Abstract
Polycomb complexes regulate cell type–specific gene expression programs through heritable silencing of target genes. Trimethylation of histone H3 lysine 27 (H3K27me3) is essential for this process. Perturbation of H3K36 is thought to interfere with H3K27me3. We show that mutants of Drosophila replication-dependent (H3.2K36R) or replication-independent (H3.3K36R) histone H3 genes generally maintain Polycomb silencing and reach later stages of development. In contrast, combined (H3.3K36RH3.2K36R) mutants display widespread Hox gene misexpression and fail to develop past the first larval stage. Chromatin profiling revealed that the H3.2K36R mutation disrupts H3K27me3 levels broadly throughout silenced domains, whereas these regions are mostly unaffected in H3.3K36R animals. Analysis of H3.3 distributions showed that this histone is enriched at presumptive Polycomb response elements located outside of silenced domains but relatively depleted from those inside. We conclude that H3.2 and H3.3 K36 residues collaborate to repress Hox genes using different mechanisms.
Histone H3.2 and H3.3 K36 residues ensure Hox gene silencing and enable development by different, but synergistic mechanisms.
INTRODUCTION
A fundamental question in developmental biology is to understand how diverse cell types are generated from undifferentiated precursor cells. Once established, cellular identities must be maintained over time. The failure to do so can result in a wide spectrum of human diseases (1–4). Covalent posttranslational modifications (PTMs) of the histone proteins that package eukaryotic genomes are thought to encode epigenetic information that is passed from one cell generation to the next (5–7), but the mechanisms by which this process occurs remain incompletely understood.
In animal cells, histone PTM functions have largely been inferred from genetic analyses of histone modifying factors (readers, writers, and erasers) rather than from studying the histone residues themselves. To help decipher the metazoan “histone code” (8), we developed an experimental system in Drosophila that allows for sophisticated phenotypic analysis following loss of a specific site of histone modification (9, 10). We found that histone missense mutants often exhibit a subset of the phenotypes caused by mutations in their cognate chromatin-modifying enzymes (9, 11–14). Here, we take advantage of this system to focus on the role of histone H3 lysine-36 (H3K36) in antagonizing the developmentally regulated gene silencing activity of the Polycomb Repressive Complex 2 (PRC2).
A large body of evidence demonstrates that trimethylation of H3 lysine 27 (H3K27me3) is deposited by PRC2 and is critical for formation of silent chromatin (15, 16). Recent work has conclusively shown that the H3K27 residue is essential for maintaining repression of homeobox (Hox) genes that control cell fate decisions in Drosophila and mice (9, 17, 18). Furthermore, allosteric interactions within the PRC2 enzyme complex serve to facilitate the spreading of H3K27me3 into neighboring chromatin domains (19–21). Thus, the H3K27me3 writer is also a reader—a finding that has profound consequences for understanding the regulation of heterochromatin (22, 23). The H3K27me3 mark is also read by PRC1, containing the Polycomb (Pc) protein, which further condenses and represses H3K27me3-marked genomic regions (24).
To counteract the spreading activity of PRC2, other chromatin marks including H3K4me3, H3K36me2, and H3K36me3 are thought to antagonize Pc silencing (25, 26). Elegant work from Müller and colleagues (27) recently elucidated the structural basis whereby modification of H3K36 inhibits the activity of EZH2, the catalytic subunit of mammalian PRC2. Cryo–electron microscopy (cryo-EM) analysis of in vitro–reconstituted nucleosomes showed that the N-terminal tail of histone H3 is threaded into the active site of EZH2 by a network of interactions that is disrupted by covalent modification of H3K36 (27). However, despite the compelling cryo-EM data, histone gene replacement studies in vivo in Drosophila showed that replication-dependent histone H3.2K36R mutants exhibit reduced H3K27me3 levels but comparatively modest Pc derepression phenotypes (12, 27, 28). In contrast, H3.2K27R mutants exhibit more widespread derepression (27). Given the exquisite ability of cells to sense changes in the levels of H3K27-modifiable nucleosomes (9, 17), the relatively mild Pc phenotypes observed in H3.2K36R larval tissues are puzzling. If the presence of an unmodified H3K36 residue really is necessary for efficient trimethylation of H3K27 in vivo, then perhaps there is some redundant histone function that serves to mask the K36R mutant phenotype.
One clear candidate for such a role is the replication-independent histone, H3.3, which differs from H3.2 by only four amino acids. The H3.3 and H3.2 N-terminal tails differ by only a single amino acid, at position 31, and thus, both can be similarly modified at their respective H3K36 and H3K27 residues (29, 30). Accordingly, functional redundancies between H3.2 and H3.3 have been described. One study showed that H3.2 can compensate for loss of H3.3 (31). Another example is highlighted by the H3K9 residue (14) as the combination of H3.2K9R and H3.3K9R mutations resulted in more severe developmental and transcriptional defects than did either mutation alone.
We therefore hypothesized that lysine-36 of H3.3 might functionally compensate for loss of H3.2K36 with respect to directly promoting enhancer of zeste [E(z)] activity, H3K27 trimethylation, and appropriate repression of Hox genes. To test this notion, we generated H3.3K36R mutants and examined them for homeotic phenotypes indicative of faulty Hox gene repression. We also compared the effects of H3.2K36R and H3.3K36R mutations on the levels and genome-wide distribution of H3K27me3. These experiments revealed that loss of H3.2K36 causes widespread disruption of H3K27 trimethylation across broad domains (e.g., the Hox gene clusters), whereas loss of H3.3K36 does not. In control genomes, we found that H3.3 preferentially accumulates at presumptive PRC2 recruitment sites, called Polycomb response elements, or PREs (32, 33). H3.3 accumulates to a lesser degree at PRE-like sites located inside broad domains of H3K27me3 silent chromatin than it does to those outside. Last, we created an H3.2K36R/H3.3K36R double mutant and found that combining these mutations synergistically derepresses Hox genes. These findings support a model wherein H3.2K36 and H3.3K36 residues are both important for proper Hox gene repression but that they carry out this function from distinct genomic subcompartments.
RESULTS
Arginine substitutions at K36 and K27 in H3.2 synergistically impair development
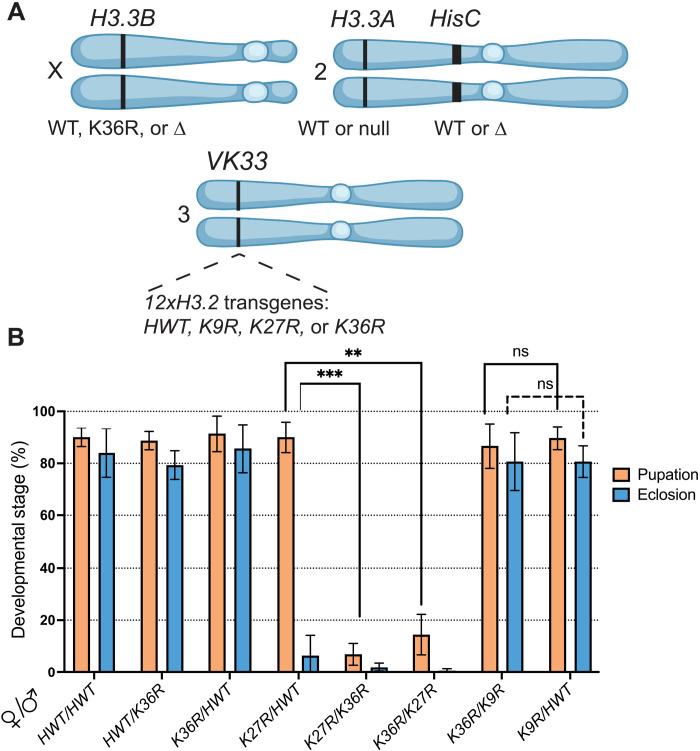
The antagonistic relationship between PRC2, which carries out H3K27 trimethylation, and complexes that methylate H3K36 is well established (25, 26, 34, 35). However, to date, there is no direct evidence demonstrating a developmental biological connection between H3K27 and H3K36 residues. We therefore analyzed animals expressing various bacterial artificial chromosome (BAC) transgenes carrying two homologous copies of a 12x tandemly arrayed 5-kb Drosophila histone gene repeat element in the background of a homozygous deletion of the endogenous histone gene cluster (∆HisC); for details, see fig. S1 and Materials and Methods. This scheme (Fig. 1A) allowed us to perform genetic complementation analyses by combining 12x transgenes of different genotypes, thereby assessing the likelihood that different H3.2 residues might participate in a common function. Adults homozygous or hemizygous for the 12xH3.2HWT (histone wild type, HWT) tandem array are viable and fertile (9); see Fig. 1B. We found that one copy of the HWT transgene rescues the larval and pupal lethality previously reported (9, 12) for 12xH3.2K36R (K36R) hemizygotes (Fig. 1B). One copy of the HWT transgene was unable to fully rescue 12xH3.2K27R (K27R) hemizygotes. These K27R/HWT animals pupate normally, but very few eclose as viable adults (Fig. 1B).
Fig. 1. Intragenic complementation analysis within a multigene family.
(A) Cartoon of chromosomal loci used in (B) and in subsequent experiments. For complete genotypes, see figs. S1, S2, and S4. The H3.3B gene (chr. X) is either WT, K36R, or ∆ (null). The H3.3A gene (chr. 2L) is either WT or ∆ (null). The endogenous replication-dependent histone gene cluster HisC (chr. 2L) is either intact (WT) or ∆ (null). The transgenic insertion site VK33 (chr. 3L, band 65B2) was used for histone gene replacement analysis. 12xH3.2 transgenes contain 12 copies of the histone repeat unit, each of which contains all five replication-dependent histone genes. Transgenes used in this study carry the following alleles of H3.2: HWT, K9R, K27R, or K36R. Panel was created using BioRender.com. (B) Developmental viability assay for complementation analysis of 12xH3.2 transgenes. All genotypes are HisC∆ and carry two 12x histone transgenes in trans (24x total). Pairs of transgenes are represented on the x axis for each set of bars. For each genotype, % pupation and % eclosion of four to six biological replicates (50 larvae/replicate vial) were calculated, and means and SD of these percentages were plotted. Statistical significance for % pupation was calculated with GraphPad Prism software using a mixed-effects analysis (can accommodate missing values) on the four genotype pairs indicated by brackets, followed by Šidák’s multiple comparisons test. **P < 0.01; ***P < 0.001. ns, not significant.
Using this assay, we observed a strong genetic interaction between H3.2K36R and H3.2K27R; nearly all the K27R/K36R animals die as larvae before pupation (Fig. 1B). As mentioned above, the control crosses displayed significantly milder phenotypes (Fig. 1B). Notably, the K27R/K36R complementation failure is not simply due to an overabundance of mutant histones as crosses between K36R and 12xH3.2K9R (K9R) produced viable adults at similar frequencies to those with HWT (Fig. 1B). Previously, this sort of intragenic complementation analysis within a large multigene family has not been possible. Our results strongly suggest that H3.2K36 and H3.2K27 residues share common pathways or mechanisms necessary for proper development to adulthood, whereas H3.2K36 and H3.2K9 do not.
H3.3K36R mutants are viable and fertile
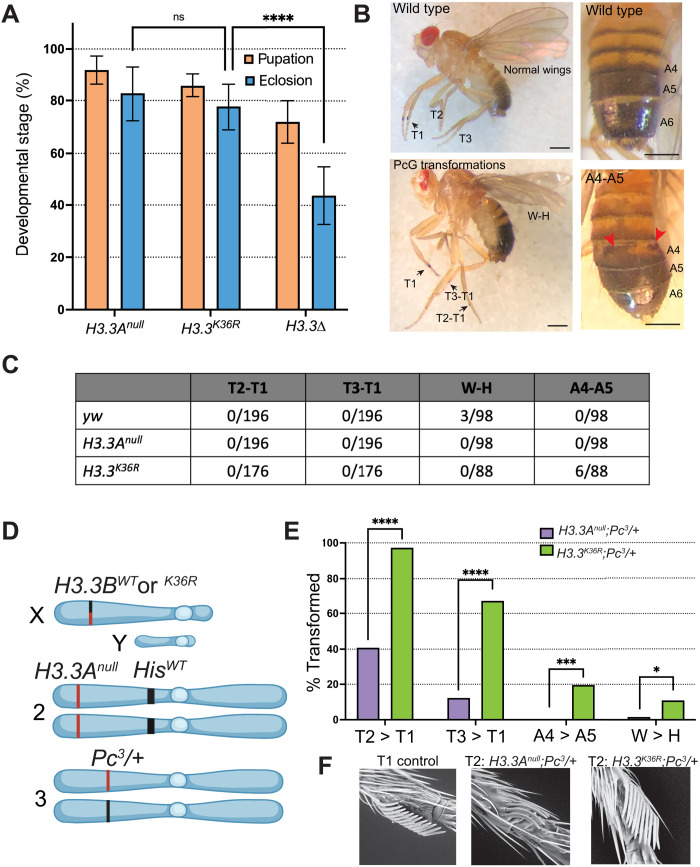
The relative failure of replication-dependent H3.2K36R mutants to elicit strong Pc phenotypes (12, 27, 28) suggests the existence of a redundant H3 function. Outside of S phase and in postreplicative cells, nucleosome turnover is largely carried out using the replication-independent histones, including H3.3 (36). In Drosophila melanogaster, there are two H3.3 genes, H3.3A and H3.3B (37). To test for genetic redundancies at H3.3 lysine-36, we generated a K-to-R missense mutation in H3.3B using Cas9-mediated homologous recombination and introgressed it onto an H3.3A null mutant (H3.3Anull) background (Fig. 1A). Complete genotypes and crossing schemes for generating these animals and all others used in this study can be found in figs. S1 to S4 and table S1. H3.3BK36R;H3.3Anull double-mutant animals (hereafter H3.3K36R) pupate and eclose at similar frequencies to H3.3Anull mutants (Fig. 2A). Thus, the viability of H3.3BK36R animals was unaffected by deletion of H3.3A. Given that animals lacking both H3.3A and H3.3B (H3.3∆) had previously been shown to complete development (31, 38), and those lacking only H3.3A are fully viable and fertile (31), this result was expected. However, unlike H3.3K9R or H3.3K27R mutants (14, 39), H3.3K36R males are fertile. Furthermore, these data reveal that the H3.3K36R protein is incorporated into chromatin and is at least partially functional as H3.3K36R mutants eclose at significantly higher frequencies than H3.3∆ animals (Fig. 2A).
Fig. 2. Homeotic transformation analysis of H3.3K36R mutants.
(A) Developmental viability of H3.3K36R mutant and controls. H3.3K36R indicates H3.3Anull combined with H3.3BK36R. Assay conditions and statistical analyses are as in Fig. 1B, with the following modifications. Eight replicates were scored for all genotypes. Statistical significance was calculated on % eclosion with one-way analysis of variance (ANOVA). Šidák’s multiple comparisons test was used as above, with the following additional P values; ****P < 0.0001. (B) Top left, a WT fly with normal legs and wings. Bottom left: A fly with three homeotic transformations scored in (C): T2-T1 (thoracic segment leg 2 to leg 1), T3-T1 (leg 3 to leg 1), and W-H (wing to haltere). Top right: A fly with a WT abdomen. Bottom right: A fly exhibiting a typical A4-A5 (abdominal segments 4 to 5) transformation scored in this assay. Individual A4-A6 segments are labeled. Red arrows highlight abnormal pigmentation indicating a partial A4-A5 transformation. Scale bar, ~0.5 mm. (C) Table showing number of transformations per scored events for each H3.3K36R mutant and control genotype. (D) H3.3K36R (and control genotypes) were combined with a heterozygous Pc3 mutation and scored for the four PcG homeotic transformations depicted in (B). For full genetic scheme, see fig. S3. Panel was created using BioRender.com. (E) The fraction transformed (%) for these phenotypes is plotted for each genotype. N value for number of flies scored for the H3.2K36R;Pc3/+ genotype (n = 55) and for the control (n = 62). Note that for T2-T1 and T3-T1, each appendage was scored separately, effectively doubling the n value for these transformations. GraphPad Prism was used to calculate a χ2 value for each transformation. Significance is abbreviated as follows: *P < 0.05, ***P < 0.001, ****P < 0.0001. (F) Image of a typical T2-T1 transformation for each genotype collected by scanning electron microscopy at ×250 magnification.
The H3.3K36R mutation enhances Pc phenotypes in adults
The H3.3WTH3.2K36R mutants exhibit delayed development, with a broad lethal phase that extends throughout larval and pupal stages; very rarely (< 0.2%), animals eclose as adults (9, 12). However, H3.3WTH3.2K36R pharate adult (uneclosed) males occasionally demonstrate homeotic transformations that are indicative of impaired regulation of Hox genes, including ectopic sex combs and antenna to leg transformations (27). To evaluate H3.3K36R, we scored adult mutant and control animals for homeotic transformations including: thoracic T2 to T1 (T2-T1) and T3 to T1 (T3-T1), abdominal segments 4 to 5 (A4-A5), and wing to haltere (W-H) transformations. Examples of these phenotypes are illustrated in Fig. 2B. The T2-T1 and T3-T1 leg transformations display ectopic sex comb bristles, the A4-A5 transformations show abnormal pigmentation of segment A4, and the W-H transformations feature abnormal wing morphology, manifesting as fully or partially crumpled wings.
Control yw flies (n = 98) with WT histone loci completely lacked leg and abdominal transformations, although 3 of 98 flies displayed wing morphology consistent with mild W-H transformations. Inspection of H3.3K36R adult males (n = 88) for homeotic transformations revealed few overt PcG phenotypes compared to controls (Fig. 2C) (40, 41). However, introducing H3.3K36R into a sensitized genetic background revealed a significant increase in homeotic transformation. Flies heterozygous for a null mutation of Pc (Pc3/+) are viable as adults but exhibit a baseline frequency of homeotic transformations, which can be modified by other mutations present in the genotype (42–44). This assay has been used reliably to implicate genes regulating Pc-dependent gene silencing in many other studies (42–44). We constructed two pairs of control and mutant genotypes for analysis (Fig. 2, D to F). As the full genotypes are quite lengthy, we have abbreviated them for clarity. See fig. S3 for complete genotypes.
First, to validate the role of H3.2K36 in Hox gene repression in this assay, and to provide a benchmark for subsequent analysis of H3.3, we scored T2-T1, T3-T1, W-H, and A4-A5 transformations in His∆/+;H3.2K36R/Pc3 mutants and His∆/+;H3.2HWT/Pc3 controls (fig. S5). Despite mutant H3.2K36R genes comprising only about 10% of the total number of H3.2 genes (9, 45), we observed significant increases in T2–T1, T3-T1, and W-H transformations in the His∆/+;H3.2K36R/Pc3 mutants compared to His∆/+;H3.2HWT/Pc3 control animals (fig. S5). These data validate this assay in the context of histone N-terminal tail residue mutation, provide further evidence that the H3.2K36 residue promotes Pc-mediated silencing of Hox genes, and provide a benchmark for the sensitivity of this assay.
Second, we further investigated whether H3.3K36 plays a role in Pc-mediated Hox gene repression by scoring homeotic transformations in H3.3K36R;Pc3/+ mutants and H3.3Anull;Pc3/+ controls (Fig. 2, D and E). Notably, these flies had a fully WT HisC locus, and thus, all replication-dependent H3.2 genes were present at WT copy number. We observed a significant increase in all four categories of homeotic transformations in the H3.3K36R;Pc3/+ mutants relative to the control group (Fig. 2E). We observed a sizeable increase in A4-A5 transformations (19.6%, P < 0.001) when H3.3 was mutated compared to when H3.2 was mutated (3.6%, P = not significant). This observation implies that the A4-A5 transformation is particularly sensitive to H3.3K36 mutation. Such relative differences in the severity of homeotic transformation between H3.2K36R and H3.3K36R mutants suggest that H3.2K36 and H3.3K36 might promote Hox gene repression by nonidentical means.
In the above assay, we quantitatively scored homeotic transformations as differences in frequency. However, we also noted qualitative differences in phenotypic severity that mirrored or exceeded these changes in frequency. For example, the H3.3BK36R;Pc3/+ mutants and H3.3Anull;Pc3/+ controls both exhibit T2-T1 sex comb transformations. However, the H3.3BK36R;Pc3/+ mutant generally demonstrated a more severe phenotype, as indicated by the number of bristles on T2. To capture these qualitative differences in sex comb transformations, we used scanning electron microscopy to image T2 sex combs of each genotype in the previous sets of experiments (Fig. 2F). We consistently observed a greater number of bristles in the T2 sex combs of both His∆/+;H3.2K36R/Pc3 and H3.3K36R;Pc3/+ mutant genotypes relative to controls (Fig. 2F). These same relative differences in severity also applied to the other transformations that we scored quantitatively. In summary, these analyses of adult animals suggest roles for both H3.2K36 and H3.3K36 in repression of Hox genes at late developmental stages.
H3.3K36 and H3.2K36 differentially affect H3K27me3 late in development
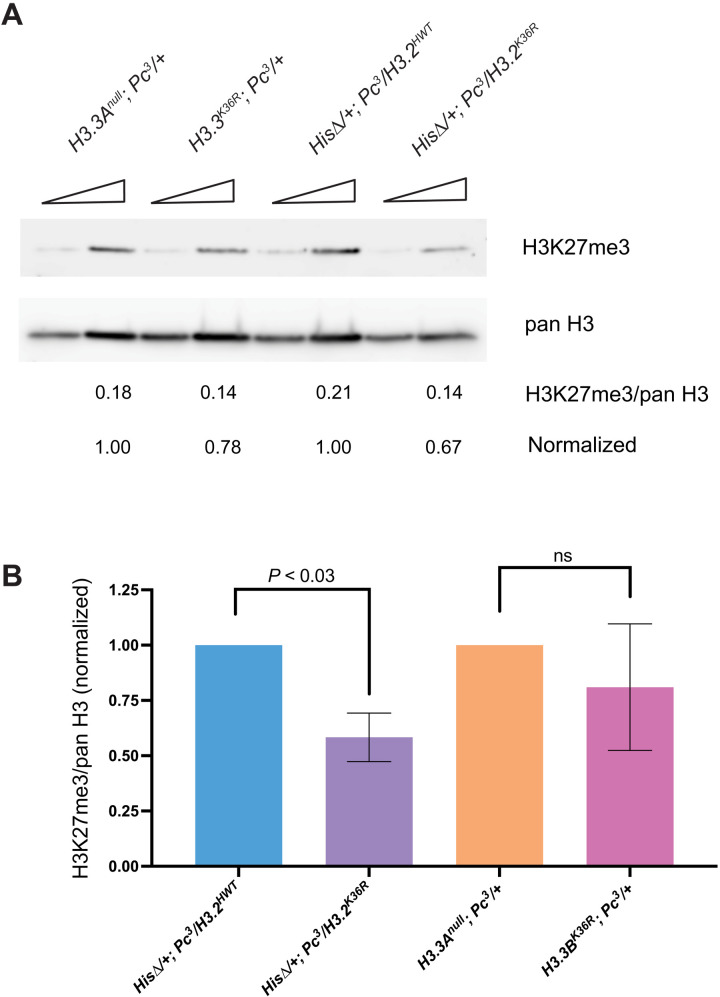
Given that PcG phenotypes arise in H3.3K36R adults, we wondered whether an effect on H3K27me3 might become evident at this later developmental time point. Therefore, we next sought to measure the relative impact of H3.2 and H3.3 K36R on H3K27me3 by Western blotting from extracts of adult heads. If unmethylated H3.3K36 were redundantly stimulating E(z) at later developmental stages, we would expect to observe reduced H3K27me3 in H3.3K36R;Pc3/+ mutants relative to H3.3Anull;Pc3/+ controls and relative to the His∆/+;H3.2K36R/Pc3 genotype. However, we did not observe any difference in global H3K27me3 levels in the H3.3K36R;Pc3/+ animals compared to control despite a clear effect on homeotic transformation frequency (Figs. 3 and 2E). In contrast, we observed an ~40% decrease in H3K27me3 in His∆/+;H3.2K36R/Pc3 mutants relative to His∆/+;H3.2HWT/Pc3 controls despite the fact that only 10% of H3.2 histone genes carry the H3.2K36R mutation in this genotype (Fig. 3 and fig. S5). This large difference held true in three separate experiments (P < 0.05; Fig. 3B). Overall, these data are consistent with the idea that H3.2K36 is more important than H3.3K36 in promoting global levels of H3K27me3 even at a late developmental time point.
Fig. 3. Western blot analysis of H3K27me3 levels in adult heads.
H3.2 and H3.3 K36R mutants were maintaned in a sensitized Pc3 genetic background. (A) Western blotting with anti-H3K27me3 and anti–pan H3 were performed using adult head extracts, loading either 40 or 80 μg of protein per lane. Three independent experiments were performed and imaged; a representative blot is shown. Mean pixel intensity was calculated for each band using equal-sized areas, and a ratio of H3K27me3/pan H3 was calculated for each biological replicate from the 80-μg lane. (B) For each independent experiment, the H3K27me3/pan H3 ratio of mutant genotypes was normalized to its control (His∆/+;H3.2K36R/Pc3 to His∆/+;H3.2HWT/Pc3 and H3.3BK36R;Pc3/+ to H3.3Anull;Pc3/+). For each genotype, the normalized mean and SD of H3K27me3/pan H3 values of all three experiments was plotted. Statistical significance between raw H3K27me3/H3 ratios was assessed by paired one-way ANOVA (within experiments), followed by Friedman tests individually comparing mutant genotypes to controls. P values are directly noted on the graph.
Mutation of H3.2K36, but not H3.3K36, causes defects in H3K27me3 spreading
Previous studies in various organisms have revealed antagonism between factors that carry out H3K36me and H3K27me. In particular, H3K36me is thought to be important for demarcating Pc domain boundaries (34, 46–48). To investigate changes in H3K27me3 patterns in the H3.2K36R and H3.3K36R mutants, we used CUT&RUN chromatin profiling in wing imaginal discs of wandering L3 (WL3) larvae (49). To directly compare H3K27me3 levels between genotypes, we analyzed the H3.3K36R mutation in the same genetic background used for the histone gene replacement platform. The resultant H3.3K36RH3.2HWT animals were compared with H3.3AnullH3.2HWT controls, each of which contains a deletion of the H3.3A gene (see fig. S4 for details). For each mutant and control, we performed three independent biological replicates and sequenced both the supernatant and pellet fractions (see Materials and Methods). The data in both fractions were consistent with respect to signal distribution; however, correlations between samples of the same genotype were superior in the pellet fraction (fig. S7), Thus, for all subsequent analyses with the anti-H3K27me3 antibody, we used the pellet fraction except where specifically noted.
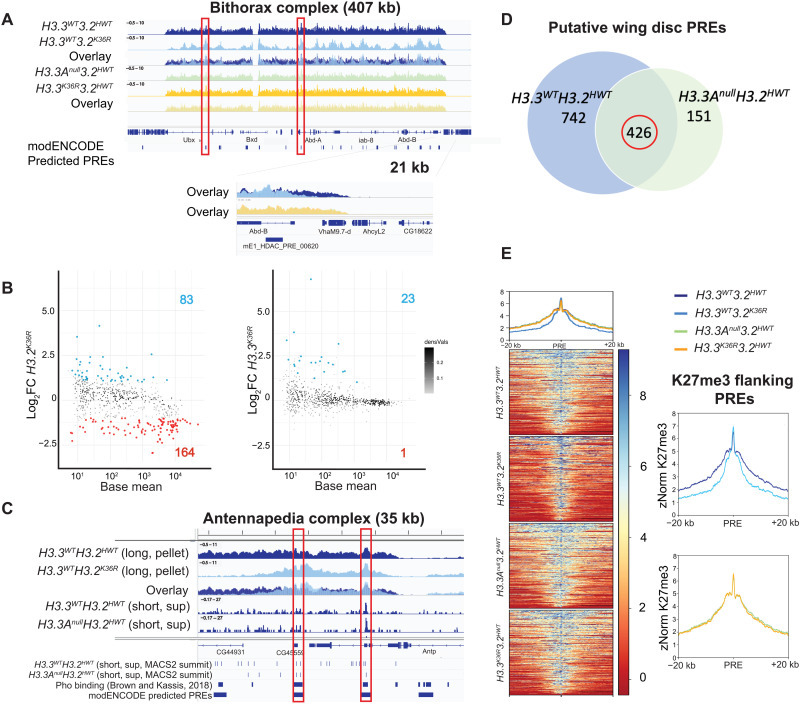
As expected, H3K27me3 was highly enriched across known Pc domains in all genotypes, exemplified by the browser shot of the Bithorax complex (BX-C) (Fig. 4A). We noted no discernable difference between the two control genotypes. Genome-wide, we also observed little difference between the H3.3K36RH3.2HWT mutant and the H3.3AnullH3.2HWT control (Fig. 4A). In contrast, H3K27me3 levels in the H3.3WTH3.2K36R mutant were markedly depleted relative to the H3.3WTH3.2 HWT control (Fig. 4A). To quantify the number of broad H3K27me3 domains that changed for each mutant, we performed a differential peak analysis using DESeq2 (Fig. 4B). These domains spanned a range of sizes and H3K27me3 signal intensities, from very large and heavily methylated regions, such as the Bithorax and Antennapedia complexes, to much smaller and less intensely methylated ones (Fig. 4B). In the H3.3K36RH3.2HWT mutants, there were very few (24 of 629) differential domains with an adjusted P value < 0.05 and a log2 fold-change threshold of > |1| (Fig. 4B). Moreover, nearly every differential peak identified in the H3.3K36RH3.2HWT animals displayed increased H3K27me3 signal intensity in the mutant, which is the opposite of what would occur if K36R were inhibiting PRC2. In contrast, the H3.3WTH3.2K36R mutants exhibited significantly more differentially methylated domains (247 of 629), the majority of which (164 of 247) decreased in intensity (Fig. 4B) (27). The most notable changes are evident in domains bearing the highest levels of methylation (base mean > 103), where nearly all of them decreased in the presence of the mutation (Fig. 4B). Overall, these data provide no evidence that H3.3K36 contributes to methylation within broad H3K27me3 domains in the wing disc, whereas H3.2K36 has a widespread effect.
Fig. 4. Mutation of H3.2K36, but not H3.3K36, impairs H3K27me3 in Pc domains.
H3K27me3 CUT&RUN profiling in WL3 wing discs (three replicates per genotype). All browser tracks, metaplots, and heatmaps used z score normalization (A) IGV (Integrative Genomics Viewer) track of the BX-C from pellet. Individual and overlay tracks for H3.3WTH3.2K36R with H3.3WTH3.2HWT control, and H3.3K36RH3.2HWT with H3.3AnullH3.2HWT control. Below the genome annotation are modENCODE predicted PREs. Red boxes indicate PREs exhibiting WT H3K27me3, with H3K27me3 declining in the H3.3WTH3.2K36R mutant in flanking regions. The Abd-B promoter is shown at higher magnification. (B) Differential peak analysis of broad H3K27me3 domains using DESeq2 (n = 629). MACS2 broad peak intervals within 10 kb were merged before DESeq2 (see fig. S8). Points with adjusted P value > 0.05 and a log2 fold change (Log2FC) > |1| colored (red, down-regulated; blue, up-regulated). Log2FC is computed relative to appropriate control. (C) IGV track of H3K27me3 from the Antennapedia complex using long-fragment reads (150 to 700 bp). Tracks are shown for the H3.3WTH3.2K36R mutant and H3.3WTH3.2HWT control, including overlay. Below overlay are subnucleosomal (20 to 120 bp) tracks from control genotypes. BED tracks represent individual MACS2 narrow peak summits, Pho-binding sites as determined in (59), and modENCODE-predicted PREs. Red boxes indicate correspondence between features. (D) To identify putative wing disc PREs, intervals containing MACS2 peak summits ± 150-bp overlapping Pho-binding regions (see Materials and Methods and fig. S8) were ascertained for each control. The number of intervals for each control is indicated in each circle. The intersection of PRE intervals identified in both controls (circled in red) was used for further analysis. (E) Heatmap and metaplots of H3K27me3 signal at putative PREs and 20-kb flanking regions determined in (D) (n = 426), calculated from merged bigwig files from each genotype. To the right, each mutant genotype is compared directly to its respective control.
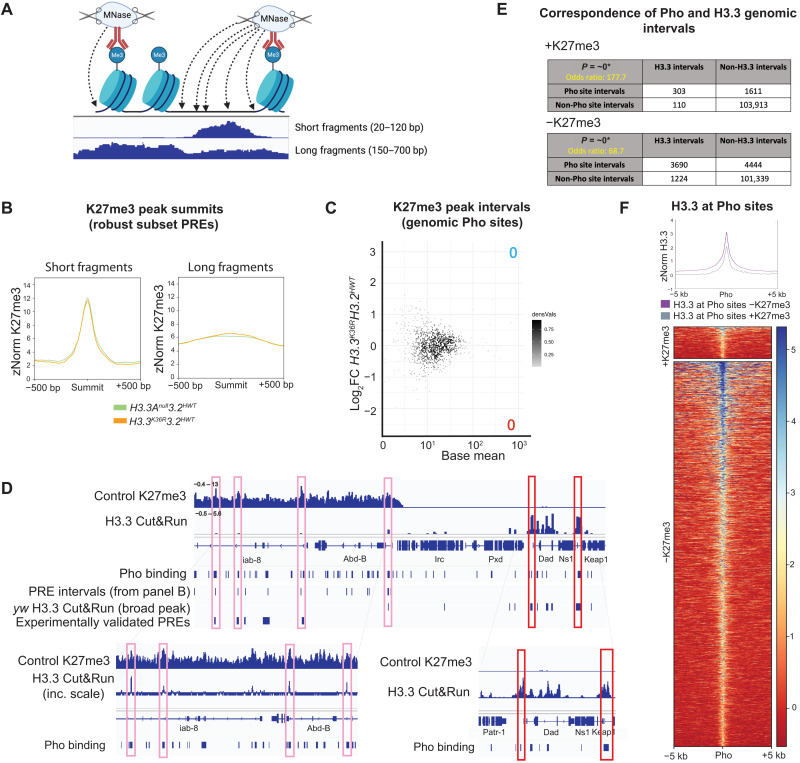
The pattern of depletion in the H3.3WTH3.2K36R mutant was also notable. That is, the “spikes” of K27 trimethylation in the mutant were of comparable magnitude to those in the controls, but the signal was severely depleted in the troughs and at Pc domain boundaries (Fig. 4A). One might expect a spiky pattern such as this if PRC2 could efficiently nucleate at specific sites (e.g., PREs) but was unable to spread effectively between them. Although PREs are generally thought to be nucleosome depleted (32, 50), DNA binding factors located at these sites are known to recruit PRC2 to adjacent nucleosomes. During the CUT&RUN assay, DNA cleavage by micrococcal nuclease (MNase) is directed by an antibody. In this case, anti-H3K27me3 is expected to recruit MNase to cleave chromatin that is near PREs into short, subnucleosomal fragments [Fig. 5A (50–52)]. Accordingly, the spikes located within broader domains of H3K27me3 signal in our data align well with PREs that have been predicted by other methods (Fig. 4A) (53).
Fig. 5. H3K27me3-directed cleavage at presumptive PREs is unchanged in H3.3K36R mutants.
(A) MNase cleavage near histone PTMs during CUT&RUN. Antibodies direct MNase localization, cleaving nearby accessible DNA to generate long (nucleosomal) and short (subnucleosomal) fragments. Image was created using BioRender.com. (B) Metaplots of K27me3 directed CUT&RUN signal ± 500 bp around peaks from all four genotypes overlapping wing disc PRE intervals identified in Fig. 4D (n = 426) for short and long fragments, respectively. The H3.3K36RH3.2HWT mutant alongside H3.3AnullH3.2HWT control. (C) DESeq2 analysis of short-fragment H3K27me3 peak intervals that overlap with Pho-binding sites identified by Kassis and colleagues (n = 985). M/A (log ratio/mean average) plot of these intervals for H3.3K36RH3.2HWT mutant versus H3.3AnullH3.2HWT control (details in fig. S8). No significant peaks were identified. (D) IGV track of H3.3 CUT&RUN at the BX-C from supernatant, merged, z score–normalized bigwig files. Two replicates each of a yw control bearing no histone mutations were performed to determine H3.3 signal, and an H3.3∆ mutant used to subtract nonspecific antibody binding. An H3K27me3 track is included for identification of H3K27me3 domains. BED tracks show positions of the following: Pho-binding sites identified in (59); PRE intervals derived in (B); MACS2 peak calls from H3.3 CUT&RUN from yw genotype; and experimentally validated PREs curated in (54). Pink boxes illustrate subthreshold H3.3 peaks within the BX-C. Red boxes show H3.3 peaks near the Dad locus. (E) Fisher’s exact tests measuring correspondence between genomic intervals encompassing Pho sites and H3.3 binding. Separate tests were performed for regions inside and outside H3K27me3 domains. Correspondence was highly significant in both cases. *Precise P value is below minimum computable value for BEDtools and is approximated as “0.” (F) Metaplot and heatmap comparing z-normalized H3.3 CUT&RUN signal in yw controls at Pho sites intersecting H3.3 peaks located inside (+K27me3, n = 303) and outside (−K27me3, n = 3690) of H3K27me3 domains.
The depletion of H3K27me3 signal between presumptive PREs in the Hox gene clusters suggests that the H3.3WTH3.2K36R mutation causes a defect in the spreading of Pc-silencing factors. PREs are tissue-specific cis-regulatory modules, so to determine whether the spreading defect is a general phenomenon, we first needed to identify a robust set of PREs that are used in the WL3 wing disc. The gold-standard definition of a PRE is a functional one, requiring experimental characterization of individual sequences in vivo. Thus, a relatively small number of PREs have been validated in this manner, and most of the experiments have not been performed in wing discs (32, 54). Bioinformatic predictions based on the presence of DNA binding protein motifs (55–58) predict elements with potential PRE activity but do not define which sequences are used in a particular developmental scenario. To circumvent these problems, we set out to identify PREs used in the WL3 wing disc in a manner that is both functional and predictive.
As functional PREs encompass sequences where DNA binding proteins assemble at open regions of chromatin, we exploited the propensity for accessible chromatin to be cleaved into subnucleosomal fragments (Fig. 5A) during the CUT&RUN assay (52) by centering our analysis on peaks derived from short fragments [20 to 120 base pairs (bp)] located within broader H3K27me3 domains (Fig. 4C). For this purpose, we used MACS2 narrow peak summit calls from short fragments in the supernatant fraction as the supernatant yielded fewer spurious peak calls due to localized noise. To improve accuracy, we also capitalized on the availability of chromatin immunoprecipitation sequencing data that were generated from a mixed sample of larval brain and imaginal discs using antibodies targeting pleiohomeotic [Pho; (59)]. In Drosophila, the presence of Pho is one of the best predictors of functional PRC activity at experimentally verified PREs (32, 60). Using these two datasets, we bioinformatically identified genomic intervals of short-fragment H3K27me3 peak summits (±150 bp) that overlapped with known Pho-binding sites identified by Kassis and colleagues (Fig. 4C). These regions correspond well with PREs predicted by modENCODE (Fig. 4C). As shown in Fig. 4D, this analysis identified 1168 and 577 candidate wing disc PREs in the H3.3WTH3.2HWT and H3.3AnullH3.2HWT genotypes, respectively. We further narrowed this list to a common set of 426 putative PREs that were identified in both control genotypes (Fig. 4D).
To ascertain whether the K36R mutation impedes spreading of H3K27me3, we generated heatmaps and metaplots of H3K27me3 signal in the regions flanking these presumptive PREs (Fig. 4E). Although the short-fragment reads were useful for identifying regions likely to harbor DNA binding proteins at PREs, we used reads from pooled bigwig files of supranucleosomal fragment length (150 to 700 bp) to compare H3K27me3 signal in the regions ±20 kb flanking the presumptive PREs for each genotype. Notably, metaplots for the two control genotypes and the H3.3K36RH3.2HWT mutant were nearly identical, demonstrating that mutation of H3.3K36 alone had no effect on spreading across broad Pc domains (Fig. 4E). In contrast, the H3.3WTH3.2K36R mutant displayed a marked depletion of H3K27me3 signal in the flanking regions but not directly over the PREs themselves (Fig. 4E). These data not only confirm and extend previous findings (27) but also demonstrate that in vivo, H3.2K36 plays a much more important role than H3.3K36 in facilitating the spreading of H3K27me3 across broad domains, suggesting that H3.3K36 promotes proper Hox gene regulation by a different means.
Loss of H3.3K36 has no effect on H3K27 trimethylation at PREs
Previous reports in mouse embryonic stem cells and Drosophila showed that H3.3 is enriched at CpG islands and PREs, regions that most consistently function as PRC2 nucleation sites, respectively (61, 62). Furthermore, GAGA factor, which can bind PREs and promote repression of associated genes (63–66), interacts with HIRA, an H3.3-specific histone chaperone (67). Although the analysis of long-fragment reads in Fig. 4 failed to identify a loss of signal within broad domains, we wondered whether the H3.3K36RH3.2HWT animals might display a more localized effect at PREs.
In CUT&RUN, the distribution and overall signal from different fragment sizes are functions of both the relative presence of the epitope of interest and the relative accessibility of the surrounding chromatin (Fig. 5A) (52). To interrogate the H3K27me3 profiling data more specifically, we generated metaplots ±500 bp around 365 short-fragment peak summits from all four genotypes that overlapped with the set of robust PREs established in Fig. 4. As shown in Fig. 5B, we observed no appreciable difference in signal between the H3.3K36RH3.2HWT mutant and its control (Fig. 5B). Despite reported colocalization of H3.2 and H3.3 at PREs in S2 cells (62), we also saw no effect at PREs in the H3.3WTH3.2K36R mutant (fig. S10). These data show that individual loss of H3.2K36 or H3.3K36 has little effect on H3K27 methylation immediately flanking presumptive PREs.
Because PREs are heterogeneous with respect to DNA sequence, binding factor occupancy, and functional properties (32, 59, 68), we wondered whether certain PREs might be preferentially affected by the H3.3K36 mutation, though we detected no general effect. To address this question, we performed DESeq2 differential expression analysis on short-fragment read counts from the K27me3 pellet fraction in all four genotypes (Fig. 5C). We broadened the approach to examine intervals that overlapped with a more general set of short fragment peak summits (fig. S11) and with Pho-binding sites (Fig. 5C and fig. S10). We identified no differential peaks (0 of 985) overlapping with Pho sites in the H3.3K36RH3.2HWT mutant (Fig. 5C). Similarly, there were only a few differential peaks (81 of 985) in the H3.3WTH3.2K36R mutant (fig. S10). These data show that mutation of H3.2 or H3.3 lysine-36 has little effect on the accessibility of PREs to MNase cutting or on H3K27 trimethylation of nucleosomes that flank Pho-binding regions.
H3.3 is enriched at Pho-binding sites inside and outside of Pc domains
Chromatin profiling for H3K27me3 in the H3.3WTH3.2K36R mutant supports a clear role for H3.2 in propagation of H3K27me3 between PREs (Fig. 4). Given that H3.3 is reportedly enriched at PREs in S2 cells (62), the failure to observe a reduction in H3K27me3 signal at these sites in H3.3K36RH3.2HWT mutant wing discs was unexpected (Fig. 5, B and C). We therefore determined whether H3.3 is also enriched at PREs in the wing disc by CUT&RUN profiling using anti-H3.3 antibodies on yw (positive) and H3.3∆ mutant (negative) control genotypes. As shown in Fig. 5 (D to F), analysis of the anti-H3.3 and anti-H3K27me3 CUT&RUN data alongside sets of previously validated PREs (54) and Pho-binding sites (59) revealed a number of interesting features.
We detected an enrichment of H3.3 at Pho-positive PREs located in the BX-C and in other H3K27me3 domains (Fig. 5D). Note that the presence of H3.3 at these sites was obvious when visualized on a genome browser (high signal, low noise), but the peak summit intensities often did not meet the threshold of the MACS2 peak caller. Consistent with the notion that H3.3 is enriched within active chromatin (61, 69–71), we observed much higher levels of H3.3 in “euchromatic” regions located outside of repressive H3K27me3 domains (Fig. 5D). The H3.3 peaks in euchromatic regions also coincided with Pho-binding sites, and visual inspection suggested that they were typically of greater intensity than those located inside of H3K27me3 domains (Fig. 5D).
To determine the significance of the association between H3.3 and Pho sites, we performed Fisher’s exact test on intervals known to have Pho-binding potential (59) and those enriched for H3.3. As shown in Fig. 5E, the correlation between H3.3 and Pho peaks was large and highly significant, regardless of H3K27me3 status [odds ratio (OR) = 177.67, P ~ 0 inside versus OR = 68.75, P ~ 0 outside]. To quantify the difference in H3.3 signal at Pho sites inside versus those outside of Pc domains, we generated heatmaps and metaplots for H3.3 signal at sites of overlapping H3.3 and Pho-binding potential (Fig. 5F). We found that H3.3 enrichment was roughly 1.6-fold greater at Pho sites located outside K27me3 domains compared to those inside (Fig. 5F). Together, our data show that H3.3 is indeed enriched at wing disc PREs; however, mutation of H3.2 or H3.3 K36 has very little effect on H3K27 trimethylation at these sites.
Complete loss of H3K36 results in profound developmental defects
Our genetic experiments demonstrate that both H3.2K36 and H3.3K36 contribute to accurate repression of Hox genes. However, Western blotting and CUT&RUN experiments suggest that they contribute to this process by different mechanisms. Although H3.3WTH3.2K36R animals clearly have a defect in H3K27 trimethylation, H3.3K36RH3.2HWT mutants do not. Furthermore, both H3.2K36R and H3.3K36R mutant animals exhibit relatively mild homeotic phenotypes compared to animals with mutations in other genes required for this process (43, 44, 72, 73).
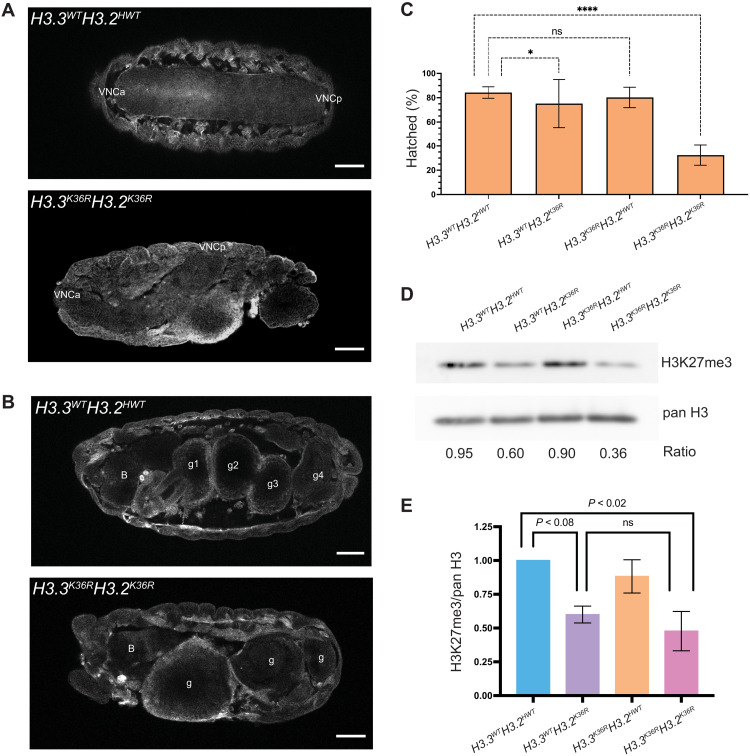
To investigate potential redundancy between H3.3K36 and H3.2K36, we engineered a combined H3.3K36RH3.2K36R mutant (fig. S4). When H3K36 is completely removed from the genome (H3.3K36RH3.2K36R), we observed a much stronger developmental phenotype than either the H3.2K36R or H3.3K36R alone as none of the H3.3K36RH3.2K36R mutants progressed beyond the L1 stage (n > 500). Furthermore, H3.3K36RH3.2K36R embryos frequently exhibited gross morphological defects, such as ventral nerve cord (VNC) and gut defects (Fig. 6, A and B). Only ~30% of the H3.3K36RH3.2K36R embryos hatched (Fig. 6C). Together, the results reveal that H3K36 is critical for proper embryogenesis and that H3.2K36 and H3.3K36 can compensate for one another during these early stages of development.
Fig. 6. Analysis of combined H3.3K36RH3.2K36R mutants during early development.
(A) Stage 16 embryos of H3.3K36RH3.2K36R mutants and H3.3WTH3.2HWT controls stained with anti-green fluorescent protein (GFP) antibody, outlining gross morphology. Top: A control with a symmetrical VNC. Anterior and posterior VNC (VNCa, VNCp). Bottom: A mutant exhibiting severe VNC defects. We observed a similar degree of VNC twisting in 4 of 44 (~9%) H3.3K36RH3.2K36R mutant embryos and 0 of 73 in controls. (B) Embryos, as in (A), but highlighting severe defects in gut development. B, brain; g1, g2, g3, and g4 correspond to four gut lobes characteristic of this stage. This phenotype was observed in combined mutants but not in other genotypes. Scale bar, 50 μm. Embryos displayed with anterior at left. (C) Embryonic viability assay (for each genotype, n = 250 to 400 embryos). Fraction of embryos progressing from embryonic to L1 stages (% hatched) was calculated. For H3.3WTH3.2K36R and H3.3K36RH3.2K36R genotypes, this value reflects adjustment for the presence of a balancer chromosome in 50% of the embryos. Significance was calculated with GraphPad Prism software using Fisher’s exact test comparing H3K36R mutant genotypes with the H3.3WTH3.2HWT control. P values abbreviated as in Fig. 1. (D) Western blotting for H3K27me3 and pan H3 on L1 larvae. (E) Quantification of four independent replicates. Mean pixel intensity for each band was measured, and ratio of H3K27me3/pan H3 was computed per replicate. For each experiment, the H3K27me3/pan H3 ratio of mutant genotypes was normalized to H3.3WTH3.2HWT controls. For each genotype, normalized means and SD of H3K27me3/pan H3 values of four replicates were plotted. Significance between raw H3K27me3/H3 ratios was assessed by paired one-way ANOVA, followed by Friedman tests comparing mutant genotypes to H3.3WTH3.2HWT controls. P values noted on graph. H3.3K36RH3.2HWT mutants are no different from H3.3WTH3.2HWT controls (P value = ns, not shown on graph), whereas, H3.3WTH3.2K36R and H3.3K36RH3.2K36R exhibit decreased H3K27me3.
Total loss of H3K36 does not alter global H3K27me3 levels but causes synergistic misexpression of Hox proteins
Because H3K27 trimethylation at PREs is believed to be a prerequisite for the spread of this PTM into flanking regions [reviewed in (74)], one might expect that if H3.2 and H3.3 functioned redundantly to enable PRC2 activity at PREs, then levels of H3K27me3 in the combined H3.3K36RH3.2K36R would be further diminished compared to what is observed in H3.3WTH3.2K36R animals. To test this idea, we performed Western blotting of L1 lysates from H3.3WTH3.2HWT (control), H3.3WTH3.2K36R, H3.3K36RH3.2HWT, and H3.3K36RH3.2K36R genotypes using anti-H3K27me3 antibodies, along with a pan-H3 loading control (Fig. 6, D and E). In total, we performed and quantified four biological replicates. In the H3.3WTH3.2K36R genotype, we observed an ~40% reduction (P < 0.08) in H3K27me3 compared to the H3.3WTH3.2HWT control (Fig. 6, D and E). This finding is consistent with previous reports showing similarly reduced H3K27me3 levels in WL3 H3.3WTH3.2K36R (12, 27). As expected, we observed no change in H3K27me3 levels in the H3.3K36RH3.2HWT mutants. Notably, we observed no significant change of H3K27me3 in the combined H3.3K36RH3.2K36R genotype compared to the H3.3WTH3.2K36R mutant, with H3K27me3 at ~40% of the H3.3WTH3.2HWT control (Fig. 6, D and E). These data suggest that H3.2 and H3.3 K36R mutations are unlikely to function redundantly at PREs by directly inhibiting PRC2.
Mutation of either residue alone confers relatively mild defects in Hox gene expression and homeotic transformation. To further explore the possibility of genetic redundancy between H3.2K36 and H3.3K36, we carried out immunostaining of control, single, and combined K36R mutations and determined the extent to which Hox proteins were mis-expressed. In WT embryos, Abdominal-B protein (AbdB) is limited to embryonic parasegments PS10 to PS14, with the highest expression levels near the posterior of this region. These parasegments roughly correspond to adult abdominal segments A4 to A9 (75). Immunostaining of stage 16 H3.3WTH3.2HWT embryos recapitulated the established pattern of AbdB expression in the VNC (Fig. 7A, bracket). We also observed the mild, stochastic derepression of AbdB in H3.3WTH3.2K36R embryos noted previously (27), although the penetrance of this phenotype was incomplete (Fig. 7A, arrowheads, and fig. S13). The H3.3K36RH3.2HWT phenotype was similar to that of the control, in that we never observed AbdB derepression in anterior parasegments (Fig. 7A and fig. S13). In contrast, H3.3K36RH3.2K36R mutants displayed extensive derepression of AbdB in anterior parasegments, comparable to that of H3.3WTH3.2K27R mutants (Fig. 7A, arrowheads, and fig. S13). We conclude that H3.3K36 and H3.2K36 can functionally compensate for one another to fully repress AbdB expression in the embryonic VNC.
Fig. 7. Combined mutation of H3.3 and H3.2 lysine 36 causes synergistic derepression of Hox genes.
(A) Stage 16 embryos of various histone H3 mutant genotypes and controls were fixed and stained with anti-AbdB antibodies. Embryos were stained with anti-GFP antibodies to detect yellow fluorescent protein (YFP) for staging and genotype selection. Images used for staging these embryos can be found in fig. S12. Brackets indicate the expected boundary of AbdB expression in WT embryos. Filled arrows highlight individual cells exhibiting anterior derepression of AbdB. Scale bar, 50 μm. Overall, the individual H3.3WTH3.2K36R and H3.3K36RH3.2HWT mutants closely resemble the H3.3WTH3.2HWT-negative controls, whereas the double-mutant H3.3K36RH3.2K36R embryos are like the H3.3WTH3.2K27R-positive control. For phenotypic scoring, 10 to 25 embryos were imaged per genotype (see fig. S13). (B) The same as (A), except stained with anti-Ubx (see fig. S14). Empty arrows highlight a small patch of Ubx-positive cells consistently observed in WT controls and thus do not indicate H3K36-dependent derepression.
To further assess redundancies between H3.2K36 and H3.3K36 in Hox gene repression, we analyzed expression of another Hox protein, Ubx. In stage 16 embryos, control H3.3WTH3.2HWT embryos recapitulated the known Ubx staining pattern (75), although we consistently observed a small cluster of Ubx expressing cells at the midline, slightly anterior to the known boundary at PS5 (Fig. 7B, arrowheads). Because these Ubx-positive cells were always present in the controls, we did not score them as derepression events in any mutant genotype. Notably, whereas Finogenova et al. (27) observed ectopic expression of Ubx in H3.3WTH3.2K36R L3 wing discs, they failed to detect derepression of Ubx in stage 16 embryos (27). Likewise, H3.3WTH3.2K36R and H3.3K36RH3.2HWT embryos resembled the H3.3WTH3.2HWT control (Fig. 7B and fig. S14). In contrast, we found extensive Ubx expression in anterior parasegments of the VNC of H3.3K36RH3.2K36R embryos, which was comparable to that of H3.3WTH3.2K27R embryos (Fig. 7B and fig. S14). As with AbdB, we conclude that H3.2K36 and H3.3K36 can functionally compensate for each other in the repression of Ubx expression in the embryonic VNC.
Together with abd-A, the Abd-B and Ubx genes are clustered within the BX-C. In addition to BX-C, the Antennapedia complex is located roughly 10 Mb away and contains the Antp, Scr, Dfd, pb, and lab genes (76, 77). We also performed immunostaining for the Antp protein (fig. S15). As with AbdB and Ubx, we also observed incompletely penetrant stochastic derepression of Antp in H3.3WTH3.2K36R embryos, WT staining in the H3.3K36RH3.2HWT embryos, and more extensive and penetrant derepression in H3.3K36RH3.2K36R mutant embryos. In general, Antp derepression in H3.3K36RH3.2K36R was less extensive than that of H3.3WTH3.2K27R embryos. These results reveal that H3.2K36 and H3.3K36 can also functionally compensate for one another to carry out repression of Antp. Together, the data are consistent with the hypothesis that lysine-36 of H3 is necessary for maintaining Hox gene repression in vivo and that the ability of H3K36 to enable this repression can be provided by either H3.2 or H3.3 (12, 27).
DISCUSSION
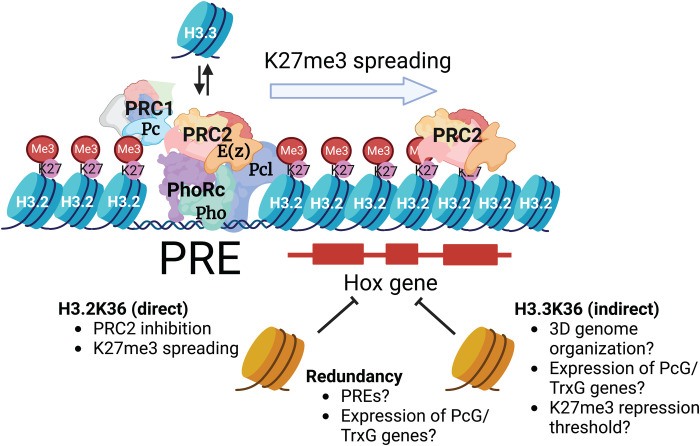
This study demonstrates that H3.2K36 and H3.3K36 collectively mediate proper repression of Hox genes throughout development. At embryonic stages, combined mutation of these two H3 variants causes profound dysregulation of Hox genes. Later in development, the individual H3.2 and H3.3 K36R mutants each exhibit mild homeotic transformations and enhance PcG homeotic phenotypes. As summarized in Fig. 8, our study strongly suggests that H3.2K36 and H3.3K36 promote proper Hox gene repression by distinct mechanisms.
Fig. 8. Working model for function of H3K36 in Pc silencing.
H3.2K36 and H3.3K36 ensure Hox gene repression by different but synergistic mechanisms. PREs nucleate binding of silencing factors including the Pho repressor complex (PhoRc). Replication-independent H3.3 containing nucleosomes are preferentially deposited at PREs, and establishment of silencing is thought to involve their turnover. Once bound to the PRE, PhoRc then recruits PRC2 and PRC1, including Pc and Pcl. Pcl stimulates the activity of the methyltransferase component of PRC2 and E(z). Mutation of replication-dependent H3.2 K36 inhibits E(z) and thereby prevents spreading of H3K27me3 across adjacent nucleosomes between PREs. Replication-independent H3.3 is excluded from PREs located within Pc domains, and mutation of H3.3K36 does not affect H3K27me3 levels. Extensive derepression of Hox genes occurs only when both H3.2 and H3.3 K36 are mutated. This genetic redundancy indicates that H3.3 participates in Pc silencing indirectly, through a variety of potential mechanisms. See text for details. Created with BioRender.com.
Previous studies suggested a critical role for unmodified H3.2K36 in E(z)-mediated H3K27 trimethylation (27). This work corroborates and extends those observations, demonstrating that H3.2K36 is critical for spreading and maintaining H3K27me3 across numerous Pc domains genome-wide (Fig. 8). We also show in vivo that this role in sustaining PRC2 function is specific to H3.2 as we observed no change in global levels or distribution of H3K27me3 in H3.3K36R mutants, and there was no additional impact when combined with an H3.2K36R mutation. Even in a sensitized Pc3/+ genetic background, where H3.3K36R mutants exhibit extensive homeotic transformations, global H3K27me3 levels are not significantly changed. In addition to having no effect on the overall distribution of H3K27me3, H3.3K36R mutation also elicits no specific effect at PREs (Fig. 5, B and C). Perhaps unexpectedly, the same is true for H3.2K36R mutation. Despite a clear defect in the H3.2K36R mutant in inter-PRE regions, H3K27me3 levels are the same as the control at putative PREs. Moreover, Western blotting suggests that global H3K27me3 levels are not significantly reduced when both H3.2 and H3.3 K36R mutations are combined. To more definitively address the possibility that localized loss of H3K27 trimethylation is responsible for Hox gene derepression, chromatin profiling of the combined mutant in L1 larvae from a relatively homogeneous cell population would be necessary. Such an experiment presents numerous technical hurdles but may be possible in the future.
Another possibility for why loss of H3K36 might fail to affect H3K27me3 levels at PREs could be because PRC2 activity is greatly enhanced at these locations by other factors (Fig. 8). Vertebrate Pc-like (PCL) proteins interact with H3K36me3 (78–80) and function as enhancers of PRC2 and EZH2 activity (81–83). Notably, structural analysis of the fly PCL ortholog (Pcl) suggests that this protein is unlikely to bind H3K36me3 (84). However, Pcl retains the ability to bind E(z) in the context of a large complex containing PRC2 (85, 86), and Pcl mutants exhibit reduced H3K27me3 (87). Future studies addressing the role of Pcl at Drosophila PREs in the context of H3K36R mutations will be of great interest.
During early stages of development, we have shown that arginine substitutions in histone H3.3 at either K27 (39) or at K36 (this work) have little effect on overall levels of H3K27 trimethylation. Similarly, overexpression of H3.3K36R histones in cultured human cells had no effect on H3K27me3 (47). These results are consistent with a model put forth by the Allis laboratory, suggesting that H3.3 does not provide a methyl-substrate nucleosome at PREs (88). Instead, increased turnover of H3.3-containing nucleosomes (Fig. 8) is thought to be a key feature in PRC2 recruitment to PREs (88). By extension, higher nucleosome turnover at PREs would be more important for establishing a silent chromatin domain than it would be for maintaining one. Thus, our finding that H3.3 occupancy was much higher at Pho sites located outside of H3K27me3 domains compared to those inside (Fig. 5, E and F) supports this model.
Given that PREs are thought to be multifaceted regulatory modules that can function as repressors or enhancers in different developmental contexts (54, 89, 90), it is intriguing to speculate on other roles that H3.3K36 might play in Hox gene regulation. Our earlier work demonstrates that H3.2K36R mutants exhibit widespread transcriptomic defects (12), and mutants for all three H3K36 lysine methyltransferases in Drosophila also produce large changes in gene expression in the larval brain (91). Given the enrichment of H3.3 in active areas of the genome, one would expect similar dysregulation in H3.3K36R mutants. Therefore, it is possible that H3.3K36 is key to maintaining proper expression levels of one or more genes involved in Pc gene silencing (Fig. 8). Transcriptomic studies in the H3.3K36R, H3.2K36R, and combined mutants would address this possibility.
A role in three-dimensional (3D) genome organization is another potential mechanism for how H3.3K36 might promote Pc silencing (Fig. 8). Several lines of evidence point to the connection between Pc domains, H3K36me, and genome organization (92, 93). PcG proteins bound at different chromosomal sites coalesce into foci known as “Polycomb Bodies” (94–97). Contacts between Hox gene clusters appear to be functionally important to gene silencing as mutation of the Fab-7 element in the BX-C, which strengthens association between the two HOX gene clusters, weakens silencing of genes in the ANTP complex (94). Notably, 3D contacts within Pc domains correspond preferentially to PREs, where H3.3 is enriched (62). In mouse, knockdown of H3.3 (but not H3.1) leads to increased chromatin compaction (98, 99). Given that spatial proximity increases the efficiency of subcellular processes (100–102), a role for H3.3K36 in genome architecture might explain why the H3.3K36R mutation enhances Hox gene derepression in two different genetically sensitized backgrounds (i.e., H3.2K36R versus Pc mutations). Reduced levels of Pc or H3K27me3 might be sufficient to maintain gene repression if these factors are concentrated spatially but insufficient when genome organization is disrupted, thus changing the threshold for eliciting a phenotype. Hence, studies probing the effect of an H3.3K36 mutation on 3D genome organization could also be of interest.
Despite several intriguing findings and conclusions, our histone gene replacement approach has several limitations. The K36R mutation effectively abolishes all K36 PTMs, including acetylation and individual states of lysine methylation (me0, me1, me2, and me3). The simultaneous loss of me0 and me2 is particularly relevant to a discussion of Pc target loci. The Ash1 methyltransferase dimethylates H3K36 (26, 103, 104). In Drosophila, mutation of ash1 impairs the expression of Hox genes, including Abd-B and Ubx, in compartments where they are normally expressed (28, 105), and in vitro studies confirm that the H3K36me2 mark inhibits E(z) function (26, 105). In this context, an H3K36R mutation is analogous to an ash1/E(z) double mutant. Previous studies showed that ash1/E(z) double-mutant clones exhibited a Hox gene derepression phenotype similar to that of an E(z) single mutation (106). Thus, E(z) is epistatic to ash1. This observation is consistent with our results in the combined K36R mutant embryos.
In summary, this study implicates the lysine-36 residue of noncentromeric H3 variants in promoting Hox gene repression in flies. Despite their nearly identical amino acid sequences and potential to permit E(z)-mediated H3K27me3, they contribute to Hox gene repression by nonidentical means. Future work should elucidate the roles played by H3K36 PTMs in different parts of the genome, and how they contribute to chromatin organization, and to the various steps of pre-mRNA processing.
MATERIALS AND METHODS
Drosophila lines and husbandry
To obtain progeny of crosses, parental flies were housed in cages plugged with grape juice agar plates containing supplemental yeast paste. Plates were changed at least daily. Embryos and L1 larvae were harvested directly from the grape juice plates. Older animals were handpicked at the L2 stage, 50 per vial, and raised on standard cornmeal-molasses food. All experimental animals were raised at 25°C.
Details regarding construction of BAC transgenes containing the 12xH3.2 histone gene arrays can be found in (10). The 12x HWT, K27R, K36R, and K9R alleles were generated previously (9, 13). His∆ corresponds to Df(HisCED1429); flies carrying the His∆, twGal4, and His∆, UAS:2xYFP chromosomes (107) were gifts from A. Herzig. The H3.3A2x1 (H3.3Anull) and H3.3B0 (31), along with the Df(2 L)Bsc110 deficiency and Pc3 allele were obtained from Bloomington Stock Center (nos. 68240, 8835, and 1730).
Generation of mutant genotypes
For detailed crossing schemes, see fig. S1. Animals of the His∆ genotype were obtained by selection for yellow fluorescent protein (YFP). Other H3.3 genotypes were selected for absence of a CyO, twGFP balancer chromosome. When not universal in progeny, selection for 12xH3.2 transgenes were identified in embryos by selecting animals that progressed beyond cellularization and by picking YFP-positive larvae at later stages. Adults scored for homeotic transformations were selected by the presence or absence of adult body markers.
CRISPR-Cas9 mutagenesis of H3.3B locus
Cas9-mediated homologous recombination of H3.3B was carried out as described previously (14, 39), with the following differences. A single-guide RNA targeting the H3.3B gene near the K36 residue was inserted into pCFD3 and coinjected along with a 2-kb repair template containing the H3.3BK36R substitution. Constructs were injected into embryos expressing Cas9 from the nanos promoter [nanos-cas9; (108)]. Recovered H3.3BK36R alleles were subsequently crossed into a H3.3A null background [H3.3A2x1 over deficiency Df(2 L)BSC110].
Pupal and adult viability assays
For each genotype, 50 L2 were picked from grape juice agar plates and transferred to vials containing molasses-cornmeal food. Pupal cases and eclosed adults were counted until 13 and 18 days after egg laying, respectively. Pupal and adult viability percentages were calculated on a per-vial basis by dividing the number of pupal cases or eclosed adults per 50 input larvae and multiplying by 100. Each vial constituted one biological replicate for statistical purposes. Between 200 and 400 total animals were analyzed per genotype.
Embryonic hatching assay
For each genotype, His∆, YFP+ embryos were moved onto a clean grape juice plate with a pick into lines of 50. At days 1 and 2 after selection, empty eggshells were counted and recorded as “hatched.” Hatching frequency (%) was calculated by dividing the total number of empty eggshells after 2 days by either the number of total embryos (for 12xH3.2HWT genotypes) or by 0.5× the total number of embryos (for 12xH3.2K36R genotypes) and multiplying by 100. The 0.5× adjustment is due to the necessity of carrying the 12xH3.2K36R transgene over TM6B and the inability to distinguish those two genotypes at the embryonic stage. Between 250 and 400 animals were analyzed per genotype.
Immunofluorescence
Embryos were collected for 3 hours on grape juice agar plates and then aged for 12 hours at 25°C. Embryos of appropriate age were collected in mesh baskets, dechorionated for 5 min in 50% bleach, and rinsed thoroughly in deionized water, followed by embryo wash buffer [1× phosphate-buffered saline (PBS) and 0.03% Triton X-100]. Embryos were transferred to a glass vial, where first 0.5 ml of fixative (1× PBS and 4% formaldehyde) was added, followed by 0.5 ml of heptane, and nutated for 20 min. The bottom layer of fixative was completely removed and replaced with 0.5 ml of 100% methanol. Embryos were devitellinized by vigorous shaking for 30 s. After removal of methanol and heptane, embryos were washed for 5 min, twice with 100% methanol, once with 1:1 methanol/PBS-T (1× PBS and 0.15% Triton X-100), and twice with 1× PBS-T. This was followed by two additional 30-min PBS-T washes. All washes were performed with nutation during the incubation period. Between the two 100% methanol washes, samples were transferred from the glass vial to an Eppendorf tube.
Embryos were blocked in PBS-T (Phosphate buffered saline, Tween 20) with 2% normal goat serum (NGS) for 1 hour at room temperature (RT). Primary antibody incubations were performed overnight at 4°C in PBS-T with 2% NGS plus one or more of the following antibodies: polyclonal rabbit anti–green fluorescent protein (GFP, 1:800; Abcam, no. ab290), monoclonal mouse anti-AbdB (1:500; Developmental Studies Hybridoma Bank, no. 1A2E9), monoclonal mouse anti-Ubx (1:500; Developmental Studies Hybridoma Bank, no. FP3.38), and monoclonal mouse anti-Antp (1:250; Developmental Studies Hybridoma Bank, no. 4C3). Embryos were washed 3× for 10 min in PBS-T. Secondary antibody incubations were performed for 45 min at RT with both anti-rabbit Alexa Fluor 546 (Invitrogen, no. A11035) and anti-mouse Alexa Fluor 488 (Invitrogen, no. A21202) in PBS-T, followed by 2-min 4′,6-diamidino-2-phenylindole stain and three additional PBS-T washes. Embryos were mounted in Vectashield mounting media and imaged on a Leica SP8 confocal microscope with 20× oil immersion objective. Images were viewed and analyzed using ImageJ. For each embryo examined, a maximum intensity Z projection through the VNC was inspected for the number of derepressed cells. For each genotype, 10 to 25 embryos were analyzed in detail.
Western blotting
Protein lysates from L1 or adult heads were obtained by homogenization with a micropestle in SUTEB buffer [1% SDS, 8 M urea, 10 mM EDTA (pH 8.0), and 5% β-mercaptoethanol, with 1:20 Halt protease inhibitor cocktail (Thermo Fisher Scientific, no. 78429)]. Chromatin was further disrupted by sonication with a Bioruptor Pico (Diagenode).
Samples were run on 4 to 15% Mini-PROTEAN TGX Stain-Free protein gels (Bio-Rad, #4568084) for 60 min at 100 V. Western blotting was performed using the Bio-Rad Trans Blot Turbo transfer system using the provided buffer (Bio-Rad, no. 10026938) onto a nitrocellulose membrane at 1.3A/25 V for 7 min. Membranes were blocked at RT in 5% milk in TBS-T (Tris buffered saline, Tween 20). Primary antibody incubation occurred overnight at 4°C in TBS-T with 5% milk with one of the following antibodies: polyclonal rabbit anti-H3K27me3 (1:1000; Activemotif, no. 39055) and polyclonal rabbit anti-H3 (Abcam, #1791). For each primary antibody, an anti-rabbit secondary (Sigma-Aldrich, no. 12-348) was used at 1:5000. Blots were incubated with chemiluminescent detection reagent (Amersham ECL Prime Western Blotting Detection Reagents, GE Healthcare, no. RPN2236) and imaged on an Amersham Imager 600 (GE Healthcare). Between primary antibody antibodies, blots were stripped, rinsed in TBS-T, and incubated with detection reagent to verify removal of antibody before reprobing.
Relative band intensity ratios were calculated on ImageJ. Briefly, a box of equal size and dimension was drawn around each band, and integrated density (IntDen) inside the box was recorded. For each blot, a ratio of H3K27me3/H3 IntDen was calculated per sample lane. In addition, a normalized value for each mutant genotype was calculated by dividing the mutant ratio by that of the H3.3WTH3.2HWT genotype (for L1 experiments) or by individual control indicated in figure legend (adult heads).
CUT&RUN chromatin profiling
We performed CUT&RUN in wing disc tissue, as described in (109), modified from (49). For each replicate, 20 WL3 imaginal wing discs were used. Two wing discs of Drosophila yakuba were also included with each replicate but were not used in downstream analyses. For the H3K27me3 experiment, αH3K27 (Cell Signaling Technology, no. 9733, 1:100) and protein AG-MNase (1:100; UNC core) were used. For the H3.3 experiment, αH3.3 (H3F33B, 1:100; Abnova, no. H00003021-M01) and protein A-MNase (gift of S. Henikoff) were used.
The ThruPLEX DNA sequencing kit was used for the library preparation. The manufacturer’s protocol was followed until the amplification step. After the addition of indexes, 16 to 21 cycles of 98˚C, 20 s and 67˚C, 10 s were run. DNA library purification was done using AMPure XP beads. Libraries were sequenced on an Illumina NextSeqP2.
Bioinformatic analyses
A chart with details of our bioinformatic workflow can be found in fig. S6. CUT&RUN data were processed using coding resources deposited in https://doi.org/10.5281/zenodo.7554890. Differential peak analysis was performed using featureCounts (110) and DESeq2, v1.34.0 (111). Details on how each list of peaks or intervals was determined during a given step of the analysis are shown in fig. S8. For broad domains (Fig. 4B), pellet reads of all fragment sizes were used for DESeq2 analysis. For PRE-based analyses in Fig. 5C and figs. S10B and S11, only short fragment (20 to 120 bp) pellet reads were analyzed with DESeq2.
All heatmaps and metaplots were generated with deepTools, v3.2.0 (112). The Y chromosome was excluded from analysis. For details on how reference points were obtained for each analysis, see fig. S8. For broad domain analysis (Fig. 4D), deepTools, v3.2.0 was used to calculate signal intensity from K27me3 z score–normalized pellet large fragment–pooled bigWig files at “robust putative PREs” (see fig. S8) and for the heatmaps and metaplots of 20-kb flanking regions. For PRE-based heatmaps and metaplots in Fig. 5B and fig. S10A, deepTools, v.3.2.0 was used to calculate and plot signal intensity from K27me3 z score–normalized pellet large fragment– and small fragment–pooled bigWig files at these “concatenated PRE intervals” and for 500-bp flanking regions. For Fig. 5F heatmaps and metaplots, deepTools, v.3.2.0 was used to calculate signal intensity from H3.3 z score–normalized supernatant all fragment–pooled yw bigWig files at Pho-binding sites (see fig. S8) and 5-kb flanking regions inside and outside broad H3K27me3 domains.
Statistical analysis
Statistical analyses for nongenomic experiments were performed and guided by GraphPad Prism according to the characteristics of each dataset. Details of each statistical analysis can be found on each corresponding figure legend. Adjusted P values obtained to call differential peaks in the CUT&RUN experiment were determined with the DESeq2 analysis package (111). The two-tailed Fisher’s exact test in Fig. 5E was performed between Pho-binding intervals and H3.3 MACS2 narrow peak summits ±150-bp intervals from the yw genotype with H3.3∆ peaks blacklisted separately by H3K27me3 status using BEDtools, v.2.3.0 (113).
Homeotic transformation assay
Individual crosses for each genotype (see fig. S3 for details) were set up in cages and fed daily with grape juice agar and yeast paste. Groups of 50 L2 were collected per vial of cornmeal molasses food, where they grew to adulthood. Adult males of the correct genotype were separated by body markers and scored for homeotic transformations under a Leica M60 stereomicroscope. Images were obtained using an iPhone camera mounted on the Leica M60.
Scanning electron microscopy
Flies collected and stored in 70% ethanol were sequentially deyhydrated in 100% ethanol. Fly bodies were mounted, and images were collected using a Hitachi 392 TM4000Plus tabletop scanning electron microscope using ×250 magnification.
Acknowledgments
We thank the Bioinformatics Analytics Research Collaborative at the University of North Carolina (UNC) School of Medicine for facilitating access to bioinformatics education and training (for H.R.S. and B.D.M.) as well as staff support (for J.M.S.). Additional educational support was provided by the UNC Chancellor’s Science Scholars program (to K.M.A.) and by the UNC SOLAR (Summer of Learning and Research) program (to S.A.B.).
Funding: This work was supported by the following: National Institutes of Health grant R35-GM136435 (A.G.M., H.R.S., V.V., and J.C.B.); National Institutes of Health grant R35-GM145258 (R.J.D.); National Institutes of Health grant R35-GM128851 (D.J.M.); and National Institutes of Health training stipend T32-GM007092 (M.P.M.).
Author contributions: Conceptualization: A.G.M. and H.R.S. Methodology: H.R.S. and M.P.M. Investigation: H.R.S., V.V., S.A.B., J.C.B., M.P.L.-J., M.P.M., and K.M.A. Software and bioinformatic analysis: B.D.M., J.M.S., and H.R.S. Visualization: H.R.S. and B.D.M. Supervision: A.G.M. and H.R.S. Writing—original draft: H.R.S. and A.G.M. Writing—review and editing: A.G.M., H.R.S., R.J.D., D.J.M., and J.M.S. Funding acquisition: A.G.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw and processed RNA-seq data were deposited to Gene Expression Omnibus (GEO) under accession GSE215017. BED files from the following previously published ChIP-seq datasets (GSE102339) were used (59). BED files derived from Pho_WT_1 (GSM2634951), Pho_WT_2 (GSM2634952), and Pho_WT_3 (GSM2634953) were also used. As described in Materials and Methods, coding resources used for data processing can be found at DOI: 10.5281/zenodo.7554890. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Fig. S1 to S15
Table S1
Legends for tables S2 to S4
References
Other Supplementary Material for this : manuscript includes the following:
Tables S2 to S4
REFERENCES AND NOTES
- 1.N. Camacho-Ordonez, E. Ballestar, H. T. M. Timmers, B. Grimbacher, What can clinical immunology learn from inborn errors of epigenetic regulators? J. Allergy Clin. Immunol. 147, 1602–1618 (2021). [DOI] [PubMed] [Google Scholar]
- 2.E. Conway, E. Healy, A. P. Bracken, PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 37, 42–48 (2015). [DOI] [PubMed] [Google Scholar]
- 3.J. H. Lee, J.-H. Kim, S. Kim, K. S. Cho, S. B. Lee, Chromatin changes associated with neuronal maintenance and their pharmacological application. Curr. Neuropharmacol. 16, 118–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. Piunti, A. Shilatifard, The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 22, 326–345 (2021). [DOI] [PubMed] [Google Scholar]
- 5.A. Elsherbiny, G. Dobreva, Epigenetic memory of cell fate commitment. Curr. Opin. Cell Biol. 69, 80–87 (2021). [DOI] [PubMed] [Google Scholar]
- 6.J.-Y. Hwang, K. A. Aromolaran, R. S. Zukin, The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 18, 347–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.K. R. Stewart-Morgan, N. Petryk, A. Groth, Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 22, 361–371 (2020). [DOI] [PubMed] [Google Scholar]
- 8.B. D. Strahl, C. D. Allis, The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 9.D. J. McKay, S. Klusza, T. J. Penke, M. P. Meers, K. P. Curry, S. L. McDaniel, P. Y. Malek, S. W. Cooper, D. C. Tatomer, J. D. Lieb, B. D. Strahl, R. J. Duronio, A. G. Matera, Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. P. Meers, M. Leatham-Jensen, T. J. R. Penke, D. J. McKay, R. J. Duronio, A. G. Matera, An animal model for genetic analysis of multi-gene families: Cloning and transgenesis of large tandemly repeated histone gene clusters. Methods Mol. Biol. 1832, 309–325 (2018). [DOI] [PubMed] [Google Scholar]
- 11.R. L. Armstrong, T. J. R. Penke, B. D. Strahl, A. G. Matera, D. J. McKay, D. M. MacAlpine, R. J. Duronio, Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila. Genome Res. 28, 1688–1700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. P. Meers, T. Henriques, C. A. Lavender, D. J. McKay, B. D. Strahl, R. J. Duronio, K. Adelman, A. G. Matera, Histone gene replacement reveals a post-transcriptional role for H3K36 in maintaining metazoan transcriptome fidelity. eLife 6, e23249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.T. J. Penke, D. J. McKay, B. D. Strahl, A. G. Matera, R. J. Duronio, Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev. 30, 1866–1880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.T. J. R. Penke, D. J. McKay, B. D. Strahl, A. G. Matera, R. J. Duronio, Functional redundancy of variant and canonical histone H3 lysine 9 modification in drosophila. Genetics 208, 229–244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B. Schuettengruber, H. M. Bourbon, L. Di Croce, G. Cavalli, Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57 (2017). [DOI] [PubMed] [Google Scholar]
- 16.J. A. Simon, R. E. Kingston, Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R. T. Coleman, G. Struhl, Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356, eaai8236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. R. Pengelly, Ö. Copur, H. Jäckle, A. Herzig, J. Müller, A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 19.K. H. Hansen, A. P. Bracken, D. Pasini, N. Dietrich, S. S. Gehani, A. Monrad, J. Rappsilber, M. Lerdrup, K. Helin, A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 (2008). [DOI] [PubMed] [Google Scholar]
- 20.L. Jiao, X. Liu, Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383-aac4383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R. Margueron, N. Justin, K. Ohno, M. L. Sharpe, J. Son, W. J. Drury III, P. Voigt, S. R. Martin, W. R. Taylor, V. De Marco, V. Pirrotta, D. Reinberg, S. J. Gamblin, Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Uckelmann, C. Davidovich, Not just a writer: PRC2 as a chromatin reader. Biochem. Soc. Trans. 49, 1159–1170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.T. Zhang, S. Cooper, N. Brockdorff, The interplay of histone modifications - writers that read. EMBO Rep. 16, 1467–1481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N. P. Blackledge, R. J. Klose, The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.F. W. Schmitges, A. B. Prusty, M. Faty, A. Stutzer, G. M. Lingaraju, J. Aiwazian, R. Sack, D. Hess, L. Li, S. Zhou, R. D. Bunker, U. Wirth, T. Bouwmeester, A. Bauer, N. Ly-Hartig, K. Zhao, H. Chan, J. Gu, H. Gut, W. Fischle, J. Muller, N. H. Thoma, Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011). [DOI] [PubMed] [Google Scholar]
- 26.W. Yuan, M. Xu, C. Huang, N. Liu, S. Chen, B. Zhu, H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.K. Finogenova, J. Bonnet, S. Poepsel, I. B. Schafer, K. Finkl, K. Schmid, C. Litz, M. Strauss, C. Benda, J. Müller, Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. eLife 9, e61964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. Dorafshan, T. G. Kahn, A. Glotov, M. Savitsky, M. Walther, G. Reuter, Y. B. Schwartz, Ash1 counteracts Polycomb repression independent of histone H3 lysine 36 methylation. EMBO Rep. 20, e46762, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.S. J. Elsaesser, A. D. Goldberg, C. D. Allis, New functions for an old variant: No substitute for histone H3.3. Curr. Opin. Genet. Dev. 20, 110–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B. Loppin, F. Berger, Histone variants: The nexus of developmental decisions and epigenetic memory. Annu. Rev. Genet. 54, 121–149 (2020). [DOI] [PubMed] [Google Scholar]
- 31.A. Sakai, B. E. Schwartz, S. Goldstein, K. Ahmad, Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19, 1816–1820 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. A. Kassis, J. L. Brown, Polycomb group response elements in Drosophila and vertebrates, in Advances in Genetics, T. Friedmann, J. C. Dunlap, S. F. Goodwin, Eds. (Elsevier, 2013), pp. 83–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Muller, J. A. Kassis, Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16, 476–484 (2006). [DOI] [PubMed] [Google Scholar]
- 34.G. Streubel, A. Watson, S. G. Jammula, A. Scelfo, D. J. Fitzpatrick, G. Oliviero, R. McCole, E. Conway, E. Glancy, G. L. Negri, E. Dillon, K. Wynne, D. Pasini, N. J. Krogan, A. P. Bracken, G. Cagney, The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol. Cell 70, 371–379.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Y. Zheng, S. M. Sweet, R. Popovic, E. Martinez-Garcia, J. D. Tipton, P. M. Thomas, J. D. Licht, N. L. Kelleher, Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc. Natl. Acad. Sci. U.S.A. 109, 13549–13554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.P. B. Talbert, S. Henikoff, Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126 (2017). [DOI] [PubMed] [Google Scholar]
- 37.A. S. Akhmanova, P. C. T. Bindels, J. Xu, K. Miedema, H. Kremer, W. Hennig, Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome 38, 586–600 (1995). [DOI] [PubMed] [Google Scholar]
- 38.M. Hodl, K. Basler, Transcription in the absence of histone H3.3. Curr. Biol. 19, 1221–1226 (2009). [DOI] [PubMed] [Google Scholar]
- 39.M. Leatham-Jensen, C. M. Uyehara, B. D. Strahl, A. G. Matera, R. J. Duronio, D. J. McKay, Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLOS Genet. 15, e1007932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S. E. Celniker, S. Sharma, D. J. Keelan, E. B. Lewis, The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9, 4277–4286 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.N. Javeed, N. J. Tardi, M. Maher, S. Singari, K. A. Edwards, Controlled expression of Drosophila homeobox loci using theHostile takeoversystem. Dev. Dyn. 244, 808–825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U. Fresán, M. A. Rodríguez-Sánchez, O. Reina, V. G. Corces, M. L. Espinàs, Haspin kinase modulates nuclear architecture and Polycomb-dependent gene silencing. PLOS Genet. 16, e1008962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Furuyama, R. Banerjee, T. R. Breen, P. J. Harte, SIR2 is required for Polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr. Biol. 14, 1812–1821 (2004). [DOI] [PubMed] [Google Scholar]
- 44.A. Lagarou, A. Mohd-Sarip, Y. M. Moshkin, G. E. Chalkley, K. Bezstarosti, J. A. Demmers, C. P. Verrijzer, dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 22, 2799–2810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. P. Lifton, M. L. Goldberg, R. W. Karp, D. S. Hogness, The organization of the histone genes in Drosophila melanogaster: Functional and evolutionary implications. Cold Spring Harb. Symp. Quant. Biol. 42 (Pt 2), 1047–1051 (1978). [DOI] [PubMed] [Google Scholar]
- 46.A. Chaouch, J. Berlandi, C. C. L. Chen, F. Frey, S. Badini, A. S. Harutyunyan, X. Chen, B. Krug, S. Hébert, A. Jeibmann, C. Lu, C. L. Kleinman, M. Hasselblatt, P. Lasko, M. Shirinian, N. Jabado, Histone H3.3 K27M and K36M mutations de-repress transposable elements through perturbation of antagonistic chromatin marks. Mol. Cell 81, 4876–4890.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Lu, S. U. Jain, D. Hoelper, D. Bechet, R. C. Molden, L. Ran, D. Murphy, S. Venneti, M. Hameed, B. R. Pawel, J. S. Wunder, B. C. Dickson, S. M. Lundgren, K. S. Jani, N. De Jay, S. Papillon-Cavanagh, I. L. Andrulis, S. L. Sawyer, D. Grynspan, R. E. Turcotte, J. Nadaf, S. Fahiminiyah, T. W. Muir, J. Majewski, C. B. Thompson, P. Chi, B. A. Garcia, C. D. Allis, N. Jabado, P. W. Lewis, Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R. Popovic, E. Martinez-Garcia, E. G. Giannopoulou, Q. Zhang, Q. Zhang, T. Ezponda, M. Y. Shah, Y. Zheng, C. M. Will, E. C. Small, Y. Hua, M. Bulic, Y. Jiang, M. Carrara, R. A. Calogero, W. L. Kath, N. L. Kelleher, J. P. Wang, O. Elemento, J. D. Licht, Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLOS Genet. 10, e1004566 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.P. J. Skene, J. G. Henikoff, S. Henikoff, Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 50.G. A. Orsi, S. Kasinathan, K. T. Hughes, S. Saminadin-Peter, S. Henikoff, K. Ahmad, High-resolution mapping defines the cooperative architecture of Polycomb response elements. Genome Res. 24, 809–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.J. G. Henikoff, J. A. Belsky, K. Krassovsky, D. M. Macalpine, S. Henikoff, Epigenome characterization at single base-pair resolution. Proc. Natl. Acad. Sci. U.S.A. 108, 18318–18323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.P. J. Skene, S. Henikoff, An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.N. Nègre, C. D. Brown, L. Ma, C. A. Bristow, S. W. Miller, U. Wagner, P. Kheradpour, M. L. Eaton, P. Loriaux, R. Sealfon, Z. Li, H. Ishii, R. F. Spokony, J. Chen, L. Hwang, C. Cheng, R. P. Auburn, M. B. Davis, M. Domanus, P. K. Shah, C. A. Morrison, J. Zieba, S. Suchy, L. Senderowicz, A. Victorsen, N. A. Bild, A. J. Grundstad, D. Hanley, D. M. MacAlpine, M. Mannervik, K. Venken, H. Bellen, R. White, M. Gerstein, S. Russell, R. L. Grossman, B. Ren, J. W. Posakony, M. Kellis, K. P. White, A cis-regulatory map of the Drosophila genome. Nature 471, 527–531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. Erceg, T. Pakozdi, R. Marco-Ferreres, Y. Ghavi-Helm, C. Girardot, A. P. Bracken, E. E. Furlong, Dual functionality of cis-regulatory elements as developmental enhancers and Polycomb response elements. Genes Dev. 31, 590–602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.B. A. Bredesen, M. Rehmsmeier, DNA sequence models of genome-wide Drosophila melanogaster Polycomb binding sites improve generalization to independent Polycomb Response Elements. Nucleic Acids Res. 47, 7781–7797 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.T. Fiedler, M. Rehmsmeier, jPREdictor: A versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Res. 34, W546–W550 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.L. Ringrose, M. Rehmsmeier, J.-M. Dura, R. Paro, Genome-wide prediction of Polycomb/trithorax response elements in Drosophila melanogaster. Dev. Cell 5, 759–771 (2003). [DOI] [PubMed] [Google Scholar]
- 58.J. Zeng, B. D. Kirk, Y. Gou, Q. Wang, J. Ma, Genome-wide polycomb target gene prediction in Drosophila melanogaster. Nucleic Acids Res. 40, 5848–5863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. L. Brown, M.-A. Sun, J. A. Kassis, Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho. Proc. Natl. Acad. Sci. U.S.A. 115, E1839–E1848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. I. Kuroda, H. Kang, S. De, J. A. Kassis, Dynamic competition of polycomb and trithorax in transcriptional programming. Annu. Rev. Biochem. 89, 235–253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. M. Deaton, M. Gómez-Rodríguez, J. Mieczkowski, M. Y. Tolstorukov, S. Kundu, R. I. Sadreyev, L. E. Jansen, R. E. Kingston, Enhancer regions show high histone H3.3 turnover that changes during differentiation. eLife 5, e15316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Y. Mito, J. G. Henikoff, S. Henikoff, Histone replacement marks the boundaries of cis-regulatory domains. Science 315, 1408–1411 (2007). [DOI] [PubMed] [Google Scholar]
- 63.A. Busturia, A. Lloyd, F. Bejarano, M. Zavortink, H. Xin, S. Sakonju, The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 128, 2163–2173 (2001). [DOI] [PubMed] [Google Scholar]
- 64.B. A. Horard, C. Tatout, S. Poux, V. Pirrotta, Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20, 3187–3197 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R. K. Mishra, J. Mihaly, S. P. Barges, A. Spierer, F. O. Karch, K. Hagstrom, S. E. Schweinsberg, P. Schedl, The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 21, 1311–1318 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. Schwendemann, M. Lehmann, Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. U.S.A. 99, 12883–12888 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.T. Nakayama, K. Nishioka, Y.-X. Dong, T. Shimojima, S. Hirose, Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 21, 552–561 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.H. Okulski, B. Druck, S. Bhalerao, L. Ringrose, Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics Chromatin 4, 4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.K. Ahmad, S. Henikoff, Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. U.S.A. 99 (Suppl 4), 16477–16484 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.H.-T. Fang, C. A. El Farran, Q. R. Xing, L.-F. Zhang, H. Li, B. Lim, Y.-H. Loh, Global H3.3 dynamic deposition defines its bimodal role in cell fate transition. Nat. Commun. 9, 1537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.E. McKittrick, P. R. Gafken, K. Ahmad, S. Henikoff, Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. U.S.A. 101, 1525–1530 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.I. Bajusz, L. Sipos, Z. Gyorgypal, E. A. Carrington, R. S. Jones, J. Gausz, H. Gyurkovics, The Trithorax-mimic allele of Enhancer of zeste renders active domains of target genes accessible to polycomb-group-dependent silencing in Drosophila melanogaster. Genetics 159, 1135–1150 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.C. T. Wu, M. Howe, A genetic analysis of the Suppressor 2 of zeste complex of Drosophila melanogaster. Genetics 140, 139–181 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.I. A. Hernández-Romero, V. J. Valdes, De novo polycomb recruitment and repressive domain formation. Epigenomes 6, 25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R. K. Maeda, F. Karch, The ABC of the BX-C: The bithorax complex explained. Development 133, 1413–1422 (2006). [DOI] [PubMed] [Google Scholar]
- 76.R. J. Diederich, V. K. Merrill, M. A. Pultz, T. C. Kaufman, Isolation, structure, and expression of labial, a homeotic gene of the Antennapedia Complex involved in Drosophila head development. Genes Dev. 3, 399–414 (1989). [DOI] [PubMed] [Google Scholar]
- 77.M. P. Scott, A. J. Weiner, T. I. Hazelrigg, B. A. Polisky, V. Pirrotta, F. Scalenghe, T. C. Kaufman, The molecular organization of the Antennapedia locus of Drosophila. Cell 35, 763–776 (1983). [DOI] [PubMed] [Google Scholar]
- 78.C. Ballare, M. Lange, A. Lapinaite, G. M. Martin, L. Morey, G. Pascual, R. Liefke, B. Simon, Y. Shi, O. Gozani, T. Carlomagno, S. A. Benitah, L. Di Croce, Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 19, 1257–1265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.L. Cai, S. B. Rothbart, R. Lu, B. Xu, W. Y. Chen, A. Tripathy, S. Rockowitz, D. Zheng, D. J. Patel, C. D. Allis, B. D. Strahl, J. Song, G. G. Wang, An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell 49, 571–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.C. A. Musselman, N. Avvakumov, R. Watanabe, C. G. Abraham, M. E. Lalonde, Z. Hong, C. Allen, S. Roy, J. K. Nunez, J. Nickoloff, C. A. Kulesza, A. Yasui, J. Cote, T. G. Kutateladze, Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 19, 1266–1272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R. Cao, H. Wang, J. He, H. Erdjument-Bromage, P. Tempst, Y. Zhang, Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol. Cell. Biol. 28, 1862–1872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.J. Choi, A. L. Bachmann, K. Tauscher, C. Benda, B. Fierz, J. Muller, DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat. Struct. Mol. Biol. 24, 1039–1047 (2017). [DOI] [PubMed] [Google Scholar]
- 83.K. Sarma, R. Margueron, A. Ivanov, V. Pirrotta, D. Reinberg, Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell. Biol. 28, 2718–2731 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.A. Friberg, A. Oddone, T. Klymenko, J. Muller, M. Sattler, Structure of an atypical Tudor domain in the Drosophila Polycomb-like protein. Protein Sci. 19, 1906–1916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.S. O’Connell, L. Wang, S. Robert, C. A. Jones, R. Saint, R. S. Jones, Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J. Biol. Chem. 276, 43065–43073 (2001). [DOI] [PubMed] [Google Scholar]
- 86.F. Tie, J. Prasad-Sinha, A. Birve, A. Rasmuson-Lestander, P. J. Harte, A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol. Cell. Biol. 23, 3352–3362 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.M. Nekrasov, T. Klymenko, S. Fraterman, B. Papp, K. Oktaba, T. Köcher, A. Cohen, H. G. Stunnenberg, M. Wilm, J. Müller, Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 26, 4078–4088 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.L. A. Banaszynski, D. Wen, S. Dewell, S. J. Whitcomb, M. Lin, N. Diaz, S. J. Elsasser, A. Chapgier, A. D. Goldberg, E. Canaani, S. Rafii, D. Zheng, C. D. Allis, Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.B. Papp, J. Muller, Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 20, 2041–2054 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.J. van Arensbergen, S. Dussaud, C. Pardanaud-Glavieux, J. García-Hurtado, C. Sauty, A. Guerci, J. Ferrer, P. Ravassard, A distal intergenic region controls pancreatic endocrine differentiation by acting as a transcriptional enhancer and as a polycomb response element. PLOS ONE 12, e0171508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.H. Lindehell, A. Glotov, E. Dorafshan, Y. B. Schwartz, J. Larsson, The role of H3K36 methylation and associated methyltransferases in chromosome-specific gene regulation. Sci. Adv. 7, eabh4390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.K. J. Evans, N. Huang, P. Stempor, M. A. Chesney, T. A. Down, J. Ahringer, Stable Caenorhabditis elegans chromatin domains separate broadly expressed and developmentally regulated genes. Proc. Natl. Acad. Sci. U.S.A. 113, E7020–E7029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.M. A. Willcockson, S. E. Healton, C. N. Weiss, B. A. Bartholdy, Y. Botbol, L. N. Mishra, D. S. Sidhwani, T. J. Wilson, H. B. Pinto, M. I. Maron, K. A. Skalina, L. N. Toro, J. Zhao, C.-H. Lee, H. Hou, N. Yusufova, C. Meydan, A. Osunsade, Y. David, E. Cesarman, A. M. Melnick, S. Sidoli, B. A. Garcia, W. Edelmann, F. Macian, A. I. Skoultchi, H1 histones control the epigenetic landscape by local chromatin compaction. Nature 589, 293–298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.F. Bantignies, V. Roure, I. Comet, B. Leblanc, B. Schuettengruber, J. Bonnet, V. Tixier, A. Mas, G. Cavalli, Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144, 214–226 (2011). [DOI] [PubMed] [Google Scholar]
- 95.P. Buchenau, J. Hodgson, H. Strutt, D. J. Arndt-Jovin, The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: Impact on models for silencing. J. Cell Biol. 141, 469–481 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.C. Grimaud, F. Bantignies, M. Pal-Bhadra, P. Ghana, U. Bhadra, G. Cavalli, RNAi components are required for nuclear clustering of polycomb group response elements. Cell 124, 957–971 (2006). [DOI] [PubMed] [Google Scholar]
- 97.A. J. Saurin, C. Shiels, J. Williamson, D. P. Satijn, A. P. Otte, D. Sheer, P. S. Freemont, The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142, 887–898 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.C. J. Lin, M. Conti, M. Ramalho-Santos, Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140, 3624–3634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Y. Wang, H. Long, J. Yu, L. Dong, M. Wassef, B. Zhuo, X. Li, J. Zhao, M. Wang, C. Liu, Z. Wen, L. Chang, P. Chen, Q.-F. Wang, X. Xu, R. Margueron, G. Li, Histone variants H2A.Z and H3.3 coordinately regulate PRC2-dependent H3K27me3 deposition and gene expression regulation in mES cells. BMC Biol. 16, 107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.D. Arias Escayola, K. M. Neugebauer, Dynamics and function of nuclear bodies during embryogenesis. Biochemistry 57, 2462–2469 (2018). [DOI] [PubMed] [Google Scholar]
- 101.R. J. Duronio, W. F. Marzluff, Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 14, 726–738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.J. E. Sleeman, L. Trinkle-Mulcahy, Nuclear bodies: New insights into assembly/dynamics and disease relevance. Curr. Opin. Cell Biol. 28, 76–83 (2014). [DOI] [PubMed] [Google Scholar]
- 103.K. M. Dorighi, J. W. Tamkun, The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 140, 4182–4192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Y. Tanaka, Z. Katagiri, K. Kawahashi, D. Kioussis, S. Kitajima, Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397, 161–168 (2007). [DOI] [PubMed] [Google Scholar]
- 105.S. Schmahling, A. Meiler, Y. Lee, A. Mohammed, K. Finkl, K. Tauscher, L. Israel, M. Wirth, J. Philippou-Massier, H. Blum, B. Habermann, A. Imhof, J. J. Song, J. Muller, Regulation and function of H3K36 di-methylation by the trithorax-group protein complex AMC. Development 145, dev163808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.T. Klymenko, J. Muller, The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5, 373–377 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.U. Gunesdogan, H. Jackle, A. Herzig, A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 11, 772–776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.S. Kondo, R. Ueda, Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.K. Ahmad, CUT&RUN with Drosophila tissues V.1., protocols.io (2018); 10.17504/protocols.io.umfeu3n. [DOI]
- 110.Y. Liao, G. K. Smyth, W. Shi, featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 111.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.F. Ramírez, D. P. Ryan, B. Grüning, V. Bhardwaj, F. Kilpert, A. S. Richter, S. Heyne, F. Dündar, T. Manke, deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.A. R. Quinlan, I. M. Hall, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.S. Andrews, FastQC: A quality control tool for high throughput sequence data (Babraham Institute, 2010). [Google Scholar]
- 115.S. W. Wingett, S. Andrews, FastQ screen: A tool for multi-genome mapping and quality control. F1000Research 7, 1338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.P. Danecek, J. K. Bonfield, J. Liddle, J. Marshall, V. Ohan, M. O. Pollard, A. Whitwham, T. Keane, S. A. McCarthy, R. M. Davies, H. Li, Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.W. J. Kent, A. S. Zweig, G. Barber, A. S. Hinrichs, D. Karolchik, BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu, Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.M. N. Patwardhan, C. D. Wenger, E. S. Davis, D. H. Phanstiel, Bedtoolsr: An R package for genomic data analysis and manipulation. J. Open Source Softw. 4, 1742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S15
Table S1
Legends for tables S2 to S4
References
Tables S2 to S4