by butyl butyrate, and ASJ was activated by 1-octanol. Similarly, isoamyl alcohol not only activated AWA, AWB, AWC, and ASH at different concentrations, as previously reported (15), but also activated ASE and ASG (Fig. 2, A to C). At high concentrations, every odorant elicited responses from multiple sensory neurons. We observed substantial overlap in the sets of responding neurons for different odorants (Fig. 2, A to D).
Most chemosensory neurons exhibited ON responses to most odorants—changes in calcium levels upon odorant onset. We also observed OFF responses—changes in calcium levels upon odorant removal. For example, AWB has been reported to exhibit ON and OFF responses at different isoamyl alcohol concentrations (15). We confirmed this result and also found that AWB had ON responses to some odorants, such as diacetyl at high concentration, and OFF responses to 1-hexanol and 1-octanol (Fig. 1D and fig. S2E).
Most chemosensory neurons exhibited excitatory responses—increases in intracellular calcium levels during stimulus presentation. Some neurons exhibited inhibitory responses—decreases in intracellular calcium levels below the baseline level. A previous work showed that AWC is inhibited by several odorants in our panel, including diacetyl, benzaldehyde, and 2-butanone (16, 17, 21, 39). In our stimulus conditions, AWC is inhibited by every odorant in our panel (Fig. 2A). We also found that ASK is inhibited by many odorants including ethyl butyrate and 2-nonanone (fig. S2F). Some neurons are inhibited by certain odorants but excited by others. For example, ASJ is strongly inhibited by 2-butanone but strongly excited by 1-nonanol (fig. S2G).
The left and right ASE neurons exhibited strong asymmetry in their responses to two odorants—heptanoate and butyl butyrate. Both activated ASEL and inactivated ASER (fig. S2H). AWC, another pair of neurons with known structural asymmetry (20), might exhibit moderate differences in their response dynamics when presented with short odorant pulses (21). Because all other left and right sensory neurons respond symmetrically to all odorants and because the left and right ASE and AWC neurons also respond symmetrically to many odorants, we grouped left and right sensory neurons in all analyses unless otherwise noted.
To compare the temporal dynamics of chemosensory neurons across odorants, we computed pairwise cross-correlations of their activity time courses across odorants (fig. S4, A and B). We found that matrices of pairwise cross-correlations are distinct for different odorants. Measured in terms of peak responses or dynamics, the diversity of ensemble-level dynamics is as large as the number of tested odorants. The compact sensory neuron ensemble of C. elegans may be able to encode the identities of numerous odorants by using this combinatorially large space of distinct activity patterns.
Sensory representations are not dependent on chemical synaptic connections
The C. elegans connectomes have revealed consistent axo-axonic chemical synapses between some sensory neurons and from some interneurons to sensory neurons (Fig. 1A) (55, 56). These connections raise the possibility that ensemble representations might not solely reflect independent responses from individual neurons.
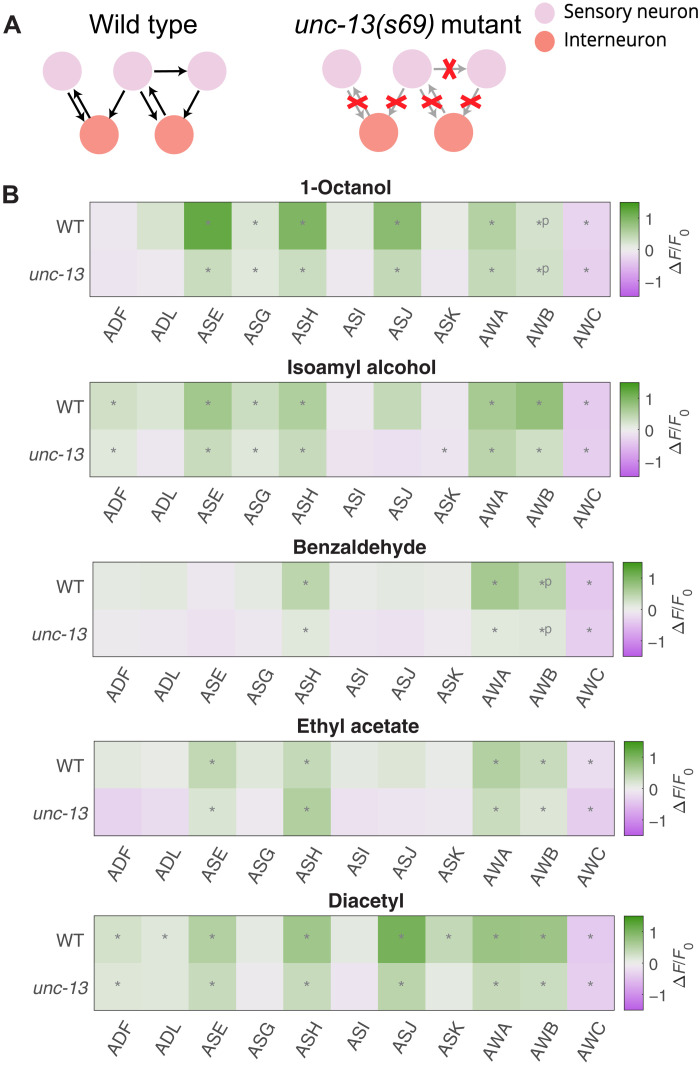
We examined this possibility by analyzing ensemble responses in an unc-13(s69) mutant where synaptic vesicle fusion is mostly blocked (57) (Fig. 3A). We sampled five odorants that represent different chemical classes. In all cases, nearly identical groups of neurons significantly responded (q ≤ 0.01) in wild-type and unc-13 mutants (Fig. 3B). Therefore, chemical synaptic transmission does not appear to be the dominant factor in ensemble responses. The tuning of each neuron to an odorant is likely to be cell intrinsic, a function of the receptors expressed in each neuron.
Fig. 3. Odorant representations in synaptic transmission mutants.
(A) Most of the chemical synapses in unc-13(s69) synaptic transmission mutants are nonfunctional. We recorded neural activity in these mutants during odor presentation. (B) When presented with the same odorants, similar sets of neurons significantly (q ≤ 0.01) responded in wild-type (WT) and unc-13 mutants. Significant OFF responses are indicated with *P.
It is possible that gap junctions or neuromodulation, which are not affected in unc-13 mutants, play roles in shaping sensory responses. We note that there are no direct gap junctions between different chemosensory neurons (Fig. 1A). We also note that response magnitudes in unc-13 mutants, on average across neurons and stimuli, were ∼60% the size of response magnitudes in wild-type animals. Chemical synaptic connections might play a role in amplifying response magnitudes.
Olfactory representations broaden with increasing concentrations
We compared the response properties of different neurons in response to odorants from our panel over three to five orders of magnitude in concentration (Fig. 2, A to C, and fig. S3, C and D). For most odorants and neurons, response magnitudes increased monotonically with odorant concentration—neurons activated at low concentrations were also activated at all higher concentrations. Every odorant is associated with the activation of a characteristic set of neurons at all concentrations above detection threshold. Across all concentrations, for example, 1-pentanol activates AWA and AWC; 1-octanol activates ASE, ASH, AWA, AWB, and AWC; and benzaldehyde activates AWA, AWB, and AWC. Each set of activated neurons may constitute a unique olfactory representation associated with each odorant identity.
For many odorants, increasing concentration spatially broadens olfactory representation by activating more sensory neurons. Different neurons exhibit different thresholds for different odorants. For example, AWB is only activated by 1-pentanol at concentrations above 10−5 dilution, and ADF, ADL, and ASG are only significantly activated by 1-pentanol at 10−4 dilution, the highest tested concentration (fig. S3E). Thus, odorant intensity is represented partly by the magnitude of responses of activated neurons and partly by the number and identities of activated neurons (Fig. 2D).
We used phase-trajectory analysis to illustrate the temporal dynamics of ensemble-level odorant representations. In a low-dimensional principal component (PC) space, these representations follow closed trajectories as they evolve over time following odor presentation (fig. S4C). Along each trajectory, neurons become activated, reach their peak responses, and return to baseline. In this space, the responses to different odorants follow trajectories with different headings from the origin. Trajectories for responses to the same odorant at different concentrations are aligned in direction but differ in magnitude.
Diversity in dose responses across neurons and odorants
We constructed dose-response curves for all 11 chemosensory neurons in response to select odorants from our panel over five orders of magnitude in concentration. The dose-response curves of the 11 chemosensory neurons exhibit substantial diversity (Fig. 2E and fig. S3F). Each odorant can evoke dose-response curves with different thresholds and steepnesses in different neurons. Conversely, each sensory neuron can exhibit dose-response curves with different thresholds and steepnesses for different odorants.
In some cases, neurons detected an odorant with slowly graded responses over a broad dynamic range. Graded responses include AWA’s response to 1-pentanol and ASG’s response to 1-octanol (Fig. 2E and fig. S3F). In other cases, neurons exhibited steep response functions, becoming fully activated or fully inhibited above a sharply defined threshold. Step-like responses include ASE’s response to 1-pentanol and AWB’s response to 1-octanol.
To estimate differences in response steepness across odorants and neurons, we performed log-linear fits on peak responses r as a function of odorant dilution c: r(c) ≈ mlog10c + I and determined the slope m through linear regression. Response steepnesses were diverse, whether for a given neuron across odorants or for a given odorant across neurons (fig. S3G).
Diversity in dose-response curves contrasts with insects and mammals, where olfactory sensory neurons exhibit similarly shaped dose-response curves across neurons and across odorants (4, 58, 59). In insects and mammals, each sensory neuron is generally equipped with one receptor type, whereas in C. elegans, each neuron likely expresses multiple receptors (12, 13). For a nematode neuron, the presence of receptors to multiple odorants, each with different binding affinities, may explain dose-response diversity.
Comparing ensemble-level representations of chemically similar odorants
Different odorants activate distinct but overlapping subsets of the chemosensory ensemble (Fig. 2, A to C). Quantitative differences in the sensitivity of chemosensory neurons to odorants will depend on cell-specific patterns of receptor expression. In most olfactory systems, a typical olfactory receptor is activated by a range of structurally similar odorant molecules with common chemical features. This leads to a systematic dependence of ensemble-level olfactory representations on odorant chemistry. To assess this dependence in C. elegans, we performed hierarchical clustering of odorants from our panel based on ensemble-level responses evoked at high concentrations (fig. S3H). The representations of some molecular classes clustered together. For example, ensemble-level responses to a set of straight-chain alcohols (1-hexanol, 1-heptanol, 1-octanol, and 1-nonanol) were similar to one another, and the ensemble-level responses to a set of ketones (2-butanone, 2,3-pentanedione, and 2-heptanone) were likewise similar. On the other hand, the esters in our panel, a group more diverse in their chemical structure, produced a broader set of representations.
Principal components analysis (PCA) is a quantitative means of comparing high-dimensional ensemble-level representations. We constructed a PC space from all average ensemble-level peak responses. We found that the first two PCs contain 65% of the variance (fig. S3I). The loading of the first two PCs allows us to assess the relative contribution of each sensory neuron to ensemble representations (fig. S3I). We observed a broad distribution of PC loading, which indicates a distributed contribution from all neurons to the separability of odorant representations.
We then asked how different odorants are distributed in the reduced PC space. Overall, the PC space appears compact without distinct clusters (Fig. 2F). Consistent with observations from hierarchical clustering, responses to certain classes of odorants, such as the straight-chain alcohols and two thiazoles, are close to each other in PC space. Responses to members of other classes, such as esters, are distributed more broadly.
Sensory neurons are either broadly or narrowly tuned in the chemical space
Olfactory sensory neurons are tuned to odorants by the relative binding affinities of receptors for different ligands (7). In animals where sensory neurons express single-receptor types, this leads to a systematic dependence of ensemble representation on the chemical properties of odorants and receptors (4, 7, 60, 61). In C. elegans, the tuning of a sensory neuron may also be shaped by the expression of multiple different receptors. To explore the tuning of sensory neurons in chemical space, we projected the activity of each neuron into a PC space based on the chemical structure of odorants (fig. S3A) (52).
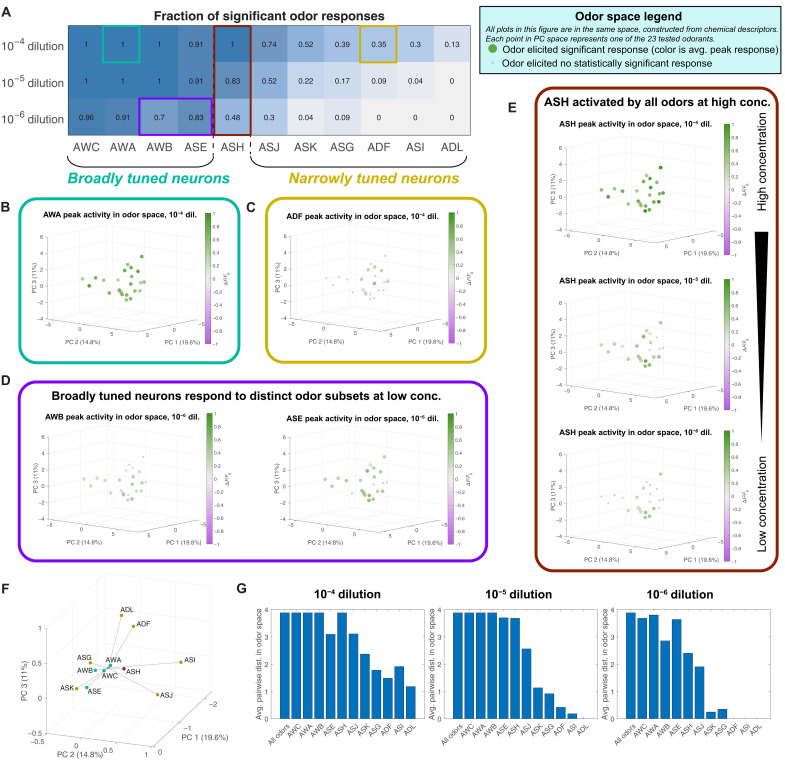
We observed a qualitative distinction in neuron tuning. Neurons AWA, AWB, AWC, and ASE each respond to most tested odorants at high concentrations, so we call them “broadly tuned” (Figs. 2, A to C, and 4A). In contrast, ADF, ADL, ASG, ASI, ASJ, and ASK each respond to a smaller set of odorants even at the highest tested concentrations, so we call them “narrowly tuned.”
Fig. 4. Chemosensory neuron tuning.
(A) Fraction of odorants in our 23-odor panel, which elicited significant responses (q ≤ 0.01) in each neuron, at three different concentrations. We class neurons that responded to the majority of presented odors at high concentration as “broadly tuned” and neurons that responded to a small number of odors as “narrowly tuned.” For each neuron, we plot peak responses to odorants in a space constructed from chemical descriptors (fig. S3A). (B) The activity of broadly tuned neurons (ex: AWA) spans this space, while (C) the activity of narrowly tuned neurons (ex: ADF) is confined to a subset of chemically similar odorants. (D) At low concentrations, broadly tuned neurons respond to distinct subsets of odorants. (E) ASH, a polymodal nociceptor, is activated by all tested odorants at high concentration but is only activated by a small set of repulsive odorants at low concentration. See fig. S5 for these plots for all neurons. (F) Centroids of the significant responses at 10–4 dilution of each neuron in odor space, weighted by the strength of each response. These centroids project in different directions from the center, suggesting that each neuron is most sensitive to a particular region of odor space. (G) Average pairwise distance between the odors that activate each neuron at high (left), medium (center), and low (right) concentrations, compared to the average pairwise distance between all 23 odors as a baseline. The broadly tuned neurons (AWC, AWA, AWB, ASE, and ASH) span most of the space, while the odors that activate narrowly tuned neurons (ASJ, ASK, ASG, ADF, ASI, and ADL) tend to be closer to each other on average and thus more similar in the chemical space.
ASH is broadly tuned at high concentrations and narrowly tuned at low concentrations (Fig. 4A), a pattern that might reflect its role as a nociceptor, mediating avoidance of any odorant when delivered at a sufficiently high concentration (Fig. 2, A to C). In previous behavioral experiments, most odorants in our panel were shown to be attractive at low concentrations and repulsive at high concentrations. A few odorants—1-heptanol, 1-octanol, and 1-nonanol—are repulsive at any tested concentration (table S2) (35). The odorants to which ASH is most sensitive are generally those that are repulsive at all concentrations.
The responses of each sensory neuron appear to occupy contiguous domains in chemical odor space. Each domain encompasses chemically similar odorant molecules that are effective stimuli for each sensory neuron (Fig. 4, B to E, and fig. S5). At high concentrations, broadly tuned neurons, such as AWA, extend responses throughout the chemical structure space. Even at high odorant concentrations, narrowly tuned neurons, such as ADF, extend responses over a smaller contiguous region of chemical space. This can be quantified as the average pairwise distance between the odors in odor space that significantly activate each neuron (Fig. 4G). Broadly tuned neurons span the entire space and therefore have the same average pairwise distance between odors as the baseline average pairwise distance of the entire odor panel. Narrowly tuned neurons have lower average pairwise distances—odors that activate narrowly tuned neurons are close to each other in odor space and are thus likely to be chemically similar.
Different sensory neurons can have overlapping response domains. However, each sensory neuron tends to be activated most strongly by a different part of odor space. We calculated the centroid of each neuron’s activity in odor space, weighted by the strength of response to each odor and plotted relative to the center of our 23-odor panel (Fig. 4F). These activity centroids project in different directions from the center, suggesting that each neuron is most sensitive to a different region of odor space. The centroids of broadly tuned neurons are closer to center, as expected because they are activated by most odorants, and it is the weighting by response magnitude that pulls them off center. The centroids of narrowly tuned neurons are further from the center, as their significantly responding neurons occupy only one patch of odor space.
Most broadly tuned neurons extend responses over a smaller region of chemical space at lower concentrations than at high concentrations. Responses at low concentrations reveal the structural characteristics of molecules to which sensory neurons are most sensitive. AWA is most strongly activated by ketones, AWB is most strongly activated by esters, and ASE is most activated by alcohols (fig. S5). These regions of odor space are consistent with the directions to which the activity centroids of these neurons project at high concentration. ASH responds to odorants throughout chemical space, perhaps allowing ASH to contribute to the repellent response of any odorant delivered at sufficiently high concentration. The observation that each sensory neuron extends its sensitivity range across a contiguous region of chemical structural space suggests that each neuron is tuned to shared molecular properties of a set of odorant stimuli, as opposed to being tuned to exclusively detect specific odorants.
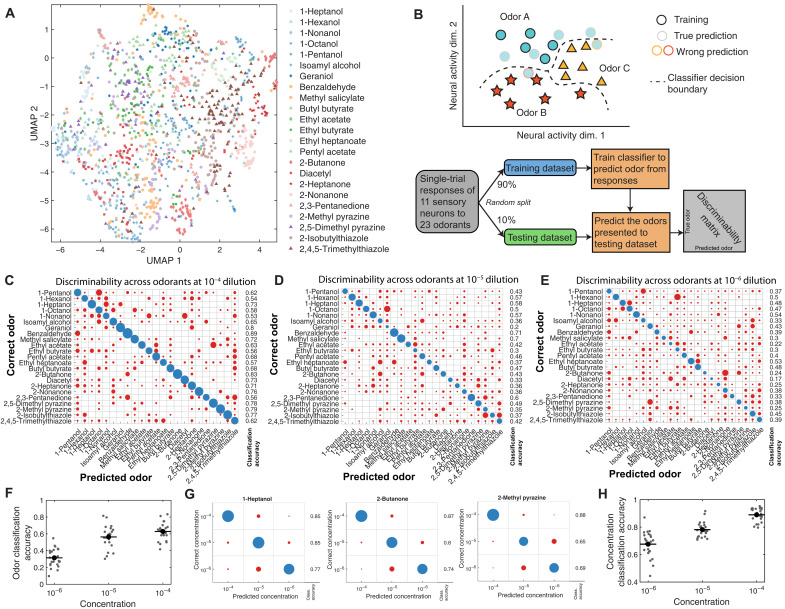
Single-trial responses suffice for discriminating odorant pairs
We observed trial-to-trial variability in odorant responses, both across animals and across odor presentations to the same animal. However, ensemble-level coding might confer robustness when discriminating odorants. We compiled all single-trial responses to each odorant across all datasets. In some recordings where data from individual neurons were missing, we imputed missing activity patterns using the rest of the ensemble (Appendix D and fig. S6, A to D). We used two dimensionality reduction methods to visualize the space spanned by single-trial responses—PCA and Uniform Manifold Approximation and Projection (UMAP). In a PC space constructed from the peak responses of all single trials, chemically similar odorants exhibit more similar representations (fig. S6D) and chemically dissimilar odorants exhibit dissimilar representations (fig. S6E). Overlap in a low-dimensional PC space is an imperfect measure for odorant discrimination because <60% of variance is explained by the first three principal components. Plotting all single-trial responses to all 23 odorants in UMAP space, trials for the same odorant also cluster together, although it is difficult to segregate trials for different odorants in this two-dimensional representation (Fig. 5A). Both PCA and UMAP analyses indicate that ensemble-level responses for the same odorant are similar. Both analyses also indicate that ensemble representations are high dimensional, as reduction to two or three dimensions removes a substantial fraction of the variance.
Fig. 5. Representative comparisons of single-trial odorant responses.
(A) Low-dimensional UMAP representation of single-trial neural responses to all 23 odorants at 10−4 dilution. Responses to any given odorant generally cluster together. (B) Schematic of the multiclass classifier used for theoretical discriminability analysis of single-trial responses. The classifier was trained to predict odor identity from the peak responses of the ensemble of sensory neurons, generating a discriminability matrix. (C) Linear discriminability analysis of single-trial peak responses to high-concentration (10−4 dilution) odorants, with the presented odorant on the y axis and the classified odorant on the x axis. Circle size indicates the number of trials, with correct classifications colored blue and incorrect classifications colored red. The fraction of correctly classified trials for each odorant is to the right. Most of the single trials are correctly classified for each odorant. At lower concentrations, 10−5 dilution (D) and 10−6 dilution (E), classification accuracy diminishes. This is summarized in (F), a scatterplot of multiclass classification accuracy at different concentrations (C to E). (G) Within a given odorant (three examples shown), the concentration of the given odorant can be correctly classified on the basis of individual peak responses. (H) Across all odorants, concentration classification accuracy at different concentrations is shown.
We asked whether olfactory representations were sufficiently dissimilar for reliable odorant discrimination based on single odorant presentations. To estimate the theoretical discriminability of odorant pairs, we computed errors in binary classification based on the pooled single-trial responses of each odorant pair using logistic regression (fig. S6F) and a support vector machine (fig. S6G). In all cases, binary classification succeeded with low error rate. Thus, any two odorants in our panel can be distinguished from each other on the basis of single-trial ensemble responses.
Odorant identification with single-trial responses
We asked whether odorant identity could reliably be decoded from single-trial ensemble responses, a task more challenging than binary classification of an odorant pair. We trained a multiclass classifier to perform linear discrimination (Fig. 5B). We randomly divided all single-trial measurements into a training set (90%) and validation (testing) set (10%). After we trained the classifier with the training set, we tested its performance in predicting odorant identities from single-trial measurements with the validation set (see Appendix E for details). This classifier successfully identified odorants in most of the single-trial measurements at high concentrations (Fig. 5C). Classification accuracy declined at lower concentrations but succeeded in the plurality of measurements (Fig. 5, D to F).
We used a similar approach to determine whether odorant intensity could be estimated from single-trial measurements. With trained multiclass classifiers, we were able to predict the concentration of a given odorant using single-trial measurements, although accuracy declined at lower concentrations (Fig. 5, G and H). In principle, the ensemble-level spatial map of sensory neuron activity contains sufficient information to determine odorant identity and intensity from single-stimulus presentations.
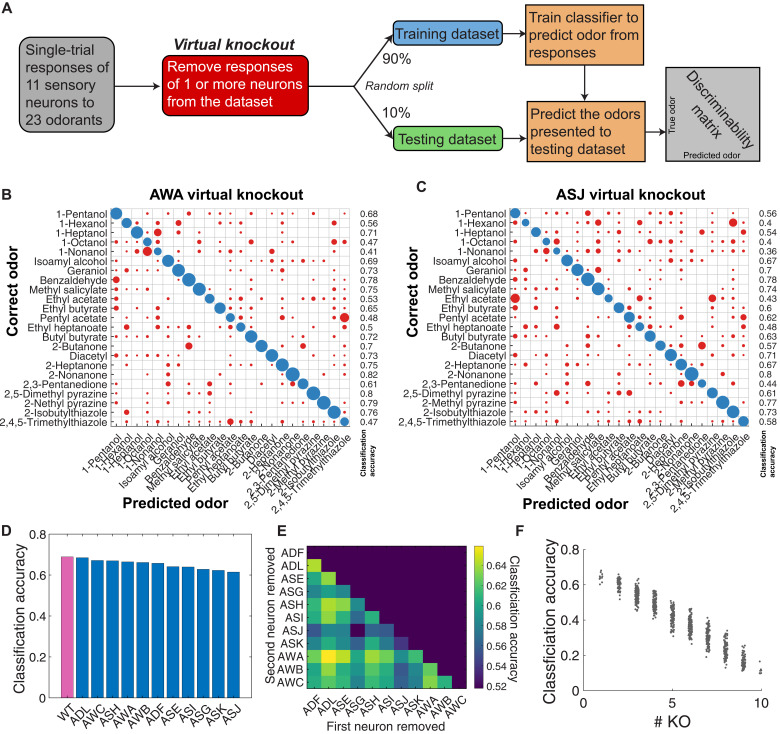
Virtual neuron knockouts degrade classifier accuracy
To quantify the relative contribution of each sensory neuron to ensemble-level discriminability, we performed virtual knockouts. We performed virtual knockouts by removing (masking) specific sensory neurons from the dataset and retraining the multiclass classifier on the remaining data. Removing any single sensory neuron led to small decreases in classification accuracy compared to wild type (Fig. 6, B to D). Classification accuracy was lower after masking narrowly tuned neurons (ASI, ASK, ASJ, or ASG) than broadly tuned neurons (AWA, ASH, or AWC).
Fig. 6. Odorant discriminability is robust to virtual knockouts.
(A) By removing the responses of one or more neurons from the dataset fed into the multiclass classifier, we assess the relative importance of different neurons to the theoretical discriminability of single-trial neural responses. Linear discriminability analysis of single-trial data, with (B) AWA or (C) ASJ virtually removed from the dataset. Removing different neurons changes the discriminability matrix in different ways. (D) We virtually removed each neuron from the dataset and computed the average classification accuracy for each virtual knockout (KO). Classification accuracy remains close to wild type (all 11 neurons) but is degraded more severely by removal of narrowly tuned neurons (ASI, ASK, ASJ, and ASG) than by removal of broadly tuned neurons. (E) Virtually removing pairs of neurons produces further reductions in average classification accuracy. (F) Plotting average classification accuracy of different sets of virtual knockouts reveals a linear relationship between theoretical classification accuracy and the number of chemosensory neurons.
Masking different neurons degrades the classification accuracy of a given odorant to different degrees. For instance, pentyl acetate is correctly classified 68% of the time when all 11 chemosensory neurons are included. ASJ masking reduces this accuracy to 62%, but AWA masking reduces accuracy to 48%. Masking any two neurons further decreases average classification accuracy (Fig. 6E). We computed the average classification accuracy when randomly removing different combinations of multiple neurons. We observed an inverse linear relationship between the number of masked neurons and classification accuracy (Fig. 6F). Odor identity across olfactory space is thus encoded in a distributed manner across all 11 chemosensory neurons.
Responses to pheromone stimuli are distinct from those of volatile odorants
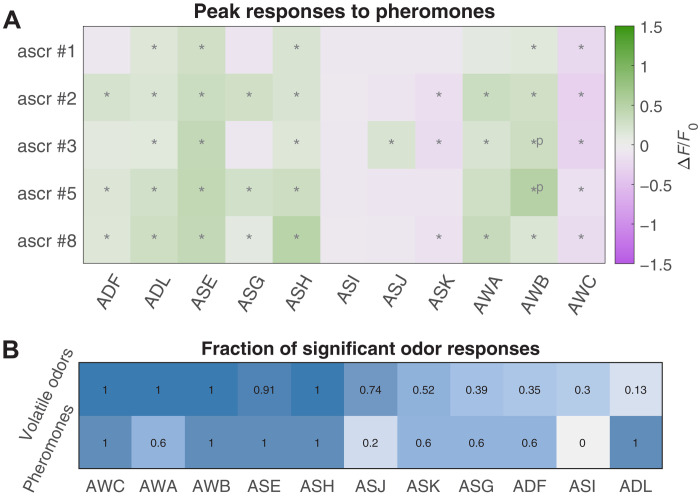
All amphid chemosensory neurons respond to volatile odorants. We asked whether ensemble-level responses extend to other stimuli. C. elegans communicate using pheromones, a mixed group of glycolipid molecules called ascarosides (24, 29). We presented young adult hermaphrodites with a panel of five single ascarosides (#1, #2, #3, #5, and #8) (25).
Similarly to volatile odorants, ascarosides activated multiple sensory neurons (Fig. 7A). Some neurons—known to respond to ascarosides but narrowly tuned with respect to our volatile-odorant panel, such as ADL, ADF, and ASK—were strongly activated across our five-pheromone panel (Fig. 7B). Pheromones also evoked some activity in neurons that are broadly tuned to volatile odorants. For example, AWA was activated less often by the pheromone panel than by the odorant panel. Thus, pheromone detection may also involve an ensemble-level code, but a code that relies more heavily on those neurons that are narrowly tuned to volatile odorants.
Fig. 7. Odorant representations of pheromones.
(A) Average peak responses of the 11 chemosensory neurons to ascaroside pheromones #1, #2, #3, #5, and #8 at a concentration of 200 nM. Responses are reported as ∆F/F0, and significant responses (q ≤ 0.01) are indicated with stars. (B) Fraction of volatile odorants (out of 23 odorants total), which elicited significant responses in each neuron at high concentration (first row), compared with the fraction of pheromone stimuli (out of five stimuli total), which elicited significant responses (second row). Many neurons (such as ADF and ADL) that are narrowly tuned with respect to volatile odorants appear to be activated more often by the ascaroside pheromones.
DISCUSSION
In insects and vertebrates, the integrated activity of chemosensory neuron ensembles is often presumed to enhance odorant discrimination and broaden the space of olfactory perceptions with ensemble-level codes (1–6). The C. elegans olfactory system contains only 11 pairs of chemosensory neurons. Each nematode chemosensory neuron is considered a unique class distinguished by dendrite morphologies, wiring partners, and stimulus selectivity (12, 34). Does the integrated activity of the C. elegans chemosensory ensemble contain information that might enhance and broaden olfactory discrimination?
We have simultaneously recorded calcium dynamics in all chemosensory neurons in nematodes exposed to a chemically diverse odorant panel. Nearly every distinct odorant stimulus evoked a distinct ensemble-level activity pattern among chemosensory neurons. We show that these highly reproducible ensemble-level patterns can, in principle, robustly encode odorant identity and intensity throughout a large chemical space.
Characterizing the ensemble-level olfactory code at the sensory periphery sets the stage for future studies aimed at their relevance for behavior and decision-making. Recording the activity of downstream interneurons will determine whether olfactory representations in circuits for behavior are similarly high dimensional, perhaps facilitating olfactory discrimination and diverse patterns of decision-making in complex environments.
Diverse sensory neuron tuning properties
The unique response properties of the chemosensory neurons allow each to contribute information to the spatial activity map that encodes olfactory stimuli. Ensemble-level activity appears to be largely independent of synaptic communication between chemosensory neurons (Fig. 3B). Moreover, the tuning of each chemosensory neuron is shaped by the expression and properties of multiple receptors, not by the sensitivity of a single receptor, as is typical in larger animals. Removing any chemosensory neuron lowers the accuracy of stimulus classification based on ensemble-level activity (Fig. 6).
We lack comprehensive information about the expression patterns and odorant sensitivity of most olfactory receptors in C. elegans. ODR-10, highly expressed in AWA, remains the only characterized olfactory receptor for diacetyl (38, 53). However, AWA also responds to many other odorants in a manner that is independent of ODR-10, evidence that AWA expresses additional receptors (15–17, 35). Other sensory neurons that do not express ODR-10 are also activated by diacetyl at higher threshold concentrations. We uncovered a diversity of odorant dose-response curves to diacetyl and other odorants (Fig. 2E and fig. S3, F and G). This diversity is consistent with the expression of multiple receptors in each chemosensory neuron. Variable dose-response curves may reflect the cumulative activities of different sets of receptors with different binding affinities for each odorant across chemosensory neurons. Every chemosensory neuron tends to be sensitive to structurally similar odorants, suggesting that the receptors expressed by each neuron might be correlated in their chemical binding affinities (Fig. 4 and fig. S5).
Because we do not know the repertoire of odorant receptors that are expressed in each chemosensory neuron, we cannot easily characterize receptor-ligand interactions based on dose-response curves, as has been done in other animals (4, 62, 63). We note that the breadth of tuning to olfactory stimuli (defined as the fraction of the odor panel that elicit significant responses) is not strongly correlated with the number of expressed GPCRs (13). For example, ADL expresses the most GPCR genes of any chemosensory neuron but is sensitive to only three odorants in our panel. ASH, ASK, and ASJ express many GPCR genes, but only ASH is broadly tuned to our odorant panel. ASE, another broadly tuned neuron, expresses the smallest number of GPCR genes. Our inability to correlate the number of expressed GPCRs with the breadth of tuning might be because many GPCRs might not function as odorant receptors. For example, ADL, narrowly tuned for odorants but broadly tuned for pheromones, might use many GPCRs as ascaroside receptors.
Comparisons with olfactory systems in larger animals
In larger animals, chemosensory cells typically express single receptor types. In these animals, when domains of sensory neuron activity are represented in a chemical odor space, response domains tend to be clustered. Olfactory neuron ensembles span odor space by connecting the clustered response domains of different olfactory sensory neurons (1–6).
In C. elegans, each sensory neuron is sensitive to a contiguous region of chemical space (Fig. 4). This suggests that each neuron is tuned to shared molecular properties, as opposed to being faithful detectors of unique odorant molecules. The broad tuning of many C. elegans sensory neurons is probably caused by the combined activities of different receptors. Each receptor may be tuned to a given region of chemical structural space. Connecting the regions of chemical space corresponding to each receptor could produce the broad region of chemical space sensed by each neuron. The tendency for even the most broadly tuned neurons to be most strongly activated by certain chemical classes suggests correlations in the cell-specific expression of receptor types. The multireceptor nature of C. elegans sensory neurons may also contribute to their graded responses over broad concentration ranges. As additional receptor types with higher thresholds are recruited at higher concentrations of a given odorant, a sensory neuron gradually and cumulatively becomes more active.
The C. elegans chemosensory neuron ensemble-level activity encodes odorant identity
Previous studies in C. elegans largely dissected the properties of individual chemosensory neurons in response to selected odorants (15–21). For example, single olfactory sensory neurons can exhibit complex temporal activity patterns in response to odorant stimulation (16, 17, 38, 46–49). Many previous studies have mapped the activities of single sensory neurons to behavioral outputs. However, using selected odorants to stimulate single sensory neurons and evoke behavioral responses does not reveal how olfactory inputs might be encoded by the sensory neuron ensemble.
We found that most olfactory stimuli activate multiple chemosensory neurons in C. elegans (Fig. 2). Chemosensory neurons that have been principally studied for roles in olfactory learning and navigation—AWA, AWB, AWC, and ASE—are the most broadly tuned, having high sensitivity to many different types of molecules. AWA is comparatively more strongly activated by ketones, AWB by some esters, and ASE by alcohols. AWC is inhibited by every odorant that we tested. Other olfactory neurons—such as ASK, ASJ, or ASG—are more narrowly tuned, activated by a small number of structurally similar odorants (Fig. 4 and fig. S5). When ensemble-level responses of broadly and narrowly tuned chemosensory neurons are taken together, a reproducible and distinct spatial activity map emerges for each stimulus. This map encodes both odorant identity and intensity across the space spanned by our panel of 23 diverse chemicals at multiple concentrations (Fig. 5).
How might C. elegans use an ensemble-level code for olfaction? Broadly tuned neurons permit coarse identification of odorants. Each narrowly tuned neuron is sensitive to a smaller region of olfactory space. When a narrowly tuned neuron is active, the possible identities of each olfactory stimulus are limited to those odorant molecules inside its region of sensitivity. When a neuron is inactive, molecules inside its region of sensitivity are ruled out. Combinatorial activity patterns among chemosensory neurons with different regions of sensitivity can provide enough information to pinpoint the identity and concentration of an odorant stimulus. Because these ensemble-level patterns are highly reproducible, accurate discrimination can be performed with single-stimulus presentations. Ensemble-level codes may also improve robustness, compensating for trial-to-trial variability in the responses of individual chemosensory neurons.
We stress that showing that ensemble-level neuron activity is capable of encoding odorant identity and intensity throughout olfactory space does not necessarily mean that downstream circuits use this olfactory information in full when making behavioral decisions. Demonstrating that information is encoded at the sensory periphery sets the stage for future experiments to determine what part of this information is decoded for animal behavior.
Pheromone detection engages the chemosensory ensemble in distinct ways
We found that chemosensory neurons that are more narrowly tuned to volatile odorants are more broadly tuned to pheromones (Fig. 7). Activation of pheromone-sensing neurons by volatile odorants might reflect cross-reactivity of pheromone receptors to small organic molecules. These narrowly tuned neurons might also express different receptors with high odorant specificity. We also do not know whether the activation of broadly tuned olfactory neurons by pheromones reflects cross-reactivity of olfactory receptors to pheromone molecules. In any case, widespread ensemble-level activity across all chemosensory neurons in response to odorants and pheromones encodes substantial information that can be used to accurately identify any chemical stimulus.
Discrepancies with previously reported chemosensory responses
We have characterized >900 neuron-stimulus pairings, including many previously undescribed responses. Where our measurements overlapped with previous studies, we found general agreement with previously reported neuronal responses. However, we observed some discrepancies.
We did not observe previously reported OFF responses in AWC. There may be two reasons for this. First, to map the tuning properties of chemosensory neurons, we used stimulus conditions that would minimize adaptation. We presented odorants in short 10-s pulses with long intervening blank periods between presentations. Previously reported OFF responses in AWC were obtained with longer odor stimulus presentations (15, 37). Second, some previously reported OFF responses were observed in one of the two asymmetric AWC neurons. Here, we did not separate the responses of AWCON and AWCOFF neurons, and so any asymmetric AWC response would be lost in the population average.
We also did not observe some previously recorded sensory neuron responses to ascarosides (26–29). This might be due to differences in the age and sex of tested animals. To be consistent with our own volatile odorant experiments, we recorded from young adult hermaphrodites. Different ascaroside responses in previous reports were obtained using males and juvenile hermaphrodites.
Limitations and future studies
Calcium imaging is a coarse-grained measure of neuronal activity. We primarily quantified peak calcium responses, omitting differences in dynamics, spiking, or asymmetric responses, all of which likely encode additional information. Thus, our estimates of the information encoded in ensemble-level activity represent conservative lower bounds.
Our analysis of synaptic transmission mutants suggests that synaptic transmission is not the primary driver of ensemble-level responses (Fig. 3). Synaptic connections and feedback might shape the magnitude and dynamics of neuronal responses in important ways. For example, it has been suggested that feedback by neuropeptide signaling causes ASE to respond when benzaldehyde is detected by other sensory neurons (17). Nonsynaptic neurotransmitter signaling and electrical synapses might also coordinate activity among chemosensory neurons.
How does ensemble-level information relate to behavior? Many odorants studied in C. elegans have known behavioral valence: either attractive or repulsive to the animal (35). At high concentrations, many odorants become behaviorally repulsive. Do these switches in behavioral valence correlate with a change in the ensemble-level code? A simple prediction is that ASH activity increases as a stimulus becomes more repulsive. To understand changes in behavioral valence, it is necessary to simultaneously measure the ensemble-level olfactory code during decision-making in freely behaving animals. This is because it is difficult to calibrate previous experiments—where the behavioral valence of volatile odorants was determined with crawling animals on agar plates—with olfactory stimulation of immobilized worms using microfluidics.
Downstream from the chemosensory ensemble, interneuron networks resemble both a reflexive avoidance circuit (consisting of the premotor interneurons AVA, AVB, and AVD that primarily receive inputs from ASH) and a circuit for learning and navigation (consisting of the interneurons AIA, AIB, AIY, and AIZ that integrate the activity of the entire chemosensory ensemble) (Fig. 1A) (10, 30, 31, 33, 36, 39, 64, 65). The activity of some of these interneurons is known to be modulated by differences in ensemble-level sensory neuron activity (41). ASH might be the start of a nociceptive reflex arc that maps the detection of noxious stimuli to rapid escape responses. The output of the entire chemosensory ensemble also appears to be integrated and decoded by another more complex interneuron network. Recently developed multineuronal recording methods (21, 66, 67) that extend from the chemosensory neurons to downstream interneurons might reveal how much olfactory information that is encoded at the sensory periphery is decoded in olfactory discrimination and behavioral decision-making.
We studied a broad panel of pure volatile odorants, all of which are known to elicit behavioral responses in the worm. In the natural environment of C. elegans, most olfactory cues will be mixtures of odorants, signifying diverse food sources or pathogens. Understanding how the chemosensory ensemble responds to these mixtures would illuminate ethologically relevant decision-making in downstream circuits.
The extent to which any animal exploits the collective activity of chemosensory neurons to decode olfactory inputs remains poorly understood. On one hand, the “dimensionality” of the olfactory code is often presumed to be as large as the number of distinct chemosensory neurons that contribute to the code (68). If so, the ability to detect even small numbers of different molecules, each with specificity to different chemosensory neurons, can create the potential to discriminate astronomical numbers of olfactory stimuli (69). On the other hand, animals might discard much of the high-dimensional olfactory information at the sensory periphery if it only needs to perform coarse categorizations of odorants such as “attractive” versus “repulsive.” Rapid and efficient olfactory coding can be accomplished using only a small number of the earliest responding (or primary) olfactory receptors and neurons, as in recent experiments that explore “primacy models” of the olfactory code in rodents (70).
With advances in microfluidics and imaging technologies, it is becoming possible to combine high-throughput odorant stimulation with brain-wide imaging and tracking in behaving animals (50, 71–74). With these tools, it will become possible to measure how much odorant information is decoded in behavioral discrimination tasks throughout the olfactory space that we characterized in this study. While the range of olfactory discrimination tasks and the combinatorial possibilities of the olfactory code are still large in C. elegans, its relatively small size makes it feasible to quantitatively assess the behavioral relevance of ensemble-level olfactory codes.
MATERIALS AND METHODS
Experimental design
The primary objective of the study was to understand how the 11 chemosensory neuron pairs in C. elegans encode odorant identity and intensity. We developed a transgenic nematode in which all of the ciliated sensory neurons were fluorescently labeled with GCaMP, allowing the activity of the 11 chemosensory neuron pairs to be recorded from simultaneously. We assembled a broad panel of 23 volatile odorants and five pheromones and used a microfluidic device to present these stimuli to nematodes. We used confocal microscopy to record from chemosensory neurons as odorant stimuli at multiple concentrations were presented.
Worm maintenance
All C. elegans lines used in this project were grown at 22°C on nematode growth medium plates seeded with the Escherichia coli strain OP50. All animal lines were allowed to recover from starvation or freezing for at least two generations before being used in experiments. All animals used in experiments were young adults.
Plasmids and crosses
To construct the ZM10104 imaging strain, we created and then crossed two integrated lines, one expressing GCaMP6s and one expressing the wCherry landmark. The first of these lines, ADS700, was made by coinjecting lin-15(n765) animals with pJH4039 (ift-20 GCaMP6s::3xNLS) and a lin-15–rescuing plasmid. A stable transgenic line (hpEx3942) with consistent GCaMP expression in the chemosensory neurons was selected for integration, and transgenic animals were irradiated with ultraviolet (UV) light to integrate the transgenes into the genome. The resulting integrated line (aeaIs008) was backcrossed four times against N2 wild type. The second line, ADS701, was similarly made by coinjecting lin-15(n765) animals with pJH4040 (gpc-1 wCherry) and a lin-15–rescuing plasmid. A stable transgenic line with good wCherry expression was selected for integration, and transgenic animals were irradiated with UV light to integrate the transgenes into the genome. The resulting integrated line (hpIs728) was backcrossed four times against N2 wild type. To make ZM10104, ADS700 hermaphrodites were crossed with N2 males. Heterozygous aeaIs008/+ male progeny were then crossed with ADS701 hermaphrodites. F1 progeny were picked for wCherry expression, and F2 progeny were picked for both GCaMP6s and wCherry expression. The line was then homozygozed in the F3 generation.
The ADS707 mutant imaging line was created by crossing the ZM10104 line with EG9631, an unc-13(s69) mutant obtained from the Caenorhabditis Genetics Center (CGC) (53). EG9631 hermaphrodites were crossed with ZM10104 males. Heterozygous (aeaIs008/+; hpIs728/+; +/unc-13) F1 hermaphrodite progeny were selected by GCaMP6s and wCherry expression and wild-type locomotion (unc-13 is recessive). F2 progeny were picked for fluorescence and the unc-13 uncoordinated phenotype. The line was homozygozed for fluorescence in the F3 generation.
Microfluidics
We used a modified version of a microfluidic system capable of delivering multiple odors to Drosophila larvae (4). The microfluidic chip is designed with an arbor containing delivery points for multiple stimuli, together with a buffer delivery point and two control switches, one for buffer and one for odor (Fig. 1B). At any given time, three flows are active: one of the control switches, the buffer blank, and one odor stimulus. The chip is designed to maintain laminar flow of each fluid, and the flow is split between a waste channel and an odor channel, which flows past the animal’s nose. The chip described here is designed to switch rapidly from one stimulus to the buffer. After the flows pass the animal, they exit the chip via a waste port at atmospheric pressure. Waste is removed with a vacuum.
We grafted the odorant delivery arbor to a C. elegans loading chamber similar to those designed by Chronis et al. (55). We designed a loading chamber suitable for adult C. elegans, a narrow channel 62 μm wide and 30 μm high, with a gently tapered end. The tapered end serves as a guide to help hold the animal’s nose in place without distorting the animal. The microfluidic device pattern was designed in AutoCAD, and the design was translated to silicon wafer using photolithography. The photomasks of the design were printed using CAD/Art Services Inc. The silicon wafer was then used as a mold for polydimethylsiloxane (PDMS) to fabricate microfluidic devices. The PDMS components were then removed from the silicon wafer, cut to size, and had access channels made with a biopsy punch. The completed PDMS components were then plasma-bonded to no. 1 glass cover slips. To minimize contamination from dust, all microfluidics assembly was done in a cleanroom.
Preparation of odorant and buffer solutions
Odorants were diluted in CTX buffer (5 mM KH2PO4/K2HPO4 at pH 6, 1 mM CaCl2, 1 mM MgSO4, and 50 mM NaCl, adjusted to 350 mOsm/liter with sorbitol). To prevent contamination, each odor condition was mixed and stored in its own glass bottle and delivered through its own glass syringe and tubing. Furthermore, a new microfluidic device was used for a single consistent panel of odors. The single ascarosides (25) were diluted in CTX buffer to 200 mM concentration for presentation to the animals.
Stimulus delivery protocol
We chose to deliver 10-s odorant pulses separated by 30-s buffer blanks. These pulse and blank lengths were sufficiently spaced to elicit similar neuronal responses across pulses, with no indication of adaptation (fig. S2, A and B). We carried out control experiments by presenting each animal in the microfluidic device with multiple conditions (odorants and concentrations) in a single trial (fig. S2, C and D). Randomizing the order of odorant delivery, we did not observe any effects of odorant order on the responses of the sensory neurons.
We previously used a similar stimulus protocol by Yemini et al. (21), presenting three chemosensory stimuli separated by buffer blanks in a randomized order. There, we similarly observed no differences in average odorant-evoked responses that were correlated with odorant delivery order.
Thus, to reduce the risk of odorant cross-contamination, we elected to conduct the remaining experiments by presenting each animal with multiple presentations of one stimulus condition (fig. S2, A and B) and averaged across the population of at least 10 animals per condition, treating the response to each odorant pulse response as an independent trial.
Imaging setup
We used a single-photon, spinning-disk confocal microscope to capture fluorescent images from intact C. elegans. The microscope was inverted to allow for easy access to the microfluidic device mounted on the stage. We used a 488-nm laser to excite GCaMP in vivo and used a 561-nm laser to excite the wCherry landmark. To minimize cross-talk between channels, lasers were fired sequentially during multicolor recordings. We captured images with a 60× water-immersion objective with a numerical aperture of 1.2. Volumes were acquired using unidirectional scans of a piezo objective scanner. All fluorescence microscopy is a trade-off between spatial resolution, temporal resolution, laser power, and signal strength. We optimized two sets of imaging conditions, one set for activity imaging and another set for landmark imaging. Both sets of imaging conditions capture the region containing most of the neurons in the head of C. elegans, a volume of 112 μm by 56 μm by 30 μm.
In any given experiment, acquisition of a landmark volume precedes acquisition of an activity movie. This volume, which contains both green and red channels, allows us to identify neurons of interest. The spatial resolution of these volumes is 0.5 μm by 0.5 μm by 1.5 μm per voxel, with the z-resolution of 1.5-μm set by the point spread function.
The activity movies were acquired at a high speed in the green channel only, with lower spatial resolution (1 μm by 1 μm by 1.5 μm per voxel). At this resolution, we could acquire volumes at 2.5 Hz in standard acquisition mode.
Analyzing multineuronal recordings
The neurons in each activity recording were identified and then tracked through time using a neighborhood correlation tracking method. The criteria for identifying each neuron class are described in the Supplementary Methods. Neurons that could not be unambiguously identified were excluded from the dataset. All neuron tracks were then manually proofread to exclude mistracked neurons. Activity traces were bleach-corrected and reported in ∆F/F0 = [F(t) − F0]/F0. Normalization by baseline fluorescence F0 allowed for direct comparisons within a given neuron class across left and right sides and across individuals. The baseline F0 value was determined individually for every recorded neuron, set at the fifth percentile of the distribution of bleach-corrected fluorescence values, with the opportunity for manual correction.
Statistical analysis
We used two-tailed, paired t tests to compare the mean signal during stimulus presentation with an unstimulated period of identical length within the same neuron. Neurons were tested for both ON and OFF responses. The P values were corrected for multiple testing using false discovery rate (76). To test for asymmetric neuron responses, we used two-tailed, two-sample t tests (unpaired). Sensory neuron responses to all conditions are publicly available at this data repository, together with plots of average responses, phase trajectories, and time trace correlation matrices.
Acknowledgments
We thank G. Si and J. Kanwal for advice on microfluidics design and operation and M. Seyedolmohadesin and M. Torkashvand for discussions on ascaroside responses. We also thank S. R. Datta, S. Flavell, and the members of the Zhen, Pehlevan, and Samuel laboratories for advice on the project and comments on the manuscript. The EG9631 strain was obtained from the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Microfluidic devices were manufactured using the Soft Materials Cleanroom facility of the Harvard MRSEC (DMR-1420570).
Funding: This work was supported by NSF Brain Eager NSF IOS-1452593 (to A.D.T.S.), NSF Physics of Living Systems NSF 1806818 (to A.L. and A.D.T.S.), NSF Ideas NSF IOS-1555914 (to A.D.T.S.), NIH Brain Initiative NIH 1UF1NS111697-01 (to C.P., A.D.T.S., and M.Z.), Burroughs Wellcome Fund Career Award at the Scientific Interface (to V.V.), and Canadian Institutes of Health Research CIHR Foundation Scheme 154274 (to M.Z.).
Author contributions: Conceptualization: A.L., V.V., M.Z., and A.D.T.S. Genetics and reagents: A.L., M.W., L.L., W.H., and M.Z. Experimental setup design and build: A.L. and V.V. Software: A.L. and V.V. Imaging experiments: A.L., H.C., G.C., N.Z.T., and R.V. Analysis and modeling: A.L. and S.Q. Supervision: C.P., M.Z., and A.D.T.S. Writing—original draft: A.L., V.V., M.Z., and A.D.T.S. Writing—review and editing: S.Q., H.C., and C.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The animal strains used in this study are available at the CGC (https://cgc.umn.edu/). Please search by strain name or genotype. The neural activity data from this project, together with analysis code, plots of activity traces, phase trajectories, and time trace correlations for all conditions, are available at this Zenodo repository (https://doi.org/10.5281/zenodo.7563053). The code is also available at this GitHub repository (https://github.com/samuellab/celegans-sensory-code).
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S6
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.B. Malnic, J. Hirono, T. Sato, L. B. Buck, Combinatorial receptor codes for odors. Cell 96, 713–723 (1999). [DOI] [PubMed] [Google Scholar]
- 2.K. Nara, L. R. Saraiva, X. Ye, L. B. Buck, A large-scale analysis of odor coding in the olfactory epithelium. J. Neurosci. 31, 9179–9191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. A. Kreher, D. Mathew, J. Kim, J. R. Carlson, Translation of sensory input into behavioral output via an olfactory system. Neuron 59, 110–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.G. Si, J. K. Kanwal, Y. Hu, C. J. Tabone, J. Baron, M. Berck, G. Vignoud, A. D. T. Samuel, Structured odorant response patterns across a complete olfactory receptor neuron population. Neuron 101, 950–962.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.J. W. Wang, A. M. Wong, J. Flores, L. B. Vosshall, R. Axel, Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282 (2003). [DOI] [PubMed] [Google Scholar]
- 6.E. A. Hallem, J. R. Carlson, Coding of odors by a receptor repertoire. Cell 125, 143–160 (2006). [DOI] [PubMed] [Google Scholar]
- 7.J. del Marmol, M. A. Yedlin, V. Ruta, The structural basis of odorant recognition in insect olfactory receptors. Nature 597, 126–131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. De Bruyne, T. C. Baker, Odor detection in insects: Volatile codes. J. Chem. Ecol. 34, 882–897 (2008). [DOI] [PubMed] [Google Scholar]
- 9.P. S. Grewal, D. J. Wright, Migration of Caenorhabditis elegans larvae towards bacteria and the nature of the bacterial stimulus. Fundam. Appl. Nematol. 15, 159–166 (1992). [Google Scholar]
- 10.C. I. Bargmann, H. Robert Horvitz, Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729–742 (1991). [DOI] [PubMed] [Google Scholar]
- 11.C. I. Bargmann, Comparative chemosensation from receptors to ecology. Nature 444, 295–301 (2006). [DOI] [PubMed] [Google Scholar]
- 12.C. I. Bargmann, Chemosensation in C. elegans, WormBook: The Online Review of C. elegans Biology (2006), pp. 1–29; 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed]
- 13.B. Vidal, U. Aghayeva, H. Sun, C. Wang, L. Glenwinkel, E. A. Bayer, O. Hobert, An atlas of Caenorhabditis elegans chemoreceptor expression. PLOS Biol. 16, 1–34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L. A. Perkins, E. M. Hedgecock, J. N. Thomson, J. G. Culotti, Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487 (1986). [DOI] [PubMed] [Google Scholar]
- 15.K. Yoshida, T. Hirotsu, T. Tagawa, S. Oda, T. Wakabayashi, Y. Iino, T. Ishihara, Odour concentration-dependent olfactory preference change in C. elegans. Nat. Commun. 3, 739 (2012). [DOI] [PubMed] [Google Scholar]
- 16.J. Larsch, D. Ventimiglia, C. I. Bargmann, D. R. Albrecht, High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 110, E4266–E4273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. G. Leinwand, C. J. Yang, D. Bazopoulou, N. Chronis, J. Srinivasan, S. H. Chalasani, Circuit mechanisms encoding odors and driving aging-associated behavioral declines in Caenorhabditis elegans. eLife 4, e10181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. Zaslaver, I. Liani, O. Shtangel, S. Ginzburg, L. Yee, P. W. Sternberg, Hierarchical sparse coding in the sensory system of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 112, 1185–1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Yu, L. Avery, E. Baude, D. L. Garbers, Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc. Natl. Acad. Sci. U. S. A. 94, 3384–3387 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.P. D. Wes, C. I. Bargmann, C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410, 698–701 (2001). [DOI] [PubMed] [Google Scholar]
- 21.E. Yemini, A. Lin, A. Nejatbakhsh, E. Varol, R. Sun, G. E. Mena, A. D. T. Samuel, L. Paninski, V. Venkatachalam, O. Hobert, NeuroPAL: A multicolor atlas for whole-brain neuronal identification in C. elegans. Cell 184, 272–288, 288.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.T. R. Thiele, S. Faumont, S. R. Lockery, The neural network for chemotaxis to tastants in Caenorhabditis elegans is specialized for temporal differentiation. J. Neurosci. 23, 11904–11911 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.H. Suzuki, T. R. Thiele, S. Faumont, M. Ezcurra, S. R. Lockery, W. R. Schafer, Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 454, 114–117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.E. Z. MacOsko, N. Pokala, E. H. Feinberg, S. H. Chalasani, R. A. Butcher, J. Clardy, C. I. Bargmann, A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458, 1171–1175 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. Narayan, V. Venkatachalam, O. Durak, D. K. Reilly, N. Bose, F. C. Schroeder, A. D. T. Samuel, J. Srinivasan, P. W. Sternberg, Contrasting responses within a single neuron class enable sex-specific attraction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 113, E1392–E1401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. S. Greene, M. Brown, M. Dobosiewicz, I. G. Ishida, E. Z. Macosko, X. Zhang, R. A. Butcher, D. J. Cline, P. T. McGrath, C. I. Bargmann, Balancing selection shapes density-dependent foraging behaviour. Nature 539, 254–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.E. Z. Aprison, I. Ruvinsky, Counteracting ascarosides act through distinct neurons to determine the sexual identity of C. elegans pheromones. Curr. Biol. 27, 2589–2599.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 28.K. A. Fagan, J. Luo, R. C. Lagoy, F. C. Schroeder, D. R. Albrecht, D. S. Portman, A single-neuron chemosensory switch determines the valence of a sexually dimorphic sensory behavior. Curr. Biol. 28, 902–914.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. T. McGrath, I. Ruvinsky, A primer on pheromone signaling in Caenorhabditis elegans for systems biologists. Curr. Opin. Syst. Biol. 13, 23–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. A. Hilliard, C. I. Bargmann, P. Bazzicalupo, C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr. Biol. 12, 730–734 (2002). [DOI] [PubMed] [Google Scholar]
- 31.M. A. Hilliard, C. Bergamasco, S. Arbucci, R. H. A. Plasterk, P. Bazzicalupo, Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. EMBO J. 23, 1101–1111 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. M. Kaplan, H. R. Horvitz, A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 90, 2227–2231 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Y. Sambongi, T. Nagae, Y. Liu, T. Yoshimizu, K. Takeda, Y. Wada, M. Futai, Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport 10, 753–757 (1999). [DOI] [PubMed] [Google Scholar]
- 34.A. Metaxakis, D. Petratou, N. Tavernarakis, Multimodal sensory processing in Caenorhabditis elegans. Open Biol. 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. I. Bargmann, E. Hartwieg, H. R. Horvitz, Odorant-selective genes and neurons mediate olfaction in C. elegans. Neuron 74, 515–527 (1993). [DOI] [PubMed] [Google Scholar]
- 36.S. H. Chalasani, N. Chronis, M. Tsunozaki, J. M. Gray, D. Ramot, M. B. Goodman, C. I. Bargmann, Erratum: Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 451, 63–70 (2008). [DOI] [PubMed] [Google Scholar]
- 37.M. Tsunozaki, S. H. Chalasani, C. I. Bargmann, A behavioral switch: cgmp and pkc signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 59, 959–971 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.E. R. Troemel, B. E. Kimmel, C. I. Bargmann, Reprogramming chemotaxis responses: Sensory neurons define olfactory preferences in C. elegans. Cell 91, 161–169 (1997). [DOI] [PubMed] [Google Scholar]
- 39.H.-i. Ha, M. Hendricks, Y. Shen, C. V. Gabel, C. Fang-Yen, Y. Qin, D. Colón-Ramos, K. Shen, A. D. T. Samuel, Y. Zhang, Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron 68, 1173–1186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D. Cheng, J. S. Lee, M. Brown, M. S. Ebert, P. T. McGrath, M. Tomioka, Y. Iino, C. I. Bargmann, Insulin/igf signaling regulates presynaptic glutamate release in aversive olfactory learning. Cell Rep. 41, 111685 (2022). [DOI] [PubMed] [Google Scholar]
- 41.M. Dobosiewicz, Q. Liu, C. I. Bargmann, Reliability of an interneuron response depends on an integrated sensory state. eLife 8, e50566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. A. Colbert, C. I. Bargmann, Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron 14, 803–812 (1995). [DOI] [PubMed] [Google Scholar]
- 43.N. D. L’Etoile, C. M. Coburn, J. Eastham, A. Kistler, G. Gallegos, C. I. Bargmann, The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36, 1079–1089 (2002). [DOI] [PubMed] [Google Scholar]
- 44.G. Jansen, D. Weinkove, R. H. A. Plasterk, The G-protein γ subunit gpc-1 of the nematode C. elegans is involved in taste adaptation. EMBO J. 21, 986–994 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. A. Hilliard, A. J. Apicella, R. Kerr, H. Suzuki, P. Bazzicalupo, W. R. Schafer, In vivo imaging of C. elegans ASH neurons: Cellular response and adaptation to chemical repellents. EMBO J. 24, 63–72 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. Levy, C. I. Bargmann, An adaptive-threshold mechanism for odor sensation and animal navigation. Neuron 105, 534–548.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 47.M. Khan, A. H. Hartmann, M. P. O’Donnell, M. Piccione, A. Pandey, P.-H. Chao, N. D. Dwyer, C. I. Bargmann, P. Sengupta, Context-dependent reversal of odorant preference is driven by inversion of the response in a single sensory neuron type. PLOS Biol. 20, e3001677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Q. Liu, P. B. Kidd, M. Dobosiewicz, C. I. Bargmann, C. elegans AWA olfactory neurons fire calcium-mediated all-or-none action potentials. Cell 175, 57–70.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 49.I. G McLachlan, T. S. Kramer, M. Dua, E. M. Diloreto, U. Dag, J. Srinivasan, S. W. Flavell, Diverse states and stimuli tune olfactory receptor expression levels to modulate food-seeking behavior. bioRxiv 2022.04.27.489714 [Preprint]. 28 April 2022. 10.1101/2022.04.27.489714. [DOI] [PMC free article] [PubMed]
- 50.N. Chronis, M. Zimmer, C. I. Bargmann, Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods 4, 727–731 (2007). [DOI] [PubMed] [Google Scholar]
- 51.S. E. Worthy, L. Haynes, M. Chambers, D. Bethune, E. Kan, K. Chung, R. Ota, C. J. Taylor, E. E. Glater, Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PLOS ONE 13, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R. Haddad, R. Khan, Y. K. Takahashi, K. Mori, D. Harel, N. Sobel, A metric for odorant comparison. Nat. Methods 5, 425–429 (2008). [DOI] [PubMed] [Google Scholar]
- 53.P. Sengupta, J. H. Chou, C. I. Bargmann, odr-10 Encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84, 899–909 (1996). [DOI] [PubMed] [Google Scholar]
- 54.P. J. Summers, R. M. Layne, A. C. Ortega, G. P. Harris, B. A. Bamber, R. W. Komuniecki, Multiple sensory inputs are extensively integrated to modulate nociception in C. elegans. J. Neurosci. 35, 10331–10342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. G. White, E. Southgate, J. N. Thomson, S. Brenner, The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 56.D. Witvliet, B. Mulcahy, J. K. Mitchell, Y. Meirovitch, D. R. Berger, Y. Wu, Y. Liu, W. X. Koh, R. Parvathala, D. Holmyard, R. L. Schalek, N. Shavit, A. D. Chisholm, J. W. Lichtman, A. D. T. Samuel, M. Zhen, Connectomes across development reveal principles of brain maturation. Nature 596, 257, 261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. E. Richmond, W. S. Davis, E. M. Jorgensen, UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2, 959–964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.H. Sass, Sensory encoding of odor stimuli in Periplaneta americana. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 107, 49–65 (1976). [Google Scholar]
- 59.M. Meister, T. Bonhoeffer, Tuning and topography in an odor map on the rat olfactory bulb. J. Neurosci. 21, 1351–1360 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.H. Saito, Q. Chi, H. Zhuang, H. Matsunami, J. D. Mainland, Odor coding by a mammalian receptor repertoire. Sci. Signal. 2, ra9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D. Zwicker, A. Murugan, M. P. Brenner, Receptor arrays optimized for natural odor statistics. Proc. Natl. Acad. Sci. U.S.A. 113, 5570–5575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. Qin, Q. Li, C. Tang, Y. Tu, Optimal compressed sensing strategies for an array of nonlinear olfactory receptor neurons with and without spontaneous activity. Proc. Natl. Acad. Sci. U.S.A. 116, 20286–20295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.K. Kajiya, K. Inaki, M. Tanaka, T. Haga, H. Kataoka, K. Touhara, Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J. Neurosci. 21, 6018–6025 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.H. Spors, M. Wachowiak, L. B. Cohen, R. W. Friedrich, Temporal dynamics and latency patterns of receptor neuron input to the olfactory bulb. J. Neurosci. 26, 1247–1259 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.T. Wakabayashi, I. Kitagawa, R. Shingai, Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci. Res. 50, 103–111 (2004). [DOI] [PubMed] [Google Scholar]
- 66.P. A. Garrity, M. B. Goodman, A. D. Samuel, P. Sengupta, Running hot and cold: Behavioral strategies, neural circuits, and the molecular machinery for thermotaxis in C. elegans and Drosophila. Genes Dev. 24, 2365–2382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.H. S. Kaplan, M. Zimmer, Brain-wide representations of ongoing behavior: A universal principle? Curr. Opin. Neurobiol. 64, 60–69 (2020). [DOI] [PubMed] [Google Scholar]
- 68.A. Lin, D. Witvliet, L. Hernandez-Nunez, S. W. Linderman, A. D. T. Samuel, V. Venkatachalam, Imaging whole-brain activity to understand behaviour. Nat. Rev. Phys. 4, 292–305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.M. Meister, On the dimensionality of odor space. eLife 4, e07865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.C. Bushdid, M. O. Magnasco, L. B. Vosshall, A. Keller, Humans can discriminate more than 1 trillion olfactory stimuli. Science 343, 1370–1372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.C. D. Wilson, G. O. Serrano, A. A. Koulakov, D. Rinberg, A primacy code for odor identity. Nat. Commun. 8, 1477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D. R. Albrecht, C. I. Bargmann, High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments. Nat. Methods 8, 599–605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.J. P. Nguyen, A. N. Linder, G. S. Plummer, J. W. Shaevitz, A. M. Leifer, Automatically tracking neurons in a moving and deforming brain. PLOS Comput. Biol. 13, e1005517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.V. Venkatachalam, N. Ji, X. Wang, C. Clark, J. K. Mitchell, M. Klein, C. J. Tabone, J. Florman, H. Ji, J. Greenwood, A. D. Chisholm, J. Srinivasan, M. Alkema, M. Zhen, A. D. T. Samuel, Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 113, E1082–E1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.K. M. Hallinen, R. Dempsey, M. Scholz, X. Yu, A. Linder, F. Randi, A. K. Sharma, J. W. Shaevitz, A. M. Leifer, Decoding locomotion from population neural activity in moving C. elegans. eLife 10, e66135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.J. D. Storey, A direct approach to false discovery rates. J. R. Stat. Soc. B 64, 479–498 (2002). [Google Scholar]
- 77.E. Candès, B. Recht, Exact matrix completion via convex optimization. Commun. ACM 55, 111–119 (2012). [Google Scholar]
- 78.L. T. Nguyen, J. Kim, B. Shim, Low-rank matrix completion: A contemporary survey. IEEE Access 7, 94215–94237 (2019). [Google Scholar]
- 79.J. Fan, L. Ding, Y. Chen, M. Udell, Factor group-sparse regularization for efficient low-rank matrix recovery. Adv. Neural Inf. Process. Syst. 32, (2019). [Google Scholar]
- 80.M. Linkert, C. T. Rueden, C. Allan, J. M. Burel, W. Moore, A. Patterson, B. Loranger, J. Moore, C. Neves, D. M. Donald, A. Tarkowska, C. Sticco, E. Hill, M. Rossner, K. W. Eliceiri, J. R. Swedlow, Metadata matters: Access to image data in the real world. J. Cell Biol. 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.P. Kovesi, Good colour maps: How to design them. arXiv:1509.03700 [cs.GR] (12 September 2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S6
Tables S1 and S2
References